- 1Facultad de Medicina Veterinaria y Zootecnia, Universidad Central del Ecuador (UC), Quito, Ecuador
- 2Centro Internacional de Zoonosis, Universidad Central del Ecuador (UC), Quito, Ecuador
- 3Dirección General de Postgrado, Universidad Tecnológica Equinoccial, Santo Domingo de los Tsáchilas, Ecuador
- 4Departamento de Ciencias de la Vida y la Agricultura, Universidad de las Fuerzas Armadas – ESPE, Carrera de Ingeniería en Ciencias Agropecuarias, Sangolqui, Ecuador
- 5Facultad de Ciencias Agrícolas, Universidad Central del Ecuador (UC), Quito, Ecuador
The Province of Santo Domingo de los Tsáchilas in Ecuador represents the largest informal cattle market. Because of its strategic position, cattle movement is very high and therefore we selected this region, to determine the strain variation of Brucella sp. Part of the study aimed at the isolation, biotyping, and genotyping of Brucella species from milk and supra-mammary lymph nodes of sero-positive bovines, using selective Farrell medium, biochemical assays, and IS711-PCR, AMOS-PCR, and HOOF-Prints techniques. In total, 656 animals from 12 sero-positive dairy herds and from the provincial slaughterhouse were diagnosed by Rose Bengal and Wright’s Slow Agglutination test with EDTA. Amongst these animals, 50 animals were sero-positive for brucellosis. Twenty-five lymph nodes and 25 milk samples from each group of positive reactors were transferred to culture medium. Isolation was possible from 4 (16%) lymph nodes and 9 (36%) milk samples; out of these, 10 isolates were diagnosed as Brucella sp. All four isolates of lymphatic tissue corresponded to Brucella abortus biotype 1, confirmed as field strains by molecular analysis. Milk isolations, showed biochemically a more dispersed pattern in which B. abortus biotypes 1 and 4 were found; yet four samples gave a pattern similar to B. abortus biotype 2; however, only biotypes 1 and 4 were confirmed by molecular analysis. The concordance between biochemical and molecular diagnostic tests reached 76.9%.
Introduction
Brucellosis is a widespread zoonotic disease, affecting cattle, sheep, goats, pigs, and humans (1). From a total of nine species of Brucella reported so far, four species are zoonoses: Brucella abortus, B. canis, B. melitensis, and B. suis which have been typically related to cattle, dogs, sheep goats, and pigs, respectively. Other species such as B. microti, B. neotomae, B. ovis, B. pinipedialis, and B. inopinata are supposed to be host specific (2, 3).
In cattle, the main symptoms associated with brucellosis include abortion and poor health in newborn calves. Epididymitis and infertility have been also reported in bulls (4, 5). In Ecuador, annual losses due to brucellosis in cattle are estimated to be around 5.5 million USD due to abortions, reduced milk yield, and mortality (6). In addition, in several municipalities in Ecuador, the presence of brucellosis in humans has been directly related to its presence in the cattle population (7), with, so far, only, B. abortus as the causative agent of human brucellosis (8, 9), contrary to neighboring Colombia and Peru, were in addition to B. abortus, B. melitensis, and B. suis have equally been reported in man (8, 10).
Determining the strain variability of Brucella can be helpful to understand the geographical and epidemiological dispersion of the disease as shown in the United States where molecular techniques have been used to evaluate strain diversity of B. abortus to define foci of transmission between cattle and wildlife, i.e., elk and bison, and also to identify infections related to the use of vaccines (11). In northern Ecuador, previous studies have reported B. abortus biotype 1 and 4 in human samples (9, 12), yet the diversity of Brucella sp. in cattle has not been investigated previously.
