- 1Northern Forestry Centre, Natural Resources Canada, Canadian Forest Service, Edmonton, AB, Canada
- 2Pacific Forestry Centre, Natural Resources Canada, Canadian Forest Service, Victoria, BC, Canada
- 3Laurentian Forestry Centre, Natural Resources Canada, Canadian Forest Service, Quebec, QC, Canada
- 4Forest Sciences Centre, Faculty of Forestry, University of British Columbia, Vancouver, BC, Canada
- 5ATTS Group, Edmonton, AB, Canada
In June, 2021, laurel willow (Salix pentandra) near Slave Lake, Alberta, was found to be infected by a Melampsora sp. that produced bright yellow urediniospores in uredia that were present on catkins, leaves, and stems. All Melampsora species previously reported in Canada are recorded as infecting leaves; therefore, further investigation was undertaken to ascertain the identity of this pathogen. To assess the relationship between this specimen and other Melampsora spp. previously collected from Canada, samples of willow leaves infected by Melampsora spp. were sourced from mycological herbariums located at the Laurentian Forestry Centre (QFB) and the Northern Forestry Centre (CFB, WINF(M)). DNA sequence data from the internal transcribed spacer (ITS) ribosomal RNA region of the fresh specimen, herbarium specimens, and DNA sequence data deposited within GenBank, were used to conduct a phylogenetic analysis. Sequencing and BLAST analysis of the material from the sample resulted in a 99.3% sequence identity match to Melampsora epitea “Mel J” collected from Larix laricina in New York State. The ITS sequence from the herbarium sample WINF(M)7356 (described as M. abieti-capraearum from Manitoba) had 100.0% identity with the Alberta sample. Additionally, specimens WINF(M)11892 (Melampsora sp. from Manitoba) and CFB8931 (Melampsora sp. from the Yukon) had 99.0% sequence identity with the Alberta sample. From these results we applied the identity of M. epitea to the rust discovered in Slave Lake, AB. With the current emphasis on willows for bioenergy production in Canada, growers must remain vigilant for this pathogen and the damages it could cause to willow plantations.
Introduction
This study was initiated as a result of a public inquiry regarding Salix pentandra L. (laurel willow) infected by a rust pathogen in Alberta, Canada, in 2021. Laurel willow is native to Europe and Asia, although it has been introduced to Canada where its cold hardiness has facilitated its establishment (Farrar, 1995). The specimens that were received were subjected to further examination because uredinia were present in cankers found on stems, a symptom that is not listed in any of the descriptions of Melampsora spp., which infect willows in Western Canada, as outlined by Ziller (1974), leading to concerns that a new pathogen may have been introduced to Canada.
The taxonomy of rust pathogens infecting willows is complicated due to the morphological similarity of the uredinial states of Melampsora spp. on willows, resulting in Melampsora epitea Thümen becoming a catch-all for Melampsora sp. infecting willow (Ziller, 1974). Recent studies based on morphology and ribosomal DNA sequences have revealed that M. epitea is a species complex (Smith et al., 2004; Bennett et al., 2011; Kenaley et al., 2014; Zhao et al., 2017). In total, 12–14 phylotypes were recognized in North America (Smith et al., 2004), and more recently, Zhao et al. (2017) have described 13 species based on specimens collected from East Asia. These groups are differentiated based on aecial hosts, subtle morphological differences in the urediniospores, uredinia and telia, and ribosomal DNA sequences (Smith et al., 2004; Bennett et al., 2011; Kenaley et al., 2014; Zhao et al., 2017). Morphology and host associations are important characters in making an identification, but within this group, this information must be supplemented with DNA sequence data to arrive at the best possible identification.
The objective of this study was to determine the identity of the Melampsora sp. causing disease on S. pentandra in Alberta to investigate its potential as a new introduction in Canada. This was achieved by comparing the ribosomal DNA sequence data from this specimen with the ribosomal DNA extracted from herbarium specimens collected from Salix spp. in Canada and ribosomal DNA sequence data present in GenBank.
Materials and methods
Collection information
The sample, which included infected catkins, leaves, and stems, was collected fresh from Salix pentandra growing on private property in the Slave Lake region of Alberta and received at the Northern Forestry Centre on 16 June 2021.
