- 1Department of Craniofacial Biology, Pathology, & Radiology, Faculty of Dentistry, University of the Western Cape, Cape Town, South Africa
- 2SAMRC/CPUT/Cardiometabolic Health Research Unit, Department of Biomedical Sciences, Faculty of Health & Wellness Sciences, Cape Peninsula University of Technology, Cape Town, South Africa
- 3Department of Oral Medicine & Periodontology, Faculty of Dentistry, University of the Western Cape, Cape Town, South Africa
Introduction: Periodontitis is a significant health challenge caused by a complex interaction between bacterial infection, host immune response, and environmental factors, leading to tooth loss, bone loss, and potential associations with major systemic diseases and conditions. While the determinants of periodontitis have been extensively investigated in other populations, such studies are lacking in South Africa, which represents a high-risk population. Therefore, this study was conducted to characterize the subgingival bacterial biodiversity in the periodontal pockets of patients with periodontitis in a Western Cape population.
Materials & methods: Pooled subgingival plaque samples were collected from the deepest pocket/crevices of five periodontitis cases and five controls using sterile paper points. Illumina MiSeq paired-end sequencing and QIIME2 software were employed for sequence filtration and analysis. Several alpha and beta-diversity metrics assessed biodiversity within-sample and population structure between different microbiota datasets, respectively. Statistical significance for alpha diversity was tested using the Kruskal–Wallis H test (p < 0.05), and beta diversity differences were evaluated using PERMANOVA. Data visualization, including beta diversity plots, was conducted with the Phyloseq package in R.
Results: Beta-diversity measures revealed significant differences between periodontitis cases and controls (p-value = 0.04), whereas alpha-diversity was higher in cases, though without statistical significance (p-value ≥ 0.05). Cases group showed high relative abundance of Fusobacterium (16%), Porphyromonas (10%), and Treponema (9%), while the periodontally healthy controls were dominated by Streptococcus (20%), Fusobacterium (15%), and Veillonella (10%), with g_Streptococcus showing a significant difference (p-value = 0.008). Differential abundance analysis revealed distinct bacterial genera enriched in cases (Bulleidia, Peptoanaerobacter, Phocaeiola, W5053) and controls (Abiotrophia, Haemophilus, Lautropia, Rothia, Streptococcus). Sample-specific variations included higher levels of Porphyromonas (15%) in grade B and Fusobacterium (20%) in grade C.
Conclusion: This exploratory study highlights distinct bacterial communities associated with periodontitis in a South African population. The findings emphasize the need for larger, population-based cohorts to validate these results and lay a foundation for future research into region-specific microbial profiles and their implications for personalized treatment strategies.
1 Introduction
According to the WHO's Global Oral Health Status Report (2022), 19% of the global population aged 15 years and older is affected by severe periodontitis, with the highest prevalence in Africa (23%) (1). Periodontitis is a multifactorial disease triggered by dysbiosis in dental biofilm that promotes inflammation with both protective and destructive effects on periodontal tissues (2). This dysbiosis negatively impacts oral and general health, reducing the quality of life (3) and contributing to a global healthcare burden and social inequality (4).
Advances in culture-independent molecular techniques have revealed distinct bacterial communities in periodontal health and disease (5). Despite limitations such as short sequencing reads, limited species-level resolution for certain bacterial genera (6), lack of standardization in selecting hypervariable regions for sequencing (7), and an inability to provide functional insights (8), 16S rRNA sequencing remains the gold standard for bacterial profiling (9). These techniques highlight the diagnostic and prognostic potential of microbiome biodiversity in periodontitis, where changes in microbial composition signal early disease, and biodiversity loss is linked to disease progression (10). Restoring microbial diversity could be a key therapeutic goal in periodontitis, promoting a balanced oral microbiome essential for achieving and maintaining periodontal stability (11).
While the current classification system for periodontitis incorporates pathophysiological and host immune factors that justified the consolidation of periodontitis into a unified category (12), it does not include microbial criteria to differentiate between stages and grades. This limitation arises primarily from insufficient evidence-based data on microbiological diagnostics in periodontitis (13).
Emerging evidence suggests that the subgingival microbial composition in periodontitis patients varies significantly based on population, demographic factors, and environmental conditions (14, 15). These findings highlight the potential relevance of microbial profiling in enhancing and refining periodontal classification systems.
However, much of our understanding of periodontitis microbiota is derived from studies conducted in non-African populations (16). Recent insights highlight the need for population-specific investigations. For instance, Aggregatibacter actinomycetemcomitans, strongly linked to Grade C Molar-Incisor periodontitis, appears to behighly prevalent among Africans, compared to other ethnic groups" (17–20).
Therefore, investigating periodontitis determinants in high-risk, diverse populations, such as South Africans (SAs), is imperative for developing a successful precision dentistry health system. Recent work established the microbial profile of the SA population who smoke (21). However, no study has been conducted to establish the baseline bacterial profile of SAs with periodontitis compared to those with a healthy periodontium.
Given these considerations, the primary objective of this study was to characterize the subgingival bacterial communities within periodontal pockets of periodontitis patients from the Western Cape, SA, and to assess whether microbial composition differs across the various grades of periodontitis. This pilot study aims to lay the foundation for future, more comprehensive research by evaluating both the feasibility and potential benefits of the proposed study within a diverse population.
2 Material and methods
2.1 Study population
Ethical approval was obtained from the Biomedical Research Ethics Committee (BMREC) of the University of the Western Cape (UWC), reference number BM20/10/9. Twenty-three consecutive South African participants, aged 18 years or older and with at least 10 teeth, were recruited from the Oral Medicine and Periodontology Department at Tygerberg Oral Health Centre between August 2021 and October 2022. Participants were excluded if they had any systemic disease, were current smokers, pregnant or lactating, edentulous, had used antiseptics or anti-inflammatory drugs for more than one week, antibiotics, antimicrobials, undergone any periodontal management in the past three months. Therefore, a final sample size of 23 patients was included in this study.
All potential participants underwent Basic Periodontal Examination (BPE) to assess their periodontal status. Eligible participants then received a comprehensive periodontal examination for detailed clinical data collection. Personal and demographic information, including age, gender, and self-reported ethnicity, were also collected.
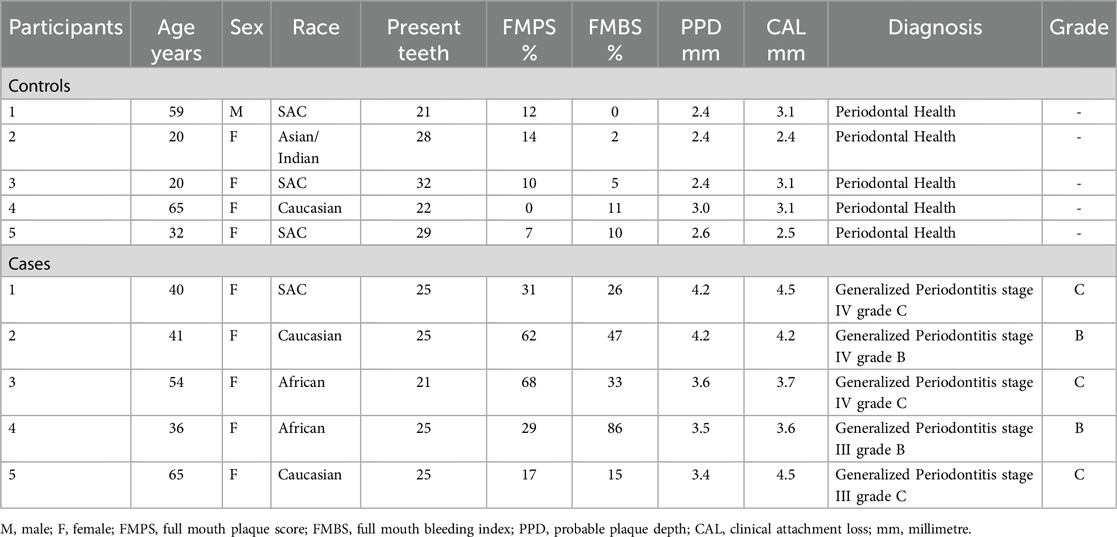
Panoramic radiographs were taken, and a calibrated periodontist with high intra-reliability (kappa > 0.81) conducted clinical examinations, documenting periodontal parameters such as full mouth plaque score (FMPS), full mouth bleeding score (FMBS), probable pocket depth (PPD), and clinical attachment level (CAL). Participants were categorized into periodontitis cases and periodontally healthy controls based on the case definitions outlined in the 2018 classification scheme of periodontal diseases (13, 22). Following this, a complete diagnosis of periodontitis cases was established using the multidimensional staging and grading systems, in which disease severity was categorized into four stages (I–IV), ranging from mild clinical attachment loss, Stage I, to severe periodontitis with extensive bone and tooth loss, Stage IV. Staging was determined based on CAL, radiographic bone loss, disease complexity, and its extent and distribution; localized or generalized. Progression was further graded as A (slow progression), B (moderate progression), or C (rapid progression). Grading was based on CAL and bone loss in relation to age, case phenotype reflected through FMPS, and the presence of grade modifiers such as diabetes mellitus (DM) and smoking (13, 22).
