- 1Teagasc Food Research Centre, Moorepark, Cork, Ireland
- 2Institute for Global Food Security, School of Biological Sciences, Queen's University Belfast, Belfast, United Kingdom
- 3IUT Dijon-Auxerre, University of Burgundy, Dijon, France
- 4Department of Microbiology and Biotechnology, Max Rubner-Institut, Federal Research Institute of Nutrition and Food, Kiel, Germany
- 5VistaMilk SFI Research Centre, Cork, Ireland
The high mortality rate associated with Listeria monocytogenes as well as its ability to adapt to the harsh conditions employed in food processing have ensured that this pathogen has become a significant concern in the ready-to-eat food industry. Lytic bacteriophages are viruses that hijack the metabolic mechanisms of their bacterial host as a means to grow and replicate, subsequently leading to host cell death due to lysis. This study reports the biological and genomic characterization of L. monocytogenes phage vB_LmoH_P61 (P61) and its potential application for the reduction of L. monocytogenes in artificially contaminated foods. Phage P61 is a virulent bacteriophage belonging to the family Herelleviridae and has a genome size of 136,485bp. The lytic spectrum of phage P61 was investigated and it was shown to infect serotypes 1/2a, 1/2b, 1/2c, 4b, 4e, and 6a. Treatment of artificially contaminated milk stored at 8 and 12°C with phage P61 resulted in a significant reduction in L. monocytogenes numbers over the product shelf life. Similarly, phage P61 reduced L. monocytogenes numbers on artificially contaminated baby spinach stored at 8, 12, and 25°C. The research findings indicate that biocontrol of L. monocytogenes with phage P61 may offer a safe and environmentally friendly approach for the reduction of L. monocytogenes numbers in certain ready-to-eat foods.
Introduction
On an annual basis, it is estimated that 600 million people globally (~1 in 10) fall ill following consumption of contaminated food, and of these, 420,000 die (World Health Organization, 2020). Contamination of food with pathogenic bacteria poses a severe threat to our socio-economic balance and healthcare systems. Bacteria such as Campylobacter, Salmonella, Shiga toxin-producing Escherichia coli (STEC), and Listeria monocytogenes are responsible for the majority of outbreaks related to bacterial foodborne illness occurring in the EU (EFSA, 2018). Compared to the other bacterial species, the numbers of cases of illness caused by L. monocytogenes is low. In 2018, there were 2,549 cases of listeriosis in the EU compared to 246,571 cases of campylobacteriosis, 92,857 cases of salmonellosis and 8,161 cases of STEC (EFSA, 2018). However, listeriosis accounts for the highest proportion of hospitalized cases (97%) and the highest number of deaths (229) in Europe, making it one of the most serious foodborne diseases (EFSA, 2018). In addition, the trend in case numbers in Europe is continuing upward over the past 10 years (EFSA, 2018). Gastroenteritis is the most common manifestation of listeriosis and although the occurrence in healthy individuals is relatively rare, the disease can progress to septicemia and death in immunocompromised individuals, pregnant women, neonates and the elderly (Vázquez-Boland et al., 2001; Smith et al., 2018).
Issues of food contamination with L. monocytogenes are primarily associated with ready-to-eat (RTE) foods, which are foods intended by the producer or the manufacturer for direct human consumption without the need for cooking or other processing effective to eliminate or reduce to an acceptable level micro-organisms of concern [Regulation (EC) No. 2073/2005]. L. monocytogenes is found ubiquitously in the environment, present in soil, water, animal feed, vegetation, industrial plants, and on farms. It can enter the RTE food chain via a number of routes, soil and water being the most common, and can persist along the food chain (Ferreira et al., 2014). The control of L. monocytogenes in the food processing setting has proven difficult due to its ability to adapt to and survive in a wide range of environmental conditions. To minimize the risk associated with L. monocytogenes, processing of RTE foods may include the addition of preservatives (sodium nitrite, sorbic acid), decontamination (water, acid), curing, smoking, fermentation, heating (pasteurizing, cooking, baking, boiling, and steaming), and drying (Ricci et al., 2018). For RTE foods that undergo mild processing treatments (e.g., washing), such as fresh fruit and vegetables, the pathogen may still survive. Therefore, antimicrobials or bacteriostatic agents may be applied to minimize the growth. An antimicrobial is defined by the USDA Food Safety and Inspection Service as a substance in or added to an RTE product that has the effect of suppressing or limiting the growth of L. monocytogenes throughout the shelf life of a product (Savage, 2004). Recent reports on bacteriophages (phages) as natural and environmentally friendly antimicrobial agents have shown effective, specific targeting of bacterial pathogens of interest in different food matrices (Taylor, 2018). The specificity of phage-host interactions ensures the targeting of pathogenic bacteria in food without disturbing the normal microbiota of the food. There is no risk of the formation of secondary components upon their application; a reported disadvantage of using chemical agents in the decontamination process of foods (Shen et al., 2016). Commercially available phage products are usually water-based solutions consisting of purified phages, with low levels of salt and little or no preservatives or additives, making them an attractive alternative to chemicals for producers of “clean label” foods and consumers (Moye et al., 2018). Research also suggests that the application of phages does not affect the organoleptic properties of foods. In comparison to other antimicrobials and food safety interventions, the cost of application of phage preparations is relatively low, reportedly costing 1–4 cents per pound of food treated (Moye et al., 2018). These biological properties and qualities of commercially available phages make them an attractive proposition for improving the safety of RTE foods (Moye et al., 2018).
To date, over 500 Listeria phages have been isolated from environmental sources adding to the knowledge of the phenotypes and genotypes of these phages (Lee et al., 2017; Vongkamjan et al., 2017; Mutlu and Şahin, 2018; Song et al., 2019). All of these phages belong to the order Caudovirales, either featuring the long non-contractile tail of the Siphoviridae family or the contractile tail of the Herelleviridae family (Klumpp and Loessner, 2013; Adriaenssens et al., 2020). Commercial listeriophage preparations have been available to the food industry for some time, comprising either single broad-host range phages or a cocktail of phages for application either on food matrices or on food processing plant surfaces (Golkar et al., 2014). In a research setting, the efficacy of phages as biocontrol agents in various food matrices have been tested including lettuce, cheese, smoked salmon, and frozen entrees (Perera et al., 2015), and melon, pear, and apple products (juices and slices) (Oliveira et al., 2014).
The aim of this study was to phenotypically and genotypically characterize a L. monocytogenes phage, vB_LmoH_P61 (P61), isolated from grass silage. In addition, we evaluated the potential of P61 as a biocontrol agent against L. monocytogenes in both liquid and solid food matrices. In particular, given the increase in the consumption of fresh and minimally processed vegetables in recent years, we chose to evaluate the efficacy of phage P61 on baby spinach, a product on which the occurrence of L. monocytogenes has been reported.