Herbarium samples
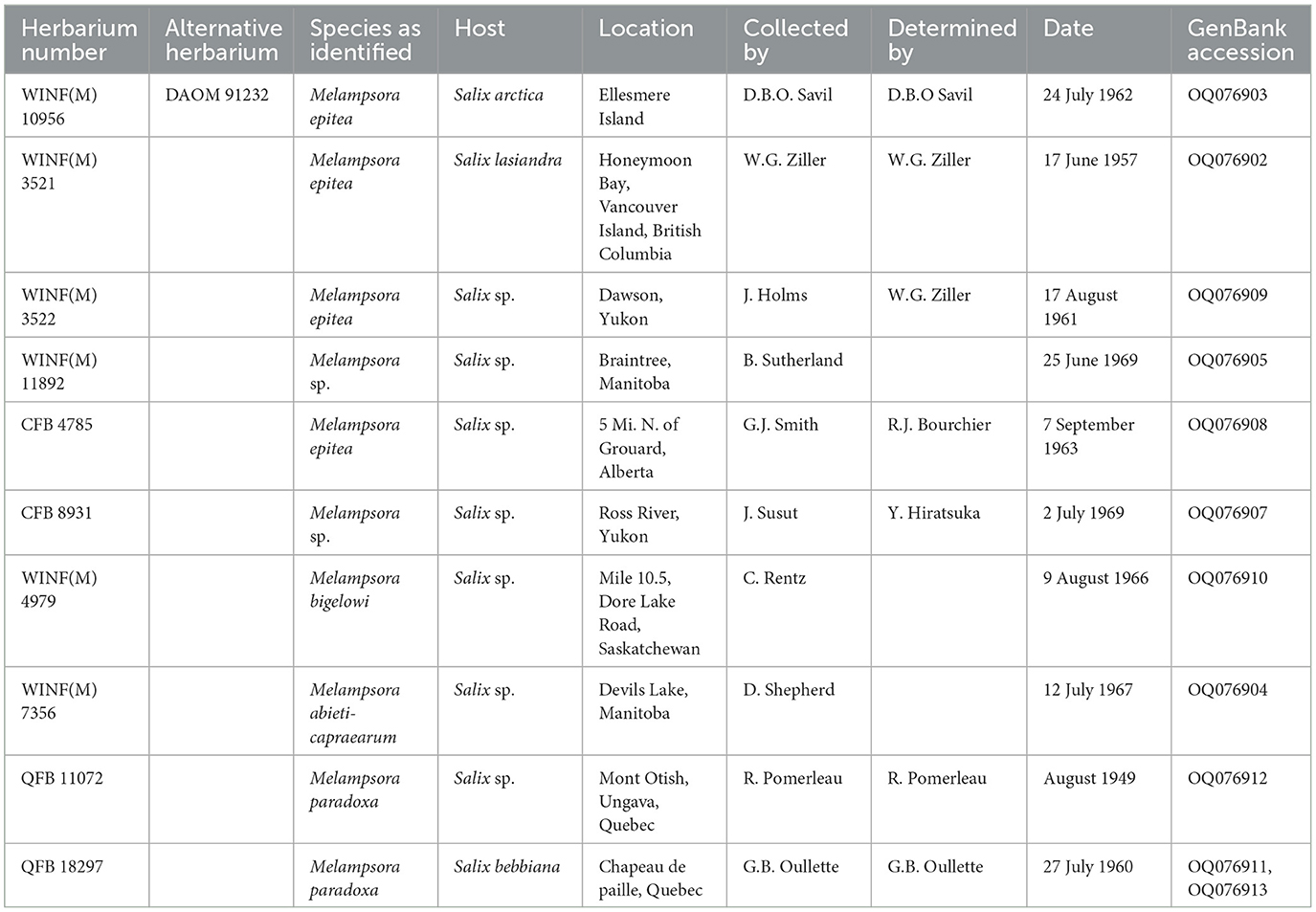
Specimens were retrieved from the QFB herbarium at the Laurentian Forestry Centre (Quebec City, Canada) and the CFB and WIN(F) herbaria at the Northern Forestry Centre (Edmonton, Canada) based on the host and proximity to the collection site (Table 1).
