Abstract
The majority of multi-cellular terrestrial life is found in tropical forests and is either an invertebrate or a plant: for decades ecologists have sought to understand why. As global change erodes the list of extant species on our planet quantifying what species remain, along with their origins and ecology, contributes to our ability to preserve ecosystem functioning and resilience. Here we study three feeding guilds of caterpillars (Lepidoptera) and seek to understand the drivers of their diet breadth across four diverse tropical plant genera in Papua New Guinea. Host specificity is central to biodiversity estimates and the resilience of ecological networks. Specifically, we calculate distance-based host specificity in relation to plant phylogenetic relationships alongside chemical and mechanical traits of leaves. In terms of chemical defenses, we focus on the major polyphenol groups, a compound class shared across many plant species. We refine our data exploration using food webs and ordinations to pick out specific traits of relevance to insect host specificity. Our results showed that the degree of specialization for caterpillars took the following order: phylogenetic>polyphenol>mechanical, such that insect specificity was explained best by host phylogeny and polyphenol chemistry in our study system. Leaf mining insects had higher host specificity than those feeding externally. Of the traits studied hexahydroxydiphenoyl derivatives, galloyl derivatives, trichome density, quinic acid derivatives, myricetins and successional index explained the most variation in overall insect community structure. Our findings build on earlier studies of New Guinean rainforest communities and add a mechanistic explanation to previous findings that host genera are functional islands for insect herbivores. Further, we demonstrate that different plant genera combine different defensive traits that appear to drive associated insect diversity. Our approach integrates trait data and phylogeny to explore dimensions of specialization and we welcome metabolomic studies that will provide more detailed explanations for insect-herbivore host use. Finally, chemical diversity is directly linked to organismal diversity and by studying a range of insect herbivore guilds we make a connection between feeding ecology and specialization that will help to predict species interactions and, potentially, the persistence of ecological networks.
1 Introduction
Plants and their associated insect communities represent 75% of terrestrial macro-diversity (Price, 2002). By understanding the ecology and evolution of herbivorous insects we can shed light on the origins and distribution of myriad extant species. A large share of woody-plant diversity in tropical forests is represented by a relatively small number of species-rich plant genera (Gentry, 1982; Foster and Hubbell, 1990). These genera, such as Ficus, Inga, Piper, Psychotria, and others have fascinated ecologists for decades (Gentry, 1982; Basset et al., 1997; Sedio et al., 2012). They can form abundant and diverse assemblages composed of multiple congeneric species occurring in sympatry, or ‘species swarms’ (Gentry, 1982). Due to their abundance and diversity, species-rich woody-plant genera serve as particularly important hosts of tropical insects (Basset et al., 1997; Novotny et al., 2010). For example, a single Ficus tree can harbor dozens of insect herbivore species from various taxonomic groups and feeding guilds (Novotny et al., 2010). Understanding what factors allow for the coexistence of so many closely related plant species facing diverse herbivore communities can reveal important rules of plant community assembly. In turn this can contribute toward understanding why tropical forests are so diverse (Sedio and Ostling, 2013).
Most insect herbivores show some level of specialization. Typically, insect herbivores are specialized at the level of host-plant genus, with both monophagous and extremely polyphagous species being rather rare (Forister et al., 2015). This is because the functional and defensive traits that govern insect host-choice show high variation among tropical plants (Cardenas et al., 2014; Sedio et al., 2017; Sedio et al., 2018). This variation in traits can be attributed to different evolutionary history and habitat preferences among the species from different genera. Plant genera frequently occur at distinct successionary stages (as gap specialists, understory shrubs or canopy trees, for example) and share adaptations to these habitats. In some cases, diversity and identity of neighboring trees may even explain insect specialization better than leaf traits (Jia et al., 2023). Similar processes also act among congeneric plants (Sedio et al., 2017). In addition, there seems to be a trend toward divergence in chemical traits among related sympatric species of tropical plants that further helps them to avoid sharing their insect herbivores (Kursar et al., 2009; Endara et al., 2017; Salazar et al., 2018; Endara et al., 2023). Indeed, chemical variation seems to increase toward the wet tropics where specialized herbivores are more prevalent (Sedio et al., 2018; Sedio et al., 2021). In turn, variation among congeneric species can be of a similar extent or even larger than between species from different genera (Sedio et al., 2017).
Various herbivores show differential responses to host-plant traits (Volf et al., 2018; Leong et al., 2022). These responses depend on the feeding guild and dietary specialization of the given herbivore species (Ali and Agrawal, 2012; Leong et al., 2022). Our study considers the larvae of Lepidoptera (caterpillars) that show different levels of specialization and belong to various feeding guilds. Generalist free-living caterpillars can be deterred by the toxic chemistry of their hosts. In contrast, specialized caterpillars can prefer to feed on toxic plants and may even benefit from their specialized metabolites (Volf et al., 2018). For example, Asota (Erebidae: Aganainae) caterpillars preferentially feed on alkaloidal Ficus species, sequestering host alkaloids and likely using them for anti-predator defense (Volf et al., 2018; Fontanilla et al., 2022). Other specialized herbivores are found among concealed caterpillars, such as those mining leaves. By feeding on leaf parenchymal tissue, leaf miners can avoid some leaf defenses associated with the leaf cuticle (Connor and Taverner, 1997). Additionally, many mining and non-mining insects are able to manipulate leaf quality and chemical composition of their hosts, which further contributes to the intimacy of their interactions (Giron et al., 2016).
In this study, we analyze host-specialization in free-living caterpillars, semi-concealed caterpillars (leaf-rollers and leaf-tiers), and leaf miners associated with speciose tree genera in the tropical forest in Papua New Guinea. Papua New Guinea is a diversity hotspot and a center of endemism for both plants and insects (Novotny and Toko, 2015). Despite covering only ca 5% of the dry land surface of our planet, it is home to at least 13,634 plant species (Höft, 1992; Novotny and Toko, 2015; Cámara-Leret et al., 2020), which represents 3.5% of the global total (Antonelli et al., 2023). Unlike many other tropical islands, it is still partially covered by vast areas of primary forest mixed with secondary forests. These habitats contain diverse plant communities, including some particularly species-rich woody plant genera (Novotny and Toko, 2015). We focus on four of them – Ficus, Macaranga, Syzygium and Psychotria (Whitfeld et al., 2012a): together these genera include 6% of all New Guinean plant species (Conn, 2008). While Syzygium and Psychotria are typically climax species Macaranga is generally a pioneer (Whitmore, 1989). Succesionary status varies among species within Ficus. We combine plant defensive traits and phylogenetic data with insect community data to explore what factors drive host specialization of caterpillars associated with Ficus, Macaranga, Syzygium and Psychotria. In terms of chemical defenses, we focus particularly on polyphenols, including various types of tannins and flavonoids. Polyphenols are broadly distributed among the plant kingdom and represent a group of defensive metabolites found in all four genera we studied. Polyphenols affect insect herbivores through various mechanisms, such as causing oxidative stress or precipitating proteins in the insect gut (Salminen and Karonen, 2011; Salminen, 2014). Due to their universal presence in plants, they have been shown to be important predictors herbivore host-choice in tropical forests of Papua New Guinea when considering host-plants from various lineages (Segar et al., 2017). It is important to acknowledge that differences across methods and habitats can determine the role of polyphenols in deterring insect herbivores, here we consider all major groups of polyphenols (as well as the well-studied condensed tannins) and include measures of biological activity. While some studies in tropical habitats have demonstrated mixed findings (Coley, 1983; Kurokawa and Nakashizuka, 2008; Cardenas et al., 2014; López-Carretero et al., 2016), a consideration of individual groups, compounds or biological activities has demonstrated a pronounced effect of polyphenols on insect herbivore damage (Moctezuma et al., 2014) or host use (Segar et al., 2017). We note that colometric assays for condensed tannins and quantification of total polyphenol content can provide meaningful results in the right context, but that high resolution data (quantification of major groups or even individual compounds) are more suitable for studying insect chemical ecology, especially when combined with measures of biological activity.
We first hypothesize that the four tree genera will harbor insect assemblages dissimilar from each other, while congeneric hosts will share most of their herbivores. This expectation is based on phylogenetic conservatism in plant defensive traits and existing evidence that insect herbivores are generally specific at the level of host genus (Novotny et al., 2010). Second, we expect that insect specialization will reflect host phylogeny and habitat preferences rather than polyphenols and physical traits (because phylogenetically conserved host traits determine insect preference). And third, we expect that when partitioned into feeding guilds, the level of specialization will decrease from miners, through semi-concealed caterpillars, to free-living caterpillars. We aim to explore how sympatric species-rich genera of tropical trees share caterpillars from different lepidopteran guilds and identify the factors that govern the patterns in host-use through combining defensive trait and phylogenetic data.
