- 1Division of Biomedical Information Analysis, Iwate Tohoku Medical Megabank Organization, Iwate Medical University, Yahaba, Japan
- 2Department of Advanced Genomic and Laboratory Medicine, Graduate School of Medicine, University of the Ryukyus, Nishihara, Japan
- 3Department of Biology, National Museum of Natural Science, National Taiwan Normal University, Taipei, Taiwan
- 4Department of Biological Sciences, Macquarie University, Sydney, NSW, Australia
- 5Division of Zoology, Endemic Species Research Institute, National Taiwan Normal University, Taipei, Taiwan
- 6Department of Life Science, National Taiwan Normal University, Taipei, Taiwan
- 7Division of Bioresource Science, Amphibian Research Center, Hiroshima University, Higashihiroshima, Japan
- 8Program of Basic Biology and Program of Biomedical Science, Graduate School of Integrated Sciences for Life, Hiroshima University, Higashihiroshima, Japan
Buergeria frog tadpoles exhibit high thermal tolerance and are occasionally found in water pools that temporarily exceed 40°C. With the aim of understanding how they can cope with the severe heat stress, we performed RNA-seq of three heat-treated (38°C) and three control (25°C) tadpoles and compared their transcriptomic profiles. We identified 382 differentially expressed transcripts. A protein-protein interaction (PPI) network analysis of these transcripts further identified hub proteins involved in protein degradation, stress granule assembly, and global suppression of DNA transcription and mRNA translation. Along with the avoidance behavior against high water temperature, these endurance mechanisms potentially support tadpoles to survive in high temperatures for short periods of time. Similar mechanisms may exist in many other amphibian species whose habitats are prone to high temperatures.
Introduction
In their natural environment, organisms face various environmental stressors such as ultraviolet light, low and high pH, osmotic pressure, freezing, and heat. For survival, organisms exhibit various responses to such stressors at multiple biological levels, including changes in morphology, physiology, behavior, and life history (Crespi et al., 2013; Wingfield, 2013; Gormally and Romero, 2020). Most of these responses are initially caused by intracellular regulations mediated by the neural and hormonal system (Mastrangelo et al., 2012; Crespi et al., 2013; Wingfield, 2013; Gormally and Romero, 2020). Therefore, these stress responses become the paradigm of how molecular changes influence individual survival under environmental challenges (Boonstra, 2013). Thus, a better understanding of how organisms cope with extreme stressors can reveal important molecular mechanisms enabling them to survive in such environments. In particular, thermal conditions pose significant challenges to ectotherms. Without thermoregulatory mechanisms, the survival and distribution of organisms are largely determined by temperature, whereby an appropriate response to thermal stress is essential for their survival.
Tree frog species of the genus Buergeria are suitable for studying how organisms cope with extreme thermal stress because several populations live in extreme hot environments. Buergeria frogs are distributed in a wide range of habitats, from mountain streams to coastal areas across continental islands in Eastern and Southeastern Asia (Haramura, 2004; Matsui and Maeda, 2018). Currently, six species have been identified, and several populations of three species, B. otai, B. choui, and B. japonica, are further found in geothermal hot springs (Morita, 1988; Chen et al., 2001; Wu and Kam, 2005; Komaki et al., 2016b).
In geothermal hot springs, the water temperature is non-uniform and varies temporally and spatially due to rainfall, water depth, air temperature, and distance from the hot spring source. Field and laboratory observations revealed that Buergeria tadpoles and adults from hot spring populations tended to avoid inhabiting and reproducing in water temperatures above ∼35°C (Wu and Kam, 2005; Komaki et al., 2020). Avoidance behaviors against heat stress are probably essential for populations to survive in hot spring environments. However, due to the nature of hot springs, the water temperature can fluctuate, and tadpoles must face inevitable temperature spikes in this fully aquatic life stage. Indeed, Buergeria frogs living in hot springs can tolerate hot water: a previous study measured 30 Buergeria tadpoles collected from a hot spring habitat and revealed tolerance of temperatures up to 46°C, whereas above this temperature, tadpoles lost balance and swimming ability or died (Komaki et al., 2016b). Such high thermal tolerance has been observed not only in hot spring populations but also in freshwater Buergeria populations across their distribution (Komaki et al., 2016a). The high thermal tolerance necessary for survival at these temperatures demands protective transcriptional and subsequent cellular responses to such stress. These responses, together with behavioral strategies, are expected to play essential roles in Buergeria frogs inhabiting hot spring environments.
Transcriptional and proteomic responses of anurans to cold or freezing temperatures have been previously characterized, and genes and proteins related to heat shock, ubiquitin-proteasome, antioxidation, and glycogenolysis were identified in response to low-temperature stress (Kiss et al., 2011; Nagasawa et al., 2013; do Amaral et al., 2020). However, to the best of our knowledge, transcriptome analysis related to heat stress has not been reported, presumably due to the lack of well-known heat-tolerant frog species. Therefore, Buergeria frogs, which are known to inhabit hot springs and exhibit high thermal tolerance, are suitable subjects for studying the transcriptional response to explicit heat stress.
Given their avoidance of high-temperature waters (Wu and Kam, 2005; Komaki et al., 2020), Buergeria frogs may be less exposed to long-term heat stress. Rather, it may be more important for these frogs to endure temporary heat stress while moving away from the stressor. In this study, to further understand the molecular mechanisms and strategies that function in response to heat stress, we performed a comparative transcriptome analysis of heat-treated and untreated B. otai tadpoles.
Materials and Methods
Sample Preparations
We collected B. otai adult females and males in Hualien, Taiwan in August 2013. The adult individuals were kept in a laboratory where they then laid eggs. It is unclear whether the eggs were laid by a single female-male pair or not. Eggs were transferred to approximately 25°C water tanks (∼50 L) with filtration systems and 9:15 light-dark cycle. The water temperature was chosen according to previous studies (Chen et al., 2001; Wu and Kam, 2005). Hatched tadpoles were kept under the same condition until use in thermal experiments. The water was not replaced. Tadpoles were fed boiled spinach once a day.