Morphology
Fresh urediniospores were mounted in water and the length (μm) and width (μm) of 10 urediniospores were measured under 400× and 1,000× magnification (n = 20) using a Zeiss Axiophot compound microscope using differential interference contrast (Carl Zeiss AG, Jena, Germany). One-way analysis of variance (α = 0.05) was performed using Microsoft Excel to compare the size of urediniospores measured under 400× and 1,000× magnification. As there was no significant difference in the length or width of urediniospores observed under 400× and 1,000× magnification (length F1,18 = 0.04, p = 0.839; width F1,18 = 2.48, p = 0.133), urediniospore size was calculated based on the pooled sample (n = 20).
To compare urediniospore size between uredia present on catkins, stems, and leaves, a total of 30 urediniospores from each tissue type (n = 90) from specimen CFB 22329 were mounted in water, and the urediniospore length (μm) and width (μm) were measured under 400× magnification using a Zeiss Axiophot compound microscope (Carl Zeiss AG, Jena, Germany). One-way analysis of variance (α = 0.05) was performed using Microsoft Excel to compare the size of urediniospores from uredia present in these different tissues. As there was no significant difference in the length or width of urediniospores collected from catkins, stems, or leaves (length F2,87 = 1.24, p = 0.294; width F2,87 = 2.19, p = 0.119), the size of urediniospore was calculated based on the pooled sample (n = 90).
DNA extraction: willow stems and seed catkins, herbarium samples
Urediniospores were collected from both fresh samples of willow stem and seed catkins and also herbarium samples of willow leaves. Plant materials were examined under a dissecting microscope, and urediniospores were aseptically harvested into 2 ml tubes with two 3.2 mm diameter stainless steel beads per tube. Due to the difficulty in harvesting from herbarium leaf samples, urediniospores were collected along with minute amounts of leaf, taking every care to minimize leaf material (Table 1). Afterward, the urediniospore samples were ground to a fine powder using the MM 300 TissueLyser (Retsch®, PA, USA) at 30 Hz for 2 min. The total genomic DNA of all urediniospore samples was extracted using the Qiagen DNeasy Plant mini kit (Qiagen, CA, USA) according to the manufacturer's instructions. DNA concentration of samples was measured using the Nanodrop ND-1000 spectrophotometer (ver. 3.8; Thermo Fisher Scientific Inc., Ottawa, ON, Canada).
Polymerase chain reaction (PCR) assay
For species identification, the ITS region of the ribosomal DNA was amplified by PCR using the universal fungal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′; Gardes and Bruns, 1993)/ITS4 (5′-TCCTCCGCTTATTGATATGC-3′; White et al., 1990) following published protocols (White et al., 1990). Each PCR reaction (25 μl total volume) contained 1X buffer (Invitrogen, Carlsbad, CA, USA), 1.5 mM MgCl2 (Invitrogen, Carlsbad, CA, USA), 0.2 mM each dNTP (Invitrogen, Carlsbad, CA, USA), 0.5 U Platinum Taq polymerase (Invitrogen, Carlsbad, CA, USA), each primer (forward and reverse) at 1 μM, and 2 μl of template DNA. The PCR protocol was performed in a Bio-Rad T100™ Thermal Cycler (Hercules, CA, USA) and included an initial denaturation step at 94°C for 3 min. This was followed by 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 55°C), extension (1 min at 72°C), and final extension at 72°C for 10 min.
The PCR products were mixed with safe green dye (Applied Biological Materials, Canada) at a 1:5 ratio and were analyzed by gel electrophoresis using 1% agarose in 1X TBE (10X: Tris base 108 g, Boric acid 55 g, and EDTA 7.5 g in 1 L of distilled water). The PCR products were visualized using the Prepone Sapphire Blue LED Illuminator system (Embi Tec, CA, USA). Subsequently, the PCR products were sent for purification and Sanger sequencing to the CHUL Research Center Sequencing and Genotyping Platform (Québec City, Québec, Canada).