Materials and Methods
Bacterial Strains and Culture Conditions
Listeria monocytogenes strains used in this study were obtained from a collection of isolates housed at the Teagasc Food Research Center, Moorepark, Cork, Ireland, unless otherwise indicated (Table 1). Overnight cultures of each strain were prepared following 18 h of incubation in tryptic soy broth (TSB) (Oxoid Ltd., Basingstoke, UK) at 25°C under aerobic conditions. Solid agar and soft agar TSB overlays contained 1.5% agar and 0.4% agarose (both Sigma-Aldrich, St. Louis, USA), respectively.
Bacteriophage Isolation and Propagation
Phage P61 was isolated from grass silage sourced from the Teagasc Moorepark farm, Fermoy, Cork following the protocol outlined by Alemayehu et al. (2009). Subsequently, individual phage plaques were subjected to four successive rounds of purification. To propagate the phage, 1 ml of TSB (Becton Dickinson and Company, Le Pont de Claix, France) contained 10% inoculum of an overnight culture of the host organism (L. monocytogenes strain 3053) and 18.5 mM CaCl2 (Merck, Darmstadt, Germany) and was incubated for 3 h at 25°C. Following this incubation, a single plaque was aseptically removed from an overlay plate using a 5 ml pipette and added to the 1 ml of the bacterial host-cell (strain 3053). The volume was increased to 10 ml with TSB containing 18.5 mM CaCl2 and 10% inoculum of strain 3053. The mixture was incubated for 18 h at 25°C. Following overnight incubation, the sample was filtered using a 0.45 μm pore filter (Sarstedt, Wexford, Ireland). Phage titer (Plaque forming unit /ml) (PFU/ml) was determined using the host sensitive strain 3053, 1 ml of phage lysate was diluted 1:10 in maximum recovery diluent (MRD) (Oxoid Ltd. Basingstoke, UK) to dilution 10−8. A plaque assay was then performed using each dilution whereby a 5 ml sloppy agar tube (TSA, 0.4% agarose) contained 2% of strain 3053 and 20% of the phage lysate dilution to be tested and 3.7 mM CaCl2. The mixture was overlaid onto TSA plates (Becton Dickinson and Company, Le pont de Claix, France) and incubated at 25°C for 18 h.
Bacteriophage Lytic Spectrum
The lytic spectrum of phage P61 was determined by plaque assays against a range of L. monocytogenes strains (listed in Table 1) using the protocol outlined by Casey et al. (2015). Efficiency of plaquing (EOP) figures were calculated by dividing the phage titer (PFU/ ml) of a given test strain, by the phage titer (PFU/ ml) of the host sensitive strain (strain 3053).
One Step Growth Curve to Determine the Growth Kinetics of P61
To determine the growth kinetics of phage P61, a one-step growth curve was conducted according to Denes et al. (2015). Briefly, a 5 ml culture of L. monocytogenes was grown in TSB to a concentration of ~1 × 109 Colony forming unit/ml (CFU/ml) and 20 mM of CaCl2 was added. P61 (1 × 108 PFU/ml) was then added to the solution [multiplicity of infection (MOI) of 0.1]. The solution was incubated at 30°C while shaking (120 rpm). Two samples were taken every 10 min, one 100 μl was transferred into a tube containing 4 drops of chloroform and the other sample was immediately diluted and enumerated using strain 3053 as the host bacterial cell. At the end of the timeline, chloroform-treated phages were enumerated yielding the total concentration of viable phage particles in the sample, including intracellular phages. The average burst size was calculated by dividing the average concentration of infected cells and free viable phages by three timepoints following the first step of lysis (time point 80, 90, and 100 min) by the average concentration of infected cells and free viable phage from the first free time points post-infection (Hyman and Abedon, 2009; Denes et al., 2015). The experiment was independently repeated three times.
Determination of Bacteriophage-Insensitive Mutant Frequency
To determine the frequency of cases of bacteriophage-insensitive mutants (BIMs), a protocol was adapted from Filippov et al. (2011). A total of 24 colonies were selected to test the frequency of BIMs. A single colony of strain 3053 was inoculated into TSB and incubated overnight at 25°C. Following the overnight incubation, the sample was serially diluted in MRD and 100 μl spread onto Listeria Chromogenic agar (Neogen) with or without a double agarose overlay (0.4%) containing 1 × 109 PFU/ml of phage P61. Plates were incubated for 48 h at 37°C. Resulting colonies were counted and BIM frequency was determined using the formula (surviving viable counts divided by initial viable counts).
Ammonium Acetate Precipitation of Phage Lysate
Phage P61 was concentrated using a protocol adapted from Casey et al. (2015). A volume of 1.5 L of P61 was propagated to a titer of ~1 × 1010 PFU/ml. The lysate was filtered through a 0.45 μm filter (Starstedt) and centrifuged at 6,000 × g at 4°C to remove any cell debris. Filtrates were then centrifuged at 25,000 × g at 4°C for 1.5 h and the resulting pellet was resuspended in 10 ml of 0.1 M ammonium acetate and centrifuged at 25,000 × g for 1.5 h at 4°C. The resulting pellet was then resuspended in 1 ml of 0.1 M ammonium acetate. The 1 ml solution was centrifuged again at 25,000 × g for another 1.5 h at 4°C and pellet resuspended in 1 ml of 0.1 M of ammonium acetate.
Transmission Electron Microscopy
Phage lysates were dialyzed for 20 min against SM-buffer buffer (20 mM Tris-HCl [pH 7.2], 10 mM NaCl, 20 mM MgSO4). After 15-min adsorption to ultrathin carbon films, 10-min fixation with 1% glutaraldehyde and negative staining with 1% (wt/vol) uranyl acetate, transmission electron microscopy was performed at an accelerating voltage of 80 kV (Tecnai 10; FEI Thermo Fisher Scientific, Eindhoven, The Netherlands). Micrographs were captured with a MegaView G2 CCD camera (Emsis, Muenster, Germany).
