- 1Department of Earth and Planetary Sciences, EDGE Institute, University of California, Riverside, Riverside, CA, United States
- 2Department of Biology, University of New Mexico, Albuquerque, NM, United States
- 3Department of Vertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 4Center for Macroecology, Evolution, and Climate, Globe Institute, University of Copenhagen, Copenhagen, Denmark
Studies of animal movement and migration over large geospatial scales have long relied on natural continental-scale hydrogen isotope (δ2H) gradients in precipitation, yet the physiological processes that govern incorporation of δ2H from precipitation into plant and then herbivore tissues remain poorly understood, especially at the molecular level. Establishing a biochemical framework for the propagation of δ2H through food webs would enable us to resolve more complicated regional-scale animal movements and potentially unlock new applications for δ2H data in animal ecology and eco-physiology. Amino acid δ2H analysis offers a promising new avenue by which to establish this framework. We report bulk tissue δ2H, δ13C, and δ15N data as well as amino acid δ2H and δ13C data from three Pipevine swallowtail (Battus philenor) tissues—caterpillars, butterfly bodies, and wings—as well as their obligate plant source: pipevine leaves (Aristolochia macrophylla). Insects are often dominant herbivores in terrestrial food webs and a major food source for many higher-level consumers, so it is particularly important to understand the mechanisms that influence insect tissue δ2H values. Our data reveal extensive δ2H variation within and among individuals of a relatively simple plant-herbivore system that cannot be explained by temporal or geospatial gradients of precipitation δ2H or dietary differences. Variations in essential amino acid δ2H and δ13C indicate that B. philenor acquire these compounds from an additional source that is isotopically distinct from pipevine leaves, potentially gut microbes. We also found multiple isotopic carryover effects associated with metamorphosis. This study emphasizes the strong influence of physiology on consumer-diet δ2H discrimination in a local population of pipevines and swallowtails and provides a template that can be broadly applied to Lepidoptera—the second most diverse insect order—and other holometabolous insects. Understanding these physiological mechanisms is critical to interpreting the large degree of δ2H variation in consumer tissues often observed at a single collection site, which has implications for using δ2H isoscapes to study animal movement. Further investigation into amino acid δ2H holds promise to elucidate how subsets of amino acids may be best utilized to address specific ecological and physiological questions for which bulk tissue δ2H is insufficient.
Introduction
For decades, numerous studies have traced terrestrial animal migrations and movements by relying on natural continental-scale gradients in hydrogen isotopes of precipitation and groundwater that intrinsically label animal tissues (Wassenaar and Hobson, 1998; Kelly et al., 2002; Altizer et al., 2015). Because of the logistical and technological challenges associated with tracking, capturing, and sampling highly mobile animals that undergo large migrations, molecular techniques like hydrogen isotope (δ2H) analysis are incredibly valuable to studies of movement ecology (Kelly et al., 2002; Hobson and Wassenaar, 2008). Yet, it remains difficult to interpret these isotope data, especially when local site-level isotopic variation limits inferences of geographic affinity. First, bulk tissue (e.g., feather) isotope values are influenced by both the isotope signature at the base of the food web and biochemical modifications that occur within consumers during resource assimilation and tissue synthesis (McMahon and Newsome, 2019). Second, the isotopic composition of a consumer’s tissue represents a weighted average of all macromolecules within the sample. Lipids and carbohydrates are known to have vastly different δ13C and δ2H values than proteins (Estep and Hoering, 1981; Terwilliger et al., 2002; Post et al., 2007); lipids also vary in concentration among different tissues and often fluctuate temporally with body condition and reproductive status, especially in invertebrates (Cherel et al., 2005; Wolf et al., 2009). Moreover, migrations are often associated with significant physiological shifts including changes in metabolic rate, energy expenditure, and fasting, depletion of fat reserves, and variation in hormones—any of which could potentially affect bulk tissue isotope values (Gannes et al., 1998; Gwinner, 2012). Consequently, it can be difficult to determine how baseline isotope values, diet, tissue type, physiology, or metabolism each contribute to consumer bulk tissue isotope data.
Despite these obstacles, bulk tissue δ2H data can be successfully implemented in movement and migration studies at large spatial scales if δ2H isoscapes are well-constrained and δ2H gradients are large enough (Hobson et al., 2003; Bowen et al., 2005). Many of these studies have relied on the feathers of migratory birds because they are metabolically inert once grown, and feather δ2H values have repeatedly been shown to correlate with those of the local precipitation where they are synthesized (Chamberlain et al., 1997; Rubenstein et al., 2002; Hobson et al., 2004). However, δ2H in animal tissues is influenced by a combination of drinking/food water and diet, and the latter source of hydrogen accounts for the majority used to synthesize proteinaceous tissues like feathers (Hobson, 1999; Wolf et al., 2011, 2012). For this reason, and because hydrogen in a molecule has many opportunities to exchange with other pools of hydrogen during metabolic reactions, hydrogen cycling is more complex than that of more conventional biomarkers such as carbon or nitrogen isotopes. In fact, nearly all the research into the biochemical processes that govern incorporation of δ2H into consumer tissues stems from controlled feeding experiments in the lab (Estep and Dabrowski, 1980; Peters et al., 2012; Fogel et al., 2016; Newsome et al., 2017; Rodriguez Curras et al., 2018), with relatively little being done in natural ecosystems across trophic levels. Establishing a biochemical framework for the propagation of δ2H through food webs would expand our ability to resolve smaller regional-scale animal movements and potentially unlock new applications for δ2H data in dietary and metabolic studies.
Amino acid hydrogen isotope analysis offers a promising new avenue by which to establish this framework, without the confounding factors associated with bulk tissue analysis. Amino acids are the building blocks of most tissues (e.g., feather, muscle, liver, blood) analyzed by ecologists, making them ideal compounds to study when examining how isotopes propagate through food webs. Although amino acid δ2H datasets are currently limited in the literature, a controlled feeding study on E. coli found that the δ2H of individual amino acids within a single sample can vary by ∼200‰ (Fogel et al., 2016)—an extreme amount of intermolecular variation that would be masked by a composite bulk tissue δ2H value. Similar patterns among amino acid δ2H values were recently reported for muscle tissue of house mice (Mus musculus) reared in controlled laboratory conditions (Newsome et al., 2020), suggesting these δ2H values are primarily controlled by biochemical processes governing amino acid synthesis that are shared among prokaryotes and eukaryotes. While this extensive variation can present challenges for ecological studies, it also provides an opportunity to examine the mechanisms driving the transfer of δ2H signals up food chains (e.g., metabolic routing) at the molecular level and on a scale that is an order of magnitude larger than the other light isotopes.
Unlike δ2H, amino acid δ13C analysis is well established in ecology. Amino acids are classified into two groups with respect to δ13C—essential and non-essential. Essential amino acids (AAEss) are particularly useful tracers in diet and food web studies because their carbon skeletons cannot be synthesized by animals, and so are directly routed into a consumer’s tissues from the diet with minimal isotopic alteration (O’Brien et al., 2002; McMahon et al., 2010). Thus, the δ13C values of AAEss in consumer tissues closely reflect those of the primary producers, bacteria, and fungi at the base of their food web, which have unique amino acid δ13C patterns or “fingerprints” that likely reflect variation in the biochemical pathways these groups use to synthesize these compounds (Scott et al., 2006; Larsen et al., 2009). In contrast, non-essential amino acids (AANEss) can either be directly routed from dietary protein or synthesized de novo by animals from dietary macromolecules (e.g., carbohydrates or lipids) or catabolized from endogenous stores (e.g., muscle or adipose tissue). The AAEss and AANEss classification scheme used in δ13C studies appears to hold up for δ2H, probably because most of the hydrogen (85–90%) in amino acids is bonded to carbon and theoretically does not exchange with body water (Wassenaar and Hobson, 2000; Fogel et al., 2016; Newsome et al., 2017). A controlled feeding study on E. coli found that a higher proportion of environmental water was used to synthesize AANEss, especially glycolytic amino acids, relative to AAEss in both protein-rich (tryptone) and protein-free (glucose) media treatments (Fogel et al., 2016). This finding illustrates how amino acid δ2H analysis can be employed to identify the sources of hydrogen from both diet (AAEss) and drinking or environmental water (AANEss) used by consumers to synthesize tissues.