Sequences and phylogenetic analysis
ITS sequence data from the fresh specimen and herbarium specimens were aligned with ITS sequences from M. epitea and closely related Melampsora spp. retrieved from GenBank. Sequence chromatograms were trimmed, manually assessed for quality, and aligned for consensus using BioEdit ver 7.2 (Hall, 1999). Sequence identity was assigned using the BLASTn (Altschul et al., 1990) on the GenBank-NCBI nucleotide collection (nt) with homolog sequences. Maximum likelihood (ML) phylogenetic trees were constructed with RaxML (Stamatakis, 2014) using the GTR + GAMMA nucleotide evolution model. ModelTest-NG (Darriba et al., 2020) was used to identify the best-fitting nucleotide evolution model for this dataset. The maximum parsimony (MP) tree was identified by conducting a heuristic search and 100 random additions of sequences (RAS) with PAUP ver. 4.0b10 (Swofford, 2003). Statistical support for nodes on the ML and MP phylogenetic trees were obtained with 100 bootstrap replicates, saved in a Newick format, and then edited using TreeGraph 2 (Stöver and Müller, 2010), collapsing nodes for the minimum 70% consensus rule.
Results
Examination of the specimens revealed bright yellow uredinia present on leaves, catkins, and stems (Figure 1). Fresh urediniospores were yellow, echinulate, and globoid to broadly ellipsoid and measured [(length × width) (min-) x̄ ± SD (-max)]: (20-) 25.4 ± 3.17 (−35) × (17.5-) 20.6 ± 1.62 (−23) μm. Further examination of dried material revealed no significant difference in urediniospore size produced in uredia on catkins, stems, or leaves (length F2,87 = 1.24, p = 0.294; width F2,87 = 2.19, p = 0.119), suggesting that all tissues were infected by the same pathogen. Urediniospores from the dried material were no longer yellow and measured (20-) 23.8 ± 1.89 (−30) × (12.5-) 15.9 ± 1.41 (−20) μm.

Figure 1. Melampsora epitae-infected Salix pentranda: (A) Uredinia on the stem (white arrow) and seed catkins (black arrow). (B) Urediniospores under 1,000× magnification in water, scale bar 20 μm. (C) Uredinia on seed catkin. (D) Uredinia on a stem. (E) Uredinia on leaves and catkins. (F) Uredinia on branch.
When the ITS sequence data were aligned with sequence data from close relatives that were deposited in GenBank, it was found that the rust collected in this study was part of the Melampsora epitea complex. Comparison of the ITS sequence data from the fresh material with ITS sequence data generated from herbarium material resulted in a 100% identity with the specimen WINF(M)7356, identified as Melampsora abietis-caprearum, which was collected in 1967 from Devils Lake, Manitoba. The sequence was also 100% identical with the GenBank sequence GQ479229, M. epitea, collected from Edmonton, Alberta, in 2006 (Figure 2), and had a 99% identity match with the herbarium specimens CFB8931, Melampsora sp. collected from the Ross River in the Yukon in 1969 and WINF(M)11892, Melampsora sp. collected from Braintree, Manitoba, in 1969.
