- 1Department of Food and Nutrition, Chung-Ang University, Anseong-si, Gyeonggi-do, Republic of Korea
- 2Department of Food and Nutrition, Gwangju University, Gwangju, Republic of Korea
- 3Division of Food Bioscience and Technology, College of Life Sciences and Biotechnology, Korea University, Seoul, Republic of Korea
- 4Bio and Environmental Technology Research Institute, Chung-Ang University, Seoul, Republic of Korea
Introduction: Foodborne viruses are a serious concern in public health. This study investigated the prevalence of eight foodborne viruses norovirus (NoV), adenovirus (AdV), sapovirus (SapoV), astrovirus, hepatitis A virus (HAV), hepatitis E virus (HEV), rotavirus, aichivirus, and influenza A virus (IAV).
Material and method: A total of 316 chicken samples were collected from three poultry processing plants to commercial markets (local and online). RT-qPCR- and PCR-positive amplicons obtained from monitoring were confirmed by sequence analysis.
Results: Foodborne viruses and IAV were not found in poultry processing plants. Of the 100 chickens purchased from the local and online markets, 19 (19.0%) AdV and 2 (2.0%) SapoV were detected. NoV, astrovirus, HAV, HEV, rotavirus, aichivirus, and IAV were not detected in the retailed chickens. Phylogenetic analysis identified 18 human AdV-41, one porcine AdV, and two SapoV-GI.1. It was the first case of the discovery of the SapoV gene in chicken. The average contamination level of detected AdV was 2.4 log DNA copies/g, but there were cases where the highest level was 5.35 log DNA copies/g.
Discussion: This study highlights the importance of chicken's contribution to the transmission of AdV with the possibility of annual variability with emerging symptoms. The prevention of AdV contamination in the food chain from slaughterhouses to retail markets should be monitored and controlled in further study.
1. Introduction
Meat consumption is increasing worldwide. Poultry consumption is expected to increase in high-income countries, driven by a growing preference for white meat, which is more convenient to prepare and perceived as a better food choice (OECD-FAO, 2017, 2022). Furthermore, consumption continues to increase in low- and middle-income countries as poultry is cheaper than other meats. According to the OECD and FAO reports in 2022, poultry meat is expected to account for 47% of the protein consumed from meat sources globally by 2031 due to a favorable meat-to-feed price ratio compared to other ruminants combined with short production cycles (OECD-FAO, 2022). Among the poultry, chicken dominates meat consumption as it is generally affordable, low in fat, and faces few religious and cultural barriers (OECD-FAO, 2017).
Foodborne illnesses from chicken consumption were often caused by Campylobacter and Salmonella spp. bacteria (Mwangi et al., 2019; Frosth et al., 2020; Golden and Mishra, 2020; Zhuge et al., 2021). There were fewer relations between foodborne viruses and chicken in the viral metagenomic research on chicken (Zhang et al., 2014). However, since the 2019 SARS-CoV-2 pandemic, there has been an ever-growing list of emerging viral pathogens that could threaten the food supply (Tetro, 2014; Ceylan et al., 2020). These include well-known foodborne viruses such as norovirus (NoV), adenovirus (AdV), sapovirus (SapoV), astrovirus, hepatitis A virus (HAV), hepatitis E virus (HEV), rotavirus, and avian influenza, as well as the less well-known aichivirus (Tetro, 2014).
Foodborne viruses were transmitted to humans primarily through the consumption of contaminated food or water (Tetro, 2014). Common symptoms of foodborne viruses are fever, vomiting, diarrhea, and headache (Pal and Ayele, 2020). Furthermore, depending on the severity of the infection, HAV causes jaundice and acute hepatitis (Gholizadeh et al., 2023). The symptoms of HEV vary from mild to severe, with jaundice appearing rapidly and acute liver failure occurring in <1% of patients (Webb et al., 2020). In South Korea, HAV and HEV were designated as class 2 infectious diseases, and NoV, AdV, SapoV, astrovirus, and rotavirus A were defined as class 4 intestinal infectious diseases according to the Waterborne and Foodborne Infectious Diseases Control Guidelines (KCDC, 2022). Symptomatic cases of HAV were reported: 17,598 cases in 2019 and 3,989 cases in 2020 (KCDC, 2022). One hundred and ninety-one HEV symptomatic cases have been reported since it was added to legal infectious diseases in 2020 (KCDC, 2022). Rotavirus A, astrovirus, AdV, NoV, and SapoV were reported in 2020 as 1,416, 201, 192, 3,219, and 70 cases, respectively (KCDC, 2022). Cases of outbreaks occurred in the order of NoV (121 cases, 27.0%), Campylobacter spp. (24 cases, 5.1%), and Salmonella spp. (22 cases, 4.7%) as the causative pathogens in 2021 (KCDC, 2022). Foodborne viral illness is primarily caused by NoV and HAV through the consumption of shellfish (oyster, clam) (Cho et al., 2021; Hyun et al., 2022; Yoo et al., 2022).
Outbreaks of foodborne virus disease caused by chicken consumption resulted in 59 outbreaks, 1,548 illnesses, 28 hospitalizations, and one death from 2009 to 2020 in the USA (Dewey-Mattia et al., 2018; White et al., 2022). In the past 15 years, the highest number of reported illness was 337 cases, and in 2018, there were reports of NoV GII cases linked to consumption of chicken wraps (White et al., 2022). One death from NoV occurred after eating chicken dishes from a restaurant in California in 2014 (White et al., 2022). Other than NoV cases, one of them involved eating chicken coconut curry due to SapoV infection in 2015 (Dewey-Mattia et al., 2018). In addition, there were 108 cases of rotavirus A outbreak caused by eating chicken or tuna salad sandwiches in 2000 (CDC, 2000).
Since 2009, due to the swine flu pandemic (also known as the “novel flu virus”) caused by highly pathogenic influenza A virus (IAV) H1N1 (strain pandemic H1N1/09 virus), there have been 700 million to 1.4 billion estimated suspension cases worldwide, and 18,449 people have died (WHO, 2009, 2011; CIDRAP, 2013). Since the large-scale pandemic, there has been a continuous awareness of IAV among consumers and farmers (Cui et al., 2022). In Taiwan, the IAV H5N1 virus was detected in live birds, and surface samples were collected from the food market in 2004–2005 (Amonsin et al., 2008). In 2003, the IAV H5N1 was isolated from imported poultry meat in Japan (Mase et al., 2005). Due to these risks and consumer awareness, IAV infection through food intake is also a concern in food safety.
Most foodborne diseases are under-reported or not reported at all, leaving many gaps in our understanding of how viruses affect food (Tetro, 2014). In this study, the foodborne viruses (NoV, AdV, SapoV, astrovirus, HAV, HEV, rotavirus, and aichivirus) and IAV prevalence from poultry processing plants to commercial markets (local and online) were studied through multilateral cooperation according to the food distribution channels. Phylogenetic analysis was used to investigate the relationship between the detected viruses.
2. Materials and methods
2.1. Ethical statement
This study only used chicken meat samples without animal or patient material and thus did not require approval from the ethics committee.