Phage DNA Extraction
DNA was extracted from phage P61 using the method adapted from Casey et al. (2015). Briefly, 1 μl of RNAse (Thermo Fisher Scientific, Leicestershire, UK) (1 mg/ml) and 12 U/μl of DNAse I (Roche Diagnostics, Mannheim, Germany) (6 μl of 2,000 U/ml) were added to 1 ml of P61 lysate (1 × 109 PFU/ml) and incubated at 37°C for 30 min. The sample was then centrifuged at 18,407 g for 5 min and the supernatant was transferred to a fresh 1.5 ml sterile Eppendorf tube. Lysis buffer (100 μl) (0.5 M Tris-HCl pH9, 0.25 M EDTA, 2.5% SDS) was then added to the sample, vortexed and incubated at 65°C for 5 min. One hundred twenty-five microliter of 8 M potassium acetate (Sigma-Aldrich, Poole, UK) was then added and mixed by inverting the tube 4–6 times. The sample was placed at −20°C for 15 min. Following this, the samples were centrifuged at 18,407 g for 5 min and the supernatant was aliquoted equally into two fresh Eppendorf tubes. The sample was extracted twice with phenol choloroform-isoamyl alcohol (Sigma-Aldrich). The top layer was again placed into a fresh Eppendorf tube where an equal volume of isopropanol (Sigma-Aldrich) was added and mixed by inverting. The sample was placed at −20°C for 40 min. The sample was centrifuged at 18,407 g for 5 min and the isopropanol was removed. The pellet was washed twice with 70% ethanol (Scharlau, Barcelona, Spain) and left to dry at 37°C. Twenty microliter of dsH2O was added to the pellet and was incubated for 30 min at 37°C. DNA was quantified using the Invitrogen Qubit 4 fluorometer (Thermo Fisher Scientific, Renfrew, UK) using the protocol outlined by the manufacturer, and visualized on a 0.8% agarose gel.
Genome Sequencing and Comparative Genomics
The whole genome sequence of phage P61 was elucidated using the Illumina MiSeq Next Generation Sequencing platform at Microbes NG (Birmingham Research Park, UK). Prior to assembly, the trimmed reads were passed through FLASH (Fast Length Adjustment of Short reads) to eliminate overlapping paired end reads. The genome of P61 was assembled using the SeqMan NGen application of DNAStar Lasergene Genomics Suite (DNAStar, Inc., USA). Once assembled, the genome was uploaded onto the RAST server for annotation. Annotations were confirmed through analysis using BLASTp (https://blast.ncbi.nlm.nih.gov/), HMMER (http://hmmer.org/), and Artemis (https://www.sanger.ac.uk/science/tools/artemis). The genome sequence of P61 has been uploaded to the NCBI database with the accession no: MT438761 and the illumina paired reads have been uploaded onto SRA NCBI database reference PRJNA599330.
Determination of the Efficacy of Phage P61 for the Reduction of L. monocytogenes in Milk
Pasteurized milk (3.5% fat) was purchased from a local retail outlet and was screened for the presence of L. monocytogenes and Listeria phages prior to commencing the trial. An overnight culture of L. monocytogenes strain 3053 was diluted 1:5 in TSB and incubated for 3 h at 25°C. Cells were then diluted in maximum recovery diluent (Oxoid Ltd.) to the desired cell numbers. Target cell numbers in the milk were 103 CFU/ml. 18.5 mM CaCl2 (Merck) was also added to the milk. Phage P61 was propagated using the protocol outlined above to a final concentration of 2 × 109 PFU/ml. The phage was added to the test samples at a concentration of 2 × 106 PFU/ml. Samples were then stored at 12 and 8°C for a total of 7 days (shelf-life of milk). Bacterial viable counts (CFU/ml) and phage counts (PFU/ml) were determined immediately after phage addition (T0) and subsequently at days 1, 3, 5, and 7. For enumeration of L. monocytogenes, samples were serially diluted and plated onto Listeria Chromogenic Agar. Plates were incubated at 37°C for 48 h. For enumeration of P61, samples were diluted, and plaque assays performed using the protocol outlined above. Technical and biological replicates were performed in duplicate for this experiment.
Determination of the Efficacy of Phage P61 for the Reduction of L. monocytogenes in Baby Spinach
L. monocytogenes strain 3053 and phage P61 were prepared following the protocol described above. An overnight culture of strain 3053 which had been grown at 25°C was diluted 1:5 in TSB at 25°C for 3 h and the optical density determined at 600 nm and cells were diluted to desired cell numbers. Target cell numbers in spinach were 2 × 103 CFU/g and the inoculum volume was 500 μl. Baby spinach was purchased from a local retail outlet and weighed out into 10 g portions and placed into sterile bags. Bacteria and phages were then separately applied by pipetting onto the surface of the baby spinach and shaking the bag vigorously for 30 s. L. monocytogenes was allowed to acclimatize to the spinach samples for ~1 h prior to the addition of phages. To the samples to be treated with phages, 500 μl aliquots of P61 were added to achieve a concentration of ~2 × 108 PFU/g of spinach. Samples were then stored at 25, 12, and 8°C for a total of 6 days (shelf life of baby spinach). Samples were diluted 1:10 in full Fraser broth (Sigma-Aldrich, St Louis, USA) and homogenized at 230 rpm for 1 min and plated as described above. Technical and biological replicates were performed in duplicate for this experiment. For enumeration of P61, samples were serially diluted to 10−7 and plaque assays performed using the protocol outlined above. Technical and biological replicates were performed in duplicate for this experiment.
Results and Discussion
Phage P61 Infects Six Serotypes of L. monocytogenes
The host range of phage P61 was determined by performing efficiency of plaquing assays against a series of L. monocytogenes strains. The strains selected for analysis represent isolates from a range of environmental niches, sample types and were of nine different serogroups (Table 1). P61 was shown to be capable of infecting 6 of the 9 serogroups of L. monocytogenes tested (1/2a, 1/2b, 1/2c, 4b, 4e, and 6a). An EOP value of 0.56 was obtained for a single strain of L. marthii that was tested (S4120) indicating that phage P61 can infect another species of Listeria. Phage P61 was unable to infect strains of serotype 4ab (Listeria innocua), 4a/c and 7. Further testing revealed that P61 infected all three L. monocytogenes strains tested belonging to the serotypes 1/2a and 1/2b, but only infected one out of two strains of the serotype 1/2c and 4b, and three out of four strains of serotype 4e. Strains tested are representatives from a given serotype, there is considerable variation between strains within a serotype, accounting for the differences shown in susceptibility to phage P61 here. Vongkamjan et al. (2012), isolated 114 Listeria phages from two dairy farms and tested the host range of these phages against 13 strains representing 9 major serotypes. This revealed that 12.3% of phage isolates showed a narrow host range and 28.9% of phages were broad host range phages (Vongkamjan et al., 2012). Vongkamjan showed that the broad host range phages in this study infected 84.6% of strains tested while the narrow host range phages infected between 7.7% and 38.5% of strains tested. In this study 17 strains representing 9 major serotypes were used to test the host range of P61 and 64.5% of strains tested were susceptible to phage infection. P61 has a limited host range in comparison to broad host range Listeria phages that have been characterized (Loessner et al., 1994; Carlton et al., 2005; Vongkamjan et al., 2017) and can infect a wider range of serotypes than narrow host range Listeria phages that have been characterized (Casey et al., 2015; Sumrall et al., 2019). Thus, P61 cannot be defined as a narrow or broad host range phage but has a moderate host range.