2.2 Subgingival sampling
After the removal of the supra-gingival biofilm, the teeth targeted for sampling were isolated with cotton rolls and dried. Sub-gingival dental biofilm samples were acquired from the meso-buccal surface of teeth with the deepest pathological pocket (or cervices in the healthy group) of each quadrant by gently introducing a sterile paper point #35 (23–30).
If a deeper pathological pocket (or cervices in the healthy group) was identified on another surface, it was used for sample collection. Paper points were immediately transferred from the participant's mouth and pooled into a sterile 2 ml Eppendorf tube containing 500 μl of phosphate-buffered saline (PBS), then immediately placed on ice. As per the recommendation of the best storage temperature for microbiome material, samples were frozen at −80°C at IMBM until DNA extraction was conducted.
2.3 DNA extraction and sequencing
DNA extraction from the dental biofilm samples was performed using the PureLink™ Microbiome DNA Purification Kit (#A29790; Thermo Fisher Scientific, Waltham, Massachusetts, USA), following the manufacturer's recommendations under sterilized conditions. The extracted DNA quality and concentration were evaluated using the NanoDrop® ND 1,000 Spectrophotometer and the Qubit® 2.0 fluorometer (Invitrogen, Thermo Fisher Scientific, Waltham, Massachusetts, USA), respectively. Samples with DNA quality metrics meeting the Illumina 16S metagenomics workflow requirements for optimal outcomes (≥20 μl of 10 ng/μl, A260/280: 1.8–2.0, A260/230: 1.5–2.2) were selected for downstream analysis. While the majority of the samples met the criteria, only the top ten with the highest quality and quantity were included in downstream analysis due to funding limitations.
For PCR amplification of the hypervariable V3-V4 regions of the16S ribosomal RNA gene (16S rRNA), the following primer sequences were used:
• Forward primer (341F primer + Illumina overhang adapter underlined): TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCA
• Reverse primer (805R primer + Illumina overhang adapter underlined): GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC
Following this, the Nextera XT v2 Indexes were used for amplicon barcoding, which were then multiplexed, spiked with 10% of a 6 pM PhiX, and sequenced using the MiSeq Reagent Kit v2 (500 cycles) on the Illumina MiSeq platform (#15044223, Illumina, San Diego, CA, USA) at the Centre for Proteomic and Genomic Research.
2.4 Data analysis
Raw sequencing reads were quality filtered and trimmed, retaining only a Q-score > 20 and overlapping regions were allowed up to two of the ambiguous bases. Analysis of raw sequence data was mainly performed using Quantitative Insights into Microbial Ecology 2 (QIIME2) 2022.2 (https://qiime2.org/, RRID:SCR_021258), with DADA2 plugin to ccorrect sequencing errors and cluster the sequences into amplicon sequence variants (ASVs). Each ASV was aligned and classified at the genus level using the Greengenes database, V.2024.09 (https://qiime2.org/, RRID:SCR_002830).
Differential abundance testing was conducted by the Wald test with Benjamin-Hochberg adjustment for false discovery (31) using an adjusted p-value < 0.05, as implemented in the DESeq2-package in R. Features appearing in <10% of the samples and a relative abundance of <5% were filtered out.
Further statistical analysis and plot generation were conducted using R package, V4.2 (http://www.r-project.org/, RRID:SCR_001905).
Alpha-diversity was assessed using various metrics, including Observed richness, Chao1, Abundance Coverage Estimator (ACE), Simpson and Shannon indices, to estimate richness, abundance, and diversity within the sample (32). The Kruskal–Wallis H test was applied to all alpha-diversity metrics to determine statistical significance, with a threshold set at p-value < 0.05.
Beta-diversity was evaluated using the Bray-Curtis, Jaccard, Weighted UniFrac, and Unweighted UniFrac distance metrics on the rarefied datasets to assess differences in microbial community structure between comparison groups. The Phyloseq package in R was used to visualize these distances through principal coordinate analysis (PCoA) plots. Distance-based permutation multivariate analysis of variance (PERMANOVA) was conducted using Adonis 2 from the vegan R package (33). This was followed by a test for homogeneity.
3 Results
3.1 Demographics and clinical parameters
The demographics and clinical parameters are detailed in Table 1. The majority of the participants in the study were females, with only one male in the control group. Within the control group, three participants self-identified as SA Coloured (SAC), while the case group had an equal distribution of Caucasian and African participants, with only one participant from the SAC ethnic group. Among the periodontitis cases examined, Stage IV was the most prevalent, observed in three out of five cases, while Stage III accounted for the remaining two. For grading, the cases were classified as either Grade B or C, with Grade C indicating the highest severity, also found in three out of five cases. Additionally, all periodontitis cases exhibited a generalized distribution, as detailed in Table 1. P-values for the participant's quantitative and qualitative variables were not included as this was a preliminary descriptive study with limited sample size.
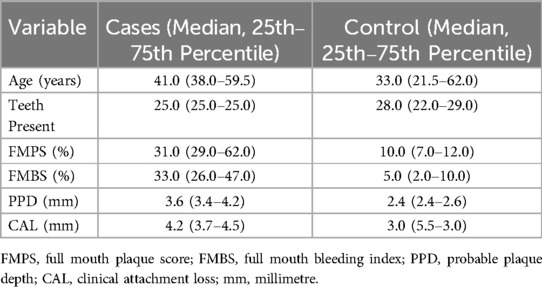
The observed distribution of the continuous variables between cases and controls is tabulated in Table 2. Based on the full-mouth clinical examination, periodontitis cases exhibited higher FMBS, FMPS, PPD, and CAL values, and fewer present teeth, when compared to the periodontally healthy controls. The differences between the median estimates all seem minimal. The control subjects seemed to be about 8 years younger on average but with larger variation between them (SD ≈ 21.49).
3.2 Sequence data
A total of 3.2 million V3–V4 16S rDNA paired-end reads were generated from the 10 pooled samples. After filtering, 2.7 million reads were left. The average length of the filtered reads was 599 bp. A total of 1,482 features were resolved, with a total frequency of 680,617 after quality filtering.
3.3 Subgingival bacterial communities
3.3.1 Bacterial compositions
The top 10 genera with the highest relative abundance in the periodontitis cases group were Fusobacterium (16%), Porphyromonas (10%), Treponema (9%), Prevotella (8%), Prevotella_7 (5%), Tannerella (5%), Filifactor (4%), F0058 (4%), Streptococcus (3%), and Veillonella (2%) (Figure 1A). In contrast, the control group was dominated by Streptococcus, accounting for 20% of all bacteria at the genus level. This was followed by Fusobacterium (15%), Veillonella (10%), Prevotella (7%), Porphyromonas (3%), Treponema (3%), Prevotella_7 (3%), Tannerella (2%), Filifactor (2%), and F0058 (2%) (Figure 1A).
