- 1Jiangxi Province Key Laboratory of Watershed Ecosystem Change and Biodiversity, Institute of Life Science and School of Life Sciences, Nanchang University, Nanchang, China
- 2State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China
East Asia is one of the plant diversity and endemism centers in the world, and the temporal and spatial patterns and processes of vascular plants attracted ecologists’ and biogeographers’ attention. However, the biogeographic patterns of endemic epiphytic plants in East Asia are still unclear. Here, we investigated the historical biogeography of an East Asian endemic epiphytic genus Holcoglossum Schltr. (Orchidaceae). Using DNA sequences of eleven chloroplast genes and one nuclear gene, we reconstructed a robust phylogenetic framework for Holcoglossum and used a relaxed-clock method to estimate divergent times for the genus. We inferred the ancestral range of lineages under the statistical dispersal-extinction cladogenesis (S-DEC) and statistical dispersal-vicariance analysis (S-DIVA), respectively. Biogeographical analysis suggested that the most recent common ancestor of Holcoglossum occurred in the Palaeotropical region in the late Miocene (6.33 Ma). Four dispersal events were inferred to explain the Holcoglossum expansion to Sino-Himalayan, Sino-Japanese, and Taiwan regions from the latest Miocene to Quaternary. The episodes of these events were associated with intensification of East Asian monsoon around 3.6–2.6 Ma and global cooling since the latest Pliocene. The disjunct distribution between mainland China and Taiwan was attributed by the sea-level fluctuations and climate changes during the late Pliocene. This study shed light on the biogeographic processes of endemic epiphytic plants in East Asia.
Introduction
East Asia is well-known for its vascular plant diversity and endemic (Qian et al., 2005). The flora of East Asia harbors ca. 258 families and more than 3000 genera, of which approximately 8.2% are endemic (Wu and Wu, 1998; Chen et al., 2018). Understanding the historical origins and processes of the plant richness of East Asia has been of major interest for ecologists and biogeographers. In the Cenozoic, East Asia underwent the uplift of the Himalaya–Tibetan Plateau (An et al., 2001) and the establishment and intensification of the Asian monsoon (Sun and Wang, 2005). A great deal of phytogeographic research on plants in East Asia has been published. East Asia is suggested as both a “Cradle” and a “Museum” for vascular plants since the Cretaceous (e.g., Wu and Wu, 1998; Wen, 1999; Jiang et al., 2019). Xiang et al. (2019) combined molecular and fossil data and inferred that the ancestors of Hamamelidaceae occurred in tropical Asia during the mid-Cretaceous and subsequently migrated to Europe, Africa, and America during the Late Cretaceous and Early Tertiary. Luo et al. (2015) carried out phylogeographic analysis of four perennial herbs in Hengduan Mountains and showed that these species originated during the Late Pliocene or early–mid-Pleistocene and diverged by climate-induced habitat fragmentation. Further, during the glacial periods of Quaternary, East Asia acted as refugia for vascular plants’ survival and evolution, especially for more recent divergent lineages (Tiffney, 1985; Ying, 2001; Qiu et al., 2011; Chen et al., 2018). However, the biogeographic patterns and processes of endemic epiphytic plants in East Asia are still poorly known.
Aeridinae is the largest and horticulturally important subtribe in Orchidaceae, which consisted of 83 genera and 1550 species (Pridgeon et al., 2014; Chase et al., 2015). The subtribe is mainly composed of epiphytes and distributed in tropical and subtropical Asia and northern Australia with some extended to northeast Asia (Hidayat et al., 2012; Chase et al., 2015). Among these, nearly 33 genera occurred across tropical Asia to temperate Asia. Holcoglossum Schltr. (Aeridinae, Orchidaceae) is used here as a typical case to illustrate the biogeographic processes of endemic epiphytic plants in East Asia. This genus encompasses ca. 16 currently accepted species (Xiang et al., 2012), mainly distributed in southwestern China and neighboring regions (Schlechter, 1919; Christenson, 1998; Jin and Wood, 2009; Xiang et al., 2012). These species are diverse in morphology, especially the structure of the flower (Figure 1). Xiang et al. (2012) employed three DNA markers (ITS, matK, and trnH-psbA) to resolve the relationship of 16 Holcoglossum species and found that Holcoglossum is monophyletic, which contains three clades, alpine clade (AC), tropical clade (TC), and the intermediate group (HC). Li et al. (2019) used the whole chloroplast genomes of 12 Holcoglossum species and obtained the same result of Xiang et al. (2012). Of Holcoglossum species, TC includes six species that occur in Southeast Asia, and AC contains five species that are mainly found in the Hengduan Mountains (Figure 2). Within HC, two species are restricted to Taiwan, and another two species are distributed in the mainland China (Figure 2). Previous studies show that the stem age of Holcoglossum was 7.71 Ma [95% highest posterior density (HPD): 4.12–12.04] (Xiang et al., 2016) with two Holcoglossum samplings. Without time estimation and biogeographic analyses, Fan et al. (2009) postulated that Holcoglossum originated in tropical Asia and migrated from the tropics to the temperate regions. Therefore, when and how the genus Holcoglossum formed current biogeographic patterns is still unclear.

Figure 1. Habitat and diversity of Holcoglossum. (A) H. rupestre; (B) H. sinicum; (C) H. flavescens; (D) H. nujiangense; (E) H. lingulatum; (F) H. omeiense; (G) H. pumilum; (H) H. quasipinifolium; (I) H. himalaicum; (J) H. kimballianum; (K) H. amesianum; (L) H. subulifolium. Photo by Xiaohua Jin.
