- 1Department of Environmental Biology, La Sapienza University of Rome, Rome, Italy
- 2Polytechnic Institute of Castelo Branco, Polytechnic University, School of Agriculture, Castelo Branco, Portugal
- 3CERNAS, Research Center for Natural Resources, Environment and Society, Polytechnic Institute of Castelo Branco, Castelo Branco, Portugal
- 4CEF, Forest Research Centre, TERRA Associated Laboratory, Superior Institute of Agronomy, Lisbon University, Lisbon, Portugal
- 5Biotech Plant Lab of Beira Interior, School of Agriculture, Castelo Branco, Portugal
- 6Department of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany
- 7Department of Genetics and Ecology, Uppsala University, Uppsala, Sweden
- 8Laboratory of Systematic Botany and Phytogeography, School of Biology, Aristotle University of Thessaloniki, Thessaloniki, Greece
- 91st General Lyceum of Kamatero, Athens, Greece
- 10Field Science Centre, Graduate School of Agricultural Science, Tohoku University, Osaki, Miyagi, Japan
- 11Graduate School of Symbiotic Systems Science and Technology, Fukushima University, Fukushima, Japan
Introduction: The woody angiosperm Styrax officinalis L., primarily occurring in the Near East and South-eastern Europe, has been historically considered a human introduction in the Italian Peninsula.
Methods: To challenge this assumption, we conducted a genetic analysis on a comprehensive sample of individuals across its range, utilizing chloroplast and nuclear microsatellites as well as a genome-wide single-nucleotide polymorphism (MIG-seq) sequencing approach.
Results: Analysis of 351 individuals revealed clear genetic structure across the species’ range. Most Italian populations form a distinct nuclear genetic cluster, suggesting long-term isolation, while three populations show signs of admixture with Cypriot individuals. Although one rare chloroplast haplotype was unique to Italy, widespread eastern haplotypes were entirely absent from the peninsula, which does not support the hypothesis of a recent human-mediated introduction.
Discussion: The results largely support the indigenous nature of the species in the Italian Peninsula, rejecting the notion of recent human introduction, and elevate S. officinalis to the status of a local relict, probably representing a component of the Late Neogene warm-temperate vegetation in Southern Paleo Europe.
1 Introduction
Styrax officinalis L. (Styracaceae) is a perennial, deciduous small or low-growing polycormic tree native to western Eurasia. It exhibits a fragmented distribution, ranging from the Near East to southeastern Europe, with its westernmost occurrences in southwestern Italy and southeastern France (Tutin et al., 1964–1993; Dimopoulos et al., 2016; POWO, 2025) (Figure 1). The species is commonly found in the warm temperate and arid regions of the Eastern Mediterranean basin and thrives under Mediterranean climatic conditions. It typically inhabits evergreen or semi-evergreen open oak and pine forests, shrublands, and rocky slopes. Styrax officinalis prospers in diverse environments and remains an integral component of Mediterranean ecosystems due to its adaptability and distinct morphology. It is known to grow on various substrates, including tuff, basalt, sedimentary rocks, conglomerate, and eolianite, often on dry, rocky slopes, at elevations ranging from sea level in Italy at Mondragone in southern Italy (Terracciano, 1875) to 1500 m a.s.l. in Lebanon (Zohary, 1973).
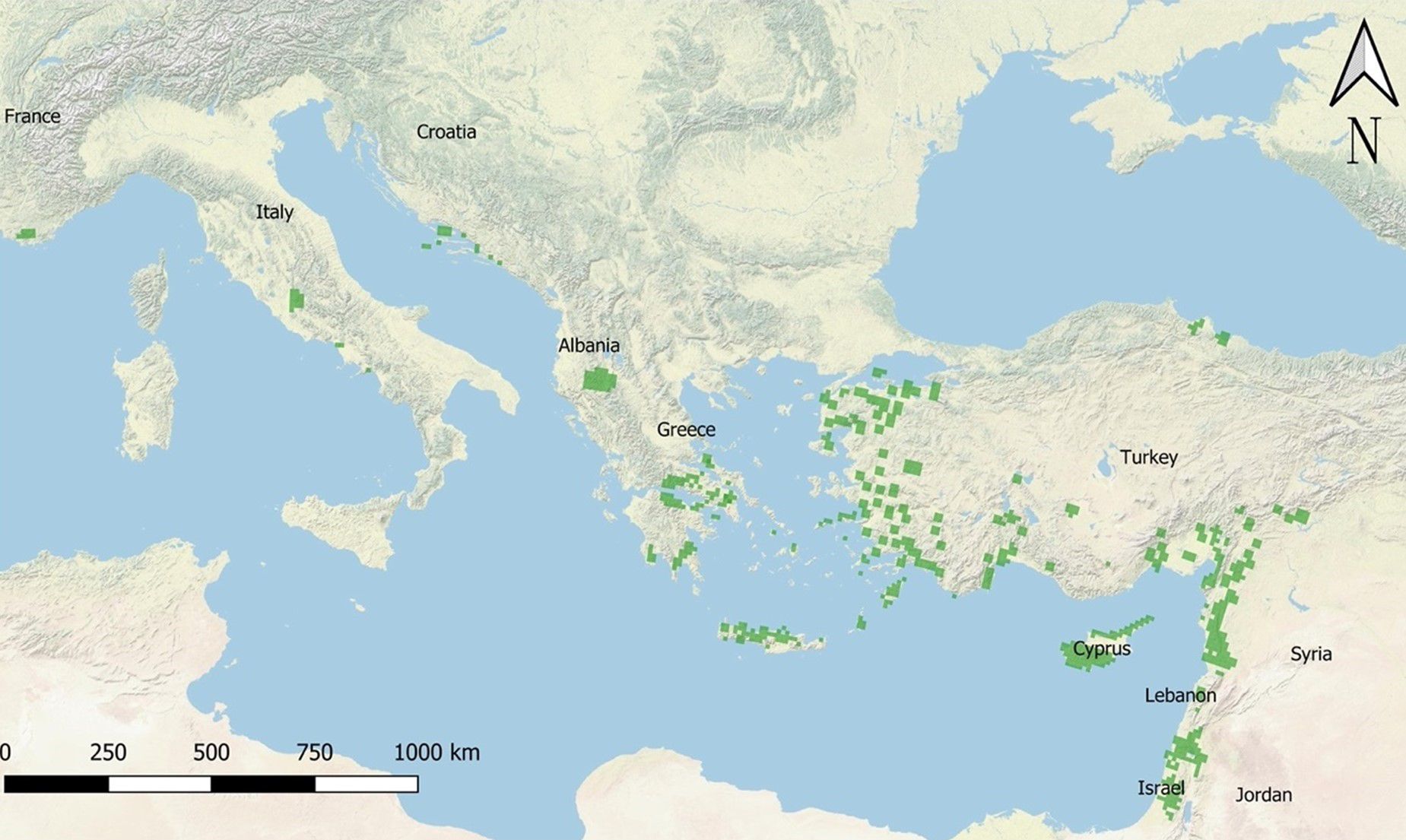
Figure 1. Map showing the natural distribution of Styrax officinalis across the Mediterranean basin. The distribution points were derived from literature data, covering regions including Italy, Greece (continental and islands), Turkey, Dalmatia, Israel, France, Croatia, and Cyprus.
