- 1School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2Institute of Quality and Safety Testing Center for Agro-Products, Xining, China
- 3College of Horticulture, Shanxi Agricultural University, Jinzhong, China
- 4Department of Plant and Environmental Sciences, Edisto Research and Education Center, Clemson University, Blackville, SC, United States
- 5Biology Program, Division of Science, New York University Abu Dhabi, Abu Dhabi, United Arab Emirates
The non-host specific Pseudomonas syringae phytotoxin Coronatine (COR) causes chlorosis and promotes toxicity by inducing physiological changes in plants. We performed transcriptome analysis to better understand plants' transcriptional and metabolic response to COR. Toward this end, mock-treated and COR-treated soybean plants were analyzed by RNA-Seq. A total of 4,545 genes were differentially expressed between the two treatments, of which 2,170 were up-regulated whereas 2,375 were down-regulated in COR treated samples. Gene annotation and pathway analysis conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases revealed that the differential genes were involved in photosynthesis, jasmonic acid (JA) synthesis, signal transduction, and phenylpropane metabolism. This study will provide new insights into COR mediated responses and extend our understanding of COR function in plants.
Introduction
Soybean [Glycine max (L.) Merr.] is one of the most important economic crops in the world but is vulnerable to various pests and diseases which drastically affect yield and quality. Soybean speck caused by Pseudomonas syringae pv. glycinea (Pgy) is a common disease, threatening soybean yield worldwide (Keen and Buzzell, 1991; Wrather et al., 1997). Pathogenic bacteria inject type three effectors (TTEs) through the type three secretion system (TTSS) in order to inhibit the plant immune response and cause virulence in compatible hosts. Besides effector proteins, pathogens also secrete phytotoxins to increase pathogenicity on plants. Phytotoxins promote pathogen growth by modifying host cell processes and by altering host metabolism (Galán, 2009). Coronatine (COR) is a non-host specific phytotoxin secreted by Pseudomonas syringae pathovars atropurpurea, glycinea, maculicola, morsprunorum, and tomato (Bender et al., 1999). Coronatine can be detected in healthy tissues adjacent to the bacterial lesions, indicating that COR could move systemically (Zhao et al., 2001). The main symptom caused by COR is chlorosis, however, COR does not destroy organelles or membrane systems (Palmer and Bender, 1995). In addition to causing chlorosis in plants, the compound can reopen stomata, which can promote bacterial invasion, proliferation, and development of plant disease symptoms (Geng et al., 2012).
Coronatine (C18H25NO4) has a relative molecular weight of 319 g/mol and is a structural and functional analog of JA-Ile. It is formed by linking the α-amino acid Coronamic Acid (CMA) with the polyketone Coronafacic Acid (CFA) through an amide linkage (Brooks et al., 2004). Coronafacic Acid structure partially mimics the plant hormone jasmonic acid (JAs), and similar to Jasmonate, its activity is mediated by binding to the Coranatine Insensitive1 (COI1) receptor (Feys et al., 1994). The affinity of COR for COI1 is more than 1,000 times that of JA-Ile (Katsir et al., 2008). Coronatine induces JA biosynthesis and inhibits SA signaling, thereby dampening SA-dependent disease resistance (Zheng et al., 2012). The treatment of COR in plants also induces ethylene production and the release rate of ethylene is proportional to the concentration of COR in tobacco leaves (Ferguson and Mitchell, 1985; Kenyon and Turner, 1992). Although CMA is structurally similar to the ethylene precursor 1-aminocyclopropene 1-carboxylic acid (ACC), it does not induce the production of ethylene responsive genes (Uppalapati et al., 2005). Furthermore, treatment with CFA, CMA, MeJA, or ACC does not induce chlorosis, indicating that COR has additional functions than those attributed to either CFA or CMA (Uppalapati et al., 2005).
High-throughput RNA-Seq is a powerful tool to study gene expression changes in plants (Gao et al., 2014). In order to better understand the function of COR in soybean, we treated soybean plant leaves with COR which was followed by transcriptome analysis. The biological annotation of differentially expressed genes (DEGs) under treatment of COR provided new insights into COR mediated plant responses on soybeans, thereby extending our understanding of COR function in plants.
Materials and Methods
Plant Material and Treatment
Soybean [Glycine max (L.) Merrill cv. Williams 82] was grown under a 12 h light /12 h dark period with light and dark temperatures of 25 and 20°C, respectively. Four-week-old soybean plants were used in the study. Coronatine purchased from Sigma-Aldrich (St. Louis, MO) was dissolved in methanol to make a stock solution (600 μM, stored at −20°C) which was subsequently diluted with water to a final concentration of 3 μM. After the addition of 0.004% Silwet L-77, the working solution was sprayed on soybean leaves and leaf samples were collected 10 h post spray. For control treatment, only water containing 0.004% Silwet L-77 solution was sprayed onto the plant leaves. Harvested samples were immediately frozen in liquid nitrogen and stored at −80°C.
Library Construction and RNA-Seq
Each treatment (COR or mock) was carried out in triplicate. Total RNA was extracted from leaves of COR-treated and control plants using Trizol reagent (Invitrogen, Carlsbad, CA, USA; Rio et al., 2010), and the concentration of RNA was quantified using a NanoDrop-2000 nucleic acid spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Nucleic acid integrity was assessed by resolving RNA on a 1% (w/v) agarose gel. Quality control and library construction were entrusted to Huada Gene Technology Company (Wuhan, Hubei, China). In brief, from a pool of total RNA, the mRNA fraction was enriched with the help of Oligo-dT magnetic beads (Vazyme Biotech Company, Jiangsu, China). This was followed by RNA fragmentation and first strand cDNA synthesis using the random hexamer primers (Illumina, San Diego, CA, USA). Following second strand cDNA synthesis, the fragments were ligated with adapters and PCR amplified to construct cDNA libraries. BGISEQ-500 platform from Huada Gene Technology Co., Ltd. (Wuhan, Hubei, China) was used to generate raw library reads. The base quaility of the raw data was the ASCII (American standard code for information interchange) value of each character in the fourth line of the FASTQ format minus 64. Reads where more than half of the component bases had a quality score below 15 was considered a low-quality read and removed by Soapnuk. Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) was used to align the filtered data to the reference genome (GCF_000004515.4_Glycine_max_v2.0; Kim et al., 2015). The RNA-seq data has been uploaded to NCBI gene expression omnibus server (accession number: PRJNA690873, Biosample ID: SAMN17266875, COR SRA ID: SRR13389957, and Mock SRA ID: SRR13389956).
