- Department of Medicine, Tufts University School of Medicine, Boston, MA, United States
Introduction: Life’s macroevolutionary patterns—rapid post-extinction recoveries and bursts of novelty—are not fully explained by mutation and vertical descent alone. I introduce Biological Recycling Theory (BRT), a cyclic, network-based framework.
Methods: An agent-based model compared four scenarios (classical, cryptic-only, HGT-only, full BRT), with extinction pulses and explicit constraints on DNA uptake/compatibility; code and runs are archived.
Results: Under 50% extinction, BRT restored ~90% of pre-event diversity in ~⅓ fewer generations than classical models and yielded ~3,600 novel genotype combinations (vs. ~2,800 cryptic-only; ~700 HGT-only; ~0 mutation-only). Longer eDNA half-life increased diversity retention and innovation.
Discussion: BRT integrates balancing selection, cryptic genetic variation, and genetic recycling via HGT/eDNA to expand the effective genetic search space across time, offering a testable framework for macroevolutionary resilience.
Conclusion: Evolution is better modeled as a cyclic network, where alleles circulate across populations, environments, and time, complementing Darwinian microevolution.
1 Introduction
Darwinian evolution has provided a robust framework for understanding biological change for more than 150 years. Natural selection acting on heritable variation remains one of the most powerful and empirically validated explanations for microevolutionary dynamics—the emergence of small-scale adaptations within species (Endler, 1986; Futuyma, 2013). Classic cases such as antibiotic resistance in bacteria or beak diversification in Darwin’s finches confirm the power of mutation and selection to shape traits within populations over relatively short timescales (Grant and Grant, 2008; Davies and Davies, 2010).
However, macroevolutionary transitions—such as the origin of new lineages, adaptive radiations after mass extinctions, and the persistence of ancient alleles across deep time—often unfold faster and at scales that mutation and vertical inheritance alone cannot fully explain (Gould and Eldredge, 1977; Lynch, 2007). Metagenomic surveys reveal large pools of uncharacterized genes circulating in environmental samples (Louca et al., 2018). Paleogenomic evidence shows that DNA can persist for long periods under favorable conditions (e.g., permafrost and sediments) (Kjær et al., 2022; Murchie et al., 2022). Genomic analyses also report horizontal gene transfer (HGT) in eukaryotes once thought resistant to it (Redmond and McLysaght, 2023). Together, these findings suggest that evolution cannot be fully captured by a one-way, branching “tree of life” alone.
Here I propose the Biological Recycling Theory (BRT): a cyclic, network-based view in which genetic material is passed vertically and recycled across temporal and taxonomic boundaries. BRT integrates three empirically grounded pillars—balancing selection (Andrés et al., 2009), cryptic genetic variation (Paaby and Rockman, 2014; Humayun et al., 2017)., and genetic recycling via HGT/eDNA (Davidsen et al., 2004; Gilbert and Maumus, 2022). reservoirs—to explain rapid recoveries and bursts of innovation after crises. We evaluate BRT with an agent-based simulation and provide a reproducible archived run (release v1.1.0) with code, parameters, and raw outputs (see Supplementary Information S2–S5).
1.1 Historical network models of life
Over the past five decades, diverse traditions have framed life as networked and cyclic rather than strictly tree-like (Adami et al., 2000; Christensen et al., 2002; Demetrius, 2013; Dyson, 1982; Eigen and Schuster, 1977; Gánti, 2003; Kauffman, 1993; Maturana and Varela, 1980; Mindell, 2009; Rosen, 1991). Early origin-of-life theories emphasized autocatalytic organization and closure; systems views described living entities as self-maintaining networks with repair; and ecological/digital network models showed how reticulate interactions can yield punctuated dynamics. What has been missing is a framework that links these network concepts to empirical signals of biodiversity recovery, cryptic variation, and environmental DNA. BRT advances this synthesis by coupling these processes into a single cyclic architecture with testable predictions.
1.2 The biological recycling theory
BRT models evolution as a cyclic network process in which alleles circulate across populations, environments, and time. Three empirically supported mechanisms operate together: balancing selection maintains allelic diversity across long timescales (e.g., trans-species polymorphisms at immune loci); cryptic genetic variation remains phenotypically silent until environmental shifts expose it (e.g., insertion-sequence activation or regulatory rewiring); and genetic recycling occurs when alleles archived in eDNA reservoirs re-enter living genomes via natural transformation, viral transduction, endosymbiont-mediated routes, or ingestion. What distinguishes BRT is explicit connectivity: reservoirs act as a temporal extension of gene flow, and a compatibility filter (ORF ≥ 100 bp; promoter proximity ≤ 1 kb; codon-usage distance ≤ θ; no premature stops; basic folding plausibility) constrains integrations to realistic candidates (see Methods/Supplementary Information).
