- 1Environmental Studies and Research Unit, Department of Microbiology, Faculty of Agriculture, Cairo University, Giza, Egypt
- 2Algorithms in Bioinformatics, Center for Bioinformatics, University of Tübingen, Tübingen, Germany
- 3Department of Horticultural Genetics and Biotechnology, Mediterranean Agronomic Institute of Chania, Chania, Greece
- 4Department of Biotechnology, Hungarian University of Life Science, Gödöllő, Hungary
- 5Department of Microbiology, Faculty of Agriculture & Natural Resources, Aswan University, Aswan, Egypt
- 6Department of Botany, Faculty of Agriculture, Cairo University, Giza, Egypt
- 7Institute for National and International Plant Health, Julius Kühn-Institute – Federal Research Centre for Cultivated Plants, Braunschweig, Germany
- 8Department of Plant Microbe Interactions, Leibniz Institute of Vegetable and Ornamental Crops, Großbeeren, Germany
Plant microbiota have co-evolved with their associated plants in the entire holobiont, and their assemblages support diversity and productivity on our planet. Of importance is in vitro cultivation and identification of their hub taxa for possible core microbiome modification. Recently, we introduced the in situ-similis culturing strategy, based on the use of plant leaves as a platform for in vitro growth of plant microbiota. Here, the strategy is further extended by exploring plant organ compatible cultivation of plant microbiota when grown on corresponding leaf/root-based culture media. Pooling the advantages of MPN enrichment methodology together with natural plant-only-based culture media, the introduced method efficiently constructed a nutritional milieu governed by vegan nutrients of plant origin, i.e., leaf strips/root segments, immersed in plain semi-solid water agar. MPN estimates exceeded log 7.0 and 4.0 g−1 of endo-rhizosphere and endo-phyllosphere, respectively, of maize and sunflower; being proportionate to those obtained for standard culture media. With sunflower, PCR-DGGE analyses indicated divergence in community composition of cultivable endophytes primarily attributed to culture media, signaling a certain degree of plant organ affinity/compatibility. Based on 16S rRNA gene sequencing of bacterial isolates, 20 genera comprising 32 potential species were enriched; belonged to Bacteroidetes, Firmicutes, and Alpha-/Gammaproteobacteria. The described cultivation strategy furnished diversified nutritive platform in terms of homologous/heterologous plant organ-based medium and ambient/limited oxygenic cultivation procedure. Duly, cultivability extended to > 8 genera: Bosea, Brevundimonas, Chitinophaga, Pseudoxanthomonas, Sphingobacterium Caulobacter, Scandinavium, and Starkeya; the latter three genera were not yet reported for Sunflower, and possible unknown species or even one new putative genus. Thus, both potential members of the major microbiome and rare isolates of satellite microbiomes can be isolated using the presented method. It is a feasible addition to traditional cultivation methods to explore new potential resources of PGPB for future biotechnological applications.
Introduction
The plant microbiome comprises highly associated keystone microbial taxa, the core microbiome, being essential for the fitness of the plant holobiont (Toju et al., 2018; Compant et al., 2019). Similar to other environmental microbiomes, the majority of its members fails to grow under in vitro laboratory conditions and still under shadow (Libby and Silver, 2019). They are, therefore, often considered as the “microbial dark matter” (Lok, 2015). Especially, the low abundant taxa, satellite/rare taxa, that are increasingly recognized as contributors to community stability by acting as a reservoir that can rapidly respond to environmental changes, are often missed in isolation studies (Shade et al., 2014).
A major breakthrough in exploring environmental microbiota has been achieved with the development of DNA sequencing technologies, i.e., culture-independent techniques, and big data handling and analysis that have revolutionized our capacity to realize the complexity of microbial diversity and function. However, the absence of cultured representatives of many lineages hinders the ability to discover the roles of numerous lineages of Bacteria and Archaea present in different environments and to study how they interact with each other (Nichols et al., 2008; Stewart, 2012; Cross et al., 2019). Therefore, recent studies have revived extensive isolation efforts combined with genome sequencing and phenotypic characterization (Bai et al., 2015; Mauchline et al., 2015; Levy et al., 2018). And, several techniques have been proposed for bacterial cultivation by employing genomic data to understand either microbial interactions (network-directed targeted bacterial isolation) or ecosystem engineering [reverse genomics] (Salam et al., 2020). Such reverse-genomics, genome-informed antibody engineering, enabled cultivation of target specific groups of as yet-uncultured microbes, especially those present at low abundance or have no selectable phenotypes (Cross et al., 2019).
The reasons behind microbial “uncultivability” are numerous, and have been associated with a requirement for factors produced by other microbes, strict interspecies interactions, slow growth, competition/inhibition and dormancy (Cross et al., 2019). Here, extensive efforts are devoted to improve the cultivability of different environmental microbiota based on simulating the in situ environments to include less abundant but functional members. This included the optimization of substrate compositions and concentrations, gelling agents, incubation time, cell density, trace additives of syntrophic growth factors, as well as signaling molecules. Successfully, such efforts improved cultivation of rarely isolated bacterial phyla (Nichols et al., 2008, 2010). Further, high-throughput methods were innovated to improve cultivation capabilities and recover not yet cultivated microorganisms; e.g., diffusion chamber (Kaeberlein et al., 2002; Bollmann et al., 2007), gel microdroplet-based microfluidic systems (Manome et al., 2001; Zengler et al., 2002), microfluidic streak plates (Jiang et al., 2016), combined with a number of micro-devices (Ingham et al., 2005, 2007). These tools allowed the recovery and characterization of new ecologically important isolates (Morris et al., 2002; Zengler et al., 2002; Ferrari et al., 2005; Nichols et al., 2008).
The paradigm shift in the study of the human gut microbiome happened through the application of multiple culture conditions (microbial culturomics). Such strategy involves the use of multiple isolation culture media, a range of culture conditions, prolonged incubation and rapid identification of developed colonies with MALDI-TOF MS and/or 16S rRNA gene sequencing (Lagier et al., 2016). Taking into consideration that the combinations of all culture media used throughout such strategy are basically “compatible to the humans/animals” in respect of composition, being bovine-based broth cultures together with various supplements of blood, rumen fluid, stool extract. Along that path, the plant-only-based culture media, based on juices, saps, homogenates and/or dehydrated powders of tested host plants were developed to be more compliant with the plant microbiome culturability (Nour et al., 2012; Sarhan et al., 2016; Youssef et al., 2016). In fact, such plant-only-based culture media proved to be competitive to replace several synthetic culture media, and to increase the cultivability of the plant microbiota. With the aid of culture-dependent and independent techniques, we were able to enrich previously not yet cultured bacteria (Sarhan et al., 2016, 2018, 2020; Hegazi et al., 2017; Nemr et al., 2020). Further, we later introduced the intact leaf-based culturing strategy that allowed the direct inoculation and cultivation of microorganisms on the leaf surfaces in the form of culture pads, following the rationale of “natural environments envelop multiplex of nutrients necessary for the growth of their inherent microbiota” (Nemr et al., 2020). Here, majority of plant nutrients are supplied to the growing microorganisms in their natural/proportionate concentrations and gradients, (Nemr et al., 2020). Exemplarily for sunflower (Helianthus annuus L.) such strategy, named “leaf in situ similis cultivation,” allowed the recovery of microorganisms reluctant for cultivation and extended the cultivable diversity from the endo-phyllosphere and the endo-rhizosphere to members of Firmicutes, Proteobacteria and Actinobacteria. As well, culturing strains of genera not commonly reported for sunflower, e.g., Aureimonas, Sphingomonas, Paracoccus, Kosakonia, and Erwinia, was achieved using such in situ similis cultivation technique (Nemr et al., 2020). However, there are still numerous genera (158), which already were detected using 16S rRNA metagenomic sequencing techniques in sunflower, but were not yet cultured in vitro (Tamošiune et al., 2020).
The “Most Probable Number, MPN” method, based on the use of liquid growth medium, is commonly used and recommended to enrich and estimate the concentration of viable microorganisms in environmental samples (Sutton, 2010; http://www.microbiologynetwork.com/content/jgxp_v14n4_most-probable-number-method-use-in-qc-microbiology.pdf; https://microbeonline.com/probable-number-mpn-test-principle-procedure-results). Although it is laborious and requires longer times for incubation, the method is effective in enrichment and detection of low abundant organisms, especially those present in highly turbid/particulate samples, e.g., sediments, sludge, mud, etc., that cannot be analyzed by plate count and/or membrane filtration. In principle, the replicated dilution series of samples in selective enrichment liquid broth are prepared, and then growth is confirmed by conventional phenotypic and biochemical assays. It is reported and recommended in literature (Sutton, 2010; Russo et al., 2014) that MPN enrichment can be followed by plating microorganisms from positive tubes on selective agar plates and subsequent biochemical and molecular biology assays for isolates genera/species identification. Often the procedure is subjected to improvement in respect of developing new methods of growth measurements as well as novel selective media suitable for target organisms. Lately, MPN was successfully integrated with qPCR, in order to considerably reduce the time required for growth confirmation and detection of pathogens, e.g., L. monocytogenes and E. coli O157:H7 with lower concentration of as low as 1 CFU g−1 in the corresponding selective media (Russo et al., 2014). As well to improve cultivability and performance of the method, variety of liquid media were developed for specific environments, where source material is included in the enumeration/isolation culture media, e.g., celluloses, activated sludge, marine sediments, and samples from the oil drilling industry, clarified rumen, soil extract and bacterial extracts (Mah and Smith, 1981; Wais, 1988; Tanner, 1989; Wirth and Wolf, 1990; Vester and Ingvorsen, 1998). Extracts from blood, serum, animal tissues, fecal material, sewage sludge supernatant, and clay particles were used as supplements to promote growth of microorganisms with fastidious and/or unknown requirements (Laanbroek and Geerligs, 1983). The principle of using natural media is expected to improve MPN enumeration of microorganisms in diverse ecosystems, especially those physio types which can be detected by chemical and molecular methods. So far, and to our knowledge, plant materials have not yet been used for MPN cultivation and enrichment of plant endophytic bacteria.
The major objective of the present study is to take advantage of the merits of the MPN method to apply our approach of “in situ similis cultivation” and to enrich the cultivable community of endophytic bacteria residing in the endo-phyllosphere and endo-rhizosphere compartments of sunflower plants. The developed MPN culturing strategy is based on leaf strips and/or root segments of tested host plants, as sole sources of nutrients, immersed in semi-solid water agar tubes. It is postulated that such semi-solid set up is creating compatible gradients of natural nutrients and gas phases in close proximity and along the immersed plant tissues to satisfy growth and enrich a wide array of plant microbiota. Furthermore, we aim to explore affinity of homologous/heterologous cultivation of plant endopytes on respective plant organs, i.e., preference of development of endophytes of eno-phyllosphere on homologous leaf strips-based culture media and of endo-rhizosphere on homologous root segments-based culture media. MPN enrichment was followed by subjecting positive tubes to: (a) derive MPN estimates, (b) analyze composition/diversity of the culturable bacterial community by Denaturing Gradient Gel Electrophoresis (DGGE), (c) isolate on agar plates/semi-solid tubes of respective plant/organ culture media representative isolates, followed by subsequent 16S rRNA gene sequencing for identification.
Materials and Methods
Experimental Approach and Design
We postulated that plant leaves and root segments immersed in plain semi-solid water agar deliver compatible plant nutrients in their factual diversity, complexity and concentrations, and create a natural milieu for enrichment of tested bacterial plant endophytes. For this purpose, a number of preliminary experiments were carried out to initially assess the suitability of leaf strips of a number of host plants in the constructed semi-solid MPN tubes to support in vitro growth of corresponding plant endophytes. The developed macroscopic and microscopic growth was monitored and evaluated. Followed, two main experiments were conducted to test: (a) MPN enrichment and estimation of endophytes of maize plants on the expense of diffused nutrients of plant leaves compared to the standard culture media of R2A and CCM, and (b) the compatibility of leaf strips compared to root segments to support cross cultivation of homologous/heterologous endo-phyllosphere and endo-rhizosphere bacterial communities of sunflower plants: growth was measured in terms of population densities, MPN estimates, and community composition of the cultivable sunflower microbiome as measured by PCR-DGGE analysis and by 16S rRNA gene sequencing of representative isolates (Figure 1).
