- 1Neonatology, Thomas Jefferson University/Nemours, Philadelphia, PA, United States
- 2Department of Pathology, Thomas Jefferson University, Philadelphia, PA, United States
- 3Laboratory of Cancer Genomics, Thomas Jefferson University, Philadelphia, PA, United States
- 4Section of Neonatology, Department of Pediatrics, Cooper University Hospital, Camden, NJ, United States
- 5Department of Environmental Medicine, University of Rochester Medical Center, Rochester, NY, United States
Background: Histological chorioamnionitis (HCA) is an infection/inflammation of fetal membranes and complicates 5.2–28.5% of all live births. Exposure to HCA can have long-term consequences including abnormal neurodevelopment and an increased risk for allergic disorders and asthma later in childhood. HCA may incite epigenetic changes, which have the potential to modulate both the immune and neurological systems as well as increase the risk of related disorders later in life. However, there is limited data on the impact of HCA on epigenetics, in particular DNA methylation, and changes to immune and neurological systems in full-term human neonates.
Objective: To determine differential DNA methylation in cord blood mononuclear leukocytes from neonates exposed to HCA.
Methods: Cord blood was collected from 10 term neonates (5 with HCA and 5 controls without HCA) and mononuclear leukocytes were isolated. Genome-wide DNA methylation screening was performed on Genomic DNA extracted from mononuclear leukocytes.
Results: Mononuclear leukocytes from cord blood of HCA-exposed neonates showed differential DNA methylation of 68 probe sets compared to the control group (44 hypermethylated, 24 hypomethylated) with a p ≤ 0.0001. Several genes involved in immune modulation and nervous system development were found to be differentially methylated. Important canonical pathways as revealed by Ingenuity Pathway Analysis (IPA) were CREB Signaling in Neurons, FcγRIIB Signaling in B Lymphocytes, Cell Cycle: G1/S Checkpoint Regulation, Interleukin-1, 2, 3, 6, 8, 10, 17, and 17A signaling, p53 signaling, dopamine degradation, and serotonin degradation. The diseases and disorders picked up by IPA were nervous system development and function, neurological disease, respiratory disease, immune cell trafficking, inflammatory response, and immunological disease.
Conclusions: HCA induces differential DNA methylation in cord blood mononuclear leukocytes. The differentially methylated genes may contribute to inflammatory, immunological and neurodevelopmental disorders in neonates exposed to HCA.
Background
Chorioamnionitis (CA) is an infection/inflammation of the placenta and fetal membranes; it complicates up to 5.2% of live births (1) with histological CA (HCA) being found in 23.6% to 28.7% of term births with spontaneous labor (2, 3). HCA has been linked to several common chronic diseases of childhood: cerebral palsy, developmental delay, asthma and allergic disorders (4–13). The exact mechanism of these associations is unclear. Exposure to perinatal inflammation may stimulate epigenetic changes that alter immune function and subsequently contribute to these pathologies.
Prior studies have shown an association between perinatal environment, including epigenetic changes, and disease later in life (14, 15). Epigenetic changes can occur through multiple mechanisms including histone modification, DNA methylation, and RNA interference. DNA methylation is an enzyme-mediated change to DNA in which a methyl group is added to cytosine at CpG sites, an important mechanism of transcriptional regulation (16). Increased methylation results in reduced transcription of adjacent genes while decreased methylation results in increased transcription of adjacent genes. HCA is known to be associated with epigenetic changes via DNA methylation of key developmental genes in preterm neonates (17). Additionally, DNA methylation has been shown to be altered in placentas and fetal membranes with HCA (18).
The effects of HCA on DNA methylation in full term neonates are not yet known and may be an important mechanism in increasing susceptibility to disease later in life. Our objective was to evaluate the DNA methylation patterns of cord blood mononuclear leukocytes from neonates exposed to HCA compared with healthy controls.
Materials and Methods
Ethical Approval: Human Study Protocol and Institutional Biosafety Approvals
All human protocols and procedures described in this study were approved by the Institutional Review Board of Thomas Jefferson University Hospital. All experiments performed in this study were approved by the Nemours Institutional Biosafety Committee. The Institutional Review Board waived informed consent as the study was performed on discarded blood and placental tissue samples.
Study Design
This is a prospective observational study to examine differential DNA methylation in mononuclear leukocytes isolated from cord blood of term neonates born to mothers with HCA compared to those without HCA. Samples of cord blood and fetal membranes were collected from term neonates (37–40 weeks of gestation). Exclusion criteria included maternal infections other than HCA, gestational diabetes, hypertension, major congenital/chromosomal anomalies, and intrauterine growth restriction.
Cord Blood Collection and Isolation of Leukocytes
The umbilical cord was wiped with 70% alcohol immediately after delivery and cut at the placental side of the clamp. Cord blood was collected in sterile EDTA tubes, mixed thoroughly, and evaluated for blood clots. Mononuclear leukocytes were isolated by Ficoll-Paque Plus density gradient (GE Healthcare Biosciences, Pittsburgh, PA), following the manufacturer's protocol and the method described previously by our group (19). Two million packed cells per vial were saved at −80°C for DNA isolation and global DNA methylation analysis.
Fetal Membrane Collection, Processing, Staining, and Diagnosis of HCA
Fetal membrane tissues were washed with cold PBS and fixed in 10% neutral buffered formalin for 24–48 h. Membrane pieces were then processed and paraffin embedded in Histoplast LP (Thermo Fisher Scientific, Fremont, CA) following the detailed method of Gayen nee' Betal et al. (19). Tissue samples were processed using standard operating procedures at the Nemours Histochemistry and Tissue Processing Core (Nemours, Wilmington, DE). The tissue samples were then examined by a blinded pathologist and classified either HCA (placental membranes score ≥ stage 1) or no HCA (no histological inflammatory changes in fetal membranes) (20) (Supplemental Figure 1).
DNA Isolation and Global DNA Methylation Analysis
Genomic DNA was isolated using QIAamp DNA Mini kit (Qiagen, Germantown, MD). DNA was quantified on a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and the DNA quality was assessed by an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). Analysis of genome-wide DNA methylation was performed using the Infinium HumanMethylationEPIC BeadChip array (Illumina, San Diego, CA) at the Children's Hospital of Philadelphia following the standard protocol provided by Illumina. In brief, 500 ng of genomic DNA was bisulfite-converted using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. The Illumina iScan Reader was used to analyze the image and data from EPIC BeadChip. Data processing was performed with Illumina GenomeStudio software.
Quality Control and Statistical Approaches for DNA Methylation Analysis
Quality control (QC) was performed on DNA methylation data to filter out probes with missing methylation values in any samples and probes with at least 75% of samples having detected a p > 0.05. 866,836 probes remained for further analysis after QC filtering. DNA methylation β-values were normalized using the SWAN normalization method in minfi package in R/Bioconductor (21, 22). After normalization, DNA methylation sites with maximum methylation level below 0.15 were further filtered out to remove probes with low DNA methylation levels from next step differential analysis. A general linear model approach was then used to detect the difference in methylation levels between healthy neonates and neonates born to mothers with HCA using the limma package in R/Bioconductor (23). The moderated t-statistics with empirical Bayes approach was used to estimate the differences in methylation levels between healthy neonates and those born to mothers with HCA. Top DNA methylation sites were selected by test statistics and p-values (with raw p < 0.0001) from the two group comparisons. A Manhattan plot was used to show the selected top DNA methylation sites and their chromosome positions. A volcano plot of all DNA methylation sites was generated to show both their p-values and fold changes from the two group comparisons. A heatmap of methylation levels from top DNA methylation sites was generated to show the DNA methylation pattern differences between groups. All plots were generated using the program “R.”
