Abstract
Tissue regeneration has been in the spotlight of research for its fascinating nature and potential applications in human diseases. The trait of regenerative capacity occurs diversely across species and tissue contexts, while it seems to decline over evolution. Organisms with variable regenerative capacity are usually distinct in phylogeny, anatomy, and physiology. This phenomenon hinders the feasibility of studying tissue regeneration by directly comparing regenerative with non-regenerative animals, such as zebrafish (Danio rerio) and mice (Mus musculus). Medaka (Oryzias latipes) is a fish model with a complete reference genome and shares a common ancestor with zebrafish approximately 110–200 million years ago (compared to 650 million years with mice). Medaka shares similar features with zebrafish, including size, diet, organ system, gross anatomy, and living environment. However, while zebrafish regenerate almost every organ upon experimental injury, medaka shows uneven regenerative capacity. Their common and distinct biological features make them a unique platform for reciprocal analyses to understand the mechanisms of tissue regeneration. Here we summarize current knowledge about tissue regeneration in these fish models in terms of injured tissues, repairing mechanisms, available materials, and established technologies. We further highlight the concept of inter-species and inter-organ comparisons, which may reveal mechanistic insights and hint at therapeutic strategies for human diseases.
Introduction: Tissue Regeneration in Model Organisms
Reparative regeneration refers to replacing damaged or lost body parts with new tissue, an injury response that restores the tissue homeostasis and function in the optimal scenario (Iismaa et al., 2018). By studying regeneration, scientists can devise biological concepts for tissue repairing and apply them to traumatic injury and degenerative diseases in humans/patients. The studies encompass the strategy to stimulate the repair mechanism to replace the damaged tissues and organs, involve cross-discipline practices, and serves as a bridge between developmental biology and clinical study. A common way to study regeneration is to introduce experimental injury to the model animals and observe how they repair the tissue and recover from the injury.
In the Kingdom Animalia, the loss of regenerative ability coincides with the evolution of new and complex cell and tissue types (Brockes et al., 2001; Maginnis, 2006; Bely and Nyberg, 2010; Elchaninov et al., 2021). Anatomically simple organisms, such as hydra and planarians, can regenerate their entire body when cut into multiple pieces (Reddien and Alvarado, 2004; Reddy et al., 2019). Considerable members of early branching vertebrate lineages (like bony fish and amphibians) can also regrow various organs upon experimental injuries (Yun, 2015; Khyeam et al., 2021). In striking contrast, mammals (like mice and humans), retain a limited regenerative capacity only in some tissues and organs (Iismaa et al., 2018; Figure 1). The differences in regeneration capacity solely rely on the cellular source for replenishing lost or damaged tissue, which may come from one of three mechanisms, including the proliferation of progenitor/stem cells, dedifferentiation of mature cells into progenitors, and transdifferentiation from one cell type to another (Jopling et al., 2011). Regenerative species usually possess the pool of progenitor cells or the potential of dedifferentiation and transdifferentiation upon activating the regenerative program within the injured tissue. Taking the heart for example, most cardiomyocytes stop proliferating soon after birth, and there is no stem-like or progenitor cell population identified in the adult hearts (Bely and Nyberg, 2010; Steinhauser and Lee, 2011; Mollova et al., 2013; Bergmann et al., 2015). Instead, mature cardiomyocytes were stimulated to dedifferentiate, proliferate, and re-differentiate to replenish the lost tissue upon injury (Eschenhagen et al., 2017). Over the years, investigations across the animal kingdom have led us to compile a list of masters in tissue regeneration who also have comparable organ systems to humans, including zebrafish (Marques et al., 2019), newts (Laube et al., 2006), and Axolotl (Cano-Martinez et al., 2010; Simon and Tanaka, 2013; Figure 1).
FIGURE 1
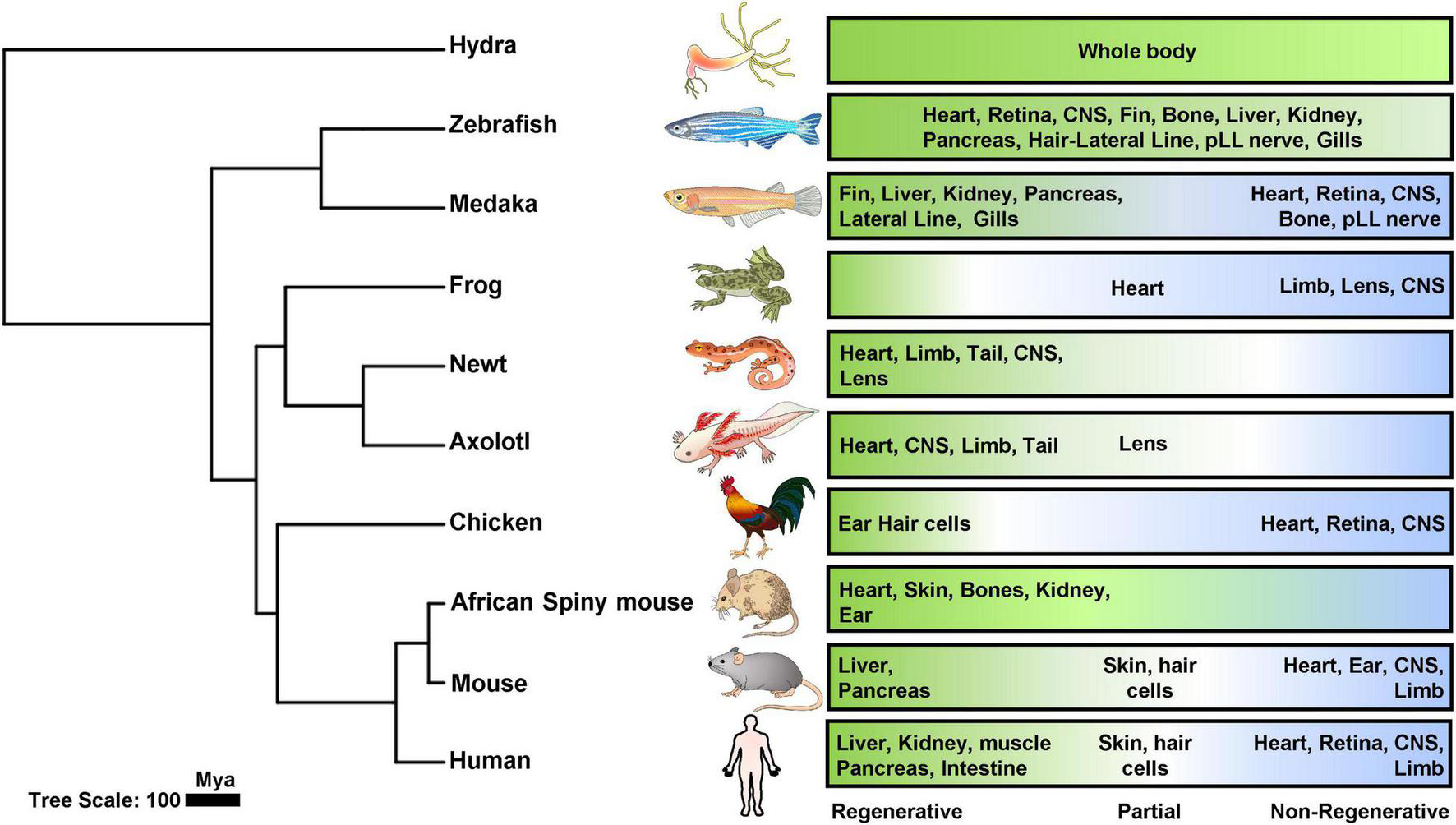
Phylogenetic analysis of tissue regeneration. Phylogenetic tree of animal models tested for regenerative capacities including cnidaria, teleost’s, amphibians, aves and mammals were generated using TimeTree (TimeTree::The Timescale of Life) (Kumar et al., 2017) and later organized by iTOL (Letunic and Bork, 2021). Regenerative (green), partial (white), and non-regenerative (blue) organs were depicted in the panels for each category.
Since regenerative capacity exists unevenly among species and their respective organs, an exciting way to uncover the mechanisms of tissue regeneration is by comparing the repair processes in animals with differential regenerative capacities. Such comparisons have been carried out in two ways: “Inter-species” comparing the repair of the same tissue/organ that is regenerative in one species (could also be age or living condition) but non-regenerative in another; Or “inter-organ” comparing two regenerative tissues within the same species to identify a central regenerative program (Potts et al., 2021). However, the regenerative species are usually quite distant in phylogeny, anatomy, and physiology from those non-regenerative ones, such as zebrafish and mice. It is thus essential to find more comparable species to overcome these shortcomings. In search of such comparative systems that can justify the disadvantages of comparing fish with mammals, zebrafish and medaka represent a more simplistic and feasible platform for comparing tissue repair and regeneration.
Zebrafish and medaka are two commonly used vertebrate models in biomedical research, given the homology with mammals and the availability of a wide range of research tools (Furutani-Seiki and Wittbrodt, 2004). Despite diverged 115–200 Mya, zebrafish and medaka are similar in anatomy, physiology, and genetics, with many conserved gene regulatory elements. In addition, zebrafish shares more than 70% of homologous genes with humans, and conserved signaling pathways and metabolic networks, making it a valuable model for biomedical research (Howe et al., 2013). Interestingly, medaka possesses regenerative capacity in fin (Katogi et al., 2004), kidney (Watanabe et al., 2009), liver (Van Wettere et al., 2013), pancreas (Otsuka and Takeda, 2017), lateral line neuromasts (Seleit et al., 2017b), and gills (Stolper et al., 2019) but is impaired to regenerate the heart (Ito et al., 2014; Lai et al., 2017), retina (Lust and Wittbrodt, 2018), brain (Shimizu and Kawasaki, 2021), and posterior lateral line (pLL) nerve cells (Seleit et al., 2022). This uneven regenerative capacity across organs is in sharp contrast with zebrafish, which can regenerate almost all organs, including the heart (Poss et al., 2002), retina (Vihtelic and Hyde, 2000; Sherpa et al., 2008), brain (Kroehne et al., 2011; Marz et al., 2011; Kishimoto et al., 2012), spinal cord (Becker et al., 1997; Ghosh and Hui, 2018), notochord (Garcia et al., 2017; Lopez-Baez et al., 2018), fin (Poss et al., 2003), kidney (Diep et al., 2011), liver (Sadler et al., 2007), pancreas (Moss et al., 2009), gills (Mierzwa et al., 2020), and lateral line (Hair cells) (Lush and Piotrowski, 2014; Cruz et al., 2015). These features make them great models for studying tissue regeneration by inter-species comparisons. Even for the inter-organ comparisons, common vs. tissue-specific regenerative programs could be revealed in zebrafish, while tissue-specific injury responses relevant to regeneration may be explored in medaka, which will be further elaborated in this review.
Here, we highlight the potential of this comparative platform by summarizing the current knowledge from published work, available tools and techniques, and elaborate on current limitations and future outlooks. This platform may provide a new opportunity for investigating the intrinsic mechanisms of tissue regeneration at the organism level and in an unbiased manner. The constraints and triggers of tissue regeneration may further translate toward novel therapeutics for related human diseases.
Evolution of the Regenerative Capacity
Deciphering the underlying mechanisms of tissue regeneration across phylogeny requires the integrative knowledge of evolutionary biology since the trait (regeneration) changes over the course of evolution (Zattara et al., 2019). This phenomenon is due to the fact that maintaining regenerative capacities requires selective pressures, in terms of the frequency and severity of major damages in an extreme living environment (Morgan, 1901; Lin et al., 2017; Elchaninov et al., 2021). These damages may compromise the fitness of the organisms, but they are not always detrimental for survival and propagation as a selective pressure (Fox and McCoy, 2000; Bernardo and Agosta, 2005). As a result, a critical phylogenetic trend identified across the animal phyla reveals declined regenerative capacity instead of preservation (Bely, 2010; Bely and Nyberg, 2010).
Various theories have been proposed to explain the declined regenerative capacity, including low damage intensity over the evolution mentioned above and the changes in adaptive value of organs (Elchaninov et al., 2021). In addition, the loss of capacity in myocardial regeneration in adult mammals could be an evolutionary trade-off related to energy metabolism (Elhelaly et al., 2016). In a different context, the regenerative capacity of limbs in amphibians but lost in other tetrapods may result from the semiautonomous module of limb development, so their limb may regenerate as a separate organ in adults without the interactions with other transient structures during development (Galis et al., 2003). Another interesting observation is that the declined regenerative capacity seems to inversely correlate with complex immune systems during development and evolution (Mescher and Neff, 2005). For example, the development and maturation of the immune system strongly correlate with the decline of regenerative capacity during frog metamorphosis (Robert and Ohta, 2009; Godwin and Rosenthal, 2014) and mammalian cardiac maturation (Porrello et al., 2011; Vivien et al., 2016). Thus, the selection pressure of the immune system may have underlying influences on the regenerative capacity that reflect at the tissue or organismic levels. However, the immune system does not always obstruct regeneration. It even acts as a critical tissue regeneration component as a coordinated innate immune response is indispensable for regenerating the axolotl limb and neonatal mouse heart (Godwin et al., 2013; Aurora et al., 2014). Moreover, the current knowledge indicates the capacity to regenerate is not only confined to organ-specific or tissue-specific levels but a coordinated involvement of systemic responses (Aurora et al., 2014; Lai et al., 2017; Sanz-Morejon et al., 2019; Bevan et al., 2020).
It is known that the loss/gain of tissue regenerative ability has evolved independently several times over the course of evolutionary history (Zattara et al., 2019). Given this, it is of paramount importance to examine and compare regeneration in a lineage-specific context (Dwaraka and Voss, 2021). Despite the availability of systematic reviews on the evolutionary origin of regeneration, only a handful of studies have addressed the regenerative potential in a lineage-specific context and have reconstructed routes of the ancestral states with the organ of interests (Zattara et al., 2019; Dwaraka and Voss, 2021). Fortunately, growing research groups are proposing comparative analyses of tissue regeneration across animal phylogeny. Comparative phylogenetic studies investigating the regenerative capacity of diverse animal taxa bring invaluable insights into the origin and preservation of regeneration throughout evolution. Novel inferences may be drawn only by comparing a wide range of organisms covering major branches/lineages of interest.
Such a study was recently reported by Hirose et al. (2019) who used cardiomyocyte ploidy as an indicator of heart regeneration and assessed the ploidy of cardiomyocytes in 41 vertebrate species. They found that the diploid cardiomyocyte frequency inversely correlated with the energy metabolism process modulated by the thyroid signaling, an evolutionary trade-off for acquiring endothermy in mammals compared to fish (Hirose et al., 2019). Evolutionary trade-offs are the manifestation of loss or gain of a particular functional trait caused by opposing selections resulting from different environments at an apparent cost (Agrawal et al., 2010). More examples of the trade-off between regenerative capacity and metabolism can be observed in Mexican cavefish (Stockdale et al., 2018). Stockdale et al. (2018) reported that the surface- and cavefish possessed similar levels of cardiomyocyte proliferative capacity, but the cave-fish showed differential upregulation of immune and scarring responses with downregulated metabolic genes compared to their surface-dwelling counterparts. These switch in metabolic regulation might play an essential role in the regenerative capacity of the cavefish when it fails to regenerate its heart and instead forms a fibrotic scar overtime. Furthermore, this intra-species comparative study nicely depicts that successful heart regeneration relies on the interplay of cardiomyocytes (CM) proliferation and scarring, which is absent in the cave-dwelling species (Stockdale et al., 2018). Moreover, the current evidence for heart regeneration further suggests that the loss of mammalian regenerative capacity is a one-trait evolutionary trade-off for higher energy metabolism in cardiac output and failure in cardiomyocyte proliferation as a capacity for heart regeneration (Elhelaly et al., 2016). The switch in metabolic reprogramming can be further correlated with cardiomyocyte proliferation observed during zebrafish heart regeneration switching from oxidative phosphorylation to glycolysis (Honkoop et al., 2019). These events, in turn, if activated in mouse hearts by ErbB2 signaling, can induce cardiomyocyte proliferation and improve functional recovery post-ischemic injury (Honkoop et al., 2019). Interestingly, medaka possesses the potential for testing these theories and makes people wonder the mechanisms underlying their uneven regenerative capacity amongst different organs, especially when compared to zebrafish.
Zebrafish and Medaka: Powerful Models for Comparative Study
Among various model systems used to study regeneration, fish species are extensively investigated. Fish is a phylogenetically “inclusive” term that encompasses four major vertebrate lineages: Sarcopterygii (lobe-finned fish), Actinopterygii (ray-finned fish), Chondrichthyes (cartilaginous fish), and Agnatha (jawless fish). Interestingly, these primitive vertebrates exhibit uneven regenerative capacity among different organs, living conditions, and between phylogenetically close species, making them perfect models to reveal how regeneration works and how to preserve or rehabilitate it in other vertebrates that have lost the capacity. Among actinopterygian fish, zebrafish are natives of the river basins in India and a well-established animal model used extensively for scientific research since the 1980s (Streisinger et al., 1981, 1986). Over the decades, zebrafish has stood out as a powerful tool for studying developmental biology, evolution, human genetics, and diseases. The advantages of the zebrafish model include a small size for manipulation, short reproductive cycle, large clutches of embryos, rapid development, cheap maintenance, comparable organs to mammals, and fully sequenced genome with well-annotated genes (Gemberling et al., 2013; Howe et al., 2013; Beffagna, 2019). Most importantly, zebrafish regenerate almost all organs upon experimental injury (Marques et al., 2019). On the other hand, medaka species are small egg-laying freshwater teleost fish home to Asia with native diversity from Japan, Korea, Taiwan, and China (Hilgers and Schwarzer, 2019). They live in rice paddy fields, rivers, and creeks in Japan, thus also named “Japanese Rice fish.” As a resident of the temperate zone, medaka can tolerate a temperature range from 4 to 40°C for both embryos and adults in the wild. In particular, Japanese medaka (Oryzias latipes) is highly tolerant to inbreeding, ideal for laboratory conditions with 14 h light and 10 h dark circles for mating conditions, with simple dietary and habitat requirements (Kirchmaier et al., 2015). They were established as a genetic model as early as 1975 (Yamamoto, 1975) and were one of the first model organisms for genetic manipulations (Ozato et al., 1986). Additionally, medaka is an ideal model organism owning to short development (7–9 days) and reproduction cycle (2–3 months), fully sequenced genome (three strains), and a transparent body throughout the juvenile stage (Ishikawa, 2000; Wittbrodt et al., 2002; Kirchmaier et al., 2015). Although zebrafish and medaka are distant relatives that got separated around 110-200 million years ago during evolution (Wittbrodt et al., 2002), they are similar in size, anatomy, and physiology, allowing them to be raised in the same laboratory conditions in terms of feeding, light-dark cycle, water temperature/quality, and propagation (Furutani-Seiki and Wittbrodt, 2004). In addition to orthologous gene-sets for genome-wide profiling and reciprocal analyses, many materials and methods can be applied equally to both zebrafish and medaka, making them ideal for comparative studies than more distantly related species (Figure 2).
FIGURE 2
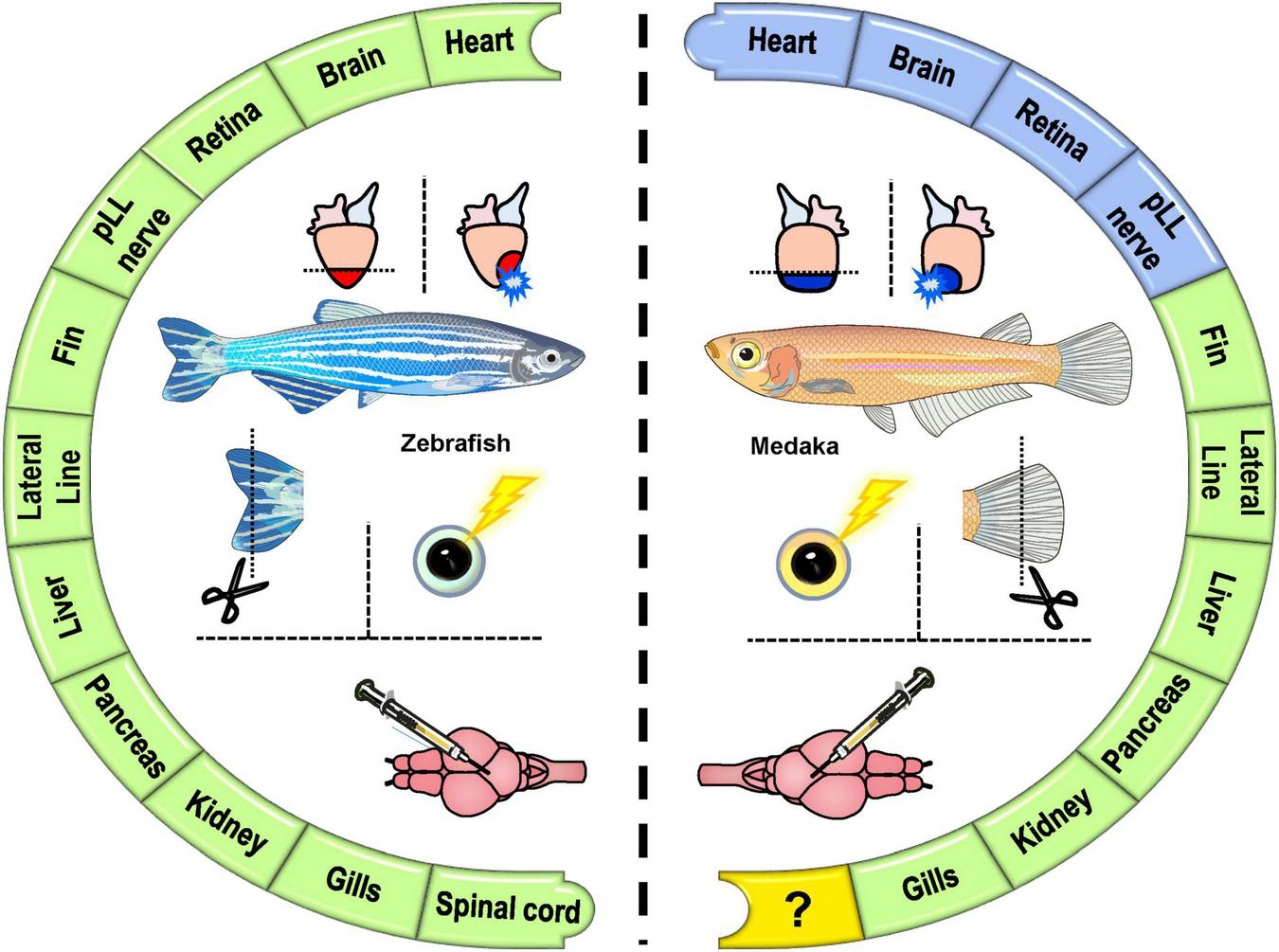
The comparative analyses of tissue regeneration in zebrafish and medaka. A summary of the regenerative (green) vs. non-regenerative (blue) organs and the comparisons (mostly inter-species) of tissue regeneration in zebrafish and medaka.