General Characteristics of the Genome of Phage P61
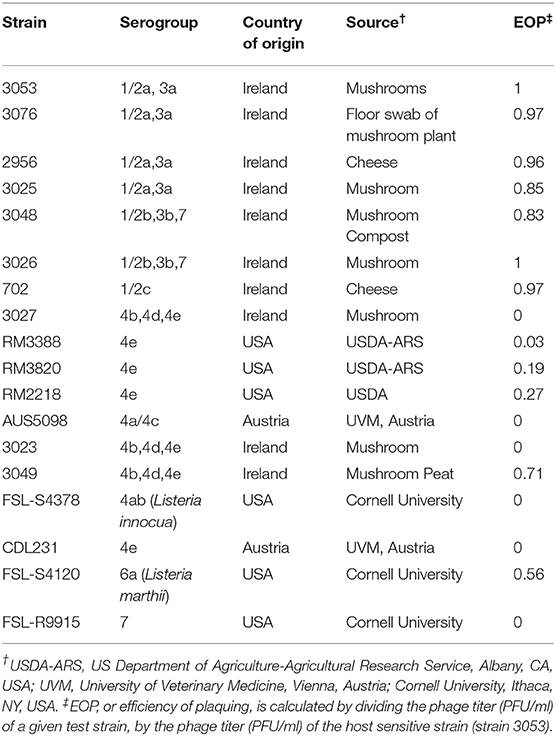
The assembled genome of P61 presented here is a draft assembly with collasped terminal repeat regions positioned approximately at 10,000–13,700 bp. Figure 1 shows the terminal repeat regions of phage A511 mapped to the draft assembly of phage P61 indicating the position of the collasped terminal repeats in the genome of phage P61. This draft assembly of P61 has a genome size of 136,485 bp and a G +C content of 35.92%, which correlates well with values for other Listeria phages, e.g., the Herelleviridae LP-125 (135,281 bp) with a GC content of 35.9%, the Herelleviridae LP-124 (135,817 bp) with a GC content of 35.9%, and the Herelleviridae LP-048 (133,096 bp) with a GC content of 36.0% (Denes et al., 2014). The total number of reads generated was 291,863 and mean coverage was 880.637x. A total of 192 open reading frames (ORFs) and a cluster of 17 tRNA genes were identified in the draft assembly of phage P61 (Supplementary Table 1). Blastp and Blastn analysis of the nucleotide sequences and amino acid sequences revealed the genome of phage P61 is similar in size and sequence to that of the Listeria phage P100 (131,384 bp) which encodes 174 gene products and 18 tRNAs (Carlton et al., 2005) and that of A511 (134,494 bp), encoding 190 gene products and 16 tRNAs. The absence of genes or homologs related to the lysogenic life cycle, e.g., integrases, excisionases, the two repressor proteins Cro repressor and cI repressor (Ohlendorf et al., 1998), attachment sites, in the genome of phage P61 suggests that it follows a strictly lytic lifecycle, a vital characteristic if phages are to be exploited as a biocontrol agent to avoid integration of phage DNA in the host cell genome (Casjens and Hendrix, 2015). Genes playing a role in host cell lysis were also identified. ORF 61 appears to encode the endolysin of phage P61, an N-acetylmuramoyl-L-alanine amidase which shares 100% sequence similarity with the N-acetylmuramoyl-L-alanine amidase of Listeria phage LP-125 (Denes et al., 2014).
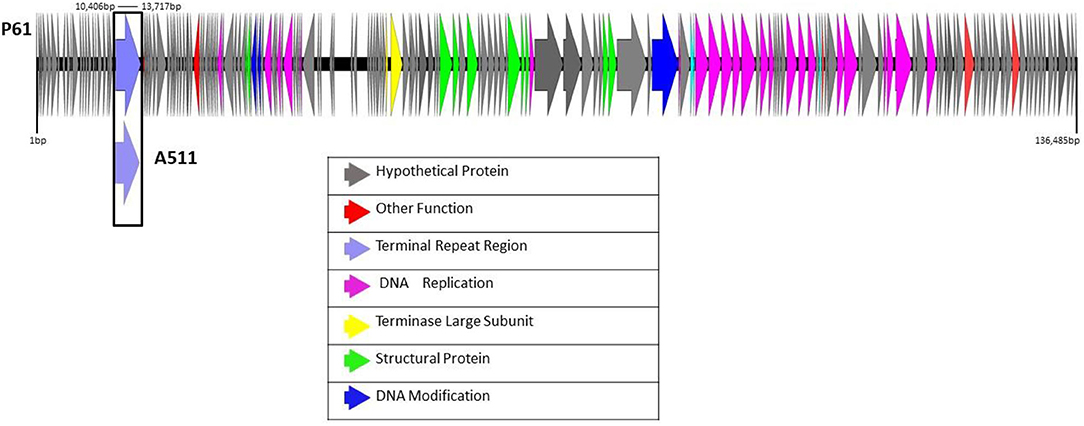
Figure 1. Linear view of the annotated draft genome of phage P61 visualized using EasyFig 2.2.4. The area 10,406–13,717 bp is marked with a box to indicate where the terminal repeat region of phage A511 maps to the draft genome of phage P61.
Transmission EM Confirms Phage P61 as a Member of the Family Herelleviridae
Electron microscopy analysis was performed to further characterize phage P61 in terms of morphology (Figure 2). Measurements of 12 phage P61 particles was shown to possess an isometric capsid of 85.5 ± 2.5 nm in diameter, with a thin collar structure evident beneath the capsid and blackberry-like surface structures on the capsid surface. A tail length of 218.5 ± 2.7 nm and width 22.4 ± 1.0 nm was measured. Thin tail fibers extending from the upper region of the baseplate structure were observed. A double disc baseplate structure was observed, with the baseplate height measured at 30.8 ± 2.3 nm and width of 50.7 ± 3.7 nm, with the collar shown to be 6.4 ± 0.4 nm (n = 7) in height and 13.8 ± 0.8 nm (n = 7) in width. The baseplate of phage P61 is complex with conformational changes occuring when the tail contracts. The baseplate appendages seem to be highly flexible, either with a brush-like appearance or forming globular structures. Morphologically, phage P61 is similar to A511 and P100 (Klumpp et al., 2008).