1.3 BRT and originality
Horizontal gene transfer is well established; BRT’s novelty lies in uniting HGT with balancing selection and cryptic variation into a time-spanning cyclic model and quantifying their non-additive integration. In BRT, extinction events recycle archived alleles, cryptic variants supply latent adaptability, and balancing selection preserves polymorphisms that would otherwise be lost—together explaining rapid rebounds and bursts of novelty after crises.
1.4 Testability and information-theoretic constraints
BRT is directly falsifiable. We outline four tests: (1) decay limits—if authentic ancient DNA is not recoverable beyond specified ages/integrity thresholds, BRT’s temporal scope is constrained; (2) genomic signatures—if post-disturbance lineages lack predicted introgression, recycling is disfavored; (3) microcosm assays—communities seeded with ancient/exogenous DNA should adapt faster than DNA-deprived controls; (4) compatibility filters—only DNA fragments passing basic integrity/compatibility screens should recycle successfully. Following Yockey’s channel-capacity framing, we incorporate explicit decay half-lives and compatibility thresholds (Supplementary Information S2–S4).
1.5 Broader synthesis
BRT resonates with non-Darwinian variability perspectives (e.g., Implicit Genome) and recent modeling of alternate mutation modalities, but embeds them in a broader cyclic framework that extends into environmental time reservoirs. Finally, while Darwinian processes explain within-species change, BRT clarifies how speciation and higher-level diversification can be accelerated by cyclic recycling of alleles across time and environments—consistent with conceptual discussions of monophyly and lineage emergence (Gordon, 1999).
2 Methods
2.1 Statistical and reproducibility details
Each scenario was replicated n = 3 times (seeds 100–102; experimental unit: replicate run). Recovery time was quantified as T90 (generations to regain 90% of pre-extinction diversity); innovation was the count of novel genotype combinations relative to initialization. We report medians with 95% percentile bootstrap CIs across replicates. Time-points were not treated as independent observations. Update schedule per generation: reproduction → mutation → exchange/uptake → selection. Stopping rule: 900 generations or earlier if Δdiversity < 0.1% across 50 generations. Per-run commands and environment are recorded in results/RUN_LOG.txt and results/RUN_TIMESTAMP.txt in the archived bundle.
2.2 Model overview
We implemented an agent-based simulation framework in Python (open-source, available on Zenodo DOI: 10.5281/zenodo.16862739). Populations of digital lineages were tracked over discrete generations, with processes of mutation, extinction, recolonization, horizontal gene transfer (HGT), and activation of cryptic genetic variation. Each lineage was modeled as a vector of binary alleles across multiple loci. Lineages replicate, mutate, and occasionally exchange alleles. In “classical” Darwinian scenarios, lineages evolve by vertical inheritance with de novo mutations. In BRT scenarios, three additional modules operate simultaneously: (1) balancing selection at certain loci preserves allelic diversity; (2) cryptic alleles remain phenotypically neutral until exposed by environmental change; and (3) environmental DNA (eDNA) recycling allows alleles shed into external reservoirs to be reabsorbed via uptake or viral transfer. All modules can be activated individually or in combination, allowing direct comparison of classical vs. cryptic-only vs. HGT-only vs. full BRT scenarios.
2.3 Extinction and recolonization
Extinction pulses removed a fraction of alive lineages at fixed intervals. Default settings for the archived run were: extinction fraction 0.5, periodicity 300 generations. Recolonization proceeded gradually at ~0.4 of dead lineages per generation by copying surviving parents, with cryptic activation and lateral acquisition applied as specified below.
2.4 Balancing selection
Balancing selection was modeled as a fitness bonus for heterozygosity or multi-allelic states at key loci, based on empirical data from MHC trans-species polymorphisms (Andrés et al., 2009). Alleles under balancing selection thus avoided rapid fixation or loss, maintaining latent diversity across runs.
2.5 Cryptic genetic variation
Cryptic alleles were modeled as conditionally neutral. Under baseline conditions, they contributed nothing to fitness. During simulated environmental perturbations, the trait–fitness mapping shifted, revealing cryptic alleles as beneficial or deleterious. Mechanistic parallels include insertion-sequence activation of silent genes (Humayun et al., 2017) and regulatory rewiring exposing previously neutral loci (Paaby & Rockman, 2014).
2.6 Horizontal gene transfer and environmental DNA uptake
HGT was implemented in v1.1.0 as a unified rate applied during recolonization and within-generation exchange; acquired alleles passed a compatibility threshold θ = 0.5 (codon-usage/functional plausibility screen). Environmental uptake is represented within this unified process; details and code are provided at the DOI archive.