To investigate the biochemical mechanisms by which hydrogen is incorporated into consumer tissues in a natural setting, we rely on a combination of bulk tissue and amino acid isotope data from a simple plant-herbivore trophic transfer in a single watershed. We exploit an obligate caterpillar-host plant association between pipevine plants in the genus Aristolochia and the Pipevine swallowtail (Battus philenor) (Figure 1; Rausher, 1980). Both the caterpillars and adult butterflies rely on flowering C3 plants in the same geographical region and should retain the isotopic fingerprint of that primary producer group in their own AAEss. Our approach eliminates the confounding factor of large latitudinal or elevational δ2H gradients in meteoric water. Furthermore, because adults of this species feed on nectar primarily composed of sugars (Scott, 1986; Lotts and Naberhaus, 2020), they can be protein-limited. This provides the unique opportunity to investigate isotopic changes associated with metamorphosis and a limited nitrogen pool, with little influence from the adult diet. In theory, therefore, any isotopic offsets in AAEss or AANEss among plants, caterpillars, and adults are the result of physiological effects associated with metamorphosis. Butterfly body tissue continues to turn over throughout the adult’s short lifetime. However, because little additional nitrogen is consumed during this time, any isotopic offsets in amino acids between butterfly body tissue and wing tissue should either reflect tissue-specific discrimination during metamorphosis or metabolic processing during the adult’s lifetime. Butterfly wing chitin is metabolically inert once formed, such that its isotopic signature reflects only larval resources (Wassenaar and Hobson, 1998). Wings also contain metabolically active hemolymph, but it represents a small component of the wing and is unlikely to significantly affect our isotope data or interpretations. See Supporting Information—Text for details.

Figure 1. Photos of Pipevine (Aristolochia macrophylla), Pipevine swallowtail caterpillar (Battus philenor), and Pipevine swallowtail butterfly, by Will Cook, Duke University.
A complicating influence on insect tissue isotope values may be gut microbiomes that have been identified in several Lepidopteran larvae (Robinson et al., 2010; Hammer et al., 2014) and adults (Paniagua Voirol et al., 2018; Hammer et al., 2020). Unlike insects, gut microbes are capable of synthesizing AAEss de novo using all dietary macromolecules, which could impart a unique isotopic signature if microbially-synthesized amino acids are used by the host to build tissues (Christensen and Fogel, 2011; Newsome et al., 2020). Such isotope effects are poorly understood even though insects are often the dominant herbivore group in terrestrial food webs (Price et al., 1980; Chikaraishi et al., 2011). Alternatively, some AAEss could originate from nectar (Levin et al., 2017). If nectar-sourced amino acids supplied a significant proportion of amino acids for insect tissue synthesis, we would expect that the isotopic compositions of plant and insect tissues to be similar.
Our results reveal a high degree of intra- and inter-individual δ2H variation in caterpillars and butterflies despite the absence of geographical variation in the δ2H of water and food sources. This variation emphasizes the complexity inherent in natural systems, even ones as simple as an obligate caterpillar-host plant interaction. The influence of metabolism on consumer-diet δ2H (and δ13C) discrimination in our study provides a template for how δ2H might fractionate in response to physiological shifts in metamorphosing insects.
Materials and Methods
Study Site and Species
Battus philenor L. is found in warm climates throughout North America, mainly across the southern and eastern portions of the United States and in Mexico (Scott, 1986). This species is non-migratory (Hall, 2020) and females feed on a variety of flowering plants (e.g., thistles, phlox, ironweed) but only oviposit on pipevine species in the genus Aristolochia (Scott, 1986; Lotts and Naberhaus, 2020). In our study system, Battus larvae fed on the foliage of Aristolochia macrophylla Lam., a large liana with ropy stems that ascends into the forest canopy. Large vines can bear hundreds or even thousands of large heart-shaped leaves. Little is known about the nectar sources exploited by Battus in the Santeetlah Creek watershed, the amino acid concentrations of those nectars, or the many other protein sources exploited opportunistically. These unknowns generate potential uncertainty when interpreting results and are addressed in the discussion.
Sample Collection
Specimens were collected throughout a narrow elevational band (872–1,191 m above sea level) from the watersheds of Big Santeetlah Creek (5350 ha) and Little Santeetlah Creek, Graham County, North Carolina, United States from 13 to 15 June 2013. These watersheds are heavily forested, dominated by C3 plants, and situated on the eastern slope of the Unicoi Mountains, a subdivision of the southern Appalachians (Graves and Romanek, 2009). Caterpillars (n = 8) were collected directly from pipevine leaves, which were also collected, along with butterfly samples (n = 8); all insects were stored in cryovials in liquid nitrogen in the field. Specimens were subsequently freeze-dried and stored at room temperature in the lab until isotope analysis. Only a subset of the leaves collected were analyzed (n = 6), each from a different plant along a ∼4 km long transect parallel to Santeetlah Creek. For amino acid analysis, we sampled internal tissues, avoiding the gut. Water samples were also collected from the Santeetlah Creek Watershed and analyzed for bulk δ2H (average = –42‰; see Supporting Information—Text for details). Butterflies were sexed in the lab on the basis of dorsal hindwing iridescence, which is brighter in males (Supplementary Figure 1; Hall, 2020). All were males, except G-14. Variation in isotope data were unrelated to butterfly sex and elevation of sample collection (see Supporting Information—Text for details).
Sample Preparation
Approximately 10–15 mg of each dry sample was hydrolyzed for 20 h at 110°C in 1 mL of 6 N HCl, with a N2 headspace to remove oxygen. Hydrolysates were evaporated to dryness under a gentle N2 stream at room temperature then prepared as TFAA derivatives (Silfer et al., 1991). The total free amino acids in the hydrolysates were esterified with acetyl chloride/2-propanol (1:4, v/v) at 110°C for 1 h then acylated with trifluoroacetic anhydride/dichloromethane (1:1, v/v; DCM) at 110°C for 10 min. The resultant derivatives were completely dried then brought up in DCM and stored at –25°C until analysis. A standard mixture of 13 amino acids purchased from Sigma Aldrich (Saint Louis, MO) with known isotope values was derivatized alongside every batch of samples. See Supporting Information for details on bulk tissue analysis and findings.
Amino Acid Analysis
All samples were analyzed at the EDGE Institute at the University of California Riverside. The δ13C and δ2H values of individual amino acids were determined by gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS) using a Trace 1310 gas chromatograph (GC) coupled with a GC-Isolink ll routed through a Conflo IV interface into a Delta V Plus isotope ratio mass spectrometer (Thermo Fisher Scientific, United States). Amino acids were either routed through a combustion reactor at 1,000°C for δ13C or an HTC reactor at 1,420°C for δ2H. Using a Triplus RSH autosampler, samples were injected splitless at 250°C onto a BPX-5 capillary column (60 m × 0.32 mm inner diameter, 1.0 um film thickness; SGE Analytical Science, United States). The GC oven temperature program was as follows: initial temperature 50°C for 2 min, followed by a ramp of 15°C/min to 125°C, a ramp of 3°C/min to 160°C, a ramp of 4°C/min to 190°C, a ramp of 6°C/min to 275°C, and a ramp of 15°C/min to 320°C. Carrier gas (He) flow was 2 mL min–1.
Stable isotope values are expressed in per mil (‰) using delta notation: δX = [(Rsample/Rstandard) − 1] × 103 where X is 13C or 2H, R is the corresponding ratio of 13C:12C or 2H:1H, and Rstandard is Vienna Pee-Dee Belemnite or VSMOW for δ13C and δ2H, respectively. We analyzed the δ13C and δ2H values of 12 individual amino acids, 6 essential—valine (Val), threonine (Thr), leucine (Leu), isoleucine (Ile), phenylalanine (Phe), lysine (Lys)—and 6 non-essential—alanine (Ala), glycine (Gly), serine (Ser), proline (Pro), aspartic acid (Asp), glutamic acid (Glu)—listed in chromatographical order. Because acid hydrolysis converts glutamine (Gln) and asparagine (Asn) into Glu and Asp, respectively, by cleaving the terminal amine group, the measurements we report are combined Gln + Glu and Asn + Asp, hereby referred to as just Glu and Asp, to be consistent with previous studies. Tyr and Arg were excluded from analyses because of missing measurements caused by low concentrations near or below detection limits.
Amino acid standards of known isotopic composition were derivatized alongside and analyzed between unknown samples that were analyzed in triplicate. Reproducibility of replicate sample injections ranged from 0.1 to 0.6‰ (average < 0.3‰) for δ13C and 1.2–5.2‰ (average 2.5‰) for δ2H. Reproducibility of the amino acid standards used for sample corrections averaged < 0.6‰ for δ13C and 2.6‰ for δ2H. Because derivatization reagents add carbon atoms to the amino acids, the standards prepared alongside the samples are used to correct for the reagent carbon added. Similarly, exchangeable hydrogen atoms are removed from the carboxyl and amine groups of the amino acids during derivatization, and an additional hydrogen atom from the amine group is likely exchanged during hydrolysis. So, our measured δ2H data are corrected using amino acid-specific mass balance equations that adjust for the percentage of hydrogen atoms removed during derivatization (Fogel et al., 2016). Thus, the amino acid δ2H values we report reflect the nonexchangeable (intrinsic) hydrogen atoms only, and not hydrogen contributed by reagents during derivatization.
