- 1Integrated and Urban Plant Pathology Laboratory, TERRA Teaching and Research Center, University of Liège, Gembloux Agro-Bio Tech, Gembloux, Belgium
- 2Plant Protection Department, Agroscope, Nyon, Switzerland
- 3School of Natural and Environmental Sciences, Newcastle University, Newcastle, United Kingdom
- 4Crops and Forest Health Unit, Walloon Agricultural Research Center (CRA-W), Gembloux, Belgium
- 5Modelization and Development Department, University of Liège, Gembloux Agro-Bio Tech, Gembloux, Belgium
The number of small-scale diversified vegetable growers in industrialized countries has risen sharply over the last 10 years. The risks associated with plant viruses in these systems have been barely studied in Europe, yet dramatic virus emergence events, such as tomato brown fruit rugose virus (ToBRFV), sometimes occur. We developed a methodology that aimed to understand better the implications related to viruses for tomato production in Belgian’s vegetable farms by comparing growers’ perception and the presence of plant-viral-like symptoms (visual inspection) with non-targeting detection of nearly all viruses present in the plants by high throughput sequencing technologies (HTS). Virus presence and impact were interpreted considering the farm’s typology and cultural practices, and the grower’s professional profiles. Overall, the data indicated that most growers have limited understanding of tomato viruses and are not concerned about them. Field observations were correlated to this perception as the prevalence of symptomatic plants was usually lower than 1%. However, important and potentially emergent viruses, mainly transmitted by insects, were detected in several farms. Notably, the presence of these viruses tended to be associated with the number of plant species grown per site (diversity) but not with a higher awareness of the growers regarding plant viral diseases, or a higher number of symptomatic plants. In addition, both HTS and perception analysis underlined the rising incidence and importance of an emergent virus: Physostegia chlorotic mottle virus. This study also revealed a notable lack of knowledge among producers regarding the highly contagious quarantine virus ToBRFV. Overall, the original methodology developed here, involving the integration of two separate fields of study (social science with phytopathology using HTS technologies), could be applied to other crops in other systems to identify emergent risks associated with plant viruses, and can highlight the communication needed with growers to mitigate epidemics. This exploratory investigation provides relevant insights, which, ideally, would be further tested on wider samples to allow finer statistical treatment to be performed.
1. Introduction
Tomato (Solanum lycopersicum L.) is one of the most popular and valuable cultivated vegetables grown worldwide, with a gross production of 102.6 billion US dollars and yield estimated at 186.8 million tons (MT) in 2020 (FAOSTAT, 2020). Tomatoes are grown in diverse production systems (open fields, under plastic tunnels, hydroponics, and high-tech greenhouses) for the fresh market or food industry. It is Europe’s main produced vegetable, with 18 MT in 2021 (Eurostat, 2021). During 2021, the major part of the supply was in Italy (6.6 MT) and Spain (4.7 MT), where production occurs in open fields and tunnels, mainly for processing and export. In northern Europe, Poland (1.1 MT), the Nederlands (0.9MT), and Belgium (0.3MT) are the largest tomato producers, mainly for the production of fresh edible tomatoes in high-tech greenhouses (Eurostat, 2021).
Tomatoes are also grown by small-scale growers, and in gardens for local consumption (Benton, 2007). In industrialized countries where small-scale growers almost disappeared during the green revolution, these production systems represent a recently expanding niche market (Morel and Léger, 2016; Laforge et al., 2018; Dumont et al., 2020). These small-scale growers promote human values and ecosystem welfare rather than profit maximization (Morel and Léger, 2016). Combining multiple logic and aspirations is indeed typical of agroecology-inspired growers (Plateau et al., 2021). Regarding farming practices, most of these growers aim to sustainably produce an extensive range of vegetables on soil, leaning on eco-system services, crop diversification, and rotations. Studies have shown that these systems have many advantages over conventional agriculture, especially for the environment and workers, as it reduces chemical and polluting inputs. In addition, these systems are supposed to have better resilience to climate change and plant diseases (King and Lively, 2012; Kremen and Miles, 2012; Mori et al., 2013). It has also been shown that multi-cropping and crop rotations increase yields in both organic and conventional cropping systems (Ponisio et al., 2015), encouraging the need for research on these agricultural practices to improve the productivity of sustainable agriculture methods.
In Belgium, there are two distinct sectors of tomato production. The most significant part of tomato production is dedicated to export and mass retailing. It is cultivated mainly in the northern part of the country (Flanders) by specialized tomato growers under high-tech greenhouses. A minor part of the production is achieved by small-scale growers producing tomatoes among other vegetables for local consumption. The number of these small-scale (< 2 ha) growers has risen sharply over the last 10 years in the southern part of Belgium, Wallonia (Dumont et al., 2020). A sociologic survey underlined that ethical and sociological factors were considered in growers’ decision processes and that many growers are not from the agricultural sector (Dumont, 2017). Most of these growers aimed to produce tomatoes on soil under tunnels or greenhouses and alternated tomatoes with other vegetables over a year (Dumont, 2017).
Tomatoes are a sensitive crop: in all growing systems, the presence of pests and diseases (fungi, bacteria, viruses…), can jeopardize tomato crops, leading to important yield losses (Blancard, 2009). The characteristics of each pathosystem (the subset of an ecosystem in which the components include a host organism and an associated pathogen or parasite) determine specific strategies for plant pathogen control (Aranda and Freitas-Astúa, 2017). Thus, viral outbreaks are often related to unknown emerging diseases, for which diagnosis is the first step in disease management (Hanssen et al., 2010; Rubio et al., 2020). Viral diseases represent nearly half of the emerging plant diseases (Anderson et al., 2004) and tomato is the plant for which the most viruses are recorded (312 viral species in 2021, Rivarez et al., 2021). Plant viruses can be spread through insects, seeds, plant-to-plant contact, fungal spores, and other means.
Many environmental factors drive the emergence of plant viruses and their outbreaks by altering interactions between viruses, hosts, and vectors. For plant viruses, climate change and human activity, such as agriculture and trade, are the main factors influencing the outcome of these interactions (Jones, 2009; Elena et al., 2014). Elena et al. (2014) decoupled the emergence of new viruses into three phases. The first one requires a virus to jump from a host (“reservoir”) to the same host in another ecological environment or to a new host (“spillover”). The second phase involves the adaptation of the virus in its new host/environment in which it develops the ability to be transmitted independently from the reservoir. The last phase is characterized by optimizing the virus transmission in this new host/environment and establishing the pathogen in the host population (Elena et al., 2014).
Although many viruses are known to infect tomatoes, all the interactions between the different actors of the pathosystem (vector, host, and virus) must occur in a favorable environment in order for a virus to lead to an epidemic and subsequent consequences on the production. For annual crops, viruses can remain largely benign if their horizontal transmission is inefficient, resulting in low prevalence in the crop during a growing season. Therefore, the presence or absence of a virus in a given environment does not necessarily reflect the health of a field, and is not always equivalent to the disease impact (“viral disease risk”). Nevertheless, it is the first step in understanding and anticipating possible risks (MacDiarmid et al., 2013).
The development of high throughput sequencing (HTS) technologies significantly improved the detection of new and potentially emergent viruses in the last decade (Massart et al., 2014). For example, it helped to carry out surveillance studies for tomatoes without a priori knowledge of viruses (Xu et al., 2017; Villamor et al., 2019; Desbiez et al., 2020; Rivarez et al., 2021; Vučurović et al., 2021). This enabled the identification of emergent new viruses such as tomato brown rugose fruit virus (ToBRFV; Salem et al., 2016), Physostegia chlorotic mottle virus (PhCMoV, Menzel et al., 2018) and facilitated the study of their evolution and epidemiology (Lefeuvre et al., 2019).
Of these emerging viral diseases, ToBRFV, which belongs to the Tombamovirus genus, has recently received the most attention from European scientists, policymakers and regulators and has sparked waves of regulatory action (Oladokun et al., 2019). ToBRFV is recommended to be regulated as a quarantine pest by EPPO.1 This phenomenon is due to the association of ToBRFV with severe yield losses on tomato and pepper crops, combined with very high transmissibility (transmission by contact: tools, hands, clothes…) and stability it can remain active in the environment for months (Oladokun et al., 2019; Zhang et al., 2022).
Physostegia chlorotic mottle virus also raised concerns as it is associated with extreme symptoms in tomato fruits. However, the low prevalence of the virus reported in the field so far makes it a lower threat. This virus has a vast host range spanning across nine families and infecting crops (eggplant, cucumber, and crosne), weeds (galinsoga), and ornementals (hellebore, etc.; Temple et al., 2022). It is likely transmitted by leafhoppers such as its close relative Alphanuclorhabdovirus, eggplant mottled dwarf virus (EMDV), and potato yellow dwarf virus (PYDV; Black, 1940; Babaie and Izadpanah, 2003).
Since viruses cannot be cured, their control mainly relies on (i) their accurate detection and (ii) the use of resistant varieties and/or limitation of their transmission, which can be either horizontal or/and vertical (seeds) and depends on the biological properties of each virus (Hull, 2014; Nicaise, 2014). Vega et al. (2019) propose to classify pathogens based on their dispersal and survival strategies, regardless of the taxonomic group to which they belong. This classification facilitates the interpretation of the occurrence of a viral disease in response to cultural practices.
The cost of plant testing may cause growers to rely mainly on their observations and knowledge to control virus infection in the field. In this context, it is crucial to determine virus perception by growers to understand the global virus-associated risks because their actions can affect the spread of viral diseases (Murray-Watson et al., 2022). In addition, growers are the first to observe the crops and to be conscious of their loss. Still, their perception of virus infection can sometimes be disconnected from reality, leading to inappropriate practices (Schreinemachers et al., 2015). Growers’ perceptions and actions depend on several factors, including their knowledge of the disease, their virus-related experience and their production systems per se. Furthermore, the growers’ actions are constrained by their financial means. In connection with the chosen production systems, growers’ aspirations can also influence how they deal with viruses. Some producers may value growing vegetables more sustainably than maximizing their profit (Morel and Léger, 2016) and would tend to display different cultural practices than “conventional growers.” These practices may play a role in virus presence and disease transmission. For example, growers who emphasize ecosystem welfare will be more reluctant to use insecticides because of their impact on non-targeted insects that might be important for other ecological functions (pollination, auxiliaires…). They may also be more likely to grow various tomato varieties, including old varieties that are not resistant to certain viruses such as tomato mosaic virus (Hanssen et al., 2010) or, re-use their own seeds, which can promote the spread of seed-transmitted viruses.
Considering the importance of studying plant viruses (emergent or not) before they become a problem, the recent threat of ToBRFV in Belgium, and the context of climate change, and sustainable agricultural challenges, this study aims to evaluate and compare the diversity of viruses in tomatoes grown on soil in diversified production systems with the associated risk perception of growers. Therefore, the first objective is to identify and understand the potential risks of viruses (“viral disease risk”) in these production systems in Wallonia, Belgium.
Duong et al. (2019) emphasize that although biosecurity is the second largest risk mentioned by producers, there is a lack of research on socio-economic factors that explain risk perceptions, especially those that influence risk perceptions related to biosecurity. In addition, the role of cultural practices in virus presence and disease risk is critical to understand plant virus epidemiology (Jones et al., 2014), especially in sustainable agriculture where options for handling viral diseases are restricted. Therefore, the secondary objective is to interpret the results considering the farm typology and cultural practices, the grower’s professional profiles, and the visual inspection of plant-viral-like symptoms, and to potentially evaluate what drives the presence of viruses and their impact. In this study, HTS will be used to assess the presence of viruses without a priori. Growers’ perceptions will be compared to the presence of viruses and to the observations on the field to understand better the disease risk associated with these viruses within the different farms.
