Abstract
Microalgae are potential sources for the sustainable production of valuable chemicals including polyphenols, pigments, and ω-3 PUFAs. However, successful exploitation of these high value compounds in the food, healthcare and pharmaceutical sectors depends greatly on their effective separation, identification, and analysis after recovery from the biomass. The findings of this review paper illustrated that chromatographic methods coupled to different types of detectors have been used as a crucial part of research on microalgal polyphenols, Omega-3 Polyunsaturated Fatty Acids (ω-3 PUFAs), and pigments production through identification, measurement, sample preparation, and purification practices. Therefore, it is important to provide a comprehensive review regarding the current research in the field. The basic operating principles, parametric optimisation and detection units of common (liquid chromatography and gas chromatography) and novel chromatographic techniques (counter current chromatography, expanded bed adsorption chromatography and supercritical fluid chromatography) used to separate, identify, and quantify polyphenols, PUFAs and pigments from microalgae matrices are comprehensively reviewed.
1 Introduction: Chromatographic Separation of Microalgal Bioproducts
Microalgae are eukaryotic photosynthetic microorganisms with a variety of species living in a diverse range of ecological circumstances such as freshwater, seawater, soil, hot spring, etc. Over 50,000 species of microalgae are present, but only around 30,000 of them have been identified and analyzed (Richmond and Hu, 2013). Microalgae have attracted significant industry and academic attention over the past few decades because of their wide range of potential commercial applications. However, there are still major limitations for the large-scale production of microalgae and their derivatives (Dębowski et al., 2020; Mohsenpour et al., 2021). Some industrially relevant algae species include Chlorella sp, Dunaliella sp., Nannochloropsis sp. and Haematococcus sp, and the filamentous cyanobacteria Arthrospira sp (traditional name: Spirulina sp.) (Buono et al., 2014). Microalgae can be used as a primary source of feed/food, biofuels, fertilizer, plant growth promoters, biopesticides, antibiotics and medicines (Halim and Danquah, 2012; Halim et al., 2012, 2019; Adetunji et al., 2022; Fernandes et al., 2022). As biological factories, microalgae can also purify wastewaters and fix CO2 in a sustainable manner (Adetunji et al., 2022; Fernandes et al., 2022; Goswami et al., 2022; Xie et al., 2022). In addition, microalgae are a good source of plant-based proteins and high-value nutraceutical compounds that can promote human health (Miranda et al., 1998; Herrero et al., 2006; Rea et al., 2011; Schwenzfeier et al., 2011; Caporgno and Mathys, 2018; Olasehinde et al., 2019; Gill et al., 2020; Ashaolu et al., 2021).
The surge in demand for natural health promoting ingredients from the pharmaceuticals and food industries has driven the recent increase in the exploration for powerful bioactive ingredients in microalgae (Sansone and Brunet, 2019). The manifold range of different healthy products, such as polyunsaturated fatty acids, polyphenols, proteins, carotenoids, pigments, sterols, and vitamins accessible from different microalgal species, clearly demonstrates a wide range of applications and importance of these versatile microbes as cellular factories in biotechnology (Gill et al., 2020; Singh et al., 2020). A summary of different categories of valuable ingredients produced by microalgae cells is depicted in Figure 1. The bioactivity of a compound (e.g., antioxidative, antihypertensive or anticarcinogenic activities) can be evaluated in vivo, ex vivo, and in vitro (Caporgno and Mathys, 2018). But in order to fully assess the health benefits of these compounds, effective extraction, purification, and separation methods are needed. Quantification of microalgal metabolites has often been carried out spectrophotometrically, but these methods lack specificity and should only be used for preliminary testing. Chromatographic methods are regarded as the gold standard for the separation and quantification of microalgal metabolites. Despite this, few studies have exclusively investigated the development of chromatographic methods for high-resolution separation of microalgal products. In most microalgal studies, little consideration has been given to the development and optimization of chromatographic separation, with most simply adapting known methods from plant and microbial matrices for their analysis. This lack of attention on chromatography can often result in poor-quality analysis which is not only restrictive to the scope of the concerned microalgal studies but also harmful to the advancement of microalgae product development as future studies emulate methods which have not been specifically tailored/optimised for microalgae.