2.2. Sample collection from poultry processing plants and commercial markets (local and online)
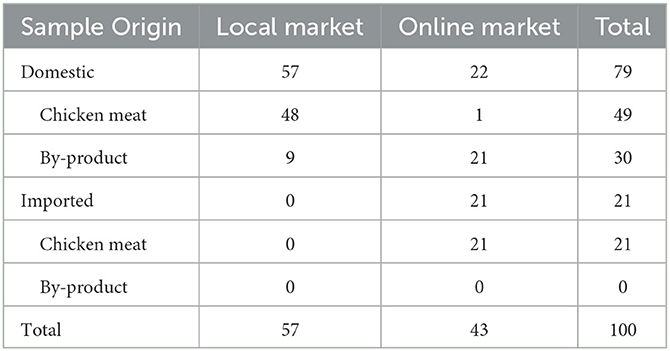
We sampled 317 samples of chicken from poultry processing plants and commercial markets (local and online markets). Local markets are physical marketplaces for buying and selling goods and services in a specific area, while online markets are digital marketplaces that allow for buying and selling goods and services over the internet, from anywhere.
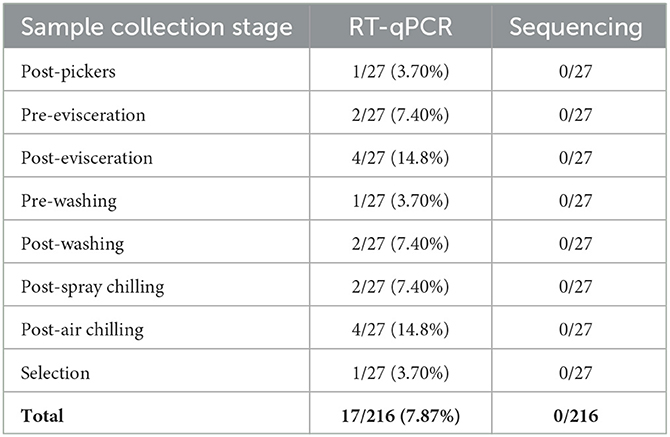
The schematic flow of the poultry processing line is shown in Supplementary Figure 1. Eight red arrows indicated the collection parts: post-pickers, pre-evisceration, post-evisceration, pre-washing, post-washing, post-spray chilling, post-air chilling, and selection. A total of 216 samples were collected by dividing three chickens into three parts (legs, wings, and breasts) at each of the eight stages in three units.
Chicken samples were collected depending on the commercial market type. A total of 57 and 43 chickens were purchased from the local and online markets, respectively. Among the 43 chickens from the online markets, 22 were domestic and 21 were imported. All the chickens purchased in local markets were domestic (Table 1).
2.3. Sample preparation and nucleic acid extraction
Nucleic acid extraction was used to prepare the sample after elution and concentration (Son et al., 2014). Chicken legs and wings were deboned using sterilized scissors and a knife, and only the meat parts were collected. Consequently, it was homogenized to take 10 g. For chicken breast and gizzard, 2 g was taken for each spot and an appropriate amount (over 10 g) was randomly sampled. After homogenization, 10 g was taken. The chicken meat homogenate was eluted in phosphate-buffered saline (PBS, pH 7.4) elution buffer at a ratio of 1:10. After elution, the elute was centrifuged at 8,000 × g for 30 min at 4°C. The first supernatant was collected in a new tube, while the remnant pellet was mixed with the same amount of elution buffer. After a second centrifugation using the same conditions, the second supernatant was pooled with the first supernatant (Son et al., 2014). The sample elution was used for concentration using vivaspin-20 ultrafiltration with a nominal molecular weight limit of 10 kDa (Sartorius, Göttingen, Germany). The tissue homogenate was ultra-filtered with centrifugation at 8,000 × g for 30 min at 4°C (Son et al., 2014). The final concentration was extracted nucleic acid using an RNeasy mini kit (Qiagen, Hilden, Germany), following the manufacturer's protocol. The final elution was performed once with a 50 μl elution buffer. The nucleic acid was stored at −80°C until analysis (Tian et al., 2010).
2.4. RT-qPCR or qPCR for virus monitoring
One-step RT-qPCR or qPCR was used for investigating eight foodborne viruses (NoV, AdV, SapoV, astrovirus, HAV, HEV, rotavirus, and aichivirus) and IAV. Primers and probes for one-step RT-qPCR or qPCR are presented in Table 2 (Spackman et al., 2002; Tian et al., 2010; Nielsen et al., 2013; Shin et al., 2019; Wang et al., 2020; Lee al., 2021; Yeo et al., 2022). One-step RT-qPCR for each RNA virus was performed using a one-step RT-PCR kit (Qiagen, Hilden, Germany). AdV was identified by qPCR using Premix Ex Taq (2X)TM (Takara, Shiga, Japan) performed on the CFX96TM Real-Time PCR system (Bio-Rad, California, United States). Titration of AdV was used with quantitative genomic DNA of human AdV-41 (ATCC® VR-930DQ, Virginia, United States) (Yeo et al., 2022).
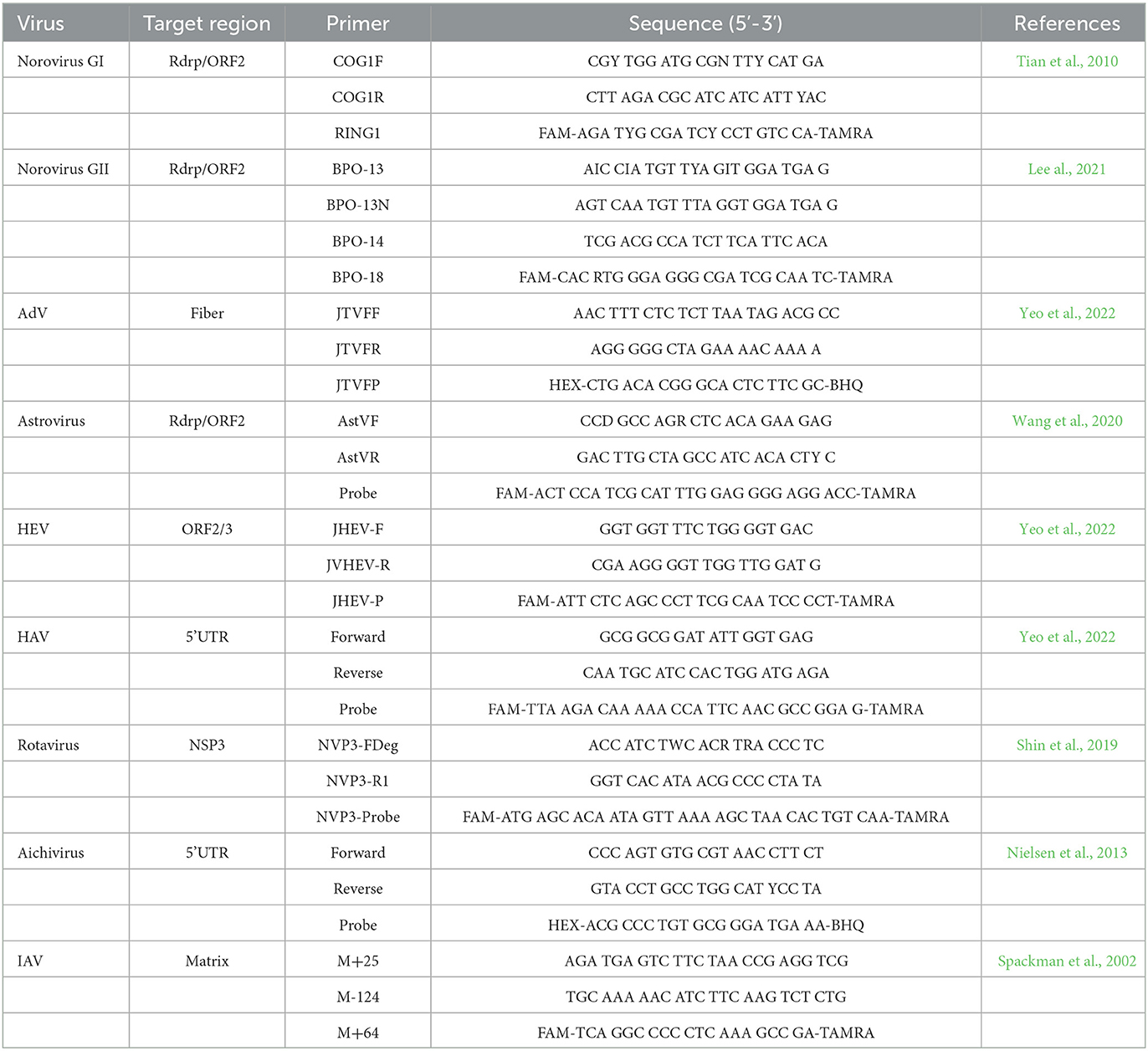
Table 2. Primers and probes for foodborne viruses and influenza A virus monitoring in RT-qPCR and qPCR.