2 Methods
2.1 Study system
Ficus (Moraceae) is a pantropical genus including ca 800 species (Cruaud et al., 2012). It is considered to be one of the key-stone tree genera in tropical forests as it supports a high diversity of herbivores, frugivores, pollinators, and their natural enemies (Berg and Corner, 2005; Cruaud et al., 2012; Volf et al., 2020). The genus occurs in both primary and secondary forests and it possesses diverse chemical defenses that show high variability among species (Berg and Corner, 2005; Volf et al., 2018). Macaranga (Euphorbiaceae) is distributed primarily in the secondary forests of the Old-World tropics. Some of its ca 300 species are myrmecophilous and rely on defense by ants (Davies, 2001; Davies et al., 2001). Such strict myrmecophily is rare among the New Guinean species. Additionally, Macaranga species produce chemical defenses including various phenolics (Eck et al., 2001). Syzygium (Myrtcaceae) includes economically important species. The genus is known for being taxonomically challenging. Currently it is estimated at ca 1,200 species (Govaerts et al., 2008; Low et al., 2022). In Papua New Guinea, it occurs mainly in primary forests (Novotny et al., 2010). Being a member of the Myrtaceae family, the genus is known for potent chemical defenses (Dixon et al., 2005; Mohamed et al., 2013). Finally, the genus Psychotria (Rubiaceae) contains over 1,500 species and represents one of the World’s largest flowering plant genera; it is an important component of tropical forests (Sedio et al., 2017). While Psychotria is primarily neotropical there are almost 150 species in New Guinea, 95% of them being endemic. This understory genus is well known for its secondary metabolites, including, among others, the entheogen dimethyltryptamine (Callaway et al., 2005; Cámara-Leret et al., 2020).
2.2 Insect community and phylogenetic data
The insect data were taken from Novotny et al. (2010), these authors sampled all caterpillars from a locally representative selection of 88 host plants in a 10 x 20 km area matrix of primary and secondary lowland rainforest in Madang province, Papua New Guinea. Specifically, the original dataset includes trees and shrubs from plant lineages that comprise a majority of forest woody species in the area. It comprises multiple representatives of locally occurring large genera including Ficus, Macaranga, Psychotria, and Syzygium. In terms of species selection, the dataset focuses on species common in the area that allowed to sample insect herbivores from 1500m2. The insect data include only reared individuals, with host associations confirmed by feeding trials. We focused on Lepidoptera as a large insect order with multiple and well represented feeding guilds. Guilds were defined as follows: leaf miners, semi-concealed caterpillars and free-living caterpillars. Leaf miners feed on the leaf mesophyll, semi-concealed caterpillars include leaf rollers and tiers while free-living caterpillars are exposed (Hrcek et al., 2013). We included recent taxonomic revisions for Choreutidae (Volf et al., 2018) and mining caterpillars (O. Kaman, unpublished data). Singleton and doubleton species were removed from all statistical analyzes because it is impossible to assess host-specificity in cases where one individual is found only on one plant and challenging for doubletons (Novotný and Basset, 2000).
We generated multigene molecular phylogenies for hosts using sequence data generated in previous studies (Novotny et al., 2010; Whitfeld et al., 2012b). We used rbcL sequences in combination with additional sequence data for the following groups: Euphorbiaceae (ndhF), Ficus (ITS) and Rubiaceae (RpS16). We used the resultant topologies as constraints in a larger rbcL only analysis for all 88 plant species from 31 families. We also used the ordinal and family level relationships as derived from the APGIII (2009) tree for constraining the higher-level angiosperm relationships Szefer et al. (2017). We estimated ultrametric phylogenies using BEAST v2.3.1 (Bouckaert et al., 2014) using substitution models derived from jMODEL Test 2 (Darriba et al., 2012) and a relaxed log-normal molecular clock (set to a rate of 1.0). We partitioned our five gene data set, grouping codons one and two (modeling codon three separately) and defined a separate substitution model for each partition. We ran two separate MCMC chains for 20,000,000 generations and assessed the effective sample sizes (ESS) of the combined chains using Tracer v1.6 (Rambaut et al., 2014) to ensure adequate sampling and convergence of the analysis (ESS values over 200 for all parameters sampled). We used the CIPRES portal v3.3 for all phylogenetic analyzes (Miller et al., 2010) and pruned this larger tree to include only our focal species.
2.3 Trait data
Our methods followed Volf et al. (2018), but we give an overview here for clarity. Plant tissue was collected in the field over 251 days between 2013 and 2014, and we sampled leaf discs of 2.4 cm in diameter from 10 young but fully expanded leaves per individual tree for between three to six individuals per species. We avoided sampling from plants heavily damaged by herbivores or pathogens to avoid measuring individuals with strongly induced levels of their defenses. We also avoided sampling from saplings and young trees as we expect that their chemical traits can differ from mature trees. Half of the leaf discs were stored in HPLC grade acetone in order to prevent enzymatic degradation and oxidization of the studied metabolites in the field. They were transferred to a dark -20°C freezer on return to the New Guinea Binatang Research Centre. These discs were later used for secondary metabolite analysis. The other half of leaf discs were weighed fresh and dry in order to estimate both the percentage of water per leaf disc, and the dry weight contained in each tube of acetone. Leaf discs collected for polyphenol analysis (ca 0.6 g of dry leaf tissue in total for each individual: 0.55 g ± 0.22 SD) were stored in 40 ml of HLPC grade acetone. Concentrations were calculated in mg/g dry weight using individual values for each sample because field collected tissue mass varied.
In the laboratory, this first acetone extract was transferred into a 50 ml falcon tube. Five ml of ultrapure water was added, and the solution was concentrated to water phase under a flow of nitrogen at room temperature. The leaf disks were cut into smaller blades and transferred into grinding tubes (DT-50, IKA-Werke GmbH & Co. KG, Germany) containing 35 ml acetone/water (80:20, v/v). The polyphenols were extracted from the leaves by grinding them for 30 min using tube dispensers at room temperature (Ultra-Turrax Tube Drive, IKA-Werke GmbH & Co. KG, Germany). The leaf material was removed and the extract was combined with the water phase obtained from the first acetone extraction above. The combined extract was diluted with acetone to a uniform volume of 50 ml. Polyphenols were extracted from the samples using 50 ml of acetone/water (80:20, v/v). This volume of extract was split, with 10 ml being taken for polyphenol analysis.
To quantify polyphenols, we ran two separate sets of assays. Firstly, we quantified total concentrations of eight main polyphenol sub-groups (in mg/g dry weight) by UPLC-QqQ-MS/MS with the methods of Engström et al. (Engström et al., 2014; Engström et al., 2015) as described in e.g. Malisch et al. (2016). The measured polyphenol sub-groups included hydrolysable tannins that were divided into (1) galloyl derivatives and (2) hexahydroxydiphenoyl derivatives (HDDP, ellagitannins), proanthocyanidins that were divided into (3) procyanidin and (4) prodelphinidin subunits, flavonol glycosides that were divided into (5) kaempferol, (6) quercetin and (7) myricetin derivatives, and (8) quinic acid derivatives. In addition, we ran two activity assays to quantify two major functions of polyphenols in anti-herbivore protection – oxidative activity and protein precipitation capacity. Polyphenol oxidative activity was measured following Salminen and Karonen (2011) using gallic acid as the standard. Protein precipitation capacity was measured following Hagerman’s radial diffusion assay (Hagerman and Butler, 1978) using pentagalloylglucose as the standard. Both assays provided activities in mg/g dry weight.
The total number of trichomes per 10 mm2 and their average length was measured on five leaf discs per individual tree (from five leaves per tree), avoiding the central vein. We used a DinoLite USB microscope to draw on a circle of the appropriate diameter and to quantify trichome density and length. Values for dorsal and ventral sides of the discs were averaged. Specific Leaf Area (SLA) was measured as the area per mass using ten dried leaf discs (twenty for Ficus, due to tissue being collected for other studies) from ten leaves per individual tree, which were cut avoiding the central vein. Total carbon and nitrogen content were determined by dry combustion using ca 0.6 g of homogenised dry leaf material across the same leaves sampled for polyphenols.
2.4 Statistical analysis
We calculated the rescaled distance-based specialization index (DSI*) (Jorge et al., 2014; Jorge et al., 2017) for each herbivore guild using host phylogenetic distance, polyphenol distance (Euclidean distance derived from a matrix quantifying the concentration of eight polyphenol sub-groups and the two types of activities) and mechanical distance (Euclidean distance derived from C:N, SLA, mean trichomes per disc and mean trichome length). DSI weights the degree of specialization by the phylogenetic and trait similarity of hosts and their availability, rather than using counts of host species, or higher taxonomic categories. In this sense, DSI measures distance-based specialization accounting for differences in the pool of available hosts, such variation in their polyphenol chemistry or phylogenetic relatedness. The rescaled DSI index (DSI*), accounts for differences in abundances and sampling intensities. Rescaled DSI has upper bounds of 1 (monophagy) and the lower bounds –1 (maximum generalization).
We used generalized linear mixed models with a beta error term and logit link function to test for differences in phylogenetic, polyphenol and mechanical specialization among guilds. The response variable in each model was DSI* scaled to be within 0.00001 and 0.99999 (to satisfy the requirements of beta-regression) five models were compared using AIC, each model included species as a random effect. The simplest model had an intercept of one while the next model included only guild as an explanatory variable, the third model included only the DSI metric (phylogenetic, polyphenol or mechanical) and the fourth included both previous explanatory variables together. The fifth model included the interaction term between these two variables. All models were fitted in the R package ‘glmmTMB’ (Magnusson et al., 2017) and models were compared using an AIC table generated using the R package ‘bbmle’ (Bolker and Bolker, 2016). Testing of model fit was conducted using the R package ‘DHARMa’ (Hartig and Hartig, 2017) (specifically: dispersion, homogeneity of variance in the residuals, correct distribution test and distribution of outliers). The model with the lowest AIC was examined further with type II ANOVA: Wald Chi-square tests were used to the summarize overall significance of the variables and pair wise tests using a Tukey adjustment were used to compare significance across both categorical variables simultaneously (implemented using the R package ‘emmeans’ (Lenth et al., 2020).