The tree typically reaches a height of about 6 m, with soft, simple, alternate leaves that are rounded or oval-shaped and greenish white in colour, featuring hairy lower surfaces. The species is monoecious, homogamous and xenogamous and its white, bell-shaped flowers have a superior ovary and 8–16 stamens, double the number of petals. The fruits are greenish-yellow drupes with a ball-like shape, a soft, elongated apex, and a single shiny, brown, stony seed inside (Venditti et al., 2018). According to the IUCN Red List of Threatened Species, the species is classified as Least Concern (Wilson, 2018). While the undisputed core of its zonal range lies in the Eastern Mediterranean region (including Palestine, Cyprus, Crete, southern continental Greece, the Aegean islands, and southern and western Turkey (Rikli, 1942; Browicz, 1983; Fritsch, 1996), all occurrences west of continental Greece have long been suspected of being non-native. In Italy the species has traditionally been considered introduced by humans since classical times, and some authors have even described its presence in both Italy and France as the result of naturalization (e.g., Fritsch, 1996). Conversely, recent literature suggests that the native range of this species extends from west-central Italy to northwestern Jordan, and that its presence in France is due to human introduction (POWO, 2025). The disputed origin of the species west of continental Greece, particularly along the Balkan coasts and Italy, dates to early botanical accounts. Fiori (1923) acknowledged natural populations in the Balkans and Italy but omitted occurrences in France. Rikli (1942), in the first comprehensive contour map, excluded East Adriatic populations, aligning with De Candolle’s view of non-native Italian and French populations (De Candolle, 1855). This perspective of non-native populations in Italy partially persisted through subsequent classifications (Tutin et al., 1964–1993; Pignatti, 1982; Browicz, 1983; Fritsch, 1996). The agreement about the non-native status of the westernmost stands of S. officinalis stems from reliance on references from classical antiquity, particularly influential among pre-Linnean botanists who emphasized cultivation in gardens over wild occurrences. In Italy, S. officinalis exhibits a restricted range between 41° and 42° 10’ N. The major enclave encompasses populations clustered on the western edge of the Italian Peninsula, including areas on Mesozoic limestone (Monti Lucretili, Monti Cornicolani, Monti Tiburtini, Monti Prenestini, Monti Ruffi), pyroclastic outcrops and tephra deposits of the Latium Volcano (Monti Albani), up to an elevation of 600 m a.s.l (Montelucci, 1949). Discontinuous stands of limited extent, covering a few hectares, are found further south on the foothills of a limestone coastal ridge near the village of Mondragone in Northern Campania. These stands thrive in mostly secondary thickets within the remnants of a disrupted mixed evergreen-deciduous thermophilic forest, characterized by the dominance of Quercus ilex and Quercus pubescens. Initially documented by Terracciano (1875), these stands have since then been subject to misinterpretation, primarily due to toponymical homonymy with the luxuriant populations growing in the estate Villa Mondragone in Frascati, near Rome (Pignatti, 1982). Additionally, a smaller enclave has been recently identified on the steep slopes of a coastal ridge (Monti Lattari) near Amalfi in Campania, positioned at 41° N (Salerno et al., 2007). Several populations north of Rome have not yet been verified. Possible errors in taxonomic identification may explain records from Monte Soratte, north of Rome, regarding Cydonia sp. and near Spoleto pertaining to Pyrus pyraster.
It is important to note that in Italy S. officinalis occurs in natural habitats and shows no preference for proximity to gardens or human settlements. Its abundance in certain rural landscapes of the Roman Campagna (Tivoli) is likely a consequence of previous woodland being replaced by olive orchards. In these areas, Styrax persists near rocky outcrops, fully demonstrating its capability to regenerate after fire events. This pattern may mistakenly suggest human-mediated expansion through artificial hedgerows. Moreover, in Italy, S. officinalis grows within plant communities that include same associated species found in East Mediterranean analogues (Spada, F. personal communication). Despite the species’ apparent resilience to human influence, localized population declines have been observed, likely due to anthropogenic pressure (Di Pietro and Germani, 2007, and references therein). Terracciano (1875) documented populations reaching the seashore at Mondragone, in contrast to the current upslope distribution. Moreover, urban sprawl has contributed to local extinctions, as seen in the southwestern periphery of Rome and along the Tiber River. Historical claims of S. officinalis cultivation in Italy are not supported by any documentation and likely originated from pre-Linnean traditions that attributed ornamental or medicinal properties to the species. Ancient authors, including Pliny the Elder, occasionally mentioned the extraction of balsam from Styrax, which contributed to the long-standing misconception of its eastern origin and cultivation in Italy for balsam production. In reality, the species has never been used on large scale for balsam extraction. Although the idea that S. officinalis was introduced in Italy is frequently repeated in the literature, we argue that this interpretation is not supported by current ecological and historical evidence. This confusion largely stems from the historical conflation of Styrax with the balsam produced by Liquidambar orientalis, commonly known as “storax” (Meikle, 1977; Bhojvaid and Chaudhari, 2004) a species native to Anatolia, and widely used in traditional medicine and perfumery (Duke, 2008; Birney and Koh, 2018). Initially, Pignatti (1982), author of the officially recognized Flora d’Italia, definitively labelled S. officinalis as non-native in Italy, contrasting Fiori’s earlier stance (Fiori, 1923). However, in the updated Flora d’Italia, Pignatti et al. (2019) presents an alternative view, echoing Montelucci’s earlier proposition that the species might be a relic from the “Riss-Würm interglacial” (Eemian), suggesting a native status in central Italy. This notion, revived by other authors (Spada, 1988), is also supported by coenological similarities between Italian S. officinalis stands and arboreal communities in Anatolia, continental Greece, Aegean Islands, Crete, and Cyprus (Zohary, 1973; Walter, 1974). Past coexistence of S. officinalis with tertiary relict species in Italy, such as Zelkova sp (Follieri et al., 1986), reinforces its relict nature. Today, few extant individuals of Zelkova sicula facing extinction occur in Sicily (Di Pasquale et al., 1992). In Crete, Zelkova abelicea considered a relict representative of the late tertiary warm-temperate flora, as well (Kozlowski et al., 2014), is associated to S. officinalis along the topographic gradient. Similarly, in the Roman district where dense populations of S. officinalis occur (Monti Cornicolani), the rare occurrence of the macro-mesothermic forb Dracunculus vulgaris (Araceae) is recorded. Additionally, the southernmost small stands of S. officinalis in Italy (Monti Lattari in Campania) are found along with the tertiary relict fern Woodwardia radicans (Blechnaceae), implying a shared relict character for S. officinalis, consistent with Paleo-European Late Neogene fossil records (Martinetto, 2001; Cancellieri et al., 2017). Consistent with this, Fritsch (2001) noted that species within the section Styrax are distributed across nearly all mixed-mesophytic forest refugia from the Tertiary in the Northern Hemisphere. Furthermore, many examples of European deciduous taxa have become extinct due to prolonged cold conditions from 70,000 to 16,000 years ago, compounded by the presence of latitudinally oriented mountain ranges. Styrax, along with some of its sclerophyllous associates, appears to have migrated southward from its central European range during the Miocene (Palamarev, 1989) to the Mediterranean region, thereby avoiding extinction. Today, the area occupied by Styrax in the Mediterranean Basin is recognized as a Tertiary refugium, also home to species such as Arbutus andrachne, Cercis siliquastrum, Liquidambar orientalis, and Platanus orientalis (Fritsch, 1996).
Given the diverse and sometimes conflicting information, we initiated a comprehensive genetic study employing chloroplast, as well as genome-wide nuclear markers across numerous individuals within S. officinalis populations spanning its current distribution. The primary objective was to investigate the genetic connections between Italian stands and those in unequivocally native regions, thereby testing the hypothesis of the native status of Italian populations.
2 Materials and methods
2.1 Sampling and distribution mapping
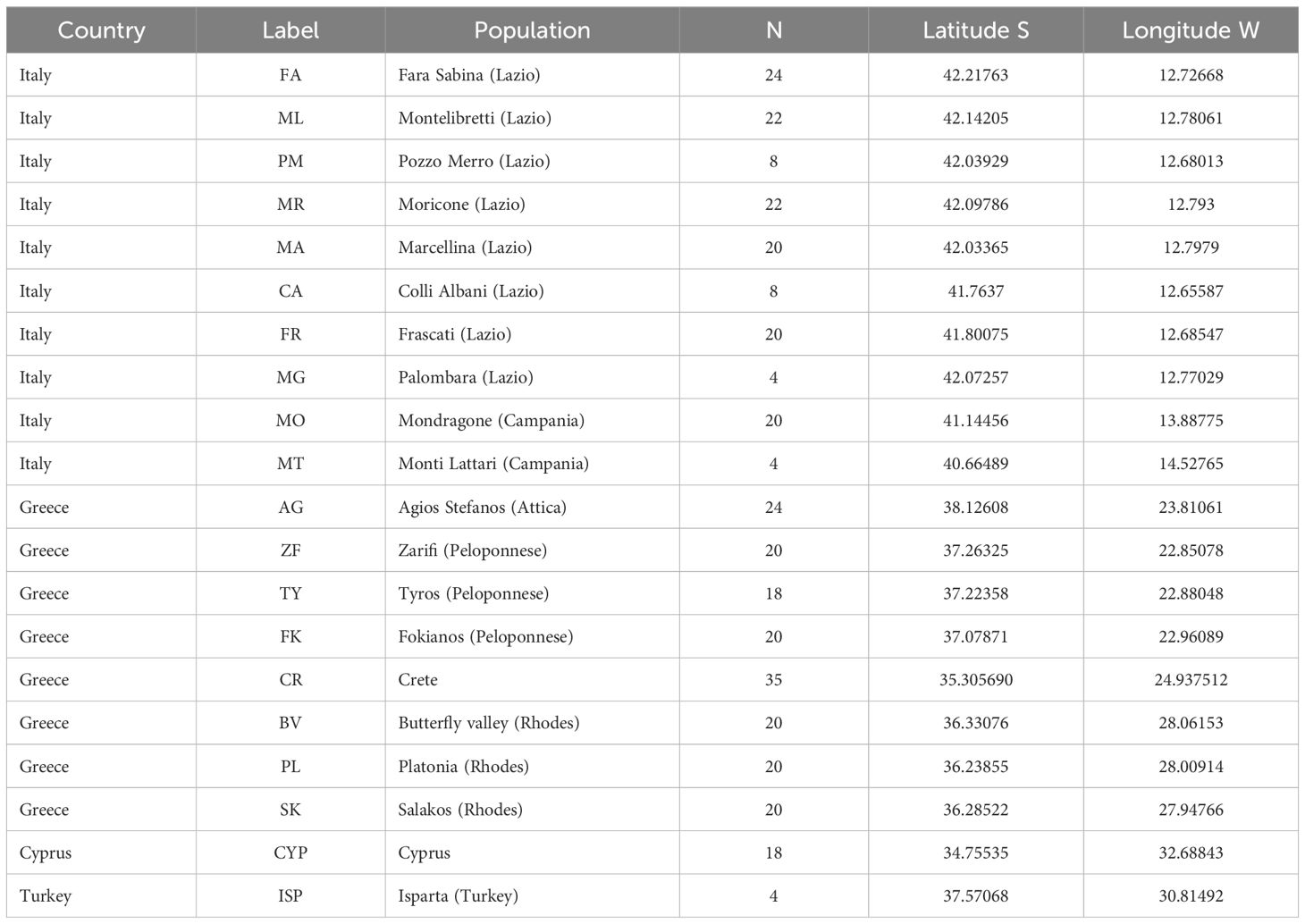
We collected 351 samples of S. officinalis from 20 populations across the species’ range, spanning from Italy to the southeastern Mediterranean region, including continental Greece (Attica and Peloponnesus), Crete, western Turkey (Isparta province), Rhodes, and Cyprus (Table 1). Although S. officinalis has been reported from Albania and France, the record for Albania is unconfirmed and lacks voucher specimens, and populations were not found during a previous expedition by one of the authors, while the French population consists of a few possibly introduced individuals (e.g. Tutin et al., 1964-1993). For this reason, we focused our sampling on areas representing the continuous and central part of the species’ Mediterranean range, which are more informative for inferring its biogeographic history. When the species was growing in protected areas the sampling was conducted after obtaining the necessary permissions from the relevant authorities and, for the populations from Greece, sampling was conducted according to the national legislation. In addition to specimens gathered directly from the wild, a small number of samples were incorporated from herbarium materials originating from Turkey, Dalmatia, Israel, France and Croatia. These herbarium samples were acquired through collaborations with colleagues and Botanical Gardens, particularly in cases where direct sampling was hindered by conflicts or accessibility challenges.