2.5. Sequencing analysis and phylogenetic tree
For genotype determination, we applied sequencing analysis, and nested RT-PCR or PCR was performed to obtain an amplicon. The AdV was amplified by nested PCR using primer: hex1deg (17,602–17,626: 5′-GCC SCA RTG GKC WTA CAT GCA CAT C-3′) and hex2deg (17,891–17,921: 5′-CAG CAC SCC ICG RAT GTC AAA-3′) for the amplification of the outer hexon gene. Nehex3deg (16,452–16,4796: 5′-GCC CHY GCM ACI GAI ACS TAC TTC-3′) and nehex4deg (16,594–16,623: 5′-CCY ACR GCC AGI GTR WAI CGM RCY TTG TA-3′) were used for inner gene amplification and direct sequencing (Lee al., 2021).
SapoV was performed using the one-step RT-PCR kit (Bioneer, Daejeon, South Korea) using primers: SV-F11 (5,098–5,117: 5′-GCY TGG TTY ATA GGT GGT AC-3′), SV-R1 (5,878–5,857: 5′-CWG GTG AMA CMC CAT TKT CCA T-3′), SV-F21 (5,157–5,177: 5′- ANT AGT GTT TGA RAT GGA GGG-3′), and SV-R2 (5,591–5,572: 5′- GWG GGR TCA ACM CCW GGT GG-3′). SV-F21 and SV-R2 were used for direct sequencing primers (Okada et al., 2002).
Influenza A virus-positive samples were amplified by a one-step RT-PCR kit (Bioneer, Daejeon, South Korea) with two sets of pan-primers for the Haemagglutinin (HA) and Neuraminidase (NA) genes. For HA gene amplification, Bm-HA-1 (5′-TAT TCG TCT CAG GGA GCA AAA GCA GGG G-3′) and Bm-NS-890R (5′-ATT CGT CTC GTA TTA GTA GAA ACA AGG GTG TTT T-3′) were used as a primer to obtain a PCR product of 1,800 bp (Hoffmann et al., 2001). NA gene was amplified using primers Bm-NA-1 (5′-TAT TGG TCT CAG GGA GCA AAA GCA GGG G-3′) and Ba-NA-1413R (5′-ATT GGT CTC GTA TTA GTA GAA ACA AGG AGT TTT T-3′) as 1,500 bp (Hoffmann et al., 2001).
The PCR products were purified using ExoSAP-IT™ (Applied Biosystems, Waltham, United States). Purified samples were directly sequenced on a SeqStudio using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Waltham, United States). All nucleotide sequences were first edited with the SeqMan program (DNASTAR, Madison, WI, United States), and the sequences were analyzed with different viral genotype sequences using BLAST (Anaclerio et al., 2021). Multiple alignments of different nucleotide sequences of the AdV and SaopV reference sequences were performed using DNASTAR Lasergene MegAlign Pro (DNASTAR Inc., WI, United States). Phylogenetic analysis of aligned sequences was performed in DNASTAR Lasergene MegAlign Pro (DNASTAR Inc., WI, United States), using RAxML v8 under the GTR GAMMA model with 500 rapid bootstrapping replicates and a search for the best-scoring maximum likelihood tree (Stamatakis, 2014). The reference sequences to build the phylogenetic tree were based on BLAST results with a high E-value (0.001) depending on the genotype. The GenBank accession number was presented in Supplementary Table S1.
2.6. Statistical analysis
The differences in detection rates of foodborne viruses and IAV between the poultry processing plants and market samples were analyzed using one-way Student's t-tests. P-values of <0.05 were considered statistically significant. All statistical analyses were performed using R version 4.1.0.
3. Results
3.1. Prevalence of foodborne viruses and IAV from poultry processing plants to retailed chickens
As a result of the investigation, there were no foodborne viruses, and IAVs were identified through sequencing analysis in each processing unit of the three poultry processing plants (A, B, and C). In each step of the processing unit, the IAV was detected by RT-qPCR. IAV-suspected samples were identified in each step of sampling (Table 3). However, none of the IAVs were confirmed by sequencing.