Results
Genome-wide DNA methylation was performed on mononuclear leukocytes from cord blood of 10 term neonates. Five neonates had evidence of HCA by placental histopathology (referred as “HCA Group”) and five neonates had no evidence of HCA (referred as “control group”).
Differential DNA Methylation
DNA methylation levels are represented by β-values. The β-value is the ratio of the methylated probe signal intensity to the total locus intensity. The β-values range from 0 to 1 where 0 indicates unmethylated and 1 indicates fully methylated. A boxplot (Figure 1) was generated from β-values of all probe sets from the 10 samples to describe distribution of the pre-processed DNA methylation level. The interquartile range of the β-values was 0.2–0.9. Although the spread of β-values varied slightly within each group, no significant differences between groups were observed.
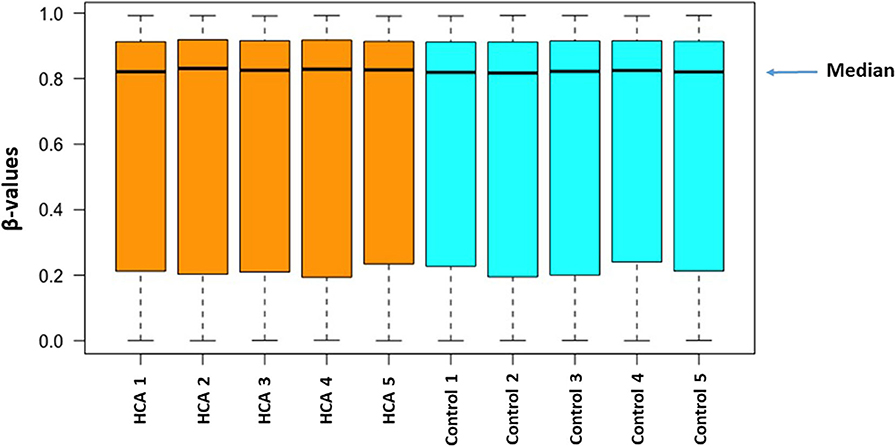
Figure 1. The distribution of methylation level (β-values) among all of the samples from the two groups.
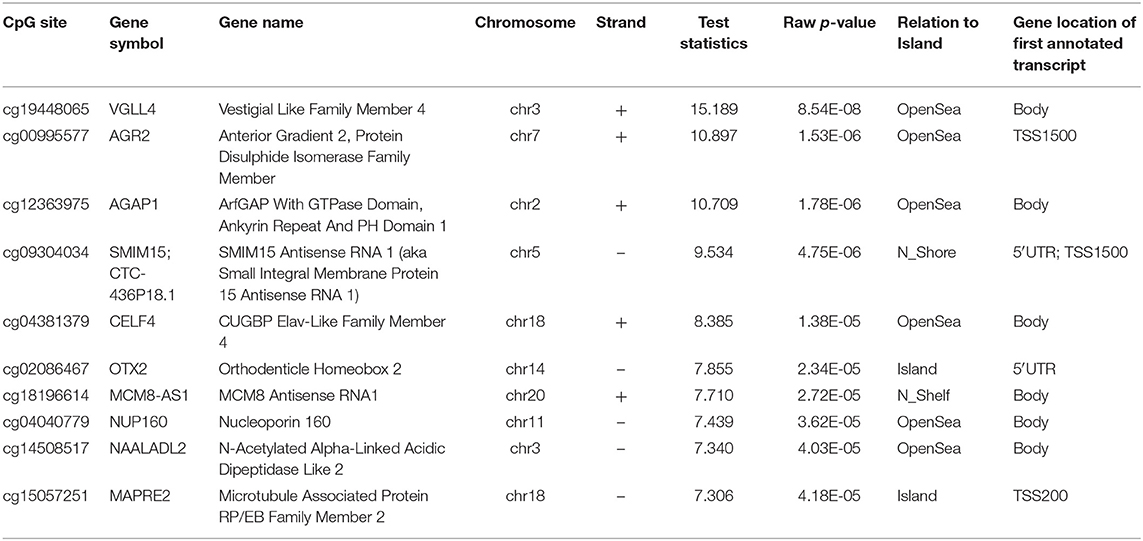
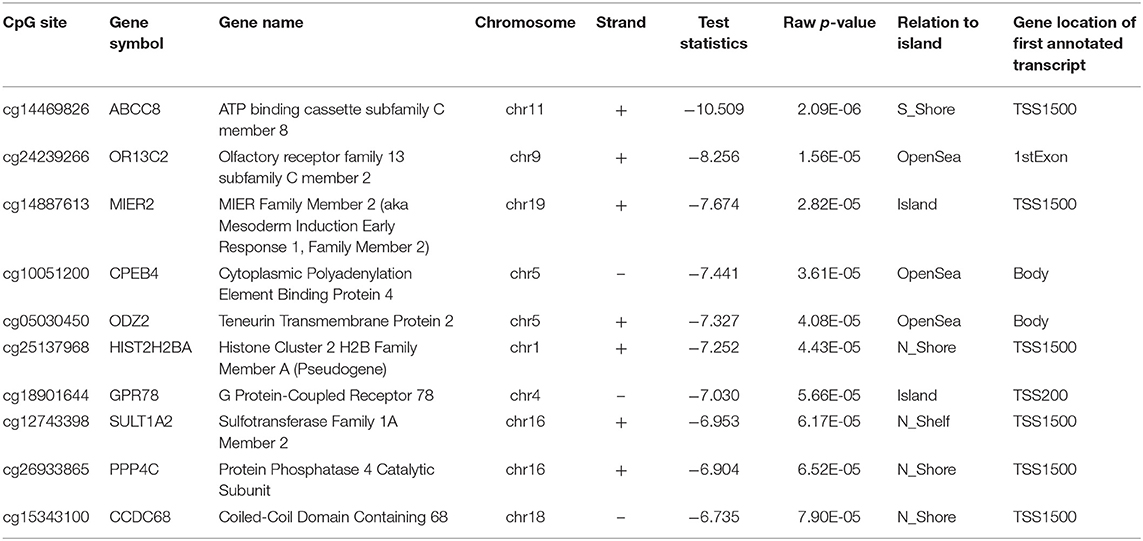
Comparison of the HCA group genome-wide DNA methylation data with the control group using Illumina GenomeStudio software identified 68 differentially methylated probe-IDs with a p ≤ 0.0001. Of these, 44 probe-IDs were significantly hypermethylated and 24 probe-IDs were significantly hypomethylated (Table 1). The top 10 hyper and hypomethylated probe-IDs along with their corresponding gene names based on p-values are listed in Tables 2, 3. Notable hypermethylated probe-IDs with gene names included AGR2 (Anterior Gradient Protein 2), which plays a role in asthma and mucin production (24), and OTX2 (Orthodenticle Homeobox 2), which interacts with nitric oxide (25). The top hypomethylated probe-IDs with gene names included MIER2 (Mesoderm Induction Early Response Protein 2), which regulates histone deacetylases (26), and PPP4C (Protein Phosphatase 4 Catalytic Subunit), a regulator of T-regulatory cells (27).