Six tadpoles at Gosner stages 26–28 (Gosner, 1960) were moved into two 1000-mL glass beakers containing 500-mL of 25°C water and were maintained for a day in incubators. After an additional 12 h fasting, three tadpoles were directly moved into a glass beaker with 38°C water which is slightly cooler than their possible thermal limit (∼40°C) (Komaki et al., 2016a). We confirmed that all three tadpoles did not exhibit any abnormal behavior after a sudden temperature change. The other three tadpoles were moved into a beaker with 25°C water. We maintained the water temperatures of each beaker for approximately 4 h using incubators, and then tadpoles were euthanized by pithing immediately after they were taken out from the water. Bodies were transferred into a RNAlater Stabilization Solution (Thermo Fisher Scientific, MA, United States).
Total RNA was extracted from the whole-body using a RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. RNA samples were shipped to BGI (Beijing, China), where RNA-seq was performed using Illumina Hi-Seq 2000 (Illumina, CA, United States).
Since amphibian tadpoles were not subject to ethical review in Hiroshima University where the experiment was conducted at that time, no institutional ethical approval was obtained. This study was carried out in compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines.1 In the experimental procedures using living tadpoles, we made an effort to minimize stress of individuals following Guidelines for Live Amphibians and Reptiles in Field and Laboratory Research developed by the American Society of Ichthyologists and Herpetologists in 2004.
Transcriptome Analyses
Raw RNA-seq data went through read filtering based on base quality and read length using Trim Galore v0.6.4 with default settings (phred score threshold of 20 and length threshold of 20 bp). Then, de novo assembly was performed using Trinity v2.8.3 (Grabherr et al., 2011), and summary statistics of the assembly were calculated by SeqKit v0.13.2 (Shen et al., 2016). Completeness of the assembly was assessed using BUSCO version 5.0.0 (Seppey et al., 2019) specifying tetrapoda_obd10 as a reference. Pseudoalignment and transcript quantification was performed using kallisto v0.45.1 (Bray et al., 2016) with 100 bootstraps. Output files of kallisto were imported into the R environment using the R package “tximport” v1.10.1 (Soneson et al., 2016), and counts of reads mapped to each contig were normalized by the trimmed means of M (TMM) values method. A differential expression analysis was then performed using the R package “edgeR” v3.24.3 (Robinson et al., 2010) in R 3.6.3 (R Foundation). A likelihood ratio test based on TMM values was applied in the analysis. Transcripts with an FDR-corrected p (q-value) ≤ 0.05 and an absolute value of log2 fold change ≥ 3 were defined as differentially expressed transcripts.
Annotation of Differentially Expressed Transcripts
To annotate differentially expressed transcripts, we first searched candidate coding regions in each contig of differentially expressed transcripts using Transdecoder v5.5.0 with default settings and the “–single_best_only” option so that only the single best candidate region per transcript was considered (Haas et al., 2013). We then scanned all predicted protein sequences for homology in the Ensembl reference protein sequence of Xenopus tropicalis (v9.1) using local BLASTP v2.10.1 + (Altschul et al., 1990). We used the default settings and set the e-value threshold to 10–5, and only the top hits for each contig were considered in the downstream analyses.
Functional Analyses
For an overview of the transcriptomic change in response to the heat stress, we performed a protein-protein interaction (PPI) analysis and functional enrichment analyses based on transcripts that were differentially expressed and annotated by BLASTP search.
A web database STRING (Szklarczyk et al., 2019) was used for the PPI analysis, specifying X. tropicalis as the target organism with default settings. Annotations of both upregulated and downregulated transcripts obtained from BLASTP annotation using X. tropicalis as a reference were analyzed together in a single PPI analysis. The obtained pairwise PPIs were further analyzed with the R package “igraph” v1.2.5 (Csardi and Nepusz, 2006) to detect hub proteins in the PPI networks. We adopted two different criteria of measurements, including degree centrality and betweenness centrality, to identify hub proteins as applied in Chen et al. (2019) and Mao et al. (2020). The degree centrality method measures the number of interactions each protein participates in, and betweenness centrality measures how often the shortest paths connecting two other proteins pass through the given protein. Proteins with a higher score of degree centrality or betweenness centrality were regarded as more important hubs in the networks.
We conducted enrichment analyses implemented in STRING database to identify gene ontologies, pathways, keywords, domains, and features overrepresented by differentially expressed and BLASTP-annotated transcripts. The available frameworks in the STRING analysis were GO (Gene Ontology) annotations, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, Uniprot keywords, Reactome pathways, Pfam protein domains, SMART (Simple Modular Architecture Research Tool) protein domains, and InterPro protein features. We performed three independent analyses: upregulated transcripts, downregulated transcripts, and all differentially expressed transcripts (upregulated and downregulated transcripts). In the analyses, strength and Fisher’s exact test p-values were calculated, and the significance threshold was set to FDR-corrected p-value < 0.05. Strength is the log10-transformed number of proteins belonging to a certain category divided by the expected number of proteins belonging to the category when proteins were randomly extracted from the database and indicates the degree of enrichment.
Results
De novo Assembly and Expression Analysis
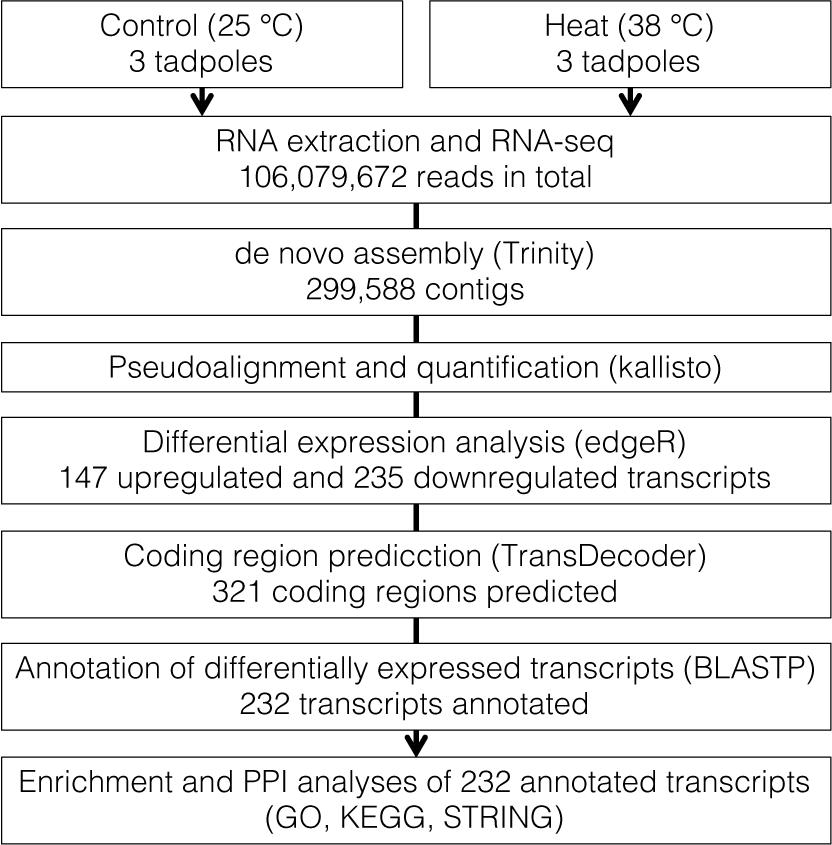
In total, 106,079,672 reads were obtained from RNA-seq of six tadpoles’ whole body and were applied for de novo assembly (Figure 1 and Supplementary Table 1). We assembled 299,588 contigs with an average length of 979.6 bp (range: 165–60,421 bp) and N50 of 2,272 bp. Estimated completeness of the assembly was 92.1% (40.7% single-copy and 51.4% duplicated BUSCOs). Fragmented and missing BUSCOs were 2.2 and 5.7%, respectively (Supplementary Table 1).