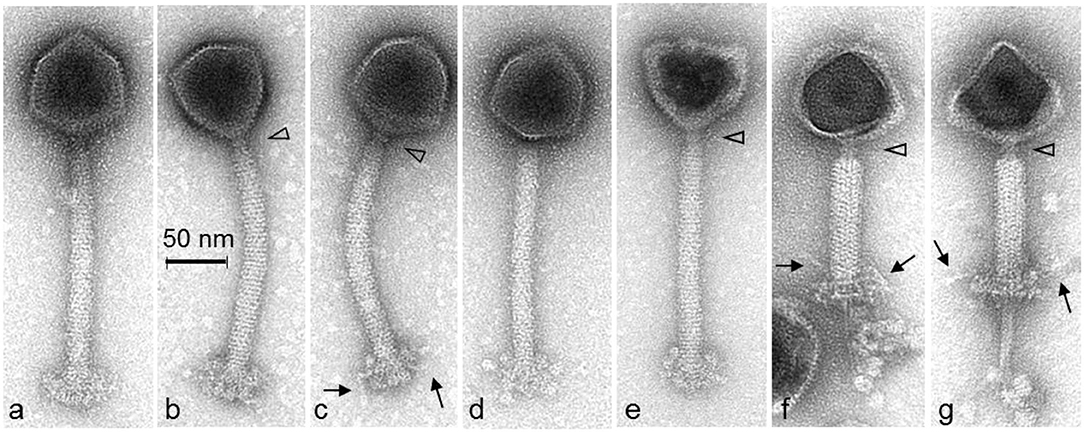
Figure 2. Transmission electron micrographs of L. monocytogenes phage P61 stained with 1% (w/v) uranyl acetate. The open triangles indicate the thin collar structure beneath the capsids. The blackberry-like surface structure of the capsid surface is visible on intact phage particles (a–d). The arrows show thin tail fibers extending from the upper region of the baseplate structure of intact (c) or contracted phage particles (f,g). The double disc structure of the baseplate is visible on phage particles with contracted tail sheaths and damaged capsids (f,g). The baseplate appendages seem to be highly flexible with a brush-like appearance (a,b) or forming globular structures (d,e).
One-Step Growth Curve and Bacteriophage Insensitive Mutant Frequency
To determine the infection kinetics of phage P61, a one-step growth curve was performed on the host strain 3053 (Figure 3). Following 30 min of incubation, 99.99% of phage particles adsorbed to the bacterial host cell. Having a high percentage of adsorption suggests that there may be a strong concentration of available host bacterial cell receptors and phage P61 has a strong affinity for these receptors. The burst size of phage P61 was calculated to be 11.03. The eclipse period, defined as the time interval between viral penetration and the production of progeny virions, was calculated as 40–50 min, and the latent period, defined as the time taken for the infected cell to lyse post-infection, was calculated as 60–70 min. The frequency of BIMs against host strain 3053 was calculated to be 4.36 × 10−5 (n = 24).
Figure 3. One-step growth curve of phage P61. Blue circles represent samples that were directly plated, representing the cumulative concentration of absorbed phages and unabsorbed phages. Red triangles represent samples that were treated with chloroform prior to plating, representing the total concentration of phages including intracellular phage. All values are means of three independent experiments.
Phage P61 Reduces L. monocytogenes Numbers in Liquid Milk and Baby Spinach
Phage P61 was evaluated as a potential biocontrol agent for the reduction of L. monocytogenes in both liquid (pasteurized milk) and solid food matrices (baby spinach). Preliminary experiments in TSB at 8 and 12°C showed that the application of phage P61 at an MOI of 100 resulted in inhibition of the growth of L. monocytogenes strain 3053 at both temperatures, with counts of L. monocytogenes reaching 2 × 108 CFU/ml in untreated samples vs. 1 × 105 CFU/ml in phage-treated samples at 8°C, and 2 × 109 CFU/ml in untreated samples vs. 3 × 108 CFU/ml in phage-treated samples at 12°C (Figure 4). Experiments at 8 and 12°C were the conducted using pasteurized milk (purchased at a local retail outlet) where samples were spiked with L. monocytogenes strain 3053 at a concentration of 1 x 103 CFU/ml, following which phage P61 was added at an MOI of 10,000. At 12°C, a significant decrease (p = 0.05) in CFU/ml of L. monocytogenes strain 3053 was observed compared to the phage free control (Figure 5), with a reduction in L. monocytogenes counts of log 2.36 (99.57% reduction) in treated vs. untreated samples at day 7. When stored at 8°C a reduction of 1.2 log (93.75% reduction) was observed at day 7 (shelf-life of milk) (p = 0.0173, Figure 5). Prior to these experiments it was shown that P61 survives well in the absence of L. monocytogenes with the inoculated concentration of P61 staying consistent throughout the 7 days.
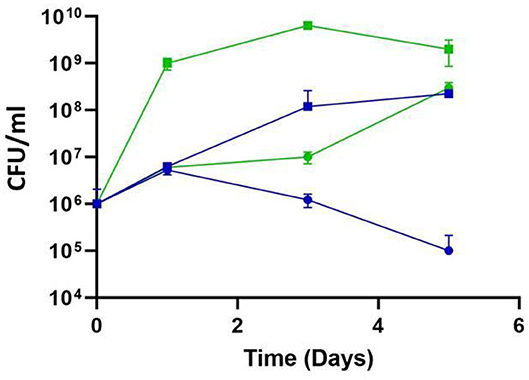
Figure 4. Impact of temperature on the efficacy of phage P61 infection in TSB. L. monocytogenes cells were added to TSB broth at a final concentration of 106 CFU/ml and phage added at 108 PFU/ml (MOI = 100). Samples were incubated at 8°C (blue symbols) and 12°C (green symbols) and L. monocytogenes enumerated daily. Squares represent untreated samples; circles represent phage-treated samples.
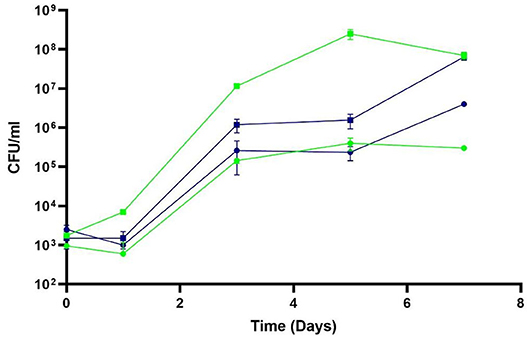
Figure 5. Efficacy of phage P61 against deliberately-inoculated L. monocytogenes strain 3,053 in pasteurized liquid milk at 8 and 12°C. Samples were incubated at 8°C (blue symbols) and 12°C (green symbols) and L. monocytogenes enumerated daily. Squares represent untreated samples; circles represent phage-treated samples. Results are presented as mean values, and error bars represent standard deviations of the means.