FIGURE 1
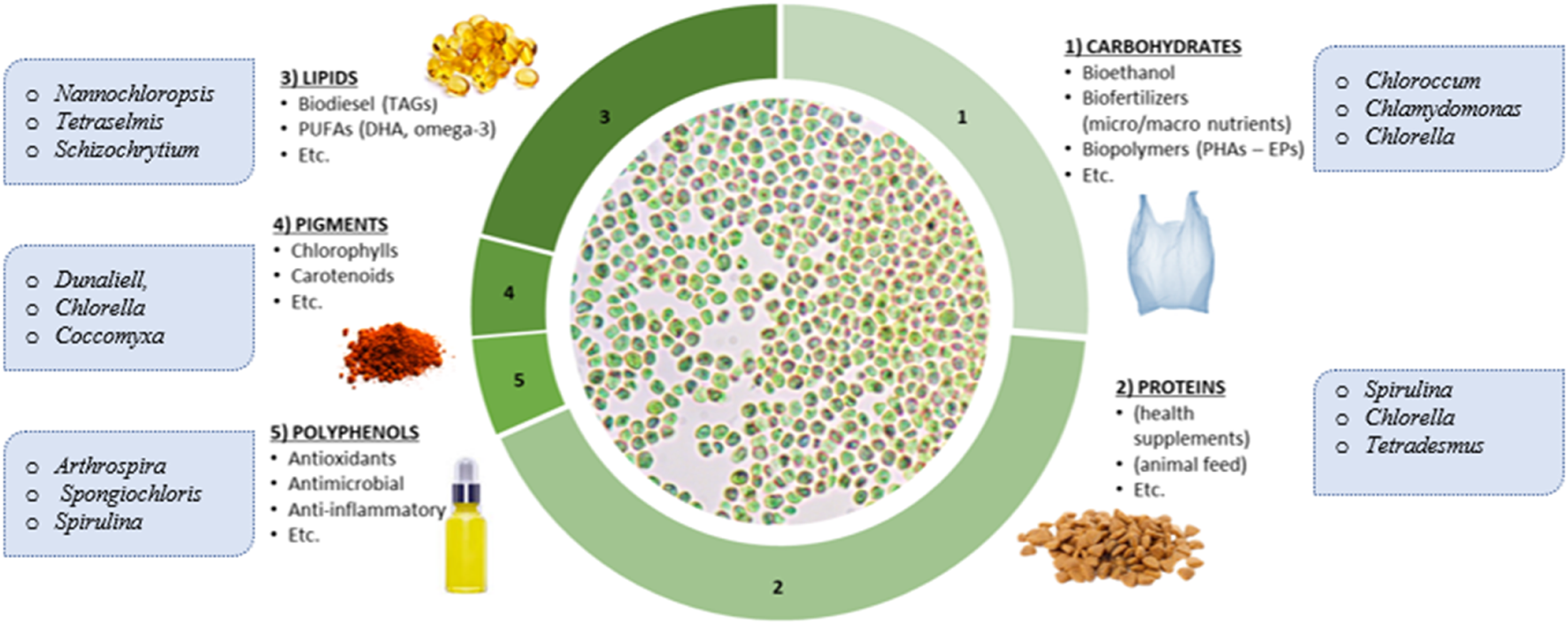
The range of valuable ingredients that can be derived from microalgae.
Therefore, in this paper, a comprehensive review of different chromatographic techniques employed for the separation and analysis of microalgal bioactive ingredients is provided. The three groups of compounds reviewed in the review (polyphenols, ω-3 PUFAs, and pigments) are well-known health-promoting agents and have mainly been selected for their, commercial significance as higher-value products from microalgae, current commercial demand and limited supply sources, and their ubiquitous presence across different industrially promising microalgae genera (e.g., Spirulina, Chlorella, Phaeodactylum, Dunaliella, Nannochloropsis, Scenedesmus, and Schizochrytium). For each group of compounds, this review provided a brief description of their composition in microalgae, biochemical properties, extraction affinity and recovery methods before discussing appropriate chromatographic techniques for their identification and quantification. The discussions will include choice of stationary and mobile phases, detection units, and chromatographic parameters.
1.1 Main Principles for the Separation and Identification of Different Algal Bioproducts
Chromatography is a separation technique widely used for the identification and/or purification purposes in different research and industrial sectors. The main principle behind this separation technique is the difference between compounds in terms of their interaction with two phases, one fixed stationary phase and another moving mobile phase (Tarafder and Miller, 2021; Kumari et al., 2022). Chromatography is classified based on the type of the stationary and mobile phases used as well as the separation apparatus working mode. A summary of different chromatography methods are summarized in Table 1.
