Abstract
For protecting endangered species, precise understanding of their distribution is crucial. However, it is often very difficult to estimate at a large scale with conventional methods (e.g., casting nets or electrofishing for aquatic species) because of their low densities in the wild. Sakhalin taimen (Parahucho perryi) is one of the largest and most critically endangered freshwater salmonid fishes in the world. In this study, we applied an environmental DNA (eDNA) detection system for this species to 120 rivers in Hokkaido, the second largest main island of Japan. We successfully detected eDNA from Sakhalin taimen in seven rivers (5.8%). Although these rivers were widely distributed across the island, > 95% of the total amounts of eDNA were detected from region-A and -I, indicating that local populations in the other regions of Hokkaido are very small and on the brink of extinction. In addition, principal component analyses based on the eDNA-based estimation of Sakhalin taimen distribution and GIS revealed their distribution determinants including limited topographic relief of watershed as well as presence of wetlands and lagoons. Our results suggest that eDNA-based detection systems are an efficient means of monitoring the population status of endangered freshwater species at large scales.
Introduction
Precise information about the current distribution and biomass of threatened species is essential for their conservation and appropriate management. Over 30,000 species are currently listed as threatened species in the Red List by the International Union for Conservation of Nature (IUCN), and the number is increasing (International Union for Conservation of Nature and Natural Resources, 2020). While many kinds of threats are considered as factors for extinction (e.g., loss or degradation of habitat, illegal trade, invasive species, or human activity), 85% of the endangered species are facing threats of habitat loss or degradation (Wilcove et al., 1998). These threats potentially accelerate further reduction of population sizes through a process known as an ‘extinction vortex” (Gilpin and Soulé, 1986). However, vast sampling efforts are often required for field surveys with conventional methods (e.g., visual observation or physical capture). In addition, specialized skills are typically needed for species identification, especially in the case of juveniles. Due to these requirements, studies on current distribution of endangered species are often incomplete and may be limited to small parts of a species’ distribution.
The Sakhalin taimen (Parahucho perryi) has been listed as a critically endangered species (CR) in the IUCN Red List since 2006 (Rand, 2006). They are a typical example of an endangered species threatened by habitat loss or degradation due to human activities (Fukushima, 2006; Rand, 2006; Fukushima et al., 2007; Zolotukhin et al., 2013). Historically, they were distributed in the Russian Far East and northern Japan (Rand, 2006; Zolotukhin et al., 2013). Hokkaido is the second largest main island of Japan (83,456 km2), and currently the only island hosting Sakhalin taimen in the country (Fukushima et al., 2011). According to a previous research (Fukushima et al., 2011), Sakhalin taimen populations require various habitats including upstream habitats, estuarine and/or coastal habitats, and lagoons and wetlands through their life history. However, even the best available scientific report on the distribution of this species above suffers from spotty and sometimes anecdotal records across large time spans, bringing some conclusions about the species’ ecology and distribution into doubt.
Environmental DNA (eDNA) techniques have been developed as a new assessment tool for aquatic organisms since 2008 (Ficetola et al., 2008; Fukumoto et al., 2015; Pfleger et al., 2016). These techniques have many advantages for detecting target species in the field, such as objectivity, high detectability, and reduced sampling effort. They have been shown to be especially useful for monitoring endangered species that are otherwise difficult to capture or observe (Fukumoto et al., 2015; Laramie et al., 2015; Pfleger et al., 2016; Carlsson et al., 2017; Maruyama et al., 2018; Atkinson et al., 2019; Iwai et al., 2019). However, the previous studies tended to apply the eDNA techniques to specific freshwater/saltwater systems or to relatively small geographical regions. For comprehensive understanding of distribution of endangered species and its determinants, an applicability of this technique to an investigation in large geographical regions with a variety of environments is crucial.
In this study, we aimed to estimate the distribution and population status of Sakhalin taimen in 120 rivers covering the entirety of Hokkaido, Japan, using an eDNA detection system established during a previous study (Mizumoto et al., 2018). Furthermore, we sought to understand key environmental determinants for the presence/absence of Sakhalin taimen using Geographic Information System (GIS), so that the current distribution could be estimated together with limiting factors for this endangered species. Given the size of Hokkaido (83,450 km2) and heterogeneity in river environments in it, this study provides a textbook example of an eDNA application to understand a wide-range distribution, population status and their environmental determinants of endangered species.
Materials and Methods
Sample Collections
Field sampling was conducted at 120 rivers in Hokkaido, Japan, from 2015 to 2018 (Figure 1 and Supplementary Table A1). Sampling sites were selected around estuaries on each river for the sake of convenience and uniformity of the sampling effort. Because of the Sakhalin taimen’s conservation status and risk of increased fishing pressure on them due to this study, eDNA detection results were projected on a map of the 14 subprefectures (A–N) of Hokkaido.