The livestock market in Santo Domingo de los Tsáchilas province is the largest in the country because of its strategic geographical location (13). This cattle market is very informal, facilitating the movement and exchange of animals and meat to large cities. It is also an important center for the trade of animals from the dairy areas of the Sierra region to different areas in the coastal region for fattening bull calves, as such it is hardly surprising that many of the outbreaks of foot-and-mouth disease started in this region (13). Thus, the sanitary condition of animals in this region might offer a reflection of the health status of cattle from different zones of the country. In this context, and given the zoonotic risk related to cattle brucellosis, the evaluation of the disease prevalence supported by a study of strain variability in cattle passing through this region will be an important epidemiological tool, including the assessment of the importance of food-borne brucellosis.
Materials and Methods
Study Design
The study area was located at Santo Domingo de los Tsáchilas Province (0.14°: −0.70°N, −78.73°: −79.62°E). In total, 656 blood samples were collected from 12 sero-positive dairy farms, previously identified during a large-scale national survey (data not published) and at the provincial abattoir between May and June 2013. Samples were analyzed by Rose Bengal plate (RB) and Wright’s Slow Agglutination Test with EDTA (SAT-EDTA). Equally, milk and supra-mammary lymph nodes were carefully sampled avoiding contamination and stored at 4°C until screening by RB and/or SAT-EDTA. Samples from positive reactors were processed for bacterial growth in the specific growth medium.
Serological Tests
All blood samples were tested by Rose Bengal (Bengatest antigen® 4% v/v suspension) and Wright’s SAT-EDTA (antigen SAW®, Synbiotics ASAW code). For RB, the slightest trace of agglutination was considered as positive. For SAT-EDTA, 100 μl of antigen was added to a doubling serum dilution from 1/12.5 up to 1/25.600. Data were recorded as international agglutination units (international units per milliliter) with values equal or greater than 30 IU/ml, corresponding to a transparency of 25% of a 1/25 dilution, considered as a positive reactions as described by Godfroid and Boelaert (14).
Microbiological Isolation
In a microbiology laboratory (biosafety type III), lymph nodes were macerated using the Stomacher®, milk samples were centrifuged at 3000 g for 10 min. Both macerated nodes and cream were tested for bacterial growth in selective Farrell medium [Columbia blood agar base CM0331 (Oxoid) + horse serum (reference: 16050-130 Gibco) + modified Brucella Selective Supplement SR0083A (Oxoid)]. Cultures were kept at 37°C and 5% CO2 for 5 days (15). Then, isolates were transferred to agar base [Columbia blood agar base CM0331 (Oxoid)] to obtain distinct Brucella sp. colonies. Finally, part of the colonies was used for DNA extraction and another part was stored at −70°C for further analysis.
Biotyping and Molecular Identification
Isolated colonies were biotypified by macroscopic observation and biochemical assays, i.e., urease, catalase, oxidase, and hydrogen sulfide production. Additionally, bacterial cultures were grown on media with stained safranin, thionin, and fuchsin at different concentrations, and tested for agglutination with Anti-A and Anti-M mono-specific sera (15).