Inter-Species Comparisons: Cardiac Regeneration
As one of the most vital organs, mammalian hearts have a minimal capacity for regeneration upon disease or injury in the post-natal period, especially for replenishing cardiac muscle cells (cardiomyocytes, CMs). Instead, the infarcted hearts undergo fibrotic repair, which in turn deteriorates tissue contractility and function, eventually leading to heart failure and organismal death (Kong et al., 2014). In contrast to the limited regenerative capacity of adult mammals, certain fishes and amphibians, and even neonatal mice can regenerate their hearts after injury (Vivien et al., 2016). This is an excellent example of how the regenerative capacity of hearts exists unevenly across species and developmental stages, as mammals possess the regenerative capacity only for a short time window after birth (Porrello et al., 2011; Haubner et al., 2012, 2016). These mammalian and non-mammalian models provide unique opportunities to study the intrinsic capacity and mechanisms of heart regeneration. Amongst, extensive knowledge was gained from the zebrafish studies. Since the ground-breaking discovery of zebrafish heart regeneration by Poss et al. (2002), researchers have made in-depth investigations to understand the mechanisms of heart regeneration in zebrafish (Marques et al., 2019; Jaźwińska and Blanchoud, 2020; Potts et al., 2021). Briefly, zebrafish hearts mount a robust immune response in the recruitment of macrophages and neutrophils immediately after injury (Lai et al., 2017) and fast revascularization that expands superficially and intraventricularly and serve as the scaffold for CM repopulation (Marín-Juez et al., 2016, 2019). Almost concurrently, epicardium activates and expands by proliferation to cover the injured area and serve as a signaling hub to stimulate CM de-differentiation and proliferation in the border zone of the injured area (Kikuchi et al., 2010, 2011a,b; Jopling et al., 2011; González-Rosa et al., 2012; Cao and Poss, 2018). Newly formed CMs gradually replace scar tissue coincident with ECM remodeling (Sanchez-Iranzo et al., 2018) and scar resolution (Bevan et al., 2020; Simoes et al., 2020), eventually restoring the morphology and function of the heart. Conserved processes have been shown in neonatal mouse heart regeneration, particularly the source of regenerated CMs (Porrello et al., 2011; Vivien et al., 2016). Furthermore, hints gained from zebrafish studies have been applied to mice models to accelerate cardiac repair (Chen et al., 2016; Honkoop et al., 2019). A comparative study in zebrafish and mouse injured hearts even revealed microRNA dynamics that may regulate CM proliferation and cardiac repair (Crippa et al., 2016). However, considering the taxonomy distance between zebrafish and mice, a comparative approach may be more feasible for more closely related species with similar physiology and structure.
Unlike zebrafish, medaka showed impaired heart regeneration, indicated by a lack of revascularization, low CM proliferation, and a permanent fibrotic scar in the injured area after resection (Ito et al., 2014). To understand the differences in cardiac repair in zebrafish and medaka, Lai and colleagues performed a global transcriptomic analysis and revealed a robust immune response and angiogenic revascularization exist preferentially in zebrafish (Lai et al., 2017). Coincidently, they observed a reduced macrophage infiltration and prolonged neutrophil recruitment/retention in medaka hearts compared to zebrafish. The blunted immune response in medaka encouraged the investigation of the acute immune response and timely macrophage recruitment in heart regeneration. Indeed, in a loss-of-function setting, delayed macrophage recruitment by clodronate liposome pre-depletion abolished the regenerative capacity in zebrafish (Lai et al., 2017), which correspond nicely with findings in neonatal mice (Aurora et al., 2014). These results support an essential role of macrophage function in heart regeneration across species and encore the requirement of the macrophage function in the regeneration of other organs, including fin, retina, optic tectum, brain, and spinal cord reviewed elsewhere (Var and Byrd-Jacobs, 2020). They further identified the immunostimulant poly I:C as one of the upstream candidates that may trigger the differential transcriptomic response found between zebrafish and medaka. Indeed, they further showed that stimulating immune response by poly I:C administration promotes heart regeneration in medaka in a gain-of-function setting (Lai et al., 2017). These results support the strength of comparative analysis using fish models to gain knowledge conserved across species and identify pro-regenerative factors. However, it remains unclear how poly I:C (or other immunostimulants) promote heart regeneration in medaka and whether the same principle may apply to mammals, awaiting further investigation.
Inter-Species Comparisons: Retina Regeneration
The capacity to regenerate retinal neurons after injury also varies drastically among vertebrate species. While mammalian Müller glia (MG) do not spontaneously regenerate lost retinal neurons, zebrafish MG cells possess a robust capacity to regenerate all retinal cell types and recover their visual ability (Vihtelic and Hyde, 2000; Sherpa et al., 2008; Goldman, 2014; Gorsuch and Hyde, 2014; Lenkowski and Raymond, 2014). Several pluripotent factors, including ascl1a, lin-28, and sox2, regulate the dedifferentiation, reprogramming, and proliferation of MG cells into various retinal cell types during retina regeneration in zebrafish (Ramachandran et al., 2010a; Gorsuch et al., 2017). Sox2 is also one of the four Yamanaka factors that induced pluripotent stem cell status (Takahashi et al., 2007). In addition to retina, retinal pigment epithelium (RPE) regeneration was also recently described in zebrafish (Leach et al., 2021). Similar to heart regeneration, the immune response, particularly the macrophages and microglia cells, responds to injury and plays a critical role in retina and RPE regeneration, potentially associated with phagocytotic debris clearance and cytokine secretion (Mitchell et al., 2019; Leach et al., 2021).
During retina development, medaka neural stem cells behave similarly to those in zebrafish (Martinez-Morales et al., 2009; Centanin et al., 2011, 2014). However, Lust and Wittbrodt discovered that medaka showed limited regenerative capacity in the retina. The MG cells proliferate but fail to self-renew and reprogram, eventually giving rise to only photoreceptor cells (Lust and Wittbrodt, 2018). Moreover, by comparing medaka with zebrafish, they identified that medaka MG cells fail to maintain sox2 expression after injury and demonstrated that sustained sox2 expression in medaka MGs confers regenerative response (Lust and Wittbrodt, 2018). Similar to the above-mentioned reciprocal analyses in heart regeneration, Lust and Wittbrodt were able to identify the critical factor and demonstrated the functional relevance of sox2 expression in promoting retina regeneration.
More recently, Hoang et al. (2020) identified the evolutionarily conserved and species-specific gene regulatory networks that control the quiescent, reactive, and proliferative MG transition after retinal injury in another cross-species comparison between mice, chick, and zebrafish. They further demonstrated that deleting the factors maintaining the quiescent state may promote MG reprogramming into regeneration-competent cells in adult mice (Hoang et al., 2020).
Inter-Species Comparisons: Central Nervous System Regeneration
Unlike mammals, zebrafish respond to injury or degeneration by inducing specific neurogenic programs and constitutive neurogenesis for tissue regeneration (Diotel et al., 2020). Learning the regenerative mechanisms occurring in zebrafish will be invaluable for developing therapeutics for brain injury and degenerative diseases. In contrast to target-oriented studies, new knowledge may come from side-by-side and unbiased comparisons of animal models with divergent regenerative capacities. Unfortunately, mice brains and zebrafish brains show distinct features other than regenerative capacity, including the overall anatomy and neurogenic niches, thus preventing direct comparisons (Diotel et al., 2020; Labusch et al., 2020). Therefore, it is relevant to explore the mechanisms of brain regeneration in more closely related models.
The regenerative capacity of zebrafish central nervous system (CNS) has been investigated in the optic tectum (Ito et al., 2010; Shimizu et al., 2018; Lindsey et al., 2019) and telencephalon (Kroehne et al., 2011; Marz et al., 2011; Kishimoto et al., 2012). In the adult zebrafish CNS, both MG and the radial glia (RG) cells are activated to proliferate and differentiate into new neuronal cells following injury (Raymond et al., 2006; Ito et al., 2010; Shimizu et al., 2018). These regenerative responses seem to be induced and facilitated by immune responses (Kyritsis et al., 2012; Caldwell et al., 2019), while a specific inflammatory signaling cascade is stimulated by microglia during zebrafish brain repair (Kanagaraj et al., 2020).
A comparative study of brain regeneration in medaka and zebrafish has also been reported very recently (Shimizu and Kawasaki, 2021). Medaka shares a similar brain structure with zebrafish and neural stem cells (NSCs) niche for brain development and growth (Adolf et al., 2006; Grandel et al., 2006; Alunni et al., 2010; Kuroyanagi et al., 2010). Main NSCs exist in the optic tectum of both zebrafish and medaka, including the proliferative neuroepithelial-like stem (NE) cells and the quiescent RG cells (Alunni et al., 2010; Ito et al., 2010; Takeuchi and Okubo, 2013; Dambroise et al., 2017). However, medaka could not regenerate their optic tectum after stab injury and thus leaving a permanent scar (Shimizu and Kawasaki, 2021). In medaka, RG cells were similarly activated for proliferation upon tectum injury, but they failed to differentiate into neuron cells. Unlike the scenario in the retina, sox2 is substantially expressed in both zebrafish and medaka optic tectum and does not associate with the differential regenerative ability. Instead, the expression of pro-regenerative transcriptional factors ascl1a and oct4 were missing in the medaka. As a result, glial scar-like structures composed of GFAP+ radial fibers filled the injured area of the medaka optical tectum. Follow-up studies might be required to test the functional relevance of the ectopic expression of ascl1a and oct4 in promoting RG differentiation and optic tectum regeneration in medaka.
Inter-Species Comparisons: Fin Regeneration
Among various tissues and organs, appendage regeneration draws major attention early on in the field as teleost fish, urodeles, and amphibians all can regenerate their fins, arms, and legs following amputation (Daponte et al., 2021). Compared to limbs of urodeles and amphibians, fish fin structure is simpler and consists of bony fin rays covered by thin epidermal cells (Grandel and Schulte-Merker, 1998). Nevertheless, limb and fin are homologous tissue across vertebrate species (Yano and Tamura, 2013). A forward-genetic screen done in zebrafish identified a novel and conserved regulator of appendage patterning. When mutated, zebrafish formed limb-like bones in fins, suggesting the conservation in skeleton development and the potential of fin-to-limb transition (Hawkins et al., 2021). The regenerative capacity of the fish fin was examined as early as the 1700s to understand appendage regeneration with the first reports by French naturalist Broussonet (Broussonet, 1786; Broussonet, 1789). Like the limb regeneration in urodeles and amphibians, zebrafish repair their caudal fin by blastema-mediated epimorphic regeneration (Poss et al., 2003). In this context, blastema originates from dedifferentiated mesenchymal cells and is the primary source for growing new tissues, including bone, nerve, and vessel (Poss et al., 2003; Straube and Tanaka, 2006; Pfefferli and Jazwinska, 2015). Conserved pathways, including the Wnt/β-catenin pathway, were shown to regulate appendage regeneration across different vertebrate species, including zebrafish, Xenopus, and axolotl (Kawakami et al., 2006; Yokoyama et al., 2007).
Like zebrafish, medaka also regenerates their fin after amputation via blastema-mediated epimorphic regeneration (Katogi et al., 2004; Nakatani et al., 2007). It will be interesting to learn if medaka fin regeneration shares a conserved regenerative program with zebrafish in a cross-species study. In addition, inflammation and macrophages play a central role in both heart and fin regeneration in zebrafish but seem deficient/blunt in medaka hearts upon injury (Petrie et al., 2014; Lai et al., 2017). One may wonder whether systemic inflammation and immune response contribute differently upon fin and heart injury in medaka, which will be further discussed and await future investigation.
Overall, all these studies highlight the strength of inter-species comparisons between zebrafish (regenerative) and medaka (non-regenerative) organs to identify (and in some cases also to validate) the potential triggers of tissue regeneration. It is worth mentioning that other comparisons of organisms/conditions exhibiting diverse regenerative capacity have also been reported. For example, Stockdale and colleagues identified genes fundamental to heart regeneration by comparing the injury response of regenerative Astyanax mexicanus surface fish with their non-regenerative counterparts Pachón cave-dwelling fish (Stockdale et al., 2018). Following such regenerative traits among other species, researchers have also explored the evolutionary concept of regeneration amongst other teleost species (Table 1). In addition, comparing the transcriptomes and open chromatin landscapes of the cardiac cells isolated from the regenerative neonatal vs. non-regenerative adult mice hearts, Wang Z. et al. (2020) revealed the gene regulatory networks in diverse cardiac cell types and extracellular mediators for cardiomyocyte proliferation, angiogenesis, and fibroblast activation. Furthermore, mammals like the African spiny mouse (Acomys) can regenerate their ear, skin, heart, and bones in contrast to the house mouse (Mus musculus), providing more opportunities for cross-species analyses (Seifert et al., 2012; Matias Santos et al., 2016; Simkin et al., 2017; Qi et al., 2021).
TABLE 1
Fish models and organs for regeneration research.
Comparing organisms with similar regenerative properties may also identify the conserved regenerative programs. For example, from an inter-species comparison of zebrafish and African killifish (Nothobranchius furzeri) following fin amputation, Wang and colleagues identified the evolutionary conserved regenerative response elements (RRE) (Wang W. et al., 2020). Activation of inhba, a gene downstream of the RRE, is essential for both fin and heart regeneration and requires the binding motifs of activator protein 1 (AP-1) complex. Such enhancer is also present in mammals, shares Ap-1 binding motifs, and responds to injury, although it cannot promote regeneration. These results suggest that RREs might have been repurposed in regeneration-incompetent animals during evolution and only promote tissue repair but not regeneration (Yang and Kang, 2019; Wang W. et al., 2020). Of note, killifish possess the regenerative capacity in multiple organs, including the heart (Wang W. et al., 2020), fin (Wendler et al., 2015), and brain (Van Houcke et al., 2021) while being phylogenetically closer to medaka (Terzibasi et al., 2007), represent an alternative model for inter-species comparison.
Inter-Organ Comparisons in Medaka and Zebrafish (Regenerative Programs, Regulatory Elements, and Systemic Immune Responses)
In addition to the availability of progenitor/stem cell populations in each tissue/organ, we wonder how systemic responses, including immune response, neural innervation, hormonal regulation, metabolic shift, contribute differently to the respective injured tissues/organs of the same organism and lead to uneven regenerative capacity. The problem is especially apparent when one teleost zebrafish can regenerate organs such as the heart, retina, and brain, while another teleost medaka cannot. Taking the heart for example, the blunt immune response seems to be the major obstacle for medaka to initiate regenerative programs, but how does the same systemic (immune) response sustain the regeneration of other organs, for example, the fin? Are there tissue-specific contributions/responses, for example, residential immune cells, tissue-specific injury response elements, or even changes in the epigenomic landscape? These questions may be best addressed in medaka where the organ-specific regenerative capacities are uneven and well studied, including the regenerative fin (Katogi et al., 2004), kidney (Watanabe et al., 2009), liver (Van Wettere et al., 2013), and pancreas (Otsuka and Takeda, 2017), and non-regenerative heart (Ito et al., 2014; Lai et al., 2017), retina (Lust and Wittbrodt, 2018), and brain (Shimizu and Kawasaki, 2021). The potential findings can be cross-species compared and further validated in zebrafish loss-of-function and medaka gain-of-function experiments. A similar concept could also apply to other species. Differential regenerative capacity can be observed in lower vertebrates where some lizards can replace their tail but not their limb (Alibardi and Toni, 2005). Even in mice, the uneven regenerative capacity exists ranging from active (intestine and skin), partial (liver, pancreas, muscle), to none (CNS, heart, and most other internal organs) (Iismaa et al., 2018; Figure 1).
Even between two regenerative organs, researchers have identified common and tissue-specific regeneration responsive elements/enhancers by inter-organ comparisons. For example, Kang et al. (2016) have performed such a comparative study identifying a tissue regeneration enhancer element (TREE) from zebrafish heart and fin, which locates upstream of lepb gene and activates following injury. This element could also be activated in neonatal mouse tissues upon injury and may be engineered to modulate the regenerative potential of vertebrate organs. Another study by Pfefferli and Jaźwińska (2017) identified a 3.18 kb regulatory element upstream of ctgfa gene, named as careg element that drives the regenerative response in both zebrafish fin and heart via TGFβ/Activin-β signaling pathway. This type of study paved the way for identifying evolutionarily conserved RREs, which can also be analyzed in medaka to decipher how these RREs exist and regulate the regenerative programs in a tissue-specific manner.
Materials and Methods Available for Comparative Study in Zebrafish and Medaka
Here, we highlight the materials and methods selected from published studies to accelerate comparative studies in tissue regeneration using zebrafish and medaka, including the injury methods, visualization of gene expression and specific cell types, and functional assays by drug delivery and genetic modifications. Online resources and new experimental models will also be summarized in this section.
Injury Models
Various injury models have been established in fish models to introduce tissue injury and investigate the reparative process. In addition to the feasibility and reproducibility, these methods were often established based on the similarity to the human diseases or trauma conditions to gain translational value (Table 2). The resection or amputation model is one of the most commonly used injury models, involving surgical removal of a part of the tissue for observing the restoration of size, morphology/structure, and function of the injured tissue. Resection is straightforward, cheap, and reproducible and has been widely adopted to fin (Poss et al., 2003; Nakatani et al., 2007) and heart (Poss et al., 2002; Ito et al., 2014) regeneration studies in both zebrafish and medaka. However, resection is often accompanied by excessive bleeding and open wounds, which leads to infection and high mortality. Also, resection is sometimes infeasible for internal organs or tissues those unexposed or too small. Similar models include stabbing and transection, which works by surgically disrupting the integrity without removing any tissue and are well established in the retina, brain, and spinal cord regeneration in both zebrafish (Becker et al., 1997; Marz et al., 2011; Shimizu et al., 2018) and medaka (Shimizu and Kawasaki, 2021).
TABLE 2
Injury models developed in zebrafish and medaka.
Slightly different from resection, researchers adapted the cauterization method in fish models to mimic the fibrotic repair in mammalian organs, which is highly associated with the inflammatory response (Strungs et al., 2013; Polizzotti et al., 2016). The cauterization technique involves burning or freezing the target tissue with electric or metal probes and introducing necrotic and apoptotic cell death. Cryoinjury is popular in heart regeneration studies in both zebrafish (Gonzalez-Rosa et al., 2011; Schnabel et al., 2011; Dyck et al., 2020) and medaka (Lai et al., 2017) as it mimics the myocardial infarction in mammals better than resection model (Chablais et al., 2011; Darehzereshki et al., 2015). This technique can also be applied to external organs, such as the fin (Chassot et al., 2016) and gills (Ramel et al., 2021). However, cauterization is technically challenging and less reproducible compared to resection. Also, the wound usually takes a longer time to recover as the cell debris needs to be cleared before regeneration occurs (Schnabel et al., 2011). To increase the accuracy and reproducibility, cauterization can also be performed by using high-powered lasers in fish tissues, including the retina (Conedera et al., 2017; Lust and Wittbrodt, 2018) and skin (Richardson et al., 2013). The target tissue has to be exposed or transparent for laser penetration in this case. Overall, most physical injuries are invasive to cause high mortality and take practice to be consistent.
Apart from physical injury models, genetic cell ablation models in fish were established by expressing enzymes that catabolize cytotoxic products added to the system or induce cell death directly (Table 2). The former method was developed by expressing the bacterial enzyme Nitroreductase (NTR), which alone is not toxic but can catabolize the prodrug metronidazole (Mtz) to induce cytotoxicity (Lindmark and Müller, 1976). This system can achieve spatial (tissue-specific expression of NTR) and temporal control (the timing of adding Mtz), and labeling the target cells (co-expression with reporter system) at the same time. NTR/Mtz system has been used to tease out the functions of specific cell types in a complex process of organ regeneration in zebrafish, including the heart (Curado et al., 2007; Wang et al., 2013; Zhang et al., 2013), fin (Petrie et al., 2014), pancreatic β-cells (Pisharath et al., 2007), bone (Willems et al., 2012) and RPE regeneration (Hanovice et al., 2019). This system is also applicable to medaka, demonstrated by accessing regeneration capacity using NTR/Mtz mediated genetic ablation of the pancreatic β-cell population (Otsuka and Takeda, 2017), osteoblasts (Willems et al., 2012), and bone progenitor cells (Dasyani et al., 2019) in fin regeneration.
The latter genetic ablation tool to study zebrafish development and regeneration involves diphtheria toxin A (DTA) expression under a tissue-specific promoter, exampled by crystallin promoter-driven DTA expression in lens (Kurita et al., 2003), elastase A promoter-driven DTA expression in exocrine pancreas (Wan et al., 2006), and myl7 promoter-driven DTA expression in cardiomyocytes (Wang J. et al., 2011). Though the DTA approach lacks temporal control for activation, it is highly toxic for killing the target cells efficiently. In a modified method, the temporal control can be achieved by expressing the human diphtheria toxin receptor (DTR) and further activate cytotoxicity by diphtheria toxin injection (Jimenez et al., 2021). The genetic ablation models are technically simpler with faster recovery. These models can be used combined with physical injury to tease the role of a specific cell type during complex organ regeneration. For example, Sanchez-Iranzo et al. (2018) depleted fibroblast cells after cardiac cryoinjury and demonstrated how fibroblasts contribute to heart regeneration. Overall, most of these injury models developed in zebrafish may also apply to medaka in comparative studies of tissue regeneration.
Visualization Tools
One of the best attributes of using zebrafish and medaka as research models is the tools and techniques available for visualizing specific cell types and biological processes in vivo. Here, we summarize the visualization tools that have been established in fish models, especially in zebrafish and correspondingly in medaka, with a focus on the transgenic reporter lines (Table 3) and alternative approches.
