- 1U.S. Geological Survey, National Wildlife Health Center, Madison, WI, United States
- 2Department of Natural Resource Management, South Dakota State University, Brookings, SD, United States
- 3Idaho Department of Fish and Game, Lewiston, ID, United States
- 4Department of Veterinary Microbiology and Pathology, Washington State University, Pullman, WA, United States
Introduction: Throughout their range, bighorn sheep (Ovis canadensis) populations have seen significant disease-associated declines. Unfortunately, understanding of the underlying epidemiological processes driving the disease dynamics in this species has hindered conservation efforts aimed at improving the health and long-term viability of these populations. Individual response to pathogen exposure emerges from dynamic interactions between competing evolutionary processes within the host and pathogen. The host’s adaptive immune system recognizes pathogens and mounts a defensive response. Pathogens have evolved strategies to overcome adaptive immune defenses including maintaining high genetic diversity through rapid evolution. The outcomes of this evolutionary warfare determine the success of pathogen invasion of the host and ultimately the success of conservation efforts.
Methods: During an epizootic dominated by a single strain, we explore these host-pathogen dynamics by examining the variation in effects of pathogen invasion on captive bighorn sheep with differing histories of exposure to genetically diverse strains of Mycoplasma ovipneumoniae (Movi). We monitored clinical signs of disease and sampled animals and their environment to detect spread of Movi among 37 bighorn sheep separated into nine pens based on known exposure histories.
Results: We documented Movi transmission within and across pens and we detected Movi DNA in air, water, and invertebrate samples. Higher levels of antibody to Movi prior to the epizootic were associated with a lower likelihood of presenting clinical signs of pneumonia. Nonetheless, higher antibody levels in symptomatic individuals were associated with more severe progressive disease, increased probability and speed of pneumonia-induced mortality, and reduced likelihood of returning to a healthy state. Bighorn sheep with previous exposure to a strain other than the predominant epizootic strain were more likely to recover.
Discussion: Our results indicate that Movi-strain variability was sufficient to overwhelm the adaptive host immunological defenses. This outcome indicates, in free-ranging herds, past exposure is likely insufficient to protect bighorn sheep from infection by new Movi strains, although it influences the progression of disease and recovery within the herd. Therefore, given Movi-strain variability and the lack of immunological protection from past exposure, focusing management efforts on minimizing the introduction of Movi into bighorn herds, through separation of domestic and bighorn sheep and avoidance of management activities that create commingling of bighorn sheep carrying differing Movi strains, will likely be the most effective approach for reducing the effects of disease and achieving bighorn sheep conservation goals.
Introduction
Pathogens and their hosts are involved in on-going evolutionary warfare. Many hosts have evolved defenses including the innate and adaptive immune systems, which the host uses to recognize and respond to infections with new pathogens as well as to mount rapid responses upon re-exposure to prevent re-infection with previously encountered pathogens. Successful pathogens have evolved various complex and efficient methods to tolerate, evade, circumvent, or overcome innate and adaptive host immune defenses, resulting in increased disease severity or duration of infection of the host (Finlay and McFadden, 2006). One subtle but highly successful mechanism, employed by pathogens to specifically evade adaptive immunity is rapid evolution resulting in alteration of surface exposed antigenic epitopes, which leads to multiple and genetically diverse strain types (Bloom, 1979). Pathogen genetic diversity has implications for individual host response to infection and ultimately determines the pathogen’s virulence, transmissibility, and severity of epidemiological outbreaks (Coscolla and Gagneux, 2010; Chae and Shin, 2018). Extensive literature describes antigenic variation within Mycoplasma spp. (class Mollicutes), the smallest and simplest self-replicating organisms (Christiansen et al., 1997; Citti et al., 2010; Betlach et al., 2019; Qin et al., 2019). Antigenic variation is of fundamental importance in determining the underlying dynamics of host-pathogen interactions. We investigated these host-pathogen interactions by examining the impacts of pathogen genetic diversity on individual bighorn sheep (Ovis canadensis).
Mycoplasma ovipneumoniae (Movi) is a common pathogen of domestic sheep and goats that exhibits a high degree of genetic (strain) and phenotypic heterogeneity (Ionas et al., 1991a,b, Parham et al., 2006, Maksimović et al., 2017, Kamath et al., 2019). Spillover of Movi into bighorn sheep is often followed by epizootic transmission with high morbidity and variable mortality (Besser et al., 2012, 2013, 2014; Cassirer et al., 2018). The long-term population-level effect of exposure to Movi varies from full recovery to functional or local extinction (i.e., mortality of ≥90% of the population; Singer et al., 2000; Sells et al., 2015). Most surviving bighorn sheep clear Movi infections with immunity restricted to that strain (Plowright et al., 2013; Cassirer et al., 2017). Nonetheless, some become chronic carriers, and despite apparent immunity from clinical disease, do not clear the infection, consistently test positive for Movi carriage and pose infection risk to other individuals (Plowright et al., 2017; Garwood et al., 2020). As a result of these acute and chronic effects, respiratory disease remains one of the major factors impeding the conservation of bighorn sheep herds (Cassirer et al., 2018).
Individual host response, resulting dynamics and negative effects of disease within bighorn sheep populations is shaped by heterogeneities in Movi strain-specific virulence, exposure dosages, and prior Movi exposure histories (Cassirer et al., 2017, 2018). Describing the effects of exposure history and adaptive immune responses on disease severity and persistence, addresses an important knowledge gap regarding host-pathogen dynamics in wildlife. To address this gap, we used information collected prior to and during an unplanned Movi-associated pneumonia epizootic in a captive bighorn sheep facility containing individuals with different pathogen strain exposure histories to investigate the heterogeneity in responses to infection. We hypothesized during the epizootic that individuals with previous exposure to the dominant infecting strain would be protected whereas prior exposure to other Movi strains would provide limited protection from disease. We also predicted that disease would be less severe in individuals that mounted a greater antibody response and higher antibody levels would improve the odds of recovery. Lastly, we investigated potential environmental routes of Movi exposure that may have facilitated the epizootic. Filling these knowledge gaps cannot be easily done with free-ranging ungulates but is essential for the conservation of bighorn sheep. Therefore, this study represented a unique opportunity to increase understanding of respiratory disease processes in bighorn sheep and ultimately inform disease prevention and mitigation actions to improve the health of bighorn sheep populations and enhance conservation efforts.
Materials and methods
Study area/animals
Thirty-seven free-ranging adult bighorn sheep were tested 1 to 7 times to classify Movi infection status over a period of up to 4 years by state wildlife agencies prior to being transported to the South Dakota State University (SDSU) Captive Wildlife Research Facility in Brookings, South Dakota (44°20′ N, 96°47′ W; Figure A in Supplementary Appendix A; Table 1). Free-ranging animals were exposed to at least 1 of 4 genetic strains (Kamath et al., 2019) of Movi. Specifically, sheep from the Hells Canyon subpopulations: Asotin (n = 9), Lostine (n = 4), and Sheep Mountain (n = 2) of Washington, Oregon, and Idaho, respectively, had been exposed to the HC-404 strain of Movi. Sheep transported in October 2014 from the Black Butte herd (n = 8) of Washington within Hells Canyon carried the HC-404 strain from 1995 until a novel Movi strain, BB-393, was detected in 2014 (Cassirer et al., 2017). We refer to this strain exposure as BB-393/HC-404. Rapid Creek (number of sheep: n = 1) and Badlands (n = 2) herds within South Dakota were exposed to the SD-398 strain, and the Snowstorm herd (n = 11) from Nevada entered the study with exposure to the NV-400 strain (Table 1). All strains were associated with all-age epizootics of pneumonia in the source herds (Cassirer et al., 2013, 2018; Smith et al., 2014; Kamath et al., 2019). The BB-393 strain faded out of the Black Butte herd 2 years following the epizootic and the transfer of sheep to the SDSU captive facility. The other strains persisted in the field and were associated with recurring severe pneumonia outbreaks in lambs.
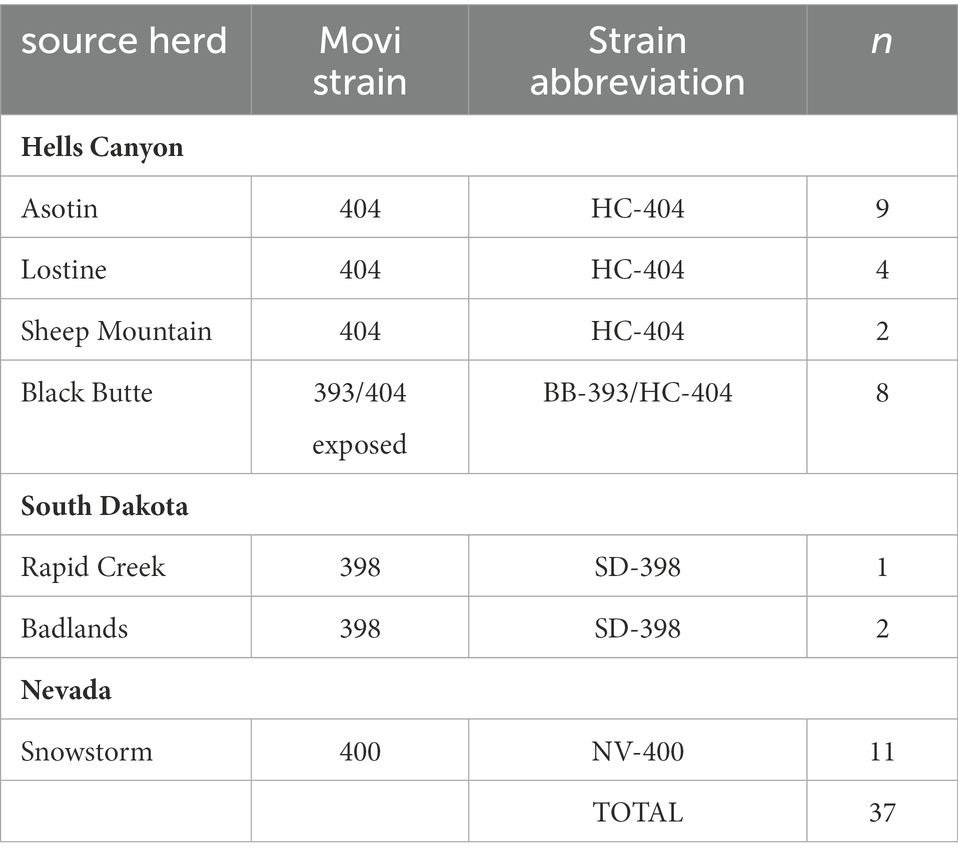
Table 1. Source herds for 37 bighorn sheep in the study at the South Dakota State University Captive Wildlife Facility, and Mycoplasma ovipneumoniae genotypes (strains) detected in those herds.