Further, we calculated the mean insect specialization per host plant (e.g., the sum of DSI* for each insect herbivore divided by the number of species on that host). Overlap of insect communities across host genera was visualized by plotting the mean of shared species among congeners and among Ficus, Macaranga, Psychotria and Syzygium. Here, 0 means that not a single species overlaps with another species or genus, whereas 1.0 means that all members of a guild are shared with other hosts. We constructed food-webs for each guild using the R package ‘bipartite’ (Dormann et al., 2008). We also calculated connectedness and modularity for each food web.
We used Principal Components Analysis (PCA) to summarize all 14 defensive traits measured across the 34 host plant species (Supplementary Table 1) as implemented in the R package ‘vegan’ (Oksanen et al., 2019). We included an additional variable, successionary index, to account for host habitat. Succesionary index ranges from 0 to 100 and represented the % of basal area in primary forest plots (data derived from Lepš et al., 2001)). Community data from all three guilds were combined for this analysis. Species were scaled to unit variance. Further, we performed Canonical Correspondence Analysis (CCA) to test for correlations between insect community composition across host species and the defensive variables. Model simplification proceeded in both directions, used 999 permutations and specified R2 scope as true and was implemented using the ‘ordistep’ function in the R package ‘vegan’.
3 Results
The mixed model with the lowest AIC included both guild and specialization metric (phylogenetic, polyphenol or mechanical) as explanatory variables describing the deviance in DSI*.
No interaction term was included. Both guild (χ2 = 45.3432,454, p<0.001) and specialization metric (χ2 = 80.2032,454, p<0.0001) explained significant amounts of deviance in DSI*. Miners were significantly more specialized than either semi-concealed (z-ratio= 6.651, p<0.0001) or free-living chewers (z-ratio=-5.347, p<0.0001) but semi-concealed and free-living chewers did not differ significantly in their specialization (z-ratio= 0.791, p= 0.7083). Specialization metric differed significantly in each pairwise comparison (polyphenol-mechanical: z-ratio= 4.317, p=0.0001, polyphenol-phylogenetic: z-ratio= -5.049, p<0.0001 and mechanical-phylogenetic: z-ratio=-8.955, p<0.0001). Miners were more specialized than the other guilds for all metrics and specialization metric takes the following order: phylogenetic>polyphenol>mechanical (Figure 1). The best model was diagnosed as having goodness of fit, indices indicated no significant difference between observed and measured dispersion, had uniform variance and there were no outliers with extreme influence. The quantile-quantile plot demonstrated visual normality of residuals, although there was significant deviation in the correct distribution test. The same model with a Gaussian error term had extreme deviation in variance and normality of residuals as well as outliers with extreme influence.
Figure 1
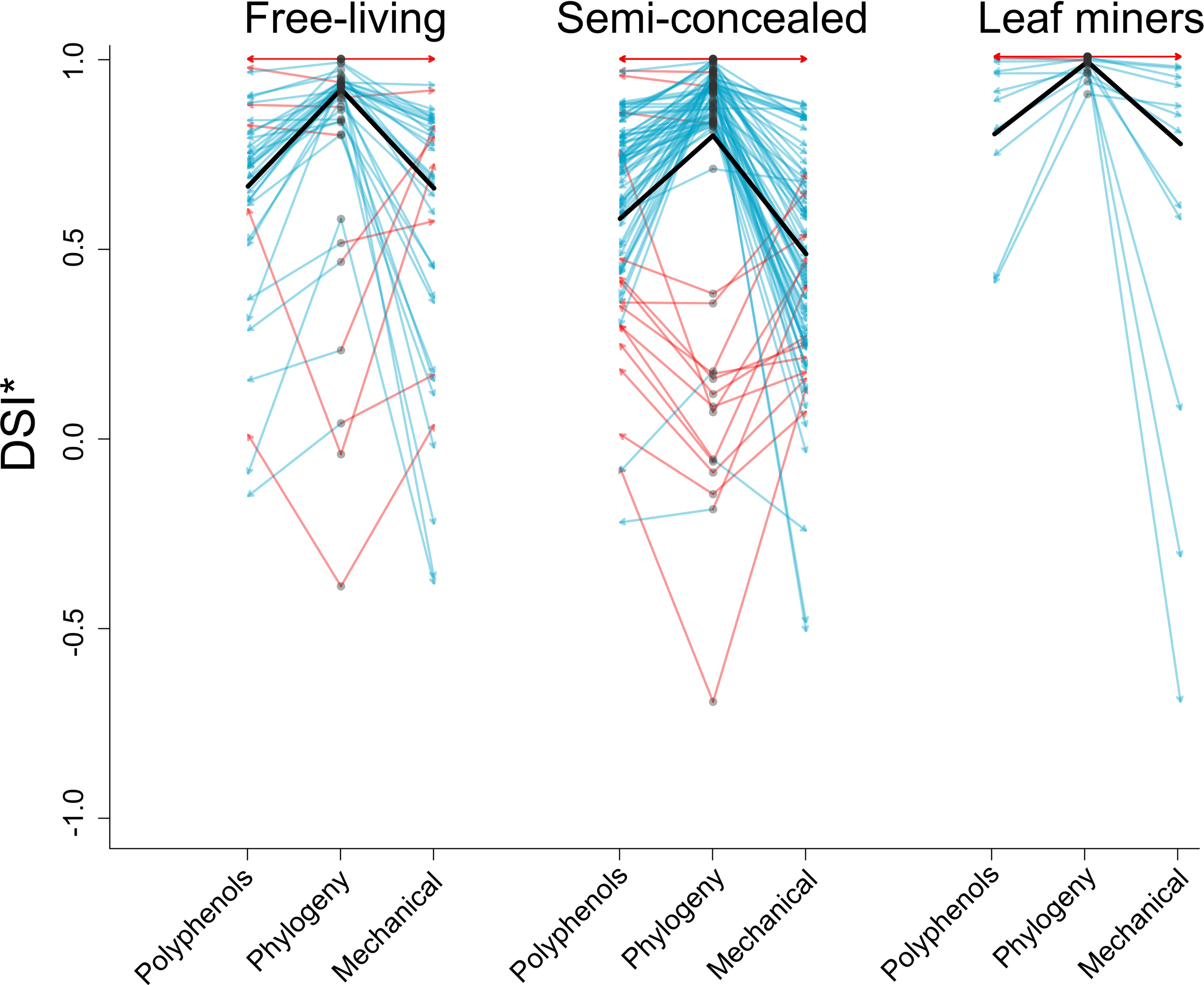
Plot showing rescaled specialization DSI* of leaf mining, free-living and semi-concealed Lepidopteran larval leaf-chewing guilds. Grey circles represent a species’ phylogenetic specialization, arrows to the left and right indicate specialization toward polyphenols and specialization toward mechanical defense. Blue arrows represent cases where either polyphenol or mechanical specialization are lower than phylogenetic specialization and red arrows represent the inverse scenario. Black bars represent means.
Plant mean specialization varied across guild and metric. Macaranga herbivores were notable for their low specialization toward polyphenols and mechanical defenses. In contrast, Ficus herbivores showed the highest mean phylogenetic and polyphenol specialization of all plant genera studied. Syzygium insect herbivores were most specialized toward mechanical defenses (Figure 2).
Figure 2
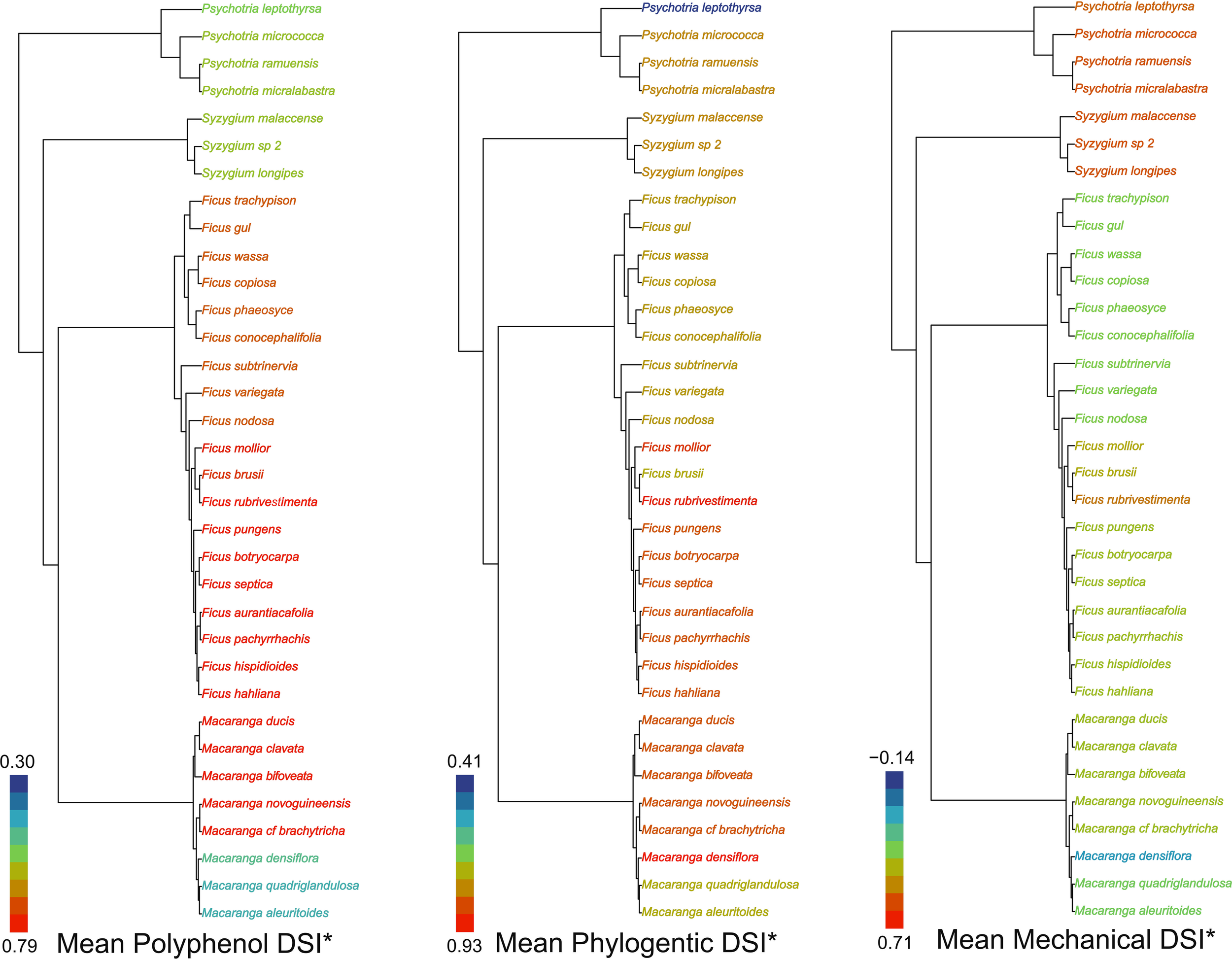
Plant mean specialization: i) polyphenol chemistry, ii) phylogeny and iii) mechanical traits. The tips of the phylogeny are colored such that warm (orange red to yellow green) colours represent high relative specialization of the herbivore community on each plant species and cooler ones (royal blue to light sky blue) low relative specialization (values indicate maximum and minimum DSI*).