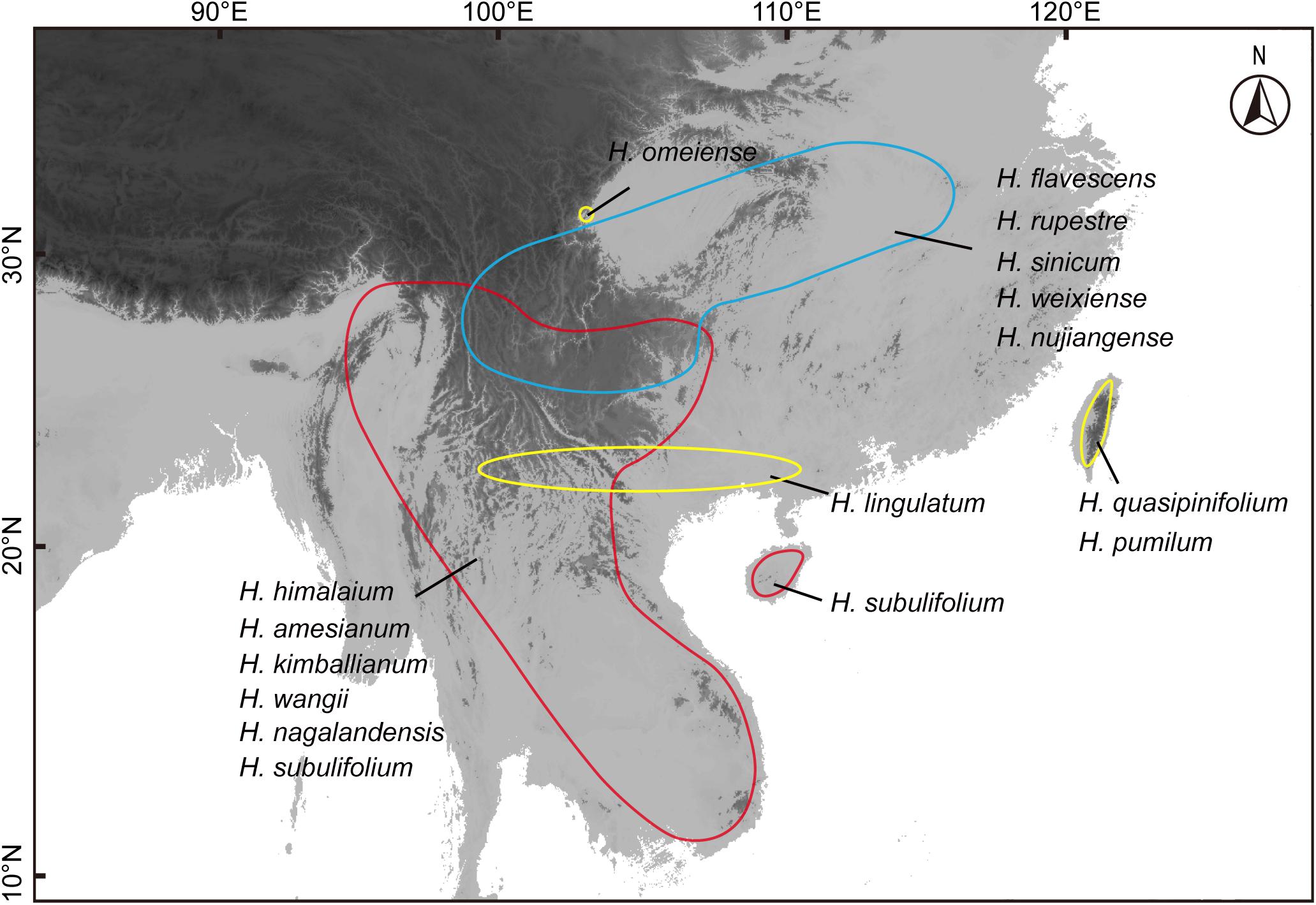
Figure 2. Geographical distribution of Holcoglossum. The red region represents the range of tropical clade (TC), the blue region is for temperate alpine clade (AC), and the yellow region is for the group between the first two clades (HC).
In this study, we reconstruct a time-calibrated phylogenetic tree with samples of nearly all species. We also reconstruct the biogeographic history of Holcoglossum. Our specific objectives were to infer (1) the spatial and temporal pattern of Holcoglossum and (2) the possible impacts of past geological and climatic oscillations on the distribution of these species.
Materials and Methods
Taxon Sampling
In this study, we sampled 15 species of Holcoglossum, representing the three clades of this genus. Based on Topik et al. (2005) and Xiang et al. (2012), the phylogenetic relationships of Aerides alliance is not all resolved, so ten species from Aerides alliance were used as outgroups (Aerangis calligera, Aeranthes ramosus, Aerides multiflora, Angraecum eburneum, Ascocentrum ampullaceum, Diaphananthe odoratissima, Neobenthamia gracilis, Neofinetia falcata, Phalaenopsis aphrodite, Vanda coerulescens). Voucher information and GenBank accession numbers are listed in Supplementary Table 1.
Molecular Data
Total genomic DNA was extracted from silica gel-dried leaves of a living plant using total DNA extraction kit (CWBIO, Beijing). One nuclear markers (ITS) and eleven chloroplast DNA markers (atpH-I, matK, psbA-trnH, psbK-I, rbcL, rpoB, rpoC1, rpS12-rpL20, trnL-F, trnS-fM, and trnS-G) were employed in this study. The primers for amplification and sequencing are listed in Supplementary Table 2. The PCR procedure of atpH-I is followed by Shaw et al. (2007); procedures of psbK-I, rpoB, and rpoC1 are followed by CBOL Plant Working Group (2009); procedures of rpS12-rpL20 and trnS-G are followed by Shaw et al. (2005); the procedure of rbcL is followed by Goldman et al. (2001); and the procedure of trnS-fM is followed by Demesure et al. (1995).