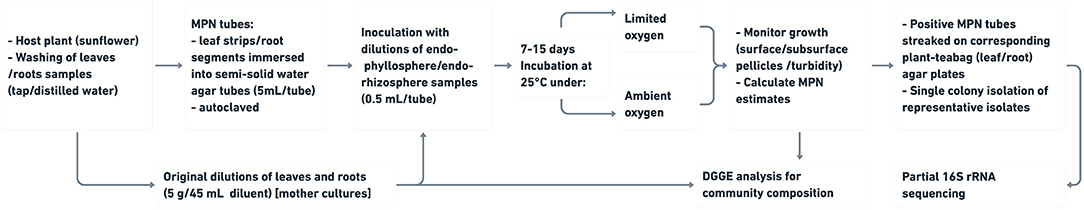
Figure 1. Workflow of compatible plant organ (leaf strips/root segments)-culturing strategy of bacterial endophytes residing endo-phyllosphere and endo-rhizosphere of plants.
Plant Materials and Sampling
Preliminary experiments tested the host plants of sunflower, maize, berseem clover, faba bean, lupine and mango. Extended experiments focused on maize (Zea mays L., cv. Cairo-1) and sunflower (Helianthus annus L., Giza landrace 13). All tested host plants were grown in the experimental fields of the Faculty of Agriculture, Cairo University, Giza, Egypt (30.0131°N, 31.2089°E), and sampled at the stage of 50% flowering. For sampling, three healthy plants were randomly-sampled, where the vegetative parts were first collected, and then the root systems (intact roots with closely adhering soil) were carefully uprooted. Samples were transferred to the lab and stored at 4°C until microbiological analysis was conducted within 24 h after sampling.
Culture Media
Plant-Teabag Culture Medium
The plant powder teabags were prepared according to Sarhan et al. (2016), with concentration of 0.5 g L−1 dehydrated powder prepared from the tested plant leaves/roots. For convenience, such plant teabag culture media were particularly used in the form of: a) agar plates (2% agar-agar, w/v) for single colony isolation of representative isolates enriched in positive MPN culture tubes, and b) semi-solid agar tubes (1.8 g agar-agar L−1) for further subculturing of secured isolates.
Standard Chemically-Synthetic Culture Media
The standard culture media of R2A (Reasoner and Geldreich, 1985), and N-deficient combined carbon sources medium, CCM (Hegazi et al., 1998), were prepared as previously described by Elsawey et al. (2020). Agar was added to prepare semi-solid agar tubes (1.8 g agar-agar L−1) and agar culture media (2% w/v). The pH of both culture media was adjusted to 7.2, and then diluted to 1:10 (w/v) and half strength, respectively.
Preparation of Semi-solid Culture Media Based on Leaf Strips and Root Segments
Fresh plant leaves/roots were carefully washed with tap and distilled water; then leaves were cut into strips (1 cm × 6 cm), while intact tap roots with adhering secondary roots were cut into segments of 6 cm length. Leaf strips (one piece) or root segments (2–3 segments) were transferred into each of test tubes containing 5 mL semi-solid (1.8 g agar-agar L−1) water agar. Then, tubes after being sterilized were inoculated with the prepared endo-rhizosphere and endo-phyllosphere samples (s. below Endo-Rhizosphere/Endo-Phyllosphere Sample Preparation and MPN-Culturing for the Recovery of Bacterial Endophytes). The present culturing method depends on the sole use of strips/segments of leaves/roots to prepare natural plant semi-solid culture media to enrich and enumerate endophytes using the MPN method (Figure 1).
All prepared culture media were autoclaved for 20 min at 121°C, and then kept overnight at 25°C for sterility check before use.
Incubation Conditions at Ambient and Limited Oxygen Conditions
For MPN estimation, two sets of MPN culture tubes were prepared for incubation at 25°C, under either ambient oxygen or limited oxygen conditions. Under ambient oxygen conditions, MPN tubes with conventional cotton plugs were incubated in traditional incubators, with ambient O2 concentration, ca. 20.95%. While for limited/deficient oxygen conditions, under aseptic conditions, cotton plugs of MPN tubes were replaced by sterilized suba-seal rubber plugs after inoculation, and then the overhead gas phase inside the tubes was replaced with nitrogen gas. Next, MPN tubes were kept in tight glass jar, with a diameter 9.5 cm and height 22.5 cm, fitted with two glass valves at the top to allow in/out gas exchange. Once a week, the gas phase inside the jars was flushed/replaced with nitrogen gas to maintain limited oxygen conditions within the jar. A candle was placed in the jar to ensure that upon flame extinction, oxygen is exhausted.
Endo-Rhizosphere/Endo-Phyllosphere Sample Preparation and MPN-Culturing for the Recovery of Bacterial Endophytes
Samples of plant leaves and roots, of maize and sunflower plants, were surface sterilized. Aliquots of 5 g of fresh, full grown and healthy leaves were carefully washed with tap and distilled water, surface-sterilized with ethanol (70% for 1 min), sodium hypochlorite (3% for 5 min) and ethanol (70% for 1 min) and then carefully washed with sterilized distilled water (de Oliveira Costa et al., 2012; Jackson et al., 2013). For roots, 5 g of almost intact roots (taproot with adhering secondary roots) were surface- sterilized with 95% ethanol for 5–10 s; followed by 3% sodium hypochlorite for 30 min and then carefully washed with sterilized distilled water (Youssef et al., 2004).
Original suspensions, named mother culture, of both endo-phyllosphere and endo-rhizosphere, were prepared under sterile conditions by crushing ca. 5 g surface-sterilized leaves or roots in a Waring blender with ca. 45 mL of basal salts of CCM as diluent. Further, 10-fold serial dilutions were prepared in the same diluent, then dry weights (at 70°C) were determined for suspended leaves and roots (Youssef et al., 2016). Aliquots of 0.5 mL of each of suitable dilutions (10−2:10−5) of plant endo-rhizosphere and (10−1:10−4) of endo-phyllosphere samples were used to inoculate the above prepared semi-solid MPN tubes of leaf strips and root segments. Two sets of MPN tubes of either plant spheres were prepared, and incubated under ambient oxygen and limited oxygen conditions. Macroscopic growth, in terms of bacterial surface/subsurface pellicles and/or developed turbidity, and microscopic observation of growing cells, was monitored throughout 7–15 days of incubation at 25°C. MPN estimates were derived using Cochran's Table (Cochran, 1950). To assure sterility of culture media used as well as to obtain appropriate growth index required for MPN estimation, it is imperative to obtain no-growth in a number of MPN culture tubes of the inoculated higher dilutions.
Positive MPN culture tubes of sunflower plants were subjected to further plating on corresponding plant agar plates for the purpose of single colony isolation of representative isolates. After 7–5 days of incubation under ambient and limited oxygen conditions, respectively, aliquots (20 μL) from homogenized growth in representative positive MPN tubes were surface-plated on agar culture media prepared from corresponding sunflower leaves or roots-based culture media. Then, agar plates were incubated for 7–15 days under corresponding incubation conditions. For limited oxygen conditions, agar plates were kept in tight plastic box, 25 × 37 × 18 cm (16.65 L capacity), fitted with two valves at the top/bottom to allow in/out gas exchange. Every 2 days, the gas phase inside the box was flushed/replaced with nitrogen gas to create limited oxygen conditions within the box. A candle was placed in the box to ensure that upon flame extinction, oxygen is exhausted. More than 1,068 colonies were randomly picked to represent all CFUs proportional to their abundance for further sub-culturing on corresponding plant agar plates, and isolates recovered were subjected to 16S rRNA gene analysis (Supplementary Table 1).
DNA Extraction and 16S rRNA Gene Analysis of Bacterial Isolates
The genomic DNA of successively-subcultured isolates (Supplementary Table 1) was extracted using the QIAGEN DNeasy Plant Mini Kit (Qiagen Inc., Hilden, Germany) according to the manufacturer's instructions (Sarhan et al., 2016). The 16S rRNA genes were amplified with the forward primer “9bfm” (GAGTTTGATYHTGGCTCAG) and reverse primer “1512R” (ACGGHTACCTTGTTACGACTT) (Mühling et al., 2008). The PCR setup in a 25 μL volume was as follows: QIAGEN TopTaq Master Mix Kit 12.5 μL, PCR water 5.5 μL, primer 9 bfm (3.1 pmol μL−1) 2.5 μL, primer 1512R (3.1 pmol μL−1) 2.5 μL, and target DNA (ca. 15 ng μL−1) 2.0 μL. The amplification of DNA was performed according to the thermal amplification cycling program: 4 min initial denaturing at 96°C, 30 thermal cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, and extension at 74°C for 1.5 min. The PCR products were purified using PCR purification kit (Qiagen Inc.,), concentration adjusted to 10 ng μL−1 and submitted for sequencing to Eurofins MWG Operon (Ebersberg, Germany).
DGGE Analysis
DNA Extraction From MPN Tubes and Root/Shoot Mother Cultures
After 7–15 days of respective ambient and limited oxygen incubation, all surface/sub-surface pellicles developed in 3 MPN positive tubes, as replicates, representing the lowest dilution of endo-phyllosphere (10−1) and endo-rhizosphere (10−2) of all tested culture media, were individually harvested in 2.0 mL Eppendorf tubes. The pellicles' harvest, as well as samples of previously prepared original leaves/root suspensions (mother cultures; in three replicates), were centrifuged at 13,000 rpm for 10 min. DNA was extracted from the resulting pellets using the genomic DNA Extraction Mini Kit (iNtRON Biotechnology, Kyungki-Do, Korea). DNA quality was checked using the BioDrop μLITE spectrophotometer (Biochrom, Holliston, MA, USA).
Amplification of the 16S rRNA Gene and DGGE Fingerprinting
The protocols for 16S rRNA gene amplification and nested PCR of the V3-16S rRNA gene by using the 341f-GC (CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGG-CCTACGGGAGGCAGCAG) and 518r (ATTACCGCGGCTGCTGG) primers (Muyzer et al., 1993; Mühling et al., 2008), adopted by Sarhan et al. (2016), were used. 16S rRNA gene amplification from DNA extracted, either from the pellicles' harvest or original leaves/root suspensions, mother cultures, was performed with the Hightech Thermocycler (SensoQuest, Göttingen, Germany).
DGGE was performed using the VS20WAVE-DGGE (Cleaver Scientific Ltd, Warwickshire, UK). Aliquots of 10 μL of each sample were mixed with 3 μL of a 6X loading dye (glycerin, xylene cyanol, bromophenol blue), then heated at 95°C for 5 min, and stored at 65°C until loading. Amplicons were electrophoresed on an 8% acrylamide gel containing a 30 to 70% denaturing gradient of formamide and urea with 1 × TAE buffer. After 3 min of initial migration at 200 V to push the sample into the gel, DGGE was conducted at 60°C for 20 h at 50 V. The gel was stained for 30 min with the 6X ethidium bromide, photographed, and analyzed for DGGE band profiles with the MicroDOC System with UV Transilluminator (Cleaver Scientific Ltd, Warwickshire, UK). A self-created standard of mixed PCR products from 4 pure bacterial strains (Arthrobacter globiformis DSM 20124 [GC content 62%], Bifidobacterium breve DSM 20213 [GC content 58.8%], Pectobacterium carotovorum DSM 30168 [GC content 49%], Lactobacillus plantarum DSM 20174 [GC content 44.3%]) was included in every DGGE run (Supplementary Figure 4). All of these strains were obtained from the “Deutsche Sammlung von Mikroorganismen und Zellkulturen” (DSMZ, dsmz.de) and revived according to the provider's instructions.
Phylogenetic Analysis
For phylogenetic analyses, the 16S rRNA gene sequences were taxonomically assigned by comparison with those available in GenBank using BlastN (Altschul et al., 1990). The 16S rRNA gene sequences of the isolates and their closely/nearest related strains were aligned together with respective type strain (T) sequences by Clustal Omega version 1.2.4 (Madeira et al., 2019). The alignment was trimmed with trimAl version 1.4.rev22 (-gt 0.8/0.7 -st 0.001 -cons 70) (Capella-Gutiérrez et al., 2009). Phylogenetic trees were constructed by using the Maximum Likelihood method under the GTRCAT model, implemented in RAxML (Stamatakis, 2014). Bootstrapping was performed on 1,000 replicates, and the inferred tree was saved in Newick format and visualized with iTol (itol.embl.de) (Letunic and Bork, 2007).
