Abstract
Accurate species delimitation is critical for biodiversity conservation. Integrative taxonomy has been advocated for a long time, yet tools allowing true integration of genetic and phenotypic data have been developed quite recently and applied to few models, especially in plants. In this study, we investigated species boundaries within a group of twelve Pseudophrys taxa from France by analyzing genetic, morphometric and chemical (i.e., floral scents) data in a Bayesian framework using the program integrated Bayesian Phylogenetics and Phylogeography (iBPP). We found that these twelve taxa were merged into four species when only genetic data were used, while most formally described species were recognized as such when only phenotypic (either morphometric or chemical) data were used. The result of the iBPP analysis performed on both genetic and phenotypic data supports the proposal to merge Ophrys bilunulata and O. marmorata on the one hand, and O. funerea and O. zonata on the other hand. Our results show that phenotypic data are particularly informative in the section Pseudophrys and that their integration in a model-based method significantly improves the accuracy of species delimitation. We are convinced that the integrative taxonomic approach proposed in this study holds great promise to conduct taxonomic revisions in other orchid groups.
Introduction
Accurately delimiting species is of critical importance for many fields of research in biology, including conservation biology. Species are commonly defined as independently evolving linages that can be delimited using various criteria (Hey, 2006; De Queiroz, 2007). As any single line of evidence may fail at detecting species boundaries (Knowles and Carstens, 2007), many authors have advocated the use of an integrative approach combining several lines of evidence, both genetic and phenotypic (Dayrat, 2005; Will et al., 2005; Padial et al., 2010; Pires and Marinoni, 2010). However, until recently, genetic and phenotypic data were almost always integrated in a purely qualitative way, as no quantitative methods were available for processing simultaneously both data types (Schlick-Steiner et al., 2010; Yeates et al., 2011). Fortunately, model-based species delimitation methods, which were originally developed for DNA sequences (Fujita et al., 2012; Naciri and Linder, 2015), were later extended to integrate quantitative traits (Guillot et al., 2012; Solís-Lemus et al., 2015), thereby improving objectivity and repeatability of integrative species delimitation. Such methods have been applied to various animal (Huang and Knowles, 2016; Pyron et al., 2016; Olave et al., 2017; Núñez et al., 2022) and plant (Yang et al., 2019; Zhang et al., 2020) clades and have proven useful in several cases. Because model-based species delimitation methods may cause oversplitting when solely based on genetic data (Sukumaran and Knowles, 2017; Mason et al., 2020), combining the latter with phenotypic data may provide more conservative estimates of species numbers (e.g., Pyron et al., 2016). Conversely, in recently radiating clades, in which species often lack clear genetic differentiation, integrating morphological or ecological data may increase the power to detect species boundaries (e.g., Edwards and Knowles, 2014; Solís-Lemus et al., 2015).
Hyperdiverse clades deserve particular conservation attention but may be taxonomically challenging. This is, for example, the case of the Orchidaceae family, which comprises more than 30,000 named species [Plants of the World Online [POWO], 2022], including some of the most threatened species in the world (Fay, 2018), but in which species boundaries are sometimes blurred (Barrett and Freudenstein, 2011; Pessoa et al., 2012). Within this family, the Mediterranean genus Ophrys L. is of particular interest, due to its high level of ecological specialization and endemism rate, but it is also considered as a textbook example of taxonomic confusion (Bertrand et al., 2021; Cuypers et al., 2022), which may affect its conservation (Agapow et al., 2004; Pillon and Chase, 2007; Vereecken et al., 2010; Schatz et al., 2014). Some of this confusion arises from conflicting views on which operational criteria should be used to delimit species in this genus. Specifically, some authors support that taxa should have achieved reciprocal monophyly (Devey et al., 2008; Bateman et al., 2011) to be considered as “good” species, while others argue that interactions between Ophrys and pollinators are more informative than neutral markers due to their key role in speciation (Schiestl and Ayasse, 2002; Ayasse et al., 2011; Vereecken et al., 2011; Baguette et al., 2020). Indeed, Ophrys species attract one or a few pollinator species (Joffard et al., 2019; Schatz et al., 2020) using sex pheromones-mimicking floral scents (Schiestl et al., 1999; Ayasse et al., 2003). In these species, changes in floral scents may cause pollinator shifts, which may in turn mediate reproductive isolation between conspecific populations and drive speciation (Sedeek et al., 2014). Distinct views on which criteria should be used to delimit species has led to the recognition of dozens (Devey et al., 2008) versus hundreds (Paulus, 2006) of Ophrys species. In addition, even authors who favor the same criteria sometimes disagree on where along the speciation continuum independently evolving lineages should be recognized as species, i.e., “splitters” (e.g., Devillers and Devillers-Terschuren, 1994; Delforge, 2016) versus “lumpers” (e.g., Pedersen and Faurholdt, 2007; Kühn et al., 2020). In this context, model-based species delimitation methods integrating genetic and phenotypic data could be particularly helpful.
In this study, we aim at delimiting species through the integration of molecular markers, morphometric characters and floral scents in a group of twelve Pseudophrys taxa. We compare species boundaries based on genetic and phenotypic data alone or in combination and we discuss the potential of integrative taxonomy in solving long-standing debates about Ophrys taxonomy.
Materials and methods
Studied species and populations
The monophyletic section Pseudophrys Godfery comprises twelve groups, each of them including one to twelve taxa (Delforge, 2016). Here, we focused on the twelve Pseudophrys taxa that are described in France (Table 1 and Figure 1; Bournérias and Prat, 2005). Among them, eight belong to the O. fusca group (namely O. bilunulata, O. delforgei subsp. “O. forestieri” sensu neotypus 1999, O. funerea, O. lupercalis, O. marmorata, O. peraiolae, O. sulcata, and O. zonata), one to the O. iricolor group (O. eleonorae), two to the O. lutea group (namely O. corsica and O. lutea) and one to the O. omegaifera group (O. vasconica). These twelve taxa differ in their geographical distribution, some of them being widely distributed (e.g., O. bilunulata, O. lupercalis, and O. lutea), while others have restricted distribution areas, e.g., in South-eastern France (O. deforgei), South-western France and Northern Spain (O. vasconica) or Corsica and Sardinia (O. corsica, O. eleonorae, O. funerea, O. marmorata, O. peraiolae, and O. zonata). By contrast, these twelve taxa do not strongly differ in their flowering phenology or habitats: except for O. sulcata and O. vasconica, they all flower in early spring and grow in open, dry habitats typical of the Mediterranean region (Bournérias and Prat, 2005). Among them, O. lupercalis, O. lutea, O. sulcata, and O. vasconica are regionally protected in France (Bournérias and Prat, 2005) and several of them are currently considered as threatened at the national or regional level, such as O. eleonorae (considered as endangered at the national level and as critically endangered in the Corsican region) and O. marmorata (considered as vulnerable in the Corsican region) (IUCN et al., 2010; Delage and Hugot, 2015).