Phylogenetic Analysis
DNA sequences were aligned using the default parameters in Clustal X v.1.83 (Thompson et al., 1997) and subsequently manually adjusted with BioEdit (Hall, 1999). Topological congruence between the chloroplast and nuclear data was evaluated using the incongruence length difference (ILD) test (Farris et al., 1994).
Phylogenetic reconstruction was performed using maximum parsimony (MP), maximum likelihood (ML) in PAUP∗ 4.0b10 (Swofford, 2003), and Bayesian inference (BI) methods in MrBayes v.3.2 (Ronquist et al., 2012), respectively. All characters were unordered and had equal weight. Gaps were treated as missing data.
For MP analyses, heuristic searches were conducted with 1000 replicates of random addition, in combination with tree bisection–reconnection (TBR) branch-swapping, Multrees in effect, and steepest descent off. Bootstrap support values were conducted with 1000 replicates with 10 random taxon additions and heuristic search options.
Before performing ML and BI analyses, the best-fit nucleotide substitution model for each DNA region was chosen based on the Akaike information criterion (AIC) as calculated using Modeltest v.3.7 (Posada and Crandall, 1998). For ML analyses, we conducted a rapid bootstrap analysis (1000 replicates) and searched for the best-scoring ML tree simultaneously. Each DNA region was assigned the best-fit model, and other parameters followed the default settings.
For BI analyses, each DNA region was assigned its own model of nucleotide substitution. Four Markov-chain Monte Carlo (MCMC) were run, sampling one tree every 1000 generations for 3,000,000 generations. Runs started with a random tree. Majority-rule (>50%) consensus trees were constructed after removing the “burn-in” samples (the first 20% of the sampled trees).
Molecular Age Estimation
We first conducted a likelihood ratio test to determine whether our sequence data were evolved in a clock-like fashion. The result rejected a constant rate (δ = 246.55, d.f. = 24, P < 0.001); we used a relaxed lognormal clock model in BEAST v.1.7.4 (Drummond and Rambaut, 2007) to generate a dated phylogeny of Holcoglossum. There are no fossil for Holcoglossum and subtribe Aeridinae, so we used the age of 33.97 Ma (95% HPD: 25.67–42.33) for the tree root prior to our analysis based on our recent broader study of Orchidaceae (Xiang et al., 2016). Following the suggestion of Ho (2007), we assigned a prior normal distribution for the calibration, with a standard deviation of 4. The YULE process was chosen as the speciation prior, and the BEAST analysis was run on the GTR + I + Γ model for each DNA region, respectively. MCMC searches were run for 100,000,000 generations, sampled every 5000 generations. Convergence was monitored using Tracer v.1.5 (Rambaut and Drummond, 2007). The effective sampling sizes (ESSs) for all parameters were more than 200. The maximum clade credibility tree was computed using TreeAnnotator v.1.7.4 (Drummond and Rambaut, 2007).
Ancestral Range Reconstruction
According to the extant distribution of Holcoglossum and outgroups, four main regions were categorized based on the floristic regions of Wu and Wu (1998) and Wu et al. (2003): A, Sino-Japanese region; B, Sino-Himalayan region; C, Palaeotropical region; and D, Taiwan. The ancestral range reconstruction was inferred using the statistical dispersal-extinction cladogenesis (S-DEC) model and statistical dispersal-vicariance analysis (S-DIVA) as implemented in RASP (Yu et al., 2015), respectively. The maximum clade credibility tree obtained from BEAST was chosen as the input tree. The random 1000 trees from BEAST trees after burn-in were input to estimate the probabilities of the ancestral range at each node.
Results
Phylogenetic Relationships Within Holcoglossum
A total of 103 new sequences for atpH-I, rbcL, psbK-I, rpoB, rpoC1, rpS12-rpL20, trnS-fM, and trnS-G were generated in this study (Supplementary Table 2). The total length of the combined chloroplast and nuclear DNA sequences was 11,176 bp, of which 1117 characters were variable, and 433 were parsimony informative (Supplementary Table 3). The result of ILD suggested that there is congruence between chloroplast and nuclear data (P = 1.00). The monophyly of Holcoglossum and the three clades previously proposed (Fan et al., 2009; Xiang et al., 2012) was strongly supported (Figure 3). The TC was basal to Holcoglossum (BI-PP: 1.00; ML-BS: 79%; MP-BS: 82%), and the AC and the group between the first two clades (HC) formed a sister group (BI-PP: 1.00; ML-BS: 100%; MP-BS: 100%). The most nodes within Holcoglossum were supported by moderate to high supporting values, except for interrelationships in the AC clade (Figure 3).
