- Department of Biomedical Engineering, Duke University, Durham, NC, United States
Organoids and microphysiological systems represent two current approaches to reproduce organ function in vitro. These systems can potentially provide unbiased assays of function which are needed to understand the mechanism of action of environmental toxins. Culture models that replicate organ function and interactions among cell types and tissues move beyond existing screens that target individual pathways and provide a means to assay context-dependent function. The current state of organoid cultures and microphysiological systems is reviewed and applications discussed. While few studies have examined environmental pollutants, studies with drugs demonstrate the power of these systems to assess toxicity as well as mechanism of action. Strengths and limitations of organoids and microphysiological systems are reviewed and challenges are identified to produce suitable high capacity functional assays.
Introduction
Animal toxicological studies provide important information about the effects of a range of drugs, toxins and pollutants. Animal models of disease often replicate some key symptoms but do not reproduce the pathology and thus do not accurately predict the response in humans. While scaling relationships between species and physiologically-based pharmacokinetic models provide important information to predict safety and efficacy in humans (1), the expression levels and activities of key liver enzymes (2) and renal transporters (3) differ between humans and rodents, which are used in many drug distribution studies. In many cases, a transgenic mouse may be the only pre-clinical model. However, a transgenic mouse model often does not match the severity or phenotype or share the same genetic basis of the related human disease. Further, rodent physiology is drastically different from that in humans and other large animals. For example, humans and mice exhibit different transcriptional responses to inflammatory diseases and variations among mouse strains are significant and unrelated (4). Animal models are of limited use when studying the response to environmental toxins in individuals who may be more susceptible than the general population, such as in the developing fetus, children, the elderly and those with diseases.
Environmental pollutants act by modulating or interfering with biochemical pathways or modifying DNA. These changes then activate the immune system, induce cancer, cause neurotoxicity, or produce birth defects or developmental disorders. The number of environmental pollutants is quite extensive and growing, while only a few have been adequately studied. To cope with the large number of unevaluated chemicals in the environment, US and European agencies have initiated a concerted effort by to develop better tools to screen compounds for potentially harmful biological effects. While carefully planned high throughput screening can produce important insights about the model of action of pollutants (5), the lack of success of targeted screens to identify new drug candidates suggest that phenotypic approaches are needed to develop a complete understanding of the mechanism by which pollutants induce pathology (6).
A number of in vitro two-dimensional cell culture systems have been used to study toxicological responses and three-dimensional systems are used for specialized applications including spheroids, hanging drop (7), and Transwell systems (8). In general, most of these simpler systems have limited utility for drug discovery and toxicology due to several limitations (9). The culture media is often not optimized to promote differentiation. Cells do not differentiate well due to the mechanical rigidity of tissue culture polystyrene and the absence of mechanical forces (9). In tissues, multiple cell types are present and coculture conditions are needed to promote optimal function of cells (7, 9).
Several features are critical to developing useful cell culture models to assess disease mechanisms of drugs as well as environmental pollutants (6). While cell lines have the advantage of reproducibility and ease of access, cell lines are transformed, and cell signaling pathways and function differ from those in the native cells. Thus, primary cells in early passage should be used and their extent of differentiation should be validated. Further, functional assays that measure one or more important physiological variables should be used in addition to gene or protein expression (6). The signaling pathways and responses activated in response to drugs, toxins, or pollutants are often very dependent on the local environment, strongly affecting the manner in which a drug or environmental chemical affects the response (10).
To address these limitations, recent advances in cell culture technology, stem cell biology, biomaterials, microfluidics and biosensors have been applied to the development of human organoids (11) and human microphysiological systems (or organs-on-a-chip) (12). While still in the early stages of development these technologies offer the potential to provide better in vitro models to perform toxicological studies by more closely modeling the in vivo environment. Advances in methods to produce differentiated cells and tissues from human pluripotent stem (hPS) cells or human induced pluripotent stem (iPS) cells, as well as transdifferentiation from one cell type to another offer new opportunities to develop in vitro disease models to study mechanisms and develop new therapies.
In this paper, we review and compare human organoid and microphysiological systems, summarize their application in toxicity studies and discuss advances needed to be used in a predictive manner for toxicological studies of environmental pollutants.
Organoid Systems
Organoids are in vitro tissue models that consist of “a collection of organ-specific cell types that develops from stem cells or organ progenitors and self-organizes through cell sorting and spatially restricted lineage commitment in a manner similar to in vivo” (13). Organoids have been produced using adult, embryonic and induced pluripotent stem cells. The stem or progenitor cells undergo division, often in a biological hydrogel matrix, such as Matrigel, and through a process of genetically-encoded self-organization (14), cells with different adhesion receptors, associate with each other and produce a three-dimensional structure often representing the functional unit of a tissue (Figure 1). Organoids can be produced by directed or undirected differentiation (15). In directed differentiation, hPS induced to a specific lineage (endoderm, mesoderm, or ectoderm) are then driven to a specific tissue phenotype using small molecular regulators of transcription factors in tissue-specific media, patterning, three-dimensional culture in hydrogels or suspension. For brain organoids, directed differentiation produced specific brain regions (15). In undirected differentiation, the hPS cells are incubated with culture media containing tissue-specific growth factors and hormones, but without small molecules that activate specific lineage pathways. The latter approach works best with tissues that are relatively uniform containing one or two different cell types; otherwise more complex tissue, like brain (15), exhibit reduced specialization and reduced organization and require more precise differentiation. When properly differentiated, the organoid reproduces the structure of the tissue or a functional unit of the tissue, with tissue-specific molecules being produced, and some limited organ function can be produced. Often organoids can be maintained in culture for months when cultured with activators of specific pathways (16).