Statistical Analysis
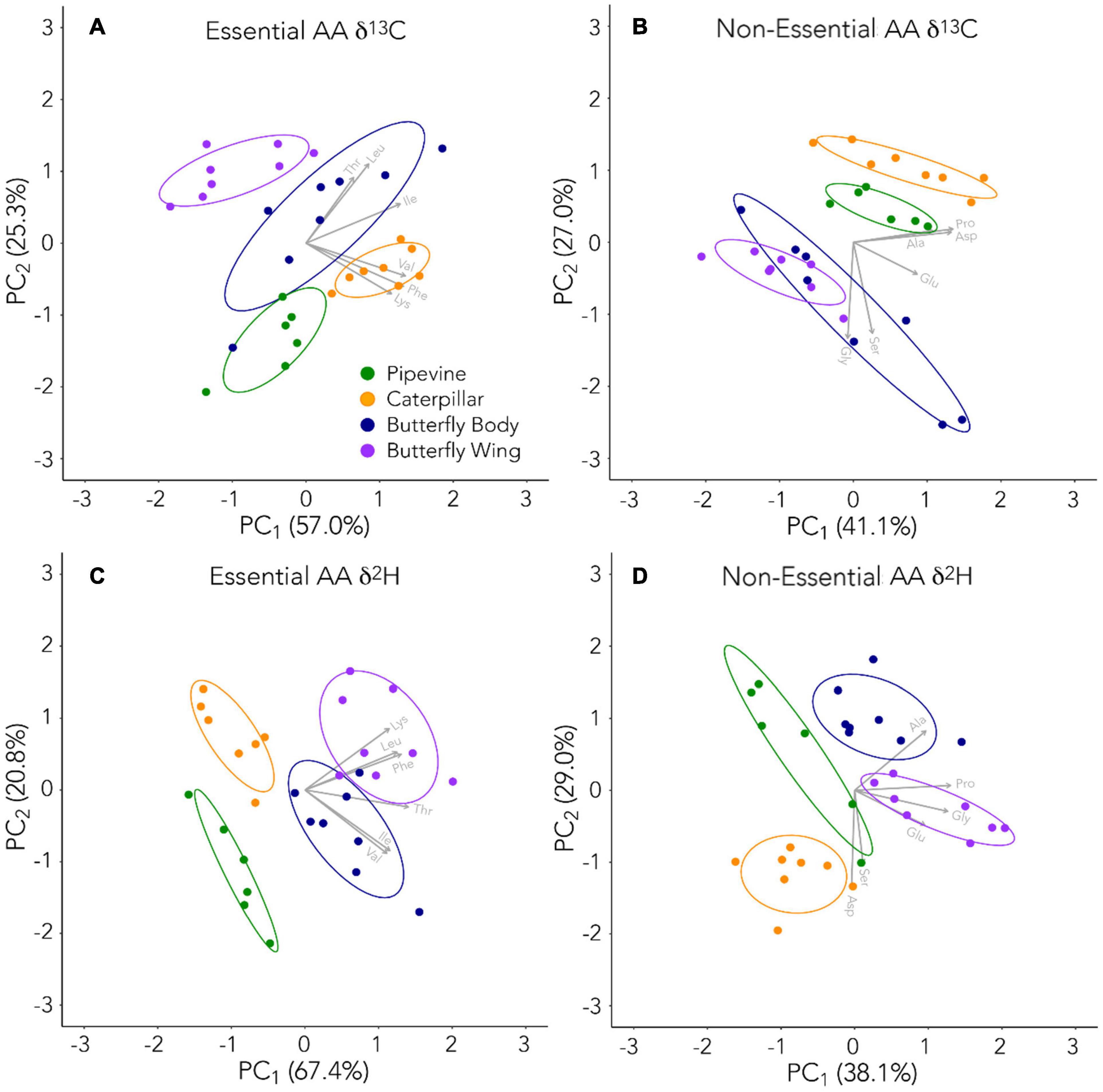
We used Shapiro-Wilks and Levene’s tests to confirm normality and homoscedasticity, respectively, and considered nonparametric equivalents before selecting ANOVA with Tukey HSD post-hoc tests to assess differences between tissue types in each individual amino acid. We further explored patterns in our amino acid δ2H and δ13C datasets with principal component analysis (PCA). AAEss and AANEss were evaluated separately. These statistical analyses were performed in R (version 4.0.3) with RStudio interface (version 1.3.1093; R Development Core Team, 2020) using the stats package.
Results
Amino Acid δ13C
The AAEss δ13C of the four tissue types were distinct from one another, a finding very different from O’Brien et al. (2002, 2003, 2005)’s laboratory studies (Figures 2A,B). PCA of AAEss δ13C clustered the samples according to their tissue type fairly well, with caterpillars the least variable and butterfly bodies the most variable (Figure 3A). Caterpillars separated from pipevines and butterfly wings along the first (PC1) and second (PC2) principal component axes. Differences between pipevines and caterpillars were driven by Thr, Leu, and Ile, while differences between caterpillars and adult butterflies (especially wings) were driven by Val, Phe, and Lys. Higher Thr and Leu also distinguished butterfly bodies from pipevines. In the ANOVA, all AAEss showed significant differences between some of the tissue types (Supplementary Table 5). Thr differed significantly between butterfly bodies and each of the other three tissues. Val differed for every pair except pipevines and butterfly bodies. Leu differed between pipevines and each of the other tissues. Ile was significantly different for half of the pairings (pipevines-caterpillars, pipevines-butterfly bodies, caterpillars-butterfly wings). Phe and Lys differed between butterfly wings and every other tissue type.
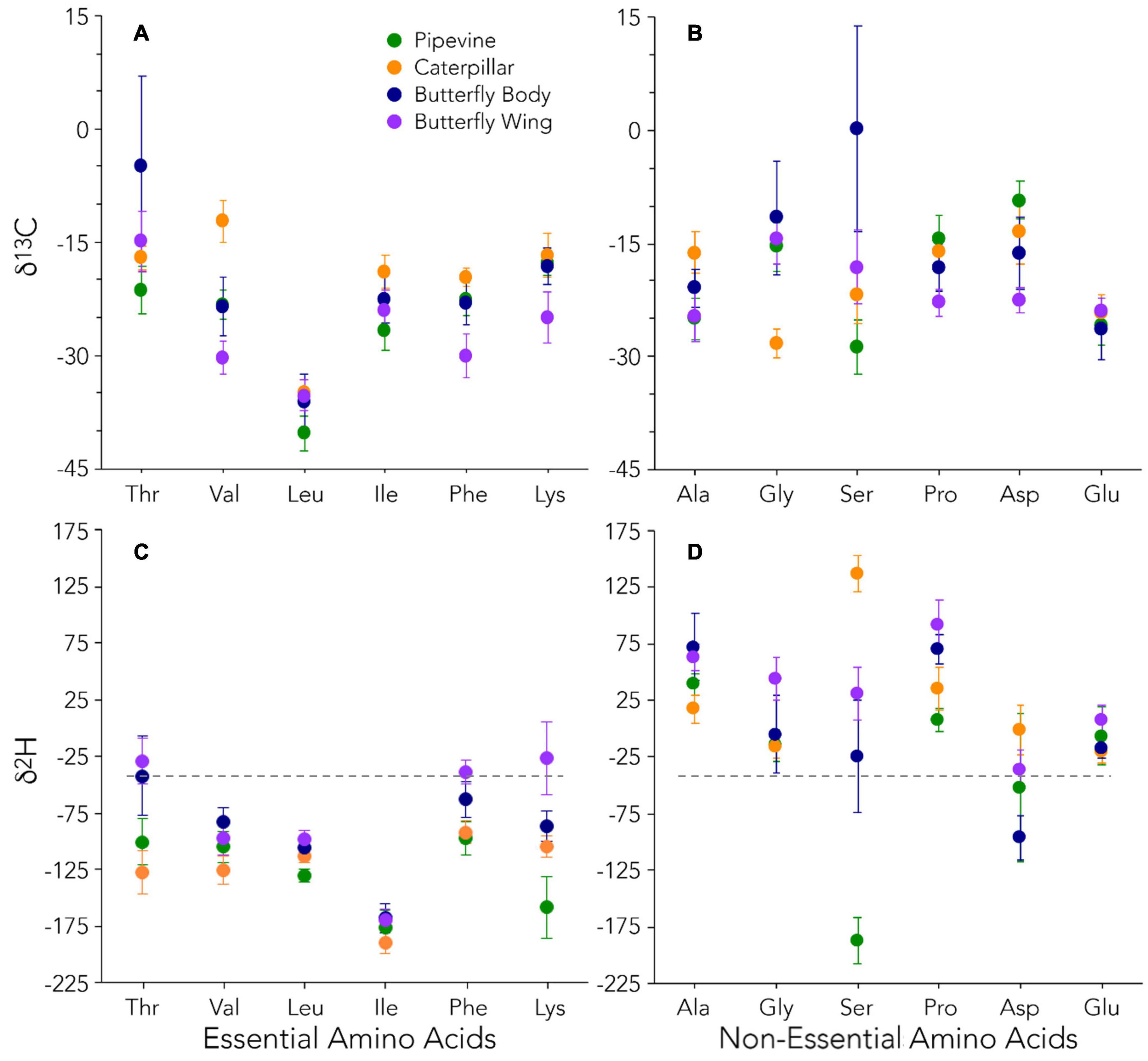
Figure 2. δ13C (A,B) and δ2H (C,D) data for essential and non-essential amino acids. Our average δ2H value (–42‰) for local water at the time of sample collection is indicated by the dashed gray line.