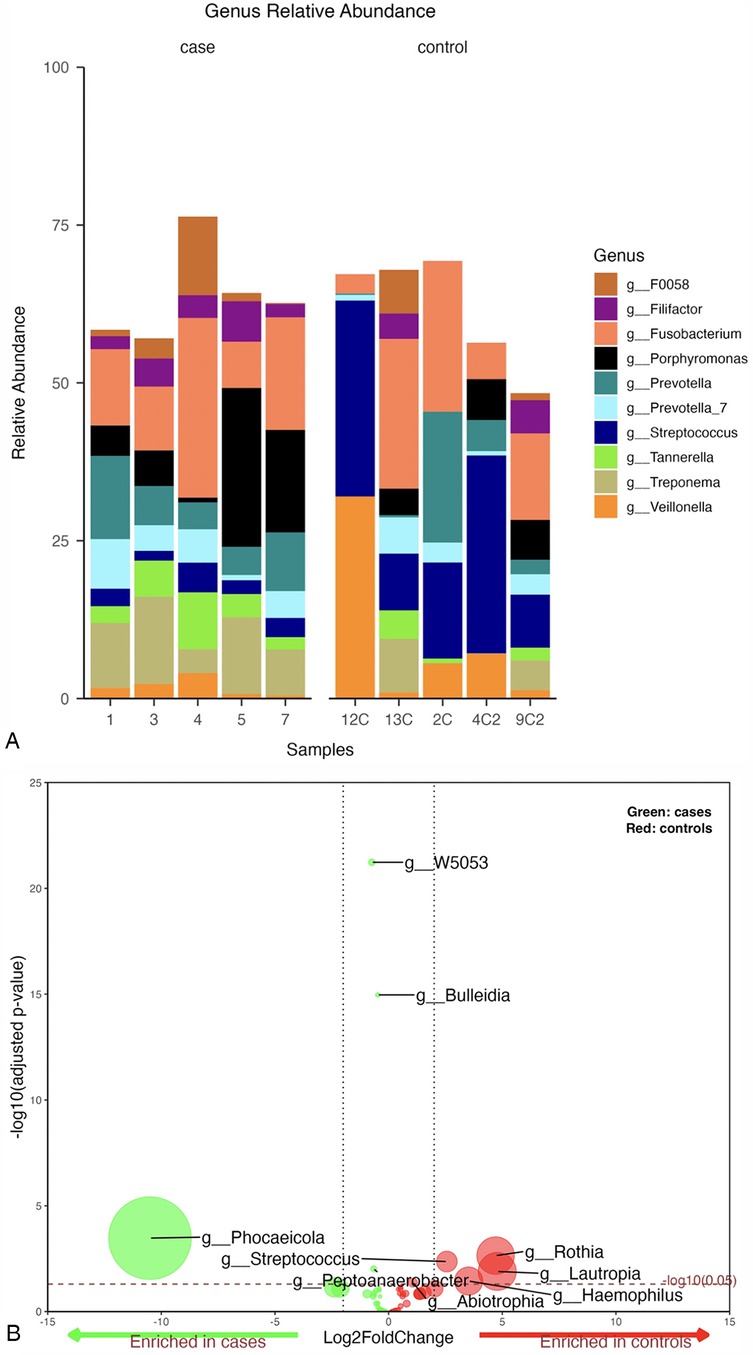
Figure 1. (A) Mean relative abundance of the top 10 genera generated from periodontitis cases vs. control samples, (B) Volcano plot depicting differentially abundant taxa as determined by DESeq2 in cases vs. controls at genus level. Significantly more discriminatory taxa appear above the threshold (red dotted line, FDR = 1). The relative abundance of taxa is indicated by circle size.
A pairwise differential abundance analysis at the genus level showed that four genera, Bulleidia, Peptoanaerobacter, Phocaeiola, and W5053, were enriched in the cases group samples, while five genera, Abiotrophia, Haemophilus, Lautropia, Rothia, and Streptococcus, were enriched in the controls (Figure 1B).
A statistically significant difference was only noted for the g_Streptococcus when comparing the top 10 genera between periodontitis cases and periodontally healthy controls at genus level (p-value = 0.008) (Supplementary Figure S1).
Among periodontitis samples, Fusobacterium was the most abundant genus (20%) in grade C, while Porphyromonas (15%) and Treponema (13%) were more dominant in grade B. In grade B, sample 2 showed a relatively equal abundance of all detected genera, with slightly higher levels of Treponema and Fusobacterium. Sample 4 was unique in having a distinctively higher abundance of Porphyromonas among all samples. All grade C samples were dominated by Fusobacterium. However, sample 3 had higher abundance of Tannerella and F0058, followed by Lentimicrobium. Samples 1 and 5 had high levels of Prevotella and Treponema, with sample 5 particularly rich in Porphyromonas and depleted in Lentimicrobium (Figure 2A).
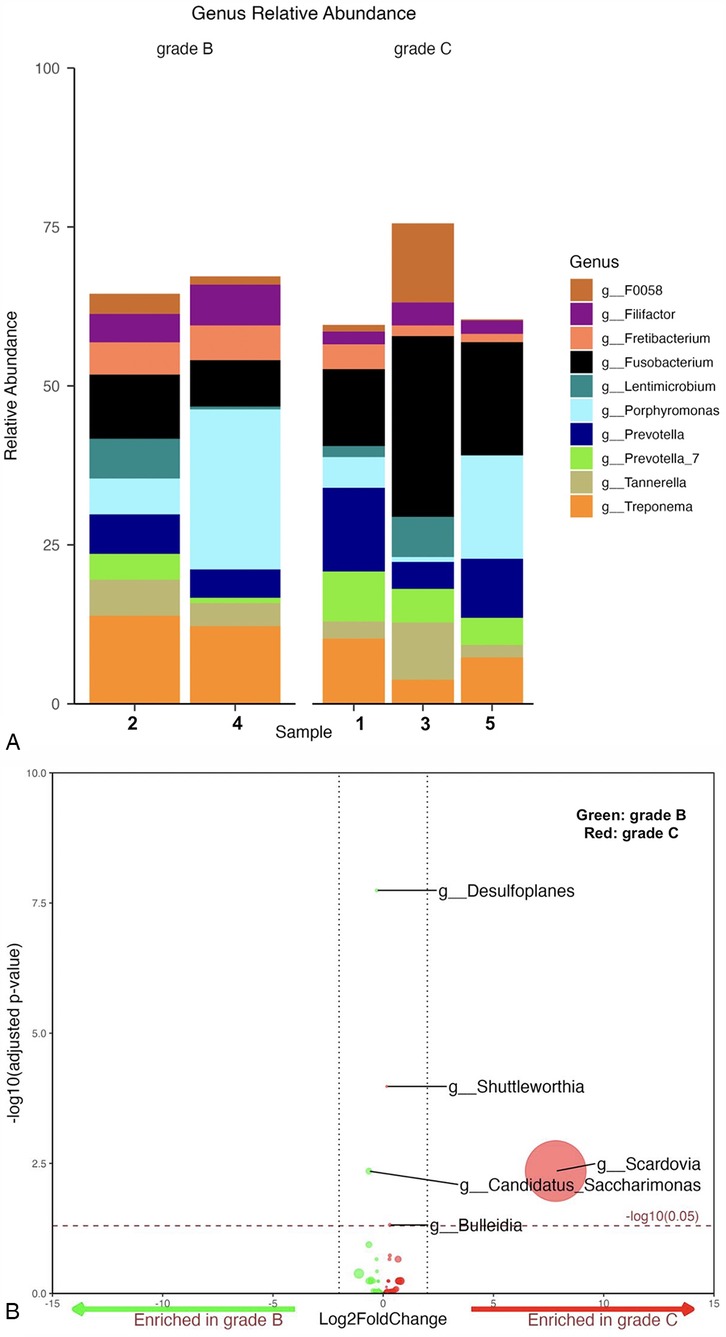
Figure 2. (A) Mean relative abundance of the top 10 genera generated from grade B vs. C samples. (B) Volcano plot depicting differentially abundance taxa as determined by DESeq2 in grade B vs. C at genus level. Significantly more discriminatory taxa appear above the threshold (red dotted line, FDR = 1). Relative abundance of taxa is indicated by circle size.