In the quantification of transcripts, 17,679,945 reads, on average, were processed per sample, 92% of which were pseudoaligned onto any contigs. Based on the log-likelihood ratio test q-value and log2 fold change thresholds from differential expression analyses, we found 147 and 235 significantly upregulated and downregulated transcripts in response to the heat stress, respectively (Figure 2 and Supplementary Table 2). There were 131 and 190 coding regions predicted by Transdecoder from 147 upregulated and 235 downregulated transcript sequences, respectively.
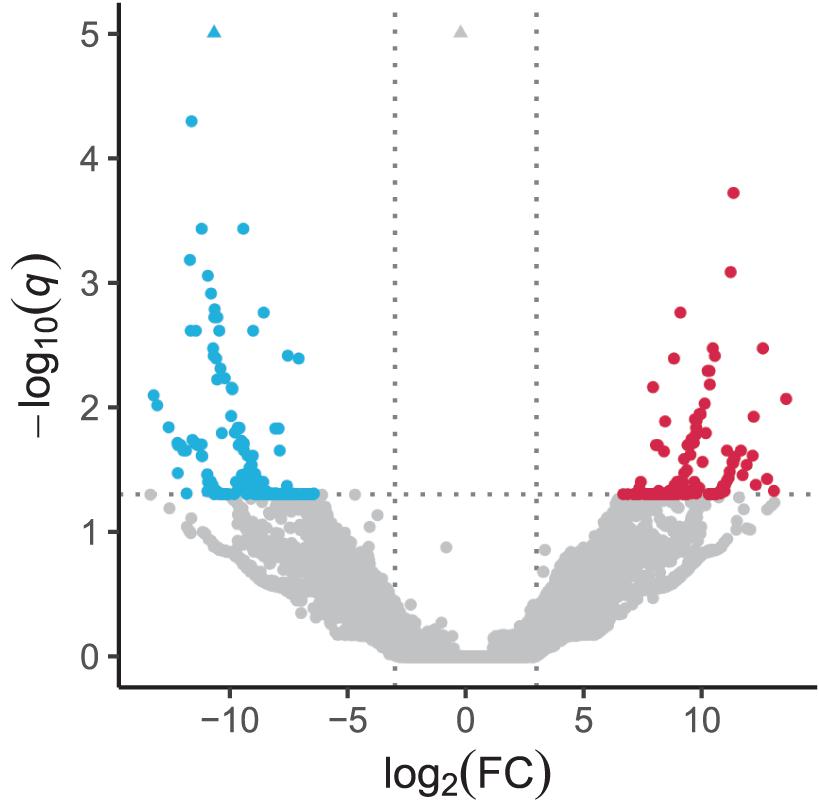
Figure 2. Volcano plot depicting transcriptomic change in response to heat stress. Significantly upregulated and downregulated transcripts are indicated by red and blue dots, respectively. Horizontal dotted line represents the FDR-adjusted p-value (q-value) = 0.05, and vertical dotted lines represent the absolute value of log2(fold change) = 3. Triangles represent transcripts with q-value < 10–5.
Annotations and Protein-Protein Interactions
The local BLASTP using X. tropicalis as a reference protein sequence annotated 86.7% (113 upregulated and 165 downregulated transcripts) of Transdecoder-predicted coding sequences. There were overlapped annotations among transcripts, and the unique number of annotations was 232.
In the STRING PPI analysis, among 232 unique genes with differentially expressed levels and BLASTP annotations, 64 had protein interactions (Figure 3). The number of protein interactions was significantly larger than expected according to the STRING network stats (PPI enrichment p = 2.2 × 10–3). In total, 13 networks were formed. Members of proteins with the highest degree centrality and betweenness centrality were similar to each other; GAK, DDX3X, and KMT2B exhibited degree centrality and betweenness centrality scores within the top five (Supplementary Table 3) and belonged to the largest network. The second-largest PPI network consisted of 10 proteins, and FBXW4 served as a hub protein.
