- 1Regional Centre of Agricultural Research of Sidi Bouzid, Sidi Bouzid, Tunisia
- 2Laboratory of Agriculture Production Systems and Sustainable Development (LR03AGR02), Department of Agricultural Production, Higher School of Agriculture of Mograne, University of Carthage, Mograne-Zaghouan, Tunisia
- 3Department of Plant Protection, College of Agriculture Engineering Science, University of Baghdad, Baghdad, Iraq
- 4Faculty of Biotechnology, October University for Modern Sciences and Arts, 6th of October, Egypt
- 5LR14AGR02, Laboratory of Bio-aggressors and Integrated Pest Management in Agriculture, National Agronomic Institute of Tunisia, University of Carthage, Tunis, Tunisia
- 6Higher Institute of the Sciences and Techniques of Waters of Gabes, Gabes University, Gabes, Tunisia
- 7Department of Botany and Microbiology, Faculty of Science, University of Suez Canal, Ismailia, Egypt
- 8Research Institute of University of the Bucharest (ICUB), Bucharest, Romania
- 9The Centre for Mineral Biogeochemistry, Faculty of Natural and Agricultural Sciences, The University of the Free State, Bloemfontein, Free State, South Africa
- 10Department of Environmental Management, Institute of Environmental Engineering, RUDN University, Moscow, Russia
Beneficial rhizobacteria play a crucial role in promoting plant growth and enhancing soil health by producing key enzymes, facilitating nutrient cycling, and suppressing phytopathogens. This study investigated the physicochemical properties of soil from 32 grapevine sites in the Sidi Bouzid region of Tunisia and evaluated the plant growth-promoting and biocontrol potential of bacterial isolates against Botrytis cinerea, the causative agent of gray mold. Soil analysis revealed significant variation in pH (6.78 to 8.07), organic matter (0.44%−1.59%), and nutrient content, while electrical conductivity remained stable. A total of 107 bacterial isolates were isolated from soil samples and recovered, of which 97 were non-pathogenic and tested for multiple plant growth-promoting traits. Enzymatic screening revealed production of catalase, pectinase, cellulase, and chitinase among others. In vitro assays identified four isolates (H3Rh1, ZRh5, GRh5, and SRh2) with strong antifungal activity, achieving up to 99.3% growth inhibition of B. cinerea. In detached leaf assays, the isolates H3Rh1, ZRh5, GRh5, and SRh2 achieved inhibitory growth potential values of 92.33%, 93.73%, 93.02%, and 96.99% against B. cinerea, respectively. Molecular analysis confirmed the isolates as Arthrobacter globiformis, Priestia megaterium, Bacillus cabrialesii, and Bacillus mojavensis, with >99% sequence identity and deposited in GenBank. These strains also demonstrated strong plant growth-promoting attributes, including nitrogen fixation, phosphate and potassium solubilization, and indole acetic acid and siderophore production. This study highlights the biocontrol potential of native rhizobacteria as eco-friendly alternatives to chemical fungicides for managing gray mold in grapevines and promoting sustainable viticulture.
1 Introduction
Vitis vinifera (L.) is a globally cultivated fruit crop with origins in the Caucasus region. Its cultivation extends across a vast geographical area, encompassing countries such as Turkey, Georgia, Spain, France, Italy, and many others. As of 2018, the global vineyard area under cultivation reached approximately 7.4 million hectares, with a corresponding production of 77.8 million tons of grapes. This widespread cultivation and significant production highlight the economic and agricultural importance of grapevines (Tsantila et al., 2024).
V. vinifera has a long history of cultivation in Tunisia, dating back to the medieval period (Snoussi et al., 2004). Grapevines are cultivated across diverse climatic and edaphic conditions throughout the country (Zoghlami et al., 2003). According to the General Directorate of Agricultural Studies and Development, districts such as Ben Arous, Nabeul, and Sidi Bouzid (the current study location) contribute significantly, accounting for approximately 44% of the total national grapevine production (Habib et al., 2020). Interestingly, in southern Tunisia's arid regions, grape cultivation thrives within oases (Gabes, Gafsa, Kebili, and Tozeur) where vines are grown under the shade of date palms (Habib et al., 2020). This unique cultivation practice highlights the adaptability of V. vinifera in Tunisia. With a national production reaching 133,500 tons in 2016, grapevine cultivation plays a substantial role in the country's socio-economic landscape (Habib et al., 2020; Saddoud Debbabi et al., 2024).
Plant pathogens pose significant threats to global food security, causing economic hardship, food spoilage (e.g., ergotism), and even historical famines such as the Irish Potato Famine (1845–1852). Pathogen infection disrupts host plant physiology through various mechanisms by secreting toxins and enzymes (Rebouh et al., 2020). The fungus Botrytis cinerea exhibits limited host specificity similar to a necrotrophic pathogen and infects a wide range of plant species (Hajji-Hedfi et al., 2023). This pathogen caused gray mold disease that significantly impacts numerous important crops economically such as grapes, strawberries, and tomatoes (Barka et al., 2002; Dean et al., 2012). Annual global losses due to B. cinerea are estimated to range from USD 10 billion to USD 100 billion (Weiberg et al., 2013; Amarouchi et al., 2021). In Tunisia, B. cinerea is the main fungal disease currently infecting vineyards. In the northeastern region of the country, infection rates reported up to 70% in grapevine bunches, approximately 20% in grapes, and between 5% and 27% in floral buds (Melki Ben Fredj et al., 2007; Chebil et al., 2004).
Various strategies are implemented to manage B. cinerea infections. Currently, chemical fungicides remain effective against this pathogen but cause negative effects on ecosystem and increase fungicide resistance. These include the emergence of B. cinerea resistance, leading to reduced or even failed disease control (Fernández-Ortuño et al., 2016; Rupp et al., 2017; Yin et al., 2018; Sautua et al., 2019). In addition, consumer demand for pesticide-free produce, growing concerns about environmental pollution, and stricter regulations on synthetic fungicides are driving the need for alternative control methods (Reeves et al., 2019). To address these limitations, research on effective and eco-friendly disease management strategies is crucial (Rebouh et al., 2019). Among these strategies, the uses of plant-based essential oils demonstrated a significant promise alternative to chemicals as natural antifungal agents. B. cinerea growth was severely inhibited by essential oils extracted from origanum, lavender, and rosemary. At extremely low doses, origanum oil showed complete inhibition. According to these results, essential oils may be useful as a bio fungicide for preventing gray mold in crops (Soylu et al., 2010). Furthermore, the application of beneficial microbes as biocontrol agents gained significant interest in recent agricultural practices. This approach offers a promising alternative for sustainable crop protection (Rebouh et al., 2022; Hajji-Hedfi et al., 2023).
Plant growth-promoting rhizobacteria (PGPR) are a group of bacteria that colonize the rhizosphere (the zone surrounding plant roots) and can influence plant growth positively through direct and indirect mechanisms (Soumare et al., 2021). PGPR can affect plant pathogen directly and indirectly. Regarding direct mechanisms, they include producing antibiotics, enzymes of hydrolysis, siderophores, competition, and volatile and non-volatile compounds. On the other hand, indirect mechanisms refer to enhance the plant growth and plant defenses (Soumare et al., 2021).
Many bacterial species identified as potential PGPR candidates, including members of the genera Arthrobacter, Azospirillum, Azotobacter, Bacillus, Burkholderia, Enterobacter, Klebsiella, Pseudomonas, and Serratia. These bacteria offer promising avenues for sustainable agriculture by enhancing plant fitness through various mechanisms (Al-Ani et al., 2020).
Plant growth-promoting rhizobacteria can exhibit various mechanisms of antagonism against plant pathogens such as B. cinerea. PGPR can utilize different mechanisms in controlling plant pathogens that may include competition for the niche-space, solubilization of phosphate, production of antibiotics, siderophores, non-volatile and volatile compounds, phytohormones, nitrogen fixation, and hydrolytic enzymes (Hasan et al., 2024). PGPR can stimulate plant defenses by inducing the production of pathogenesis-related (PR) proteins (Santoyo et al., 2012). These PR proteins play a crucial role in plant resistance against B. cinerea infection in grapes, highlighting the intricate relationship between plant metabolism and disease response (Kumar et al., 2010). However, the primary inhibitory action of most antagonistic bacteria is believed to be the production of antifungal metabolites and antibiotics, directly targeting and suppressing the growth of the pathogen (Peian et al., 2021; Al-Ani et al., 2020). Among beneficial bacteria, the genus Bacillus used as biological control agents, regarding their large secondary metabolites (Shafi et al., 2017), including lipopeptides such as iturins, fengycins, and surfactins. By introducing their hydrophobic tails into the cytoplasmic membrane and causing autoaggregation to produce pores in the cellular membrane, the iturins cause cell leakage. Furthermore, these species able to produce polyketides and volatile organic compounds with antifungal activities (Ongena and Jacques, 2008; Caulier et al., 2019). In Tunisia, previous studies have demonstrated the efficacy of two strains of Bacillus subtilis group, namely, B27 and B29, to control the gray mold in grapevine, where they reduced the development on grape leaf by 77% and 99%, respectively (Maachia et al., 2015). The widespread occurrence and significant economic impact of Botrytis cinerea-induced gray mold disease on various economic plants as grapevine required effective control strategies. To investigate biocontrol options against B. cinerea, this study aimed to identify local rhizobacteria that were isolated from Tunisian vineyard soils and that have not been previously reported for biocontrol of gray mold in Tunisian vineyards. They were evaluated for their plant growth-promoting (PGP) activities as potential biocontrol agents against B. cinerea-infested Vitis vinifera in Tunisia. The study presents regionally adapted strains of bacterial isolates from 32 vineyard sites, suggesting promising potential for sustainable biocontrol approaches.
2 Materials and methods
2.1 Experimental sites and sampling
This study was conducted at V. vinifera (grapevine) cv. Victoria plantation located in the Regueb delegation of Sidi Bouzid region, Tunisia (Supplementary Table S1). A total number of 32 grapevine fields were prospected between 2021 and 2022. To assess the rhizobacteria, bulk soil samples were collected from a depth of 20–40 cm directly beneath the canopy of mature grapevines exhibiting the same phenological state. Five independent soil samples were collected from each field in sterile and polyethylene bags and transferred directly to the lab until they were plated out.
2.2 Soil characterization
The physicochemical parameters, such as pH, electrical conductivity (EC), organic carbon content (OC), total nitrogen (N), carbon (C)/total nitrate (N) ratio, soil texture, total limestone (TL), available phosphorus (P2O5), and exchangeable potassium (K2O), were evaluated using a variety of titration methods. The Robinson pipette method (Jackson, 1971; Mathieu and Pieltain, 2003) was employed to evaluate the particle size texture. The soil pH was measured using a digital pH meter (Consort C1010) after being vortexed for 5–6 min in a 1:5 (soil: water) suspension. In accordance with reference (Jackson, 1971), the conductivity of the soil supernatant liquid was determined by employing potassium chloride (KCl) and Equiptronic's digital electrical conductivity meter (Consort C1010). The Walkley and Black method (Jackson, 1971) was employed to evaluate the organic matter content. The available nitrogen in soil samples was evaluated using the Kjeldahl method (Kjeldahl, 1883). The carbon-to-nitrogen ratio of soil mass (C/N) was ascertained in accordance with the method (Zhang et al., 2011).
Nevertheless, the optical density was estimated as the available phosphorus by measuring it with a UV spectrometer at a wavelength of 660 nm. The flame photometer was employed to determine the available potassium in the soil, which was carried out using a chemical extraction method. Calibration was conducted following standardization with a solution containing potassium at concentrations of 10, 15, and 40 ppm. The Bernard's calcimeter was used to estimate the total limestone available for each subsample after the addition of HCl (50%). The entire limestone analysis was monitored by measuring the CO2 released from the contact of HCl with a precise weight of soil containing pure and dry CaCO3 in a known quantity.
2.3 Isolation of Botrytis cinerea
The occurrence of B. cinerea at different positions on grapes, showing characteristic gray mold infections, was recorded and assessed in selected vineyards. The isolation technique involved isolating grape skins at bunch closure, onset ripening, and harvest stages. Two grapes were randomly cut at the pedicel from each of the 10 bunches per vineyard. The grapes were then divided into two groups, each containing 10 grapes. Grapes of one group were left untreated, and those of the other group were surface-sterilized (30 s in 70% ethanol, 2 min in 0.35% sodium hypochlorite, and 30 s in 70% ethanol) according to Abdel-Azeem and Salem (Abdel-Azeem and Salem, 2012) and air-dried. The isolation was carried out by plating grapes directly on Botrytis selective medium (BSM) according to Edwards and Seddon (Edwards and Seddon, 2001). Isolates were maintained on potato dextrose agar slants and kept at 4°C.
2.4 Identification and pathogenicity test of Botrytis cinerea
Phenotypic identification of B. cinerea was based on established morphological features, including colony structure, growth rate [calculated as growth at 48 h minus that at 24 h (Zhou et al., 2014)], mycelial shape and color, days to sporulation and sclerotia formation, as well as the number, shape, and color of sclerotia. These criteria align with standard taxonomic references (Tanovic et al., 2009, 2014).