Statistical and Numerical Analyses
The analysis of variance (ANOVA, Tukey-test) and the honestly significant differences (HSD) were calculated using MSTAT-C software (Michigan State University, MI, United States). The R-package “ggplot2” was used for the construction of barplots.
The band profiling of DGGE gels was analyzed using CLIQS (TotalLab, Newcastle upon Tyne, UK) and binary data were exported and further used for α-diversity indices calculations using PAST v.3.22 (Hammer et al., 2001).
Results
Semi-solid Culture Media Based on Leaf Strips of Various Host Plants Equipped in vitro Enrichment of Plant Endophytes
Preliminary experiments demonstrated that nutrients available in leaf strips of a variety of host plants immersed in plain semi-solid water agar are sufficient enough to support and enrich in vitro growth of plant endophytes. The semi solid tubes prepared from leaf strips of berseem-clover, faba bean; lupine and mango created rich nutritional matrix that supported profuse growth of plant endophytes. Microscopic examination of developed surface/subsurface pellicles resolved diverse cell morphologies, e.g., short rods, cocci cells, long rods, spore-forming bacilli, free spores and yeast cells (Supplementary Figure 1).
Comparable to Standard R2A and CCM, Leaf Strips-Based Culture Media Supported Good Growth of Bacterial Endophytes of Maize Endo-Phyllosphere
Growth/enrichment of bacterial entophytes of maize endophyllosphere was assayed in semi-solid agar tubes prepared from standard culture media, CCM and R2A, and strips of both young and old leaves of maize plants. Upon incubation, the visual/macroscopic and confirmed microscopic observation of bacterial growth was monitored in the prepared semi-solid culture tubes. It appeared that leaf strips furnished a rich nutritional milieu that supported dense MPN estimates of bacterial endophytes of maize endo-phyllosphere, being > log 7.0 g−1 leaf and very much proportionate to those derived for standard R2A and CCM culture media (Table 1; Supplementary Figure 2). Strips of young leaves were relatively more nutritive, and significantly supported higher MPN estimates compared to old leaves.

Table 1. MPN estimates of bacterial endophytes of endo-phyllosphere of maize as developed on semi-solid culture media of leaf-strip-based culture media and diluted standard CCM and R2A culture media: One-way ANOVA analysis for culture media effect.
Not Only Leaf Strips but Also Root Segments Promoted in vitro Cultivation of Bacterial Endophytes of Sunflower Plant Compartments
With sunflower plants, preliminary tests indicated that not only leaf strips but also root segments supported good growth of bacterial endophytes of endo-phyllosphere and endo-rhizosphere. Microscopic examination confirmed the presence of dense and diverse cell morphologies, e.g., short rods, long rods, spore-forming bacilli, free spores and cocci cells (Supplementary Figure 3). Therefore, experimentation was enlarged to include semi-solid culture media based on both leaf strips and root segments, as well as incubation under ambient and limited oxygen conditions.
The most probable number (MPN) method was used to estimate the population densities of bacteria in both compartments of the sunflower endo-rhizosphere and endo-phyllosphere. MPN estimates were based on visual and microscopic observation/confirmation of bacterial growth developed in semi-solid agar culture media based on leaf strips and root segments under ambient or limited oxygen conditions. In general, bacterial populations of either plant compartments amply enriched in MPN culture tubes based on both plant leaf strips and root segments.
Under ambient oxygen atmosphere, significant differences were attributed to the single effects of plant spheres and culture media. Higher MPN estimates were reported for the endo-rhizosphere (6.9 log MPN estimates g−1) compared to the endo-phyllosphere (only 3.8 log MPN estimates g−1) (Table 2). Notably, the root segments-based culture medium supported somewhat lower bacterial growth (5.3 log MPN estimates g−1) in comparison to the leaf strips-based culture medium (5.4 log MPN estimates g−1). Of interest is that higher MPN estimates were achieved for homologous vs. heterologous culturing; i.e., endo-rhizosphere bacteria favored culture media based on root segments with 7.2 log MPN estimates per g root related to only 6.7 log MPN estimates g−1 when grown on leaf strips-based culture medium. Likewise, endo-phyllosphere bacteria preferred the culture medium based on leaf strips with 4.1 log MPN estimates g−1 contrast to 3.5 log MPN estimates g−1 on root segments-based culture medium (Table 2).
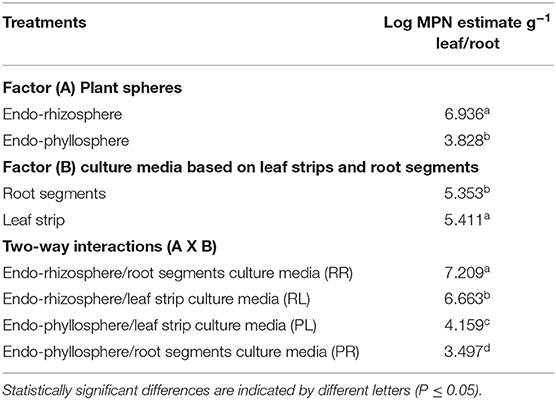
Table 2. MPN estimates of endo-rhizosphere and endo-phyllosphere bacteria of sunflower plants as developed on leaf strips/root segments-based semi-solid culture media, under ambient oxygen conditions: two-way ANOVA analysis.
Under limited oxygen conditions, the effects of plant spheres and culture media were not as distinctive as under ambient oxygen conditions. But, likewise, the endo-rhizosphere MPN estimates were higher compared to those of the endo-phyllosphere (5.9 and 5.2 log MPN estimates g−1, respectively). The culture medium effect was not significant (Table 3). Compared to heterologous cultivation, the homologous cultivation on corresponding leaf strips/root segments culture media were more advantageous; however, differences did not approach significant levels.
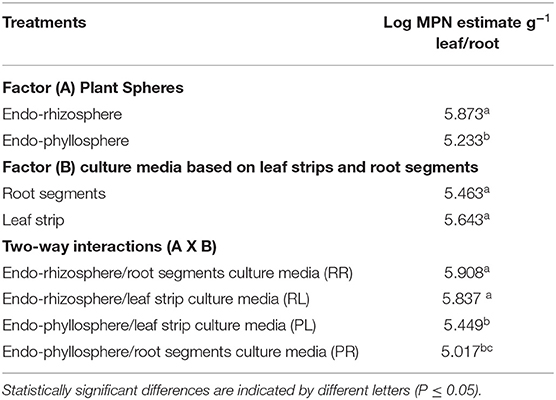
Table 3. MPN estimates of bacteria of endo-rhizosphere/endo-phyllosphere of sunflower plants as developed on leaf-strips/root segments- based semi-solid culture media, under limited oxygen conditions: two-way ANOVA analysis.
DGGE Analysis Indicated Divergence in Community Composition of Cultivable Bacterial Endophytes of Sunflower Attributed to Homologous/Heterologous Cultivation
PCR-DGGE fingerprinting of the 16S rRNA gene segment recovered from in vitro growth developed in MPN culture tubes, representing all combinations of tested culture media and growth conditions, was used to compare the composition of enriched cultivable endophytic communities, in both endo-rhizosphere and endo-phyllosphere of sunflower. We primarily aimed at checking possible differential enrichment of endophytic bacteria of endo-phyllosphere when developed in homologous leaf strips-based culture media compared to the heterologous root strips-based culture media, and vice versa. Included in the analysis as well were the genomic 16S rRNA samples that were extracted from the initial root and leaf suspensions, originally prepared for MPN culturing. DGGE analysis resulted in clear banding patterns (Supplementary Figure 4), and cluster analyses of produced DGGE bands of bacterial endophytes of both plant compartments are illustrated in Figures 2 and 3, respectively.
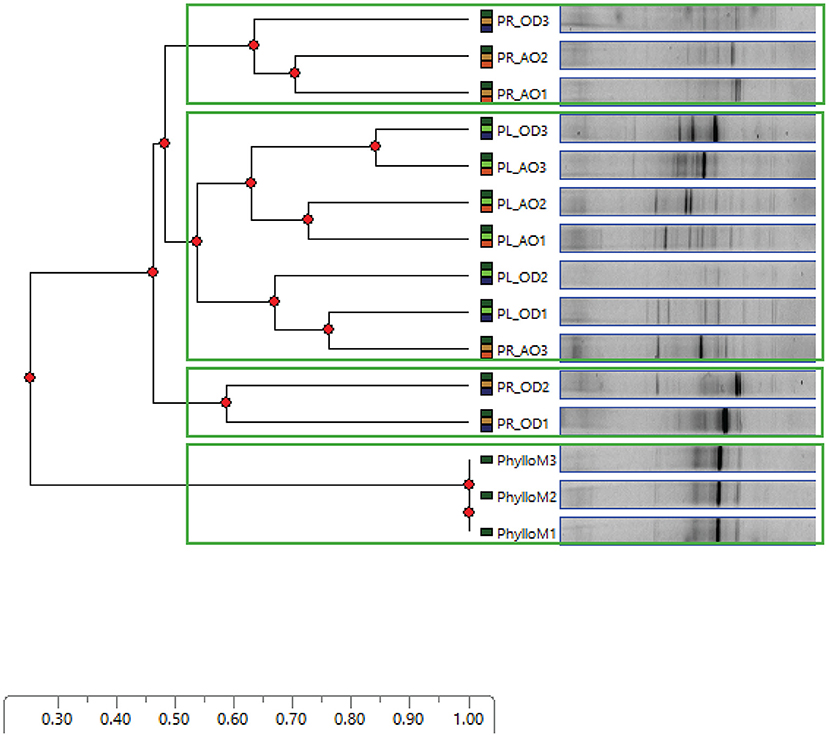
Figure 2. DGGE bands obtained for sunflower endo-phyllosphere bacterial community, developed in MPN culture tubes of leaf strips (PL;  ) or root segments-based (PR;
) or root segments-based (PR;  ) culture media; UPGMA cluster analysis based on Euclidean distances of DGGE lanes. Each lane represents the bacterial growth harvested from one MPN culture tube, and each culture medium is represented by three MPN tubes (1–3) incubated under ambient oxygen (AO;
) culture media; UPGMA cluster analysis based on Euclidean distances of DGGE lanes. Each lane represents the bacterial growth harvested from one MPN culture tube, and each culture medium is represented by three MPN tubes (1–3) incubated under ambient oxygen (AO;  ) or limited oxygen (OD;
) or limited oxygen (OD;  ) conditions. PhylloM (
) conditions. PhylloM ( ), DNA extracted from original suspension of plant leaves (1–3).
), DNA extracted from original suspension of plant leaves (1–3).
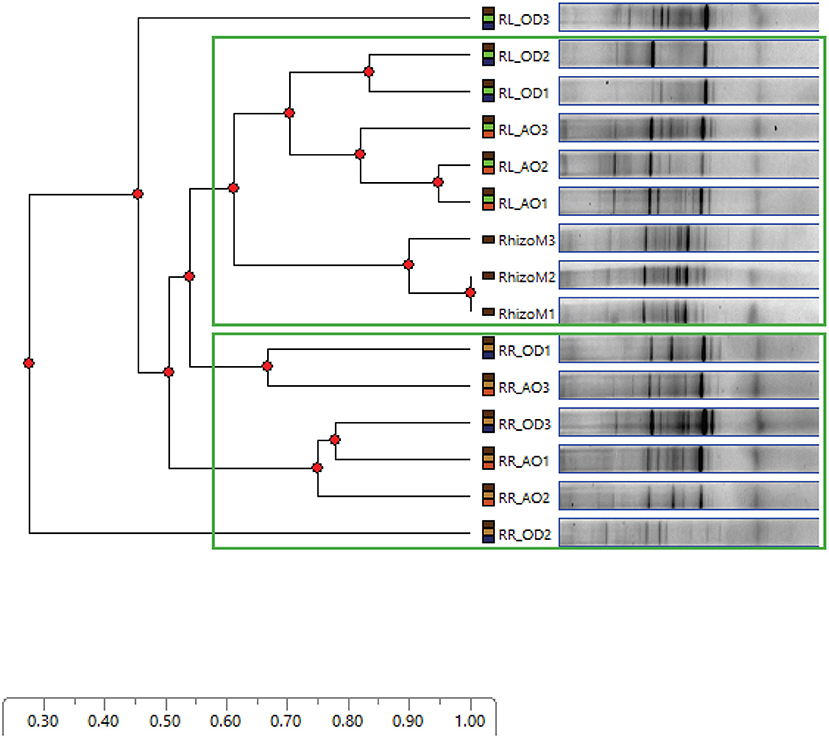
Figure 3. DGGE bands obtained for sunflower endo-rhizosphere bacterial community, developed in MPN culture tubes of root segments (RR;  ) or leaf strips-based (RL;
) or leaf strips-based (RL;  ) culture media; UPGMA cluster analysis based on Euclidean distances of DGGE lanes. Each lane represents the growth harvested from one MPN culture tube, and each culture medium is represented by three MPN tubes (1–3), incubated under ambient oxygen (AO;
) culture media; UPGMA cluster analysis based on Euclidean distances of DGGE lanes. Each lane represents the growth harvested from one MPN culture tube, and each culture medium is represented by three MPN tubes (1–3), incubated under ambient oxygen (AO;  ) or limited oxygen (OD;
) or limited oxygen (OD;  ) conditions. RhizoM (
) conditions. RhizoM ( ), DNA extracted from original suspension of plant roots (1–3).