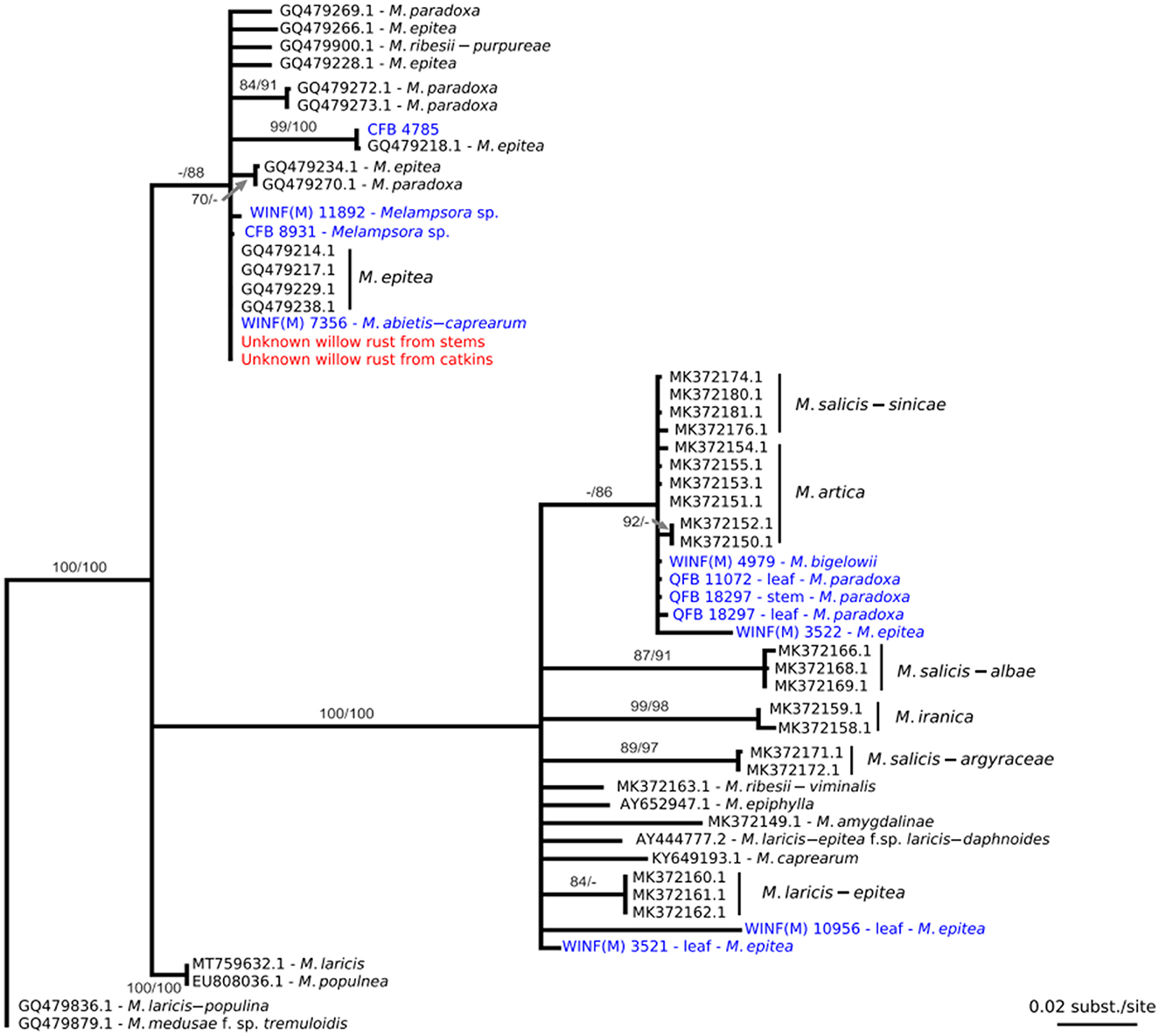
Figure 2. Maximum likelihood phylogenetic tree constructed from a nucleotide alignment of 613 bp of the internal transcribed spacer regions (ITS) of the nuclear rDNA including sequence data collected from the willow rust specimen identified in Alberta (in red), various herbaria specimens (in blue), and sequence data from GenBank (in black). The numbers above the nodes are statistical support values obtained from 100 bootstrap samples in maximum likelihood/maximum parsimony; statistical support under 70% is not represented. Scale bars indicate the number of nucleotide substitutions per site.
A representative sample from the specimen has been entered into the CFB herbarium under the accession number CFB22329. The ITS sequence data from this specimen has been accessioned in GenBank under the numbers OQ076914 for urediniospores from the stem and OQ076915 for urediniospores from catkins.
Discussion
Diseased S. pentandra with uredinia on stems, catkins, and leaves were collected from northern Alberta, Canada. As Ziller (1974) states that M. epitea is responsible for yellow leaf spots or blight, further examination of this material was undertaken. Urediniospores were larger than that reported by Ziller (1959) but otherwise matched the description. Further examination using DNA sequence data to determine the identity of this pathogen was performed. When the ITS sequence data from this specimen was compared with sequence data in GenBank, it was revealed that the specimen was part of the Melampsora epitea species complex. Phylogenetic analysis of the ITS data from this specimen and sequence data from herbarium specimens and GenBank accessions resulted in a 100% match with the specimen WINF(M)7356, which is morphologically identified as M. abieti-capraearum [which Ziller (1974) states as indistinguishable from M. epitea in the aecial, uredinial, and telial stages] collected in 1967 from Manitoba, and GenBank accession GQ479229, which is identified as M. epitea collected from Edmonton, Alberta, in 2006. Stem cankers have also been observed on Salix sp. in Minnesota (Ostry and Anderson, 2001) and on Salix arctica Pall. from Ellesmere Island (Smith et al., 2004). Stem cankers have been hypothesized to provide an overwintering mechanism in the closely related species M. paradoxa (Crowell et al., 2022). Altogether, these results suggest that the diseased material collected in northern Alberta in 2021 is not the product of a recent introduction. Instead, the pathogen responsible for this stem canker symptom on willows seems to be relatively widespread in Western Canada. Two additional public inquiries during the summer of 2022 from ornamental willows in the Edmonton region determined it to be the same pathogen, further supporting the conclusion that the pathogen is established in Canada.
