- 1Universidade do Algarve, Faro, Portugal
- 2Australian Institute of Marine Science, Townsville, QLD, Australia
- 3Australian Institute of Marine Science, University of Western Australia, Perth, WA, Australia
- 4Marine Ecology Research Centre, Southern Cross University, Lismore, NSW, Australia
- 5National Marine Science Centre, Faculty of Science and Engineering, Southern Cross University, Coffs Harbour, NSW, Australia
- 6Lizard Island Research Station, Australian Museum, Sydney, NSW, Australia
Coral reefs are increasingly threatened by climate change-induced stressors, including marine heatwaves, which can lead to coral mortality, reduced reproductive output, and compromised natural recovery. Successful coral reef recovery requires the settlement of coral larvae and recruitment in degraded areas, replenishing coral communities and promoting resilience. Some restoration strategies involve utilizing natural spawning slicks, composed of coral gametes and embryos, to produce larvae to reseed reefs. However, verifying the taxonomic composition of these slicks is challenging. Here, we tested the performance of two coral ITS primer sets, CoralITS2 and CoralITS2_acro, on mock communities to evaluate their ability to capture genera composition and relative abundances. Both primer sets demonstrated high accuracy (>97%) in detecting and quantifying coral taxa. Subsequently, these primers were applied to wild-collected spawning slicks from the Great Barrier Reef, revealing variation in scleractinian (reef-building) coral community composition among slicks. For the CoralITS2_acro assay, Acropora was consistently the most abundant resolved genus detected across wild slick sample sites, with the exception of samples from the Whitsundays region, where Platygyra was dominant. The CoralITS2 assay successfully differentiated reef-building (Scleractinian) corals from other co-occurring spawning taxa, such as soft corals, anemones, and sponges, and revealed that these other co-spawners dominated slicks at two sites. Our findings underscore the potential of eDNA-based monitoring as a scalable tool to confirm the presence and relative abundance of diverse coral assemblages in natural slicks, informing restoration efforts. By enabling the characterization and comparison of slick composition across large spatial and temporal scales, eDNA metabarcoding can support restoration practices that align with the ecological requirements of reef ecosystems, safeguarding biodiversity and promoting resilience against future disturbances.
1 Introduction
Marine heatwaves and other direct human impacts threaten global biodiversity, as reefs face a combination of disturbances, including coastal development, pollution, and destructive fishing practices, alongside increasingly frequent extreme warming events that affect critical foundation species such as reef-building corals (Fox and Caldwell, 2006; Smale et al., 2019). While marine heatwave events do not always cause mass coral mortality, their increasing severity and frequency are resulting in increased coral losses and reef degradation (Maynard et al., 2008; Weis, 2010; Gadoutsis et al., 2019; Intergovernmental Panel on Climate Change (IPCC), 2023; Yadav et al., 2023). For instance, marine heatwaves have triggered five mass coral bleaching events on the Great Barrier Reef (GBR) in just nine years (2016 to 2024), reducing coral cover on shallow water coral reefs by as much as 50% (Hughes et al., 2017; Dietzel et al., 2021; Henley et al., 2024). The loss of coral, particularly reef-building species, diminishes the structural complexity of these habitats and disrupts ecological functions, compromising the biodiversity and ecosystem services coral reefs provide (Gratwicke and Speight, 2005; Graham and Nash, 2013; Newman et al., 2015; Souter et al., 2020).
The recovery of coral communities following major disturbances depends on successful reproduction and recruitment, in which the dispersive larval phase replenishes populations (Gilmour et al., 2013; Holbrook et al., 2018; Gouezo et al., 2020) and maintains genetic diversity. While coral communities have an inherent capacity for natural recovery after disturbance events (Baker et al., 2008; Sheppard et al., 2008), this process is increasingly hampered by various biological (declines in fecundity of source corals) and environmental challenges (sedimentation, biofilm, and suitable substrates), and can take a significant period of time free from additional disturbances (Ortiz et al., 2018). One critical factor in determining recovery success is the dispersive larval phase, during which coral larvae supply the new recruits needed to rebuild populations (Harrison and Wallace, 1990; Richmond and Hunter, 1990; Connell et al., 1997; Harrison, 2024a); this process requires successful larval settlement in degraded areas for recovery to occur.
Coral bleaching events and other severe disturbances significantly impact larval recruitment by reducing the availability of mature, breeding corals, which are vital for sustaining reproductive output (Hughes et al., 2019). Recruitment levels often plummet to a fraction of historical averages following such disturbances, with the composition of recruits frequently shifting toward more tolerant taxa (Gilmour et al., 2013; Holbrook et al., 2018; Hughes et al., 2019). Adding to this challenge, frequent disturbances subject many surviving adults to sublethal stressors, causing impaired fecundity and reduced gamete quality, diminishing the supply of healthy larvae and undermining the ability of coral populations to sustain themselves (Michalek-Wagner and Willis, 2001; Ward et al., 2002; Cox, 2007; Gilmour et al., 2013; Piñón-González and Banaszak, 2018; Johnston et al., 2020; Briggs et al., 2024). This decline in larval supply, coupled with altered species composition, hampers natural recovery processes: fewer larvae are available to settle and repopulate degraded reefs, which extends recovery timelines and drives long-term changes in community structure and biodiversity. Moreover, the increasing frequency of mass bleaching events leaves insufficient recovery intervals, often preventing coral communities from regenerating fully before the next disturbance (Babcock et al., 2021). This shortened window for recovery exacerbates the instability of coral populations and contributes to the long-term destabilization of reef ecosystems (Gilmour et al., 2013; van Woesik et al., 2016; Ortiz et al., 2018; Babcock et al., 2021).
Critical gaps exist in monitoring coral reproductive output across assemblages. Current assessment methods are often limited to a few species and rely on resource-intensive techniques such as tissue sampling for fecundity measurements (Howells et al., 2016; Briggs et al., 2024), recruitment data from settlement devices (Mallela and Crabbe, 2009), and benthic surveys (Jonker et al., 2008). These techniques are costly, often restricting their application to small specific sites and target species, and their reliance on morphological identification of recruits can result in low taxonomic resolution; indeed, recruits typically can’t be identified beyond the family level for the first several months of life (Babcock et al., 2003). This narrow focus can overlook broader, community-wide reproductive dynamics, such as potential shifts in the taxonomic composition of spawning slicks towards more tolerant taxa. Understanding these shifts is crucial to monitoring and forecasting the response of coral communities to climate change.
Active restoration efforts are increasingly recognized as an essential strategy for sustaining coral ecosystems and their services (Bullock et al., 2011; Anthony et al., 2017; Boström-Einarsson et al., 2020; Vaughan, 2021; Banaszak et al., 2023). Among these interventions, two prominent approaches include the use of seeding devices containing settled coral recruits (Chamberland et al., 2017; Randall et al., 2021, 2023; Miller et al., 2022; Whitman et al., 2024) and the collection and deployment of wild coral spawning slicks (Heyward et al., 2002; dela Cruz and Harrison, 2017; 2020; Harrison et al., 2021). Seeding devices often involve controlled breeding and settlement of one or a few species (Chamberland et al., 2017; Randall et al., 2021, 2023; Whitman et al., 2024) to target high-value species, populations, and genotypes of interest, but cannot fully capture the extensive diversity found within broadcast-spawning communities, particularly on Indo-Pacific reefs, including the GBR. In contrast, the use of wild spawning slicks overcomes these limitations by collecting gametes and embryos from diverse coral assemblages and settling them on degraded reefs (Heyward et al., 2002; Doropoulos et al., 2019; Tabalanza et al., 2020). However, the taxonomic composition of these slicks has remained largely uncharacterized, which limits our understanding of the diversity of corals present in the slicks and highlights the risk that other potentially competitive broadcast spawner species (e.g., soft corals, anemones, and sponges) could be inadvertently captured and settled. This underscores the need for monitoring tools to ensure that restoration efforts are inclusive of diverse taxa and aligned to maintain coral ecosystem resilience, as well as for a better understanding of potential species bias within slicks.
Environmental DNA (eDNA) refers to genetic material obtained directly from the environment. Sequencing of eDNA is widely applied in terrestrial and marine ecosystems for biodiversity assessments (Willerslev et al., 2003; Thomsen et al., 2012; Everett and Park, 2018; Cilleros et al., 2019; Alexander et al., 2020; Dugal et al., 2022) and has been shown to produce coral biodiversity assessments comparable to traditional visual surveys (Dugal et al., 2022). Here, we leveraged coral eDNA sequencing to characterize the taxonomic diversity of corals and their relative abundances in natural spawning slicks. We first generated eDNA profiles of known slick mixes containing larvae of 10 coral species from the GBR to assess the accuracy of existing coral eDNA methodology when applied to coral slicks. Following this, we sequenced eDNA of wild slick samples collected from the Cooktown, Townsville, and Whitsunday Islands sectors of the GBR. We show that eDNA profiles accurately recapitulate the taxonomic composition of coral slicks, thus demonstrating the potential of incorporating eDNA assays into wild spawning slick collection activities to identify the diversity of corals in slicks and target coral species most in need of conservation.
2 Materials and methods
2.1 Creation of a mock coral slick community
To assess the detection sensitivity of existing eDNA assays on coral slicks, a mock slick mix was created using larvae from cultures of 10 known coral species belonging to the four genera Acropora, Dipsastraea (formerly Favia), Platygyra, and Mycedium from the central GBR (Table 1). Bulk monospecific larval cultures were established from colony fragments that were spawned, fertilized, and cultured in November 2018 at the National Sea Simulator Facility (SeaSim) at the Australian Institute of Marine Science (AIMS) in Townsville, Australia (see Randall et al., 2024 for details). Five replicate mocks were created, each containing three larvae of each coral species for a total of 30 larvae. Samples were fixed in 100% ethanol and stored at room temperature until further processing for DNA extraction. Sampling equipment was rinsed with 10% bleach and fully dried between use.