TABLE 1
| Taxon | Population | Molecular data | Morphometric data | Chemical data |
| Ophrys bilunulata Risso (1844) | Gruissan, 11, France | 3 (1) | 15 | 10 |
| Clapier, 34, France | 3 (1) | 10 | 5 | |
| La Gaude, 06, France | 2 (1) | − | − | |
| Ophrys corsica Soleirol ex Foelsche and Foelsche (2002) | Bonifacio, 2A, France* | 1 | 25 | 15 |
| Ophrys delforgei Devillers-Terschuren and Devillers (2006) | Martigues, 13, France* | 2 | 25 | 20 |
| Ophrys eleonorae Paulus and Gack (2004) | Antisanti, 2B, France | 1 | − | 2 |
| Ophrys funerea Viviani (1824) | Palasca, 2B, France | 3 (2) | 20 | 9 |
| Corte, 2B, France | 3 (2) | 15 | 9 | |
| Laconi, Sardinia, Italy | 2 | − | − | |
| Ophrys lupercalis Devillers and Devillers-Terschuren (1994) | Armissan, 11, France* | 2 | 0 | 0 |
| Saint Bauzille de Montmel, 34, France | 2 (1) | 20 | 15 | |
| Saint-Florent, 2B, France | − | 5 | 5 | |
| Ophrys lutea Cavanilles (1793) | Montferrier sur Lez, 34, France | 3 (1) | 10 | 10 |
| Montarnaud, 34, France | 3 (1) | 20 | 10 | |
| Cassis, 13, France | 1 | − | − | |
| Maala, Kabylia, Algeria | 1 | − | − | |
| Benicolet, Valencian community, Spain | 1 | − | − | |
| Sempere, Valencian community, Spain | 1 | − | − | |
| Ophrys marmorata Foelsche and Foelsche (1998) | Bonifacio, 2A, France* | 5 (3) | 20 | 8 |
| Ophrys peraiolae Foelsche et al. (2000) | Palasca, 2B, France* | 3 | 15 | 8 |
| Ophrys sulcata Devillers and Devillers-Terschuren (1994) | Lapanouse, 12, France | 3 (2) | 25 | 10 |
| Oléron, 17, France* | 2 | − | − | |
| Vence, 06, France | 1 | − | − | |
| Ophrys vasconica Delforge (1991) | Belpech, 11, France | 1 | 20 | 15 |
| Ophrys zonata Devillers and Devillers-Terschuren (1994) | Saint-Florent, 2B, France | 3 (1) | 25 | 15 |
Number of populations and individuals sampled for molecular, morphometric, and chemical data.
*: Populations located at the locus classicus. (): Number of newly-published sequences.
FIGURE 1

Photographs of the 12 French Pseudophrys taxa sampled for molecular, morphometric, and chemical data. From left to right and top to bottom: Ophrys bilunulata, O. delforgei, O. funerea, O. lupercalis, O. marmorata, O. peraiolae, O. sulcata and O. zonata(O. fusca group), O. eleonorae(O. iricolor group), O. corsica, O. lutea(O. lutea group), and O. vasconica(O. omegaifera group). © N. Joffard and B. Schatz.
Four hundred ninety individuals belonging to one to six populations per taxon were selected and sampled for molecular, morphometric, or chemical analysis between 2013 and 2016 (Table 1). Within populations, molecular, morphometric, and chemical data were not collected on the same individuals as the iBPP program (see below) requires independence of genetic and phenotypic data. Molecular data were collected in one to six populations per taxon, distributed over most of their geographic range, in up to five individuals per population. Morphometric and chemical data were collected in one or two of these populations only, but on up to 25 individuals per population. One population was sampled at the locus classicus (i.e., site where the species was described for the first time) for six of these twelve taxa.
Genetic data collection and analysis
One leaf of one to five individual(s) per population were collected between 2014 and 2017 and dried in silica gel for a few days. DNA extraction was performed with a Plant Minikit (®Quiagen). Three genes were amplified and sequenced in 52 individuals: the internal transcribed spacers (ITS) 1 and 2, the first intron of the beta-galactosidase-like (BGP) gene and the first intron of the LEAFY/FLORICULA (LFY) gene. For 36 individuals, sequences have been published in Joffard et al. (2020), while for 16 individuals, sequences are published for the first time in this study (Supplementary Table 1).
Polymerase chain reaction (PCR) and sequencing were carried out as described in Joffard et al. (2020). Sequences were edited using CodonCode Aligner v.4.2.7 (CodonCode Corporation). Uncertainties and alleles from heterozygous individuals were merged into consensus sequences using International Union for Pure and Applied Chemistry (IUPAC) coding. Consensus sequences were aligned using the Muscle algorithm (Edgar, 2004) as implemented in SeaView v.4.4.2 (Gouy et al., 2010) prior to concatenation.
A phylogenetic analysis was performed on the concatenated alignment using MrBayes v.3.1.2 (Ronquist and Huelsenbeck, 2003). Ophrys cinerophila from Samos (Greece) was used as outgroup based on Joffard et al. (2020). The best partitioning scheme and the best model for each partition was chosen using the Bayesian Information Criterion (BIC) as estimated by PartitionFinder v.1.1.1 (Lanfear et al., 2012). Bayesian analysis was conducted with two separate runs of four Markov chain Monte Carlo (MCMC) chains for 10 million generations with tree sampling every 1,000 generations. 25% of the sampled trees were discarded as burn-in, and the 75% best scoring trees were used to calculate the consensus tree.
A DNA barcoding analysis was performed on the concatenated alignment using the Automatic Barcode Gap Detection (ABGD) website (Puillandre et al., 2012). ABGD is a tool designed to infer species hypotheses based on automatized identification of barcode gaps between intra- and interspecific pairwise distances. It aims at revealing a significant barcoding gap in the distribution of pairwise genetic distances, reflecting a discontinuity between intra- and interspecific distances among individuals. ABGD partitions individuals into groups in a recursive manner until no further splits are possible, while integrating priors on maximum and minimum intraspecific differentiation and barcode gap width. In this study, pairwise distances were computed as K2P-corrected distances. We left the default values of 10 steps from Pmin = 0.001 to Pmax = 0.1 for number of steps and intraspecific differentiation, and the default value of 1.5 for barcode gap width.
Phenotypic data collection and analysis
For morphometric data, fifteen to thirty-five individuals per taxon were sampled in 2015 and 2016 (Table 1). Ophrys eleonorae was not sampled for morphometric data as no flowering individuals were found in 2015 nor in 2016. However, this species is known to be morphologically distinct from the eleven other taxa in that it has a particularly long (∼15 to 25 mm) and wide (∼10 to 20 mm) labellum (Bournérias and Prat, 2005). In each individual, twelve morphometric characters were measured to the nearest 0.01 mm in the field using a digital caliper. Four of these characters concerned the labellum, whose size and shape are important because they must match those of the pollinator, but the length and/or width of the stigmatic cavity, lateral petals, and sepals were also measured (Supplementary Figure 1).
For chemical data, eight to twenty individuals per taxon were sampled for floral scents in 2014 and 2015 (Table 1) using solid phase microextraction (SPME) (except in the case of O. eleonorae in which only two individuals were sampled) as described in Joffard et al. (2016). Floral scents were then analyzed by GC-MS using a Shimadzu QP2010 Plus gas chromatograph-mass spectrometer with an OPTIMA ® 5-MS capillary column (30 m × 0.25 mm × 0.25 μm, Macherey-Nagel, Düren, Germany) and helium as carrier gas with the method described in Joffard et al. (2016). Retention times of a series of n-alkanes (Qualitative retention time mix, ASTM, Sigma Aldrich®) were used to convert retention times into retention index. Compounds were identified based on retention index and mass spectra which were compared to those recorded in databases (NIST, 2007, Wiley Registry 9th) and in the literature (Adams, 2007) and, for some of them, to retention index and mass spectra of analytical standards. Peak areas were measured with the software GCMSsolution (4.11) (Shimazu®).
Two partial least square discriminant analyses (PLS-DA) were performed, one for morphometric characters and one for floral scents. PLS-DA was chosen over Principal Component Analysis (PCA) because it is suitable for data that are non-independent (due to allometry in the case of morphometric characters and shared biosynthetic pathways in the case of floral scents). Because variances were non-homogenous among compounds, floral scents data were centred log-ratio-transformed prior to analysis. Statistical analyses were performed in R version 3.1.2 (R Development Core Team, 2008).