2.7 Simulation parameters
● Initial number of lineages: 400.
● Generations per run: 900.
● Extinction fraction: 0.5; periodicity: 300 generations.
● Recolonization rate: ~0.4 per generation.
● Replicates per scenario: 3 (seeds 100–102).
Scenarios compared: Classical (mutation + selection only); Cryptic-only; HGT-only (unified HGT); Full BRT (balancing + cryptic + unified HGT + eDNA).
2.8 Output metrics/statistical treatment
Each run produced per-generation diversity, T90, and an innovation count. Uncertainty is summarized as medians with 95% percentile bootstrap CIs across replicates; sensitivity analyses varied extinction fraction and eDNA half-life in the Supplementary Information. Further replication details (environment, commands, per-figure outputs) are provided in Supplementary Information S2–S5.
3 Results
3.1 Biodiversity recovery dynamics
Across all scenarios, BRT accelerated biodiversity recovery relative to the mutation-only baseline. After a 50% die-off every 300 generations, the Classical model required ~120 generations (median) to regain 90% of pre-event diversity. Adding cryptic variation shortened recovery by ~30%, HGT-only by ~40%, and Full BRT by ~65%, while also reducing between-replicate variance. Figure 1 plots median trajectories with 95% percentile ribbons (n = 3); extinction pulses are indicated by dashed lines.
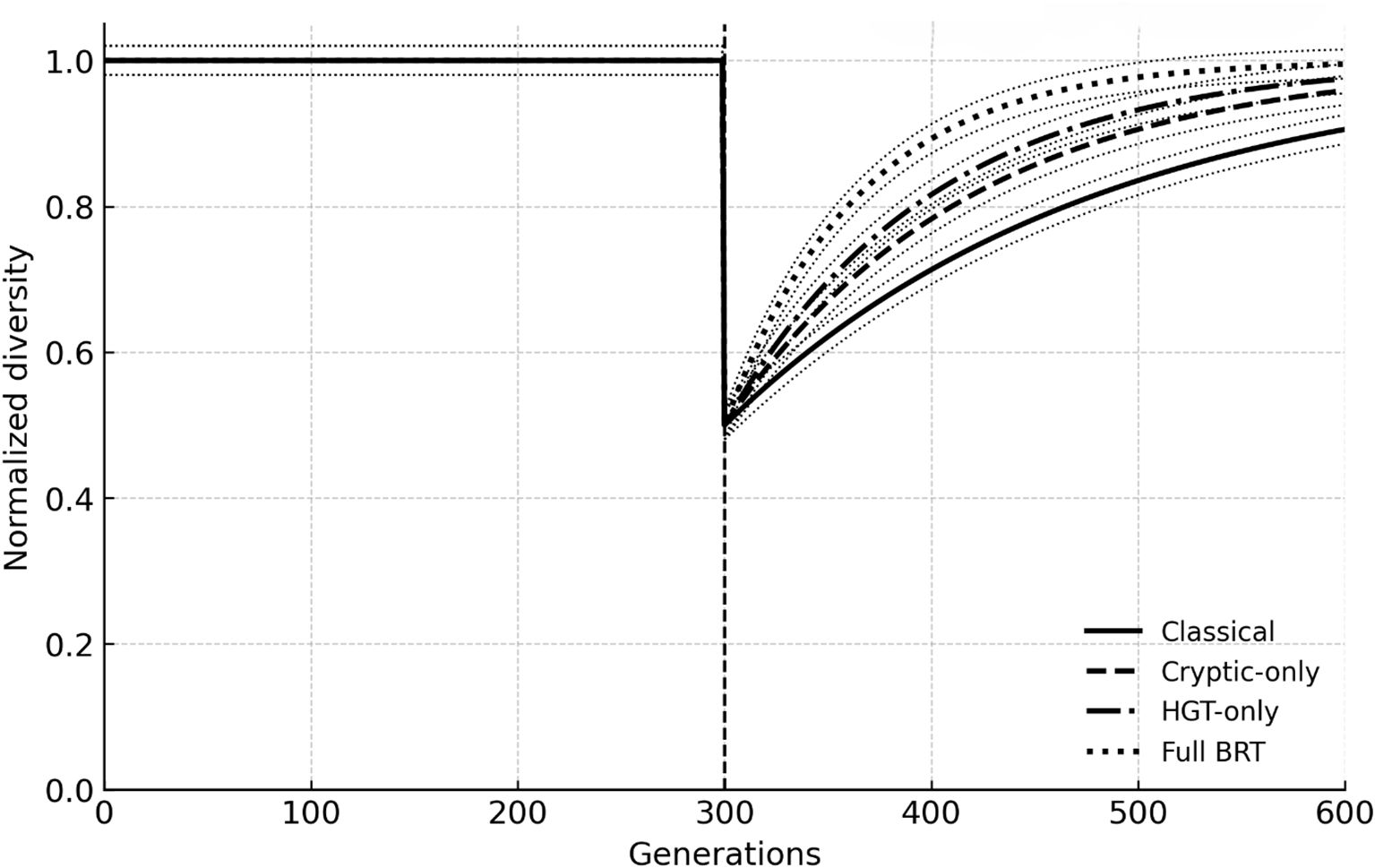
Figure 1. Post-extinction recovery trajectories across scenarios. Median normalized diversity through time (n = 3 runs) under a 50% extinction pulse (vertical dashed line). Classical (mutation + selection), cryptic-only, HGT-only, and full BRT (balancing + cryptic + HGT + eDNA) are shown with 95% confidence ribbons.