TABLE 3
| Tissue type | Cell-type | Zebrafish transgenic lines (References) | Medaka transgenic lines (References) |
| Blood vessels | Pan-endothelial cells |
Tg(tie2:EGFP) (Motoike et al., 2000) Tg(fli1a:EGFP)y1 (Lawson and Weinstein, 2002) |
Tg(tie2:GFP) (Nakatani et al., 2008) Tg(fli1:GFP) (Moriyama et al., 2010) |
| Arterial- endothelial cells | Tg(kdrl:mCherry)is5 (Wang et al., 2010) | Tg(kdrl:DsRed2) (TG1252, NBRP) | |
| Lymphatic vessels | Pan-lymphatic cells |
Tg(lyve1:DsRed2)nz101 (Okuda et al., 2012) TgBAC(flt4:Citrine)hu7135 (Gordon et al., 2013) Tg(mrc1a:egfp)y251 (Jung et al., 2017) |
Tg(flt4-EGFP) (Deguchi et al., 2012) |
| Heart cells | Pan- cardiomyocytes |
Tg(cmlc2:DsRed2-Nuc) (Rottbauer et al., 2002) Tg(myl7:EGFP)twu26 (Huang et al., 2003) |
Cab-Tg(zfmlc2-5.1k:DsRed2-nuc) (Taneda et al., 2010); Anti-MyHC (MF20) (Ito et al., 2014) |
| Dedifferentiating-cardiomyocytes |
Tg(gata4:EGFP)ae1 (Kikuchi et al., 2010) TgBAC(nppa:mCitrine) (Honkoop et al., 2019) |
Not available | |
| Epicardium |
Tg(wt1b:GFP) (Perner et al., 2007) Tg(tcf21:nucEGFP)pd41 (Wang J. et al., 2011) |
Not available | |
| Endocardium |
Tg(flt1:YFP)hu4624 (Hogan et al., 2009) Anti-Raldh2 (Kikuchi et al., 2011b) |
Tg(raldh2-GFP) (Ito et al., 2014) | |
| Muscles and whole body | Skeletal muscles | Tg(mylz2:gfp) (Ju et al., 2003) | Tg(mylz2:gfp) (Zeng et al., 2005) |
| Ubiquitous |
Tg(actc1b:GFP) (Higashijima et al., 1997) Tg(bactin2:switch) (Bertrand et al., 2010) Tg(-3.5ubi:EGFP) (Mosimann et al., 2011) |
Tg(pOBA-GFP) (Hamada et al., 1998) Tg(EF-1α-A-GFP) (Kinoshita et al., 2000) Tg(CMV-EGFP-ITR) (Chou et al., 2001) Tg(β-actin-EGFP-ITR) (Chou et al., 2001) Wimbledon (Centanin et al., 2011) Gaudi Toolkit (Centanin et al., 2014) |
|
| Skin | Epithelial cells | Tg(krt4:nlsEGFP)cy34 (Chen et al., 2011) |
Tg(krt8:rfp) (Zeng et al., 2005) Tg(K15:H2B-EGFP) (Seleit et al., 2017a,b) Tg(K15:H2B-EGFP) and Tg(K15:LifeAct-tRFP) (Seleit et al., 2022) |
| Fibroblasts | Activated fibroblasts | Tg(postnb:citrine)cn6 (Sanchez-Iranzo et al., 2018) | Not available |
| Collagen producing fibroblasts | Tg(col1a2:loxP-mCherry-NTR)cn11 (Sanchez-Iranzo et al., 2018) | Not available | |
| Fibroblasts | Tg1(-6.8wt1a:EGFP)li7Tg (Bollig et al., 2009) | Not available | |
| Immune cells | Macrophages |
Tg(mpeg1:EGFP)gl22 (Ellett et al., 2011) Tg(mpeg1.4:mCherry-F)ump2 (Bernut et al., 2014) Tg(mfap4:tdTomato-CAAX)xt6 (Walton et al., 2015) |
Tg(mpeg1:mCherry) (Phan et al., 2020) Isolectin B4 (Lai et al., 2017) |
| Mononuclear phagocyte system |
Tg(ptprc:DsRed)sd3 (Bertrand et al., 2008) Tg(mhc2dab:GFP)sd6 (Wittamer et al., 2011) |
Tg(Cxcr3a:GFP) (Aghaallaei et al., 2010) | |
| Pro-inflammatory cells |
Tg(tnfa:EGFP-F)ump5Tg (Nguyen-Chi et al., 2015) Tg(irg1:EGFP) (Sanderson et al., 2015) |
Not available | |
| Neutrophils | TgBAC(mpx:GFP)i114 (Renshaw et al., 2006) |
Tg(FmpoP::EB3-EGFP/FmpoP::RFP-Lifeact) (Crespo et al., 2014) Tg(FmpoP::mCherry) (TG1044, NBRP) |
|
| T-cells |
Tg(lck:lck-EGFP)cz2 (Langenau et al., 2004) Tg(ikzf1:GFP)fr24 (Bajoghli et al., 2009) |
Tg(lck:gfp) (Bajoghli et al., 2015) | |
| T-regulatory cells | TgBAC(foxp3a:EGFP) (Hui et al., 2017) | Not available | |
| Progenitors and thymocytes |
Tg(rag1:GFP) (Jessen et al., 1999) Tg(rag2:GFP) (Jessen et al., 2001) |
Tg(rag1-egfp) (Li et al., 2007) Tg(ccr9a:gfp) and Tg(rag2:gfp-pest) (Bajoghli et al., 2015) |
|
| B-cells |
Tg(Cau.Ighv-ighm:EGFP)sd19 (Page et al., 2013) Tg(cd79a:GFP) and Tg(cd79b:GFP) (Liu et al., 2017) |
Not available | |
| Blood | Erythrocyte |
Tg(gata1:DsRed)sd2 (Traver et al., 2003) Tg(runx1P1:EGFP) and Tg(runx1P2:EGFP) (Yi Ni Lam et al., 2009) |
Tg(fli::GFP;gata1::GFP) (Schaafhausen et al., 2013) |
| Thrombocytes | Tg(CD41:GFP) (Lin et al., 2005) | Not available | |
| Eyes | Rod cells | Tg(XlRho:EGFP)fl1 (Fadool, 2003) | Not available |
| Müller glia cells | Tg(gfap:EGFP)nt11 (Thummel et al., 2008) |
Tg(rx2:H2B-RFP) (Inoue and Wittbrodt, 2011) Tg(rx2:lifeact-EGFP) and Tg(rx2:H2B-EGFP) (Lust and Wittbrodt, 2018) |
|
| Photoreceptor | Tg(-5.5opn1sw1:EGFP)kj9 (Takechi et al., 2003) | ||
| Retinal pigment epithelium (RPE) | Tg(rpe65a:EGFP) (Collery et al., 2016) | ||
| CNS | Pan-neurons | Tg(huC:GFP) (Park et al., 2000) | Tg(kif5a:gfp) (Kawasaki et al., 2012) |
| Radial glial cells |
Tg(gfap:GFP)mi200–1 (Raymond et al., 2006) Tg(cyp19a1b:cyp19a1b-GFP) (Tong et al., 2009) |
Anti-Gfap immunostaining, Tg(cyp19a1b-GFP) (Takeuchi and Okubo, 2013) | |
| Neural stem cells | Tg(-1.7Cau.Tuba1:GFP) (Goldman et al., 2001) |
Tg(rx2::H2B-RFP) (Inoue and Wittbrodt, 2011; Reinhardt et al., 2015) Tg(wdr12:GFP) (Dambroise et al., 2017) Tg(cndp::eGFP-caax) (Becker et al., 2021) |
|
| Oligodendrocyte | Tg(olig2:DsRed2) and Tg(sox10:mRFP) (Kucenas et al., 2008) | Not available | |
| Notochord | Sheath cells | Tg(col9a2:GFPCaaX)pd1151 (Garcia et al., 2017) | Tg(desmogon:EGFP) (Seleit et al., 2020) |
| Vacuolated cells | Tg(col8a1a:GFPCaaX) (Garcia et al., 2017) | ||
| Intervertebral disk | Tg(twhh:gfp) (Du and Dienhart, 2001) | Not available | |
| Lateral Line | Neuromast | Tg(brn3c:GAP43-GFP)s356t (Xiao et al., 2005) |
Tg(eya1:EGFP), Tg(eya1:mECFP), Tg(K15:H2B-EGFP), and Tg(K15:H2B-RFP) (Seleit et al., 2017a,b) Tg(K15:LifeAct-tRFP) (Seleit et al., 2022) |
| Fin and Bone | Osteoblasts and precursors | Tg(sp7:EGFP)b1212 (DeLaurier et al., 2010) |
Tg(osx-mCherry) (Renn and Winkler, 2009) Tg(col10a1:nlGFP) (Renn et al., 2013) |
| Osteoclasts | TgBAC(ctsk:Citrine) (Bussmann and Schulte-Merker, 2011) |
Tg(ctsk:mEGFP) (To et al., 2012) Tg(TRAP:GFP) (Chatani et al., 2011) |
|
| Pancreas | Pancreatic endocrine cells |
Tg(-6.5pdx1:GFP) (Huang et al., 2001) Tg(-8.5nkx2.2a:GFP) (Zecchin et al., 2007) Tg(-4.0ins:GFP) (Huang et al., 2001) |
Tg(pdx1-EGFP) (Otsuka et al., 2015) Tg(insulin-EGFP-NTR) (Otsuka et al., 2015) |
| Pancreatic exocrine cells | Tg(elaA:gfp) (Wan et al., 2006) | Tg(ptf1a-mCherry) (Otsuka et al., 2015) | |
| Liver | Hepatocytes |
Tg(-2.8fabp10a:EGFP) (Her et al., 2003) Tg(-1.7apoa2:GFP) (Wang R. et al., 2011) |
Tg(chg-L1.5 kb/GFP-emgb/RFP) (Ueno et al., 2004) |
Visualization tools developed in zebrafish and medaka.
Since fluorescence reporter driven by tissue-specific gene promoter is applicable and efficient in zebrafish and medaka, many transgenic lines have been generated to study specific tissues/organs in development and diseases. Here we summarize tissue-specific reporters described in major tissue regeneration studies previously mentioned in Table 3. In addition to tissue-specific reporter lines, researchers can also use antibodies against cell-specific transcription factors or cytosolic proteins to assess cellular dynamics in growth, development, and regeneration. For example, Mef-2 (sc-313), nkx2.5 (GTX128357) or MF20 (Fischman, D.A., DSHB) antibody can be used in combination with proliferation/cell cycle markers PCNA (GTX124496), Anti-phospho-Histone H3 (Ser10) (06-670, Merck), and BrdU/EdU (C10086, ThermoFisher, Eugene, OR, United States) to label the proliferating CMs (Chablais et al., 2011; Chablais and Jaźwińska, 2012a). Similarly, transgenic medaka lines have also been generated to facilitate cardiac research using zebrafish cmlc2 regulatory elements for myocardial expression (Taneda et al., 2010). In addition to labeling cardiac tissues, researchers have successfully developed medaka reporters utilizing zebrafish skeletal specific mylz2 promoter to label skeletal muscles (Zeng et al., 2005). Vice versa, medaka mylz2 promoter can also recapitulate GFP expression in zebrafish (Zeng et al., 2005). On the same note, medaka β-actin promoter can drive ubiquitous gene expression in both medaka and zebrafish (Yoshinari et al., 2012), while zebrafish krt8 promoter can label both skin and intestinal epithelium in medaka, as almost identical to zebrafish (Zeng et al., 2005). These examples showed that the transcriptional regulation of many genes is highly conserved in both species, with many tissue-specific transgenes developed to facilitate research, as summarized in Table 3.
Restoring vasculature and circulation is one of the first steps during tissue regeneration (Jung and Kleinheinz, 2013). Both these blood vessels and lymphatic vessels that regulate tissue homeostasis and immune cell trafficking can be visualized by reporters and have been used extensively for regenerative studies in zebrafish (Table 3). To name a few of the most commonly used reporter lines, tie2 reporter for pan-endothelial cells (Motoike et al., 2000), fli1a reporter for endothelial and endocardial cells (Lawson and Weinstein, 2002), kdrl reporter for arterial vessels (Wang et al., 2010), and lyve1 (Okuda et al., 2012) reporter for lymphatic vessels in zebrafish. Correspondingly, a wide range of transgenic reporter strains has been generated in medaka based on orthologous gene promoters (Table 3). Moreover, one can also use staining methods to label the vasculature when transgenic animals are inaccessible. One of the widely adopted vasculature staining methods involves alkaline phosphatase (AP) staining, which relies on the endogenous AP activity to convert NBT/BCIP into purple precipitates in endothelial cells for rapid visualization in larvae (Childs et al., 2002) and heart (Lai et al., 2017). In addition, the Fli1 antibody (ab133485) can be used to mark endothelial cell nuclei in zebrafish by immunostaining (Bensimon-Brito et al., 2020). Aside from post-fixation staining, angiography can be achieved by fluorescent dextran injection into the circulation of larval (Hoeppner et al., 2015; Takanezawa et al., 2021) and adult fish (Pugach et al., 2009).
Immune response, especially inflammatory cell infiltration and resolution, is a critical component of tissue regeneration to prevent infection, clear damaged tissue, maintain tissue integrity, and sometimes even is associated with the fibrotic response and cell proliferation (Julier et al., 2017). On top of the tremendous capacity in regeneration, zebrafish possess both innate and adaptive immunity comparable to mammals (Trede et al., 2004), making it a powerful model to study the role of immune response in tissue repair and regeneration (Var and Byrd-Jacobs, 2020). Inflammatory cells, including neutrophils and macrophages, are among the first responders recruited to the injured tissue by chemokines and damage-associated molecular patterns (DAMPs) (McDonald et al., 2010; Soehnlein and Lindbom, 2010). Some of the zebrafish reporter lines have been used to visualize these innate immune cells, including fluorescent genes expression driven by neutrophil-specific mpx promoter (Renshaw et al., 2006) and macrophages specific mpeg1.1/mpeg1.4 (Ellett et al., 2011) and mfap4 (Walton et al., 2015) promoters. Taking advantage of the transparent tissue at the larval stage, zebrafish have been extensively used for studying the dynamic and function of these inflammatory cells in tissue repair/regeneration (Li et al., 2012). Apart from the innate immune system, some adaptive immune cell reporters were also established in zebrafish (Table 3). Materials for zebrafish immune research have also been previously reviewed (Martins et al., 2019). The immune system in medaka is less studied compared to zebrafish. Still, some immune cell reporters have been generated in medaka based on zebrafish orthologous genes (Table 3), including neutrophil-specific mpo reporter (alias to mpx) (Grabher et al., 2007; Crespo et al., 2014), macrophages specific mpeg1.1 reporter (Phan et al., 2020), and pan mononuclear phagocytes cxcr3.2 reporter (Aghaallaei et al., 2010). For the adaptive immune cells, medaka gained interest for studying T-cell development where Tg lines were developed respectively (Bajoghli et al., 2019). Given the importance of immune response in tissue repair/regeneration and the amount of knowledge gained in zebrafish, it is pretty evident that corresponding transgenic reporter lines in medaka await future development for comparative studies. Specifically, it would be interesting to learn more about the critical roles of immune cells that plays similarly or differently in these two model systems.
Due to the limited resource of antibodies against fish proteins and reporters for labeling immune cells, other approaches can be applied to label and even isolate immune cells. For example, isolectin B4 (IB4) and liposome-uptake may label macrophage and other phagocytes in both zebrafish and medaka. In a comparative study, IB4 labels mainly macrophages in zebrafish and medaka and show colocalized signals with zebrafish mpeg1 reporter signals (Lai et al., 2017). On the other hand, DiI liposomes can label the phagocytes efficiently in both zebrafish and medaka based on their properties of macrophage ablation when loaded with clodronate (Lai et al., 2017). Despite in limited numbers, some antibodies work in both fish models in labeling the immune cells, including Lcp1 (GTX124420) (Redd et al., 2006), Lyz (GTX132379) for leukocytes, Spi/Pu.1 for myeloid cells (GTX128266), Mpx (GTX128379) for neutrophils (Lai et al., 2017), Mpeg1 (GTX54246) for macrophages (Simoes et al., 2020), and anti-4C4 for microglia (Becker and Becker, 2001).
Here, we have tabulated the most widely used fish reporter and transgenic lines in Table 3. More transgenic lines that label different tissues can be easily looked up in the Zebrafish Information Network (ZFIN1) and accessed from the Zebrafish International Resource Center (ZIRC2) and the European Zebrafish Resource Center (EZRC3), or the NBRP Medaka website4. Overall, common regulatory elements between zebrafish and medaka support that the activation of zebrafish-specific factors can be well recapitulated in the medaka and vice-versa. This evidence further highlights the conserved gene-regulatory networks between zebrafish and medaka, making them excellent models to perform comparative studies in tissue regeneration.
Genetic Manipulations in Zebrafish and Medaka
Taking advantage of being model organisms and a broad research community, zebrafish and medaka are well-equipped with tools for genetic manipulations for generating animal models for specific cell ablations, visualization, and functional manipulations introduced previously. Here, we summarize these toolsets established in zebrafish and medaka (Table 4) to investigate the cellular and molecular mechanisms of tissue regeneration.
TABLE 4
Genetic manipulation tools in zebrafish and medaka.
Genetic manipulation in forward genetic screening has been established in fish models early on (Driever et al., 1996; Wienholds et al., 2003). As a standard method, ENU (ethylnitrosourea) treatment introduced point mutations via base alkylation that give rise to single base mutations (often called ENU mutants) in zebrafish (Driever et al., 1996) and medaka (Loosli et al., 2000; Furutani-Seiki et al., 2004). Lacking efficient methods for targeted gene mutagenesis used to be a weak spot of zebrafish reverse genetics, but was recently overcome by the invention of zinc-finger nucleases (ZFN) (Doyon et al., 2008), transcription activator-like effector nucleases (TALENs) (Huang et al., 2011; Bedell et al., 2012), and the Clustered, Regularly Interspaced, Short Palindromic Repeat (CRISPR)/CRISPR-associated 9 (Cas9) technology (Hruscha et al., 2013; Hwang et al., 2013). CRISPR became the favorable genetic manipulation strategy given its ease to generate and assemble, and application in almost all eukaryotic cells. In addition to gene knockout/mutagenesis, CRISPR technology was further modified for generating knock-in/transgenic zebrafish carrying reporter/functional genes under endogenous gene regulation (Kimura et al., 2014). Like zebrafish, genome editing using TALEN (Ansai et al., 2013, 2014) and CRISPR/Cas9 (Ansai and Kinoshita, 2014) have been established in medaka. CRISPR/Cas9 mediated gene knock-in via NHEJ was used to generate transgenic and mutant medaka with a high germline transmission rate (Watakabe et al., 2018). In addition, the knock-in method using CRISPR was also applied to generate conditional knockout zebrafish by targeted insertion of loxP sites (Burg et al., 2018). Instead of NHEJ mediated knock-in which is error-prone, Wierson et al. (2020) have further developed knock-in method based on homology mediated end joining (HMEJ) repair for a more efficient and precise genome editing in zebrafish known as the GENEWELD method. Likewise, homology-directed repair (HDR) mediated knock-in strategies were also feasible in medaka (Murakami et al., 2017).
This concurrent development of technologies in zebrafish and medaka highlights the reciprocal nature of exchanging tools and methods between these model systems, facilitating the advancement of scientific research. Besides mutagenesis, genetic manipulation via stable transgenesis was first demonstrated in medaka (Ozato et al., 1986). Later, Kawakami and Shima (1999) identified the Tol2 transposon system in Medaka and adapted this system in zebrafish for transgenesis which revolutionized the field. To better recapitulate the endogenous gene expression patterns, insertion of BAC constructs by Tol2 transposase has been widely used in generating zebrafish reporter lines (Suster et al., 2011). Despite the fact that Tol2 was originally identified in medaka, it is more efficient in zebrafish (Kawakami, 2007). Therefore, another method for insertional transgenesis was developed in fish models using I-SceI meganuclease and greatly facilitated transgenesis in medaka (Thermes et al., 2002; Grabher and Wittbrodt, 2008).
As applications, spatial (e.g., specific tissue) and temporal regulation of ectopic gene expression can be achieved by combining transgenic lines generated by the above-mentioned methods (Tables 4, 5). For example, the Cre/lox system is widely used to perform reverse genetics, ectopic gene expression, and lineage tracing experiments in multicellular organisms, including zebrafish (Felker and Mosimann, 2016). To achieve spatial-temporal control, the Cre recombinase gene is fused with a human estrogen receptor (ER) domain and expressed under tissue-specific promoter, resulting in recombination of lox sequences in specific tissue upon estrogen stimulation (Metzger et al., 1995; Feil et al., 1996). Since then, there have been growing numbers of tissue-specific Cre lines generated in zebrafish (Jungke et al., 2013, 2015). Within the scope of tissue repair and regeneration, we have summarized a list of Cre driver and switch lines applied in previously described studies (Table 5). Additionally, robust co-expression of multiple genes following switch cassette can be accomplished by placing polycistronic ORFs separated by short viral 2A peptides (Provost et al., 2007). In contrast to zebrafish, there is way fewer medaka Cre/lox transgenic lines that we found and summarized with respect to the comprehensive list in zebrafish with available databases like CreZoo (Jungke et al., 2013, 2015). Fortunately, the advances of CRISPR technology allow genetic manipulation in both fish models and generate powerful tools for fate mapping and functional experiments (Liu et al., 2019). Overall, our review encompasses a portion of tools and strategies commonly used in zebrafish and medaka, which were extensively reviewed elsewhere for medaka (Kirchmaier et al., 2015) and zebrafish (Sassen and Köster, 2015), respectively.