With PCA for AANEss δ13C, we found very tight clustering and excellent separation among pipevines, caterpillars, and butterfly wings. However, the two ellipses for butterfly tissues overlap (Figure 3B) because bodies show a high degree of variation, producing an ellipse that stretches across the entire PC1 axis. Consequently, half of the butterfly body samples plot near the wings, while the other half plot below pipevines and caterpillars. PC1, and the separation between butterfly wings on one end and caterpillars and pipevines on the other end, is strongly influenced by Ala, Pro, and Asp. PC2 is strongly influenced by Gly and Ser, and accounts for even more of the separation among pipevines, caterpillars, and butterfly wings. Interestingly, the half of the butterfly body samples that plotted most closely with pipevines and caterpillars along PC1 plot the farthest from them along PC2. ANOVA indicated that every AANEss except Glu showed significant differences among some of the tissue types (Supplementary Table 5). Ala and Gly differed between caterpillars and each of the butterfly tissues. Ser differed between butterfly bodies and the other tissues. Pro and Asp behaved similarly and only differed significantly between two pairings—-pipevines-caterpillars, caterpillars-butterfly bodies.
Amino Acid δ2H
Similar to the δ13C data, AAEss δ2H were distinct from one another, supporting the above finding that essential amino acids were not simply routed from plants to caterpillars to adult butterflies (Figures 2C,D, 4). PCA of AAEss δ2H shows the tightest clustering of samples by their tissue types, with four non-overlapping ellipses (Figure 3C). The two butterfly tissues show the greatest variability and the greatest similarity to each other. PC1 provides excellent separation of pipevines and caterpillars from both butterfly tissues. PC2 similarly provides substantial separation between pipevines and caterpillars. Differences between pipevines and adult butterfly tissues were mainly due to Lys, Leu, and Phe; Thr was most responsible for separating adult butterfly tissues from the caterpillars and plant samples, and Val and Ile drove the separation between caterpillars and butterfly bodies. ANOVA revealed that Thr differed significantly between all but two pairs—pipevines-caterpillars and the two butterfly tissues (Supplementary Table 5). Leu was significantly different between all pipevine pairs and between caterpillars and butterfly wings. For Ile, caterpillars differed from both butterfly tissues. Phe was significantly different for all pairings involving butterfly tissues. Lys was significantly different for all pipevine and butterfly wing pairs.
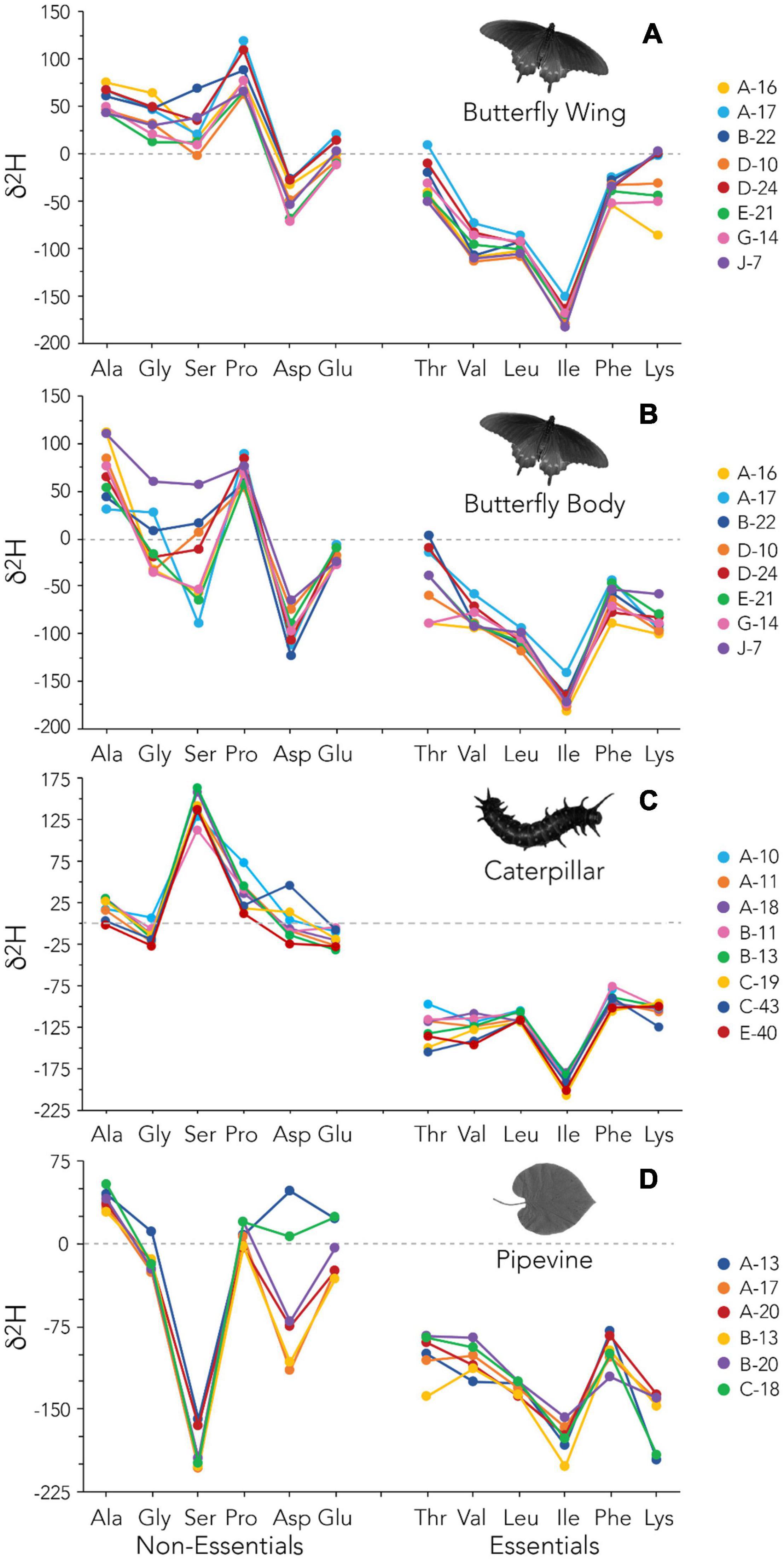
Figure 4. Amino acid δ2H data for individual butterfly wings (A), butterfly bodies (B), caterpillars (C), and pipevine leaves (D).
Like AAEss δ2H, all PCA ellipses for AANEss δ2H are non-overlapping (Figure 3D). In contrast to δ13C, AANEss δ2H varies most among pipevines and least among butterfly wings. PC1 mainly separates butterfly wings from pipevines and caterpillars and is controlled by Pro, Gly, and Glu. PC2 is governed primarily by Asp and Ser, and to a lesser extent, Ala. These three amino acids create the separation of caterpillars from the other three tissue types. Asp and Ser are also responsible for the separation between the two butterfly tissues. The pipevine ellipse is oblong, such that it stretches across most of PC2. Within this ellipse space, the pipevine samples form two clusters—four samples group at the top of PC2 to the left of the butterfly body samples while the other two samples are centrally located between caterpillars and butterfly wings. This suggests high variability among pipevine Asp and Ser. In ANOVA, Ala was significantly different in half of the pairings—pipevines-butterfly bodies, caterpillars-butterfly bodies, and caterpillars-butterfly wings (Supplementary Table 5). Gly differed for every butterfly wing pairing. Ser was significantly different for all six pairs. Pro differed for every pairing except that of the two butterfly tissues. Like Ala, Asp differed significantly in half of the pairs—pipevines-caterpillars, caterpillars-butterfly bodies, and the two butterfly tissues. Glu only differed between caterpillars-butterfly wings, and the two butterfly tissues.