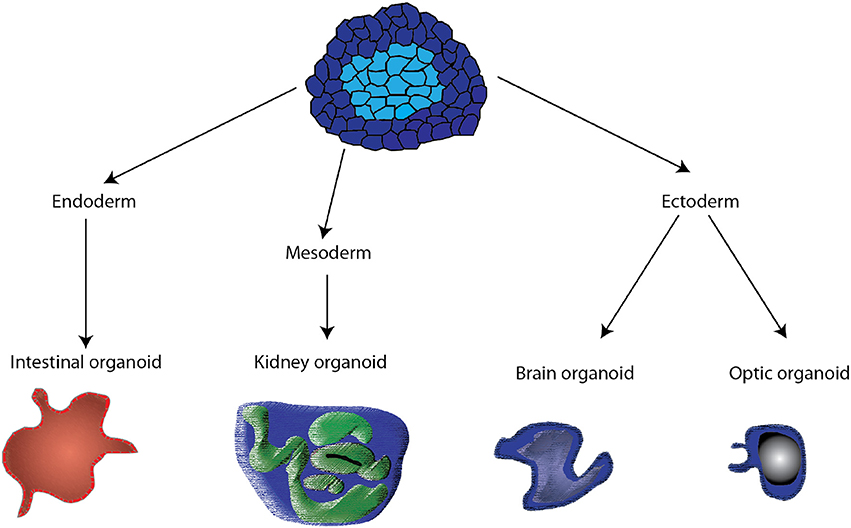
Figure 1. Schematic of the different pathways by which organoids are derived from pluripotent stem cells. Control of media and the addition of small molecules activing specific transcription factors drive the stem cells to endoderm, mesoderm, and ectoderm. Subsequent treatments are then used to produce specific organoids.
Examples of Organoid Systems
Murine organoids have been obtained for a large number of tissues (13). Human organoid systems have been developed for the fallopian tubes (16), small intestine (17), retina (18), cornea (19), brain (15), liver (20, 21), pancreas (22), prostate (23), salivary glands (24), lung (8), kidney (25), and endometrium (26). Using adult stem cells, organ progenitor cells or iPS cells, organoids can be developed that represent the diversity of a population to examine various mutations that increase susceptibility to disease. Human iPS cells can be differentiated into specific tissues to study disease development (13), cancer (27), infections of brain and gut-microbiome organoids (28), and generate models of cystic fibrosis (29). CRISPR technology can be used to test models of cancer development or test the effectiveness of proposed corrections of genetic diseases (30).
Current Limitations of Organoid Systems
Several challenges exist in using organoid cultures to assess environmental toxins. The organoids tend to form clumps or spherical structures with internal cavities, often altering the anatomic structure in vivo. This limits the ability to make functional measurements when the native tissue structure involves perfusion and/or secretion, a challenging issue for epithelial transport in organoids derived from the intestine and kidney. One approach to address this limitation is to culture organoids in a two-dimensional format on Transwell membranes as has been done with gut enteroids(17) or lung organoids (8), or to differentiate the organoids in microfluidic channels, replicating the normal tissue organization (31). The addition of endothelium and mesenchymal stem cells within a developing organoid (32) or to small numbers of established organoids (33) has improved function in vitro and led to rapid vascularization and function in vivo (32, 33).
Many organoids consist of one or two cell types and don't reflect the complete organization of the tissue. Due to the limited number of cell types arising in the organoids, resident immune cells are often missing (11), limiting studies of inflammation. Although organoids follow a developmental pathway, their size and organization tend to be variable, affecting functional measures.
Several systematic approaches have identified fabrication and culture conditions to result in more uniform structures and reduce the overall heterogeneity of the organoid population (11), while sorting gut epithelial spheroids by size, produced more uniform intestinal organoids (34). While the organoids display some mature tissue functions, they represent an early developmental stage. For instance, gut enteroids do not differentiate into the intestinal segments, do not display peristalsis and have a limited ability to form villus structures (17). As such, organoids may be a suitable model for studies of environmental toxins during early development.
