- 1Department of Clinical Laboratory Medicine, The First Affiliated Hospital of Shandong First Medical University and Shandong Provincial Qianfoshan Hospital, Jinan, China
- 2Department of Pathogen Biology, School of Clinical and Basic Medical Sciences, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
Influenza pandemics occur annually, highlighting the importance of understanding the influenza A virus (IAV) in the context of orthomyxoviruses and virology as a whole. However, many undergraduate students find the complex knowledge surrounding virology challenging to grasp., particularly the three-dimensional structure and dynamic processes of IAV. To address this issue, we developed a tangible, interactive, and cost-effective model using ultra-lightweight paper clay. This model visualizes the three-dimensional structure of IAV, its life cycle, antigenic variation, and the mechanisms underlying vaccines and antiviral drugs. Key features include a magnet-enhanced design that replicates molecular interactions, a reflective inner wall simulating viral infection dynamics, and reversible components illustrating processes such as replication and antigenic transitions. By engaging multiple senses, this hands-on model enhances comprehension, facilitates memory retention, and provides an accessible learning experience. Through improved virology literacy, students can better understand IAV and even disseminate accurate information to the general public. This model has received positive student feedback, demonstrating its potential for integration into virology curricula.
1 Introduction
Influenza pandemics occur annually, underscoring the need for a comprehensive understanding of influenza A virus (IAV), a key member of the Orthomyxoviridae family (Krammer et al., 2018). Knowledge of IAV is essential not only for studying this viral family but also for advancing virology, a field increasingly pivotal to modern science (Kushner and Pekosz, 2021). However, the complexity of virology concepts, particularly the intricate microscopic structures of viruses, often poses challenges in educational settings. Traditional lectures typically rely on static visuals, which are insufficient for demonstrating the dynamic and three-dimensional nature of viruses.
To enhance microbiology education, tools such as visual aids, animations, and virtual lab simulations have been introduced, showing promise in improving engagement and comprehension (Stuckey and Stuckey, 2007; Sweeney et al., 2022). Yet, these tools are not without limitations, as some students struggle to adapt to new technology-based formats, encounter a lack of session continuity, or face technological constraints (Wood, 2009). To address these issues, we developed a hands-on, interactive, and cost-effective teaching model constructed from ultra-lightweight paper clay. This tangible IAV model integrates visual, tactile, and interactive elements, fostering a multi-modal approach that promotes effective learning and longer-lasting memory retention. Constructed with two hemispheres and interchangeable accessories, the model incorporates strategically placed magnets to simulate molecular interactions, enabling demonstrations of key virology concepts, including viral morphology, replication cycles, mutation processes, and mechanisms of antiviral drugs. By combining interactive and visual elements, this model provides a hands-on learning experience, helping students grasp complex concepts, deepen their understanding of virology, and foster stronger engagement with the subject matter.
2 Method and results
2.1 Overview
The outer casing, an integral component of the model, features an inner wall coated with a mirror-reflective film (Figures 1A–C). This film visually amplifies a single virus (Figure 1D) and a small patch of plasma membrane into multiple virions and a large continuous plasma membrane, creating a realistic scenario that mimics virus-cell interactions. The bottom tray of the model represents the plasma membrane and, when flipped over, reveals a detailed visualization of the viral replication cycle (Figure 1E) (Samji, 2009; Kumar and Sakharam, 2024) and antigenic shift processes (Figure 1E) (Zhang et al., 2023; Luczo and Spackman, 2024).

Figure 1. Overview of the tangible IAV model. (A) The full model in its protective casing. (B,C) The mirror-reflective coating represents the interaction of a virus with a localized patch of the plasma membrane. This mirrors a single virus interacting with a membrane and visualizes it as numerous virions interacting with a continuous and infinite plasma membrane surface, simulating the scenario during cellular infection. (D) A detailed close-up view of the IAV model. (E) Beneath the tray, a schematic diagram illustrating the stages of the IAV life cycle, which was adapted from the reference (Kumar and Sakharam, 2024), along with a depiction of the antigenic shift process in the H7N9 avian influenza virus, was displayed.