Capture, transport, daily care, and animal sampling protocols were reviewed and approved by the SDSU Institutional Animal Care and Use Committee (Number 14-076A). With the exception of individuals from the Snowstorm herd (Table 1), sheep were penned together (2–4 adults per pen; Figure 1) with other bighorn sheep with similar exposure and carriage status, which were known for each individual based on prior sampling as described above. In contrast, the carrier status for sheep from the Snowstorm herd (n = 11) was unknown. Thus, sheep were housed together in pen 9 (Figure 1) while their carrier status was being determined; however, the epizootic occurred before their carrier status could be ascertained.
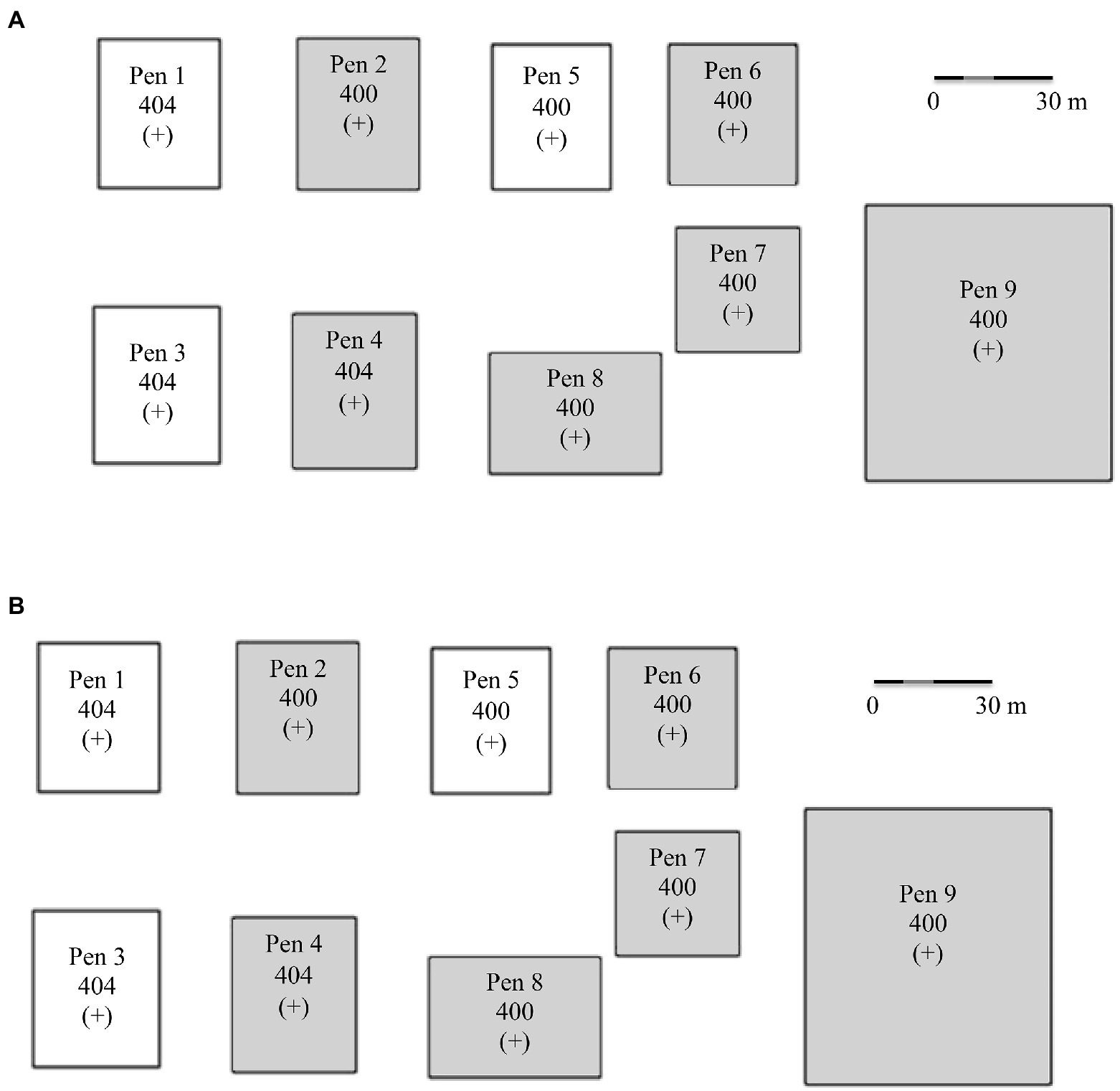
Figure 1. (A) Captive bighorn sheep research facility design and pen assignments based on known Mycoplasma ovipneumoniae history and current carriage as of 1 January 2015. (B) Unintentional indirect novel Mycoplasma ovipneumoniae strain infections as of 1 January 2016. Strain types: 404 = HC-404; 400 = NV-400; 398 = SD-398; 393 = BB-393/HC-404. A (+) indicates Movi detected in pen and (−) indicates Movi not detected in pen. Shaded pens indicate pneumonia-induced mortality occurred in that pen.
The distance over which Movi might be transmitted was unknown, therefore, we established a minimum distance of 15 m between carrier and other pens to minimize the potential for pathogen transmission between pens under the assumption most transmission is the result of close contact between individuals. Additionally, considering the prevailing winds, chronic Movi carriers were housed in pens in the eastern\downwind edge of the research facility (Figure 1). Further, personnel strictly followed biosecurity protocols including: (1) the installation of disinfecting foot baths at each pen gate for use immediately prior to entering and exiting each pen; (2) use of pen-specific feed and water pails; (3) changing protective clothing when handling possible Movi-positive sheep; and (4) use of order-of-pen-entry from west (Movi-negatives) to east (Movi-positives; Figure 1).
Microbiological sampling
Starting the autumn after arrival at SDSU, we collected serial microbiological samples from all sheep during 1 October–15 March annually, and periodically throughout the year from sheep without lambs at heel using chemical immobilizing agents (BAM; 0.43 mg/kg butorphanol, 0.29 mg/kg azaperone, 0.17 mg/kg medetomidine, Wildlife Pharmaceuticals, CO, United States) and a CO2 powered dart projector (Pneu-dart, Williamsport, PA, United States). Once under anesthesia, we fully inserted single polyurethane culture swabs (BD CultureSwab™ EZ System) into both nares and slowly rotated the swab shaft while gently contacting the mucosal tissue of the nasal wall and withdrawing the swab with circular motions. Duplicate swabs were collected and stored at −20°C after replacement in the sterile sheath prior to submission to the lab. We also collected 8–10 ml of blood via jugular venipuncture and extracted 0.5–1 ml serum for detection of antibodies to Movi via competitive enzyme-linked immunosorbent assay (ELISA) performed by the Washington Animal Disease Diagnostic Laboratory. We shipped all samples overnight to Washington State University (Pullman, WA, United States) for PCR analyses (Ziegler et al., 2014) and strain-typing using multi-locus sequence typing (MLST; Cassirer et al., 2017). If a mortality occurred, we collected the same samples, as well as bronchial swabs to detect and strain-type Movi from the lower respiratory tract. The ELISA values were used to assess the effect of the individual’s immune response during active infection on disease transition probabilities as described below.
Clinical observations
To maintain consistency and minimize disturbance, the same observer conducted daily 20-min vehicle-based observations using binoculars at distances of ≥27 m throughout the study (1 April 2015 to 1 April 2017). We observed and ranked signs of respiratory disease from 0 to 10 to indicate severity ranging from absent to extremely severe. Signs recorded for each individual included lethargy, sternal recumbency, ear paresis, nose licking, nasal discharge, and coughing (Supplementary Appendix B). To track disease progression during the epizootic event, we used daily clinical scores for all adults from 16 July 2015 to 1 April 2017.
Environmental samples
To identify environmental factors potentially contributing to transmission of Movi across pens, we sampled air and water from August 2015 to September 2016. We conducted aerosol sampling once every other week at each of 5 pens (Sartorius MD8 Airport Portable Air Sampler, Fisher Scientific, Waltham, MA, United States) for 20-min at 3 defined locations: immediately outside and down-wind of the enclosure being tested (<1 m from pen fence), halfway between the enclosure being tested and the immediately adjacent down-wind pen (halfway from source pen fence), and immediately outside the nearest adjacent down-wind pen (15–30 m from source pen fence) of the pen being sampled. We collected weekly drinking water samples from the permanent 94.6-L trough in each pen. We collected 50-ml of surface water and then thoroughly mixed the water and collected another 50-ml sample from the center of the trough. We also opportunistically conducted sampling of flying insects, bird nests in pens, and soil. We conducted invertebrate (e.g., Musca spp.) contamination assessments using fly tape traps and collecting replicate swabs of the external surface of trapped flying invertebrates. Finally, twice during the sampling period, we tested observer-fomite transmission by securing gauze to the bottom of work boots and traversing the pen for 5-min focusing on areas heavily used by sheep (e.g., feed, water stations, and shelters). Immediately after collection, air sample filters were cut in half; one half was dissolved in 20 ml sterile Hayflick’s broth media and the other half was placed dry in a sterile envelope. All environmental samples were overnight shipped to Dr. Tom Besser’s laboratory at Washington State University for analyses. Figure B in Supplementary Appendix A portrays the timeline for each of the major activities associated with this study.
Laboratory methods varied by sample type. We varied the volumes for resuspension and for culture in response to different amounts of debris or dirt in the sample types (i.e., air sample filters the cleanest, water samples intermediate, and boot wash samples the dirtiest). Specifically, we transferred aliquots of water samples (25–80 ml, based on 50% of the volume collected) to centrifuge tubes (Product# 3119-0050PK, Thermo-Fisher Scientific, Waltham MA) and pelleted [20 min, 4°C, 18,500 × g (RCF average, J-25.50 rotor)]. We resuspended pellets in 1 ml phosphate buffered saline (PBS), which was divided into two 500 μl aliquots, one of which was retained for PCR detection of Movi, and the other was cultured to detect viable Movi. For air samples, the Hayflick’s broth containing the dissolved half filter cultured to detect viable Movi. The dry half air filter was dissolved in PBS, pelleted similarly, and the pellet was resuspended in a 500 μl aliquot of PBS, which was retained for PCR detection of Movi. We agitated the boot wash gauze samples in 100 ml PBS in new Ziploc bags (1 gal). We then removed 30 ml and pelleted as described above for water samples. We resuspended the pellets in 5 ml PBS and removed a 500 μl aliquot for PCR detection of Movi. Another 1 ml aliquot was then removed and cultured for detection of viable Movi. Swab samples from arthropods and other miscellaneous environmental (e.g., air) samples were processed for Movi detection by realtime PCR.
For detection of Movi by realtime PCR, we weighed pellets obtained from the 500 μl aliquots described for water and boot samples, and limited the amount retained for DNA extraction to 25 mg. For air samples, since the pellets were invariably <25 mg, we extracted the entire pellet. Swab samples were eluted directly into 500 μl aliquots of PBS. We obtained DNA extracts from these specimen and aliquots using QIAmp DNA Kit (Qiagen United States, Germantown MD) according to the manufacturer’s directions. Realtime PCR was then used to analyze extracted DNA (Ziegler et al., 2014).