IBPP species delimitation
A joint Bayesian inference based on genetic and phenotypic data was used to delimit species using the program iBPP v.2.1.3 (Solís-Lemus et al., 2015). This program is an extension of the multispecies coalescent model-based program BPP (Rannala and Yang, 2003; Yang, 2015) which includes models of evolution for phenotypic data under a Brownian Motion (BM) process. Because the program assumes independence of phenotypic data, scores on the first two (for morphometric characters) and five (for floral scents) components resulting from two preliminary PCA were included in the analysis. Note that given the role of floral scents in pollinator attraction, these scents may not have evolved according to the assumptions of a BM process, but the results of the program iBPP have been shown to be robust to such a violation (Solís-Lemus et al., 2015). The program begins with a strictly bifurcating guide tree, which in our case was constructed with the software MrBayes (see above), and collapses internal nodes sequentially. We used a prior gamma distribution G (2, 2,000) for τ (branch lengths) and θ (product of Ne the population size and μ the mutation rate) for genetic data and left the default values of 0 for σ2 (variance) and λ (within/between species ratio) for phenotypic data (non-informative priors). A reversible MCMC analysis was ran over 1,00,000 generations, sampled every ten generations, with 1,000 generations (10%) discarded as burn-in. Seven analyses were performed: (i) with genetic data only, (ii) with morphometric data only, (iii) with chemical data only, (iv) with both genetic and morphometric data, (v) with both genetic and chemical data, (vi) with both morphometric and chemical data, and (vii) with the entire dataset. Because phylogenetic relationships between the taxa O. bilunulata, O. delforgei, and O. marmorata, as well as between the taxa O. funerea, O. sulcata, and O. zonata could not be resolved, several alternative topologies were tested for the guide tree and the topology that gave the most conservative species delimitation model for these two triplets was retained. The robustness of the results was tested by analyzing the data with both the fine tune settings of zero and one (Yang and Rannala, 2010), and by repeating each analysis five times.
Results
Genetic data
ITS, BGP, and LFY sequences were obtained for 52, 49, and 52 individuals, respectively (153 sequences, including 45 that are newly published). Sequences were obtained for at least three individuals per taxon, except for O. corsica, O. eleonorae, and O. vasconica (sequences obtained for one individual only). ITS, BGP, and LFY sequences contained 73, 562, and 603 parsimony-informative sites on 809, 948, and 2,210 sites, respectively. The phylogenetic tree (Figure 2) was congruent with the one described in Joffard et al. (2020), with two well-supported clades, one comprising the taxa O. lutea, O. corsica, O. lupercalis, O. peraiolae, O. delforgei, O. bilunulata, and O. marmorata and one comprising the taxa O. eleonorae, O. vasconica, O. sulcata, O. funerea, and O. zonata. On the nine taxa for which several individuals were sampled for molecular analyses, only two - namely O. lutea and O. peraiolae - were found to be monophyletic with a posterior probability of 0.98 and 1, respectively. The AGBD method detected three species only: it recognized O. eleonorae as a species but merged O. lutea, O. corsica, O. lupercalis, O. peraiolae, O. delforgei, O. bilunulata, and O. marmorata on the one hand, and O. vasconica, O. sulcata, O. funerea, and O. zonata on the other hand. Mean K2P-corrected distances between individuals were of 2.38 × 10–3 substitutions per site within and 7.09 × 10–3 substitutions per site between these species. The barcoding gap was located between 3.00 × 10–3 and 4.00 × 10–3 substitutions per site (Supplementary Figure 2).
FIGURE 2
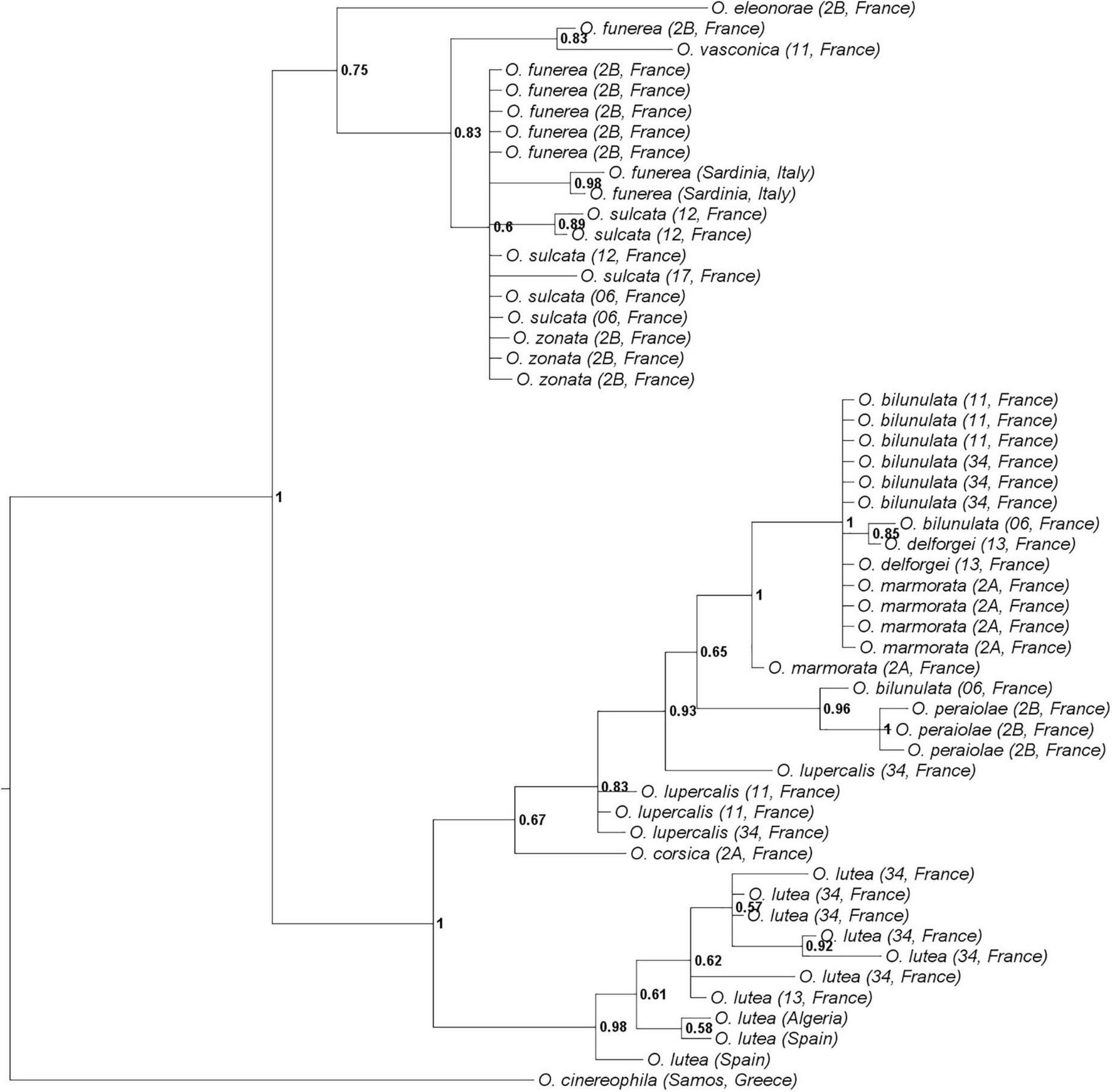
Phylogenetic relationships between the twelve French Pseudophrys taxa represented by the 50-majority rule consensus tree from the MrBayes analysis. Posterior probabilities are indicated at each node.
Morphometric data
Labellum length ranged from 6.11 to 14.35 mm, with a mean of 9.11 (±1.45) mm, and labellum width from 5.88 to 14.23 mm, with a mean of 8.98 (±1.51) mm. Ophrys lupercalis and O. vasconica were characterized by large sepals, petals, and labella compared to other species, while O. sulcata, O. funerea, and O. zonata were characterized by long petals and sepals but a relatively small labellum, with a high length/width ratio. By contrast, O. bilunulata, O. marmorata, and O. peraiolae were characterized by larger labella with a lower length/width ratio. Finally, the yellow-flowered O. corsica and O. lutea were characterized by short sepals and petals and a short but wide labellum (Figure 3 and Supplementary Table 2).