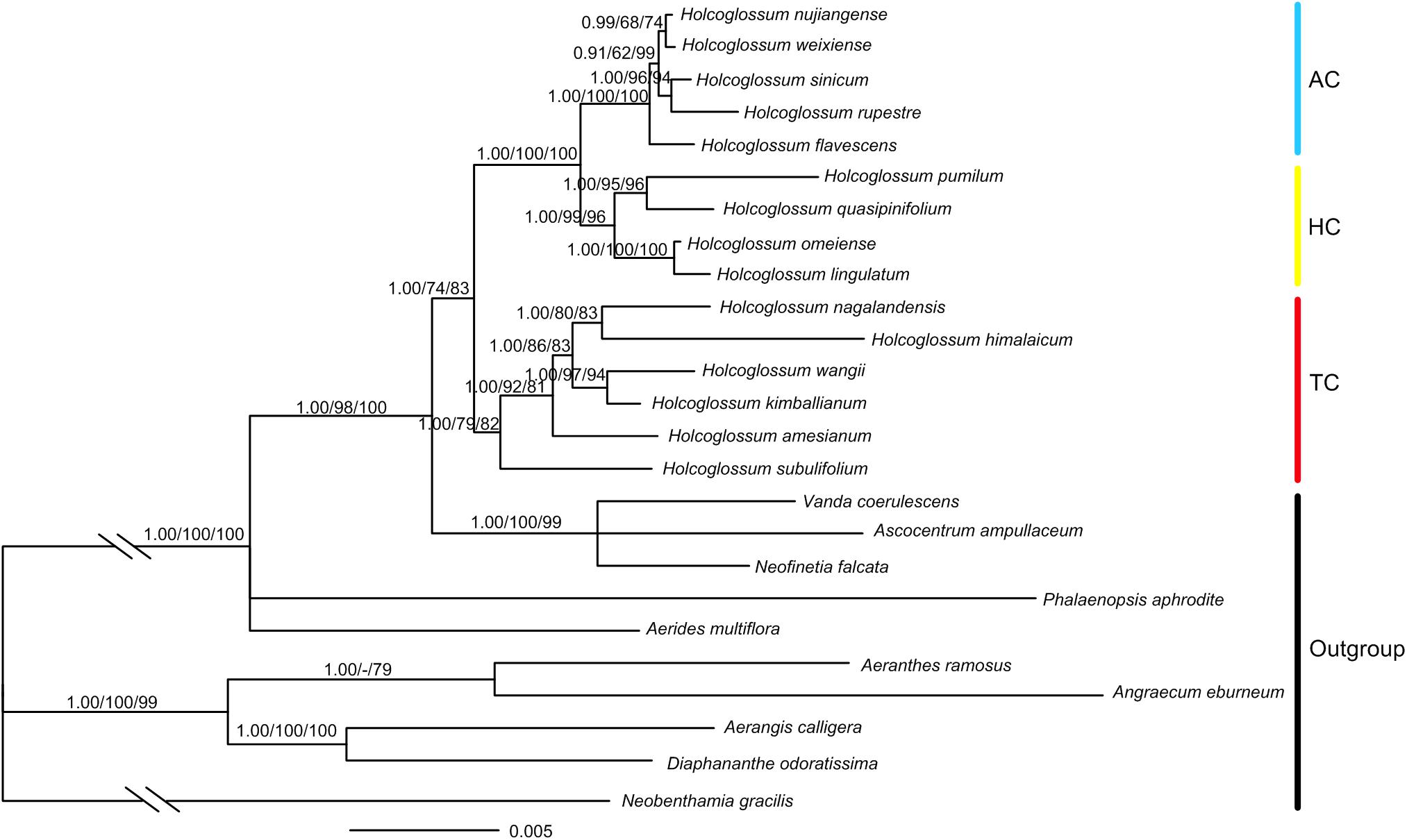
Figure 3. Chronogram obtained from Bayesian inference analysis based on combined nrDNA and cpDNA data. Numbers on the branches are supporting values of Bayesian inference, maximum likelihood, and maximum parsimony, respectively. Dash indicates that the node was not supported.
Divergent Time Estimation
The likelihood ratio test showed that our sequence data were evolving in a non-clock-like model, and a chronogram of Holcoglossum based on the relaxed-clock model is presented in Figure 4. Our time estimates suggested that Holcoglossum might have originated during the Late Tertiary (8.53 Ma, 95% HPD: 4.19–14.02, node 1). The crown age of Holcoglossum was 6.33 Ma (95% HPD: 3.3–10.6, node 2). The TC clade, containing Holcoglossum himalaicum, H. nagalandensis, H. kimballianum, H. wangii, H. amesianum, and H. subulifolium, diverged first during the Early Pliocene (5.31 Ma, 95% HPD: 2.5–8.81, node 3). Then, the HC clade and AC clade diverged during the Late Pliocene (3.63 Ma, 95% HPD: 1.64–6.24, node 4). The crown age of the HC clade, containing Holcoglossum ligulatum, H. omeiense, H. pumilum, and H. quasipinifolium, was at latest Pliocene (2.7 Ma, 95% HPD: 1.16–4.78, node 5), and then the two Taiwan species (H. pumilum, H. quasipinifolium) were divergent from their sister group. The AC clade might have started to diversify around Middle Pleistocene (1.22 Ma, 95% HPD: 0.5–2.39, node 8), and its five species (Holcoglossum nujiangense, H. weixiense, H. rupestre, H. sinicum, H. flavescens) rapidly differentiated since then.