To culture viable Movi, if samples were not already suspended in a Mycoplasma broth culture media, we transferred samples to Mycoplasma broth tubes (Product# R102, Hardy Diagnostics, Santa Maria CA) and mixed well. One milliliter pre-incubation aliquots were removed from both Hayflick’s broth (air filter samples) and Mycoplasma broth, and stored at −20°C. The remainder of the inoculated broths were incubated at 37°C with 5% CO2 for 72 h, after which we removed and stored at −20°C a second 1 ml post-incubation aliquot. The pre-and post-incubation aliquots were thawed, DNA extracted (QIAmp DNA Kit), and analyzed by realtime PCR (Ziegler et al., 2014). We used relative cycle threshold (CT) values to identify samples with stronger post-incubation realtime PCR detection consistent with growth of viable Movi.
Statistical analyses
To capture the initial transmission and incubation prior to the epizootic, which was first observed 15 July 2015, we initiated our model 1 January 2015 and concluded it on 1 April 2017. Using Kermack and McKendrick’s (1927) classic compartmental SIR model structure, we developed a model with three main individual disease states: susceptible (S; i.e., susceptible to infection), infected (I; i.e., those that were currently displaying clinical symptoms of respiratory disease and were presumed infectious), and recovered (R; i.e., those who have had the disease but were no longer symptomatic; Anderson and May, 1991). To model an individual’s disease progression during the epizootic between the SIR compartments, we used a Bayesian mixture model for competing risks (Larson and Dinse, 1985; Figure 2). This approach assumed all individuals will eventually transition from their current state to a new state (i.e., probability of staying in its current state as time , and this new state was determined by some stochastic mechanism when they entered their current state (Larson and Dinse, 1985). Individual host response resulted in heterogeneous transition times into each new state. We also assumed that the day when an individual entered a state was when the daily hazard of transitioning from that state began.
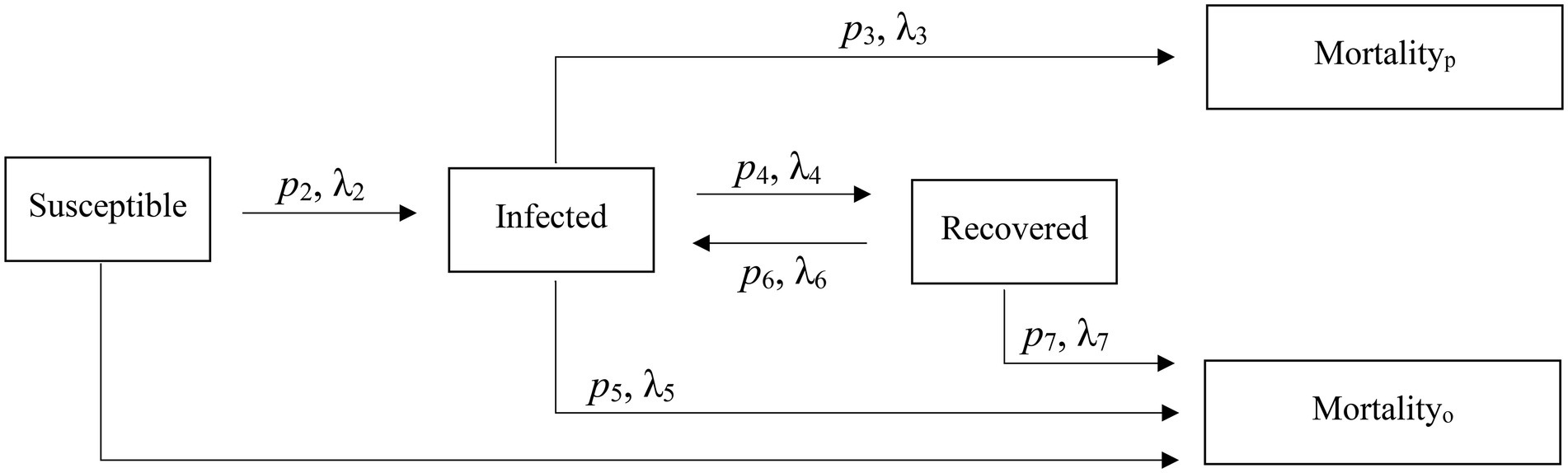
Figure 2. Reversible epidemiological SIR model and associated parameters used to characterize an epizootic in captive bighorn sheep.
We modeled the overall probability an individual in state i transitioned to state j, and then, conditional on this transition, we estimated the associated daily hazard rate of making the transition. Thus, the transition probabilities acted as mixing parameters for the various hazard rates (Larson and Dinse, 1985). We initially classified all adults as S because no individuals were initially displaying clinical signs and a general lack of Movi cross-strain immunity (Cassirer et al., 2017). We used cough and nasal discharge scores to define disease states. We defined the start of disease state I as 3 weeks prior to the date of the onset of cough (score ≥ 1) or nasal discharge (score of >2; Besser et al., 2014, Supplementary Appendix B). We classified an individual as being in the R state if they did not present any indication of coughing or nasal discharge (score ≤ 2) during daily clinical symptom observations for a minimum of 60 days (Besser et al., 2014).
The model was structured so I individuals could not return to the S state; however, R individuals could return to the I state. We also included two absorptive states (i.e., probability of transitioning from an absorptive state = 0): (1) death unassociated with disease (mortalityo), and individuals could die and enter this state from any of the states; and (2) death due to pneumonia (mortalityp). Only I individuals could enter this latter state. Lastly, individuals that did not die during the study we considered right-censored in the state they occupied at the study’s conclusion. The model structure is shown in Figure 2.
Transition Probabilities-We estimated most transition probabilities as a function of covariates using a logit link function as follows:
where is the probability of transitioning from state i to j, X is a covariate matrix, which varied between transition probabilities, and are the parameters for the covariate effects. However, individuals transitioning from state I could move to 3 different potential states and therefore, we used a multinomial logit model:
where are the parameters for the covariates associated with transitioning to state j from I.
When estimating the probabilities of transitioning from S, (p2), we included an effect for the (1) individual’s immune response (Movi cELISA % inhibition value; %II hereafter) at the nearest sampling event (4–7 months) prior to the epizootic, and (2) the initial Movi strain detected in each individual at the study start. For the probabilities of transitioning from I, (p3, p4), for each individual we included (1) the %II effect, (2) the individual’s immune response during active infection (i.e., averaged Movi cELISA % inhibition values for all tests conducted while an individual was alive and had an active infection based on clinical signs; %IA hereafter), (3) the individual’s initial Movi strain, and (4) an indicator of whether the NV-400 strain was detected in an individual during the epizootic (CST NV-400 hereafter). Lastly, we specified the probability of transitioning back to I from R (disease recurrence; p6) as a function of the individual’s initial Movi strain (Table 2).
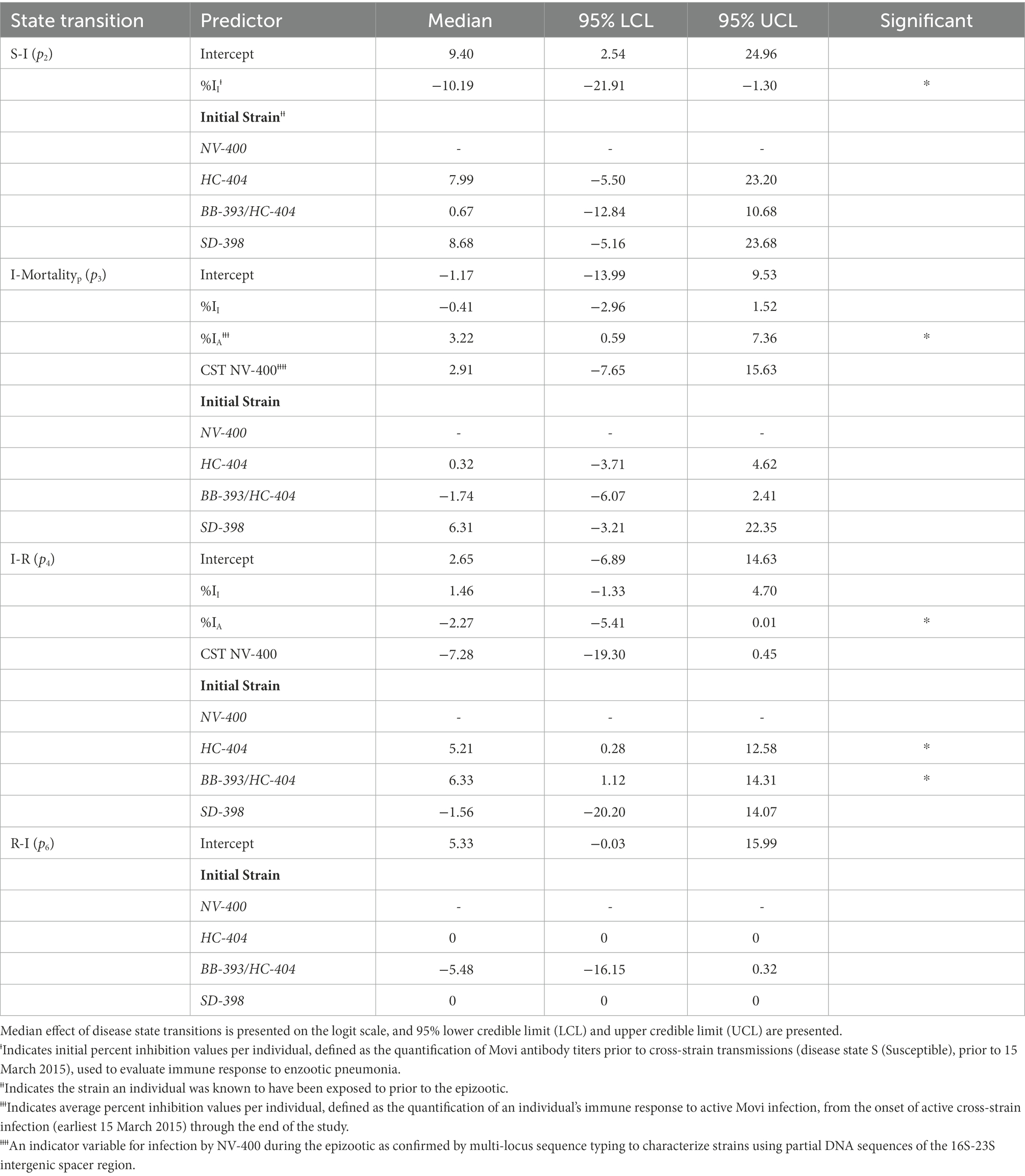
Table 2. Estimated posterior distribution of disease state transition probabilities (pn) for bighorn sheep as a function of covariates using a logit link function for state transitions from susceptible to infected (S-I) and disease recurrence (R-I) and a multinomial logit model for individuals transitioning from the infected state: infected to pneumonia-related mortality (I-Mortalityp) and infected to recovered (I-R).
Transition Hazard Rates-We used a piece-wise constant function to model each daily transition hazard as a function of covariates of interest using a proportional hazards assumption:
where is the daily hazard of transitioning from state i to j during interval t, are the parameters for the covariate effects and is a regularizing term for daily interval t, which is used to account for temporal autocorrelation and provide temporal smoothing. We used a constant model for where j is mortalityo regardless of the current state i because this transition hazard rate was independent of disease state.