Given the absence of a map illustrating the natural distribution of S. officinalis in the literature, we constructed one using QGIS® Desktop v3.10.4. This map was developed by compiling all available data from the literature regarding the presence of the species in the Mediterranean region (Figure 1).
2.2 DNA extraction
DNA extraction from fresh or silica gel dried leaves was conducted using the Dneasy® Plant Mini kit (QIAGEN, Hilden, Germany). Approximately 20 mg (silica gel dried) or 50 mg (fresh) material was rapidly frozen by flash-freezing with liquid nitrogen and subsequently powdered with Tissue Lyser II. Subsequently, the extraction procedure was carried out following the manufacturer’s instructions, with 30 minutes’ incubation time to boost the cell wall lysis and increase the DNA final yield. The resultant DNA was then stored at -20°C in 2 ml plastic tubes until further use. Although we were able to extract DNA of relatively good quality and carry out molecular analyses on the herbarium samples, the resulting data contained a too high proportion of missing SNPs (~95%). This limited their usability in downstream analyses, and therefore, these samples were ultimately excluded from the study.
2.3 Chloroplast markers
The complete chloroplast genome of Styrax grandiflorus obtained from the National Center for Biotechnology Information Database Resources (https://www.ncbi.nlm.nih.gov/nuccore/) was used as a reference for primer design, as no sequences for S. officinalis were available. Using SciroKo 3.4 software (Kofler et al., 2007), we identified short sequence repeats (cpSSRs) and primer design was done via the online program Primer3Plus version 2.4.2 (Untergasser et al., 2012). Several designs were created to optimize resources and allow multiplexing in fragment analysis, considering the estimated fragment sizes. Design parameters included primer length (18–23 bp, with 20 bp being optimal), PCR product sizes (100–450 bp, within the 55°C - 60°C melting temperature range), and a GC content > 40% for enhanced stability. Eight chloroplast microsatellite primers were selected and used for analysis (Table 2). Chloroplast analyses were conducted on 343 samples from 18 populations. The MT and MG populations were not included in the cpDNA analyses because sampling in these localities was conducted after these analyses were completed.
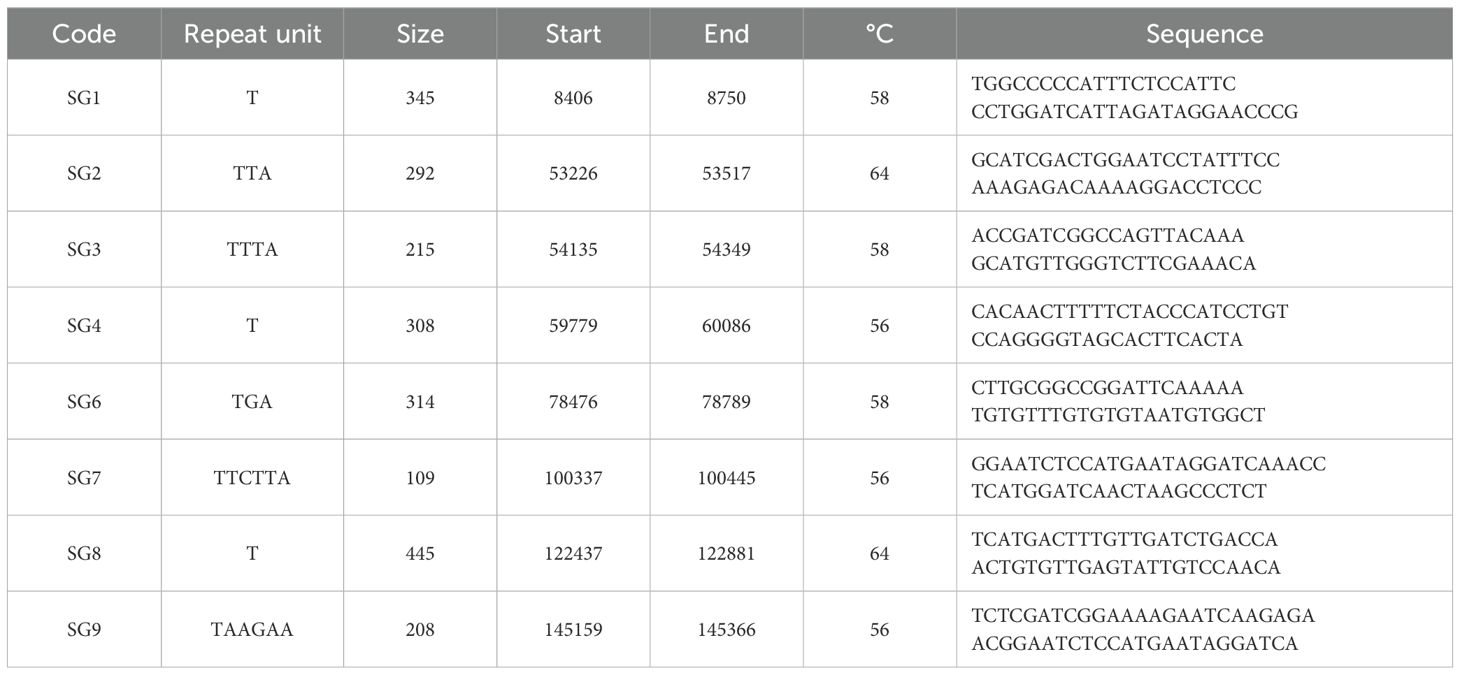
Table 2. Code, repeat unit, size (bp, base pairs), fragment start and end position on the Styrax grandiflorus chloroplast complete genome, annealing temperature (°C) and sequence of the eight cpSSR primers used in this study.
Three multiplex PCRs were conducted each tailored to the specific annealing temperature of its primer set (Table 2). For fluorescence labelling, the forward sequence of each primer pair was utilized: SG1, SG2, and SG8 were 6-FAM labelled; SG3 and SG4 were VIC labelled; and SG6, SG7, and SG9 were NED labelled.