Gene Annotation and Classification
Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) was used to evaluate gene expression levels, and DEGseq R package (Anders and Huber, 2010) was used to determine the DEGs between the two samples. The screening threshold for differential genes was | log2 Fold Change | ≥ 1, FDR < 0.01, FPKM ≥ 1.
Based on Gene Ontology (GO) (http://www.geneontology.org/) analysis, DEGs were divided into three major functional categories including molecular function, cellular component, and biological process. These genes were further classified into biological pathways based on Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) annotation and the phyper function in R was used for enrichment analysis.
Quantitative Real Time PCR (qRT-PCR) Validation
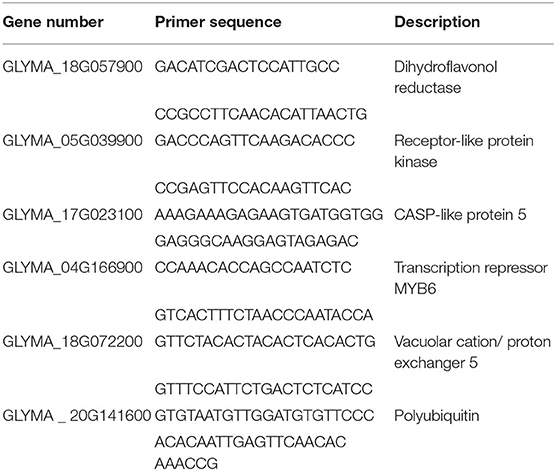
In order to assess the reliability of the data obtained from RNA-Seq, we measured the relative fold change of five genes by qRT-PCR analysis (Yu et al., 2014). RNA was extracted from frozen leaf tissue using Trizol reagent. In order to determine RNA integrity, the RNA samples were resolved on a 1% (w/v) agarose gel. For first strand cDNA synthesis, the PrimeScript™ RT (TaKaRa, Mountain View, CA), and the gDNA Eraser (TaKaRa, Mountain View, CA) kits were used. For qPCR analysis, the TB GreenTM Premix Ex Taq™ (TaKaRa, Mountain View, CA) kit was used under the following set of conditions. A pre-denaturation step of 95°C for 30 s, 40 cycles with each cycle employing a denaturation temperature of 95°C for 5 s, and an annealing/extension step with a temperature of 60°C for 30 s followed by melt curve analysis. For each treatment, we analyzed a total of three biological replicates, each of which included three technical repeats. The 2−ΔΔCT method was used for the relative quantification of the five genes. The primers used for the qRT-PCR analysis are listed in Table 1. The GLYMA_20G141600 gene that encodes for polyubiquitin served as an internal control.
Results
Overview of the Soybean Transcriptome
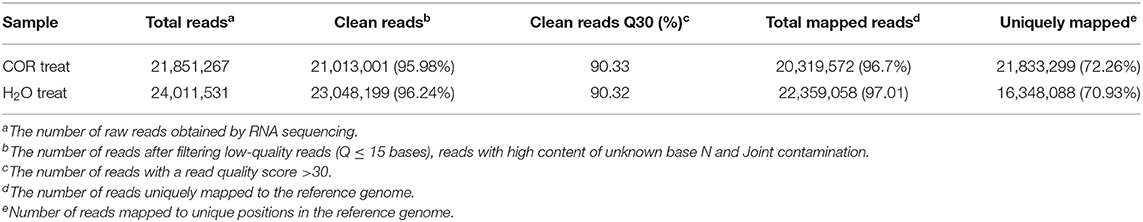
Transcriptome sequencing was performed on samples of COR and mock treated soybean leaves. A high proportion of clean reads (>96%) were obtained after filtration of the raw reads and more than 90% of the total reads had a quality value of 30, suggesting the sequencing reads were of high quality. Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) was used to align the filtered data to the reference genome, and the mapping result statistics are shown in Table 2 (Kim et al., 2015).
Gene expression based on fragments per kilobase per million mapped reads (FPKM), generated sequence information for 41,290 and 40,505 reference genes from the COR and mock samples, respectively. Most of the expressed genes were distributed between 10 and 100 FPKM (Table 3). Transcriptome sequencing analysis was carried out on soybean leaves from the COR and the mock groups. The overall transcriptome data analysis showed that the transcriptome sequencing data was of high quality and could meet the basic requirements necessary for downstream analysis.

Table 3. Distribution of FPKM-quantified expression level of detected genes in COR and mock treated samples.
Analysis of Differentially Expressed Genes
Our results showed that the COR-vs.-mock comparison yielded 4,531 DEGs. In order to summarize the results obtained from our analysis, and to make visualization of the regulatory patterns more intuitive, we have drawn a volcanic map (Figure 1). The volcano diagram clearly and intuitively shows the expression profile of differential genes in soybeans under different treatments. Although COR-treatment resulted in a greater number of down-regulated genes (2,370), most of the genes with large fold differences were up-regulated (2,161; Figure 1).
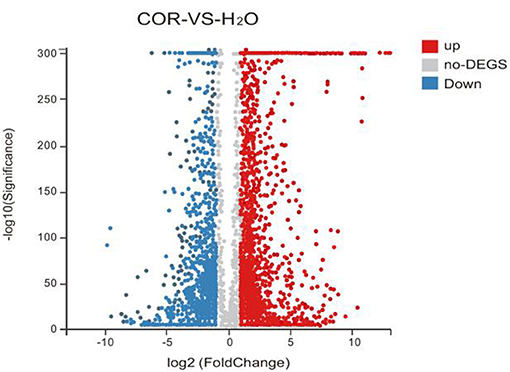
Figure 1. Volcano plot for DEGs. The X-axis represents the log2-transformed expression changes between COR-treated and mock samples, and the Y-axis represents the –log10-transformed p-value denoting the significance of differential expression. The red dots, blue dots, and gray dots denote up-regulated, down-regulated, and non-differential genes, respectively.