2. Materials and methods
2.1. Study design
To better understand the implications of plant viruses for tomato production in Walloon vegetable farms, a three-tiered survey was designed: (1) interviews with the owner or manager of the tomato production, (2) field observations, and (3) analysis of the tomato virome through HTS technologies.
In 2020, a pilot survey was carried out with five growers and three members of the Interprofessional Center of vegetable growers (CIM, Regional extension services supervising vegetable production in Wallonia) to test and improve the study design to homogenize the questionnaire. Members of the CIM mentioned that they barely encountered outbreaks due to viral diseases in vegetable crops, including tomatoes: “most of the time, there are few virus-infected plants here and there, but viral epidemics are uncommon.” For them, significant problems encountered during tomato culture are related to cryptogamic diseases.
A year after this pilot survey, the study was conducted with a standardized questionnaire with 21 tomato-growers in the province of Namur and Walloon Brabant at the end of the growing season (from August 18 to October 1, 2021) because the prevalence of viral diseases is usually highest at this time since the viral infection has been building up throughout the season.
2.2. Semi-structured interviews
2.2.1. Data collection
Growers’ contact details were collected through the CIM, informal growers’ network and by word of mouth. WalOnMap2 was used to determine the agricultural area where the different farms were located, and growers were grouped based on agricultural area and geographical relevance (growers located in the adjacent area of sandy-silty and silty were grouped and the ones in the adjacent Condroz and Famenne areas as well).
Interviews were conducted face to-face with the grower, informing the survey objective before the visit. During the exchanges, notes were taken, and interviews were audio recorded with the grower’s approval. The questionnaire had two main objectives: (1) to describe the typology of the farms, grower’s profiles, and cultural practices of tomato growing and, (2) to evaluate the perception of growers regarding tomato viral diseases. First, information about the farm [six variables: farm age, area, localization, number of vegetable species grown, organic label, and number of employees (full-time equivalent)], tomato culture (six variables: number of plants and varieties grown, seedling origin, number of production years, disinfection of tool, and usage of homemade seeds), and professional background of the interviewed person (four variables: registered at the CIM, number of years’ experience in the field, reconverted after another job, and have relatives in the agricultural sector) were collected. The questionnaire is described in Supplementary Data, and the answers for each farm are presented in Supplementary Table 1. To obtain a global view of the answers, median, average, and min and max values were calculated for the quantitative data and the sum for the binary data.
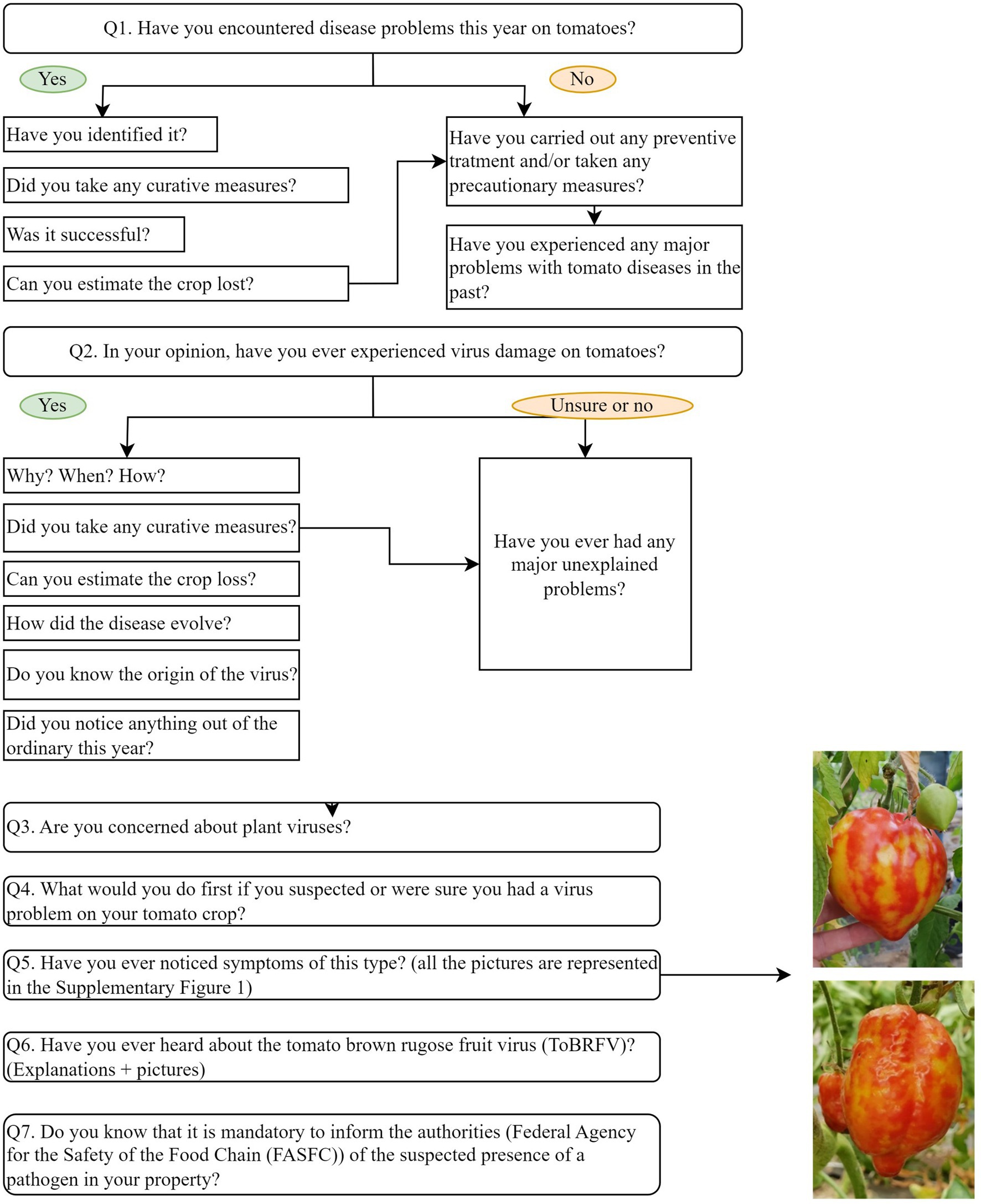
The second part of the questionnaire evaluated how growers perceived tomato viral diseases and which control measures were applied or envisioned. A mix of “open-ended” questions encouraging discussion and closed questions were asked in a specific order (Figure 1).
At question Q5, pictures of viral symptoms induced by PhCMoV on different host plants were shown to assess if the growers recognized the symptoms (Supplementary Figure 1). Since viral symptoms are difficult to notice and sometimes resemble other stresses to the plant, respondents were able to validate or correct their answers to Q2, “In your opinion, have you ever experienced virus damage on tomatoes?”
Physostegia chlorotic mottle virus symptoms were chosen because they are severe and can be easily identified in tomato fruits. They can also be mistaken with other important plant viruses known to be present in Belgium, such as ToBRFV or PePMV (Hanssen et al., 2010; EPPO Bulletin, 2020; Temple et al., 2022). In addition, it was the most frequently detected virus-causing symptom during the pilot survey in 2020.
In Q6, whether growers were aware of ToBRFV was investigated, as this virus was recently widely publicized by different stakeholders involved in the tomato production chain.
At the end of the interview, information on the biology of these two viruses (PhCMoV and ToBRFV), which require different control measures, were given to the growers.
2.2.2. Data interpretation
The second part of the questionnaire investigated the perceptions and worries of the growers and allowed them to express themselves spontaneously. Interviews were first transcribed word by word, and answers were grouped based on the content of the replies. For instance, concerning Q2, responses were classified into four categories: (1) those who thought they already had viruses, (2) those who did not say “yes” clearly but who “did not rule out the possibility,” (3) those who “did not know,” and (4) those who “did not think” they already had viruses.
2.3. Observations and sampling
Prior to sample collection, each grower was explicitly told that if a quarantine virus (e.g., ToBRFV) was detected, it was mandatory to notify Belgian’s NPPO (Federal Agency for the Safety of Food chain). Thus, oral consent to sample was sought from each grower.
In each farm, 100 asymptomatic tomato leaves were systemically collected following a W-shaped transect. When there were several tunnels on a farm, an equal number of plants was sampled per tunnel to reach 100 plants per farm. The tomato plants that showed viral-like symptoms (fruits: deformations, anomalies of coloration; leaves: vein clearing, deformation, mosaic; plant: reduced size) were pictured, counted, and collected in a separate bag.
Since the symptoms of PhCMoV can be easier to spot on eggplant than on tomato, eggplants were also examined when present on the farm. When the symptoms of PhCMoV were noticed on eggplants or tomatoes, the number of symptomatic plants was recorded, and at least three symptomatic plants per farm were collected.
In Belgium, 2021 was not optimal for outdoor tomato production due to very wet, cloudy, and cool weather (also exacerbated by storms and floods). These conditions favored the development of fungal diseases, asphyxiated the root systems, and slowed down the ripening of the fruit. Consequently, some growers removed a part of the planting before the visit. These growers were listed (Supplementary Table 1).
2.4. Virus analysis
2.4.1. High throughput sequencing
After the collection, 100 mg of fresh tomato asymptomatic leaves from the same farm (i.e., 10 g for 100 plants) were pooled in a filter bag and stored at −80°C. The symptomatic plants were also pooled per farm. The weight of material per plant varied according to the number of plants in total (5 g in total). After that, the samples were analyzed for viruses using a virion-associated nucleic acids enrichment protocol (VANA) before HTS on Illumina. The VANA protocol and library preparation used for the samples followed the method described by Maclot et al. (2021) adapted from Filloux et al. (2015).
In brief, 5 or 10 g of tissue was ground, respectively, in 25 or 50 mL of a cold Hanks’ buffered salt solution (HBSS, composed of 0.137 M NaCl, 5.4 mM KCl, 0.25 mM Na₂HPO₄, 0.07 g glucose, 0.44 mM KH₂PO₄, 1.3 mM CaCl₂, 1.0 mM MgSO₄, and 4.2 mM NaHCO₃) using a tissue homogenizer. In a 50 mL falcon tube, the clarification was obtained from a centrifugation run of 10,000 g for 10 min at 4°C. Supernatant was then filtered through a 0.45 μm sterile syringe filter and 10.4 mL of supernatant was put into an ultracentrifuge tube [Beckman Ultra clear 14 mL tubes (#344085)]. In each tube, 0.1 mL (1:100) of a clarified banana sample infected with banana bract mosaic virus (BBrMV) was added as an internal positive control to evaluate the analytical sensitivity of the test (Massart et al., 2014). Then, a sucrose cushion, made of 1 mL of 30% sucrose in 0.2 M potassium phosphate at pH 7.0, was deposited at the bottom of the tube. The extract was then centrifuged at 40,000 rpm for 2 h at 4°C using the 50Ti rotor (Beckman). A protocol previously used in the laboratory and described in Maclot et al. (2021) was used for the library preparation.
PCR products were pooled by 6–12 according to the linkers and cleaned using the Nucleospin Gel and PCR cleanup (Macherey-Nagel). Samples containing asymptomatic plants were pooled separately from those containing symptomatic leaves to limit potential cross-contaminations of highly concentrated viruses in symptomatic pools. A positive external alien control containing infected beans with Endornavirus was processed simultaneously as the asymptomatic samples to monitor potential cross-contaminations as recommended in Massart et al. (2014).
