Abstract
Numb is an intracellular protein with multiple functions. The two prevalent isoforms, Numb p66 and Numb p72, are regulators of differentiation and proliferation in neuronal development. Additionally, Numb functions as cell fate determinant of stem cells and cancer stem cells and its abnormal expression has been described in several types of cancer. Involvement of deregulated Numb expression has been described in the malignant childhood brain tumor medulloblastoma, while Numb isoforms in these tumors and in cancer stem-like cells derived from them, have not been studied to date. Here we show that medulloblastoma stem-like cells and cerebellar neuronal stem cells (NSCs) express Numb p66 where its expression tampers stemness features. Furthermore, medulloblastoma samples evaluated in this study express decreased levels of Numb p66 while overexpressed Numb p72 compared with normal tissues. Our results uncover different roles for the two major Numb isoforms examined in medulloblastoma and a critical role for Numb p66 in regulating stem-like cells and NSCs maintenance.
Introduction
Medulloblastoma (MB) arises in the cerebellum and is the most common malignant pediatric brain tumor (1, 2), originating from both granule cell progenitor (GCPs) and neural stem cells (3). Human medulloblastoma is divided in at least 4 molecular subgroups (WNT, SHH, Group 3, and Group 4) which are characterized by different patterns of gene expression, genetic aberrations, and clinical outcomes (4). SHH MBs account for roughly 27% of tumors and are characterized by aberrant activation of the Sonic hedgehog (Shh) pathway. This aberrant activation is mostly achieved by genetic loss of negative regulators (i.e., Ptch) or amplification of positive regulators (i.e., Gli2) (4–6).
Cancer subpopulations with stemness features (stem-like cells, SLCs) are considered the ultimate reservoir of cancer cells and have been isolated and characterized in several solid tumors, including brain tumors (7). In our previous studies we isolated and characterized cerebellar NSCs and SLCs of human and murine origin and we showed that Shh is a major driver of stemness in this context, through the transcriptional activity of the transcription factor Gli1 and the post-transcriptional regulation of Gli1 activity (8–12).
Numb is an adaptor protein, evolutionary conserved from flies to mammals, (13), which is involved in many cellular processes (14), and is able to behave as a cell fate determinant, being responsible for daughter cell polarization in asymmetric division (14–16). Numb has been described in cortical ventricular zone cells (17) and neural crest lineages (18), where it segregates preferentially in neural daughter cell during asymmetric division (16). Mammalian Numb is transcribed in four isoforms, namely p65, p66, p71, and p72, produced by alternative splicing (19), of which the p66 and p72 have been the main focus of research. Several studies described the role of Numb in stem cells compartment's maintenance, acting as intrinsic determinant through the interaction with signaling pathways such as Notch, p53 and Shh (20–23). To this regard, Numb was shown to control Gli1 function by inducing Gli1 ubiquitination and degradation (23, 24). Interestingly, Numb is abnormally expressed in many cancer types (25–29), and has been demonstrated to play a role in cancer stem cell subpopulation of colorectal cancer (30) and gliomas (31).
The role of Numb as fate determinant in different types of stem cells, the important role of Shh in the maintenance of NSCs and MB SLCs and the molecular relationship between Numb and Gli prompted us to investigate the role of Numb in the mouse cerebellar neural stem cells (NSCs) as well as in medulloblastoma stem-like cells (MB-SLCs). Moreover, we aimed to investigate the expression of Numb isoforms in MB subgroups, since previous studies were conducted before the definition of the molecular subgroups and also the differential expression of Numb isoforms was not explored (23).
Materials and methods
Cell cultures
Stem Cells. Cerebellar NSCs were obtained from cerebella of postnatal 4-day-old wild-type black 6 /C56 (C57BL/6) mice (Charles River). NSCs were derived as previously described (12).
Mouse Medulloblastoma stem-like cells (mMB-SLCs) were derived from spontaneous tumors arisen in Ptc+/– mice, as previously described (9, 32).
Human Medulloblastoma stem-like cells (hMB-SLCs) were derived from primary human MB during surgical resection as previously described (9). hMB-SLCs were immunostained with APC-conjugated anti-CD133 (Miltenyi Biotec) according to manufacturer's protocol and sorted using a FACSAriaIII (BD Biosciences) prior to experiments (32).
NSCs and MB-SLCs were cultured in Selective medium (SM) for stem cells enrichment, containing DMEM/F12 (Gibco) supplemented with 0.6% glucose, 25 mg/ml insulin, 60 mg/ml N-acetyl-L-cysteine, 2 mg/ml heparin, 20 ng/ml EGF, 20 ng/ml bFGF (Peprotech, Rocky Hill, NJ), 1X penicillin-streptomycin, and B27 supplement without vitamin A (Gibco).
For neurosphere/oncosphere forming assay NSCs and MB-SLCs were disaggregated to single cell and plated at clonal density (1–2 cells/mm2) into 96-well plates, in selective medium. After 10–14 days, the number of neurospheres or oncospheres was divided by the number of cells plated to determine the percentage of neurosphere forming cells and oncosphere forming cells, respectively.
To induce differentiation, NSCs were disaggregated and plated on poly-lysine coated dishes in differentiation medium containing platelet-derived growth factor (PDGF; 10 ng/ml) (Sigma, P3076), for 48 h (9).
Animal experiments were approved by local ethic authorities and conducted in accordance with Italian Governing Law (D.lgs 26/2014; Prot. no. 03/2013).
P19 were purchased from ATCC and maintained in Alpha Minimum Essential Medium with ribonucleosides and deoxyribonucleosides supplemented with 7.5% bovine calf serum, 2.5% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Thermo scientific).
Treatments
Transduced NSCs were treated with a Smoothened antagonist cyclopamine-KAAD, (Calbiochem), at the 1 μM, and with Smo-agonist SAG (200 nM, Alexis), for 48 h. For differentiation experiment, cells were treated with platelet derived growth factor (PDGF, Sigma-Aldrich) for 48 h.
Immunofluorescence
To detect Gli1 and Numb, neurospheres were blandly disaggregated and plated on poly-lysine-coated Lab-Tek chamber slides (cover slips) for 2 h. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, incubated in blocking solution (5% normal goat serum, 1% BSA, 0.1% Triton X-100) and stained overnight with primary antibodies diluted in blocking solution and for 2 h with secondary antibodies. Primary antibodies were mouse anti-Gli1 and rabbit anti-Numb (Cell Signaling Technology Inc). 594- or 488-conjugated anti-mouse and anti- rabbit secondary antibodies were purchased from Molecular Probes (Invitrogen, Eugene, OR). Nuclei were counterstained with Hoechst reagent. Cover slips were mounted with fluorescence mounting medium (Dako, Carpinteria, CA). Images were acquired with Carl Zeiss microscope (Axio Observer Z1) and AxioVision Digital Image Processing Software.
Lentiviral transduction
Neurospheres were transduced with pGreenZeo Lentiviral Reporter Vectors containing specific promoters for NANOG (Nanog-GFP) or CMV (Zeo-GFP) (9) and infected cells were selected with Zeocin (Thermo Fisher) treatment.