Gene Ontology Enrichment Analysis
The GO database was used to functionally annotate DEGs into the following three major categories: Biological process, Cellular component, and Molecular function. There were 4,545 significant DEGs between COR and mock treated samples. Twelve GO subclasses belonged to biological processes sub-category, 12 subclasses were found within the cellular components category while 9 subclasses were enriched in the molecular function category (Figure 2). Within the biological process category, cellular processes, and metabolic processes accounted for a relatively high proportion of hits. In the cell components category, cell, organelle, membrane, and membrane part were the four subclasses with higher percentage representation whereas within the molecular function category, binding, and catalytic activity were the two majorly represented subclasses.
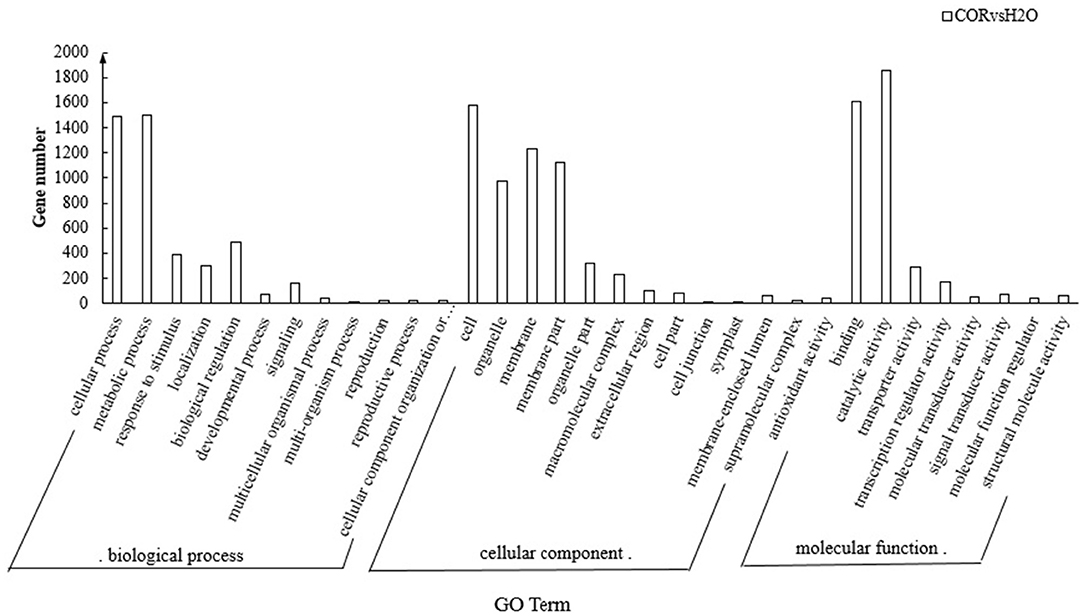
Figure 2. GO functional categories of DEGs in different sub-groups. The GO terms are sorted according to their P-values in the cellular component, molecular functions, and biological process categories. The bars represent the number of enriched genes in major sub-categories within each GO category.
KEGG Annotation of DEGs
The KEGG database integrates gene catalogs obtained from a completely sequenced genome with higher-level system functions at the cell, species, and ecosystem level. The integrative function of the database transforms the expressed gene data into gene networks. By mapping DEGs to the reference genes in the KEGG database, we identified 2,100 DEGs involved in 131 KEGG pathways. Enrichment analysis showed that KEGG enrichment with a Q-value < 0.05 was reliable, and on the basis of that threshold, a total of 30 KEGGs were found to be significantly enriched (Figure 3, Supplementary Table 3). The KEGG pathway with the largest number of enriched genes was related to “plant hormone signal transduction” (ko04075), with 181 genes annotated in this pathway. Although there were only 26 genes in the “Photosynthesis—antenna proteins” (ko00196) pathway, the enrichment factor reached 0.764, which was the highest of all annotated pathways. Analysis of the enriched KEGG pathway showed that the vast majority of KEGG genes belonged to the “metabolic branch” and were related to photosynthesis, hormone transduction, and secondary metabolism (Figure 3, Supplementary Table 3).
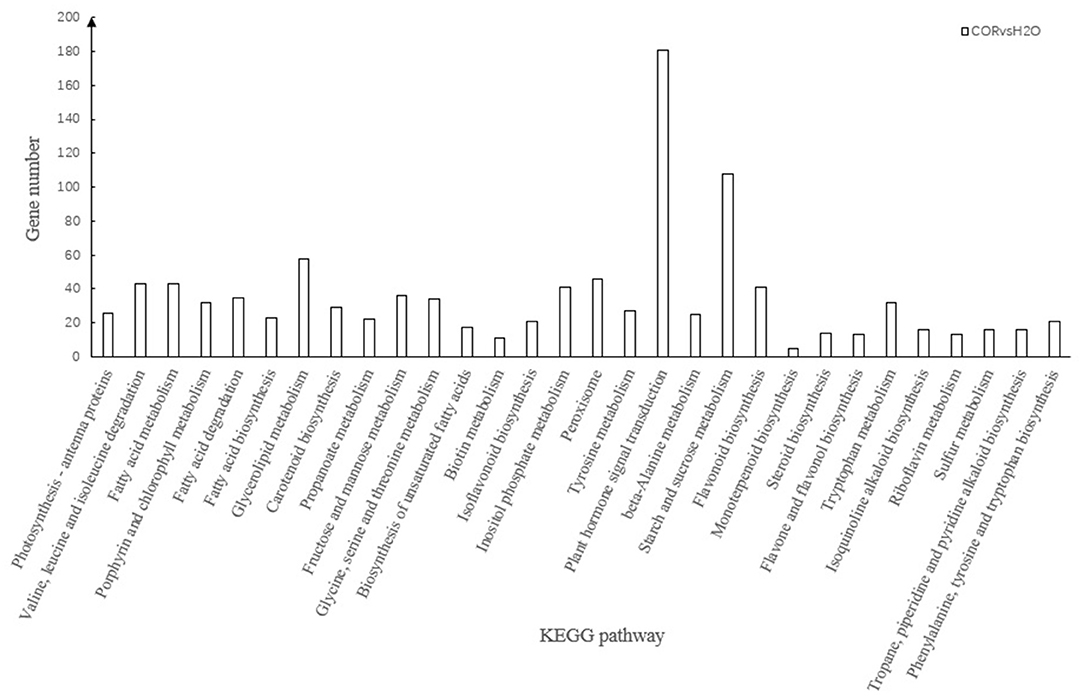
Figure 3. Over-representation of DEGs in KEGG pathways. The X-axis shows different KEGG pathways, and the Y-axis denotes the number of DEGs involved in each category.