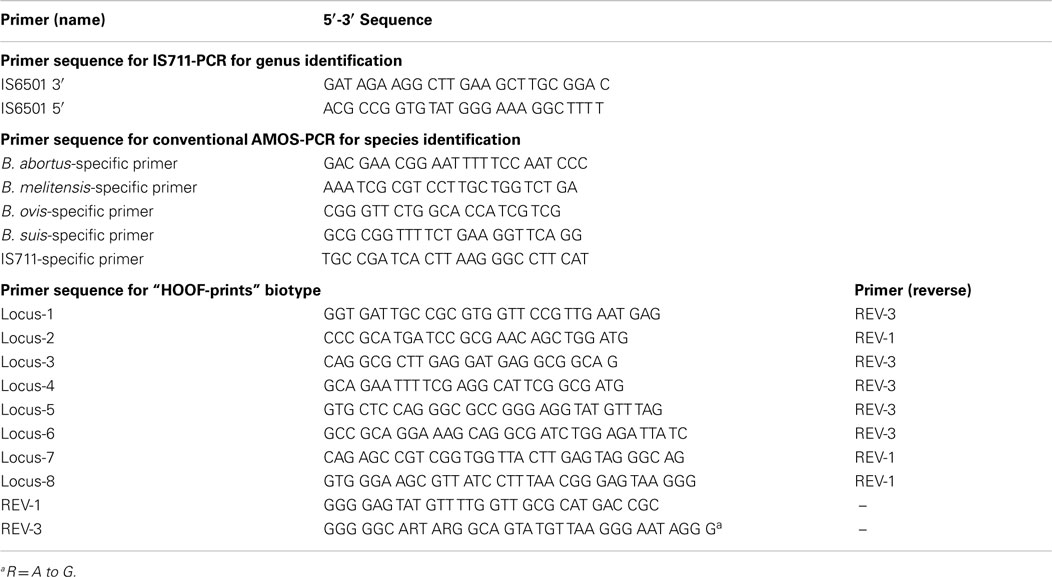
For molecular identification, genomic DNA was extracted according to Marmur and Kirby [phenol–chloroform–isoamyl alcohol (16)]. DNA amplification was performed using protocols IS711-PCR and AMOS-PCR as described by Ref. (17, 18) to identify genera and species, respectively. Primers for DNA amplification are presented in Table 1. Each PCR-reaction had a final volume of 20 μl. Master mix was made with 1 U/45 μl of Taq Polymerase, 1X buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 mM of each primer, and approximately 10 ng of DNA. To characterize the Brucella biotype, the “HOOF-Print” technique was used as described by Bricker et al. (19) and Bricker and Ewalt (20) for eight loci; all VNTR were amplified separately using primers described in Table 1; each PCR-reaction had a final volume of 15 μl and the master mix was composed with 0.6 U of Taq Polymerase, 1X buffer, 1.5 mM MgCl2, 0.25 mM dNTPs, 0.2 mM of each primer, and approximately 10 ng of DNA.
Data Analysis
The proportions of isolation of Brucella sp. were contrasted by Fisher exact test with 5% statistical significance. Additionally, an estimation of the test concordance was measured in terms of positive and negative agreements over the total isolations. Data were analyzed in “R” software version 3.1.0.
Results
Serology
Out of 656 blood samples, 50 were sero-positive, i.e., 25 were from the slaughterhouse and 25 were from sero-positive dairy farms of Santo Domingo.
Microbiological Isolation
Twenty-five milk and 25 lymph node samples were processed and isolated in a specific microbiological medium. The bacterial growth of Brucella spp. was evidenced in nine (36%) and four cases (16%), respectively. No statistical difference was found between the types of sample used for the isolation (p-value = 0.1085); yet isolation from milk appeared to be better than from tissues.
Bio-Typification
Table 2 shows the biochemical features of the microbiological isolations from sero-positive animals and from those where Brucella was isolated (milk or supra-mammary lymph nodes). Out of nine milk isolations, six were biochemically compatible with B. abortus biotype and three were “not determined” isolations (ND, samples: 8, 10, and 13) because they did not present urease activity, nor growth in CO2 and no H2S production. Isolations from lymphatic nodes (samples 1–4) were also biochemically compatible with B. abortus. In total, nine isolates were sensitive to inhibition by basic fuchsin, four were insensitive but agglutinated with anti-A sera. Nine isolates agglutinated with anti-A sera (i.e., samples 1–5, 6, 7, 9, and sample 11) and only one agglutinated with anti-M sera (sample 12) hence corresponding to B. abortus biotype 4. As described by Corbel and Brinley Morgan (21), Mayfield et al. (22), and Rodríguez Torres et al. (23), growth in basic fuchsin medium and agglutination with anti-A sera, is indicative for B. abortus biotype 1; however, lack of bacterial growth in basic fuchsin and agglutination with anti-A sera is indicative for B. abortus biotype 2. Yet, as shown in Table 2, by molecular analysis, all isolates were B. abortus biotype 1. All milk isolates were identified as B. abortus biotypes 1 and 4.