Each PCR reaction was carried out in a 10 µL final volume, comprising 50–60 ng of template DNA, 1.0 U Supreme NZYTaq 2x Colourless Master Mix® with separate MgCl2 (Nzytech, Lisbon, Portugal), 2.5 mM MgCl2, and 0.2 µM of each primer. Amplifications were executed on a UNO96 Gradient thermocycler (VWR®, Leuven, Belgium). The PCR protocol initiated with a 10-minute denaturation step at 95°C, followed by 30 cycles of denaturation (30 s at 95°C), annealing (30 s at the optimal temperature for each primer pair, see Table 2), and amplification (30 s at 72°C). A final extension step of 10 minutes at 72°C concluded the amplification cycles. Post-amplification, the PCR products were diluted in 100 µL Milli-Q water, and 0.5 µL of the diluted product was combined with 10 µL of formamide and 0.3 µL of LIZ-600 size standard. Genotyping was performed using an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Fragment analysis and binning were conducted using GeneMapper 4.0 software (Applied Biosystems, Foster City, CA, USA).
2.3.1 CpSSR genetic diversity and genetic variation
The cpSSR fragments analysed were combined to infer the chloroplast haplotype of each sample. The program CONTRIB v. 1.4 (Petit et al., 1998) was employed to compute key metrics for each population, including the number of haplotypes (Nh), unbiased haplotypic diversity based on haplotype frequencies (He) (Nei, 1987), and haplotypic richness (AR). The latter was determined as the number of different haplotypes found when a specific sample size is drawn from the population, and it was computed to account for uneven population sampling using a rarefaction method, with the population size fixed at four. In addition, GenAlEx v. 6.501 (Peakall and Smouse, 2012) was utilized to calculate the effective number of haplotypes (Ae) and the number of private haplotypes (Ph). CONTRIB also furnished insights into the contribution of each population to total diversity (CT%) and total haplotypic richness (CTR%). These contributions were divided into a first component attributed to population diversity, and a second component resulting from differentiation from other populations (Petit et al., 1998). An haplotype network was created using the mst function by using a minimum spanning tree based on the differences in number of repeats between haplotypes, and visualized through the plot.mst function in the pegas package in R (Paradis, 2010; R-Core-Team, 2020).
To investigate the genetic variation, a non-hierarchical analyses of molecular variance (AMOVA) was conducted using Arlequin 3.5 software (Excoffier and Lischer, 2010). The infinite allele mutation model (IAM) was used by considering the distances between haplotypes as the number of different alleles, and the stepwise mutation model (SMM) in view of those distances as the number of repeat units for each locus. This comprehensive test estimated variance components among and within populations. Significance values were derived from 1,000 permutations.
2.4 Genome-wide nuclear markers
To better investigate the genetic connections between Italian stands and those in the eastern Mediterranean region, we conducted simultaneous amplification and sequencing of genome-wide regions of the nuclear genome, utilizing the MIG-seq approach (Suyama and Matsuki, 2015). This method involves the amplification and sequencing of single nucleotide polymorphic regions of the genome (SNPs), providing a robust dataset for population demographic analysis.
To assess MIG-seq performance on S. officinalis, we adjusted template DNA concentrations to 10–50 ng/μl using NanoDrop One. We used MIG-seq primer set-1 developed by Suyama and Matsuki (2015) and a MIG-seq library was prepared following the protocol by Suyama et al. (2022) and sequenced using the MiSeq system (Illumina, San Diego, CA, USA) and MiSeq Reagent Kit v3 (150 cycles).
2.4.1 Bioinformatic and population structure analyses
Raw forward and reverse demultiplex reads were merged and processed together. Since the insert size of the sequenced molecules were longer than the number of cycles sequenced, no overlap between the forward and reverse reads was expected. STACKS v. 2.55 (Catchen et al., 2013) was used to call genotypes using default settings. In short, cstacks was employed to create a catalogue, and sstacks was further employed to create stacks from the previously created catalogue. The function tsv2bam was used to transpose the genotype data by locus into a bam file. The bam files were then analysed with gstacks in “De novo mode” to call genotypes. Finally, an unfiltered vcf-file was created using Populations. Vcftools (v 0.1.15) (Danecek et al., 2011) was used to calculate the total number of loci covered. A total of 13 samples were removed due to overall low number of SNPs (<3000) (Supplementary Figure 1). The remaining samples where further filtered by excluding all loci with minor allele frequency below 0.02%. Two missing data thresholds (20%, 80%) were used to assess its effect. Loci with high LD were pruned with plink2 (v2.00a3) using the –indep-pairwise 10000 0.2. The main analysis was performed with maximal 80% missing data to capitalize the number of loci in the analysis. Principal Component Analysis (PCA) was preformed using the SmartPCA function from Eigensoft (Patterson et al., 2006). Admixture analysis was performed with Admixture (v1.3.0) for k=3 to k=10 and cross-validation values were used to select the most probable K. Identity by descent (IBD) was calculated using plink2 –genome. All results were visualised in R (R-Core-Team, 2020) using ggplot (Wickham, 2016). Missing data was not correlated with the observed genetic structure (Supplementary Figure 2).
3 Results
3.1 Chloroplast analysis
The distribution of S. officinalis chloroplast haplotypes, based on the analysis of the eight cpSSR primers is illustrated in Figure 2. The predominant haplotype H5 (red) exhibits a widespread presence across the entire distribution, with notable prevalence in Greece and the Near East. Conversely, haplotype H7 (blue) is also common, but observed mainly in the northern and the eastern regions of the Italian Peninsula. The H2 haplotype (yellow) is only identified in the southern part of the range (Greece and the Near East), while the haplotype H4 (orange) is only found in Crete. Finally, the haplotype H8 (purple) is confined to Italy. Notably, Cyprus, which is the most differentiated and diverse population in the studied region, stands out with its unique haplotype H3 (light blue). Also, in Greece (Attica), the AG population displayed a unique haplotype H1 (white). The Cypriot populations revealed some affinity with the Italian populations, despite the geographical proximity with populations from Greece and the Near East as shown by the mutual presence of the haplotype H6 (green), though the Cypriot populations lack the common H7 (blue) Italian haplotype.
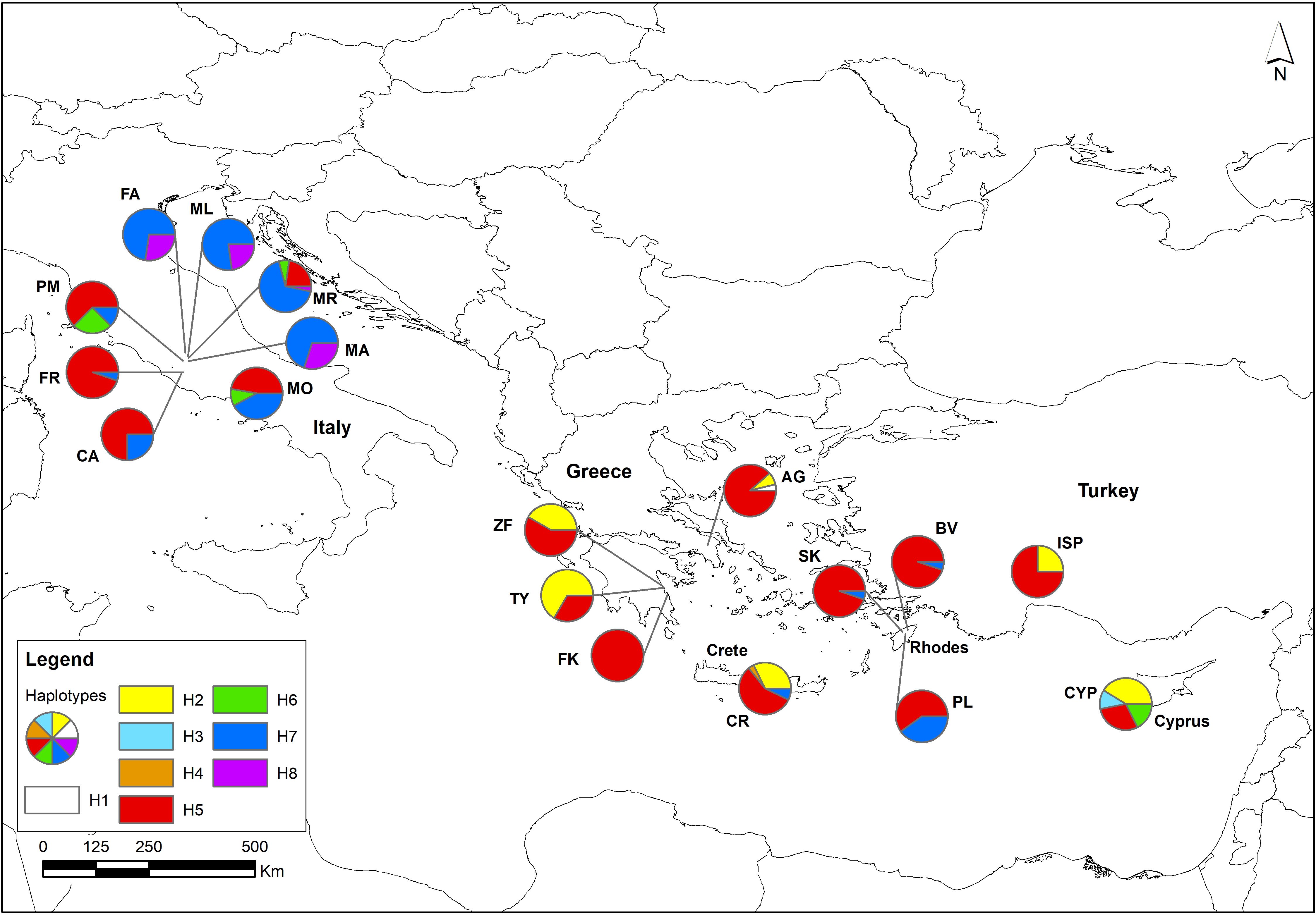
Figure 2. Haplotype distribution among populations, shown as pie charts placed at their respective geographical locations. Population abbreviations are provided in Table 1.