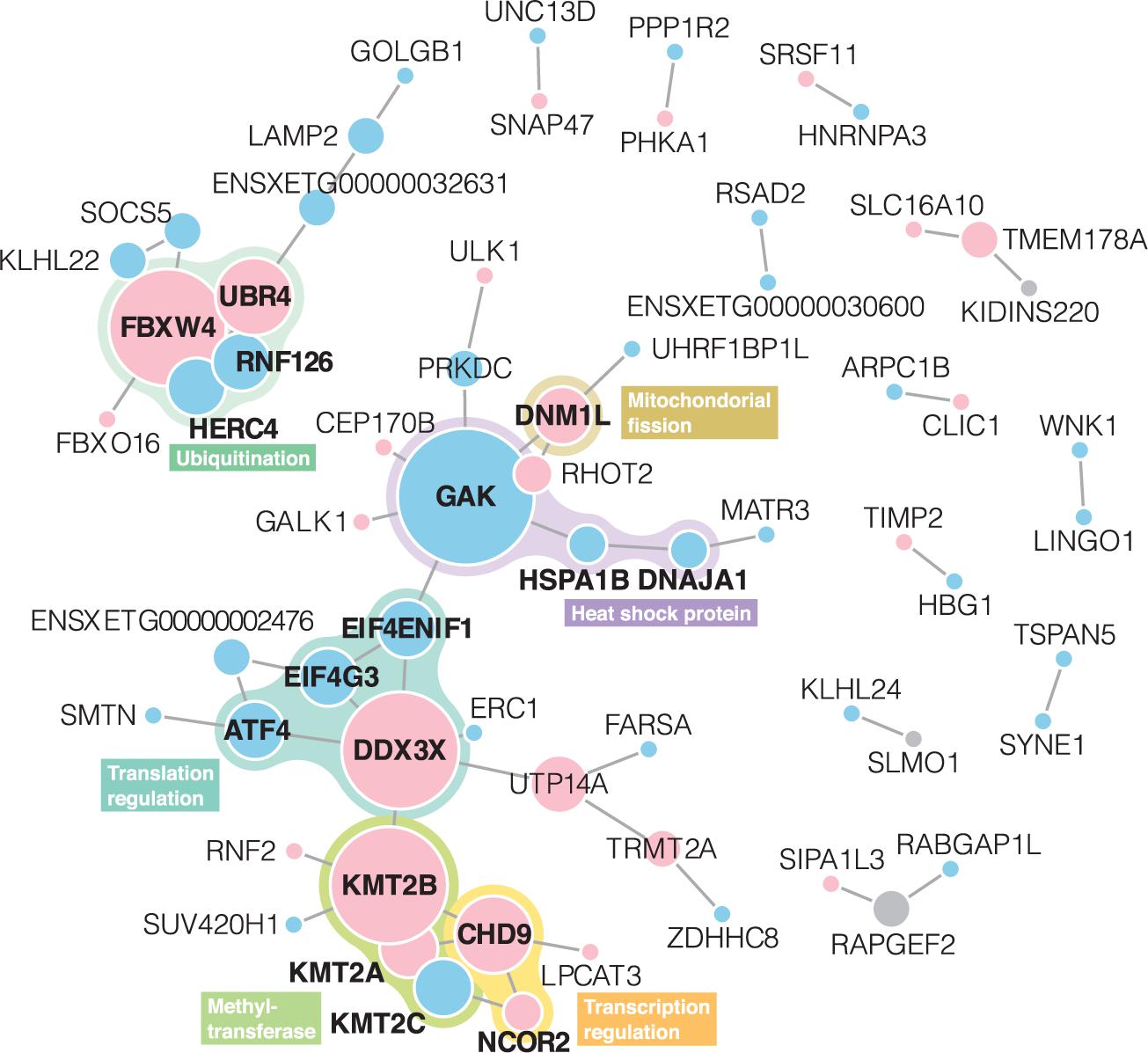
Figure 3. Protein-protein interaction networks derived from the STRING web database. Node size represents degree centrality (Supplementary Table 2). Gene symbols obtained in BLASTP annotation using X. tropicalis as a reference are presented in each node. Red and blue nodes represent proteins with upregulated and downregulated expressions, respectively. Gray nodes indicate that both upregulated and downregulated transcripts shared the annotations. Proteins written in bold letters are those mentioned in the main text.
Function of Hub and Related Proteins
Integrating the gene functions, six subgroups were identified from the PPI networks: heat shock protein (HSP), mitochondrial fission, translation regulation (×2), methyltransferase, and ubiquitination.
Heat shock protein subgroup contained three proteins of GAK, HSPA1B, DNAJA1. GAK, a hub protein in the network, is a member of the DnaJ/HSP40 family. DNAJA1, closely located to GAK on the network, is also a member of the family. HSPA1B, which had PPI interactions with GAK and DNAJA1, belongs to the HSP70. mRNA expression levels of these proteins were downregulated in response to the heat stress.
DNM1L, which had an interaction with GAK, mediates mitochondrial fission. Upregulation of this gene was also reported in a cow and mouse in response to heat stress (Yu et al., 2018; Chen et al., 2020). Under the heat stress, DNM1L-induced mitochondrial fission does not play a protective role for the cell but leads to apoptosis (Yu et al., 2016).
The translation regulation subgroups were found in two non-directly linked regions. One subgroup contained DDX3X, ATF4, eIF4G3, and eIF4ENIF1. DDX3X which was identified as a hub protein of this subgroup is an important component for stress granule assembly and molecular dynamics (Hilliker et al., 2011). Stress granules are dense aggregations of RNAs and RNA-binding proteins. It is formed in the cytosol when the cell is exposed to stress and is expected to have a protective function for the cell through mRNA translation regulation including, sequestration, degradation, and translation re-initiation of mRNA (Lake et al., 2016). ATF4 is a master transcription factor during the stress response. For the mRNA expression of ATF4, the binding of DDX3X to the eIF4F complex is required (Adjibade et al., 2017). The downregulated expression level of ATF4 detected in this study thus can be involved in the possible suppression of the eIF4F complex formation under the heat stress. eIF4F (eukaryotic initiation factor 4F) is a translation initiation factor, and consists of eIF4E, eIF4G, and eIF4A. eIF4G3 is an isoform of eIF4G, and its expression level was downregulated in B. otai tadpoles. eIF4ENIF1, a transporter of eIF4E, also exhibited a downregulated expression level in response to the heat stress. It was demonstrated that eIF4F formation is disrupted, and subsequently, mRNA translation is inhibited in mice under heat stress so that protein synthesis and subsequent possible misfolding can be prevented (Cuesta et al., 2000). The other translation regulation subgroup contained CHD9 and NCOR2, which were closely located to the KMT2 family in the PPI network, are known as transcriptional regulators.
In the methyltransferase subgroup, three proteins (KMT2A–C) of the lysine methyltransferase 2 (KMT2) family were included. This family mainly functions as a histone methyltransferase and is thus known as a transcriptional regulator. In response to the heat stress, KMT2A and KMT2B were upregulated whereas KMT2C was downregulated.
Ubiquitination subgroups including FBXW4, UBR4, RNF126, and HERC4 formed an independent PPI network. They are ubiquitin-related proteins and known to promote ubiquitination and degradation of proteins (O’Leary et al., 2016; UniProt Consortium, 2019). All of these proteins are E3 ubiquitin-protein ligases, however, expression patterns were different: FBXW4 and UBR4 were upregulated and HERC4 and RNF126 were downregulated in response to the heat stress.