Discussion
We found extensive isotopic variation within and between individuals and among tissue types despite an obligate caterpillar-host plant relationship, a C3 herbivorous diet for both larval and adult B. philenor, and sampling from a single watershed (Figures 2, 3 and Supplementary Table 4). Our data reveal the extent to which physiological processes—particularly those associated with metamorphosis—contribute to isotopic variation and persist into the adult lifestage as carryover effects. Additionally, our results suggest that the gut microbiome contributes to the amino acid pool used by B. philenor for tissue synthesis throughout their life cycle. Understanding what drives δ2H variation among primary producers and herbivores is critical for isoscape development and δ2H-based animal migration studies.
Essential Amino Acid δ13C
Because AAEss cannot be synthesized by B. philenor, they should be obtained directly from dietary amino acids. Direct routing from dietary protein is assumed to result in minimal isotopic fractionation such that isotope values of AAEss in caterpillars/butterflies and pipevines would be similar. In contrast, the use of AAEss synthesized by microbes from hydrogen or carbon in dietary biochemicals would yield a mismatch in the isotopic composition of AAEss between consumer and dietary protein. Adult B. philenor consume very little—if any—protein, which means their AAEss primarily derive from larval resources (Scott, 1986; O’Brien et al., 2005; Lotts and Naberhaus, 2020). Thus, we expected the AAEss δ13C values of B. philenor tissues to closely resemble each other and that of pipevines. However, our data show significant variation in AAEss δ13C values among pipevines, caterpillars, and butterfly tissues (Figure 2A and Supplementary Table 3), which must be driven by foraging behavior and/or gut microbial contributions. One such foraging behavior is compensatory feeding, wherein insects increase food consumption—and frass production—to compensate for a low-protein diet (Lavoie and Oberhauser, 2004; Le Gall and Behmer, 2014). This foraging strategy could potentially lessen the severity of protein-limitation in B. philenor caterpillars but the extent to which it might have a carryover effect on adult fitness is unknown.
Alternatively, some nectarivorous butterflies combat protein limitation by supplementing their adult diet with amino acids, which can increase their fecundity or even lifespan (O’Brien et al., 2005; Otis et al., 2006; Cahenzli and Erhardt, 2013). These supplements can potentially be acquired from nectar (Cahenzli and Erhardt, 2013; Levin et al., 2017), pollen (Erhardt and Baker, 1990; O’Brien et al., 2003), or mud-puddling, a behavior of aggregating at moist soil, tears, animal scat, or carcasses that is typically employed by males (Otis et al., 2006). However, amino acids are only present in trace amounts in nectar (Baker and Baker, 1973; Baker, 1975), B. philenor does not consume pollen (Konagaya et al., 2015), and mud-puddling is likely used to acquire sodium rather than protein (Otis et al., 2006). Moreover, even if amino acid supplements were contributing to the AAEss pool in butterfly body tissues, this would not explain differences in δ13C values among pipevines, caterpillars, and butterfly wings. In four out of six AAEss, it is either the caterpillar or the wing tissue that deviates most strongly from the δ13C fingerprint of pipevine leaves (Figure 2A). Thus, while B. philenor butterflies may opportunistically consume some amino acids from nectar, our data suggest that amino acid supplements do not substantially contribute to their AAEss budgets.
A more likely explanation for observed differences in the δ13C of AAEss between plant and consumer is subsidization by the gut microbiome. Although it remains unknown whether or not most Lepidopteran species host a gut microbiome, our data suggest that it contributes to the AAEss pool used by both caterpillars and butterflies for tissue synthesis. How the microbial signature deviates from that of dietary AAEss depends on whether gut microbes use carbohydrates or lipids—which are 13C-enriched or depleted, respectively, relative to dietary proteins—as a carbon source to synthesize AAEss. For example, gut microbes appear to synthesize Val, Leu, and Ile in caterpillars and Thr in butterflies from carbohydrates as the δ13C of these particular amino acids is substantially more positive (∼10‰) than those in leaf tissue (Figure 2A). Conversely, Val, Phe, and Lys in wing tissues seem to reflect larval gut microbial contributions from lipid stores during metamorphosis because these amino acids have more negative δ13C (∼7‰) than those in leaf tissue. We also found evidence of direct routing of AAEss between pipevines and caterpillars and during metamorphosis. δ13C values of Leu are identical among the insect tissues and only ∼5‰ lower in pipevines (Supplementary Table 3).
Non-essential Amino Acid δ13C
AANEss δ13C values are known to reflect a combination of direct routing from dietary protein and de novo synthesis by consumers from dietary carbohydrates and lipids (Jim et al., 2006; Newsome et al., 2014). Because pipevine leaves and nectar have less protein content than animal tissue, we expected that caterpillars and butterflies would use other dietary macromolecules (e.g., nectar carbohydrates) and limited endogenous nitrogen stores to construct some of the AANEss they need for metabolism and reproduction. In support of this prediction, we did not find a common AANEss δ13C fingerprint among tissue types, indicating extensive metabolic remodeling of amino acids during each insect life history stage (Figure 2B and Supplementary Figure 4B). Glu was the only AANEss that had a similar δ13C composition among tissue types (Supplementary Table 3); we hypothesize that this pattern results from Glu being the most abundant amino acid in animals, so its isotopic composition is relatively stable in comparison to amino acids that have smaller pool sizes. In contrast, Pro and Asp in wings are 13C-depleted relative to caterpillars, suggesting that pupae, or their gut microbes, draw from lipid stores to synthesize these AANEss for wing synthesis. Lipids are the most abundant energy reserve available during metamorphosis (Arrese and Soulages, 2010), and lipid carbon is easily introduced as acetyl CoA into the TCA cycle, from which Pro and Asp are derived (Horton et al., 2002).
Because adult B. philenor consume carbohydrates almost exclusively, it follows that amino acids synthesized from intermediaries in glycolysis (Ala, Gly, Ser) should reflect the strongest carbohydrate δ13C signature of all the tissue types. In support of this prediction, Ser δ13C is on average ∼20‰ higher in butterfly bodies than in the other three tissues. Of all the AANEss measured, Ser has the most variable δ13C values (Figure 2B and Supplementary Table 3), which range > 50‰ among pipevines and butterflies and almost 40‰ in butterfly bodies alone (Supplementary Figure 3B). This is a substantial amount of δ13C variation for a single plant-herbivore system and indicates extensive de novo synthesis, particularly by the nectarivorous adult butterflies. Ala δ13C values are significantly higher by ∼9‰ in caterpillars relative to pipevine leaves (Supplementary Table 5), also indicative of synthesis from carbohydrates. Interestingly, caterpillars have uniquely low Gly δ13C values, suggesting either a mass balance effect associated with metamorphosis or synthesis of this AANEss with lipid carbon. Overall, isotope data for glycolytic amino acids demonstrate that the magnitude of 13C-enrichment imparted by carbohydrate carbon is not universal. Rather, it likely depends on the interaction between the pool sizes and fluxes of each amino acid dictated by supply and demand of these metabolites, as well as the individual fractionation factors of the enzymes used to synthesize them.
Essential Amino Acid δ2H
AAEss δ2H data further show that the hydrogen isotope composition of individual amino acids is systematically modified according to the biochemical pathways used for synthesis and that eukaryotes and many prokaryotes utilize the same basic metabolic machinery to do so (Rodriguez Curras et al., 2018; Newsome et al., 2020). As was the case with E. coli (Fogel et al., 2016) and house mice (Newsome et al., 2020) reared in controlled feeding experiments, Ile has the lowest δ2H values of all amino acids in our dataset. Ile also has the most uniform δ2H values, suggesting it was mainly routed directly from the larval diet into the insect tissues. Furthermore, the extreme variation we found in AAEss δ2H within individuals and among tissue types (Figure 2C and Supplementary Table 4) mirrors that seen previously in E. coli and mice. AAEss δ2H varied over 300‰ in a single caterpillar (Figure 4C) and variation was similar in the other tissues measured, even within a single pipevine leaf (∼230‰).
In every AAEss other than Ile, δ2H was highest in either butterfly wings (Phe, Lys) or both wings and butterfly bodies (Thr, Val, Leu) relative to pipevines and caterpillars. This 2H-enrichment in adult insect tissues suggests that gut microbes, rather than direct incorporation from nectar, are contributing AAEss to their hosts using some combination of hydrogen sourced from the metabolic breakdown of carbohydrates, and possibly from drinking water. Environmental water typically has higher δ2H values than all dietary hydrogen sources because of the large negative discrimination during the synthesis of organic compounds in photosynthesis (Estep and Hoering, 1981). The fact that AAEss in wings are similar to or more enriched than the butterfly bodies points to body water as the predominant hydrogen source over carbohydrates because although stored glycogen can be accessed for glycolytic fuel during metamorphosis, without frequent feeding these carbohydrates are far less abundant than fat reserves and are quickly depleted (Arrese and Soulages, 2010). Although the adult gut microbiome may also play a role in butterfly body δ2H, carryover effects from the larval stage appear to persist throughout the butterfly’s lifetime and are explored further below.