A total of five taxa were found to be significantly differentially abundant (Figure 2B). Of the five, Desulfoplanes and Candidatus Saccharimonas were enriched in grade B cases, while Shuttleworthia, Bulleidia, and Scardovia were enriched in the grade C cases. No other statistically significant differences were identified among all genera between both grades (p-value ≥ 0.059) (Supplementary Figure S2).
3.3.2 Bacterial diversity
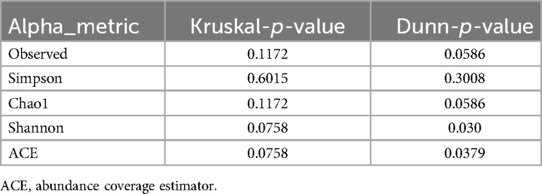
Despite observing higher median alpha-diversity in the periodontitis samples compared to the controls (Supplementary Figure S3), there was no statistically significant difference (all p-values > 0.05) in the alpha-diversity metrics used. Borderline significant p-values for Shannon and ACE suggest potential subtle differences (Dunn test, Table 3). Values of multiple alpha diversity and richness indices conducted for cases and controls group samples are presented in Supplementary Table S4.
The phylogeny-based Weighted-Unifrac distance matrix explained the highest amount of variance (69.7%) compared to Bray Curtis, Unweighted Unifrac, and Jaccard measures, which explained 39.6%, 40.4%, and 31.8% of the variance, respectively. The PCoA plot based on Weighted-Unifrac distance matrix showed that all cases samples clustered together, while the controls samples appeared more dispersed (Figure 3). PERMANOVA tests revealed significant differences in the average community composition between cases and controls (p-value = 0.04) but with a non-significant homogeneity condition test result (p-value = 0.16) (Table 4).
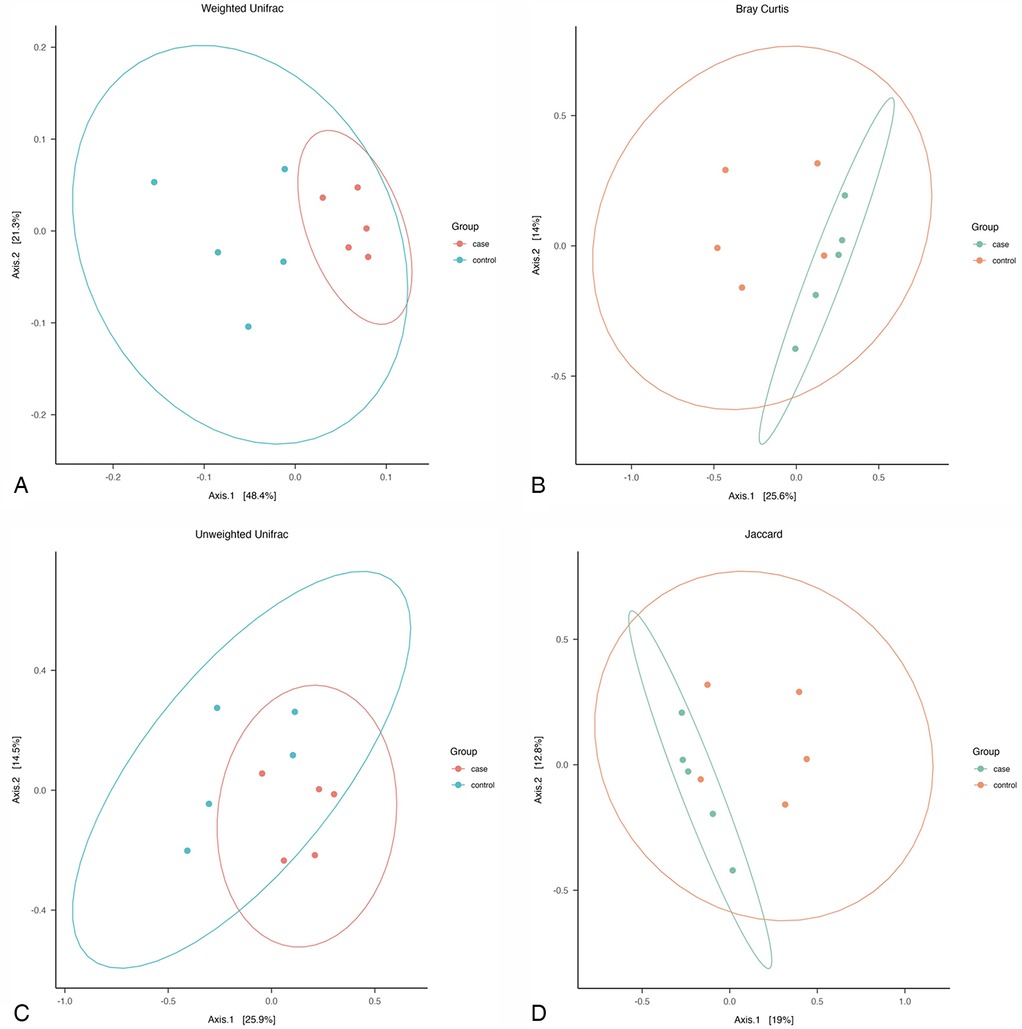
Figure 3. Pcoa plot of: (A) weighted unifarc distance, (B) bray curtiz dissimilarity distances, (C) unweighted unifrac distances, and (D) jaccard distances, across all samples.
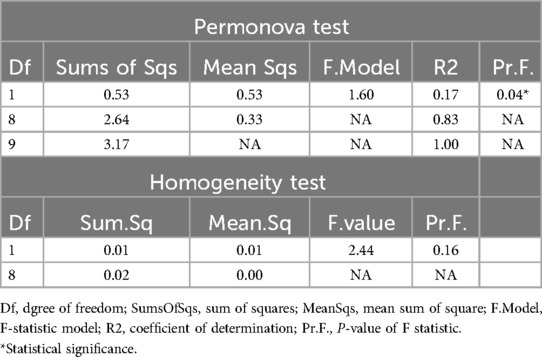
Table 4. PERMANOVA and PERMANOVA homogeneity tests for beta diversity conducted between cases and control groups.
4 Discussion
In this study, the subgingival microbiota in patients with periodontitis within a SA cohort, was characterised. The analysis demonstrated that, although the progression of periodontitis did not exhibit notable variations in bacterial diversity as assessed by alpha-diversity metrics, significant differences in community structure were identified using multiple beta-diversity indices.
The Weighted UniFrac distance analysis confirmed distinct clustering within the periodontitis group, whereas the control group exhibited greater dispersion, suggesting higher heterogeneity in microbiota composition and structure among healthy individuals.
These findings are consistent with those of Shi et al. (34), who utilized the Unweighted UniFrac distance metric to evaluate microbial variation in 25 periodontally healthy Chinese individuals, and Lenartová et al. (35), who employed the Bray-Curtis index to investigate microbial composition in 151 Czech participants across different clinical statuses and age groups. Both studies identified individualized subgingival plaque structures and attributed the dissimilarity observed in healthy biofilms to the presence of shallow gingival sulci, permitting environmental factors to influence microbial composition.
The “microbial succession” model dictates that several perio-pathogens initially invade the healthy microbiota, developing a diverse community of both health and disease-associated microbiota. As disease progresses, the transitional microbial community is then replaced by predominantly disease-associated organisms, resulting in the development of a more homogenous microbiota (10).
Findings from this study are consistent with studies conducted in Chilean, English, Chinese, and French populations (14, 36–38). They also partially align with the results of Nibali et al. (39), who reported significant differences in alpha and beta diversity across ethnic groups in England. However, our findings contrast with Schulz et al.'s (40), who observed no significant diversity differences in patients with aggressive periodontitis of Caucasian descent from Central Germany.
Several taxa exhibited comparable abundances between cases and controls, making it challenging to categorize them into distinct health status groups across studies (14, 35, 41). This challenge may stem from the potential limitations in accurately assigning health status at the microbial level, as such classifications are often based on clinical criteria. This issue was evident in the present investigation, where the relative abundances of certain genera showed minimal variation between cases with periodontitis and controls, including Fusobacterium (16% vs. 15%), Prevotella (8% vs. 7%), Prevotella_7 (5% vs. 3%), Tannerella (5% vs. 2%), F0058 (4% vs. 2%), and Filifactor (4% vs. 2%). In contrast, notable differences were observed in Veillonella (2% vs. 10%), Streptococcus (3% vs. 20%), Porphyromonas (10% vs. 3%), and Treponema (9% vs. 3%), with Streptococcus being significantly more abundant in controls (p = 0.008).