Chlorophyll and Photosynthesis Related Genes Were Down-Regulated by COR Treatment
Coronatine treatment resulted in the down-regulation of a large number of genes involved in chloroplast metabolism (Supplementary Table 3), indicating that COR had a significant influence on plant health. Chlorophyll a/b binding protein was significantly down-regulated, whereas genes related to magnesium removal, such as magnesium dechelatase, chlorophyllase, uroporphyrin-III C-methyltransferase, pheophorbidase, pheophorbide an oxygenase, and glucuronosyltransferase were significantly upregulated (Supplementary Table 3). Furthermore, COR induced the expression of chlorophyllase, an enzyme that degrades chlorophyll (Takamiya et al., 2000; Xie et al., 2008). In the three photosynthetic pathways: Photosynthesis-antenna proteins (ko00196), Carbon fixation in photosynthetic organisms (ko00710), Photosynthesis (ko00195), there were 129 genes with significant gene expression changes (Supplementary Table 1). Out of these, only 15 genes were up-regulated while 114 genes were down-regulated, indicating that photosynthesis was significantly downregulated upon COR treatment.
Genes Related to ROS Generation Were Upregulated in Response to COR Treatment
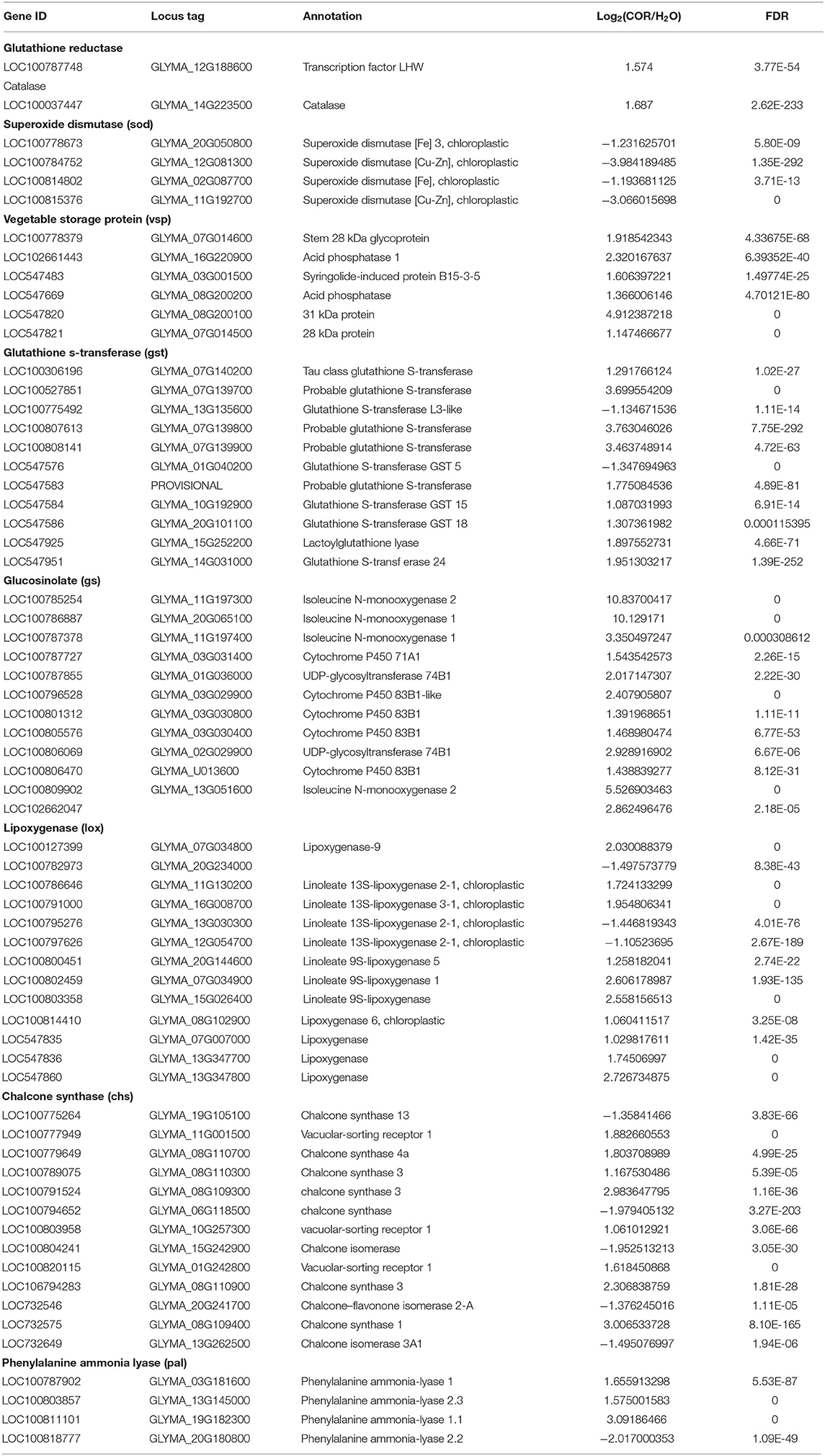
When plants are under biological stress, rapid generation of reactive oxygen species (ROS) can be observed (Wojakowska et al., 2013; Gao et al., 2017). While ROS cannot prevent the development of macroscopic symptoms, its production limits the invasiveness of pathogens (Zou et al., 2005). Reactive oxygen species strengthens cell walls through oxidative crosslinking of glycoproteins, and acts as a second messenger in some cell signaling pathways (Lamb and Dixon, 1997). However, ROS also exhibits potential toxicity as its increased accumulation can accelerate membrane lipid peroxidation (Mittler, 2002; Mittler et al., 2004). In order to increase their resistance to oxidative damage, plants require high levels of antioxidant enzymes (Sudhakar et al., 2001). The antioxidant enzymes or ROS scavengers can reduce ROS levels (Keppler and Baker, 1989). Various ROS scavengers, including ascorbic acid peroxidase (PER), glutathione, superoxide dismutase (SOD), and catalase (CAT) maintain ROS homeostasis in different compartments of plant cells (Mittler et al., 2004). Most of the enzymes involved in ROS detoxification, such as glutathione S-transferase (GST), CAT, PER, and glutathione reductase (GR), were up-regulated in COR treated samples, while SOD-related enzymes were down-regulated (Table 4).