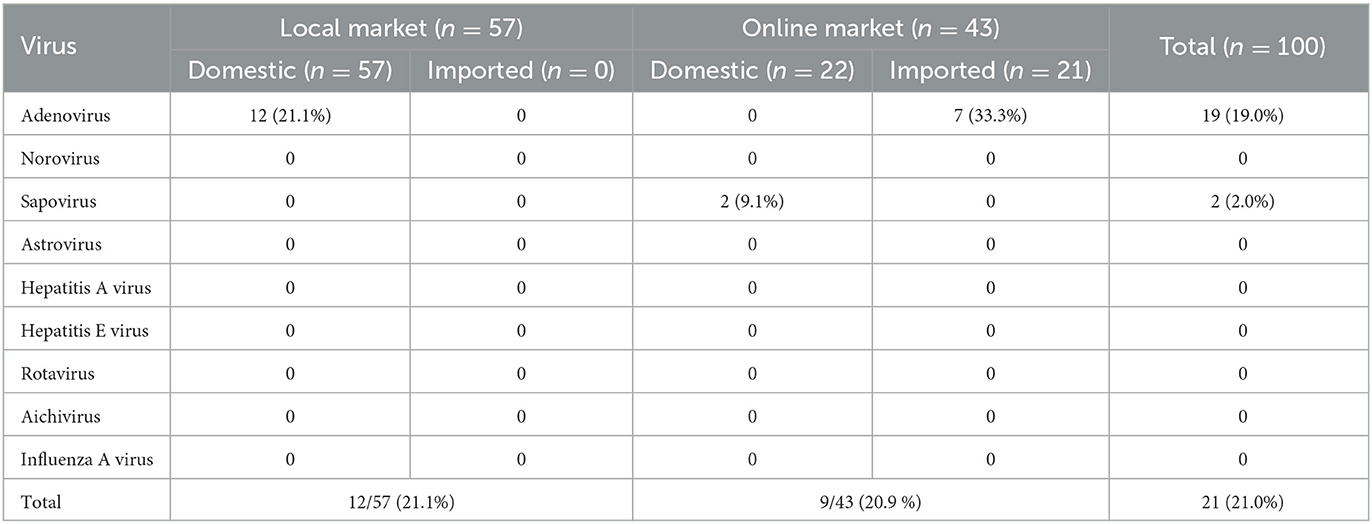
Of the 100 chickens from the purchased local and online markets, 19 (19.0%) of AdV and 2 (2.0%) of SapoV were positive for foodborne viruses (Table 4). NoV GI, NoV GII, astrovirus, HAV, HEV, rotavirus, aichivirus, and IAV were not identified in the retailed chickens. Among 18 human AdV-41 positive samples, six were identified in imported chicken legs purchased from the online market, and 12 were domestic chicken purchased from the local market. Human AdV-41 was identified in two, one, eight, and one cases of chicken breast, wing, leg, and gizzard from the chicken meat purchased from the local market. One porcine AdV was identified from an imported chicken leg purchased from the online market. All samples contaminated with SapoV were domestic gizzards purchased from the online market (Supplementary Table S1). In this group, detection rates of foodborne viruses in the local and online markets were similar at 21.1% (12/57) and 20.9% (9/43) which was not a statistically significant difference (p = 0.563). The detection rate of the foodborne virus by country of origin was 33.3% (7/21) for imported chicken and 17.7% (14/79) for domestic chicken. Based on the statistical analysis, the detection rate of foodborne viruses from the imported chicken was significantly higher than that from domestic chicken (p = 0.045).
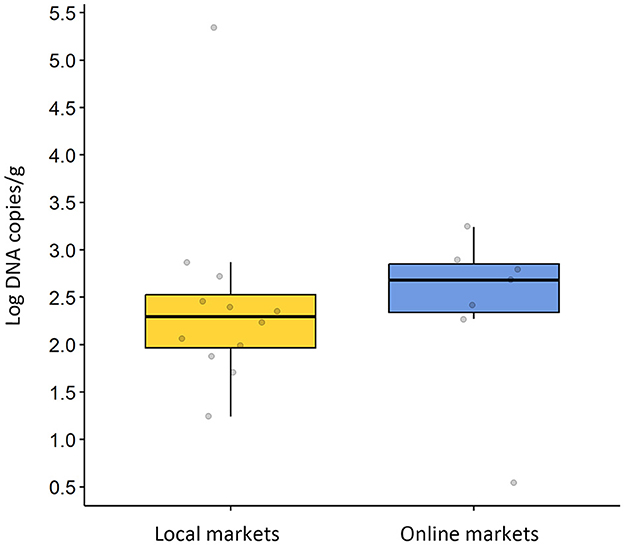
The average AdV viral copy number was similar to 2.43 log DNA copies/g at the local market and 2.40 log RNA copies/g at the online market, as shown in Figure 1. There was no statistically significant difference in the average AdV viral copy number (p = 0.941). There were distinct differences in AdV titer (genome copy number) for each sampling, with a maximum of 3.24 log DNA copies/g and a minimum of 0.54 log DNA copies/g when purchased from the online market. For chickens purchased from the local market, the highest AdV contamination level was 5.35 log DNA copies/g and the minimum value was 1.24 log DNA copies/g. The information on virus-detected samples is presented in Supplementary Table S1.
3.2. Phylogenetic analysis of AdV in chicken samples
The AdV-positive samples were analyzed for the hexon gene using a phylogenetic tree, which was shown in Figure 2. Eleven AdV-41, twelve AdV-40, and eleven porcine AdV strain sequences were used as reference strains. Nineteen AdV genotypes were detected for the hexon gene, 18 were human AdV-41, and one was porcine AdV.
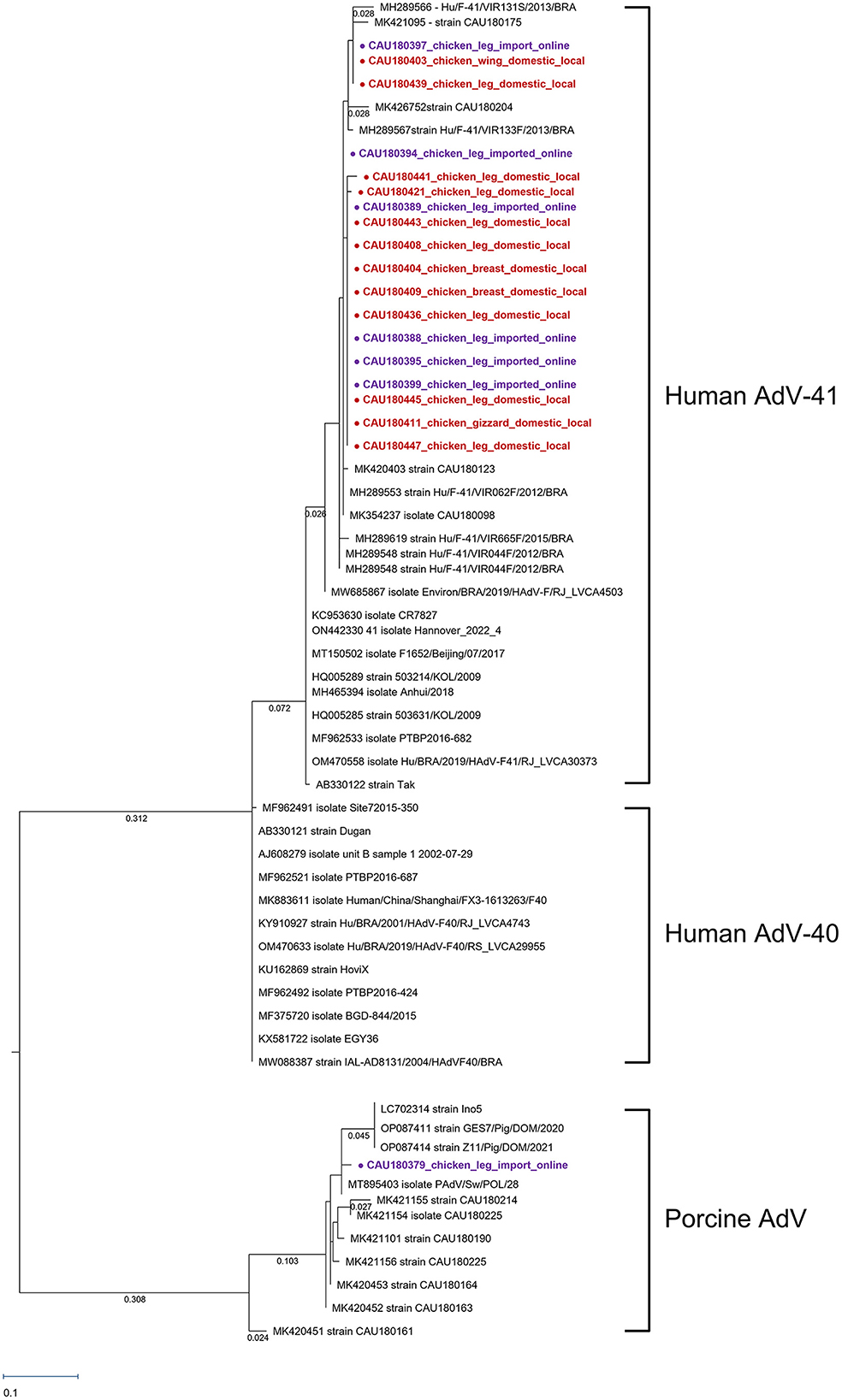
Figure 2. Phylogenetic tree of AdV detected in chickens. Sequences detected in this study are represented as circles ( ). The red circles (
). The red circles ( ) were AdV samples purchased in local markets, and the purple circles (
) were AdV samples purchased in local markets, and the purple circles ( ) were AdV samples purchased in online markets. Phylogenetic tree analysis for the AdV hexon gene using the maximum likelihood method and based on the 171-bp sequence. The numbers of compressed sequence distances were shown in parentheses.