Enrichment Analyses
Enrichment analyses performed on upregulated proteins demonstrated that three Pfam, four InterPro, and four SMART categories were enriched, which are involved in methylation and other expression regulations, and no GO term and KEGG pathway were enriched (Supplementary Table 4). Methyltransferase genes, such as KMT2A/B, were involved in the enriched categories. In the downregulated protein analyses, only two UniProt categories (coiled coil and kelch repeat), which were involved in expression regulation and ubiquitination, were enriched (Supplementary Table 4). Three proteins, ENC1.2, KLHL22, and KLHL24, and 32 proteins (e.g., EIF4G3, KMT2C, and HSPA1B), were involved in the coiled-coil and kelch repeat, respectively. There was no overlapping category between the upregulated and downregulated protein analyses. The enrichment analyses performed on all differentially expressed proteins demonstrated 16 overrepresented pathway, keywords, domains, and features while no enriched GO term was detected (Table 1). All of them, except for the coiled-coil and kelch repeat, can be directly or indirectly linked to methyltransferase activity.
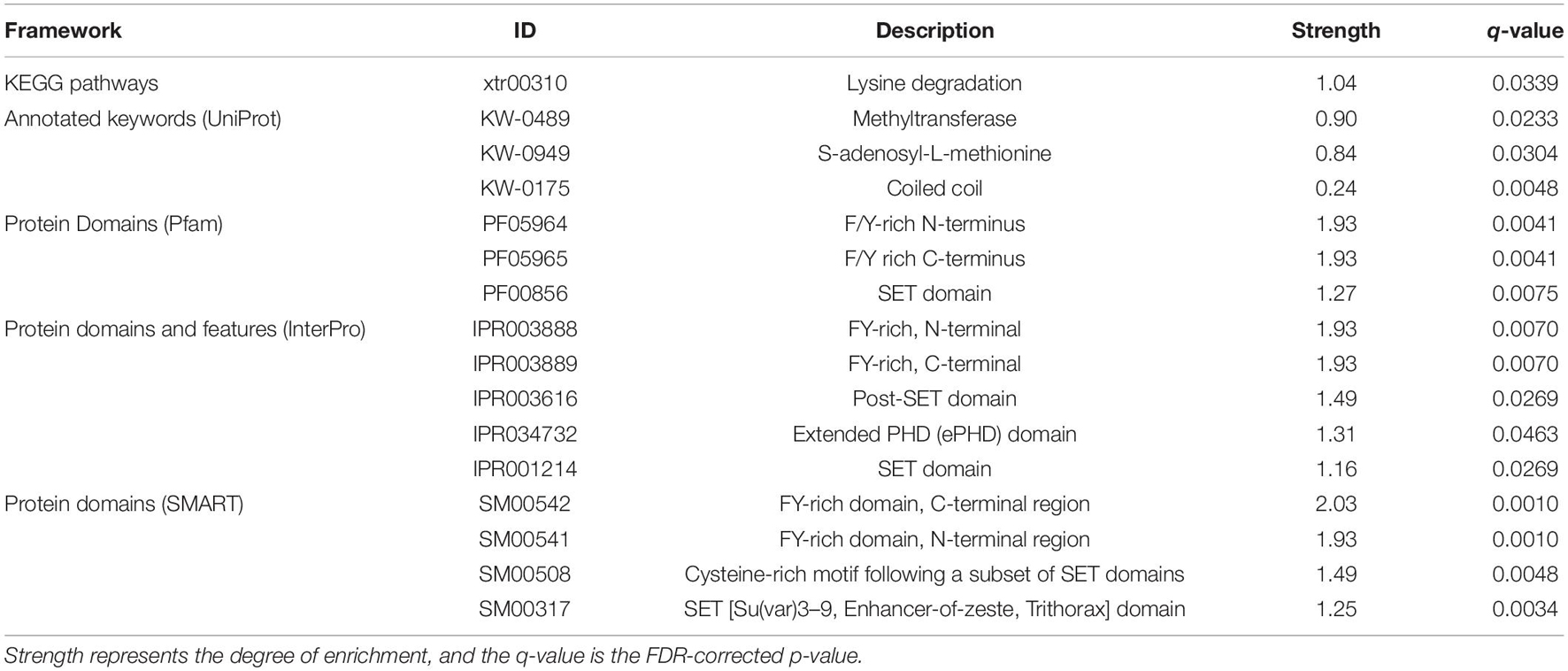
Table 1. Results of STRING functional enrichment analyses based on combined dataset of upregulated and downregulated proteins.
Discussion
Through a comparative transcriptomic analysis, we identified hundreds of transcripts differentially expressed between heat-treated and control tadpoles of B. otai. Knowledge-based analyses imply that our transcriptome analyses detected sets of genes with important functions in coping with the heat stress. Specifically, the PPI network analysis identified hub proteins with functions such as HSPs, stress granule assembly, methylation, and ubiquitination.
Transcriptional and Translational Regulation Mechanisms
First, the transcriptional response to the environmental stimuli is led by epigenetic regulation mechanisms. This study of comparative transcriptional analyses identified genes, pathways, domains, and features involved in different epigenetic mechanisms like DNA and histone methylation. Methyltransferase itself does not have a specific target of genes but is involved in the global change of DNA and histone methylation status. Subsequently, it can cause the transcriptional change of genes. It is not clear how many differentially expressed genes identified in this study are regulated by the methyltransferase activity. However, expression levels of all genes mentioned in the results section, except for DDX3X on the X chromosome, are associated with DNA methylation levels of neighboring DNA methylation sites in humans (Komaki et al., 2018).
Expression levels of KMT2A and KMT2B were upregulated in response to heat stress, whereas that of KMT2C was downregulated. The contrasting expression patterns may be explained by the structural and functional differences between them. For example, the KMT2 enzymes can be distinguished by the number of PHD fingers: KMT2A/B contain four PHD fingers and have a similar overall domain structure. KMT2C, on the other hand, contains eight PHD fingers, which is the largest number among the known KMT2 enzymes (Ali et al., 2014). These structural differences will lead to functional differences. It is known that KMT2A/B and KMT2C have unique subunits, which are known to be associated with differences of binding regions: KMT2A/B are suggested to bind to the promoter and enhancer regions, while KMT2C binds to the enhancer region (Rao and Dou, 2015). The CXXC domain, unique to KMT2A/B, also contributes to the binding of the enzymes to CpG sites (Rao and Dou, 2015). Taken together, the contrasting expression pattern of KMT2 genes implies that specific regulatory mechanisms, such as DNA methylation, may be responsive to heat stress.