Numb p66 lentiviral infection was performed using a lentiviral vector pRRL–CPPT–CMV–PGK–GFP–WPRE (TWEEN) containing Numb p66 coding sequence (23). NSCs and MB-SLCs were infected for 48 h prior to analyses.
Knockdown experiments
Silencing of endogenous Numb was performed using ON-TARGET plus Human NUMB siRNA (L-015902-00-0005) for hMB-SLCs and ON-TARGET plus Mouse Numb siRNA (L-046935-01-0005) for NSCs and mMB-SLCs. Dharmacon. Hiperfect reagent (Qiagen) was used for siRNA transfections according to manual instructions. After 72 h, cells were harvested and subjected to mRNA expression analysis.
RNA extraction and gene expression analysis
Total RNA was isolated from cells and human tissue using Trireagent (Ambion) and reverse transcribed in cDNA as previously described (12). cDNA was used for quantitative RT-PCR (qRT-PCR) analysis using ViiA ™ 7 Real-Time PCR System and SensiFAST™ Probe Lo-ROX (Bioline).
For each mRNA analysis 10 ng of cDNA were used. We selected best coverage TaqMan gene expression assay from Applied Biosystems and used according to the manufacturer's instructions. To analyze Numb isoforms, the following assay IDs were used: murine Numb p66 Mm01302754_m1; murine Numb p72 Mm01304901_m1, human Numb p66 Hs01105435_m1; human Numb p72 Hs01105426_m1.
mRNA quantification was expressed in arbitrary units and each amplification reaction was performed in triplicate. All Results were evaluated using the 2-ΔΔCT method and values were normalized to three endogenous controls: ß-actin, ß2-microglobulin, and Gapdh.
Western blot assay
Western blot was performed as previously described (33). Cellular pellets were lysed using lysis buffer: Tris-HCl pH 7.6 50 mM, deoxycholic acid sodium salt 0.5%, NaCl 140 mM, NP40 1%, EDTA 5 mM, NaF 100 mM, sodium pyrophosphate 2 mM, and protease inhibitors. Cellular lysates were separated on 8% acrylamide gel and western blot analysis was performed using standard procedures. Membranes were incubated overnight with the following antibodies: anti-Numb (ab4147; Abcam), anti-mouse Nanog (Cosmo Bio Co, Japan), anti-GAPDH (ab8245; Abcam), anti-Actin I-19 (sc-1616; Santa Cruz Biotechnology), anti-mouse Gli1 (#2643; Cell signaling), anti-NeuN (MAB377 Millipore), anti-βIII-tubulin (MAB 1637 Millipore). HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) were applied on membranes and signals were visualized by enhanced chemiluminescence (ECL Advansta). Densitometry was performed using ImageJ software and protein levels were normalized to the respective loading control. Error bars represent mean ± standard deviation of at least three experiments.
Human MB samples and controls
Surgical specimens of primary MBs were originated from a cohort of patients included in the present study, enrolled with Institutional Review Board approval, as previously described (34). Molecular subgroup classification was performed as described in (34). Number of MBs analyzed for each molecular subgroup: WNT n: 10; SHH n: 25; G3 n: 25; G4 n: 19.
Correlation analysis between the isoforms was measured using GraphPad Prism 6 software (La Jolla, CA, USA).
Commercial non-neoplastic cerebellum was purchased from Bio-chain Institute (n = 4: R1234039-50, Total RNA-Human Brain cerebellum Adult; n = 4: R1244041-50 and R1244040-50, Total RNA-Human Brain cerebellum Fetal).
Results
Numb has a pro-differentiation role in cerebellar neural stem cell (NSCs)
Involvement of Numb in cell determination and differentiation and in cortical neurogenesis has already been described (35), while the role of Numb in cerebellar neural stem cell (NSCs) differentiation has not been studied to date. First of all, we evaluated Numb protein expression in NSCs with respect to starting population (Figure 1A). NSCs were identified as the neurosphere forming cells after at least 30 days in selective medium (SM), and were compared to both the bulk cell population and to cerebellar cells after 5 days in SM. Notably, Numb protein level was lower at day 5 in SM with respect to both bulk population and NSCs, probably due to a selection of stem cells in medium, and its expression increased at day 30, when NSC culture was established (Figure 1A). Since only one band was revealed by western blot analysis, we compared Numb protein expression pattern of NSCs with the protein expression in murine embryonal carcinoma P19 cells after differentiation stimuli. P19 cells represent a model of neuronal differentiation which express both Numb p66 and Numb p72 isoforms (19). Interestingly, NSCs expressed high levels of the Numb p66 isoform while Numb p72 was not detectable (Supplementary Figure 1). To investigate the distribution of Numb positive cells in the heterogeneous population of neurosphere culture, we performed immunofluorescence staining of Numb and Gli1 (Figure 1B), a stemness marker in the context of cerebellar NSCs (9). Interestingly, Numb is expressed in both Gli1 positive and Gli1 negative cells.