Non-essential Amino Acid δ2H
In agreement with the analogous δ13C data, AANEss δ2H data suggest considerable de novo synthesis in B. philenor, particularly in Ser that ranges > 300‰ (–187‰–137‰) among tissue types (Figure 2D and Supplementary Table 4). Variation in Ser δ2H is not surprising given the essential function of serine proteases in insect metabolism—such as aiding digestion and development (Cao and Jiang, 2018)—which involves serine donating a hydrogen then gaining one from water (Radisky et al., 2006). Generally, structurally simple amino acids like Ser are more likely to exchange hydrogen with body water because a larger proportion of their hydrogen is bound to oxygen and nitrogen relative to more complex amino acids (Newsome et al., 2020). In controlled feeding studies on E. coli (Fogel et al., 2016) and mice (Newsome et al., 2020), drinking water appeared to have a particularly strong effect on the δ2H of glycolytic amino acids—this was especially true when organisms were fed diets containing no or low protein contents. Our data indicate that the same is true of our protein-limited herbivores.
The influence of body water on tissue δ2H is likely amplified in a holometabolous insect like B. philenor. Inside its chrysalis, which the caterpillar must construct from protein, the pupa undergoes a complete morphological remodel. While wings are an iconic part of this transformation, the pupa must also shorten the gut, grow limbs, expand the tracheal system, reshape the mouthparts, and develop a hard exoskeleton (Lowe et al., 2013). This complex process requires energy, expends water, and generates waste, and it is unclear how differential routing of amino acids might affect fractionation of carbon and hydrogen isotopes among newly synthesized tissues. What is clear, though, is that there are myriad opportunities during metamorphosis for isotopic discrimination to occur.
Evaluation of over 50 Lepidopteran species revealed that pupae typically lose a large percentage (≤80%) of their weight during metamorphosis (Molleman et al., 2011). Most of the weight lost is water (Wappner and Quesada-Allué, 1996; Molleman et al., 2011). This substantial water loss likely results in 2H-enrichment of the remaining body water pool, which is then used to synthesize 2H-enriched amino acids for adult butterfly tissues. δ2H patterns in Ala, Pro, and many microbially synthesized AAEss follow this prediction (Figures 2C,D). If wings are synthesized later in metamorphosis than adult body tissues, they may acquire higher δ2H values as a consequence of this gradual process, particularly if they are strongly influenced by hydrogen in body water. For instance, δ2H values of Gly, Ser, and Asp in butterfly wings are 40–50‰ more positive than in butterfly bodies (Supplementary Figure 4A and Supplementary Table 4). Furthermore, prior studies have identified bulk tissue 15N- and 2H-enrichments following metamorphosis in multiple species, including Lepidoptera, and primarily ascribed them to the excretion of 15N-depleted waste products and the inclusion of 2H-enriched body water for tissue synthesis (Doi et al., 2007; Tibbets et al., 2008; Peters et al., 2012; see Supporting Information—Text for additional discussion of Pro).
Inter-Individual Variation in Amino Acid Isotope Values
We focus this discussion on δ2H, though we lean on δ13C for context, because understanding inter-individual δ2H variation among primary producers and herbivores is critical for isoscape development and further elucidates the influence of physiology on tissue δ2H values. Also see Supporting Information—Text for a brief discussion of mycoheterotrophy as a potential explanation for inter-individual variation in plant isotope values. We found that bulk tissue δ2H values of caterpillars are often less variable than those of pipevines (Figures 4C,D). Relative to all other amino acids, Asp has the most variable δ2H values in both pipevine and caterpillar samples; Asp δ13C values are also variable in caterpillars (Supplementary Figure 3C). In plants, aspartate is a precursor for asparagine, which is used to transport and store nitrogen, and a precursor for Thr, Ile, Lys, and methionine synthesis (Azevedo et al., 2006). In caterpillars, Asp is abundant in a storage protein (Telfer and Kunkel, 1991) that is catabolized in adult Lepidoptera and used as a critical substrate for reproduction (O’Brien et al., 2002). These key metabolic functions may explain the unusually high inter-individual variability observed in Asp. Despite these exceptions, the similarity in AAEss δ13C and δ2H fingerprints among individual caterpillars implies relatively consistent fractionation during the plant-herbivore trophic transfer (Figure 4C and Supplementary Figure 3C). This uniformity strongly contrasts with the high degree of inter-individual variation among adult butterflies (Figures 4A,B and Supplementary Figures 3A,B). It is not surprising that caterpillars would be less variable than butterfly body tissue, given the generalist diet of the adults and the obligate host plant relationship maintained by the larvae. However, the fact that caterpillars also tend to be less variable than wings—which are largely metabolically inert—emphasizes the dominant role played by physiology during metamorphosis in determining δ2H values of adult tissues.
Concurrently assessing both δ13C and δ2H fingerprints for individual butterfly bodies reveals an additional interesting feature of our dataset that is best illustrated by the highly variable Ser and Thr: the relative rankings of individuals’ 13C-enrichment are fairly consistent independent of amino acid (Supplementary Figure 3B). That is, the three butterflies with the highest Ser δ13C also have the highest Thr δ13C. The same is not true for δ2H (Figure 4B). This is likely a reflection of the increased complexity of hydrogen cycling relative to that of carbon, but perhaps it also signifies a promising opportunity. δ2H analysis of specific subsets of amino acids may be best suited to answering different kinds of ecological or physiological questions. For example, glycolytic amino acids have previously been proposed as the most faithful proxies for δ2H of drinking water (Newsome et al., 2020) and so may be most beneficial for assigning geographic origin to migratory animals, particularly when bulk tissue δ2H is insufficient. Because caterpillars are an important food source for many higher trophic level organisms, their AAEss δ2H (and δ13C) values could be useful dietary tracers. Moreover, AAEss δ2H and δ13C provide a way to assess the presence and role of more complex metabolic processes like gut microbial input in systems where the isotopic composition of diet is well characterized. Several microbially synthesized AAEss as well as Ala, Pro, and Asp may be particularly valuable for studying physiology and biochemistry during metamorphosis, including the pupa’s use of a 2H-enriched body water pool to synthesize AANEss or AAEss (via its microbiome), amino acid routing, and the order in which adult tissues are synthesized.
Conclusion
The use of δ2H data in movement and migration studies is built on the premise that bulk animal tissue δ2H is primarily determined by precipitation δ2H. The presumption being that δ2H signatures are transferred from meteoric water to primary producers to animal consumers in a systematic fashion such that δ2H is a faithful intrinsic record of tissue synthesis location. Yet, our data show that bulk tissue δ2H values can range over 30‰ within a single population of pipevine-feeding caterpillars and amino acid δ2H values can vary by 300‰ within and among these herbivorous insects collected from a single watershed over a narrow elevation range. Given the absence of geographical, dietary, or temporal variation among our study organisms, this is a clear indication that physiology drives δ2H and δ13C fractionation during the trophic transfer from primary producers to caterpillars and especially during metamorphosis. Insects are a major herbivore group in terrestrial food webs and a common food source for consumers, including birds that are frequently of interest in δ2H-based studies of avian migration. Therefore, understanding how δ2H and δ13C values of insect tissues are biochemically mediated and vary among individuals is particularly critical for elucidating how δ2H propagates through food webs. Bulk tissue δ2H can be an invaluable tool for answering questions regarding breeding and overwintering grounds that are separated by thousands of kilometers along steep and well-established gradients in precipitation δ2H. However, further investigation into amino acid δ2H may yield a complementary tool that can be more routinely applied to studies of bird and insect movement across smaller geospatial scales or to better understand the physiological processes associated with metamorphosis in insects.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SN, GG, and MF formulated the project idea and collected samples. KM performed stable isotope and statistical analyses and led the writing of the manuscript. All authors engaged in intellectual exchange and manuscript editing and gave final approval for manuscript submission.