Fungal spores were collected from 10-day-old cultures grown at 22°C in sterile water containing 0.1% (v/v) Tween 20. The suspension was placed through two layers of lens cleaning tissue (Whatman 105) to remove any mycelial fragments. It was then centrifuged at 10,000 × g for 2 min at 4°C. The supernatant was decanted, and the spore pellet was re-suspended in 0.01% (v/v) Tween-20 to extract nutrients from the medium. Standard inoculums were adjusted to 1 × 107-2 × 107 CFU ml−1, based on 0.5 McFarland standards.
After being surface-sterilized with 1% NaOCl, three rinses in sterile distilled water, and an overnight drying period, 320 grapes (10 from each site) were then placed on wet filter paper in a plastic container measuring 29 × 22 × 15 cm. The grapes were gradually inoculated with the inoculum suspension of the disease-causing pathogen at a concentration of 104 conidia/mL. After 24 h of incubation in a humid chamber set at 20°C with 100% relative humidity, samples were allowed to cool at room temperature for further observation. To validate Koch's hypotheses, the causative agent was re-isolated from the lesions. Three separate runs of the pathogenicity test were performed.
2.5 Bacteria strain isolation
The standard serial dilution technique was used for the isolation of bacteria from soil samples collected from 32 different sites according to Prashanthi et al. method (Prashanthi et al., 2021). One gram of soil sample was mixed with l0 ml of sterile water and serially diluted up to 10−7. From the serially diluted soil sample, 100 μl was mixed with solidified nutrient agar medium (NA), King's Medium B (KB), and ISP2 medium (ISP2) plates, and natamycin (Sigma-Aldrich, USA) at 20 μg/ml was amended with a molten agar medium at 50°C to prevent fungal growth (Liao et al., 2013) and incubated at 25°C ± 2°C for optimal bacterial growth. After 48 h, the plates had a lawn of mixed bacterial colonies. The individual colonies were selected according to morphology, color, transparency, and other characteristics and then picked and streaked onto fresh nutrient agar plates to get pure cultures. The purified slants were stored in a 4°C freezer for ulterior assays.
2.6 Hypersensitive reaction
The total screened pure bacterial isolates revealed from the soil samples were subjected to pathogenicity tests as described by Xiang et al. (2022). To confirm the absence of phytopathogenic properties in the antagonistic bacterial isolates, a hypersensitivity test was conducted on tobacco (Nicotiana tabacum) and geraniums (Pelargonium hederaefolium) leaves using a standardized inoculum concentration of 200 colony-forming units per milliliter. The tobacco and geraniums plants used in the present research were obtained from the Regional Centre of Agricultural Research of Sidi Bouzid, Tunisia. The bacterial suspension was delivered via intercellular injection using a sterile syringe. Sterile distilled water served as the negative control, while Agrobacterium vitis (phytopathogenic bacteria obtained from the Laboratory of Plant Protection, CRRA, Sidi Bouzid, Tunisia), known to elicit a hypersensitive response in tobacco, acted as the positive control. The absence of a complete collapse of the inoculated tobacco leaf tissue after 24 h was interpreted as a negative reaction, indicating no phytopathogenic potential of the tested bacterial isolates (Mohamed et al., 2022; Xiang et al., 2022).
2.7 Screening of antagonistic bacteria
According to Li et al. (2020), the agar disk confrontation assay was applied to evaluate the antagonistic potential of candidate bacterial strains against B. cinerea. In brief, 5 mm-diameter fungal disks, grown for 8 days on potato dextrose agar (PDA), were placed in the center of Petri dishes containing PDA medium. Bacterial antagonists were then inoculated on the same day at the four cardinal points around the fungal disk. These bacterial inoculates consisted of 5-mm-diameter PDA disks containing 72-h-old bacterial growth. The control plates included only the fungal disk on the PDA, and then, all plates were incubated at 28°C. Every 24 h for 7 days, a systematic measure was performed to measure the colony radial growth of both B. cinerea and the antagonistic bacteria. Five replicate plates for each treatment were conducted, and the entire experiment was repeated three times to ensure data robustness. The percentage of radial growth inhibition (PRGI) formula was used to quantify each bacterial isolate's anti-fungal capacity as follows:
where R1 represents the average radial growth of the fungus control (without bacterial antagonism), and R2 represents the average radial growth of the fungus in confrontation with the bacterial antagonist.
2.8 Detached leaf assay
To assess the antifungal potential of recovered bacterial isolates against B. cinerea, in vitro bioassay was conducted using detached leaves of V. vinifera cv. Victoria, a cultivar that is susceptible to gray mold (Wu et al., 2023). Sporangial suspension of the fungal pathogen was prepared in distilled water and adjusted the concentration up to 106 spores/ml using a hemocytometer. Detached leaves were surface-sterilized with 70% ethanol, rinsed with sterile water, and then positioned on saturated filter paper in Petri dishes. Ten microliter volumes of bacterial suspensions was applied onto the abaxial (underside) surface of each leaf. After a 2-h incubation period, we directly applied the B. cinerea conidia suspension to the treated areas to facilitate bacterial establishment. The positive control leaves were treated with conidia suspension, while the negative control leaves were treated only with sterile water. Five leaves for each treatment (controls and tested bacterial strains) were assessed. We conducted the entire experiment in triplicate to account for potential variability. All Petri dishes were sealed and incubated in a growth chamber at 22°C ± 1°C during a 12-h-light/12-h-dark cycle. The disease progression on leaves was monitored daily for 14 days post-inoculation (dpi) to evaluate the efficacy of bacterial interventions in B. cinerea growth suppression (Hajji-Hedfi et al., 2023).
Disease progression of Botrytis cinerea on detached grapevine leaves was evaluated using several parameters according to Macan et al. (2022). The efficacy of each bacterial treatment against B. cinerea was then categorized based on the calculated disease severity indices (DSI) using the following classification system: Extremely effective (EE): DSI = 0%; Highly effective (HE): DSI = 0.1%−5%; Effective (E): DSI = 5.1%−25%; Ineffective (I): DSI = 25.1%−50%; Highly ineffective (HI): DSI = 50.1%−100% (Milagres et al., 1999). Finally, the protective potential (PP) of each bacterial treatment was determined by comparing the average DSI of positive controls with the average DSI of treated leaves, expressed as a percentage. This calculation provides a measure of disease reduction attributable to bacterial antagonism (Mahajan et al., 2020):
2.9 Screening of plant growth-promoting traits
The total collection of isolated non-pathogenic bacterial strains was screened for their potential PGP traits, including phosphate and potassium solubilization, production of indole acetic acid (IAA), siderophore production, and antibiotic production (HCN) and production of seven extracellular enzymes, as follows:
2.9.1 Indole acetic acid production
The bacterial isolates strains were cultured in plates on Luria-Bertani (LB) medium supplemented with 5 mM tryptophan, 0.06% SDS, and 1% glycerol. Then, a Whatman filter paper disk of 80 mm diameter was placed on the plate surface for incubation at 28°C for 48 h. After incubation, the disk was removed and treated with Salkowski reagent (2% FeCl3 in 0.5 M perchloric acid). The disks were incubated in Petri dishes for 10 to 30 min, during which the presence of indole-3-acetic acid (IAA) was manifested by the development of a pink to reddish halo. A shift from yellow to yellow-brown or brown indicated the presence of other indole derivatives, as described by Glickmann and Dessaux (1995).
2.9.2 Phosphate solubilization efficiency
The phosphate solubilization potential of the candidate bacterial strains was evaluated using Pikovskaya (1948)'s agar medium. Bacterial colonies obtained from pure cultures were inoculated onto Petri dishes containing 20 ml of solidified Pikovskaya's agar medium. The inoculated plates were then incubated at 28°C for 5 days. The formation of a clear, lighter-colored halo zone surrounding the bacterial colonies was indicative of phosphate solubilization by the candidate bacterial strains. This halo zone represents the area where insoluble phosphates in the medium have been transformed into soluble forms accessible for plant uptake (Oladapo et al., 2020).
2.9.3 Potassium solubilization
The potential for potassium solubilization was examined using the Aleksandrov agar medium, where bacterial isolates were inoculated and incubated at 28°C for 5 days. The appearance of a halo zone around the colonies was regarded as a positive result (Anwar et al., 2022).
2.9.4 Atmospheric nitrogen fixation
The nitrogen fixation potential of the candidate bacterial strains was evaluated using Ashby's nitrogen-free agar medium. Bacterial cultures were inoculated onto the solidified medium and incubated at 28°C for 5 days. The presence of visible bacterial growth on the plates after incubation served as a positive indicator of the isolate's ability to fix atmospheric nitrogen (N2). This process converts N2 gas, which is unusable by most plants, into a bioavailable form (ammonium or nitrate) that can be readily assimilated by plants, potentially promoting their growth (Hajji-Hedfi et al., 2023).
2.9.5 Siderophore production
Siderophore production was assessed using chrome azurol S (CAS) agar medium. Siderophores are iron-chelating compounds secreted by some bacteria. In CAS agar, these siderophores compete with the medium's chelator for iron, resulting in the formation of a yellow-orange halo zone surrounding colonies of siderophore-producing bacteria. The presence of such a halo zone served as a positive indicator for siderophore production by the bacterial strain (Milagres et al., 1999).
2.9.6 HCN production assay
Hydrogen cyanide (HCN) production by candidate bacterial strains was assessed using the method described by Abd Alhakim et al. (2022). Bacterial cultures were inoculated onto a solidified nutrient agar medium supplemented with glycine (4.4 g L−1). A Whatman number 1 filter paper, saturated with a solution of 0.5% picric acid and 2% sodium carbonate (w/v), was placed on top of the inoculated plates. The plates were then incubated, and the color change of the filter paper was monitored. A shift in color from yellow to light brown or reddish-brown indicated a positive test for HCN production. This color change signifies the reaction between HCN gas, potentially produced by the bacteria, and the picric acid in the filter paper (Hajji-Hedfi et al., 2023).
2.9.7 Extracellular enzyme production
The ability of bacterial isolates to produce hydrolytic enzymes was screened on basal medium supplemented with (1% w/v) different sole carbon sources (carboxymethyl cellulose [CMC], laminarin, starch, pectin of citrus peel, tween-80 as well as colloidal chitin and skimmed milk for the testing of the production of cellulase, β-1,3-glucanase, amylase, pectinase, lipase, chitinase, and proteases, respectively) (Hankin and Anagnostakis, 1975). To detect cellulase producing colonies, they were flooded with 1% Congo red solution and washed with distilled water (Méndez-Santiag et al., 2021), while amylase and pectinase activity plates were flooded with 1% iodine solution. The clear zones around colonies indicated positive enzyme activity (Hankin and Anagnostakis, 1975; Faramarzi et al., 2009).
2.10 Molecular identification of potent bacterial isolates
In this study, four isolates were identified according to the high ability inhibiting B. cinerea, as follows:
2.10.1 PCR reaction
The DNA extraction for four bacteria isolates was following the method of Griffiths et al. (2000) and Farkas et al. (2017). Polymerase chain reaction of the bacterial by amplification of 16S rRNA gene was performed using universal primers: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1525R (5′AAGGAGGTGWTCCARCC-3′) (Lane, 1991), and following the protocol of MyTaq™ Red Mix (Bioline) including 3 min initial denaturation at 95°C, followed by 35 cycles of 15 s denaturation at 95°C, 15 s annealing at 56°C, and 1 min extension at 72°C. Then, sequencing of PCR products was assumed by using 1st BASE DNA sequencing service (https://base-asia.com/services/sanger-sequencing-services/).
2.10.2 Sequence alignment and phylogenetic analysis
The sequence data were edited and quality checked by analyzing the chromatogram peaks with BioEdit 7.2.5 software (Hall, 1999). Blast was used to check the identity and similarity of the resulting sequences for the expected product on both the EzBioCloud server (Kim et al., 2012) and the NCBI database (Madden et al., 1996). The 16S rRNA gene sequences of each strain (H3Rh1, SRh2, ZRh5, and GRh5) were aligned and compared to reference sequences using the MEGA V.7 software (Kumar et al., 2016). Phylogenetic tree was constructed using the maximum likelihood (ML) method. The optimal model for ML was first determined using IQ-TREE v1.4.4 (Nguyen et al., 2015), followed by 1,000 bootstrap replicates (Minh et al., 2013). Phylogenetic trees were created and visualized with FigTree v1.4.4. All generated nucleotide sequences were deposited in GenBank with accession numbers PQ596295, PQ596293, PQ631724, and PQ631348.
2.11 Data analysis and statistics
The data were explored for normality by checking normal distribution and the Shapiro–Wilk test. Principal component analysis (PCA) was carried out to analyze the correlation between the physicochemical properties and bacterial density data. Data were analyzed using the ANOVA and Duncan tests for intergroup comparisons (p < 0.05). All figures were generated in this study using the ggplot2 package in RStudio software (Version 2024.12.0+467).