Microphysiological Systems
Advances in tissue engineering, microfabrication, microfluidics, and biosensors have enabled the in vitro fabrication of a tissue or organ by organizing primary human cells or differentiated cells from hPS cells in a spatial arrangement similar to that observed in vivo. The microphysiological system is a smaller scale recapitulation of the functional unit of an organ or tissue. The ideal human microphysiological system has the following characteristics (35): (1) multicellular architecture similar to the native tissue; (2) functional representation of normal human biology; (3) reproducible and viable operation under physiological conditions for multiple weeks; (4) key features of normal and disease phenotypes; (5) range of functional behavior that represents the population diversity; (6) capable of integrating with other organ systems; and (7) amenable to high content screening for repeated dose efficacy testing, and for toxicology [absorption, distribution, metabolism, excretion and toxicity (ADMET)] and safety screening. Some, but not all of these features have been met. Human microphysiological systems have been developed for the heart (36, 37), liver (38, 39), kidney glomerulus (40), kidney proximal tubule (41), female reproductive tract (42), placental barrier (43, 44), heart (45–47), skeletal muscle (48), blood vessel (49), white adipose tissue (50), microvasculature (51), and blood-brain barrier (36, 37).
Design Considerations in Microphysiological Systems
The functional unit of organs such as lung, liver, kidney blood vessel, placental barrier, and gastrointestinal tract consist of transport across an epithelial layer separating a cavity from capillaries. The basic design that mimics this barrier region consists of a thin, porous deformable membrane separating two channels (37, 52–54). Epithelial cells form a monolayer on one side and an endothelial monolayer on the other side, representing the plasma compartment (Figure 2). For the blood-brain barrier, a confluent layer of endothelial cells are on the luminal side in contact with blood or blood mimicking media, with astrocytes on the side in contact with brain tissue (37). The fluid composition and flow rates of each channel can be controlled separately. The walls of the channels are often made from polydimethylsiloxane (PDMS), a biocompatible, transparent, and deformable polymer used in the fabrication of microfluidic devices. Some groups have used a vacuum system to expand the channels and stretch the membrane to mimic stretch that occurs in the lung during breathing (55) or peristalsis in the intestine (54). Often, other cell types are interspersed with the epithelial layer reflecting the composition of the tissue. For example, the lung airway consists of a mixture of epithelial cells that are functionally differentiated in terms of presence of cilia, secretion of mucus (goblet cells) and glycosaminoglycans (club cells) and neuroendocrine cells, overlaying stem cells (basal cells) (56). Differentiation of the epithelial layer is promoted by using the appropriate fluid, such as air for airway epithelia (56).
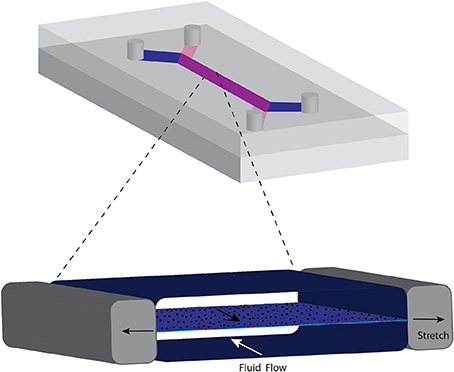
Figure 2. Common format of microphysiological chip for transport across barrier membranes such as those occurring in the lung and intestines. A porous elastic membrane separates the two flow streams and cells can be grown on either side. Negative pressure created in the side chambers stretches the membrane, mimicking mechanical deformation of the membrane. The cycle of stretching is varied to represent breathing or peristaltic motion.
The immune function in the liver has been addressed by incorporating cryopreserved human Kupfer cells (38) or the U937 human monocytic cell line (39). Exposure of the liver microphysiological system after 7 days of operation with the bacterial coat lipopolysaccharide (LPS) resulted in substantial production of TNFα (38, 39), as well as the cytokines G-CSF, RANTES, IL-8, and IL-6, which are known to be associated with LPS-induced inflammation (38). Such inflammatory responses increased the cytotoxicity of drugs (39) which may help to explain acute drug-induced liver toxicity (57) and may increase the toxicity of environmental pollutants.
Mechanical forces play a critical role in promoting differentiation of cells in microphysiological systems. In a model of the lung alveolus using the design in Figure 2, mechanical stretching of the membrane to which the endothelial cells and epithelial cells enhanced the inflammatory response and generation of reactive oxygen species (55). Following treatment of the alveolar chip model with IL-2 to model pulmonary inflammation, mechanical strain enhanced the increase in permeability due to formation of gaps between the cells and fluid accumulation on the epithelial side, leading reduced oxygen saturation (58). In a gut model that stretches the epithelial layer using an arrangement similar to that shown in Figure 2 with just gut epithelial cells, applying 10% cell strain at 0.15 Hz caused the Caco-2 human epithelial cell line to form villi with columnar epithelium and a brush border and prevented a noncommensal bacteria from disrupting the epithelial barrier (54). Microarray analysis showed that these differentiated cells cultured with gut microbes were closer to human ileum than were cells cultured on a Transwell insert (54). Likewise, exposure of mouse ovarian tissue to flow in microfluidic chambers enhanced follicle maturation of ovarian follicles and secretion of follicular hormones (42).