This study has demonstrated the importance of preserving specimens in biological collections. The CFB, DACFP, S, and WINF(M) collections maintained at the Northern Forestry Centre in Alberta contain a combined 237 accessions of Salix spp. infected by Melampsora spp. that are identified as, M. epitea, M. paradoxa, M. abieti-capaearum, and M. bigelowii. The QFB collection maintained in the Pomerleau herbarium at the Laurentian Forestry Centre in Quebec contains 87 specimens of willow infected by Melampsora rusts. Analysis of the ITS sequence data generated from a subset of these herbaria specimens was key to determining that the pathogen collected in this study was already present in Canada. The specimens housed in these herbaria were all identified based on morphology and host associations. Identifying the causal agent of rust disease on willows based on morphology is difficult due to the similarity of key characteristics (Ziller, 1974), and recent DNA-based studies have suggested that M. epitea is far more complex than revealed by morphological examination (Smith et al., 2004; Bennett et al., 2011; Kenaley et al., 2014; Zhao et al., 2017). Having these specimens available for research will be an important resource for any future studies related to understanding the composition of the M. epitea complex in Canada.
Within Canada, short-rotation willow plantations are becoming increasingly common for phytoremediation (Jerbi et al., 2023) and bioenergy production (Buss et al., 2022). These plantations rely on genotypes that have been selected for growth and yield, and some of these genotypes have been demonstrated to become infected by M. epitea (McCracken and Dawson, 2003). Losses caused by a stem-infecting form of Melampsora that caused stem cankers on Salix viminalis L. in the United Kingdom (Pei et al., 1995) also demonstrated the risk posed to Canadian willows. Surveys of phytoremediation willow plantations in the Edmonton area that were conducted as part of a project to assess willow health have not revealed the presence of any Melampsora spp. on leaves or stems to date. As these plantations become more prominent in Canada, health surveys should include M. epitea as a pathogen of concern.
Further research is necessary to understand the exact identity of the pathogen observed in this study and to determine the aecial host as all materials that were examined were from the uredinial host. The finding that the ITS sequence data from the specimen examined in this study was a 100% match with herbarium material, collected between 1967 and 2006 in Canada, suggests that it is not a recent introduction, but rather that the pathogen has been long-established in central and northwestern parts of Canada. The formation of stem cankers, however, appears to have been rarely observed due to the lack of herbarium specimens that include stem material and the descriptions included within Ziller (1974). It is unknown why stem cankers have not been observed more frequently in the past, but the sample examined in this study was collected from an exotic ornamental by a homeowner who was closely monitoring the health of the trees on their property.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ076902–OQ076913.
Author contributions
This work was conceived by TDR who led writing of the manuscript, performed microscopic analysis, and provided herbarium material from the Northern Forestry Centre. NF conducted the phylogenetic analysis and contributed to writing. PT provided herbarium material from the Laurentian Forestry Centre and contributed to writing. RCH provided funding for DNA sequence analysis and contributed to writing. PH performed DNA extractions and sequencing and contributed to writing. TB provided sample material and observations from Alberta. All authors contributed to the article and approved the submitted version.
Funding
Funding provided by the Canadian Forest Service. RCH was funded by Genome Canada's Large Scale Applied Research Program Project 10106.
Acknowledgments
Pascal Frey (INRAE, France) and Salvatore Moricca (UniFI, Italy) provided important information early in this study that helped to focus our research direction. The technical assistance of Malithi Sella Kapu and Bradley Tomm is greatly appreciated.