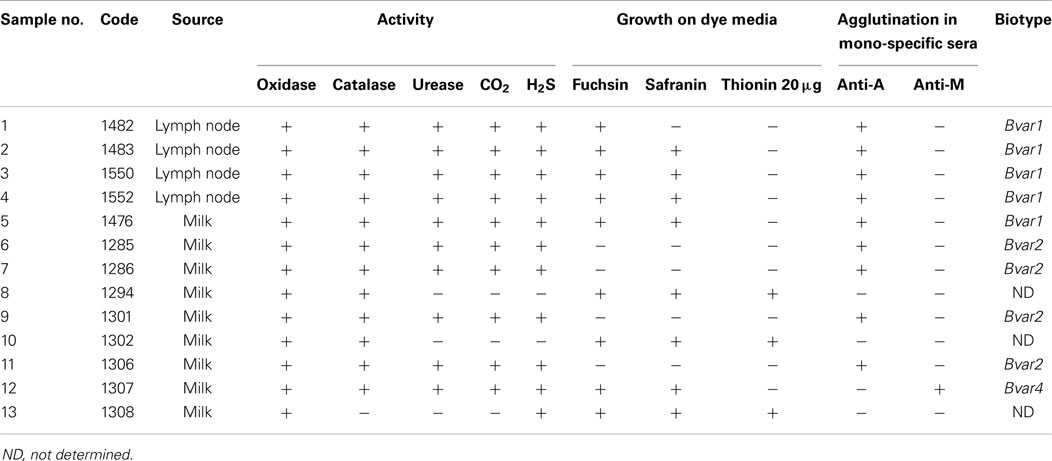
Table 2. Differential characters of B. abortus and biotypes isolated from milk and lymph nodes collected in Santo Domingo de los Tsáchilas province.
Molecular Identification
In total, 13 isolates corresponded to B. abortus identified by IS711 and AMOS-PCR (Table 3). The “HOOF-Prints” protocol allows biotype classification, as such VNTR markers evidenced the presence of B. abortus biotype 1 in 12 out of 13 isolates. All these isolates were field strains and were different from vaccine strains S19 and RB51, as confirmed by conventional AMOS-PCR. Furthermore, one isolate, from a milk sample, was confirmed to be B. abortus biotype 4 (Sample 12). The allelic diversity found in Brucella isolates from Santo Domingo Province is given in Table 4. Molecular patterns found are similar to biotype 1 and 4, reported by Bricker et al. (19). Samples, biochemically found as biotype 2 (samples 6, 7, 9, and 11), were confirmed as B. abortus biotype 1 whilst sample 12 was corroborated as biotype 4.
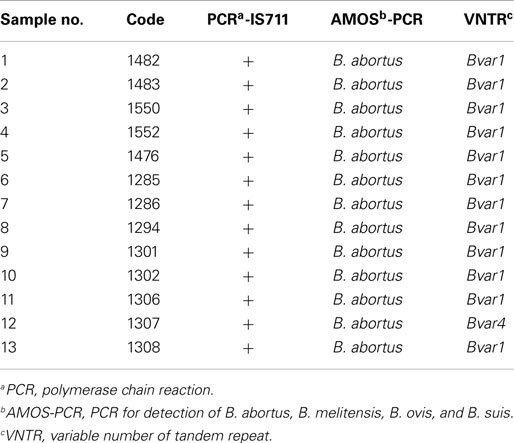
Table 3. Genotyping of Brucella spp. from isolates of milk and lymph nodes collected in Santo Domingo de los Tsáchilas province.