FIGURE 1
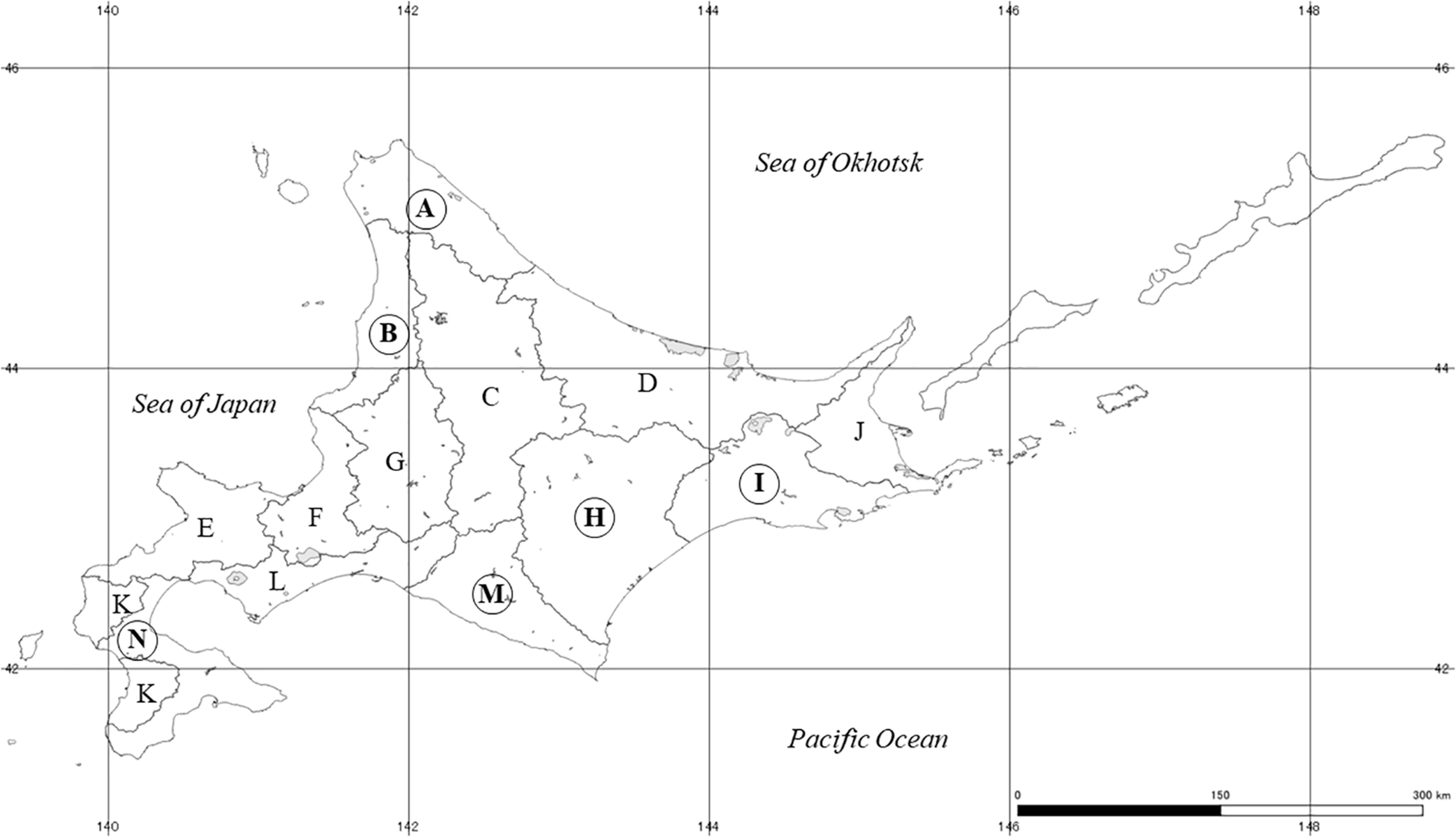
The map of our survey areas. Sampling was conducted from 2015 to 2018 at 120 rivers in total. All sampling sites were located around estuaries, however, detailed geographical information was not presented for the protection and conservation of Sakhalin taimen. Instead a larger scale review was included for the 14 subprefectures (A-N) of Hokkaido. The number of rivers located each region were 16 (region A), 8 (region B), 12 (region D), 12 (region E), 3 (region F), 6 (region H), 16 (region I), 18 (region J), 7 (region L), 8 (region M) and 14 (region N). Circled alphabets represent areas where Sakhalin taimen DNA was detected. The river and coastline data were provided by the National Land Information Division, Ministry of Land, Infrastructure, Transport and Tourism of Japan, under the CC BY 4.0 license.