Figure 1
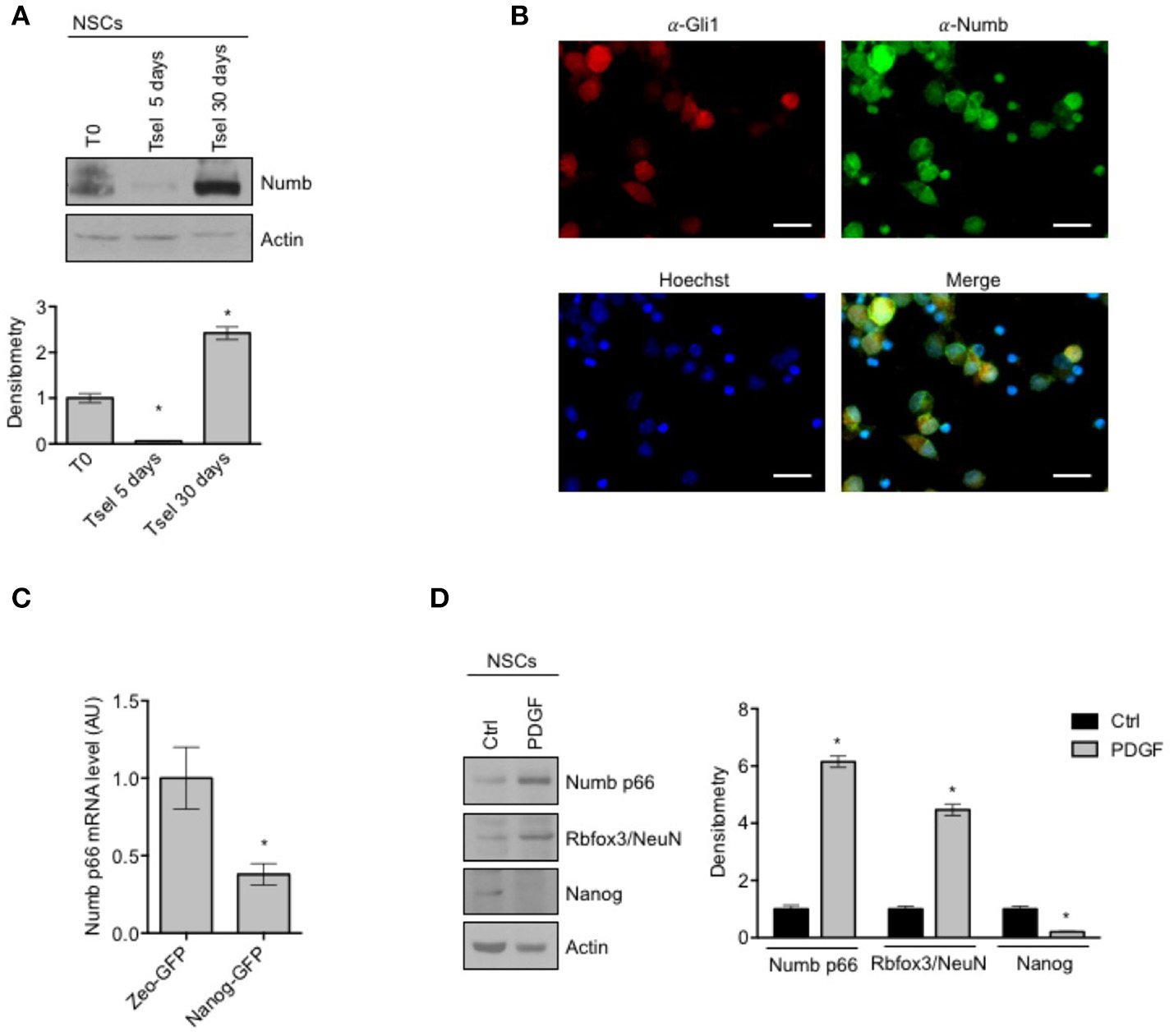
Numb expression in NSCs. (A) Representative Western Blot (WB) (up) and densitometric analysis (bottom) of endogenous Numb isoforms (p66 and p72) in NSCs grown in selective medium for 5 or 30 days, compared to bulk cells (T0). Actin was used as loading control. P-values *p < 0.05. (B) Representative images of immunofluorescence staining of NSCs for Numb (green) and Gli1 (red); nuclei are counterstained with Hoechst (blue). Scale bar: 10 μm for all panels. (C) qRT-PCR analysis showed mRNA expression of Numb p66, evaluated in NSCs infected with lentivirus carrying Nanog-GFP or Zeo-GFP as control and sorted for GFP. Data represent means ± SD from three independent experiments. P-values *p < 0.05. (D) Representative Western Blot (WB) (left) and densitometric analysis (right) of NSCs before and after in vitro differentiation for 48 h (PDGF); p66, Rbfox3/NeuN, Nanog were evaluated. Actin was used as loading control. P-values *p < 0.05. (A–D) Data are means ± SD from three independent experiments. Full-length images are presented in Supplementary Figures.
To further investigate whether Numb was associated with stemness features in the neurosphere population, we sorted cells according to their expression of the stemness factor Nanog (9), and we observed that Nanog positive cells expressed significantly lower levels of Numb p66, with respect to control (Figure 1C).
In order to explore the role of Numb in influencing the balance between stemness and neural differentiation, we evaluated Numb p66 protein level with western blot analysis, in NSCs before and after in vitro differentiation (Figure 1D). Numb p66 protein level was increased in NSCs after differentiation stimuli such as platelet-derived growth factor (PDGF), together with an enhanced expression of differentiation markers (Rbfox3/NeuN) and a reduced expression of Nanog stemness marker.
We next proceeded to investigate the role of Numb p66 in NSCs by modulating its expression. We performed lentiviral infection of NSCs with a virus encoding the ORF of Numb p66 (LvNumb) and evaluated the effects after 48 h. LvNumb-transduced NSC cells showed a differentiated phenotype with adherent morphology (Figure 2A, left), up-regulation of differentiation neural markers (Rbfox3/NeuN and βIII-tubulin) and down-regulation of the stemness markers Nanog and Gli1 (Figure 2A, right). Thus, our data support the role for Numb in promoting the “differentiated phenotype” of NSCs.
Figure 2
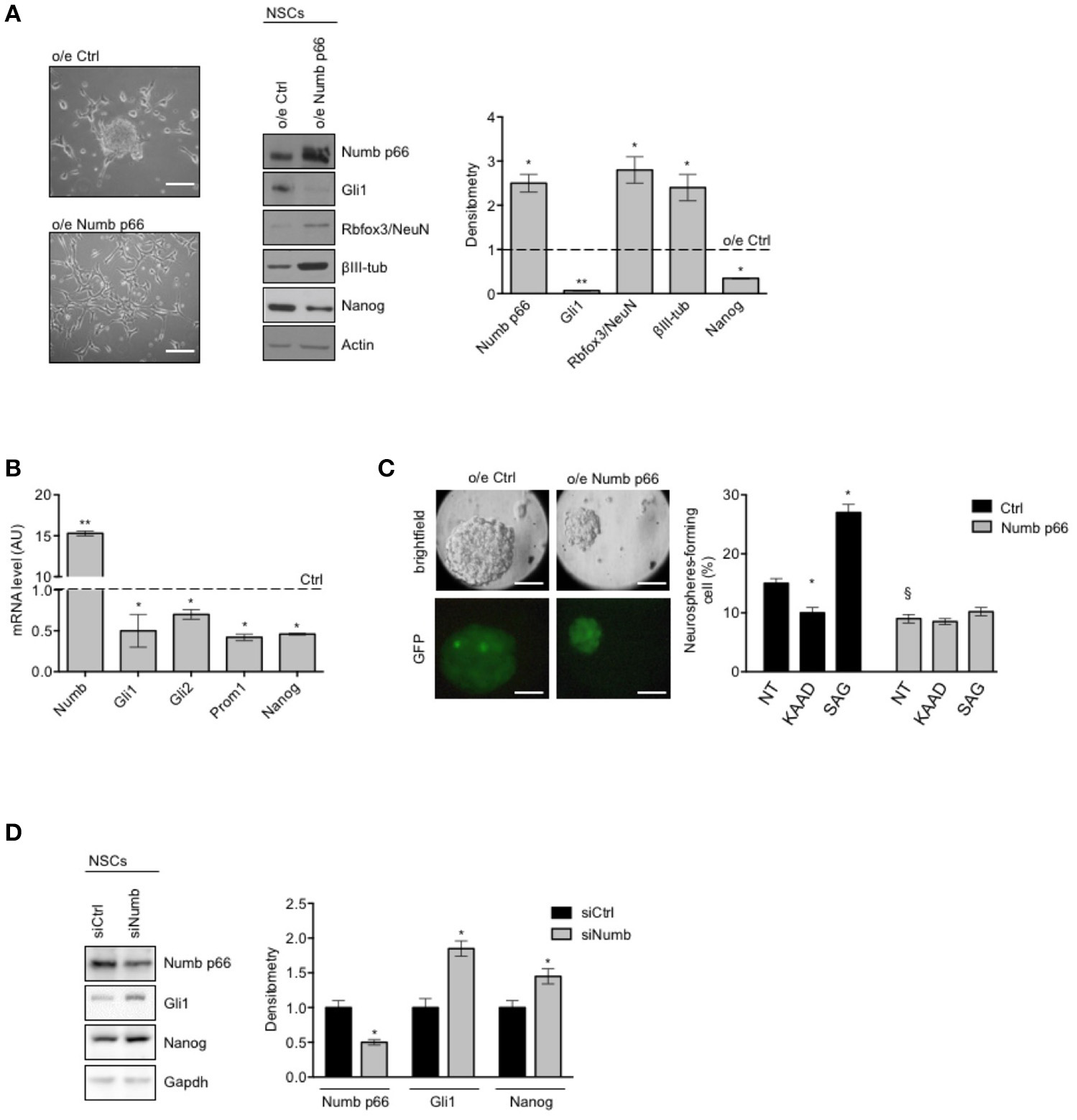
Numb antagonizes stemness and promotes differentiation of cerebellar NSCs. (A) (Left) Representative bright-field images of neurospheres after overexpression of p66 (or Ctrl). Scale bar: 100 μm. Representative western blot images (center) and densitometric analysis (right) of analysis of p66, stemness, and/or Hedgehog markers Nanog and Gli1 and differentiation markers Rbfox3/NeuN and βIII-tubulin (βIII-tub), in NSCs after overexpression of p66 vs. Ctrl. Actin was used as loading control. P-values *p < 0.05. (B) mRNA expression levels of Numb p66 and stemness and/or Hedgehog markers (Gli1, Gli2, Prom1, and Nanog), in NSCs after overexpression of p66. Dashed bars represent control cells (Ctrl). Data are means ± SD from three independent experiments. P-values *p < 0.05; **p < 