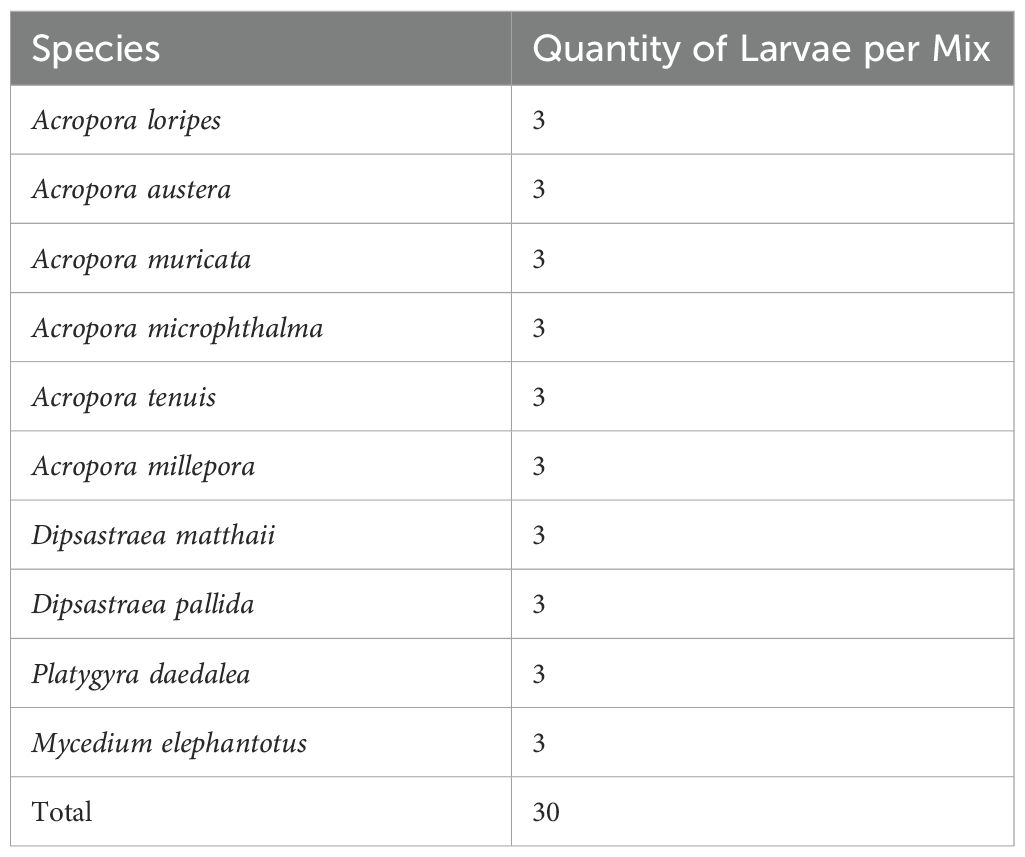
Table 1. List of Great Barrier Reef scleractinian coral species and quantities used to create known mixes.
2.2 Study sites and wild spawn slick sample collection
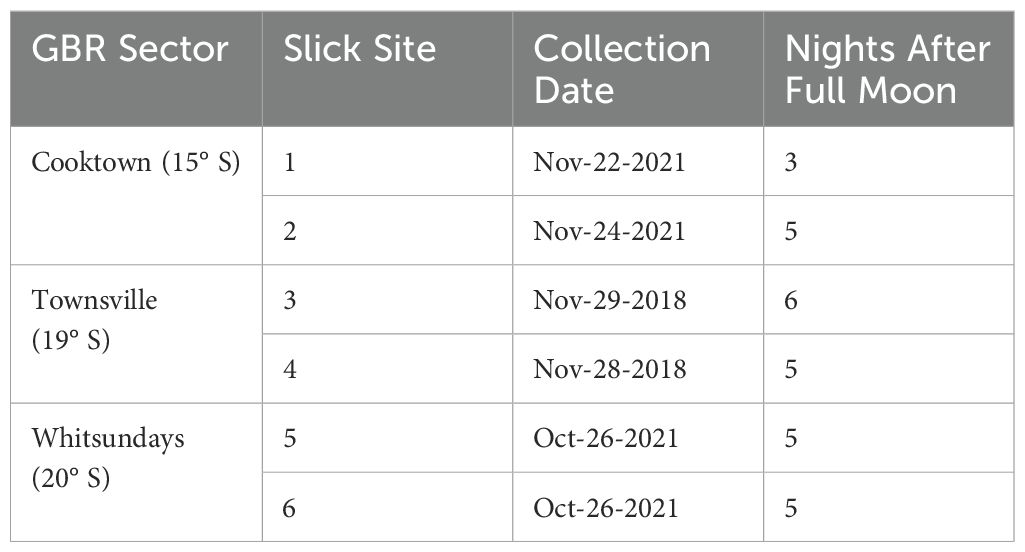
Six reef sites along the northern and central GBR were sampled opportunistically during the 2018 and 2021 spawning seasons (Figure 1). Coral spawning slick material was collected from Lizard Island (-14.6802, 145.4466) in the Cooktown area, Backnumbers Reef (-18.5087, 147.1529) and Keeper Reef (-18.7492, 147.2656) in the central mid-shelf near Townsville, and Hook Island in the Whitsunday Islands (-20.1705, 148.9225) (Table 2). Coral spawning was visually confirmed by the presence of spawning slicks at the sea surface of each reef during the predicted mass spawning period (Harrison et al., 1984; Baird et al., 2021), and samples were viewed under microscopes and imaged. Slick samples were scooped from the water surface using 1L beakers, and spawn collection nets from Lizard and Hook Island sites (see Harrison, 2024b for details), and 3–5 mL of the slick was transferred using sterile transfer pipettes to 15 mL sterile sample tubes. Samples were fixed in 100% ethanol and stored at room temperature until processing for DNA extraction.
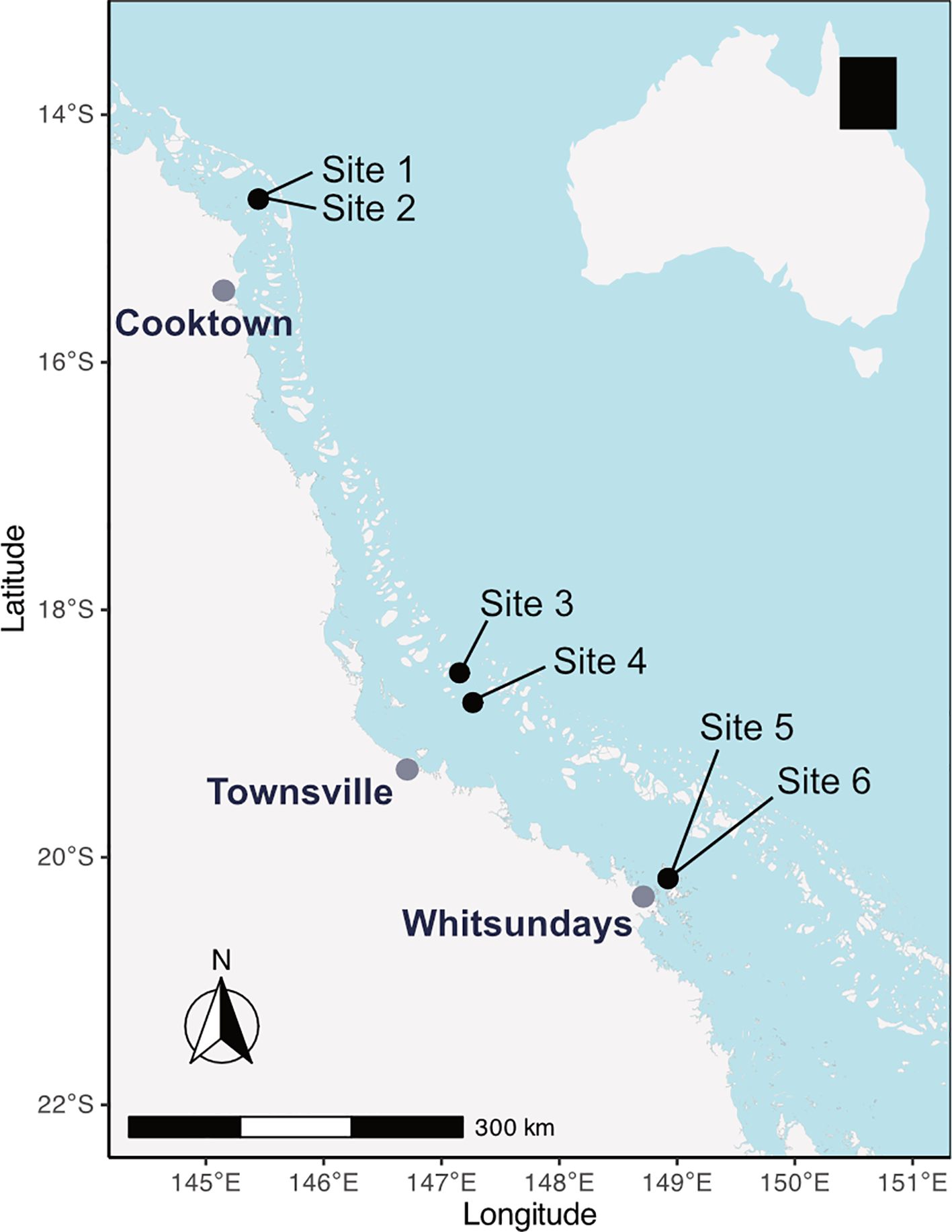
Figure 1. Locations of collection sites of wild coral spawning slicks along the Great Barrier Reef, Australia. Sites are grouped into sectors based on location: Sites 1 & 2 (Cooktown), Sites 3 & 4 (Townsville), Sites 5 & 6 (Whitsundays).
2.3 eDNA extraction and sequencing
DNA was extracted following Liew et al., 2020, with a 2.5–3 hour incubation in preheated buffer (100 mM Tris/EDTA/NaCl, 1% SDS) and RNase treatment. DNA extractions were sent to the Australian Genome Research Facility (AGRF) (Sydney, Australia) for PCR amplicon generation and DNA sequencing. Each PCR reaction mixture (25 μL) contained 10x PCR Gold buffer, 50 nM MgCl2, 25 nM dNTPs, 5x SYBR Green, 1 U AmpliTaq PCR buffer, 10 μM of forward and reverse primer (CoralITS2 and CoralITS2_acro, Supplementary Table S1), 20 μM of forward and reverse tags, and DNA template. The thermal cycling conditions were as follows: initial denaturation at 95°C for 5 min, followed by 45 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 45 seconds, with a final extension of 72°C for 10 min. Separate DNA libraries were created for the CoralITS2 samples and the CoralITS2_acro samples. The final libraries were size-selected to a range of 160–550 bp (for CoralITS2) and 175–600 bp (for CoralITS2_acro). The cleaned libraries were then quantified, pooled, and sequenced on the Illumina MiSeq platform with 500-cycle V2 chemistry for paired-end sequencing (Illumina, USA).