), DNA extracted from original suspension of plant roots (1–3).
Based on the analysis of distance scores by Phoretix 1D pro software, the constructed dendrogram of endo-phyllosphere is shown in Figure 2. Setting a Cluster Cutoff Value of 0.55 resulted in four different clusters. The original suspension of the leaf material clearly clustered apart from all other samples at earlier similarity level of 0.20. The remaining three clusters divided the MPN culture tubes in regard to the culture medium used. One independent cluster, at similarity level 0.52, contained endo-phyllosphere bacteria developed in the homologous MPN leaf strips-based culture media. While the separated endo-phyllosphere bacteria grown on the heterologous root segments-based culture medium split into two subgroups/clusters. No obvious trend/effect was related to the oxygen status during incubation. These results show a distinct enrichment of endo-phyllospheric bacteria when grown on leaf strips-based compared to root segments-based media.
As to the cultivable bacterial communities of the endo-rhizosphere (Figure 3), and at the same Cluster Cutoff Value of 0.55, two independent clusters were detectable. The first cluster separated endo-rhizosphere communities developed on the MPN culture tubes of homologous root segments away from those of the heterologous leaf strips-based MPN culture tubes. The second cluster contained sub-clusters of the community of DNA extracted from the original suspension of plant roots and of the culturable endo-rhizosphere community grown on leaf strips-based culture medium. Again, no specific separation or pattern was specifically acclaimed to oxygen availability during incubation.
Comparing the band pattern composition and density of the original plant materials to those developed in leaf strips/root segments-based MPN culture tubes, an enrichment of minor abundant groups was visible in the plant- based medium. Bacterial endophytes with higher GC content seem to be notably enriched on the plant- based semi-solid MPN culture tubes prepared for either endo-phyllosphere or the endo-rhizosphere samples.
Leaf Strips/Root Segments Setup Significantly Enriched/Extended Diversity of Bacterial Isolates Representing Culturable Bacterial Endophytes of Sunflower Plants
Due to problems known to be related to domestication of isolates (Nichols et al., 2010), out of 1,068 CFUs randomly selected on all tested culture media and under various incubation conditions, a number of 566 isolates were progressively sub-cultured. And, 163 of which produced good quality sequences (Supplementary Table 1). These isolates represented the cultivable microbiota of endo-rhizosphere (69) and endo-phyllosphere (94) of sunflower plants, being developed under ambient (116) or oxygen limited conditions (47), and enriched on leaf strips and root segments-based culture media (Figures 4, 5; Supplementary Table 2). Over all, the leaf strips/root segments cultivation strategy in terms of combinations of plant compartments, tested culture media and growth conditions create opportunities for the isolation of multiple and diverse bacterial genera of sunflower microbiota (Supplementary Figure 5). In total, such combinations facilitated the cultivability of 20 genera, belonging to the three phyla Bacteroidetes, Firmicutes and Proteobacteria, and comprising 32 potential species (Supplementary Table 2).
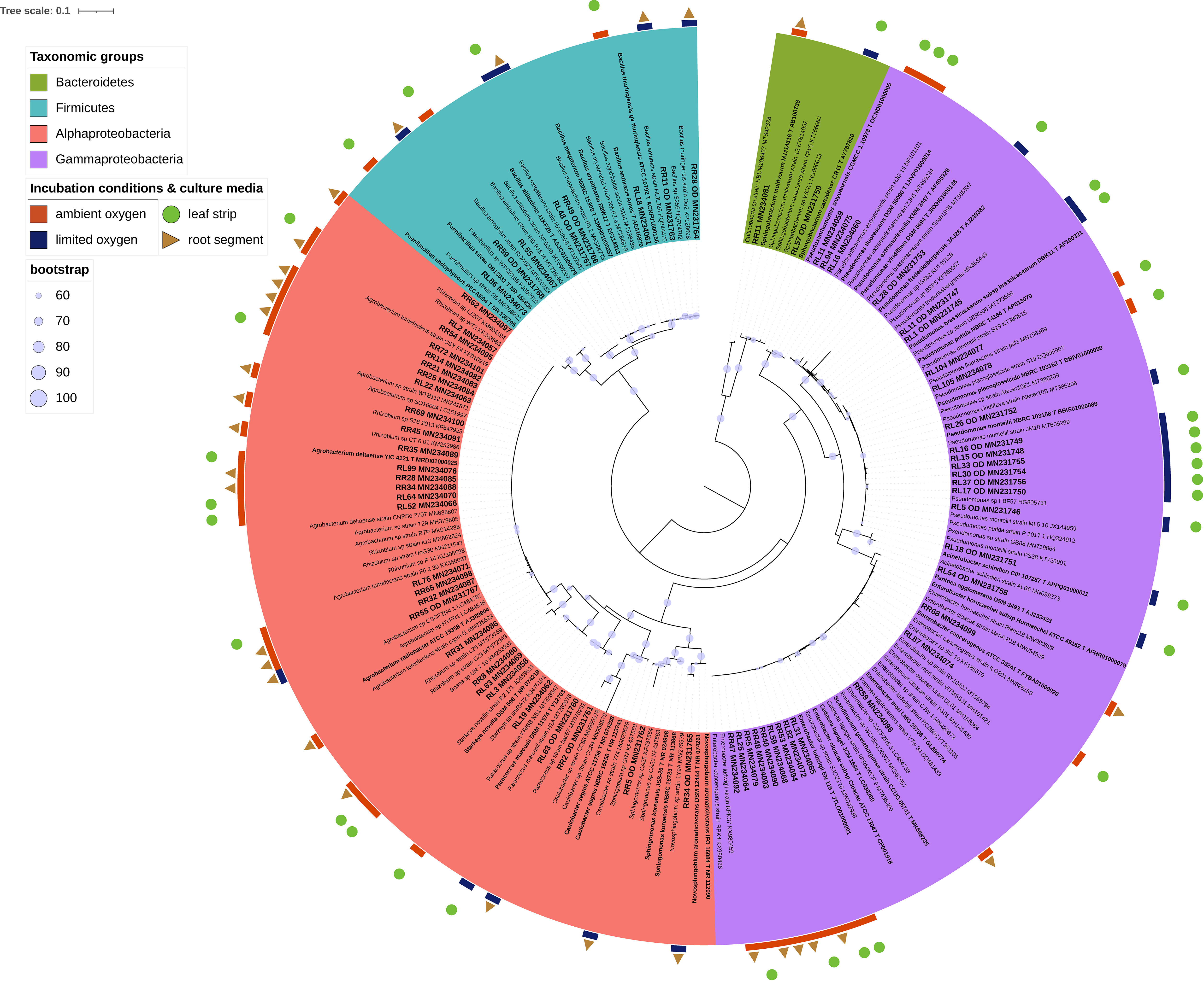
Figure 4. Phylogenetic tree of 16S rRNA gene sequences of the 69 isolates representing endo-rhizosphere of sunflower plants. The colored labels indicate the taxonomic groups (Firmicutes, Alphaproteobacteria, Gammaproteobacteria, Bacteriodetes), incubation conditions (ambient and limited oxygen), and culture media (leaf strips and root segments). Respective isolates identifier followed by their accession number are highlighted in bold and larger font style. In contrast, closely related strains are written in normal style, while type strains are displayed in bold, indicated by a T followed by the accession number. The phylogenetic tree was interfered with the Maximum Likelihood method under the GTRCAT model, using RAxML. The bootstrap values for 1,000 replicates are adorning the branches as cycles.
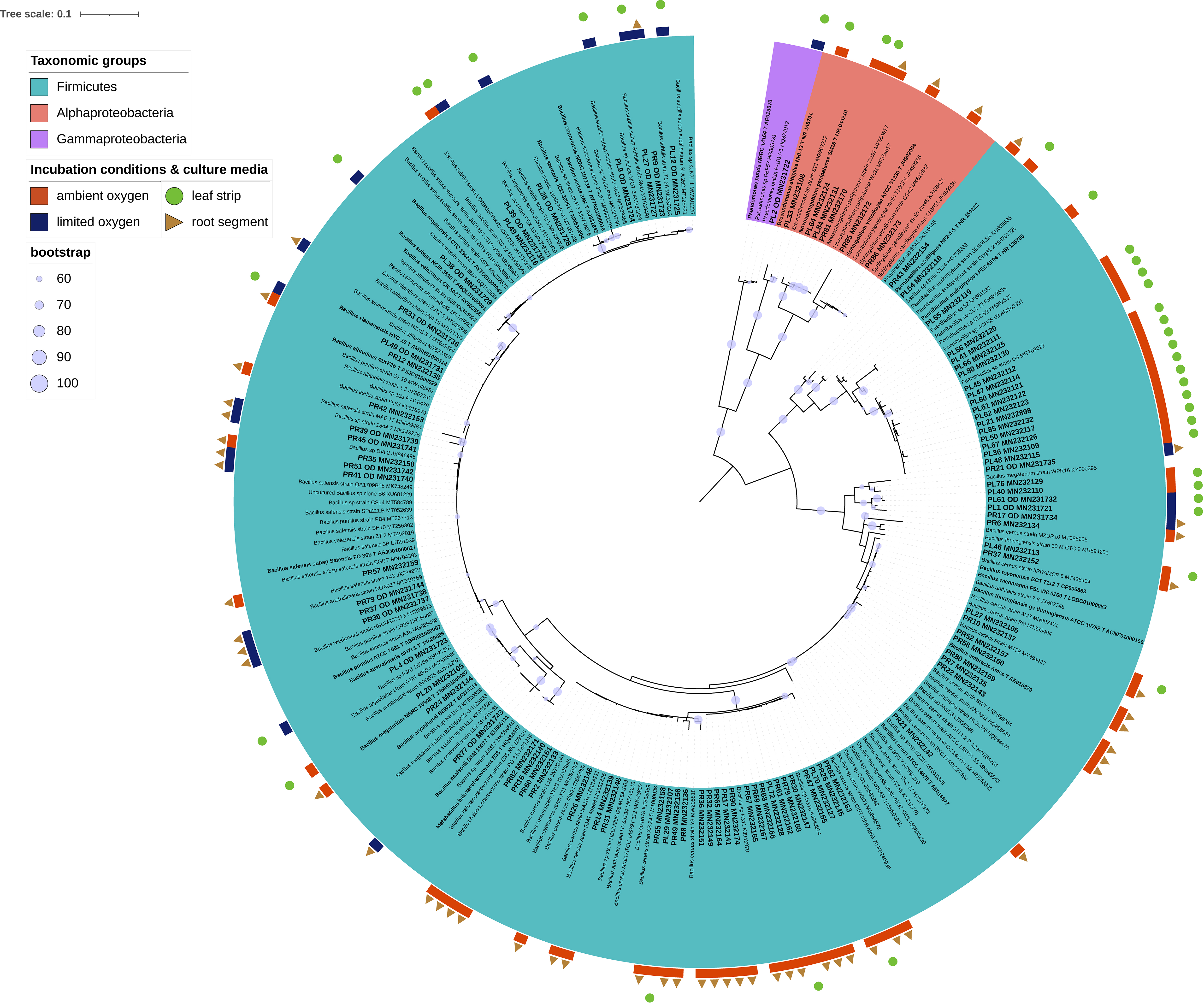
Figure 5. Phylogenetic tree of 16S rRNA gene sequences of the 94 isolates representing endo-phyllosphere of sunflower plants. The colored labels indicate the taxonomic groups (Firmicutes, Alphaproteobacteria, Gammaproteobacteria), incubation conditions (ambient and limited oxygen), and culture media (leaf strips and root segments). Respective isolates identifier followed by their accession number are highlighted in bold and larger font style. In contrast, closely related strains are written in normal style, while type strains are displayed in bold, indicated by a T followed by the accession number. The phylogenetic tree was interfered with the Maximum Likelihood method under the GTRCAT model, using RAxML. The bootstrap values for 1,000 replicates are adorning the branches as cycles.