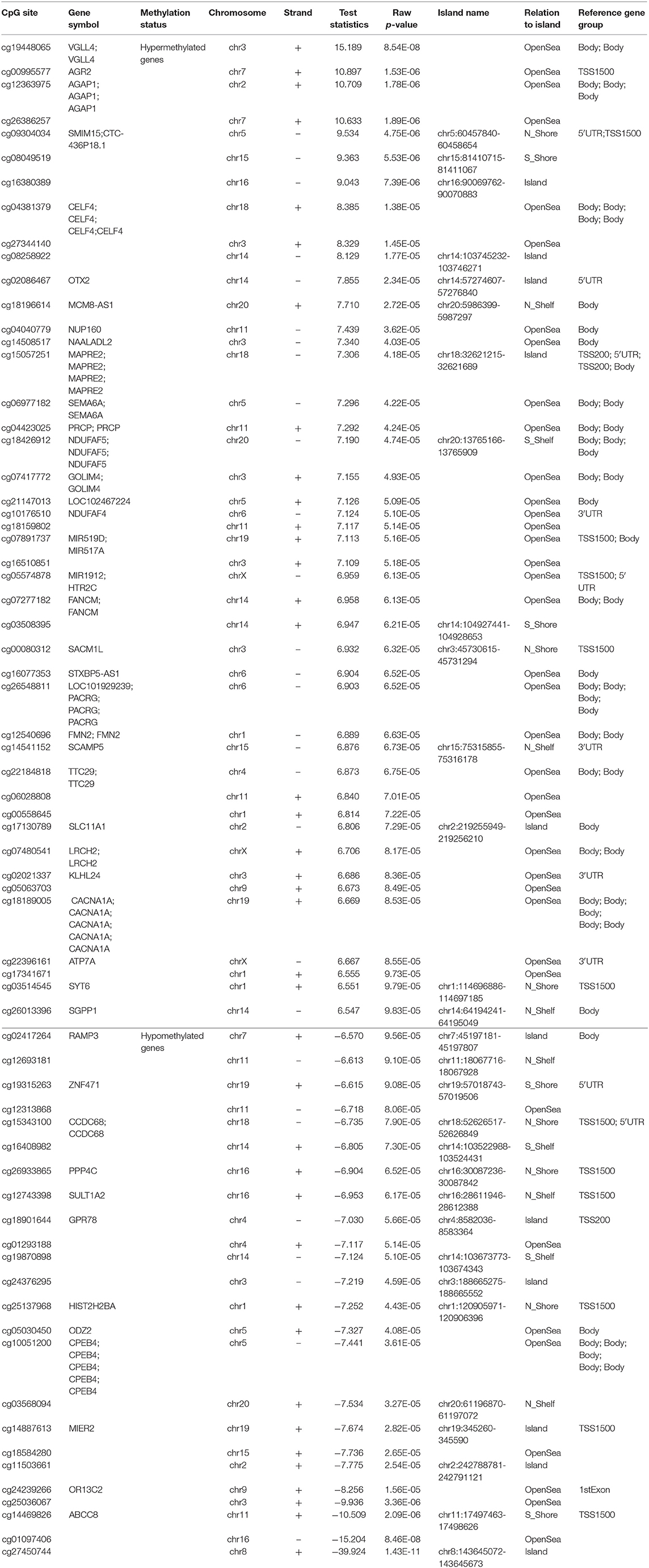
Table 1. Differentially methylated CpGs with exposure to histological chorioamnionitis (p ≤ 0.0001).
A Manhattan plot (Figure 2) shows the distribution of possible differentially methylated CpG sites identified across chromosomes. Genome-wide significance threshold of p ≤ 0.0001 was used as a cut off (red line). The top candidates in pairwise comparison were selected by test statistics and p-values for comparison. The black dotted line indicates a significance threshold of p ≤ 10−7. Three probe sets including two non-annotated probes (cg27450744, cg01097406) and VGLL4 (cg19448065) remained significant at a threshold of p ≤ 10−7. The differentially methylated CpG sites are depicted in a volcano plot (Figure 3). A volcano plot is a scatter plot of significance, the negative log of the p-value vs. the log of fold change.
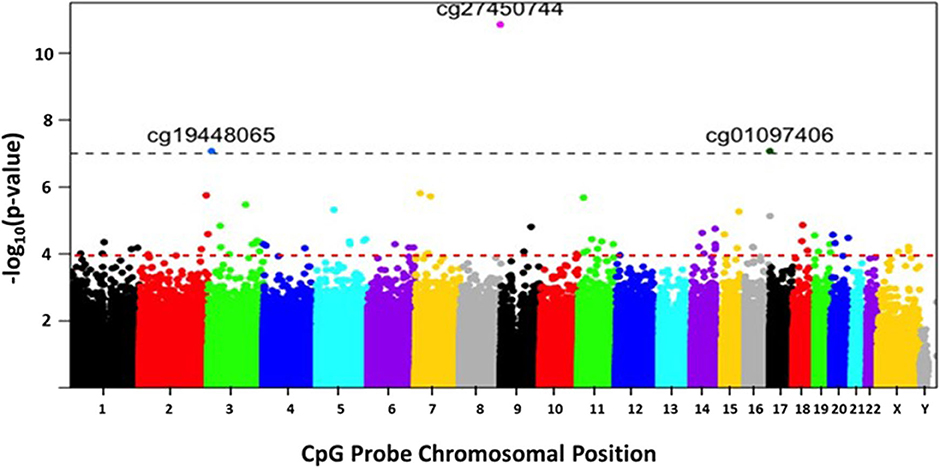
Figure 2. The distribution of differentially methylated CpG sites identified across chromosomes. The x-axis represents the chromosomal location of the CpG probes. The y-axis represents differential methylation as the negative log of the p-value of their association. Genome-wide significance threshold of p ≤ 0.0001 is used as a cut off (horizontal red line). The black dotted line indicates a significance threshold of p ≤ 10−7.