2.4 Amplicon sequence data processing
Demultiplexed reads provided by AGRF were imported into QIIME2 (v. 2024.5; Bolyen et al., 2019). DADA2 (Callahan et al., 2016) implemented in QIIME2 was used to trim adapter, primer and low-quality sequences, and then denoise, merge, and check the remaining sequences for chimeras, and finally to generate an amplicon sequence variant (ASV) counts table. The LULU algorithm was then used to merge ASVs based on sequence similarity and co-occurrence patterns (Frøslev et al., 2017). To assign taxonomy, the representative sequence of each ASV was compared against the UNITE ITS database (Abarenkov et al., 2024) v9 using BLASTN v2.13.0+ and the alignment outputs imported into MEGAN v6.24.20 (Huson et al., 2007) for assignment using the Lowest Common Ancestor (LCA) algorithm (within top 5% (CoralITS2_acro) and 3% (CoralITS2) based on BLASTN alignment score). Sequences with no matches or taxonomic assignments only at the domain level were discarded.
2.5 Statistical and ecological analysis
Relative abundances were calculated by dividing each feature count by total library size (i.e. total sum scaling). For the mock community samples, average relative abundances were calculated separately for the CoralITS2 and CoralITS2_acro community profiles and the expected versus observed outcomes of average relative abundances were assessed using a hierarchical model in a Bayesian framework (Gelman, 2003). Specifically, a beta-binomial model with weakly informative priors was constructed. The Bayesian model included three chains, each 5,000 interactions, thinned by a factor of ten, with the first 1,000 iterations used for warmup and excluded from the analysis. The model demonstrated good mixing and convergence on a stable posterior (Supplementary Figures S4-S6). Validation was performed using simulated residuals with the DHARMa package (Hartig, 2022; R Foundation for Statistical Computing, 2024). All statistical analyses were performed in the R Statistical and Graphical Environment (version 4.4.1; R Core Team, 2024) via the brms package (Bürkner, 2017). Post-hoc comparisons between observed and expected values were made based on the full posteriors before summarizing using draw-level chi-squared values, medians, and highest probability intervals.
To compare the six wild slick samples, a Bray-Curtis distance matrix was calculated for the ASVs detected, and a Principal Coordinate Analysis (PCoA) was performed to visualize the multidimensional relationships between samples in a lower-dimensional space. Each PCoA was created using the ‘cmdscale’ function in R (R Core Team, 2024) and visualized with Rstudio (v2024.4.2.764; Posit Team, 2025).
3 Results
3.1 Coral slick eDNA metabarcoding recapitulates expected profiles of mock communities
Metabarcoding of the mock communities yielded an average of 31,724 and 24,306 reads per sample for the CoralITS2_acro and CoralITS2 assays, respectively. After processing in DADA2 and merging co-occurring ASVs with high sequence similarity using LULU, this resulted in 54 and 10 ASVs in the CoralITS2_acro and CoralITS2 data sets, respectively. At the ASV level, the CoralITS2_acro and CoralITS2 primer sets were unable to resolve species taxonomies. Of the 10 known coral species present (Table 1), eight including A. loripes, A. austera, A. microphthalma, D. mathaii, D. pallida, M. elephantotus, and P. daedalea were not taxonomically identified in the CoralITS2_acro and CoralITS2 primer sets (Supplementary Figure S1). As such, ASV data were collapsed to genus or next lowest available rank.
In the CoralITS2_acro assay, we identified Platygyra ASVs, indicating that Platygyra were resolved to the genus level whereas Dipsastraea and Mycedium were likely only resolved to the suborder level (Vacatina). We therefore combined Dipsastraea and Mycedium into one expected Vacatina group with an expected 30% relative abundance. Overall, the observed average relative abundances in the known mix were comparable to the expected proportions: Acropora (59% observed vs. 60% expected), Vacatina (33% vs. 30%), and Platygyra (9% vs. 10%) (Table 3; Figure 2A). Variation between replicate samples was greatest for Acropora and Vacatina, resulting in a standard deviation of 18.9% and 15.5%, respectively. Platygyra showed average values close to the expected relative abundances and was more consistent between replicates, resulting in a low standard deviation of 3.3%.
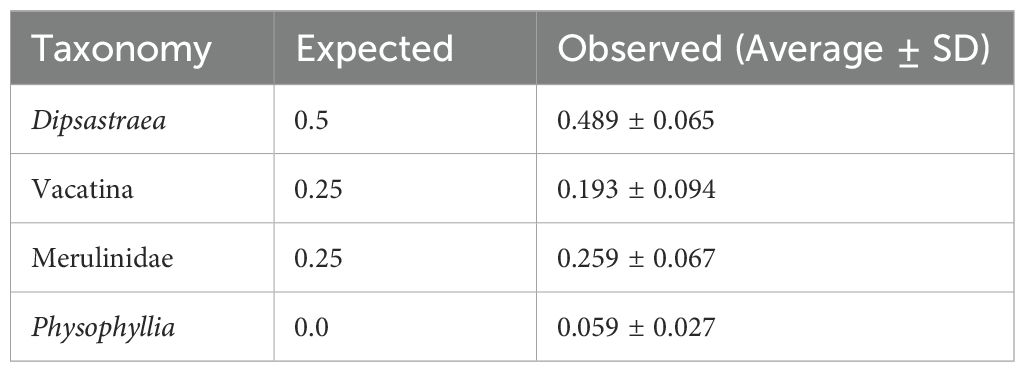
Table 3. Expected and observed (average and standard deviation) relative abundances of scleractinian coral taxa in known mixes of coral larvae for the CoralITS2 assay.
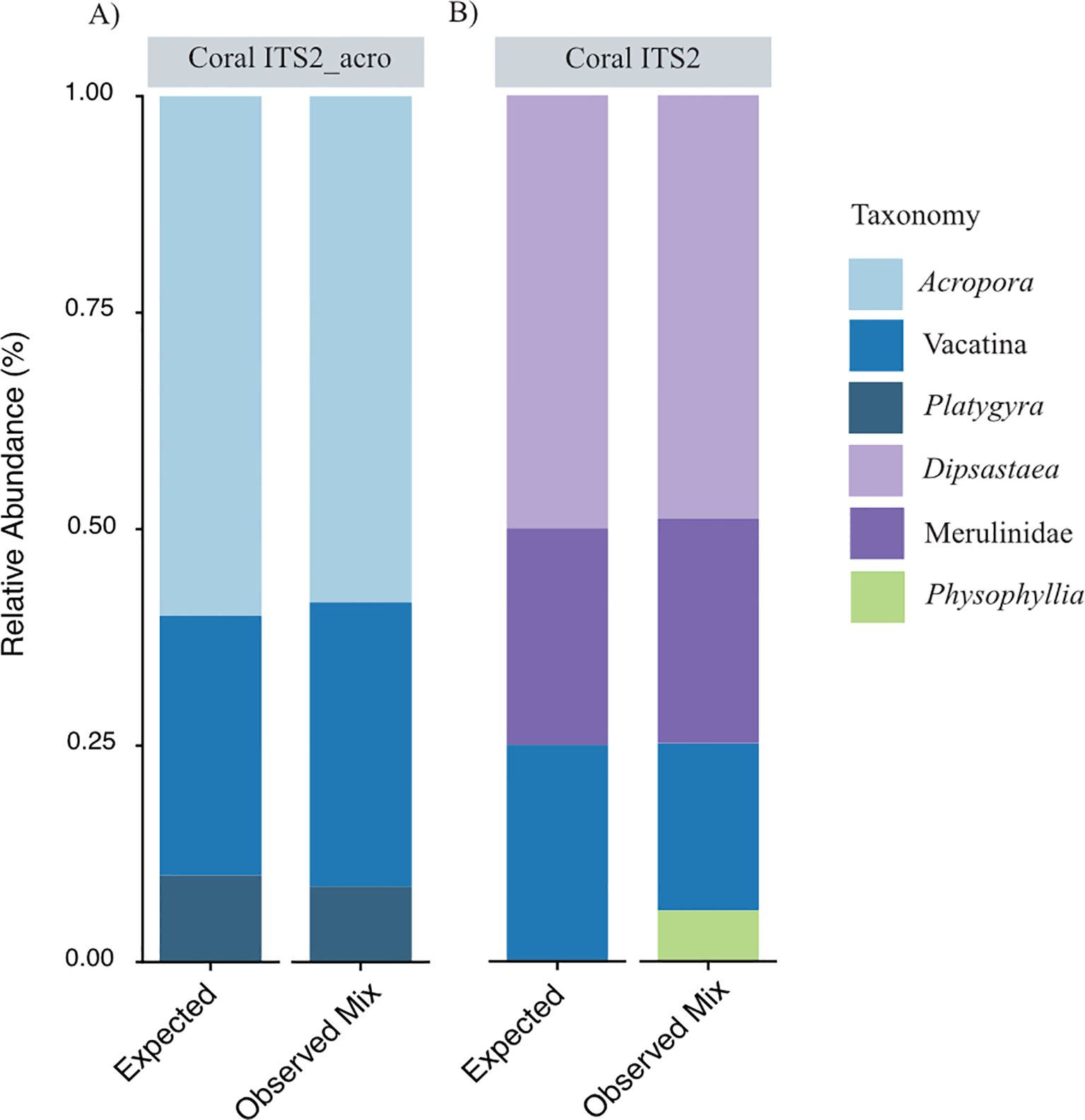
Figure 2. Bar plot showing expected versus observed relative abundance of scleractinian taxa within known mixes of coral larvae using the (A) CoralITS2_acro and (B) Coral ITS2 eDNA metabarcoding assays.
The CoralITS2 assay resulted in 10 ASV matches: eight mapping to known taxonomy in the mocks, one to a genus not present in the mock (Physophyllia), and one with no known taxonomic match. Dipsastraea was resolved to the genus level (Dipsastraea), while Platygyra was resolved to the family level (Merulinidae). The remaining ASVs resolved as suborder Vacatina likely represented Mycedium. Using this primer set, the observed vs. expected relative abundances were: Dipsastraea (49% observed vs. 50% expected), Vacatina (19% vs. 25%), Merulinidae (26% vs. 25%), and Physophyllia (6% vs. 0%) (Table 4; Figure 2B). Variation in relative abundances between replicates using this primer set was lower compared to coralITS2_acro, with standard deviations for Dipsastraea at 6.5%, Vacatina at 9.4%, Merulinidae at 6.7%, and Phosphyllia at 2.7% (Table 4).