Filtering procedures are described in Mizumoto et al. (2018). In brief, collected water samples were filtered at field stations or visitor centers near sampling sites as soon as possible. When we estimated that it had taken > 6 h from water collection to filtration, we added benzalkonium chloride (BAC) for preserving DNA at a final concentration of 0.01% (Yamanaka et al., 2017; Supplementary Table A1). At most sampling sites, 2 L of environment water were collected and filtered separately to provide 1 L duplicates, but in some samples, only 500 mL could be filtered due to clogging by fine particles in turbid water. All water samples were filtered through glass-membrane filters (Whatman GF/F, GE Healthcare Japan, Tokyo, Japan) using an aspirator. After filtration, 10 mL of 70% molecular grade ethanol was filtered for preserving DNA in the sample. Negative control samples were collected by filtering 500 mL of sterilized distilled water before filtering each river sample. All filter samples were stored around −20°C at field stations and at –25°C in the lab until further analyses. To prevent cross-contamination, all filtration equipment was bleached using a 6% sodium hypochlorite solution and carefully rinsed with sterilized distilled water after each filtration.
DNA Extraction and qPCR Procedures
DNA extractions were conducted using DNeasy Blood and Tissue kits (Qiagen, Hilden, Germany) following Mizumoto et al. (2018). The final volume of the extracted DNA was 110 μL/sample. qPCRs were carried out in 20 μL volumes with 400 nM of Sakhalin taimen specific primer and probe (Forward-primer: 5′-GCAATAGGCCTCCTATCAACAA-3′, Reverse-primer: 5′-CGAATTGTAAAATTAGTACTATCCATCCA-3′, Probe: 5′-FAM-AGCATACTCTTCAATCGCCCACCT-TAMRA-3′) designed by Mizumoto et al. (2018) in a Brilliant III Ultra-Fast qPCR Master Mix with Low ROX (Agilent Technologies, Inc.) and 2 μL of the extracted DNA as a template sample (Mizumoto et al., 2018). Triplicates of 2 μL were applied to each qPCR reaction on a Stratagene Mx3000P (Agilent Technologies, Inc.) with the same thermal-cycling regime as in the previous study (Mizumoto et al., 2018). For the quantification of environmental samples, we applied synthetic linear DNA of the target region containing 2 × 101, 2 × 102, 2 × 103, 2 × 104 and 2 × 105 copies per tube as standards in all qPCR assays (n = 21, average R2 = 0.992, SD = 0.007, and average PCR efficiency = 0.894, SD = 0.126). In addition, sterilized distilled water samples were applied in all qPCR assays as PCR negative control samples.
Estimation of Distribution and GIS analyses
We accepted sites as positive detections when we had at least one detection in six PCR replicates per site (triplicate PCR reactions x two filters per site). The lowest detection limit was set at “0.01 copy/2 μL∗site.” To adjust for the variation in water volumes we sampled (500 mL or 1,000 mL, see above), estimated DNA concentrations were represented as/1,000 mL of environmental water after averaging over the six replicates. In the following analyses, qPCR outputs (copies/1,000 mL) that were obtained from the same river system’s samples were averaged across the river system (Supplementary Table A1). Ultimately, 116 river systems were used for GIS analyses.
To investigate factors determining the settlement of Sakhalin taimen, we examined 23 environmental variables (18 for natural environment and 5 for human activity) in 116 rivers (Table 1). For this study, we looked into the same variables that were tested in Fukushima et al. (2011) to confirm the current population status of Sakhalin taimen. Among the environmental variables, information on the distribution of marshes were collected from the 5th National Survey on Natural Environment (The Ministry of the Environment, 1995), and the other variable data were provided by the Geospatial Information Authority of Japan (Ministry of Land, Infrastructure, Transport and Tourism, 2020). In general, an aggregation of variables is often used by a mesh as a division unit. However, in order to grasp the habitat of Sakhalin taimen, it is effective to use a regional classification based on basin. The basins of division unit were merged from the basin mesh data (30 × 45 s for latitude and longitude, about 1 km per side) based on information published by Geospatial Information Authority of Japan. For each unit basin created, the variable mesh was calculated as the sum (population, number of lagoons, number of wetlands), average (temperature, precipitation, altitude, slope), or ratio (low land area, land use). The degree of meandering was obtained by dividing the flow path extension from the start point to the end of the longest river in the relevant water system by the linear distance. We used ArcGIS 10.7 (ESRI) to aggregate these environment variables.
TABLE 1
| Variable type | Variable | Unit | Number of variables |
| Natural variables | Drainage area | ha | 1 |
| Elevation | m | 1 | |
| Area below 10, 20, and 30 m | % | 3 | |
| Lagoon presence, number, and area | ha | 3 | |
| Marsh presence, number, and area | ha | 3 | |
| Forest | % | 1 | |
| Slope | Degree | 1 | |
| Annual air temperature (max, min, and average) | °C | 3 | |
| Precipitation | mm | 1 | |
| Degree of meandering | 1 | ||
| Human activity | Dam number and density | 2 | |
| Human population and density | /ha | 2 | |
| Farmland | % | 1 |
Summary of the 23 environmental variables (18 for natural environment and 5 for human activity) used for evaluation of 116 rivers.
Marsh distribution was collected from the 5th National Survey on Natural Environment (The Ministry of the Environment, 1995), and the other variables were prepared by the Geospatial Information Authority of Japan (Ministry of Land, Infrastructure, Transport and Tourism, 2020).