FIGURE 3
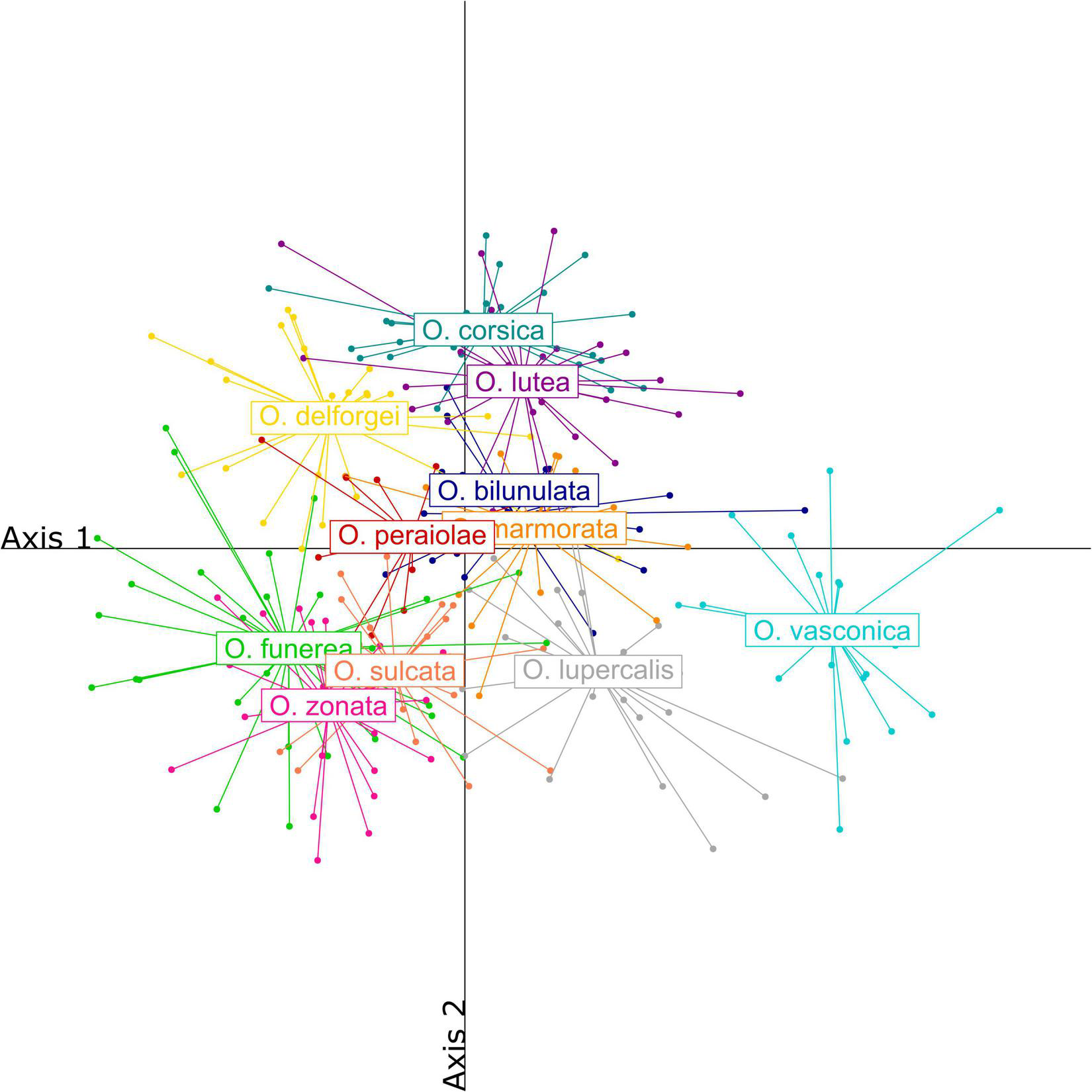
Partial least squares-discriminant analysis (PLS-DA) of morphometric characters measured on the twelve French Pseudophrys taxa.
Chemical data
Over one hundred VOCs were detected in the blends of the twelve studied taxa, mostly alkanes (19), alkenes and alkadienes (29), aldehydes (23), acids (13) and fatty acid esters (24) (Supplementary Table 3). Blends were dominated by alkenes and alkadienes (58.6%) as well as alkanes (28.2%), but aldehydes and fatty acid esters both accounted for more than 5% of the blends. The blends of the O. fusca, O. iricolor, O. omegaifera, and O. lutea groups were well differentiated, both qualitatively and quantitatively (Figure 4). More precisely, taxa from the O. fusca group generally did not produce any fatty acid esters, while taxa from the O. iricolor, O. omegaifera, and O. lutea groups produced significant amounts of nonyl, decyl, and octyl esters, respectively. Within the O. fusca group, some species also had well-differentiated blends, although this differentiation was often quantitative rather than qualitative. By contrast, several taxa, such as the O. funerea/O. zonata pair, produced very similar floral scents.
FIGURE 4
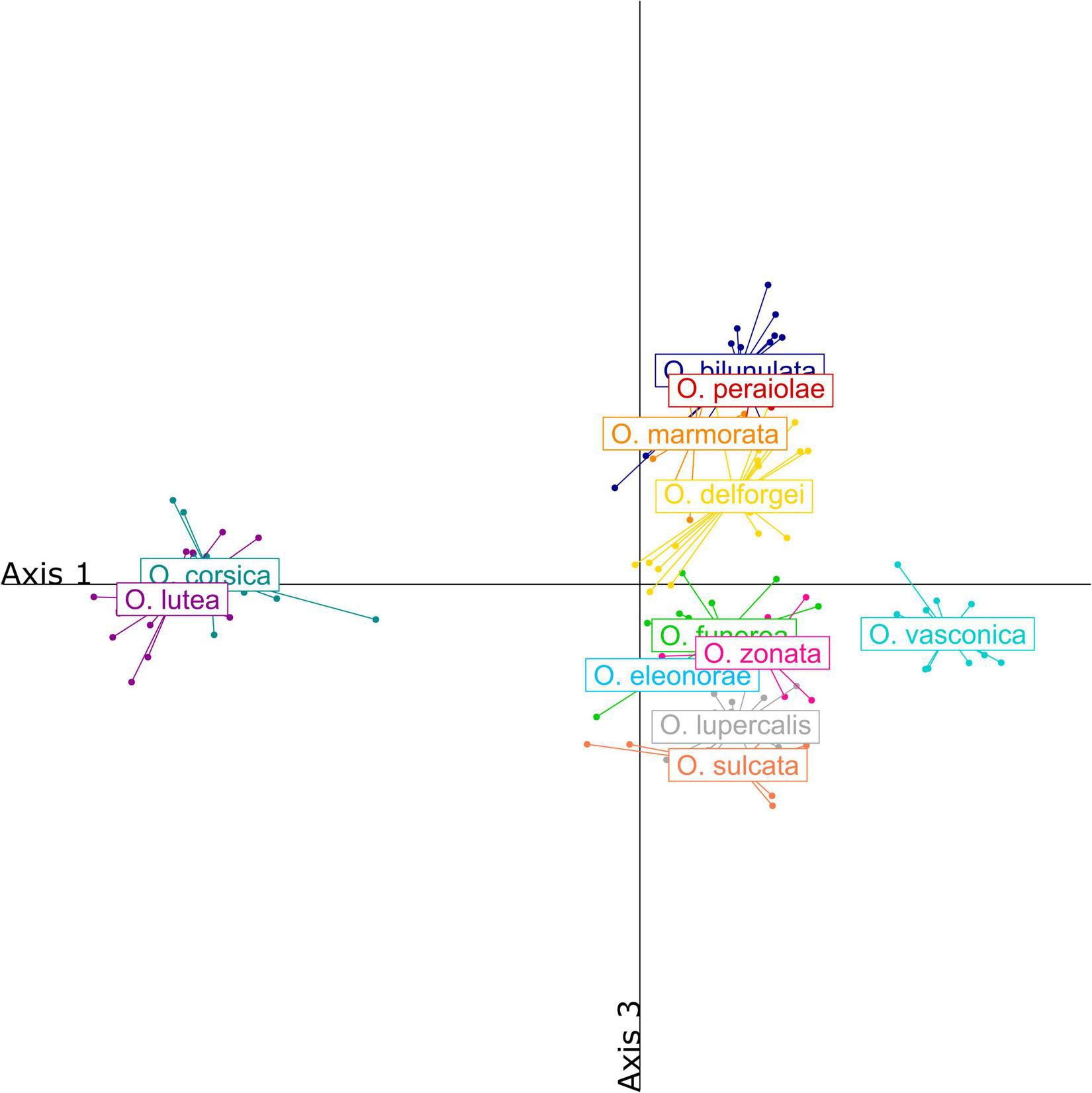
Partial least squares-discriminant analysis (PLS-DA) of floral scents detected in the blends of the 12 French Pseudophrys taxa. Individuals were represented along axes 1 and 3 to better visualize variation within the O. fusca group.