Overall, a distinct division emerges between eastern and western S. officinalis populations, with the exclusive occurrence of the purple haplotype H8 in the Italian populations, and with the yellow H2 being absent.
In Table 3 we present the genetic parameters for each population based on the observed chloroplast SSRs primers. Only eight haplotypes were detected, five of which showed low frequencies (Supplementary Figure 3). Only three cpSSRs showed polymorphic variations. Overall, the average haplotype diversity within populations is low (0.40) as well as the mean effective number of haplotypes (1.75) and the mean number of haplotypes (2.44). These results suggest that the observed genetic diversity is largely influenced by the predominance of a few common haplotypes (H5, H7, and H2), which inflates the apparent variation within populations. The highest haplotype diversity is observed in the Cyprus population (CYP), with an effective number of haplotypes (Ae) of 3.32 and a haplotype diversity (He) of 0.74, followed by Italy/Mondragone (MO) (2.42 and 0.62), Crete (2.29 and 0.59), and Italy/Pozzo Merro (PM) (2.13 and 0.71). The lowest haplotypic diversity is observed in the Greece/Fokianos (FK) (1.00 and 0.00) and in the Greece/Rhodes/Butterfly Valley (BV) (1.10 and 0.10) populations. Haplotypic richness (AR), follows a similar pattern, with the populations showing the highest and lowest values for this parameter as those observed for haplotype diversity.
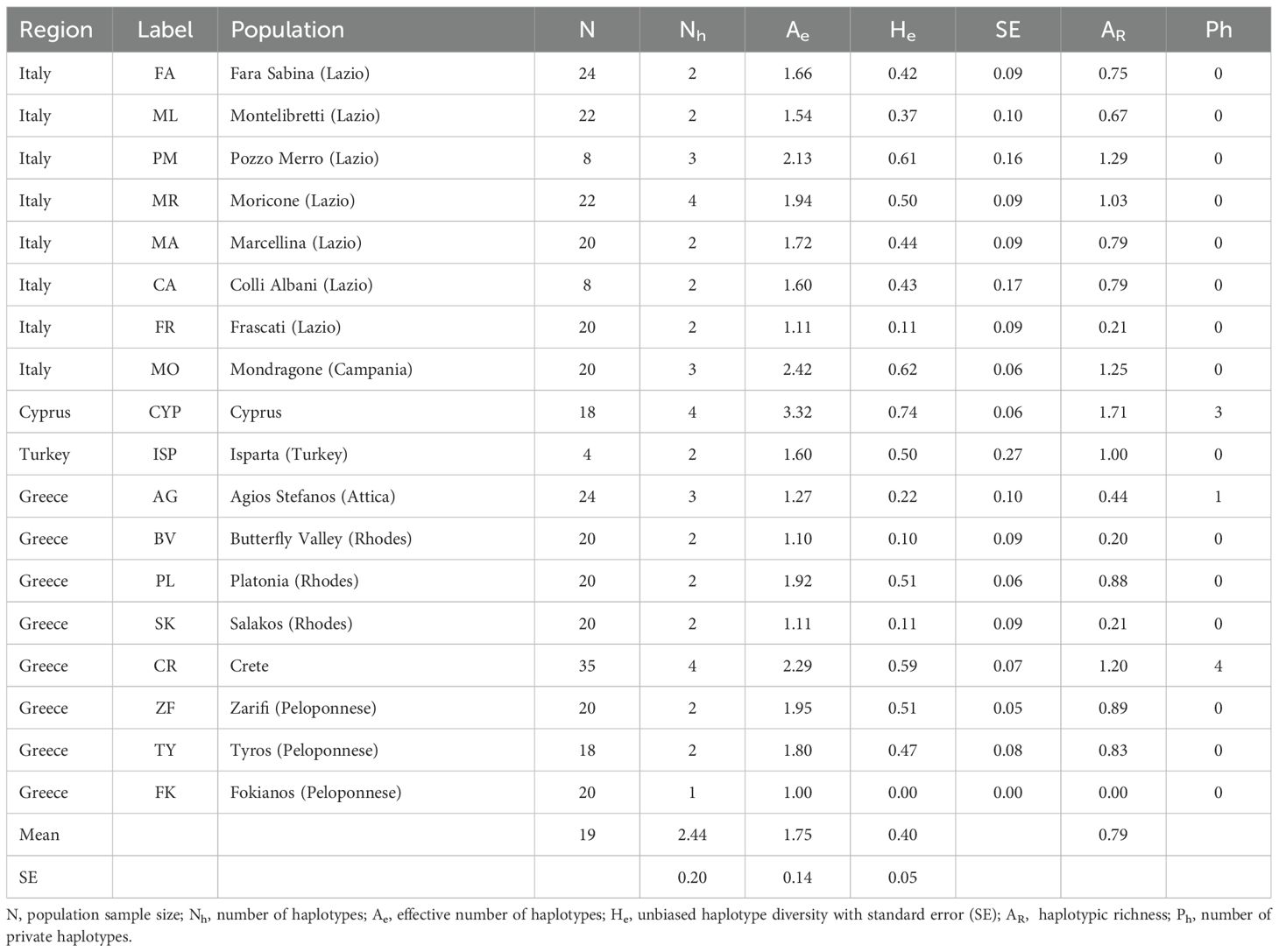
Table 3. Within-population genetic parameters for the S. officinalis populations analysed in this study.
Figure 3 shows the contribution of each population to total haplotypic diversity (CT%), haplotypic richness (CTR%) and total genetic diversity, considering both positive and negative components of diversity and differentiation. Overall, populations contribute more to haplotypic richness than to total genetic diversity. Among them, the Cyprus population emerges as the main contributor to overall genetic diversity and haplotype richness. Similarly, the Italian populations FA, ML and MA also contribute positively to both parameters, while CA and FR show negative contributions, mainly to total haplotypic richness. Most non-Italian populations show minimal contribution, mainly to genetic diversity, except for the one from Crete.
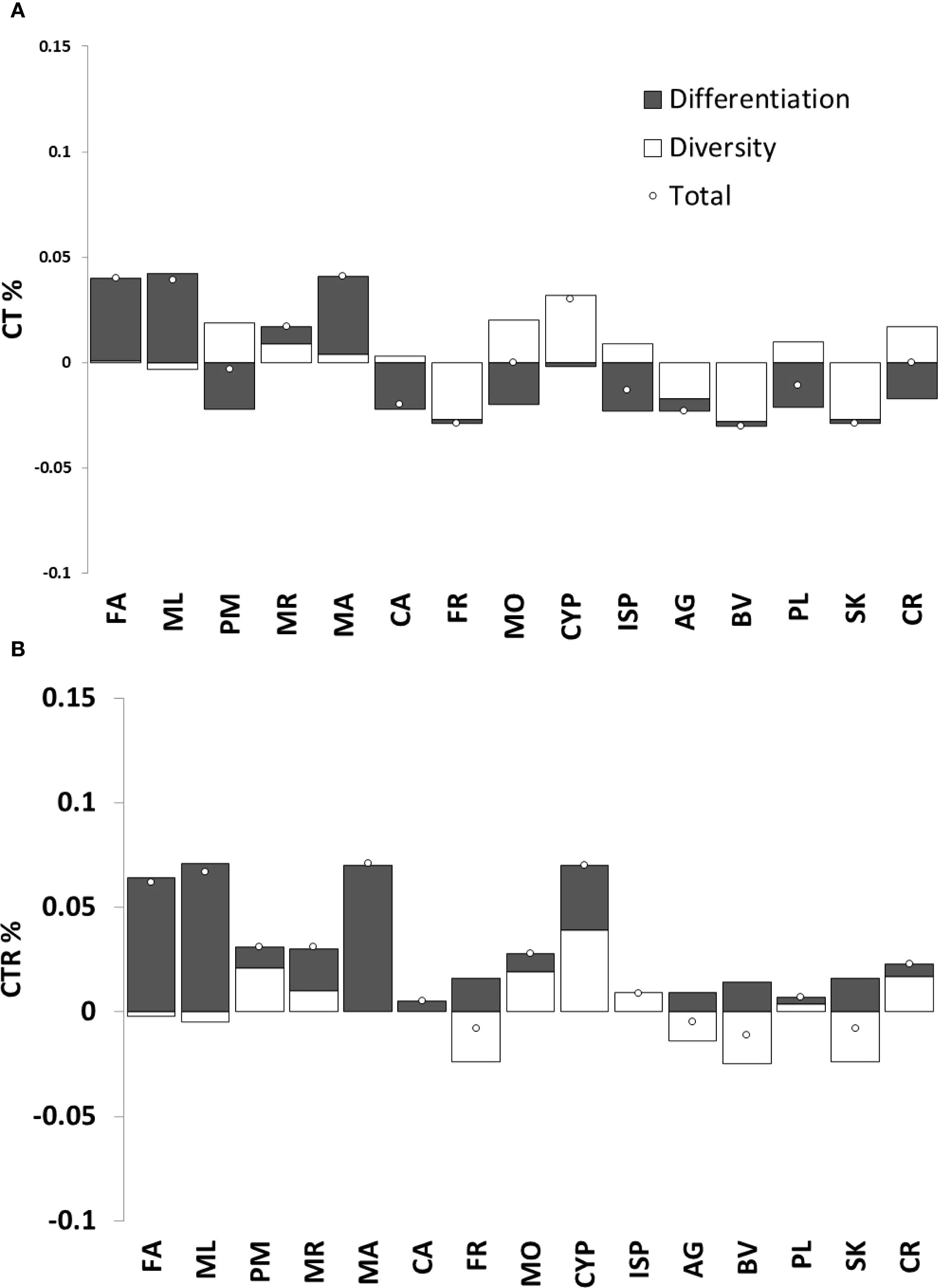
Figure 3. Population diversity components according to Petit et al. (1998): (A) Contribution of each population to total diversity (CT%), divided into own diversity (white) and divergence (dark grey) components. (B) Contribution of each population to total haplotypic richness (CTR%), also divided into own diversity (white) and divergence (dark grey) components.