Functional measures of microphysiological systems demonstrate that they can reproduce the behaviors of native tissue. Transendothelial electrical resistance values approach those occurring in vivo for the blood-brain barrier (36, 37) or gut (54). Many drugs are metabolized by cytochrome P450 (59) and measurement of its activity provides a suitable functional assessment of liver microphysiological systems. By controlling oxygen tension, cytochrome P450 activity is greater in the low oxygen concentration region of the acinus, as occurs in vivo (60).
Contractile force represents a functional measure of skeletal (48) and cardiac muscle (47) systems and oxygen consumption has been characterized in a skeletal muscle microphysiological system (61). Skeletal, smooth and cardiac muscle attached to deformable thin films have provided data on contractile stresses (62, 63) and contraction and dilation in the presence of agonists provides a measure of vasoactivity (49). Conduction velocity and action potential duration provide important information about the electrophysiological properties of cardiac muscle microphysiological systems that can be compared with in vivo behavior (47).
Organoids have been integrated into microfluidic devices to take advantage of the tissue organization that occurs with organoids and the polarity induced by membrane designs. Intestinal organoids from human duodenum biopsies were expanded and then integrated into a microfluidic channel as in Figure 2. Then the organoids were differentiated under mechanical stimulation to form villous structures that contained all of the intestinal cell types, exhibited barrier function and exhibited gene expression profiles closer to human intestine than microphysiological systems using Caco-2 cell lines (31). The advantage of this approach is that the cells in the organoid are positioned in the proper arrangement and the microfluidic system provides flow and mechanical stimulation that can promote further differentiation of the cells.
Multiple microphysiological systems have been interlinked to replicate interactions among various organs (Figure 3). System integration has ranged from two to ten organs. While a number of scaling approaches have been considered (64–66), one organ serves as a reference and tissue mass (or volume assuming a tissue density slightly greater than the density of water) is scaled based on body proportions. Flow rates to organs are based on the distribution of flow throughout the body and the tissue residence time (67, 68). Many systems used a closed format that resembles the body's blood circulation with a central mixing region that enables gas exchange (50, 68). Alternatively, an open format in which the media in each organ is exposed to sterile air ensures efficient gas exchange for each organ (67). By coupling gut and liver models, first pass metabolism was demonstrated (69, 70), and the contribution of each organ and organ-organ interactions assessed (69). Adding the kidney enables assessment of absorption, metabolism and excretion. The resulting clearance values for a number of drugs are similar to those reported in vivo (65). Linking other organs to the gut-liver-kidney modules enabled assessment of vitamin D3 transport and metabolism, blood-brain transport, skeletal muscle toxicity, and biodistribution (65). Ten (67) and thirteen (71) organ systems have been developed indicating that many of the features of the entire human body can be replicated in microfluidic systems.
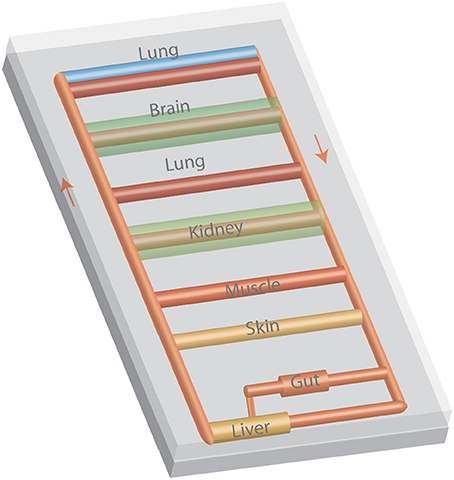
Figure 3. A multiple organ-on-a-chip with eight organ models. The tubing represents blood vessels and arrows represent the flow of culture media. Media can be moved with a variety of pneumatic or electromagnetic pumps or with a modification of design with a rocker to induce flow by gravity.
Challenges in Developing Microphysiological Systems
Several factors influence the design and development of individual organ systems and integrated multi-organ microphysiological systems. While pumps are often used to link multiple organs in microfluidic devices, tubing volumes must be limited in order to maintain the correct relationship among the fluid and tissue volumes. Pneumatic (67, 69) or electromagnetic (42) pumps built into the multiorgan units can reduce the need for additional tubing. Alternatively, rocking platforms that utilize gravity to generate flow (72) and robotic fluid handlers (73).
Hydrophobic molecules are more likely to adsorb to PDMS, with a dramatic change in adsorption for an octanol/water partition coefficient greater than 300 (74). Drug loss of molecules with high partition coefficients can be minimized if the contact time is minimized (75), although this may not be practical given other constraints on flow and system dimensions. Drug and pollutant binding to PDMS can be reduced with a variety of coatings (74, 76), although binding should be assessed for specific molecules under study. Other materials such as polysulfones (67), polycarbonates and thermoplastics (77) exhibit low drug binding and can be micromachined to form chambers for microphysiological systems.
To increase throughput, multiple replicate, functional tubular (78) or microvascular (51) systems have been developed to increase throughput for screening, but most systems are below a 96 well format.