0.01. (C) (Left) LvNumb-transduced NSCs were plated for clonogenic assay as described in Material and Methods. Upper panels show brightfield images of secondary neurospheres, lower panels show GFP of transduced cells. Scale bar: 100 μm. (Right) Neurosphere-formation capacity of LvNumb-transduced NSCs and treated with cyclopamine-KAAD (KAAD) to suppress endogenous Hedgehog signaling or with Smo-agonist (SAG). Data are means ± SD from three independent experiments. *P-values (vs. NT), *p < 0.05. §P-value (vs. LvCtrl), §p < 0.05. (D) Representative Western Blot (WB) (left) and densitometric analysis (right) of NSCs before and after in vitro differentiation for 48 h (PDGF); p66, Rbfox3/NeuN, Nanog were evaluated. Actin was used as loading control. P-values *p < 0.05. GAPDH was used as loading control. (A–D) Data are means ± SD from three independent experiments. Full-length images are presented in Supplementary Figures.
To deepen our understanding of the relationship between Numb p66 expression, stemness features and Shh signaling, we evaluated the expression of relevant markers in LvNumb-transduced NSCs. In detail, we observed a strong reduction, at transcription level, of both key components in the Shh pathway (Gli1, Gli2) and the stemness markers Prom1 and Nanog compared with control cells (Figure 2B). Consistently with mRNA level data of stemness markers, Numb overexpression also impaired the ability of NSCs to form secondary neurospheres, i.e., their clonogenicity (Figure 2C) and Numb transduced NSCs formed smaller neurospheres. We previously showed that the modulation of the Shh signaling in NSCs is able to significantly enhance or impair clonogenicity (9). Interestingly, after modulation of the Shh signaling in LvNumb-transduced NSCs, we did not observe any significant variation of self-renewal, suggesting that Numb overexpression could counteract this pathway (Figure 2C, right).
In order to further investigate the role of Numb p66 in NSCs, we performed silencing of Numb (siNumb) and observed an upregulation of Nanog and Gli1 (Figure 2D). Altogether, these data support a role for Numb p66 in NSCs where it induces neural differentiation and controls stemness by negatively regulating the Shh signaling.
NUMB p66 controls self-renewal of SHH MB stem-like cells
To investigate the role of NUMB in cancer stem-like cells (SLCs) from SHH MB, we performed the following set of experiments in SLCs derived from a human SHH MB, referred to as hMB-SLCs, as previously described (32). First of all, we sorted hMB-SLCs for the stemness marker CD133 (32), and analyzed the expression profile of CD133 positive (CD133+) and negative (CD133-) cells.
As shown in Figure 3A, expression level of NUMB p66 was significantly lower in CD133+ cells, with respect to CD133– ones, while NUMB p72 showed a positive trend in CD133 positive cells, without reaching statistical significance. Interestingly, mRNA levels of NUMB p66 resulted higher than NUMB p72 in CD133+ hMB-SLCs (Supplementary Figure 1B), in accordance with protein data. We also investigated markers of the Shh pathway (GLI1, CYCLIN D1, HIP1), that we previously showed to drive stemness in cerebellar NSCs and SHH MB SLCs (9). Shh pathway resulted more active in CD133+ (Figure 3A). These data suggest that NUMB p66 was associated with a reduced state of stemness hMB-SLCs.