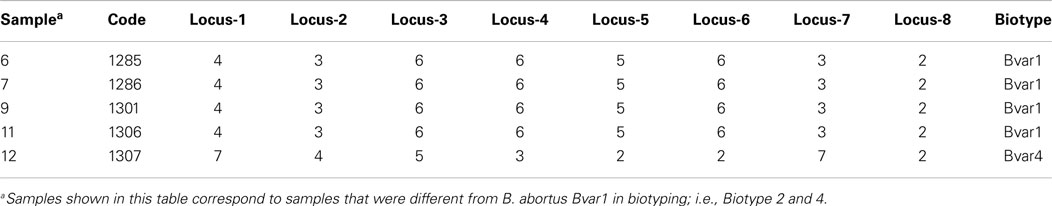
Table 4. HOOF-Prints: results of alleles configuration to identify Brucella abortus biotypes from isolates of milk collected in Santo Domingo Province in Ecuador.
On the other hand, the concordance of biochemical and molecular tests estimated a proportion of coincidences of 76.92%.
Discussion
This study demonstrated the presence of bovine brucellosis in the province of Santo Domingo de los Tsáchilas province.
Biochemical tests used for biotyping isolates allowed the identification of B. abortus biotypes 1, 2, and 4, biotypes which have been previously reported in human populations in Ecuador using biochemical and molecular techniques (9, 12). Samples 6, 7, 9, and 11, were biochemically identified as B. abortus biotype 2, yet as B. abortus biotype 1 by HOOF-Prints protocol, which is highly sensitive test (11, 19). It is known that the biochemical tests are of limited use for identifying biotypes, since the biochemical response depends on environmental conditions during the preparation of media and reagents and the amount and time for growth of the strains (24–26). In addition, the intraspecific Brucella molecular variability could have caused this biochemical response (21–23, 27). However, further studies are suggested to confirm or reject the presence of B. abortus biotype 2 in Ecuador or that the biochemical results are due to a genetic adaptation of B. abortus biovar 1.
Molecular tests indicated that all strains described in this study were field strains and not vaccine-type strains; as for B. abortus biotype 1 field strains, in spite of being genetically similar to vaccine strains, the former do not grow in thionin (2 μg/ml) in a culture medium.
The presence of B. abortus biotype 4 as previously reported in humans by Ron-Román et al. (9), was confirmed in this study. The biochemical characteristics of B. abortus biotype 4 differ from B. abortus biotype 1 and 2 because the former is agglutinated by anti-A instead of anti-M sera. In the same way, the allelic configuration allowed differentiating between biotypes 1 and 4 in HOOF-Prints technics.
The type strains of all classical Brucella species and biovars were surveyed to assess the discriminating power of microsatellite fingerprint technique. This technique was used to assess the level of divergence amongst and within populations of naturally infected cattle and wildlife (19, 20, 28, 29).
In this survey, both B. abortus biotype 1 and 4 were reported as described by Ron-Román et al. (9, 12) in humans from northern Ecuador. The presence of the two biotypes (1 and 4) in animals in Santo Domingo province shows that due to intensive cattle movement, the presence of several biotypes is possible. Finally, the study findings suggest that microbiological isolation of Brucella spp. is more successful from milk samples (44%) than from lymph nodes in slaughter cattle (16%).
In conclusion, the strain diversity of B. abortus was assessed in a region with intensive cattle movement and B. abortus biotypes 1 and 4 were found; although, some isolations of B. abortus biotype 1 presented phenotypic variability according to biochemical tests. These findings were correlated with results found in humans in northern Ecuador. Further research is needed to study intra-species variability and to investigate the possibility of other biotypes and Brucella species present in the tropical regions of Ecuador.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This survey was carried out with the financial support of the Universidad Tecnológica Equinoccial (UTE) with the V.SDO.AGR.27 project. The technical support in the development of laboratory tests from Paulina Fernandez staff member at CIZ staff is greatly appreciated.
References
1. Acha P, Szyfres B. “Brucellosis” in Zoonoses and Communicable Disease Common to Man and Animals. Third ed. (Vol. 1). Scientific and Technical Publication No. 580. Washington, DC: Pan America Health Organization (2001). p. 40–67.