Illumina library was prepared at GIGA Genomics (University of Liege, Belgium) using NEBNext Ultra II DNA library prep kit (New England BioLabs, United States), and libraries were sequenced on the Illumina NextSeq500 sequencer for the generation 10 M of paired-end reads (2 × 150 base pair) per library. Resulting sequence reads were first demultiplexed according to the linker and trimmed from the adaptor, then quality trimming, pairing, and merging were performed using the Geneious R11 software platform3 before de novo assembly with (RNA) SPAdes assembler 3.10.0 (Bankevich et al., 2012). Next, contigs were compared using tBlastx against a database of viruses and viroids sequences downloaded from NCBI in November 2021 (RefSeq virus database). Then, reads were mapped on the closest reference sequences using geneious parameters Medium-Low Sensitivity/ Fast. The presence of viruses was considered positive when the coverage (% of reference genome) was superior to 50% for most viruses and when the number of mapped reads on the closest genome reference was >90. On another hand, the presence of tombusviruses and alphanecroviruses was assessed at a different threshold (12% of ref. seq) since the number of reads which map on reference genomes, were very low compared to other viruses, such as the internal control BBrMV. This threshold was determined after a manual expertise assessment of the difference between contamination and low concentration of each virus (Rong et al., 2022).
2.4.2. RT-PCR for PhCMoV detection in eggplant
Eggplant leaves showing symptoms of PhCMoV were subjected to RNA extraction and RT-PCR for testing the presence of PhCMoV. RNA extraction followed the method described by Oñate-Sánchez and Vicente-Carbajosa (2008). Then, the extracts were reverse transcribed using random hexamers and Tetro RT enzyme (Bioline). The obtained cDNA was amplified with the MangoTaq™ DNA Polymerase and the primers described by Gaafar et al. (2017). Thermal cycling corresponded to: 94°C for 1 min, 35 cycles at 94°C for 15 s, 60°C for 20 s, and 72°C for 45 s, with a final 72°C extension for 3 min. Amplicons were analyzed by electrophoresis on a 1% agarose gel in Tris-acetate-EDTA (TAE) buffer stained with GelRed® Nucleic Acid Gel Stain (Biotium) and visualized under UV light.
2.4.3. RT-PCR and sanger sequencing for confirmation of challenging HTS results
To confirm the detection of strawberry latent ringspot virus (SLRV) and carnation Italian ringspot virus (CIRV), RNA from the original 100 frozen leaves of the positive sample were re-extracted in pools of 25 with the Spectrum plant total RNA kit (Sigma-Aldrich) and tested by RT-PCR using the Titan One Tube RT-PCR kit. For the detection of SLRV, the primers SLRSV1/2 described by Bertolini et al. (2003) were used and for the detection of CIRV, primers were designed based on the consensus sequence using Geneious designing primer tool: CIRV-F: “CGTGGCAGTTACCAGACAGT,” CIRV-R: “CTCCA TCCCAACGTTCACCA” (product length: ~1 kb). Amplicons were Sanger-sequenced, and the obtained sequences were searched against the NCBI database using BLASTn to confirm the presence of the virus.
The status of tomato mosaic virus (ToMV) was assessed in six samples where HTS yielded a small number of reads mapping the reference genome. The 100 frozen leaves for each of these sample and the positive sample were extracted in pools of 25 following the Spectrum plant total RNA kit (Sigma-Aldrich). RNA extracts were tested by RT-PCR using the Titan One Tube RT-PCR kit (Roche) and the primers of Li et al., 2018.
2.5. Data analyses
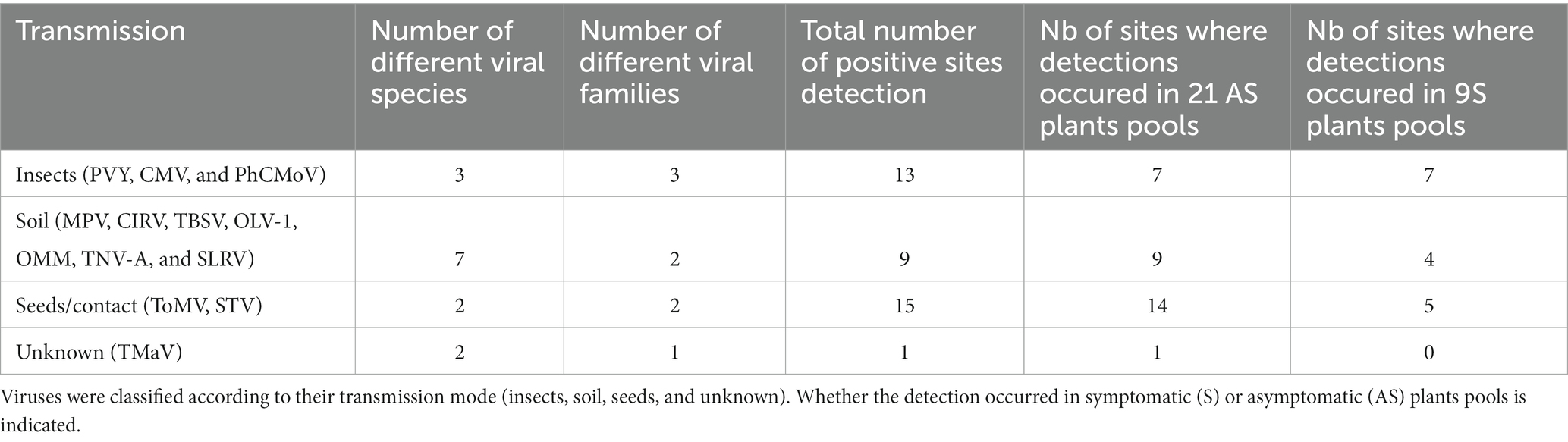
In this study, detected viruses were classified based on their transmission mode (insect-vector, soil, seeds, fungi, and unknown; Table 1). Then, sites were grouped in categories based on the (transmission mode) class of viruses detected (Table 2; Supplementary Table 1).
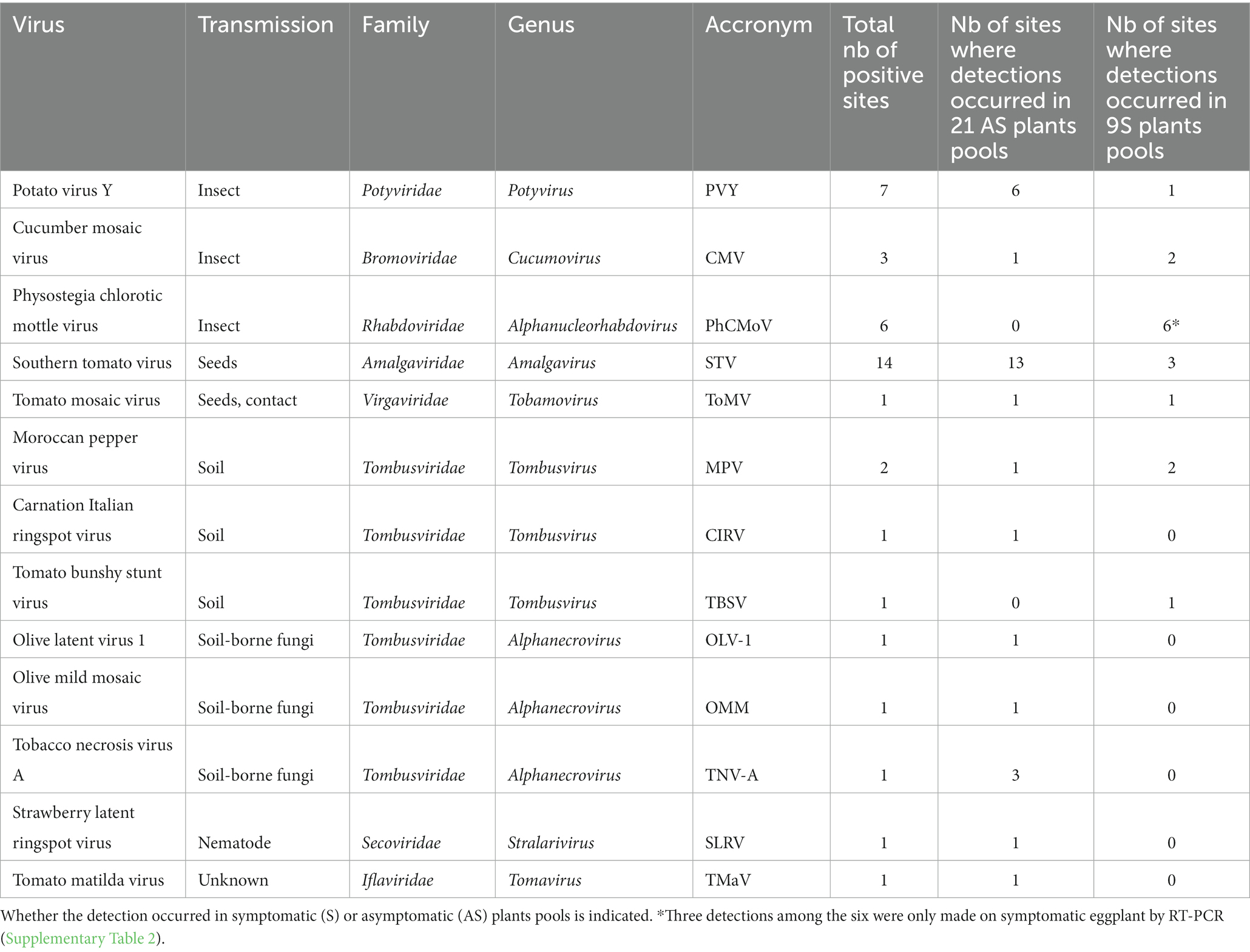
Table 1. Taxonomic characteristics of detected viruses and the number of sites where they were detected.
To evaluate the significance of the associations between the different variables, all the generated raw data in Supplementary Table 1 were tested two-by-two without a priori using Orange Mining’s Sieve Plot diagrams widget (https://orangedatamining.com; Demsar et al., 2013). A Sieve Plot Diagram is a graphical method for visualizing the results of a chi-squared test of independence in a two-way contingency table. The chi-squared test is a statistical method that evaluates whether two categorical variables are independent of each other or whether they are related in some way. In the contingency table generated by this display, the area of each rectangle is proportional to the expected frequency, while the observed frequency is shown by the number of squares in each rectangle. The difference between observed and expected frequency (proportional to the standard Pearson residual) appears as the density of shading, using color to indicate whether the deviation from independence is positive (blue) or negative (red). The area of each rectangle is proportional to the expected frequency, while the observed frequency is shown by the number of squares in each rectangle. Thereafter, using the relevance scoring options and attributes research of the Sieve Plot Diagram widget, the variables that were the most associated with interesting results were identified.
In addition, selected features related to the sites where viruses were transmitted through the most prevalent mode in symptomatic plants were compared to other sites. For that purpose, quantitative data were transformed into qualitative data based on the median value of each feature. In addition, all the data were normalized according to the number of growers with or without the viruses transmitted the same way.
3. Results
3.1. General description of the farms, professional profile of the growers, and tomato culture
During the first step of the questionnaire, information was gathered on the general characteristics of the farm, the professional profile of the growers, and tomato cultivation practices. The detailed data are presented for each farm in Supplementary Table 1.
The 21 surveyed growers sold their products locally (e.g., shop in the farm, markets, baskets, and “pick your own”). The median time they had worked in vegetable farming was 7 years, with only 4 years (median) of tomato production on the studied site (Table 3). This difference is because most of the growers worked on another vegetable farm before starting their own production. These growers stated that they had always grown tomatoes on their own farms. Less than half of the growers (9/21) had only worked in vegetable cultivation, while the others (12/21) switched careers after having worked in other sectors (Table 4). Only five respondents have relatives in the agricultural sector (Table 4).