COR Regulation of JA Biosynthesis and Related Hormone Pathways
Plant hormones not only play an essential role in growth and development, but they also contribute to how plants respond to both biotic and abiotic stressors. For instance, Jasmonate Acid (JA) modulates developmental as well as innate immune responses. Jasmonate Acid belongs to a small class of lipid-derived molecules and acts as a salicylic acid (SA) antagonist and an ethylene (ET) synergist (Gfeller et al., 2010). The first step in JA synthesis is accomplished by lipoxygenase (LOX)-catalyzed oxidation of linolenic acid (Vick and Zimmerman, 1984). In the JA synthesis pathway, 12 genes were up-regulated and only 2 genes were down-regulated. Among the genes encoding LOX, two genes were down-regulated, while five genes were up-regulated. In particular, the genes encoding allene oxide synthase (AOS) and allene oxide cyclase (AOC) were all up-regulated, suggesting that COR induced the activation of the JA biosynthetic pathway (Supplementary Table 2). Coronatine treatment also affected the expression of ethylene (ET) synthesis genes (Supplementary Table 2). In this study, a total of 14 DEGs involved in ethylene synthesis were identified, of which 13 genes were up-regulated and 1 gene was down-regulated (Supplementary Table 2), indicating that COR could promote ethylene biosynthesis.
COR Induced the Expression of Genes Involved in the Production of Stress Associated Proteins and Plant Secondary Metabolites
The accumulation of secondary metabolites constitutes an important component of plant stress response (Dixon et al., 2002). Some secondary metabolites play important defense roles during pathogen invasion (Erb and Kliebenstein, 2020). Vegetable Storage Protein (VSP) can be rapidly synthesized or degraded and plays an important role in the process of plant stress adaptation (Staswick et al., 1991). In our study VSP was up-regulated upon COR treatment (Table 4). Glucosinolates (GS) are also defense-related secondary metabolites, and related genes were also up-regulated in the presence of COR (Table 4). Coronatine also induced the expression of key enzymes in the phenylpropane pathway such as phenylalanineammonialyase (PAL), chalcone synthase (CHS), and phenylalanineammonialyase (PAL) (Table 4, Supplementary Table 3). These results suggest that COR may affect plant defense responses by regulating plant secondary metabolism, which is known to play important roles during plant pathogenesis.
qRT-PCR Validation of RNA-Seq Results
A total of 5 DEGs (GLYMA_18G057900, GLYMA_05G039900, GLYMA_17G023100, GLYMA_04G166900, and GLYMA_18G072200) identified through RNA-Seq analysis were selected for qRT-PCR verification. Our comparison results (Figure 4) showed that the relative expression levels of these genes in RNA-seq and qRT-PCR followed the same trend, suggesting that the results of RNA-seq were reliable.
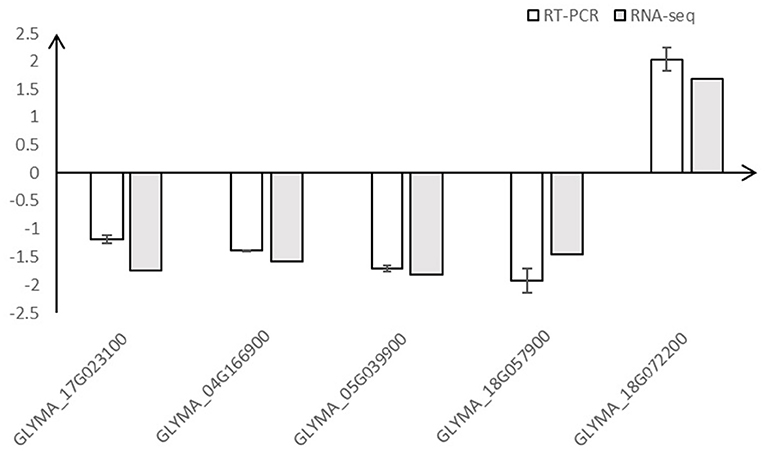
Figure 4. qRT-PCR validation of DEGs identified by RNA-seq. Five differentially expressed genes (GLYMA_18G057900, GLYMA_05G039900, GLYMA_17G023100, GLYMA_04G166900, and GLYMA_18G072200) identified through RNA-Seq analysis were selected for qRT-PCR validation, and the gene expression changes between COR-treated and mock samples detected by RNA-seq and qRT-PCR were compared. The white columns represent the mean fold change of the five genes through qRT-PCR analysis (error bars represent standard error of mean) while the gray bars represent fold changes obtained through RNA-Seq analysis. Pearson correlation coefficient used to measure the strength of the correlation between both methods showed a significant positive correlation (r = 0.974, p = 0.0053).