) were AdV samples purchased in online markets. Phylogenetic tree analysis for the AdV hexon gene using the maximum likelihood method and based on the 171-bp sequence. The numbers of compressed sequence distances were shown in parentheses.
One human AdV-41 clade was formed with accession numbers MH289566–MK354237. The 18 human AdV-41 sequences detected had a nucleotide sequence identity of 98.54% with the reference AB330122 (strain Tak). Furthermore, the nucleotide sequence difference among the 18 human AdV-41 sequences was only 0.14%, indicating a high level of similarity between these sequences.
Through phylogenetic analysis, CAU180379 was found to be related to porcine AdV. There was a high nucleotide sequence similarity of 99.27% with reference strain MT895403. The detected CAU180379 sequence had the same clade with reference strain LC70231 identified as porcine AdV type 5, but a different root from reference strain MK420451 identified as porcine AdV type 3. Phylogenetic analyses suggest that the CAU180379 strain sequences detected in this study could be of porcine AdV type 5 origin.
3.3. Phylogenetic analysis of SapoV in chicken samples
We analyzed the SapoV VP1 capsid region, as presented in Figure 3. As reference strains—15, 5, 3, 2, 3, and 2—SapoV-GI.1, SapoV-GI.2, SapoV-GI.3, SapoV-GII, SapoV-GV, and SapoV-GIII were used, respectively. As a result of phylogenetic analysis, two detected SapoV were of the GI.1 type. A single clade composed of SapoV-GI has three sub-genotypes. Both detected SapoV-GI (CAU180457 and CAU180464) have high identities with MK450330 (99.5% nucleotide sequence identity). In addition, the CAU180457 and CAU180464 detected were 100% identical. The nucleotide sequence identity with the SapoV-GI.1, KP298674 reported from South Korea strain (Hu/GI.1/Seoul/ROK62/2013/KOR), was 96.1%. Both discovered viruses were found in samples of chicken gizzards, however, the samples were obtained from separate companies.
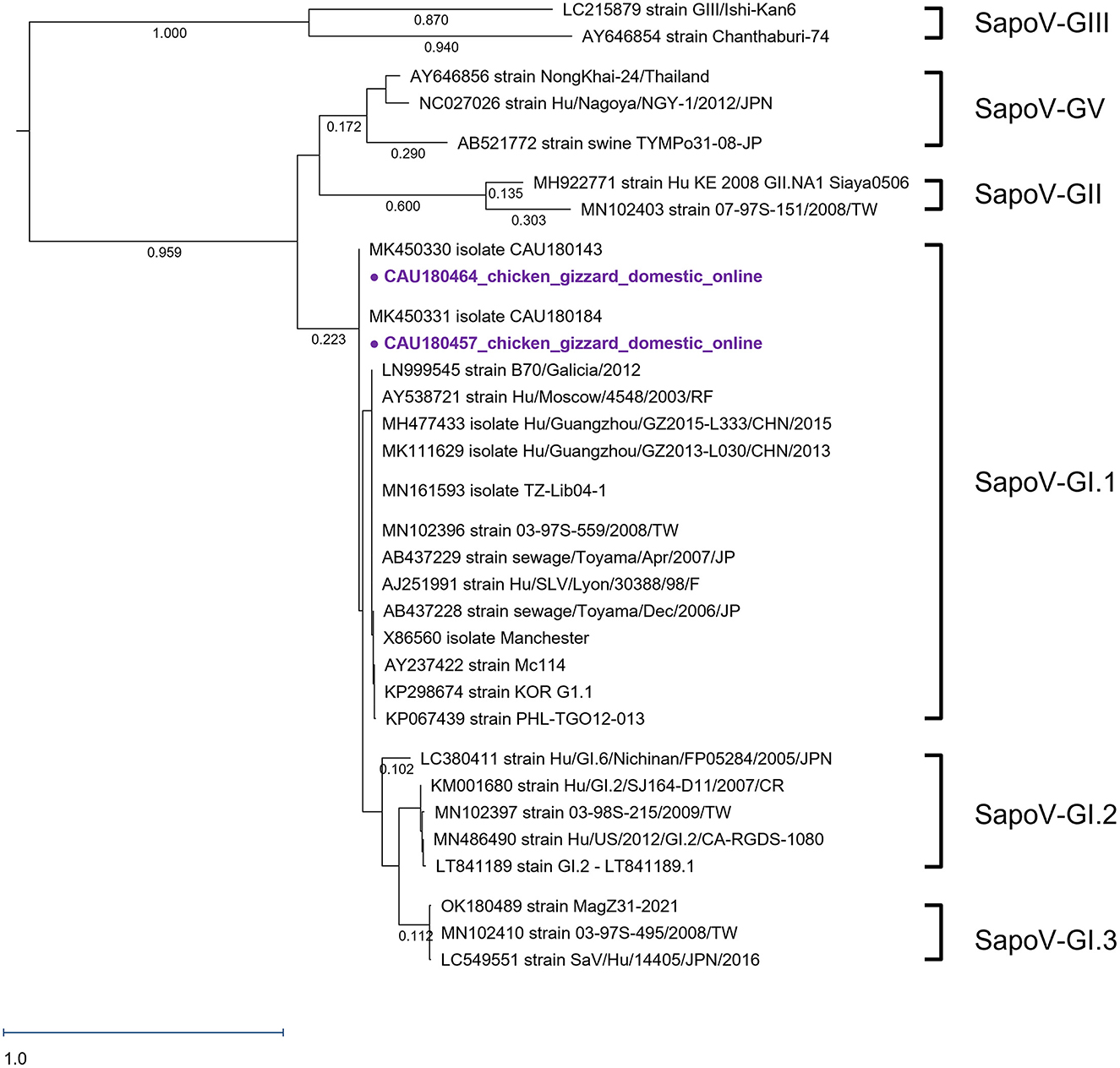
Figure 3. Phylogenetic analysis of SapoV detected in chickens. The SapoV phylogenetic tree was constructed following the maximum likelihood method and based on 416-bp sequences of the VP1 region. The purple circles ( ) were AdV samples purchased in online markets. The numbers at the nodes of the tree indicate sequence distance.
) were AdV samples purchased in online markets. The numbers at the nodes of the tree indicate sequence distance.