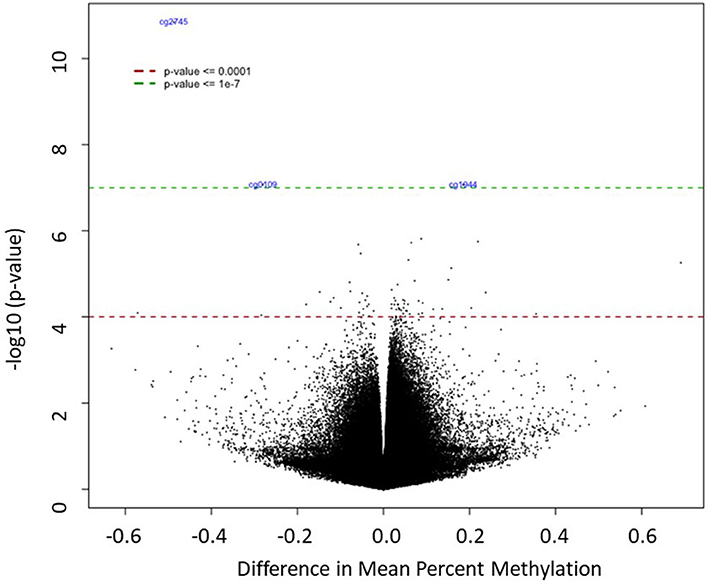
Figure 3. The differentially methylated CpG sites between the HCA and control groups. The x-axis shows difference in mean percent of DNA methylation and the y-axis shows the significance of differential methylation of each probe, represented as the negative log of the p-value. The red and green line indicate genome-wide significance thresholds of p ≤ 0.0001 and p ≤ 10−7, respectively.
Cluster Analysis – Heatmap
Cluster analysis was performed on the 68 differentially methylated loci for the two sample groups using the hierarchical clustering method. A heatmap of the methylation levels for the 68 DNA methylation loci illustrates the differences between the two groups (Figure 4). The hypermethylated loci are depicted in green, and the hypomethylated loci are depicted in red. Each column in the heatmap represents a sample.
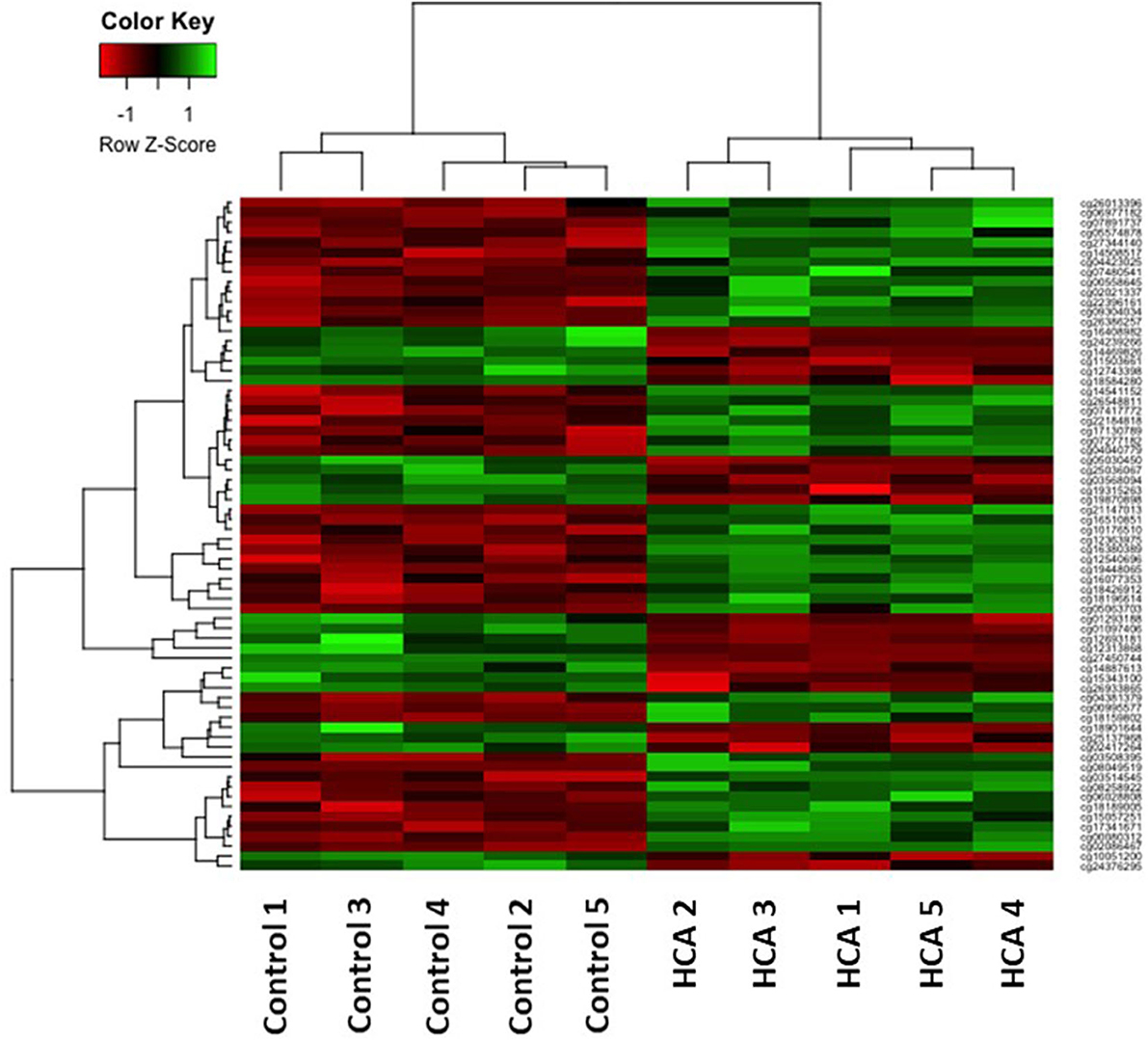
Figure 4. The 68 differentially methylated probe sets (p ≤ 0.0001) in the individual HCA samples (right) compared to control samples (left). The green and red colors indicate hyper-methylated and hypo-methylated loci, respectively.
Differential DNA Methylation Related to Immune Response
Eleven genes related to immune response and regulation of inflammatory pathways were identified (Table 4). Seven genes were hypermethylated and 4 genes were hypomethylated. The hypermethylated genes have roles in asthma (VGLL4, AGR2) (24, 28), neurologic development (AGAP1, OTX2) (29, 30), and immune regulation (SCAMP5, SLC11A1) (31, 32). The hypomethylated genes have roles in immune regulation (PPP4C) (27), transcription regulation (MIER2) (26), and inflammation (ABCC8) (33).
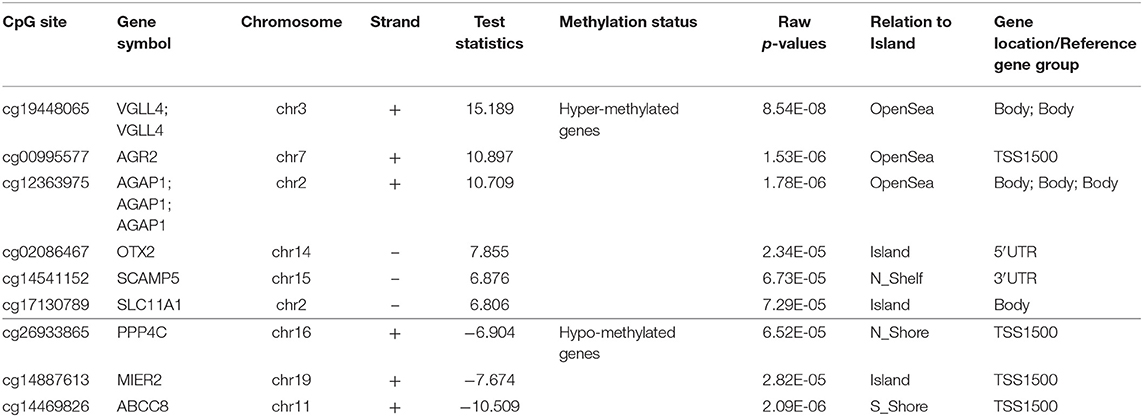
Table 4. Key genes related to immune response with exposure to histological chorioamnionitis (p ≤ 0.0001).