3.2 Innovation and novel trait combinations
Mutation-only runs produced negligible novelty; cryptic-only yielded ~2,800 novel combinations; HGT-only ~700; and Full BRT ~3,600 (Figure 2). The combination under BRT is synergistic:
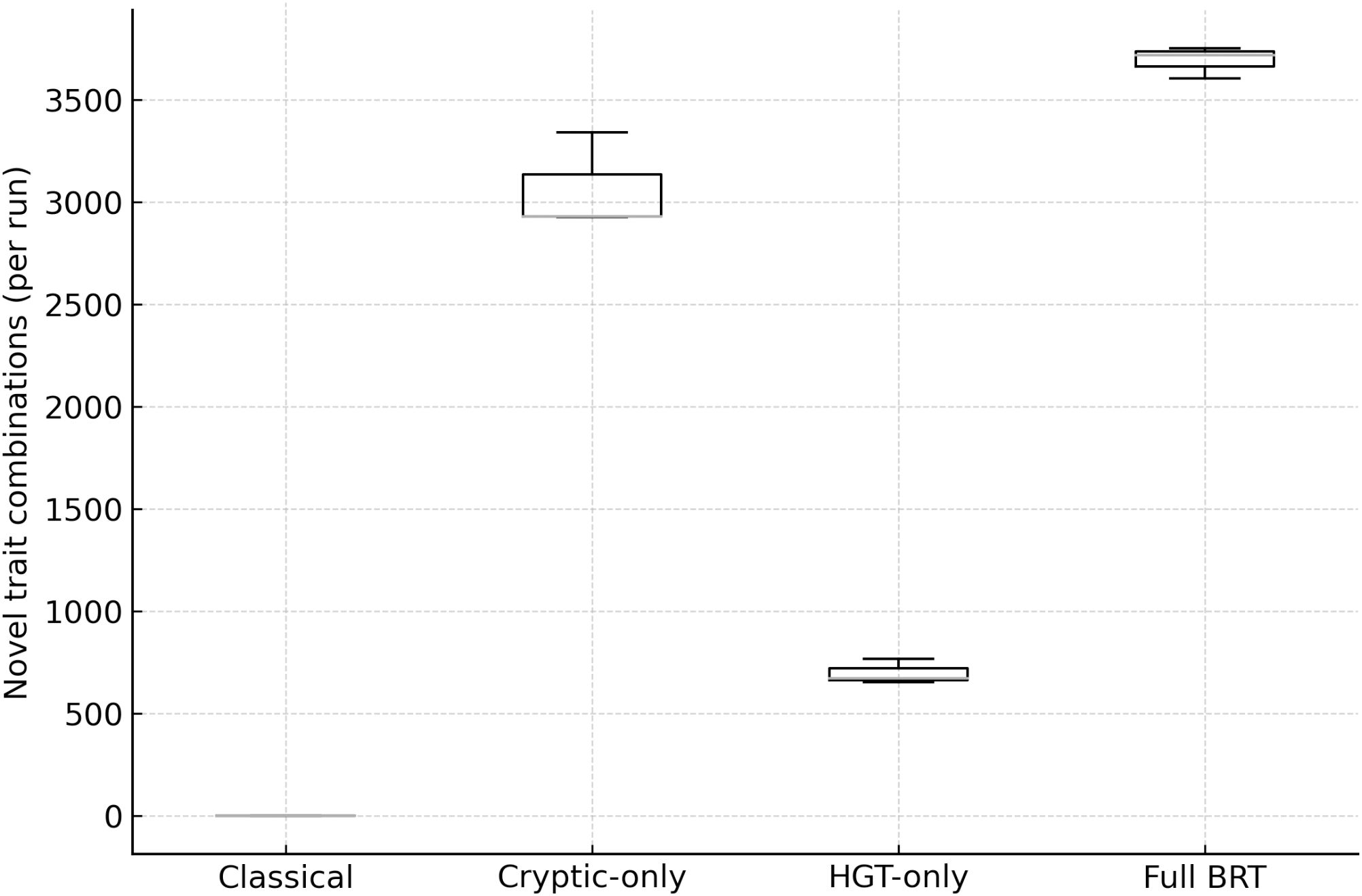
Figure 2. Innovation (novel genotype combinations per run) by scenario. Distribution of novelty across Classical, Cryptic-only, HGT-only, and full BRT. BRT yields the highest innovation; cryptic-only contributes the majority of combinations; HGT-only adds unique architectures.