We used a kernel convolution model (Higdon, 2002) for the parameters to regularize across days of the study when transitioning from S to I and from I to mortalityp. This modeling approach provided a flexible means of accounting for temporal autocorrelation and permitted the estimation of the level of smoothing supported by our data (Higdon, 2002). For the S to I transition hazard with knot locations set at each day of the study, the model was:
which uses a Normal density kernel truncated at 120 days (i.e., days beyond 120 days from the t do not influence ) with a variance of , is the squared distance between t and the nth knot location, and is the latent random day effect at the nth knot. Knots were created for each day. We specified a Gamma (1, 1) for and a Normal ( prior for the vector of effects. We used a Gamma (1, 1) prior on the precision, . We used the same kernel convolution model for smoothing the temporal effects, , for the hazard of transitioning from I to disease-associated death state and once again we used a Gamma (1, 1) prior on and a Gamma (1, 1) prior on .
When estimating the log hazard rate for transitioning from S to I, (λ2), we examined the effect of individual immune response and historical strain exposure. Specifically, we include aa (1) %II effect and (2) an effect for distance from NV-400 strain pen. To model the log hazard of transitioning from I to the mortalityp state (λ3), we used a (1) %II effect and (2) %IA effect in the model. For the transition from I to the R state (λ4), we modeled the log hazard using a (1) %II effect, a (2) %IA effect, and the individual’s initial Movi strain. Finally, we modeled the hazard of transitioning back to I from R (i.e., disease recurrence; λ6) as a constant. Table 3 contains the covariates used in each hazard model.
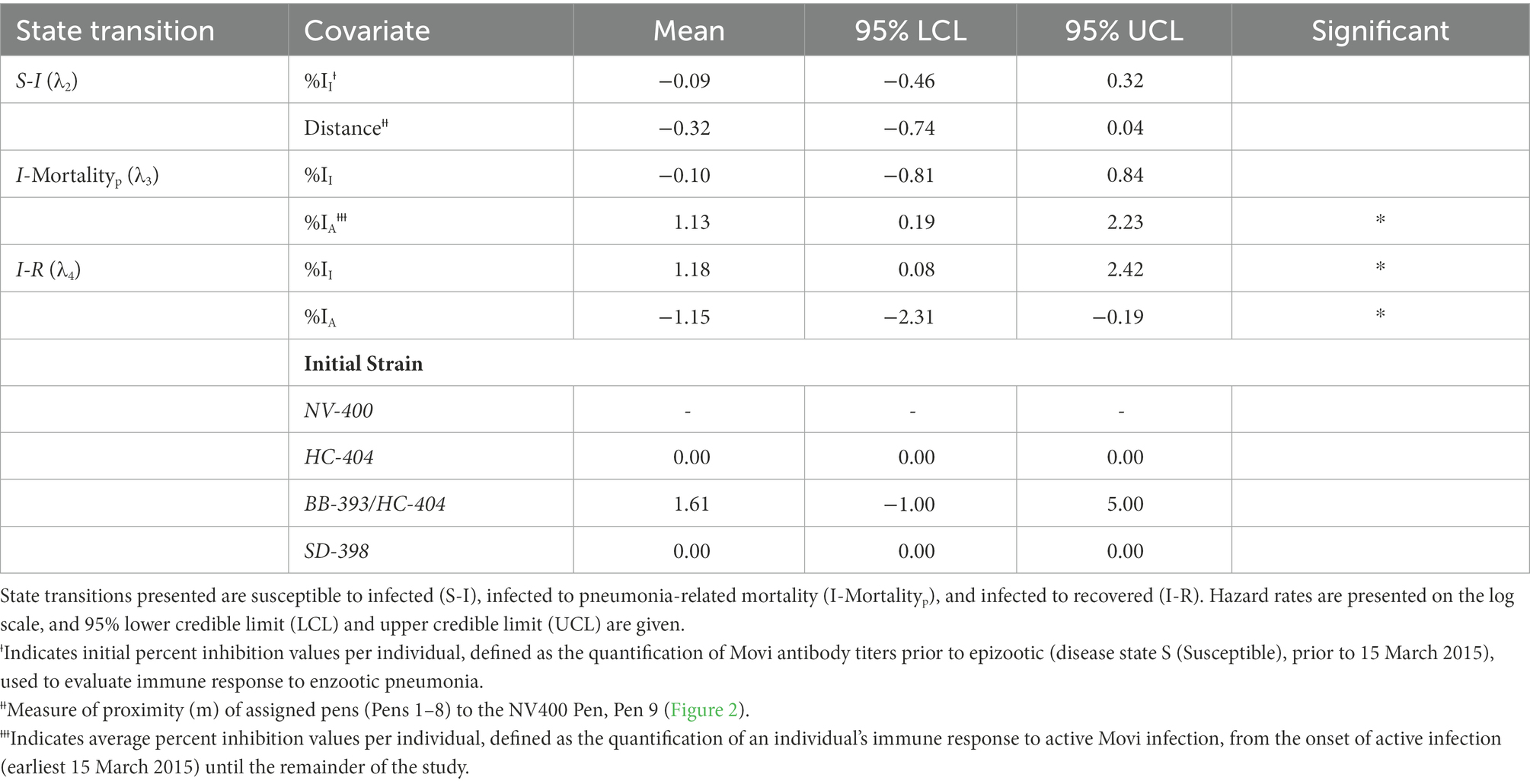
Table 3. Estimated posterior distribution of daily disease state transition hazard (λn) for bighorn sheep as a function of antibody levels and prior exposure history.
Posterior Distribution-Given these probabilities and hazards, the kth individual’s transition from state i to j makes the following contribution to the log likelihood, :
where ei,k is the kth individual’s entry time into state i and ri,k is the last time the kth individual was known to be in state i. Note, this model allows for interval censoring of the transition time (e.g., the transition time is only known to have occurred between ri,k and ej,k). Individuals who are in a state other than death at the end of the study (i.e., right censored) contribute the following:
where the first summation is over the J possible states that can be transitioned to from state i, and T is the day the study ended. To complete our model, we specified diffuse Uniform (−100, 100) priors on the intercept/base-line log hazard rate, and Normal (µ=0, σ2=100) priors for all covariate parameters used in estimating the transition probabilities and the daily hazard rates. The posterior distribution is then proportional to the sum of the log of the prior distributions and the log likelihood.
Estimation-We employed JAGS (Plummer, 2003) in Program R (R Core Team, 2018) via the R2JAGS package (Su and Yajima, 2015) to estimate the posterior distributions of our parameters of interest using Markov Chain Monte Carlo (MCMC) methods. Because our likelihood is in a non-standard format, we used the “zeros trick” (Lunn et al., 2013) to permit its use in JAGS. We ran 3 chains for 100,000 repetitions, and removed the first 25,000 iterations for burn-in. Each chain was started with dispersed starting values, and graphical checks were used to monitor for evidence of non-convergence.
Results
Epizootic dynamics
We first observed pneumonia in lambs, followed by adults and eventually confirmed pneumonia-induced deaths in 6 of 9 pens in the study (Figure 1). These disease events were often associated with infection by Movi strains novel to the bighorn sheep within the affected pens (Supplementary Appendix C).
Timing of Pneumonia by Age Class, Pen, and Strain Type – In 2015, parturition occurred from 24 March to 21 June and all lambs died from pneumonia or other causes by early August (Supplementary Appendix C). The onset of clinical signs of pneumonia in lambs preceded the detection of pneumonia in adults. Over the course of the epizootic, two strains of Movi spread across pens to individuals previously exposed to different strains (i.e., cross-strain transmission, Figure 1).
Clinical signs of pneumonia were first detected in the Snowstorm lambs in pen 9 (Figure 1B) on 28 April 2015. The Snowstorm lamb deaths occurred from May through June 28 at a mean age of 43 days (range = 22–87 days; n = 7). Comparatively, births and onset of clinical signs in lambs were observed later in the other pens. In an adjacent pen (pen 7; Figure 1B), the first observation of clinical signs was 27 May 2015 and subsequent mean age at mortality was 38 days (range = 27–49; n = 2). The presence of the Snowstorms NV-400 strain type detected in the mortality samples for both pen 7 lambs was not expected based on the HC-404 strain carried by ewes present in pen 7 (Figure 1A) and represented the first genetic confirmation of cross-strain transmission. The final lamb in pen 7 died 14 July 2015 from a confirmed cross-strain infection, immediately prior to clinical detection of cross-strain infection in adults (Supplementary Appendix C).
Near the end of July 2015, we witnessed the unexpected mortality of two Snowstorm ewes (pen 9; Figure 1B). Cause of death was attributed to a severe acute pneumonia associated with infection by strain NV-400. Simultaneously, we noticed adults in pen 7, which were previously asymptomatic, exhibiting severe clinical indications of pneumonia immediately after the last lamb expired in that pen (Supplementary Appendix C). Subsequent to the index case of clinical pneumonia attributed to cross-strain infections in each pen, the median number of days for all adults in the pen to display signs of clinical disease was 15 days (range = 9–47 days). Median time between onset of symptoms and mortalityp was 126 days (range = 45–489 days).
Clinical observations summary-Observation of clinical symptoms of respiratory disease was the basis of the morbidity and infection calculation used in our study. We detected 21 instances of transmissions of the NV-400 strain to sheep previously exposed to HC-404, BB-393/HC-404, and SD-398 and 4 transmissions of HC-404 to the uninfected sheep previously exposed to BB-393/HC-404 and SD-398. After 1 November 2015, Movi strains NV-400 and HC-404 were the only strains detected. We documented signs of respiratory disease in 95% (n = 35) of study animals; however only 68% of the individuals were documented as infected with a novel Movi strain. Thus, 27% of affected individuals were symptomatic and infected with their initial strain. Most (82%, 9/11) individuals entering the study with NV-400 (the Snowstorm bighorn sheep herd) showed clinical signs when only their initial strain was detectable. Cross-strain infections were detected from July to October 2015, and time to detection varied by pen assignment but generally moved from east to west, opposite of the prevailing winds, within the research facility (Figure 1).
Health sampling
Between February 2015 and December 2016 we collected 2–10 nasal swab samples from the 37 bighorn sheep in our study. The proportion of Movi-positive individuals detected within each month increased from 0.19 to 0.83 during the epizootic. We confirmed pneumonia as the cause of death in 43% (n = 16) of the bighorn sheep in the study. Other sources of mortality included darting complications (n = 2), birthing complications (n = 1), West Nile Virus (n = 1), liver hemorrhage (n = 1), flystrike (n = 1), gastric abscess (n = 1), euthanasia due to emaciation (n = 1), and injury (n = 1). Survival to the end of the study was 33%, of which half (n = 6) remained symptomatic and in the I state.