Discussion
Bacterial COR has been shown to induce the activation of LOXs, which catalyze the synthesis of JA and MeJA. Through a positive feedback loop, JA can also activate LOXs and promote JA accumulation even further (Bell and Mullet, 1991). Both COR and JA induce ethylene biosynthesis (Czapski and Saniewski, 1992; Uppalapati et al., 2005) which plays an important role in the development of chlorotic symptoms associated with soybean speck disease (Lund et al., 1998). During plant–pathogen interactions, SA signaling activates resistance against biotrophic and hemi-biotrophic pathogens, whereas JA and ethylene (ET) activate resistance against necrotrophic pathogens while suppressing SA dependent signaling (Glazebrook, 2005). As bacteria expressing COR induce the production of JA and ET, we would expect a dampening of SA-dependent responses and enhanced growth of biotrophic pathogens. Coronatine-induced ethylene has also been shown to accelerate plant senescence and inhibit photosynthesis in a dose-dependent manner (Ueda and Kato, 1980; Stall and Hall, 1985; Kenyon and Turner, 1990; Bleecker and Kende, 2000; He et al., 2002; Uppalapati et al., 2005). Consistent with previous studies, we showed that in COR-treated soybean leaves, photosynthetic related genes were down-regulated, suggesting that COR is involved in inducing chlorotic symptoms through the inhibition of the photosynthetic machinery.
Reactive oxygen species generation is one of the initial responses to pathogen invasion (Lamb and Dixon, 1997). High levels of ROS are produced in plant cells undergoing stress. Superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (OH−) are the main forms of ROS produced during photosynthesis (Karpinski et al., 2003; Apel and Hirt, 2004). Pseudomonas syringae infection is reported to result in COR-induced decomposition of chlorophyll, which in turn promotes ROS production (Mur et al., 2010). As high level of ROS leads to cell toxicity (Mittler, 2002), its overproduction usually results in the generation of free radical scavenging enzymes. GST is one such enzyme, which is known to be involved in regulating redox potential of infected tissues (Zhang et al., 2015). Consistent with these findings, COR has been reported to cause GST mRNA accumulation 4–9 h after bacterial inoculation (Greulichi et al., 1995). However, not all antioxidant enzymes are up-regulated upon ROS generation. As some studies show that COR inhibits the expression of thylakoid-localized Cu/Zn SOD (Ishiga et al., 2009). These results may explain why we observed a down-regulation of SOD expression and an upregulation of GSTs upon COR treatment.
In Arabidopsis, COR also induces the expression of genes related to phenylpropane metabolism (Attaran et al., 2014). Flavonoids are secondary metabolites that are synthesized via the phenylpropane pathway and are involved in imparting resistance against necrotrophic pathogens. Lamb and Dixon (1997) proposed that the isoflavone phytoalexin pathway may also be the main participant involved in the modulation of oxidative stress responses (Lamb and Dixon, 1997; Winkel-Shirley, 2001). The up-regulation of PAL and CHS indicated that COR targets the phenylpropane metabolic pathway and promotes the synthesis of flavonoids which in turn induce the production of ROS-scavenging enzymes. Of note, ROS scavengers may be beneficial to the host as well as the pathogen. To increase the resistance to oxidative damage induced upon bacterial infection and to improve plant health, plants need high levels of these antioxidants. However, COR can inhibit the early onset of pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) and induce disease-related necrotic cell death at later stages of disease progression. While COR has not been shown to function as a ROS scavenger, tomato plants infected with COR-producing bacterial strains have reduced ROS levels through the activity of ROS scavenging enzymes (Ishiga et al., 2009). It is therefore not surprising that most ROS scavenger-related genes were up-regulated in our study.
In addition to ROS scavengers, we focused on other genes that were specifically regulated by COR. These included the vegetative storage proteins (VSP) and glucosinolate (GS). Vegetative storage proteins is known to be upregulated during insect herbivory and is specifically induced by JA (Benedetti et al., 1995). Other plant hormones below a certain concentration threshold do not modulate VSP levels in plant cells (Anderson et al., 1989). In Arabidopsis, VSP expression was not detected in the Arabidopsis coi1 mutant seedlings, and VSP in wild-type Arabidopsis seedlings was induced upon MeJA and COR treatments (Benedetti et al., 1995). Thioglucoside and its hydrolyzed metabolites are involved in multiple functions including antioxidant activity, chemical protection, and resistance to biotic and abiotic stressors (Guo et al., 2013). The ability of coi1 mutants to induce indole-GS through MeJA was severely impaired, suggesting that COI1 plays a positive role in inducing GS production (Mewis et al., 2006; Brader et al., 2007). It is worth noting that GS, VSP, and ethylene expression can be induced by both COR and JA, indicating that these genes may be induced by COR and JA in a similar manner. Interestingly, the ability of COR to induce chlorosis is also achieved through COI1 (Mecey et al., 2011). The above results indicate that COI1 plays a major role in the regulation of COR-induced responses in plants.