3 Results
3.1 Soil characterization
Across all studied sites, a variation in soil physicochemical properties showed in the different investigated sites, except for the electrical conductivity (EC) (Supplementary Tables S2, S3). Soil pH ranged from 6.78 to 8.07. There were substantial differences in water content (WC), where S15 recorded the highest content with 23.89% and S22 showed the lowest content with 6.05%.
All soil samples showed a low content of organic soil matter (OM) and organic carbon (OC), S15 showed the highest value with 1.59%, while S21 recorded the lowest value with 0.44%. Regarding the nitrate content (NO3), S5 recorded the highest value with 36.73 ppm, while S26 and S18 showed the lowest content with 4.89 and 4.63 ppm, respectively. Sodium (Na) concentrations varied considerably, with S30 having the highest (57.25 ppm) and S12 having the lowest (24.09 ppm). S24 and S30 had exceptionally high K2O levels with values of 1,109.67 and 960 ppm, respectively. Regarding the P2O5 values, S25 and S27 exhibited the highest levels with 15.48 and 11.53 ppm, respectively. The total limestone values ranged between 2.47 and 2.95. The analysis of sand, clay, and silt ratios shows that the soils were mainly coarse, with a sandy composition; S17 showed the highest levels of TN with 0.663%, and S27 and S28 showed the lowest values with 0.065% for each. Sites S16 and S29 showed the highest C/N ratio values with 11.51 and 8.35, respectively (Supplementary Table S2).
3.2 Collection of bacterial isolates
The analysis focused on quantifying bacterial populations using three different media types: nutrient agar, Kings B, and ISP2. The bacterial populations exhibited significant variations (p < 0.05) across the sites, and a total of 107 bacterial isolates were obtained. Some sites, such as S4, S15, S16, and S20, displayed high bacterial counts on all media, suggesting a thriving microbial community (5.67 × 105 – 4.03 × 105 CFU/g for Nutrient Agar; 2.84 × 105 – 2.62 × 105 CFU/g for King's Medium B; 2.42 × 105 – 1.22 × 105 CFU/g for ISP2 Medium). However, sites S21, S22 S23, S24, and S30 had consistently low bacterial counts (<1 × 105 CFU/g) across all media, indicating a less diverse and abundant microbial population (Supplementary Table S3). Statistically significant correlations were observed between the physicochemical characters of the soil and the total bacterial counts. The water content, organic matter content, and clay content showed a positive correlation with the total bacterial count from the three selected media types (CFU_Na, CFU_KB, and CFU_ISP2) with values of r = 0.592, 0.669, 0.725, 0.654, 0.705, 0.808, 0.508, 0.522, and 0.683, respectively (Table 1).
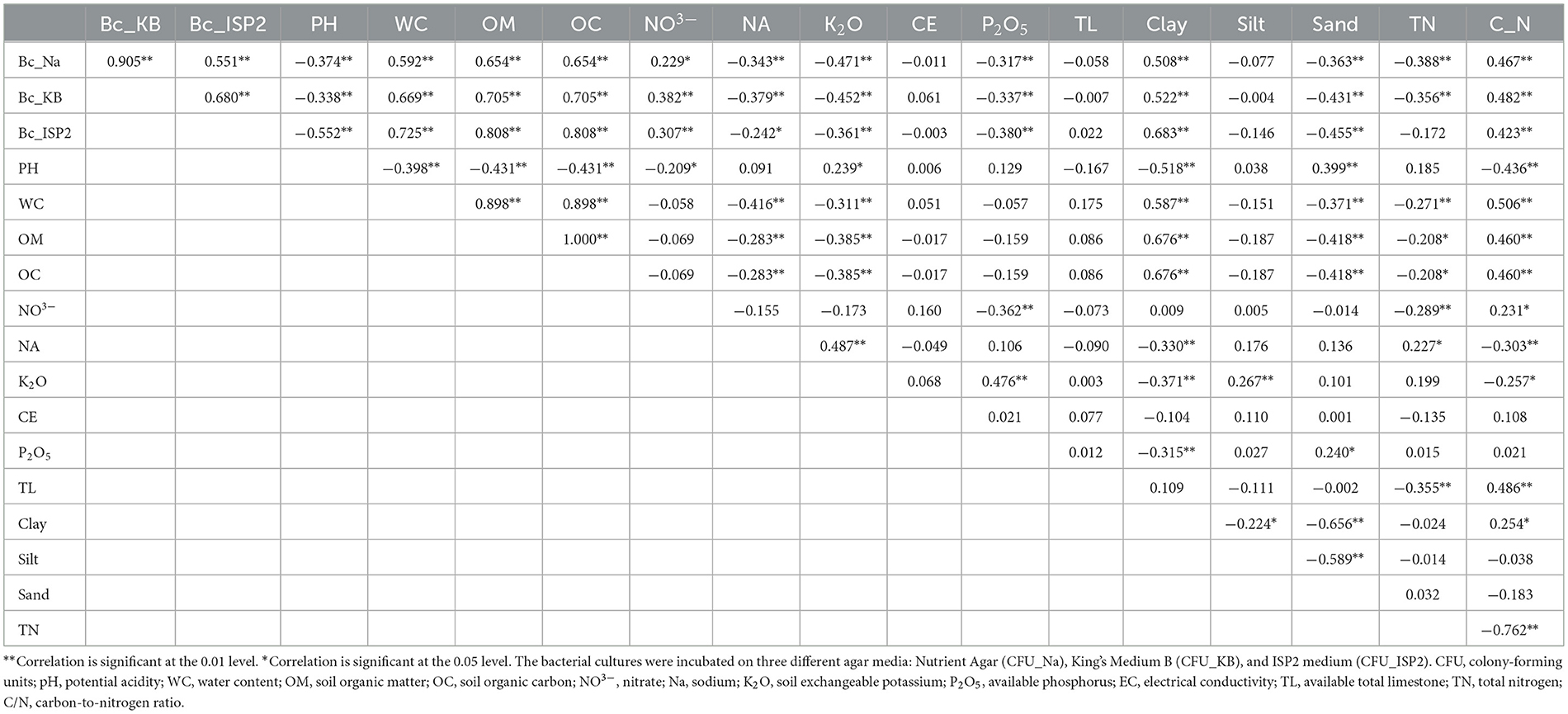
Table 1. Pearson correlation coefficients between pH, WC, OM, OC, NO3−, Na, K2O, CE, P2O5, TL, percentage of clay, percentage of silt, percentage of sand, TN, C/N, and bacterial density in 32 vineyard sites.
Principal component analysis (PCA) was employed to investigate the relationships between soil physicochemical properties collected from different environments and bacterial communities' growth measured on three distinct culturing media (CFU_Na, CFU_KB, and CFU_ISP2). The first (CCA1 = 48.933) and second (CCA 2 = 12.095) principal components explain 60.028% of the variance. These principal components represent underlying factors that capture the most significant variation within the data set of soil properties (Figure 1)
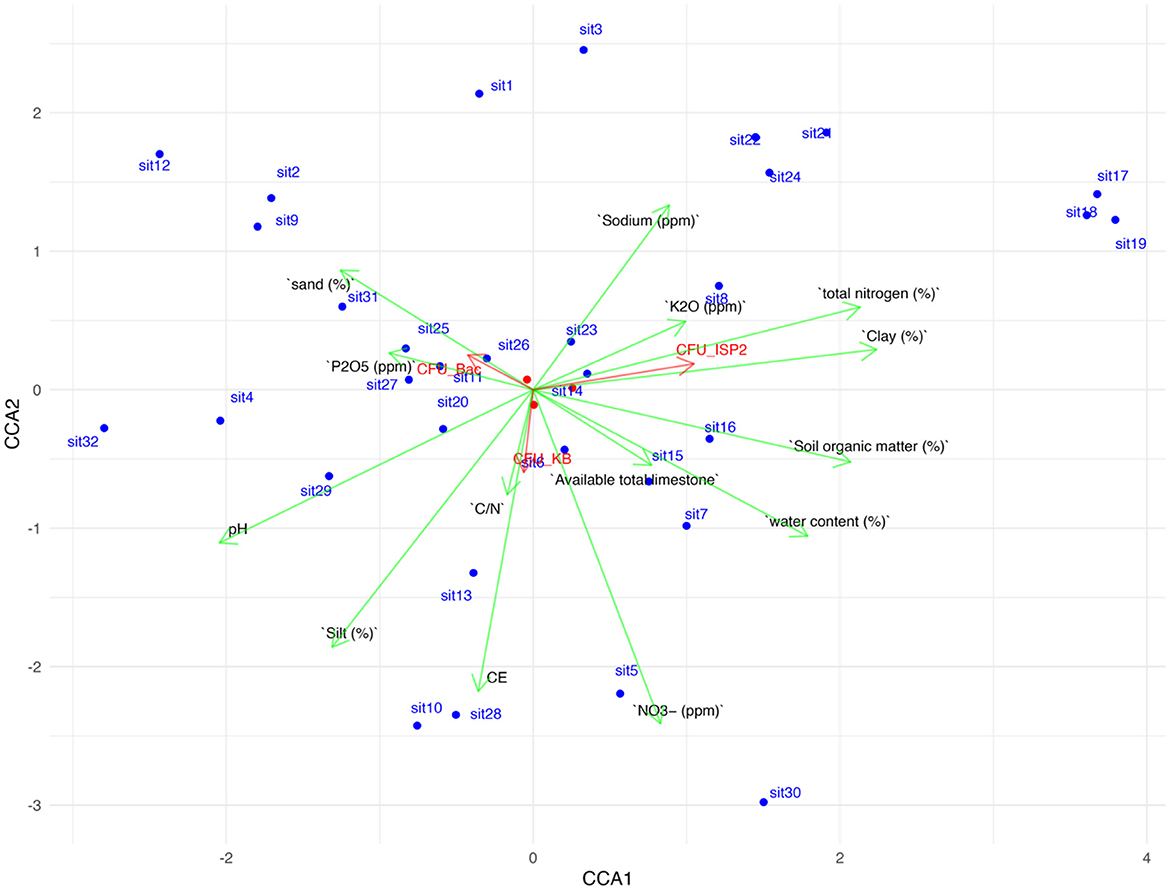
Figure 1. Principle component analysis (CCA) of the first three principal components, depicting relationship among soil physicochemical properties (EC, electrical conductivity; BD, bulk density; OC, organic carbon; OM, organic matter; TN, total nitrogen; C/N, carbon/total nitrate ratio; TL, total limestone; P2O5, available phosphorus; K2O, exchangeable potassium; sand; clay; and silt), soil bacterial grow on three media (CFU_Na, CFU_KB, and CFU_ISP2). The first (CCA1 = 48.933) and second (CCA 2 = 12.095) principal components explain 60.028% of the variance.
3.3 Identification and pathogenicity assessment of Botrytis cinerea
B. cinerea colonies produced cottony, gray-white aerial mycelia that turned gray and formed conidia after 7 days of incubation, followed by the development of black sclerotia after 14 days on PDA. Sclerotia measured 1.1 to 4.4 mm in length and 1.0 to 3.6 mm in width (average = 3.45 by 2.4 mm). Conidiophores were erect, solitary, or clustered, with brown-branched tops and lengths and widths ranging from 500 to 2,210 by 6 to 17 mm (average: 715 by 13 mm). The conidia were unicellular, hyaline to light brown in color, and ellipsoid to ovoid in shape, measuring 6.65 to 13.42 mm long and 3.99 to 6.19 mm wide (average = 8.17 by 5.79 mm) (Supplementary Figure 1). Recovered taxon was deposited at the Fungarium of Suez Canal University (https://ccinfo.wdcm.org/details?wdcmnumber=1180) under the accession numbers SFUF00001882.
3.4 Phytopathogenicity potential of bacterial isolates
Hypersensitivity reaction (HR) assay revealed 10 of the total bacterial isolates caused tissue collapse within 24 h of inoculation, indicating a positive HR. These bacterial isolates with phytopathogenic effect were excluded, and the remained 97 bacterial isolates with a negative HR were maintained for antagonistic activity assay (Supplementary Table S4).
3.5 In vitro antagonistic activity
Among the screened bacterial isolates, 16 showed more than 50% radial growth inhibition, indicating moderate-to-strong antagonistic activity against B. cinerea (Figure 2). In contrast, 58 isolates exhibited low inhibitory activity, ranging from 0.11% to 9.62%. In addition, 45 isolates demonstrated moderate inhibition, with values ranging from 11.58% to 85%, which indicates more than 50% of screened bacteria in soil having a low ability in inhibiting some important plant pathogens such as B. cinerea. Consequently, four bacteria isolates, namely, H3Rh1, ZRh5, GRh5, and SRh2, showed high in vitro inhibitory activity against B. cinerea, with inhibition rates of 91.11%, 91.11%, 92.22%, and 99.3%, respectively (Figure 3).