2.2 Key components of IAV
The core of the IAV model features a genome comprising eight RNA segments and associated proteins, including nucleoproteins (NPs) and RNA-dependent RNA polymerases (RdRps)—the latter comprising PA, PB1, and PB2 proteins (Zhu et al., 2023). Together, these proteins which are symbolized by kneaded super-light clay (Linyi Yilanzi Stationery Co., Ltd.), a type of paper clay, with designated colors, bind to the RNAs, symbolized by rolled formable iron wires, to form ribonucleoprotein (RNP) complexes (Figure 2A). The viral envelope is depicted as a two-layered structure: an inner layer of matrix protein 1 (M1) (Peukes et al., 2021) and an outer lipoprotein layer (Figure 2B). The two structures that comprise the hemisphere are both crafted from clay. There are several methods for preparing the hemispherical viral envelope, with the molding method being the most straightforward. To begin, select a sphere, such as a bowl, to serve as the mold and wrap its exterior in plastic wrap. Next, drape a rolled-out sheet of paper clay evenly over the form. Press the clay to conform to the shape of the mold, trimming any excess material along the rim. Allow the formed hemisphere to air-dry completely before gently removing it from the mold. Alternatively, if a suitable mold is not available, the assembly method can be employed. Start by cutting the flat paper clay into petal shapes (Figure 2B). The edges of these shapes are then raised and carefully joined together. Smooth the seams to create a hemisphere, which is then left to air-dry. Appropriate magnets, such as 5-mm-diameter Neodymium beads (from Pan’an County Leilei Buck Ball Toys Co., Ltd.), are embedded within the docking edges of the two hemispheres. This allows the two hemispheres to be securely fixed together using magnetic attraction, forming a spherical viral particle. Embedded within the envelope are two types of spikes: hemagglutinin (HA) (Russell et al., 2018) and neuraminidase (NA) (Wen and Wan, 2019) (Figures 2C,D). Specific holes are punched in the hemispherical envelope to facilitate the insertion of both the HA and NA proteins into their corresponding transmembrane regions, allowing for the possibility of replacing them with mutant forms. Each encoded protein spike may undergo structural changes due to viral mutations. For example, in studying the shift and drift processes of the H7N9 virus, we have created multiple models of the H9N2 IAV NA, H7N9 IAV NA, H9N2 IAV HA, mutant H9N2 IAV HA, and H7N9 IAV HA proteins using kneaded clay.