Environmental samples
During the epizootic, we detected Movi DNA in air, water, and invertebrate samples (Table 4). We detected aerosolized Movi DNA via PCR downwind of the target pen (directly outside of target pen: n = 9 detected (CT ≤ 35), n = 2 indeterminate (CT = 35.01–39.9); between target pen and nearest occupied pen downwind from target pen: n = 1 detected, n = 4 indeterminate; and outside nearest occupied pen downwind from target pen: n = 1 detected, n = 2 indeterminate). We detected Movi DNA in 22 water, 5 invertebrate, and 1 permanent fly trap samples. We did not detect Movi DNA on the soil surface sampled via the gauze on our boots or in a bird nest constructed in an occupied pen’s shelter (CT ≥40; Table 4). Viable Movi, based on Movi growth in broth culture, were not detected in any environmental sample.
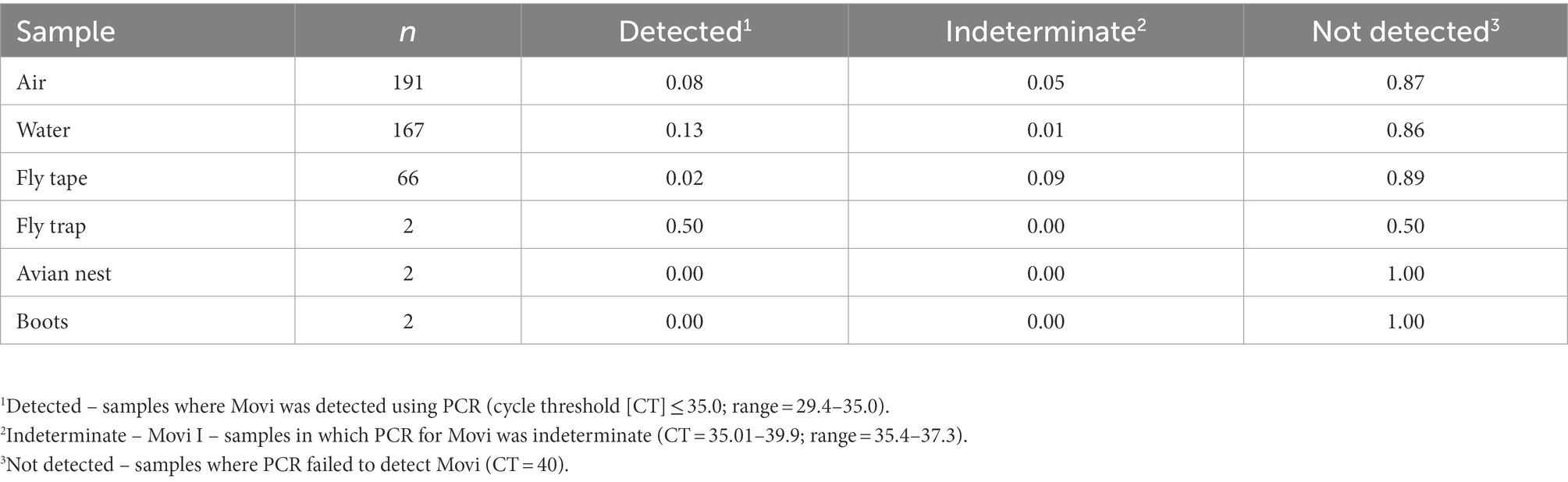
Table 4. Prevalence of Mycoplasma ovipneumoniae detections in environmental samples collected to identify possible modes of cross pen transmission for bighorn sheep at the South Dakota State University Captive Wildlife Facility from 2015 to 2016.
Disease state transition analyses
Right Censored Susceptible Adults-One chronic carrier ewe with HC-404 Movi from the Lostine herd (e.g., always positive by PCR; n = 8) failed to display clinical symptoms of pneumonia during our study and was right censored in the S state. A second ewe from the Snowstorm herd was neither documented carrying Movi nor did it display clinical symptoms and therefore, was right censored in the S state. All other adults in our study transitioned out of the S state as documented by clinical signs.
Transition from the Susceptible State to the Infected State-A high probability existed that a previously exposed bighorn sheep will become clinically infected when exposed to a novel Movi strain (Table 2). The probability of cross-strain infection was not influenced by previous strain type exposure, whereas pre-infection antibody level (%II defined as the Movi cELISA % inhibition value) had a significant negative (protective) effect on cross-strain transmission (median effect size = −10.19; 95% credible interval [CI] = −21.91 – −1.30; effect sizes of covariates on probabilities and daily hazards are presented on the logit and log scale, respectively; Table 2), with individuals having higher pre-existing %II values being less likely to become infected with a novel strain (Figure 3A).
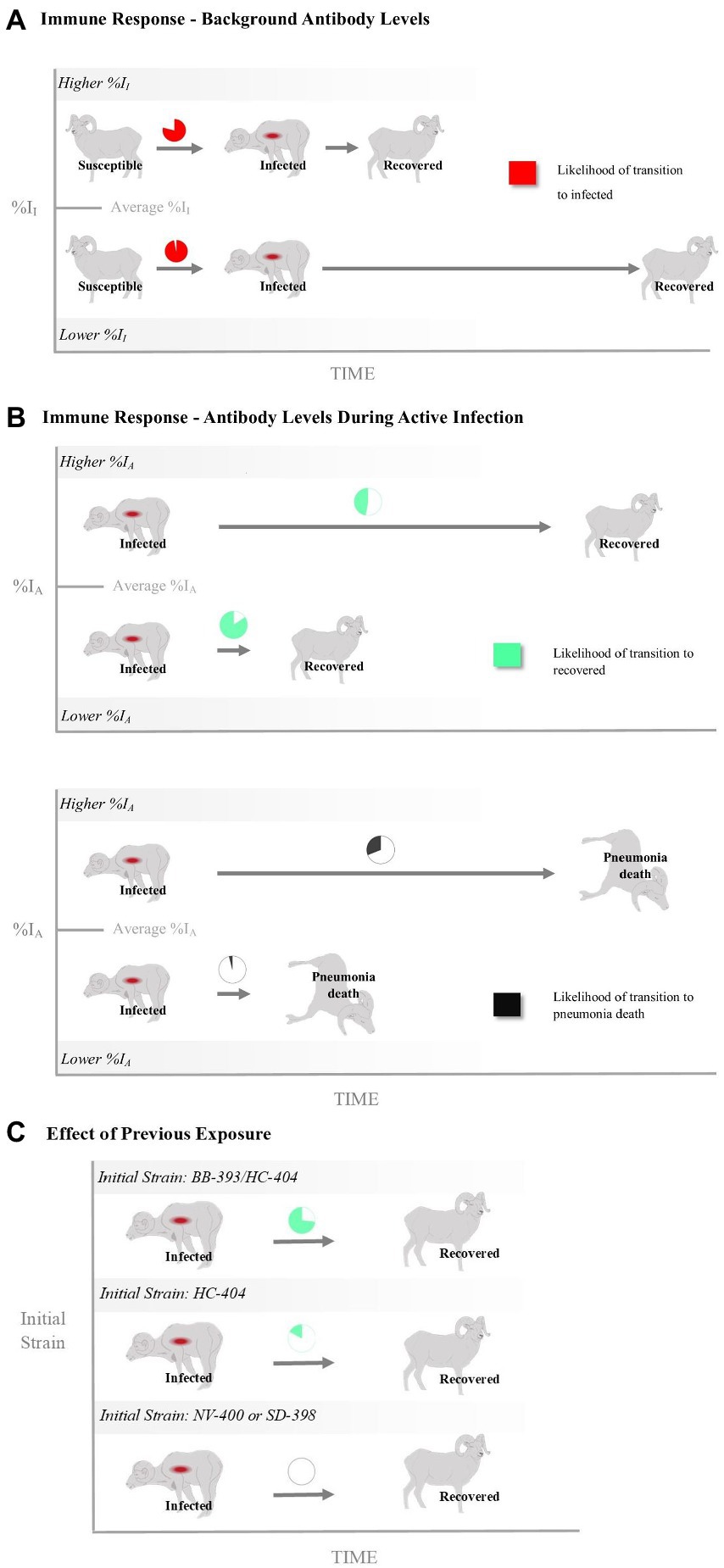
Figure 3. The relationship of bighorn sheep immune responses with infection and death as represented by the probability and rate of transitioning between disease states (i.e., susceptible, infected, recovered and disease-induced mortality). Shown are effects of antibody levels measured (A) prior to and (B) during a Mycoplasma ovipneumoniae (Movi) epizootic and (C) as a function of past exposure history. The %II effect is an individual’s Movi cELISA percent inhibition (%I) value at the nearest sampling event prior to the epizootic. The %IA effect is the averaged Movi cELISA % inhibition values for all tests conducted while an individual was alive and had an active infection based on clinical signs. The initial strain is the strain the individual was known to have been exposed to prior to the epizootic. Pie charts for %II and %IA depict the mean probabilities above (Higher) or below (Lower) the mean covariate values. Times to transition are depicted as the relative difference in rates between 1 standard deviation above and below the mean covariate values.
The rate at which individual bighorn sheep became infected exhibited a bimodal distribution over time. Most individuals exhibited disease onset approximately day 200 (late July 2015) or 275 [early October 2015 (λ2; Figure 4A)]. An individual’s %II value did not affect the rate of becoming infected (Table 3). Proximity to the NV-400 pen/pen 9 (mean effect size = −0.32; SD = 0.20; 95% CI = −0.74 – 0.04) had a marginal effect on the infection hazard, with individuals penned closer to the NV-400 pen becoming infected sooner than individuals penned farther away; however, the effect was not statistically significant.