IBPP species delimitation
Results of iBPP analyses (best species delimitation models, with their respective species numbers and posterior probabilities) are summarized Figure 5 and detailed Supplementary Table 4.
FIGURE 5

Results from the iBPP analysis based on molecular (Mol.), morphometric (Mor.), and chemical (Che.) data analyzed alone or in combination.
Whatever the type of data included in the analysis, the posterior probability of the best model never exceeded 70%, showing relatively weak support for this model compared to the next best ones. When only genetic data were considered, the three best models (i.e., those for which the sum of posterior probabilities exceeded 80%) all recognized O. lutea, O. corsica, O. lupercalis, O. peraiolae, O. deforgei, O. bilunulata, and O. marmorata as a single species. They also all considered O. eleonorae as a genuine species and merged O. funerea and O. zonata. The best model (PP = 61.86%) was a four-species model recognizing O. vasconica as a species but merging O. sulcata with the O. funerea/O. zonata pair. By contrast, when only morphometric data were considered, the two best models recognized most taxa as genuine species: the first one (PP = 67.10%) merged O. bilunulata and O. marmorata, while the second one (PP = 21.98%) delimited twelve species. The same results were obtained when molecular data were included in the analysis, but the posterior probabilities of these two best models decreased, while that of a ten-species model merging both O. marmorata with O. bilunulata and O. zonata with O. funerea increased (Supplementary Table 4). Likewise, when only chemical data were considered, the two best models were the ten-species model merging both pairs, and an eleven-species model merging O. zonata with O. funerea only, with comparable posterior probabilities (PP = 47.63 and 44.84%, respectively). Including molecular data in the analysis increased support for the first model (PP = 53.59%) compared to the second one (PP = 39.50%). When only phenotypic (morphometric + chemical) data were considered, the three best models either suggested to merge both the O. bilunulata/O. marmorata and O. funerea/O. zonata pairs (PP = 44.83%) or one only (PP = 29.04% for the model merging O. marmorata with O. bilunulata and 16.02% for the one merging O. zonata with O. funerea). Finally, the same results were obtained when genetic and phenotypic data were combined, with only slight differences in the posterior probability attributed to each of these three best models compared to the previous analysis (PP = 49.88, 27.63, and 14.19%, respectively).
These results were robust to the algorithm that was used to collapse internal nodes, and repetitions of each analysis gave similar results. By contrast, these results varied depending on the topology of the guide tree. More specifically, when O. bilunulata and O. marmorata on the one hand, and O. funerea and O. zonata on the other hand were not considered as sister-species in the guide tree, the best model was the twelve-species model, because these two taxa were distinct from O. delforgei and O. sulcata, respectively.
Discussion
Our study aimed at comparing species boundaries drawn from molecular, morphometric, and chemical data alone or in combination in a group of twelve Pseudophrys taxa. Our results showed that including phenotypic data in the analysis helped being more accurate when delimiting species in this group. Based on this integrative taxonomic approach, eight formally described species were recognized as such, while the best model suggested merging two pairs of taxa into one species each.
Integration of genetic and phenotypic data in the section Pseudophrys
Our results showed that genetic differentiation between the twelve studied taxa was often limited and that species delimitations drawn from genetic data only (using either ABGD or iBPP) were thus very conservative, with only a few taxa recognized as genuine species. Such a limited genetic differentiation could result from incomplete lineage sorting or hybridization (Soliva et al., 2001; Soliva and Widmer, 2003), that are both likely given the recent diversification of the section Pseudophrys (Breitkopf et al., 2014; Baguette et al., 2020) and the weakness of post-zygotic barriers between sympatric species (Cortis et al., 2009). Although the markers used in this study were selected because of their high resolution at the scale of the genus, they may not be informative enough to discriminate between such closely related taxa. Just like other non-model organisms, the Ophrys genus will likely benefit from the democratization of high-throughput sequencing technics allowing to develop more resolutive markers (e.g., Bateman et al., 2018).
By contrast, phenotypic differentiation between the twelve studied taxa was often significant, perhaps because morphometric characters and floral scents are selected by pollinators and may thus evolve faster and be less affected by hybridization than neutral markers (Sedeek et al., 2014). Indeed, the size and shape of the labellum are likely to be selected to match those of the pollinator’s body (Triponez et al., 2013), and floral scents to match sex pheromones of female insects (Schiestl et al., 1999; Ayasse et al., 2003). Interestingly, our results show that morphometric characters are as informative as floral scents to discriminate between Pseudophrys species. Both are classically used as criteria to delimit Ophrys species (Bernardos et al., 2005; Mant et al., 2005), but in the past decades, much more emphasis has been put on chemical signals (Schiestl et al., 1999; Ayasse et al., 2003; Stökl et al., 2005; Véla et al., 2007). However, our results suggest that using morphometric characters for taxonomic purposes is relevant in the section Pseudophrys and emphasize the potential role of orchid enthusiasts in providing valuable data for taxonomic research (Véla et al., 2015). As in the case of molecular markers, more informative morphometric or chemical markers could be developed using more sophisticated techniques, such as geometric morphometrics (Rakosy et al., 2017; Gibert et al., 2022). More importantly, it would be interesting to distinguish between selected (i.e., functionally significant) and neutral phenotypic traits through electrophysiological and/or behavioral studies (Schiestl et al., 1999; Rakosy et al., 2017). The distinction between biologically active and non-active floral scents, in particular, would likely provide further insights into the taxonomy of the section Pseudophrys (Stökl et al., 2005, 2009), as shown in the section Euophrys (e.g., Mant et al., 2005).
Because of these heterogeneous levels of resolution between molecular, morphometric and chemical data, species boundaries drawn from genetic versus phenotypic data were not congruent. Such an incongruence mirrors disagreements between authors favoring phylogenetic distinctness versus reproductive isolation through attraction of distinct pollinator species as a criterion to delimit Ophrys species (Paulus, 2006; Devey et al., 2008; Bateman et al., 2010). Methods based on genetic data are often judged more reliable than methods based on phenotypic data because they are not subject to investigator bias, nor affected by environmental or maternal conditions (Fujita et al., 2012). However, speciation sometimes leaves no signature at the level of neutral markers, especially when it is recent and when only a few loci mediate reproductive isolation, as it is assumed to be the case in the genus Ophrys (Xu and Schlüter, 2015). In this case, neutral markers-based methods may fail at detecting species boundaries by putting aside the data that are the most informative. Our results also show that integrating several phenotypic traits (in our case, morphometric and chemical) in the analysis may be helpful. For example, in our study, two taxa were slightly distinct morphologically, but strictly similar from a chemical point of view. When only morphometric characters were analysed, these two taxa were recognized as species, whereas when both morphometric characters and floral scents were analysed, they were merged. This shows that integrating new data types – either new molecular markers or new phenotypic traits – may challenge previous taxonomic inferences, species boundaries being hypotheses which should be tested using many data types to increase their robustness (Padial et al., 2010).