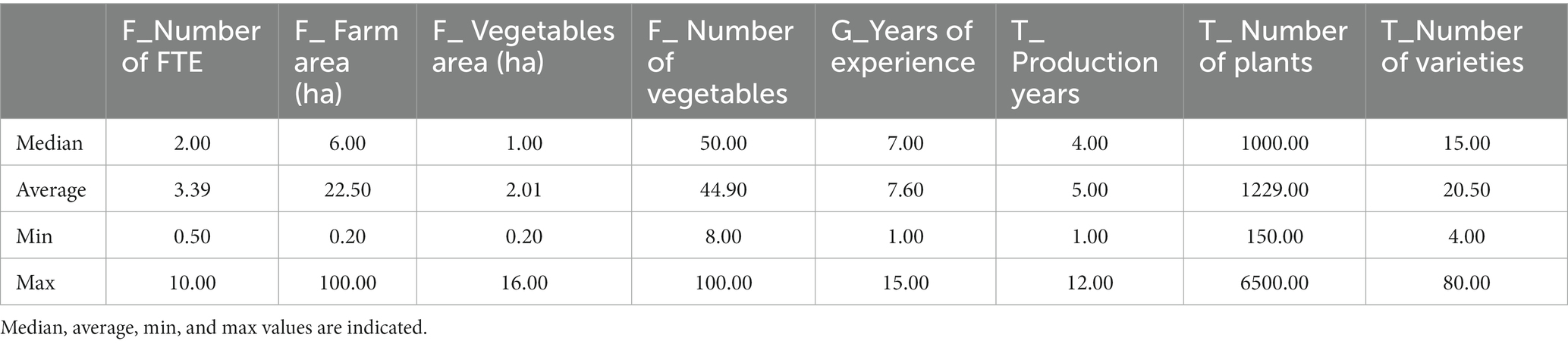
Table 3. Description of the main quantitative characteristics of the farms (F); grower’s profiles (G) and, tomato cultural practices (T) of the respondents (n = 21).

Table 4. Description of the main qualitative characteristics related to the farms (F); grower’s profiles (G) and, tomato cultural practices (T) of the respondents (n = 21).
Regarding growing systems, half of the respondents (11/21) grew vegetables under the organic label or were in the process of obtaining it. However, among the other half, most growers followed organic or agroecological farming practices without certification. Moreover, some of them explained that they do not need the organic label while following the practices because they are “close to their consumers” (notably in the case of pick-your-own farms). Therefore, it is challenging to mobilize this feature in the analyses.
Most of the surveyed growers produced many different vegetable species per year (median = 50) on a small surface area (median = 1.1 ha; Table 3). The different crops (root vegetables, fruit vegetables, leafy green, cruciferous, marrow, aromatics…) were alternated on the same piece of land throughout the year. Only two growers stood out from the others by growing vegetables on a larger surface (INX-29: 4.5 ha and INX-37: 16 ha; Supplementary Table 1).
Regarding tomato production, the median number of tomato plants grown per year, per farm, was 1,000, with a median of 15 different cultivars. The growers bought their plants from nurseries or made seedlings from commercial or home-made seeds from the previous year (Table 3).
3.2. General description of the grower perception, observations of viral-like symptoms, and virus detected in the 21 farms
3.2.1. Grower perception
During the interviews, 20 out of 21 growers had faced cryptogamic diseases during the current year (Figure 2—Q1). One grower (INX-40) mentioned encountering problems with viruses (at the beginning of the interview process).
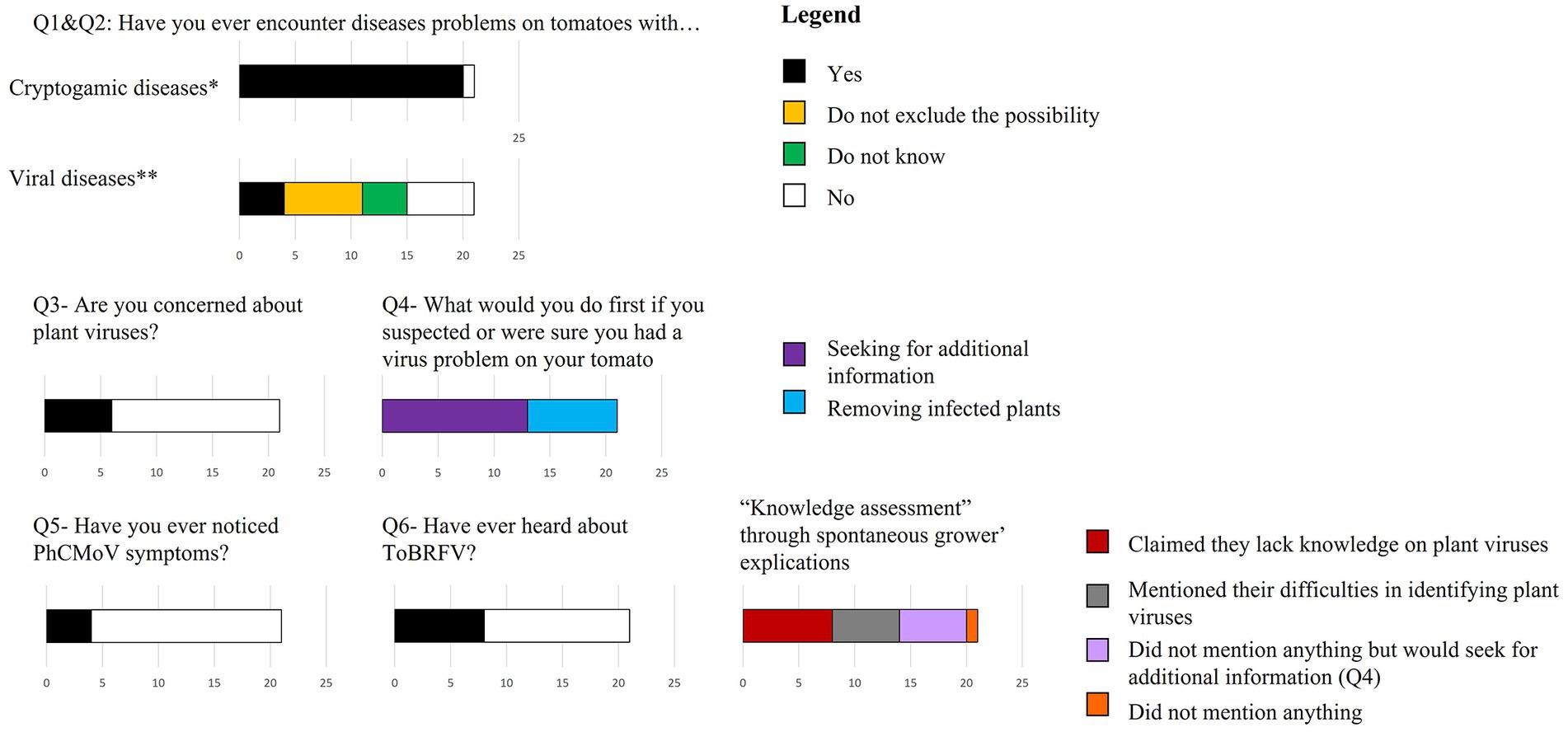
Figure 2. Perception of growers on viral diseases affecting tomato plants. *During the current year, **Since ever, n = 21.
In the second question, specific to viral diseases, only four growers responded that they had faced virus infection, but it had never been problematic (except for the grower INX-40). Seven other growers did not rule out the possibility of having viruses, but were unsure. The rest of the growers “did not know” or “did not think” they ever had tomatoes infected by viruses (Figure 2—Q2). Interestingly, among the growers who did not think they had faced plant viruses or were unsure, no growers highlighted important unexplained troubles with tomato production at Q2. In addition, most growers (15/21) responded that they were not concerned about viruses (Figure 2—Q3).
Subsequently, the interviews revealed that many growers had little knowledge of viruses. Many (14/21) respondents naturally mentioned that they “did not know about viruses,” that they were “unaware of them,” or that they “did not know how to recognize them.” The only grower that demonstrated his knowledge about plant viruses was the one with the most significant tomato production (INX-37). The lack of knowledge was exposed by question Q6 where less than half of the growers were aware of the potential danger posed by ToBRFV. They also mentioned that they did not know the list of quarantine pathogens or how to recognize them. In contrast, the growers seemed aware of fungal diseases as they all mentioned one disease name in Q1, and none highlighted their lack of knowledge about fungal diseases.
3.2.2. Observations of viral-like symptoms
In the parcels, 122 viral-like symptomatic tomato plants were collected in nine farms. In most cases, the proportion of symptomatic tomato plants was lower than 1% (Supplementary Table 1).
Only one exception was noticed with the grower INX-40, who was aware of putatively virus-induced symptoms in his tomato crop (Q1, Q2). In this farm, 40/300 tomato plants (13%) showed strong viral symptoms like the ones associated with PhCMoV (Supplementary Figure 3). In addition, one of the three tunnels from this farm was particularly impacted with 38/85 tomato plants showing these typical symptoms. In this tunnel, other plants were grown (cucumber, capsicum, mint, strawberry…), and the grower mentioned that he saw the symptoms related to PhCMoV in cucumber early in the season. In addition, three eggplants near the greenhouse also showed the symptoms of PhCMoV and tested positive.
In total, 30 eggplants showing PhCMoV symptoms (vein clearing on the leaves) were observed across five farms (including two farms where symptomatic tomatoes were observed, Supplementary Table 2). The maximum number of symptomatic eggplants per farm was 11 (Supplementary Table 2).
3.2.3. Virus detection
First, internal spike BBrMV was identified in all the asymptomatic pools with a number of reads mapped on the reference genome NC_009745 ranging between 5 and 99. Additionally, the external alien control (endornavirus) was detected in the two samples where it was expected with 27,690 and 43,744 reads mapped to the NC_038422 genome. There were no reads of endornavirus found in any other samples, indicating that the extent of cross-contamination was minimal after the filtering steps.
In total, 13 different viral species belonging to eight viral families were identified during this survey (Table 1). The number of virus species detected per farm varied between 1 and 4 (Supplementary Table 2).
These viruses were classified into four categories based on their transmission mode: transmitted by insects, by the soil (including virus transmitted by soil-borne fungi or nematodes), by seeds and/or contact, and the viruses for which the transmission is not known to date (Table 2). In total, three viral species transmitted by insects were detected across 16 different farms, seven viral species transmitted through the soil across nine farms, two viral species transmitted through seeds and/or contact across 14 farms, and one species for which biological data on its transmission was lacking, on one farm. In the symptomatic tomato plant pools, insect-borne viruses were the most prevalent, as they were detected across seven farms out of nine (Table 2).
The most frequently detected virus (n = 14) was southern tomato virus (STV), a persistent virus transmitted by seeds. STV was detected more frequently in asymptomatic plant pools than in symptomatic pools (Table 1).
Then, the most prevalent viruses were the ones transmitted by insects: potato virus Y (PVY), PhCMoV, and cucumber mosaic virus (CMV), which were detected in, respectively, seven, six, and three farms (Table 1).
High throughput sequencing identified PhCMoV in three farms on symptomatic tomato showing fruit deformations and anomalies of coloration and by RT-PCR and on three additional farms on eggplants showing vein clearing on the leaves (Table 1). Overall, during the study, all the tested plants that showed PhCMoV symptoms were positive for this virus. PhCMoV was only detected in plants with typical PhCMoV symptoms and not in asymptomatic plants, whereas PVY was primarily identified in asymptomatic plants. Leafhoppers transmit PhCMoV, while aphids transmit PVY. These two viruses appeared on a farm where many symptomatic plants were found, including cucumber and eggplant and where the growers complained of tomato virus disease (INX-40). The symptoms observed were very characteristic of PhCMoV (Supplementary Figure 2C), which was detected by HTS on the pool of symptomatic tomatoes and by RT-PCR in four separate symptomatic tomato plants.
Cucumber mosaic virus was associated with symptoms in two symptomatic pools and in one asymptomatic pool (Supplementary Table 2A; Blancard, 2009).