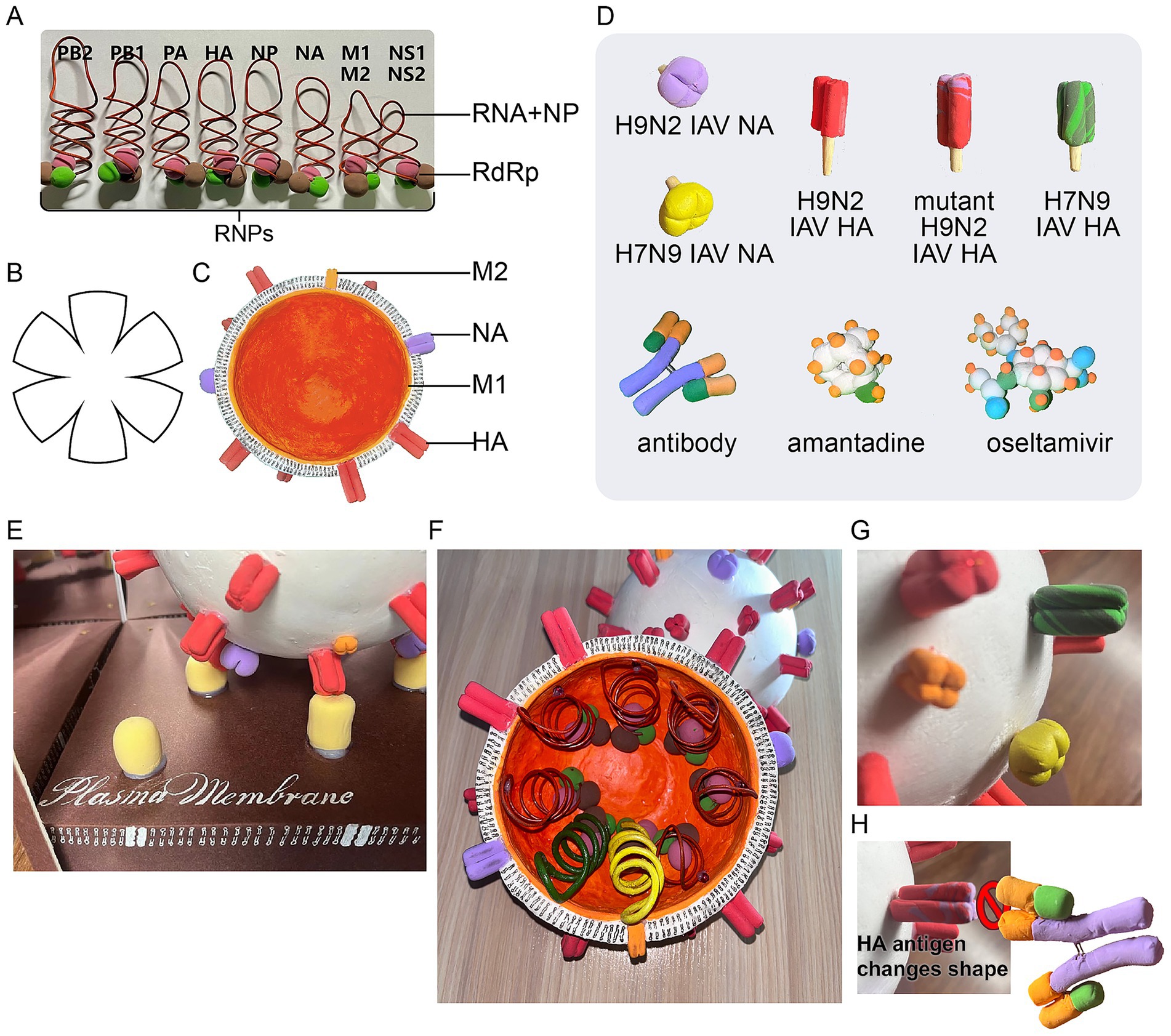
Figure 2. Key components of the IAV. (A) The RNP complex, composed of RNA segments, NPs, and RdRps. (B) The shape of the petals can serve as a template to assist in cutting out the paper clay and shaping it into a hemisphere. (C) The viral envelope, which consists of an M layer and an outer lipoprotein bilayer containing HA and NA glycoproteins. (D) Accessories included with the model: H9N2 IAV NA, H7N9 IAV NA, H9N2 IAV HA, mutant H9N2 IAV HA, H7N9 IAV HA, antibodies, amantadine, and oseltamivir. (E) The full assembled model with its protective casing. (F) Detailed components of the RNP complex in the H7N9 IAV. (G) Changes in HA and NA antigens during IAV antigenic shift. (H) Conformational changes in the HA antigen that prevent its binding to antibodies.
Beyond structural insights, the model also replicates the viral life cycle (Samji, 2009). By bringing the simulated virion close to the tray, the HA spike is shown to interact with and bind to the sialic acid receptor on the plasma membrane (via magnetic attraction), simulating the initial infection process of IAV (Figure 2E). Subsequent stages of the infection process are illuminated by diagrams displayed beneath the tray (Figure 1C). After HA binds to the sialic acid receptor, the virus undergoes invagination and is internalized into the cell. The viral matrix protein 2 (M2) ion channel then lowers the pH within the viral envelope, triggering a conformational change in HA, which facilitates fusion between the viral envelope and the plasma membrane, ultimately releasing the viral nucleocapsid into the cytoplasm (Manzoor et al., 2017). The nucleocapsid is transported to the nucleus via nuclear pores, where transcription and replication of viral RNA occur. The resulting mRNA moves to the cytoplasm, enabling the synthesis of viral proteins. These newly synthesized components then assemble into progeny viruses, which are subsequently released from the cell.