3. Scholz HC, Nöckler K, Göllner C, Bahn P, Vergnaud G, Tomaso H, et al. Brucella inopinata sp. nov., isolated from a breast implant infection. Int J Syst Evol Microbiol (2010) 60:801–8. doi:10.1099/ijs.0.011148-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. England T, Kelly L, Jones R, MacMillan A, Wooldrige M. A simulation model of brucellosis spread in British cattle under several testing regimes. Prev Vet Med (2004) 63:63–73. doi:10.1016/j.prevetmed.2004.01.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Yamamoto T, Toshiyuki T, Nishiguchi A, Kobayashi S. Evaluation of surveillance strategies for bovine brucellosis in Japan using a simulation model. Prev Vet Med (2008) 86:57–74. doi:10.1016/j.prevetmed.2008.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Agrocalidad. Programa Nacional de Control de la Brucelosis Bovina. (2009). Available from: http://www.agrocalidad.gob.ec/agrocalidad/images/pdfs/sanidadanimal/programa_nacional_brucelosis_bovina.pdf
7. Ron L, Benítez W, Speybroeck N, Ron J, Saegerman C, Berkvens D, et al. Spatio-temporal clusters of incident human brucellosis cases in Ecuador. Spat Spatiotemporal Epidemiol (2013) 5:1–10. doi:10.1016/j.sste.2013.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Lucero NE, Ayala SM, Escobar GI, Jacob NR. Brucella isolated in humans and animals in Latin America from 1968 to 2006. Epidemiol Infect (2008) 136:496–503. doi:10.1017/S0950268807008795
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Ron-Román J, Ron-Garrido L, Abatih E, Celi-Erazo M, Vizcaíno-Ordóñez L, Calva-Pacheco J, et al. Human brucellosis in Northwest Ecuador: typifying Brucella spp., seroprevalence, and associated risk factors. Vector Borne Zoonotic Dis. (2014) 14(2):1–10. doi:10.1089/vbz.2012.1191
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Tique V, Daza E, Álvarez J, Mattar S. Seroprevalencia de Brucella abortus y ocurrencia de Brucella melitensis en caprinos y ovinos de Cesar y Sucre. Revista UDCA Actual de Divulgación Científica (2010) 13(2):133–9.
11. Higgins J, Stuber T, Quance C, Edwards W, Tiller R, Linfield T, et al. Molecular epidemiology of Brucella abortus isolates from cattle, elk and bison in the United States, 1998 to 2011. Appl Environ Microbiol (2012) 78:3674–84. doi:10.1128/AEM.00045-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Ron-Román J, Saegerman C, Minda E, Benítez W, Brandt J, Douce R. Case report; first report of orchitis in man caused by Brucella abortus biovar 1 in Ecuador. Am J Trop Med Hyg. (2012) 87:534–8. doi:10.4269/ajtmh.2012.11-0341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Lindholm A, Hewitt E, Torres P, Lasso M, Echeverria C, Shaw J, et al. Epidemiologic aspects of a foot-and-mouth disease epidemic in cattle in Ecuador. Int J Appl Res Vet Med (2007) 5(1):17–24.
14. Godfroid J, Boelaert F. Prescriptions Pour le Diagnostic Serologique de la Brucellose. Belgium: CODA-CERVA (Ed.) (1995). 47 p.
15. Alton G, Jones L, Angus R, Verger J. Techniques for the Brucellosis Laboratory. Paris: INRA (1988).
17. Bricker B, Halling S. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol (1994) 32:2660–6.
18. Bricker B, Halling S. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J Clin Microbiol (1995) 33:1640–2.
19. Bricker B, Ewalt D, Halling S. Brucella ‘HOOF-Prints’: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol (2003) 3:15. doi:10.1186/1471-2180-3-15
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Bricker B, Ewalt D. Evaluation of the HOOF-print assay for typing Brucella abortus strains isolated from cattle in the United States: results with four performance criteria. BMC Microbiol (2005) 5:37. doi:10.1186/1471-2180-5-37
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Corbel MJ, Brinley Morgan WJ. Proposal for minimal standards for description of new species and biotypes of the genus Brucella. Int J Syst Bacteriol (1975) 25(1):83–9. doi:10.1099/00207713-25-1-83
22. Mayfield J, Bantle J, Ewalt D, Meador V, Tabatabai L. Detection of Brucella cells and cell components. In: Nielsen K, Duncan R editors. Animal Brucellosis. Florida: CRC Press (1992). p. 97–101.