Another virus transmitted by seeds, tomato mosaic virus (ToMV, Supplementary Figure 2B), was detected in one farm on asymptomatic and symptomatic plants. ToMV is a Tobamovirus which has been widely studied and is also transmitted by contact (Jones et al., 2014). RT-PCR confirmed the presence of the virus in the sample where the highest number of reads was found and not detected in all the other samples.
In addition, six different viral species belonging to the Tombusvidae family (tomato bushy stunt virus, moroccan pepper virus, olive latent virus 1, olive mild mosaic virus, tobacco necrosis virus A, and CIRV) were primarily detected in asymptomatic plants (Table 1). These viruses are transmitted through the soil, mainly by soil-borne fungi and are not considered economically significant tomato pathogens (Yamamura and Scholthof, 2005).
Finally, SLRV (Secoviridae) and tomato matilda virus (TMaV; Ilflaviridae) were detected on asymptomatic plants only. SLRV is transmitted by a nematode, and the transmission mode of TMaV is unknown. This reports the first detection of SLRV and CIRV on tomatoes. RT-PCR and sanger sequencing was performed on the original plant samples and confirmed the presence of these viruses in tomatoes.
3.3. Associations
To investigate whether the presence of insect-borne viruses (PhCMoV, PVY, and CMV) was associated with any specific metric related to the farms characteristics, cultural practices, grower’s profiles or perception, and the widget sieve diagram on orange mining was used on Supplementary Table 1, and associations with a value of p < 0.1 were noted.
Regarding cultural practices, the presence of these three viruses was associated with the increased number of different vegetable species grown per farm (“diversity”). Furthermore, these insect-borne viruses were also associated with the farms situated in the silty or sandy-silty area. Finally, regarding perception and actions, the presence of PhCMoV, CMV, and PVY was associated with the growers who would remove the virus-infected plants as a first reflex; with the ones who were not concerned about viruses; and with the ones who recognized PhCMoV symptoms (Figure 3).
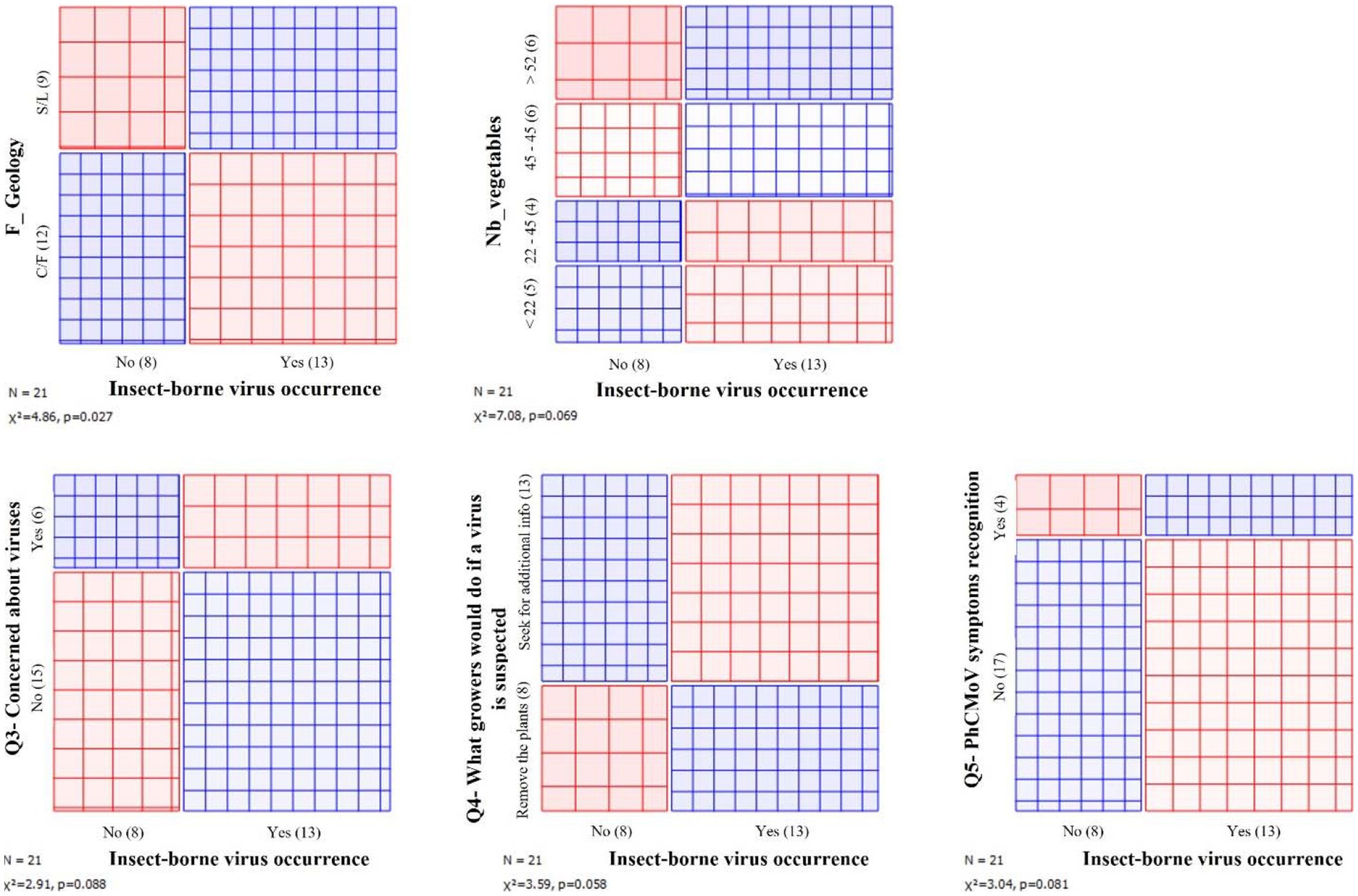
Figure 3. Associations between the presence of insect-borne viruses and different metrics. The area of each rectangle is proportional to the expected frequency, while the observed frequency is shown by the number of squares in each rectangle. The difference between observed and expected frequency (proportional to the standard Pearson residual) appears as the density of shading, using color to indicate whether the deviation from independence is positive (blue) or negative (red). S/S, silty and sandy-silty area; C/F, condroz and famenne area.
In a second step, selected features of interest related to the grower’s perception, observations, or other farms characteristics were converted into qualitative data if needed and evaluated against the presence of insect-borne viruses. This analysis showed that the farms with insect-borne viruses tended to have fewer tomato plants, which could be related to a higher number of different plant species per year, and with the growers who produce vegetables on smaller surfaces.
In addition, the four growers who thought they already had virus problems, and the one who recognized the symptoms of PhCMoV had insect-borne viruses (Figure 4). Interestingly, most of these growers were not concerned about viruses (Figure 4, Supplementary Table 1). This perception can be related to their understanding that there was an absence of any significant problem of viruses on their farms, and highlights that the presence of PVY, CMV, or PhCMoV is, in most cases, not associated with important yield losses. Also, three out of four growers who recognized the PhCMoV symptoms had PhCMoV in their farms. In addition, the growers who thought they had more than 30% losses during the current year due to fungal diseases on tomato were less likely than others to have insect-borne viruses, suggesting that yield losses from viral diseases, if any, were negligible in 2021 (Figure 4).
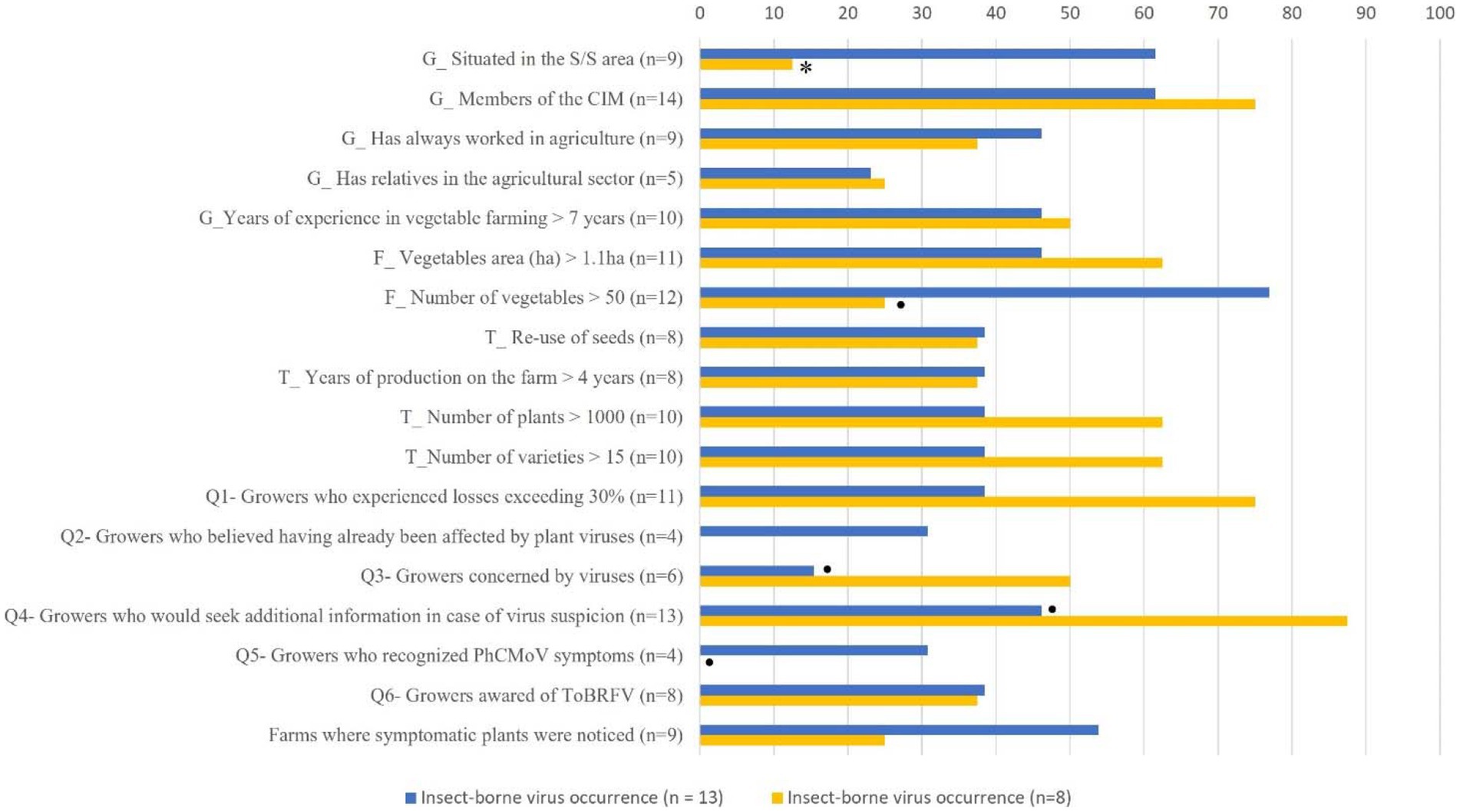
Figure 4. Percentage of growers with insect-borne viruses (blue) or not (yellow) vs. selected characteristics related to the farms (F); grower’s profiles (G) and, tomato cultural practices (T). S/S, silty and sandy-silty area; CIM, regional extension services supervising vegetable production in Wallonia; n represents the number of growers in each feature on which the analysis was based. The difference statistically highlighted with orange mining was indicated when p < 0.1 = ●, p < 0.05 = *.
Since there were no significant variations in how growers perceived viral diseases, it was difficult to compare the perception to the farms’ characteristics and growers’ profiles, and to understand if socio-economic factors can explain plant viruses risk perception. Only one grower claimed that he had problems with plant viruses. Using the widget sieve diagram on orange mining, it was tested whether being awared of ToBRFV (n = 8) or being concerned by tomato viruses (n = 6) was associated to any factors, and no significant associations were identified.