The model is especially useful for explaining the antigenic variation of IAV, which is classified into subtypes based on the unique characteristics of its HA and NA antigens. Due to the segmented nature of its genome (comprising eight RNA segments), IAV can undergo “reassortment” when two viruses from different host species co-infect the same cell. This process can produce antigenically novel strains, a phenomenon termed antigenic shift (Scholtissek, 1995). For instance, the model simulates the creation of the H7N9 avian influenza virus: a domestic duck transmits an H7N3 virus to an intermediate host (a chicken) carrying the H9N2 subtype, while a wild bird transmits a different H7N9 virus to the same chicken. The reassortment of genes from these strains results in the novel H7N9 virus (Figures 2F,G). Conversely, antigenic drift involves continuous or minor variations within a specific subtype, which can alter the structure of an antigen, subsequently reducing antibody-binding affinity and weakening the immune response (Yewdell, 2011). Both antigenic shift and drift can lead to changes in the shape of the HA antigen, which can be simulated in the model by altering the type of antigen. This modification renders pre-existing antibodies ineffective (Figure 2H) (Treanor, 2004).
2.3 Vaccines and antiviral mechanisms
Preventing influenza requires a proactive, science-based approach (Cowling and Okoli, 2024). Vaccination remains the most effective strategy. Each year, flu vaccines are tailored to include three or four strains of the virus, which may vary annually. Upon vaccination, the body generates antibodies targeting the HA antigens of the virus contained in the vaccine. These antibodies block the virus from binding to and infecting host cells (Figure 3A). The model illustrates how a viral antigen and a specific antibody attract each other through magnetic forces. However, if the HA gene mutates, the encoded antigen may also change shape, rendering previously effective antibodies unable to bind and allowing the mutated virus to infect host cells. As demonstrated in the model, the antibody stimulated by the vaccine is unable to bind to the mutant antigens, as these antigens do not incorporate the magnetic beads (Figure 2H).
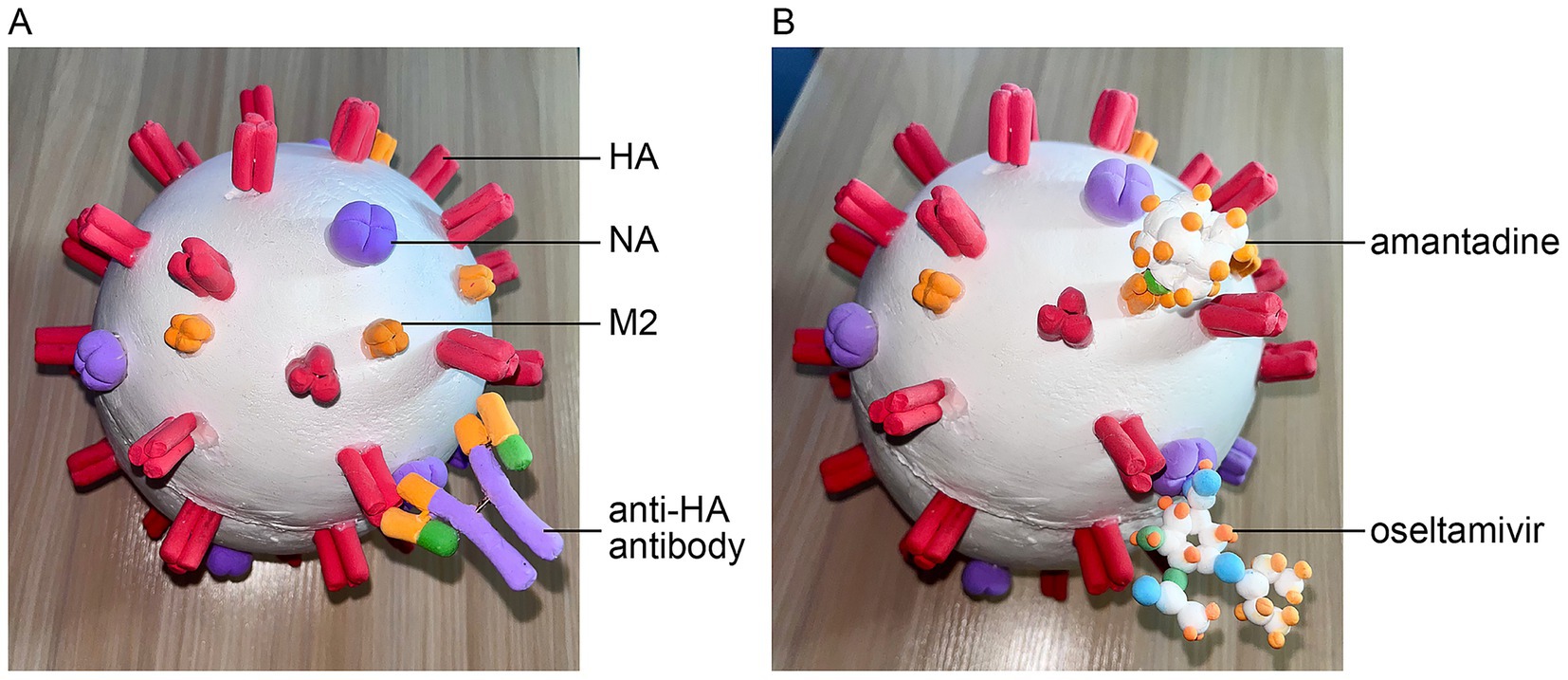
Figure 3. Antiviral mechanisms of the IAV. (A) Visualization of antibodies binding to the HA antigen of the IAV. (B) The mechanism of action for antiviral drugs: oseltamivir carboxylic acid binding to NA and amantadine blocking the M2 ion channel.