Host specificity varied across guilds when considering the overlap of insect species (Figure 3). For example, there was not a single miner species shared between host species from different genera. In contrast, the congeneric host species on average shared ca 75% of their miner species and more than 90% of their free-living and semi-concealed caterpillars. Food webs were generally modular, with miners being restricted to one host genus (modularity of 0.88, connectedness of 0.07) while the food webs for free-living (modularity of 0.58, connectedness of 0.11) and semi-concealed (modularity of 0.66, connectedness of 0.16) caterpillars were more connected (Figure 4).
Figure 3
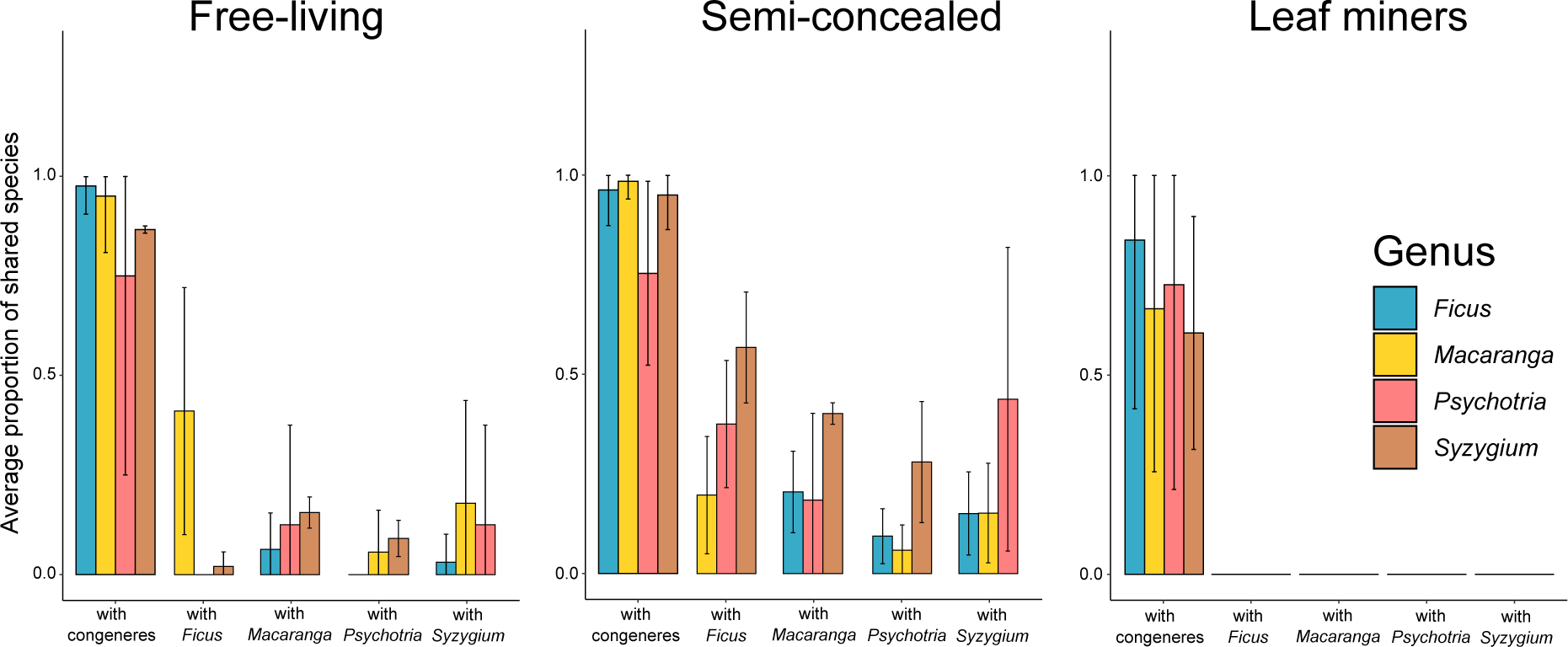
Overlap of free-living caterpillars, semi-concealed caterpillars, and leaf miners among Ficus, Macaranga, Psychotria and Syzygium. A value of 1.0 indicates that all of the herbivore species on the given plant species are shared with at least one other host species either within or among genera.
Figure 4
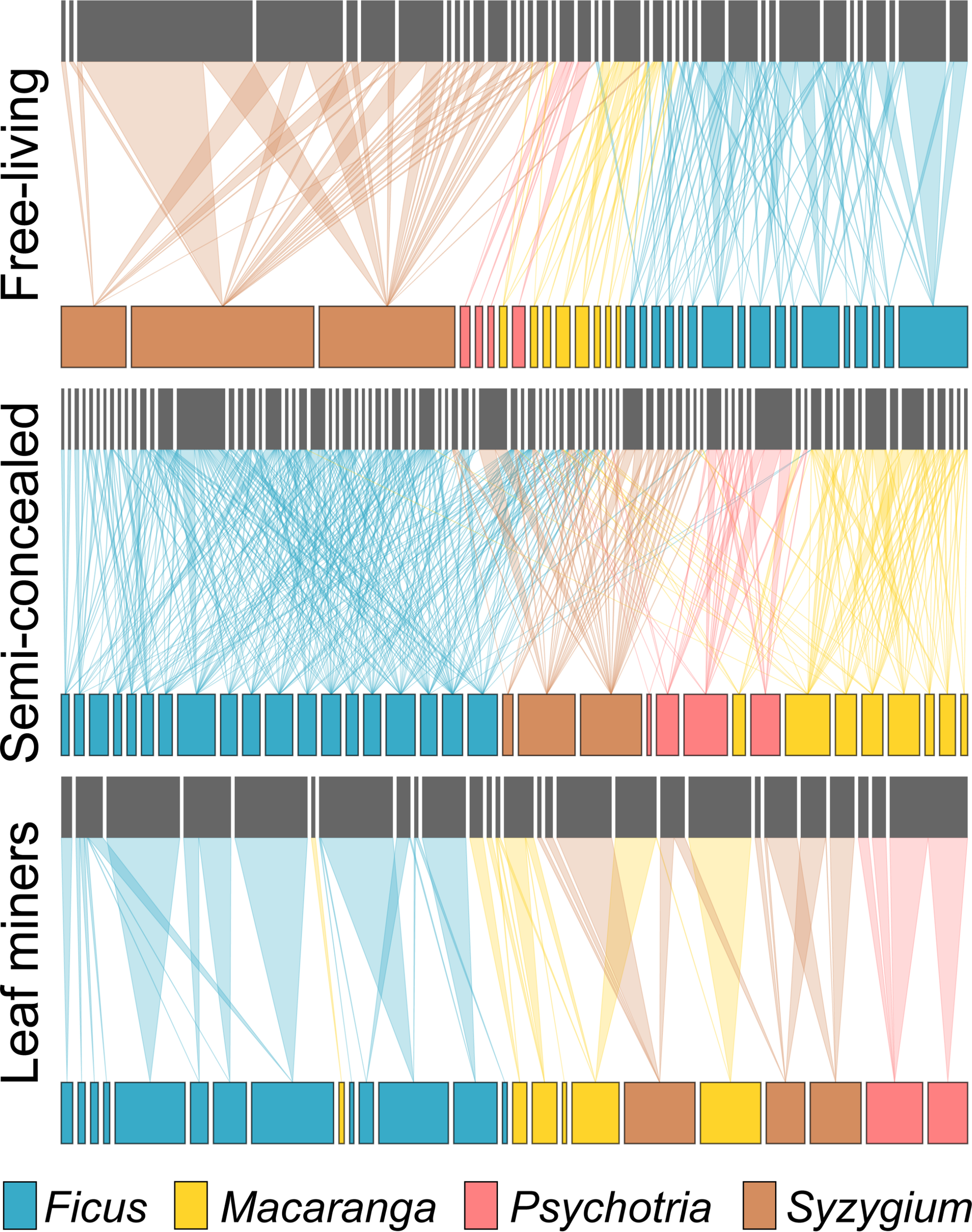
Food webs free-living caterpillars, semi-concealed caterpillars, and leaf miners.