Taxonomy and conservation of the section Pseudophrys
The section Pseudophrys is known to be taxonomically challenging, due to the lack of resolution of classic molecular markers in this section (Schlüter et al., 2007; Devey et al., 2008) and to the striking morphological similarity between its members (Bernardos et al., 2005). Ninety-seven Pseudophrys species are described across the Mediterranean region (Delforge, 2016), but the taxonomic rank of many of them has been questioned when confronted to molecular, morphometric or chemical evidence (e.g., O. arnoldii: Bernardos et al., 2005; O. vallesiana: Gögler et al., 2016). Our analysis supports most Pseudophrys species that are described in France, with two remarkable exceptions: on the one hand, the first and second best models suggested to merge O. marmorata with the previously described species O. bilunulata, and on the other hand, the first and third best models suggested to merge O. zonata with the previously described species O. funerea. The similarity within these two pairs of taxa has been emphasized before (Bournérias and Prat, 2005; Tison and de Foucault, 2014), O. marmorata being sometimes called “O. bilunulata from Corsica” (Bournérias and Prat, 2005). Tison and de Foucault (2014) also suggested merging O. delforgei with the O. bilunulata/O. marmorata pair, and O. sulcata with the O. funerea/O. zonata pair, but our analysis does not support these proposals, since we found that both O. delforgei and O. sulcata were morphologically and chemically distinct from their closest relatives. The continental O. bilunulata and the Corsican O. marmorata were found to be genetically and phenotypically similar, whereas the Cyrno-Sardinian O. funerea and O. zonata were found to be slightly distinct morphologically but similar both genetically and chemically. Interestingly, recent records suggest that both O. bilunulata and O. marmorata are pollinated by Andrena flavipes (Schatz et al., 2021), which supports our proposal to merge these two taxa. Likewise, both O. funerea and O. zonata are pollinated by this species (Foelsche et al., 2000; Schatz et al., 2021), suggesting that these two taxa are not reproductively isolated and should be considered as conspecifics. The proposal to merge O. marmorata with O. bilunulata does not imply that this taxon should not be considered as vulnerable in Corsica anymore; however, it implies that it should not be considered as threatened at the national level. Another important conclusion of our study is the fact that O. peraiolae – which is sometimes described as a morph of O. marmorata (Delforge, 2005) and sometimes merged with O. funerea and O. zonata (Delage and Hugot, 2015) – likely corresponds to a genuine species, although it may be of hybrid origin (Tison and de Foucault, 2014). More generally, our analysis supports many species that are not recognized in the latest version of the European Red List (e.g., O. lupercalis, O. bilunulata, etc.), in which they are all referred to as “O. fusca sensu lato” (Rankou, 2011). We hope that this study will prompt the reassessment of their UICN status and the implementation of appropriate conservation actions, especially for species with extremely restricted distribution areas and declining population sizes such as O. peraiolae. We encourage the use of the integrative taxonomic approach proposed in this study to other orchid groups in which species boundaries are blurred, as it provides a framework to interpret patterns of genetic and phenotypic divergence among taxa and would speed up taxonomic revisions that are urgently needed for defining conservation priorities.
Statements
Data availability statement
The data presented in the study are deposited in the NCBI GenBank repository, and accession numbers are provided in the Supplementary material.
Author contributions
NJ, EV, CM, and BS conceived and designed the analysis. NJ, EV, and BS collected the data. NJ, BB, VA, and CM performed the analysis. All authors discussed the results and contributed to the final manuscript.
Funding
This study was funded by heSam Université and its Paris Nouveaux Mondes program, the Conservatoire Botanique National de Corse (Contract N°15/006, OEC-CBNC), and the Observatoire de REcherche Méditerranéen de l’Environnement (SO Ocove, OSU OREME).
Acknowledgments
We thank the Service des Marqueurs Génétiques en Ecologie (SMGE - CEFE) and the Plateforme d’Analyses Chimiques en Ecologie (PACE - CEFE) of the LABoratoire d’EXcellence Centre Méditerranéen de l’Environnement et de la Biodiversité for technical support; M. Busi, P. Cortis, A. Delage, P. Escudié, P. Fouquet, P. Geniez, T. Guillausson, L. Hugot, and J. Oltra for their help with fieldwork and C. Ané, M. Hervé, and C. Solís-Lemus for statistical advice.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1058550/full#supplementary-material
References
1
Adams R. (2007). Identification of essential oil components by gas chromatography/mass spectrometry, 4th Edn. Carol Stream, IL: Allured Publishing.
2
Agapow P. Bininda-Emonds O. Crandall K. Gittleman J. Mace G. Marshall J. et al (2004). The impact of species concept on biodiversity studies.Q. Rev. Biol.79161–179. 10.1086/383542
3
Ayasse M. Schiestl F. Paulus H. Ibarra F. Francke W. (2003). Pollinator attraction in a sexually deceptive orchid by means of unconventional chemicals.Proc. R. Soc. Lond. B270517–522. 10.1098/rspb.2002.2271
4
Ayasse M. Stökl J. Francke W. (2011). Chemical ecology and pollinator-driven speciation in sexually deceptive orchids.Phytochemistry721667–1677. 10.1016/j.phytochem.2011.03.023
5
Baguette M. Bertrand J. Stevens V. Schatz B. (2020). Why are there so many bee-orchid species? Adaptive radiation by intraspecific competition for mnesic pollinators.Biol. Rev.951630–1663. 10.1111/brv.12633
6
Barrett C. Freudenstein J. (2011). An integrative approach to delimiting species in a rare but widespread mycoheterotrophic orchid.Mol. Ecol.202771–2786. 10.1111/j.1365-294X.2011.05124.x
7
Bateman R. Bradshaw E. Devey D. Glover B. Malmgren S. Sramkó G. et al (2011). Species arguments: Clarifying competing concepts of species delimitation in the pseudo-copulatory orchid genus Ophrys.Bot. J. Linn. Soc.165336–347. 10.1111/j.1095-8339.2011.01121.x
8
Bateman R. Devey D. Malmgren S. Bradshaw E. Rudall P. (2010). Conflicting species concepts underlie perennial taxonomic controversies in Ophrys.Cah. Société Française Orchidophilie787–104.
9
Bateman R. Murphy A. Hollingsworth P. Hart M. Denholm I. Rudall P. (2018). Molecular and morphological phylogenetics of the digitate-tubered clade within subtribe Orchidinae s.s. (Orchidaceae: Orchideae).Kew Bull.731–30. 10.1007/s12225-018-9782-1
10
Bernardos S. Crespí A. Del Rey F. Amich F. (2005). The section pseudophrys (Ophrys, Orchidaceae) in the Iberian Peninsula: A morphometric and molecular analysis.Bot. J. Linn. Soc.148359–375. 10.1111/j.1095-8339.2005.00403.x
11
Bertrand J. Baguette M. Joffard N. Schatz B. (2021). “Challenges inherent in the systematics and taxonomy of genera that have recently experienced explosive radiation: The case of orchids of the genus Ophrys,” in Systematics and the exploration of life, edsGrandcolasP.MaurelM. C. (London: ISTE CNRS), 10.1002/9781119476870.c
12
Bournérias M. Prat D. (2005). Les orchidées de France, Belgique et Luxembourg, deuxième édition. Biotope editions, ed.Société Françaised’Orchidophilie (Mèze: Biotope Editions), 504.
13
Breitkopf H. Onstein R. Cafasso D. Schlüter P. M. Cozzolino S. (2014). Multiple shifts to different pollinators fuelled rapid diversification in sexually deceptive Ophrys orchids.N. Phytol.207377–389. 10.1111/nph.13219
14
Cortis P. Vereecken N. Schiestl F. Barone Lumaga M. Scrugli A. Cozzolino S. (2009). Pollinator convergence and the nature of species’ boundaries in sympatric Sardinian Ophrys (Orchidaceae).Ann. Bot.104497–506. 10.1093/aob/mcn219
15
Cuypers V. Reydon T. Artois T. (2022). Deceiving insects, deceiving taxonomists? Making theoretical sense of taxonomic disagreement in the European orchid genus Ophrys.Perspect. Plant Ecol. Evol. Syst56:125686. 10.1016/j.ppees.2022.125686
16
Dayrat B. (2005). Towards integrative taxonomy.Biol. J. Linn. Soc.85407–415. 10.1111/j.1095-8312.2005.00503.x
17
De Queiroz K. (2007). Species concepts and species delimitation.Syst. Biol.56879–886. 10.1080/10635150701701083
18
Delage A. Hugot L. (2015). Liste rouge régionale de la flore vasculaire de Corse.Corte: Conservatoire Botanique National de Corse, Office de l’Environnement de la Corse, 72.