AMOVA analysis with the IAM model indicate that the total genetic diversity among populations (Φst) is very high (40%) and the remaining 60% is attributable to differences within populations. When the stepwise mutation model (SMM) is used, the total genetic diversity among populations rises to 62% (Supplementary Table 1).
3.2 Genome-wide nuclear analysis
After the exclusion of 1,134 SNPs showing strong linkage-disequilibrium, the final dataset included 2,811 unlinked SNPs. We applied a missing data threshold of 80% to maximize sample inclusion. A comparative test between thresholds of 20% and 80% missing data showed no significant differences in the genetic structure results, supporting the decision to use the more inclusive 80% threshold to retain a larger number of loci in the analysis.
In Figure 4A, a PCA conducted on the SNP dataset identified four major genetic clusters: (i) Italy, (ii) Italy combined with Cyprus, (iii) Turkey and mainland Greece (Attica), and (iv) Greek islands, including the Peloponnese, Rhodes, and Crete. Within the Italy-Cyprus cluster, three distinct subgroups are evident: one from Cyprus and two separate groups within Italy, suggesting some regional genetic differentiation.
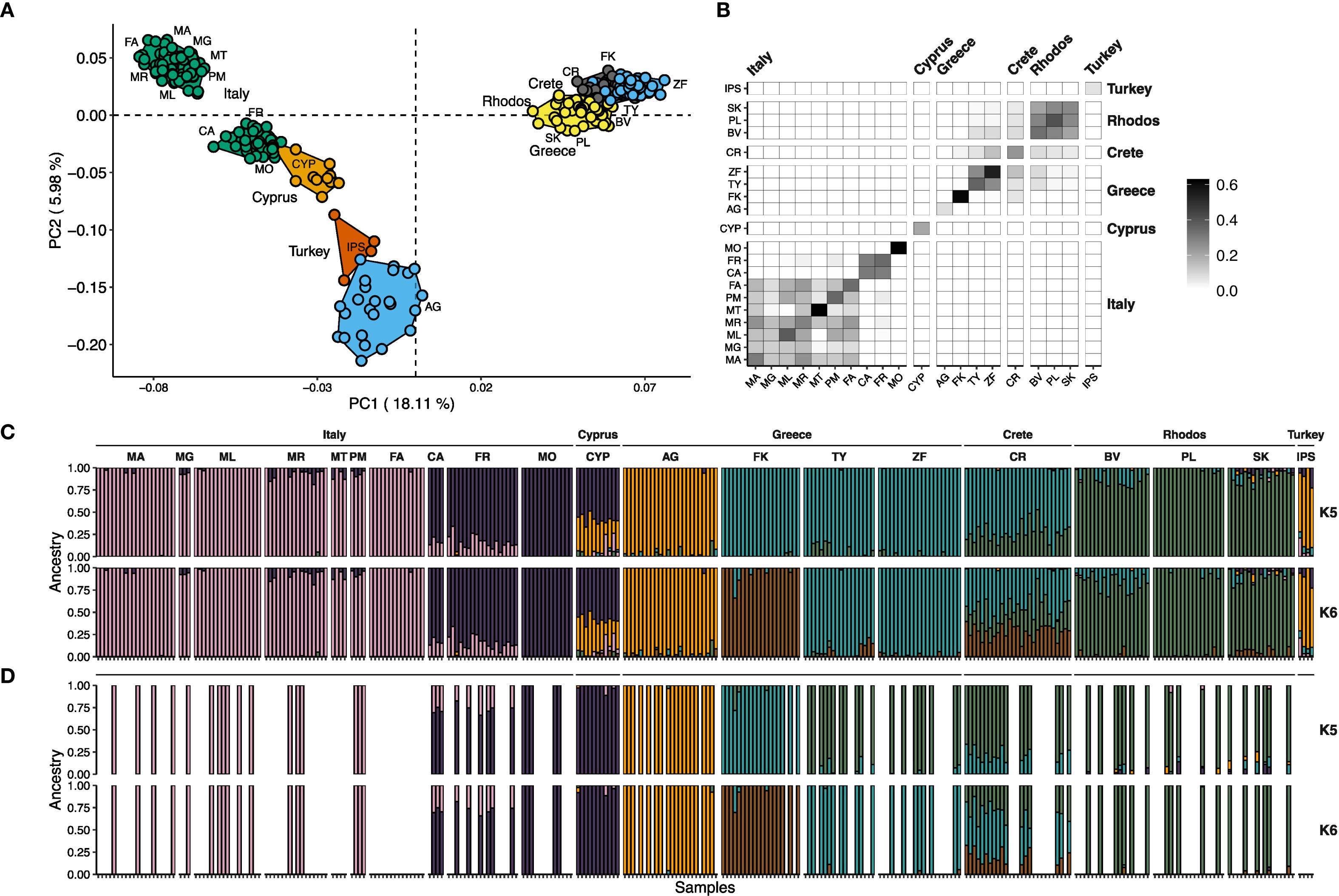
Figure 4. Population genetic analysis using 2,811 unlinked SNPs (maximum 80% missing data). (A) Principal Component Analysis (PCA) reveals four distinct genetic clusters: (1) Italy, (2) Italy + Cyprus, (3) Turkey + Greece, and (4) Greece + Crete + Rhodes. (B) Heatmap of mean relatedness shows high relatedness within populations and across geographic regions. Notable exceptions include Crete, which shares some genetic relatedness with populations from Greece and Rhodes. (C) Admixture analysis for K=5 and K=6, showing clustering patterns consistent with the PCA and relatedness analyses. Admixture is evident between Cypriot and Italian populations, as well as between Crete, Greece, and Rhodes. Populations from Cyprus and Turkey remain genetically indistinguishable. The main difference between K=5 and K=6 is the emergence of population FK (Greece) as a distinct cluster, exhibiting greater admixture with Crete. (D) Admixture analysis for K=5 and K=6 with a down-sampled set of individuals.
Mean relatedness analysis (Figure 4B) supports the PCA results, showing high genetic relatedness both within populations and between geographically close populations, consistent with limited gene flow over larger distances. For example, the Crete population is closely related to those from Peloponnese and Rhodes reflecting geographic proximity and potential historical connectivity. The highest within-population relatedness was observed in Italian populations MT and MO, and Greek populations FK and ZF, possibly indicating local isolation or bottlenecks. Interestingly, Italian populations CA and FR exhibit high relatedness to each other but not to other Italian populations, which themselves form a distinct related group. A similar pattern is observed between the Greek populations TY and ZF.