The first two principal components in the PCA analysis explained 48.7% of the total inertia in the studied traits across the host species (Figure 5A). The best CCA model explaining the inertia in herbivore communities consisted of the variables HHDPs (F=1.3825, p=0.062), galloyl derivatives (F=1.5191, p=0.023), trichomes per disc (F=1.5763, p=0.010), quinic acid derivatives (F=1.4402, p=0.019), CN (F=1.4509, p=0.025), myricetins (F=2.3237, p=0.001) and successional index (F=1.3319, p=0.074). These variables together explained 34.8% of the total inertia in the model (χ2 = 4.35626,27, F=1.9783, p<0.001). The first two CCA axes explained 14.8% of the total inertia (Figure 5B).
Figure 5
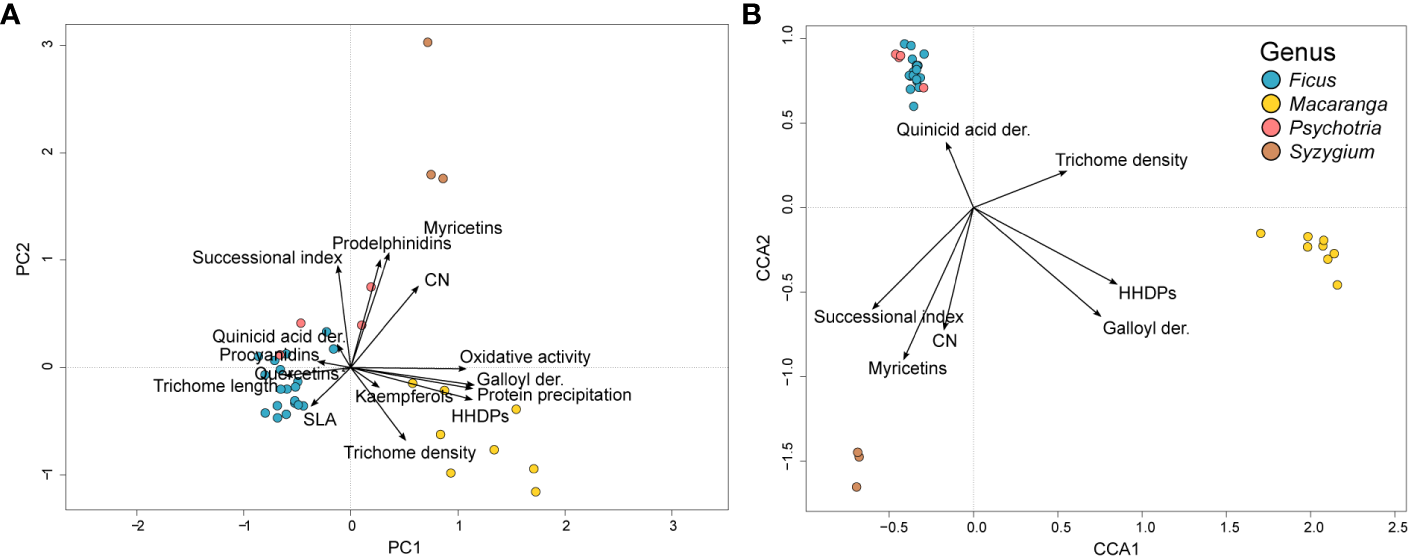
Trait variation among the focal tree species as analyzed with PCA (A) and the correlation between traits and insect communities as analyzed with CCA (B). The first two principal components explained 48.7% of the total inertia in trait variation across the tree species when analyzed with PCA. The CCA ordination diagram shows the traits best explaining the variation in herbivore communities. Together, these traits explained 34.8% of the total inertia in the model. Traits are shown as black arrows. Tree species appear as circles colored according to the genus.
4 Discussion
Species-rich plant genera represent an important part of woody plant diversity in tropical forests (Gentry, 1982; Foster and Hubbell, 1990). These ‘species swarms’ provide resources for a non-trivial component of terrestrial diversity: herbivorous insects. Fundamental to estimating insect species richness is their host-specificity, understanding their diversification requires placing this into a phylogenetic context. In this study we explored phylogenetic and functional patterns of specialization in insect herbivores associated with such hosts, and analyzed what host traits govern these patterns. Overall, our study shows that Ficus, Macaranga, Psychotria, and Syzygium harbor largely distinct herbivore assemblages, with the four genera serving as distinct host islands that support insect diversity in the forest (Novotny et al., 2004). Novotny and Basset (2005) found that only 27% of insect herbivores appear to be monophagous in tropical forest communities where several congeneric hosts are present. Here the recovered numbers of caterpillar species shared among the congeneric hosts are even higher. This suggests that host-shifts may be particularly frequent in caterpillars using species-rich plant genera, as may be the case for other even more generalist herbivores such as adult Coleoptera and Orthoptera.
Insect specialization was most strongly associated with host phylogeny and polyphenol traits conserved at the level of genus. Mechanical traits we measured played a smaller role, apart from in Syzygium. Polyphenols affect insect herbivores through protein precipitation and oxidative stress. In caterpillars that have high pH in their mid-gut, the protein precipitation is likely limited. Polyphenol oxidative activity is thus supposed to play a more important role (Salminen and Karonen, 2011; Salminen, 2014). Indeed, the multivariate analyzes picked hydrolysable tannins (HHDPs) with high oxidative activity as one of the traits affecting assemblages of caterpillars associated with hosts from the four genera we studied. A distinct polyphenol profile also characterized Syzygium, which was largely driven by the presence of myricetins. The dominance of myricetins has previously been reported in Syzygium cumini (Chagas et al., 2018) and these likely influence insect host use and shape food web modularity. Yet, the fact that insect specialization was more closely related to host phylogeny than to chemical differences quantified based on polyphenols or physical traits suggests that phylogenetically conserved, but unmeasured, traits are important predictors of host use.
We focused our chemical analyzes on polyphenols that are known to structure insect communities and provide anti-herbivore defense in all four genera we studied (Sagers and Coley, 1995; Segar et al., 2017; Volf et al., 2018). While our targeted approach works well when considering a subset of (well-studied) chemical diversity for which commercial standards are widely available, it fails to capture the higher order complexity of the sample. Our focal plant hosts also possess characteristic specialized metabolites. For example, Ficus produces phenanthroindolizidine alkaloids and leaf proteases that affect its interactions with insect herbivores (Volf et al., 2018; Volf et al., 2020). Phenanthroidolizidine alkaloids are even sequestered by some of Ficus-specialized herbivores and largely govern their host choice (Fontanilla et al., 2022). Specialized metabolites showing some level of lineage-specificity can be also found in the other studied genera, such as indole alkaloids in Psychotria (Gregianini et al., 2004). Unfortunately, we do not have data to analyze the effect of these metabolites on the differences in insect specialization and assemblage structure between Ficus, Macaranga, Psychotria, and Syzygium. However, results from other studies suggest a key role of lineage specific metabolites in promoting turnover of insect species among host-plants (Richards et al., 2015; Volf et al., 2020). Together with the variation that occurs among congeneric species, these trends make chemical variation among tropical plants one of the key factors supporting diversity of insect herbivores in tropical forests (Richards et al., 2015; Endara et al., 2017; Salazar et al., 2018). By quantifying polyphenol groups, we were able to detect correlations among them, this allowed us to explain the distribution of insects across host plants with a higher degree of accuracy than using measures of total content. Different compound groups influence insect feeding in different ways.
We expected that host-plant habitat preferences could explain significant amount of variation in insect assemblages in addition to the variation explained by the chemical and physical traits of the hosts. The studied tree species differ in their preference for primary and secondary forests where both biotic and abiotic can be profoundly different (Fibich et al., 2016). Habitat preferences are also correlated with variation in leaf chemistry, which could be another factor affecting the herbivores (Sedio et al., 2017). Our multivariate analyzes did find an effect of host-plant preference for primary vs. secondary forest on the associated insect assemblages, explained largely by the distinct assemblage on Syzygium. This relationship, however was not strong. This suggests that, when accounting for host-plant traits, habitat differences between primary and secondary forests per se may play a smaller role in structuring caterpillar assemblages than in other insect groups such as ants (Holloway et al., 1992; Klimes et al., 2012). This can be due to the low correlation between host palatability and successional status in tropical forests (Lepš et al., 2001). Here, the observed differences in insect herbivores between hosts preferring either primary or secondary forests were mainly associated with host species from the genera Macaranga and Syzygium. Species from Macaranga harbored insects with low polyphenol and mechanical specialization, those feeding on Syzygium showed specialization toward mechanical defenses. The presence of such host lineages rather than systematic underlying effects can be responsible for a substantial share of differences in specialization and species composition of herbivorous insects between primary and secondary forests (Lepš et al., 2001).