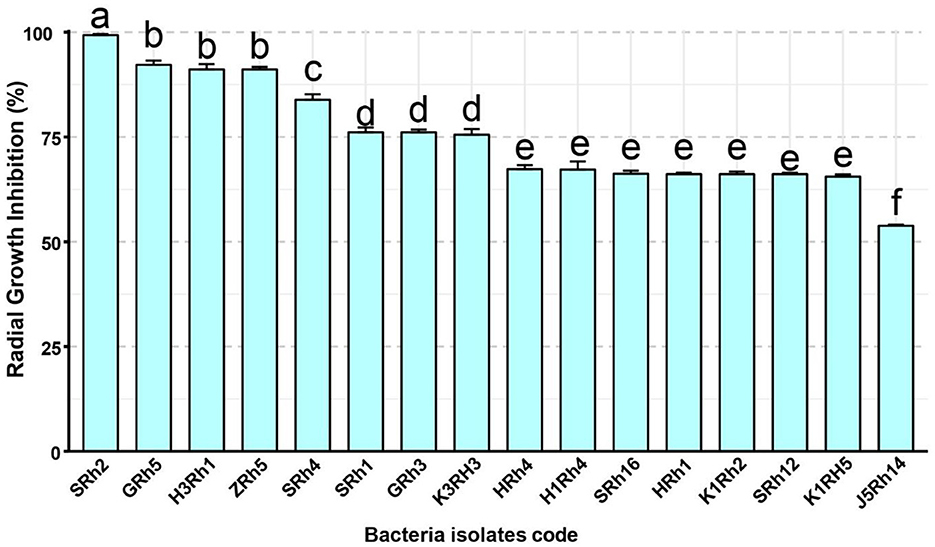
Figure 2. Antifungal activity of bacterial isolates against the radical growth of the gray mold Botrytis cinerea (%; Mean ± SE). Different letters indicate statistically significant differences (P ≤ 0.05).
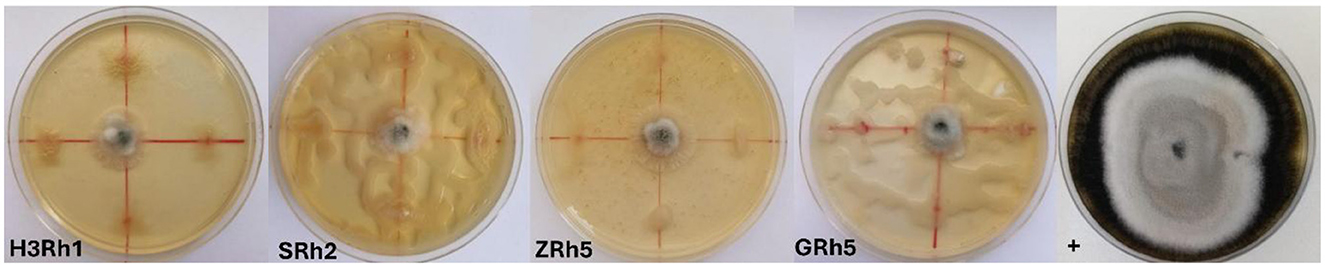
Figure 3. In vitro direct confrontation effect of bacterial isolates H3Rh1, SRh2, ZRh5, and GRh5 against Botrytis cinerea, in 7 days of incubation, +, positive control.
3.6 Detached leaf assay
The 16 bacterial isolates were evaluated for their potential in controlling B. cinerea using a detached leaf assay. Four bacteria isolates (H3Rh1, ZRh5, GRh5, and SRh2) showed high significant (p ≤ 0.05) in reduction of the disease development after 14 dpi. These results suggest the high efficacy of these specific bacteria isolates in controlling B. cinerea colonization. Reduction for mold growth (PLA) by H3Rh1, ZRh5, GRh5, and SRh2 was significant at 7.29, 5.96, 6.64, and 2.86, respectively, compared to the control (Figure 4). In addition, these bacterial isolates showed a high reduction in disease severity index (DSI) of 3.85, 1.99, 2.11, and 1.49%, respectively. These isolates were categorized as highly effective (HE), achieving remarkably low DSI values (Table 2). Therefore, it was suggested that these isolates highly effectively suppressed fungal growth on grapevine leaves.
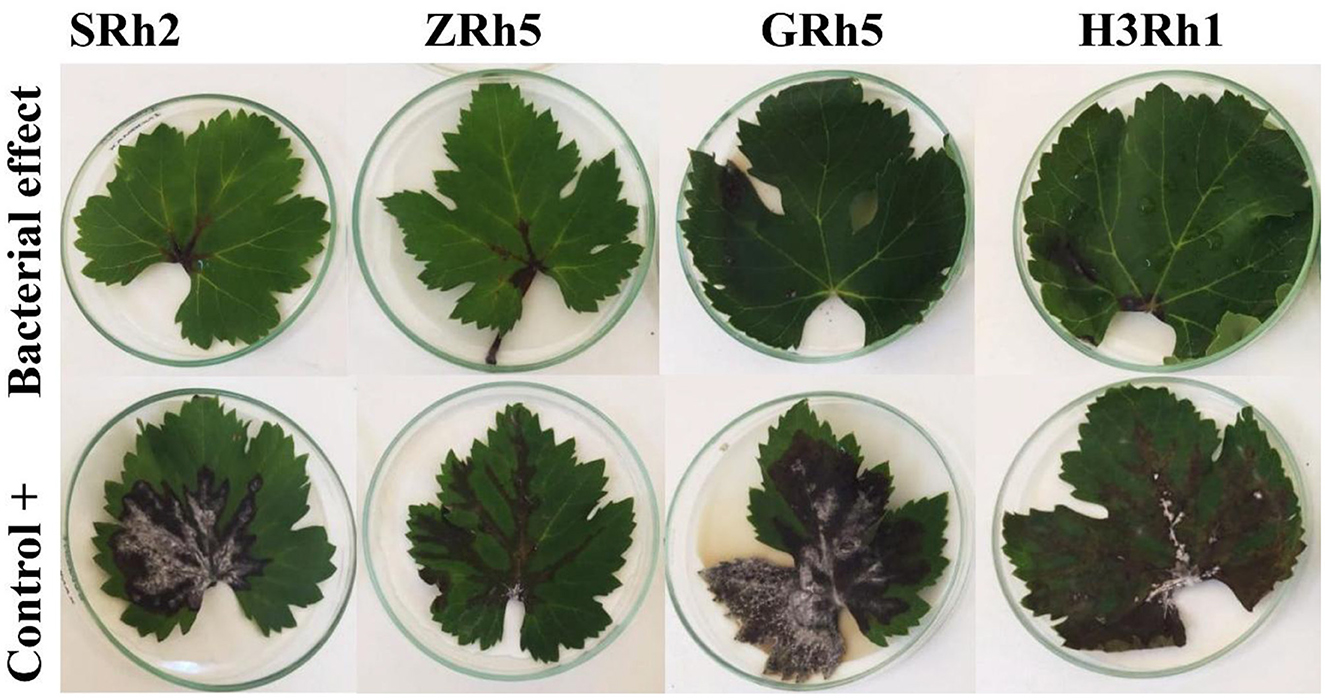
Figure 4. Impact of bacterial strains SRh2, ZRh5, GRh5, and H3Rh1 on detached leaf assay: inhibition of Botrytis cinerea infection compared to positive control.
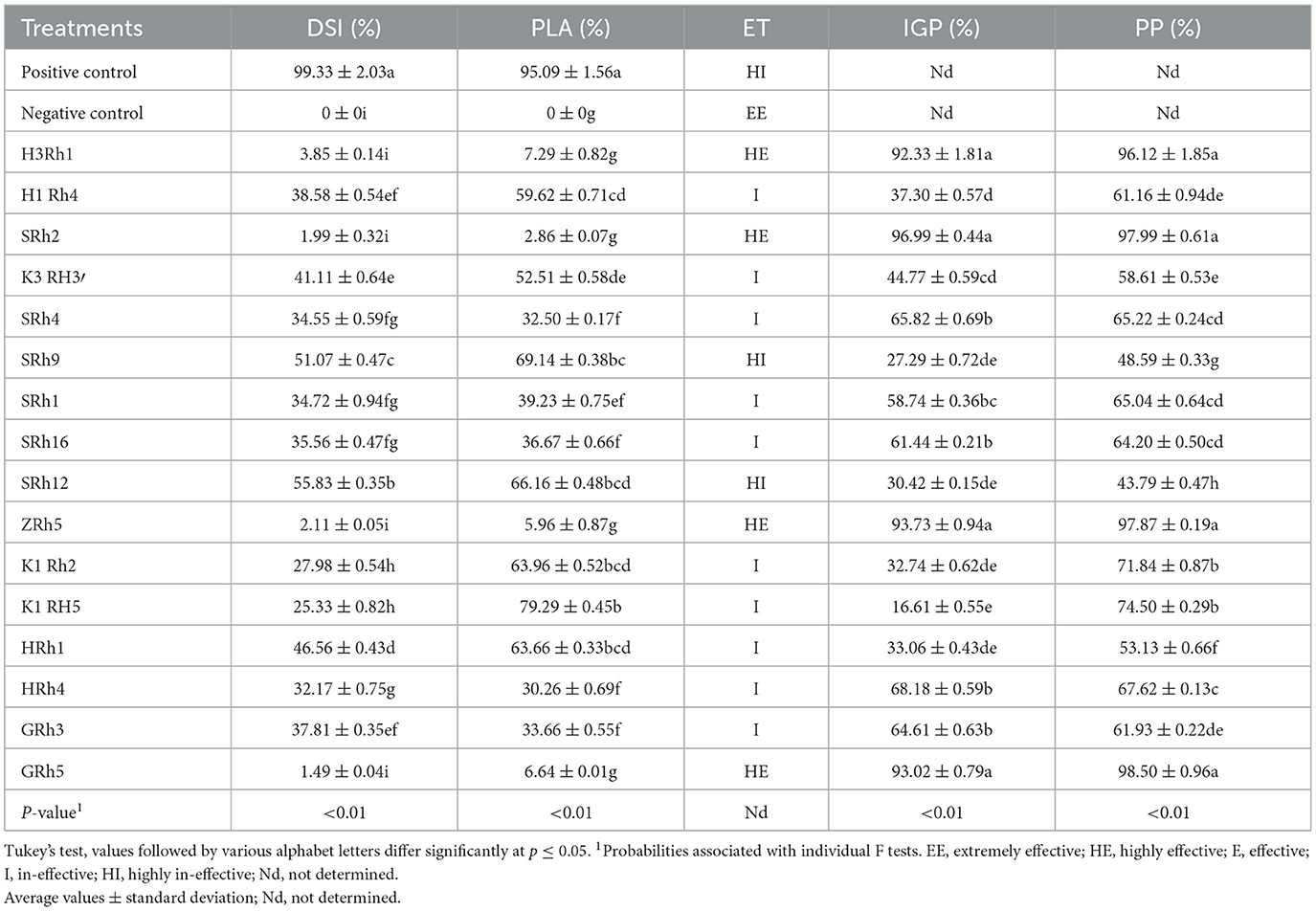
Table 2. Effect of bacterial isolates against Botrytis cinerea on the relative leaf area covered by gray mold (PLA), disease severity index (DSI), the protective potential of treatments (ET), inhibitory growth potential (IGP), and protective potential (PP) after 14 days of post-inoculation.
The results pointed in Table 2 revealed that the inhibitory growth potential was found to be highly effective in isolates H3Rh1, ZRh5, GRh5, and SRh2, with inhibitory rates of 92.33%, 93.73%, 93.02%, and 96.99%, respectively. In addition, these bacterial isolates also showed significantly high protective potential (PP) values of 96.12%, 97.87%, 98.50%, and 97.99%, respectively. Their overall disease protection exceeded 96%, indicating excellent control against gray mold caused by B. cinerea. In contrast, most of the tested bacterial strains were categorized as ineffective (I), with disease severity index (DSI) values ranging from 25.33% to 51.07%, indicating minimal to no protection against B. cinerea (Table 2). The SRh12 isolate showed a relatively low DSI of 55.83%, suggesting limited effectiveness. The K1Rh5 isolate exhibited the lowest protective leaf area (PLA) effectiveness at 79.29%, and it also showed very low IGP activity at 16.61%. Furthermore, the SRh9 isolate had low protective potential (PP) at 48.59%. Both SRh12 and SRh9 were classified as highly ineffective (HI), with DSI values exceeding 50%, reflecting minimal direct antifungal activity and limited disease suppression (Table 2).
3.7 In vitro plant growth-promoting activities
The analysis of the remaining 97 non-phytopathogenic bacterial isolates revealed varying positive results in the screening of plant growth-promoting (PGP) traits (Supplementary Table S5). The 16 bacterial isolates displayed a wide variety of characteristics that promote plant growth. All isolates were able to fix nitrogen, produce indole acetic acid (IAA), solubilize potassium and phosphate, and synthesize hydrogen cyanide (HCN) and siderophores. These isolates also showed a range of enzyme activities, such as lipase, chitinase, β-1,3-glucanase, amylase, pectinase, cellulase, and catalase, which are essential for nutrition cycling and pathogen suppression (Figure 5). Bacterial isolates H3Rh1, SRh2, and GRh5 showed positive responses for all investigated attributes, indicating their strong potential for use in biocontrol and plant growth promotion (Figure 6). Although ZRh5 exhibited significant PGP and enzymatic properties as well, it was unable to produce β-1,3-glucanase, cellulase, or chitinase (Figure 7).