19
Delforge P. (2005). Guide des orchidées d’Europe, d’afrique du nord et du proche-orient.Lonay: Delachaux et Niestlé, 640.
20
Delforge P. (2016). Orchidées d’Europe, d’Afrique du Nord et du Proche-Orient.Delachaux et Niestlé, Neuchâtel-Paris, Switzerland, p. 544.
21
Devey D. Bateman R. Fay M. Hawkins J. (2008). Friends or relatives? Phylogenetics and species delimitation in the controversial European orchid genus Ophrys.Ann. Bot.101385–402. 10.1093/aob/mcm299
22
Devillers P. Devillers-Terschuren J. (1994). Essai d’analyse systématique du genre Ophrys.Les Naturalistes Belg.75273–400.
23
Edgar R. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Res.321792–1797. 10.1093/nar/gkh340
24
Edwards D. Knowles L. (2014). Species detection and individual assignment in species delimitation: Can integrative data increase efficacy?Proc. R. Soc. Lond. B28120132765–20132765. 10.1098/rspb.2013.2765
25
Fay M. (2018). Orchid conservation: How can we meet the challenges in the twenty-first century?Bot. Stud.591–6. 10.1186/s40529-018-0232-z
26
Foelsche G. Foelsche W. Gerbaud M. Gerbaud O. (2000). Ophrys peraiolae spec. nov. und die taxa der Ophrys fusca gruppe in Korsika (Ophrys peraiolae spec. nov. and the taxa of the Ophrys fusca group in Corsica.).J. Eur. Orchideen32403–455.
27
Fujita M. Leaché A. Burbrink F. McGuire J. Moritz C. (2012). Coalescent-based species delimitation in an integrative taxonomy.Trends Ecol. Evol.27480–488. 10.1016/j.tree.2012.04.012
28
Gibert A. Louty F. Buscail R. Baguette M. Schatz B. Bertrand J. (2022). Extracting quantitative information from images taken in the wild: A case study of two vicariants of the Ophrys aveyronensis species complex.Diversity14:400. 10.3390/d14050400
29
Gögler J. Zitari A. Paulus H. Cozzolino S. Ayasse M. (2016). Species boundaries in the Ophrys iricolor group in Tunisia: Do local endemics always matter?Plant Syst. Evol.302481–489. 10.1007/s00606-015-1271-5
30
Gouy M. Guindon S. Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol. Biol. Evol.27221–224. 10.1093/molbev/msp259
31
Guillot G. Renaud S. Ledevin R. Michaux J. Claude J. (2012). A unifying model for the analysis of phenotypic, genetic, and geographic data.Syst. Biol.61897–911. 10.1093/sysbio/sys038
32
Hey J. (2006). On the failure of modern species concepts.Trends Ecol. Evol.21447–450. 10.1016/j.tree.2006.05.011
33
Huang J. Knowles L. (2016). The species versus subspecies conundrum: Quantitative delimitation from integrating multiple data types within a single Bayesian approach in hercules beetles.Syst. Biol.65685–699. 10.1093/sysbio/syv119
34
IUCN, MNHN, FCBN, and SFO. (2010). “Chapitre orchidées de france métropolitaine,” in La liste rouge des espèces menacées de France (Paris: IUCN).
35
Joffard N. Arnal V. Buatois B. Montgelard C. Schatz B. (2020). Floral scent evolution in the section Pseudophrys: Phylogenetic constraints or pollinator-mediated selection?Plant Biol.22881–889. 10.1111/plb.13104
36
Joffard N. Buatois B. Schatz B. (2016). Integrative taxonomy of the fly orchid group: Insights from chemical ecology.Sci. Nat.1031–5. 10.1007/s00114-016-1403-y
37
Joffard N. Massol F. Grenié M. Montgelard C. Schatz B. (2019). Effect of pollination strategy, phylogeny and distribution on pollination niches of Euro-Mediterranean orchids.J. Ecol.107478–490. 10.1111/1365-2745.13013
38
Knowles L. Carstens B. (2007). Delimiting species without monophyletic gene trees.Syst. Biol.56887–895. 10.1080/10635150701701091
39
Kühn R. Perdersen H. Cribb P. (2020). Orchidées d’europe et de méditerranée.Mèze: Biotope editions, 430.
40
Lanfear R. Calcott B. Ho S. Guindon S. (2012). PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses.Mol. Biol. Evol.291695–1701. 10.1093/molbev/mss020
41
Mant J. Peakall R. Schiestl F. P. (2005). Does selection on floral odor promote differentiation among populations and species of the sexually deceptive orchid genus Ophrys?Evolution591449–1463. 10.1111/j.0014-3820.2005.tb01795.x
42
Mason N. Fletcher N. Gill B. Funk W. Zamudio K. (2020). Coalescent-based species delimitation is sensitive to geographic sampling and isolation by distance.Syst. Biodivers.18269–280. 10.1080/14772000.2020.1730475
43
Naciri Y. Linder H. P. (2015). Species delimitation and relationships: The dance of the seven veils.Taxon643–16. 10.12705/641.24
44
Núñez R. Willmott K. Álvarez Y. Genaro J. Pérez-Asso A. Quejereta M. et al (2022). Integrative taxonomy clarifies species limits in the hitherto monotypic passion-vine butterfly genera Agraulis and Dryas (Lepidoptera, Nymphalidae, Heliconiinae).Syst. Entomol.47152–178. 10.1111/syen.12523
45
Olave M. Avila L. Sites J. Morando M. (2017). Hidden diversity within the lizard genus Liolaemus: Genetic versus morphological divergence in the L. rothi complex (Squamata: Liolaeminae).Mol. Phylogenet. Evol.10756–63. 10.1016/j.ympev.2016.09.009
46
Padial J. Miralles A. De la Riva I. Vences M. (2010). The integrative future of taxonomy.Front. Zool7:16. 10.1186/1742-9994-7-16
47
Paulus H. (2006). Deceived males–pollination biology of the mediterranean orchid genus Ophrys (Orchidaceae).J. Eur. Orchideen38303–353.
48
Pedersen H. Faurholdt N. (2007). Ophrys: The bee orchids of Europe.London: Royal Botanic Gardens.
49
Pessoa E. Alves M. Alves-Araújo A. Palma-Silva C. Pinheiro F. (2012). Integrating different tools to disentangle species complexes: A case study in Epidendrum (Orchidaceae).Taxon61721–734. 10.1002/tax.614002
50
Pillon Y. Chase M. (2007). Taxonomic exaggeration and its effects on orchid conservation: Research note.Conserv. Biol.21263–265. 10.1111/j.1523-1739.2006.00573.x
51
Pires A. Marinoni L. (2010). DNA barcoding and traditional taxonomy unified through integrative taxonomy: A view that challenges the debate questioning both methodologies.Biota Neotrop.10339–346. 10.1590/S1676-06032010000200035
52
Plants of the World Online [POWO] (2022). Facilitated by the royal botanic gardens. Available online at: http://www.plantsoftheworldonline.org/(accessed September 22, 2022).
53
Puillandre N. Lambert A. Brouillet S. Achaz G. (2012). ABGD, Automatic barcode gap discovery for primary species delimitation.Mol. Ecol.211864–1877. 10.1111/j.1365-294X.2011.05239.x
54
Pyron R. Hsieh F. Lemmon A. Lemmon E. Hendry C. (2016). Integrating phylogenomic and morphological data to assess candidate species-delimitation models in brown and red-bellied snakes (Storeria).Zool. J. Linn. Soc.177937–949. 10.1111/zoj.12392
55
R Development Core Team. (2008). R: A language and environment for statistical computing.Vienna: R Foundation for Statistical Computing.
56
Rakosy D. Cuervo M. Paulus H. Ayasse M. (2017). Looks matter: Changes in flower form affect pollination effectiveness in a sexually-deceptive orchid.J. Evol. Biol.301978–1993. 10.1111/jeb.13153
57
Rankou H. (2011). Ophrys fusca. The IUCN red list of threatened species 2011: e.T175999A7166962.Gland: IUCN.