As expected, our results also show important differences in the specialization of various insect guilds associated with the four large genera. The miners were most specialized in all regards, while there was little difference in mean DSI* indices between free-living and semi-concealed caterpillars. These patterns follow the overall guild specialization patterns recovered in tropical forests (Novotny et al., 2010). However, there was also variation within guilds. For example, a sizeable subset of semi-concealed caterpillars was more polyphenol specialized than phylogenetically specialized, illustrating potential convergence of caterpillars onto chemically similar plants. Certain caterpillar lineages seem to be tracking hosts with similar polyphenol profiles. For example, Segar et al. (2017) showed that geometrids predominantly occurred on hosts with high oxidative activity of polyphenols. We observed that caterpillars from the family Tortricidae are frequently associated Syzygium while Dichomeris spp. (Gelechiidae) are typically associated with Macaranga (within the context of this study at least). While chemical turnover among congeneric hosts (chemical β-diversity) is more pronounced in some genera (e.g., Ficus) than others (e.g., Macaranga), restricted metabolites drive insect herbivore host use. For example, Macaranga is distinct among the four genera studied here yet each Macaranga species harbours a similar insect community and has similar chemical composition. DSI* measures the deviation of the diet of a given herbivore from the pool of available hosts and it accounts for any imbalance in taxonomic composition of the sampled species. Despite our findings are thus robust, we advocate wider and more balanced sampling of these genera. While inter-generic variation largely exceeds inter-generic variation in our samples, we acknowledge that more balanced sampling would improve the reliability of our multivariate analyzes and strengthen conclusions relating to chemical β-diversity.
In conclusion, we show how speciose genera of tropical woody plants harbor largely different caterpillar communities. As proposed by Janzen (1968) such plant lineages can be seen as islands for their herbivore fauna. Host shifts among congeneric hosts from the Ficus, Macaranaga, Psychotria, and Syzygium seem to be frequent even in highly specialized guilds, such as miners. The overall diversity of insects harbored by individual genera may be particularly affected by their species diversity and variation in functional traits that govern herbivore host specialization (Richards et al., 2015; Volf et al., 2019; Narango et al., 2020). Here, caterpillar specialization seems to be more closely related to phylogenetic relationships between the hosts than to the host traits or successional preference. In particular, while traits such as polyphenols or trichomes that occur across the studied host genera explained some variation in the composition of caterpillar assemblages, we expect that most of the effect of phylogeny we recovered was driven by traits that we did not measure. In miners that are intimately associated with their hosts, such traits can include leaf morphology or host susceptibility to manipulation (Zhang et al., 2016). Indeed, many insect species can manipulate plant defenses (Giron et al., 2016). While our field survey was not designed to detect induced defenses (but rather to avoid manipulated leaves and hosts), we have to expect that some variation can be attributed to up- or downregulated polyphenol defenses (Eyles et al., 2010). We suggest that experiments in more controlled conditions would help to unravel these effects further. Another large share of the variation can be likely attributed to lineage specific specialized metabolites. Rapidly developing fields of metabolomics and bioinformatics offer new possibilities for measuring concentrations of thousands of metabolites in non-model plant species and assessing their structural diversity (Sedio, 2017). Approaches that consider structural or spectral diversity of metabolites are particularly promising in this regard. This is because they allow to quantify chemical variation among distantly related plants that show only a minimal overlap in the content of individual metabolites (Sedio et al., 2017; Philbin et al., 2022). Combining metabolomic data with information on other types of data, such as other plant traits and habitat preferences that we studied here, can help us to understand the foundational role of speciose plant genera in tropical forest food webs. Greater mechanistic understanding of ecological network structure will ultimately lead to more scientifically informed conservation practices as well as insights into the origins of plant ‘species swarms’ and their associated insect communities.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
SS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. LRJ: Conceptualization, Formal Analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. LN: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. YB: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – review & editing. JR: Data curation, Investigation, Writing – review & editing. OK: Formal Analysis, Investigation, Methodology, Writing – review & editing. MS: Data curation, Investigation, Methodology, Writing – review & editing. BG: Data curation, Investigation, Methodology, Writing – review & editing. CD: Data curation, Investigation, Methodology, Writing – review & editing. PB: Formal Analysis, Investigation, Methodology, Software, Writing – review & editing. PM: Visualization, Writing – review & editing. SM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing. GW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing – review & editing. J-PS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing. VN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. MV: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding VN, MV, and SS acknowledge the Grant Agency of the Czech Republic (grant number 19-28126X). VN acknowledges support from the European Research Council grant no. 669609. SS acknowledges departmental support from Harper Adams University. J-PS acknowledges funding from the Academy of Finland. This material is based upon work supported by the U.S. National Science Foundation under DEB 9707928, 0211591 and 0515678, 0816749 and 0841885. DNA barcoding was provided by the Biodiversity Institute of Ontario, University of Guelph, with funding from Genome Canada and the Ontario Genomics Institute to the International Barcode of Life Project.
Acknowledgments
We thank the assistants, taxonomists, and the staff of the New Guinea Binatang Research Centre in Papua New Guinea. We also thank the Papua New Guinea Forest Research Institute and the Department of Environment and Conservation for their help in getting plant and insect export permits, and the lab members of the Natural Chemistry Research Group at the University of Turku, Finland for their assistance with chemical analyzes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1308608/full#supplementary-material
SUPPLEMENTARY TABLE 1Segar, Simon (2023). Interaction_matrix_large_genera_310723.xlsx. figshare. Dataset. https://doi.org/10.6084/m9.figshare.24191136.v1SUPPLEMENTARY TABLE 2Segar, Simon (2023). Large Genera_171123.R. figshare. Software. https://doi.org/10.6084/m9.figshare.24191184.v1; Segar, Simon (2023). Means_PNG results 280618_FORMATTED_MV.xlsx. figshare. Dataset. https://doi.org/10.6084/m9.figshare.24191133.v1SUPPLEMENTARY TABLE 3Segar, Simon (2023). PNG_Plants_2_SUBST.tre. figshare. Dataset. https://doi.org/10.6084/m9.figshare.24191187.v1References
1
AliJ. G.AgrawalA. A. (2012). Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci.17, 293–302. doi: 10.1016/j.tplants.2012.02.006
2
Angiosperm Phylogeny Group. (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical J. Linn. Soc.161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
3
AntonelliA.FryC.SmithR. J.EdenJ.GovaertsR. H.A.KerseyP.et al. (2023). State of the world’s plants and fungi 2023 (Kew, UK: Royal Botanic Gardens). doi: 10.34885/wnwn-6s63
4
BassetY.NovotnyV.WeiblenG. (1997). “Ficus: a resource for arthropods in the tropics, with particular reference to New Guinea,” in Forests and insects. Eds. WattD. A.StorkN. E.HunterM. D. (London: Chapman & Hall), 341–361.
5
BergC.CornerE. (2005). Moraceae (Ficus). Flora malesiana, series I (Seed plants) Vol. 17 (Leiden: National Herbarium of the Netherlands).
6
BolkerB.BolkerM. (2016). Package “bbmle”: tools for general maximum likelihood estimation. R package version 1. Available at: https://cran.r-project.org/web/packages/bbmle/index.html.
7
BouckaertR.HeledJ.KühnertD.VaughanT.WuC. H.XieD.et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PloS Comput. Biol.10, e1003537. doi: 10.1371/journal.pcbi.1003537
8
CallawayJ. C.BritoG. S.NevesE. S. (2005). Phytochemical analyses of Banisteriopsis caapi and Psychotria viridis. J. Psychoactive Drugs37, 145–150. doi: 10.1080/02791072.2005.10399795
9
Cámara-LeretR.FrodinD. G.AdemaF.AndersonC.AppelhansM. S.ArgentG.et al. (2020). New Guinea has the world’s richest island flora. Nature584, 579–583. doi: 10.1038/s41586-020-2549-5
10
CardenasR. E.ValenciaR.KraftN. J.ArgotiA.DanglesO. (2014). Plant traits predict inter-and intraspecific variation in susceptibility to herbivory in a hyperdiverse Neotropical rain forest tree community. J. Ecol.102, 939–952. doi: 10.1111/1365-2745.12255
11
ChagasV. T.CoelhoR.GasparR. S.Da SilvaS. A.MastrogiovanniM.MendoncaC. J.et al. (2018). Protective Effects of a Polyphenol-Rich Extract from Syzygium cumini (L.) Skeels Leaf on Oxidative Stress-Induced Diabetic Rats. Oxid. Med. Cell Longev.2018, 5386079. doi: 10.1155/2018/5386079
12
ColeyP. D. (1983). Herbivory and defensive characteristics of tree species in a lowland tropical forest. Ecol. Monogr.53, 209–234. doi: 10.2307/1942495
13
ConnB. (2008) Census of vascular plants of papua new Guinea. Available at: http://www.pngplants.org/PNGCensus.
14
ConnorE. F.TavernerM. P. (1997). The evolution and adaptive significance of the leaf-mining habit. Oikos79, 6–25. doi: 10.2307/3546085
15
CruaudA.RønstedN.ChantarasuwanB.ChouL. S.ClementW. L.CoulouxA.et al. (2012). An extreme case of plant-insect codiversification: Figs and fig-pollinating wasps. Systematic Biol.61, 1029–1047. doi: 10.1093/sysbio/sys068
16
DarribaD.TaboadaG. L.DoalloR.PosadaD. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods9, 772–772. doi: 10.1038/nmeth.2109
17
DaviesS. J. (2001). Systematics of macaranga sects. Pachystemon and pruinosae (Euphorbiaceae). Harvard Papers Bot.6 (2), 371–448.