Ingenuity Pathway Analysis
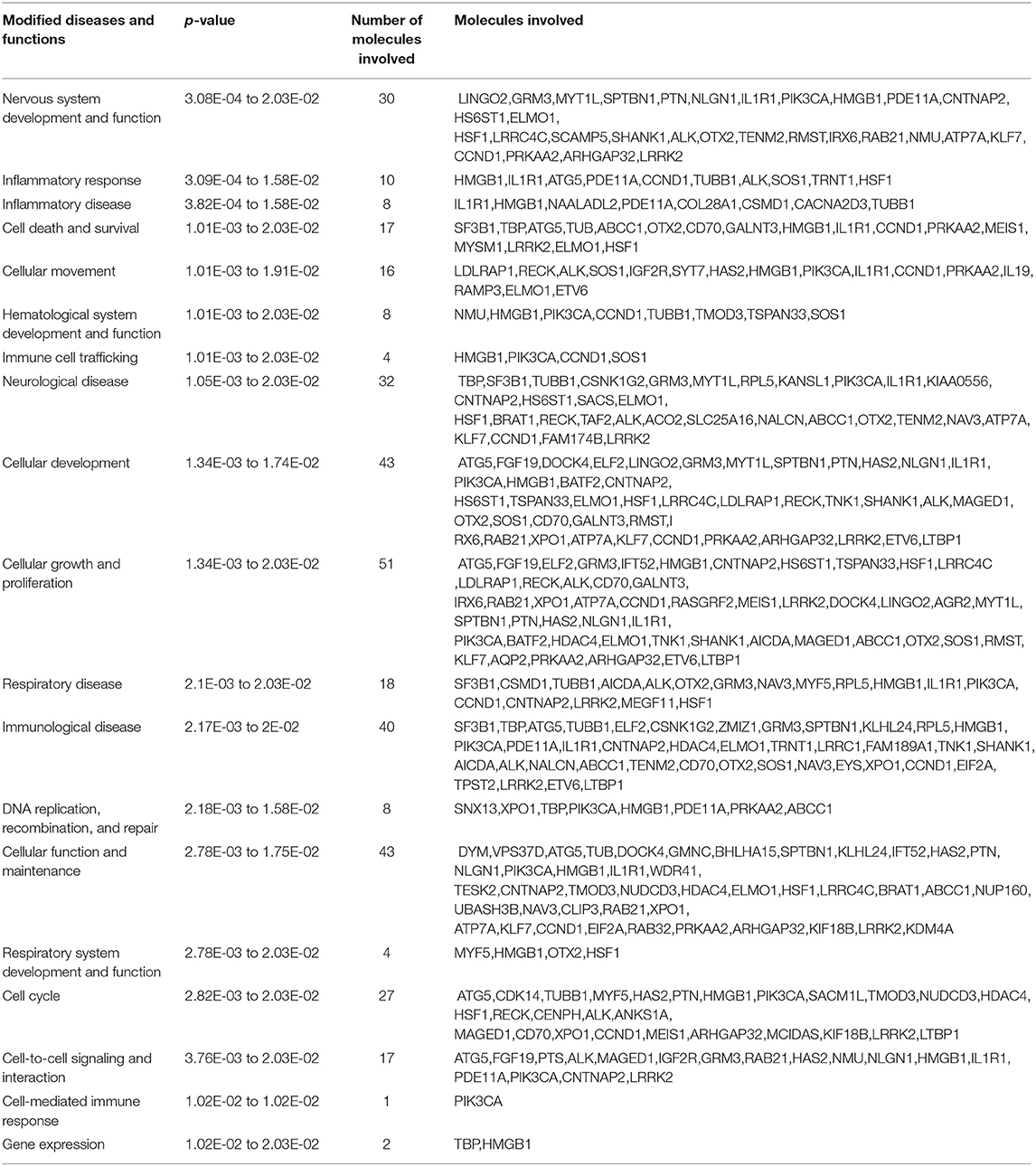
Ingenuity Pathway Analysis (IPA) software (Qiagen Inc., Germantown, MD) was used to perform pathway analysis by loading 448 probe sets that were found to be differentially methylated with exposure to HCA for a p ≤ 0.001. A total of 76 diseases and functions were altered with exposure to HCA. Selected key diseases and functions altered with HCA are listed in Table 5. The modification of functions with HCA included genes related to nervous system development and function, inflammatory response and disease, cellular movement, death and survival, hematological system development and function, and respiratory disease, development and function. IPA identified 224 canonical pathways that were modified with exposure to HCA. Selected key pathways important in immune regulation and inflammatory responses are shown in Table 6.
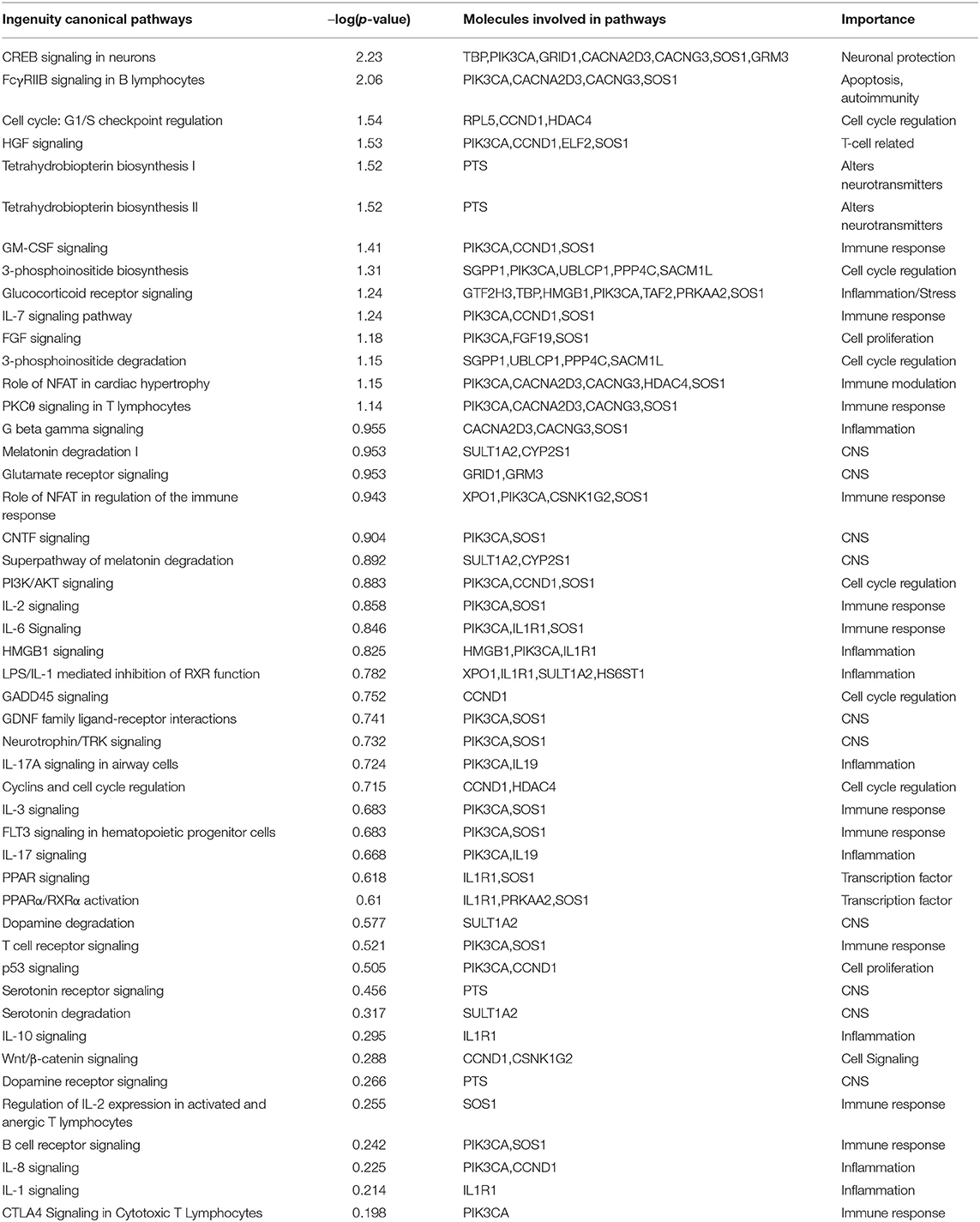
Table 6. Canonical pathways in Ingenuity Pathway Analysis associated with exposure to histological chorioamnionitis.