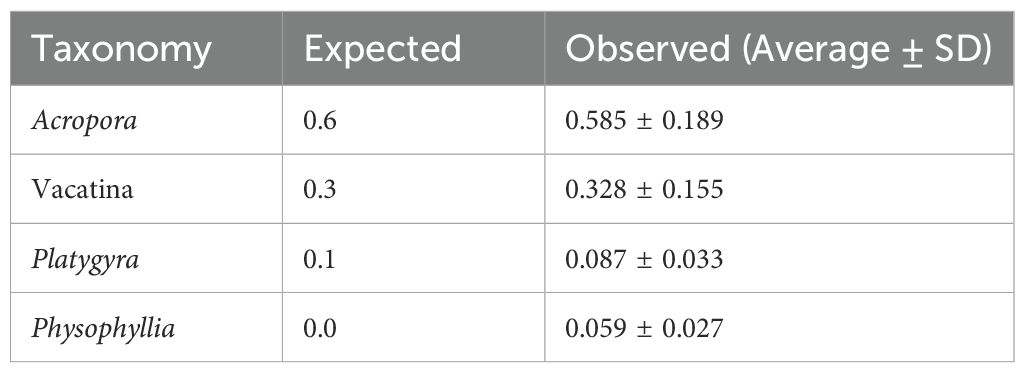
Table 4. Expected and observed (average and standard deviation) relative abundances of scleractinian coral taxa in known mixes of coral larvae for the CoralITS2_acro assay.
We applied a Bayesian beta-binomial model to evaluate whether the observed relative abundances deviated from the expected values. The model showed no evidence that the observed relative abundances differed from the expected values for both the CoralITS2_acro assay (median deviation = 0.065, 95% CI [-0.27 – 0.5]; x < 0 = 0.329, x > 0 = 0.671; Supplementary Figure S2) and the CoralITS2 assay (median deviation = 0.0622, 95% CI [-0.32 – 0.47]; x < 0 = 0.351, x > 0 = 0.649; Supplementary Figure S3). The variations observed were consistent with database quality, DNA quality, and random fluctuations versus a systematic effect.
3.2 Community composition of wild coral spawning slicks
Metabarcoding of the wild coral spawning slick samples yielded a total of 632,909 and 382,586 demultiplexed sequences, with an average of 37,230 and 23,911 reads per sample for the CoralITS2_acro and CoralITS2 assays, respectively. The CoralITS2_acro assay resulted in a total of 129 ASVs among coral spawning slicks, with an average of 35 ASVs per replicate sample. The CoralITS2 assay resulted in fewer ASVs, 52 in total, with an average of 7 ASVs per sample. In a few instances, one of the three replicates was excluded from downstream analyses of CoralITS2_acro (sites 5, 6) and CoralITS2 (site 4) communities due to failed PCR amplification or low yield with no taxonomic match. A further replicate from site 6 was included in the CoralITS2 relative abundance analysis but removed from the PCoA due to the recovery of a single ASV which made it incomparable with other samples.Taxonomic assignments for the majority of ASVs (89%) were resolved to at least the family level (Figures 3A, B). With the CoralITS2_acro assay, spawning slicks were dominated by Acroporidae at sites 1 (98-99% relative abundance), 3 (37-96%) and 4 (99-100%), and Vacatina (a suborder encompassing Merulinidae and Mussidae) at sites 2 (81-87%), 5 (55-90%) and 6 (98-100%). Poritidae was detected at sites 1 (0-<1%) and 2 (<1%) at low relative abundances (Figure 3A). Nearly all assigned taxonomic detections with the CoralITS2_acro assay were reef-building corals (Scleractinia), with the order Zoantharia detected only at site 1 (Cooktown, <1%). With the CoralITS2 assay, a greater diversity of families was recovered with similar differences in community composition among slicks. Acroporidae remained dominant in the slicks from site 3 (2-53%), while Merulinidae, and Vacatina were prevalent in varying proportions within slicks at sites 2 (2-97%), 5 (0-100%), and 6 (0-100%). However, Alcyoniidae (soft corals) was the dominant family in the slicks at site 1 (75-90%), and Actiniidae (sea anemones) was the dominant family within the slick at site 4 (78-82%). Other scleractinian (Pectiniidae, Poritidae) and non-scleractinian (Aplysinidae, Clionaidae) families were also detected in low quantities (<1%) in one or more slick samples, except Pectinidae detection at site 5 (0-18%).
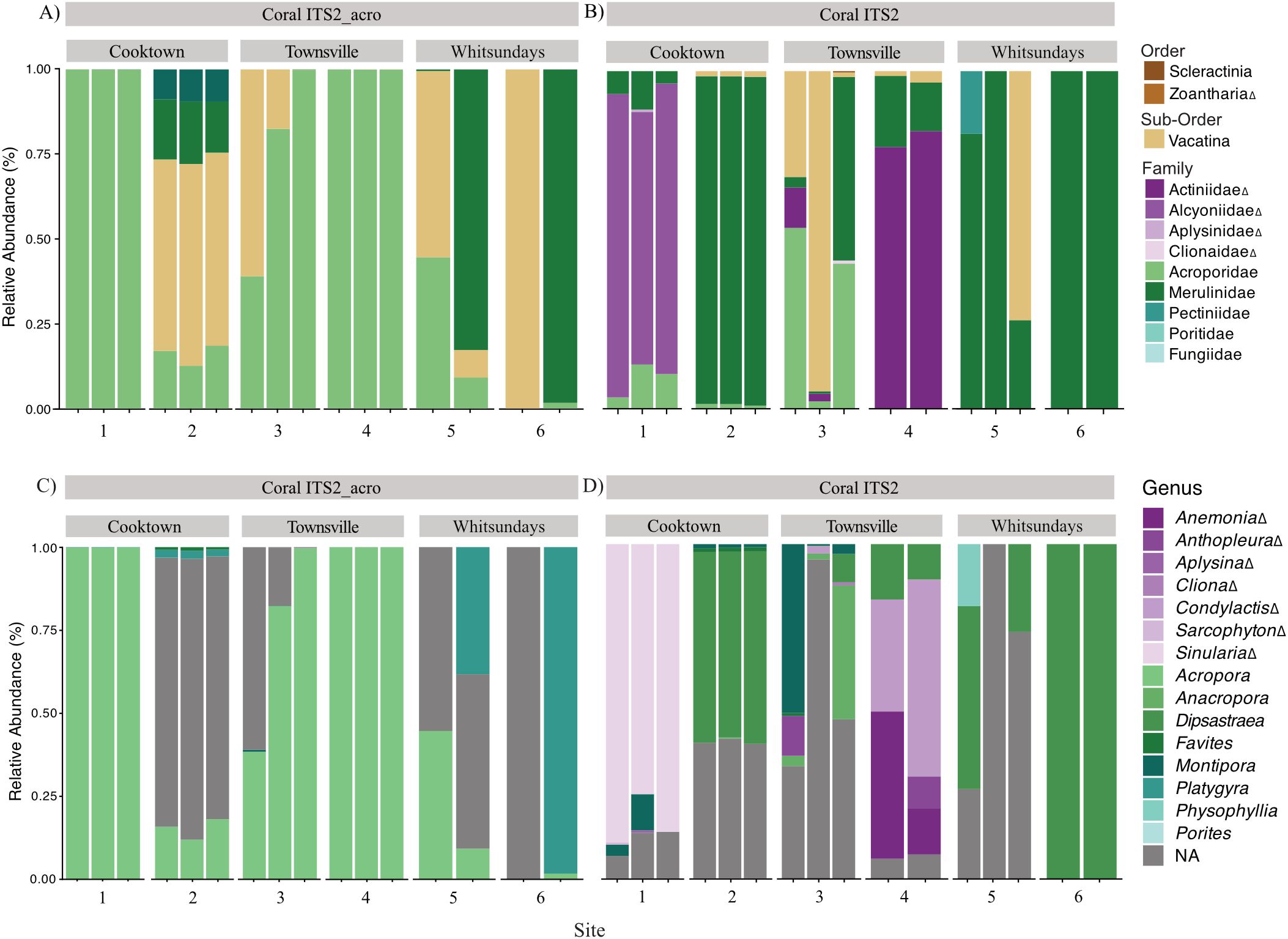
Figure 3. Community composition of wild coral spawning slicks on the Great Barrier Reef. Bar plots show the relative abundance of taxa detected using the (A) CoralITS2_acro and (B) CoralITS2 eDNA metabarcoding assays, resolved to the family level where possible; taxa that could not be resolved to a family are shown at the most resolved taxonomic level possible. (C, D) show the same data resolved to the genus level where possible, with grey bars (NA) indicating ASVs that could not be assigned at the genus level. Δ denotes detections of non-scleractinian (non-reef-building) corals.
At the genus level, there was a substantial reduction in explained diversity as 42 ASVs could not be resolved, particularly within the family Merulinindae (3 ASVs, CoralITS2_acro; 6 ASVs, CoralITS2) and suborder Vacatina (10, CoralITS2_acro; 8, CoralITS2). This reduction varies widely from 0% to 100% within replicate samples, with an average decrease of 31% (CoralITS2_acro) and 37% (CoralITS2) (Figures 3C, D). For the CoralITS2_acro assay, Acropora was consistently the most abundant resolved genus detected across slicks, except for two samples from sites 5 and 6 (Whitsundays) where Platygyra was dominant (Figure 3C). For the CoralITS2 assay, Dipsastraea was the most abundant resolved genus at sites 2, 5, and 6, and Anacropora and Montipora were most abundant at site 3. Soft coral (Sinularia) and anemone (Condylactis, Anemonia) genera were dominant at sites 1 and 4, respectively (Figure 3D).
The relative abundance data revealed notable variability across reef sites regarding taxa composition. At sites 1, 3, and 4, Acropora emerged as the most prevalent group, with the highest variation observed at site 3 (SD = 31.6%), while the other sites showed consistent ratios across replicates (SD < 0.01%). The site 2 spawning slick contained the most diverse taxa, with consistent relative abundance across replicates. While the standard deviation for Acropora was comparatively higher at 3.1%, other taxa showed generally low variation (SD < 1%). In contrast, the two Whitsundays sites (5 and 6) exhibited substantial variability, with large standard deviations for Acropora (25.1%), Vacatina (33.1%), and Merulinidae (31.1%) at Whitsundays site 6, as well as a notable detection of Platygyra in just one replicate (27.1%). The variability in these sites complicates the interpretation of their community composition.