18
DaviesS. J.LumS. K.ChanR.WangL. (2001). Evolution of myrmecophytism in western Malesian Macaranga (Euphorbiaceae). Evolution55, 1542–1559. doi: 10.1111/j.0014-3820.2001.tb00674.x
19
DixonR. A.XieD. Y.SharmaS. B. (2005). Proanthocyanidins–a final frontier in flavonoid research? New Phytol.165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
20
DormannC. F.GruberB.FründJ. (2008). Introducing the bipartite package: analysing ecological networks. R News8, 8–11.
21
EckG.FialaB.LinsenmairK. E.Bin HashimR.ProkschP. (2001). Trade-off between chemical and biotic antiherbivore defense in the South East Asian plant genus Macaranga. J. Chem. Ecol.27, 1979–1996. doi: 10.1023/A:1012234702403
22
EndaraM.-J.ColeyP. D.GhabashG.NichollsJ. A.DexterK. G.DonosoD. A.et al. (2017). Coevolutionary arms race versus host defense chase in a tropical herbivore–plant system. Proc. Natl. Acad. Sci. U.S.A.114, E7499–E7505. doi: 10.1073/pnas.1707727114
23
EndaraM.-J.ForristerD. L.ColeyP. D. (2023). The evolutionary ecology of plant chemical defenses: from molecules to communities. Annu. Rev. Ecol. Evolution Systematics54, 107–127. doi: 10.1146/annurev-ecolsys-102221-045254
24
EngströmM. T.PälijärviM.FryganasC.GrabberJ. H.Mueller-HarveyI.SalminenJ.-P. (2014). Rapid qualitative and quantitative analyses of proanthocyanidin oligomers and polymers by UPLC-MS/MS. J. Agric. Food Chem.62, 3390–3399. doi: 10.1021/jf500745y
25
EngströmM. T.PälijärviM.SalminenJ.-P. (2015). Rapid fingerprint analysis of plant extracts for ellagitannins, gallic acid, and quinic acid derivatives and quercetin-, kaempferol- and myricetin-based flavonol glycosides by UPLC-QqQ-MS/MS. J. Agric. Food Chem.63, 4068–4079. doi: 10.1021/acs.jafc.5b00595
26
EylesA.BonelloP.GanleyR.MohammedC. (2010). Induced resistance to pests and pathogens in trees. New Phytol.185, 893–908. doi: 10.1111/j.1469-8137.2009.03127.x
27
FibichP.LepšJ.NovotnýV.KlimešP.TěšitelJ.MolemK.et al. (2016). Spatial patterns of tree species distribution in New Guinea primary and secondary lowland rain forest. J. Vegetation Sci.27, 328–339. doi: 10.1111/jvs.12363
28
FontanillaA. M.AubonaG.SisolM.KuukkanenI.SalminenJ. P.MillerS. E.et al. (2022). What goes in must come out? The metabolic profile of plants and caterpillars, frass, and adults of Asota (Erebidae: Aganainae) feeding on Ficus (Moraceae) in New Guinea. J. Chem. Ecol.48, 718–729. doi: 10.1007/s10886-022-01379-x
29
ForisterM. L.NovotnyV.PanorskaA. K.BajeL.BassetY.ButterillP. T.et al. (2015). The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. U.S.A.112, 442–447. doi: 10.1073/pnas.1423042112
30
FosterR. B.HubbellS. P. (1990). “The floristic composition of the Barro Colorado Island forest,” in Four neotropical rainforests. Ed. GentryA. H. (New Haven, Connecticut, USA: Yale University Press), 85–98.
31
GentryA. H. (1982). Neotropical floristic diversity: phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Ann. Missouri Botanical Garden69, 557–593. doi: 10.2307/2399084
32
GironD.HuguetE.StoneG. N.BodyM. (2016). Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J. Insect Physiol.84, 70–89. doi: 10.1016/j.jinsphys.2015.12.009
33
GovaertsR.SobralM.AshtonP.BarrieF.HolstB. K.LandrumL. L.et al. (2008). World checklist of myrtaceae (Royal Botanic Gardens: University of Chicago Press, Chicago).
34
GregianiniT. S.PortoD. D.NascimentoN. C. D.FettJ. P.HenriquesA. T.Fett-NetoA. G. (2004). Environmental and ontogenetic control of accumulation of brachycerine, a bioactive indole alkaloid from Psychotria brachyceras. J. Chem. Ecol.30, 2023–2036. doi: 10.1023/B:JOEC.0000045592.24785.33
35
HagermanA. E.ButlerL. G. (1978). Protein precipitation method for the quantitative determination of tannins. J. Agric. Food Chem.26, 809–812. doi: 10.1021/jf60218a027
36
HartigF.HartigM. F. (2017). Package ‘DHARMa’. R package. Available at: https://cran.r-project.org/web/packages/DHARMa/index.html.
37
HöftR. (1992). Plants of New Guinea and the Solomon Islands: dictionary of the genera and families of flowering plants and ferns Vol. 3 (Port Moresby, Papua New Guinea: University of Papua New Guinea Press).
38
HollowayJ.Kirk-SpriggsA.KhenC. V. (1992). The response of some rain forest insect groups to logging and conversion to plantation. Philos. Trans. R. Soc. London Ser. B: Biol. Sci.335, 425–436. doi: 10.1098/rstb.1992.0034
39
HrcekJ.MillerS. E.WhitfieldJ. B.ShimaH.NovotnyV. (2013). Parasitism rate, parasitoid community composition and host specificity on exposed and semi-concealed caterpillars from a tropical rainforest. Oecologia173, 521–532. doi: 10.1007/s00442-013-2619-6
40
JanzenD. H. (1968). Host plants as islands in evolutionary and contemporary time. Am. Nat.102, 592–595. doi: 10.1086/282574
41
JiaS.YangX.CastagneyrolB.YangL.YinQ.HeC.et al. (2023). Neighbouring tree effects on leaf herbivory: Insect specialisation matters more than host plant leaf traits. J. Ecol. 00, 1–11. doi: 10.1111/1365-2745.14228
42
JorgeL. R.NovotnyV.SegarS. T.WeiblenG. D.MillerS. E.BassetY.et al. (2017). Phylogenetic trophic specialization: a robust comparison of herbivorous guilds. Oecologia185, 551–559. doi: 10.1007/s00442-017-3980-7
43
JorgeL. R.PradoP. I.Almeida-NetoM.LewinsohnT. M. (2014). An integrated framework to improve the concept of resource specialisation. Ecol. Lett.17, 1341–1350. doi: 10.1111/ele.12347
44
KlimesP.IdigelC.RimandaiM.FayleT. M.JandaM.WeiblenG. D.et al. (2012). Why are there more arboreal ant species in primary than in secondary tropical forests? J. Anim. Ecol.81, 1103–1112. doi: 10.1111/j.1365-2656.2012.02002.x
45
KurokawaH.NakashizukaT. (2008). Leaf herbivory and decomposability in a Malaysian tropical rain forest. Ecology89, 2645–2656. doi: 10.1890/07-1352.1
46
KursarT. A.DexterK. G.LokvamJ.PenningtonR. T.RichardsonJ. E.WeberM. G.et al. (2009). The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc. Natl. Acad. Sci. U.S.A.106, 18073–18078. doi: 10.1073/pnas.0904786106
47
LenthR.SingmannH.LoveJ.BuerknerP.HerveM. (2020). Package ‘emmeans’ R topics documented: R Package Version 1. 15, Vol. 15, 1. Available at: https://cran.r-project.org/web/packages/emmeans/index.html.
48
LeongJ. V.JorgeL. R.SeifertC. L.VolfM. (2022). Quantity and specialisation matter: Effects of quantitative and qualitative variation in willow chemistry on resource preference in leaf-chewing insects. Insect Conserv. Diversity15, 453–460. doi: 10.1111/icad.12559
49
LepšJ.NovotnýV.BassetY. (2001). Habitat and successional status of plants in relation to the communities of their leaf-chewing herbivores in Papua New Guinea. J. Ecol.89, 186–199. doi: 10.1046/j.1365-2745.2001.00540.x
50
López-CarreteroA.BoegeK.Díaz-CastelazoC.DomínguezZ.Rico-GrayV. (2016). Influence of plant resistance traits in selectiveness and species strength in a tropical plant-herbivore network. Am. J. Bot.103, 1436–1448. doi: 10.3732/ajb.1600045
51
LowY. W.RajaramanS.TomlinC. M.AhmadJ. A.ArdiW. H.ArmstrongK.et al. (2022). Genomic insights into rapid speciation within the world’s largest tree genus Syzygium. Nat. Commun.13, 5031. doi: 10.1038/s41467-022-32637-x
52
MagnussonA.SkaugH.NielsenA.BergC.KristensenK.MaechlerM.et al. (2017). Package ‘glmmtmb’. R package version. Available at: https://cran.r-project.org/web/packages/glmmTMB/index.html.
53
MalischC. S.SalminenJ.-P.KoüllikerR.EngströmM. T.SuterD.StuderB.et al. (2016). Drought effects on proanthocyanidins in sainfoin (Onobrychis viciifolia Scop.) are dependent on the plant’s ontogenetic stage. J. Agric. Food Chem.64, 9307–9316. doi: 10.1021/acs.jafc.6b02342
54
MillerM. A.PfeifferW.SchwartzT. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in 2010 gateway computing environments workshop (GCE) (New Orleans, LA, USA: Ieee), 1–8.