TABLE 5
| Tissue type | Zebrafish transgenic lines (References) | Medaka transgenic lines (References) |
| Cardiomyocytes |
Tg(myl7:creERT2) (Kikuchi et al., 2010) Tg(gata4:creERT2) (Kikuchi et al., 2011a) |
Not available |
| Skeletal muscles | Tg(cry:mCherry;-1.9mylz2:CreERT2) (Mukherjee and Liao, 2018) | Tg(myl2::nlsCreCherry) (TG938, NBRP) |
| Endothelial cells | Tg(fli1a:CreERT2)cn9 (Sanchez-Iranzo et al., 2018) | Not available |
| Epicardial cells | Tg(tcf21:CreERT2)pd42Tg (Kikuchi et al., 2011a) | Not available |
| Neuronal cells | Tg(Cau.tuba1a:CreERT2, Cau.tuba1a:CFP)mi19/+ (Ramachandran et al., 2010b) |
Tg(rx2::CreERT2) (Reinhardt et al., 2015) Tg(K15:Ert2-Cre) (Seleit et al., 2017b) Tg(cndp::CreERT2) (Becker et al., 2021) |
| Macrophages |
Tg(mpeg:Cre)fh506 (Roh-Johnson et al., 2017) Tg(mfap4:iCre:p2A-tdTomato)xt8 (Walton et al., 2015) |
Not available |
| Fibroblast and collagen producing cells | Tg(periostin:CreERT2)cn7 and Tg(wt1a:CreERT2)cn10 (Sanchez-Iranzo et al., 2018) | Not available |
| Bone | Tg(Ola.Sp7:CreERT2-P2A-mCherry)tud8 (Knopf et al., 2011) | Not available |
| Heat-shock (temporal) | Tg(hsp70l:mCherry, Cre-ERT2)tud104 (Hans et al., 2011) | GaudíHspCRE.A (Centanin et al., 2014) |
| Pan-cells switch type |
(ubi:Switch) and Tg(-3.5ubb:CreErT2, myl7:EGFP)cz1702 (Mosimann et al., 2011) Tg(bactin2:loxp-DsRed-STOP-loxp-EGFP) (Kikuchi et al., 2010) |
GaudiRSG toolkit (Centanin et al., 2014) Tg(Olactb:loxP-dsR2-loxP-EGFP) (Yoshinari et al., 2012) |
| Switch ablation line | Tg(bactin2:loxP-mCherry-STOP-loxP-DTA176)pd36 (Wang J. et al., 2011) | Not available |
The Cre drivers and switch lines for studying tissue regeneration.
Delivery of Pharmaceutical Reagents
Since zebrafish and medaka are routinely used for drug screening and validations, developing various routes of pharmaceutic administration is essential (Table 6). As these fish are tiny compared to mice, the development of administration methods requires further optimization to mimic the delivery route in mice.
TABLE 6
| Description | Website | References |
| Dissection of different organs from the Adult Zebrafish | Dissection of Organs from the Adult Zebrafish | Protocol (jove.com) | Gupta and Mullins, 2010 |
| Dissection of the Adult Zebrafish Kidney | Dissection of the Adult Zebrafish Kidney | Protocol (jove.com) | Gerlach et al., 2011 |
| Induction of myocardial infarction in adult zebrafish using cryoinjury | Induction of Myocardial Infarction in Adult Zebrafish Using Cryoinjury | Protocol (jove.com) | Chablais and Jaźwińska, 2012a |
| Brain injury model by stabbing in Adult Zebrafish | Stab Wound Injury of the Zebrafish Adult Telencephalon: A Method to Investigate Vertebrate Brain Neurogenesis and Regeneration | Protocol (jove.com) | Schmidt et al., 2014 |
| Spinal cord injury by transection in larval zebrafish | Spinal Cord Transection in the Larval Zebrafish | Protocol (jove.com) Zebrafish In Situ Spinal Cord Preparation for Electrophysiological Recordings from Spinal Sensory and Motor Neurons | Protocol (jove.com) |
Briona and Dorsky, 2014; Moreno et al., 2017 |
| Laser-induced retinal injury model in zebrafish | Müller Glia Cell Activation in a Laser-induced Retinal Degeneration and Regeneration Model in Zebrafish | Protocol (jove.com) | Conedera et al., 2017 |
| Examining muscle regeneration in zebrafish models of muscle disease | Examining Muscle Regeneration in Zebrafish Models of Muscle Disease | Protocol (jove.com) | Au - Montandon et al., 2021 |
| Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration | Hepatocyte-specific Ablation in Zebrafish to Study Biliary-driven Liver Regeneration | Protocol (jove.com) | Choi et al., 2015 |
| Intraperitoneal injection in zebrafish | Intraperitoneal Injection: A Method of Solution Delivery into the Abdominal Cavity of an Adult Zebrafish | Protocol (jove.com) | Kinkel et al., 2010 |
| Intrathoracic injection for the study of adult zebrafish heart | Intrathoracic Injection for the Study of Adult Zebrafish Heart | Protocol (jove.com) | Bise and Jazwinska, 2019 |
| Retro-orbital injection in adult zebrafish | Retro-orbital Injection in Adult Zebrafish | Protocol (jove.com) | Pugach et al., 2009 |
| Nanoparticle-mediated siRNA gene-silencing in adult zebrafish heart | Nanoparticle-mediated siRNA Gene-silencing in Adult Zebrafish Heart | Protocol (jove.com) | Xiao et al., 2018 |
| CRISPR/Cas9-generated gene knockouts in zebrafish | Efficient Production and Identification of CRISPR/Cas9-generated Gene Knockouts in the Model System Danio rerio | Protocol (jove.com) | Sorlien et al., 2018 |
| Imaging blood vessels and lymphatic vessels in the zebrafish | Methods in Cell Biology | The Zebrafish - Cellular and Developmental Biology, Part A Cellular Biology | ScienceDirect.com by Elsevier | Jung et al., 2016 |
| Microinjection of medaka embryos for use as a model genetic organism | Microinjection of Medaka Embryos for use as a Model Genetic Organism | Protocol (jove.com) | Porazinski et al., 2010b |
| Dechorionation of medaka embryos and cell transplantation for the generation of chimeras | Dechorionation of Medaka Embryos and Cell Transplantation for the Generation of Chimeras | Protocol (jove.com) | Porazinski et al., 2010a |
| Medaka: Biology, Management, and Experimental protocols | Volume 1 and 2 (https://onlinelibrary.wiley.com/doi/book/10.1002/9781119575399#) | Kinoshita et al., 2009; Murata et al., 2019 |
Tutorials/protocols for injury and drug delivery for zebrafish and medaka.
Intraperitoneal (IP) injection was one of the first delivery methods introduced to deliver reagents in adult zebrafish and is adapted from veterinary practice in bigger fishes (Kinkel et al., 2010). The injection is performed using a 31G needle with a small injection volume (usually < 10–15 μl) into the abdominal cavity posterior to the pelvic girdle and midline to the pelvic fins in zebrafish (Kinkel et al., 2010). The reagents administrated by IP injection will distribute majorly to the spleen and liver within 72 h and later into the circulation, making it a favorable method for studying biochemical modulation in zebrafish over multiple injections or an extended period (Ruyra et al., 2014). IP is widely used for systemic administration of drugs, small-molecule inhibitors, nanoparticles, reagents in regeneration studies, including clodronate liposomes for ablating macrophages (de Preux Charles et al., 2016), poly I:C for immune-stimulation in medaka (Lai et al., 2017), mTOR-inhibitor rapamycin for autophagy inhibition (Chavez et al., 2020), IWR-1-endo for Wnt inhibition (Chen et al., 2009; Zhao et al., 2019), and tamoxifen for Cre-mediated recombination (Hans et al., 2009).
On the other hand, intravenous (IV) injection is effective for drug delivery in mice but has been technically challenging for zebrafish, due to the small vessel diameter and poor vasculature visibility. To improve vessel visibility, transparent casper fishes can be injected intravenously through their cardinal vein described in cancer research in zebrafish (Benjamin and Hynes, 2017). As alternative methods for direct access to the circulation, the intra-cardiac injection has been tested but showed high mortality (White et al., 2008), while the retro-orbital (RO) injection is suitable for delivering both reagents and cells into the blood circulation (Pugach et al., 2009; Simoes et al., 2020).
For more tissue/organ-localized delivery methods, intra-tissue injections may be applied. For example, intrathoracic (IT) injections were developed in zebrafish to test the effects of exogenous factors on adult heart regeneration, including nanoparticles encapsulated siRNAs (Xiao et al., 2018; Bise and Jazwinska, 2019). Cerebroventricular microinjection (CVMI) was also developed to deliver reagents to the adult zebrafish brain (Kizil and Brand, 2011). Intravitreal injections for the targeted delivery to the vitreous space of the retina (Fimbel et al., 2007) and intraspinal injection to the spinal cord were also developed in zebrafish (Wehner et al., 2018).
In contrast to invasive methods, which sometimes lead to tissue damage and mortality, a straightforward and convenient way for pharmaceutic delivery is immersion/incubation. Although immersion/incubation can be applied to both larvae and adults, it is more costly for incubating adults due to a large amount of reagent needed to reach the same dose. The biodistribution from immersion varies among reagents and different stages. In adults, the reagent is mainly intake through gills and ingestion, then absorbed/digested in the intestine and liver, which resembles oral uptake in the mice model (Ruyra et al., 2014). Examples of incubation experiments include tamoxifen treatment for Cre activation and Erbb2 inhibitor AG1478 in heart regeneration of both zebrafish larvae and adults (de Koning et al., 2015; Gemberling et al., 2015). Incubation of Alk5/4 inhibitor SB431542 has also been used in adult fin (Jaźwińska et al., 2007) and heart (Chablais and Jaźwińska, 2012b) to understand the dynamics of fibrosis in tissue regeneration.
Overall, based on the size and anatomic similarities between zebrafish and medaka, most of these delivery methods (Table 6) can be adapted in medaka and are regularly used in our laboratory as well as in other studies (Maekawa et al., 2016; Lai et al., 2017; Marín-Juez et al., 2019).
Genomics Resources for Zebrafish and Medaka
Due to the interest in exploring cellular and molecular mechanisms throughout the last few decades, both zebrafish (∼1,412 Mb) (Howe et al., 2013) and medaka genome (∼800 Mb) (Kasahara et al., 2007; Kobayashi and Takeda, 2008) have been sequenced and are publicly available in the databases, including Ensembl for zebrafish5 (see footnote 1) and medaka6 (Table 7). Like zebrafish, medaka has emerged as one of the most popular and influential animal models to investigate development and disease. At the genomic level, the regenerative capacity of zebrafish might rely on gene regulatory networks, which might be repressed in other non-regenerative animal models (Yang and Kang, 2019). Early findings in zebrafish suggest epigenetic modifications ranging from histone modifications to initiation of enhancer-induced activation of regenerative programs (Kang et al., 2016; Pfefferli and Jaźwińska, 2017). Growing evidence support the compatibility of zebrafish and medaka for comparative transcriptomic analyses (Tena et al., 2014; Lai et al., 2017). Moreover, with the advances of epigenetic profiling, medaka has also been explored for epigenetic changes associated with embryogenesis, development, and evolution (Nakamura et al., 2014; Tena et al., 2014; Ichikawa et al., 2017; Marletaz et al., 2018; Uesaka et al., 2019; Li et al., 2020). These properties may bring medaka as a new teleost model in epigenetics for comparative studies in tissue regeneration.
TABLE 7
| Fish | Resource | Description | Website and references |
| Zebrafish | The Zebrafish Information Network (ZFIN) | Central database of zebrafish resources, studies and protocols | http://zfin.org/ (Ruzicka et al., 2019) |
| Ensembl: Danio rerio | Genome assembly, GRCz11 | http://asia.ensembl.org/Danio_rerio/Info/Index | |
| zfRegeneration | Dataset for zebrafish associated regeneration studies | http://www.zfregeneration.org/ (Nieto-Arellano and Sanchez-Iranzo, 2019) | |
| CreZoo | Database of Zebrafish Cre driver lines | Zebrafish CreZoo (Jungke et al., 2013) | |
| Zebrafish International Resource Center | Zebrafish stock center, United States | https://zebrafish.org/home/guide.php | |
| European Zebrafish Resource Center (EZRC) | Zebrafish stock center, KIT-Europe | https://www.ezrc.kit.edu/index.php | |
| The Taiwan Zebrafish Core Facility (TZCF) | Core facility for Zebrafish stock | http://www.tzcf-tzenh.org/ (You et al., 2016) | |
| NBRP-Zebrafish | Japan stock center for zebrafish resource | https://shigen.nig.ac.jp/zebra/index_en.html | |
| China Zebrafish Resource Center (CZRC) | Zebrafish resources, developing new lines and technology | http://en.zfish.cn/ | |
| Medaka | NBRP medaka | Central repository and achieve for medaka resources | https://shigen.nig.ac.jp/medaka/ (Sasado et al., 2010) |
| NBRP strains | Repository for strains and transgenics | https://shigen.nig.ac.jp/medaka/strain/strainTop.jsp | |
| Ensembl: Japanese medaka HdrR | Gene assembly and gene annotation (ASM223467v1) | http://asia.ensembl.org/Oryzias_latipes/Info/Index | |
| NBRP genome tools | Genome Mapping | http://viewer.shigen.info/medakavw/mapview/ | |
| MEPD | Gene expression data by in situ hybridization | http://mepd.cos.uni-heidelberg.de/mepd/ (Alonso-Barba et al., 2016) | |
| mODP | OMICs data and epigenetic modification database | http://tulab.genetics.ac.cn/modp/#/Browser?species=medaka (Li et al., 2020) | |
| Both Models | CCTop | CRISPR/Cas9 design tool | https://cctop.cos.uni-heidelberg.de:8043/index.html (Stemmer et al., 2015) |
| CRISPRscan | CRISPR/Cas9 design tool | https://www.crisprscan.org/ (Moreno-Mateos et al., 2015) |
Resources for zebrafish and medaka research.
Hybrid/Chimera Fish and Cell Transplantations
Among various teleost fishes that come in different shapes and sizes, zebrafish and medaka have a similar developmental process overall. However, medaka shows a slower pace and hatch at 9 days compared to 3 days in zebrafish. Considering the similarities and differences between zebrafish and medaka, inter-species blastula transplantation was explored to study the genetic developmental timing during organogenesis (Hong et al., 2012; Fuhrmann et al., 2020). Generating chimeric organisms named “Zebraka” or “Medrafish” involves cell transplantation at the blastula stage and ectopic formation of chimeric organs like retina from zebrafish donor cells in the medaka host. This approach helps researchers determine the transcriptional dynamics of retinal organogenesis and further state the existence of organ-intrinsic mechanisms independent of the development pace of the host (Fuhrmann et al., 2020). It will be intriguing to consider the differential regenerative capacity of each organ in these hybrid animals. For example, if the heart was contributed by zebrafish donor cells and grows in medaka, would it still be regenerative (and why)? Hybrid animals may help researchers dissect the intrinsic and extrinsic properties of tissue regeneration, and zebrafish and medaka chimeras may provide a unique opportunity for such study.
Similar ideas have been exploited in the intra-species blastula transplantation in embryonic and adoptive transfer in adult zebrafish to determine the cellular contribution of specific biological processes (cell-autonomous vs. non-cell-autonomous actions). The concept was recently adopted in a heart regeneration study where macrophages were isolated from zebrafish larvae donors and adoptively transferred into adult hosts. Larval macrophages were found to infiltrate the injured hearts and contribute to scar formation by directly secreting collagens in adults (Simoes et al., 2020). Similar strategies may pave the way for future research, in which zebrafish donor cells might be transferred in medaka host or vice-versa and determine the cellular contributions in tissue regeneration.
Limitations, Future Directions, and Concluding Remarks
In this review, we illustrated the concept of comparative study in tissue regeneration and highlighted the examples in zebrafish and medaka models. Zebrafish and medaka are phylogenetically close model organisms compared to animals from different phyla across the animal kingdom yet possess a distinct tissue regeneration capacity. Various tools and techniques commonly used in zebrafish and medaka were summarized here, supporting the unique strength of conducting comparative tissue regeneration research. Nevertheless, we cannot ignore some technical challenges and conceptual oversight that must be addressed or overcome.
On a technical aspect, orthologous gene annotation and identification are critical for inter-species comparisons. The Zebrafish genome has been well annotated and referenced due to a broad research interest with orthologous genes identified and translatable to the mammalian system (Howe et al., 2013). However, the medaka genome is less annotated than zebrafish. The limitation may sometimes be overcome by mapping the medaka genome against zebrafish to identify the orthologous genes. Though this might not be the optimal approach for genome annotation, it is currently the most feasible method to translate genetic information to the zebrafish and the mammalian context. Using such a method, Lai et al. (2017) have identified more than 15,000 orthologous genes across zebrafish and medaka in a comparative transcriptomic analysis.
On a conceptual aspect, it is still unclear how the trait of regeneration evolved along with all the other physiological and anatomical differences among species under the pressure of natural selection. Following the same logic, the similarities and differences between zebrafish and medaka might not necessarily associate with their regenerative capacity, which confounds the causal relationship between candidate factors and tissue regeneration. In addition, the concept of zebrafish and medaka comparison is based on the existence of conserved mechanisms underly tissue regeneration between these organisms, which might not be accurate, and the same blindside also exists for other inter-species comparisons. In light of this, potential factors identified from the comparative analyses must be further examined in both gain-of-function and loss-of-function experiments to determine their exact role in tissue regeneration. Comparative studies in the heart and retina provide nice examples of such practice (Lai et al., 2017; Lust and Wittbrodt, 2018). To gain translational opportunities, candidate factors (biological processes/signaling pathways) should be further tested to regulate regenerative capacity in mammals. Overall, the comparative approach in different models may gain basic knowledge in tissue regeneration and hint at new therapeutic strategies.
Since the pioneer studies in zebrafish tissue regeneration, broad interests have been invested in the regenerative program and other mechanistic insights underlying various tissue regeneration, hoping that the knowledge will hint or translate into therapeutics in regenerative medicine. Although the medaka has only begun to enter the stage of regenerative biology, their similar characteristics to zebrafish and abundant resources as a model animal draw more and more attention for inter-species and inter-organ comparisons in tissue regeneration and await further exploration.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
KC, SL, and S-LL contributed to the conception, writing and revision of the manuscript. KC drafted the manuscript and prepared the tables and figures. S-LL reviewed, edited, and oversaw the manuscript preparation. All the authors read and approved the submitted version.
Funding
The research in the S-LL group was supported by the Ministry of Science and Technology (MOST 108-2320-B-001-032-MY2), the IBMS/Academia Sinica (IBMS-CRC108-P03), and the Grand Challenge Project, Academia Sinica (AS-GC-110-L06) in Taiwan. KC is the recipient of the Academia Sinica-Taiwan International Graduate Program (AS-TIGP) Research Progress Fellowship.
Acknowledgments
We thank Tsung-Hung Hung from the Medical Art Room, IBMS for the artwork, and I-Ting Lin, Khai-Lone Lim, Wei-Han Lang, and Ke-Hsuan Wei for their critical comments and suggestions during the manuscript preparation. We apologize to our colleagues whose articles we could not cite due to space limitations. We also thank the reviewers’ time and efforts during the manuscript revision.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
2.^ https://zebrafish.org/home/guide.php
4.^ https://shigen.nig.ac.jp/medaka/
5.^ http://asia.ensembl.org/Danio_rerio/Info/Index
6.^ https://asia.ensembl.org/Oryzias_latipes/Info/Annotation
References
1
Adolf B. Chapouton P. Lam C. S. Topp S. Tannhauser B. Strahle U. et al (2006). Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon.Dev. Biol.295278–293. 10.1016/j.ydbio.2006.03.023
2
Aghaallaei N. Bajoghli B. Schwarz H. Schorpp M. Boehm T. (2010). Characterization of mononuclear phagocytic cells in medaka fish transgenic for a cxcr3a:gfp reporter.Proc. Natl. Acad. Sci. U.S.A.10718079–18084. 10.1073/pnas.1000467107
3
Agrawal A. A. Conner J. K. Rasmann S. (2010). “Tradeoffs and negative correlations in evolutionary ecology,” in Evolution Since Darwin: The First,edsBellM. A.EanesW. F.FutuymaD. J.LevintonJ. S. (Sunderland, MA: Sinauer Associates), 243–268.
4
Alibardi L. Toni M. (2005). Wound keratins in the regenerating epidermis of lizard suggest that the wound reaction is similar in the tail and limb.J. Exp. Zool. A. Comp. Exp. Biol.303845–860. 10.1002/jez.a.213
5
Alonso-Barba J. I. Rahman R. U. Wittbrodt J. Mateo J. L. (2016). MEPD: medaka expression pattern database, genes and more.Nucleic Acids Res.44D819–D821. 10.1093/nar/gkv1029
6
Alunni A. Hermel J. M. Heuze A. Bourrat F. Jamen F. Joly J. S. (2010). Evidence for neural stem cells in the medaka optic tectum proliferation zones.Dev. Neurobiol.70693–713. 10.1002/dneu.20799
7
Ansai S. Inohaya K. Yoshiura Y. Schartl M. Uemura N. Takahashi R. et al (2014). Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka.Dev. Growth Differ.5698–107. 10.1111/dgd.12104
8
Ansai S. Kinoshita M. (2014). Targeted mutagenesis using CRISPR/Cas system in medaka.Biol. Open3362–371. 10.1242/bio.20148177
9
Ansai S. Ochiai H. Kanie Y. Kamei Y. Gou Y. Kitano T. et al (2012). Targeted disruption of exogenous EGFP gene in medaka using zinc-finger nucleases.Dev. Growth Differ.54546–556. 10.1111/j.1440-169X.2012.01357.x
10
Ansai S. Sakuma T. Yamamoto T. Ariga H. Uemura N. Takahashi R. et al (2013). Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases.Genetics193739–749. 10.1534/genetics.112.147645
11
Asakawa K. Kawakami K. (2008). Targeted gene expression by the Gal4-UAS system in zebrafish.Dev. Growth Differ.50391–399. 10.1111/j.1440-169X.2008.01044.x
12
Au - Montandon M. Au - Currie P. D. Au - Ruparelia A. A. (2021). Examining muscle regeneration in Zebrafish Models of Muscle Disease.J. Vis. Exp.167:e62071. 10.3791/62071
13
Aurora A. B. Porrello E. R. Tan W. Mahmoud A. I. Hill J. A. Bassel-Duby R. et al (2014). Macrophages are required for neonatal heart regeneration.J. Clin. Invest.1241382–1392. 10.1172/JCI72181
14
Bajoghli B. Aghaallaei N. Hess I. Rode I. Netuschil N. Tay B. H. et al (2009). Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates.Cell138186–197. 10.1016/j.cell.2009.04.017
15
Bajoghli B. Dick A. M. Claasen A. Doll L. Aghaallaei N. (2019). Zebrafish and Medaka: two Teleost Models of T-Cell and Thymic Development.Int. J. Mol. Sci.20:4179. 10.3390/ijms20174179
16
Bajoghli B. Kuri P. Inoue D. Aghaallaei N. Hanelt M. Thumberger T. et al (2015). Noninvasive in toto imaging of the thymus reveals heterogeneous migratory behavior of developing T cells.J. Immunol.1952177–2186. 10.4049/jimmunol.1500361
17
Becker C. Lust K. Wittbrodt J. (2021). Igf signaling couples retina growth with body growth by modulating progenitor cell division.Development148:dev199133. 10.1242/dev.199133
18
Becker T. Becker C. G. (2001). Regenerating descending axons preferentially reroute to the gray matter in the presence of a general macrophage/microglial reaction caudal to a spinal transection in adult zebrafish.J. Comp. Neurol.433131–147. 10.1002/cne.1131
19
Becker T. Wullimann M. F. Becker C. G. Bernhardt R. R. Schachner M. (1997). Axonal regrowth after spinal cord transection in adult zebrafish.J. Comp. Neurol.377577–595. 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8gt;3.0.co;2-#
20
Bedell V. M. Wang Y. Campbell J. M. Poshusta T. L. Starker C. G. Krug R. G. II et al (2012). In vivo genome editing using a high-efficiency TALEN system.Nature491114–118.