The Admixture analysis (Figure 4C) largely agree with the PCA and relatedness findings. Unexpectedly, evidence of admixture was detected in the Cyprus population, which shows genetic contributions from both a subgroup of Italian populations and the Attica population (AG), possibly reflecting historical gene flow. A similar admixture pattern is seen in the Turkish population. At K=6, the Crete population appears admixed with populations from Rhodes and Peloponnese (FK), the latter forming a separate cluster. However, this admixture signal disappears at K values between 5 and 8 when the sample size is balanced through down-sampling (Figure 4D). Under these conditions, the Cyprus population is genetically indistinguishable from the Italian MO population, but shows admixture with Italian populations CA and FR, supporting complex population dynamics between these regions.
4 Discussion
While our sampling was extensive, covering most of the species’ distribution, populations from the Middle East (Lebanon, Syria, Israel) could not be included due to political instability in those regions. Although this represents a limitation, given that these areas constitute part of the species’ present-day core range, our analyses of chloroplast DNA and nuclear SNPs from 351 individuals across the remaining range (Figure 1) still offer valuable insights and revealed a clear population structure, addressing the native status of this taxon in Italy.
Chloroplast haplotype H8 (purple) is unique to three of the eight sampled populations on the Italian peninsula, with a frequency of 5%, suggesting it may represent a remnant of historical haplotypic diversity in this region. Interestingly, H8 is positioned at the opposite end of the cpDNA network from H7 (Supplementary Figure 4), which is more frequent in populations from the East. H8 also appears close to H6, a relatively rare haplotype found both in Italy and Cyprus. Meanwhile, H2 and H5 dominate the non-Italian samples and cluster closely together. Haplotype H2 (yellow) is found in all eastern sampling locations except the island of Rhodes but is absent in Italy, which contradicts the hypothesis of large-scale human-mediated movement of S. officinalis into Italy, as a widespread haplotype would likely have been introduced through such dispersal (Hartl and Clark, 2007). While the network suggests that H8 is unique and distant from the more common haplotypes, potentially indicating a distinct lineage for Italian populations, it is important to note that the haplotypes differ only by single nucleotides. Therefore, the visual separation in the network may exaggerate the true genetic distances, and we remain cautious about overinterpreting its structure.
Nuclear DNA analysis identified four major genetic clusters consisting of individuals from: (i) Italy, (ii) Italy and Cyprus, (iii) Turkey and mainland Greece (Attica), and (iv) Greek islands, including Peloponnese, Rhodes, and Crete (Figure 4A). Seven Italian populations form a distinct, well-defined cluster, showing no signs of genetic admixture with populations outside Italy (Figure 4C). These results suggest that the S. officinalis populations from these seven Italian sites are genetically distinct and do not have an evident external origin, supporting the hypothesis that they are relic populations, possibly dating back to the early Miocene. According to macrofossils, this was a time when S. maximus group, the closest relative to the modern S. officinalis, was widespread all over central and southern Paleo-Europe, from the Atlantic coast to Caucasus (Palamarev, 1989; Fritsch, 1996).
While most Italian populations appear to be native relics based on current results, three Italian populations (CA, FR, MO) behave differently and are genetically distinct from the other seven Italian populations and show closest affinity with samples from Cyprus, Turkey, and Attica. Chloroplast haplotype H6 (green) is restricted to Italy and Cyprus but differs by only a single nucleotide from haplotype H5 (red), which is in turn widespread.
Populations PM, FR, CA, and MO exhibit haplotype H5 (red) at a frequency of nearly 50%, similar to most eastern populations. A stronger connection emerges in the nuclear DNA data, where these populations group with Cyprus in the PCA analysis, though they do not fully overlap, except for one sample. Admixture analysis, depending on the dataset used (all samples vs. a balanced subset), reveals either Italian populations admixed with Cyprus individuals (balanced dataset) or Cyprus individuals appearing genetically intermediate between distinct Italian and Greek populations from Attica. The two admixed populations (CA and FR) are located on Pleistocene volcanic outcrops and tephra deposits south of Rome, while MO is situated on a modern coastal limestone ridge. In contrast, all other Italian populations are found along a limestone ridge that emerged in the early Miocene, marking a Pliocene coastline northeast of Rome. IBD analysis (Figure 4B) indicates that the Miocene limestone populations experience gene flow with high IBD values, reflecting high levels of first- and second-degree relationships, but share little to no IBD with the three admixed populations (CA, FR, MO). Notably, populations CA and FR are strongly connected via IBD but lack connectivity with MO, a pattern also evident in the admixture plot, where MO exhibits only minor contributions from the “pure” Italian cluster.
Considering the fossil record (Palamarev, 1989), the similarity patterns found in this study may reflect shared ancient lineages surviving in two disjunct regions (Italy and Cyprus). A possible scenario would be that S. maximus, morphologically identical to S. officinalis (Palamarev, 1989), migrated southward during the Late Tertiary, due to progressive climatic cooling, from the Atlantic Brittany through Central Europe, Poland, and into the Caucasus. The current Italian populations would represent a relic of this ancient distribution, rather than the result of peripheral isolation. These are the northernmost surviving populations, retaining geographical and genetic continuity with Tertiary Paleo-Europe. Their marked genetic diversity suggests relic status from now-extinct northern populations, rather than independent genomic development. In southern Italy, Styrax probably did not expand due to either geographic disconnection from the emerging southern lands or harsh Quaternary droughts.
In the East, Styrax reached Cyprus before the land connection with Anatolia was severed (over 5 million years ago). Cypriot populations show genomic components similar to the Italian Mondragone population, suggesting they preserved ancient Central European genomic elements, lost elsewhere in Western Asia. Considering the fossil record (Palamarev, 1989), this similarity may reflect the persistence of ancient, shared lineages in two disjunct regions. Cyprus’s long isolation, similar to that observed in Cedrus brevifolia, may have allowed the conservation of an archaic southeastern Paleo-European genomic structure (Eliades et al., 2011). Both chloroplast and nuclear data would support this descriptive-relational model. However, alternative scenarios, including long-distance dispersal, or the possibility of ancient human-mediated introduction, cannot be entirely excluded and require further investigation.
In conclusion, these findings strongly support S. officinalis as a native species of the Italian flora. Macrofossil records (Palamarev, 1989; Martinetto, 2001), and that it should not be considered merely an eastern element at the westernmost edge of its range, but rather a remnant of a northwest-to-southeast retreat of a warm-temperate Tertiary flora that once characterized Paleo-Europe in the late Neogene. This study highlights the importance of integrating paleogeographical data into phylogeographical analyses, as genetic similarities inferred solely from present-day geographical proximity may be misleading over long timescales.
4.1 Future research and open questions
The absence of populations from the Middle East regions remains a major limitation. Sampling in these areas would clarify the core range of S. officinalis and help determine whether peripheral populations (e.g., in France and Croatia) are native, for example by detecting haplotype H8, which is currently found only in the Italian Peninsula.
Further studies using a larger number of SNPs, genotype calling based on a reference genome, and new mitochondrial DNA data could improve our understanding of the population dynamics and evolutionary history of S. officinalis across the Mediterranean, which appears to be more complex than previously thought.
Data availability statement
The original contributions presented in the study are publicly available. All raw MIG-seq data were deposited at the DDBJ Sequence Read Archive (DRA) with accession number PRJDB37664.
Author contributions
LP: Writing – original draft, Methodology, Supervision, Conceptualization, Investigation, Funding acquisition, Resources, Writing – review & editing, Project administration. MR: Conceptualization, Visualization, Validation, Writing – review & editing, Formal analysis, Supervision, Writing – original draft, Data curation, Methodology. KN: Supervision, Writing – review & editing, Validation, Software, Methodology, Resources, Formal analysis, Visualization, Data curation. AN: Investigation, Formal analysis, Writing – review & editing, Visualization, Data curation. SS: Formal analysis, Visualization, Writing – review & editing, Investigation. TD: Investigation, Writing – review & editing. AD: Supervision, Resources, Writing – review & editing, Investigation. EA: Resources, Writing – review & editing. SH: Validation, Visualization, Data curation, Software, Formal analysis, Methodology, Resources, Writing – review & editing. YS: Resources, Formal analysis, Software, Validation, Data curation, Supervision, Methodology, Writing – review & editing. FS: Visualization, Investigation, Resources, Validation, Funding acquisition, Conceptualization, Supervision, Formal analysis, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Swedish Phytogeographical Society (LP, FS and KN), Lazioadisu, “TORNO SUBITO 2017 (AN), the Fundação para a Ciência e Tecnologia, I.P. through project references: CERNAS (UIDB/00681), CEF (UID/00239; DOI 10.54499/UIDB/00239/2020), and TERRA (LA/P/0092/2020) (MMR).
Acknowledgments
We thank Ayumi Matsuo for assistance with the MIG-seq analysis, Dr. T. Konstantinidis, Dr. E. Kalpoutzakis and Dr. E. Mpaliousis for sharing information on the Styrax officinalis distribution in Greece. This work was carried out during Laura Parducci and Kevin Nota´s time at Uppsala University. We are grateful to Prof. Nicklas Jansson from Linköping University for providing S. officinalis samples from Turkey.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. To improve language style in very few parts of the main text and in the title.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2025.1598113/full#supplementary-material
References
Bhojvaid P. P. and Chaudhari D. C. (2004). “Non-wood products: resins, latex and palm oil,” in Encyclopedia of Forest Sciences. Ed. Burley J. (Elsevier, Oxford), 620–627.