Figure 3
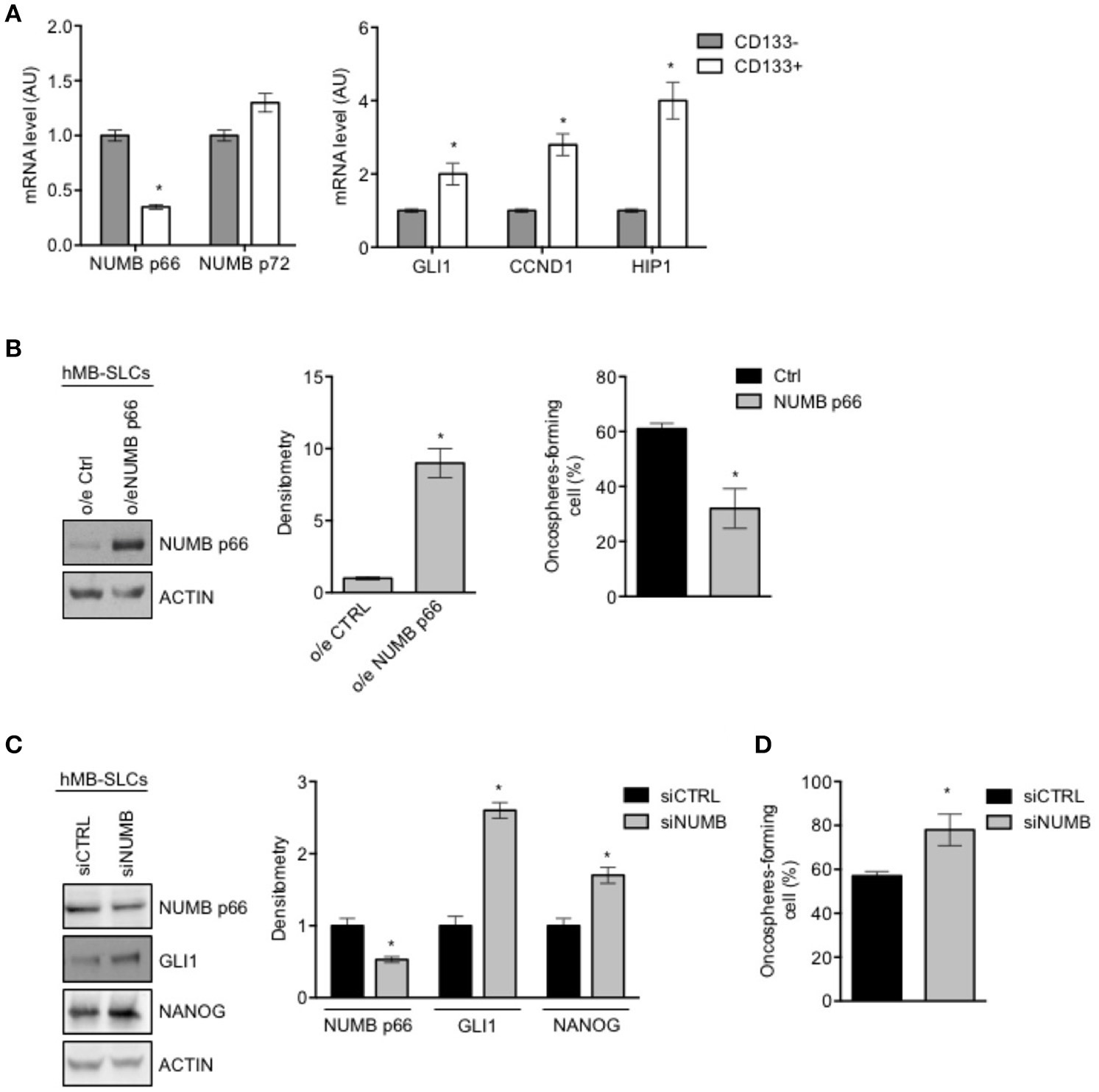
Modulation of NUMB in hMB-SLCs. (A) qRT-PCR analysis showed mRNA expression of NUMB isoforms, p66, and p72, and of Hedgehog markers (GLI1, CCND1, HIP1) evaluated in hMB-SLCs CD133+ vs. CD133−. Data represent means ± SD from 3 independent experiments. P-values *p < 0.05. (B) Representative western blot images (left) and densitometric analysis (center) of p66 in hMB-SLCs, after overexpression of p66 vs. Ctrl. ACTIN was used as loading control. (Right) Oncospheres-formation capacity of LvNumb-transduced hMB-SLCs vs. LvCtrl. P-values *p < 0.05. (C) Representative images of western blot (left) and densitometric analysis (right) analysis of p66, NANOG, and GLI1, in hMB-SLCs transfected with siRNA against NUMB (siNUMB) or non-targeting siRNA controls (siCTRL). ACTIN was used as loading control. P-values *p < 0.05. (D) Oncospheres-formation capacity of siNUMB hMB-SLCs vs. siCTRL. P-values vs. siCTRL. Data represent means ± SD from three independent experiments. P-values *p < 0.05. (B–C) Data are means ± SD from three independent experiments. Full-length images are presented in Supplementary Figures.
We then proceeded to evaluate the role of NUMB in hMB-SLCs in vitro. As shown in Supplementary Figure 1A, hMB-SLCs express high protein levels of NUMB p66 isoform. To explore the role of NUMB p66 in hMB-SLCs, we performed lentivirus-mediated overexpression of NUMB p66 (Figure 3B, left). We then evaluated the clonogenicity of LvNumb-transduced hMB-SLCs cells, a significant decrease of the oncospheres' forming capability was observed in hMB-SLCs overexpressing NUMB vs. control cells (Figure 3B, right).
To further understand the role of p66 in regulating stemness in MB-SLCs, we depleted the expression of NUMB using small interfering RNA (siNUMB) in hMB-SLCs. As shown in Figure 2C, following NUMB silencing, hMB-SLCs showed an increase of GLI1 protein level respect to control siRNA-transfected cells. Moreover, siNUMB hMB-SLCs showed an increase of NANOG stemness marker (Figure 3C) and oncosphere-forming ability (Figure 3D). These results suggest that NUMB may have a role in negatively regulating stemness features in hMB-SLCs. Additionally, we performed silencing of Numb in stem like cells isolated from murine SHH MB spontaneously arisen in Ptch +/− mice (9, 32). Numb silencing caused an increase in Gli1 and Nanog protein levels (Supplementary Figure 2A) and enhanced clonogenicity (Supplementary Figure 2B).
Our evidence show that NUMB plays a critical role in influencing MB-SLCs behavior, blocking the Sonic Hedgehog signaling and stemness features.
Numb alternative splicing in medulloblastoma
We then investigated NUMB expression in a cohort of MB using the MB dataset provided by Cavalli (Tumor Medulloblastoma–Cavalli−763 -rma_sketch—hugene11t) in the R2 platform (36). MB samples were divided into molecular subgroups and according to total NUMB expression level (low or high). As shown in Supplementary Figure 3, low expression of NUMB was associated with shorter overall survival (OS) in SHH and G3 subgroups.
Unfortunately, in the publicly available datasets the present probes do not discriminate the different isoforms of NUMB.
The expression level of NUMB 66 (p66) [Isoform 2, identifier: P49757-2] and NUMB 72 (p72) [Isoform 1, identifier: P49757-1] was evaluated in a cohort of human MB patients' samples characterized and divided in molecular subgroups.
The transcript level of p66 resulted significantly reduced only in SHH subgroup, whereas the p72 mRNA level was significantly up-regulated in all tumor molecular subgroups, as shown in Figure 4A, compared with normal adult cerebellar tissue (control). Interestingly, the up-regulation of p72 was stronger and more significant in G3, G4, and WNT with respect to SHH, although not reaching statistical significance in the comparison among subgroups. Furthermore, the expression of Numb p66 transcript was significantly higher in non-SHH tumors with respect to SHH. We also investigated the mRNA expression of NUMB isoforms in normal fetal cerebellum. With respect to adult tissues, fetal cerebellum showed a trend toward down-regulation of p66 and a trend of up-regulation of p72, even though it didn't reach statistical significance (Figure 4A). NUMB p66 expression in MBs failed to show statistical significance in the comparison with fetal cerebellum, and NUMB p72 resulted statistically up-regulated only in G4 MBs and WNT MBs. These data support previous findings that showed that SHH MB derive from of proliferating granule cell progenitors in the external granular layer (EGL) (37), that physiologically dissipates during the first year of postnatal life (38).