Conflict of interest
TB was employed by ATTS Group Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bennett, C., Aime, M. C., and Newcombe, G. (2011). Molecular and pathogenic variation within Melampsora on Salix in western North America reveals numerous cryptic species. Mycologia 103, 1004–1018. doi: 10.3852/10-289
Buss, J., Mansuy, N., Laganière, J., and Persson, D. (2022). Greenhouse gas mitigation potential of replacing diesel fuel with wood-based bioenergy in an arctic Indigenous community: A pilot study in Fort McPherson, Canada. Biomass Bioenergy 159, 106367. doi: 10.1016/j.biombioe.2022.106367
Crowell, C. R., Wilkerson, D. G., Smart, L. B., and Smart, C. D. (2022). Evidence of asexual overwintering of Melampsora paradoxa and mapping of stem rust host resistance in Salix. Plants 11, 2385. doi: 10.3390/plants11182385
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2020). ModelTest-NG: A new and scalable tool for ethe selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi: 10.1093/molbev/msz189
Farrar, J. L. (1995). Trees in Canada. Ottawa: Fitzhenry & Whiteshade Limited and the Canadian Forest Service, Natural Resources Canada. Canada Communication Group – Publishing Supply and Services Canada.
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Jerbi, A., Kalwahali-Muissa, M., Krygier, R., Johnston, C., Blank, M., Sarrazin, M., et al. (2023). Comparative wood anatomy, composition and saccharification yields of wastewater irrigated willow cultivars at three plantation in Canada and Northern Ireland. Biomass Bioenergy 170, 106683. doi: 10.1016/j.biombioe.2022.106683
Kenaley, S. C., Smart, L. B., and Hudler, G. W. (2014). Genetic evidence for three discrete taxa of Melampsora (Pucciniales) affecting willows (Salix spp.) in New York State. Fungal Biol. 118, 704–720. doi: 10.1016/j.funbio.2014.05.001
McCracken, A. R., and Dawson, W. M. (2003). Rust disease (Melampsora epitea) of willow (Salix spp.) grown as short rotation coppice (SRC) in inter- and intra-species mixtures. Ann. App. Biol. 143, 381–393. doi: 10.1111/j.1744-7348.2003.tb00308.x
Ostry, M. E., and Anderson, N. A. (2001). Melampsora leaf rust of willow causing a stem canker in Minnesota. Plant Dis. 85, 229. doi: 10.1094/PDIS.2001.85.2.229C
Pei, M. H., Royle, D. J., and Hunter, T. (1995). A comparative study of stem- and leaf-infecting forms of Melampsora rust on Salix viminalis in the U.K. Mycol. Res. 99, 357–363. doi: 10.1016/S0953-7562(09)80913-1
Smith, J. A., Blanchette, R. A., and Newcombe, G. (2004). Molecular and morphological characterization of the willow rust fungus, Melampsora epitea, from arctic and temperate hosts in North America. Mycologia 96, 1330–1338. doi: 10.1080/15572536.2005.11832882
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post—analysis of large phylogenies. J. Bioinform. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stöver, B. C., and Müller, K. F. (2010). TreeGraph 2: Combinin and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 11, 7. doi: 10.1186/1471-2105-11-7
Swofford, D. L. (2003). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates Inc.
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. Innis, D. Gelfand, J. Shinsky, and T. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Zhao, P., Kakishima, M., Wang, Q., and Cai, L. (2017). Resolving the Melampsora epitea complex. Mycologia 109, 391–407. doi: 10.1080/00275514.2017.1326791
Ziller, W. G. (1959). Studies of western tree rusts V. The rusts of hemlock and fir caused by Melampsora epitea. Can. J. Bot. 37, 109–119. doi: 10.1139/b59-010
Keywords: Melampsora, Salix, rust disease, willow health, canker
Citation: Ramsfield TD, Feau N, Tanguay P, Hamelin RC, Herath P and Bozic T (2023) First report of Melampsora epitea causing stem cankers on Salix pentandra in Alberta, Canada. Front. For. Glob. Change 6:1172889. doi: 10.3389/ffgc.2023.1172889
Received: 23 February 2023; Accepted: 09 May 2023;
Published: 06 June 2023.
Edited by:
Julio Javier Diez Casero, University of Valladolid, SpainReviewed by:
Trudy Paap, University of Pretoria, South AfricaJane E. Stewart, Colorado State University, United States
Copyright © 2023 Ramsfield, Feau, Tanguay, Hamelin, Herath and Bozic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tod D. Ramsfield, dG9kLnJhbXNmaWVsZEBOUkNhbi1STkNhbi5nYy5jYQ==
 Tod D. Ramsfield
Tod D. Ramsfield Nicolas Feau
Nicolas Feau Philippe Tanguay
Philippe Tanguay Richard C. Hamelin
Richard C. Hamelin Padmini Herath4
Padmini Herath4