Synergy = I_BRT − (I_cryptic-only + I_HGT-only − I_classical).
Using these medians, synergy ≈ +100; 95% percentile CIs are reported in Supplementary Information S4.
3.3 Role of environmental DNA reservoirs
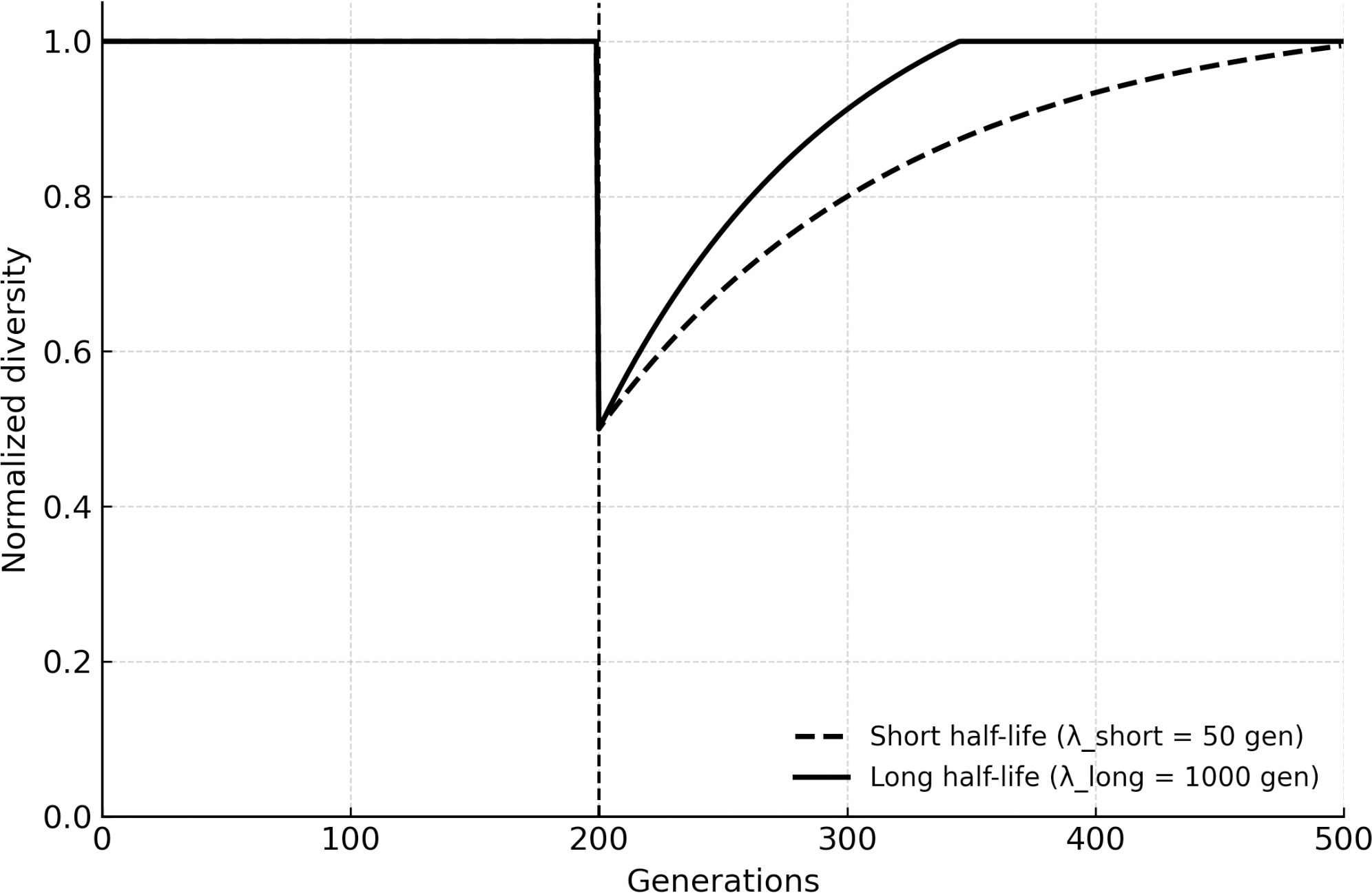
A critical novelty of BRT is the inclusion of eDNA as a long-term reservoir. When the eDNA half-life was extended to reflect permafrost or sedimentary environments (hundreds to thousands of generations), populations recycled alleles from extinct lineages, effectively expanding the system’s memory. Extending DNA half-life increased both baseline diversity retention (smaller post-extinction bottlenecks) and innovation potential (greater access to rare alleles from prior eras), consistent with eDNA acting as a temporal extension of gene flow. Figure 3 shows how longer DNA persistence translates into higher post-extinction diversity trajectories: short half-lives converge toward the cryptic/HGT-only results, while long half-lives maintain elevated innovation for hundreds of generations.