These patterns of variability and consistency across sites are further supported by the Bray-Curtis dissimilarity analysis, which visualized the relationship between samples based on their taxonomic composition and abundance. The PCoA plot (Figure 4) illustrates that replicate samples from each reef site generally clustered together, suggesting that sites shared similar community compositions. However, certain sites showed closer relationships within and among their slicks. For example, sites 1, 2, 3, and 4 showed greater similarity among their replicate samples, while sites 5 and 6 demonstrated more divergent patterns (Figure 4). This trend is also evident in the CoralITS2 assay (Figure 4B), where site 2 consistently separated from other groups, and site 1 showed divergence within the clustering. Notably, the replicates from Whitsundays sites (5 and 6) were positioned farther apart, aligning with the observed discrepancies in relative abundance, particularly for Acropora. Shannon diversity varied across sampling locations within each primer set (Figure 4C). For the ITS2 assay, diversity was lowest at Site 6 (0) and highest at Sites 1, 3, and 4 (1.3-1.5). Similarly, in the ITS2_acro assay, diversity was lowest at Site 6 (0.7) and highest at sites 1, 3, and 4 (2.7-2.8).
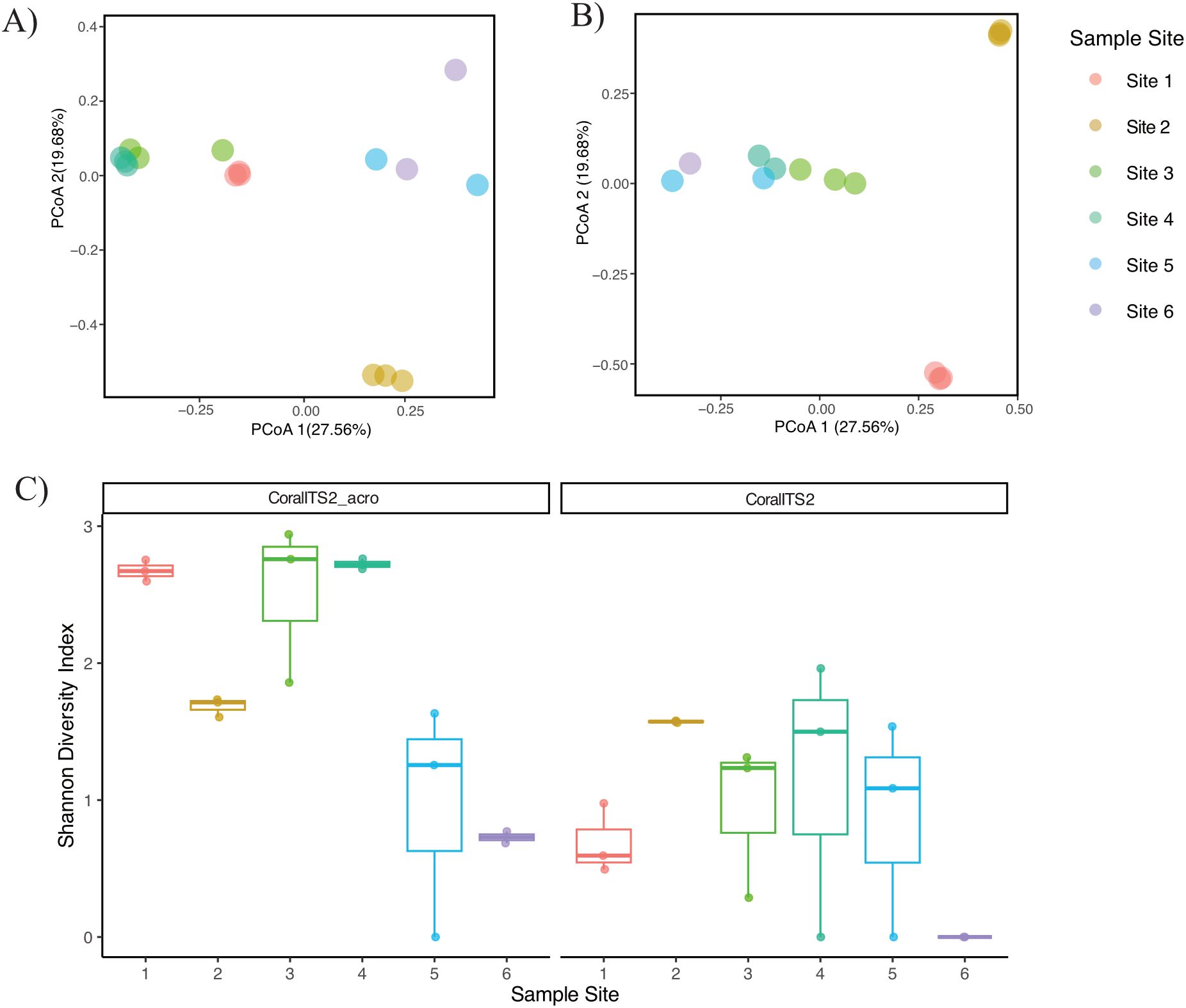
Figure 4. Dissimilarity and diversity plots of the community composition of wild coral spawning slicks from the Great Barrier Reef. Principal coordinates analysis was based on the Bray-Curtis dissimilarity metric for ASVs recovered from the (A) CoralITS2_acro and (B) CoralITS2 eDNA metabarcoding assays. (C) Shannon diversity plots by sample site for each primer set.
3.3 Assay detection
The assay results showed a variety of identified taxonomic groups, with some taxa not being resolved beyond the family and sub-order level. There was considerable cross-amplification of taxonomic groups across the two assays, and each assay identified unique families and genera (Figure 5). CoralITS2_acro identified a total of five families and six genera, while CoralITS2 identified seven families and 13 genera. As expected, the majority of ASVs recovered by the CoralITS2_acro (135) and CoralITS2 (52) assays were assigned to reef-building coral taxa (order Scleractinia). Several ASVs from the CoralITS2 assay were assigned to non-scleractinian coral taxa, including soft corals (5), anemones (8), sponges (2), and zoanthids (2). While CoralITS2 detected more unique taxonomic groups, of the 13 genera identified by the CoralITS2 assay, five were not corals but rather sea anemones and sponges. Additionally, at the genus level, Acropora was exclusively detected with the CoralITS2_acro assay and represented 54% of the ASVs identified in this study, highlighting the importance of this revised primer set in detecting this reef-dominant species. Lastly, 23% of the ASVs identified to the family level could not be resolved to genus, indicating that improved reference database resolution could lead to more precise identification (Figure 5).
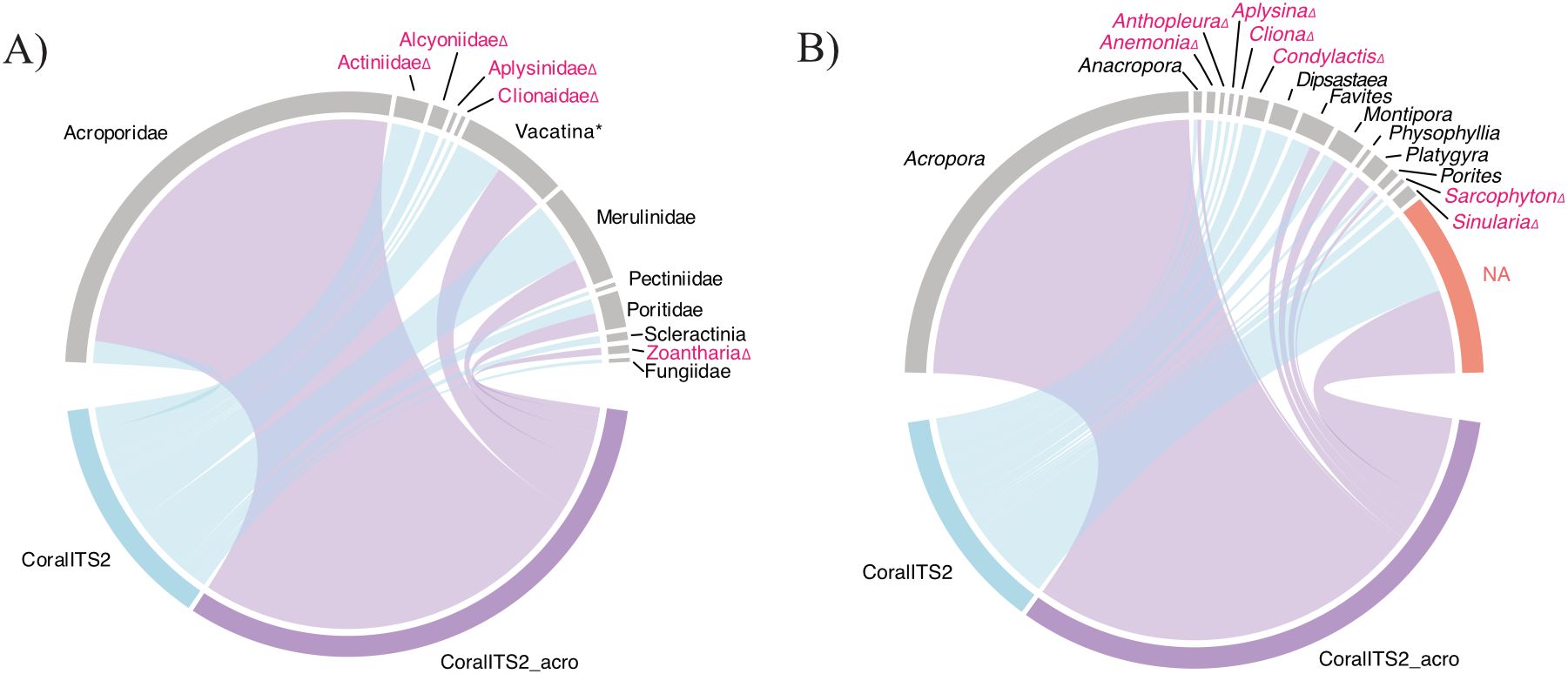
Figure 5. Taxonomic chord diagram of detections across CoralITS2_acro and CoralITS2 eDNA metabarcoding assays in wild coral spawning slicks from the Great Barrier Reef and known mixes of coral larvae. (A) Taxonomy is resolved to the family level where possible and is otherwise noted with an *. (B) Taxonomy is refined to the genus level where possible and is otherwise represented by NA. Non-scleractinian coral detections are noted with Δ and the names are colored pink. The size of endpoints reflects the number of unique ASVs attributed to each group.
4 Discussion
This study revealed the community composition of coral spawning slicks for the first time and demonstrated the applicability of using eDNA metabarcoding technology for this purpose. Analysis using the ITS2 region (CoralITS2 and CoralITS2_acro assays) identified 191 ASVs comprising 9 families and 15 genera from wild-collected slick samples on the Great Barrier Reef. The detection capabilities of both assays were validated with samples of known taxonomic diversity, confirming that observed relative abundance values aligned with expected profiles. These findings support further development of eDNA metabarcoding as a cost-effective approach for surveying the community composition of wild-captured spawning slicks, providing valuable insights for monitoring and restoration efforts.