A similar experiment was conducted with baby spinach artificially inoculated with L. monocytogenes strain 3053 and treated with phage P61 at an MOI of 10,000. However, the results were not promising, with no significant difference observed between the untreated and phage-treated samples during the trial, other than at a single time point on day 3 where a reduction of 0.8 log (p = 0.0034) was observed (data not shown). Further investigation revealed that L. monocytogenes strain 3053 was not actively growing on the surface of the spinach, thus potentially hindering the phage action. A number of strains were tested on the spinach surface and L. monocytogenes strain 702 was chosen for further analysis as this strain was found to grow well on spinach and was also shown to be susceptible to phage P61 (Table 1). The experiment on spinach was repeated with strain 702 and on this occasion, the L. monocytogenes was left to acclimatize to the surface of the spinach prior to the application of phage P61 (MOI of 100,000). A reduction of 1.93 log (98.78%) was seen on day 5 at 8°C (p = 0.0024), 2.06 log (99.12%) at 12°C (p = 0.002) and 3.3 log (99.95%) at 25°C (p = 0.0083; Figure 6).
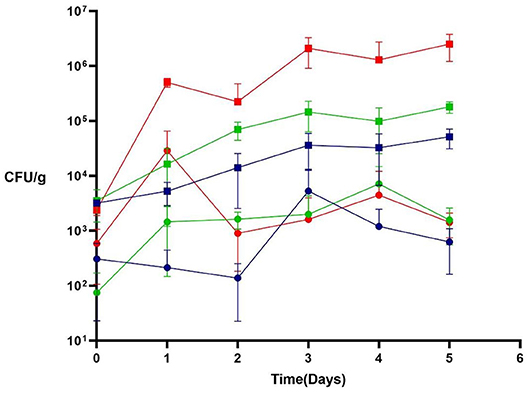
Figure 6. Efficacy of phage P61 against deliberately-inoculated L. monocytogenes strain 702 on baby spinach leaves at 8°C (blue symbols), 12°C (green symbols), and 25°C (red symbols). L. monocytogenes were enumerated daily; squares represent untreated samples; circles represent phage-treated samples. Results are presented as mean values, and error bars represent standard deviations of the means.
Our results suggest that there are a number of determining factors as to whether the application of phage P61 will successfully reduce or inhibit the growth of L. monocytogenes. These include the food matrix, the storage temperature, the stage of growth of the bacterial pathogen and the MOI. In addition to liquid milk and baby spinach, we had previously tested a number of other semi-solid and solid food products, including smoked salmon, hummus and cream cheese. Phage P61 did not inhibit or reduce the growth of L. monocytogenes in these matrices, despite the fact that the organism grew well and the phage survived well in these environments (data not shown). Marcó et al. (2010) suggest that the diffusion of phages could be hindered or favored depending on the structure and composition of the matrix and the environmental conditions. To exploit phages as a biocontrol agent on different food matrices, the application must be specifically optimized for individual food systems. The storage temperature is also a key determinant of phage activity. In our study, notable differences were seen when phage P61 was applied at 8 and 12°C in pasteurized milk, and 8, 12, and 25°C on baby spinach. In the case of pasteurized milk, L. monocytogenes counts decreased by 1.20 log in the phage-treated samples stored at 8°C, in comparison to the 2.36 log reduction in the phage-treated samples stored at 12°C after 7 days. There was little difference between the growth of L. monocytogenes at the two temperatures (0.03 log difference), and thus, this may indicate that strain 3053 used in this experiment is less susceptible to phage P61 at lower temperatures in a liquid medium. Tokman et al. (2016), discuss the effect of temperature on the susceptibility of L. monocytogenes to phages. The team highlight that L. monocytogenes can gain physiological refuge from phage infection indicating that adsorption rates of phage can be affected by the physiological state of the host (Tokman et al., 2016). In the case of baby spinach, a reduction of 1.93 logs, 2.06 log, and 3.3 log was seen in phage treated samples at the end of the storage period at 8, 12, and 25°C, respectively. As expected, in samples that were not treated with phages, L. monocytogenes grew best at 25°C and it was at this temperature where the highest reduction in CFU/ml was observed in the phage-treated samples. The increase in the concentration of actively growing host cells may increase the likelihood of phages to come into contact with these cells and begin the infection cycle. Henderson et al. (2019) recently discussed the effect of temperature and surrounding environmental conditions on the efficiency of phage application to reduce the growth of L. monocytogenes in a laboratory cheese model. These authors reported that treatment with phage P100 is more effective when the food is stored at higher temperatures, showing that the average L. monocytogenes counts on phage-treated samples were significantly lower when samples were stored at 22°C as opposed to samples stored at 6 or 14°C (Henderson et al., 2019). This correlates with what we see for the solid matrix tested in our study, baby spinach. Henderson et al. (2019) also concluded that the pH of the food and serotype of L. monocytogenes used influences the ability of phage P100 to reduce the growth of L. monocytogenes. This has also been observed for a number of other foodborne pathogens and their phages in food system experiments (O'Flynn et al., 2004; Hong et al., 2016). In the O'Flynn et al. (2004) study, the authors noted that significant reductions in E. coli cell numbers were obtained at 30 and 37°C; however, the lytic ability of the phage cocktail was greatly reduced at 12°C. Radford et al. (2016) described the efficient production of a high concentration of Listeria phage under refrigeration conditions, a method that may be adapted to optimize the application of phages as a biocontrol on food matrices in future experiments (Radford et al., 2016).
The MOI is also a determining factor as to whether the application of phages as a biocontrol agent will be successful. Tomat et al. (2013) suggested that the application of higher numbers of phages and higher MOI values will result in a greater reduction of the numbers of the foodborne pathogen. Guenther et al. (2009) also report the significance of phage concentration. When applying Listeria phage A511 to hot dogs, chocolate milk, and cabbage, these authors noted that lower doses of phages resulted in a less significant growth suppression of L. monocytogenes, with the higher titer of phage increasing the likelihood of phage making contact with its bacterial host cell (Guenther et al., 2009). Seo et al. (2016) used varying MOI values (1,000–100,000) for the inhibition of E. coli O157:H7 in beef, pork and chicken meat and indicated using a higher MOI value of 100,000 had a greater inhibitory effect compared to lower values used. In these set of experiments using a MOI of 100,000 resulted in greater log reductions of L. monocytogenes than when lower MOIs were used in preliminary studies in broth and pasteurized milk (Seo et al., 2016).