IPA identified 15 networks, of which network 2 (cellular growth and proliferation, cellular development, connective tissue development and function) and network 13 (cellular movement, hematological system development and function, immune cell trafficking) are closely related to the focus of this study.
Discussion
Exposure to HCA is associated with an increased risk of medical problems later in life including cerebral palsy, developmental delay, asthma, and allergic disorders (6–13). Although the mechanism of this increased risk is not yet known, alteration in gene expression secondary to differential DNA methylation may contribute to this increased risk. To our knowledge, this is the first study to report differential DNA methylation in human cord blood mononuclear leukocytes in term neonates exposed to HCA. Our data suggest that HCA altered the methylation status of genes involved in inflammation, immune regulation, respiratory development, and neurologic development in cord blood mononuclear leukocytes from term neonates.
Infants born to mothers with HCA are at increased risk for asthma but the exact mechanism is unknown (6, 7, 11, 12). Our results show that exposure to HCA was associated with differential methylation of several genes involved in signaling pathways related to the lung development, lung inflammation, and asthma. Vestigial Like Family Member 4 (VGLL4) is a gene related to Wnt/β-catenin signaling pathway regulation, cell cycle regulation, and immune regulation via T-cell mediated tumor regression (28, 34, 35). The Wnt/β-catenin signaling pathway plays a pivotal role in lung development, lung injury, and repair (28). Aberrant expression of Wnt/β-catenin signaling can lead to asthmatic airway remodeling with hyperplasia of airway smooth muscle cells, goblet cell metaplasia, and extracellular matrix deposition (28). Down-regulation of VGLL4 due to hypermethylation after exposure to HCA could lead to dysregulation of Wnt/β-catenin signaling, abnormal lung development, and an increased risk for asthma.
Cyclin D-1 gene (CCND1), another regulator of the Wnt/β-catenin signaling pathway, is differentially methylated after exposure to HCA and associated with airway remodeling and smooth muscle proliferation in asthma (36, 37). Mutation in the ATP Binding Cassette Subfamily C Member 8 (ABCC8) gene is associated with pulmonary arterial hypertension (38). Receptor Activity-Modifying Protein 3 (RAMP3), the top hypomethylated gene with exposure to HCA, plays a role in human lung development (39). An additional top hypermethylated gene is Anterior Gradient 2 (AGR2), a protein disulfide isomerase family member known to be involved in asthma and allergen-induced mucin production (24).
Previous studies have also shown that HCA is associated with neurodevelopmental impairment and cerebral palsy (8–10, 13). Several genes involved in nervous system development and function, neurological diseases, and neurological pathways were found to be differentially methylated in cord blood mononuclear leukocytes after exposure to HCA. Hypermethylated genes involved in neurological disorders include AGAP1, CACNA1A, and OTX2. AGAP1 is known to be involved in dopamine regulation and has been shown to increase endocytic recycling of M5 muscarinic receptors involved in dopamine release in the midbrain (29). Recent work by Pacault et al. showed that deletion of AGAP1 is reported in a patient with autism (40). Genetic variation in the CACNA1A gene is associated with cerebral palsy (41). OTX2 mutations are associated with severe ocular phenotypes, seizures, and developmental delay (30).
Examples of hypomethylated genes in our study include ABCC8, MIER2, and PPP4C. ABCC8 has been shown to be associated with inflammation regulation in the nervous system in autoimmune encephalomyelitis (33). MIER2 is involved in recruitment of histone deacetylase complexes (26). PPP4C plays a key role in cortical neurodevelopment by ensuring symmetric division of early cortical cells (42). Absence of PPP4C has been shown to result in early asymmetric division and premature neurogenesis. Hypomethylation of PPP4C may result in abnormal cortical development, increasing the risk of neurodevelopmental disorders. PPP4C has also been shown to be involved in immune regulation through proliferation and expansion of regulatory T cells and subsequent adaptive immunity (27). We speculate that the fetal systemic inflammatory response from HCA leads to changes in methylation pattern in leukocytes. The systemic inflammatory response outside the fetal brain can induce neuroinflammation and neuronal injury (43, 44). The inflammatory response in fetal brain due to HCA may change the DNA methylation pattern in nervous system cells.
We report differential DNA methylation in genes involved in lung development, neurodevelopment, immune regulation, and inflammation after exposure to HCA. Changes in DNA methylation may result in differential gene expression and alter these processes throughout life, being a potential mechanism for the increased risk of disorders seen with exposure to HCA. Our hierarchical clustering method analysis showed extremely consistent results across the samples from each group suggesting high reliability of the differential methylation patterns that we found. Furthermore, IPA identified diseases and functions that were consistent with the known associations of HCA, including modification of nervous system development and function, inflammatory response and disease, as well as respiratory disease, development, and function.
Our study has multiple strengths. This is the first study reporting differential DNA methylation of cord blood mononuclear leukocytes in term neonates born to mothers with HCA. Genome-wide DNA methylation was performed on the mononuclear leukocytes rather than whole blood. We chose umbilical cord blood which allowed us to obtain a significant volume of blood with no invasive risk to the neonate. We also recognize several limitations in our study. The sample size of 10 neonates is small, although similar sample sizes have been used in prior studies of differential DNA methylation (45). There is a chance of finding differential gene expression when comparing such a high number of genes but that was mitigated by using a very stringent p-value. Another weakness of the study is a lack a validation cohort. A larger study to confirm our finding is needed.