Across the Cooktown and Townsville wild slick collection sites, Acropora was consistently the most abundant genus detected using the Coral ITS2_acro assay, aligning with survey data that shows Acropora as one of the most, and often the most, abundant broadcast spawning hard coral genera in these regions during the collection years (Australian Institute of Marine Science Long Term Monitoring Program1). In contrast, slicks from the Whitsundays collection sites were dominated by non-acroporid hard corals, particularly members of the Merulinidae family, which is consistent with benthic hard coral composition in this region during the collection years. These results underscore the ability of eDNA to capture region-specific reproductive outputs that mirror known benthic assemblages.
Leveraging eDNA methods for monitoring the community composition of coral spawning slicks offers several significant advantages for understanding coral spawning patterns and coral restoration applications. Spawning slicks represent a critical life-history stage for corals, acting as a source of larval dispersal and recruitment that ultimately shapes reef population dynamics (Harrison and Wallace, 1990; Jones et al., 2009). Understanding the taxonomic composition of these slicks allows for improved monitoring of reproductive success, assessment of species-specific contributions, and evaluation of how environmental stressors may influence reproductive output. This study makes key advancements in this area by providing the first taxonomic characterization of wild-collected spawning slicks, validating the use of eDNA for slick monitoring, and highlighting the potential for applying this approach in restoration efforts.
eDNA enhances detection capabilities by allowing for the simultaneous identification of multiple taxa, including those that are difficult to observe directly, such as cryptic or rare species. This approach also enables differentiation between reef-building corals and other marine mass synchronized spawners—such as soft corals, anemones, worms, and sponges (see Babcock et al., 1986, 1992) that may have contributed to the spawning slick and might otherwise be mistaken for corals based on visual observations alone. Moreover, eDNA reduces the need for specialized training and extensive night diving to accurately record observations of coral spawning on the reef during nocturnal spawning periods (Harrison et al., 1984; Babcock et al., 1986). This makes it an accessible and scalable option for monitoring coral reefs, especially for sites that are expensive and/or difficult to reach, facilitating frequent and widespread surveys at a lower cost. Additionally, eDNA-based methods provide a valuable tool for tracking changes in spawning patterns in response to environmental stressors, such as bleaching events, and offer insights into the health and reproductive output of coral spawning communities over time. Furthermore, emerging eRNA approaches, such as those described by Ye et al. (2025), may offer additional benefits by providing more immediate assessments of metabolically active organisms. These methods could enhance monitoring efforts during the culture of slicks for restoration, offering insights into coral larval viability and early-stage development.
Despite its advantages, the resolution of eDNA-based identification is currently constrained by inherent challenges in taxonomic assignment. The ITS2 region captures both inter- and intra-specific variation, describing diversity within and across coral species. Historical and contemporary hybridization events contribute to the intragenomic variation observed in the ITS2 region (Coleman and Van Oppen, 2008; Chan et al., 2018, 2019; Lamb et al., 2024) and can lead to ambiguous or incorrect taxonomic assignments, resulting in errors in sequence-based identification (Willis et al., 2006). Additionally, multiple potential matches may arise, leading to lower resolution in taxonomic assignments. Here we observed a reduction in the overall explained diversity when resolving taxa from the family to the genus level, likely due to these complexities.
Taxonomic resolution is further constrained by limitations in available sequence databases, which may lack comprehensive or up-to-date references for coral taxa (Dugal et al., 2022). These database gaps introduce uncertainties in eDNA-based identifications, increase the likelihood of unresolved classifications, and result in trade-offs between detailed resolution (genus-level) and the completeness of diversity representation (family-level). To address these issues, future work could enhance reference databases with better-curated and updated taxonomy and/or include internal spiked datasets containing known samples to improve detection depth and classification accuracy (Dugal et al., 2022). Moreover, differences in the coral taxa recovered between the CoralITS2 and CoralITS2_acro assays underscore the necessity of integrating multiple or alternative DNA markers, especially to capture Acropora and non-Acropora taxa.
ITS2 community profiling could be biased by variation in DNA copy number and fragment length, with shorter amplicons preferentially amplified during PCR, and species with more ITS2 copies potentially overestimated. To account for this, mock communities composed of known coral taxa were included in this study. A Bayesian beta-binomial model was applied to compare observed and expected taxonomic compositions within these mock communities. The model showed no evidence that expected and recovered relative abundances differed. This quality control step lends support that the taxonomic profiles recovered from wild slick samples reflect true biological community composition rather than technical artifacts introduced during amplification or sequencing.
Here we used three technical replicates per sample of briefly mixed samples. This recommended approach (Ficetola et al., 2015; Fonseca, 2018) produced consistent averages in our curated mix, however, more variable relative abundance ratios were observed in the wild slick samples. We suspect this is due to biological sample heterogeneity as gametes and larvae may clump together, particularly for slicks collected shortly after spawning when not all gamete bundles have dissociated. Yet, we cannot exclude the inconsistencies in DNA extraction efficiency and PCR amplification as contributors to variation among technical replicates. In future studies, we recommend implementing a more thorough mixing process to fully homogenize samples and exploring the effect of various levels of replication in quantifying the abundance of taxa in wild samples.
To further enhance the robustness of future analyses, we suggest that biological replicates from the same spawning slick are also collected to provide more precise estimates of community composition. Pairing this information with comprehensive environmental data such as hydrodynamic models (Gouezo et al., 2025), water temperature, and other relevant conditions during spawning (Keith et al., 2016; Lin and Nozawa, 2023; Nozawa, 2012; van Woesik, 2010) and with the context of disturbance history (e.g. bleaching events) would enable a deeper understanding of how environmental factors influence community-level reproductive output. Resampling the same regions over multiple spawning seasons can also shed light on the trajectories of community recovery following bleaching events, where surviving corals are expected to experience reduced fecundity (Ward et al., 2002). Once temporal variability of taxa in slicks at a given reef site has been characterized, shifts in the relative abundance of taxa within spawning slicks can be linked to changes on the reef benthos; these data would provide important insights into the long-term dynamics of coral populations and assemblages.
Importantly, sampling limitations arise from the timing of surface-collected spawning slick material, which may not capture the full diversity of coral taxa. For example, certain groups, such as taxa that release negatively or neutrally buoyant gamete bundles, and species that spawn during the day rather than at night, and at different times after sunset or on different lunar phases may be missed (Babcock et al., 1986; Harrison and Wallace, 1990; Harrison, 2024a). The timing of sampling relative to the full moon also influences what species may be captured due to differences in reproductive timing across lunar phases and months (Baird et al., 2021). Furthermore, brooding coral species, which release fully formed swimming larvae will not be captured in spawning slicks. Improving the temporal resolution of eDNA sampling around the timing of spawning to collect over multiple days and nights around the full moon would increase our understanding of the diversity of species reproducing during these periods (Ip et al., 2023).
Lastly, in the context of restoration activities that use wild slicks as material for restoring degraded reefs (Heyward et al., 2002; Harrison et al., 2021; Banaszak et al., 2023; Harrison, 2024a), the current processing times for PCR and sequencing of eDNA samples can exceed the time available before settlement begins. Understanding the species composition and diversity within larval cohorts is essential for predicting settlement and recruitment dynamics. More importantly, identifying potential contamination by soft corals, which are major competitors for reef-building corals, is crucial before larvae are introduced to target reef sites. A potential solution to this challenge is the use of lateral flow assay (LFA) dipstick methods, as demonstrated by Doyle and Uthicke (2020). LFA enables rapid, field-based detection of eDNA within minutes, rather than the days required for traditional sequencing-based workflows. Incorporating LFA into spawning slick monitoring could facilitate real-time taxa verification, ensuring that the collected spawn produces larval cohorts that are appropriate for larval restoration, including target taxa before larval enhancement occurs.
This study provides significant advances in our understanding of the taxonomic composition of wild coral spawning slicks and demonstrates the feasibility of using eDNA metabarcoding for their characterization. While current limitations in taxonomic resolution and database completeness remain challenges, the integration of additional markers, database improvements, and methodological refinements can further enhance eDNA-based monitoring. The successful validation of this approach in both mock communities and wild slicks establishes eDNA as a valuable tool for assessing coral community reproductive output, monitoring ecosystem health, and informing restoration strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1230602.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because Proper permits obtained (see manuscript), no other ethical approval required.
Author contributions
BM: Visualization, Formal analysis, Writing – review & editing, Writing – original draft. HE: Supervision, Writing – review & editing. YY: Data curation, Writing – review & editing, Supervision. CR: Resources, Writing – review & editing. AH: Resources, Writing – review & editing. PH: Funding acquisition, Resources, Writing – review & editing. ML: Formal analysis, Writing – review & editing. EH: Conceptualization, Methodology, Writing – review & editing, Investigation, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by The Reef Restoration and Adaptation Program (RRAP), a partnership between the Australian Governments Reef Trust and the Great Barrier Reef Foundation, and a Paul G Allen Family Foundation Grant to PH.
Acknowledgments
We acknowledge the Dingaal, Bindal, and Ngaro Peoples as the Traditional Custodians of the sea Countries where this research took place, and we acknowledge their Elder’s past, present, and emerging, and their continuing spiritual connection to sea Country. We thank Christina Langley for collecting the Whitsundays samples and the SCU and CSIRO coral larval restoration team members for field support at Lizard Island and in the Whitsundays. This study was funded by The Reef Restoration and Adaptation Program (RRAP), a partnership between the Australian Governments Reef Trust and the Great Barrier Reef Foundation, and a Paul G Allen Family Foundation Grant to PH. Broodstock coral colonies for mock slick creation were collected under permits G12/35236.1 and G19/43024.1, and spawn slick samples were collected under permit G21/46077.1, issued by the Great Barrier Reef Marine Park Authority (GBRMPA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. AI tool ChatGPT 4o was utilized to improve the manuscript flow.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2025.1602543/full#supplementary-material
Footnotes
References
Abarenkov K., Nilsson R. H., Larsson K. H., Taylor A. F. S., May T. W., Frøslev T. G., et al. (2024). The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Res. 52, D791–D797. doi: 10.1093/nar/gkad1039
Alexander J. B., Bunce M., White N., Wilkinson S. P., Adam A. A. S., Berry T., et al. (2020). Development of a multi-assay approach for monitoring coral diversity using eDNA metabarcoding. Coral Reefs 39, 159–171. doi: 10.1007/s00338-019-01875-9
Anthony K., Bay L. K., Costanza R., Firn J., Gunn J., Harrison P., et al. (2017). New interventions are needed to save coral reefs. Nat. Ecol. Evol. 1, 1420–1422. doi: 10.1038/s41559-017-0313-5
Babcock R. C., Baird A. H., Piromvaragorn S., Thomson D. P., and Willis B. L. (2003). Identification of scleractinian coral recruits from Indo-Pacific reefs. Zool. Stud. 42, 211–226.