21
Beffagna G. (2019). Zebrafish as a smart model to understand regeneration after heart injury: how fish could help humans.Front. Cardiovasc. Med.6:107. 10.3389/fcvm.2019.00107
22
Bely A. E. (2010). Evolutionary loss of animal regeneration: pattern and process.Integr. Comp. Biol.50515–527. 10.1093/icb/icq118
23
Bely A. E. Nyberg K. G. (2010). Evolution of animal regeneration: re-emergence of a field.Trends Ecol. Evol.25161–170. 10.1016/j.tree.2009.08.005
24
Benjamin D. C. Hynes R. O. (2017). Intravital imaging of metastasis in adult Zebrafish.BMC Cancer17:660. 10.1186/s12885-017-3647-0
25
Bensimon-Brito A. Ramkumar S. Boezio G. L. Guenther S. Kuenne C. Helker C. S. et al (2020). TGF-β signaling promotes tissue formation during cardiac valve regeneration in adult zebrafish.Dev. Cell529–20.e7. 10.1016/j.devcel.2019.10.027
26
Bergmann O. Zdunek S. Felker A. Salehpour M. Alkass K. Bernard S. et al (2015). Dynamics of cell generation and turnover in the human heart.Cell1611566–1575. 10.1016/j.cell.2015.05.026
27
Bernardo J. Agosta S. J. (2005). Evolutionary implications of hierarchical impacts of nonlethal injury on reproduction, including maternal effects.Biol. J. Linn. Soc.86309–331.
28
Bernstein J. J. (1964). Relation of spinal cord regeneration to age in adult goldfish.Exp. Neurol.9161–174. 10.1016/0014-4886(64)90014-7
29
Bernut A. Herrmann J. L. Kissa K. Dubremetz J. F. Gaillard J. L. Lutfalla G. et al (2014). Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation.Proc. Natl. Acad. Sci. U.S.A.111E943–E952. 10.1073/pnas.1321390111
30
Bertrand J. Y. Chi N. C. Santoso B. Teng S. Stainier D. Y. Traver D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development.Nature464108–111. 10.1038/nature08738
31
Bertrand J. Y. Kim A. D. Teng S. Traver D. (2008). CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis.Development1351853–1862. 10.1242/dev.015297
32
Bevan L. Lim Z. W. Venkatesh B. Riley P. R. Martin P. Richardson R. J. (2020). Specific macrophage populations promote both cardiac scar deposition and subsequent resolution in adult zebrafish.Cardiovasc. Res.1161357–1371. 10.1093/cvr/cvz221
33
Bise T. Jazwinska A. (2019). Intrathoracic Injection for the Study of Adult Zebrafish Heart.J. Vis. Exp.147:e59724. 10.3791/59724
34
Bollig F. Perner B. Besenbeck B. Kothe S. Ebert C. Taudien S. et al (2009). A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros.Development1362883–2892. 10.1242/dev.031773
35
Briona L. K. Dorsky R. I. (2014). Spinal cord transection in the larval zebrafish.J. Vis. Exp.87:51479. 10.3791/51479
36
Brockes J. P. Kumar A. Velloso C. P. (2001). Regeneration as an evolutionary variable.J. Anat.199(Pt 1–2), 3–11. 10.1046/j.1469-7580.2001.19910003.x
37
Broussonet M. (1786). Observations sur la régénérations de quelques parties du corps des poissons.Hist. Acad. R. Des. Sci.105625–641.
38
Broussonet M. (1789). Memoir on the Regeneration of Certain Parts of the Bodies of Fishes. (London: Printed for the Proprietors and Sold by C. Forster), 111–113.
39
Burg L. Palmer N. Kikhi K. Miroshnik E. S. Rueckert H. Gaddy E. et al (2018). Conditional mutagenesis by oligonucleotide-mediated integration of loxP sites in zebrafish.PLoS Genet.14:e1007754. 10.1371/journal.pgen.1007754
40
Bussmann J. Schulte-Merker S. (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish.Development1384327–4332. 10.1242/dev.068080
41
Caldwell L. J. Davies N. O. Cavone L. Mysiak K. S. Semenova S. A. Panula P. et al (2019). Regeneration of Dopaminergic Neurons in Adult Zebrafish depends on immune system activation and differs for distinct populations.J. Neurosci.394694–4713. 10.1523/JNEUROSCI.2706-18.2019
42
Cano-Martinez A. Vargas-Gonzalez A. Guarner-Lans V. Prado-Zayago E. Leon-Oleda M. Nieto-Lima B. (2010). Functional and structural regeneration in the axolotl heart (Ambystoma mexicanum) after partial ventricular amputation.Arch. Cardiol. Mex.8079–86.
43
Cao J. Poss K. D. (2018). The epicardium as a hub for heart regeneration.Nat. Rev. Cardiol.15631–647. 10.1038/s41569-018-0046-4
44
Carl M. Loosli F. Wittbrodt J. (2002). Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye.Development1294057–4063.
45
Centanin L. Ander J. J. Hoeckendorf B. Lust K. Kellner T. Kraemer I. et al (2014). Exclusive multipotency and preferential asymmetric divisions in post-embryonic neural stem cells of the fish retina.Development1413472–3482. 10.1242/dev.109892
46
Centanin L. Hoeckendorf B. Wittbrodt J. (2011). Fate restriction and multipotency in retinal stem cells.Cell Stem Cell9553–562. 10.1016/j.stem.2011.11.004
47
Chablais F. Jaźwińska A. (2012a). Induction of myocardial infarction in adult zebrafish using cryoinjury.J. Vis. Exp.62:3666. 10.3791/3666
48
Chablais F. Jaźwińska A. (2012b). The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling.Development1391921–1930. 10.1242/dev.078543
49
Chablais F. Veit J. Rainer G. Jaźwińska A. (2011). The zebrafish heart regenerates after cryoinjury-induced myocardial infarction.BMC Dev. Biol.11:21. 10.1186/1471-213X-11-21
50
Chassot B. Pury D. Jazwinska A. (2016). Zebrafish fin regeneration after cryoinjury-induced tissue damage.Biol. Open5819–828. 10.1242/bio.016865
51
Chatani M. Takano Y. Kudo A. (2011). Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish.Dev. Biol.36096–109. 10.1016/j.ydbio.2011.09.013
52
Chavez M. N. Morales R. A. Lopez-Crisosto C. Roa J. C. Allende M. L. Lavandero S. (2020). Autophagy activation in Zebrafish heart regeneration.Sci. Rep.10:2191. 10.1038/s41598-020-59106-z
53
Chen B. Dodge M. E. Tang W. Lu J. Ma Z. Fan C. W. et al (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer.Nat. Chem. Biol.5100–107. 10.1038/nchembio.137
54
Chen C. F. Chu C. Y. Chen T. H. Lee S. J. Shen C. N. Hsiao C. D. (2011). Establishment of a transgenic zebrafish line for superficial skin ablation and functional validation of apoptosis modulators in vivo.PLoS One6:e20654. 10.1371/journal.pone.0020654
55
Chen W. C. Wang Z. Missinato M. A. Park D. W. Long D. W. Liu H. J. et al (2016). Decellularized zebrafish cardiac extracellular matrix induces mammalian heart regeneration.Sci. Adv.2:e1600844. 10.1126/sciadv.1600844
56
Childs S. Chen J. N. Garrity D. M. Fishman M. C. (2002). Patterning of angiogenesis in the zebrafish embryo.Development129973–982.
57
Choi T.-Y. Khaliq M. Ko S. So J. Shin D. (2015). Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration.J. Vis. Exp.99:e52785. 10.3791/52785
58
Chou C.-Y. Horng L.-S. Tsai H.-J. (2001). Uniform GFP-expression in transgenic medaka (Oryzias latipes) at the F0 generation.Transgenic Res.10303–315. 10.1023/a:1016671513425
59
Collery R. F. Volberding P. J. Bostrom J. R. Link B. A. Besharse J. C. (2016). Loss of Zebrafish Mfrp causes nanophthalmia, hyperopia, and accumulation of subretinal macrophages.Invest. Ophthalmol. Vis. Sci.576805–6814. 10.1167/iovs.16-19593
60
Conedera F. M. Arendt P. Trepp C. Tschopp M. Enzmann V. (2017). Muller Glia cell activation in a laser-induced retinal degeneration and regeneration model in Zebrafish.J. Vis. Exp.128:e56249. 10.3791/56249
61
Cox A. G. Saunders D. C. Kelsey P. B. Jr. Conway A. A. Tesmenitsky Y. Marchini J. F. et al (2014). S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury.Cell Rep.656–69. 10.1016/j.celrep.2013.12.007
62
Crespo C. L. Vernieri C. Keller P. J. Garre M. Bender J. R. Wittbrodt J. et al (2014). The PAR complex controls the spatiotemporal dynamics of F-actin and the MTOC in directionally migrating leukocytes.J. Cell Sci.127(Pt 20), 4381–4395. 10.1242/jcs.146217
63
Crippa S. Nemir M. Ounzain S. Ibberson M. Berthonneche C. Sarre A. et al (2016). Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways.Cardiovasc. Res.11073–84. 10.1093/cvr/cvw031
64
Cruz I. A. Kappedal R. Mackenzie S. M. Hailey D. W. Hoffman T. L. Schilling T. F. et al (2015). Robust regeneration of adult zebrafish lateral line hair cells reflects continued precursor pool maintenance.Dev. Biol.402229–238. 10.1016/j.ydbio.2015.03.019
65
Cuervo R. Hernandez-Martinez R. Chimal-Monroy J. Merchant-Larios H. Covarrubias L. (2012). Full regeneration of the tribasal Polypterus fin.Proc. Natl. Acad. Sci. U.S.A.1093838–3843. 10.1073/pnas.1006619109
66
Curado S. Anderson R. M. Jungblut B. Mumm J. Schroeter E. Stainier D. Y. (2007). Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies.Dev. Dyn.2361025–1035. 10.1002/dvdy.21100
67
Dambroise E. Simion M. Bourquard T. Bouffard S. Rizzi B. Jaszczyszyn Y. et al (2017). Postembryonic fish brain proliferation zones exhibit neuroepithelial-type gene expression profile.Stem Cells351505–1518. 10.1002/stem.2588
68
Daponte V. Tylzanowski P. Forlino A. (2021). Appendage Regeneration in Vertebrates: What Makes This Possible?Cells10:242. 10.3390/cells10020242
69
Darehzereshki A. Rubin N. Gamba L. Kim J. Fraser J. Huang Y. et al (2015). Differential regenerative capacity of neonatal mouse hearts after cryoinjury.Dev. Biol.39991–99. 10.1016/j.ydbio.2014.12.018
70
Darnet S. Dragalzew A. C. Amaral D. B. Sousa J. F. Thompson A. W. Cass A. N. et al (2019). Deep evolutionary origin of limb and fin regeneration.Proc. Natl. Acad. Sci. U.S.A.11615106–15115. 10.1073/pnas.1900475116
71
Dasyani M. Tan W. H. Sundaram S. Imangali N. Centanin L. Wittbrodt J. et al (2019). Lineage tracing of col10a1 cells identifies distinct progenitor populations for osteoblasts and joint cells in the regenerating fin of medaka (Oryzias latipes).Dev. Biol.45585–99. 10.1016/j.ydbio.2019.07.012
72
Davidson A. E. Balciunas D. Mohn D. Shaffer J. Hermanson S. Sivasubbu S. et al (2003). Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon.Dev. Biol.263191–202. 10.1016/j.ydbio.2003.07.013
73
de Koning C. Beekhuijzen M. Tobor-Kaplon M. de Vries-Buitenweg S. Schoutsen D. Leeijen N. et al (2015). Visualizing Compound Distribution during Zebrafish Embryo Development: the Effects of Lipophilicity and DMSO.Birth Defects Res. B. Dev. Reprod. Toxicol.104253–272. 10.1002/bdrb.21166
74
de Preux Charles A. S. Bise T. Baier F. Marro J. Jazwinska A. (2016). Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart.Open Biol.6:160102. 10.1098/rsob.160102
75
Deguchi T. Fujimori K. E. Kawasaki T. Maruyama K. Yuba S. (2012). In vivo visualization of the lymphatic vessels in pFLT4-EGFP transgenic medaka.Genesis50625–634. 10.1002/dvg.22018
76
DeLaurier A. Eames B. F. Blanco-Sanchez B. Peng G. He X. Swartz M. E. et al (2010). Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration.Genesis48505–511. 10.1002/dvg.20639
77
DiCicco R. M. Bell B. A. Kaul C. Hollyfield J. G. Anand-Apte B. Perkins B. D. et al (2014). Retinal regeneration following OCT-guided laser injury in zebrafish.Invest. Ophthalmol. Vis. Sci.556281–6288. 10.1167/iovs.14-14724
78
Diep C. Q. Ma D. Deo R. C. Holm T. M. Naylor R. W. Arora N. et al (2011). Identification of adult nephron progenitors capable of kidney regeneration in zebrafish.Nature47095–100. 10.1038/nature09669
79
Diotel N. Lubke L. Strahle U. Rastegar S. (2020). Common and Distinct Features of Adult Neurogenesis and Regeneration in the Telencephalon of Zebrafish and Mammals.Front. Neurosci.14:568930. 10.3389/fnins.2020.568930
80
Doyon Y. McCammon J. M. Miller J. C. Faraji F. Ngo C. Katibah G. E. et al (2008). Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases.Nat. Biotechnol.26702–708. 10.1038/nbt1409
81
Driever W. Solnica-Krezel L. Schier A. F. Neuhauss S. C. Malicki J. Stemple D. L. et al (1996). A genetic screen for mutations affecting embryogenesis in zebrafish.Development12337–46.
82
Du S. J. Dienhart M. (2001). Zebrafish tiggy-winkle hedgehog promoter directs notochord and floor plate green fluorescence protein expression in transgenic zebrafish embryos.Dev. Dyn.222655–666. 10.1002/dvdy.1219
83
Dwaraka V. B. Voss S. R. (2021). Towards comparative analyses of salamander limb regeneration.J. Exp. Zool. B Mol. Dev. Evol.336129–144. 10.1002/jez.b.22902
84
Dyck P. K. V. Hockaden N. Nelson E. C. Koch A. R. Hester K. L. Pillai N. et al (2020). Cauterization as a simple method for regeneration studies in the zebrafish heart.J. Cardiovasc. Dev. Dis.7:41. 10.3390/jcdd7040041
85
Elbaz I. Yelin-Bekerman L. Nicenboim J. Vatine G. Appelbaum L. (2012). Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish.J. Neurosci.3212961–12972. 10.1523/JNEUROSCI.1284-12.2012
86
Elchaninov A. Sukhikh G. Fatkhudinov T. (2021). Evolution of regeneration in animals: a tangled story.Front. Ecol. Evol.9:621686. 10.3389/fevo.2021.621686
87
Elhelaly W. M. Lam N. T. Hamza M. Xia S. Sadek H. A. (2016). Redox regulation of heart regeneration: an evolutionary Tradeoff.Front. Cell Dev. Biol.4:137. 10.3389/fcell.2016.00137
88
Ellett F. Pase L. Hayman J. Andrianopoulos A. Lieschke G. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish.Blood117E49–E56. 10.1182/blood-2010-10-314120
89
Emelyanov A. Gao Y. Naqvi N. I. Parinov S. (2006). Trans-kingdom transposition of the maize dissociation element.Genetics1741095–1104. 10.1534/genetics.106.061184
90
Eschenhagen T. Bolli R. Braun T. Field L. J. Fleischmann B. K. Frisén J. et al (2017). Cardiomyocyte regeneration: a consensus statement.Circulation136680–686. 10.1161/circulationaha.117.029343
91
Fadool J. M. (2003). Development of a rod photoreceptor mosaic revealed in transgenic zebrafish.Dev. Biol.258277–290. 10.1016/s0012-1606(03)00125-8
92
Feil R. Brocard J. Mascrez B. LeMeur M. Metzger D. Chambon P. (1996). Ligand-activated site-specific recombination in mice.Proc. Natl. Acad. Sci. U.S.A.9310887–10890. 10.1073/pnas.93.20.10887
93
Felker A. Mosimann C. (2016). “Chapter 11 - Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments,” in Methods in Cell Biology, edsWilliam DetrichH.WesterfieldM.ZonL. I. (Amsterdam: Elsevier), 219–244. 10.1016/bs.mcb.2016.01.009
94
Ferguson H. Kongtorp R. Taksdal T. Graham D. Falk K. (2005). An outbreak of disease resembling heart and skeletal muscle inflammation in Scottish farmed salmon, Salmo salar L., with observations on myocardial regeneration.J. Fish Dis.28119–123. 10.1111/j.1365-2761.2004.00602.x
95
Fimbel S. M. Montgomery J. E. Burket C. T. Hyde D. R. (2007). Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish.J. Neurosci.271712–1724. 10.1523/JNEUROSCI.5317-06.2007
96
Fox S. F. McCoy J. K. (2000). The effects of tail loss on survival, growth, reproduction, and sex ratio of offspring in the lizard Uta stansburiana in the field.Oecologia122327–334. 10.1007/s004420050038
97
Froschauer A. Sprott D. Gerwien F. Henker Y. Rudolph F. Pfennig F. et al (2012). Effective generation of transgenic reporter and gene trap lines of the medaka (Oryzias latipes) using the Ac/Ds transposon system.Transgenic Res.21149–162. 10.1007/s11248-011-9514-x
98
Fuhrmann J. F. Buono L. Adelmann L. Martinez-Morales J. R. Centanin L. (2020). Genetic developmental timing revealed by inter-species transplantations in fish.Development147:dev192500. 10.1242/dev.192500
99
Furutani-Seiki M. Sasado T. Morinaga C. Suwa H. Niwa K. Yoda H. et al (2004). A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes.Mech. Dev.121647–658. 10.1016/j.mod.2004.04.016
100
Furutani-Seiki M. Wittbrodt J. (2004). Medaka and zebrafish, an evolutionary twin study.Mech. Dev.121629–637. 10.1016/j.mod.2004.05.010
101
Galis F. Wagner G. P. Jockusch E. L. (2003). Why is limb regeneration possible in amphibians but not in reptiles, birds, and mammals?Evol. Dev.5208–220. 10.1046/j.1525-142x.2003.03028.x
102
Garcia J. Bagwell J. Njaine B. Norman J. Levic D. S. Wopat S. et al (2017). Sheath cell invasion and trans-differentiation repair mechanical damage caused by Loss of Caveolae in the Zebrafish Notochord.Curr. Biol.271982–1989.e3. 10.1016/j.cub.2017.05.035
103
Gemberling M. Bailey T. J. Hyde D. R. Poss K. D. (2013). The zebrafish as a model for complex tissue regeneration.Trends Genet.29611–620. 10.1016/j.tig.2013.07.003
104
Gemberling M. Karra R. Dickson A. L. Poss K. D. (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish.eLife4:e05871. 10.7554/eLife.05871
105
Géraudie J. Brulfert A. Monnot M. J. Ferretti P. (1994). Teratogenic and morphogenetic effects of retinoic acid on the regenerating pectoral fin in zebrafish.J. Exp. Zool.26912–22.