Conclusions
Exposure to HCA results in differential DNA methylation of cord blood mononuclear leukocytes in term neonates. This differential DNA methylation was present in genes involved in immune modulation, inflammation, lung development, and nervous system development. Future studies are needed to further validate these results in a larger group of neonates and by examining functional differences related to these genes. Our data may lead to an improved understanding of the genes and pathways involved in exposure to HCA. Identifying and understanding the role of novel candidate genes can lead to the development of prognostic biomarkers and therapies to mitigate the long-term consequences of abnormal DNA methylation induced by HCA.
Data Availability Statement
The raw data are available at the Gene Expression Omnibus (GEO) database of the NIH, under accession number GSE153668.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of Thomas Jefferson University Hospital and the Nemours Institutional Biosafety Committee. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author's Note
Presented in part at the Eastern Society for Pediatric Research Annual Meeting; Philadelphia, PA; March 2018 and the Annual Meeting of the Pediatric Academic Societies; Toronto, Canada; May 2018.
Author Contributions
GF, SG, and SM contributed equally to concept and design, sample collection and processing, acquisition and assembly of data, data analysis and interpretation, and manuscript writing. MF contributed to sample collection, acquisition and analysis of data, and manuscript writing. JC and SA contributed to data analysis and interpretation. VB, TS, and JG contributed to concept and design and data analysis and interpretation. IR contributed to concept and design, interpretation, and manuscript editing. ZA contributed to concept and design, data analysis and interpretation, and manuscript writing and editing. All authors have approved the version of the submitted manuscript.
Funding
This work was supported in part by the Institutional Development Award (IDeA) (Aghai) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number U54-GM104941 (PI: Binder-Macleod) and NIH COBRE P30GM114736 (PI: TS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2020.00437/full#supplementary-material
References
1. Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol. (2015) 213:S29–52. doi: 10.1016/j.ajog.2015.08.040
2. Mi Lee S, Romero R, Lee KA, Jin Yang H, Joon Oh K, Park CW, et al. The frequency and risk factors of funisitis and histologic chorioamnionitis in pregnant women at term who delivered after the spontaneous onset of labor. J Matern Fetal Neonatal Med. (2011) 24:37–42. doi: 10.3109/14767058.2010.482622
3. Torricelli M, Voltolini C, Conti N, Vellucci FL, Orlandini C, Bocchi C, et al. Histologic chorioamnionitis at term: implications for the progress of labor and neonatal wellbeing. J Mater Fetal Neonatal Med. (2013) 26:188–92. doi: 10.3109/14767058.2012.722724
4. Arneson CL, Durkin MS, Benedict RE, Kirby RS, Yeargin-Allsopp M, Van Naarden Braun K, et al. Prevalence of cerebral palsy: autism and developmental disabilities monitoring network, three sites, United States, 2004. Disabil Health J. (2009) 2:45–8. doi: 10.1016/j.dhjo.2008.08.001
5. Boulet SL, Boyle CA, Schieve LA. Health care use and health and functional impact of developmental disabilities among US children, 1997-2005. Arch Pediatr Adolesc Med. (2009) 163:19–26. doi: 10.1001/archpediatrics.2008.506
6. De Luca G, Olivieri F, Melotti G, Aiello G, Lubrano L, Boner AL. Fetal and early postnatal life roots of asthma. J Mater-Fetal Neonatal Med. (2010) 23:80–3. doi: 10.3109/14767058.2010.509931
7. Getahun D, Strickland D, Zeiger RS, Fassett MJ, Chen W, Rhoads GG, et al. Effect of chorioamnionitis on early childhood asthma. Arch Pediatr Adolesc Med. (2010) 164:187–92. doi: 10.1001/archpediatrics.2009.238
8. Shatrov JG, Birch SC, Lam LT, Quinlivan JA, McIntyre S, Mendz GL. Chorioamnionitis and cerebral palsy: a meta-analysis. Obstet Gynecol. (2010) 116:387–92. doi: 10.1097/AOG.0b013e3181e90046
9. Pappas A, Kendrick DE, Shankaran S, Stoll BJ, Bell EF, Laptook AR, et al. Chorioamnionitis and early childhood outcomes among extremely low-gestational-age neonates. JAMA Pediatr. (2014) 168:137–47. doi: 10.1001/jamapediatrics.2013.4248
10. Shevell A, Wintermark P, Benini R, Shevell M, Oskoui M. Chorioamnionitis and cerebral palsy: lessons from a patient registry. Eur J Paediatr Neurol. (2014) 18:301–7. doi: 10.1016/j.ejpn.2013.12.005
11. Akinbami LJ, Simon AE, Rossen LM. Changing trends in asthma prevalence among children. Pediatrics. (2016) 137:1–7. doi: 10.1542/peds.2015-2354
12. McDowell KM, Jobe AH, Fenchel M, Hardie WD, Gisslen T, Young LR, et al. Pulmonary morbidity in infancy after exposure to chorioamnionitis in late preterm infants. Ann Am Thorac Soc. (2016) 13:867–76. doi: 10.1513/AnnalsATS.201507-411OC
13. Shi Z, Ma L, Luo K, Bajaj M, Chawla S, Natarajan G, et al. Chorioamnionitis in the development of cerebral palsy: a meta-analysis and systematic review. Pediatrics. (2017) 139:e20163781. doi: 10.1542/peds.2016-3781
14. Barker DJP. The origins of the developmental origins theory. J Internal Med. (2007) 261:412–7. doi: 10.1111/j.1365-2796.2007.01809.x
15. Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod Toxicol. (2007) 23:297–307. doi: 10.1016/j.reprotox.2006.08.012
16. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38:23–38. doi: 10.1038/npp.2012.112
17. Liu Y, Hoyo C, Murphy S, Huang Z, Overcash F, Thompson J, et al. DNA methylation at imprint regulatory regions in preterm birth and infection. Am J Obstet Gynecol. (2013) 208:395. e1–7. doi: 10.1016/j.ajog.2013.02.006
18. Konwar C, Price EM, Wang LQ, Wilson SL, Terry J, Robinson WP. DNA methylation profiling of acute chorioamnionitis-associated placentas and fetal membranes: insights into epigenetic variation in spontaneous preterm births. Epigene Chromatin. (2018) 11:63. doi: 10.1186/s13072-018-0234-9
19. Gayen Nee' Betal S, Murthy S, Favara M, Fong G, Chan JSY, Addya S, et al. Histological chorioamnionitis induces differential gene expression in human cord blood mononuclear leukocytes from term neonates. Sci Rep. (2019) 9:5862. doi: 10.1038/s41598-019-42205-x
20. Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, Vogler C. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. (2003) 6:435–48. doi: 10.1007/s10024-003-7070-y
21. Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium humanmethylation450 beadchips. Genome Biol. (2012) 13:R44. doi: 10.1186/gb-2012-13-6-r44
22. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. (2014) 30:1363–9. doi: 10.1093/bioinformatics/btu049
23. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
24. Schroeder BW, Verhaeghe C, Park SW, Nguyenvu LT, Huang X, Zhen G, et al. AGR2 is induced in asthma and promotes allergen-induced mucin overproduction. Am J Respir Cell Mol Biol. (2012) 47:178–85. doi: 10.1165/rcmb.2011-0421OC
25. Filpa V, Carpanese E, Marchet S, Pirrone C, Conti A, Rainero A, et al. Nitric oxide regulates homeoprotein OTX1 and OTX2 expression in the rat myenteric plexus after intestinal ischemia-reperfusion injury. Am J Physiol Gastrointest Liver Physiol. (2017) 312:G374–89. doi: 10.1152/ajpgi.00386.2016
26. Derwish R, Paterno GD, Gillespie LL. Differential HDAC1 and 2 recruitment by members of the MIER family. PLoS ONE. (2017) 12:e0169338. doi: 10.1371/journal.pone.0169338
27. Liao FH, Hsiao WY, Lin YC, Chan YC, Huang CY. T cell proliferation and adaptive immune responses are critically regulated by protein phosphatase 4. Cell Cycle. (2016) 15:1073–83. doi: 10.1080/15384101.2016.1156267
28. Hussain M, Xu C, Lu M, Wu X, Tang L, Wu X. Wnt/beta-catenin signaling links embryonic lung development and asthmatic airway remodeling. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:3226–42. doi: 10.1016/j.bbadis.2017.08.031
29. Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HCJr, Sulzer D, et al. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. Embo J. (2010) 29:2813–26. doi: 10.1038/emboj.2010.154
30. Gat-Yablonski G. Brain development is a multi-level regulated process–the case of the OTX2 gene. Pediatr Endocrinol Rev. (2011) 9:422–30.