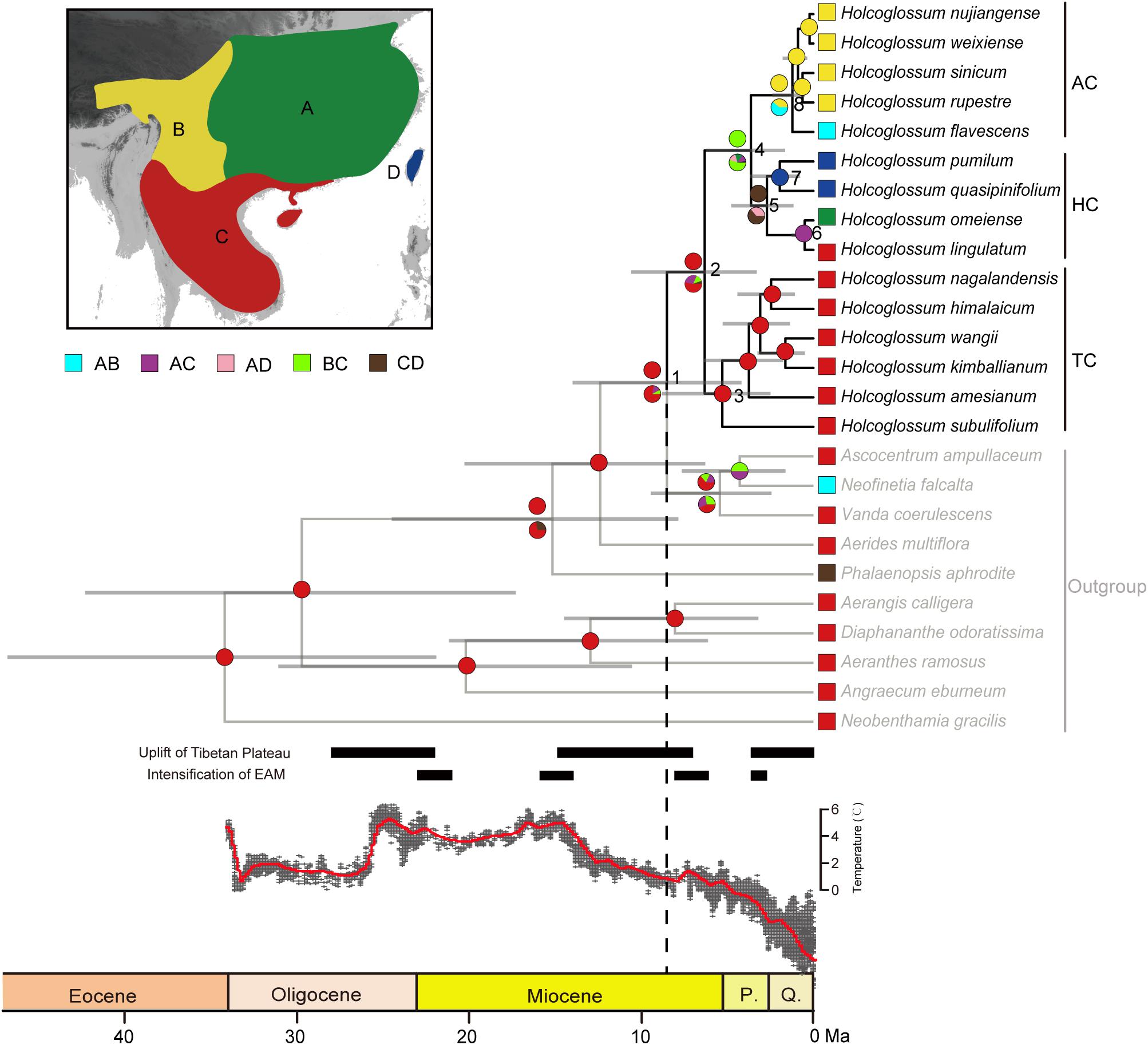
Figure 4. Ancestral range reconstruction of Holcoglossum in RASP. The same results of S-DEC and S-DIVA are marked by one circle at the node. Colored circles at nodes above branches for S-DIVA and below branches for S-DEC. A, Sino-Japanese region; B, Sino-Himalayan region; C, Palaeotropical region; D, Taiwan. The global temperature curve is modified from Zachos et al. (2008).
Ancestral Range Reconstruction
The ancestral range reconstruction of Holcoglossum based on S-DEC and S-DIVA is shown in Figure 4. Both S-DEC and S-DIVA inferred that the most recent common ancestor (MRCA) of Holcoglossum was probably distributed in the Palaeotropical region (node 2). The MRCA of TC was also in the Palaeotropical region (node 3). The MRCA of HC distributed in the Palaeotropical region and Taiwan (node 5), which were later split into mainland China and Taiwan. The MRCA of AC was inferred at the Sino-Japanese region and Sino-Himalayan region by S-DEC, and Sino-Himalayan by S-DIVA (node 8), which subsequently diversified in the Sino-Himalayan region.
Discussion
Phylogenetic Relationship of Holcoglossum
A robust phylogenetic framework is needed for biogeographical analyses. In this study, the phylogenetic result based on combined chloroplast and nuclear data strongly supported Holcoglossum as monophyletic, which included three clades, i.e., TC, HC, and TC, which is consistent with Xiang et al. (2012) and Li et al. (2019). However, the phylogenetic relationships within TC and AC are slightly different. For example, Holcoglossum naglandensis and H. himalaicum among TC are sisters in this study and Xiang et al. (2012), while H. naglandensis is close to H. amesianum and H. himalaicum is close to H. wangii due to different DNA sequences. Nevertheless, these species of TC and AC are distributed in the same floristic region, respectively, and there is no influence on subsequently biogeographic analyses.
Temporal and Spatial Patterns of Holcoglossum
Both S-DEC and S-DIVA analyses detected that the modern distribution of Holcoglossum is attributed to four dispersal events (Figure 4). Here, we adopted the S-DEC result to explain the temporal and spatial patterns of Holcoglossum. The biogeographical analyses indicate that the stem and crown of Holcoglossum most likely occurred in the Palaeotropical region, partly in agreement with the conclusion from Fan et al. (2009) that Holcoglossum was dispersed from the tropical region.
Within TC, all six currently recognized species distribute in the Asian tropical region. The crown group of TC happened in the Palaeotropical region since 5.31 Ma (95% HPD: 4.19–14.02, node 3) and then six species diversified there among 5.31–1.63 Ma. The weather in Southeast Asia had no distinct seasonality, and the climate is constantly moist and warm since Pliocene (Clements et al., 2006), which is beneficial for Holcoglossum colonization in tropical forests. Further, a putative abrupt intensification of the East Asian monsoon system during 3.6–2.6 Ma (Zhang et al., 2012) brought abundant precipitation, which provided favorable climatic conditions for the survival of epiphytic species in East Asia.