The Response of Microphysiological Systems and Organoids to Xenobiotics and Pollutants
Most studies with microphysiological systems have focused on examining drugs for toxicity and, in some cases, efficacy. Human primary of hPS cells are necessary to assess toxicity, given the shortcomings of animal models. Various drugs that affect cardiac contractility strength and duration exerted similar effects on cardiac microphysiological systems formed using iPS-derived cardiomyocytes as determined by beat frequency (46, 79) or contractile stress (80); EC50 values were within a factor of two of values obtained with cardiac muscle. A number of drugs with known liver toxicity have been examined in several liver microphysiological systems. In a four-organ system, the gut and liver metabolized the toxic terfenadine to its nontoxic and vasoactive form fexofenadine which was unable to cross the blood-brain barrier (70). A four organ system, consisting of human cardiac, liver, skeletal muscle and neuronal cultures using a common defined medium for 2 weeks, demonstrated that differential responsiveness to a 48 h exposure to 5 μM doxorubicin (68). Consistent with reports for humans, the viability of cardiac and liver cells was reduced whereas for skeletal muscle contraction was affected, but not viability (68). Skeletal muscle systems exhibited toxicity to cerivastatin below 1 nM, which was removed the market, whereas doses above pharmacological levels were needed before toxicity to lovastatin was observed (81).
Only a few studies have examined the effect of environmental pollutants. An airway model was used to model smoking by linking the microphysiological system to a device that mimicked inhaling and exhaling cigarette smoke (52). Cigarette smoking reduces mucus clearance from the lung, due to altered function of cilia. In this model, smoking did not affect the average cilia beat frequency, but altered the distribution of beat frequencies which may alter mucus flow. Further, epithelial from patients with chronic obstructive pulmonary disease exhibited large increases in IL-8 secretion after exposure to cigarette smoke in this system, whereas epithelium from healthy nonsmoking donors did not exhibit IL-8 secretion. Interestingly, the device detected differences in the response of the airway epithelium between conventional cigarettes and e-cigarettes (52).
Brain organoids were cultured on microfluidic channels to enable perfusion and then the response to nicotine was tested (82). Nicotine exposure at levels occurring with heavy smokers inhibited differentiation and organization of the organoids which the authors concluded was similar to impaired neurogenesis observed in the fetal brain of mothers who were heavy smokers (82). Recently, dose-response toxicity studies with organoids formed with hPSCs differentiated to cardiac myocytes or primary liver cells indicated that toxicity to lead, mercury, glyphosate, and mercury, but at higher doses than observed with cultured cells or animal toxicity studies (83). This difference in toxicity may reflect a combination of the assays used, the extent of differentiation of the organoids, and the short duration of exposure (24-48 h).
Aristolochic acids (AA) are widely used in herbal teas but are implicated in kidney disease and kidney and liver cancers. Combining liver and kidney proximal tubule microphysiological systems showed that the liver modified AA-1 to produce a metabolite that was more toxic to kidney epithelial cells (84). Enzymes that modify AA-1 were identified and organic ion transporters were shown to be involved in uptake by kidney proximal tubules (84). The study showed the usefulness of microphysiological systems to identify the manner in which a naturally occurring toxin is metabolized and transported into target cells.
Perspective
Organoids and microphysiological systems offer the potential to identify and characterize the biological impact of environmental pollutants. These systems move beyond existing screens that target individual pathways and provide a means to assay context-dependent function.
Critical issues include developing a common media, maintaining the relevant size relationship among tissues, producing the differentiated state of cells, replicating the in vivo organization of tissues and organs, developing more robust organoid culture systems, increasing throughput, and testing population variability. Isolation of primary cells can be challenging and limiting when doing scale-up for higher throughput screening (85). While hPS cells provide a ready source of cells, the differentiation protocols need further improvement to ensure a well-differentiated state that can maintain function following subculture for a wide range of tissues. Linking organoids and micromachined devices represents an opportunity to produce an organ system that more closely resembles the in vivo organ. To effectively screen large numbers of environmental pollutants, protocols or systems are needed to enable higher throughput testing. This involves identifying critical features that measure key functions of the tissue and the development of low cost sensor systems, establishing uniform methods to access and perfuse different organ systems, and integration with existing lab automation systems (85).
While multi-organ systems can replicate the entire range of absorption, disposition, metabolism and excretion (77), current microphysiological systems are complex, limiting scaling for high throughput assays. There is not likely to be a universal system useful for all applications. Rather, for the particular topic of study, the key function output need to be identified and the system designed accordingly.
Promising solutions developed to create individual and multi-organ systems with human cells suggest that the field is likely to develop rapidly over the next decade, making available new approaches to assess environmental pollutants.
Author Contributions
GT conceived of this topic and wrote the review.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants HL138252 from NHLBI, NIH grant R21AR072283, from NIAMS, and NIH grant UG3TR002142 from NCATS and NIAMS.