4. Discussion
Overall, this study has provided knowledge of growers’ perceptions regarding viruses, and the occurrence of tomato viruses, in small and diversified farms.
First, our selection process was representative because it was aligned with the classification of Dumont (2017): most of the interviewed vegetable growers produced a large diversity of vegetables sustainably on a surface area up to 2 ha.
Second, their perception of viruses was somewhat unclear: most interviewed growers found it challenging to say whether they had problems linked to viral diseases. Hence, growers usually agreed that viruses were not a major problem in their tomato production. Observations of tomato plants in the field correlated with this perception, as a small number of symptomatic plants were found. However, some viruses known to affect tomato production were identified, such as ToMV, CMV, and PhCMoV (Hanssen et al., 2010; Mahjabeen Akhtar et al., 2012; Ullah et al., 2017; Temple et al., 2022).
Regarding the origin of the difference between perception, observations, and the presence of viruses, a first hypothesis (H1) is that the detected viruses do not cause significant problems in the agricultural system under study (low viral disease risk) and, perception and observations correlate well with reality. A second hypothesis (H2) might be that growers are unaware of the problem, and field observations are not representative because symptoms of other diseases (fungal, bacterial, and abiotic stress) might mask viral symptoms. This hypothesis can be supported by the fact that some growers admitted their lack of understanding of plant viruses or difficulties for identifying them.
While H2 cannot be entirely set aside, many elements suggest that H1 better explains the difference between perception and HTS results. Firstly, during the survey, all respondents admitted that they had a fungal disease problem and developed a control strategy for this issue. Many fungal disease problems in 2021 were observed due to wet weather conditions. The results related to the general tomato diseases perception suggested that growers seemed to be aware of the disease when they were severely impacted. Another reason supporting H1 is that the sole grower who reported a viral infection in their tomato crop experienced a significant virus outbreak. The observations and laboratory analysis were consistent with the growers’ perception: many plants with typical PhCMoV symptoms were noticed, and the presence of the virus was confirmed. Finally, in the pilot survey, the regional extension services mentioned that viruses were not a significant problem in vegetable production, and that they rarely get requests on this subject. This information suggested that viral diseases were, at the time of study, not the most important diseases for tomato production in Wallonia, even if pathogenic viruses were detected. Many reasons can explain why a virus does not necessarily cause problems in a cropping system. For instance, its transmission from plant to plant might be inefficient resulting in a low number of plants infected, or the plants might get infected late, and the virus not have time to cause damage (Jeger, 2020; Trebicki, 2020).
Overall, these results contrast with the situation in the province of Alméria in Spain, where viruses represent a crucial threat to tomato production (Panno et al., 2021), and novel viruses are detected at a rate of 0.9/year (Velasco et al., 2020). Consequently, the growers in Alméria recognize the significant risk of plant viruses (Velasco et al., 2020). It shows that growers’ perception is also correlated to viral disease risk. Agricultural systems and tomato culture history may explain the difference. For example, in Alméria, tomatoes are grown intensively under plastic greenhouses at a very high density over 8,423 ha (in 2021; Análisis de la campaña hortofrutícola. Campaña 2020/2021). There is almost no space between the parcels, which can facilitate the spread of viruses between fields. This is in contrast to the tomato growing systems observed in this study.
In the Nordic European countries, where most tomatoes are grown independently of seasonality in high-tech greenhouses, ToBRFV is of particular concern to intensive tomato growers because of the high risks associated with the movement of fruits, equipment, and employees in greenhouses and warehouses (Zhang et al., 2022). This results in strict phytosanitary controls and a high level of awareness for all persons allowed to enter these greenhouses. In contrast, less than half of the small-scale producers interviewed in this study were aware of the ToBRFV despite the recent high-coverage for the disease in general, and specialized media. In addition, most of the small-scale growers were also unaware that reporting quarantine pests to authorities was mandatory. This highlights the existence of a gap in the information channel between the plant health authorities and small-scale producers. Lack of awareness of harmful viruses may result from the system model in which most growers in this study operate: trade is often limited and local and other crops can compensate for low tomato yields in a given year. This can lead to a potential underlying infection that could remain unnoticed for some time. An outbreak would be very challenging to manage for a small farm, as a virus such as ToBRFV can survive in the environment over successive seasons. Whereas in a large high-tech glasshouse, a complete cleanup is costly and complex but possible. We found no socio-economic reasons or patterns to explain this low perception of ToBRFV. Nevertheless, this result demonstrates the value of addressing sociological factors that influence producers’ decision-making in the face of a pathogen outbreak.
In this study, the most common viruses detected in symptomatic plants were insect-borne viruses. Subsequently, the characteristics that could possibly explain the presence of these viruses were investigated. The low number of producers (21) limited the scope of possible analyses. However, some patterns were detected, which might warrant further investigation. In terms of perception and action, the presence of insect-borne viruses was associated with growers who reported removing suspect plants; those who were not concerned about plant viruses; and those who recognized PhCMoV. The fact that the presence of PhCMoV is associated with growers who recognize its symptoms can be explained by the striking symptoms caused by this virus. Removing plants infected with viruses is often advisable to mitigate the spread of plant viruses. Therefore, it is surprising that growers of tomatoes infected with insect-borne viruses indicated that they would remove the symptomatic plant if they were sure of the viral origin of the symptoms. Furthermore, those who were not concerned about viruses (15/21) had the most insect-borne viruses. This result is also remarkable since insect-borne viruses were frequently detected in symptomatic plants. It suggests that growers did not notice the symptoms, and/or that these viruses were not associated with a high prevalence.
In addition, our analysis suggested that insect-borne viruses were more present in the farms where numerous different plant species were cultivated, and in the farms situated in the silty and sandy-silty areas. In both farms with and without insect-borne viruses, growers had the same profile: settled and working in agriculture for the same number of years. It is challenging to state whether being diversified (numerous plant species cultivated), located in the silty and sandy-silty area, or whether a range of characteristics explain the presence of these viruses, but the different elements can be justified.
Insect-borne viruses were detected more in the silty and sandy-silty area, where field crops such as cereals, sugar beet, potatoes, or flax are common. About half of the Walloon horticulture farms are in the silty area. It is recognized that the intensification of agriculture increased the emergence of viral diseases (Pinel-Galzi et al., 2015; Roossinck and García-Arenal, 2015), and the proximity of large-scale cultivated plants next to the study plots could serve as virus reservoirs (Bernardo et al., 2018). In the Walloon context and our study case, potatoes have been primarily grown in the silty area for centuries. This crop is the primary host of PVY; thus, the presence of potatoes might partly explain the presence of PVY in the farms situated in the silty region.
Cucumber mosaic virus, PhCMoV, and PVY are insect-borne viruses with a broad host plant range which can affect tomato yield (Jones et al., 2014; Temple et al., 2022). The results tend to suggest that insect-borne viruses were more likely to be detected in the most diversified farms (which were not necessarily located in the silty/sandy silty area, data not shown). Growing a high number of vegetable plant species could increase the number of host plants harboring insect vectors (Knops et al., 1999; Cook-Patton et al., 2011) or the number of host plants enabled to host viruses (“amplification effect,” Keesing et al., 2006). Plant-cultivated diversity in small-scale production systems may also increase the first step of emergence: virus-host jump between wild and cultivated reservoirs as the number of potential new hosts is higher. In addition, these diversified farms have expanded in the past 10 years throughout industrialized countries (including Belgium), and introduced new crops in the environment, a factor known to promote virus emergence (Elena et al., 2014). In this study, an emergent virus (PhCMoV) was detected in six farms out of 21. Even though the virus was detected in Germany since 2003 in a diversified system (Temple et al., 2022), its prevalence in Belgium could potentially be associated with the development of diversified farms.
Interestingly, in most cases, the presence of insect-borne viruses was not found to be associated with greater grower concern or an important number of symptomatic plants, suggesting that their presence did not pose a high risk for the production in diversified production systems. Although these results must be taken with precautions because of the relatively small number of studied farms, some studies postulate that plant-cultivated diversity protects against the spread of viral diseases (Haddad et al., 2009; Keesing et al., 2010; Pagán et al., 2012; Roossinck and García-Arenal, 2015). Different protection mechanisms are involved, for example, creating a large genetic diversity for plants may lead to a dilution protective effect since pathogens have more chances of encountering resistant hosts in diverse habitats and more difficulties spreading (Liu et al., 2020; Keesing and Ostfeld, 2021). In addition, plant diversity might increase the diversity of insect-vector but also of the predators, which should theoretically lead to an ecological balance (Haddad et al., 2009; Cook-Patton et al., 2011). It might also disturb the movement of insect vectors and thus reduce the spread of the disease (Power, 1991). Lichtenberg et al., 2017 demonstrate that organic farming and higher in-field plant diversity enhanced arthropod abundance, particularly for rare taxa, resulting in increased richness but decreased evenness. Our results tend to align with this statement since insect-borne viruses were more present in most diversified production systems but were not associated with higher viral disease risk. Nevertheless, more in-depth studies need to be undertaken to confirm this hypothesis. Alongside its other benefits, plant diversity has been shown to reduce the impact of other pathogens, such as fungi on crops and can be used for their management (Mundt, 2002; Ratnadass et al., 2012). The agroecological paradigm states that pests and pathogens can be controlled, but not eliminated, through antagonistic ecological processes facilitated by crop genetic diversity or enhanced plant health (Van Bruggen et al., 2016). In addition, diversified farms increase the resilience of the systems to biotic, abiotic and economic threats because if one crop is affected, growers can rely on other crops (Mori et al., 2013; Petersen-Rockney et al., 2021). These systems minimize the negative impact of agriculture on ecosystems and are less dependent on unstable geopolitical contexts, which can hamper food distribution from producers to consumers (Wezel et al., 2014; Hertel et al., 2021). Altogether, and in complement to the documented economic potential of diversified production systems (Van der Ploeg et al., 2019), these considerations suggest that additional research efforts on small-scale systems are needed to better optimize them.
Plant viruses constantly evolve, and monitoring them is essential to forecast emergence and outbreaks due to viral diseases. This study highlighted that some detected viruses would require more attention than others. For example, PhCMoV was widely distributed, associated with severe symptoms on few plants, and considered problematic by one grower. In addition, knowledge on the biology of this virus was limited (Temple et al., 2022). Thus, interactions of the virus with plants and vectors within the environment (reservoirs plant, transmission…) needs to be investigated more in-depth to understand how the disease can be developed, and to set up managment strategies.
Other not-well-known viruses were detected in tomatoes, among which two species (CIRV, SLRV) had not been detected in tomatoes before. However, since their presence was not associated with specific symptoms when no co-infection was noticed with other pathogenic viruses and growers did not notice it, their characterization might not be the priority for the moment. This study also reports, to our knowledge, the first detection of TMaV in Belgium. This virus was described for the first time in 2015 on tomato and did not seem to be associated with symptoms (Saqib et al., 2015). ToMV was detected on symptomatic plants showing typical ToMV symptoms (mosaic) and asymptomatic plants on a farm. ToMV was considered a severe threat to tomato production worldwide before the use of resistant cultivars and is currently re-emerging with the increased use of older, non-resistant cultivars (Hanssen et al., 2010). The virus is spread by seed and contact, and the grower affected with ToMV in this study was re-using their seeds from 1 year to another. Therefore, more information can be communicated to avoid practices that increase the likelihood of certain viruses spreading. Finally, STV was detected in 2/3rd of the farms (14/21) predominantly in asymptomatic plant pools, suggesting a potential minor role in plant pathogenicity.