Of common presence and prevalence was the genus Bacillus with 76 isolates (46.6% of total isolates), with potential species of Bacillus sp. (46 isolates), B. cereus (10), B. anthracis (7), B. subtilis (4), B. safensis (4), B. altitudinis (2), B. thuringiensis (1), B. megaterium (1), and B. pumilus (1). The following genera were Paenibacillus (19 isolates, 11.7%); Rhizobium (18 isolates, 11%); Pseudomonas (15 isolates, 9%) and Enterobacter (8 isolates, 5%) (Supplementary Figure 5). The remaining genera, representing 53.4% of the isolates, were of low incidence, represented with ≤ 4 isolates each, such as Novosphingobium (4), Agrobacterium (3), Pseudoxanthomonas (3), Scandinavium. (3), Starkeya (3), Sphingobium (2), and with single isolate of Acinetobacter, Bosea, Brevundimonas, Caulobacter, Chitinophaga, Paracoccus, Pantoea, Sphingobacterium, and Sphingomonas. Interestingly, the taxonomic and phylogenetic analysis revealed significantly lower identity values for the isolates PR85 (MN232172, Sphingobium sp.) and PR21_OD (MN231735, Bacillus sp.) of 98.85 and 98.49% identity for their partial 16S rRNA gene fragments of 435 and 797 nt length, respectively (Supplementary Table 2). Moreover, isolate PR21_OD showed a unique split that resulted into a long branch, indicating an earlier divergence from the Sphingobium isolates (Figure 5).
Homologous/Heterologous Culturing on Leaf Strips/Root Segments-Based Culture Media of Significant Impact and Signaling Potential Host Organ Compatible Cultivation
Irrespective of culture media and growth conditions, the endo-rhizosphere isolates were of higher diversity and belonged to 18 genera, associated to three different phyla, dominated by Proteobacteria (Alphaproteobacteria), rather than by Firmicutes and Bacteroidetes (Figure 6). In contrast, only 6 genera were recovered from the endo-phyllosphere, which are assigned to two phyla, dominated by Firmicutes more than Proteobacteria (mostly Alphaproteobacteria).
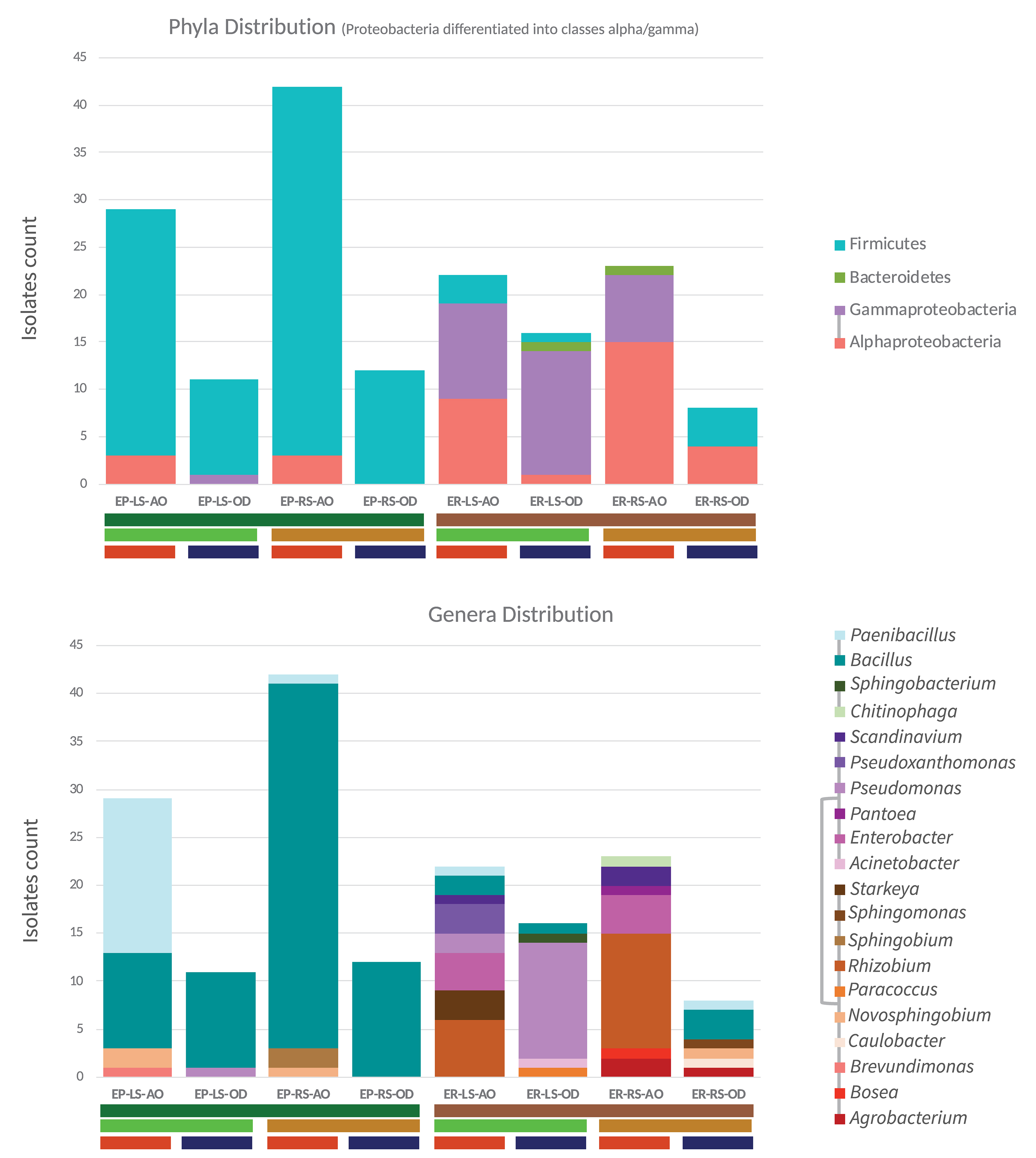
Figure 6. Based on 16S rRNA gene sequences, the taxonomic status of endo-phyllosphere (EP) and endo-rhizosphere (ER) bacterial isolates enriched on leaf strips (LS) and root segments (RS) based culture media under ambient (AO) and limited oxygen (OD) conditions. Respective isolate counts are shown on phylum and genus levels. The phyla affiliation of each genus is indicated by the phyla-specific coloring and connected lines in the legend.
In detail, 50 of the Bacillus isolates from the endo-phyllosphere (in total 70 of 76 isolates) were cultivated on the root segments based medium and among them the majority of the isolates grew under ambient oxygen condition (38/50). And, 17 out of 19 Paenibacillus isolates were obtained from the endo-phyllosphere, being more abundant (16/17) on leaf strips-based medium under ambient oxygen level. Endo-rhizospheric isolates of the genus Rhizobium prevailed under ambient oxygen level in both plant compartment sources with higher presence on root specific media (12/18). Noteworthy, all 15 Pseudomonas isolates, mostly isolated from the endo-rhizosphere (14/15), exhibited preferences for cultivation on leaf strip media only, mostly under limited oxygen conditions (13/15). Enterobacter isolates, all of which isolated from the endo-rhizosphere, obtained exclusively under ambient oxygen and on both leaf strips/root segments culture media.
Regarding the plant spheres, 2 genera of Brevundimonas and Sphingobium were solely isolated from the endo-phyllosphere, while 14 genera were obtained uniquely from the endo-rhizosphere (e.g., Rhizobium, Enterobacter, Agrobacterium, Starkeya, Caulobacter and Scandinavium). Interestingly an overlap of only 4 genera, Bacillus, Paenibacillus, Novosphinobium, and Pseudomonas, was detected for both plant compartments (Figure 7A).
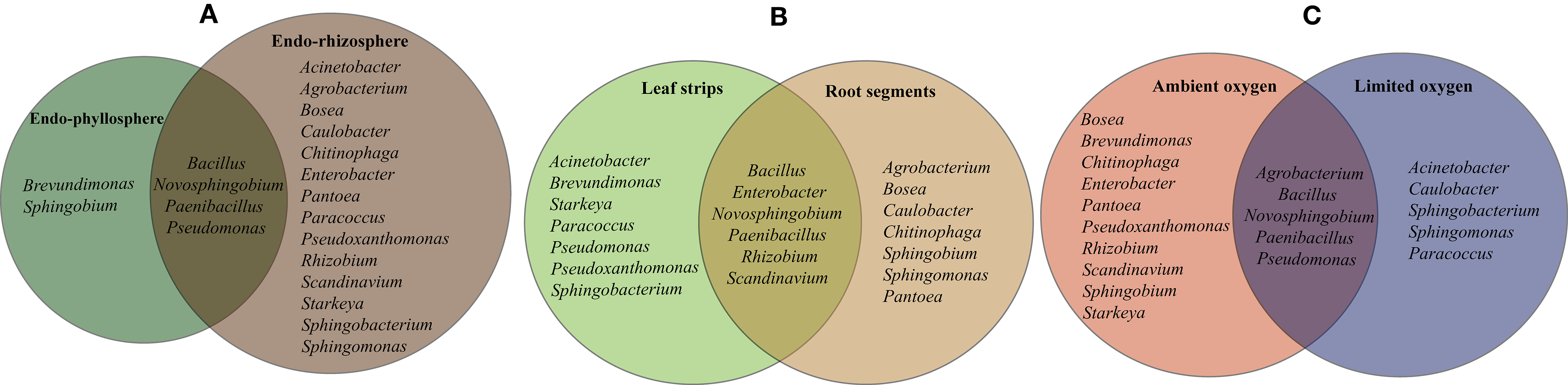
Figure 7. Based on 16S rRNA gene sequences and genera identification, the diversity of sequenced isolates is significantly influenced by the single effect of plant compartments, endo-rhizosphere or endo-phyllosphere (A), culture media based on leaf strips or root segments (B), and incubation conditions under ambient or limited oxygen conditions (C).
Comparing the cultivability on both culture media, a number of bacterial genera were confined to either culture media, where 7 genera were recovered on leaf strips-based culture medium (e. g. Brevundimonas, Pseudoxanthomonas, Starkeya, Sphingobacterium) and 7 genera exclusively on root segments-based medium (e. g. Agrobacterium, Sphingobium, Bosea, Caulobacter, Chitinophaga). The remaining 6 genera, Bacillus, Paenibacillus, Rhizobium, Enterobacter, Novosphingobium, and Scandinavium, exhibited good growth in both culture media (Figure 7B).
Representatives of the cultivable genera varied in their oxygen requirements, where 10 genera favored ambient oxygen (e.g., Rhizobium, Enterobacter, Scandinavium, Starkeya, Pseudoxanthomonas, Bosea, Chitinophaga, Brevundimonas) and 5 genera dominated the limited oxygen conditions, such as Sphingobacterium and Caulobacter among others (Figure 7C). The representative isolates of Bacillus, Paenibacillus, Novosphingobium, Pseudomonas, and Agrobacterium were able to grow and develop under both ambient and limited oxygen conditions.
Characteristically and under ambient oxygen, a number of genera were exclusively developed in the homologous combinations of endo-rhizosphere/root segments (Bosea, Pantoea, and Chitinophaga) and of endo-phyllosphere/leaf strips (Brevundimonas) culture media. Heterologous combinations allowed the growth of representative of a few numbers of genera, e.g., endo-rhizosphere/leaf strips (Starkeya, Pseudoxanthomonas), endo-phyllosphere/root segments (Sphingobium). Under limited oxygen conditions, isolates representing Caulobacter and Sphingomonas were particularly enriched under homologous conditions of endo-rhizosphere/root segments. Among other examples of heterologous combinations are Acinetobacter, Sphingobacterium, and Paracoccus that were enriched from the endo-rhizophere on leaf strips media.