Funding
Funding for this project was provided by an NSF award to SN (DEB-1343015) and MF (DEB-1437845).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ying Lin, manager of the University of California Riverside EDGE lab, for bulk tissue analysis and maintaining the EA. The EDGE Institute also supported KM. GG thanks the Alexander Wetmore Fund (Smithsonian Institution) and the Smoketree Trust for support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.729258/full#supplementary-material
References
Altizer, S., Hobson, K. A., Davis, A. K., De Roode, J. C., and Wassenaar, L. I. (2015). Do healthy monarchs migrate farther? Tracking natal origins of parasitized vs. uninfected monarch butterflies overwintering in Mexico. PLoS One 10:e0141371. doi: 10.1371/journal.pone.0141371
Arrese, E. L., and Soulages, J. L. (2010). Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225. doi: 10.1146/annurev-ento-112408-085356
Azevedo, R. A., Lancien, M., and Lea, P. J. (2006). The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 30, 143–162. doi: 10.1007/s00726-005-0245-2
Baker, H. G. (1975). Sugar concentrations in nectars from hummingbird flowers. Biotropica 7, 37–41. doi: 10.2307/2989798
Baker, H. G., and Baker, I. (1973). Amino-acids in nectar and their evolutionary significance. Nature 241, 543–545. doi: 10.1038/241543b0
Bowen, G. J., Wassenaar, L. I., and Hobson, K. A. (2005). Global application of stable hydrogen and oxygen isotopes to wildlife forensics. Oecologia 143, 337–348. doi: 10.1007/s00442-004-1813-y
Cahenzli, F., and Erhardt, A. (2013). Nectar amino acids enhance reproduction in male butterflies. Oecologia 171, 197–205. doi: 10.1007/s00442-012-2395-8
Cao, X., and Jiang, H. (2018). Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects. Insect Biochem. Mol. Biol. 103, 53–69. doi: 10.1016/j.ibmb.2018.10.006
Chamberlain, C. P., Blum, J. D., Holmes, R. T., Feng, X. H., Sherry, T. W., and Graves, G. R. (1997). The use of isotope tracers for identifying populations of migratory birds. Oecologia 109, 132–141. doi: 10.1007/s004420050067
Cherel, Y., Hobson, K. A., Bailleul, F., and Groscolas, R. (2005). Nutrition, physiology, and stable isotopes: new information from fasting and molting penguins. Ecology 86, 2881–2888. doi: 10.1890/05-0562
Chikaraishi, Y., Ogawa, N. O., Doi, H., and Ohkouchi, N. (2011). 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: a case study of terrestrial insects (bees, wasps, and hornets). Ecol. Res. 26, 835–844. doi: 10.1007/s11284-011-0844-1
Christensen, H., and Fogel, M. L. (2011). Feeding ecology and evidence for amino acid synthesis in the periodical cicada (Magicicada). J. Insect Physiol. 57, 211–219. doi: 10.1016/j.jinsphys.2010.11.005
Doi, H., Kikuchi, E., Takagi, S., and Shikano, S. (2007). Changes in carbon and nitrogen stable isotopes of chironomid larvae during growth, starvation and metamorphosis. Rapid Commun. Mass Spectr. 21, 997–1002. doi: 10.1002/rcm.2925
Erhardt, A., and Baker, I. (1990). Pollen amino acids—an additional diet for a nectar feeding butterfly? Plant Syst. Evol. 169, 111–121. doi: 10.1007/BF00935989
Estep, M. F., and Dabrowski, H. (1980). Tracing food webs with stable hydrogen isotopes. Science 209, 1537–1538. doi: 10.1126/science.209.4464.1537
Estep, M. F., and Hoering, T. C. (1981). Stable hydrogen isotope fractionations during autotrophic and mixotrophic growth of microalgae. Plant Physiol. 67, 474–477. doi: 10.1104/pp.67.3.474
Fogel, M. L., Griffin, P. L., and Newsome, S. D. (2016). Hydrogen isotopes in individual amino acids reflect differentiated pools of hydrogen from food and water in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, E4648–E4653. doi: 10.1073/pnas.1525703113
Gannes, L. Z., Martinez del Rio, C., and Koch, P. (1998). Natural abundance variations in stable isotopes and their potential uses in animal physiological ecology. Comp. Biochem. Physiol. Part A 119, 725–737. doi: 10.1016/S1095-6433(98)01016-2
Graves, G. R., and Romanek, C. S. (2009). Mesoscale patterns of altitudinal tenancy in migratory wood warblers inferred from stable carbon isotopes. Ecol. Appl. 19, 1264–1273. doi: 10.1890/08-0934.1
Gwinner, E. (Ed) (2012). Bird migration: Physiology and Ecophysiology. Berlin: Springer Science and Business Media, doi: 10.1007/978-3-642-74542-3
Hall, D. W. (2020). University of Florida Institute of Food and Agricultural Sciences Entomology and Nematology Featured Creatures. Available online at: http://entnemdept.ufl.edu/creatures/bfly/pipevine_swallowtail.htm, [Accessed Jan 12, 2021].
Hammer, T. J., Dickerson, J. C., McMillan, W. O., and Fierer, N. (2020). Heliconius butterflies host characteristic and phylogenetically structured adult-stage microbiomes. Appl. Environ. Microbiol. 86, e02007–e02020. doi: 10.1128/AEM.02007-20
Hammer, T. J., McMillan, W. O., and Fierer, N. (2014). Metamorphosis of a butterfly-associated bacterial community. PLoS One 9:e86995. doi: 10.1371/journal.pone.0086995
Hobson, K. A. (1999). Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia 120, 314–326.
Hobson, K. A., and Wassenaar, L. I. (Eds) (2008). Tracking Animal Migration With Stable Isotopes. Amsterdam: Elsevier, doi: 10.1016/S1936-7961(07)00002-4
Hobson, K. A., Bowen, G. J., Wassenaar, L. I., Ferrand, Y., and Lormee, H. (2004). Using stable hydrogen and oxygen isotope measurements of feathers to infer geographical origins of migrating European birds. Oecologia 141, 477–488. doi: 10.1007/s00442-004-1671-7
Hobson, K. A., Wassenaar, L. I., Milá, B., Lovette, I., Dingle, C., and Smith, T. B. (2003). Stable isotopes as indicators of altitudinal distributions and movements in an Ecuadorean hummingbird community. Oecologia 136, 302–308. doi: 10.1007/s00442-003-1271-y
Horton, R., Moran, L. A., Scrimgeour, G., Perry, M., and Rawn, D. (2002). Principles of Biochemistry. New York, NY: Prentice Hall.
Jim, S., Jones, V., Ambrose, S. H., and Evershed, R. P. (2006). Quantifying dietary macronutrient sources of carbon for bone collagen biosynthesis using natural abundance stable carbon isotope analysis. Br. J. Nutr. 95, 1055–1062. doi: 10.1079/BJN20051685
Kelly, J. F., Atudorei, V., Sharp, Z. D., and Finch, D. M. (2002). Insights into Wilson’s warbler migration from analyses of hydrogen stable-isotope ratios. Oecologia 130, 216–221. doi: 10.1007/s004420100789
Konagaya, T., Mutoh, N., Suzuki, M., Rutowski, R. L., and Watanabae, M. (2015). Estimates of female lifetime fecundity and changes in the number and types of sperm stored with age and time since mating in the monandrous swallowtail butterfly, Battus philenor (Lepidoptera: Papilionidae) in the Arizona desert. Appl. Entomol. Zool. 50, 311–316. doi: 10.1007/s13355-015-0336-9
Larsen, T., Taylor, D. L., Leigh, M. B., and O’Brien, D. M. (2009). Stable isotope fingerprinting: a novel method for identifying plant, fungal, or bacterial origins of amino acids. Ecology 90, 3526–3535. doi: 10.1890/08-1695.1
Lavoie, B., and Oberhauser, K. S. (2004). Compensatory feeding in Danaus plexippus (Lepidoptera: Nymphalidae) in response to variation in host plant quality. Environ. Entomol. 33, 1062–1069. doi: 10.1603/0046-225X-33.4.1062
Le Gall, M., and Behmer, S. T. (2014). Effects of protein and carbohydrate on an insect herbivore: the vista from a fitness landscape. Integr. Comp. Biol. 54, 942–954. doi: 10.1093/icb/icu102
Levin, E., McCue, M. D., and Davidowitz, G. (2017). More than just sugar: allocation of nectar amino acids and fatty acids in a Lepidopteran. Proc. R. Soc. B 284, 20162126. doi: 10.1098/rspb.2016.2126
Lotts, K., and Naberhaus, T. (2020). Butterflies and Moths of North America. Available online at:http://www.butterfliesandmoths.org/ [Accessed Jan 13, 2021].