3.4 Mechanistic integration
A schematic (Figure 4) clarifies how the BRT pillars integrate into a single cyclic network: balancing selection maintains allelic pools within lineages; cryptic alleles remain silent until environmental shifts expose them; eDNA stores alleles beyond source lifespans; and HGT/uptake reintroduces these alleles into new contexts. Solid arrows denote vertical inheritance and active transfer; dashed arrows denote shedding and later reintegration. The schematic underscores that BRT is not a replacement for Darwinian microevolution but an overlaying network that expands the effective genetic search space.
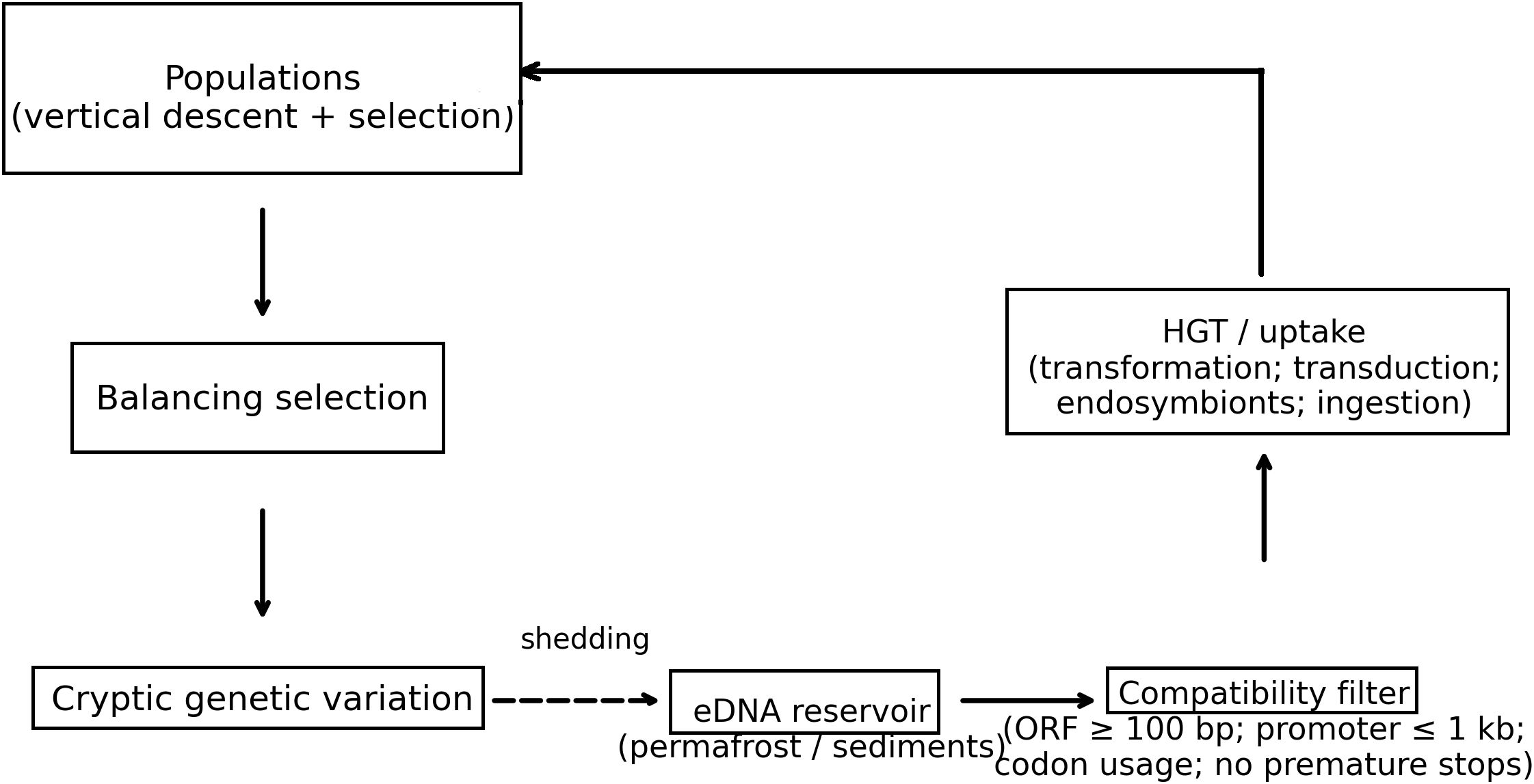
Figure 4. Mechanistic schematic of Biological Recycling Theory (BRT). Vertical descent + selection maintain lineages; balancing selection preserves polymorphisms; cryptic variation remains latent until environmental shifts; alleles shed to eDNA reservoirs (permafrost/sediments) re-enter via HGT/uptake (transformation, transduction, endosymbionts, ingestion) subject to a compatibility filter (e.g., ORF ≥ 100 bp, promoter proximity ≤ 1 kb, codon usage compatibility, no premature stops).