However, we must bear in mind, that representative isolates of the other genera were commonly grown under a variety of combinations of tested growth conditions (Supplementary Figure 6). Exemplarily, under limited oxygen growth conditions, and common to the ambient oxygen conditions, representatives of Bacillus, Paenibacillus, Pseudomonas, Agrobacterium, and Novosphingobium were enriched. Hereby, under ambient conditions, mostly all Paenibacillus isolates were enriched from endo-phyllosphere on leaf strips-media, while phyllospheric Bacillus isolates were more often found on root segments- media (Supplementary Figure 6B). Regarding the Proteobacteria isolates, among 18 Rhizobium isolates that were enriched from endo-rhizosphere under ambient oxygenic conditions, 6 were preferably grown on leaf strips; and out of the 8 rhizobacterial Enterobacter recovered under oxygen availability, 4 isolates were enriched on leaf strips-based media (Supplementary Figure 6A). Notwithstanding these commonalities, the cultivation and recovery of other multiple genera were found to be allied to one or more of the specific cultivation conditions provided, in terms of plant compartments, plant-organ-culture media and/or oxygen availability.
Discussion
Cultivation-dependent approaches are becoming a forefront of microbiological research to explore the potential of environmental microbiomes, and to shed light on their ecology, physiology and biotechnological application. In fact, traditional methods for in vitro cultivation recover <1% of bacteria from all sorts of natural environments. Here, better understanding of the bacterial environment allowed the development of new culture media and new culture conditions required by the hidden fastidious bacteria. The subject was broadly treated and reviewed by quite a number of publications (e.g., Burmølle et al., 2009; Sarhan et al., 2019; Bonnet et al., 2020; Elsawey et al., 2020; Nemr et al., 2020).
As a breakthrough, we provided sufficient evidences that plant materials “as such” without any supplement, are compatible vegan substrates, very competitively support in vitro cultivability and explore diversity of plant microbiota (Sarhan et al., 2019). Further, we successfully advanced the intact plant leaf-based culturing method as “in situ similis strategy” for exploring the plant microbiota (Nemr et al., 2020). The direct inoculation/cultivation of microorganisms on the top of leaf surfaces/pads (Figure 8) furnish in vitro growing microorganisms with nutrients in their natural composition, concentrations and gradients (Watve et al., 2000; Nemr et al., 2020). In the present study we extend the concept of such in situ-similis cultivation by exploring plant organ compatible cultivation of bacterial community of endo-phyllospheric/endo-rhizosphere when grown on corresponding leaf strips/root segments-based culture media (see Figure 8 illustrating the two different methods). Efforts were made to pooling the advantages of MPN enrichment methodology together with the natural plant-only-based culture media. Where the MPN culture tubes efficiently constructed a nutritional milieu governed solely by vegan nutrients of plant origin, i.e., leaf strips/root segments, immersed in plain semi-solid water agar. The profuse growth of endophytic bacteria developed in the form of distinguishable surface and sub surface pellicles and/or diffused turbidity of viable cells of diverse morphologies. As to population's density of maize endophytes, the derived MPN estimates of bacterial endophytes in the endo-phyllosphere exceeded log 7.0 g−1 root, and were proportionate to those reported with standard R2A and CCM culture media. Further nutrients' compatibility was distinguished for sunflower endophyes, where a certain degree of plant organ specificity/preference was reported for culture media based on homologous plant organ, i.e., endo-rhizosphere bacteria favored culture media based on root segments while those of endo-phyllosphere preferred culture media based on leaf strips (Table 2). With 16S rRNA gene sequences of representative isolates, it appeared that homologous cultivation of endo-rhizosphere samples in root segments-based culture medium favored species of the genera of Bosea, Chitinophaga, Pantoea, Caulobacter, Scandinavium, and Agrobacterium, where Scandinavium and Caulobacter were isolated for the first time among sunflower microbiota. With leaf strips-based culture media, a number of potential rare species of the genera of Bacillus and Sphingobium were enriched from the endo-phyllosphere community. Surprisingly isolates of the genus Starkeya were isolated for the first time from the sunflower endo-rhizosphere when cultivated on leaf strips-based culture media under ambient oxygen conditions. Under restricted oxygen concentrations, preference of better growth of the endo-phyllospere community was reported in comparison to that of endo-rhizosphere.
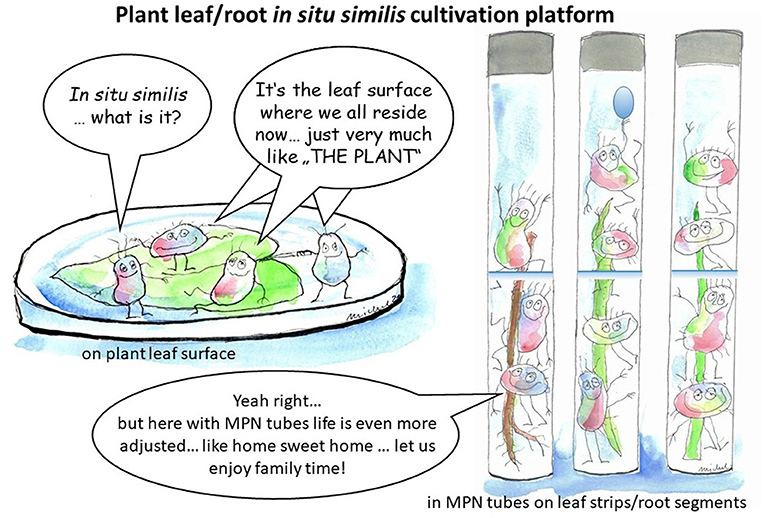
Figure 8. Plant leaf/root in situ similis cultivation platform: On the left, direct inoculation on the top of leaf surfaces/pads placed in water agar plates; on the right, inoculation of leaf strips/root segments-based MPN culture tubes.
A number of arguments have been put forward to justify the exceptional power of bacterial enrichment and selection provided in the set-up of MPN-semi solid culture tubes. It is reported that the environmental conditions at the solid (e.g., leaf/root)-liquid (e.g., semi-solid water agar) interface, that is constructed in the semi-solid MPN tubes, differ from those in conventional culture media in the form of the bulk aqueous phase of liquid culture media and/or solid phase of agar plates (Fletcher, 1984). Accordingly, the growth and physiological activity of bacteria attached to leaf/root surfaces may differ from that of free-living cells in respect of: (a) nutrient concentration and/or accessibility may be different at the interface because of adsorption/absorption/desorption of low molecular weight or macromolecular substrates; (b) direct contact that allows the surfaces of the natural plant substratum to provide a site for colonization and the development of a bacterial biofilm. This particular microenvironment of growth allows normal/natural interactions between multiple residents of various functions in the plant compartments.
In spite of the preference reported for culturability of Bacillus sp., the applied MPN method provided a unique nutritional platform and culturing conditions for the growth of not–yet-cultured genera of sunflower microbiota. As we obtained 163 representative isolates, belonging to 20 genera, expanding the diversity of cultivable sunflower microbiota to include multiple genera representing Alphaproteobacteria (Bosea sp.; Brevundomonas sp.; Caulobacter sp.; Starkeya sp.); Gammaproteobacteria (Pseudoxanthomonas sp.; Scandinavium sp.) and Bacteriodetes (Chitinophaga sp.; Sphingobacterium sp.) (Table 4; Supplementary Figure 5). Recently, Tamošiune et al. (2020) reported the occurrence of 158 different genera in sunflower phyllosphere and rhizosphere using 16S rRNA metagenomics sequencing method. Adding up all genera described for sunflower plants, including those of our study, a total of 176 different genera are until now in vitro recaptured out of the sunflower microbiota (Supplementary Table 3). Interestingly, the difference of 18 genera comprises cultivated isolates that did not show up in the previous molecular 16S rRNA metagenomics study. Furthermore, out of the above-mentioned 8 genera, that were genuinely isolated on our MPN-leaf/strips and root/segments-based media (Table 4; Supplementary Table 3), isolates of 3 genera were brought into cultivation, Caulobacter, Scandinavium, and Starkeya, that describe novel members of the sunflower microbiome, as they were not reported before (Supplementary Table 3, marked in bold letters). Additionally, our results strongly suggest the novel plant organ specific cultivation of at least two isolates, namely PR21_OD (MN231735, Bacillus sp.) and PR85 (MN232172, Sphingobium sp.). Very likely they are putative novel species, and even the latter as a putative novel genus, due to their significantly lower identity value and early long branching from other closely related Sphingobium isolates currently found in the NCBI nr database. Unfortunately, the 16S rRNA gene fragment size is quite short for both isolates, that might interfere with the phylogenetic tree building algorithm and, thus, needs further proof in future.
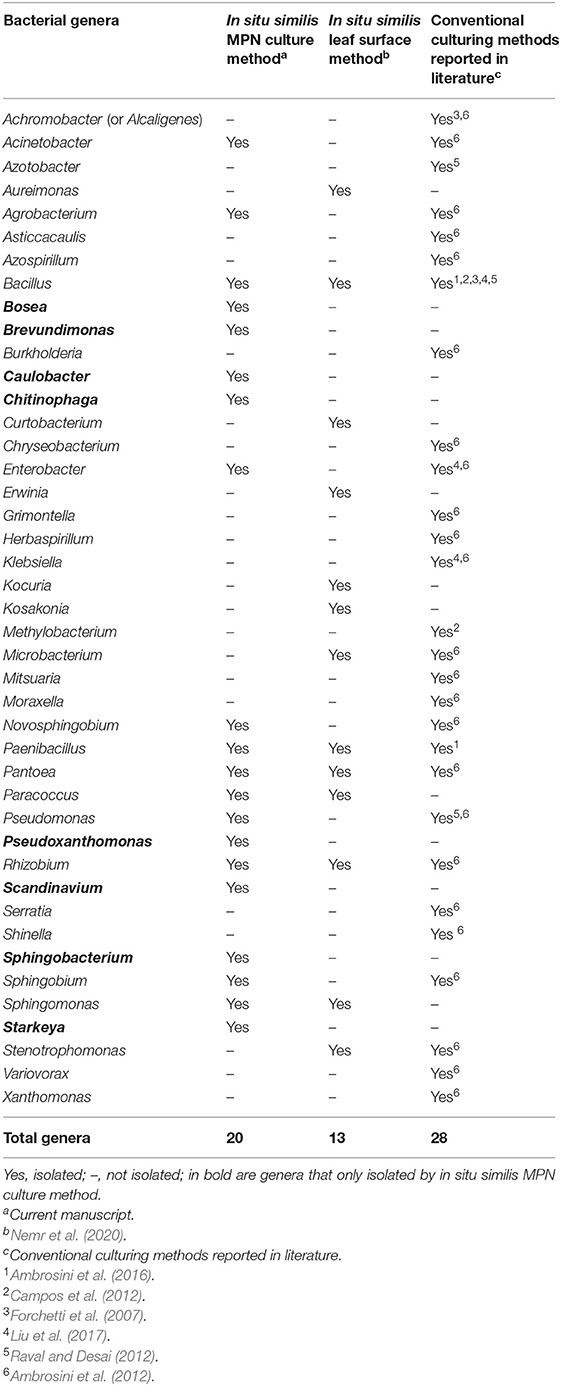
Table 4. Genera of cultivable endophytic bacteria of sunflower isolated by in situ similis culturomic strategies compared to other conventional culturing methods reported in literature.
Generally, the tested endo-rhizosphere isolates were of higher diversity, represented by 18 genera (belonged to Gammaproteobacteria, Alphaproteobacteria, Firmicutes, and Bacteroidetes) compared to those of the endo-phyllosphere (represented by only 7 genera) of the phyla Firmicutes and Proteobacteria (Alphaproteobacteria and Gammaproteobacteria). This is in accordance to what was reported in literature about the greater microbial diversity in belowground (roots, root zone, and bulk soil) than aboveground (leaves, flowers, and grapes) samples (Zarraonaindia et al., 2015). It is established that among other factors, organ (leaf vs. root) and habitat (epiphytes vs. endophytes) define the community of plant microbiota (Bodenhausen et al., 2013). Our results indicated some overlap at the rank of bacterial genera, namely Novosphingobium, Pseudomonas, Bacillus, and Paenibacillus, between leaf- and root-derived bacteria (4 genera, Figure 7A). This is in consistence with previous reports on different host plants, which further advance that soil may serve as a common bacterial reservoir for belowground and aboveground (Zarraonaindia et al., 2015; Wagner et al., 2016). Although the communities associated of both plant compartments share some bacterial species, they still differ in structure/composition (Bodenhausen et al., 2013). In the present study 15 genera (75%) were detected in either the endo-rhizosphere or the endo-phyllosphere.