55
MoctezumaC.HammerbacherA.HeilM.GershenzonJ.Méndez-AlonzoR.OyamaK. (2014). Specific polyphenols and tannins are associated with defense against insect herbivores in the tropical oak Quercus oleoides. J. Chem. Ecol.40, 458–467. doi: 10.1007/s10886-014-0431-3
56
MohamedA. A.AliS. I.El-BazF. K. (2013). Antioxidant and antibacterial activities of crude extracts and essential oils of Syzygium cumini leaves. PloS One8, e60269. doi: 10.1371/journal.pone.0060269
57
NarangoD. L.TallamyD. W.ShropshireK. J. (2020). Few keystone plant genera support the majority of Lepidoptera species. Nat. Commun.11, 1–8. doi: 10.1038/s41467-020-19565-4
58
NovotnýV.BassetY. (2000). Rare species in communities of tropical insect herbivores: pondering the mystery of singletons. Oikos89, 564–572. doi: 10.1034/j.1600-0706.2000.890316.x
59
NovotnyV.BassetY. (2005). Host specificity of insect herbivores in tropical forests. Proc. R. Soc. London B: Biol. Sci.272, 1083–1090. doi: 10.1098/rspb.2004.3023
60
NovotnyV.MillerS. E.LepsJ.BassetY.BitoD.JandaM.et al. (2004). No tree an island: the plant–caterpillar food web of a secondary rain forest in New Guinea. Ecol. Lett.7, 1090–1100. doi: 10.1111/j.1461-0248.2004.00666.x
61
NovotnyV.MillerS. E.BajeL.BalagawiS.BassetY.CizekL.et al. (2010). Guild-specific patterns of species richness and host specialization in plant-herbivore food webs from a tropical forest. J. Anim. Ecol.79, 1193–1203. doi: 10.1111/j.1365-2656.2010.01728.x
62
NovotnyV.TokoP. (2015). “Ecological research in Papua New Guinean rainforests: insects, plants and people,” in The state of forests of papua new Guinea: measuring change over the period 2002-2014. Eds. BryanJ. E.ShearmanP. L. (Port Moresby: University of Papua New Guinea), 71–85.
63
OksanenJ.et al. (2019). vegan: Community Ecology Package. R package version 2.5-6. Available at: https://CRAN.R-project.org/package=vegan.
64
PhilbinC. S.DyerL. A.JeffreyC. S.GlassmireA. E.RichardsL. A. (2022). Structural and compositional dimensions of phytochemical diversity in the genus Piper reflect distinct ecological modes of action. J. Ecol.110, 57–67. doi: 10.1111/1365-2745.13691
65
PriceP. W. (2002). Resource-driven terrestrial interaction webs. Ecol. Res.17, 241–247. doi: 10.1046/j.1440-1703.2002.00483.x
66
RambautA.SuchardM.XieD. (2014) Drummond A Tracer v1. 6. Available at: http://beast.bio.ed.ac.uk.Tracer (Accessed 27th July 2014).
67
RichardsL. A.DyerL. A.ForisterM. L.SmilanichA. M.DodsonC. D.LeonardM. D.et al. (2015). Phytochemical diversity drives plant–insect community diversity. Proc. Natl. Acad. Sci. U.S.A.112, 10973–10978. doi: 10.1073/pnas.1504977112
68
SagersC. L.ColeyP. D. (1995). Benefits and costs of defense in a neotropical shrub. Ecology76, 1835–1843. doi: 10.2307/1940715
69
SalazarD.LokvamJ.MesonesI.VásquezM.AyarzaJ.FineP. (2018). Origin and maintenance of chemical diversity in a species-rich tropical tree lineage. Nat. Ecol. Evol.2, 983–990. doi: 10.1038/s41559-018-0552-0
70
SalminenJ. P. (2014). The chemistry and chemical ecology of ellagitannins in plant–insect interactions: from underestimated molecules to bioactive plant constituents. Recent Adv. Polyphenol Res.4, 83–113. doi: 10.1002/9781118329634.ch4
71
SalminenJ. P.KaronenM. (2011). Chemical ecology of tannins and other phenolics: we need a change in approach. Funct. Ecol.25, 325–338. doi: 10.1111/j.1365-2435.2010.01826.x
72
SedioB. E. (2017). Recent breakthroughs in metabolomics promise to reveal the cryptic chemical traits that mediate plant community composition, character evolution and lineage diversification. New Phytol.214, 952–958. doi: 10.1111/nph.14438
73
SedioB. E.SpasojevicM. J.MyersJ. A.WrightS. J.PersonM. D.ChandrasekaranH.et al. (2021). Chemical similarity of co-occurring trees decreases with precipitation and temperature in North American forests. Front. Ecol. Evol.9, 679638. doi: 10.3389/fevo.2021.679638
74
SedioB. E.OstlingA. M. (2013). How specialised must natural enemies be to facilitate coexistence among plants? Ecol. Lett.16, 995–1003. doi: 10.1111/ele.12130
75
SedioB. E.ParkerJ. D.McMahonS. M.WrightS. J. (2018). Comparative foliar metabolomics of a tropical and a temperate forest community. Ecology99, 2647–2653. doi: 10.1002/ecy.2533
76
SedioB. E.Rojas EcheverriJ. C.BoyaP.CristopherA.WrightS. J. (2017). Sources of variation in foliar secondary chemistry in a tropical forest tree community. Ecology98, 616–623. doi: 10.1002/ecy.1689
77
SedioB. E.WrightS. J.DickC. W. (2012). Trait evolution and the coexistence of a species swarm in the tropical forest understorey. J. Ecol.100, 1183–1193. doi: 10.1111/j.1365-2745.2012.01993.x
78
SegarS. T.VolfM.IsuaB.SisolM.RedmondC. M.RosatiM. E.et al. (2017). Variably hungry caterpillars: predictive models and foliar chemistry suggest how to eat a rainforest. Proc. R. Soc. London B: Biol. Sci.284, 20171803. doi: 10.1098/rspb.2017.1803
79
SzeferP.CarmonaC. P.ChmelK.KonečnáM.LibraM.MolemK.et al. (2017). Determinants of litter decomposition rates in a tropical forest: functional traits, phylogeny and ecological succession. Oikos126, 1101–1111. doi: 10.1111/oik.03670
80
VolfM.SegarS. T.MillerS. E.IsuaB.SisolM.AubonaG.et al. (2018). Community structure of insect herbivores is driven by conservatism, escalation and divergence of defensive traits in Ficus. Ecol. Lett.21, 83–92. doi: 10.1111/ele.12875
81
VolfM.LaitilaJ. E.KimJ.SamL.SamK.IsuaB.et al. (2020). Compound specific trends of chemical defences in Ficus along an elevational gradient reflect a complex selective landscape. J. Chem. Ecol.46, 442–454. doi: 10.1007/s10886-020-01173-7
82
VolfM.SalminenJ.-P.SegarS. T. (2019). Evolution of defences in large tropical plant genera: perspectives for exploring insect diversity in a tri-trophic context. Curr. Opin. Insect Sci.32, 91–97. doi: 10.1016/j.cois.2018.12.005
83
WhitfeldT.KressW.EricksonD.WeiblenG. (2012a). Change in community phylogenetic structure during tropical forest succession: evidence from New Guinea. Ecography35, 821–830. doi: 10.1111/j.1600-0587.2011.07181.x
84
WhitfeldT. J. S.NovotnyV.MillerS. E.HrcekJ.KlimesP.WeiblenG. D. (2012b). Predicting tropical insect herbivore abundance from host plant traits and phylogeny. Ecology93, S211–S222. doi: 10.1890/11-0503.1
85
WhitmoreT. (1989). Canopy gaps and the two major groups of forest trees. Ecology70, 536–538. doi: 10.2307/1940195
86
ZhangH.de BernonvilleT. D.BodyM.GlevarecG.ReicheltM.UnsickerS.et al. (2016). Leaf-mining by Phyllonorycter blancardella reprograms the host-leaf transcriptome to modulate phytohormones associated with nutrient mobilization and plant defense. J. Insect Physiol.84, 114–127. doi: 10.1016/j.jinsphys.2015.06.003
Summary
Keywords
Ficus, Macaranga, Syzygium, Psychotria, leaf miner, rainforest, phytochemical, biodiversity
Citation
Segar ST, Re Jorge L, Nicholls L, Basset Y, Rota J, Kaman O, Sisol M, Gewa B, Dahl C, Butterill P, Mezzomo P, Miller SE, Weiblen G, Salminen J-P, Novotny V and Volf M (2024) Species swarms and their caterpillar colonisers: phylogeny and polyphenols determine host plant specificity in New Guinean Lepidoptera. Front. Ecol. Evol. 11:1308608. doi: 10.3389/fevo.2023.1308608
Received
06 October 2023
Accepted
14 December 2023
Published
08 January 2024
Volume
11 - 2023
Edited by
Rafael E. Cárdenas, Pontificia Universidad Católica del Ecuador, Ecuador
Reviewed by
Lora A. Richards, University of Nevada, Reno, United States
Melissa R. L. Whitaker, ETH Zürich, Switzerland
Updates
Copyright
© 2024 Segar, Re Jorge, Nicholls, Basset, Rota, Kaman, Sisol, Gewa, Dahl, Butterill, Mezzomo, Miller, Weiblen, Salminen, Novotny and Volf.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simon T. Segar, ssegar@harper-adams.ac.uk
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.