This investigation found Fusobacterium, Porphyromonas, Treponema, Prevotella, Provetella_7, Tannerella, Filifactor, and F0058 genera to be closely associated with periodontitis, while Streptococcus and Veillonella, typically linked to healthy periodontium, showed decreased abundance.
These findings align with previous research and reaffirm the typical taxa associated with periodontitis, as demonstrated by decades of studies across diverse populations (37, 42–45). Notably, Fusobacterium emerged as the most dominant genus in this study, surpassing Porphyromonas, which remained highly abundant. Similar results were reported in Chilean and American populations using 454-pyrosequencing (14, 42), and in a Chinese cohort where Fusobacterium was detected in all chronic periodontitis (CP) patients and in over 86% of healthy controls using real-time PCR (46). In contrast, Kumar et al. (47) found Fusobacterium to be rare in healthy controls and nearly absent in individuals with shallow or deep pockets. Although Fusobacterium is generally elevated in periodontitis patients who smoke (21), the participants in this study were non-smokers.
One plausible explanation for this study finding is that, although Porphyromonas gingivalis is widely recognized as the key pathogen for periodontitis development, it is significantly less invasive to host cells than Fusobacterium species, as noted by De Andrade et al. (48). Fusobacterium demonstrates a stronger adhesion capacity to human gingival cells relative to P. gingivalis, thereby facilitating superior colonization (49). Furthermore, Zhang et al. (50) showed that co-infection with Fusobacterium nucleatum enhances the invasive potential of P. gingivalis in oral epithelial cells. This “promoter” capability of F. nucleatum underscores its critical role in mixed infections. Additionally, the high prevalence of Fusobacterium has been linked to various diseases, including colorectal and gastric cancers, across different populations (51–53). Consequently, the abundance of Fusobacterium in the SA population with periodontitis may suggest a population-specific predisposition.
In this study, Filifactor ranked among the top 10 genera in both periodontitis cases and controls. Filifactor alocis displays virulence traits that facilitate its persistence in periodontal pockets, thereby contributing to disease progression (54). The abundance of Filifactor varies across populations with CP, with prevalence rates of 30% in Swedish population (55), 66.7% in German population (56), and 83% in deep pockets and 36% in shallow pockets among Koreans (57).
The F0058 genus, constituting 4% of cases and 2% of controls, was previously reported as abundant in Down syndrome patients with periodontitis (58), and in the salivary microbiome of Papillon–Lefèvre syndrome patients with neutrophil impairment (59). Its role in periodontitis warrants further investigation.
In the healthy group, Streptococcus and Veillonella were the predominant genera, consistent with findings from previous studies (35, 60–62). Streptococcus, a primary colonizer on clean tooth surfaces, plays a key role in promoting microbial adhesion (63), while Veillonella, considered a biomarker of health, consumes lactic acid produced by Streptococcus mutans and is associated with successful periodontal therapy (64). P. Zhou et al. (65) proposed that Veillonella functions as an “accessory pathogen”, facilitating the growth of pathogenic species within dysbiotic biofilms.
Fusobacterium was detected in 16% of healthy controls. While commonly present in a healthy periodontium, studies by Han (49) and Șurlin et al. (53) have also implicated Fusobacterium in the transition from gingivitis to periodontitis and its progression to more severe stages. Fusobacterium serves as a bridge between gram-positive symbionts and gram-negative pathogens in dysbiosis (66). Additionally, it has been shown to inhibit neutrophil proteases, thereby controlling tissue damage and providing defense against P. gingivalis (67).
Prevotella was found in comparable abundance (8% vs. 7%), consistent with prior studies (37, 42–45).Notably, sample 4 from the healthy controls exhibited a unique microbial profile, predominantly composed of Veillonella and Streptococcus, with periodontal pathogens such as Treponema, Tannerella, Filifactor, and F0058 either absent or present in insufficient abundance. This atypical composition may be attributed to factors such as diet or the patient's advanced age, both of which can reduce microbial diversity (68). Further studies with larger sample sizes are needed.
Peptoanaerobacter showed higher differential abundance in the periodontitis group, highlighting its recently recognized role in exacerbating inflammation and its prevalence in periodontitis biofilms (69). Less common genera, including Phocaeiola (gut-associated), W5053 (Firmicutes phylum), and Bulleidia (fecal origin), were also observed. While these genera have occasionally been detected in subgingival plaque of periodontitis patients (63–65, 70–72), their roles remain uncertain. However, these findings support the principle of multiple organisms contributing to biofilm-related disease, as previously described (73).
The control group showed a high differential abundance of genera linked to periodontal health including Streptococcus, Rothia, and Abiotrophia (35, 47, 74). Haemophilus, associated with Streptococcus in healthy periodontium (61, 75), and Lautropia, more common in younger individuals (35), were also observed.
To assess the risk of periodontitis progression based on microbial composition, bacterial abundance was compared between grade B and grade C cases. In moderate-risk grade B cases, Porphyromonas (15%) and Treponema (13%) were the dominant genera, consistent with their roles as keystone pathogens of the red complex, which induce dysbiosis and exhibit synergistic virulence in subgingival plaque (76, 77). In contrast, in grade C, associated with rapid disease progression, the abundance of the previously mentioned genera decreased (7% for each), while Fusobacterium (20%) and Prevotella (9%) became more dominant. The abundance of Prevotella was similar in both grades, as species such as P. intermedia and P. nigrescens from the orange complex are strongly linked to periodontitis (35, 44, 78) and are found in high abundance in both moderate and severe stages of periodontitis (79).
This study found that as periodontitis progresses from grade B to C, Fusobacterium abundance significantly increases. Unlike P. gingivalis, Fusobacterium potentiates inflammation by stimulating IL-8 and other virulence factors (49), explaining its persistence. However, the findings of this study contrast with those of a monozygotic twin study, where Fusobacterium was dominant in the grade B twin, while Treponema and Fretibacterium were more abundant in the grade C twin (80). The differences in severity between the twins, as well as the discrepancy with the present study's findings, may be attributed to the small sample size and non-shared environmental factors, such as long-term smoking in the grade C twin and corticosteroid use in the grade B twin (80).Additionally, a study in Brazil found higher P. gingivalis prevalence in grade C, suggesting ethnic and geographic variability (81).
Scardovia was differentially abundant in grade C patients, aligning with an animal study (82), though a pilot study found Scardovia wiggsiae, more common in periodontal health (83). This discrepancy may result from differences in sequencing techniques used (84). Candidatus saccharimonas typically present in low abundance, was differentially abundant in grade B samples, with elevated levels linked to severe periodontitis (37, 61, 85), though its role remains unclear (86). Shuttleworthia and Bulleidia, enriched in grade C, are associated with periodontitis progression (34, 87).
This pilot study had several limitations, primarily due to financial and resources constraints, which restricted the sample size to ten participants. These participants were selected based solely on the highest DNA purity and quantity measures, regardless of demographic matching. This introduced statistical limitations (e.g.,: the absence of p-values for quantitative and qualitative variables), and prevented comparisons based on gender or ethnicity. While previous research (88) was unable to identify sex-specific oral bacteria using 16S rRNA, the small sample size limits generalization. Age may impact oral microbial composition by reducing bacterial diversity (89). Future studies should use age-matched participants, a larger sample, and explore bacterial profiles across ethnic groups in SA to enhance generalizability and clinical reference. Finally, the recent development of periodontitis staging and grading systems limited the ability to draw definitive comparisons and conclusions.
However, this study has strengths in supporting the polymicrobial synergy and dysbiosis model by identifying shared bacteria like Fusobacterium between both groups, as well as underappreciated bacteria such as F0058 and Peptoanaerobacter, which mediate inflammation. Additionally, the study cohort, consisting of SA with diverse ethnic admixtures, may further influence microbial composition due to genetic and environmental factors (14), highlighting population-specific patterns. By analyzing bacterial abundance across periodontitis grades (B and C), the study provides insights into the microbiota's role in disease progression.