Cross cultivation of rhizobacteria on homologous/heterologous host plan-based culture media, not plant organ specific media, was investigated by Mourad et al. (2018). They indicated that despite the promiscuous nature of the plant materials tested to culture rhizobacteria of different host plants, results indicated that plant materials of a homologous nature to the tested host plant, at least at the family level, and/or of the same environment were more likely to be selected. The present study further highlights cross cultivation of plant microbiota, of endo-rhizosphere and endo-phyllosphere, on homologous/heterologous leaf and root organ specific culture media of the same host plant, sunflower. In other words, to check if the leaf strips-based culture medium will enrich the homologous endo-phyllosphere specific community different to that developed on the heterologous root segment based medium, and vice versa with the endo-rhizosphere populations? Results of CFUs counts reflected such a plant organ compatible culturability. In addition, the DGGE profiles obtained for endo-phyllosphere showed that bacterial communities developed in MPN culture tubes of homologous leaf strips clustered away from those of MPN culture tubes of heterologous root segments. Comparably, with endophytes of the endo-rhizosphere, endophytes developed on the homologous root segments separated away from those of the heterologous leaf strips-based MPN culture tubes.
Analyzing the 16S rRNA based community composition of representative isolates underlined as well such plant organ affinity/compatibility. In general, the leaf strips-based culture medium enriched a distinct bacterial community compared to the root segments-based culture medium for the tested samples of both endo-phyllosphere and endo-rhizosphere of sunflower plants (Figure 6; Genera Distribution). An additional impact was demonstrated for incubation under different oxygen availability that further increased the richness of cultivable organisms (Supplementary Figure 6). As to the endo-phyllosphere community, the homologous leaf strips-based culture medium generally enriched Novosphingobium sp. and Brevundimonas sp. under ambient oxygen conditions, while the heterologous root segments- based medium favored the growth of Sphingobium sp. (Figure 6; Supplementary Figure 6B). On both culture media and under ambient or limited conditions common were isolates of different Bacillus species. Bacillus safensis and Bacillus pumilus were only recovered from endo-phyllosphere samples when they were grown on root segments-based culture medium. For endo-rhizosphere community, common genera that developed on both leaf strips/root segments-based medium were Rhizobium, Enterobacter, and Scandinavium. The root segments-based medium additionally enriched Agrobacterium, Pantoea, Chitinophaga, and Boesa. While the leaf strips-based medium enriched Pseudomonas growth from endo-rhizosphere sunflower samples, particularly under limited conditions (Figure 6; Supplementary Figure 6A). Additionally, isolates of Starkeya sp. and Pseudoxanthomonas sp. were detected from the endo-rhizosphere on leaf strips-based culture medium.
Currently, thorough assessment of genetic diversity of microbial communities is gained by high-throughput techniques such as NGS; however, PCR-DGGE (Muyzer et al., 1993) is a routine and commonly used technique in plant-soil ecosystem studies. Admitting that the technique has some limitations (Orlewska et al., 2018; Vischetti et al., 2020), recently a number of protocols are optimized for use with plant and soil DNA. They are confirmed to be reliable, do not require complex bioinformatics for the analyses of results, and present an overall picture on the main differences in microbial community composition when coupled with culture-dependent microbiological approaches (Sarhan et al., 2016; Vischetti et al., 2020). And, common to the NGS methods, different primer sets can be used in PCR-DGGE, to addressing microbial communities at the phylogenetic level (e.g., 16S primers for bacteria and archaea), or at the functional level, depending on functional selected genes (Valášková and Baldrian, 2009). The obtained DGGE profiles during this study were indicative to reflect differences in community composition attributed to the type of culture media, not the oxygen status during incubation. For endo-phyllosphere, bacterial communities developed in MPN culture tubes of homologous leaf strips clustered away from those of MPN culture tubes of heterologous root segments. As well, endophytes of the endo-rhizosphere produced two independent clusters separating endophytes developed on the homologous root segments distant from those of the heterologous MPN leaf strip culture tubes. Such differences in DGGE patterns points to a certain degree of plant-organ affinity and compatibility, that architect the community composition of culturable bacterial endophytes of endo-phyllosphere/endo-rhizosphere in response to growth in leaf strips/root -based culture media. An effect that is directly allied to the nutritional make-up of the plant leaves or roots used in the preparation of the plant-only-based culture media. In this respect, it is reported that the mean C, N, and P concentrations and C/N, C/P, and N/P ratios differed among leaf, stem and root of plants, and that stoichiometric patterns in different plant organs had different responses to environmental variables (He et al., 2015).
Conclusion
The presented strategy of in vitro cultivation based on strips/segments of plant leaves/roots construct an appropriate semi-solid interface together with a continuum of varying concentrations of nutrients and gas phases. Such conditions credibly meet complex nutritional requirements of the diverse taxa of plant endophytes. The strategy enhanced cultivation of plant microbiota, and significantly extended diversity of cultivable populations. As well, it shows plant organ compatibility for enrichment of plant microbiota when cultivated on the respective homologous plant organ-based culture media. This strategy is a feasible supplement to traditional cultivation to probe deeper for the yet-uncultured taxa and fill the many cultivation gaps of plant microbiota. It is presented as a significant tool to bringing additional potential resources of plant growth promoting bacteria into cultivation for further biotechnological applications.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
NH, SR, KE-S, and RN: conceptualization. RN, SA, MK, AB, MRA, and HG: methodology. SP, RN, MH, HY, and MRA: software. NH, SR, SP, KE-S, MF, and HG: validation. RN, MH, HY, HG, MRA, AM, and MA: formal analysis. RN, SA, MK, AB, and MRA: investigation. SR, NH, and MF: resources. SR, NH, SP, MF, and KE-S: data curation. RN, NH, MF, KE-S, HG, and MA: writing—original draft preparation. NH, SR, SP, MF, and MB: writing—review and editing. RN, MB, SP, AM, and MH: visualization. NH, SR, MF, and KE-S: supervision. NH, SR, MH, and MA: project administration. SR and NH: funding acquisition. All authors have read and agreed to this version of the manuscript.
Funding
The authors declare that Cairo university-Egypt and Leibniz Institute of Vegetable and Ornamental Crops, GroSSbeeren, Germany funded the research being submitted.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer EM declared a past co-authorship with several of the authors RN, SP, SA, MRA, AM, HY, MH, MA, MF, SR, NH, and MB to the handling Editor.
Acknowledgments
It is a great pleasure to dedicate this publication in the honor of our co-author late KE-S. KE-S who passed away on 28 September 2020 was a leading Egyptian Botanist at Cairo University. NH and RN acknowledge the financial support of the Alexander von Humboldt Foundation, Bonn, Germany. They are grateful to all kinds of support provided by Prof. Eckhard George in his capacity as the research director of IGZ. We thank Michael Becker for creating the graphical abstract on the in situ similis cultivation platform (Figure 8).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.660790/full#supplementary-material
References
Altschul, S. F., Gish, W., Pennsylvania, T., and Park, U. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ambrosini, A., Beneduzi, A., Stefanski, T., Pinheiro, F. G., Vargas, L. K., and Passaglia, L. M. P. (2012). Screening of plant growth promoting rhizobacteria isolated from sunflower (Helianthus annuus L.). Plant Soil 356, 245–264. doi: 10.1007/s11104-011-1079-1
Ambrosini, A., Stefanski, T., Lisboa, B. B., Beneduzi, A., Vargas, L. K., and Passaglia, L. M. P. (2016). Diazotrophic bacilli isolated from the sunflower rhizosphere and the potential of Bacillus mycoides B38V as biofertiliser. Ann. Appl. Biol. 168, 93–110. doi: 10.1111/aab.12245
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Bodenhausen, N., Horton, M. W., and Bergelson, J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8:e56329. doi: 10.1371/journal.pone.0056329
Bollmann, A., Lewis, K., and Epstein, S. S. (2007). Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73, 6386–6390. doi: 10.1128/AEM.01309-07
Bonnet, M., Lagier, J. C., Raoult, D., and Khelaifia, S. (2020). Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbe New Infect. 34:100622. doi: 10.1016/j.nmni.2019.100622
Burmølle, M., Johnsen, K., Al-soud, W. A., Hansen, L. H., and Sørensen, S. J. (2009). The presence of embedded bacterial pure cultures in agar plates stimulate the culturability of soil bacteria. J. Microbiol. Methods 79, 166–173. doi: 10.1016/j.mimet.2009.08.006
Campos, K. G., Luisa, M. C., Cattelan, A. J., Nogueira, M. A., Guilherme, C. P., and Luiz, A. M. (2012). Biochemical and molecular characterization of high population density bacteria isolated from Sunflower. J. Microbiol. Biotechnol. 22, 437–447. doi: 10.4014/jmb.1109.09007
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cochran, W. G. (1950). Estimation of bacterial densities by means of the “most-probable numbers.” Biometrics 6, 5–116. doi: 10.2307/3001491
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cross, K. L., Campbell, J. H., Balachandran, M., Campbell, A. G., Cooper, S. J., Griffen, A., et al. (2019). Targeted isolation and cultivation of uncultivatedbacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321. doi: 10.1038/s41587-019-0260-6
de Oliveira Costa, L. E., de Queiroz, M. V., Borges, A. C., de Moraes, C. A., and de Araújo, E. F. (2012). Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 43, 1562–1575. doi: 10.1590/S1517-83822012000400041
Elsawey, H., Patz, S., Nemr, R. A., Sarhan, M. S., Hamza, M. A., Youssef, H. H., et al. (2020). Plant broth- (not bovine-) based culture media provide the most compatible vegan nutrition for in vitro culturing and in situ probing of plant microbiota. Diversity 12:418. doi: 10.3390/d12110418
Ferrari, B. C., Binnerup, S. J., and Gillings, M. (2005). Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71, 8714–8720. doi: 10.1128/AEM.71.12.8714-8720.2005
Fletcher, M. (1984). “Comparative physiology of attached and free-living bacteria,” in Microbial Adhesion and Aggregation, ed K. C. Marshall (Berlin; Heidelberg: Springer), 223–232.
Forchetti, G., Masciarelli, O., Alemano, S., Alvarez, D., and Abdala, G. (2007). Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 76, 1145–1152. doi: 10.1007/s00253-007-1077-7
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software: package for education and data analysis. Palaeontol. Electr. 4:9.
He, M., Zhang, K., Tan, H., Hu, R., Su, J., Wang, J., et al. (2015). Nutrient levels within leaves, stems, and roots of the xeric species Reaumuria soongorica in relation to geographical, climatic, and soil conditions. Ecol. Evol. 5, 1494–1503. doi: 10.1002/ece3.1441
Hegazi, N. A., Hamza, M. A., Osman, A., Ali, S., and Sedik, M. Z. (1998). “Modified combined carbon N-deficient medium for isolation, enumeration and biomass production of diazotrophs,” in Nitrogen Fixation With Non-legumes, eds K. A. Malik, M. S. Mirza, and J. K. Ladha (Dordrecht: Springer), 247–253.