To summarize the aggregated environmental variables into characteristic environments, we used principal component analysis (PCA). From the obtained PCA scores, we performed cluster analyses using the k-means method and categorized the rivers. The number of clusters was determined using the largest score according to the Calinski-Harabasz score. Statistical analyses were performed using R3.6.1 (R Core Team, 2019), cluster analyses were performed using Package “cclust,” and the Calinski-Harabasz score was calculated using Package “vegan.”
Results
We detected eDNA of Sakhalin taimen in 7 of 120 rivers in Hokkaido (5.8%). No detection was observed in any negative control samples. Among the 14 subprefectures of Hokkaido (A-N), two sites were positive in region-A, and one site was positive in region-B, -H, -I, -M, and -N (Table 2). Three of the seven river systems with positive eDNA detection had no previous reports of Sakhalin taimen, whereas Sakhalin taimen in the other four river systems were previously reported (Fukushima et al., 2011). Among the eDNA detections in the seven river systems, the estimated copy numbers varied largely (Table 3). The highest number of DNA copies were obtained in river-A-11 and -I-2, being consistent with healthy populations of Sakhalin taimen reported previously in these river systems (Fukushima et al., 2011).
TABLE 2
| Region | Detection | Copies | Ratio |
| A | 2/16 | 351.69 | 56.36% |
| B | 1/8 | 11.73 | 1.88% |
| D | 0/12 | – | – |
| E | 0/12 | – | – |
| F | 0/3 | – | – |
| H | 1/6 | 0.67 | 0.11% |
| I | 1/16 | 246.40 | 39.49% |
| J | 0/18 | – | – |
| L | 0/7 | – | – |
| M | 1/8 | 8.89 | 1.42% |
| N | 1/14 | 4.62 | 0.74% |
| Total | 7/120 | 624.01 | 100% |
Summary of the results of qPCR from 120 river water samples.
Copies represent the sum of the estimated DNA copy numbers per 1,000 mL from each site. Three of the seven river systems with positive eDNA detections from region-B, -M, and -N had no previous reports of Sakhalin taimen.
TABLE 3
| River | Detection | eDNA concentration (SD) | Ratio |
| A-8 | 4/6 | 103.92 (105.18) | 16.65% |
| A-11 | 4/6 | 247.78 (190.10) | 39.71% |
| B-5 | 1/6 | 11.73 (26.24) | 1.88% |
| H-4 | 1/6 | 0.67 (1.49) | 0.11% |
| I-2 | 4/6 | 246.40 (289.71) | 39.49% |
| M-6 | 1/6 | 8.89 (19.88) | 1.42% |
| N-14 | 1/6 | 4.62 (10.34) | 0.74% |
Results of qPCR from 7 rivers where Sakhalin taimen DNA was positively detected.
River names were reassigned with alphanumeric codes (A-8, A-11, B-5, H-4, I-2, M-6, and N-14) based on the region where these rivers were located. Detection represents the number of positive detections from six qPCR replicates at each site and eDNA concentration represents the estimated DNA copy number per 1,000 mL from each site. SD represents standard deviation. Ratio represents contribution rate for the total estimated eDNA concentration.
Among the results of GIS-based PCA, percentages of the top three explained variances were 29.3, 18.2, and 10.4% (illustrated as Dimension 1, 2, and 3, respectively, in Figure 2). Regarding the three explained variances, ten parameters (slope, below 30 m, forest, below 20, elevation, wetland presence, below 10 m, lagoon presence, precipitation, and farmland) that affected geographical flatness had high variance contributions in dimension 1, six parameters (drainage area, dam number, human population, wetland number, minimum air temperature and average air temperature) affecting river scales had high variance contributions in dimension 2, and seven parameters (average air temperature, minimum air temperature, maximum air temperature, dam density, dam number, wetland number, and human population) affecting climates had high variance contributions in dimension 3 (Figure 2). Forest appeared to have a negative effect of Sakhalin taimen’s presence although it was estimated to have a positive effect in the previous study (Fukushima et al., 2011). In contrast, the number of dams and farmland appeared to have positive effects of Sakhalin taimen’s presence, whereas these factors were estimated to have negative effects in the previous study (Fukushima et al., 2011).