31. Correa MA, Canhamero T, Borrego A, Katz ISS, Jensen JR, Guerra JL, et al. Slc11a1 (Nramp-1) gene modulates immune-inflammation genes in macrophages during pristane-induced arthritis in mice. Inflamm Res. (2017) 66:969–80. doi: 10.1007/s00011-017-1077-8
32. Han C, Chen T, Yang M, Li N, Liu H, Cao X. Human SCAMP5, a novel secretory carrier membrane protein, facilitates calcium-triggered cytokine secretion by interaction with SNARE machinery. J Immunol. (2009) 182:2986–96. doi: 10.4049/jimmunol.0802002
33. Makar TK, Gerzanich V, Nimmagadda VK, Jain R, Lam K, Mubariz F, et al. Silencing of Abcc8 or inhibition of newly upregulated Sur1-Trpm4 reduce inflammation and disease progression in experimental autoimmune encephalomyelitis. J Neuroinflammation. (2015) 12:210. doi: 10.1186/s12974-015-0432-3
34. Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. (2014) 24:331–43. doi: 10.1038/cr.2014.10
35. Wu A, Wu Q, Deng Y, Liu Y, Lu J, Liu L, et al. Loss of VGLL4 suppresses tumor PD-L1 expression and immune evasion. Embo J. (2019) 38:e99506. doi: 10.15252/embj.201899506
36. Ge HY, Huang M, Yao X, Yin KS, Yang Y. [Cyclin D1 and its association with airway remodeling in a murine model of asthma]. Zhonghua Jie He He Hu Xi Za Zhi. (2006) 29:607–11.
37. Du CL, Xu YJ, Liu XS, Xie JG, Xie M, Zhang ZX, et al. Up-regulation of cyclin D1 expression in asthma serum-sensitized human airway smooth muscle promotes proliferation via protein kinase C alpha. Exp Lung Res. (2010) 36:201–10. doi: 10.3109/01902140903290022
38. Bohnen MS, Ma L, Zhu N, Qi H, McClenaghan C, Gonzaga-Jauregui C, et al. Loss-of-function ABCC8 mutations in pulmonary arterial hypertension. Circ Genom Precis Med. (2018) 11:e002087. doi: 10.1161/CIRCGEN.118.002087
39. Ramos CG, Sun X, Johnson EB, Nelson HE, Gonzalez Bosc LV. Adrenomedullin expression in the developing human fetal lung. J Investig Med. (2014) 62:49–55. doi: 10.2310/JIM.0000000000000020
40. Pacault M, Nizon M, Pichon O, Vincent M, Le Caignec C, Isidor B. A de novo 2q37.2 deletion encompassing AGAP1 and SH3BP4 in a patient with autism and intellectual disability. Eur J Med Genet. (2018) 62:103586. doi: 10.1016/j.ejmg.2018.11.020
41. Takezawa Y, Kikuchi A, Haginoya K, Niihori T, Numata-Uematsu Y, Inui T, et al. Genomic analysis identifies masqueraders of full-term cerebral palsy. Ann Clin Transl Neurol. (2018) 5:538–51. doi: 10.1002/acn3.551
42. Xie Y, Juschke C, Esk C, Hirotsune S, Knoblich JA. The phosphatase PP4c controls spindle orientation to maintain proliferative symmetric divisions in the developing neocortex. Neuron. (2013) 79:254–65. doi: 10.1016/j.neuron.2013.05.027
43. Malaeb S, Dammann O. Fetal inflammatory response and brain injury in the preterm newborn. J Child Neurol. (2009) 24:1119–26. doi: 10.1177/0883073809338066
44. Gussenhoven R, Westerlaken RJJ, Ophelders D, Jobe AH, Kemp MW, Kallapur SG, et al. Chorioamnionitis, neuroinflammation, and injury: timing is key in the preterm ovine fetus. J Neuroinflamm. (2018) 15:113. doi: 10.1186/s12974-018-1149-x
Keywords: epigenetics, leukocytes, histological chorioamnionitis, differential DNA methylation, immune modulation
Citation: Fong G, Gayen nee' Betal S, Murthy S, Favara M, Chan JSY, Addya S, Shaffer TH, Greenspan J, Bhandari V, Li D, Rahman I and Aghai ZH (2020) DNA Methylation Profile in Human Cord Blood Mononuclear Leukocytes From Term Neonates: Effects of Histological Chorioamnionitis. Front. Pediatr. 8:437. doi: 10.3389/fped.2020.00437
Received: 14 April 2020; Accepted: 23 June 2020;
Published: 04 August 2020.
Edited by:
Anup C. Katheria, Sharp Mary Birch Hospital for Women and Newborns, United StatesReviewed by:
Trent E. Tipple, University of Oklahoma Health Sciences Center, United StatesHannes Sallmon, Charité – Universitätsmedizin Berlin, Germany
Copyright © 2020 Fong, Gayen nee' Betal, Murthy, Favara, Chan, Addya, Shaffer, Greenspan, Bhandari, Li, Rahman and Aghai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zubair H. Aghai, emFnaGFpQG5lbW91cnMub3Jn
 Gina Fong1
Gina Fong1 Suhita Gayen nee' Betal
Suhita Gayen nee' Betal Michael Favara
Michael Favara Vineet Bhandari
Vineet Bhandari Irfan Rahman
Irfan Rahman Zubair H. Aghai
Zubair H. Aghai