58
Rannala B. Yang Z. (2003). Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci.Genetics1641645–1656. 10.1093/genetics/164.4.1645
59
Ronquist F. Huelsenbeck J. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models.Bioinformatics191572–1574. 10.1093/bioinformatics/btg180
60
Schatz B. Gauthier P. Debussche M. Thompson J. (2014). A decision tool for listing species for protection on different geographic scales and administrative levels.J. Nat. Conserv.2275–83. 10.1016/j.jnc.2013.09.003
61
Schatz B. Genoud D. Claessens J. Kleynen J. (2020). Orchid-pollinator network in Euro-Mediterranean region: What we know, what we think we know, and what remains to be done.Acta Oecol.107:103605. 10.1016/j.actao.2020.103605
62
Schatz B. Genoud D. Escudié P. Geniez P. Wünsch K. Joffard N. (2021). Is Ophrys pollination more opportunistic than previously thought? Insights from different field methods of pollinator observation.Bot. Lett.168333–347. 10.1080/23818107.2021.1959394
63
Schiestl F. Ayasse M. (2002). Do changes in floral odor cause speciation in sexually deceptive orchids?Plant Syst. Evol.234111–119. 10.1007/s00606-002-0187-z
64
Schiestl F. Ayasse M. Paulus H. Löfstedt C. Hansson B. Ibarra F. et al (1999). Orchid pollination by sexual swindle.Nature399421–421. 10.1038/20829
65
Schlick-Steiner B. Steiner F. Seifert B. Stauffer C. Christian E. Crozier R. (2010). Integrative taxonomy: A multisource approach to exploring biodiversity.Annu. Rev. Entomol.55421–438. 10.1146/annurev-ento-112408-085432
66
Schlüter P. Kohl G. Stuessy T. Paulus H. (2007). A screen of low-copy nuclear genes reveals the LFY gene as phylogenetically informative in closely related species of orchids (Ophrys).Taxon56493–504. 10.1002/tax.562019
67
Sedeek K. Scopece G. Staedler Y. Schönenberger J. Cozzolino S. Schiestl F. et al (2014). Genic rather than genome-wide differences between sexually deceptive Ophrys orchids with different pollinators.Mol. Ecol.236192–6205. 10.1111/mec.12992
68
Solís-Lemus C. Knowles L. Ané C. (2015). Bayesian species delimitation combining multiple genes and traits in a unified framework.Evolution69492–507. 10.1111/evo.12582
69
Soliva M. Widmer A. (2003). Gene flow across species boundaries in sympatric, sexually deceptive Ophrys (Orchidaceae) species.Evolution572252–2261. 10.1554/02-442
70
Soliva M. Kocyan A. Widmer A. (2001). Molecular phylogenetics of the sexually deceptive orchid genus Ophrys (Orchidaceae) based on nuclear and chloroplast DNA sequences.Mol. Phylogenet. Evol.2078–88. 10.1006/mpev.2001.0953
71
Stökl J. Paulus H. Dafni A. Schulz C. Francke W. Ayasse M. (2005). Pollinator attracting odour signals in sexually deceptive orchids of the Ophrys fusca group.Plant Syst. Evol.254105–120. 10.1007/s00606-005-0330-8
72
Stökl J. Schlüter P. Stuessy T. Paulus H. Fraberger R. Erdmann D. et al (2009). Speciation in sexually deceptive orchids: Pollinator-driven selection maintains discrete odour phenotypes in hybridizing species.Biol. J. Linn. Soc.98439–451. 10.1111/j.1095-8312.2009.01279.x
73
Sukumaran J. Knowles L. (2017). Multispecies coalescent delimits structure, not species.PNAS1141607–1612. 10.1073/pnas.1607921114
74
Tison J.-M. de Foucault B. (2014). Flora gallica: Flore de France.Mèze: Biotope editions, 1200.
75
Triponez Y. Arrigo N. Pellissier L. Schatz B. Alvarez N. (2013). Morphological, ecological and genetic aspects associated with endemism in the fly orchid group.Mol. Ecol.221431–1446. 10.1111/mec.12169
76
Véla E. Rebbas K. Martin R. de Premorel G. Tison J.-M. (2015). Waiting for integrative taxonomy: Morphospecies as an operational proxy for the radiative and reticulate genus Ophrys L. (Orchidaceae)?Eur. J. Environ. Sci.5153–157. 10.14712/23361964.2015.89
77
Véla E. Tirard A. Renucci M. Suehs C. Provost E. (2007). Floral chemical signatures in the genus Ophrys L. (Orchidaceae): A preliminary test of a new tool for taxonomy and evolution.Plant Mol. Biol. Rep.2583–97. 10.1007/s11105-007-0009-0
78
Vereecken N. Dafni A. Cozzolino S. (2010). Pollination syndromes in mediterranean orchids–implications for speciation, taxonomy and conservation.Bot. Rev.76220–240. 10.1007/s12229-010-9049-5
79
Vereecken N. Streinzer M. Ayasse M. Spaethe J. Paulus H. Stökl J. et al (2011). Integrating past and present studies on Ophrys pollination–a comment on Bradshaw et al.Bot. J. Linn. Soc.165329–335. 10.1111/j.1095-8339.2011.01112.x
80
Will K. Mishler B. Wheeler Q. (2005). The perils of DNA barcoding and the need for integrative taxonomy.Syst. Biol.54844–851. 10.1080/10635150500354878
81
Xu S. Schlüter P. (2015). Modelling the two-locus architecture of divergent pollinator adaptation: How variation in SAD paralogs affects fitness and evolutionary divergence in sexually deceptive orchids.Ecol. Evol.5493–502. 10.1002/ece3.1378
82
Yang L. Kong H. Huang J. Kang M. (2019). Different species or genetically divergent populations? Integrative species delimitation of the Primulina hochiensis complex from isolated karst habitats.Mol. Phylogenet. Evol.132219–231. 10.1016/j.ympev.2018.12.011
83
Yang Z. (2015). The BPP program for species tree estimation and species delimitation.Curr. Zool.61854–865. 10.1093/czoolo/61.5.854
84
Yang Z. Rannala B. (2010). Bayesian species delimitation using multilocus sequence data.Proc. Natl. Acad. Sci. U.S.A.1079264–9269. 10.1073/pnas.0913022107
85
Yeates D. Seago A. Nelson L. Cameron S. Joseph L. Trueman J. (2011). Integrative taxonomy, or iterative taxonomy?Syst. Entomol.36209–217. 10.1111/j.1365-3113.2010.00558.x
86
Zhang C. Low S. Song Y. Kozlowski G. Li L. Zhou S. et al (2020). Shining a light on species delimitation in the tree genus Engelhardia Leschenault ex Blume (Juglandaceae).Mol. Phylogenet. Evol.152:106918. 10.1016/j.ympev.2020.106918
Summary
Keywords
integrative taxonomy, species delimitation, iBPP, floral scents, orchids
Citation
Joffard N, Buatois B, Arnal V, Véla E, Montgelard C and Schatz B (2022) Delimiting species in the taxonomically challenging orchid section Pseudophrys: Bayesian analyses of genetic and phenotypic data. Front. Ecol. Evol. 10:1058550. doi: 10.3389/fevo.2022.1058550
Received
30 September 2022
Accepted
24 October 2022
Published
23 November 2022
Volume
10 - 2022
Edited by
Pavel Kindlmann, Charles University, Czechia
Reviewed by
Przemyslaw Baranow, University of Gdańsk, Poland; Tiiu Kull, Estonian University of Life Sciences, Estonia
Updates
Copyright
© 2022 Joffard, Buatois, Arnal, Véla, Montgelard and Schatz.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bertrand Schatz, bertrand.schatz@cefe.cnrs.fr
†These authors have contributed equally to this work and share last authorship
This article was submitted to Conservation and Restoration Ecology, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.