5 Conclusion
This pilot study highlights the complexity of the oral microbiome in periodontitis, providing preliminary data that may inform future analyses extending beyond 16S sequencing. These analyses could further explore the role of Fusobacterium and the overall composition of the microbiota, with particular focus on population-specific patterns.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1110749.
Ethics statement
Ethical approval was obtained from the Biomedical Research Ethics Committee (BMREC) of the University of the Western Cape (UWC), reference number BM20/10/9.
Author contributions
SK: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing – original draft. YP: Supervision, Validation, Writing – review & editing. NN: Supervision, Writing – review & editing. GD: Data curation, Supervision, Writing – review & editing. HH: Supervision, Writing – review & editing. SD: Supervision, Validation, Writing – review & editing. MC: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Research reported in this study was supported by the South African Medical Research Council under a Self-Initiated Research Grant. The views and opinions expressed are those of the author(s) and do not necessarily represent the official views of the SA-MRC. Additional financial support for the research, authorship, and/or publication of this article was provided by the UWC Research Chair for the SBDG project under the University Research Chairs Programme.
Acknowledgments
S.K. expresses profound gratitude to the Department of Craniofacial Biology, colleagues, and friends for their unwavering support. Special thanks are extended to the Oral Medicine and Periodontology registrars for their assistance in data collection, the Centre for Proteomics and Genomics Research team for their training, the IMBM team for their support, and Hassan Siddiq for his statistical guidance since 2020. Appreciation is also given to the Cape Peninsula University of Technology Cardiometabolic Unit for their support and collaboration. Finally, we express our gratitude to the authors of the studies cited in this review, whose significant contributions have served as the foundation for this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction Note
A correction has been made to this article. Details can be found at: 10.3389/froh.2025.1640915.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2025.1568393/full#supplementary-material
References
1. World Health Organization. Global oral health status report: towards universal health coverage for oral health by 2030: regional summary of the Western Pacific Region. (2022). Available online at: https://www.who.int/publications-detail-redirect/9789240070868 (cited May 27, 2024).
2. Abu Saleh T. Prevalence of interleukin-1 polymorphisms and its association with periodontal disease status in the Xhosa population of South Africa. etd.uwc.ac.za. (2007). Available online at: http://etd.uwc.ac.za/xmlui/handle/11394/2344 (cited January 1, 2020).
3. Kapila YL. Oral health’s inextricable connection to systemic health: special populations bring to bear multimodal relationships and factors connecting periodontal disease to systemic diseases and conditions. Periodontol 2000. (2021) 87(1):11–6. doi: 10.1111/prd.12398
4. Genco RJ, Sanz M. Clinical and public health implications of periodontal and systemic diseases: an overview. Periodontol 2000. (2020) 83(1):7–13. doi: 10.1111/prd.12344
5. Loos BG, Van Dyke TE. The role of inflammation and genetics in periodontal disease. Periodontol 2000. (2020) 83(1):26–39. doi: 10.1111/prd.12297
6. Bailén M, Bressa C, Larrosa M, González-Soltero R. Bioinformatic strategies to address limitations of 16rRNA short-read amplicons from different sequencing platforms. J Microbiol Methods. (2020) 169:105811. doi: 10.1016/j.mimet.2019.105811
7. Hrovat K, Dutilh BE, Medema MH, Melkonian C. Taxonomic resolution of different 16S rRNA variable regions varies strongly across plant-associated bacteria. ISME Commun. (2024) 4(1):ycae034. doi: 10.1093/ismeco/ycae034
8. Matchado MS, Rühlemann M, Reitmeier S, Kacprowski T, Frost F, Haller D, et al. On the limits of 16S rRNA gene-based metagenome prediction and functional profiling. Microb Genom. (2024) 10(2):001203. doi: 10.1099/mgen.0.001203
9. Botan A, Campisciano G, Zerbato V, Di Bella S, Simonetti O, Busetti M, et al. Performance of 16S rRNA gene next-generation sequencing and the culture method in the detection of Bacteria in clinical specimens. Diagnostics. (2024) 14(13):1318. doi: 10.3390/diagnostics14131318
10. Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol. (2015) 81(2):783–93. doi: 10.1128/AEM.02712-14
11. Cui Z, Wang P, Gao W. Microbial dysbiosis in periodontitis and peri-implantitis: pathogenesis, immune responses, and therapeutic. Front Cell Infect Microbiol. (2025) 15:1517154. doi: 10.3389/fcimb.2025.1517154
12. Caton JG, Armitage G, Berglundh T, Chapple ILC, Jepsen S, Kornman KS, et al. A new classification scheme for periodontal and peri-implant diseases and conditions—introduction and key changes from the 1999 classification. J Clin Periodontol. (2018) 45(S20):S1–8. doi: 10.1111/jcpe.12935
13. Papapanou PN, Sanz M, Buduneli N, Dietrich T, Feres M, Fine DH, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Clin Periodontol. (2018) 45(1):S162–70. doi: 10.1002/JPER.17-0721
14. Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. (2013) 7(5):1016–25. doi: 10.1038/ismej.2012.174
15. Lafaurie GI, Neuta Y, Ríos R, Pacheco-Montealegre M, Pianeta R, Castillo DM, et al. Differences in the subgingival microbiome according to stage of periodontitis: a comparison of two geographic regions. PLoS One. (2022) 17(8):e0273523. doi: 10.1371/journal.pone.0273523
16. Olujitan M, Ayanbadejo PO, Umeizudike K, Oyapero A, Okunseri C, Butali A. Periodontal diseases in Africa. Periodontol 2000. (2024). doi: 10.1111/prd.12617
17. Fine DH, Schreiner H, Velusamy SK. Aggregatibacter, a low abundance pathobiont that influences biogeography, microbial dysbiosis, and host defense capabilities in periodontitis: the history of A bug, and localization of disease. Pathogens. (2020) 9(3):179. doi: 10.3390/pathogens9030179
18. Yoshida A, Bouziane A, Erraji S, Lakhdar L, Rhissassi M, Miyazaki H, et al. Etiology of aggressive periodontitis in individuals of African descent. Jpn Dent Sci Rev. (2021) 57:20–6. doi: 10.1016/j.jdsr.2020.12.001
19. Molli VLP, Kissa J, Baraniya D, Gharibi A, Chen T, Al-Hebshi NN, et al. Bacteriome analysis of Aggregatibacter actinomycetemcomitans-JP2 genotype-associated grade C periodontitis in Moroccan adolescents. Front Oral Health. (2023) 4:1288499. doi: 10.3389/froh.2023.1288499
20. Velsko IM, Peter H, Natalia C, Jennifer B, Hong H, Ikramuddin A, et al. Grade C molar-incisor pattern periodontitis subgingival microbial profile before and after treatment. J Oral Microbiol. (2020) 12(1):1814674. doi: 10.1080/20002297.2020.1814674
21. Prince Y, Davison GM, Davids SFG, Erasmus RT, Kengne AP, Raghubeer S, et al. The effect of cigarette smoking on the oral microbiota in a South African population using subgingival plaque samples. Heliyon. (2024) 10(10):e31559. doi: 10.1016/j.heliyon.2024.e31559
22. Tonetti MS, Greenwell H, Kornman KS. Staging and grading of periodontitis: framework and proposal of a new classification and case definition. J Periodontol. (2018) 89(20):S159–72. doi: 10.1002/JPER.18-0006