Finally, the presence of actively growing bacterial host cells vs. stationary phase cells seems vital for the application of phage as a biocontrol agent. Abedon (2017) distinguishes between an “active treatment” and a “passive treatment” for phage biocontrol. In an active treatment, phages should actively produce newly formed virions in situ to create sufficient titers to eradicate the bacterial pathogen over reasonable timeframes. By contrast, passive treatment uses phages that are bactericidal but incapable of generating new phage virions during their interactions with the bacterial host cell. It could be argued that having actively growing host cells allows the phage to hijack its host's metabolic mechanisms allowing for successful completion of its lifecycle and release of newly formed phages into the surrounding environment (Abedon, 2017). L. monocytogenes strain 702, used in the second set of experiments, actively grew on the baby spinach leaves in comparison to strain 3053 used in the initial study, which did not grow during the course of the experiment. Phage P61 coming into contact with a growing host cell (strain 702) may have increased the release of newly formed progeny into the environment, thus increasing the infection rate and subsequent reduction in L. monocytogenes numbers.
Concluding Remarks
The work outlined in this study indicates that phage P61 could potentially be exploited for use as a biocontrol agent, particularly in liquid foods, for the inhibition of L. monocytogenes. Findings from this study indicate that the application of phages as a biocontrol agent against foodborne pathogens needs to be optimized for each food matrix used and the environment it is applied to. Temperature, food matrix, environment, and the growth phase of the bacterial host cell are all factors contributing to the success of application. The inhibition may also be affected by the type of application used (spraying, spreading, and pipetting). However, the positive results observed on the spinach matrix show promise for further study on this and other horticultural products, where reports of the occurrence of L. monocytogenes are increasing and where limited options are available for safe, natural antimicrobials for inhibition of L. monocytogenes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ES: original research and writing. AL and HN: research. OM: conceptualization, resourcing, writing, and editing. IG and KC: editing. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Teagasc (project ref. MDBY0027). ES was supported by a Teagasc Walsh Scholarship (ref. 2016034).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Lisa Gorski, U.S. Department of Agriculture-Agricultural Research Service, Albany, CA; Prof. Martin Wiedmann, Cornell University, Ithaca, NY; and Prof. Martin Wagner, University of Veterinary Medicine, Vienna, Austria for provision of strains to Teagasc, and Dr. Achim Schmalenberger, University of Limerick, for advice regarding spinach trials. Angela Back (Max Rubner-Institut) is acknowledged for her technical assistance with the electron microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.521645/full#supplementary-material
References
Abedon, S. (2017). Active bacteriophage biocontrol and therapy on sub-millimeter scales towards removal of unwanted bacteria from foods and microbiomes. AIMS Microbiol. 3, 649–688. doi: 10.3934/microbiol.2017.3.649
Adriaenssens, E. M., Sullivan, M. B., Knezevic, P., van Zyl, L. J., Sarkar, B. L., Dutilh, B. E., et al. (2020). Taxonomy of prokaryotic viruses: 2018-2019 update from the ICTV bacterial and archaeal viruses subcommittee. Arch. Virol. 165, 1253–1260. doi: 10.1007/s00705-020-04577-8
Alemayehu, D., Ross, R. P., O'Sullivan, O., Coffey, A., Stanton, C., Fitzgerald, G. F., et al. (2009). Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic lactobacillus paracasei cheese strain. Gene 448, 29–39. doi: 10.1016/j.gene.2009.08.008
Carlton, R. M., Noordman, W. H., Biswas, B., De Meester, E. D., and Loessner, M. J. (2005). Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43, 301–312. doi: 10.1016/j.yrtph.2005.08.005
Casey, A., Jordan, K., Neve, H., Coffey, A., and McAuliffe, O. (2015). A tail of two phages: genomic and functional analysis of listeria monocytogenes phages vB_LmoS_188 and vB_LmoS_293 reveal the receptor-binding proteins involved in host specificity. Front. Microbiol. 6:1107. doi: 10.3389/fmicb.2015.01107
Casjens, S. R., and Hendrix, R. W. (2015). Bacteriophage lambda: early pioneer and still relevant. Virology 479–480, 310–330. doi: 10.1016/j.virol.2015.02.010
Denes, T., Den Bakker, H. C., Tokman, J. I., Guldimann, C., and Wiedmann, M. (2015). Selection and characterization of phage-resistant mutant strains of listeria monocytogenes reveal host genes linked to phage adsorption. Appl. Environ. Microbiol. 81, 4295–4305. doi: 10.1128/AEM.00087-15
Denes, T., Vongkamjan, K., Ackermann, H. W., Moreno Switt, A. I., Wiedmann, M., and den Bakker, H. C. (2014). Comparative genomic and morphological analyses of listeria phages isolated from farm environments. Appl. Environ. Microbiol. 80, 4616–4625. doi: 10.1128/AEM.00720-14
EFSA (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSAJ. 16:5077. doi: 10.2903/j.efsa.2018.5500
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Protect. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Filippov, A. A., Sergueev, K. V., He, Y., Huang, X.-Z., Gnade, B. T., Mueller, A. J., et al. (2011). Bacteriophage-resistant mutants in yersinia pestis: identification of phage receptors and attenuation for mice. PLoS ONE 6:e25486. doi: 10.1371/journal.pone.0025486
Golkar, Z., Bagasra, O., and Gene Pace, D. (2014). Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J. Infect. Dev. Countries 8, 129–136. doi: 10.3855/jidc.3573
Guenther, S., Huwyler, D., Richard, S., and Loessner, M. J. (2009). Virulent bacteriophage for efficient biocontrol of listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 75, 93–100. doi: 10.1128/AEM.01711-08
Henderson, L. O., Cabrera-Villamizar, L. A., Skeens, J., Kent, D., Murphy, S., Wiedmann, M., et al. (2019). Environmental conditions and serotype affect listeria monocytogenes susceptibility to phage treatment in a laboratory cheese model. J. Dairy Sci. 102, 9674–9688. doi: 10.3168/jds.2019-16474
Hong, Y., Schmidt, K., Marks, D., Hatter, S., Marshall, A., Albino, L., and Ebner, P. (2016). Treatment of salmonella-contaminated eggs and pork with a broad-spectrum, single bacteriophage: assessment of efficacy and resistance development. Foodb. Pathog. Dis. 13, 679–688. doi: 10.1089/fpd.2016.2172