References
1. Kuepfer L, Niederalt C, Wendl T, Schlender JF, Willmann S, Lippert J, et al. Applied concepts in PBPK modeling: how to build a PBPK/PD model. CPT Pharmaco Syst Pharmacol. (2016) 5:516–31. doi: 10.1002/psp4.12134
2. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. (2013) 138:103–41. doi: 10.1016/j.pharmthera.2012.12.007
3. Morrissey KM, Stocker SL, Wittwer MB, Xu L, Giacomini KM. Renal transporters in drug development. Ann Rev Pharmacol Toxicol. (2013) 53:503–29. doi: 10.1146/annurev-pharmtox-011112-140317
4. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci. (2013) 110:3507–12. doi: 10.1073/pnas.1222878110
5. Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, et al. Update on EPA's toxcast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol. (2012) 25:1287–302. doi: 10.1021/tx3000939
6. Vincent F, Loria P, Pregel M, Stanton R, Kitching L, Nocka K, et al. Developing predictive assays: The phenotypic screening “rule of 3.” Sci Trans Med (2015) 7:293ps15. doi: 10.1126/scitranslmed.aab1201
7. Fang Y, Eglen RM. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. (2017) 22:456–72. doi: 10.1177/1087057117696795
8. Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BLM. Lung organoids: current uses and future promise. Development (2017) 144:986–97. doi: 10.1242/dev.140103
9. Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D, et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. (2016) 15:751–69. doi: 10.1038/nrd.2016.175
10. Jones DS, Jenney AP, Joughin BA, Sorger PK, Lauffenburger DA. Inflammatory but not mitogenic contexts prime synovial fibroblasts for compensatory signaling responses to p38 inhibition. Sci Signal. (2018) 11:eaal1601. doi: 10.1126/scisignal.aal1601
11. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell (2016) 18:25–38. doi: 10.1016/j.stem.2015.12.005
12. Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. (2014) 32:760. doi: 10.1038/nbt.2989
13. Lancaster MA, Knoblich JA. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science (2014) 345:1247125. doi: 10.1126/science.1247125
14. Turner DA, Baillie-Johnson P, Martinez Arias A. Organoids and the genetically encoded self-assembly of embryonic stem cells. BioEssays (2016) 38:181–91. doi: 10.1002/bies.201500111
15. Paşca SP. The rise of three-dimensional human brain cultures. Nature (2018) 553:437–45. doi: 10.1038/nature25032
16. Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. (2015) 6:8989. doi: 10.1038/ncomms9989
17. In JG, Foulke-Abel J, Estes MK, Zachos NC, Kovbasnjuk O, Donowitz M. Human mini-guts: new insights into intestinal physiology and host–pathogen interactions. Nat Rev Gastroenterol Amp Hepatol. (2016) 13:633–42. doi: 10.1038/nrgastro.2016.142
18. Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. (2014) 5:4047. doi: 10.1038/ncomms5047
19. Foster JW, Wahlin K, Adams SM, Birk DE, Zack DJ, Chakravarti S. Cornea organoids from human induced pluripotent stem cells. Sci Rep. (2017) 7:41286. doi: 10.1038/srep41286
20. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Ellis E, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell (2015) 160:299–312. doi: 10.1016/j.cell.2014.11.050
21. Leite SB, Roosens T, El Taghdouini A, Mannaerts I, Smout AJ, Najimi M, et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials (2016) 78:1–10. doi: 10.1016/j.biomaterials.2015.11.026
22. Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell (2015) 160:324–38. doi: 10.1016/j.cell.2014.12.021
23. Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell (2014) 159:163–75. doi: 10.1016/j.cell.2014.08.017
24. Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, Ben Giepmans NG, et al. Long-term in vitro expansion of salivary gland stem cells driven by wnt signals. Stem Cell Rep. (2016) 6:150–62. doi: 10.1016/j.stemcr.2015.11.009
25. Taguchi A, Nishinakamura R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell (2017) 21:730–46.e6. doi: 10.1016/j.stem.2017.10.011
26. Turco MY, Gardner L, Hughes J, Cindrova-Davies T, Gomez MJ, Farrell L, et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. (2017) 19:568. doi: 10.1038/ncb3516
27. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (2018) 359:920–6. doi: 10.1126/science.aao2774
28. Dutta D, He I, Clever H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. (2017) 23:393–410. doi: 10.1016/j.molmed.2017.02.007
29. Dekkers JF, Berker G, Kruisselbrin E, Vonk A, de Jonge HR, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Trans Med. (2016) 8:344ra84-344ra84. doi: 10.1126/scitranslmed.aad8278
30. Driehuis E, Clevers H. CRISPR/Cas 9 genome editing and its applications in organoids. Am J Physiol Gastroint Liver Physiol. (2017) 312:G257–65. doi: 10.1152/ajpgi.00410.2016
31. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep. (2018) 8:2871. doi: 10.1038/s41598-018-21201-7
32. Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature (2013) 499:481. doi: 10.1038/nature12271
33. Takahashi Y, Sekine K, Kin T, Takebe T, Taniguchi H. Self-condensation culture enables vascularization of tissue fragments for efficient therapeutic transplantation. Cell Rep. (2018) 23:1620–9. doi: 10.1016/j.celrep.2018.03.123
34. Arora N, Imran Alsous J, Guggenheim JW, Mak M, Munera J, Wells JM, et al. A process engineering approach to increase organoid yield. Development (2017) 144:1128–36. doi: 10.1242/dev.142919
35. Low LA, Tagle DA. Tissue chips - innovative tools for drug development and disease modeling. Lab Chip (2017) 17:3026–36. doi: 10.1039/C7LC00462A
36. Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics (2015) 9:054124. doi: 10.1063/1.4934713
37. Wang YI, Abaci HE, Shuler ML. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng. (2017) 114:184–94. doi: 10.1002/bit.26045
38. Sarkar U, Ravindra KC, Large E, Young CL, Rivera-Burgos D, Yu J, et al. Integrated assessment of diclofenac biotransformation, pharmacokinetics, and omics-based toxicity in a three-dimensional human liver-immunocompetent coculture system. Drug Metabol Disposition (2017) 45:855–66. doi: 10.1124/dmd.116.074005
39. Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Shun TY, Gough A, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med. (2016) 241:101–14. doi: 10.1177/1535370215592121
40. Musah S, Mammoto A, Ferrante TC, Jeanty SS, Hirano-Kobayashi F, Mammoto M, et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng. (2017) 1:0069. doi: 10.1038/s41551-017-0069
41. Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. (2016) 90:627–37. doi: 10.1016/j.kint.2016.06.011
42. Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun. (2017) 8:14584. doi: 10.1038/ncomms14584
43. Blundell C, Tess ER, Schanzer AS, Coutifaris RC, Su E, Parry JS, et al. A microphysiological model of the human placental barrier. Lab Chip (2016) 16:3065–73. doi: 10.1039/C6LC00259E
44. Lee JS, Romero R, Han YM, Kim HC, Kim CJ, Hong JS, et al. Placenta-on-a-chip: a novel platform to study the biology of the human placenta. J. Matern-Fetal Neonat Med. (2015) 29:1046–54. doi: 10.3109/14767058.2015.1038518
45. Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip (2013) 13:3599–608. doi: 10.1039/c3lc50350j
46. Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. (2015) 5:8883. doi: 10.1038/srep08883
47. Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials (2013) 34:5813–20. doi: 10.1016/j.biomaterials.2013.04.026
48. Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. eLife (2015) 4:e04885. doi: 10.7554/eLife.04885
49. Fernandez CE, Yen RW, Perez SM, Bedell HW, Povsic TJ, Reichert WM, et al. Human Vascular Microphysiological System for in vitro Drug Screening. Sci Rep. (2016) 6:21579. doi: 10.1038/srep21579
50. Loskill P, Sezhian T, Tharp KM, Lee-Montiel FT, Jeeawoody S, Reese WM, et al. WAT-on-a-chip: a physiologically relevant microfluidic system incorporating white adipose tissue. Lab Chip (2017) 17:1645–54. doi: 10.1039/C6LC01590E
51. Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, et al. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip (2017) 17:511–20. doi: 10.1039/C6LC01422D
52. Benam KH, Novak R, Nawroth J, Hirano-Kobayashi M, Ferrante TC, Choe Y, et al. Matched-Comparative Modeling of Normal and Diseased Human Airway Responses Using a Microengineered Breathing Lung Chip. Cell Syst. (2016) 3:456–66.e4. doi: 10.1016/j.cels.2016.10.003
53. Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Int Biol. (2013) 5:1119–29. doi: 10.1039/c3ib40049b
54. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci. (2016) 113:E7–15. doi: 10.1073/pnas.1522193112
55. Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting Organ-Level Lung Functions on a Chip. Science (2010) 328:1662–8. doi: 10.1126/science.1188302
56. Hiemstra PS, Grootaers GAM, van der Does C, Krul, AM, Kooter IM. Human lung epithelial cell cultures for analysis of inhaled toxicants: Lessons learned and future directions. Toxicol in vitro (2018) 47:137–46. doi: 10.1016/j.tiv.2017.11.005
57. Adams DH, Ramaiah SK, Uetrecht J, Jaeschke H. Mechanisms of Immune-Mediated Liver Injury. Toxicol Sci. (2010) 115:307–21. doi: 10.1093/toxsci/kfq009
58. Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, et al. A human disease model of drug toxicity–induced pulmonary edema in a lung-on-a-chip microdevice. Sci Trans Med. (2012) 4:159ra147-159ra147. doi: 10.1126/scitranslmed.3004249
59. Murry DJ. Pharmacogenetics of the cytochrome P450 enzyme system: review of current knowledge and clinical significance. J Pharm Pract. (2007) 20:206–18. doi: 10.1177/0897190007304821