In addition to methyltransferase activity, we identified differentially expressed genes involved in the activation and suppression of transcription and translation. NCOR2 is a transcriptional co-suppressor, and it was upregulated under heat stress. Conversely, genes involved in forming the eIF4F complex, a translation initiation factor, were downregulated. These results imply that gene transcription and translation are globally suppressed in B. otai tadpoles under heat stress. Presumably, the mechanisms contribute to the prevention of misfolding protein synthesis under heat stress. The upregulation of DDX3X, a stress granule component, is representative of the hypothesis. Stress granule formation is a well-known cellular stress response and prevents RNA from being translated.
CHD9 upregulation is also notable. The gene belongs to the chromodomain helicase DNA-binding (CHD) family. CHD is a chromatin remodeler that contributes to chromatin structural change and thus, affects chromatin accessibility. Different from the above-mentioned proteins that can change the global DNA transcription and RNA translation pattern, CHD9 is suggested to have a specific target of genes related to osteogenesis, such as RUNX2 (Newton and Pask, 2020). Indeed, it is well documented that heat stimulus promotes osteogenesis (Chen et al., 2013; Ikuta et al., 2015), and it is conceivable that CHD9 participates in the process. However, it is unclear whether the CHD9-mediated osteogenesis can be linked to the protective function against heat stress. Since water temperature is known to affect the timing of metamorphosis (Maciel and Juncá, 2009; Goldstein et al., 2017), the upregulation of CHD9 possibly be related to the body plan change associated with accelerated metamorphosis.
Protein Degradation
We also identified ubiquitination-related proteins that formed an independent PPI network. Proteins damaged by denaturation, such as heat stress are degraded by proteasomes, and the process is usually mediated by ubiquitination (Hershko, 1996). Transcriptional upregulation of ubiquitination-related genes was also identified in two comparative transcriptome studies in which amphibians were exposed to cold stress (Nagasawa et al., 2013; do Amaral et al., 2020). Thus, in combination with above-mentioned protein synthesis suppression, ubiquitination-mediated protein degradation may contribute to reducing dysfunctional proteins.
There were four differentially expressed genes of E3 ubiquitin-protein ligases, although RNF126 and HRC4 were downregulated, while FBXW4 and UBR4 were upregulated in response to the heat stress. There are hundreds of known E3 ligases and each ligase is thought to have specific substrate, however, the relationship is still not well understood, and it is also not known how ligases recognize misfolded proteins. In addition, only a limited number of E3 ligases are known to participate in the misfolded protein ubiquitination (Chhangani et al., 2013), and various other functions have been proposed for ubiquitination, such as DNA repair, signal transduction, and transcriptional regulation (Wilkinson et al., 2005). Therefore, we cannot provide a clear explanation for the up- and downregulations of each E3 ligase gene in response to the heat stress. However, the contrasting expression patterns strongly suggest that these genes play different roles under heat stress.
Heat Shock Proteins
The HSP family is the most famous protein family produced under heat stress. Three genes belonging to HSP40 and HSP70 were differentially expressed in B. otai tadpoles but were all downregulated in heat-treated individuals. Since HSP40 is a co-chaperone of HSP70, it is probable that they demonstrated the same transcriptional change pattern. However, we have no clear insights to explain why they were downregulated in response to heat stress. Contrary to the current study, several frog species exposed to cold stress exhibited upregulation of HSP70 (Kiss et al., 2011; do Amaral et al., 2020). Although the upregulation of HSP70 in those species is expected to function in the degradation of proteins misfolded due to the stress, it cannot explain the downregulation of HSP70 under the heat stress, which also leads to protein misfolding. The downregulation of HSP genes in response to heat stress (21°C compared to 11°C) was also observed in a salamander species (Clay et al., 2019). However, upregulation of the other HSP genes were simultaneously observed and the responses of these specific genes have not been discussed in the previous study. HSPs have various functions, several of which change the responsiveness according to the duration of the stress (Gormally and Romero, 2020). Furthermore, they can be controlled by negative feedback. These gene-specific functions and regulations may underlie the suppression of HSP genes in response to heat stress.
Suppressive Transcriptional Response Against Heat Stress
It has been demonstrated that tadpoles prefer cooler water (Wu and Kam, 2005; Komaki et al., 2020) and the water temperature in hot springs can drop at night due to the lower air temperature; thus, tadpoles can avoid long-term exposure to heat even in hot spring environments. Rather, it probably important for them to tolerate the short-term heat spike. The present study showed that the majority of hub proteins identified in this study had functions related to suppression of protein synthesis and subsequent prevention of protein misfolding. These responses should negatively impact tadpole development if the suppression persists. Therefore, although not verified, the mechanisms are not likely to be sustained under long-term heat stress. That is, Buergeria tadpoles may adopt a combination of suppressive transcriptional responses to tolerate occasional heat stress and avoidance behavioral response to prevent long-term heat exposure.
Suppression of protein synthesis under heat stress is not a mechanism specific to hot spring tadpoles but a widely conserved mechanism. Thus, the mechanisms identified in this study were not evolutionarily acquired in Buergeria lineages. Many anuran species lay eggs in temporally formed shallow water pools, such as paddy fields, whereby water temperatures can reach 36°C (Maruyama et al., 2017). Therefore, although the present study employed rapid and acute heat stress and did not take into account acclimation effects that may occur in the field, a wide range of anuran species may have a similar transcriptomic response to heat stress as that observed in B. otai tadpoles.
Limitations
It should be noted that the sample size in this study is small. Therefore, it is possible that the set of differentially expressed genes may change when the sample is increased. To reduce the uncertainty arising from the small sample size, we mainly focused on the summarized characteristics of upregulated and downregulated gene sets obtained from enrichment and PPI network analyses.
It is also important to mention that this study only focused on annotated transcripts. This means that even if there are novel heat stress response-related transcripts specific to Buergeria frogs, they cannot be identified in this study design. Since RNA-seq data obtained in this study is publicly available, it will possible to approach this point in future studies.