5. Conclusion
The methodological approach used here makes it possible to obtain a holistic view of issues related to tomato viruses by combining, for the first time, a survey of growers perceptions with the characterization of the tomato virome by HTS technologies. In particular, the grower’s perception enabled a more realistic understanding of the impact of the tomato virome on the field. Furthermore, it highlighted the need for better characterization of viruses detected by HTS, particularly when they can threaten production (e.g., PhCMoV). Overall, the results showed that, for this peculiar season (2021), the presence of plant viruses was not necessarily linked to a high disease risk for the production according to the perception of growers and the symptom prevalence in diversified production systems in Belgium. Our preliminary investigation on what could explain these results suggests that more insect-borne viruses tended to be detected in the most diversified field. Since the detection were associated with low a viral disease pressure and a lack of concern from the growers, this finding supports the hypothesis that plant diversity might mitigate the impact of plant viruses on crops. Although these results need to be further validated over several years with a higher number of growers, they converge with the hypothesis of Keesing et al. (2010) which postulates that high biodiversity may provide a larger potential source of novel pathogens but would reduce further transmission for both long-established and newly-established emerging diseases. One of the findings of this study was that there was very little awareness of the virus threat among small-scale producers, as evidenced by the lack of knowledge of the ToBRFV, and the legal requirements associated with its presence. The individual interviews with the producers were an opportunity to inform them about known virus threats and their phytosanitary obligations.
This novel research approach, which combines the assessment of growers perception of the presence of viruses with their holistic laboratory detection, could be applied to other crops. The measured perception is valuable for directing technical communication toward knowledge gaps and for addressing phytosanitary risks. In this study, the detected viruses and their descriptions were provided to the interviewed growers and extension services. This is a starting point for raising farmers’ awareness of viruses, which, combined with technical support on how to report viruses and control them safely, should lead to better preparedness and therefore mitigation of the impact of future viral disease outbreaks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA927265.
Ethics statement
Oral consent was given from all the participants before the questionnaire. They were all informed and consented that there was a mandatory notification to the local authorities if a quarantine pathogen was detected in their crops. The university’s ethical committee is mainly focused on medical studies and was therefore unsuitable for evaluating this study.
Author contributions
CT, AB, SM, and KM contributed to the conception and design of the study and the methodology design. CT, AB, SM, KM, and ST validated the study and revised and edited the manuscript. CT realized the interviews, field observations, sample collection, and bioinformatics analyzes, and wrote the first original draft. CT and SS performed the laboratory analysis. KM provided a strong support for analyzing the interviews. SM and KM provided resources. SM and AB provided supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by European Union’s Horizon 2020 Research and Innovation program under the Marie Sklodowska-Curie, Grant Agreement no. 813542 and Federal public service, public health, Belgium, Grant Agreement no. RT 18/3 SEVIPLANT 55.
Acknowledgments
The authors would like to thank the National Institute of Biology of Slovenia and especially Denis Kutnjak and Mark Paul Selda Rivarez for their help in bioinformatic analysis. We also acknowledge the Phytopathology team of ULiege Gembloux Agro-biotech (Gbx) Laurent Minet (Hortiforum asbl/Center Technique Horticole de Gembloux), and the members of the extension services (Center Interprofessionel Maraicher) for their support and brainstorming effort in designing the survey and the questionnaire. Elisabeth Demonty and Pierre Hellin (Plant Virology Lab, CRA-W) are also thanks for their support in the laboratory analyzes. We are also very grateful to the growers who took the time to reply to the questions, allowed us to access their properties and collect samples. We thank Johan Rollin, Nuria Fontdevila, François Maclot (Gbx), and Sandrine Dury (CIRAD) for their support on analyzing the data and their helped in improving the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1139090/full#supplementary-material
Footnotes
1. ^https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list
References
Anderson, P. K., Cunningham, A. A., Patel, N. G., Morales, F. J., Epstein, P. R., and Daszak, P. (2004). Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. doi: 10.1016/j.tree.2004.07.021
Aranda, M. A., and Freitas-Astúa, J. (2017). Ecology and diversity of plant viruses, and epidemiology of plant virus-induced diseases. Ann. Appl. Biol. 171, 1–4. doi: 10.1111/aab.12361
Babaie, G., and Izadpanah, K. (2003). Vector transmission of eggplant mottled dwarf virus in Iran. J. Phytopathol. 151, 679–682. doi: 10.1046/j.1439-0434.2003.00788.x
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bernardo, P., Charles-Dominique, T., Barakat, M., Ortet, P., Fernandez, E., Filloux, D., et al. (2018). Geometagenomics illuminates the impact of agriculture on the distribution and prevalence of plant viruses at the ecosystem scale. ISME J 12, 173–184. doi: 10.1038/ismej.2017.155
Bertolini, E., Olmos, A., López, M. M., and Cambra, M. (2003). Multiplex nested reverse transcription-polymerase chain reaction in a single tube for sensitive and simultaneous detection of four RNA viruses and Pseudomonas savastanoi pv. Savastanoi in olive trees. Phytopathology 93, 286–292. doi: 10.1094/PHYTO.2003.93.3.286
Blancard, D. (2009). Les maladies de la tomate—Identifier, connaître, maîtriser. Librairie Quae. Available at: https://www.quae.com/produit/844/9782759213627/les-maladies-de-la-tomate
Cook-Patton, S. C., McArt, S. H., Parachnowitsch, A. L., Thaler, J. S., and Agrawal, A. A. (2011). A direct comparison of the consequences of plant genotypic and species diversity on communities and ecosystem function. Ecology 92, 915–923. doi: 10.1890/10-0999.1
Demsar, J., Curk, T., Erjavec, A., Gorup, C., Hocevar, T., Milutinovic, M., et al. (2013). Orange: data mining toolbox in Python. J. Mach. Learn. Res. 14, 2349–2353.
Desbiez, C., Wipf-Scheibel, C., Millot, P., Berthier, K., Girardot, G., Gognalons, P., et al. (2020). Distribution and evolution of the major viruses infecting cucurbitaceous and solanaceous crops in the French Mediterranean area. Virus Res. 286:198042. doi: 10.1016/j.virusres.2020.198042
Dumont, A.M. (2017). Analyse systémique des conditions de travail et d’emploi dans la production de légumes pour le marché du frais en Région wallonne (Belgique), dans une perspective de transition agroécologique. PhD Dissertation, UCLouvain. Louvain-la-Neuve.
Dumont, A. M., Gasselin, P., and Baret, P. V. (2020). Transitions in agriculture: three frameworks highlighting coexistence between a new agroecological configuration and an old, organic and conventional configuration of vegetable production in Wallonia (Belgium). Geoforum 108, 98–109. doi: 10.1016/j.geoforum.2019.11.018
Duong, T. T., Brewer, T., Luck, J., and Zander, K. (2019). A global review of farmers’ perceptions of agricultural risks and risk management strategies. Agriculture 9:10. doi: 10.3390/agriculture9010010
EPPO Bulletin (2020). Tomato brown rugose fruit virus (2021). EPPO Bulletin 51, 178–197. doi: 10.1111/epp.12723
Elena, S. F., Fraile, A., and García-Arenal, F. (2014). “Chapter three—evolution and emergence of plant viruses” in Advances in Virus Research. eds. K. Maramorosch and F. A. Murphy (Advances in Virus Research, Elsevier: Academic Press), 161–191.
Eurostat (2021). Statistics on tomatoes production, prices and trade. Available at: https://agriculture.ec.europa.eu/data-and-analysis/markets/overviews/market-observatories/fruit-and-vegetables/tomatoes-statistics_en#statistics/ (Accessed November 30, 2022).
FAOSTAT (2020). Statistics Division of the Food and Agricultural Organization of the United Nations. Available at: http://www.fao.org/faostat/en/ (Accessed November 30, 2022).
Filloux, D., Dallot, S., Delaunay, A., Galzi, S., Jacquot, E., and Roumagnac, P. (2015). Metagenomics approaches based on Virion-associated nucleic acids (VANA): an innovative tool for assessing without a priori viral diversity of plants. Methods Mol. Biol. 1302, 249–257. doi: 10.1007/978-1-4939-2620-6_18
Gaafar, Y. Z. A., Abdelgalil, M. A. M., Knierim, D., Richert-Pöggeler, K. R., Menzel, W., Winter, S., et al. (2017). First report of physostegia chlorotic mottle virus on tomato (Solanum lycopersicum) in Germany. Plant Dis. 102:255. doi: 10.1094/PDIS-05-17-0737-PDN
Haddad, N. M., Crutsinger, G. M., Gross, K., Haarstad, J., Knops, J. M. H., and Tilman, D. (2009). Plant species loss decreases arthropod diversity and shifts trophic structure. Ecol. Lett. 12, 1029–1039. doi: 10.1111/j.1461-0248.2009.01356.x
Hanssen, I. M., Lapidot, M., and Thomma, B. P. H. J. (2010). Emerging viral diseases of tomato crops. Mol. Plant-Microbe Interact. 23, 539–548. doi: 10.1094/MPMI-23-5-0539
Hertel, T., Elouafi, I., Tanticharoen, M., and Ewert, F. (2021). Diversification for enhanced food systems resilience. Nat. Food 2, 832–834. doi: 10.1038/s43016-021-00403-9
Jeger, M. J. (2020). The epidemiology of plant virus disease: towards a new synthesis. Plan. Theory 9:1768. doi: 10.3390/plants9121768
Jones, R. A. C. (2009). Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 141, 113–130. doi: 10.1016/j.virusres.2008.07.028
Jones, J. J., Zitter, T. A., Momol, T. M., and Miller, S. A. (eds.) (2014). Compendium of Tomato Diseases and Pests 2nd Edn. St-Paul, Minnesota: The American Phytopathological Society Press, 86–88
Keesing, F., Belden, L. K., Daszak, P., Dobson, A., Harvell, C. D., Holt, R. D., et al. (2010). Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652. doi: 10.1038/nature09575
Keesing, F., Holt, R. D., and Ostfeld, R. S. (2006). Effects of species diversity on disease risk. Ecol. Lett. 9, 485–498. doi: 10.1111/j.1461-0248.2006.00885.x
Keesing, F., and Ostfeld, R. S. (2021). Dilution effects in disease ecology. Ecol. Lett. 24, 2490–2505. doi: 10.1111/ele.13875
King, K. C., and Lively, C. M. (2012). Does genetic diversity limit disease spread in natural host populations? Heredity 109, 199–203. doi: 10.1038/hdy.2012.33
Knops, J. M. H., Tilman, D., Haddad, N. M., Naeem, S., Mitchell, C. E., Haarstad, J., et al. (1999). Effects of plant species richness on invasion dynamics, disease outbreaks, insect abundances and diversity. Ecol. Lett. 2, 286–293. doi: 10.1046/j.1461-0248.1999.00083.x
Kremen, C., and Miles, A. (2012). Ecosystem Services in Biologically Diversified versus conventional farming systems: benefits, externalities, and trade-offs. Ecol. Soc. 17. doi: 10.5751/ES-05035-170440
Laforge, J., Fenton, A., Lavalée-Picard, V., and McLachlan, S. (2018). New farmers and food policies in Canada. Can. Food Stud. 5, 128–152. doi: 10.15353/cfs-rcea.v5i3.288
Lefeuvre, P., Martin, D. P., Elena, S. F., Shepherd, D. N., Roumagnac, P., and Varsani, A. (2019). Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 17, 632–644. doi: 10.1038/s41579-019-0232-3
Li, Y., Tan, G., Lan, P., Zhang, A., Liu, Y., Li, R., et al. (2018). Detection of tobamoviruses by RT-PCR using a novel pair of degenerate primers. J. Virol. Methods 259, 122–128. doi: 10.1016/j.jviromet.2018.06.012
Lichtenberg, E. M., Kennedy, C. M., Kremen, C., Batáry, P., Berendse, F., Bommarco, R., et al. (2017). A global synthesis of the effects of diversified farming systems on arthropod diversity within fields and across agricultural landscapes. Glob. Chang. Biol. 23, 4946–4957. doi: 10.1111/gcb.13714
Liu, X., Chen, L., Liu, M., García-Guzmán, G., Gilbert, G. S., and Zhou, S. (2020). Dilution effect of plant diversity on infectious diseases: latitudinal trend and biological context dependence. Oikos 129, 457–465. doi: 10.1111/oik.07027
MacDiarmid, R., Rodoni, B., Melcher, U., Ochoa-Corona, F., and Roossinck, M. (2013). Biosecurity implications of new technology and discovery in plant virus research. PLoS Pathog. 9:e1003337. doi: 10.1371/journal.ppat.1003337
Maclot, F. J., Debue, V., Blouin, A. G., Fontdevila Pareta, N., Tamisier, L., Filloux, D., et al. (2021). Identification, molecular and biological characterization of two novel secovirids in wild grass species in Belgium. Virus Res. 298:198397. doi: 10.1016/j.virusres.2021.198397
Mahjabeen Akhtar, K. P., Sarwar, N., Saleem, M. Y., Asghar, M., Iqbal, Q., and Jamil, F. F. (2012). Effect of cucumber mosaic virus infection on morphology, yield and phenolic contents of tomato. Arch. Phytopathol. Plant Protect. 45, 766–782. doi: 10.1080/03235408.2011.595965
Massart, S., Adams, I., Al Rwahnih, M., Baeyen, S., Bilodeau, G. J., Blouin, A. G., et al. (2014). Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Research 188, 90–96. doi: 10.1016/j.virusres.2014.03.029
Menzel, W., Richert-Pöggeler, K., Winter, S., and Knierim, D. (2018). Characterization of a nucleorhabdovirus from Physostegia. Acta Hortic. 1193, 29–38. doi: 10.17660/ActaHortic.2018.1193.5
Morel, K., and Léger, F. (2016). A conceptual framework for alternative farmers’ strategic choices: the case of French organic market gardening microfarms. Agroecol. Sustain. Food Syst. 40, 466–492. doi: 10.1080/21683565.2016.1140695
Mori, A. S., Furukawa, T., and Sasaki, T. (2013). Response diversity determines the resilience of ecosystems to environmental change. Biol. Rev. 88, 349–364. doi: 10.1111/brv.12004
Mundt, C. C. (2002). Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 40, 381–410. doi: 10.1146/annurev.phyto.40.011402.113723
Murray-Watson, R. E., Hamelin, F. M., and Cunniffe, N. J. (2022). How growers make decisions impacts plant disease control. PLoS Comput. Biol. 18:e1010309. doi: 10.1371/journal.pcbi.1010309
Nicaise, V. (2014). Crop immunity against viruses: outcomes and future challenges. Frontiers in Plant Science 5. Available at: https://www.frontiersin.org/articles/10.3389/fpls.2014.00660 (Accessed February 17, 2023).