60. Lee-Montiel FT, George SM, Gough A.H, Sharma A. D, Wu J, DeBiasio R, Vernetti L. A, Taylor D. L. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Experimental Biology and Medicine, (2017). 242(16): p. 1617-1632. doi: 10.1177/1535370217703978
61. Davis BNJ, Santoso J, Walker M, Koves T, Kraus W, Truskey G. Human engineered skeletal muscle platform to detect mitochondrial drug-induced toxicity. Tissue Eng Part C (2017) 23:189–99. doi: 10.1089/ten.tec.2016.0264
62. Nesmith AP, Agarwal A, McCain ML, Parker KK. Human airway musculature on a chip: an in vitro model of allergic asthmatic bronchoconstriction and bronchodilation. Lab Chip (2014) 14:3925–36. doi: 10.1039/C4LC00688G
63. Nesmith AP, Wagner MA, Pasqualini FS, O'Connor BB, Pincus MJ, August PR, et al. A human in vitro model of Duchenne muscular dystrophy muscle formation and contractility. J Cell Biol. (2016) 215:47–56. doi: 10.1083/jcb.201603111
64. Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Int Biol. (2015) 7:383–91. doi: 10.1039/C4IB00292J
65. Maass C, Stokes CL, Griffith LG, Cirit M. Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS). Int Biol. (2017) 9:290–302. doi: 10.1039/C6IB00243A
66. Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip (2013) 13:3496–511. doi: 10.1039/c3lc50243k
67. Edington CD, Chen WL, Geishecker KE, Kassis T, Soenksen LR, Bhushan BM, et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci Rep. (2018) 8:4530. doi: 10.1038/s41598-018-22749-0
68. Oleaga C, Smith AST, Srinivasan B, Jackson M, McLamb W, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. (2016) 6:20030. doi: 10.1038/srep20030
69. Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS J. (2017) 19:1499–512. doi: 10.1208/s12248-017-0122-4
70. Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, et al. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep. (2017) 7:42296. doi: 10.1038/srep42296
71. Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol Bioeng. (2016) 113:2213–27. doi: 10.1002/bit.25989
72. Esch MB, Prot JM, Wang YI, Miller P, Llamas-Vidales JR, Naughton BA, et al. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip (2015) 15:2269–77. doi: 10.1039/C5LC00237K
73. Prantil-Baun R, Novak R, Das D, Somayaji MR, Przekwas A, Ingber DE. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. Annu Rev Pharmacol Toxicol. (2018) 58:37–64. doi: 10.1146/annurev-pharmtox-010716-104748
74. Wang JD, Douville NJ, Takayama S, ElSayed M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann Biomed Eng. (2012) 40:1862–73. doi: 10.1007/s10439-012-0562-z
75. Shirure VS, George SC. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip (2017) 17:681–90. doi: 10.1039/C6LC01401A
76. van Meer BJ, de Vries HK, Firth SA, van Weerd J, Tertoolen LGJ, Karperien HBJ, et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun. (2017) 482:323–8. doi: 10.1016/j.bbrc.2016.11.062
77. Wang YI, Carmona C, Hickman JJ, Shuler ML. Multiorgan microphysiological systems for drug development: strategies, advances, and challenges. Adv Healthcare Mater. (2018) 7:1701000. doi: 10.1002/adhm.201701000
78. Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat Commun. (2017) 8:262. doi: 10.1038/s41467-017-00259-3
79. Huebsch N, Loskill P, Deveshwar N, Spencer CI, Judge LM, Mandegar MA, et al. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci Rep. (2016) 6:24726. doi: 10.1038/srep24726
80. Lind JU, Yadid M, Perkins I, O'Connor BB, Eweje F, Chantre CO, et al. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip (2017) 17:3692–703. doi: 10.1039/C7LC00740J
81. Backman JT, Filppula AM, Niemi M, Neuvonen PJ. Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol Rev. (2016) 68:168–241. doi: 10.1124/pr.115.011411
82. Wang Y, Wang L, Zhu Y, Qin J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip (2018) 18:851–60. doi: 10.1039/C7LC01084B
83. Forsythe SD, Devarasetty M, Shupe T, Bishop C, Atala A, Soker S, et al. Environmental toxin screening using human-derived 3D bioengineered liver and cardiac organoids. Front Public Health (2018) 6:103. doi: 10.3389/fpubh.2018.00103
84. Chang SY, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight (2017) 2:95978. doi: 10.1172/jci.insight.95978
Keywords: microphysiological systems, organoids, toxicity, environmental pollutants, cell culture, stem cells
Citation: Truskey GA (2018) Human Microphysiological Systems and Organoids as in Vitro Models for Toxicological Studies Front. Public Health 6:185. doi: 10.3389/fpubh.2018.00185
Received: 20 March 2018; Accepted: 11 June 2018;
Published: 10 July 2018.
Edited by:
Alexander Kinev, Creative Scientist, United StatesReviewed by:
Xiangzhu Zhu, Vanderbilt University, United StatesMichael J. Van Kanegan, ZenBio, Inc., United States
Copyright © 2018 Truskey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George A. Truskey, Z3RydXNrZXlAZHVrZS5lZHU=
 George A. Truskey
George A. Truskey