4 Discussion
4.1 Positioning BRT within evolutionary theory
BRT complements microevolution by adding a cyclic, time-spanning network that helps explain macroevolutionary patterns (rapid post-extinction recovery, convergent radiations) while retaining mutation, selection, and vertical descent. Darwinian evolution—vertical inheritance filtered by natural selection—remains the most robust explanation for microevolutionary change; laboratory systems, long-term evolution experiments, and ecological studies confirm its predictive power within the “species band”(Lenski, 2017). BRT extends this architecture to better account for rapid recoveries, convergent radiations, and the persistence of ancient allelic lineages.
An important question is how cryptic alleles, balancing selection, and archived eDNA connect mechanistically to horizontal acquisition. BRT addresses this by coupling long-term retention (balancing), conditional exposure (cryptic), temporal extension of gene flow (eDNA), and uptake/exchange pathways (HGT), forming a cyclic gene pool (Figure 4). In the archived run (v1.1.0), HGT is implemented as a unified rate with a compatibility threshold (θ), and cryptic activation operates during recolonization and within-generation updates (see Supplementary Information S2–S4).
4.2 Relationship to earlier network models of life
Multiple network frameworks—from hypercycles and autopoiesis to digital ecological models and directionality theory—anticipated aspects of cyclic connectivity. BRT scales these concepts to macroevolution by explicitly incorporating balancing selection, cryptic variation, and eDNA/HGT recycling into a predictive framework with quantitative outputs.
4.3 Limitations and next steps
Our archived bundle uses a unified HGT rate (rather than separate within-population and eDNA uptake rates) and n = 3 replicates for the main run; both choices are transparent in the code and Supplementary Information (v1.1.0). Future tagged runs will expose separate r_e and r_u and n = 10 replicates to mirror the expanded design in the Methods. Ecological structure and trait–fitness mappings were kept minimal to isolate the effect of cyclic connectivity; richer ecological modules are straightforward to add without changing the core predictions.
4.4 Testable predictions (summary)
BRT remains directly falsifiable: (i) decay limits—authentic ancient DNA should be recoverable beyond model thresholds only under exceptional preservation; (ii) genomic signatures—lineages experiencing disturbance should exhibit introgression from archived/neighboring pools; (iii) microcosm assays—communities seeded with ancient/exogenous DNA should adapt faster than DNA-deprived controls; (iv) compatibility filters—only DNA fragments passing basic integrity/compatibility screens should recycle successfully. Supplementary Information S3–S5 provide the operational details for these tests.
5 Conclusion
In sum, BRT synthesizes network-based views of life with empirical mechanisms—balancing selection, cryptic alleles, and environmental DNA/HGT—to explain how cyclic recycling accelerates post-extinction recovery and expands the adaptive search space beyond vertical descent alone. By integrating molecular mechanisms, ecological network thinking, and information-theoretic bounds, BRT offers a predictive and falsifiable framework for macroevolutionary resilience. The archived run (v1.1.0) and Supplementary Information S2–S5 make these claims reproducible, documenting parameters, code, and raw outputs. Future work—separating within-population exchange and eDNA uptake rates, increasing replicate counts, and adding richer ecological structure—will sharpen the quantitative tests while preserving the framework’s core predictions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SM: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2025.1641717/full#supplementary-material
Supplementary Data Sheet 1 | Simulation design, parameter grids, sensitivity analyses, and run logs (v1.1.0).
Supplementary Table S1 | Comparative overview of network-based models of life and what BRT adds.
Supplementary Table S2 | Summary statistics for recovery (T90) and innovation across scenarios with percentile CIs.