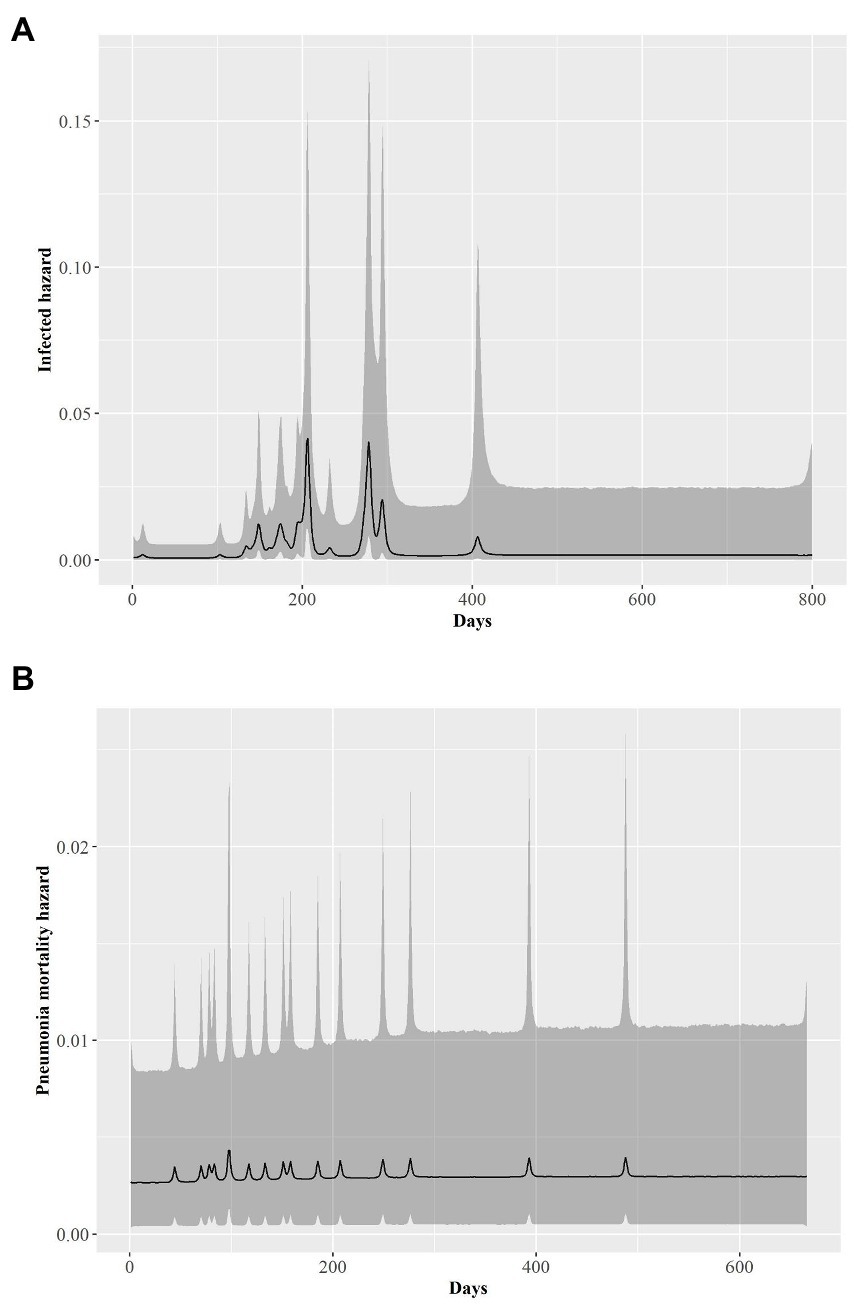
Figure 4. Time series of the estimated posterior distributions on the log scale of daily hazard rates (black line) for bighorn sheep (A) transitioning from being healthy to infected by pneumonia and (B) dying of pneumonia-related causes from 1 January 2015 to 1 April 2017. Gray shading is associated the 95% credible envelopes.
Transition from the Infected State to the Mortalityp State–There was a large degree of uncertainty when estimating the long-term probability of bighorn sheep transitioning out of the infected state (Table 2). The effects of %II, initial Movi strain type exposure, and CST NV-400 did not have a significant effect on the probability of an infected bighorn transitioning to mortalityp (Table 2). Nonetheless, individuals that mounted a higher antibody response to active symptomatic infection (%IA, defined as the average Movi cELISA % inhibition values for all tests conducted while an individual was alive and had an active infection based on clinical signs) were significantly more likely to transition to pneumonia-induced mortalityp (median effect size = 3.22; 95% CI = 0.59–7.36; Figure 3B).
Daily hazard rate of mortalityp was relatively constant with most mortality events occurring by day 300 of our study (λ3; Figure 4B). Mortalityp hazard was not associated with %II (median effect size = −0.10; SD = 0.42; 95% CI = −0.81–0.84; Table 3). In contrast, there was an effect for %IA (median effect size = 1.13; SD = 0.52; 95% CI = 0.19–2.23; Table 3) on mortalityp hazard, with individuals with a larger %IA experiencing mortalityp faster than individuals with a lower %IA (Figure 3B).
Transition from the Infected State to the Recovered State–Our model predicts the probability that an individual remains in the infected state is relatively high (Table 2). Probability of recovery was not affected by %II (Table 2); however, the initial strain an individual was exposed to prior to the epizootic did influence recovery. The individuals exposed to the BB-393/HC-404 or the HC-404 strain had higher recovery probabilities compared with those exposed to NV-400 or the SD-398 strains (Figure 3C; Table 2). In addition, bighorn sheep with a larger %IA were less likely to recover from novel Movi strain infection (median = −2.27; 95% CI = −5.41–0.01; Figure 3B).
The effect of immune response may drive recovery hazards (λ4), but with varying influences. If an individual were to recover, recovery occurred faster for bighorn sheep with increased prior immunity (%II; mean effect size = 1.18; SD = 0.59; 95% CI = 0.08–2.42; Figure 3A; Table 3) but slower for bighorn sheep experiencing a higher immune response during the active cross-strain infection (%IA; mean effect size = −1.15; SD = 0.53; 95% CI = −2.31 – −0.19; Figure 3B; Table 3).
Transition from the Recovered State to the Infected State–Very few individuals experienced disease recurrence in our study (n = 3). Although our sample size is small, our model does not indicate differences in the probability of clinical disease recurrence based on initial Movi strain exposure histories (p6; Table 2).
Discussion
Our findings support our hypothesis and conclusions from previous work that suggest that Movi strains vary in pathogenicity and that naturally acquired immunity in the bighorn sheep host is strain-specific (Justice-Allen et al., 2016; Cassirer et al., 2017). Previous exposures may influence survival following novel strain infection although in contrast to our hypothesis, even previous exposure to the outbreak strain did not provide complete protection. Immunity from previous exposure, as indexed by serologic antibody scores prior to the outbreak, did not prevent infection but was associated with resistance to outbreak-associated disease, and a faster rate of recovery in the rare cases in which it occurred. Counter to our hypothesis, however, higher immune responses during the outbreak corresponded to both increased probability and rate of pneumonia-induced mortality, and if it occurred, a slower rate of recovery; associations likely driven by disease progression and an inability to control infection.
Surprisingly, we observed that the most pathogenic, predominant strain in the study, which was detected in 81% of deaths, was associated with high mortality (45%) of sheep with previous exposure to and current infection with this strain. This result contrasts with observations of relatively low levels of sporadic pneumonia deaths of adults in chronically infected, free-ranging, bighorn sheep populations attributed to protective immunity (Cassirer et al., 2013; Plowright et al., 2013; Smith et al., 2015). Generally, immunity is developed during infection of a host, and acts to reduce pathogen establishment, survival, reproduction, or maturation (Wilson et al., 2002). Nevertheless, immune responses may also be non-protective, ranging from benign non-neutralizing responses that simply serve as a marker of antigenic exposures associated with infection to immunopathological responses associated with adverse reactions and increased disease severity (Hornef et al., 2002; Monack et al., 2004; Quinton and Mizgerd, 2015). An incomplete immunity in the host observed in this study might have been aggravated by simultaneous exposure to more than one strain, infectious dose, and undetected genetic variation in the pathogen.
Host immune response
Our findings indicate that adaptive immunity is an important factor driving individual heterogeneity in the response of bighorn sheep to disease. It is generally accepted that immune responses are genotype-specific, with previously unseen genotypes providing those pathogens with a growth advantage in semi-immune hosts (Simenka, 2005). Our finding reflects the complex notion that individual bighorn sheep with stronger immune responses following previous Movi exposure were less likely to exhibit disease symptoms and experienced shorter recovery times (Table 3; Figure 3). Only their initial strain or a strain that they had previously been exposed to in the wild (NV-400 or BB-393/HC-404, respectively) was detected in most (67%; n = 6) of bighorn sheep that recovered. In contrast, only 13% (n = 3) of bighorn sheep that experienced a novel cross-strain transmission event recovered in our study.
Pathogen strain type and competition
Some investigators have reported that simultaneous Movi infections with multiple strains in domestic hosts can result in more severe pneumonia (Parham et al., 2006; Rifatbegović et al., 2011). Our strain typing method was unable to detect more than one strain unless both strains were at similar concentrations in the sample and were amplified for sequencing at similar efficiency. We did not detect a cross-strain infection in any of the sheep in pens with current infection and with prior exposure to the predominant strain, although they were in close proximity to and downwind of the prevailing wind direction to the pens with other strain types. This outcome indicates that the predominant strain was more prolific and outcompeted other strains making it unlikely that we would detect co-infection with MLST. The question as to whether co-infection with a subdominant strain contributed to disease and mortality in this study is unclear.
While most mortality in this study was attributed to a single strain of Movi, any intrinsic reasons for this virulence could not be determined. The MLST strain-typing method used in our study has been widely applied in epidemiologic and evolutionary studies. Kamath et al. (2019) validated the strengths of employing this method to describe Movi strain diversity and bighorn sheep spillover events from 1984 to 2017 throughout the western United States. The MLST, however, characterizes approximately 0.15% of the genome (Cassirer et al., 2017) and provides no information about variation in presence or expression of specific virulence genes. Therefore, our strain-typing method fails to distinguish within-strain variants that may differ in virulence or neutralizing surface epitopes. Consequently, another possible explanation for unexpected lack of protection of prior exposure to a strain is that a new, undetected, variant developed during the outbreak. The likelihood and effect of multiple simultaneous Movi strain infections and antigenic or other variation within strains in this host is unknown and warrants investigation.
Modes of transmission
We detected instances of Movi transmission based on invasion of new strain types across distances of up to 30 m from infected bighorn sheep. In spite of careful planning and consideration of prevailing winds, we hypothesize that spread occurred through aerosolized droplets produced by symptomatic bighorn sheep in adjacent pens. Closely related Mycoplasma spp. infectious agents have been recognized as viable and transmissible through aerosolization. Mycoplasma hyopneumoniae, which causes atypical pneumoniae in swine (Stärk et al., 1998; Desrosiers, 2011), has been documented to be transmissible via aerosol droplets 9.2 km from the infected source pen (Otake et al., 2010). In addition, M. bovis, the primary agent in cattle pneumonia epizootics, and M. synoviae and M. gallisepticum, which cause acute or chronic respiratory disease in poultry, respectively, can infect livestock by airborne pathogen transmissions (Landman et al., 2010; Kanci et al., 2017). Furthermore, secondary Pasteurellaceae agents are transmissible through aersolization up to 18 m (Dixon et al., 2002) as well as by fomite contamination (Burriel, 1997; Clifford et al., 2009).
We detected Movi DNA in aerosol, fly, and water samples although we were unable to culture viable organisms from any samples that were cultured. This is likely due, at least in part, to the fastidious nature and rapid death of Movi outside the host and we might have been more successful if we had immediately inoculated aerosolized droplets into culture broth. Besser et al. (2014) reported Movi transmission within and between pens up to 12 m distant. Although average wind direction was generally opposite to pen-to-pen transmissions (Supplementary Appendix C), wind direction frequently varied and wind speeds of >60 km/h were common; wind gusts >100 km/h that occurred in late June and early August may have facilitated rapid movement of droplets containing viable bacteria across the facility (Figure 1). We detected aerosolized Movi DNA at the boundary of the nearest downwind pen at the maximum range tested (30 m) from infected bighorn sheep.