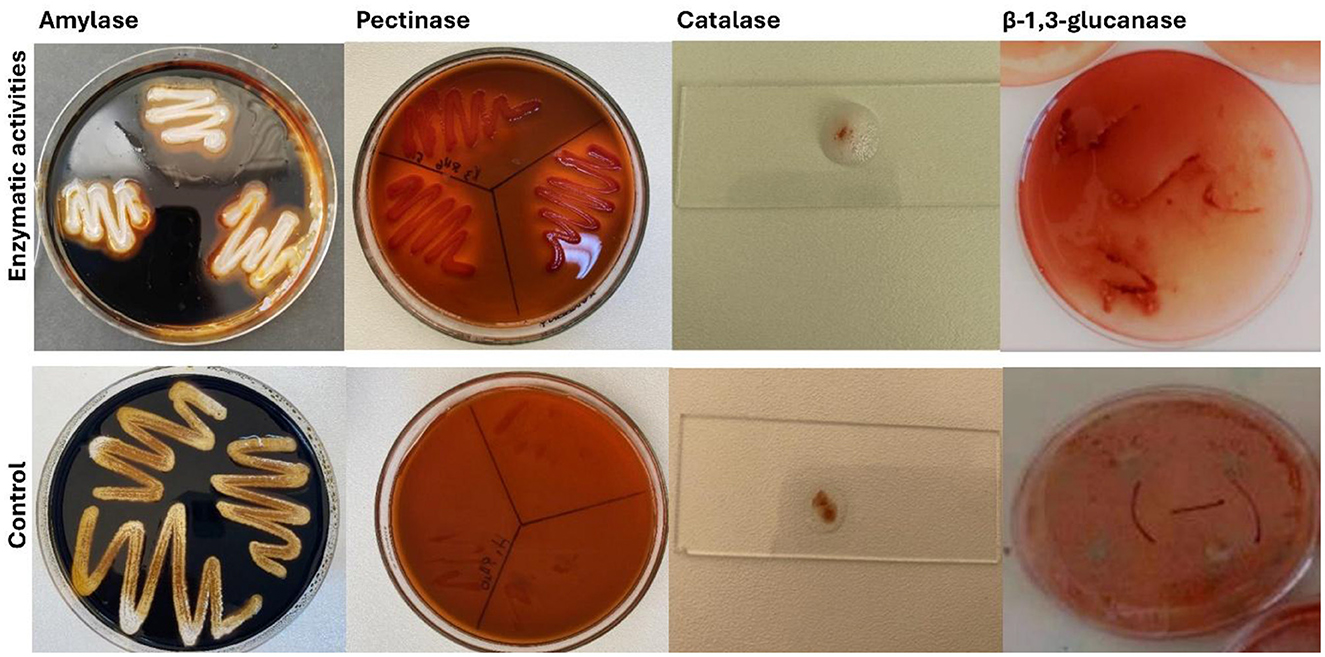
Figure 5. Representative positive extracellular enzymatic activities (amylase, pectinase, catalase, and β-1,3-glucanase) detected among the bacterial isolates (H3Rh1, ZRh5, GRh5, and SRh2) and control.
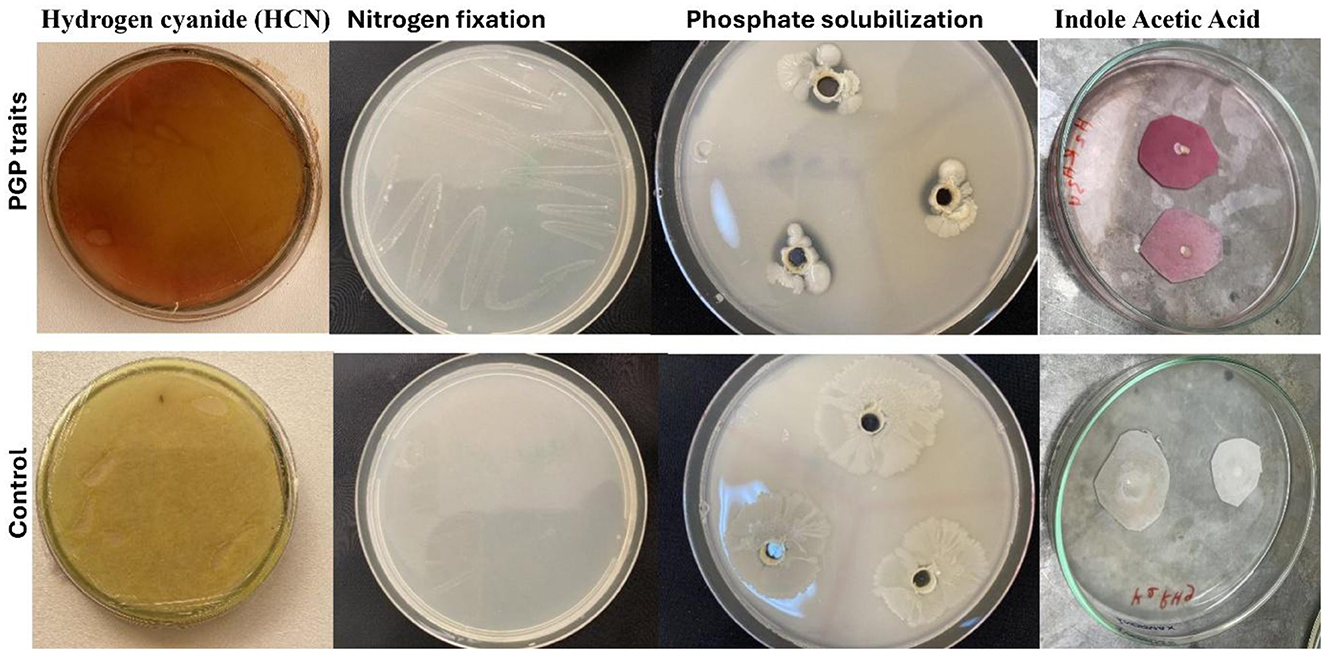
Figure 6. Plant growth-promoting (PGP) activities (HCN production, nitrogen fixation, phosphate solubilization, and indole acetic acid production) observed in bacterial isolates (H3Rh1, ZRh5, GRh5, and SRh2) and control.
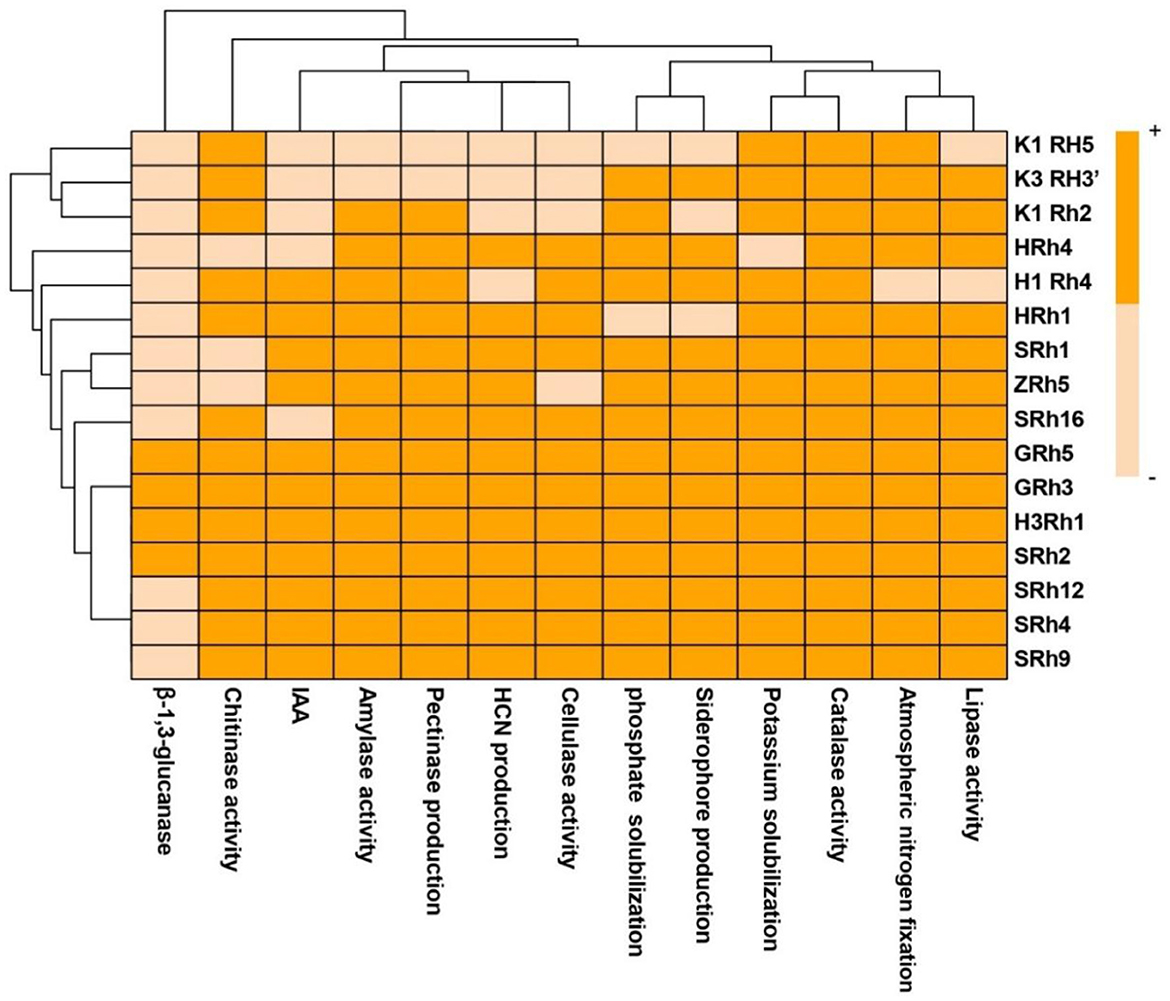
Figure 7. Heatmap of plant growth-promoting traits and extracellular enzyme of 16 bacterial isolates.
3.8 Preservation of fungal and bacterial taxa
Recovered taxa of Botrytis cinerea, Arthrobacter globiformis, Priestia megaterium, Bacillus cabrialesii, and Bacillus mojavensis were deposited in the Fungarium of Suez Canal University (https://ccinfo.wdcm.org/collection/by_id/1180), at Botany and Microbiology Department, Faculty of Science, Ismailia 41,522, Egypt, under the accession numbers from SCUF0000311 to SCUF0000315, respectively. The sequences of the identified bacterial isolate of A. globiformis, P. megaterium, B. cabrialesii, and B. mojavensis have been submitted to GenBank under accession numbers PQ596293, PQ596295, PQ631724, and PQ631348, respectively.
3.9 Molecular identification
The 16S rRNA gene of the four bacterial strains, namely, A. globiformis (H3Rh1), P. megaterium (SRh2), B. cabrialesii (ZRh5), and B. mojavensis (GRh5), was compared with reference sequences that are known by using BLAST in the GenBank database (NCBI) and is available at EzBioCloud server. Analysis of phylogeny tree showed that the strain H3Rh1 was clustered in Arthrobacter genus, the strain SRh2 clustered in Priestia genus, while both ZRh5 and GRh5 strains were grouped within Bacillus genus (Figure 8).