After TC diverged from AC and HC, a subsequent dispersal from the Palaeotropical region to the Sino-Himalayan region occurred since the Late Miocene (3.63 Ma, 95% HPD: 1.64–6.24, node 4). Within HC, the ancestor of HC dispersed from the Palaeotropical region and Sino-Himalayan region into Taiwan (2.7 Ma, 95% HPD: 1.16–4.78, node 5). Taiwan island formed at 5–6 Ma and had been long linked to the Chinese mainland via Fujian-Taiwan land bridge (Teng, 1990; Zeng, 1993), which led to the close relationship between flora of Taiwan and mainland China (e.g., Zeng, 1993; Hsieh, 2002; Nie et al., 2007; Ying and Chen, 2011; Xiang et al., 2017). Wang (1992) proposed that plant lineages could extend from Southwest China eastward along the Nanling Corridor and along other mountain chains in Central China to East China or Taiwan. The biogeography of Cycas taitungensis (Huang et al., 2001), Sassafras tzumu (Nie et al., 2007), and Dichocarpum basilare (Xiang et al., 2017) showed that dispersal is attributed to the disjunct distribution between Asian mainland and Taiwan. Holcoglossum lineages may also have dispersed eastward along the same route to east China and then across the Fujian-Taiwan land bridge to Taiwan. Conversely, the global cooling and the subsequent rise of sea level caused the interruption of the continuous distribution of Holcoglossum since the latest Pliocene. The Palaeotropical Holcoglossum lingulatum and Sino-Japanese H. omeiense split at 0.56 Ma (95%: 0.11–1.17, node 6) (Figure 4). The global cooling and frequently glacial periods happened at Quaternary, which resulted in H. omeiense being restricted in Omei Mountains in China and H. lingulatum in Southeast Asia.
Within AC, the ancestor of AC migrated from the Palaeotropical region and Sino-Himalayan region to the Sino-Japanese region (1.22 Ma, 95% HPD: 0.5–2.36, node 8). All five currently recognized species distribute in Hengduan Mountains, except for H. flavescens, which extend to the Sino-Japanese region. These species are inferred to diverge rapidly during a short time interval (<1 Ma; Figure 4). The time is markedly later than the time of the Hengduan Mountains Region uplift (around 4–3 Ma; Chen, 1992, 1996; Shi et al., 1998). However, the uplift of the Hengduan Mountains formed a complex range of topographies, climates, and habitats (Hoorn et al., 2013), and many areas of this region are ice-free during Quaternary glaciations (Zheng et al., 1998), which can allow immigrations and survivals of plant lineages. Holcoglossum species of AC clade grow on the trees of evergreen broad-leaved forests, especially Quercus section Heterobalanus (Jin, 2003). The continuous uplift of Himalaya-Hengduan Mountains in the Late Miocene to Early Pliocene triggered the diversification of Quercus section Heterobalanus (Meng et al., 2017), which provided suitable habits for Holcoglossum in the Hengduan Mountains. Extensive biogeographical researches proved that the uplift of the Hengduan Mountains directly or indirectly plays vital roles in the species diversification in multiple lineages, such as Rhodiola (Crassulaceae; Zhang et al., 2014), Rhododendron (Eriaceae; Xing and Ree, 2017), Saxifraga (Saxifragaceae; Ebersbach et al., 2017), and alpine bamboo (Ye et al., 2019). The divergence of Holcoglossum still supported that the Hengduan Mountains Region is an adaptive plant diversification center in the Quaternary climate oscillations (Boufford, 2014; Jian et al., 2015; Xing and Ree, 2017).
Conclusion
In this study, we reconstructed a phylogenetic tree and estimated divergent times of Holcoglossum, a genus endemic to East Asia. Our biogeographic inference indicated that the MRCA of Holcoglossum occurred in the Palaeotropical region. Four dispersal events happened from the Palaeotropical region to the Sino-Himalayan region at ca. 3.63 Ma, from Chinese Mainland to Taiwan at ca. 2.7 Ma, from the Sino-Himalayan and Palaeotropical to Sino-Japanese regions at ca. 1.22 Ma, and from the Palaeotropical to Sino-Japanese regions at ca. 0.5 Ma. During Quaternary, the global cooling climate caused the H. pumilum and H. quasipinifolium endemic in Taiwan and the restriction H. omeiense in the Omei Mountains of China. The temporal and spatial patterns of Holcoglossum shed light on the divergence and diversification of recently evolved epiphytic plants within East Asia.
Data Availability Statement
The datasets analyzed for this study can be found in the GenBank Database under the accession numbers listed in the Supplementary Material.
Author Contributions
XX and XJ conceived and designed the experiments. XX performed the experiments. XX, JZ, XL, PZ, and LZ analyzed the data. JZ, XX, XL, PZ, and LZ wrote the manuscript. XX, JZ, XL, PZ, LZ, and XJ revised the draft. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (31300181 and 31670212) and the National Natural Science Foundation of China–Yunnan Joint Fund Project (U1802242). XX is supported by the Youth Innovation Promotion Association Foundation of the Chinese Academy of Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00245/full#supplementary-material
References
An, Z. S., Kutzbach, J. E., Prell, W. L., and Porter, W. L. (2001). Evolution of Asian monsoons and phased uplift of the himalaya-tibetan plateau since late miocene times. Nature 411, 62–66. doi: 10.1038/35075035
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797.
Chase, M. K., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Chen, F. B. (1992). Hengduan event: an important tectonic event of the late Cenozoic in Eastern Asia. Mountain Res. 10, 195–202.
Chen, Y. S., Deng, T., Zhou, Z., and Sun, H. (2018). Is the East Asian flora ancient or not? Natl. Sci. Rev. 5, 920–932. doi: 10.1093/nsx/nwx156
Christenson, E. A. (1998). Two new species of Holcoglossum Schltr. (Orchidaceae: Aeridinae) from China. Lindleyana 13, 121–124.