Our study is the first to document flies (Musca spp.) as a possible vector of transmission for Movi in bighorn sheep infections. Some fly species feed on nasal and oral discharge and have been implicated in the rapid spread of similar agents, including M. conjunctivae, which causes infectious keratoconjunctivitis in wild and domestic Caprinae (Degiorgis et al., 1999; Giacometti et al., 2002; Fernández Aguilar et al., 2019). Permanent fly traps were secured to the roof of shelters where symptomatic bighorn sheep spent a considerable amount of time, particularly as disease progressed. The positive Movi detection of the permanent fly trap could be the result of airborne particles expelled during coughing. Nonetheless, the Movi-positive fly tape was a direct sample of flies and offers strong evidence as a possible route of transmission in captive studies. Nonetheless, the strain-type that was identified directly from flies (SD-398) was never detected in bighorn sheep carriage in that pen (i.e., pen 1). Pens 4 and 6, which were approximately 35 m and 115 m away, were the pens containing bighorn sheep with the previously detected SD-398 strain. Additional investigations aimed at detecting transmissible and viable Movi from flies that feed on oronasal secretions are necessary to better understand the effect of vector-borne Movi transmission in bighorn sheep epizootics.
Study limitations
We did not intend to evaluate bighorn sheep responses to novel Movi strain invasions, and we purposefully designed the pens to prevent this from happening. Because we were not prepared for the outbreak, our data are largely limited to observation of clinical signs. Close observation of clinical signs in habituated animals or animals in captive facilities, however, has been used to classify health status and pathogen transmission in previous studies (Lonsdorf et al., 2018; Sandel et al., 2021), and when capture and testing were feasible our disease state classifications based on observations concurred with 94% of all PCR and serological analyses. Another weakness of the opportunistic nature of the study is that the first adult infections may have pre-dated the start of our intensive monitoring of clinical signs on 15 July. Therefore, duration of infection may have been longer than we assumed in the model.
Secondly, we could not directly measure dose effects. Pen 9 contained the most adults (11) of which 7 gave birth to lambs that subsequently became infected and developed respiratory disease, whereas other pens contained from 2 to 4 adults and 0–3 infected lambs. Therefore, pen 9 had the highest disease burden and adults in pen 9 were likely exposed to the highest cumulative doses of Movi from each other and from their offspring. We attempted to account for dose effects using an index, the distance to pen 9 (Figure 1), the index pen. We did not document a significant effect of this index on the rate at which individuals became infected (Table 3); however, we were not able to completely disentangle the effect of distance and strain exposure histories. Thus, a better understanding is needed of how dose might overcome immunity obtained from prior exposure and influence response to infection and conversely, whether reducing dose could decrease virulence.
Lastly, our measure of immune response to Movi is a cELISA test, which detects antibodies based on their ability to inhibit binding of a Movi-specific monoclonal antibody to Movi antigen in vitro. The monoclonal antibody used in the assay was selected on the basis of immunodominance of its epitope across multiple bighorn sheep populations infected by diverse Movi strains, but this epitope has not been shown to be involved in protective immunity. As a result, immune responses as measured by percent inhibition (%I) should be considered to primarily reflect the intensity of current or past Movi exposure but not the effectiveness of the immune response. Both beneficial and adverse effects of immune responses to Movi infection are plausible: while immune responses may be associated with decreased pathogen carriage (Niang et al., 1998; Plowright et al., 2013), strong humoral immune responses may induce autoimmune responses in domestic sheep and have been hypothesized in wild sheep (Niang et al., 1998; Cassirer et al., 2018). Lacking a better understanding of the interactions between the host’s immune system and Movi virulence factors, the cELISA may fail to distinguish protective from ineffective or harmful immune responses, complicating interpretation of immunity in the study.
Conclusion
Despite limitations associated with an unplanned outbreak, this study provided the rare opportunity to examine the effect of exposure history on individual response of animals from multiple free-ranging populations to a pathogen with high genetic diversity. The results offer insight into patterns of repeated respiratory disease outbreaks observed in various bighorn sheep populations, demonstrating cross-strain infections may produce similar morbidity and mortality patterns as initial exposure of naïve bighorn sheep populations to Movi. Additionally, some Movi strains appear to cause more disease and outcompete others. Therefore, in free-ranging herds past exposure is likely insufficient to protect bighorn sheep from infection by new Movi strains, although it may influence the progression of disease and recovery within the herd.
This study illustrates the complexity of interactions and outcomes arising from the host-pathogen evolutionary arms race and underscores the need for ecologists, when investigating wildlife systems where pathogens are directly affecting conservation efforts, to account for dynamic evolutionary processes that give rise to heterogeneity in individual responses because they ultimately determine the negative effects of disease on a population. Finally, our findings emphasize that although exposure history might influence the progression of disease and recovery, bighorn sheep conservation would benefit from focusing on preventing pathogen introduction; reliance on past exposure and bighorn sheep adaptive immunological defenses to protect populations from pathogen invasion is unlikely to protect the health of the herd. Therefore, maintaining separation of domestic and bighorn sheep and avoiding management efforts that commingle bighorn sheep with differing Movi strains (e.g., translocations) will likely be the most effective management strategy for minimizing respiratory disease effects to bighorn sheep herds and achieving conservation goals.
Data availability statement
Data and code collected and used in this study is available at: https://doi.org/10.5066/P9Q69IAL.
Ethics statement
The animal study was reviewed and approved by South Dakota State University Institutional Animal Care and Use Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
DW provided funding, statistical support, and contributed to manuscript development. FC provided experimental animals and contributed to manuscript development. TB provided laboratory support and contributed to manuscript development. JJ provided funding, support for animal care and handling, and contributed to manuscript development. BF led animal care and handling, led data collection, assisted with analysis and contributed to manuscript development. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the U.S. Geological Survey (animal support and laboratory testing), the South Dakota Agricultural Experiment Station (Salaries and facilities), South Dakota Department of Game, Fish and Parks (Facilities), Washington State University Rocky Crate Endowment and National Wild Sheep Foundation (Salaries and Laboratory support), Federal Aid to Wildlife Restoration (Animals, travel, laboratory support), and the U.S. Department of Agriculture Forest Service (Laboratory support).
Acknowledgments
We thank the Idaho Department of Fish and Game, Washington Department of Fish and Wildlife, Oregon Fish and Wildlife, South Dakota Game, Fish and Parks, and Nevada Department of Wildlife for providing bighorn sheep for our study. We thank Katy Baker for laboratory assistance, and Michelle Mucciante and Larry Holler for general health and necropsy assistance. We recognize Justin Jensen, Austin Wieseler, Spencer Carstens, Cassie Auxt, and Corey Lee for their assistance, and Matthew Mumma and two reviewers for improving this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1039234/abstract#supplementary-material
References
Anderson, R. M., and May, R. M. (1991). Infectious diseases in humans: Dynamics and control. Oxford University Press Inc., New York, USA.
Besser, T. E., Cassirer, E. F., Highland, M. A., Wolff, P., Justice-Allen, A., Mansfield, K. M., et al. (2013). Bighorn sheep pneumonia: sorting out the etiology of a polymicrobial disease. J. Prev. Vet. Med. 108, 85–93. doi: 10.1016/j.prevetmed.2012.11.018
Besser, T. E., Cassirer, E. F., Potter, K. A., Lahmers, K., Oaks, J. L., Shanthalingam, S., et al. (2014). Epizootic pneumonia of bighorn sheep following experimental exposure to Mycoplasma ovipneumoniae. PLoS One 9:e110039. doi: 10.1371/journal.pone.0110039
Besser, T. E., Highland, M., Baker, K., Cassirer, E. F., Anderson, N. J., Ramsey, J. M., et al. (2012). Causes of pneumonia epizootics among bighorn sheep, western United States, 2008–2010. Emerg. Infect. Dis. 18, 406–414. doi: 10.3201/eid1803.111554
Betlach, A. M., Maes, D., Garza-Moreno, L., Tamiozzo, P., Sibila, M., Haesebrouck, F., et al. (2019). Mycoplasma hyopneumoniae variability: current trends and proposed terminology for genomic classification. Transbound. Emerg. Dis. 66, 1840–1854. doi: 10.1111/tbed.13233
Bloom, B. R. (1979). Games parasites play: How parasites evade immune surveillance. Nature 279, 21–26. doi: 10.1038/279021a0
Burriel, A. R. (1997). Isolation of Pasturella haemolytica from grass, drinking water, and straw bedding used by sheep. Curr. Microbiol. 35, 316–318. doi: 10.1007/s002849900261
Cassirer, E. F., Manlove, K. R., Almberg, E. S., Kamath, P. L., Cox, M., Wolff, P., et al. (2018). Pneumonia in bighorn sheep: risk and resilience. J. Wildl. Manag. 82, 32–45. doi: 10.10002/jwmg.21309
Cassirer, E. F., Manlove, K. R., Plowright, R. K., and Besser, T. E. (2017). Evidence for strain-specific immunity to pneumonia in bighorn sheep. J. Wildl. Manag. 81, 133–143. doi: 10.1002/jwmg.21172
Cassirer, E. F., Plowright, R. K., Manlove, K. R., Cross, P. C., Dobson, A. P., Potter, K. A., et al. (2013). Spatio-temporal dynamics of pneumonia in bighorn sheep. J. Anim. Ecol. 82, 518–528. doi: 10.1111/1365-2656.12031
Chae, H., and Shin, S. J. (2018). Importance of differential identification of Mycobacterium tuberculosis strains for understanding differences in their prevalence, treatment efficacy, and vaccine development. J. Microbiol. 56, 300–311. doi: 10.1007/s12275-018-8041-3
Christiansen, G., Jensen, L. T., Boesen, T., Emmersen, J., Ladefoged, S. A., Schiøtz, L. K., et al. (1997). Molecular biology of mycoplasma. Wien. Klin. Wochenschr. 109, 557–561.
Citti, C., Nouvel, L.-X., and Baranowski, E. (2010). Phase and antigenic variation in microplasmas. Future Microbiol. 5, 1073–1085. doi: 10.2217/fmb.10.71
Clifford, D. L., Schumaker, B. A., Stephenson, T. R., Bleich, V. C., Cahn, M. L., Gonzales, B. J., et al. (2009). Assessing disease risk at the wildlife-livestock interface: a study of the Sierra Nevada bighorn sheep. Biol. Conserv. 142, 2559–2568. doi: 10.1016/j.biocon.2009.06.001
Coscolla, M., and Gagneux, S. (2010). Does M. tuberculosis genomic diversity explain disease severity? Drug Discov. Today: Dis. Mech. 7, e43–e59. doi: 10.1016/j.ddmec.2010.09.004
Degiorgis, M.-P., Obrecht, E., Ryser, A., and Giacometti, M. (1999). The possible role of eye-frequenting flies in the transmission of Mycoplasma conjunctivae. Mitt. Schweiz. Entomol. Ges. 72, 189–194.
Desrosiers, R. (2011). Transmission of swine pathogens: different means, different needs. Anim. Health Res. Rev. 12, 1–13. doi: 10.1017/S1466252310000204
Dixon, D. M., Rudolph, K. M., Kinsel, M. L., Cowan, L. M., Hunter, D. L., and Ward, A. C. S. (2002). Viability of airborne Pasteurella spp. Biennial Sym. Northern Wild Sheep Goat Coun. 13, 6–13.