23. Borilova Linhartova P, Bartova J, Poskerova H, Machal J, Vokurka J, Fassmann A, et al. Apolipoprotein E gene polymorphisms in relation to chronic periodontitis, periodontopathic bacteria, and lipid levels. Arch Oral Biol. (2015) 60(3):456–62. doi: 10.1016/j.archoralbio.2014.10.003
24. Borilova Linhartova P, Vokurka J, Poskerova H, Fassmann A, Zakovicova Holla L. Haplotype analysis of interleukin-8 gene polymorphisms in chronic and aggressive periodontitis. Mediators Inflamm. (2013) 2013:342351. doi: 10.1155/2013/342351
25. Nibali L, Tonetti MS, Ready D, Parkar M, Brett PM, Donos N, et al. Interleukin-6 polymorphisms are associated with pathogenic Bacteria in subjects with periodontitis. J Periodontol. (2008) 79(4):677–83. doi: 10.1902/jop.2008.070453
26. Reichert S, Stein JM, Klapproth J, Zimmermann U, Reichert Y, Gläser C, et al. The genetic impact of the Q551R interleukin-4 receptor alpha polymorphism for aggressive or chronic periodontitis and the occurrence of periodontopathic bacteria. Arch Oral Biol. (2011) 56(12):1485–93. doi: 10.1016/j.archoralbio.2011.06.003
27. Reichert S, Machulla HKG, Klapproth J, Zimmermann U, Reichert Y, Gläser C, et al. Interleukin-2 −330 and 166 gene polymorphisms in relation to aggressive or chronic periodontitis and the presence of periodontopathic bacteria. J Periodontal Res. (2009) 44(5):628–35. doi: 10.1111/j.1600-0765.2008.01173.x
28. Reichert S, Machulla HKG, Klapproth J, Zimmermann U, Reichert Y, Gläser CH, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. (2007) 43(1):40–7. doi: 10.1111/j.1600-0765.2007.00992.x
29. Schulz S, Stein JM, Altermann W, Klapproth J, Zimmermann U, Reichert Y, et al. Single nucleotide polymorphisms in interleukin-1gene cluster and subgingival colonization with Aggregatibacter actinomycetemcomitans in patients with aggressive periodontitis. Hum Immunol. (2011) 72(10):940–6. doi: 10.1016/j.humimm.2011.05.009
30. Schulz S, Zissler N, Altermann W, Klapproth J, Zimmermann U, Gläser C, et al. Impact of genetic variants of CD14 and TLR4 on subgingival periodontopathogens. Int J Immunogenet. (2008) 35(6):457–64. doi: 10.1111/j.1744-313X.2008.00811.x
31. Noble WS. How does multiple testing correction work? Nat Biotechnol. (2009) 27(12):1135–7. doi: 10.1038/nbt1209-1135
32. Finotello F, Mastrorilli E, Di Camillo B. Measuring the diversity of the human microbiota with targeted next-generation sequencing. Brief Bioinform. (2018) 19(4):679–92. doi: 10.1093/bib/bbw119
33. Anderson MJ. permutational multivariate analysis of variance (PERMANOVA). In: Colton T, Everitt B, Piegorsch WW, Ruggeri F, Teugels JL, editors. Wiley StatsRef: Statistics Reference Online. Hoboken, NJ: John Wiley & Sons, Ltd (2017). p. 1–15. Available online at: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118445112.stat07841 (cited February 10, 2023).
34. Shi W, Tian J, Xu H, Zhou Q, Qin M. Distinctions and associations between the microbiota of saliva and supragingival plaque of permanent and deciduous teeth. PLoS One. (2018) 13(7):e0200337. doi: 10.1371/journal.pone.0200337
35. Lenartova M, Tesinska B, Janatova T, Hrebicek O, Mysak J, Janata J, et al. The oral microbiome in periodontal health. Front Cell Infect Microbiol. (2021) 11:629723. doi: 10.3389/fcimb.2021.629723
36. Esparbès P, Legrand A, Bandiaky ON, Chéraud-Carpentier M, Martin H, Montassier E, et al. Subgingival Microbiota and cytokines profile changes in patients with periodontitis: a pilot study comparing healthy and diseased sites in the same oral cavities. Microorganisms. (2021) 9(11):2364. doi: 10.3390/microorganisms9112364
37. Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PLoS One. (2013) 8(8):e71227. doi: 10.1371/journal.pone.0071227
38. Zhou X, Nanayakkara S, Gao JL, Nguyen KA, Adler CJ. Storage media and not extraction method has the biggest impact on recovery of bacteria from the oral microbiome. Sci Rep. (2019) 9(1):14968. doi: 10.1038/s41598-019-51448-7
39. Nibali L, Sousa V, Davrandi M, Liu L, Spratt D, Donos N. Patterns of subgingival microbiota in different periodontal phenotypes. J Dent. (2022) 117:103912. doi: 10.1016/j.jdent.2021.103912
40. Schulz S, Pütz N, Jurianz E, Schaller HG, Reichert S. Are there any common genetic risk markers for rheumatoid arthritis and periodontal diseases? A case-control study. Mediators Inflamm. (2019) 2019:2907062. doi: 10.1155/2019/2907062
41. Tsai CY, Tang CY, Tan TS, Chen KH, Liao KH, Liou ML. Subgingival microbiota in individuals with severe chronic periodontitis. J Microbiol Immunol Infect. (2018) 51(2):226–34. doi: 10.1016/j.jmii.2016.04.007
42. Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. (2012) 6(6):1176–85. doi: 10.1038/ismej.2011.191
43. Curtis MA, Diaz PI, Van Dyke TE. The role of the microbiota in periodontal disease. Periodontol 2000. (2020) 83(1):14–25. doi: 10.1111/prd.12296
44. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. (1998) 25(2):134–44. doi: 10.1111/j.1600-051X.1998.tb02419.x
45. Szafranski SP, Wos-Oxley ML, Vilchez-Vargas R, Jáuregui R, Plumeier I, Klawonn F, et al. High-Resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Appl Environ Microbiol. (2015) 81(3):1047–58. doi: 10.1128/AEM.03534-14
46. Wang P, Duan D, Zhou X, Li X, Yang J, Deng M, et al. Relationship between expression of human gingival beta-defensins and levels of periodontopathogens in subgingival plaque. J Periodontal Res. (2015) 50(1):113–22. doi: 10.1111/jre.12187
47. Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. (2005) 43(8):3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005
48. De Andrade KQ, Almeida-da-Silva CLC, Ojcius DM, Coutinho-Silva R. Differential involvement of the canonical and noncanonical inflammasomes in the immune response against infection by the periodontal bacteria Porphyromonas gingivalis and Fusobacterium nucleatum. Curr Res Microb Sci. (2021) 2:100023. doi: 10.1016/j.crmicr.2021.100023
49. Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. (2015) 23:141–7. doi: 10.1016/j.mib.2014.11.013
50. Zhang L, Chen F, Zeng Z, Xu M, Sun F, Yang L, et al. Advances in metagenomics and its application in environmental microorganisms. Front Microbiol. (2021) 12:766364. doi: 10.3389/fmicb.2021.766364
51. Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. (2018) 8(1):158. doi: 10.1038/s41598-017-18596-0
52. Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, et al. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. (2020) 69(11):998–2007. doi: 10.1136/gutjnl-2019-319635
53. Șurlin P, Nicolae FM, Șurlin VM, Pătrașcu Ș, Ungureanu BS, Didilescu AC, et al. Could periodontal disease through periopathogen Fusobacterium nucleatum be an aggravating factor for gastric cancer? J Clin Med. (2020) 9(12):3885. doi: 10.3390/jcm9123885
54. Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis. Infect Immun. (2011) 79(10):3872–86. doi: 10.1128/IAI.05631-11
55. Cato EP, Moore LVH, Moore WEC. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. Nov. From the human gingival sulcus. Int J Syst Bacteriol. (1985) 35(4):475–7. doi: 10.1099/00207713-35-4-475
56. Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, et al. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. (2010) 10(1):66. doi: 10.1186/1471-2180-10-66
57. Yun JH, Park JE, Kim DI, Lee SI, Choi SH, Cho KS, et al. Identification of putative periodontal pathogens in Korean chronic periodontitis patients. J Korean Acad Periodontol. (2013) 38(2):143–52. doi: 10.5051/jkape.2008.38.2.143