Babcock R. C., Bull G. D., Harrison P. L., Heyward A. J., Oliver J. K., Wallace C. C., et al. (1986). Synchronous spawnings of 105 scleractinian coral species on the Great Barrier Reef. Mar. Biol. 90, 379–394. doi: 10.1007/BF00428562
Babcock R. C., Mundy C., Keesing J., and Oliver J. (1992). Predictable and unpredictable spawning events: In situ behavioural data from free-spawning coral reef invertebrates. Invertebrate Reprod. Dev. 22, 213–227. doi: 10.1080/07924259.1992.9672274
Babcock R. C., Thomson D. P., Haywood M. D. E., Vanderklift M. A., Pillans R., Rochester W. A., et al. (2021). Recurrent coral bleaching in north-western Australia and associated declines in coral cover. Mar. Freshw. Res. 72, 620–632. doi: 10.1071/MF19378
Baird A. H., Guest J. R., Edwards A. J., Bauman A. G., Bouwmeester J., Mera H., et al. (2021). An Indo-Pacific coral spawning database. Sci. Data 8. doi: 10.1038/s41597-020-00793-8
Baker A. C., Glynn P. W., and Riegl B. (2008). Climate change and coral reef bleaching: An ecological assessment of long-term impacts, recovery trends and future outlook. Estuar. Coast. Shelf Sci. 80, 435–471. doi: 10.1016/j.ecss.2008.09.003
Banaszak A. T., Marhaver K. L., Miller M. W., Hartmann A. C., Albright R., Hagedorn M., et al. (2023). Applying coral breeding to reef restoration: best practices, knowledge gaps, and priority actions in a rapidly-evolving field. Restor. Ecol. 31, e13913. doi: 10.1111/rec.13913
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boström-Einarsson L., Babcock R. C., Bayraktarov E., Ceccarelli D., Cook N., Ferse S. C. A., et al. (2020). Coral restoration – A systematic review of current methods, successes, failures and future directions. PloS One 15, e0226631. doi: 10.1371/journal.pone.0226631
Briggs N. D., Page C. A., Giuliano C., Alessi C., Hoogenboom M., Bay L. K., et al. (2024). Dissecting coral recovery: bleaching reduces reproductive output in Acropora millepora. Coral Reefs 43, 557–569. doi: 10.1007/s00338-024-02483-y
Bullock J. M., Aronson J., Newton A. C., Pywell R. F., and Rey-Benayas J. M. (2011). Restoration of ecosystem services and biodiversity: Conflicts and opportunities. Trends Ecol. Evol. 26, 541–549. doi: 10.1016/j.tree.2011.06.011
Bürkner P. C. (2017). brms: An R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, 1–28. doi: 10.18637/jss.v080.i01
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chamberland V. F., Petersen D., Guest J. R., Petersen U., Brittsan M., and Vermeij M. J. A. (2017). New seeding approach reduces costs and time to outplant sexually propagated corals for reef restoration. Sci. Rep. 7, 18076. doi: 10.1038/s41598-017-17555-z
Chan W. Y., Hoffmann A. A., and van Oppen M. J. H. (2019). Hybridization as a conservation management tool. Conserv. Lett. 12, e12652. doi: 10.1111/conl.12652
Chan W. Y., Peplow L. M., Menéndez P., Hoffmann A. A., and van Oppen M. J. H. (2018). Interspecific hybridization may provide novel opportunities for coral reef restoration. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00160
Cilleros K., Valentini A., Allard L., Dejean T., Etienne R., Grenouillet G., et al. (2019). Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 19, 27–46. doi: 10.1111/1755-0998.12900
Coleman A. W. and Van Oppen M. J. H. (2008). Secondary structure of the rRNA ITS2 region reveals key evolutionary patterns in acroporid corals. J. Mol. Evol. 67, 389–396. doi: 10.1007/s00239-008-9160-y
Connell J. H., Hughes T. P., and Wallace C. C. (1997). A 30-year study of coral abundance, recruitment, and disturbance at several scales in space and time. Ecol. Monogr. 67, 461–488. doi: 10.1890/0012-9615(1997)067[0461:AYSOCA]2.0.CO;2
Cox E. F. (2007). Continuation of sexual reproduction in Montipora capitata following bleaching. Coral Reefs 26, 721–724. doi: 10.1007/s00338-007-0251-9
dela Cruz D. W. and Harrison P. L. (2017). Enhanced larval supply and recruitment can replenish reef corals on degraded reefs. Sci. Rep. 7. doi: 10.1038/s41598-017-14546-y
dela Cruz D. W. and Harrison P. L. (2020). Enhancing coral recruitment through assisted mass settlement of cultured coral larvae. PloS One 15, 13985. doi: 10.1371/journal.pone.0242847
Dietzel A., Connolly S. R., Hughes T. P., and Bode M. (2021). The spatial footprint and patchiness of large-scale disturbances on coral reefs. Global Change Biol. 27, 4825–4838. doi: 10.1111/gcb.15805
Doropoulos C., Vons F., Elzinga J., ter Hofstede R., Salee K., van Koningsveld M., et al. (2019). Testing industrial-scale coral restoration techniques: harvesting and culturing wild coral-spawn slicks. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00658
Doyle J. and Uthicke S. (2020). Sensitive environmental DNA detection via lateral flow assay (dipstick)—A case study on corallivorous crown-of-thorns sea star (Acanthaster cf. solaris) detection. Environ. DNA 3, 323–342. doi: 10.1002/edn3.123
Dugal L., Thomas L., Wilkinson S. P., Richards Z. T., Alexander J. B., Adam A. A. S., et al. (2022). Coral monitoring in northwest Australia with environmental DNA metabarcoding using a curated reference database for optimized detection. Environ. DNA 4, 63–76. doi: 10.1002/edn3.199
Everett M. V. and Park L. K. (2018). Exploring deep-water coral communities using environmental DNA. Deep-Sea Res. Part II: Topical Stud. Oceanogr. 150, 229–241. doi: 10.1016/j.dsr2.2017.09.008
Ficetola G. F., Pansu J., Bonin A., Coissac E., Giguet-Covex C., De Barba M., et al. (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 15, 543–556. doi: 10.1111/1755-0998.12338
Fonseca V. G. (2018). Pitfalls in relative abundance estimation using edna metabarcoding. Mol. Ecol. Resour. 18, 923–926. doi: 10.1111/1755-0998.12902
Fox H. E. and Caldwell R. L. (2006). Recovery from blast fishing on coral reefs: A tale of two scales. Ecol. Appl. 16, 1631–1635. doi: 10.1890/1051-0761(2006)016[1631:RFBFOC]2.0.CO;2
Frøslev T. G., Kjøller R., Bruun H. H., Ejrnæs R., Brunbjerg A. K., Pietroni C., et al. (2017). Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188. doi: 10.1038/s41467-017-01312-x
Gadoutsis E., Daly C. A. K., Hawkins J. P., and Daly R. (2019). Post-bleaching mortality of a remote coral reef community in Seychelles, Western Indian Ocean. Western Indian Ocean J. Mar. Sci. 18. doi: 10.4314/wiojms.v18i1.2
Gelman A. (2003). A Bayesian formulation of exploratory data analysis and goodness-of-fit testing. Int. Stat. Rev. 71, 369–382. doi: 10.1111/j.1751-5823.2003.tb00203.x
Gilmour J. P., Smith L. D., Heyward A. J., Baird A. H., and Pratchett M. S. (2013). Recovery of an isolated coral reef system following severe disturbance. Science 340, 69–71. doi: 10.1126/science.1232310
Gouezo M., Langlais C., Beardlsey J., Roff G., Harrison P., Thomson D., et al. (2025). Going with the flow: leveraging reef-scale hydrodynamics for upscaling larval-based restoration. bioRxiv. 35(3), e70020. doi: 10.1002/eap.70020
Gouezo M., Olsudong D., Fabricius K., Harrison P., Golbuu Y., and Doropoulos C. (2020). Relative roles of biological and physical processes influencing coral recruitment during the lag phase of reef community recovery. Sci. Rep. 10, 2471. doi: 10.1038/s41598-020-59111-2
Graham N. A. J. and Nash K. L. (2013). The importance of structural complexity in coral reef ecosystems. Coral Reefs 32, 315–326. doi: 10.1007/s00338-012-0984-y
Gratwicke B. and Speight M. R. (2005). The relationship between fish species richness, abundance and habitat complexity in a range of shallow tropical marine habitats. J. Fish Biol. 66, 650–667. doi: 10.1111/j.0022-1112.2005.00629.x
Harrison P. L. (2024a). “Sexual reproduction of reef corals and application to coral restoration,” in Oceanographic Processes of Coral Reefs. (CRC Press). doi: 10.1201/9781003320425-32
Harrison P. L. (2024b). Reef-based mass coral larval culture and restoration methods [Technical Report] (Lismore: Southern Cross University). doi: 10.25918/report.434
Harrison R. L., Babcock R. C., Bull G. D., Oliver J. K., Wallace C. C., and Willis B. L. (1984). Mass spawning in tropical reef corals. Science 223, 1186–1189. doi: 10.1126/science.223.4641.1186
Harrison P. L., dela Cruz D. W., Cameron K. A., and Cabaitan P. C. (2021). Increased coral larval supply enhances recruitment for coral and fish habitat restoration. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.750210
Harrison P. L. and Wallace C. (1990). Reproduction, dispersal and recruitment of scleractinian corals Ecosystems of the world. 25: Coral Reefs. Ecosyst. World 25: Coral Reefs 133–207.
Hartig F. (2022). DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. Available online at: https://CRAN.R-project.org/package=DHARMa. (Accessed July 9, 2024).