Clements, R., Sodhi, N. S., Schilthuizen, M., and Ng, P. K. (2006). Limestone karsts of southeast Asia: imperiled arks of biodiverisity. BioScience 56, 733–742.
Demesure, B., Sodzi, N., and Pettit, R. J. (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol. Ecol. 4, 129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x
Drummond, A. J., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Ebersbach, J., Schnitzler, J., Favre, A., and Muellner-Riehl, A. N. (2017). Evolutionary radiations in the species-rich mountain genus Saxifraga L. BMC Evol. Biol. 17:119. doi: 10.1186/s12862-017-0967-2
Fan, J., Qin, H. N., Li, D. Z., and Jin, X. H. (2009). Molecular phylogeny and biogeography of Holcoglossum (Orchidaceae: Aeridinae) based on nuclear ITS, and chloroplast trnL-F and matK. Taxon 58, 849–861. doi: 10.1371/journal.pone.0024864
Farris, J. S., Källersjö, M., Kluge, A. G., and Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319. doi: 10.1006/clad.1994.1021
Goldman, D. H., Freudenstein, J. V., Kores, P. J., Molvray, M., Jarrell, D. C., Whitten, W. M., et al. (2001). Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Syst. Bot. 2, 670–695. doi: 10.1043/0363-6445-26.3.670
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid. S. 41, 95–98.
Hidayat, T., Weston, P. H., Yukawa, T., Ito, M., and Rice, R. (2012). Phylogeny of subtribe Aeridinae (Orchidaceae) inferred from DNA sequences data: advanced analyses including Australasian genera. J. Teeknol. 59, 87–95. doi: 10.11113/jt.v59.1591
Ho, S. Y. W. (2007). Calibrating molecular estimates of substitution rates and divergence times in bird. J. Avian Biol. 38, 409–414. doi: 10.1111/j.0908-8857.2007.04168.x
Hoorn, C., Mosbrugger, V., Mulch, A., and Antonelli, A. (2013). Biodiversity from mountain building. Nat. Geosci. 6, 154–154. doi: 10.1038/ngeo1742
Hsieh, C. F. (2002). Composition, endemism and phytogeographical affinities of the Taiwan flora. Taiwannia 47, 298–310.
Huang, S., Chiang, Y. C., Schaal, B. A., Chou, C. H., and Chiang, T. Y. (2001). Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol. Ecol. 10, 2669–2681. doi: 10.1046/j.0962-1083.2001.01395.x
Jian, H. Y., Tang, K. X., and Sun, H. (2015). Phylogeography of Rosa soulieana (Rosaceae) in the Hengduan mountains: refugia and ‘melting’ pots in the Quaternary climate oscillations. Plant. Syst. Evol. 301, 1819–1830. doi: 10.1007/s00606-015-1195-0
Jiang, D. C., Klaus, S., Zhang, Y. P., Hillis, D. M., and Li, J. T. (2019). Asymmetric biotic interchange across the Bering land bridge between Eurasia and North America. Nat. Sci. Rev. 6, 739–754. doi: 10.1093/nsr/nwz035
Jin, X. H. (2003). Systematics on the Genus Holcgolossum Schltr. (Orchidaceae). Thesis, Chinese Academy of Sciences, Beijing.
Jin, X. H., and Wood, J. J. (2009). “Holcoglossum,” in Flora of China, eds Z. Y. Wu and P. H. Raven (Beijing: Missouri Botanical Garden Press), 499–502.
Li, Z. H., Ma, X., Wang, D. Y., Li, Y. X., Wang, C. W., and Jin, X. H. (2019). Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Plant Biol. 19:63. doi: 10.1186/s12862-019-1384-5
Luo, D., Yue, J. P., Sun, W. G., Xu, B., Li, Z. M., Comes, H. P., et al. (2015). Evolutionary history of the subnival flora of the Himalaya-Hengduan mountains: first insights from comparative phylogeography of four perennial herbs. J. Biogeogr. 43, 31–43. doi: 10.1111/jbi.12610
Meng, H. H., Su, T., Gao, X. Y., Li, J., Jiang, X. L., Sun, H., et al. (2017). Warm-cold colonization: response of oaks to uplift of the Himalaya-Hengduan Mountains. Mol. Ecol. 26, 3276–3294. doi: 10.1111/mec.14092
Nie, Z. L., Wen, J., and Sun, H. (2007). Phylogeny and biogeography of Sassafras (Lauraceae) disjunct between eastern Asia and eastern North America. Pl. Syst. Evol. 267, 191–203. doi: 10.1007/s00606-007-0550-1
Posada, D. L., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pridgeon, A. M., Cribb, P. J., Chase, M. W., and Rasmussen, F. N. (2014). Genera Orchidacearum Epidendroideae, Part III. Vol. 6. Oxford: Oxford University Press.
Qian, H., Ricklefs, R. E., and White, P. S. (2005). Beta diversity of angiosperms in temperate floras of eastern Asia and eastern North America. Ecol. Lett. 8, 15–22. doi: 10.1111/j.1461-0248.2004.00682.x
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Rambaut, A., and Drummond, A. J. (2007). Tracer v1.5 [online]. Available online at: http://beast.bio.ed.ac.uk/Tracer (accessed November 30, 2009).
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schlechter, R. (1919). Orchideologiae Sino-Japonicae Prodromus. Feddes. Repert. Spec. Nov. Regni. Veg. Beih. 4:285.
Shaw, J., Lickey, E. B., Beck, J. T., Farmer, S. B., Liu, W. S., Miller, J., et al. (2005). The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer. J. Bot. 92, 142–166. doi: 10.3732/ajb.92.1.142
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Shi, Y. F., Li, J. J., and Li, B. Y. (1998). Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the Late Cenozoic. Guangzhou: Guangdong Science and Technology Press.