FIGURE 2
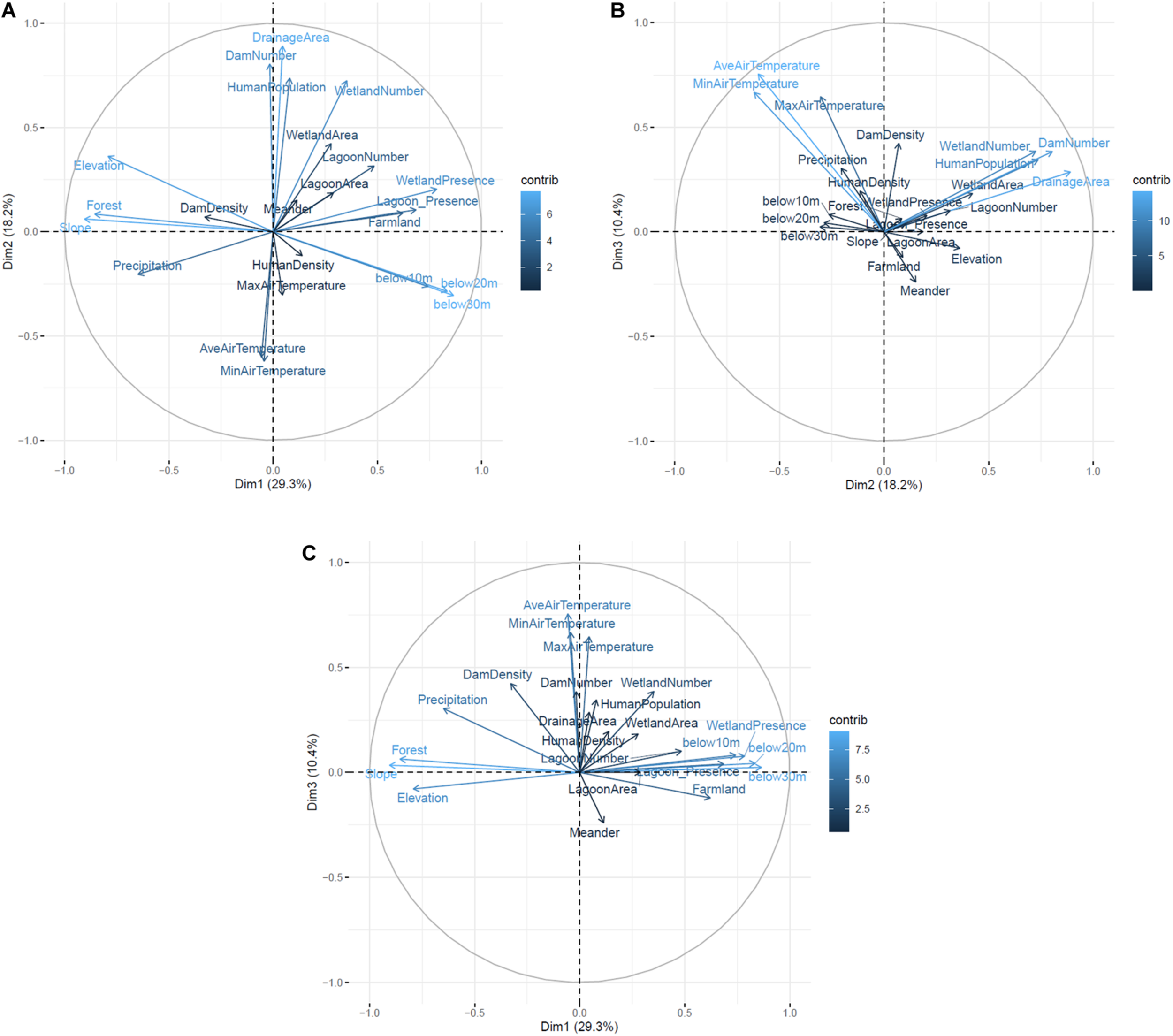
Results of the GIS-based PCA. The top three explained variances (Dimensions 1–3) were described bidimensionally in each combination [(A) Dimension 1 vs. 2, (B) Dimension 2 vs. 3, (C) Dimension 1 vs. 3]. Blue colored gradation and arrow length represent the contribution percentages and arrow direction represents directions of effects onto each explained variance.
Discussion
In this study, we applied an eDNA detection system to estimate the distribution of a critically endangered salmonid species, Sakhalin taimen (P. perryi), across Hokkaido, Japan. As a result, we detected Sakhalin taimen eDNA in seven out of 120 rivers in Hokkaido. Of the seven rivers where Sakhalin taimen’s eDNA were successfully detected, three rivers had no previous reports (river-B-5, -M-6, and -N-14, Tables 2, 3).
Tank experiments in our previous study (Mizumoto et al., 2018) revealed a high sensitivity of the Sakhalin taimen eDNA detection system and its potential for biomass estimation in flowing waters. In general, however, the detectability of eDNA from aquatic organisms is affected by both biotic and abiotic factors such as the range from the target species, sizes and density of them, water temperature, pH, water current, flow volume and so on (e.g., Barnes et al., 2014; Song et al., 2017; Maruyama et al., 2018; Kasai et al., 2020). Therefore, actual detectability of Sakhalin taimen’s eDNA in the natural river systems would rather be limited, and we are almost certain that we missed eDNA detections from some river systems, especially from large rivers or areas with low density of Sakhalin taimen.
For the biomass estimation, eDNA concentration is considered as a relatively good proxy of biomass according to previous studies (Takahara et al., 2012; Doi et al., 2017). If we assumed that the eDNA was evenly distributed within each river and that the estimated eDNA concentrations reflected the relative biomass of this species among rivers, the distributions of Sakhalin taimen in this island should be considered to have a serious bias (> 95% in region-A and -I, Table 3). Interestingly, these two regions were previously suggested to support the most stable Sakhalin taimen populations in Japan (Fukushima et al., 2011). For better understanding of fine-scale distribution within/among river systems, however, further studies with more eDNA samplings and/or with conventional sampling methods would be required.