Fernández Aguilar, X., Lopez-Olvera, J. R., Puig Ribas, M., Begovoeva, M., Velarde, R., Cardells, J., et al. (2019). Mycoplasma conjunctivae in insect vectors and anatomic locations related to transmission and persistence. Vet. Microbiol. 228, 7–11. doi: 10.1016/j.vetmic.2018.11.004
Finlay, B. B., and McFadden, G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cells 124, 767–782. doi: 10.1016/j.cell.2006.01.034
Garwood, T., Lehman, C. P., Walsh, D. P., Cassirer, E. F., Besser, T. E., and Jenks, J. A. (2020). Removal of chronic Mycoplasma ovipneumoniae carrier ewes eliminates pneumonia in a bighorn sheep population. Ecology and Evolution, 10, 3491–3502. doi: 10.1002/ece3.6146
Giacometti, M., Janovsky, M., Belloy, L., and Frey, J. (2002). Infectious keratoconjunctivitis of ibex, chamois and other Caprinae. Rev. Sci. Tech. 21, 335–345. doi: 10.20506/rst.21.2.1338
Higdon, D. (2002). “Space and space-time modeling using process convolutions” in Quantitative methods for current environmental issues. eds. C. W. Anderson, V. Barnett, P. C. Chatwin, and A. H. El-Shaarawi (London: Springer), 37–56.
Hornef, M. W., Wick, M. J., Rhen, M., and Normark, S. (2002). Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 3, 1033–1040. doi: 10.1038/ni1102-1033
Ionas, G., Norman, N. G., Clarke, J. K., and Marshall, R. B. (1991a). A study of the heterogeneity of isolates of Mycoplasma ovipneumoniae from sheep in New Zealand. Vet. Microbiol. 29, 339–347. doi: 10.1016/0378-1135(91)90141-2
Ionas, G., Norman, N. G., Clarke, J. K., and Marshall, R. B. (1991b). Isolation of multiple strains of Mycoplasma ovipneumoniae from individual pneumonic sheep lungs. Vet. Microbiol. 29, 349–360.
Justice-Allen, A. E., Butler, E., Pebworth, J., Munig, A., Wolff, P., and Besser, T. E. (2016). Investigation of pneumonia mortalities in Mycoplasma-Positive Desert Bighorn sheep and detection of different strain of Mycoplasma ovipneumoniae. Biennial North. Wild Sheep Goat Coun. 20, 68–72.
Kamath, P. L., Manlove, K., Cassirer, E. F., Cross, P. C., and Besser, T. E. (2019). Genetic structure of Mycoplasma ovipneumoniae informs pathogen spillover dynamics between domestic and wild Caprinae in the western United States. Sci. Rep. 9:15318. doi: 10.1038/s41598-019-51444-x
Kanci, A., Wawegama, N. K., Marenda, M. S., Mansell, P. D., Browning, G. F., and Markham, P. F. (2017). Reproduction or respiratory mycoplasmosis in calves by exposure to an aerosolized culture of Mycoplasma bovis. Vet. Microbiol. 210, 167–173. doi: 10.1016/j.vetmic.2017.09.013
Kermack, W. O., and McKendrick, A. G. (1927). A contribution to the mathematical theory of epidemics. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. 115, 700–721.
Landman, W. J. M., Corbanie, E. A., Feberwee, A., and van Eck, J. H. H. (2010). Aerosolization of Mycoplasma synoviae compared with Mycoplasma gallisepticum and Enterococcus faevalis. Avian Pathol. 33, 210–215. doi: 10.1080/0307945042000195812
Larson, M. G., and Dinse, G. E. (1985). A mixture model for the regression analysis of competing risks data. Appl. Stat. 34, 201–211. doi: 10.2307/2347464
Lonsdorf, E. V., Gillespie, T. R., Wolf, T. M., Lipende, I., Raphael, J., Bakuza, J., et al. (2018). Socioecological correlates of clinical signs in two communities of wild chimpanzees (Pan troglodytes) at Gombe National Park, Tanzania. Am. J. Primatol. 80:80. doi: 10.1002/ajp.22562
Lunn, D., Jackson, C., Best, N., Thomas, A., and Spiegelhalter, D. (2013). The BUGS book: A practical introduction to Bayesian analysis. 381, CRC Press, Boca Raton, Florida, USA.
Maksimović, Z., De la Fe, C., Amores, J., Gómez-Martín, Á., and Rifatbegović, M. (2017). Comparison of phenotypic and genotypic profiles among caprine and ovine mycoplasma ovipneumoniae strains. Vet. Rec. 180:180. doi: 10.1136/vr.103699,article179
Monack, D. M., Mueller, A., and Falkow, S. (2004). Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2, 747–765. doi: 10.1038/nrmicro955
Niang, M., Rosenbusch, R. F., Andrews, J. J., Lopez-Virella, J., and Kaeberle, M. L. (1998). Occurrence of autoantibodies to cilia in lambs with ‘coughing syndrome’. Vet. Immunol. Immunopathol. 64, 191–205. doi: 10.1016/S0165-2427(98)00133-0
Otake, S., Dee, S., Corzo, C., Oliveira, S., and Deen, J. (2010). Long-distance airborne transport of infectious PRRSV and Mycoplasma hyopneumoniae from a swine population infected with multiple viral variants. Vet. Microbiol. 145, 198–208. doi: 10.1016/j.vetmic.2010.03.028
Parham, K., Churchward, C. P., McAuliffe, L., Nicholas, R. A., and Ayling, R. D. (2006). A high level of strain variation within the Mycoplasma ovipneumoniae population of the UK has implications for disease diagnosis and management. Vet. Microbiol. 118, 83–90. doi: 10.1016/j.vetmic.2006.07.005
Plowright, R. K., Manlove, K. R., Besser, T. E., Páez, D. J., Andrews, K. R., Matthews, P. E., et al. (2017). Age-specific infectious period shapes dynamics of pneumonia in bighorn sheep. Ecol. Lett. 20, 1325–1336. doi: 10.1111/ele.12829
Plowright, R. K., Manlove, K. R., Cassirer, E. F., Cross, P. C., and Besser, T. E. (2013). Use of exposure history to identify patterns of immunity to pneumonia in bighorn sheep (Ovis canadensis). PLoS One :e61919. doi: 10.1371/journal.pone.0061919
Plummer, M. (2003). “JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling.” in Proceedings of the 3rd international workshop on distributed statistical computing. Available at: https://www.r-project.org/conferences/DSC-2003/Proceedings/Plummer.pdf [Accessed May 1, 2016].
Qin, L., Chen, Y., and You, X. (2019). Subversion of the immune response by human pathogenic mycoplasmas. Front. Microbiol. 10:1934. doi: 10.3389/fmicb.2019.01934
Quinton, L. J., and Mizgerd, J. P. (2015). Dynamics of lung defense in pneumonia: resistance, resilience, and remodeling. Annu. Rev. Physiol. 77, 407–430. doi: 10.1146/annurev-physiol-021014-071937
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/ (Accessed May 27, 2020).
Rifatbegović, M., Maksimović, Z., and Hulaj, B. (2011). Mycoplasma ovipneumoniae associated with severe respiratory disease in goats. Vet. Rec. 168:565. doi: 10.1136/vr.d886
Sandel, A. A., Rushmore, J., Negrey, J. D., Mitani, J. C., Lyons, D. M., and Caillaud, D. (2021). Social network predicts exposure to respiratory infection in a wild chimpanzee group. EcoHealth 17, 437–448. doi: 10.1007/s10393-020-01507-7
Sells, S. N., Mitchell, M. S., Nowak, J. J., Lukacs, P. M., Anderson, N. J., Ramsey, J. M., et al. (2015). Modeling risk of pneumonia epizootics in bighorn sheep. J. Wildl. Manag. 79, 195–525. doi: 10.1002/jwmg.824
Simenka, J. W. (2005). “Immune responses following mycoplasma infection” in Mycoplasma molecular biology, pathogenicity and strategies for control. eds. A. Blanchard and G. Browning (Horizon Bioscience: Norfolk, UK), 485–534.
Singer, F. J., Williams, E., Miller, M. W., and Zeigenfuss, L. C. (2000). Population growth, fecundity, and survivorship in recovering populations of bighorn sheep. Restor. Ecol. 8, 75–84. doi: 10.1046/j.1526-100x.2000.80067.x
Smith, J. B., Grovenburg, T. W., Monteith, K. L., and Jenks, J. A. (2015). Survival of female bighorn sheep (Ovis canadensis) in the Black Hills, South Dakota. Am. Midl. Nat. 174, 290–301. doi: 10.1674/0003-0031-174.2.290
Smith, J. B., Jenks, J. A., Grovenburg, T. W., and Klaver, R. W. (2014). Disease and predation: sorting out causes of a Bighorn sheep (Ovis canadensis) decline. PLoS One 9, 165–174. doi: 10.1002/wsb.360
Stärk, K. D. C., Nicolet, J., and Frey, J. (1998). Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Appl. Environ. Microbiol. 64, 543–548. doi: 10.1128/AEM.64.2.543-548.1998
Su, Y.-S., and Yajima, M. (2015). R2jags: using R to run ‘JAGS’. R package version 0.5-7. Available at: https://CRAN.R-project.org/package=R2jags (Accessed May 27, 2020).
Wilson, K., Bjørnstad, O. N., Dobson, A. P., Merler, S., Poglayen, G., Randolph, S. E., et al. (2002). in Heterogenities in macroparsite infections: Patterns and processes in the ecology of wildlife diseases. eds. P. J. Hudson, A. Dobson, B. Grenfell, H. Heesterbeek, and A. P. Dobson (New York, USA: Oxford University Press), 6–44.
Keywords: bighorn sheep, disease state, eDNA, hazard, immune response, Mycoplasma ovipneumoniae, strain, transmission
Citation: Walsh DP, Felts BL, Cassirer EF, Besser TE and Jenks JA (2023) Host vs. pathogen evolutionary arms race: Effects of exposure history on individual response to a genetically diverse pathogen. Front. Ecol. Evol. 10:1039234. doi: 10.3389/fevo.2022.1039234
Edited by:
R. Terry Bowyer, University of Alaska Fairbanks, United StatesReviewed by:
Aniruddha V. Belsare, Emory University, United StatesEmily Pascoe, Wageningen University and Research, Netherlands
Copyright © 2023 Walsh, Felts, Cassirer, Besser and Jenks. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel P. Walsh, ✉ ZHdhbHNoQHVzZ3MuZ292
‡Present address: Daniel P. Walsh and Brandi L. Felts U.S. Geological Survey, Montana Cooperative Wildlife Research Unit, University of Montana, Missoula, MT, United States
†These authors have contributed equally to this work and share co-first authorship
 Daniel P. Walsh
Daniel P. Walsh Brandi L. Felts2,3†‡
Brandi L. Felts2,3†‡ E. Frances Cassirer
E. Frances Cassirer Thomas E. Besser
Thomas E. Besser Jonathan A. Jenks
Jonathan A. Jenks