Lowe, T., Garwood, R. J., Simonsen, T. J., Bradley, R. S., and Withers, P. J. (2013). Metamorphosis revealed: time-lapse three-dimensional imaging inside a living chrysalis. J. R. Soc. Interface 10:20130304. doi: 10.1098/rsif.2013.0304
McMahon, K. W., and Newsome, S. D. (2019). “Chapter 7: Amino acid isotope analysis: a new frontier in studies of animal migration and foraging ecology,” in Tracking Animal Migration With Stable Isotopes, eds K. A. Hobson and L. I. Wassenaar (San Diego, CA: Academic Press), 173–190. doi: 10.1016/B978-0-12-814723-8.00007-6
McMahon, K. W., Fogel, M. L., Elsdon, T. S., and Thorrold, S. R. (2010). Carbon isotope fractionation of amino acids in fish muscle reflects biosynthesis and isotopic routing from dietary protein. J. Anim. Ecol. 79, 1132–1141. doi: 10.1111/j.1365-2656.2010.01722.x
Molleman, F., Javois, J., Esperk, T., Teder, T., Davis, R. B., and Tammaru, T. (2011). Sexual differences in weight loss upon eclosion are related to life history strategy in Lepidoptera. J. Insect Physiol. 57, 712–722. doi: 10.1016/j.jinsphys.2011.02.009
Newsome, S. D., Nakamoto, B. J., Rodriguez Curras, M., and Fogel, M. L. (2020). Compound-specific δ2H analysis highlights the relationship between direct assimilation and de novo synthesis of amino acids from food and water in a terrestrial mammalian omnivore. Oecologia 193, 827–842. doi: 10.1007/s00442-020-04730-9
Newsome, S. D., Wolf, N., Bradley, C. J., and Fogel, M. L. (2017). Assimilation and isotopic discrimination of hydrogen in tilapia: implications for studying animal diet with δ2H. Ecosphere 8:e01616. doi: 10.1002/ecs2.1616
Newsome, S. D., Wolf, N., Peters, J., and Fogel, M. L. (2014). Amino acid δ13C analysis shows flexibility in the routing of dietary protein and lipids to the tissue of an omnivore. Integr. Comp. Biol. 54, 890–902. doi: 10.1093/icb/icu106
O’Brien, D. M., Boggs, C. L., and Fogel, M. L. (2003). Pollen feeding in the butterfly Heliconius charitonia: isotopic evidence for essential amino acid transfer from pollen to eggs. Proc. R. Soc. Lond. B 270, 2631–2636. doi: 10.1098/rspb.2003.2552
O’Brien, D. M., Boggs, C. L., and Fogel, M. L. (2005). The amino acids used in reproduction by butterflies: a comparative study of dietary sources using compound-specific stable isotope analysis. Physiol. Biochem. Zool. 78, 819–827. doi: 10.1086/431191
O’Brien, D. M., Fogel, M. L., and Boggs, C. L. (2002). Renewable and nonrenewable resources: Amino acid turnover and allocation to reproduction in Lepidoptera. Proc. Natl. Acad. Sci. U.S.A. 99, 4413–4418. doi: 10.1073/pnas.072346699
Otis, G. W., Locke, B., McKenzie, N. G., Cheung, D., MacLeod, E., Careless, P., et al. (2006). Local enhancement in mud-puddling swallowtail butterflies (Battus philenor and Papilio glaucus). J. Insect Behav. 19, 685–698. doi: 10.1007/s10905-006-9049-9
Paniagua Voirol, L. R., Frago, E., Kaltenpoth, M., Hilker, M., and Fatouros, N. E. (2018). Bacterial symbionts in Lepidoptera: their diversity, transmission, and impact on the host. Front. Microbiol. 9:556. doi: 10.3389/fmicb.2018.00556
Peters, J. M., Wolf, N., Stricker, C. A., Collier, T. R., and Martínez del Rio, C. (2012). Effects of trophic level and metamorphosis on discrimination of hydrogen isotopes in a plant-herbivore system. PLoS One 7:e32744. doi: 10.1371/journal.pone.0032744
Post, D. M., Layman, C. A., Arrington, D. A., Takimoto, G., Quattrochi, J., and Montana, C. G. (2007). Getting to the fat of the matter: models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 152, 179–189. doi: 10.1007/s00442-006-0630-x
Price, P. W., Bouton, C. E., Gross, P., McPheron, B. A., Thompson, J. N., and Weis, A. E. (1980). Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Annu. Rev. Ecol. Syst. 11, 41–65. doi: 10.1146/ANNUREV.ES.11.110180.000353
R Development Core Team (2020). R: A Language And Environment For Statistical Computing. Vienna: R Foundation for Statistical Computing.
Radisky, E. S., Lee, J. M., Lu, C. J. K., and Koshland, D. E. (2006). Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediaries. Proc. Natl. Acad. Sci. U.S.A. 103, 6835–6840. doi: 10.1073/pnas.0601910103
Rausher, M. (1980). Host abundance, juvenile survival, and oviposition preference in Battus Philenor. Evolution 34, 342–355. doi: 10.2307/2407398
Robinson, C. J., Schloss, P., Ramos, Y., Raffa, K., and Handelsman, J. (2010). Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb. Ecol. 59, 199–211. doi: 10.1007/s00248-009-9595-8
Rodriguez Curras, M., Fogel, M. L., and Newsome, S. D. (2018). Assimilation and discrimination of hydrogen isotopes in a terrestrial mammal. Oecologia 188, 381–393. doi: 10.1007/s00442-018-4221-4
Rubenstein, D. R., Chamberlain, C. P., Holmes, R. T., Ayres, M. P., Waldbauer, J. R., Graves, G. R., et al. (2002). Linking breeding and wintering ranges of a migratory songbird using stable isotopes. Science 295, 1062–1065. doi: 10.1126/science.1067124
Scott, J. A. (1986). The Butterflies of North America: A Natural History And Field Guide. Stanford, CA: Stanford University Press, 583.
Scott, J. H., O’Brien, D. M., Emerson, D., Sun, H., McDonald, G. D., Salgado, A., et al. (2006). An examination of the carbon isotope effects associated with amino acid biosynthesis. Astrobiology 6, 867–880. doi: 10.1089/ast.2006.6.867
Silfer, J. A., Engel, M. H., Macko, S. A., and Jumeau, E. J. (1991). Stable carbon isotope analysis of amino acid enantiomers by conventional isotope ratio mass spectrometry and combined gas chromatography/isotope ratio mass spectrometry. Anal. Chem. 63, 370–374.
Telfer, W. H., and Kunkel, J. G. (1991). The function and evolution of insect storage hexamers. Annu. Rev. Entomol. 36, 205–228. doi: 10.1146/annurev.en.36.010191.001225
Terwilliger, V. J., Betancourt, J. L., Leavitt, S. W., and Van de water, P. K. (2002). Leaf cellulose δD and δ18O trends with elevation differ in direction among co-occurring, semiarid plant species. Geochim. Cosmochim. Acta 66, 3887–3900. doi: 10.1016/S0016-7037(02)00964-X
Tibbets, T. M., Wheeless, L. A., and Martinez del Rio, C. (2008). Isotopic enrichment without change in diet: an ontogenetic shift in δ15N during insect metamorphosis. Funct. Ecol. 22, 109–113. doi: 10.1111/j.1365-2435.2007.01342.x
Wappner, P., and Quesada-Allué, L. A. (1996). Water loss during cuticle sclerotization in the medfly Ceratitis capitata is independent of catecholamines. J. Insect Physiol. 42, 705–709. doi: 10.1016/0022-1910(96)00006-6
Wassenaar, L. I., and Hobson, K. A. (1998). Natal origins of migratory monarch butterflies at wintering colonies in Mexico: new isotopic evidence. Proc. Nat. Acad. Sci. U.S.A. 95, 15436–15439. doi: 10.1073/pnas.95.26.15436
Wassenaar, L. I., and Hobson, K. A. (2000). Improved method for determining the stable-hydrogen isotopic composition (δD) of complex organic materials of environmental interest. Environ. Sci. Technol. 34, 2354–2360. doi: 10.1021/es990804i
Wolf, N., Bowen, G. J., and del Rio, C. M. (2011). The influence of drinking water on the δD and δ18O values of house sparrow plasma, blood and feathers. J. Exp. Biol. 214, 98–103. doi: 10.1242/jeb.050211
Wolf, N., Carleton, S. A., and del Rio, C. M. (2009). Ten years of experimental animal isotopic ecology. Funct. Ecol. 23, 17–26. doi: 10.1111/j.1365-2435.2009.01529.x
Keywords: amino acid, hydrogen, carbon, stable isotope analysis, metamorphosis, butterfly, individual variation, physiology
Citation: Morra KE, Newsome SD, Graves GR and Fogel ML (2021) Physiology Drives Reworking of Amino Acid δ2H and δ13C in Butterfly Tissues. Front. Ecol. Evol. 9:729258. doi: 10.3389/fevo.2021.729258
Received: 22 June 2021; Accepted: 21 September 2021;
Published: 11 October 2021.
Edited by:
Todd Jason McWhorter, University of Adelaide, AustraliaReviewed by:
Gerhard Gebauer, University of Bayreuth, GermanyGoggy Davidowitz, University of Arizona, United States
Copyright © 2021 Morra, Newsome, Graves and Fogel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaycee E. Morra, a2F5Y2VlbUB1Y3IuZWR1
 Kaycee E. Morra
Kaycee E. Morra Seth D. Newsome
Seth D. Newsome Gary R. Graves
Gary R. Graves Marilyn L. Fogel
Marilyn L. Fogel