Henley B. J., McGregor H. V., King A. D., Hoegh-Guldberg O., Arzey A. K., Karoly D. J., et al. (2024). Highest ocean heat in four centuries places Great Barrier Reef in danger. Nature 632, 320–326. doi: 10.1038/s41586-024-07672-x
Heyward A. J., Smith L. D., Rees M., and Field S. N. (2002). Enhancement of coral recruitment by in situ mass culture of coral larvae. Mar. Ecol. Prog. Ser. 230, 113–118. doi: 10.3354/meps230113
Holbrook S. J., Adam T. C., Edmunds P. J., Schmitt R. J., Carpenter R. C., Brooks A. J., et al. (2018). Recruitment drives spatial variation in recovery rates of resilient coral reefs. Sci. Rep. 8, 7338. doi: 10.1038/s41598-018-25414-8
Howells E. J., Ketchum R. N., Bauman A. G., Mustafa Y., Watkins K. D., and Burt J. A. (2016). Species-specific trends in the reproductive output of corals across environmental gradients and bleaching histories. Mar. pollut. Bull. 105, 532–539. doi: 10.1016/j.marpolbul.2015.11.034
Hughes T. P., Kerry J. T., Álvarez-Noriega M., Álvarez-Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Hughes T. P., Kerry J. T., Baird A. H., Connolly S. R., Chase T. J., Dietzel A., et al. (2019). Global warming impairs stock–recruitment dynamics of corals. Nature 568, 387–390. doi: 10.1038/s41586-019-1081-y
Huson D. H., Auch A. F., Qi J., and Schuster S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Intergovernmental Panel on Climate Change (IPCC) (2023). “Climate change 2022 – impacts, adaptation and vulnerability,” in Climate Change 2022 – Impacts, Adaptation and Vulnerability. (Cambridge University Press) doi: 10.1017/9781009325844
Ip Y. C. A., Chang J. J. M., Tun K. P. P., Meier R., and Huang D. (2023). Multispecies environmental DNA metabarcoding sheds light on annual coral spawning events. Mol. Ecol. 32, 6474–6488. doi: 10.1111/mec.16621
Johnston E. C., Counsell C. W. W., Sale T. L., Burgess S. C., and Toonen R. J. (2020). The legacy of stress: Coral bleaching impacts reproduction years later. Funct. Ecol. 34, 2315–2325. doi: 10.1111/1365-2435.13653
Jones G. P., Almany G. R., Russ G. R., Sale P. F., Steneck R. S., Van Oppen M. J. H., et al. (2009). Larval retention and connectivity among populations of corals and reef fishes: History, advances and challenges. Coral Reefs 28, 307–325. doi: 10.1007/s00338-009-0469-9
Jonker M., Johns K., and Osborne K. (2008). “Surveys of benthic reef communities using underwater digital photography and counts of juvenile corals,” in Long-term Monitoring of the Great Barrier Reef. vol. 10.
Keith S. A., Maynard J. A., Edwards A. J., Guest J. R., Bauman A. G., van Hooidonk R., et al. (2016). Coral mass spawning predicted by rapid seasonal rise in ocean temperature. Proc. R. Soc. B: Biol. Sci. 283, 20160011. doi: 10.1098/rspb.2016.0011
Lamb A. M., Peplow L. M., Chan W. Y., Crane Z. J., Everson G. A., Harrison P. L., et al. (2024). Fertile hybrids could aid coral adaptation. Ecol. Evol. 14, e70570. doi: 10.1002/ECE3.70570
Liew Y. J., Howells E. J., Wang X., Michell C. T., Burt J. A., Idaghdour Y., et al. (2020). Intergenerational epigenetic inheritance in reef-building corals. Nat. Climate Change 10, 254–259. doi: 10.1038/s41558-019-0687-2
Lin C. H. and Nozawa Y. (2023). The influence of seawater temperature on the timing of coral spawning. Coral Reefs 42, 417–426. doi: 10.1007/s00338-023-02349-9
Mallela J. and Crabbe M. J. C. (2009). Hurricanes and coral bleaching linked to changes in coral recruitment in Tobago. Mar. Environ. Res. 68, 158–162. doi: 10.1016/j.marenvres.2009.06.001
Maynard J. A., Anthony K. R. N., Marshall P. A., and Masiri I. (2008). Major bleaching events can lead to increased thermal tolerance in corals. Mar. Biol. 155, 173–182. doi: 10.1007/s00227-008-1015-y
Michalek-Wagner K. and Willis B. L. (2001). Impacts of bleaching on the soft coral Lobophytum compactum. I. Fecundity, fertilization and offspring viability. Coral Reefs 19, 231–239. doi: 10.1007/s003380170003
Miller M. W., Latijnhouwers K. R. W., Bickel A., Mendoza-Quiroz S., Schick M., Burton K., et al. (2022). Settlement yields in large-scale in situ culture of Caribbean coral larvae for restoration. Restor. Ecol. 30, e13512. doi: 10.1111/rec.13512
Newman S. P., Meesters E. H., Dryden C. S., Williams S. M., Sanchez C., Mumby P. J., et al. (2015). Reef flattening effects on total richness and species responses in the Caribbean. J. Anim. Ecol. 84, 1678–1689. doi: 10.1111/1365-2656.12429
Nozawa Y. (2012). Annual variation in the timing of coral spawning in a high-latitude environment: Influence of temperature. Biol. Bull. 222, 192–202. doi: 10.1086/BBLv222n3p192
Ortiz J. C., Wolff N. H., Anthony K. R. N., Devlin M., Lewis S., and Mumby P. J. (2018). Impaired recovery of the great barrier reef under cumulative stress. Sci. Adv. 4, eaar6127. doi: 10.1126/sciadv.aar6127
Piñón-González V. M. and Banaszak A. T. (2018). Effects of partial mortality on growth, reproduction and total lipid content in the Elkhorn coral Acropora palmata. Front. Mar. Sci. 9. doi: 10.3389/fmars.2018.00396
Posit Team (2025). RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA. URL http://www.posit.co/.
Randall C. J., Giuliano C., Allen K., Bickel A., Miller M., and Negri A. P. (2023). Site mediates performance in a coral-seeding trial. Restor. Ecol. 31, e13745. doi: 10.1111/rec.13745
Randall C. J., Giuliano C., Heyward A. J., and Negri A. P. (2021). Enhancing coral survival on deployment devices with microrefugia. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.662263
Randall C. J., Giuliano C., Stephenson B., Whitman T. N., Page C. A., Treml E. A., et al. (2024). Larval precompetency and settlement behaviour in 25 Indo-Pacific coral species. Commun. Biol. 7, 142. doi: 10.1038/s42003-024-05824-3
R Foundation for Statistical Computing (2024). “R core,” in R: A language and environment for statistical computing.
Richmond R. and Hunter C. (1990). Reproduction and recruitment of corals: comparisons among the Caribbean, the Tropical Pacific, and the Red Sea. Mar. Ecol. Prog. Ser. 60, 185–203. doi: 10.3354/meps060185
Sheppard C. R. C., Harris A., and Sheppard A. L. S. (2008). Archipelago-wide coral recovery patterns since 1998 in the Chagos Archipelago, central Indian Ocean. Mar. Ecol. Prog. Ser. 362, 109–117. doi: 10.3354/meps07436
Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Climate Change 9, 306–312. doi: 10.1038/s41558-019-0412-1
Souter D., Planes S., Wicquart J., Logan M., David O., and Staub F. (2020). Status of coral reefs of the world: 2020 executive summary. Global Coral Reef Monit. Netw.
Tabalanza T. D., Jamodiong E. A., Diaz L. A., Tañedo M. C. S., Leriorato J. C., Villanueva R. D., et al. (2020). Successfully cultured and reared coral embryos from wild caught spawn slick in the Philippines. Aquaculture 525, 735354. doi: 10.1016/j.aquaculture.2020.735354
Thomsen P. F., Kielgast J., Iversen L. L., Møller P. R., Rasmussen M., and Willerslev E. (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PloS One 7, e41732. doi: 10.1371/journal.pone.0041732
van Woesik R. (2010). Calm before the spawn: Global coral spawning patterns are explained by regional wind fields. Proc. R. Soc. B: Biol. Sci. 277, 715–722. doi: 10.1098/rspb.2009.1524
van Woesik R., Cacciapaglia C., and Randall C. J. (2016). “Thermal-stress response of coral communities to climate change,” in The Cnidaria, past, present and Future: The World of Medusa and her Sisters. 545–552. doi: 10.1007/978-3-319-31305-4_33
Vaughan D. E. (2021). Active coral restoration: Techniques for a changing planet (Plantation FL USA: J. Ross Publishing), 610.
Ward S., Harrison P., and Hoegh-guldberg O. (2002). Coral bleaching reduces reproduction of scleractinian corals and increases susceptibility to future stress. Proc. 9th Int. Coral Reef Symposium 1123–1129.
Weis V. M. (2010). The susceptibility and resilience of corals to thermal stress: Adaptation, acclimatization or both?: NEWS and VIEWS. Mol. Ecol. 19, 1515–1517. doi: 10.1111/j.1365-294X.2010.04575.x
Whitman T. N., Hoogenboom M. O., Negri A. P., and Randall C. J. (2024). Coral-seeding devices with fish-exclusion features reduce mortality on the Great Barrier Reef. Sci. Rep. 14, 1–15. doi: 10.1038/s41598-024-64294-z
Willerslev E., Hansen A. J., Binladen J., Brand T. B., Gilbert M. T. P., Shapiro B., et al. (2003). Diverse plant and animal genetic records from holocene and pleistocene sediments. Science 300, 791–795. doi: 10.1126/science.1084114
Willis B. L., Van Oppen M. J. H., Miller D. J., Vollmer S. V., and Ayre D. J. (2006). The role of hybridization in the evolution of reef corals. Annu. Rev. Ecol. Evol. Syst. 37, 489–517. doi: 10.1146/annurev.ecolsys.37.091305.110136
Yadav S., Roach T. N. F., McWilliam M. J., Caruso C., de Souza M. R., Foley C., et al. (2023). Fine-scale variability in coral bleaching and mortality during a marine heatwave. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1108365
Keywords: biodiversity, EDNA, ITS2, metabarcoding, great barrier reef, coral seeding, monitoring
Citation: Marquardt B, Elder H, Yeoh YK, Heyward A, Randall CJ, Harrison PL, Logan M and Howells E (2025) Determining the diversity and relative abundance of coral taxa in wild spawning slicks for effective restoration. Front. Ecol. Evol. 13:1602543. doi: 10.3389/fevo.2025.1602543
Received: 29 March 2025; Accepted: 04 June 2025;
Published: 02 July 2025.
Edited by:
Carlos Prada, University of Rhode Island, United StatesReviewed by:
Viridiana Alvarado, Center for Research and Advanced Studies - Mérida Unit, MexicoAlicia Vollmer, Florida Fish and Wildlife Research Institute, United States
Copyright © 2025 Marquardt, Elder, Yeoh, Heyward, Randall, Harrison, Logan and Howells. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bailey Marquardt, Ym1hcnF1YXJkdDJAZ21haWwuY29t
 Bailey Marquardt
Bailey Marquardt Holland Elder2
Holland Elder2 Yun Kit Yeoh
Yun Kit Yeoh Andrew Heyward
Andrew Heyward Carly J. Randall
Carly J. Randall Peter L. Harrison
Peter L. Harrison