4. Discussion
This study investigated foodborne viruses and IAV in chickens from poultry processing plants to commercial markets (local and online markets). In order to identify the source of contamination, we analyzed the sequences of the detected viruses. However, AdV and SapoV were discovered, but other viruses including NoV, astrovirus, HAV, HEV, rotavirus, aichivirus, and IAV were not discovered from poultry processing plants to commercial markets. Through phylogenetic analysis, 18 human AdV-41, 1 porcine AdV, and 2 SapoV-GI.1 were identified.
By fecal–oral transmission, human AdV-41 mainly caused gastroenteritis and diarrhea (Shieh, 2021). Although there were no specific studies on the infectious dose of AdV, enteric viruses were known to be infected with 10–100 viral particles (Prevost et al., 2016). An equivalent comparison was difficult because the particle number of AdV was not confirmed by protein assay, but we observed an average of 2.41 log DNA copies/g of AdV in this study. Among the AdV-positive samples, a high titer of 5.35 log DNA copies/g was detected in CAU180436 which was closed with MH289553 and MK420403, as shown in Figure 3. Reference strain MH289553 and MK420403, both discovered in 2018, were in different regions of Brazil and South Korea, respectively (Portal et al., 2019; Yeo et al., 2022). As shown in Figure 3, human AdV-41 strains discovered in 2018 constitute one clade with MH289566~MK354237. In each respective node, there were construction clades depending on the collection year. Therefore, it could be inferred that the year-specific factor has a greater effect on the evolution of the human AdV-41 sequence than the regional factor, despite the short AdV sequence region. Moreover, in late 2021 and early 2022, cases of human AdV-41 caused hepatitis in children in the UK and the USA (Gutierrez Sanchez et al., 2022; Kelgeri et al., 2022). Therefore, as the yearly AdV-41 sequence mutations identified in this study have increased and new cases of AdV hepatitis have recently been reported, research on the association between AdV sequence mutations and the symptoms is needed.
This study was the first report to detect SapoV in chickens. In particular, cases found in gizzards were not the main subject of research in the field of food safety, thus, it was assumed that no reports have been made. SapoV was only detected in chicken gizzards purchased from the online market. Human SaVs were currently divided into 18 genotypes, 7 GI (GI.1–GI.7), 8 GII (GII.1–GII.8), a single GIV, and 2 GV (GV.1 and GV.2) (Okitsu et al., 2021). All SapoVs detected in this study were of the GI.1 type. The nucleotide sequence identities between the detected SapoV and the reference strains MK450330, KP298674, and LT841189 were 100.0, 96.1, and 99.8%, respectively. MK450330 and MK450331 having the same node were detected in Korean porcine stools in 2018 (Yeo et al., 2022). SapoV (CAU180457, CAU180464) detected in SapoV-GI.1 and KP298674 with high sequence identity was isolated from human stool in 2013 in South Korea (Choi et al., 2015). In previous studies, genotype GI was consistently dominant at over 70.0% during 2013–2019 in South Korea (Cho et al., 2021). SapoV detected in the feces of Korean patients in 2018 confirmed 19 cases of GI genotype and one case of GII genotype (Cho et al., 2021). In the GI group, the GI.1 genotype revealed the highest detection rate of 60.0% (Cho et al., 2021). SapoV-GI.1 (CAU180464 and CAU180457) detected in this study also had a very low homology with the GII strain (accession number: MH922771), with a nucleotide sequence identity of 55.7%.
In poultry processing plants, the IAV gene was detected as 17/216 (7.87%) by RT-qPCR. On the other hand, in the commercial market, only 1/100 (1.0%) was detected in chicken legs purchased from the online market. However, the sequence was not confirmed in all samples. In poultry processing plants, the IAV was detected by RT-qPCR at eight sample collection steps. Among them, the most frequently detected stages were post-evisceration and post-air chilling. In general, IAV was found in the caudal and cranial parts of the chicken lung (Rebel et al., 2011). Consistent with these reports, the highest detection rate in the post-evisceration stage could be attributed to cross-contamination during the extraction process. Our sample collection plant used the chilling system with chlorinated water spray cooling and air cooling. In the previous study of microbial cross-contamination experiments with chillers, in the water chillers, Escherichia coli K12 was dispersed in all directions from a single inoculated carcass, and transmission was increased by the use of chlorinated water sprays (Mead et al., 2000). Influenza viruses were highly stable and contagious in aerosols over a wide range of relative humidity (Kormuth et al., 2018). Due to these factors, it was probable that the IAV gene was detected in the chilling step.
The identified viruses (human AdV-41, porcine AdV, and SapoV-GI.1) do not use chicken as a host. Chicken contamination was not caused by an infection in the chicken itself. NoV, astrovirus, HAV, HEV, rotavirus, and aichivirus were not identified in chickens. These viruses were transmitted by the fecal–oral route, and the main infectious agent was human feces (Li et al., 2021). In particular, AdV was not only an indicator of fecal contamination but also an indicator of viral water quality (Rames et al., 2016). AdV was the only DNA virus among foodborne viruses (Sarantis et al., 2004). As DNA virus genes were more stable compared to RNA viruses (Sarantis et al., 2004), AdV had a higher probability of being detected than RNA viruses.
Although nine viruses were searched, the sample number was not enough to investigate the epidemiology. Looking at the results of the AdV phylogenetic tree, it was detected without distinction according to origin and place of purchase, but imported chicken cannot be purchased from the local market, thus, there was a limitation in that the equivalent detection rate was not compared. In addition, though the hexon gene contained a hypervariable region in which the detected virus genotype was analyzed (Sarantis et al., 2004), it was compared to a relatively short sequence of 171 bp. Further research is needed on methods of physical and chemical reduction of chicken meat that can prevent AdV contamination derived from environmental and epidemiological traces through large-scale monitoring and whole-genome analysis.
5. Conclusion
Foodborne viruses and IAV were investigated in poultry processing plants to commercial markets. The detection rate of foodborne viruses from imported chicken was significantly higher than that from domestic chicken. Poultry processing plants had no relationship to commercial market contamination viral sources, regardless of region. In the poultry processing plant, we could not identify any viruses. Contrary to the high concern of consumers against IAV contamination, IAV was not detected in any commercial chickens (local and online). As chickens are well cooked at high temperatures, the risk of IAV contamination in chickens was very low. However, 18 human AdV-41, 1 porcine AdV, and 2 SapoV-GI were identified in distributed chicken products in the local and online markets. Especially, AdV has the possibility for annual variability with emerging symptoms, thus, continuous follow-up from slaughterhouses to retail markets should be monitored and controlled.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession numbers OP846433-OP846451 and OP889258-OP889259.
Author contributions
DS, MR, and CC: conceptualization. DY, MH, and SJ: validation and visualization. DY and SJ: formal analysis, data curation, methodology, and software. DY and MS: investigation. MR and CC: supervision and funding acquisition. DY, MR, and CC: project administration. CC: resources. DY: writing—original draft preparation. DY, ZW, MH, SJ, and CC: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a Grant (17162MFDS034) from the Ministry of Food and Drug Safety in 2017-2019.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1113743/full#supplementary-material
Supplementary Figure 1. The schematic flow of poultry processing plants sampled in this study. Red arrows (→) indicate each sampled step. In each sampling step, 27 (3 × 9) samples were sampled for each step.
Supplementary Table S1. Information on the samples which was detected by the foodborne virus.