58. Nóvoa L, Sánchez MDC, Blanco J, Limeres J, Cuenca M, Marín MJ, et al. The subgingival microbiome in patients with down syndrome and periodontitis. J Clin Med. (2020) 9(8):2482. doi: 10.3390/jcm9082482
59. Lettieri GM, Santiago LM, Lettieri GC, Borges LGDA, Marconatto L, de Oliveira LA, et al. Oral phenotype and salivary microbiome of individuals with papillon–lefèvre syndrome. Front Cell Infect Microbiol. (2021) 11:720790. doi: 10.3389/fcimb.2021.720790
60. Bizzarro S, Laine ML, Buijs MJ, Brandt BW, Crielaard W, Loos BG, et al. Microbial profiles at baseline and not the use of antibiotics determine the clinical outcome of the treatment of chronic periodontitis. Sci Rep. (2016) 6(1):20205. doi: 10.1038/srep20205
61. Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, et al. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. (2015) 6:119. doi: 10.3389/fmicb.2015.00119
62. Meuric V, Le Gall-David S, Boyer E, Acuña-Amador L, Martin B, Fong SB, et al. Signature of microbial dysbiosis in periodontitis. Appl Environ Microbiol. (2017) 83(14):e00462–17. doi: 10.1128/AEM.00462-17
63. Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. (2009) 28(8):397–403. doi: 10.1089/dna.2009.0868
64. Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. (2012) 83(10):1279–87. doi: 10.1902/jop.2012.110566
65. Zhou P, Manoil D, Belibasakis GN, Kotsakis GA. Veillonellae: beyond bridging species in oral biofilm ecology. Front Oral Health. (2021) 2:774115. doi: 10.3389/froh.2021.774115
66. Chen Y, Huang Z, Tang Z, Huang Y, Huang M, Liu H, et al. More than just a periodontal pathogen –the research progress on Fusobacterium nucleatum. Front Cell Infect Microbiol. (2022) 12:815318. doi: 10.3389/fcimb.2022.815318
67. Yang NY, Zhang Q, Li JL, Yang SH, Shi Q. Progression of periodontal inflammation in adolescents is associated with increased number of Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis, and Fusobacterium nucleatum. Int J Paediatr Dent. (2014) 24(3):226–33. doi: 10.1111/ipd.12065
68. Liu T, Salguero P, Petek M, Martinez-Mira C, Balzano-Nogueira L, Ramšak Ž, et al. Paintomics 4: new tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. (2022) 50(W1):W551–9. doi: 10.1093/nar/gkac352
69. Prucsi Z, Płonczyńska A, Potempa J, Sochalska M. Uncovering the oral dysbiotic Microbiota as masters of neutrophil responses in the pathobiology of periodontitis. Front Microbiol. (2021) 12:729717. doi: 10.3389/fmicb.2021.729717
70. Chen C, Hemme C, Beleno J, Shi ZJ, Ning D, Qin Y, et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. (2018) 12(5):1210–24. doi: 10.1038/s41396-017-0037-1
71. Liu S, Xie G, Chen M, He Y, Yu W, Chen X, et al. Oral microbial dysbiosis in patients with periodontitis and chronic obstructive pulmonary disease. Front Cell Infect Microbiol. (2023) 13:1121399. doi: 10.3389/fcimb.2023.1121399
72. Vieira Lima CP, Grisi DC, Guimarães MDCM, Salles LP, de Kruly PC, Do T, et al. Enrichment of sulphate-reducers and depletion of butyrate-producers may be hyperglycaemia signatures in the diabetic oral microbiome. J Oral Microbiol. (2022) 14(1):2082727. doi: 10.1080/20002297.2022.2082727
73. Arora N, Mishra A, Chugh S. Microbial role in periodontitis: have we reached the top? Some unsung bacteria other than red complex. J Indian Soc Periodontol. (2014) 18(1):9–13. doi: 10.4103/0972-124X.128192
74. Mikkelsen L, Theilade E, Poulsen K. Abiotrophia species in early dental plaque. Oral Microbiol Immunol. (2000) 15(4):263–8. doi: 10.1034/j.1399-302x.2000.150409.x
75. Liljemark WF, Bloomquist CG, Uhl LA, Schaffer EM, Wolff LF, Pihlstrom BL, et al. Distribution of oral Haemophilus species in dental plaque from a large adult population. Infect Immun. (1984) 46(3):778–86. doi: 10.1128/iai.46.3.778-786.1984
76. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. (2012) 10(10):717–25. doi: 10.1038/nrmicro2873
77. Yamamoto K, Kobayashi T, Grossi S, Ho AW, Genco RJ, Yoshie H, et al. Association of Fcγ receptor IIa genotype with chronic periodontitis in caucasians. J Periodontol. (2004) 75(4):517–22. doi: 10.1902/jop.2004.75.4.517
78. Najmanova L, Sabova L, Lenartova M, Janatova T, Mysak J, Vetrovsky T, et al. R/G value—a numeric Index of individual periodontal health and oral microbiome dynamics. Front Cell Infect Microbiol. (2021) 11:602643. doi: 10.3389/fcimb.2021.602643
79. Ortiz AP, Acosta-Pagán KT, Oramas-Sepúlveda C, Castañeda-Avila MA, Vilanova-Cuevas B, Ramos-Cartagena JM, et al. Oral microbiota and periodontitis severity among Hispanic adults. Front Cell Infect Microbiol. (2022) 12:965159. doi: 10.3389/fcimb.2022.965159
80. Yamamoto T, Taniguchi M, Matsunaga K, Kawata Y, Kawamura M, Okubo K, et al. Analysis of subgingival microbiota in monozygotic twins with different severity and progression risk of periodontitis. Clin Case Rep. (2022) 10(4):e05725. doi: 10.1002/ccr3.5725
81. Rodrigues RS, Silveira VR, Rego RO. Analysis of Porphyromonas gingivalis fimA genotypes in severe periodontitis patients. Braz Oral Res. (2020) 34:e090. doi: 10.1590/1807-3107bor-2020.vol34.0090
82. Chen Y, Li EM, Xu LY. Guide to metabolomics analysis: a bioinformatics workflow. Metabolites. (2022) 12(4):357. doi: 10.3390/metabo12040357
83. Chakrapani AR, Ajjappa BG, Prakash S, Bhat K. Scardovia wiggsiae: role in periodontal health and disease–A pilot study. Indian J Dent Adv. (2019) 11(4):119–25.
84. Yap M, O’Sullivan O, O’Toole PW, Cotter PD. Development of sequencing-based methodologies to distinguish viable from non-viable cells in a bovine milk matrix: a pilot study. Front Microbiol. (2022) 13:1036643. doi: 10.3389/fmicb.2022.1036643/full
85. Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. (2003) 69(3):1687–94. doi: 10.1128/AEM.69.3.1687-1694.2003
86. Bor B, Bedree JK, Shi W, McLean JS, He X. Saccharibacteria (TM7) in the human oral microbiome. J Dent Res. (2019) 98(5):500–9. doi: 10.1177/0022034519831671
87. Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, et al. Newly identified pathogens associated with periodontitis. J Dent Res. (2014) 93(9):846–58. doi: 10.1177/0022034514542468
88. Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, et al. Human oral viruses are personal, persistent and gender-consistent. ISME J. (2014) 8(9):1753–67. doi: 10.1038/ismej.2014.31
Keywords: periodontitis, periodontal health, bacterial profile, 16S rRNA, Fusobacterium, diversity measures, South Africa
Citation: Kabbashi S, Prince Y, Ngwa NE, Holmes H, Davison GM, Davids SFG and Chetty M (2025) Feasibility and preliminary findings of a bacterial diversity study in periodontitis: a pilot investigation from the Western Cape. Front. Oral Health 6:1568393. doi: 10.3389/froh.2025.1568393
Received: 29 January 2025; Accepted: 28 March 2025;
Published: 23 April 2025;
Corrected: 26 June 2025.
Edited by:
Thuy Do, University of Leeds, United KingdomReviewed by:
Ahmed Hashim, King Faisal University, Saudi ArabiaSaliha Saad, University of the West of England, United Kingdom
Copyright: © 2025 Kabbashi, Prince, Ngwa, Holmes, Davison, Davids and Chetty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salma Kabbashi, c2thYmJhc2hpQHV3Yy5hYy56YQ==
 Salma Kabbashi
Salma Kabbashi Yvonne Prince
Yvonne Prince Ndonwi Elvis Ngwa2
Ndonwi Elvis Ngwa2 Haly Holmes
Haly Holmes Saarah F. G. Davids
Saarah F. G. Davids Manogari Chetty
Manogari Chetty