The phytotoxin COR plays an important role in promoting the pathogenicity of bacteria, and the virulent activity of bacteria lacking COR is significantly reduced (Geng et al., 2014). Our results showed that COR induced the expression of genes related to JA and ethylene synthesis, suggesting that COR may activate these two hormone pathways to regulate plant defenses. Coronatine also induced expression of Phenylalanine ammonia lyase (PAL), which is the key enzyme involved in the production of phenylpropane metabolites in plants. This indicated that COR could dampen plant defenses through perturbing phenylpropane metabolism. Our comprehensive transcriptional analysis of the interaction between COR and soybean through RNA-Seq would help better understand the non-host resistance (NHR) response in plants and would also provide a theoretical basis for the improvement of soybean resistance against non-adapted pathogens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XZ, BH, and XG designed the research strategy and analyzed the sequenced data. SS, ZZ, and TL conducted part of experiments. XZ, AJA, and XG wrote the manuscript. HW, ZL analyzed part of data. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Shanghai international scientific and technological cooperation foundation (No. 19390743400) and the key R&D and transformation program of Xining city (CN) (No. 2019-y-35).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Huada gene institution in China for providing technical assistance with bioinformatics analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.663238/full#supplementary-material
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, 1–12. doi: 10.1186/gb-2010-11-10-r106
Anderson, J. M., Spilatro, S. R., Klauer, S. F., and Franceschi, V. R. (1989). Jasmonic acid-dependent increase in the level of vegetative storage proteins in soybean. Plant Sci. 62, 45–52. doi: 10.1016/0168-9452(89)90188-X
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Attaran, E., Major, I. T., Cruz, J. A., Rosa, B. A., Koo, A. J., Chen, J., et al. (2014). Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 165, 1302–1314. doi: 10.1104/pp.114.239004
Bell, E., and Mullet, J. E. (1991). Lipoxygenase gene expression is modulated in plants by water deficit, wounding, and methyl jasmonate. Mol. Gen. Genet. 230, 456–462. doi: 10.1007/BF00280303
Bender, C. L., Alarconchaidez, F., and Gross, D. C. (1999). Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292. doi: 10.1128/MMBR.63.2.266-292.1999
Benedetti, C. E., Xie, D., and Turner, J. G. (1995). COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109, 567–572. doi: 10.1104/pp.109.2.567
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Brader, G., Djamei, A., Teige, M., Palva, E. T., and Hirt, H. (2007). The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol. Plant Microbe Interact. 20, 589–596. doi: 10.1094/MPMI-20-5-0589
Brooks, D. M., Hernández-Guzmán, G., Kloek, A. P., Alarcón-Chaidez, F., Sreedharan, A., Rangaswamy, V., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17, 162–174. doi: 10.1094/MPMI.2004.17.2.162
Czapski, J., and Saniewski, M. (1992). Stimulation of ethylene production and ethylene-forming enzyme activity in fruits of the non-ripening nor and rin tomato mutants by methyl jasmonate. J. Plant Physiol. 139, 265–268. doi: 10.1016/S0176-1617(11)80334-2
Dixon, D. P., Lapthorn, A., and Edwards, R. (2002). Plant glutathione transferases. Genome Biol. 3:reviews3004. doi: 10.1186/gb-2002-3-3-reviews3004
Erb, M., and Kliebenstein, D. J. (2020). Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 184, 39–52. doi: 10.1104/pp.20.00433
Ferguson, I., and Mitchell, R. (1985). Stimulation of ethylene production in bean leaf discs by the pseudomonad phytotoxin coronatine. Plant Physiol. 77, 969–973. doi: 10.1104/pp.77.4.969
Feys, B. J., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. doi: 10.2307/3869877
Galán, J. E. (2009). Common themes in the design and function of bacterial effectors. Cell Host Microbe 5, 571–579. doi: 10.1016/j.chom.2009.04.008
Gao, J.-X., Yu, C.-J., Wang, M., Sun, J.-N., Li, Y.-Q., and Chen, J. (2017). Involvement of a velvet protein ClVelB in the regulation of vegetative differentiation, oxidative stress response, secondary metabolism, and virulence in Curvularia lunata. Sci. Rep. 7, 1–13. doi: 10.1038/srep46054
Gao, S., Li, Y., Gao, J., Suo, Y., Fu, K., Li, Y., et al. (2014). Genome sequence and virulence variation-related transcriptome profiles of Curvularia lunata, an important maize pathogenic fungus. BMC Genomics 15:627. doi: 10.1186/1471-2164-15-627
Geng, X., Cheng, J., Gangadharan, A., and Mackey, D. (2012). The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24, 4763–4774. doi: 10.1105/tpc.112.105312
Geng, X., Jin, L., Shimada, M., Kim, M. G., and Mackey, D. (2014). The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 240, 1149–1165. doi: 10.1007/s00425-014-2151-x
Gfeller, A., Liechti, R., and Farmer, E. E. (2010). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3:cm4. doi: 10.1126/scisignal.3109cm4
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Greulichi, F., Yoshihara, T., and Ichihara, A. (1995). Coronatine, a bacterial phytotoxin, acts as a stereospecific analog of jasmonate type signals in tomato cells and potato tissues. J. Plant Physiol. 147, 359–366. doi: 10.1016/S0176-1617(11)82168-1
Guo, R., Shen, W., Qian, H., Zhang, M., Liu, L., and Wang, Q. (2013). Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana. J. Exp. Bot. 64, 5707–5719. doi: 10.1093/jxb/ert348
He, Y., Fukushige, H., Hildebrand, D. F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
Ishiga, Y., Uppalapati, S. R., Ishiga, T., Elavarthi, S., Martin, B., and Bender, C. L. (2009). Involvement of coronatine-inducible reactive oxygen species in bacterial speck disease of tomato. Plant Signal. Behav. 4, 237–239. doi: 10.4161/psb.4.3.7915
Karpinski, S., Gabrys, H., Mateo, A., Karpinska, B., and Mullineaux, P. M. (2003). Light perception in plant disease defence signalling. Curr. Opin. Plant Biol. 6, 390–396. doi: 10.1016/S1369-5266(03)00061-X
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105. doi: 10.1073/pnas.0802332105
Keen, N., and Buzzell, R. (1991). New disease resistance genes in soybean against Pseudomonas syringae pv glycinea: evidence that one of them interacts with a bacterial elicitor. Theor. Appl. Genet. 81, 133–138. doi: 10.1007/BF00226123
Kenyon, J., and Turner, J. (1990). Physiological changes in Nicotiana tabacum leaves during development of chlorosis caused by coronatine. Physiol. Mol. Plant Pathol. 37, 463–477. doi: 10.1016/0885-5765(90)90037-X
Kenyon, J. S., and Turner, J. G. (1992). The stimulation of ethylene synthesis in Nicotiana tabacum leaves by the phytotoxin coronatine. Plant Physiol. 100, 219–224. doi: 10.1104/pp.100.1.219
Keppler, L. D., and Baker, C. J. (1989). O2-initiated lipid peroxidation in a bacteria-induced hypersensitive reaction in tobacco cell suspensions. Phytopathology 79, 555–562. doi: 10.1094/phyto-79-555
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Lamb, C., and Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Lund, S. T., Stall, R. E., and Klee, H. J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10, 371–382. doi: 10.2307/3870595
Mecey, C., Hauck, P., Trapp, M., Pumplin, N., Plovanich, A., Yao, J., et al. (2011). A critical role of STAYGREEN/Mendel's I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 157, 1965–1974. doi: 10.1104/pp.111.181826
Mewis, I., Tokuhisa, J. G., Schultz, J. C., Appel, H. M., Ulrichs, C., and Gershenzon, J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67, 2450–2462. doi: 10.1016/j.phytochem.2006.09.004
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mur, L. A. J., Aubry, S., Mondhe, M., Kingstonsmith, A. H., Gallagher, J., Timmstaravella, E., et al. (2010). Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 188, 161–174. doi: 10.1111/j.1469-8137.2010.03377.x
Palmer, D., and Bender, C. (1995). Utrastructure of tomato leaf tissue treated with the pseudomonad phytotoxin coronatine and comparison with methyl jasmonate. Mol. Plant Microbe Interact. 8, 683–692. doi: 10.1094/MPMI-8-0683
Rio, D. C., Ares, M., Hannon, G. J., and Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010:pdb.prot5439. doi: 10.1101/pdb.prot5439
Stall, R., and Hall, C. (1985). Chlorosis and ethylene production in pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Phytopathology 74, 373–375.