The GIS-based PCA revealed that top three dimensions (dimensions 1–3) had effects on geographical flatness, river scales, and climates (Figure 2). Sakhalin taimen is known as a long-living freshwater fish, and river connectivity between upstream and estuarine habitats is essential for them to complete their life history (for spawning, growing foraging and wintering. (Fukushima, 1994; Edo et al., 2000; Esteve et al., 2009; Honda et al., 2012, 2014, 2017). According to Fukushima et al. (2011), Sakhalin taimen are more likely to persist in rivers with low and flat areas like wetlands or lagoons, and average air temperature combined with agricultural development were cited as seriously contributing to their risk of extinction. The results of our 2015–2018 eDNA survey were generally consistent with previous reports. Namely, they provided a further support for the importance of wetlands and lagoons for the protection of endangered species.
However, some parameters showed non-consistent results between the two studies (e.g., forest, the number of dams, farmland). There are two potential explanations on them; one is the temporal changes of the restrictions for their presence from the previous study (Fukushima et al., 2011) to the present one. For example, Fukushima et al. (2019) reported that there was a Sakhalin taimen population that was landlocked by a huge reservoir in northern Hokkaido, and this population potentially helped sustain the metapopulation dynamics of them at the watershed scale. In addition, these kinds of artificial barriers can be the restrictions of invasive species (Sharov, 2004; Vélez-Espino et al., 2011). In Hokkaido, Sakhalin taimen is considered to be threatened by the impacts of introduced rainbow trout (Oncorhynchus mykiss) because rainbow trout disturbed Sakhalin taimen’s spawning beds (Nomoto et al., 2010), therefore these kinds of barriers might restrict invasion of rainbow trout. Considering these potential effects of barriers on Sakhalin taimen, the effect of dams on the presence of Sakhalin taimen that were estimated in the previous study may change with the times. However, the effects of artificial barriers should be carefully considered in conservation programs because artificial barriers can also be restrictions of Sakhalin taimen’s migration (Fukushima et al., 2019). Another possible explanation is differently estimated distributions of the species between the two studies. Three rivers (river-B-5, -M-6, and -N-14) were newly suggested to hold Sakhalin taimen populations, and one river was reassessed to have avoided extinction (river-H-4). The difference might influence the determinant evaluation because of the added regional variations, for example, river-N-14 was located in southern Hokkaido where had relatively warmer air temperature (Max, Min and Ave) than the other regions in this study. In addition, river-H-4 had the largest values of drainage area and human population, and the second largest number of dams in this study. Regardless, more sensitive filtration system and/or more frequently spatio-temporal sampling are still needed to help estimating their distribution determinants more accurately.
Conclusion
In conclusion, we estimated the current distribution of Sakhalin taimen in Hokkaido and illustrated some of the population’s main vulnerabilities. While conventional capture surveys or visual observations can provide data on individuals of a target species (e.g., body size, sex, and age), our study shows that eDNA surveys have strong advantages for collecting population-level data on endangered species (e.g., distribution, population status, and threats of extinction) both non-invasively and objectively compared to traditional methods. Assuming that estimated DNA concentrations are reflective of a target species’ relative biomass, we believe that the Sakhalin taimen’s biomass distribution in Japan was heavily skewed into a few regions during our sampling. In drawing these conclusions, we believe that eDNA technologies might not only help establish species’ distribution, but may also show the spatial distribution of biomass and therefore infer population health over large scales. These findings should help to optimize conservation efforts for wildlife populations that are facing critical threats.
Statements
Data availability statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the animal study because we did not find/touch any animals.
Author contributions
HM and HA conceived ideas and collected samples. HM wrote the manuscript. HM and TM collected data and conceived models. TM conducted data analyses and visualized analyzed data. HA acquired funding and gave final approval for publication. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Asahi Glass Foundation, Environment Research and Technology Development Fund (4-1602 and JPMEERF20204004) of the Environmental Restoration and Conservation Agency, Japan, and JSPS KAKENHI grant no. 17H03623 (HA).
Acknowledgments
We thank K. Osanai, M. Kawahara, M. Fukushima, and the other members of the Sarufutsu Itou Conservation Council for supporting our study. We also thank Dr. M. Nakaoka and the other members of the Akkeshi Marine Station of the Field Science Center for Northern Biosphere Hokkaido University, Dr. K. Miyashita and the other members of the Field Science Center for Northern Biosphere Aquatic Research Station Division of Marine Bioresource and Environmental Science, and K. Takahashi and the other members of the Council of Biodiversity in Shiribeshi Region for supporting our sampling in Hokkaido. For English corrections and comments, we thank T. E. Squires. We declare that the experiments complied strictly with the current laws in Japan.