Birney K. J. and Koh A. J. (2018). Adventures in Storax (Full Spectrum: Micro-Publications from OpenARCHEM).
Browicz K. (1983). Chorology of trees and shrubs in South-West Asia and adjacent regions (Warzawa, Poznán: Polish Scientific Publishers).
Cancellieri L., Caneva G., and Cutini M. (2017). Phytosociology and ecology of the Mediterranean forests ecosystems in the Amalfi Coast (Monti Lattari, Italy). Rendiconti Lincei 28, 654–655. doi: 10.1007/s12210-017-0635-x
Catchen J., Hohenlohe P. A., Bassham S., Amores A., and Cresko W. A. (2013). Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. doi: 10.1111/mec.12354
Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
De Candolle A. (1855). Geógraphie botanique raisonneé: Exposition des faits principaux et des lois concernant la distribution geógraphique des plantes de l’epoque actuelle (Paris V: Masson).
Di Pasquale G., Garfì G., and Quézel P. (1992). Sur la présence d’un Zelkova nouveau en Sicile sud-orientale (Ulmaceae). Biocosme Mesogeen. 8-9, 401–409.
Di Pietro R. and Germani D. (2007). Preliminary phytosociological and cenological considerations about the presence of Styrax officinalis in the area of Cornicolani Mounts (central Latium). Fitosociologia 44, 219–223.
Dimopoulos P., Raus T., Bergmeier E., Constantinidis T., Gregoris I., Kokkini S., et al. (2016). Vascular plants of Greece: An annotated checklist. Supplement. Willdenowia 46, 301–347. doi: 10.3372/wi.46.46303
Duke J. A. (2008). “Storax (Liquidambar orientalis Mill. and L. styraciflua L.),” in Duke’s Handbook of Medicinal Plants of the Bible (Boca Raton London New York: Taylor & Francis), 258–259.
Eliades N.-G. H., Gailing O., Leinemann L., Fady B., and Finkeldey R. (2011). High genetic diversity and significant population structure in Cedrus brevifolia Henry, a narrow endemic Mediterranean tree from Cyprus. Plant Syst. Evol. 294, 185–198. doi: 10.1007/s00606-011-0453-z
Excoffier L. and Lischer H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Eco. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Follieri M., Magri D., and Sadori L. (1986). Late pleistocene zelkova extinction in central Italy. New Phytol. 103, 269–273. doi: 10.1111/j.1469-8137.1986.tb00613.x
Fritsch P. (1996). Isozyme analysis of intercontinental disjuncts within Styrax (Styracaceae): Implications for the Madrean-Tethyan hypothesis. Am. J. Bot. 83, 342–355. doi: 10.2307/2446169
Fritsch P. W. (2001). Phylogeny and biogeography of the flowering plant genus styrax (Styracaceae) based on chloroplast DNA restriction sites and DNA sequences of the internal transcribed spacer region. Mol. Phylogen. Evol. 19, 387–408. doi: 10.1006/mpev.2001.0933
Hartl D. and Clark A. (2007). Principles of population genetics (Sunderland, MA: Sinauer and Associates).
Kofler R., Schlötterer C., and Lelley T. (2007). SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23, 1683–1685. doi: 10.1093/bioinformatics/btm157
Kozlowski G., Frey D., Fazan L., Egli B., Bétrisey S., Gratzfeld J., et al. (2014). The Tertiary relict tree Zelkova abelicea (Ulmaceae): distribution, population structure and conservation status on Crete. Oryx 48, 80–87. doi: 10.1017/S0030605312001275
Martinetto E. (2001). The role of central Italy as a centre of refuge for thermophilous plants in the late Cenozoic. Acta Palaeobot. 41, 299–319.
Montelucci G. (1949). Cenni ecologici su alcune piante notevoli (o nuove) per la flora romana e loro attività nella costituzione della vegetazione Laziale (Firenze: Istituto Botanico).
Palamarev E. (1989). Paleobotanical evidences of the Tertiary history and origin of the Mediterranean sclerophyll dendroflora. Plant Syst. Evol. 162, 93–107. doi: 10.1007/BF00936912
Paradis E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Patterson N., Price A. L., and Reich D. (2006). Population structure and eigenanalysis. PloS Genet. 2, e190. doi: 10.1371/journal.pgen.0020190
Peakall R. and Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Petit R., El-Mousadik A., and Pons O. (1998). Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855. doi: 10.1111/j.1523-1739.1998.96489.x
Pignatti S., Guarino R., and La Rosa M. (2019). Flora d’Italia 2ª ed. Vol. 3. (Edagricole, Bologna).
POWO (2025). Styrax officinalis. Plants of the World Online (Kew: Facilitated by the Royal Botanic Gardens). Available online at: https://powo.science.kew.org/ (Accessed February, 1, 2025).
R-Core-Team (2020). R: A language and environment for statistical computing (Vienna. Austria: R Foundation for Statistical Computing).
Rikli M. (1942). Das Pflanzenkleid der Mittelmeerländer. II. Lieferung. Zweite, unveränderte Auflage (Zürich: Hans Huber Bern).
Salerno G., Ceschin S., and Cutini M. (2007). Contributo alla conoscenza floristica della Campagna Romana: l’area archeologica di Gabii-Castiglione (Roma). Inform. Bot. Ital. 39, 167–180.
Spada F. (1988). “Il paesaggio vegetale dei Monti Lucretili,” in Monti Lucretili, 3 ed. Ed. De Angelis G. (Provincia di Roma, Comitato Promotore Parco Naturale Regionale Monti Lucretili, Roma), 173–185.
Suyama Y., Hirota S. K., Matsuo A., Tsunamoto Y., Mitsuyuki C., Shimura A., et al. (2022). Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecol. Res. 37, 171–181. doi: 10.1111/1440-1703.12270
Suyama Y. and Matsuki Y. (2015). MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci. Rep. 5, 16963. doi: 10.1038/srep16963
Terracciano N. (1875). Terza relazione sulle peregrinazioni botaniche in Terra di Lavoro. Nobile e C. Caserta. Caserta, Nobile e C.o.
Tutin T., Heywood U., Burges N., Valentine D., Walters S., and Webb D. (1964–1993). Flora Europaea (Cambridge: Cambridge University Press).
Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3—new capabilities and interfaces. Nucl. Acids Res. 40, e115. doi: 10.1093/nar/gks596
Venditti A., Frezza C., Serafini I., Pulone S., Scardelletti G., Sciubba F., et al. (2018). Chemical profiling of the fruits of Styrax officinalis L. from Monti Lucretili (Latium region, Central Italy): Chemotaxonomy and nutraceutical potential. Trends Phytochem. Res. 2, 1–12. Available online at: https://oiccpress.com/tpr/article/view/11708.
Wilson B. (2018). Styrax officinalis (The IUCN Red List of Threatened Species 2018), e.T79927884A119836558.
Keywords: Styrax officinalis, chloroplast and nuclear DNA, genome-wide SNPs, quaternary climatic fluctuations, late neogene vegetation, Paleo-European relict, floristic disjunctions
Citation: Parducci L, Ribeiro MM, Nota K, Nobile A, De Santis S, Diamantino T, Drouzas AD, Aplada E, Hirota SK, Suyama Y and Spada F (2025) Genetic data support the relict and native status of Styrax officinalis L. (Styracaceae) in Italy. Front. Ecol. Evol. 13:1598113. doi: 10.3389/fevo.2025.1598113
Received: 22 March 2025; Accepted: 18 September 2025;
Published: 07 October 2025.
Edited by:
Richard John Edwards, University of Western Australia, AustraliaReviewed by:
Xiu Yan Feng, Chinese Academy of Sciences (CAS), ChinaAnna Maria Mercuri, University of Modena and Reggio Emilia, Italy
Copyright © 2025 Parducci, Ribeiro, Nota, Nobile, De Santis, Diamantino, Drouzas, Aplada, Hirota, Suyama and Spada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Parducci, bGF1cmEucGFyZHVjY2lAdW5pcm9tYTEuaXQ=
†These authors have contributed equally to this work and share first authorship
 Laura Parducci
Laura Parducci Maria Margarida Ribeiro
Maria Margarida Ribeiro Kevin Nota6
Kevin Nota6 Alessandro Nobile
Alessandro Nobile Simone De Santis
Simone De Santis Andreas D. Drouzas
Andreas D. Drouzas Eirini Aplada
Eirini Aplada Shun K. Hirota
Shun K. Hirota Francesco Spada
Francesco Spada