Staswick, P. E., Huang, J.-F., and Rhee, Y. (1991). Nitrogen and methyl jasmonate induction of soybean vegetative storage protein genes. Plant Physiol. 96, 130–136. doi: 10.1104/pp.96.1.130
Sudhakar, C., Lakshmi, A., and Giridarakumar, S. (2001). Changes in the antioxidant enzyme efficacy in two high yielding genotypes of mulberry (Morus alba L.) under NaCl salinity. Plant Sci. 161, 613–619. doi: 10.1016/S0168-9452(01)00450-2
Takamiya, K.-I., Tsuchiya, T., and Ohta, H. (2000). Degradation pathway (s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci. 5, 426–431. doi: 10.1016/S1360-1385(00)01735-0
Ueda, J., and Kato, J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66, 246–249. doi: 10.1104/pp.66.2.246
Uppalapati, S. R., Ayoubi, P., Weng, H., Palmer, D. A., Mitchell, R. E., Jones, W., et al. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42, 201–217. doi: 10.1111/j.1365-313X.2005.02366.x
Vick, B. A., and Zimmerman, D. C. (1984). Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 75, 458–461. doi: 10.1104/pp.75.2.458
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wojakowska, A., Muth, D., Narozna, D., Madrzak, C., Stobiecki, M., and Kachlicki, P. (2013). Changes of phenolic secondary metabolite profiles in the reaction of narrow leaf lupin (Lupinus angustifolius) plants to infections with Colletotrichum lupini fungus or treatment with its toxin. Metabolomics 9, 575–589. doi: 10.1007/s11306-012-0475-8
Wrather, J. A., Anderson, T., Arsyad, D., Gai, J., Ploper, L., Porta-Puglia, A., et al. (1997). Soybean disease loss estimates for the top 10 soybean producing countries in 1994. Plant Dis. 81, 107–110. doi: 10.1094/PDIS.1997.81.1.107
Xie, Z., Duan, L., Tian, X., Wang, B., Eneji, A. E., and Li, Z. (2008). Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. J. Plant Physiol. 165, 375–384. doi: 10.1016/j.jplph.2007.06.001
Yu, C., Fan, L., Wu, Q., Fu, K., Gao, S., Wang, M., et al. (2014). Biological role of Trichoderma harzianum-derived platelet-activating factor acetylhydrolase (PAF-AH) on stress response and antagonism. PLoS ONE 9:e100367. doi: 10.1371/journal.pone.0100367
Zhang, Z., Zhang, X., Hu, Z., Wang, S., Zhang, J., Wang, X., et al. (2015). Lack of K-dependent oxidative stress in cotton roots following coronatine-induced ROS accumulation. PLoS ONE 10:e0126476. doi: 10.1371/journal.pone.0126476
Zhao, Y., Jones, W., Sutherland, P., Palmer, D., Mitchell, R., Reynolds, P., et al. (2001). Detection of the phytotoxin coronatine by ELISA and localization in infected plant tissue. Physiol. Mol. Plant Pathol. 58, 247–258. doi: 10.1006/pmpp.2001.0334
Zheng, X. Y., Spivey, N. W., Zeng, W., Liu, P. P., Fu, Z. Q., Klessig, D. F., et al. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596. doi: 10.1016/j.chom.2012.04.014
Zou, J., Rodriguez-Zas, S., Aldea, M., Li, M., Zhu, J., Gonzalez, D. O., et al. (2005). Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol. Plant Microbe Interact. 18, 1161–1174. doi: 10.1094/MPMI-18-1161
Keywords: soybean, Pseudomonas syringae, coronatine, RNA-Seq, differentially expressed genes
Citation: Zhang X, He B, Sun S, Zhang Z, Li T, Wang H, Liu Z, Afzal AJ and Geng X (2021) Transcriptome Analysis Identified Gene Regulation Networks in Soybean Leaves Perturbed by the Coronatine Toxin. Front. Sustain. Food Syst. 5:663238. doi: 10.3389/fsufs.2021.663238
Received: 02 February 2021; Accepted: 18 March 2021;
Published: 19 April 2021.
Edited by:
Wei Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Gang Wei, Fudan University, ChinaJinxin Gao, New York University, United States
Songbiao Chen, Minjiang University, China
Copyright © 2021 Zhang, He, Sun, Zhang, Li, Wang, Liu, Afzal and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueqing Geng, eHFnZW5nQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Xiong Zhang1†
Xiong Zhang1† Ahmed Jawaad Afzal
Ahmed Jawaad Afzal Xueqing Geng
Xueqing Geng