Figure 4
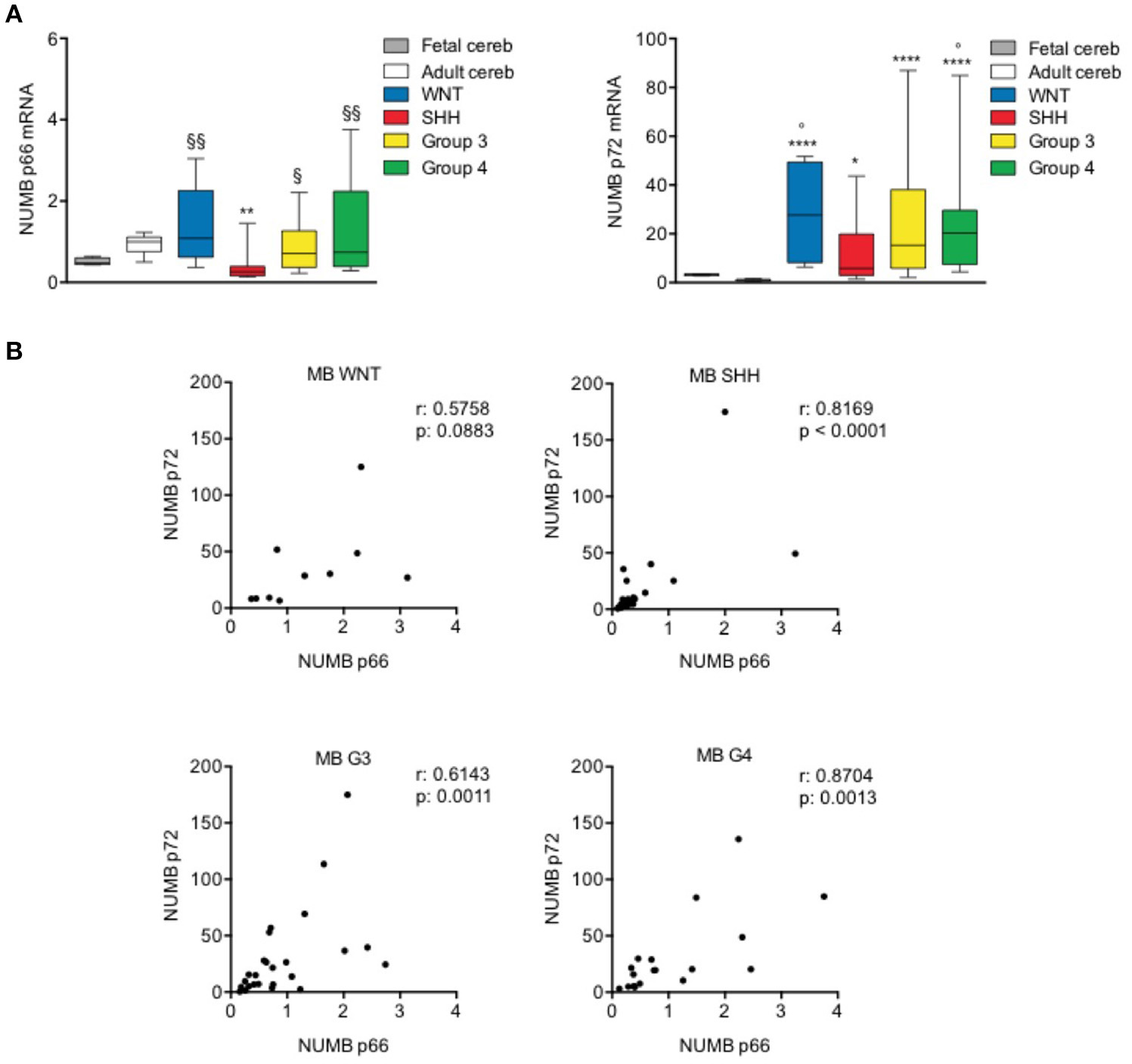
NUMB isoforms expression in human MB samples. (A) qRT-PCR analysis showed NUMB isoforms [p66 (left) and p72 (right)] mRNA expression for molecular subgroups of primary MBs (WNT n: 10; SHH n: 25; G3 n: 25; G4 n: 19), adult normal cerebellum (Adult cereb) and fetal normal cerebellum (Fetal cereb). *P-value vs. Adult cereb, *p < 0.05; **p < 0.01; ****p < 0.0001. ° = P vs. Fetal cereb, °p < 0.05. §P-value vs. SHH MBs, §p < 0.05; §§p < 0.01. (B) Pearson's correlation test of p66 vs. p72, in human MB subgroups, to evaluate the link between the expression of NUMB isoforms (r = correlation coefficient; p < 0.05 significance).
Next, we performed a correlation analysis of the expression level of the two isoforms analyzed that indicated significant positive correlation (p < 0.05) in SHH, G3, and G4 subgroups. WNT subgroup showed a trend toward positive correlation but didn't reach the statistical significance, possibly due to the small number of samples included in the study, with respect to other subgroups (Figure 4B).
Together, these data suggest a different role of NUMB p66 and p72 isoforms in human MB samples. Interestingly, we demonstrated a decreased expression of p66 only in the SHH subset, suggesting that different isoforms may exert divergent functions in the different MB subgroups.
Discussion
In this study we investigated for the first time, the expression of the two main NUMB isoforms p66 and p72 in a wide MB cohort, finding that different NUMB isoforms were differentially expressed among subgroups.
Indeed, NUMB p66 and p72 have been shown to have different/opposite roles regulating cellular functions. In murine embryonic carcinoma cells, it is described that p66 isoform is involved in differentiation but not in proliferation, whereas p72 has a role in proliferation but not in differentiation (39). This difference in the role of each isoform could explain the different functions described for NUMB in different types of cancer.
Specifically, NUMB has been described as an oncosuppressor in breast cancer (29), esophageal squamous cell carcinoma (27) and mesothelioma (40), but evidence showed also a role for NUMB as an oncogene in hepatocellular carcinoma (41), in astrocytomas (42) in cervical squamous carcinoma cells (43), and in endometrial cancer (44).
These different roles could reflect the different isoforms' expression in our cohort of human MB samples. Indeed, regardless of the molecular subgroup, NUMB p72 is up-regulated. The expression of this isoform could have a role in accelerating cell proliferation and promoting EMT features in MB as suggested by studies in other context (41, 45).
Interestingly, we found that NUMB p66 was significantly down-regulated in SHH MB only. Numb p66 is expressed during development and in adult brain (39, 46), and in murine P19 embryonic carcinoma cell line, p66 is reported to promote neural differentiation (39). Numb p66 was also shown to be down-regulated in murine models of SHH MB and to control Shh pathway activation through the regulation of Gli1 function, via its ubiquitination and proteasome dependent degradation (23, 24). Thus, our results strongly suggest that low levels of NUMB p66 in SHH MB contribute to Shh pathway deregulation keeping cancer cells in an undifferentiated state and enhancing their cancer stemness features. Altogether these results point out to different roles for NUMB isoforms in the MB subgroups possibly reflecting the diverse cell signaling pathways governing them. We believe that the dual role of NUMB as oncosuppressor or oncogene might be ascribed to the different isoform expressed, a topic that has not been fully investigated in the cited literature.