106
Gerlach G. F. Schrader L. N. Wingert R. A. (2011). Dissection of the adult zebrafish kidney.J. Vis. Exp.54:e2839. 10.3791/2839
107
Ghosh S. Hui S. P. (2018). Axonal regeneration in zebrafish spinal cord.Regeneration543–60. 10.1002/reg2.99
108
Godoy R. Noble S. Yoon K. Anisman H. Ekker M. (2015). Chemogenetic ablation of dopaminergic neurons leads to transient locomotor impairments in zebrafish larvae.J. Neurochem.135249–260. 10.1111/jnc.13214
109
Godwin J. W. Pinto A. R. Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration.Proc. Natl. Acad. Sci. U.S.A.1109415–9420. 10.1073/pnas.1300290110
110
Godwin J. W. Rosenthal N. (2014). Scar-free wound healing and regeneration in amphibians: immunological influences on regenerative success.Differentiation8766–75. 10.1016/j.diff.2014.02.002
111
Goldman D. (2014). Muller glial cell reprogramming and retina regeneration.Nat. Rev. Neurosci.15431–442. 10.1038/nrn3723
112
Goldman D. Hankin M. Li Z. Dai X. Ding J. (2001). Transgenic zebrafish for studying nervous system development and regeneration.Transgenic Res.1021–33. 10.1023/a:1008998832552
113
Goldstein A. M. Fishman M. C. (1998). Notochord regulates cardiac lineage in zebrafish embryos.Dev. Biol.201247–252. 10.1006/dbio.1998.8976
114
Gonzalez-Rosa J. M. Martín V. Peralta M. Torres M. Mercader N. (2011). Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish.Development1381663–1674. 10.1242/dev.060897
115
González-Rosa J. M. Peralta M. Mercader N. (2012). Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration.Dev. Biol.370173–186. 10.1016/j.ydbio.2012.07.007
116
Gordon K. Schulte D. Brice G. Simpson M. A. Roukens M. G. Van Impel A. et al (2013). Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant milroy-like primary lymphedema.Circ. Res.112956–960. 10.1161/CIRCRESAHA.113.300350
117
Gorsuch R. A. Hyde D. R. (2014). Regulation of muller glial dependent neuronal regeneration in the damaged adult zebrafish retina.Exp. Eye Res.123131–140. 10.1016/j.exer.2013.07.012
118
Gorsuch R. A. Lahne M. Yarka C. E. Petravick M. E. Li J. Hyde D. R. (2017). Sox2 regulates muller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a.Exp. Eye Res.161174–192. 10.1016/j.exer.2017.05.012
119
Grabher C. Cliffe A. Miura K. Hayflick J. Pepperkok R. Rorth P. et al (2007). Birth and life of tissue macrophages and their migration in embryogenesis and inflammation in medaka.J. Leukoc. Biol.81263–271. 10.1189/jlb.0806526
120
Grabher C. Henrich T. Sasado T. Arenz A. Wittbrodt J. Furutani-Seiki M. (2003). Transposon-mediated enhancer trapping in medaka.Gene32257–66. 10.1016/j.gene.2003.09.009
121
Grabher C. Joly J.-S. Wittbrodt J. (2004). Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI.Methods Cell Biol.77381–401. 10.1016/s0091-679x(04)77021-1
122
Grabher C. Wittbrodt J. (2004). Efficient activation of gene expression using a heat-shock inducible Gal4/Vp16-UAS system in medaka.BMC Biotechnol.4:26. 10.1186/1472-6750-4-26
123
Grabher C. Wittbrodt J. (2007). Meganuclease and transposon mediated transgenesis in medaka.Genome Biol.8 (Suppl 1):S10.
124
Grabher C. Wittbrodt J. (2008). Recent advances in meganuclease-and transposon-mediated transgenesis of medaka and zebrafish.Methods Mol. Biol.461521–539. 10.1007/978-1-60327-483-8_36
125
Grandel H. Kaslin J. Ganz J. Wenzel I. Brand M. (2006). Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate.Dev. Biol.295263–277. 10.1016/j.ydbio.2006.03.040
126
Grandel H. Schulte-Merker S. (1998). The development of the paired fins in the zebrafish (Danio rerio).Mech. Dev.7999–120. 10.1016/s0925-4773(98)00176-2
127
Grivas J. Haag M. Johnson A. Manalo T. Roell J. Das T. L. et al (2014). Cardiac repair and regenerative potential in the goldfish (Carassius auratus) heart.Comp. Biochem. Physiol. C Toxicol. Pharmacol.16314–23. 10.1016/j.cbpc.2014.02.002
128
Gulías P. Guerra-Varela J. Gonzalez-Aparicio M. Ricobaraza A. Vales A. Gonzalez-Aseguinolaza G. et al (2019). Danio rerio as model organism for adenoviral vector evaluation.Genes10:1053. 10.3390/genes10121053
129
Gupta T. Mullins M. C. (2010). Dissection of organs from the adult zebrafish.J. Vis. Exp.37:1717. 10.3791/1717
130
Hamada K. Tamaki K. Sasado T. Watai Y. Kani S. Wakamatsu Y. et al (1998). Usefulness of the medaka beta-actin promoter investigated using a mutant GFP reporter gene in transgenic medaka (Oryzias latipes).Mol. Mar. Biol. Biotechnol.7173–180.
131
Hanovice N. J. Leach L. L. Slater K. Gabriel A. E. Romanovicz D. Shao E. et al (2019). Regeneration of the zebrafish retinal pigment epithelium after widespread genetic ablation.PLoS Genet.15:e1007939. 10.1371/journal.pgen.1007939
132
Hans S. Freudenreich D. Geffarth M. Kaslin J. Machate A. Brand M. (2011). Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish.Dev. Dyn.240108–115. 10.1002/dvdy.22497
133
Hans S. Kaslin J. Freudenreich D. Brand M. (2009). Temporally-controlled site-specific recombination in zebrafish.PLoS One4:e4640. 10.1371/journal.pone.0004640
134
Harris J. A. Cheng A. G. Cunningham L. L. MacDonald G. Raible D. W. Rubel E. W. (2003). Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio).J. Assoc. Res. Otolaryngol.4219–234. 10.1007/s10162-002-3022-x
135
Haubner B. J. Adamowicz-Brice M. Khadayate S. Tiefenthaler V. Metzler B. Aitman T. et al (2012). Complete cardiac regeneration in a mouse model of myocardial infarction.Aging4966–977. 10.18632/aging.100526
136
Haubner B. J. Schneider J. Schweigmann U. Schuetz T. Dichtl W. Velik-Salchner C. et al (2016). Functional recovery of a human neonatal heart after severe myocardial infarction.Circ. Res.118216–221. 10.1161/CIRCRESAHA.115.307017
137
Hawkins M. B. Henke K. Harris M. P. (2021). Latent developmental potential to form limb-like skeletal structures in zebrafish.Cell184899–911.e13. 10.1016/j.cell.2021.01.003
138
Her G. M. Yeh Y. H. Wu J. L. (2003). 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish.Dev. Dyn.227347–356. 10.1002/dvdy.10324
139
Higashijima S.-I. Okamoto H. Ueno N. Hotta Y. Eguchi G. (1997). High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin.Dev. Biol.192289–299. 10.1006/dbio.1997.8779
140
Hilgers L. Schwarzer J. (2019). The natural history of model organisms: the untapped potential of medaka and its wild relatives.eLife8:e46994. 10.7554/eLife.46994
141
Hirose K. Payumo A. Y. Cutie S. Hoang A. Zhang H. Guyot R. et al (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition.Science364184–188. 10.1126/science.aar2038
142
Hoang T. Wang J. Boyd P. Wang F. Santiago C. Jiang L. et al (2020). Gene regulatory networks controlling vertebrate retinal regeneration.Science370:eabb8598. 10.1126/science.abb8598
143
Hoeppner L. H. Sinha S. Wang Y. Bhattacharya R. Dutta S. Gong X. et al (2015). RhoC maintains vascular homeostasis by regulating VEGF-induced signaling in endothelial cells.J. Cell Sci.1283556–3568.
144
Hogan B. M. Bos F. L. Bussmann J. Witte M. Chi N. C. Duckers H. J. et al (2009). Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting.Nat. Genet.41396–398. 10.1038/ng.321
145
Hong N. Chen S. Ge R. Song J. Yi M. Hong Y. (2012). Interordinal chimera formation between medaka and zebrafish for analyzing stem cell differentiation.Stem Cells Dev.212333–2341. 10.1089/scd.2011.0630
146
Honkoop H. de Bakker D. E. Aharonov A. Kruse F. Shakked A. Nguyen P. D. et al (2019). Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart.eLife8:e50163. 10.7554/eLife.50163
147
Hosoya O. Chung M. Ansai S. Takeuchi H. Miyaji M. (2021). A modified Tet-ON system minimizing leaky expression for cell-type specific gene induction in medaka fish.Dev. Growth Differ.63397–405. 10.1111/dgd.12743
148
Howe K. Clark M. D. Torroja C. F. Torrance J. Berthelot C. Muffato M. et al (2013). The zebrafish reference genome sequence and its relationship to the human genome.Nature496498–503. 10.1038/nature12111
149
Hruscha A. Krawitz P. Rechenberg A. Heinrich V. Hecht J. Haass C. et al (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish.Development1404982–4987. 10.1242/dev.099085
150
Huang C. J. Tu C. T. Hsiao C. D. Hsieh F. J. Tsai H. J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish.Dev. Dyn.22830–40. 10.1002/dvdy.10356
151
Huang H. Vogel S. S. Liu N. Melton D. A. Lin S. (2001). Analysis of pancreatic development in living transgenic zebrafish embryos.Mol. Cell Endocrinol.177117–124. 10.1016/s0303-7207(01)00408-7
152
Huang P. Xiao A. Zhou M. Zhu Z. Lin S. Zhang B. (2011). Heritable gene targeting in zebrafish using customized TALENs.Nat. Biotechnol.29699–700. 10.1038/nbt.1939
153
Hui S. P. Sheng D. Z. Sugimoto K. Gonzalez-Rajal A. Nakagawa S. Hesselson D. et al (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs.Dev. Cell43659–672.e5. 10.1016/j.devcel.2017.11.010
154
Hwang W. Y. Fu Y. Reyon D. Maeder M. L. Tsai S. Q. Sander J. D. et al (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system.Nat. Biotechnol.31227–229. 10.1038/nbt.2501
155
Ichikawa K. Tomioka S. Suzuki Y. Nakamura R. Doi K. Yoshimura J. et al (2017). Centromere evolution and CpG methylation during vertebrate speciation.Nat. Commun.8:1833. 10.1038/s41467-017-01982-7
156
Iismaa S. E. Kaidonis X. Nicks A. M. Bogush N. Kikuchi K. Naqvi N. et al (2018). Comparative regenerative mechanisms across different mammalian tissues.NPJ Regen. Med.3:6. 10.1038/s41536-018-0044-5
157
Inoue D. Wittbrodt J. (2011). One for all—a highly efficient and versatile method for fluorescent immunostaining in fish embryos.PLoS One6:e19713. 10.1371/journal.pone.0019713
158
Ishikawa Y. (2000). Medakafish as a model system for vertebrate developmental genetics.Bioessays22487–495. 10.1002/(SICI)1521-1878(200005)22:5lt;487::AID-BIES11gt;3.0.CO;2-8
159
Ito K. Morioka M. Kimura S. Tasaki M. Inohaya K. Kudo A. (2014). Differential reparative phenotypes between zebrafish and medaka after cardiac injury.Dev. Dyn.2431106–1115. 10.1002/dvdy.24154
160
Ito Y. Tanaka H. Okamoto H. Ohshima T. (2010). Characterization of neural stem cells and their progeny in the adult zebrafish optic tectum.Dev. Biol.34226–38. 10.1016/j.ydbio.2010.03.008
161
Jaźwińska A. Badakov R. Keating M. T. (2007). Activin-βA signaling is required for zebrafish fin regeneration.Curr. Biol.171390–1395. 10.1016/j.cub.2007.07.019
162
Jaźwińska A. Blanchoud S. (2020). Towards deciphering variations of heart regeneration in fish.Curr. Opin. Physiol.1421–26. 10.1016/j.cophys.2019.11.007
163
Jessen J. R. Jessen T. N. Vogel S. S. Lin S. (2001). Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons.Genesis29156–162. 10.1002/gene.1019
164
Jessen J. R. Willett C. E. Lin S. (1999). Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish.Nat. Genet.2315–16. 10.1038/12609
165
Jh B. (1947). Regeneration and transplantation of fin rays in the goldfish.Anat. Rec.99648–648.
166
Jimenez E. Slevin C. C. Colón-Cruz L. Burgess S. M. (2021). Vestibular and auditory hair cell regeneration following targeted ablation of hair cells with diphtheria toxin in zebrafish.bioRxiv [Preprint]. 10.1101/2021.06.08.447587
167
Jopling C. Boue S. Izpisua Belmonte J. C. (2011). Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration.Nat. Rev. Mol. Cell Biol.1279–89. 10.1038/nrm3043
168
Ju B. Chong S. W. He J. Wang X. Xu Y. Wan H. et al (2003). Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter.Dev. Dyn.22714–26. 10.1002/dvdy.10273
169
Julier Z. Park A. J. Briquez P. S. Martino M. M. (2017). Promoting tissue regeneration by modulating the immune system.Acta Biomater.5313–28. 10.1016/j.actbio.2017.01.056
170
Jung H. M. Castranova D. Swift M. R. Pham V. N. Venero Galanternik M. Isogai S. et al (2017). Development of the larval lymphatic system in zebrafish.Development1442070–2081. 10.1242/dev.145755
171
Jung H. M. Isogai S. Kamei M. Castranova D. Gore A. V. Weinstein B. M. (2016). Imaging blood vessels and lymphatic vessels in the zebrafish.Methods Cell Biol.13369–103. 10.1016/bs.mcb.2016.03.023
172
Jung S. Kleinheinz J. (2013). “Angiogenesis—the key to regeneration,” in Regenerative Medicine and Tissue Engineering, ed.AndradesJ. A. (London: IntechOpen). 10.5772/55542
173
Jungke P. Hammer J. Hans S. Brand M. (2015). Isolation of novel CreERT2-driver lines in zebrafish using an unbiased gene trap approach.PLoS One10:e0129072. 10.1371/journal.pone.0129072
174
Jungke P. Hans S. Brand M. (2013). The zebrafish CreZoo: an easy-to-handle database for novel CreERT2-driver lines.Zebrafish10259–263. 10.1089/zeb.2012.0834
175
Kanagaraj P. Chen J. Y. Skaggs K. Qadeer Y. Conner M. Cutler N. et al (2020). Microglia Stimulate Zebrafish Brain Repair Via a Specific Inflammatory Cascade.bioRxiv [Preprint]. 10.1101/2020.10.08.330662
176
Kang J. Hu J. Karra R. Dickson A. L. Tornini V. A. Nachtrab G. et al (2016). Modulation of tissue repair by regeneration enhancer elements.Nature532201–206. 10.1038/nature17644
177
Kasahara M. Naruse K. Sasaki S. Nakatani Y. Qu W. Ahsan B. et al (2007). The medaka draft genome and insights into vertebrate genome evolution.Nature447714–719. 10.1038/nature05846
178
Katogi R. Nakatani Y. Shin-i T. Kohara Y. Inohaya K. Kudo A. (2004). Large-scale analysis of the genes involved in fin regeneration and blastema formation in the medaka, Oryzias latipes.Mech. Dev.121861–872. 10.1016/j.mod.2004.03.015
179
Kawakami K. (2007). Tol2: a versatile gene transfer vector in vertebrates.Genome Biol.8(Suppl. 1):S7. 10.1186/gb-2007-8-s1-s7
180
Kawakami K. Shima A. (1999). Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio.Gene240239–244. 10.1016/s0378-1119(99)00444-8
181
Kawakami Y. Rodriguez Esteban C. Raya M. Kawakami H. Marti M. Dubova I. et al (2006). Wnt/beta-catenin signaling regulates vertebrate limb regeneration.Genes. Dev.203232–3237. 10.1101/gad.1475106
182
Kawasaki T. Kurauchi K. Higashihata A. Deguchi T. Ishikawa Y. Yamauchi M. et al (2012). Transgenic medaka fish which mimic the endogenous expression of neuronal kinesin, KIF5A.Brain Res.148012–21. 10.1016/j.brainres.2012.08.047
183
Kettleborough R. N. Busch-Nentwich E. M. Harvey S. A. Dooley C. M. de Bruijn E. van Eeden F. et al (2013). A systematic genome-wide analysis of zebrafish protein-coding gene function.Nature496494–497. 10.1038/nature11992
184
Khyeam S. Lee S. Huang G. N. (2021). Genetic, epigenetic, and post-transcriptional basis of divergent tissue regenerative capacities among vertebrates.Adv. Genet.7:e10042. 10.1002/ggn2.10042
185
Kikuchi K. Holdway J. E. Major R. J. Blum N. Dahn R. D. Begemann G. et al (2011b). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration.Dev. Cell20397–404. 10.1016/j.devcel.2011.01.010
186
Kikuchi K. Gupta V. Wang J. Holdway J. E. Wills A. A. Fang Y. et al (2011a). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration.Development1382895–2902. 10.1242/dev.067041
187
Kikuchi K. Holdway J. E. Werdich A. A. Anderson R. M. Fang Y. Egnaczyk G. F. et al (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes.Nature464601–605. 10.1038/nature08804
188
Kimura Y. Hisano Y. Kawahara A. Higashijima S. (2014). Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering.Sci. Rep.4:6545. 10.1038/srep06545
189
Kinkel M. D. Eames S. C. Philipson L. H. Prince V. E. (2010). Intraperitoneal injection into adult zebrafish.J. Vis. Exp.42:2126. 10.3791/2126
190
Kinoshita M. Kani S. Ozato K. Wakamatsu Y. (2000). Activity of the medaka translation elongation factor 1α-A promoter examined using the GFP gene as a reporter.Dev. Growth Differ.42469–478. 10.1046/j.1440-169x.2000.00530.x
191
Kinoshita M. Murata K. Naruse K. Tanaka M. (2009). Medaka: Biology, Management, and Experimental Protocols.Honoken, NJ: John Wiley & Sons.
192
Kirchmaier S. Höckendorf B. Möller E. K. Bornhorst D. Spitz F. Wittbrodt J. (2013). Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase.Development1404287–4295. 10.1242/dev.096081
193
Kirchmaier S. Naruse K. Wittbrodt J. Loosli F. (2015). The genomic and genetic toolbox of the teleost medaka (Oryzias latipes).Genetics199905–918. 10.1534/genetics.114.173849
194
Kishimoto N. Shimizu K. Sawamoto K. (2012). Neuronal regeneration in a zebrafish model of adult brain injury.Dis. Model. Mech.5200–209. 10.1242/dmm.007336
195
Kizil C. Brand M. (2011). Cerebroventricular microinjection (CVMI) into adult zebrafish brain is an efficient misexpression method for forebrain ventricular cells.PLoS One6:e27395. 10.1371/journal.pone.0027395
196
Knopf F. Hammond C. Chekuru A. Kurth T. Hans S. Weber C. W. et al (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin.Dev. Cell20713–724. 10.1016/j.devcel.2011.04.014
197
Knopf F. Schnabel K. Haase C. Pfeifer K. Anastassiadis K. Weidinger G. (2010). Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish.Proc. Natl. Acad. Sci. U.S.A.10719933–19938. 10.1073/pnas.1007799107
198
Kobayashi D. Takeda H. (2008). Medaka genome project.Brief Funct. Genomic. Proteomic.7415–426. 10.1093/bfgp/eln044
199
Koga A. Hori H. Sakaizumi M. (2002). Gene transfer and cloning of flanking chromosomal regions using the medaka fish Tol2 transposable element.Mar. Biotechnol.46–11. 10.1007/s10126-001-0086-2
200
Kong P. Christia P. Frangogiannis N. G. (2014). The pathogenesis of cardiac fibrosis.Cell. Mol. Life Sci.71549–574.