In addition, RNA was extracted from the whole body. Thus, genes showing tissue- or cell type-specific heat stress response patterns are unlikely to be detected in this study. The differentially expressed genes identified in this study are genes whose expression patterns do not conflict among tissues or cell types.
Data Availability Statement
RNA-seq data obtained in this study are deposited in DDBJ Sequence Read Archive (DRA) (BioProject accession number: PRJDB12187). Codes used in this study are available on GitHub (https://github.com/RolyPolyCoily/BuergeriaOtai_2021).
Ethics Statement
Ethical review and approval was not required for the animal study because amphibian tadpoles were not subject to ethical review in Hiroshima University where the experiment was conducted at that time; therefore, no institutional ethical approval was obtained.
Author Contributions
SK designed the study, carried out field and laboratory works and data analyses, and led the writing of the draft. MM carried out data analyses and participated in the writing of the draft. J-WL, K-HL, Y-PL, and YL participated in sample collection and critically revised the manuscript. S-ML coordinated the field work, participated design of the study, and critically revised the manuscript. TI designed the study, carried out laboratory works and data analyses, and revised the manuscript. All authors gave final approval for publication.
Funding
The study was supported by the JSPS KAKENHI (18K14795 and 25-5065 for SK and 23710282 and 18K06365 for TI).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.706887/full#supplementary-material
Supplementary Table 1 | Summary of RNA-seq and subsequent analyses.
Supplementary Table 2 | List of transcripts differentially expressed between heat-treated and control tadpoles with analytical results and annotations.
Supplementary Table 3 | Scores of degree centrality and betweenness centrality for each protein mapped on STRING protein-protein interaction networks.
Supplementary Table 4 | Results of STRING enrichment analyses.
Supplementary Text 1 | Methods and results of additional annotations.
Footnotes
References
Adjibade, P., Grenier St-Sauveur, V., Bergeman, J., Huot, M. E., Khandjian, E. W., Mazroui, R. (2017). DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-14262-7
Ali, M., Hom, R. A., Blakeslee, W., Ikenouye, L., and Kutateladze, T. G. (2014). Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim. Biophys. Acta Mol. Cell Res. 1843, 366–371. doi: 10.1016/j.bbamcr.2013.11.016
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Boonstra, R. (2013). The ecology of stress: a marriage of disciplines. Funct. Ecol. 27, 7–10. doi: 10.1111/1365-2435.12048
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Chen, J., Shi, Z.-D., Ji, X., Morales, J., Zhang, J., Kaur, N., et al. (2013). Enhanced osteogenesis of human mesenchymal stem cells by periodic heat shock in self-assembling peptide hydrogel. Tissue Eng. Part A 19, 716–728. doi: 10.1089/ten.tea.2012.0070
Chen, K.-L., Wang, H.-L., Jiang, L.-Z., Qian, Y., Yang, C.-X., Chang, W.-W., et al. (2020). Heat stress induces apoptosis through disruption of dynamic mitochondrial networks in dairy cow mammary epithelial cells. In Vitro Cell Dev. Biol. Anim. 56, 322–331. doi: 10.1007/s11626-020-00446-5
Chen, S.-J., Liao, D.-L., Chen, C.-H., Wang, T.-Y., and Chen, K.-C. (2019). Construction and analysis of protein-protein interaction network of heroin use disorder. Sci. Rep. 9:4980. doi: 10.1038/s41598-019-41552-z
Chen, T.-C., Kam, Y.-C., and Lin, Y.-S. (2001). Thermal Physiology and reproductive phenology of Buergeria japonica (Rhacophoridae) breeding in a stream and a geothermal hotspring in Taiwan. Zoolog. Sci. 18, 591–596. doi: 10.2108/zsj.18.591
Chhangani, D., Jana, N. R., and Mishra, A. (2013). Misfolded proteins recognition strategies of E3 ubiquitin ligases and neurodegenerative diseases. Mol. Neurobiol. 47, 302–312. doi: 10.1007/s12035-012-8351-0
Clay, T. A., Steffen, M. A., Treglia, M. L., Torres, C. D., Trujano-Alvarez, A. L., and Bonett, R. M. (2019). Multiple stressors produce differential transcriptomic patterns in a stream-dwelling salamander. BMC Genomics 20:482. doi: 10.1186/s12864-019-5814-y
Crespi, E. J., Williams, T. D., Jessop, T. S., and Delehanty, B. (2013). Life history and the ecology of stress: how do glucocorticoid hormones influence life-history variation in animals? Funct. Ecol. 27, 93–106. doi: 10.1111/1365-2435.12009
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9.
Cuesta, R., Laroia, G., and Schneider, R. J. (2000). Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 14, 1460–1470. doi: 10.1101/gad.14.12.1460
do Amaral, M. C. F., Frisbie, J., Crum, R. J., Goldstein, D. L., and Krane, C. M. (2020). Hepatic transcriptome of the freeze-tolerant Cope’s gray treefrog, Dryophytes chrysoscelis: responses to cold acclimation and freezing. BMC Genomics 21:226. doi: 10.1186/s12864-020-6602-4
Goldstein, J. A., Hoff, K., von, S., and Hillyard, S. D. (2017). The effect of temperature on development and behaviour of relict leopard frog tadpoles. Conserv. Physiol. 5:cow075. doi: 10.1093/conphys/cow075
Gormally, B. M. G., and Romero, L. M. (2020). What are you actually measuring? A review of techniques that integrate the stress response on distinct time-scales. Funct. Ecol. 34, 2030–2044. doi: 10.1111/1365-2435.13648
Gosner, K. L. (1960). A simplified table for staging anuran embryos larvae with notes on identification. Herpetologica 16, 183–190.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Haramura, T. (2004). Salinity and other abiotic characteristics of oviposition sites of the Rhacophorid Frog, Buergeria japonica, in coastal habitat. Curr. Herpetol. 23, 81–84. doi: 10.5358/hsj.23.81
Hershko, A. (1996). The ubiquitin system for protein degradation. Biochem. Soc. Trans. 24:627S. doi: 10.1042/bst024627sc
Hilliker, A., Gao, Z., Jankowsky, E., and Parker, R. (2011). The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43, 962–972. doi: 10.1016/j.molcel.2011.08.008
Ikuta, K., Urakawa, H., Kozawa, E., Hamada, S., Ota, T., Kato, R., et al. (2015). In vivo heat-stimulus-triggered osteogenesis. Int. J. Hyperth. 31, 58–66. doi: 10.3109/02656736.2014.988662
Kiss, A. J., Muir, T. J., Lee, R. E. Jr., and Costanzo, J. P. (2011). Seasonal variation in the hepatoproteome of the dehydration- and freeze-tolerant wood frog, Rana sylvatica. Int. J. Mol. Sci. 12, 8406–8414. doi: 10.3390/ijms12128406
Komaki, S., Lau, Q., and Igawa, T. (2016b). Living in a Japanese onsen: field observations and physiological measurements of hot spring amphibian tadpoles, Buergeria japonica. Amphibia Reptilia 37, 311–314. doi: 10.1163/15685381-00003052
Komaki, S., Igawa, T., Lin, S.-M., and Sumida, M. (2016a). Salinity and thermal tolerance of Japanese stream tree frog (Buergeria japonica) tadpoles from island populations. Herpetol. J. 26, 209–213.