References
Amonsin, A., Choatrakol, C., Lapkuntod, J., Tantilertcharoen, R., Thanawongnuwech, R., Suradhat, S., et al. (2008). Influenza virus (H5N1) in live bird markets and food markets, Thailand. Emerg Infect. Dis. 14, 1739. doi: 10.3201/eid1411.080683
Anaclerio, F., Ferrante, R., Mandatori, D., Antonucci, I., Capanna, M., Damiani, V., et al. (2021). Different strategies for the identification of SARS-CoV-2 variants in the laboratory practice. Genes. 12, 1428. doi: 10.3390/genes12091428
CDC. (2000). Foodborne outbreak of group A rotavirus gastroenteritis among college students-District of Columbia, March-April. MMWR Morbidity Mort Weekly Rep. 49, 1131–1133. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm4950a2.htm
Ceylan, Z., Meral, R., and Cetinkaya, T. (2020). Relevance of SARS-CoV-2 in food safety and food hygiene: potential preventive measures, suggestions and nanotechnological approaches. Virus Dis. 31, 154–160. doi: 10.1007/s13337-020-00611-0
Cho, S. R., Chae, S. J., Jung, S., Choi, W., Han, M. G., Yoo, C. K., et al. (2021). Trends in acute viral gastroenteritis among children aged ≤ 5 years through the national surveillance system in South Korea, 2013–2019. J. Med. Virol. 93, 4875–4882. doi: 10.1002/jmv.26685
Choi, H. L., Suh, C. I., Park, S. W., Jin, J. Y., Cho, H. G., Paik, S. Y., et al. (2015). Whole-genome sequencing analysis of sapovirus detected in South Korea. PLoS ONE. 10, e0132328. doi: 10.1371/journal.pone.0132328
CIDRAP. (2013). Study Puts Global 2009 Pandemic H1N1 Infection Rate at 24%. Minnesota, MN: Center for Infectious Disease Research and Policy. Available online at: https://www.cidrap.umn.edu/h1n1-2009-pandemic-influenza/study-puts-global-2009-pandemic-h1n1-infection-rate-24
Cui, B., Wang, L. D. L., Chen, X., Xu, M. Y., Ke, J., Tian, Y., et al. (2022). Chicken meat taste preferences, perceived risk of human infection with avian influenza virus, and self-reported chicken meat consumption in China. Prev. Vet. Med. 203, 105658. doi: 10.1016/j.prevetmed.2022.105658
Dewey-Mattia, D., Manikonda, K., Hall, A. J., Wise, M. E., and Crowe, S. J. (2018). Surveillance for foodborne disease outbreaks—United States, 2009–2015. Morbid. Mortal. Wkly. Rep. 67, 1. doi: 10.15585/mmwr.ss6710a1
Frosth, S., Karlsson-Lindsjö, O., Niazi, A., Fernström, L. L., and Hansson, I. (2020). Identification of transmission routes of Campylobacter and on-farm measures to reduce Campylobacter in chicken. Pathogens. 9, 363. doi: 10.3390/pathogens9050363
Gholizadeh, O., Akbarzadeh, S., Ghazanfari Hashemi, M., Gholami, M., Amini, P., Yekanipour, Z., et al. (2023). Hepatitis A: viral structure, classification, life cycle, clinical symptoms, diagnosis error, and vaccination. Can. J. Infect. Dis. Med. Microbiol. 4, 2023.4263309. doi: 10.1155/2023/4263309
Golden, C. E., and Mishra, A. (2020). Prevalence of Salmonella and Campylobacter spp. in alternative and conventionally produced chicken in the United States: a systematic review and meta-analysis. J. Food Prot. 83, 1181–1197. doi: 10.4315/JFP-19-538
Gutierrez Sanchez, L. H., Shiau, H., Baker, J. M., Saaybi, S., Buchfellner, M., Britt, W., et al. (2022). A case series of children with acute hepatitis and human adenovirus infection. N. Engl. J. Med. 387, 620–630. doi: 10.1056/NEJMoa2206294
Hoffmann, E., Stech, J., Guan, Y., Webster, R., and Perez, D. (2001). Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146, 2275–2289. doi: 10.1007/s007050170002
Hyun, J. H., Yoon, J. Y., and Lee, S. H. A. (2022). Case-control study of acute hepatitis A in South Korea, 2019. Osong Public Health Res. Perspect. 13, 352–359. doi: 10.24171/j.phrp.2022.0141
KCDC. (2022). Notification No. 11-1790387-000354-01. Waterborne and Foodborne Infectious Disease Control Guidelines. Cheongju: KCDC Publising.
Kelgeri, C., Couper, M., Gupte, G. L., Brant, A., Patel, M., Johansen, L., et al. (2022). Clinical spectrum of children with acute hepatitis of unknown cause. N. Engl. J. Med. 387, 611–619. doi: 10.1056/NEJMoa2206704
Kormuth, K. A., Lin, K., Prussin, A. J., Vejerano, E. P., Tiwari, A. J., Cox, S. S., et al. (2018). Influenza virus infectivity is retained in aerosols and droplets independent of relative humidity. J. Infect. Dis. 218, 739–747. doi: 10.1093/infdis/jiy221
Lee, D.-Y., Cho, S.-R., Chae, S.-J., Choi, W., and Han, M.-G. (2021). Evaluation of a test method to detect hepatitis A virus in salted shellfish. J. Food Saf. 41, e12883. doi: 10.1111/jfs.12883
Li, D., Zhao, M. Y., and Tan, T. H. M. (2021). What makes a foodborne virus: comparing coronaviruses with human noroviruses. Curr. Opin. Food Sci. 42, 1–7. doi: 10.1016/j.cofs.2020.04.011
Mase, M., Eto, M., Tanimura, N., Imai, K., Tsukamoto, K., Horimoto, T., et al. (2005). Isolation of a genotypically unique H5N1 influenza virus from duck meat imported into Japan from China. Virology. 339, 101–109. doi: 10.1016/j.virol.2005.05.010
Mead, G., Allen, V., Burton, C., and Corry, J. (2000). Microbial cross-contamination during air chilling of poultry. Br. Poult. Sci. 41, 158–162. doi: 10.1080/713654915
Mwangi, S., Timmons, J., Fitz-Coy, S., and Parveen, S. (2019). Characterization of Clostridium perfringens recovered from broiler chicken affected by necrotic enteritis. Poult. Sci. 98, 128–135. doi: 10.3382/ps/pey332
Nielsen, A. C. Y., Gyhrs, M. L., Nielsen, L. P., Pedersen, C., and Böttiger, B. (2013). Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J. Clin. Virol. 57, 239–242. doi: 10.1016/j.jcv.2013.03.015
OECD-FAO. (2017). Meat. In: OECD-FAO Agricultural Outlook 2017-2026. Paris: OECD Publising (2017). p. 195–211.
OECD-FAO. (2022). Meat. In: OECD-FAO Agricultural Outlook 2022-2031. Paris: OECD Publising (2022). p. 195–211.