201
Kroehne V. Freudenreich D. Hans S. Kaslin J. Brand M. (2011). Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors.Development1384831–4841. 10.1242/dev.072587
202
Kucenas S. Takada N. Park H. C. Woodruff E. Broadie K. Appel B. (2008). CNS-derived glia ensheath peripheral nerves and mediate motor root development.Nat. Neurosci.11143–151. 10.1038/nn2025
203
Kumar S. Stecher G. Suleski M. Hedges S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times.Mol. Biol. Evol.341812–1819. 10.1093/molbev/msx116
204
Kurita R. Sagara H. Aoki Y. Link B. A. Arai K.-I. Watanabe S. (2003). Suppression of lens growth by αA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish.Dev. Biol.255113–127. 10.1016/s0012-1606(02)00079-9
205
Kuroyanagi Y. Okuyama T. Suehiro Y. Imada H. Shimada A. Naruse K. et al (2010). Proliferation zones in adult medaka (Oryzias latipes) brain.Brain Res.132333–40. 10.1016/j.brainres.2010.01.045
206
Kyritsis N. Kizil C. Zocher S. Kroehne V. Kaslin J. Freudenreich D. et al (2012). Acute inflammation initiates the regenerative response in the adult zebrafish brain.Science3381353–1356. 10.1126/science.1228773
207
Labusch M. Mancini L. Morizet D. Bally-Cuif L. (2020). Conserved and Divergent Features of Adult Neurogenesis in Zebrafish.Front. Cell Dev. Biol.8:525. 10.3389/fcell.2020.00525
208
Lafontant P. J. Burns A. R. Grivas J. A. Lesch M. A. Lala T. D. Reuter S. P. et al (2012). The giant danio (D. aequipinnatus) as a model of cardiac remodeling and regeneration.Anat. Rec.295234–248. 10.1002/ar.21492
209
Lai S.-L. Marín-Juez R. Moura P. L. Kuenne C. Lai J. K. H. Tsedeke A. T. et al (2017). Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration.eLife6:e25605. 10.7554/eLife.25605
210
Langenau D. M. Ferrando A. A. Traver D. Kutok J. L. Hezel J. P. Kanki J. P. et al (2004). In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish.Proc. Natl. Acad. Sci. U.S.A.1017369–7374. 10.1073/pnas.0402248101
211
Laube F. Heister M. Scholz C. Borchardt T. Braun T. (2006). Re-programming of newt cardiomyocytes is induced by tissue regeneration.J. Cell Sci.119(Pt 22), 4719–4729. 10.1242/jcs.03252
212
Lawson N. D. Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish.Dev. Biol.248307–318. 10.1006/dbio.2002.0711
213
Le X. Langenau D. M. Keefe M. D. Kutok J. L. Neuberg D. S. Zon L. I. (2007). Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish.Proc. Natl. Acad. Sci. U.S.A.1049410–9415. 10.1073/pnas.0611302104
214
Leach L. L. Hanovice N. J. George S. M. Gabriel A. E. Gross J. M. (2021). The immune response is a critical regulator of zebrafish retinal pigment epithelium regeneration.Proc. Natl. Acad. Sci. U.S.A.118e2017198118. 10.1073/pnas.2017198118
215
Lenkowski J. R. Raymond P. A. (2014). Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish.Prog. Retin. Eye Res.4094–123. 10.1016/j.preteyeres.2013.12.007
216
Letunic I. Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation.Nucleic Acids Res.49W293–W296. 10.1093/nar/gkab301
217
Li J. Iwanami N. Hoa V. Q. Furutani-Seiki M. Takahama Y. (2007). Noninvasive intravital imaging of thymocyte dynamics in medaka.J. Immunol.1791605–1615. 10.4049/jimmunol.179.3.1605
218
Li L. Yan B. Shi Y. Q. Zhang W. Q. Wen Z. L. (2012). Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration.J. Biol. Chem.28725353–25360. 10.1074/jbc.M112.349126
219
Li Y. Liu Y. Yang H. Zhang T. Naruse K. Tu Q. (2020). Dynamic transcriptional and chromatin accessibility landscape of medaka embryogenesis.Genome Res.30924–937. 10.1101/gr.258871.119
220
Lin H.-F. Traver D. Zhu H. Dooley K. Paw B. H. Zon L. I. et al (2005). Analysis of thrombocyte development in CD41-GFP transgenic zebrafish.Blood1063803–3810. 10.1182/blood-2005-01-0179
221
Lin J.-W. Chen Y.-R. Wang Y.-H. Hung K.-C. Lin S.-M. (2017). Tail regeneration after autotomy revives survival: a case from a long-term monitored lizard population under avian predation.Proc. R. Soc. B Biol. Sci.284:20162538. 10.1098/rspb.2016.2538
222
Lindmark D. G. Müller M. (1976). Antitrichomonad action, mutagenicity, and reduction of metronidazole and other nitroimidazoles.Antimicrob. Agents Chemother.10476–482. 10.1128/AAC.10.3.476
223
Lindsey B. W. Aitken G. E. Tang J. K. Khabooshan M. Douek A. M. Vandestadt C. et al (2019). Midbrain tectal stem cells display diverse regenerative capacities in zebrafish.Sci. Rep.9:4420. 10.1038/s41598-019-40734-z
224
Liu K. Petree C. Requena T. Varshney P. Varshney G. K. (2019). Expanding the CRISPR Toolbox in Zebrafish for studying development and disease.Front. Cell Dev. Biol.7:13. 10.3389/fcell.2019.00013
225
Liu X. Li Y.-S. Shinton S. A. Rhodes J. Tang L. Feng H. et al (2017). Zebrafish B cell development without a pre–B cell stage, revealed by CD79 fluorescence reporter transgenes.J. Immunol.1991706–1715. 10.4049/jimmunol.1700552
226
Long D. W. Webb C. H. Wang Y. (2019). Persistent fibrosis and decreased cardiac function following cardiac injury in the Ctenopharyngodon idella (grass carp).bioRxiv [Preprint]. 10.1101/627752
227
Loosli F. Koster R. W. Carl M. Kuhnlein R. Henrich T. Mucke M. et al (2000). A genetic screen for mutations affecting embryonic development in medaka fish (Oryzias latipes).Mech. Dev.97133–139. 10.1016/s0925-4773(00)00406-8
228
Lopez-Baez J. C. Simpson D. J. LLeras Forero L. Zeng Z. Brunsdon H. Salzano A. et al (2018). Wilms Tumor 1b defines a wound-specific sheath cell subpopulation associated with notochord repair.eLife7:e30657. 10.7554/eLife.30657
229
Lush M. E. Piotrowski T. (2014). Sensory hair cell regeneration in the zebrafish lateral line.Dev. Dyn.2431187–1202. 10.1002/dvdy.24167
230
Lust K. Wittbrodt J. (2018). Activating the regenerative potential of Müller glia cells in a regeneration-deficient retina.eLife7:e32319. 10.7554/eLife.32319
231
Maekawa S. Chiang Y.-A. Hikima J.-I. Sakai M. Lo C.-F. Wang H.-C. et al (2016). Expression and biological activity of two types of interferon genes in medaka (Oryzias latipes).Fish Shellfish Immunol.4820–29. 10.1016/j.fsi.2015.11.036
232
Maginnis T. L. (2006). The costs of autotomy and regeneration in animals: a review and framework for future research.Behav. Ecol.17857–872. 10.1093/beheco/arl010
233
Maheras A. L. Dix B. Carmo O. M. Young A. E. Gill V. N. Sun J. L. et al (2018). Genetic pathways of neuroregeneration in a novel mild traumatic brain injury model in adult zebrafish.Eneuro5:ENEURO.0208-17.2017. 10.1523/ENEURO.0208-17
234
Marín-Juez R. El-Sammak H. Helker C. S. Kamezaki A. Mullapuli S. T. Bibli S.-I. et al (2019). Coronary revascularization during heart regeneration is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation.Dev. Cell51503–515.e4. 10.1016/j.devcel.2019.10.019
235
Marín-Juez R. Marass M. Gauvrit S. Rossi A. Lai S.-L. Materna S. C. et al (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration.Proc. Natl. Acad. Sci. U.S.A.11311237–11242. 10.1073/pnas.1605431113
236
Marletaz F. Firbas P. N. Maeso I. Tena J. J. Bogdanovic O. Perry M. et al (2018). Amphioxus functional genomics and the origins of vertebrate gene regulation.Nature56464–70. 10.1038/s41586-018-0734-6
237
Marques I. J. Lupi E. Mercader N. (2019). Model systems for regeneration: zebrafish.Development146:dev167692. 10.1242/dev.167692
238
Martinez-Morales J. R. Rembold M. Greger K. Simpson J. C. Brown K. E. Quiring R. et al (2009). Ojoplano-mediated basal constriction is essential for optic cup morphogenesis.Development1362165–2175. 10.1242/dev.033563
239
Martins R. R. Ellis P. S. MacDonald R. B. Richardson R. J. Henriques C. M. (2019). Resident immunity in tissue repair and maintenance: the zebrafish model coming of age.Front. Cell Dev. Biol.7:12. 10.3389/fcell.2019.00012
240
Marz M. Schmidt R. Rastegar S. Strahle U. (2011). Regenerative response following stab injury in the adult zebrafish telencephalon.Dev. Dyn.2402221–2231. 10.1002/dvdy.22710
241
Matias Santos D. Rita A. M. Casanellas I. Brito Ova A. Araujo I. M. Power D. et al (2016). Ear wound regeneration in the African spiny mouse Acomys cahirinus.Regeneration352–61. 10.1002/reg2.50
242
McDonald B. Pittman K. Menezes G. B. Hirota S. A. Slaba I. Waterhouse C. C. et al (2010). Intravascular danger signals guide neutrophils to sites of sterile inflammation.Science330362–366. 10.1126/science.1195491
243
Mekdara P. J. Schwalbe M. A. Coughlin L. L. Tytell E. D. (2018). The effects of lateral line ablation and regeneration in schooling giant danios.J. Exp. Biol.221:jeb175166. 10.1242/jeb.175166
244
Meng X. Noyes M. B. Zhu L. J. Lawson N. D. Wolfe S. A. (2008). Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases.Nat. Biotechnol.26695–701. 10.1038/nbt1398
245
Mescher A. L. Neff A. W. (2005). “Regenerative capacity and the developing immune system,” in Regenerative Medicine I: Theories, Models and Methods, ed.YannasI. V. (Berlin: Springer), 39–66. 10.1007/b99966
246
Metzger D. Clifford J. Chiba H. Chambon P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase.Proc. Natl. Acad. Sci. U.S.A.926991–6995. 10.1073/pnas.92.15.6991
247
Mierzwa A. S. Nguyen F. Xue M. Jonz M. G. (2020). Regeneration of the gill filaments and replacement of serotonergic neuroepithelial cells in adult zebrafish (Danio rerio).Respir. Physiol. Neurobiol.274:103366. 10.1016/j.resp.2019.103366
248
Miskey C. Izsvák Z. Plasterk R. H. Ivics Z. (2003). The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells.Nucleic Acids Res.316873–6881. 10.1093/nar/gkg910
249
Mitchell D. M. Sun C. Hunter S. S. New D. D. Stenkamp D. L. (2019). Regeneration associated transcriptional signature of retinal microglia and macrophages.Sci. Rep.9:4768. 10.1038/s41598-019-41298-8
250
Moens C. B. Donn T. M. Wolf-Saxon E. R. Ma T. P. (2008). Reverse genetics in zebrafish by TILLING.Brief Funct. Genomic Proteomic7454–459. 10.1093/bfgp/eln046
251
Mollova M. Bersell K. Walsh S. Savla J. Das L. T. Park S. Y. et al (2013). Cardiomyocyte proliferation contributes to heart growth in young humans.Proc. Natl. Acad. Sci. U.S.A.1101446–1451. 10.1073/pnas.1214608110
252
Moreno R. L. Josey M. Ribera A. B. (2017). Zebrafish in Situ spinal cord preparation for electrophysiological recordings from spinal sensory and Motor Neurons.J. Vis. Exp.122:e55507. 10.3791/55507
253
Moreno-Mateos M. A. Vejnar C. E. Beaudoin J. D. Fernandez J. P. Mis E. K. Khokha M. K. et al (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo.Nat. Methods12982–988. 10.1038/nmeth.3543
254
Morgan T. H. (1901). Regeneration and liability to injury.Science14235–248. 10.1126/science.14.346.235
255
Moriyama A. Inohaya K. Maruyama K. Kudo A. (2010). Bef medaka mutant reveals the essential role of c-myb in both primitive and definitive hematopoiesis.Dev. Biol.345133–143. 10.1016/j.ydbio.2010.06.031
256
Mosimann C. Kaufman C. K. Li P. Pugach E. K. Tamplin O. J. Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish.Development138169–177. 10.1242/dev.059345
257
Mosimann C. Puller A. C. Lawson K. L. Tschopp P. Amsterdam A. Zon L. I. (2013). Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system.Dev. Dyn.242949–963. 10.1002/dvdy.23989
258
Moss J. B. Koustubhan P. Greenman M. Parsons M. J. Walter I. Moss L. G. (2009). Regeneration of the pancreas in adult zebrafish.Diabetes581844–1851. 10.2337/db08-0628
259
Motoike T. Loughna S. Perens E. Roman B. L. Liao W. Chau T. C. et al (2000). Universal GFP reporter for the study of vascular development.Genesis2875–81. 10.1002/1526-968x(200010)28:2lt;75::aid-gene50gt;3.0.co;2-s
260
Mukherjee K. Liao E. C. (2018). Generation and characterization of a zebrafish muscle specific inducible Cre line.Transgenic Res.27559–569. 10.1007/s11248-018-0098-6
261
Murakami Y. Ansai S. Yonemura A. Kinoshita M. (2017). An efficient system for homology-dependent targeted gene integration in medaka (Oryzias latipes).Zool. Lett.3:10. 10.1186/s40851-017-0071-x
262
Murata K. Kinoshita M. Naruse K. Tanaka M. Kamei Y. (2019). Medaka: Biology, Management, and Experimental Protocols.Honoken, NJ: John Wiley & Sons, Incorporated.
263
Nakamura R. Tsukahara T. Qu W. Ichikawa K. Otsuka T. Ogoshi K. et al (2014). Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates.Development1412568–2580. 10.1242/dev.108548
264
Nakamura S. Saito D. Tanaka M. (2008). Generation of transgenic medaka using modified bacterial artificial chromosome.Dev. Growth Differ.50415–419. 10.1111/j.1440-169X.2008.01027.x
265
Nakatani Y. Kawakami A. Kudo A. (2007). Cellular and molecular processes of regeneration, with special emphasis on fish fins.Dev. Growth Differ.49145–154. 10.1111/j.1440-169X.2007.00917.x
266
Nakatani Y. Nishidate M. Fujita M. Kawakami A. Kudo A. (2008). Migration of mesenchymal cell fated to blastema is necessary for fish fin regeneration.Dev. Growth Differ.5071–83. 10.1111/j.1440-169X.2007.00977.x
267
Nasevicius A. Ekker S. C. (2000). Effective targeted gene ‘knockdown’in zebrafish.Nat. Genet.26216–220. 10.1038/79951
268
Ng G. H. B. Gong Z. (2011). Maize Ac/Ds transposon system leads to highly efficient germline transmission of transgenes in medaka (Oryzias latipes).Biochimie931858–1864. 10.1016/j.biochi.2011.07.006
269
Nguyen-Chi M. Laplace-Builhe B. Travnickova J. Luz-Crawford P. Tejedor G. Phan Q. T. et al (2015). Identification of polarized macrophage subsets in zebrafish.eLife4:e07288. 10.7554/eLife.07288
270
Nieto-Arellano R. Sanchez-Iranzo H. (2019). zfRegeneration: a database for gene expression profiling during regeneration.Bioinformatics35703–705. 10.1093/bioinformatics/bty659
271
Offen N. Blum N. Meyer A. Begemann G. (2008). Fgfr1 signalling in the development of a sexually selected trait in vertebrates, the sword of swordtail fish.BMC Dev. Biol.8:98. 10.1186/1471-213X-8-98
272
Okuda K. S. Astin J. W. Misa J. P. Flores M. V. Crosier K. E. Crosier P. S. (2012). lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish.Development1392381–2391. 10.1242/dev.077701
273
Okuyama T. Isoe Y. Hoki M. Suehiro Y. Yamagishi G. Naruse K. et al (2013). Controlled Cre/loxP site-specific recombination in the developing brain in medaka fish, Oryzias latipes.PLoS One8:e66597. 10.1371/journal.pone.0066597
274
Otsuka T. Takeda H. (2017). Targeted Ablation of Pancreatic beta Cells in Medaka.Zool. Sci.34179–184. 10.2108/zs170004
275
Otsuka T. Tsukahara T. Takeda H. (2015). Development of the pancreas in medaka, Oryzias latipes, from embryo to adult.Dev. Growth Differ.57557–569. 10.1111/dgd.12237
276
Ozato K. Kondoh H. Inohara H. Iwamatsu T. Wakamatsu Y. Okada T. S. (1986). Production of transgenic fish: introduction and expression of chicken delta-crystallin gene in medaka embryos.Cell Differ.19237–244. 10.1016/0045-6039(86)90100-4
277
Page D. M. Wittamer V. Bertrand J. Y. Lewis K. L. Pratt D. N. Delgado N. et al (2013). An evolutionarily conserved program of B-cell development and activation in zebrafish.Blood J. Am. Soc. Hematol.122e1–e11. 10.1182/blood-2012-12-471029
278
Park H. C. Kim C. H. Bae Y. K. Yeo S. Y. Kim S. H. Hong S. K. et al (2000). Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons.Dev. Biol.227279–293. 10.1006/dbio.2000.9898
279
Perner B. Englert C. Bollig F. (2007). The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros.Dev. Biol.30987–96. 10.1016/j.ydbio.2007.06.022
280
Petrie T. A. Strand N. S. Yang C. T. Rabinowitz J. S. Moon R. T. (2014). Macrophages modulate adult zebrafish tail fin regeneration.Development1412581–2591. 10.1242/dev.098459
281
Pfefferli C. Jazwinska A. (2015). The art of fin regeneration in zebrafish.Regeneration272–83. 10.1002/reg2.33
282
Pfefferli C. Jaźwińska A. (2017). The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin.Nat. Commun.8:15151. 10.1038/ncomms15151
283
Phan Q. T. Tan W. H. Liu R. Sundaram S. Buettner A. Kneitz S. et al (2020). Cxcl9l and Cxcr3.2 regulate recruitment of osteoclast progenitors to bone matrix in a medaka osteoporosis model.Proc. Natl. Acad. Sci. U.S.A.11719276–19286. 10.1073/pnas.2006093117
284
Pisharath H. Rhee J. M. Swanson M. A. Leach S. D. Parsons M. J. (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase.Mech. Dev.124218–229. 10.1016/j.mod.2006.11.005
285
Poleo G. Brown C. W. Laforest L. Akimenko M. A. (2001). Cell proliferation and movement during early fin regeneration in zebrafish.Dev. Dyn.221380–390. 10.1002/dvdy.1152
286
Polizzotti B. D. Ganapathy B. Haubner B. J. Penninger J. M. Kühn B. (2016). A cryoinjury model in neonatal mice for cardiac translational and regeneration research.Nat. Protoc.11542–552. 10.1038/nprot.2016.031
287
Porazinski S. R. Wang H. Furutani-Seiki M. (2010b). Microinjection of medaka embryos for use as a model genetic organism.J. Vis. Exp.46:e1937. 10.3791/1937
288
Porazinski S. R. Wang H. Furutani-Seiki M. (2010a). Dechorionation of medaka embryos and cell transplantation for the generation of chimeras.J. Vis. Exp.46:e2055. 10.3791/2055
289
Porrello E. R. Mahmoud A. I. Simpson E. Hill J. A. Richardson J. A. Olson E. N. et al (2011). Transient regenerative potential of the neonatal mouse heart.Science3311078–1080. 10.1126/science.1200708
290
Poss K. D. Keating M. T. Nechiporuk A. (2003). Tales of regeneration in zebrafish.Dev. Dyn.226202–210. 10.1002/dvdy.10220
291
Poss K. D. Wilson L. G. Keating M. T. (2002). Heart regeneration in zebrafish.Science2982188–2190. 10.1126/science.1077857
292
Potts H. G. Stockdale W. T. Mommersteeg M. T. M. (2021). Unlocking the secrets of the regenerating fish heart: comparing regenerative models to shed light on successful regeneration.J. Cardiovasc. Dev. Dis.8:10004. 10.3390/jcdd8010004
293
Powell C. Cornblath E. Elsaeidi F. Wan J. Goldman D. (2016). Zebrafish Müller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons.Sci. Rep.6:24851. 10.1038/srep24851
294
Provost E. Rhee J. Leach S. D. (2007). Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos.Genesis45625–629. 10.1002/dvg.20338
295
Pugach E. K. Li P. White R. Zon L. (2009). Retro-orbital injection in adult zebrafish.J. Vis. Exp.34:1645. 10.3791/1645
296
Qi Y. Dasa O. Maden M. Vohra R. Batra A. Walter G. et al (2021). Functional heart recovery in an adult mammal, the spiny mouse.Int. J. Cardiol.338196–203. 10.1016/j.ijcard.2021.06.015
297
Ramachandran R. Fausett B. V. Goldman D. (2010a). Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway.Nat. Cell Biol.121101–1107. 10.1038/ncb2115
298
Ramachandran R. Reifler A. Parent J. M. Goldman D. (2010b). Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration.J. Comp. Neurol.5184196–4212. 10.1002/cne.22448
299
Ramel M.-C. Progatzky F. Rydlova A. Wane M. Schymeinsky J. Williams C. et al (2021). Dynamics of repair and regeneration of adult zebrafish respiratory gill tissue after cryoinjury.bioRxiv [Preprint]. 10.1101/2021.05.27.445469
300
Raymond P. A. Barthel L. K. Bernardos R. L. Perkowski J. J. (2006). Molecular characterization of retinal stem cells and their niches in adult zebrafish.BMC Dev. Biol.6:36. 10.1186/1471-213X-6-36
301
Raymond P. A. Reifler M. J. Rivlin P. K. (1988). Regeneration of goldfish retina: rod precursors are a likely source of regenerated cells.J. Neurobiol.19431–463. 10.1002/neu.480190504
302
Redd M. J. Kelly G. Dunn G. Way M. Martin P. (2006). Imaging macrophage chemotaxis in vivo: studies of microtubule function in zebrafish wound inflammation.Cell Motil. Cytoskeleton63415–422. 10.1002/cm.20133
303
Reddien P. W. Alvarado A. S. (2004). Fundamentals of planarian regeneration.Annu. Rev. Cell Dev. Biol.20725–757. 10.1146/annurev.cellbio.20.010403.095114
304
Reddy P. C. Gungi A. Unni M. (2019). Cellular and molecular mechanisms of Hydra regeneration.Results Probl. Cell Differ.68259–290. 10.1007/978-3-030-23459-1_12
305
Reimschuessel R. Williams D. (1995). Development of new nephrons in adult kidneys following gentamicin-induced nephrotoxicity.Ren. Fail.17101–106. 10.3109/08860229509026246
306
Reinhardt R. Centanin L. Tavhelidse T. Inoue D. Wittbrodt B. Concordet J. P. et al (2015). Sox2, Tlx, Gli3, and Her9 converge on Rx2 to define retinal stem cells in vivo.EMBO J.341572–1588. 10.15252/embj.201490706
307
Renn J. Büttner A. To T. T. Chan S. J. H. Winkler C. (2013). A col10a1: nlGFP transgenic line displays putative osteoblast precursors at the medaka notochordal sheath prior to mineralization.Dev. Biol.381134–143. 10.1016/j.ydbio.2013.05.030
308
Renn J. Winkler C. (2009). Osterix-mCherry transgenic medaka for in vivo imaging of bone formation.Dev. Dyn.238241–248. 10.1002/dvdy.21836
309
Renshaw S. A. Loynes C. A. Trushell D. M. Elworthy S. Ingham P. W. Whyte M. K. (2006). A transgenic zebrafish model of neutrophilic inflammation.Blood1083976–3978. 10.1182/blood-2006-05-024075
310
Richardson R. Slanchev K. Kraus C. Knyphausen P. Eming S. Hammerschmidt M. (2013). Adult zebrafish as a model system for cutaneous wound-healing research.J. Invest. Dermatol.1331655–1665. 10.1038/jid.2013.16
311
Robert J. Ohta Y. (2009). Comparative and developmental study of the immune system in Xenopus.Dev. Dyn.2381249–1270. 10.1002/dvdy.21891
312
Roh-Johnson M. Shah A. N. Stonick J. A. Poudel K. R. Kargl J. Yang G. H. et al (2017). Macrophage-dependent cytoplasmic transfer during melanoma invasion in vivo.Dev. Cell43549–562.e6. 10.1016/j.devcel.2017.11.003
313
Rottbauer W. Saurin A. J. Lickert H. Shen X. Burns C. G. Wo Z. G. et al (2002). Reptin and pontin antagonistically regulate heart growth in zebrafish embryos.Cell111661–672. 10.1016/s0092-8674(02)01112-1
314
Ruyra A. Cano-Sarabia M. Garcia-Valtanen P. Yero D. Gibert I. Mackenzie S. A. et al (2014). Targeting and stimulation of the zebrafish (Danio rerio) innate immune system with LPS/dsRNA-loaded nanoliposomes.Vaccine323955–3962. 10.1016/j.vaccine.2014.05.010
315
Ruzicka L. Howe D. G. Ramachandran S. Toro S. Van Slyke C. E. Bradford Y. M. et al (2019). The Zebrafish information network: new support for non-coding genes, richer gene ontology annotations and the alliance of genome resources.Nucleic Acids Res.47D867–D873. 10.1093/nar/gky1090
316
Sadler K. C. Krahn K. N. Gaur N. A. Ukomadu C. (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1.Proc. Natl. Acad. Sci. U.S.A.1041570–1575. 10.1073/pnas.0610774104
317
Sanchez-Iranzo H. Galardi-Castilla M. Sanz-Morejon A. Gonzalez-Rosa J. M. Costa R. Ernst A. et al (2018). Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart.Proc. Natl. Acad. Sci. U.S.A.1154188–4193. 10.1073/pnas.1716713115
318
Sander J. D. Cade L. Khayter C. Reyon D. Peterson R. T. Joung J. K. et al (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs.Nat. Biotechnol.29697–698. 10.1038/nbt.1934
319
Sanderson L. E. Chien A.-T. Astin J. W. Crosier K. E. Crosier P. S. Hall C. J. (2015). An inducible transgene reports activation of macrophages in live zebrafish larvae.Dev. Comp. Immunol.5363–69. 10.1016/j.dci.2015.06.013
320
Sano S. Takashima S. Niwa H. Yokoi H. Shimada A. Arenz A. et al (2009). Characterization of teleost Mdga1 using a gene-trap approach in medaka (Oryzias latipes).Genesis47505–513. 10.1002/dvg.20528
321
Sanz-Morejon A. Garcia-Redondo A. B. Reuter H. Marques I. J. Bates T. Galardi-Castilla M. et al (2019). Wilms Tumor 1b Expression Defines a Pro-regenerative Macrophage Subtype and Is Required for Organ Regeneration in the Zebrafish.Cell Rep.281296–1306.e6. 10.1016/j.celrep.2019.06.091
322
Sasado T. Tanaka M. Kobayashi K. Sato T. Sakaizumi M. Naruse K. (2010). The National BioResource Project Medaka (NBRP Medaka): an integrated bioresource for biological and biomedical sciences.Exp. Anim.5913–23. 10.1538/expanim.59.13
323
Sassen W. A. Köster R. W. (2015). A molecular toolbox for genetic manipulation of zebrafish.Adv. Genomics Genet.5151–163. 10.2147/AGG.S57585
324
Schaafhausen M. K. Yang W.-J. Centanin L. Wittbrodt J. Bosserhoff A. Fischer A. et al (2013). Tumor angiogenesis is caused by single melanoma cells in a reactive oxygen species and NF-κB dependent manner.J. Cell Sci.1263862–3872. 10.1242/jcs.125021
325
Schall K. A. Holoyda K. A. Grant C. N. Levin D. E. Torres E. R. Maxwell A. et al (2015). Adult zebrafish intestine resection: a novel model of short bowel syndrome, adaptation, and intestinal stem cell regeneration.Am. J. Physiol. Gastrointest. Liver Physiol.309G135–G145. 10.1152/ajpgi.00311.2014
326
Schmidt R. Beil T. Strähle U. Rastegar S. (2014). Stab wound injury of the zebrafish adult telencephalon: a method to investigate vertebrate brain neurogenesis and regeneration.J. Vis. Exp.90:e51753. 10.3791/51753
327
Schnabel K. Wu C. C. Kurth T. Weidinger G. (2011). Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation.PLoS One6:e18503. 10.1371/journal.pone.0018503
328
Schuck J. B. Smith M. E. (2009). Cell proliferation follows acoustically-induced hair cell bundle loss in the zebrafish saccule.Hear. Res.25367–76. 10.1016/j.heares.2009.03.008
329
Seifert A. W. Kiama S. G. Seifert M. G. Goheen J. R. Palmer T. M. Maden M. (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys).Nature489561–565. 10.1038/nature11499
330
Seleit A. Gross K. Onistschenko J. Hoang O. P. Theelke J. Centanin L. (2022). Local tissue interactions govern pLL patterning in medaka.Dev. Biol.4811–13. 10.1016/j.ydbio.2021.09.002
331
Seleit A. Gross K. Onistschenko J. Woelk M. Autorino C. Centanin L. (2020). Development and regeneration dynamics of the Medaka notochord.Dev. Biol.46311–25. 10.1016/j.ydbio.2020.03.001
332
Seleit A. Krämer I. Riebesehl B. F. Ambrosio E. M. Stolper J. S. Lischik C. Q. et al (2017b). Neural stem cells induce the formation of their physical niche during organogenesis.eLife6:e29173. 10.7554/eLife.29173
333
Seleit A. Kramer I. Ambrosio E. Dross N. Engel U. Centanin L. (2017a). Sequential organogenesis sets two parallel sensory lines in medaka.Development144687–697. 10.1242/dev.142752
334
Senut M.-C. Gulati-Leekha A. Goldman D. (2004). An element in the α1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish.J. Neurosci.247663–7673. 10.1523/JNEUROSCI.2281-04.2004
335
Sherpa T. Fimbel S. M. Mallory D. E. Maaswinkel H. Spritzer S. D. Sand J. A. et al (2008). Ganglion cell regeneration following whole-retina destruction in zebrafish.Dev. Neurobiol.68166–181. 10.1002/dneu.20568
336
Shimizu Y. Ito Y. Tanaka H. Ohshima T. (2015). Radial glial cell-specific ablation in the adult Z ebrafish brain.Genesis53431–439. 10.1002/dvg.22865
337
Shimizu Y. Kawasaki T. (2021). Differential regenerative capacity of the optic Tectum of Adult Medaka and Zebrafish.Front. Cell Dev. Biol.9:686755. 10.3389/fcell.2021.686755
338
Shimizu Y. Ueda Y. Ohshima T. (2018). Wnt signaling regulates proliferation and differentiation of radial glia in regenerative processes after stab injury in the optic tectum of adult zebrafish.Glia661382–1394. 10.1002/glia.23311
339
Simkin J. Gawriluk T. R. Gensel J. C. Seifert A. W. (2017). Macrophages are necessary for epimorphic regeneration in African spiny mice.eLife6:e24623. 10.7554/eLife.24623
340
Simoes F. C. Cahill T. J. Kenyon A. Gavriouchkina D. Vieira J. M. Sun X. et al (2020). Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair.Nat. Commun.11:600. 10.1038/s41467-019-14263-2
341
Simon A. Tanaka E. M. (2013). Limb regeneration.Wiley Interdiscip. Rev. Dev. Biol.2291–300. 10.1002/wdev.73
342
Singh S. P. Holdway J. E. Poss K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts.Dev. Cell22879–886. 10.1016/j.devcel.2012.03.006
343
Soehnlein O. Lindbom L. (2010). Phagocyte partnership during the onset and resolution of inflammation.Nat. Rev. Immunol.10427–439. 10.1038/nri2779
344
Sorlien E. L. Witucki M. A. Ogas J. (2018). Efficient Production and Identification of CRISPR/Cas9-generated Gene Knockouts in the Model System Danio rerio.J. Vis. Exp.138:56969. 10.3791/56969
345
Steinhauser M. L. Lee R. T. (2011). Regeneration of the heart.EMBO Mol. Med.3701–712.