For antiviral treatment, oseltamivir is the drug of choice. Once metabolized in the body, it is converted to the active compound oseltamivir carboxylic acid, a potent and selective inhibitor of the influenza NA enzyme. By inhibiting NA, this drug prevents the release of progeny virions from infected cells, thereby curbing virus spread (Figures 1E, 3B) (Jackson et al., 2011). Similarly, amantadine blocks the M2 ion channel, preventing the release of viral nucleic acids into the cytoplasm and thereby exhibiting antiviral activity (Figures 1E, 3B) (Jackson et al., 2011). The interaction between drugs and their targets is also modeled using magnetic forces to simulate attraction.
3 Conclusion
This paper clay-based IAV model effectively simplifies complex virology concepts, including viral structure, replication, antigenic variation, and mechanisms of vaccines and antivirals, into tangible, approachable learning experiences. Lightweight, portable, and eco-friendly, it serves as a practical teaching tool. Strategically placed magnets enhance interactivity, promoting hands-on engagement for better comprehension and retention. The model was utilized in Medical Microbiology courses during the Spring 2025 semester at Shandong First Medical University, allowing students to observe the three-dimensional structure of the IAV and manipulate the viral mutation process repeatedly. Students responded positively to this hands-on approach, emphasizing the model’s potential to enhance virology education. When integrated with online resources, this interactive tool could reach a broader audience, addressing key challenges in microbiology education.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
JL: Writing – original draft, Software, Investigation. LZ: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research reported here was supported by grants from National Natural Science Foundation of China (82272306) and Taishan Scholars Program (tstp20221142).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cowling, B. J., and Okoli, G. N. (2024). Influenza vaccine effectiveness and Progress towards a universal influenza vaccine. Drugs 84, 1013–1023. doi: 10.1007/s40265-024-02083-8
Jackson, R. J., Cooper, K. L., Tappenden, P., Rees, A., Simpson, E. L., Read, R. C., et al. (2011). Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J. Infect. 62, 14–25. doi: 10.1016/j.jinf.2010.10.003
Krammer, F., Smith, G. J. D., Fouchier, R. A. M., Peiris, M., Kedzierska, K., Doherty, P. C., et al. (2018). Influenza. Nat. Rev. Dis. Primers 4:3. doi: 10.1038/s41572-018-0002-y
Kumar, G., and Sakharam, K. A. (2024). Tackling influenza a virus by M2 ion channel blockers: latest progress and limitations. Eur. J. Med. Chem. 267:116172. doi: 10.1016/j.ejmech.2024.116172
Kushner, D. B., and Pekosz, A. (2021). Virology in the classroom: current approaches and challenges to undergraduate- and graduate-level virology education. Annu Rev Virol 8, 537–558. doi: 10.1146/annurev-virology-091919-080047