NUMB isoforms differential expression is controlled by alternative splicing. Of note, RBFOX3 a member of RNA-binding Fox (Rbfox) family, is one of the most important regulators of NUMB alternative splicing in neuronal lineages (47). RBFOX3/NEUN is expressed in neurons (47, 48) and during spinal neuronal development, RBFOX3/NEUN regulates neuronal differentiation promoting the alternative splicing of NUMB mRNA, from the isoform p72 to p66 (47). Interestingly, we found up-regulated RBFOX3/NEUN both after in vitro differentiation of NSCs (Figure 1D) and after Numb p66 over-expression (Figure 2A), suggesting that there could be a regulatory feedback between Numb p66 and RBFOX3/NEUN. As shown in Figure 1D, RBFOX3/NEUN is up-regulated during differentiation of NSCs along with Numb. Moreover, SHH MB shows the highest expression of RBFOX3/NEUN compared to the other molecular subgroups (Supplementary Figure 4). These evidence suggest that RBFOX3/NEUN could be responsible for NUMB alternative splicing in SHH MB context, leading to isoforms shift from p72 to p66.
Notably, we identified expression of Numb p66 also in NSCs and in MB-SLCs, as a common feature of “stem cell phenotype.” We have previously demonstrated that these cellular models are characterized by Hedgehog pathway activation (9, 49). Numb p66 expression inversely correlates with stem cells features: indeed, NSCs and SHH MB-SLCs show high clonogenic potential when Numb is silenced (Figures 2C, 3C), where it cannot antagonize survival pathways, such as Hedgehog signaling (23). We demonstrated the role of the Numb p66 isoform in promoting neural differentiation, antagonizing the expression of stemness/Shh markers and inhibiting self-renewal of stem cells.
In conclusion, in this study we demonstrated for the first time the expression of Numb isoforms in Medulloblastoma, highlighting possible different roles for each isoform in MB subgroups, that we believe are worthy of further investigation in follow up studies. Moreover, we described the suppressive role of Numb p66 on stemness features of cerebellar NSC and of SHH MB-SLCs.
Statements
Author contributions
LA and AP designed experiments, analyzed the data, and wrote the manuscript. AM, EM, AC, ZB, and MM performed the experiments and analyzed the data. EDS, FG, FL, and EF assisted the study.
Acknowledgments
The work was supported by Sapienza University Research Grant 2016 to EF; Sapienza University Research Grant 2017 to EDS; Ricerca Corrente to EM; Associazione Heal ONLUS to AM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2018.00315/full#supplementary-material
- MB
Medulloblastoma
- Shh
Sonic Hedgehog
- NSC
Neural Stem Cells
- SLC
Stem-like cells
- LvNumb
Lentivirus Numb
- hMB-SLC
Human Medulloblastoma stem-like cells
- MB-SLC
Murine Medulloblastoma stem-like cells.
Abbreviations
References
1.
Ferretti E Di Marcotullio L Gessi M Mattei T Greco A Po A et al . Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene (2006) 25:7267–73. 10.1038/sj.onc.1209716
2.
Northcott P Shih DJ Peacock J Garzia L Morrissy A Zichner T et al . Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature (2012) 488:49–56. 10.1038/nature11327
3.
Fan X Eberhart CG . Medulloblastoma stem cells. J Clin Oncol. (2008) 26:2821. 10.1200/JCO.2007.15.2264
4.
Kool M Korshunov A Remke M Jones D Schlanstein M Northcott P et al . Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. (2012) 123:473–84. 10.1007/s00401-012-0958-8
5.
Ocbina PJR Anderson KV . Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev Dyn. (2008) 237:2030–8. 10.1002/dvdy.21551
6.
Kim J Kato M Beachy PA . Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. (2009) 106:21666–71. 10.1073/pnas.0912180106
7.
Singh SK Hawkins C Clarke ID Squire JA Bayani J Hide T et al . Identification of human brain tumour initiating cells. Nature (2004) 432:396–401. 10.1038/nature03128
8.
Gulino A Ferretti E De Smaele E . Hedgehog signalling in colon cancer and stem cells. EMBO Mol Med. (2009) 1:300–2. 10.1002/emmm.200900042
9.
Po A Ferretti E Miele E De Smaele E Paganelli A Canettieri G et al . Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. (2010) 29:2646–58. 10.1038/emboj.2010.131
10.
Garg N Po A Miele E Campese A Begalli F Silvano M et al . microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. (2013) 32:2819–32. 10.1038/emboj.2013.214
11.
Miele E Po A Begalli F Antonucci L Mastronuzzi A Marras CE et al . β-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer (2017) 17:488. 10.1186/s12885-017-3477-0
12.
Besharat ZM Abballe L Cicconardi F Bhutkar A Grassi L Le Pera L et al . Foxm1 controls a pro-stemness microRNA network in neural stem cells. Sci Rep. (2018) 8:3523. 10.1038/s41598-018-21876-y
13.
Uemura T Shepherd S Ackerman L Jan LY Jan YN . Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell (1989) 58:349–60. 10.1016/0092-8674(89)90849-0
14.
Gulino A Di Marcotullio L Screpanti I . The multiple functions of Numb. Exp Res. (2010) 316:900–6. 10.1016/j.yexcr.2009.11.017
15.
Petersen PH Zou K Krauss S Zhong W . Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. (2004) 7:803–11. 10.1038/nn1289
16.
Cicalese A Bonizzi G Pasi CE Faretta M Ronzoni S Giulini B et al . The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell (2009) 138:1083–95. 10.1016/j.cell.2009.06.048
17.
Zhong W Feder JN Jiang M-M Jan LY Jan YN . Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron (1996) 17:43–53. 10.1016/S0896-6273(00)80279-2
18.
Wakamatsu Y Maynard TM Weston JA . Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development (2000) 127:2811–21. Available online at: http://dev.biologists.org/content/127/13/2811
19.
Rajendran D Zhang Y Berry D McGlade C . Regulation of numb isoform expression by activated ERK signaling. Oncogene (2016) 35:5202–13. 10.1038/onc.2016.69
20.
Androutsellis-Theotokis A Leker RR Soldner F Hoeppner DJ Ravin R Poser SW et al . Notch signalling regulates stem cell numbers in vitro and in vivo. Nature (2006) 442:823–6. 10.1038/nature04940
21.
Jiang J Hui C-C . Hedgehog signaling in development and cancer. Dev Cell (2008) 15:801–12. 10.1016/j.devcel.2008.11.010
22.
Meletis K Wirta V Hede S-M Nistér M Lundeberg J Frisén J . p53 suppresses the self-renewal of adult neural stem cells. Development (2006) 133:363–9. 10.1242/dev.02208
23.