Conflict of interest
TM was employed by the company Pacific Consultants Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.569425/full#supplementary-material
References
1
Atkinson S. Carlsson J. E. Ball B. Kelly-Quinn M. Carlsson J. (2019). Field application of an eDNA assay for the threatened white-clawed crayfish Austropotamobius pallipes.Freshw. Sci.38503–509. 10.1086/704712
2
Barnes M. A. Turner C. R. Jerde C. L. Renshaw M. A. Chadderton W. L. Lodge D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems.Environ. Sci. Technol.481819–1827. 10.1021/es404734p
3
Carlsson J. E. Egan D. Collins P. C. Farrell E. D. Igoe F. Carlsson J. (2017). A qPCR MGB probe based eDNA assay for European freshwater pearl mussel (Margaritifera margaritifera L.).Aquat. Conserv. Mar. Freshw. Ecosyst.271341–1344. 10.1002/aqc.2788
4
Doi H. Inui R. Akamatsu Y. Kanno K. Yamanaka H. Takahara T. et al (2017). Environmental DNA analysis for estimating the abundance and biomass of stream fish.Freshw. Biol.6230–39. 10.1111/fwb.12846
5
Edo K. Kawamula H. Higashi S. (2000). The structure and dimensions of redds and egg pockets of the endangered salmonid, Sakhalin taimen.J. Fish. Biol.56890–904. 10.1111/j.1095-8649.2000.tb00879.x
6
Esteve M. Gilroy D. McLennan D. A. (2009). Spawning behaviour of taimen (Hucho taimen) from the Uur River, Northern Mongolia.Environ. Biol. Fish.84185–189. 10.1007/s10641-008-9407-x
7
Ficetola G. F. Miaud C. Pompanon F. Taberlet P. (2008). Species detection using environmental DNA from water samples.Biol. Lett.4423–425. 10.1098/rsbl.2008.0118
8
Fukumoto S. Ushimaru A. Minamoto T. (2015). A basin-scale application of environmental DNA assessment for rare endemic species and closely related exotic species in rivers: a case study of giant salamanders in Japan.J. Appl. Ecol.52358–365. 10.1111/1365-2664.12392
9
Fukushima M. (1994). Spawning migration and redd construction of Sakhalin taimen, Hucho perryi (Salmonidae) on northern Hokkaido Island, Japan.J. Fish. Biol.44877–888. 10.1111/j.1095-8649.1994.tb01261.x
10
Fukushima M. (2006). The effects of damming on masu salmon and the Sakhalin taimen and the assessment of their conservation areas based on predictive habitat models.Ecol. Civil Eng.8233–244.
11
Fukushima M. Harada C. Yamakawa A. Iizuka T. (2019). Anadromy sustained in the artificially land-locked population of Sakhalin taimen in northern Japan.Environ. Biol. Fish.1021219–1230. 10.1007/s10641-019-00904-4
12
Fukushima M. Kameyama S. Kaneko M. Nakao K. Ashley S. E. (2007). Modelling the effects of dams on freshwater fish distributions in Hokkaido, Japan.Freshw. Biol.521511–1524. 10.1111/j.1365-2427.2007.01783.x
13
Fukushima M. Shimazaki H. Rand P. S. Kaeriyama M. (2011). Reconstructing Sakhalin taimen Parahucho perryi historical distribution and identifying causes for local extinctions.Trans. Am. Fish. Soc.1401–13.
14
Gilpin M. E. Soulé M. E. (1986). “Minimal viable populations: processes of species extinction,” in Conservation Biology: The Science of Scarcity and Diversity, ed.SouléM. E. (Sunderland, MA: Sinauer Associates Inc), 19–34.
15
Honda K. Kagiwada H. Takahashi N. Miyashita K. (2012). Seasonal stream habitat of adult Sakhalin taimen, Parahucho perryi, in the Bekanbeushi River system, eastern Hokkaido, Japan.Ecol. Freshw. Fish.21640–657. 10.1111/j.1600-0633.2012.00585.x
16
Honda K. Kagiwada H. Takahashi N. Miyashita K. (2014). Movement patterns of adult Sakhalin taimen, Parahucho perryi, between stream habitats of the Bekanbeushi River system, eastern Hokkaido, Japan.Ichthyol. Res.61142–151. 10.1007/s10228-013-0387-2
17
Honda K. Takahashi N. Yamamoto K. Kagiwada H. Tsuda Y. Mitani Y. et al (2017). First documentation of detailed behaviors of endangered adult Sakhalin taimen Parahucho perryi in the Bekanbeushi River system, eastern Hokkaido, Japan, using bio-logging and acoustic telemetry concurrently.Ichthyol. Res.64357–364. 10.1007/s10228-016-0570-3
18
International Union for Conservation of Nature and Natural Resources (2020). The IUCN Red List of Threatened Species classified in CR, EN and VU. Available online at: http://www.iucnredlist.org/search(accessed February 20, 2020).