Hyman, P., and Abedon, S. T. (2009). Practical methods for determining phage growth parameters. Methods Mol. Biol. 501, 175–202. doi: 10.1007/978-1-60327-164-6_18
Klumpp, J., Dorscht, J., Lurz, R., Bielmann, R., Wieland, M., Zimmer, M., et al. (2008). The terminally redundant, nonpermuted genome of listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190, 5753–5765. doi: 10.1128/JB.00461-08
Klumpp, J., and Loessner, M. J. (2013). Listeria phages. Bacteriophage 3:e26861. doi: 10.4161/bact.26861
Lee, S., Kim, M. G., Lee, H. S., Heo, S., Kwon, M., and Kim, G. B. (2017). Isolation and characterization of listeria phages for control of growth of listeria monocytogenes in milk. Korean J. Food Sci. Anim. Resourc. 37, 320–328. doi: 10.5851/kosfa.2017.37.2.320
Loessner, M. J., Estela, L. A., Zink, R., and Scherer, S. (1994). Taxonomical classification of 20 newly isolated listeria bacteriophages by electron microscopy and protein analysis. Intervirology 37, 31–35. doi: 10.1159/000150353
Marcó, M. B., Reinheimer, J. A., and Quiberoni, A. (2010) Phage adsorption to lactobacillus plantarum: influence of physiological environmental factors. Int. J. Food Microbiol. 138, 270–75. doi: 10.1016/j.ijfoodmicro.2010.01.007
Moye, Z. D., Woolston, J., and Sulakvelidze, A. (2018). Bacteriophage applications for food production and processing. Viruses 10:205. doi: 10.3390/v10040205
Mutlu, N., and Şahin, M. (2018). Isolation and characterization of Listeria Monocytogenes bacteriophages from environmental sources in Kars-Turkey. Pak. Vet. J. 39, 91–95. doi: 10.29261/pakvetj/2018.090
O'Flynn, G., Ross, R. P., Fitzgerald, G. F., and Coffey, A. (2004). Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70, 3417–3424. doi: 10.1128/AEM.70.6.3417-3424.2004
Ohlendorf, D. H., Tronrud, D. E., and Matthews, B. W. (1998). Refined structure of Cro repressor protein from bacteriophage λ suggests both flexibility and plasticity. J. Mol. Biol. 280, 129–136. doi: 10.1006/jmbi.1998.1849
Oliveira, M., Viñas, I., Colàs, P., Anguera, M., Usall, J., and Abadias, M. (2014). Effectiveness of a bacteriophage in reducing Listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiol. 38, 137–142. doi: 10.1016/j.fm.2013.08.018
Perera, M. N., Abuladze, T., Li, M., Woolston, J., and Sulakvelidze, A. (2015). Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 52, 42–48. doi: 10.1016/j.fm.2015.06.006
Radford, D. R., Ahmadi, H., Leon-Velarde, C. G., and Balamurugan, S. (2016). Propagation method for persistent high yield of diverse listeria phages on permissive hosts at refrigeration temperatures. Res. Microbiol. 67, 685–691. doi: 10.1016/j.resmic.2016.05.010
Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., Fernández Escámez, P. S., et al. (2018). Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 16:5134. doi: 10.2903/j.efsa.2018.5134
Savage, R. (2004). Food Safety Magazine. Available online at: https://www.foodsafetymagazine.com/magazine-archive1/december-2003january-2004/current-issues-in-raw-and-rte-products/ (accessed June 23, 2020).
Seo, J., Seo, D. J., Oh, H., Jeon, S. B., Oh, M. H., and Choi, C. (2016). Inhibiting the growth of Escherichia coli O157:H7 in beef, pork, and chicken meat using a bacteriophage. Korean J. Food Sci. Anim. Resour. 36, 186–193. doi: 10.5851/kosfa.2016.36.2.186
Shen, C., Norris, P., Williams, O., Hagan, S., and Li, K. W. (2016). Generation of chlorine by-products in simulated wash water. Food Chem. 190, 97–102. doi: 10.1016/j.foodchem.2015.04.146
Smith, A., Moorhouse, E., Monaghan, J., Taylor, C., and Singleton, I. (2018). Sources and survival of Listeria monocytogenes on fresh, leafy produce. J. Appl. Microbiol. 125, 930–942. doi: 10.1111/jam.14025
Song, Y., Peters, T. L., Bryan, D. W., Hudson, L. K., and Denes, T. G. (2019). Homburgvirus LP-018 has a unique ability to infect phage-resistant Listeria monocytogenes. Viruses 11:1166. doi: 10.3390/v11121166
Sumrall, E. T., Shen, Y., Keller, A. P., Rismondo, J., Pavlou, M., Eugster, M. R., et al. (2019). Phage resistance at the cost of virulence: listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 15:e1008032. doi: 10.1371/journal.ppat.1008032
Taylor, T. M. (2018). Natural food antimicrobials: recent trends in their use, limitations, and opportunities for their applications in food preservation. ACS Symposium Series 1287, 25–43. doi: 10.1021/bk-2018-1287.ch002
Tokman, J. I., Kent, D. J., Wiedmann, M., and Denes, T. (2016). Temperature significantly affects the plaquing and adsorption efficiencies of listeria phages. Front. Microbiol. 7:631. doi: 10.3389/fmicb.2016.00631
Tomat, D., Migliore, L., Aquili, V., Quiberoni, A., and Balagué, C. (2013). Phage biocontrol of enteropathogenic and shiga toxin-producing Escherichia coli in meat products. Front. Cell. Infect. Microbiol. 3:20. doi: 10.3389/fcimb.2013.00020
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/CMR.14.3.584-640.2001
Vongkamjan, K., Benjakul, S., Kim Vu, H. T., and Vuddhakul, V. (2017). Longitudinal monitoring of listeria monocytogenes and listeria phages in seafood processing environments in Thailand. Food Microbiol. 66, 11–19. doi: 10.1016/j.fm.2017.03.014
Vongkamjan, K., Switt, A. M., den Bakker, H. C., Fortes, E. D., and Wiedmann, M. (2012). Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol, 78, 8666–8675. doi: 10.1128/AEM.01859-12
World Health Organization (2020). Food Safety. Available online at: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed June 29, 2020).
Keywords: bacteriophage, Listeria monocytogenes, biocontrol, broad-host range, food safety
Citation: Stone E, Lhomet A, Neve H, Grant IR, Campbell K and McAuliffe O (2020) Isolation and Characterization of Listeria monocytogenes Phage vB_LmoH_P61, a Phage With Biocontrol Potential on Different Food Matrices. Front. Sustain. Food Syst. 4:521645. doi: 10.3389/fsufs.2020.521645
Received: 19 December 2019; Accepted: 29 September 2020;
Published: 05 November 2020.
Edited by:
Avelino Alvarez-Ordóñez, Universidad de León, SpainReviewed by:
Pilar García, Consejo Superior de Investigaciones Científicas (CSIC), SpainThomas Denes, The University of Tennessee, Knoxville, United States
Copyright © 2020 Stone, Lhomet, Neve, Grant, Campbell and McAuliffe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivia McAuliffe, b2xpdmlhLm1jYXVsaWZmZUB0ZWFnYXNjLmll
 Edel Stone
Edel Stone Antoine Lhomet1,3
Antoine Lhomet1,3 Horst Neve
Horst Neve Irene R. Grant
Irene R. Grant Katrina Campbell
Katrina Campbell Olivia McAuliffe
Olivia McAuliffe