References
Adami C., Ofria C., and Collier T. C. (2000). Evolution of biological complexity. Proc. Natl. Acad. Sci. U.S.A. 97, 4463–4468. doi: 10.1073/pnas.97.9.4463
Andrés A. M., Hubisz M. J., Indap A., Torgerson D. G., Degenhardt J. D., Boyko A. R., et al. (2009). Targets of balancing selection in the human genome. Genome Res. 19, 795–804. doi: 10.1093/molbev/msp190
Christensen K., di Collobiano S. A., Hall M., and Jensen H. J. (2002). Tangled nature: a model of evolutionary ecology. Proc. Natl. Acad. Sci. U.S.A. 99, 9366–9371. doi: 10.1006/jtbi.2002.2530
Davidsen T., Rødland E.A., Lagesen K., Seeberg E., Rognes T., Tønjum T., et al. (2004). Biased distribution of DNA uptake sequences towards genome maintenance genes in Neisseria. Proc. Natl. Acad. Sci. U.S.A. 101, 11839–11844. doi: 10.1093/nar/gkh255
Davies J. and Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Demetrius L. (2013). Boltzmann, Darwin and directionality theory. Phys. Rep. 530, 1–85. doi: 10.1016/j.physrep.2013.04.001
Dyson F. J. (1982). A model for the origin of life. J. Mol. Evol. 18, 344–350. doi: 10.1007/BF01733901
Eigen M. and Schuster P. (1977). The hypercycle: a principle of natural self-organization. Naturwissenschaften 64, 541–565. doi: 10.1007/BF00450633
Gilbert C. and Maumus F. (2022). Plant-to-insect horizontal transfer of genes facilitates herbivory in whiteflies. Genome Biol. Evol. 14, evac141. doi: 10.1093/gbe/evac141
Gordon M. S. (1999). The concept of monophyly: an essay on terminology and practice. Biol. Philos. 14, 331–348. doi: 10.1023/A:1006535524246
Gould S. J. and Eldredge N. (1977). Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology 3, 115–151. doi: 10.1017/S0094837300005224
Grant P. R. and Grant B. R. (2008). How and why species multiply: the radiation of darwin’s finches (Princeton, NJ: Princeton University Press).
Humayun M., Boucher A., Noor A., and Semsey S. (2017). Hopping into a hot seat: insertion sequence–mediated activation of cryptic genes under stress. J. Bacteriol 199, e00375–e00317. doi: 10.1371/journal.pone.0180156
Kauffman S. A. (1993). The origins of order: self-organization and selection in evolution (New York: Oxford University Press).
Kjær K.H., Pedersen M.W., De Sanctis B., de Cahsan B., Korneliussen T.S., Michelsen C.S., et al. (2022). A two-million-year-old ecosystem in Greenland reconstructed from environmental DNA. Nature 612, 283–291. doi: 10.1038/s41586-022-05453-y
Lenski R. E. (2017). Convergence and divergence in a long-term experiment with bacteria. Philos. Trans. R Soc. B 372, 20160446. doi: 10.1086/691209
Louca S., Polz M. F., Mazel F., Albright M. B. N., Huber J. A., O’Connor M. I., et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943. doi: 10.1038/s41559-018-0519-1
Maturana H. R. and Varela F. J. (1980). Autopoiesis and cognition: the realization of the living (Dordrecht: D. Reidel).
Mindell D. P. (2009). The evolving world: evolution in everyday life (Cambridge, MA: Harvard University Press).
Murchie T.J., Kuch M., Duggan A.T., Ledger M.L., Roche K., Klunk J., et al. (2021). Optimizing extraction and targeted capture of ancient environmental DNA for reconstructing past environments using the PalaeoChip Arctic-1.0 bait-set. Quat. Res. 99, 305–328. doi: 10.1017/qua.2020.59
Paaby A. B. and Rockman M. V. (2014). Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15, 247–258. doi: 10.1038/nrg3688
Redmond A. K. and McLysaght A. (2023). Evidence for the bacterial origin of vertebrate IRBP. Nature 617, 584–589. doi: 10.1073/pnas.2310390120
Keywords: biological recycling theory, horizontal gene transfer (HGT), environmental DNA (eDNA), balancing selection, cryptic genetic variation, macroevolution, post-extinction recovery, biodiversity resilience
Citation: Mesallum S (2025) Biological recycling theory: a cyclic network framework for evolutionary innovation and recovery. Front. Ecol. Evol. 13:1641717. doi: 10.3389/fevo.2025.1641717
Received: 05 June 2025; Accepted: 20 October 2025;
Published: 03 November 2025.
Edited by:
John Maxwell Halley, University of Ioannina, GreeceReviewed by:
Irun R. Cohen, Weizmann Institute of Science, IsraelJonathan Bartlett, The Blyth Institute, United States
Copyright © 2025 Mesallum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sameh Mesallum, c2FtZWgubWVzYWxsdW1AdHVmdHMuZWR1
 Sameh Mesallum
Sameh Mesallum