23. Rodríguez Torres A, Abad R, Orduña A. Especies y biovars del género Brucella. Etiología de la brucelosis humana en España. Enfermedades Infecciosas y Microbiología Clínica (1992) 10:43–8.
24. Saegerman C, Berkvens D, Godfroid J, Walravens K. Bovine brucellosis. In: Lefévre P, Blancou J, Chermette R, Uilenberg G editors. Infectious and Parasitic Diseases of Livestock. London: Lavoisier (2011). p. 991–1021.
25. Godfroid J, Nielsen K, Saegerman C. Diagnosis of brucellosis in livestock and wildlife. Croat Med J (2010) 51:296–305. doi:10.3325/cmj.2010.51.296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Godoy M, Orozco L. Identificación de micobacterias no tuberculosas: comparación de métodos bioquímicos y moleculares. Revista de la Sociedad Venezolana de Microbiología (2011) 28:96–104.
27. Minharro S, Silva JP, Dorneles EMS, Pauletti RB, Neubauer H. Biotyping and genotyping (MLVA16) of Brucella abortus isolated from cattle in Brazil, 1977 to 2008. PLoS One (2013) 8(12):e81152. doi:10.1371/journal.pone.0081152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Wang Y, Chen C, Cui B, Liu J. [Comparative study on identity of B. ovis 019 strain by traditional methods and HOOF-prints technique]. Wei Sheng Wu Xue Bao (2007) 47(2):240–3.
29. Kang S-I, Her M, Heo EJ, Nam HM, Jung SC, Cho D. Molecular typing for epidemiological evaluation of Brucella abortus and Brucella canis isolated in Korea. J Microbiol Methods (2009) 78(2):144–9. doi:10.1016/j.mimet.2009.05.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Bovine brucellosis, Brucella abortus, Ecuador, Brucella abortus biotype 1, Brucella abortus biotype 4, VNTR
Citation: Rodríguez-Hidalgo RI, Contreras-Zamora J, Benitez Ortiz W, Guerrero-Viracocha K, Salcan-Guaman H, Minda E and Ron Garrido L (2015) Circulating strains of Brucella abortus in cattle in Santo Domingo de los Tsáchilas Province – Ecuador. Front. Public Health 3:45. doi: 10.3389/fpubh.2015.00045
Received: 30 June 2014; Accepted: 18 February 2015;
Published online: 10 March 2015.
Edited by:
Juan-Carlos Navarro, Universidad Central de Venezuela, VenezuelaReviewed by:
Elsa Leclerc Duarte, Universidade de Évora, PortugalAraceli Contreras-Rodriguez, Escuela Nacional de Ciencias Biológicas Instituto Politécnico Nacional, Mexico
Copyright: © 2015 Rodríguez-Hidalgo, Contreras-Zamora, Benitez Ortiz, Guerrero-Viracocha, Salcan-Guaman, Minda and Ron Garrido. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richar Ivan Rodríguez-Hidalgo, Central University of Ecuador, Faculty of Veterinary Medicine, International Centre for Zoonoses, Av. America s/n., 170517 Quito, Ecuador e-mail:cnJvZHJpZ3VlekB1Y2UuZWR1LmVj
 Richar Ivan Rodríguez-Hidalgo
Richar Ivan Rodríguez-Hidalgo Javier Contreras-Zamora
Javier Contreras-Zamora Washington Benitez Ortiz1,2
Washington Benitez Ortiz1,2 Elizabeth Minda
Elizabeth Minda Lenin Ron Garrido
Lenin Ron Garrido