346
Stemmer M. Thumberger T. Del Sol Keyer M. Wittbrodt J. Mateo J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool.PLoS One10:e0124633. 10.1371/journal.pone.0124633
347
Stockdale W. T. Lemieux M. E. Killen A. C. Zhao J. Hu Z. Riepsaame J. et al (2018). Heart regeneration in the Mexican Cavefish.Cell Rep.251997–2007.e7. 10.1016/j.celrep.2018.10.072
348
Stolper J. Ambrosio E. M. Danciu D.-P. Buono L. Elliott D. A. Naruse K. et al (2019). Stem cell topography splits growth and homeostatic functions in the fish gill.eLife8:e43747. 10.7554/eLife.43747
349
Straube W. L. Tanaka E. M. (2006). Reversibility of the differentiated state: regeneration in amphibians.Artif. Organs30743–755. 10.1111/j.1525-1594.2006.00296.x
350
Streisinger G. Singer F. Walker C. Knauber D. Dower N. (1986). Segregation analyses and gene-centromere distances in zebrafish.Genetics112311–319. 10.1093/genetics/112.2.311
351
Streisinger G. Walker C. Dower N. Knauber D. Singer F. (1981). Production of clones of homozygous diploid zebra fish (Brachydanio rerio).Nature291293–296. 10.1038/291293a0
352
Strungs E. G. Ongstad E. L. O’Quinn M. P. Palatinus J. A. Jourdan L. J. Gourdie R. G. (2013). Cryoinjury models of the adult and neonatal mouse heart for studies of scarring and regeneration.Methods Mol. Biol.1037343–353. 10.1007/978-1-62703-505-7_20
353
Suehiro Y. Kinoshita M. Okuyama T. Shimada A. Naruse K. Takeda H. et al (2010). Transient and permanent gene transfer into the brain of the teleost fish medaka (Oryzias latipes) using human adenovirus and the Cre-loxP system.FEBS Lett.5843545–3549. 10.1016/j.febslet.2010.06.047
354
Suster M. L. Abe G. Schouw A. Kawakami K. (2011). Transposon-mediated BAC transgenesis in zebrafish.Nat. Protoc.61998–2021. 10.1038/nprot.2011.416
355
Sveen L. R. Timmerhaus G. Krasnov A. Takle H. Handeland S. Ytteborg E. (2019). Wound healing in post-smolt Atlantic salmon (Salmo salar L.).Sci. Rep.9:3565. 10.1038/s41598-019-39080-x
356
Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. et al (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors.Cell131861–872. 10.1016/j.cell.2007.11.019
357
Takanezawa S. Saitou T. Imamura T. (2021). Wide field light-sheet microscopy with lens-axicon controlled two-photon Bessel beam illumination.Nat. Commun.12:2979. 10.1038/s41467-021-23249-y
358
Takechi M. Hamaoka T. Kawamura S. (2003). Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish.FEBS Lett.55390–94. 10.1016/s0014-5793(03)00977-3
359
Takeuchi A. Okubo K. (2013). Post-proliferative immature radial glial cells female-specifically express aromatase in the medaka optic tectum.PLoS One8:e73663. 10.1371/journal.pone.0073663
360
Taneda Y. Konno S. Makino S. Morioka M. Fukuda K. Imai Y. et al (2010). Epigenetic control of cardiomyocyte production in response to a stress during the medaka heart development.Dev. Biol.34030–40. 10.1016/j.ydbio.2010.01.014
361
Taniguchi Y. Takeda S. Furutani-Seiki M. Kamei Y. Todo T. Sasado T. et al (2006). Generation of medaka gene knockout models by target-selected mutagenesis.Genome Biol.7:R116. 10.1186/gb-2006-7-12-r116
362
Tena J. J. Gonzalez-Aguilera C. Fernandez-Minan A. Vazquez-Marin J. Parra-Acero H. Cross J. W. et al (2014). Comparative epigenomics in distantly related teleost species identifies conserved cis-regulatory nodes active during the vertebrate phylotypic period.Genome Res.241075–1085. 10.1101/gr.163915.113
363
Terzibasi E. Valenzano D. R. Cellerino A. (2007). The short-lived fish Nothobranchius furzeri as a new model system for aging studies.Exp. Gerontol.4281–89. 10.1016/j.exger.2006.06.039
364
Thermes V. Grabher C. Ristoratore F. Bourrat F. Choulika A. Wittbrodt J. et al (2002). I-SceI meganuclease mediates highly efficient transgenesis in fish.Mech. Dev.11891–98. 10.1016/s0925-4773(02)00218-6
365
Thummel R. Burket C. T. Brewer J. L. Sarras M. P. Jr. Li L. Perry M. et al (2005). Cre-mediated site-specific recombination in zebrafish embryos.Dev. Dyn.2331366–1377. 10.1002/dvdy.20475
366
Thummel R. Kassen S. C. Enright J. M. Nelson C. M. Montgomery J. E. Hyde D. R. (2008). Characterization of Müller glia and neuronal progenitors during adult zebrafish retinal regeneration.Exp. Eye Res.87433–444. 10.1016/j.exer.2008.07.009
367
To T. T. Witten P. E. Renn J. Bhattacharya D. Huysseune A. Winkler C. (2012). Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model.Development139141–150. 10.1242/dev.071035
368
Tong S. K. Mouriec K. Kuo M. W. Pellegrini E. Gueguen M. M. Brion F. et al (2009). A cyp19a1b-gfp (aromatase B) transgenic zebrafish line that expresses GFP in radial glial cells.Genesis4767–73. 10.1002/dvg.20459
369
Traver D. Paw B. H. Poss K. D. Penberthy W. T. Lin S. Zon L. I. (2003). Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants.Nat. Immunol.41238–1246. 10.1038/ni1007
370
Trede N. S. Langenau D. M. Traver D. Look A. T. Zon L. I. (2004). The use of zebrafish to understand immunity.Immunity20367–379. 10.1016/s1074-7613(04)00084-6
371
Ueno T. Yasumasu S. Hayashi S. Iuchi I. (2004). Identification of choriogenin cis-regulatory elements and production of estrogen-inducible, liver-specific transgenic Medaka.Mech. Dev.121803–815. 10.1016/j.mod.2004.03.027
372
Uesaka M. Kuratani S. Takeda H. Irie N. (2019). Recapitulation-like developmental transitions of chromatin accessibility in vertebrates.Zool. Lett.5:33. 10.1186/s40851-019-0148-9
373
Underwood J. L. Hestand R. S. III Thompson B. Z. (1986). Gonad regeneration in grass carp following bilateral gonadectomy.Prog. Fish Cult.4854–56. 10.1577/1548-8640(1986)48<54:grigcf>2.0.co;2
374
Van Houcke J. Mariën V. Zandecki C. Vanhunsel S. Moons L. Ayana R. et al (2021). Aging impairs the essential contributions of non-glial progenitors to neurorepair in the dorsal telencephalon of the Killifish Nothobranchius furzeri.Aging Cell20:e13464. 10.1111/acel.13464
375
Van Wettere A. J. Kullman S. W. Hinton D. E. Law J. M. (2013). Immunohistochemical characterization of the hepatic progenitor cell compartment in medaka (Oryzias latipes) following hepatic injury.J. Comp. Pathol.149434–445. 10.1016/j.jcpa.2013.03.008
376
Var S. R. Byrd-Jacobs C. A. (2020). Role of macrophages and microglia in zebrafish regeneration.Int. J. Mol. Sci.21:4768. 10.3390/ijms21134768
377
Vihtelic T. S. Hyde D. R. (2000). Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina.J. Neurobiol.44289–307. 10.1002/1097-4695(20000905)44:3lt;289::aid-neu1gt;3.0.co;2-h
378
Vivien C. J. Hudson J. E. Porrello E. R. (2016). Evolution, comparative biology and ontogeny of vertebrate heart regeneration.NPJ Regen. Med.1:16012. 10.1038/npjregenmed.2016.12
379
Walton E. M. Cronan M. R. Beerman R. W. Tobin D. M. (2015). The macrophage-specific promoter mfap4 allows live, long-term analysis of macrophage behavior during Mycobacterial Infection in Zebrafish.PLoS One10:e0138949. 10.1371/journal.pone.0138949
380
Wan H. Korzh S. Li Z. Mudumana S. P. Korzh V. Jiang Y.-J. et al (2006). Analyses of pancreas development by generation of gfp transgenic zebrafish using an exocrine pancreas-specific elastaseA gene promoter.Exp. Cell Res.3121526–1539. 10.1016/j.yexcr.2006.01.016
381
Wang J. Karra R. Dickson A. L. Poss K. D. (2013). Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration.Dev. Biol.382427–435. 10.1016/j.ydbio.2013.08.012
382
Wang J. Panakova D. Kikuchi K. Holdway J. E. Gemberling M. Burris J. S. et al (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion.Development1383421–3430. 10.1242/dev.068601
383
Wang R. Li Z. Wang Y. Gui J.-F. (2011). An Apo-14 promoter-driven transgenic zebrafish that marks liver organogenesis.PLoS One6:e22555. 10.1371/journal.pone.0022555
384
Wang Z. Cui M. Shah A. M. Tan W. Liu N. Bassel-Duby R. et al (2020). Cell-type-specific gene regulatory networks underlying murine neonatal heart regeneration at single-cell resolution.Cell Rep.33:108472.
385
Wang W. Hu C. K. Zeng A. Alegre D. Hu D. Gotting K. et al (2020). Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates.Science369:eaaz3090. 10.1126/science.aaz3090
386
Wang Y. Kaiser M. S. Larson J. D. Nasevicius A. Clark K. J. Wadman S. A. et al (2010). Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis.Development1373119–3128. 10.1242/dev.048785
387
Watakabe I. Hashimoto H. Kimura Y. Yokoi S. Naruse K. Higashijima S. I. (2018). Highly efficient generation of knock-in transgenic medaka by CRISPR/Cas9-mediated genome engineering.Zool. Lett.4:3. 10.1186/s40851-017-0086-3
388
Watanabe N. Kato M. Suzuki N. Inoue C. Fedorova S. Hashimoto H. et al (2009). Kidney regeneration through nephron neogenesis in medaka.Dev. Growth Differ.51135–143. 10.1111/j.1440-169X.2009.01090.x
389
Wehner D. Becker T. Becker C. G. (2018). Restoration of anatomical continuity after spinal cord transection depends on Wnt/β-catenin signaling in larval zebrafish.Data Brief1665–70. 10.1016/j.dib.2017.10.068
390
Wendler S. Hartmann N. Hoppe B. Englert C. (2015). Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri.Aging Cell14857–866. 10.1111/acel.12367
391
White R. M. Sessa A. Burke C. Bowman T. LeBlanc J. Ceol C. et al (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis.Cell Stem Cell2183–189. 10.1016/j.stem.2007.11.002
392
Wienholds E. Van Eeden F. Kosters M. Mudde J. Plasterk R. H. Cuppen E. (2003). Efficient target-selected mutagenesis in zebrafish.Genome Res.132700–2707. 10.1101/gr.1725103
393
Wierson W. A. Welker J. M. Almeida M. P. Mann C. M. Webster D. A. Torrie M. E. et al (2020). Efficient targeted integration directed by short homology in zebrafish and mammalian cells.eLife9:e53968. 10.7554/eLife.53968
394
Willems B. Buttner A. Huysseune A. Renn J. Witten P. E. Winkler C. (2012). Conditional ablation of osteoblasts in medaka.Dev. Biol.364128–137. 10.1016/j.ydbio.2012.01.023
395
Wittamer V. Bertrand J. Y. Gutschow P. W. Traver D. (2011). Characterization of the mononuclear phagocyte system in zebrafish.Blood J. Am. Soc. Hematol.1177126–7135. 10.1182/blood-2010-11-321448
396
Wittbrodt J. Shima A. Schartl M. (2002). Medaka—a model organism from the far East.Nat. Rev. Genet.353–64. 10.1038/nrg704
397
Xiao C. Wang F. Hou J. Zhu X. Luo Y. Xiong J. W. (2018). Nanoparticle-mediated siRNA gene-silencing in adult zebrafish heart.J. Vis. Exp.137:58054. 10.3791/58054
398
Xiao T. Roeser T. Staub W. Baier H. (2005). A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection.10.1242/dev.01861
399
Yamamoto T. (1975). Medaka (killifish): Biology and Strains.Tokyo: Keigaku Pub. Co.
400
Yang K. Kang J. (2019). Tissue regeneration enhancer elements: a way to unlock endogenous healing power.Dev. Dyn.24834–42. 10.1002/dvdy.24676
401
Yano T. Tamura K. (2013). The making of differences between fins and limbs.J. Anat.222100–113. 10.1111/j.1469-7580.2012.01491.x
402
Yi Ni Lam E. Chau J. Y. Kalev-Zylinska M. L. Fountaine T. M. Mead R. S. et al (2009). Zebrafish runx1 promoter-EGFP transgenics mark discrete sites of definitive blood progenitors.Blood J. Am. Soc. Hematol.1131241–1249. 10.1182/blood-2008-04-149898
403
Yokoyama H. Ogino H. Stoick-Cooper C. L. Grainger R. M. Moon R. T. (2007). Wnt/β-catenin signaling has an essential role in the initiation of limb regeneration.Dev. Biol.306170–178.
404
Yoshinari N. Ando K. Kudo A. Kinoshita M. Kawakami A. (2012). Colored medaka and zebrafish: transgenics with ubiquitous and strong transgene expression driven by the medaka beta-actin promoter.Dev. Growth Differ.54818–828. 10.1111/dgd.12013
405
You M. S. Jiang Y. J. Yuh C. H. Wang C. M. Tang C. H. Chuang Y. J. et al (2016). A Sketch of the Taiwan Zebrafish Core Facility.Zebrafish13(Suppl. 1), S24–S29. 10.1089/zeb.2015.1208
406
Yun M. H. (2015). Changes in regenerative capacity through lifespan.Int. J. Mol. Sci.1625392–25432. 10.3390/ijms161025392
407
Zattara E. E. Fernandez-Alvarez F. A. Hiebert T. C. Bely A. E. Norenburg J. L. (2019). A phylum-wide survey reveals multiple independent gains of head regeneration in Nemertea.Proc. Biol. Sci.286:20182524. 10.1098/rspb.2018.2524
408
Zecchin E. Filippi A. Biemar F. Tiso N. Pauls S. Ellertsdottir E. et al (2007). Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish.Dev. Biol.301192–204. 10.1016/j.ydbio.2006.09.041
409
Zeng Z. Liu X. Seebah S. Gong Z. (2005). Faithful expression of living color reporter genes in transgenic medaka under two tissue-specific zebrafish promoters.Dev. Dyn.234387–392. 10.1002/dvdy.20491
410
Zhang R. Han P. Yang H. Ouyang K. Lee D. Lin Y. F. et al (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration.Nature498497–501. 10.1038/nature12322
411
Zhao L. Ben-Yair R. Burns C. E. Burns C. G. (2019). Endocardial Notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism.Cell Rep.26546–554.e5. 10.1016/j.celrep.2018.12.048
Summary
Keywords
zebrafish, medaka (Oryzias latipes), tissue regeneration, heart, retina, fin, evolution, comparative genomics
Citation
Chowdhury K, Lin S and Lai S-L (2022) Comparative Study in Zebrafish and Medaka Unravels the Mechanisms of Tissue Regeneration. Front. Ecol. Evol. 10:783818. doi: 10.3389/fevo.2022.783818
Received
27 September 2021
Accepted
10 January 2022
Published
01 February 2022
Volume
10 - 2022
Edited by
Mathilda Mommersteeg, University of Oxford, United Kingdom
Reviewed by
Yuki Shimizu, National Institute of Advanced Industrial Science and Technology, Japan; Lazaro Centanin, Heidelberg University, Germany
Updates
Copyright
© 2022 Chowdhury, Lin and Lai.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shih-Lei Lai, ben.s.lai@ibms.sinica.edu.tw
This article was submitted to Evolutionary Developmental Biology, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.