Komaki, S., Shiwa, Y., Furukawa, R., Hachiya, T., Ohmomo, H., Otomo, R., et al. (2018). iMETHYL: an integrative database of human DNA methylation, gene expression, and genomic variation. Hum. Genome Var. 5:18008. doi: 10.1038/hgv.2018.8
Komaki, S., Sutoh, Y., Kobayashi, K., Saito, S., Saito, C. T., Igawa, T., et al. (2020). Hot spring frogs (Buergeria japonica) prefer cooler water to hot water. Ecol. Evol. 10, 9466–9473. doi: 10.1002/ece3.6637
Lake, D., Corrêa, S. A. L., and Müller, J. (2016). Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci. 73, 4397–4413. doi: 10.1007/s00018-016-2297-8
Maciel, T. A., and Juncá, F. A. (2009). Effects of temperature and volume of water on the growth and development of tadpoles of Pleurodema diplolister and Rhinella granulosa (Amphibia: Anura). Zoologica 26, 413–418. doi: 10.1590/S1984-46702009000300005
Mao, Y., Fisher, D. W., Yang, S., Keszycki, R. M., and Dong, H. (2020). Protein-protein interactions underlying the behavioral and psychological symptoms of dementia (BPSD) and Alzheimer’s disease. PLoS One 15:e0226021. doi: 10.1371/journal.pone.0226021
Maruyama, A., Nemoto, M., Hamasaki, T., Ishida, S., and Kuwagata, T. (2017). A water temperature simulation model for rice paddies with variable water depths. Water Resour. Res. 53, 10065–10084. doi: 10.1002/2017WR021019
Mastrangelo, A. M., Marone, D., Laidò, G., De Leonardis, A. M., and De Vita, P. (2012). Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 185–186, 40–49. doi: 10.1016/j.plantsci.2011.09.006
Morita, T. (1988). Terrestrial vertebrate fauna on Kuchinoshima Island of the Tokara Archipelago where wild cows live [transrated from Japanese title]. Rikabukaishi 30, 17–19.
Nagasawa, K., Tanizaki, Y., Okui, T., Watarai, A., Ueda, S., and Kato, T. (2013). Significant modulation of the hepatic proteome induced by exposure to low temperature in Xenopus laevis. Biol. Open 2, 1057–1069. doi: 10.1242/bio.20136106
Newton, A. H., and Pask, A. J. (2020). CHD9 upregulates RUNX2 and has a potential role in skeletal evolution. BMC Mol. Cell Biol. 21:27. doi: 10.1186/s12860-020-00270-5
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Rao, R. C., and Dou, Y. (2015). Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346. doi: 10.1038/nrc3929
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shen, W., Le, S., Li, Y., and Hu, F. (2016). SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11:e0163962. doi: 10.1371/journal.pone.0163962
Soneson, C., Love, M. I., and Robinson, M. D. (2016). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4:1521. doi: 10.12688/f1000research.7563.2
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
UniProt Consortium (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Wilkinson, K. D., Ventii, K. H., Friedrich, K. L., and Mullally, J. E. (2005). The ubiquitin signal: assembly, recognition and termination. EMBO Rep. 6, 815–820. doi: 10.1038/sj.embor.7400506
Wingfield, J. C. (2013). Ecological processes and the ecology of stress: the impacts of abiotic environmental factors. Funct. Ecol. 27, 37–44. doi: 10.1111/1365-2435.12039
Wu, C.-S., and Kam, Y.-C. (2005). Thermal tolerance and thermoregulation by Taiwanese rhacophorid tadpoles (Buergeria japonica) living in geothermal hot springs and streams. Herpetologica 61, 35–46. doi: 10.1655/04-50
Yu, T., Deuster, P., and Chen, Y. (2016). Role of dynamin-related protein 1-mediated mitochondrial fission in resistance of mouse C2C12 myoblasts to heat injury. J. Physiol. 594, 7419–7433. doi: 10.1113/JP272885
Keywords: amphibia, anura, differentially expressed genes, protein-protein interaction network, RNA-seq, high thermal tolerance, heat stress response
Citation: Komaki S, Matsunami M, Lin J-W, Lee K-H, Lin Y-P, Lee Y, Lin S-M and Igawa T (2021) Transcriptomic Changes in Hot Spring Frog Tadpoles (Buergeria otai) in Response to Heat Stress. Front. Ecol. Evol. 9:706887. doi: 10.3389/fevo.2021.706887
Received: 08 May 2021; Accepted: 08 September 2021;
Published: 13 October 2021.
Edited by:
Robert A. Haney, Ball State University, United StatesReviewed by:
Saurabh Sarkar, Guskara Mahavidyalaya, IndiaIvan Gomez-Mestre, Estación Biológica de Doñana (EBD), Spain
Copyright © 2021 Komaki, Matsunami, Lin, Lee, Lin, Lee, Lin and Igawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeshi Igawa, dGlnYXdhQGhpcm9zaGltYS11LmFjLmpw
 Shohei Komaki
Shohei Komaki Masatoshi Matsunami
Masatoshi Matsunami Jhan-Wei Lin
Jhan-Wei Lin Ko-Huan Lee4
Ko-Huan Lee4 Yen-Po Lin
Yen-Po Lin Takeshi Igawa
Takeshi Igawa