Oladokun, J. O., Halabi, M. H., Barua, P., and Nath, P. D. (2019). Tomato brown rugose fruit disease: current distribution, knowledge and future prospects. Plant Pathol. 68, 1579–1586. doi: 10.1111/ppa.13096
Oñate-Sánchez, L., and Vicente-Carbajosa, J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1:93. doi: 10.1186/1756-0500-1-93
Pagán, I., González-Jara, P., Moreno-Letelier, A., Rodelo-Urrego, M., Fraile, A., Piñero, D., et al. (2012). Effect of biodiversity changes in disease risk: exploring disease emergence in a plant-virus system. PLoS Pathog. 8:e1002796. doi: 10.1371/journal.ppat.1002796
Panno, S., Davino, S., Caruso, A. G., Bertacca, S., Crnogorac, A., Mandić, A., et al. (2021). A review of the Most common and economically important diseases that undermine the cultivation of tomato crop in the Mediterranean Basin. Agronomy 11:2188. doi: 10.3390/agronomy11112188
Petersen-Rockney, M., Baur, P., Guzman, A., Bender, S. F., Calo, A., Castillo, F., et al. (2021). Narrow and brittle or broad and nimble? Comparing adaptive capacity in simplifying and diversifying farming systems. Front. Sustain. Food Syst. 5:564900. doi: 10.3389/fsufs.2021.564900
Pinel-Galzi, A., Traoré, O., Séré, Y., Hébrard, E., and Fargette, D. (2015). The biogeography of viral emergence: rice yellow mottle virus as a case study. Current Opinion in Virology 10, 7–13. doi: 10.1016/j.coviro.2014.12.002
Plateau, L., Roudart, L., Hudon, M., and Maréchal, K. (2021). Opening the organisational black box to grasp the difficulties of agroecological transition. An empirical analysis of tensions in agroecological production cooperatives. Ecol. Econ. 185:107048. doi: 10.1016/j.ecolecon.2021.107048
Ponisio, L. C., M’Gonigle, L. K., Mace, K. C., Palomino, J., de Valpine, P., and Kremen, C. (2015). Diversification practices reduce organic to conventional yield gap. Proc. R. Soc. B Biol. Sci. 282:20141396. doi: 10.1098/rspb.2014.1396
Power, A. G. (1991). Virus spread and vector dynamics in genetically diverse plant populations. Ecology 72, 232–241. doi: 10.2307/1938917
Ratnadass, A., Fernandes, P., Avelino, J., and Habib, R. (2012). Plant species diversity for sustainable management of crop pests and diseases in agroecosystems: a review. Agron. Sustain. Dev. 32, 273–303. doi: 10.1007/s13593-011-0022-4
Rivarez, M. P. S., Vučurović, A., Mehle, N., Ravnikar, M., and Kutnjak, D. (2021). Global advances in tomato Virome research: current status and the impact of high-throughput sequencing. Front. Microbiol. 12:671925. doi: 10.3389/fmicb.2021.671925
Rong, W., Rollin, J., Hanafi, M., Roux, N., and Massart, S. (2022). Validation of high throughput sequencing as virus indexing test for Musa germplasm: performance criteria evaluation and contamination monitoring using an alien control. PhytoFrontiers. doi: 10.1094/PHYTOFR-03-22-0030-FI
Roossinck, M. J., and García-Arenal, F. (2015). Ecosystem simplification, biodiversity loss and plant virus emergence. Curr. Opin. Virol. 10, 56–62. doi: 10.1016/j.coviro.2015.01.005
Rubio, L., Galipienso, L., and Ferriol, I. (2020). Detection of plant viruses and disease management: relevance of genetic diversity and evolution. Front. Plant Sci. 11:1092. doi: 10.3389/fpls.2020.01092
Salem, N., Mansour, A., Ciuffo, M., Falk, B. W., and Turina, M. (2016). A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 161, 503–506. doi: 10.1007/s00705-015-2677-7
Saqib, M., Wylie, S. J., and Jones, M. G. K. (2015). Serendipitous identification of a new Iflavirus-like virus infecting tomato and its subsequent characterization. Plant Pathol. 64, 519–527. doi: 10.1111/ppa.12293
Schreinemachers, P., Balasubramaniam, S., Boopathi, N. M., Ha, C. V., Kenyon, L., Praneetvatakul, S., et al. (2015). Farmers’ perceptions and management of plant viruses in vegetables and legumes in tropical and subtropical Asia. Crop Prot. 75, 115–123. doi: 10.1016/j.cropro.2015.05.012
Temple, C., Blouin, A. G., De Jonghe, K., Foucart, Y., Botermans, M., Westenberg, M., et al. (2022). Biological and genetic characterization of Physostegia chlorotic mottle virus in Europe based on host range, location, and time. Plant Dis. 106, 2797–2807. doi: 10.1094/PDIS-12-21-2800-RE
Trebicki, P. (2020). Climate change and plant virus epidemiology. Virus Res. 286:198059. doi: 10.1016/j.virusres.2020.198059
Ullah, N., Ali, A., Ahmad, M., Fahim, M., Din, N., and Ahmad, F. (2017). Evaluation of tomato genotypes against tomato mosaic virus (ToMV) and its effect on yield contributing parameters. Pak. J. Bot. 49, 1585–1592.
van Bruggen, A. H. C., Gamliel, A., and Finckh, M. R. (2016). Plant disease management in organic farming systems. Pest Manag. Sci. 72, 30–44. doi: 10.1002/ps.4145
van der Ploeg, J. D., Barjolle, D., Bruil, J., Brunori, G., Costa Madureira, L. M., Dessein, J., et al. (2019). The economic potential of agroecology: empirical evidence from Europe. J. Rural. Stud. 71, 46–61. doi: 10.1016/j.jrurstud.2019.09.003
Vega, D., Gally, M. E., Romero, A. M., and Poggio, S. L. (2019). Functional groups of plant pathogens in agroecosystems: a review. Eur. J. Plant Pathol. 153, 695–713. doi: 10.1007/s10658-018-01616-8
Velasco, L., Ruiz, L., Galipienso, L., Rubio, L., and Janssen, D. (2020). A historical account of viruses in intensive horticultural crops in the Spanish Mediterranean arc: new challenges for a sustainable agriculture. Agronomy 10:860. doi: 10.3390/agronomy10060860
Villamor, D. E. V., Ho, T., Al Rwahnih, M., Martin, R. R., and Tzanetakis, I. E. (2019). High throughput sequencing for plant virus detection and discovery. Phytopathology 109, 716–725. doi: 10.1094/PHYTO-07-18-0257-RVW
Vučurović, A., Kutnjak, D., Mehle, N., Stanković, I., Pecman, A., Bulajić, A., et al. (2021). Detection of four new tomato viruses in Serbia using post hoc high-throughput sequencing analysis of samples from a large-scale field survey. Plant Dis. 105, 2325–2332. doi: 10.1094/PDIS-09-20-1915-RE
Wezel, A., Casagrande, M., Celette, F., Vian, J.-F., Ferrer, A., and Peigné, J. (2014). Agroecological practices for sustainable agriculture. A review. Agron. Sustain. Dev. 34, 1–20. doi: 10.1007/s13593-013-0180-7
Xu, C., Sun, X., Taylor, A., Jiao, C., Xu, Y., Cai, X., et al. (2017). Diversity, distribution, and evolution of tomato viruses in China uncovered by small RNA sequencing. J. Virol. 91. doi: 10.1128/JVI.00173-17
Yamamura, Y., and Scholthof, H. B. (2005). Tomato bushy stunt virus: a resilient model system to study virus–plant interactions. Mol. Plant Pathol. 6, 491–502. doi: 10.1111/j.1364-3703.2005.00301.x
Keywords: virome, grower’s perception, high throughput sequencing, tomato, small-scale vegetable farms, Belgium
Citation: Temple C, Blouin AG, Tindale S, Steyer S, Marechal K and Massart S (2023) High throughput sequencing technologies complemented by growers’ perceptions highlight the impact of tomato virome in diversified vegetable farms and a lack of awareness of emerging virus threats. Front. Sustain. Food Syst. 7:1139090. doi: 10.3389/fsufs.2023.1139090
Edited by:
Mohamed Ait-El-Mokhtar, University of Hassan II Casablanca, MoroccoReviewed by:
Fidele Tiendrebeogo, Institut de l'environnement et de la Recherche Agricole (INERA), Burkina FasoJacqueline Fletcher, Oklahoma State University, United States
Copyright © 2023 Temple, Blouin, Tindale, Steyer, Marechal and Massart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastien Massart, c2ViYXN0aWVuLm1hc3NhcnRAdWxpZWdlLmJl
 Coline Temple
Coline Temple Arnaud G. Blouin
Arnaud G. Blouin Sophie Tindale3
Sophie Tindale3 Stephan Steyer
Stephan Steyer Sebastien Massart
Sebastien Massart