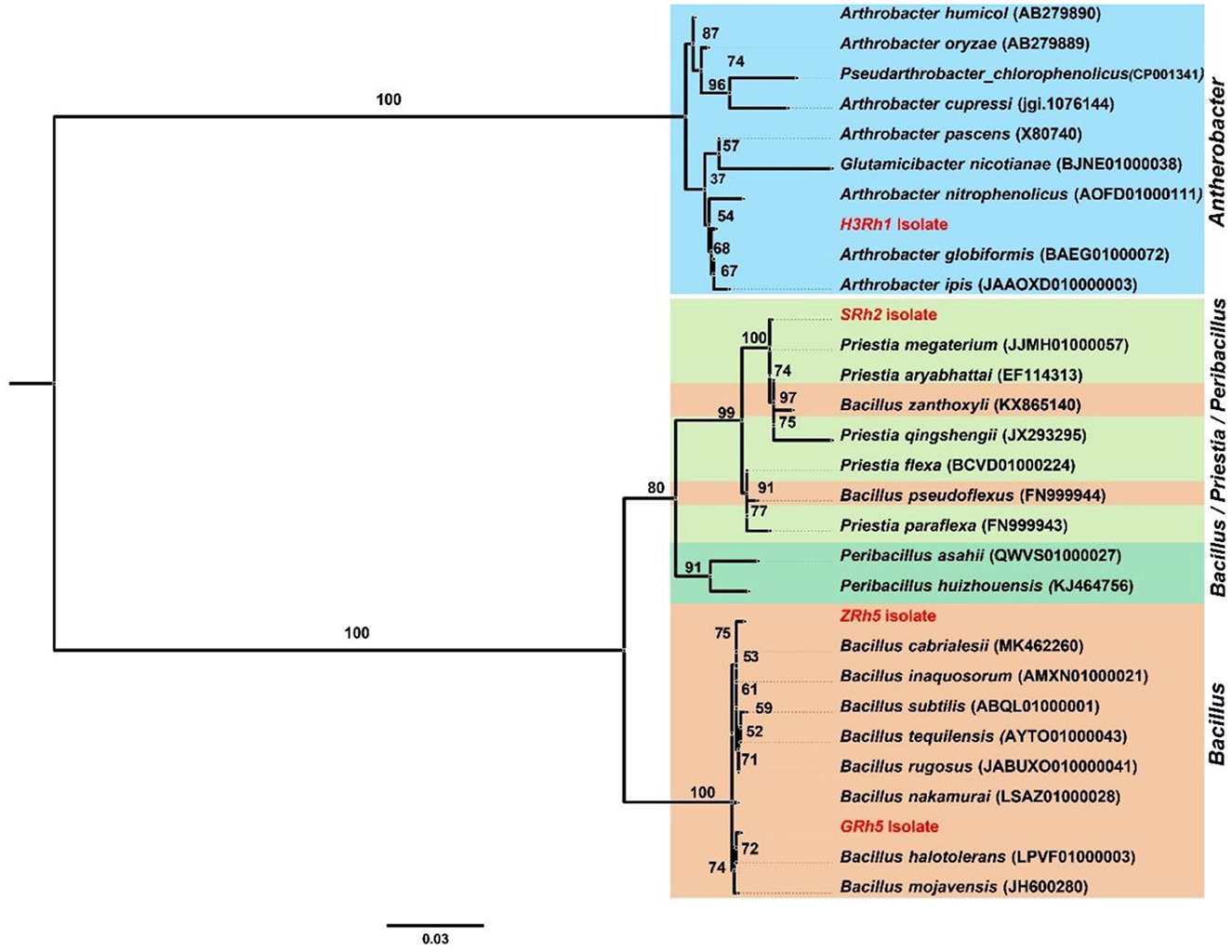
Figure 8. Maximum likelihood (ML) tree based on 16s rRNA gene sequences of four isolates (H3Rh1, SRh2, ZRh5, and GRh5). The percentage of replicate trees in which the associated species clustered together in the bootstrap test (1,000 replicates) is shown next to each branch. Identified isolates are indicated in red color.
Both server EzBioCloud and NCBI provided a high similarity percentage of 99% with known species of bacteria (Table 3). The right sentence is 'The H3Rh1 isolate identified as Arthrobacter globiformis species from both servers with a sequence length of 1180 bp and 99.49% similarity based to EzbioCloud, and 99.16% to NCBI respectively. Indeed, the highest similarity was provided via EzBioCloud (99.66%) for SRh2 isolate, to be identified as Priestia megaterium with a sequence length of 1,180 bp, as well as NCBI indicated the same species with similarity of 99.41%. Bacillus cabrialesii was identified as the most closest species matched for isolate ZRh5, with a sequence length of 1,115 bp and similarities of 99.66% and 99.41% based on EzBioCloud and NCBI, respectively. Based on EzBioCloud server, the isolate GRh5 was the closest match for Bacillus mojavensis with a sequence length of 1,144 bp and similarities of 99.74% and 99.30% based on EzBioCloud and NCBI, respectively. Sequences obtained were deposited in the GenBank database and registered with accession numbers PQ596293.1, PQ596295.1, PQ631724.1, and PQ631348.1, respectively.
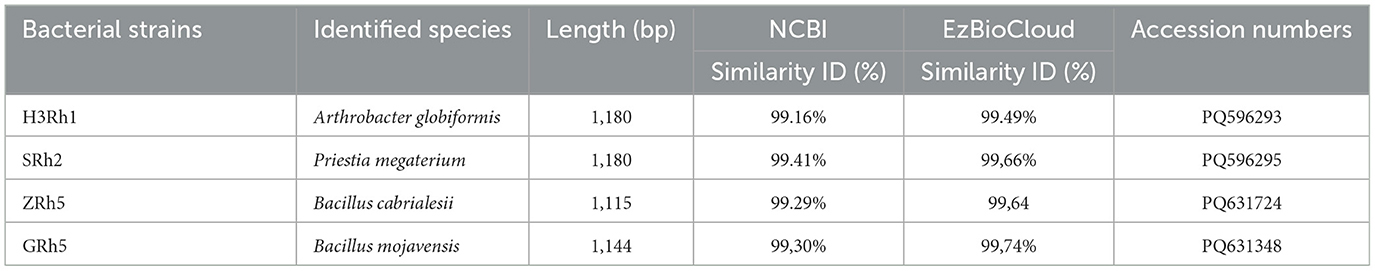
Table 3. Molecular identification of four bacterial isolates from the rhizosphere of Vitis vinifera.
4 Discussion
This study successfully isolated and characterized beneficial rhizospheric bacteria from grapevine soils in Tunisia, with a focus on their plant growth-promoting (PGP) traits and antagonistic activity against Botrytis cinerea. Soil analysis revealed heterogeneity in key properties such as organic matter content, pH, and nutrient concentrations, which are known to shape microbial community structure and function (Trivedi et al., 2013; Sun and Badgley, 2019; Wang et al., 2019). These findings align with previous research highlighting the strong influence of edaphic factors and plant genotype on rhizospheric microbial assemblages (Strano et al., 2018; Wu et al., 2019). Isolation on nutrient-specific media (NA, KB, ISP2) enabled the recovery of 107 diverse bacterial strains, with Gram-positive isolates predominating. Conducting soil analysis is necessary for determining the relationship between bacteria diversity and soil characterization. Microbial communities within soil play a critical role in maintaining healthy ecosystems. Recognizing the intricate link between soil microbial communities and the physical and chemical properties of the soil is increasingly acknowledged as crucial for long-term sustainability. In addition, special media such as NA, KB, and ISP2 medium were used for the isolation of beneficial bacteria. The isolates that showed the diversity in form and color, size, arrangement, and margin of colony were selected. The plant root exudates have the potential to change the soil microbiome (such as bacteria) and the soil environment. Plants can alter the composition of soil microbiota rhizosphere (Hu et al., 2018). The results suggest the difference in ratio of Gram-positive (GP): Gram-negative (GN) returns to the activity of grapevine root exudates. The root exudates attract beneficial bacteria to colonize the soil area around the root and the rhizosphere. There is a relationship between bacteria diversity and activity with grapevine root rhizosphere (Darriaut et al., 2022). The plant has the potential to recruit beneficial bacteria, either symbiotic or non-symbiotic, to confront the stress abiotic and biotic rhizosphere (Arif et al., 2020).
Furthermore, some of the bacterial isolates in this study exhibited the plant-growth promoting (PGP) traits including IAA, phosphate solubilization, dissolving potassium, nitrogen fixation, and siderophore, as well as HCN, and the production of different enzymes involved during biocontrol activity such as cellulase, chitinase, β-1,3-glucanase, amylase, pectinase, lipase, and catalase. It was found that all bacterial GP isolates could produce IAA (Mohite, 2013). Some studies mentioned previously GN producing IAA (Datta and Basu, 2000). The bacterial isolates can improve plant growth by fixing nitrogen, solubilizing phosphate, and dissolving potassium (Almuhawish et al., 2024). The production of siderophores and hydrogen cyanide (HCN) by both gram negative and positive bacteria is characteristic to suppress plant diseases (Sehrawat et al., 2022). Moreover, the GP bacteria can degrade complex organic compounds by producing extracellular enzymes like β-1,3-glucanase, Catalase, cellulase, lipase, amylase, pectinase, and chitinase (Tan et al., 2021).
Taking into consideration the high percentage of gram-positive bacteria isolates, they exhibit abilities in producing IAA, HCN, β-1,3-glucanase, catalase, and hydrolytic enzymes, as well as in phosphate solubilization, and potassium dissolving, compared to lower percentages among Gram-negative (GN) isolates. However, nitrogen-fixing ability was more commonly observed among Gram-negative (GN) bacterial isolates compared to Gram-positive (GP) isolates. In contrast, only four GP bacterial isolates demonstrated the ability to produce β-1,3-glucanase. These four isolates can produce all enzymes, PGP traits, siderophores, and HCN that were tested in our study.
Indeed, the differences in the abilities of bacterial isolates are probably attributed to genetics and microenvironment characteristics. The capabilities of bacteria can differ according to kind of strain or species, conditions of growth, competitiveness, water deficiency, survival, and available and type of substrate (Hartmann et al., 2017). The higher number of gram-positive bacteria in isolation compared to gram negative bacteria may be returned to the higher ability to survive and higher activity of enzymes that was confirmed through the results. Gram-positive bacteria can survive more from Gram-negative bacteria due to their robust metabolic activities, including the production of extracellular enzyme that is useful to utilize the recalcitrant compounds (Naylor and Coleman-Derr, 2018; Tocheva et al., 2016). Furthermore, this group of bacteria can have thick and strong interlinked peptidoglycan cell walls, compared with gram negative bacteria that lost these characteristics (Schimel et al., 2017), as well positive bacteria showed to be more effective than Gram-negative species in various mechanisms, including biocontrol and stress tolerance (Soylu et al., 2021). However, the determination of pathogenicity is found in Gram-positive bacterial isolates on tobacco (Nicotiana tabacum) and geraniums (Pelargonium hederaefolium) leaves, while no symptoms were observed from Gram-negative bacterial isolates. The plant bacterial pathogen can cause bacterial blight and bacterial leaf spot diseases (Francis et al., 2010; Soylu et al., 2010). According to Nasfi et al. (2018), most bacterial pathogens for plants belong to the group of Gram-positive.
Remarkably, four isolates from 107 isolates bacteria showed a high antagonistic activity against B. cinerea in vitro and on the leaf. The efficacy of these four isolates in high inhibition for B. cinerea is attributed to their antifungal properties. The strong inhibitory effect of these four isolates against B. cinerea suggests their potential to secrete antifungal compounds. Based on 16S rRNA sequence analysis, the antagonistic isolates were identified as Arthrobacter globiformis, Priestia megaterium, Bacillus cabrialesii, and Bacillus mojavensis. They reported that a greater number of prominent families were found among soil bacteria (86%) (Biedendieck et al., 2021). In this study, identified bacterial isolate ZRh5 has not previously been reported in Tunisian soils, making this an important addition to the regional microbial repertory, while the bacterial isolate SRh2 was identified as Priestia megaterium in the present study but previously was called Bacillus megaterium (Biedendieck et al., 2021).
Thus, A. globiformis, P. megaterium, and B. mojavensis could produce important enzymes such as chitinase and β-1,3-glucanase along with HCN and siderophores, which may inhibit plant fungal pathogens. In addition, B. cabrialesii was highly antagonistic by producing HCN and siderophores. Previously, these two enzymes and compounds were identified as factors influencing plant pathogens (Naylor and Coleman-Derr, 2018; Tocheva et al., 2016; Schimel et al., 2017). Strains such as A. globiformis, P. megaterium, and various Bacillus species exhibited strong antagonistic activity against several plant pathogens through the production of antifungal compounds, induction of systemic resistance, and promotion of plant growth (Naylor and Coleman-Derr, 2018; Soylu et al., 2010; Francis et al., 2010; Pylak et al., 2020; Zhou et al., 2021). Diabankana et al. (2021) noted the high efficiency of beneficial bacterial in suppressing the growth of several plant fungal pathogens, such as Fusarium oxysporum, F. graminearum, Fusarium chlamydosporum, Ascochyta pisi, Verticillium dahliae, Sclerotinia sclerotiorum, Epicoccum nigrum, and Alternaria alternate. This is due to the secretion of some important hydrolytic enzymes, including chitinase, β-glucanase, protease, lipase, and cellulase. However, B. cabrialesii showed high suppress for growth of fungal pathogens without secreting hydrolytic enzymes such as chitinase, β-glucanase, and cellulase. B. cabrialesii (ZRH5) is possibly producing some secondary metabolites, volatile and non-volatile compounds, which inhibited growth of fungal pathogen by breaking fungal cell walls. In recent studies, the production of antifungal compounds such as fengycin H, surfactin, and rhizocticin A secreted by Bacillus cabrialesii TE3 was confirmed to suppress B. cinerea. In addition, antibacterial metabolites including bacillaene, bacilysin, bacillibactin, and subtilosin were also identified (Diabankana et al., 2021; Valenzuela Ruiz et al., 2022; Ma et al., 2022; Bhargavi et al., 2024; Rahman et al., 2023). Overall, the findings support the idea that grapevine-associated rhizospheric microbiomes harbor untapped microbial resources with significant biotechnological potential. The four strains identified in this study fulfill key criteria for biocontrol agents: safety (non-pathogenic), multiple PGP traits, strong antifungal effects, and genetic identification. These strains offer promising avenues for the development of environmentally friendly alternatives to synthetic fungicides, in line with integrated pest management strategies. Future work should focus on genome sequencing, formulation stability, field trials, and potential synergism with other biocontrol agents or agricultural inputs.