19
Iwai N. Yasumiba K. Takahara T. (2019). Efficacy of environmental DNA to detect and quantify stream tadpoles of Odorrana splendida.R. Soc. Open Sci.6:181798. 10.1098/rsos.181798
20
Kasai A. Takada S. Yamazaki A. Masuda R. Yamanaka H. (2020). The effect of temperature on environmental DNA degradation of Japanese eel.Fish. Sci.86465–471. 10.1007/s12562-020-01409-1
21
Laramie M. B. Pilliod D. S. Goldberg C. S. (2015). Characterizing the distribution of an endangered salmonid using environmental DNA analysis.Biol. Conserv.18329–37. 10.1016/j.biocon.2014.11.025
22
Maruyama A. Sugatani K. Watanabe K. Yamanaka H. Imamura A. (2018). Environmental DNA analysis as a non-invasive quantitative tool for reproductive migration of a threatened endemic fish in rivers.Ecol. Evol.811964–11974. 10.1002/ece3.4653
23
Ministry of Land, Infrastructure, Transport and Tourism (2020). National Regional Planning Bureau: Digital National Land Information. Available online at: http://nlftp.mlit.go.jp/ksj-e/gml/gml_datalist.html(accessed February 20, 2020).
24
Mizumoto H. Urabe H. Kanbe T. Fukushima M. Araki H. (2018). Establishing an environmental DNA method to detect and estimate the biomass of Sakhalin taimen, a critically endangered Asian salmonid.Limnology19219–227. 10.1007/s10201-017-0535-x
25
Nomoto K. Omiya H. Sugimoto T. Akiba K. Edo K. Higashi S. (2010). Potential negative impacts of introduced rainbow trout on endangered Sakhalin taimen through redd disturbance in an agricultural stream, eastern Hokkaido.Ecol. Freshw. Fish.19116–126. 10.1111/j.1600-0633.2009.00396.x
26
Pfleger M. O. Rider S. J. Johnston C. E. Janosik A. M. (2016). Saving the doomed: using eDNA to aid in detection of rare sturgeon for conservation (Acipenseridae).Glob. Ecol. Conserv.899–107. 10.1016/j.gecco.2016.08.008
27
R Core Team (2019). R: A Language and Environment for Statistical Computing.Vienna: R Foundation for Statistical Computing.
28
Rand P. S. (2006). Hucho perryi: IUCN (International Union for the Conservation of Nature) 2020 Red List of Threatened Species, Version 2020-1. IUCN. Available online at: https://www.iucnredlist.org/species/61333/12462795(accessed February 20, 2020).
29
Sharov A. A. (2004). Bioeconomics of managing the spread of exotic pest species with barrier zones.Risk Anal.24879–892. 10.1111/j.0272-4332.2004.00486.x
30
Song J. W. Small M. J. Casman E. A. (2017). Making sense of the noise: the effect of hydrology on silver carp eDNA detection in the Chicago area waterway system.Sci. Total Environ.605713–720. 10.1016/j.scitotenv.2017.06.255
31
Takahara T. Minamoto T. Yamanaka H. Doi H. Kawabata Z. I. (2012). Estimation of fish biomass using environmental DNA.PLoS One7:e35868. 10.1371/journal.pone.0035868
32
The Ministry of the Environment (1995). Report of Wetland Survey, the 5th National Survey On The Natural Environment. Available online at: http://www.biodic.go.jp/reports3/5th/5_wetland/5_wetland.pdf in Japanese (accessed February 20, 2020).
33
Vélez-Espino L. A. McLaughlin R. L. Jones M. L. Pratt T. C. (2011). Demographic analysis of trade-offs with deliberate fragmentation of streams: control of invasive species versus protection of native species.Biol. Conserv.1441068–1080. 10.1016/j.biocon.2010.12.026
34
Wilcove D. S. Rothstein D. Dubow J. Phillips A. Losos E. (1998). Quantifying threats to imperiled species in the United States.Bioscience48607–615. 10.2307/1313420
35
Yamanaka H. Minamoto T. Matsuura J. Sakurai S. Tsuji S. Motozawa H. et al (2017). A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant.Limnology18233–241. 10.1007/s10201-016-0508-5
36
Zolotukhin S. Makeev S. Semenchenko A. (2013). Current status of the Sakhalin taimen, Parahucho perryi (Brevoort), on the mainland coast of the Sea of Japan and the Okhotsk Sea.Arch. Pol. Fish.21205–210.
Summary
Keywords
environmental DNA, endangered species, Sakhalin taimen, distribution, habitat degradation
Citation
Mizumoto H, Mitsuzuka T and Araki H (2020) An Environmental DNA Survey on Distribution of an Endangered Salmonid Species, Parahucho perryi, in Hokkaido, Japan. Front. Ecol. Evol. 8:569425. doi: 10.3389/fevo.2020.569425
Received
04 June 2020
Accepted
09 October 2020
Published
06 November 2020
Volume
8 - 2020
Edited by
Matthew A. Barnes, Texas Tech University, United States
Reviewed by
Peter Rand, Prince William Sound Science Center, United States; Andrew R. Mahon, Central Michigan University, United States
Updates
Copyright
© 2020 Mizumoto, Mitsuzuka and Araki.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hitoshi Araki, arakih@res.agr.hokudai.ac.jp
This article was submitted to Conservation and Restoration Ecology, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.