Luczo, J. M., and Spackman, E. (2024). Epitopes in the HA and NA of H5 and H7 avian influenza viruses that are important for antigenic drift. FEMS Microbiol. Rev. 48:fuae014. doi: 10.1093/femsre/fuae014
Manzoor, R., Igarashi, M., and Takada, A. (2017). Influenza a virus M2 protein: roles from ingress to egress. Int. J. Mol. Sci. 18:2649. doi: 10.3390/ijms18122649
Peukes, J., Xiong, X., and Briggs, J. A. G. (2021). New structural insights into the multifunctional influenza a matrix protein 1. FEBS Lett. 595, 2535–2543. doi: 10.1002/1873-3468.14194
Russell, C. J., Hu, M., and Okda, F. A. (2018). Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 26, 841–853. doi: 10.1016/j.tim.2018.03.005
Scholtissek, C. (1995). Molecular evolution of influenza viruses. Virus Genes 11, 209–215. doi: 10.1007/bf01728660
Stuckey, T. A., and Stuckey, B. D. (2007). Virtual labs in the online biology course: Student perceptions of effectiveness and usability. MERLOT J. Online Learn. Teach. 3, 105–111.
Sweeney, M. O., Farkas, J. E., Homan, E. P., and Raytcheva, D. D. A. (2022). Customized virtual simulations provide an interactive lab experience. J Microbiol Biol Educ 23:e00331–21. doi: 10.1128/jmbe.00331-21
Treanor, J. (2004). Influenza vaccine--outmaneuvering antigenic shift and drift. N. Engl. J. Med. 350, 218–220. doi: 10.1056/NEJMp038238
Wen, F., and Wan, X. F. (2019). Influenza neuraminidase: underrated role in receptor binding. Trends Microbiol. 27, 477–479. doi: 10.1016/j.tim.2019.03.001
Wood, W. B. (2009). Innovations in teaching undergraduate biology and why we need them. Annu. Rev. Cell Dev. Biol. 25, 93–112. doi: 10.1146/annurev.cellbio.24.110707.175306
Yewdell, J. W. (2011). Viva la revolución: rethinking influenza a virus antigenic drift. Curr. Opin. Virol. 1, 177–183. doi: 10.1016/j.coviro.2011.05.005
Zhang, N., Quan, K., Chen, Z., Hu, Q., Nie, M., Xu, N., et al. (2023). The emergence of new antigen branches of H9N2 avian influenza virus in China due to antigenic drift on hemagglutinin through antibody escape at immunodominant sites. Emerg Microbes Infect 12:2246582. doi: 10.1080/22221751.2023.2246582
Keywords: influenza A virus, interactive teaching model, antigenic variation, antiviral, microbiology education
Citation: Li J and Zhang L (2025) A handmade influenza A virus teaching model for enhancing virology education. Front. Educ. 10:1660649. doi: 10.3389/feduc.2025.1660649
Edited by:
Dina Tavares, Polytechnic University of Leiria, PortugalReviewed by:
Amanda J. Chase, Nova Southeastern University, United StatesStacy Hrizo, Slippery Rock University of Pennsylvania, United States
Copyright © 2025 Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leiliang Zhang, YXJtemhhbmdAaG90bWFpbC5jb20=
†ORCID: Leiliang Zhang, orcid.org/0000-0002-7015-9661
 Jincheng Li
Jincheng Li Leiliang Zhang
Leiliang Zhang