5 Conclusion
This study identified four native rhizospheric bacterial strains, namely, A. globiformis, P. megaterium, Bacillus cabrialesii, and B. mojavensis, with significant potential as plant growth-promoting and biocontrol agents against Botrytis cinerea, the causative agent of gray mold in grapevines. These isolates exhibited multiple beneficial traits, including nitrogen fixation, phosphate and potassium solubilization, enzyme production, and strong antifungal activity both in vitro and in planta. Their high efficacy is likely due to the combined effects of enzyme synergy, secondary metabolite production, siderophore-mediated competition, and possibly volatile compound emissions. Notably, the identification of B. cabrialesii in Tunisian grapevine soils is reported here for the first time. These findings provide a strong foundation for the development of sustainable microbial-based products aimed at enhancing viticulture health and productivity. Further studies should focus on genomic characterization, formulation, field evaluation, and the potential integration of these strains into holistic crop management strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LH-H: Validation, Conceptualization, Writing – original draft, Supervision. TW: Investigation, Writing – review & editing, Formal analysis, Data curation. LT: Writing – original draft, Data curation, Formal analysis. BB: Writing – review & editing, Formal analysis, Data curation. ST: Investigation, Writing – review & editing, Formal analysis. AM-H: Writing – review & editing, Visualization, Supervision, Formal analysis. WH: Writing – review & editing, Methodology. AA: Visualization, Writing – review & editing, Validation. NR: Funding acquisition, Writing – review & editing, Validation, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The financial support has been provided by REVINE project within the framework of PRIMA, a program supported by H2020, the European Programme for Research and Innovation and the Tunisian Ministry of Higher Education and Scientific Research (MERS). In addition, the RUDN University Strategic Academic Leadership Program has supported this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2025.1619801/full#supplementary-material
References
Abd Alhakim, A., Hashem, A., Abdelaziz, A. M., and Attia, M. S. (2022). Impact of plant growth promoting fungi on biochemical defense performance of tomato under fusarial infection. Egyptian J. Chem. 65, 291–301. doi: 10.21608/ejchem.2022.124008.5532
Abdel-Azeem, A. M., and Salem, F. M. (2012). Biodiversity of laccase producing fungi in Egypt. Mycosphere 3, 900–920. doi: 10.5943/mycosphere/3/6/4
Al-Ani, L. K. T., Franzino, T., Aguilar-Marcelino, L., Haichar, F. Z., Furtado, E. L., Razaf, W., et al. (2020). “The role of microbial signals in plant growth and development: current status and future prospects,” in New and future developments in microbial biotechnology and bioengineering, eds. A. A. Rastegari, A. N. Yadav, A. K. Awasthi, N. Yadav (Amsterdam: Elsevier), 225–42. doi: 10.1016/B978-0-12-820526-6.00015-4
Almuhawish, M. A., Kotb, E., Alkhaldi, E., and Ahmed, A. A. (2024). Production and antibacterial activity of atypical siderophore from Pseudomonas sp. QCS59 recovered from Harpachene schimperi. Pharmaceuticals (Basel) 17:1126. doi: 10.3390/ph17091126
Amarouchi, Z., Esmaeel, Q., Sanchez, L., Jacquard, C., Hafidi, M., Vaillant-Gaveau, N., et al. (2021). Beneficial microorganisms to control the gray mold of grapevine: from screening to mechanisms. Microorganisms 9:1386. doi: 10.3390/microorganisms9071386
Anwar, A. R., Ala, A., Kuswinanti, T., and Syam'Un, E. (2022). The ability of potassium-solubilizing fungi isolated from leucite potassium rock deposits. Biodiver. J. Biol. Diver. 23, 6579–6586. doi: 10.13057/biodiv/d231257
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Barka, E. A., Gognies, S., Nowak, J., Audran, J. C., and Belarbi, A. (2002). Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote grapevine growth. Biol. Control. 24, 135–142. doi: 10.1016/S1049-9644(02)00034-8
Bhargavi, G., Arya, M., Jambhulkar, P. P., Singh, A., Rout, A. K., Behera, B. K., et al. (2024). Evaluation of biocontrol efficacy of rhizosphere dwelling bacteria for management of Fusarium wilt and Botrytis gray mold of chickpea. BMC Genom Data. 25:7. doi: 10.1186/s12863-023-01178-7
Biedendieck, R., Knuuti, T., Moore, S. J., and Jahn, D. (2021). The “beauty in the beast” the multiple uses of Priestia megaterium in biotechnology. Appl. Microbiol. Biotechnol. 105, 5719–5737. doi: 10.1007/s00253-021-11424-6
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., Mahillon, J., et al. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10:302. doi: 10.3389/fmicb.2019.00302
Chebil, S., Roudet, J., Ghorbel, A., and Dubos, B. (2004). Effect of early contamination by Botrytis cinerea on the development of grey mould on Muscat d'Italie in Tunisian vineyard. OENO One 38, 131–139. doi: 10.20870/oeno-one.2004.38.2.921
Darriaut, R., Antonielli, L., Martins, G., Ballestra, P., Vivin, P., Marguerit, E., et al. (2022). Soil composition and rootstock genotype drive the root-associated microbial communities in young grapevines. Front. Microbiol. 13:1031064. doi: 10.3389/fmicb.2022.1031064
Datta, C., and Basu, P. (2000). Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanus cajan. Microbiol. Res. 155, 123–127. doi: 10.1016/S0944-5013(00)80047-6
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Diabankana, R. G. C., Afordoanyi, D. M., Safin, R. I., Nizamov, R. M., Karimova, L. Z., Validov, S. Z., et al. (2021). Antifungal properties, abiotic stress resistance, and biocontrol ability of Bacillus mojavensis PS17. Curr. Microbiol. 78, 3124–3132. doi: 10.1007/s00284-021-02578-7
Edwards, S. G., and Seddon, B. (2001). Selective media for the specific isolation and enumeration of Botrytis cinerea conidia. Lett Appl. Microbiol. 32, 63–66. doi: 10.1046/j.1472-765x.2001.00857.x
Faramarzi, M. A., Fazeli, M., Yazdi, M. T., Adrangi, S., Al-Ahmadi, K. J., Tasharrofi, N., et al. (2009). Optimization of cultural conditions for production of chitinase by a soil isolate of Massilia timonae. Biotechnology 8, 93–99. doi: 10.3923/biotech.2009.93.99
Farkas, K., Hassard, F., McDonald, J. E., Malham, S. K., and Jones, D. L. (2017). Evaluation of molecular methods for the detection and quantification of pathogen-derived nucleic acids in sediment. Front. Microbiol. 8:53. doi: 10.3389/fmicb.2017.00053
Fernández-Ortuño, D., Torés, J. A., Chamorro, M., Pérez-García, A., and de Vicente, A. (2016). Characterization of resistance to six chemical classes of site-specific fungicides registered for gray mold control on strawberry in Spain. Plant Dis. 100, 2234–2239. doi: 10.1094/PDIS-03-16-0280-RE
Francis, I., Holsters, M., and Vereecke, D. (2010). The Gram-positive side of plant-microbe interactions. Environ. Microbiol. 12, 1–12. doi: 10.1111/j.1462-2920.2009.01989.x
Glickmann, E., and Dessaux, Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61, 793–796. doi: 10.1128/aem.61.2.793-796.1995
Griffiths, R. I., Whiteley, A. S., O'Donnell, A. G., and Bailey, M. J. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000
Habib, A., Ben Maachia, S., Sahli, A., and Harbi Ben Slimane, M. (2020). Berry quality of principal grapevines in the Oasis of El Jerid, Tunisia. J. Hortic. Posthar. Res. 3, 141–150. doi: 10.22077/jhpr.2019.2753.1087
Hajji-Hedfi, L., Rhouma, A., Hajlaoui, H., Hajlaoui, F., and Rebouh, N. Y. (2023). Understanding the influence of applying two culture filtrates to control gray mold disease (Botrytis cinerea) in tomato. Agronomy 13:1774. doi: 10.3390/agronomy13071774
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hankin, L., and Anagnostakis, S. L. (1975). The use of solid media for detection of enzyme production by fungi. Mycologia 67, 597–607. doi: 10.1080/00275514.1975.12019782
Hartmann, M., Brunner, I., Hagedorn, F., Bardgett, R. D., Stierli, B., Herzog, C., et al. (2017). A decade of irrigation transforms the soil microbiome of a semi-arid pine forest. Mol. Ecol. 26, 1190–1206. doi: 10.1111/mec.13995
Hasan, A., Tabassum, B., Hashim, M., and Khan, N. (2024). Role of plant growth promoting rhizobacteria (PGPR) as a plant growth enhancer for sustainable agriculture: a review. Bacteria 3, 59–75. doi: 10.3390/bacteria3020005
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kjeldahl, J. A. (1883). New method for the determination of nitrogen in organic matter. Zeitschrift Analyt. Chem. 22, 366–382. doi: 10.1007/BF01338151
Kumar, H., Bajpai, V. K., Dubey, R. C., Maheshwari, D. K., and Kang, S. C. (2010). Wilt disease management and enhancement of growth and yield of Cajanus cajan (L) var. Manak by bacterial combinations amended with chemical fertilizer. Crop Prot. 29, 591–598. doi: 10.1016/j.cropro.2010.01.002
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds. E. Stackebrandt, M. Goodfellow (New York: Wiley), 115–75.
Li, J., Hu, M., Xue, Y., Chen, X., Lu, G., Zhang, L., et al. (2020). Screening, identification and efficacy evaluation of antagonistic bacteria for biocontrol of soft rot disease caused by Dickeyazeae. Microorganisms 8:697. doi: 10.3390/microorganisms8050697
Liao, P., Zhao, J. X., and Shaobo, L. (2013). Effects of natamycin on the elimination of fungal contamination in rice and Arabidopsis thaliana tissue cultures for Agrobacterium mediated transformation. Res. J. Biotechnol. 8, 3–9.
Ma, K., Cristina, A., María, A., Dayanna, N., and Isela, F. (2022). Bacillus cabrialesii: five years of research on a novel species of biological control and plant growth-promoting bacteria. Plants 12:2419. doi: 10.3390/plants12132419
Maachia, B., Rafik, E., Chérif, M., Nandal, P., Mohapatra, T., Bernard, P., et al. (2015). Biological control of the grapevine diseases 'grey mold' and 'powdery mildew' by Bacillus B27 and B29 strains. Indian J. Exp. Biol. 53, 109–115.