Sun, X. J., and Wang, P. X. (2005). How old is the Asian monsoon system? – Palaeobotanical records from China. Palaeogeogr. Palaeoclimat. Palaeoecol. 222, 181–222. doi: 10.1016/j.palaeo.2005.03.005
Swofford, D. L. (2003). PAUP∗: Phylogenetic Analysis Using Parsimony (∗ and related methods), Version 4.0b10. Sunderland: Sinauer Associates.
Teng, L. S. (1990). Geotectonic evolution of late Cenozoic arc-continent collision in Taiwan. Tectonophysics 183, 57–76. doi: 10.1016/0040-1951(90)90188-e
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids. Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tiffney, B. H. (1985). Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arbor. 66, 73–94. doi: 10.5962/bhl.part.13179
Topik, H., Yukawa, T., and Ito, M. (2005). Molecular phylogenetics of subtribe Aeridinae (Orchidaceae): insights from plastid matK and nuclear ribosomal ITS sequences. J. Plant. Res. 118, 271–284. doi: 10.1007/s10265-005-0217-3
Wang, W. T. (1992). On some disjunction patterns and some migration routes found in the east Asiatic region. Acta Phytotax. Sin. 30, 1–24.
Wen, J. (1999). Evolutionary of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst. 30, 421–455. doi: 10.1146/annurev.ecolsys.30.1.421
Wu, Z. Y., and Wu, S. (1998). “A proposal for a new floristic kingdom (realm) the East Asiatic Kingdom, its delineation and characteristics,” in Floristic Characteristics and diversity of East Asian Plants, eds A. L. Zhang and S. G. Wu (Beijing: Higher Education Press and Springer Verlag Press), 1–42.
Wu, Z. Y., Zhou, Z. K., Li, D. Z., Peng, H., and Sun, H. (2003). The areal types of the world families of seed plants. Acta Bot. Yunnanica 25, 245–257. doi: 10.1023/A:1022289509702
Xiang, K. L., Zhao, L., Erst, A. S., Yu, S. X., Jabbour, F., and Wang, W. (2017). A molecular phylogeny of Dichocarpum (Ranunculaceae): implications for eastern Asian biogeography. Mol. Phylogenet. Evol. 107, 594–604. doi: 10.1016/j.ympev.2016.12.026
Xiang, X. G., Li, D. Z., Jin, X. H., Hu, H., Zhou, H. L., Jin, W. T., et al. (2012). Monophyly or paraphyly-the taxonomy of Holcoglossum (Aeridinae: Orchidaceae). PLoS One 7:e52050. doi: 10.1371/journal.pone.0052050
Xiang, X. G., Mi, X. C., Zhou, H. L., Li, J. W., Chung, S. W., Li, D. Z., et al. (2016). Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr. 43, 1310–1323. doi: 10.1111/jbi.12726
Xiang, X. G., Xiang, K. L., Ortiz, R. D. C., Jabbour, F., and Wang, W. (2019). Intergrating palaeontological and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamelidaceae. J. Biogeogr. 46, 2622–2631. doi: 10.1111/jbi.13690
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.161606311
Ye, X. Y., Ma, P. F., Yang, G. Q., Guo, C., Zhang, Y. X., Chen, Y. M., et al. (2019). Rapid diversification of alpine bamboos associated with the uplift of the Hengduan Mountains. J. Biogeogr. 46, 2678–2689. doi: 10.1111/jbi.13723
Ying, T. S. (2001). Species diversity and distribution pattern of seed plants in China. Biodiver. Sci. 9, 393–398. doi: 10.17520/biods.1999021
Ying, T. S., and Chen, M. L. (2011). Plant Geography of China. Shanghai: Shanghai Scientific & Technical Publishers, 290–342.
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Zachos, J. C., Dickens, G. R., and Zeebe, R. E. (2008). An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451, 279–283. doi: 10.1038/nature06588
Zeng, W. B. (1993). The passageway of the flora migration on both sides of the Taiwan strait in Pleistocene epoch. Acta Bot. Yunnanica 16, 107–110.
Zhang, J. Q., Meng, S. Y., Allen, G. A., Wen, J., and Rao, G. Y. (2014). Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol. Phylogenet. Evol. 77, 147–158. doi: 10.1016/j.ympev.2014.04.013
Zhang, Q. Q., Ferguson, D. K., Mosbrugger, V., Wang, Y. F., and Li, C. S. (2012). Vegetation and climatic changes of SW China in response to the uplift of Tibetan Plateau. Palaeogeogr. Palaeocl. 363, 23–36. doi: 10.1016/j.palaeo.2012.08.009
Keywords: biogeography, East Asia, Holcoglossum, molecular dating, phylogeny
Citation: Zhao J, Zhou P, Li X, Zhang L, Jin X and Xiang X (2020) Temporal and Spatial Pattern of Holcoglossum Schltr. (Orchidaceae), an East Asian Endemic Genus, Based on Nuclear and Chloroplast Genes. Front. Ecol. Evol. 8:245. doi: 10.3389/fevo.2020.00245
Received: 03 February 2020; Accepted: 06 July 2020;
Published: 31 July 2020.
Edited by:
Meng Yao, Peking University, ChinaReviewed by:
Xin Zhou, China Agricultural University, ChinaXiaolei Huang, Fujian Agriculture and Forestry University, China
Copyright © 2020 Zhao, Zhou, Li, Zhang, Jin and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoguo Xiang, eGlhbmd4ZzIwMTBAMTYzLmNvbQ==
 Jiahong Zhao
Jiahong Zhao Peng Zhou
Peng Zhou Xiaoqian Li
Xiaoqian Li Liguo Zhang
Liguo Zhang Xiaohua Jin
Xiaohua Jin Xiaoguo Xiang
Xiaoguo Xiang