Hegazi, N. A., Sarhan, M. S., Fayez, M., Patz, S., Murphy, B. R., and Ruppel, S. (2017). Plant-fed versus chemicals-fed rhizobacteria of Lucerne: plant-only teabags culture media not only increase culturability of rhizobacteria but also recover a previously uncultured Lysobacter spp., Novospphingobium spp. and Pedobacter spp. PLoS ONE 12:e00180424. doi: 10.1371/journal.pone.0180424
Ingham, C. J., Spprenkels, A., Bomer, J., Molenaar, D., van den Berg, A., van Hylckama Vlieg, J. E. T., et al. (2007). The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 104, 18217–18222. doi: 10.1073/pnas.0701693104
Ingham, C. J., Van Den Ende, M., Pijnenburg, D., Wever, P. C., and Schneeberger, P. M. (2005). Growth and multiplexed analysis of microorganisms on a subdivided, highly porous, inorganic chip manufactured from anopore. Appl. Environ. Microbiol. 71, 8978–8981. doi: 10.1128/AEM.71.12.8978-8981.2005
Jackson, C. R., Randolph, K. C., Osborn, S. L., and Tyler, H. L. (2013). Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 13:274. doi: 10.1186/1471-2180-13-274
Jiang, C. Y., Dong, L., Zhao, J. K., Hu, X., Shen, C., Qiao, Y., et al. (2016). High- throughput single-cell cultivation on microfluidic streak plates. Appl. Environ. Microbiol. 82, 2210–2218. doi: 10.1128/AEM.03588-15
Kaeberlein, T., Lewis, K., and Epstein, S. S. (2002). Isolating “Uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129. doi: 10.1126/science.1070633
Laanbroek, H. J., and Geerligs, H. J. (1983). Influence of clay particles (Illite) on substrate utilization by sulfate-reducing bacteria. Arch. Microbiol. 134:161–163. doi: 10.1007/BF00407951
Lagier, J. C., Khelaifia, S., Alou, M. T., Ndongo, S., Dione, N., Hugon, P., et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1:16203. doi: 10.1038/nmicrobiol.2016.203
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Levy, A., Conway, J. M., Dangl, J. L., and Woyke, T. (2018). Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 24, 475–485. doi: 10.1016/j.chom.2018.09.005
Libby, E. A., and Silver, P. A. (2019). Harnessing undomesticated life. Nat. Microbiol. 4, 212–213. doi: 10.1038/s41564-018-0355-8
Liu, X., Li, X., Li, Y., Li, R., and Xie, Z. (2017). Plant growth promotion properties of bacterial strains isolated from the rhizosphere of the Jerusalem artichoke (Helianthus tuberosus L.) adapted to salt-alkaline soils and their effect on wheat growth. Can. J. Microbiol. 63, 228–237. doi: 10.1139/cjm-2016-0511
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mah, R. A., and Smith, M. R. (1981). “The methanogenic bacteria,” in The Prokaryotes, eds M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (New York, NY: Springer-Verlag), 948–977.
Manome, A., Zhang, H., Tani, Y., and Katsuragi, T. (2001). Application of gel microdroplet and fow cytometry techniques to selective enrichment of non- growing bacterial cells. FEMS Microbiol. Lett. 197, 29–33. doi: 10.1111/j.1574-6968.2001.tb10578.x
Mauchline, T. H., Chedom-Fotso, D., Chandra, G., Samuels, T., Greenaway, N., Backhaus, A., et al. (2015). An analysis of Pseudomonas genomic diversity in take-all infected wheat fields reveals the lasting impact of wheat cultivars on the soil microbiota. Environ. Microbiol. 17, 4764–4778. doi: 10.1111/1462-2920.13038
Morris, R. M., Rappé, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A., et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810. doi: 10.1038/nature01240
Mourad, E. F., Sarhan, M. S., Daanaa, H.-S. A., Abdou, M., Morsi, A. T., Abdelfadeel, M. R., et al. (2018). Plant materials are sustainable substrates supporting new technologies of plant-only-based culture media for in vitro culturing of the plant microbiota. Microb. Environ. 33, 40–49. doi: 10.1264/jsme2.ME17135
Mühling, M., Woolven-Allen, J., Murrell, J. C., and Joint, I. (2008). Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2, 379–392. doi: 10.1038/ismej.2007.97
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microb. 59, 695–700. doi: 10.1128/AEM.59.3.695-700.1993
Nemr, R. A., Khalil, M., Sarhan, M. S., Abbas, M., Elsawey, H., Youssef, H. H., et al. (2020). “In situ similis” culturing of plant microbiota : a novel simulated environmental method based on plant leaf blades as nutritional pads. Front. Microbiol. 11:454. doi: 10.3389/fmicb.2020.00454
Nichols, D., Cahoon, N., Trakhtenberg, E. M., Pham, L., Mehta, A., Belanger, A., et al. (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial sppecies. Appl. Environ. Microbiol. 76, 2445–2450. doi: 10.1128/AEM.01754-09
Nichols, D., Lewis, K., Orjala, J., Mo, S., Ortenberg, R., O'Connor, P., et al. (2008). Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74, 4889–4897. doi: 10.1128/AEM.00393-08
Nour, E. H., Hamza, M. A., Fayez, M., Monib, M., Ruppel, S., and Hegazi, N. A. (2012). The crude plant juices of desert plants as appropriate culture media for the cultivation of rhizospheric microorganisms. J. Adv. Res. 3, 35–43. doi: 10.1016/j.jare.2011.03.002
Orlewska, K., Piotrowska-seget, Z., and Cycoń, M. (2018). Use of the PCR-DGGE method for the analysis of the bacterial community structure in soil treated with the cephalosporin antibiotic cefuroxime and / or Inoculated with a multidrug-resistant Pseudomonas putida strain MC1. Front. Microbiol. 9:1387. doi: 10.3389/fmicb.2018.01387
Raval, A. A., and Desai, P. B. (2012). Rhizobacteria from rhizosphere of sunflower (Helianthus annuus L.) and their effect on plant growth. Res. J. Recent Sci. 1, 58–61.
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture ofbacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/AEM.49.1.1-7.1985
Russo, P., Botticella, G., Capozzi, V., Massa, S., Spano, G., and Beneduce, L. (2014). A fast, reliable, and sensitive method for detection and quantification of Listeria monocytogenes and Escherichia coli O157 : H7 in ready-to-eat fresh-cut products by MPN-qPCR. BioMed Res. Inter. 2014, 1–9. doi: 10.1155/2014/608296
Salam, N., Xian, W., Asem, M., Xiao, M., and Li, W. (2020). From ecophysiology to cultivation methodology: filling the knowledge gap between uncultured and cultured microbes. Mar. Life Sci. Tech. 3, 132–147. doi: 10.1007/s42995-020-00064-w
Sarhan, M. S., Hamza, M. A., Youssef, H. H., Patz, S., Becker, M., Elsawey, H., et al. (2019). Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media – a review. J. Adv. Res. 19, 15–27. doi: 10.1016/j.jare.2019.04.002
Sarhan, M. S., Mourad, E. F., Hamza, M. A., Youssef, H. H., Scherwinski, A. C., El- Tahan, M., et al. (2016). Plant powder teabags: a novel and practical approach to resolve culturability and diversity of rhizobacteria. Physiol. Plant 157, 403–413. doi: 10.1111/ppl.12469
Sarhan, M. S., Mourad, E. F., Nemr, R. A., Abdelfadeel, M. R., Daanaa, H. A., Youssef, H. H., et al. (2020). An inoculum-dependent culturing strategy (IDC) for the cultivation of environmental microbiomes and the isolation of novel endophytic Actinobacteria. J. Antibiot. 73, 66–71. doi: 10.1038/s41429-019-0226-4
Sarhan, M. S., Patz, S., Hamza, M. A., Youssef, H. H., Mourad, E. F., Fayez, M., et al. (2018). G3 phylochip analysis confirms the promise of plant-based culture media for unlocking the composition and diversity of the maize root microbiome and for recovering unculturable candidate divisions/phyla. Microb. Environ. 33, 317–325. doi: 10.1264/jsme2.ME18023
Shade, A., Jones, S. E., Caporaso, J. G., Handelsman, J., Knight, R., Fierer, N., et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. Am. Soc. Microbiol. 5, 1–9. doi: 10.1128/mBio.01371-14
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
Sutton, S. (2010). The Most Probable Number method and its uses in enumeration, qualification, and validation. J. Valid. Tech. 16, 35–38.
Tamošiune, I., Gelvonauskiene, D., Haimi, P., MildaŽiene, V., Koga, K., Shiratani, M., et al. (2020). Cold plasma treatment of sunflower seeds modulates plant-associated microbiome and stimulates root and lateral organ growth. Front. Plant Sci. 28, 1–13. doi: 10.35848/1882-0786/ab9712
Tanner, R. S. (1989). Monitoring sulfate-reducing bacteria: comparison of enumeration media. J. Microbiol. Methods 10, 83–90. doi: 10.1016/0167-7012(89)90004-3
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., et al. (2018). Core microbiomes for sustainable agroecosystems. Nat. Plants 4, 247–257. doi: 10.1038/s41477-018-0139-4
Valášková, V., and Baldrian, P. (2009). Denaturing gradient gel electrophoresis as a fingerprinting method for the analysis of soil microbial communities. Plant Soil Environ. 55, 413–423. doi: 10.17221/132/2009-PSE
Vester, F., and Ingvorsen, K. (1998). Improved Most-Probable-Number method to detect sulfate- reducing bacteria with natural media and a radiotracer. Appl. Environ. Microbiol. 64, 1700–1707. doi: 10.1128/AEM.64.5.1700-1707.1998
Vischetti, C., Casucci, C., Bernardi, A., De Monaci, E., Tiano, L., Marcheggiani, F., et al. (2020). Sub-lethal effects of pesticides on the DNA of soil organisms as early ecotoxicological biomarkers. Front. Microbiol. 11:1892. doi: 10.3389/fmicb.2020.01892
Wagner, M. R., Lundberg, D. S., Del Rio, T. G., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 1–15. doi: 10.1038/ncomms12151
Wais, A. C. (1988). Recovery of halophilic archaebacteria from natural environments. FEMS Microbiol. Ecol. 53, 211–216. doi: 10.1111/j.1574-6968.1988.tb02666.x
Watve, M., Shejval, V., Sonawane, C., Rahalkar, M., Matapurkar, A., Shouche, Y., et al. (2000). The “K” selected oligophilic bacteria: a key to uncultured diversity? Curr. Sci. 78, 1535–1542. doi: 10.1177/0022487110369555
Wirth, S. J., and Wolf, G. A. (1990). Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J. Microbiol. Meth. 12, 197–205. doi: 10.1016/0167-7012(90)90031-Z
Youssef, H. H., Fayez, M., Monib, M., and Hegazi, N. (2004). Gluconacetobacter diazotrophicus: a natural endophytic diazotroph ofNile Delta sugarcane capable of establishing an endophytic association with wheat. Biol. Fertil. Soils 39, 391–397. doi: 10.1007/s00374-004-0728-4
Youssef, H. H., Hamza, M. A., Fayez, M., Mourad, E. F., Saleh, M. Y., Sarhan, M. S., et al. (2016). Plant-based culture media: efficiently support culturing rhizobacteria and correctly mirror their in-situ diversity. J. Adv. Res. 7, 305–316. doi: 10.1016/j.jare.2015.07.005
Zarraonaindia, I., Owens, S. M., Weisenhorn, P., West, K., Hampton-Marcell, J., Lax, S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. MBio 6:e02527-14. doi: 10.1128/mBio.02527-14
Keywords: Plant microbiota, In situ similis cultivation, leaf strips/root segments culture media, plant organ compatible cultivation, MPN enrichment of plant microbiota, PCR-DGGE, endo-phyllosphere/endo-rhizosphere of sunflower plants
Citation: Nemr RA, Patz S, Abdelwakeel SM, Khalil M, Ben Djadid A, Abdelfadeel MR, Morsi AT, Goda HA, Youssef HH, Hamza M, Abbas M, Fayez M, El-Sahhar KF, Becker M, Ruppel S and Hegazi NA (2021) Culture Media Based on Leaf Strips/Root Segments Create Compatible Host/Organ Setup for in vitro Cultivation of Plant Microbiota. Front. Sustain. Food Syst. 5:660790. doi: 10.3389/fsufs.2021.660790
Received: 29 January 2021; Accepted: 25 May 2021;
Published: 08 July 2021.
Edited by:
Everlon Cid Rigobelo, São Paulo State University, BrazilReviewed by:
Elhussein Fouad Mourad, Politecnico di Bari, ItalyRoberta Mendes, São Paulo State University, Brazil
Copyright © 2021 Nemr, Patz, Abdelwakeel, Khalil, Ben Djadid, Abdelfadeel, Morsi, Goda, Youssef, Hamza, Abbas, Fayez, El-Sahhar, Becker, Ruppel and Hegazi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nabil A. Hegazi, aGVnYXppbmFiaWw4QGdtYWlsLmNvbQ==
 Rahma A. Nemr
Rahma A. Nemr Sascha Patz
Sascha Patz Saad M. Abdelwakeel
Saad M. Abdelwakeel Mohab Khalil
Mohab Khalil Ali Ben Djadid4
Ali Ben Djadid4 Mohamed R. Abdelfadeel
Mohamed R. Abdelfadeel Hanan A. Goda
Hanan A. Goda Mervat Hamza
Mervat Hamza Silke Ruppel
Silke Ruppel Nabil A. Hegazi
Nabil A. Hegazi