Macan, G. P. F., Khalil, S., Kalyandurg, P. B., Pareek, N., and Vetukuri, R. R. A. (2022). detached leaf assay for rapidly screening plant pathogen-biological control agents. Methods Molec. Biol. 2536, 449–458. doi: 10.1007/978-1-0716-2517-0_26
Madden, T. L., Tatusov, R. L., and Zhang, J. (1996). Applications of network BLAST server. Meth. Enzymol. 266, 131–141. doi: 10.1016/S0076-6879(96)66011-X
Mahajan, S., Kumar, D., Singh, S. K., Mahajan, D., Kumar, D., Paswal, S., et al. (2020). Evaluation of different fungicides and bioagents for the management of chickpea wilt (Fusarium oxysporum f. sp. ciceri). Curr. J. Appl. Sci. Technol. 39, 19–30. doi: 10.9734/cjast/2020/v39i1430693
Melki Ben Fredj, S., Chebil, S., Lebrihi, A., Lasram, S., Ghorbel, A., Mliki, A., et al. (2007). Occurrence of pathogenic fungal species in Tunisian vineyards. Int. J. Food Microbiol. 113, 245–250. doi: 10.1016/j.ijfoodmicro.2006.07.022
Méndez-Santiag, E. W., Gómez-Rodríguez, O., Sánchez-Cruz, R., Folch-Mallol, J. L., Hernández-Velázquez, V. M., Villar-Luna, E., et al. (2021). Serratia sp. an endophyte of Mimosa pudica nodules with nematicidal, antifungal activity and growth-promoting characteristics. Arch. Microbiol. 203, 549–559. doi: 10.1007/s00203-020-02051-2
Milagres, A. M., Machuca, A., and Napoleão, D. (1999). Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods. 37, 1–6. doi: 10.1016/S0167-7012(99)00028-7
Minh, B. Q., Nguyen, M. A., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mohamed, A. H., Abd El-Megeed, F. H., Hassanein, N. M., Youseif, S. H., Farag, P. F., Saleh, S. A., et al. (2022). Native rhizospheric and endophytic fungi as sustainable sources of plant growth promoting traits to improve wheat growth under low nitrogen input. J. Fungi. 8:94. doi: 10.3390/jof8020094
Mohite, B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13, 638–649. doi: 10.4067/S0718-95162013005000051
Nasfi, Z., Busch, H., Kehraus, S., Linares-Otoya, L., König, G. M., Schäberle, T. F., et al. (2018). Soil bacteria isolated from Tunisian arid areas show promising antimicrobial activities against gram-negatives. Front. Microbiol. 9:2742. doi: 10.3389/fmicb.2018.02742
Naylor, D., and Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oladapo, B. O., Ekundayo, E. A., Mokoolu, M. O., and Ekundayo, F. O. (2020). Phosphate solubilization potentials of rhizosphere fungi isolated from insecticide treated soil. Adv. Res. Life Sci. 4, 58–69. doi: 10.2478/arls-2020-0020
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Peian, Z., Haifeng, J., Peijie, G., Sadeghnezhad, E., Qianqian, P., Tianyu, D., et al. (2021). Chitosan induces jasmonic acid production leading to resistance of ripened fruit against Botrytis cinerea infection. Food Chem. 337:127772. doi: 10.1016/j.foodchem.2020.127772
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
Prashanthi, R., Shreevatsa, G., Krupalini, S., and Mano, L. (2021). Isolation, characterization, and molecular identification of soil bacteria showing antibacterial activity against human pathogenic bacteria. J. Genet. Eng. Biotechnol. 19:120. doi: 10.1186/s43141-021-00219-x
Pylak, M., Oszust, K., and Frac, M. (2020). Searching for new beneficial bacterial isolates of wild raspberries for biocontrol of phytopathogens antagonistic properties and functional characterization. Int. J. Mol. Sci. 21:9361. doi: 10.3390/ijms21249361
Rahman, V., Meena, K. R., Al-Ani, L. K. T., Singh, A., and Kumar, A. (2023). Bacillus velezensis strain improvement to control Helminthosporium maydis causing southern corn leaf blight disease in maize. Eur. J. Plant Pathol. 2023, 1–13. doi: 10.1007/s10658-023-02708-w
Rebouh, N. Y., Aliat, T., Polityko, P. M., Kherchouche, D., Boulelouah, N., Temirbekova, S. K., et al. (2022). Environmentally friendly wheat farming: biological and economic efficiency of three treatments to control fungal diseases in winter wheat (Triticum aestivum L.) under field conditions. Plants 11:1566. doi: 10.3390/plants11121566
Rebouh, N. Y., Latati, M., Polityko, P., Kucher, D., Hezla, L., Norezzine, A., et al. (2020). Influence of three cultivation technologies to control Fusarium spp. in winter wheat (Triticum aestivum L.) production under Moscow conditions. Res. Crops 21, 17–25. doi: 10.31830/2348-7542.2020.003
Rebouh, N. Y., Polityko, P. M., Pakina, E., Plushikov, V. G., Norezzine, A., Gadzhikurbanov, A., et al. (2019). Impact of three integrated crop protection treatments on the varieties of winter wheat (Triticum aestivum L.) in Moscow area, Russia. Res. Crops 20, 161–168. doi: 10.31830/2348-7542.2019.022
Reeves, W. R., McGuire, M. K., Stokes, M., and Vicini, J. L. (2019). Assessing the safety of pesticides in food: how current regulations protect human health. Adv. Nutr. 10, 80–88. doi: 10.1093/advances/nmy061
Rupp, S., Weber, R. W. S., Rieger, D., Detzel, P., and Hahn, M. (2017). Spread of Botrytis cinerea strains with multiple fungicide resistance in German horticulture. Front. Microbiol. 7:2075. doi: 10.3389/fmicb.2016.02075
Saddoud Debbabi, O., Ben Slimane, M., Ben Hadj Alouane, R. B., Montemurro, C., Snoussi, H., Miazzi, M. M., et al. (2024). Traditional foods as a way to preserve the genetic diversity of the grapevine (Vitis vinifera) in Tunisia. Horticulturae 10:423. doi: 10.3390/horticulturae10040423
Santoyo, G., Orozco-Mosqueda, M. D. C., and Govindappa, M. (2012). Mechanisms of biocontrol and plant growth promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci. Technol. 22, 855–872. doi: 10.1080/09583157.2012.694413
Sautua, F. J., Baron, C., Pérez-Hernández, O., and Carmona, M. A. (2019). First report of resistance to carbendazim and procymidone in Botrytis cinerea from strawberry, blueberry and tomato in Argentina. Crop Prot. 125, 2017–2010. doi: 10.1016/j.cropro.2019.104879
Schimel, J., Balser, T. C., and Wallenstein, M. (2017). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Sehrawat, A., Sindhu, S. S., and Glick, B. B. (2022). Hydrogen cyanide production by soil bacteria: biological control of pests and promotion of plant growth in sustainable agriculture. Pedosphere 32, 15–38. doi: 10.1016/S1002-0160(21)60058-9
Shafi, J., Tian, H., and Bacillus, J.i. M. (2017). species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip. 31, 446–459. doi: 10.1080/13102818.2017.1286950
Snoussi, H., Harbi Ben Slimane, M., Ruiz-García, L., Martínez-Zapater, J. M., and Arroyo-García, R. (2004). Genetic relationship among cultivated and wild grapevine accessions from Tunisia. Genome 47, 1211–1219. doi: 10.1139/g04-072
Soumare, A., Diédhiou, A. G., Arora, N. K., Al-Ani, L. K. T., Ngom, M., Fall, S., et al. (2021). Potential role and utilization of plant growth promoting microbes in plant tissue culture. Front. Microbiol. 12:649878. doi: 10.3389/fmicb.2021.649878
Soylu, E. M., Kurt, S., and Soylu, S. (2010). In vitro and in vivo antifungal activities of the essential oils of various plants against tomato grey mould disease agent Botrytis cinerea. Int. J. Food Microbiol. 143, 183–189. doi: 10.1016/j.ijfoodmicro.2010.08.015
Soylu, S., Kara, M., Uysal, A., Kurt, S., and Soylu, E. M. (2021). Determination of antagonistic potential of endophytic bacteria isolated from lettuce against lettuce white mould disease caused by Sclerotinia sclerotiorum. Zemdirbyste-Agric. 108, 317–324. doi: 10.13080/z-a.2021.108.039
Strano, C. P. A., Malacrino, O., Campolo, V., and Palmeri, V. (2018). Influence of host plant on Thaumetopoea pityocampa gut bacterial community. Microb. Ecol. 75, 487–494. doi: 10.1007/s00248-017-1019-6
Sun, S., and Badgley, B. D. (2019). Changes in microbial functional genes within the soil metagenome during forest ecosystem restoration. Soil Biol. Biochem. 135, 163–172. doi: 10.1016/j.soilbio.2019.05.004
Tan, B., Li, Y., Liu, T., Tan, X., He, Y., You, X., et al. (2021). Response of plant rhizosphere microenvironment to water management in soil- and substrate-based controlled environment agriculture (CEA) systems: a review. Front. Plant Sci. 12:691651. doi: 10.3389/fpls.2021.691651
Tanovic, B., Delibasic, G., Milivojevic, J., and Nikolic, M. (2009). Characterization of Botrytis cinerea isolates from small fruits and grapevine in Serbia. Arch. Biol. Sci. 61, 419–429. doi: 10.2298/ABS0903419T
Tanovic, B., Hrustic, J., Mihajlovic, M., Grahovac, M., and Delibasic, G. (2014). Botrytis cinerea in raspberry in Serbia I: morphological and molecular characterization. Pesticidi i fitomedicina 29, 237–247. doi: 10.2298/PIF1404237T
Tocheva, E. I., Ortega, D. R., and Jensen, G. J. (2016). Sporulation, bacterial cell envelopes and the origin of life. Nat. Rev. Microbiol. 14, 535–542. doi: 10.1038/nrmicro.2016.85
Trivedi, P., Anderson, I. C., and Singh, B. K. (2013). Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 21, 641–651. doi: 10.1016/j.tim.2013.09.005
Tsantila, E. M., Esslinger, N., Christou, M., Papageorgis, P., and Neophytou, C. M. (2024). Antioxidant and anticancer activity of Vitis vinifera extracts in breast cell lines. Life 14:228. doi: 10.3390/life14020228
Valenzuela Ruiz, V., Santoyo, G., Gómez Godínez, L. J., Cira Chávez, L. A., Parra Cota, F. I., and De los Santos Villalobos, S. (2022). Complete genome sequencing of Bacillus cabrialesii TE3T: a plant growth-promoting and biological control agent isolated from wheat (Triticum turgidum subsp. durum) in the Yaqui Valley. Curr. Res. Microb. Sci. 4:100193. doi: 10.1016/j.crmicr.2023.100193
Wang, J., Liu, G., Zhang, C., Wang, G., Fang, L., Cui, Y., et al. (2019). Higher temporal turnover of soil fungi than bacteria during long-term secondary succession in a semiarid abandoned farmland. Soil Tillage Res. 194:104305. doi: 10.1016/j.still.2019.104305
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wu, N., Li, Z. F., Wu, M., and Tang, M. (2019). Microenvironment and microbial community in the rhizosphere of dioecious Populus cathayana at Chaka Salt Lake. J. Soils Sedim. 19, 2740–2751. doi: 10.1007/s11368-019-02263-0
Wu, Z., Gao, T., Liang, Z., Hao, J., Liu, P., Liu, X., et al. (2023). Dynamic changes in plant secondary metabolites induced by Botrytis cinerea infection. Metabolites 13:654. doi: 10.3390/metabo13050654
Xiang, L. G., Wang, H. C., Wang, F., Cai, L. T., Li, W. H., Hsiang, T., et al. (2022). Analysis of phyllosphere microorganisms and potential pathogens of tobacco leaves. Front. Microbiol. 13:843389. doi: 10.3389/fmicb.2022.843389
Yin, W. X., Adnan, M., Shang, Y., Lin, Y., and Luo, C. X. (2018). Sensitivity of Botrytis cinerea from nectarine/cherry in China to six fungicides and characterization of resistant isolates. Plant Dis. 102, 2578–2585. doi: 10.1094/PDIS-02-18-0244-RE
Zhang, L., Kinkelaar, D., Huang, Y., Li, Y., Li, X., Wang, H. H., et al. (2011). Acquired antibiotic resistance: are we born with it? Appl. Environ. Microbiol. 77, 7134–7141. doi: 10.1128/AEM.05087-11
Zhou, L., Song, C., Muñoz, C. Y., and Kuipers, O. P. (2021). Bacillus cabrialesii BH5 protects tomato plants against Botrytis cinerea by production of specific antifungal compounds. Front. Microbiol. 12:707609. doi: 10.3389/fmicb.2021.707609
Zhou, Y. J., Zhang, J., Wang, X. D., Yang, L., Jiang, D. H., Li, G. Q., et al. (2014). Morphological and phylogenetic identification of Botrytis sinoviticola, a novel cryptic species causing gray mold disease of table grapes (Vitis vinifera) in China. Mycologia 106, 43–56. doi: 10.3852/13-032
Keywords: antifungal, Botrytis cinerea, enzyme activity, grapevine, soil microbiota, growth promotion, sustainable viticulture
Citation: Hajji-Hedfi L, Wannassi T, Tawfeeq Al-Ani LK, Balbool BA, Tissaoui S, Mougou-Hamdane A, Hamdi W, Azeem AMA and Rebouh NY (2025) Investigating the potential role of beneficial rhizobacteria for protecting grapevine health and promoting growth. Front. Sustain. Food Syst. 9:1619801. doi: 10.3389/fsufs.2025.1619801
Received: 28 April 2025; Accepted: 02 June 2025;
Published: 07 July 2025.
Edited by:
Mohamed Trigui, Institut Préparatoire aux Etudes d'Ingénieur de Sfax (IPEIS), TunisiaReviewed by:
Soner Soylu, Mustafa Kemal University, TürkiyeArshad Ali, Universiti Malaysia Sabah, Malaysia
Shabiha Nudrat Hazarika, Arborist Innovations, India
Copyright © 2025 Hajji-Hedfi, Wannassi, Tawfeeq Al-Ani, Balbool, Tissaoui, Mougou-Hamdane, Hamdi, Azeem and Rebouh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lobna Hajji-Hedfi, ZWxoYWpqaWxvYm5hQHlhaG9vLmZy; bG9ibmEuaGFqamlAaXJlc2EuYWdyaW5ldC50bg==; Nazih Y. Rebouh, cmVidWtoX255YUBwZnVyLnJ1; bi55YWNlcjE2QG91dGxvb2suZnI=
†ORCID: Lobna Hajji-Hedfi orcid.org/0000-0002-3587-4790
Takwa Wannassi orcid.org/0000-0002-4568-582X
Laith Khalil Tawfeeq Al-Ani orcid.org/0000-0001-5138-0224
Bassem Ayman Balbool orcid.org/0000-0002-9699-3558
Wissem Hamdi orcid.org/0000-0002-4753-0901
Ahmed M. Abdel Azeem orcid.org/0000-0003-2897-3966
Nazih Y. Rebouh orcid.org/0000-0002-8621-6595
 Lobna Hajji-Hedfi
Lobna Hajji-Hedfi Takwa Wannassi
Takwa Wannassi Laith Khalil Tawfeeq Al-Ani
Laith Khalil Tawfeeq Al-Ani Bassem Ayman Balbool
Bassem Ayman Balbool Salma Tissaoui5
Salma Tissaoui5 Wissem Hamdi
Wissem Hamdi Ahmed M. Abdel Azeem
Ahmed M. Abdel Azeem Nazih Y. Rebouh
Nazih Y. Rebouh