Di Marcotullio L Ferretti E Greco A De Smaele E Po A Sico M et al . Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. (2006) 8:1415–23. 10.1038/ncb1510
24.
Di Marcotullio L Greco A Mazza D Canettieri G Pietrosanti L Infante P et al . Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene (2011) 30:65–76. 10.1038/onc.2010.394
25.
Xie C Lu Z Liu G Fang Y Liu J Huang Z et al . Numb downregulation suppresses cell growth and is associated with a poor prognosis of human hepatocellular carcinoma. Int J Mol Med. (2015) 36:653–60. 10.3892/ijmm.2015.2279
26.
Maiorano E Favia G Pece S Resta L Maisonneuve P Di Fiore P et al . Prognostic implications of NUMB immunoreactivity in salivary gland carcinomas. Int J Immunopathol Pharmacol. (2007) 20:779–89. 10.1177/039463200702000414
27.
Hong J Liu Z Zhu H Zhang X Liang Y Yao S et al . The tumor suppressive role of NUMB isoform 1 in esophageal squamous cell carcinoma. Oncotarget (2014) 5:5602–14. 10.18632/oncotarget.2136
28.
Liu D Cui L Wang Y Yang G He J Hao R et al . Hepatitis B e antigen and its precursors promote the progress of hepatocellular carcinoma by interacting with NUMB and decreasing p53 activity. Hepatology (2016) 64:390–404. 10.1002/hep.28594
29.
Pece S Serresi M Santolini E Capra M Hulleman E Galimberti V et al . Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. (2004) 167:215–21. 10.1083/jcb.200406140
30.
Hwang W-L Yang M-H . Numb is involved in the non-random segregation of subcellular vesicles in colorectal cancer stem cells. Cell Cycle (2016) 15:2697–703. 10.1080/15384101.2016.1218101
31.
Jiang X Xing H Kim TM Jung Y Huang W Yang HW et al . Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells (2012) 30:1313–26. 10.1002/stem.1120
32.
Miele E Valente S Alfano V Silvano M Mellini P Borovika D et al . The histone methyltransferase EZH2 as a druggable target in SHH medulloblastoma cancer stem cells. Oncotarget (2017) 8:68557–70. 10.18632/oncotarget.19782
33.
Celano M Schenone S Cosco D Navarra M Puxeddu E Racanicchi L et al . Cytotoxic effects of a novel pyrazolopyrimidine derivative entrapped in liposomes in anaplastic thyroid cancer cells in vitro and in xenograft tumors in vivo. Endocrine Relat Cancer (2008) 15:499–510. 10.1677/ERC-07-0243
34.
Laneve P Po A Favia A Legnini I Alfano V Rea J et al . The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget (2017) 9:31003–15. 10.18632/oncotarget.16049
35.
Zhong W Jiang M-M Schonemann MD Meneses JJ Pedersen RA Jan LY et al . Mouse numb is an essential gene involved in cortical neurogenesis. Proc Natl Acad Sci USA. (2000) 97:6844–9. 10.1073/pnas.97.12.6844
36.
R2. R2 : Genomics Analysis and Visualization Platform. Available online at: http://r2.amc.nl
37.
Skowron P Ramaswamy V Taylor MD . Genetic and molecular alterations across medulloblastoma subgroups. J Mol Med. (2015) 93:1075–84. 10.1007/s00109-015-1333-8
38.
Volpe JJ . Cerebellum of the premature infant: rapidly developing, vulnerable, clinically important. J Child Neurol. (2009) 24:1085–104. 10.1177/0883073809338067
39.
Verdi JM Bashirullah A Goldhawk DE Kubu CJ Jamali M Meakin SO et al . Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci USA. (1999) 96:10472–6. 10.1073/pnas.96.18.10472
40.
Kang Y Ding M Tian G Guo H Wan Y Yao Z et al . Overexpression of Numb suppresses tumor cell growth and enhances sensitivity to cisplatin in epithelioid malignant pleural mesothelioma. Oncol Rep. (2013) 30:313–9. 10.3892/or.2013.2429
41.
Lu Y Xu W Ji J Feng D Sourbier C Yang Y et al . Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology (2015) 62:1122–31. 10.1002/hep.27923
42.
Yan B Omar FM Das K Ng WH Lim C Shiuan K et al . Characterization of Numb expression in astrocytomas. Neuropathology (2008) 28:479–84. 10.1111/j.1440-1789.2008.00907.x
43.
Chen H Chen X Ye F Lu W Xie X . Symmetric division and expression of its regulatory gene Numb in human cervical squamous carcinoma cells. Pathobiology (2009) 76:149–54. 10.1159/000209393
44.
Wang C Cui T Feng W Li H Hu L . Role of Numb expression and nuclear translocation in endometrial cancer. Oncol Lett. (2015) 9:1531–6. 10.3892/ol.2015.2901
45.
Misquitta-Ali CM Cheng E O'Hanlon D Liu N McGlade CJ Tsao MS et al . Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. (2011) 31:138–50. 10.1128/MCB.00709-10
46.
Dho SE French MB Woods SA McGlade CJ . Characterization of four mammalian numb protein isoforms identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. (1999) 274:33097–104. 10.1074/jbc.274.46.33097
47.
Kim KK Nam J Mukouyama Y-S Kawamoto S . Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. (2013) 200:443–58. 10.1083/jcb.201206146
48.
McKee AE Minet E Stern C Riahi S Stiles CD Silver PA . A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomicallyrestricted expression in the developing mouse brain. BMC Dev Biol. (2005) 5:14. 10.1186/1471-213X-5-14
49.
Ronci M Catanzaro G Pieroni L Po A Besharat ZM Greco V et al . Proteomic analysis of human sonic hedgehog (SHH) medulloblastoma stem-like cells. Mol Biosyst. (2015) 11:1603–11. 10.1039/C5MB00034C
Summary
Keywords
medulloblastoma, numb, NUMB isoforms, cancer stem cells, neural stem cells, sonic hedgehog signaling
Citation
Abballe L, Mastronuzzi A, Miele E, Carai A, Besharat ZM, Moretti M, De Smaele E, Giangaspero F, Locatelli F, Ferretti E and Po A (2018) Numb Isoforms Deregulation in Medulloblastoma and Role of p66 Isoform in Cancer and Neural Stem Cells. Front. Pediatr. 6:315. doi: 10.3389/fped.2018.00315
Received
03 July 2018
Accepted
04 October 2018
Published
01 November 2018
Volume
6 - 2018
Edited by
Rimas J. Orentas, Seattle Children's Research Institute, United States
Reviewed by
Mingfu Wu, Albany Medical College, United States; Anshu Malhotra, Emory University, United States
Updates
Copyright
© 2018 Abballe, Mastronuzzi, Miele, Carai, Besharat, Moretti, De Smaele, Giangaspero, Locatelli, Ferretti and Po.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnese Po agnese.po@uniroma1.it
This article was submitted to Pediatric Oncology, a section of the journal Frontiers in Pediatrics
†These authors share first authorship
‡These authors share last authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.