Okada, M., Shinozaki, K., Ogawa, T., and Kaiho, I. (2002). Molecular epidemiology and phylogenetic analysis of Sapporo-like viruses. Arch. Virol. 147, 1445–1451. doi: 10.1007/s00705-002-0821-7
Okitsu, S., Khamrin, P., Thongprachum, A., Hikita, T., Kumthip, K., Pham, N. T. K., et al. (2021). Diversity of human sapovirus genotypes detected in Japanese pediatric patients with acute gastroenteritis, 2014–2017. J. Med. Virol. 93, 4865–4874. doi: 10.1002/jmv.26934
Pal, M., and Ayele, Y. (2020). Emerging role of foodborne viruses in public health. Biomed. Res. Int. 5, 01–04. doi: 10.31579/2637-8914/021
Portal, T. M., Reymão, T. K. A., Quindere Neto, G. A., Fiuza, M. K. D. C., Teixeira, D. M., Lima, I. C. G., et al. (2019). Detection and genotyping of enteric viruses in hospitalized children with acute gastroenteritis in Belém, Brazil: Occurrence of adenovirus viremia by species F, types 40/41. J. Med. Virol. 91, 378–384. doi: 10.1002/jmv.25321
Prevost, B., Goulet, M., Lucas, F., Joyeux, M., Moulin, L., Wurtzer, S., et al. (2016). Viral persistence in surface and drinking water: suitability of PCR pre-treatment with intercalating dyes. Water Res. 91, 68–76. doi: 10.1016/j.watres.2015.12.049
Rames, E., Roiko, A., Stratton, H., and Macdonald, J. (2016). Technical aspects of using human adenovirus as a viral water quality indicator. Water Res. 96, 308–326. doi: 10.1016/j.watres.2016.03.042
Rebel, J. M., Peeters, B., Fijten, H., Post, J., Cornelissen, J., Vervelde, L., et al. (2011). Highly pathogenic or low pathogenic avian influenza virus subtype H7N1 infection in chicken lungs: small differences in general acute responses. Vet. Res. 42, 1–11. doi: 10.1186/1297-9716-42-10
Sarantis, H., Johnson, G., Brown, M., Petric, M., and Tellier, R. (2004). Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42, 3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004
Shieh, W. J. (2021). Human adenovirus infections in pediatric population-an update on clinico–pathologic correlation. Biomed. J. 45, 38–49. doi: 10.1016/j.bj.2021.08.009
Shin, H., Park, H., Seo, D. J., Jung, S., Yeo, D., Wang, Z., et al. (2019). Foodborne viruses detected sporadically in the fresh produce and its production environment in South Korea. Foodborne Pathog. Dis. 16, 411–420. doi: 10.1089/fpd.2018.2580
Son, N. R., Seo, D. J., Lee, M. H., Seo, S., Wang, X., Lee, B. H., et al. (2014). Optimization of the elution buffer and concentration method for detecting hepatitis E virus in swine liver using a nested reverse transcription-polymerase chain reaction and real-time reverse transcription-polymerase chain reaction. J. Virol. Methods. 206, 99–104. doi: 10.1016/j.jviromet.2014.05.026
Spackman, E., Senne, D. A., Myers, T., Bulaga, L. L., Garber, L. P., Perdue, M. L., et al. (2002). Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40, 3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tetro, J. (2014). Control of Foodborne Viruses at Retail. Springer: Retail food safety Springer. p. 59–79.
Tian, P., Yang, D., Jiang, W., Zhong, W., Cannon, J. L., Burkhardt, W., et al. (2010). Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J. Appl. Microbiol. 109, 1753–1762. doi: 10.1111/j.1365-2672.2010.04812.x
Wang, Z., Shin, H., Jung, S., Yeo, D., Park, H., Shin, S., et al. (2020). Effects of weather and environmental factors on the seasonal prevalence of foodborne viruses in irrigation waters in Gyeonggi Province, Korea. Microorganisms. 8, 1224. doi: 10.3390/microorganisms8081224
Webb, G. W., Kelly, S., and Dalton, H. R. (2020). Hepatitis A and Hepatitis E: Clinical and epidemiological features, diagnosis, treatment, and prevention. Clin. Microbiol. Newsl. 42, 171–179. doi: 10.1016/j.clinmicnews.2020.10.001
White, A. E., Tillman, A. R., Hedberg, C., Bruce, B. B., Batz, M., Seys, S. A., et al. (2022). Foodborne illness outbreaks reported to national surveillance, United States, 2009–2018. Emerging Infect. Dis. 28, 1117. doi: 10.3201/eid2806.211555
WHO. (2009). Report of the Review Committee on the Functioning of the International Health Regulations (2005) in Relation to Pandemic (H1N1) 2009. Geneva: World Health Organization. p. 1–180.
WHO. (2011). Standardization of Terminology of the Pandemic A (H1N1) 2009 Virus. Influenza. Available online at: https://policycommons.net/artifacts/505564/standardization-of-terminology-of-the-pandemic-ah1n1-2009-virus-normalisation-de-la-terminologie-pour-le-virus-de-la-grippe-pandemique-a-h1n1-2009/1480987/ (accessed January 18, 2023).
Yeo, D., Hossain, M., Jung, S., Wang, Z., Seo, Y., Woo, S., et al. (2022). Prevalence and phylogenetic analysis of human enteric emerging viruses in porcine stool samples in the Republic of Korea. Front. Vet. Sci. 9, 913622. doi: 10.3389/fvets.2022.913622
Yoo, Y., Oh, H., Lee, Y., Sung, M., Hwang, J., Zhao, Z., et al. (2022). Risk assessment for norovirus foodborne illness by raw oyster (Ostreidae) consumption and economic burden in Korea. Fish. Aquatic Sci. 25, 287–297. doi: 10.47853/FAS.2022.e26
Zhang, W., Li, L., Deng, X., Kapusinszky, B., and Delwart, E. (2014). What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology. 468, 303–310. doi: 10.1016/j.virol.2014.08.025
Zhuge, X., Zhou, Z., Jiang, M., Wang, Z., Sun, Y., Tang, F., et al. (2021). Chicken-source Escherichia coli within phylogroup F shares virulence genotypes and is closely related to extraintestinal pathogenic E. coli causing human infections. Transbound. Emerg. Dis. 68, 880–895. doi: 10.1111/tbed.13755
Keywords: foodborne virus, adenovirus, chicken, poultry processing plant, influenza A virus
Citation: Yeo D, Song M, Hossain MI, Jung S, Wang Z, Seo DJ, Rhee MS and Choi C (2023) Prevalence of foodborne viruses and influenza A virus from poultry processing plants to retailed chickens. Front. Sustain. Food Syst. 7:1113743. doi: 10.3389/fsufs.2023.1113743
Received: 01 December 2022; Accepted: 27 February 2023;
Published: 30 March 2023.
Edited by:
Viviana Parreño, Instituto Nacional de Tecnología Agropecuaria, ArgentinaReviewed by:
Viviana Re, National University of Cordoba, ArgentinaKalmia Kniel, University of Delaware, United States
Copyright © 2023 Yeo, Song, Hossain, Jung, Wang, Seo, Rhee and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changsun Choi, Y2Nob2lAY2F1LmFjLmty
 Daseul Yeo
Daseul Yeo Mengxiao Song1
Mengxiao Song1 Md. Iqbal Hossain
Md. Iqbal Hossain Soontag Jung
Soontag Jung Zhaoqi Wang
Zhaoqi Wang Min Suk Rhee
Min Suk Rhee Changsun Choi
Changsun Choi