TABLE 1
| Method | Sub-categ | Governing principles | Detector units | Application |
|---|---|---|---|---|
| Liquid chromatography (LC and/or HPLC) | Ion-exchange | Ionic interaction with both stationary and mobile phases. Mobile phase is changed during separation | Conductivity, Light emitting detectors, Photo diode array (PDA), Amperometric, potentiometric, Mass spectrometer (MS) | Metal elimination and water treatment; Wilde and Benemann, (1993); Sandau et al. (1996); McSheehy and Szpunar, (2000); Monteiro et al. (2012); Jiang et al. (2020); Wang et al., 2021), Protein analysis and purification (Schwenzfeier et al., 2011; Al-Zuhair et al. (2017); Silva et al. (2019) |
| Normal–phase | Polar stationary phase, non-polar mobile phase | MS, UV, Fluorescence, Refractive index, Conductivity, Evaporative light scattering | low-molecular or high-molecular polar compounds, such as peptides, proteins, oligosaccharides, and metabolites; Do Nguyen et al. (2017); Moreau et al. (2018); Differentiate between classes of lipids and sterols Donot et al. (2016); Fagundes et al. (2020) | |
| Reversed-phase | Non-polar stationary phase, polar mobile phase | Different organic compounds including acids, sugars, pigments, vitamins, polyphenols, low chain fatty acids, polar lipids, volatile compounds, amino acids, flavonoids; Stein et al. (1992); Kroumova and Wagner, (1995); Derrien et al. (1998); Goiris et al. (2014); Do Nguyen et al. (2017); Soares et al. (2019); Fischer et al. (2021) | ||
| Size-exclusion | Separation based on size or weight, flow through a packed column containing beads of specific polymers | Extracellular polysaccharides; Gómez-Ordóñez et al. (2012); Dogra et al. (2017); González-Hourcade et al. (2020); active biomolecules such as phytochelatins and phlorotannins; Scarano and Morelli, (2002); Zhou et al. (2019); and othe small proteins and peptides; Morelli and Scarano, (2004) | ||
| Affinity | Separation according to specific interaction between molecules and specific substance in the column, a highly selective method | Different polysaccharides; Fredrick, (1975); Hahn et al. (2016); lectins; Pajic et al. (2002); Hori et al. (2007); Range of proteins; Bielser et al. (2021) | ||
| Paper chromatography | A relatively old method, mostly replaced by other new techniques, colored molecules travel along with the solvent through a paper | Flavonoids; Birch et al. (1953); Markham and Porter, (1969);, Chlorophyll (Jeffrey, 1961);, cytokinin; Stirk et al. (2002);, other pigments | ||
| Thin-layer chromatography (TLC) | Thin layer of absorbent deposited onto a sheet, mobile phase travels by capillary action. Separation performed based on solubility in solvent | A wide range of chemicals including algal pigments; Sathya, (2017); Peraman and Nachimuthu, (2019);, phenolic compounds; Akbarizareh et al. (2019);, steroids; Megawati et al. (2020);, lipid analysis; Jouhet et al. (2017); Lage and Gentili, (2018); Viegas et al. (2020) | ||
| Gas chromatography (GC) | Mobile phase is a heated gas | MS, Flame ionization (FID), Flame photometric (FPD), Atomic emission, Thermal conductivity, Electron capture, Photo ionization, Olfactometric | Lipids and fat soluble compounds; Moazami et al. (2011); Halim et al. (2012); Law et al. (2018);, such as fatty acid profile, volatile compounds; Alalayah et al. (2017); Hrebien et al. (2020); Olander et al., 2021) dimethylsulphoniopropionate; Archer et al. (2011);, several other compounds; Guidetti Vendruscolo et al. (2019) | |
| Supercritical fluid chromatography (SFC) | Fluids in their supercritical region are used (Temperature and pressure controlled) | FID, FPD | Carotenoids; Matsubara et al. (2009); Abrahamsson et al. (2012); Jumaah et al. (2016);, lipid analysis and lipidomics; Bernal et al. (2013); Laboureur et al. (2015); Chollet et al. (2019);, other specific metabolite; Herrero et al. (2006) |
summary of different techniques and the governing principles of different chromatography methods used for microalgae ingredients analysis.
In general, High Performance Liquid Chromatography (HPLC) and Gas Chromatography (GC) techniques are the most widely used methods in the field of microalgae research. The chromatography method is able to deliberately separate different compounds from each other in a mixture, but a complementary technique is required for the detection and/or quantification of substances. A wide range of methods have been coupled to chromatographic separation instruments for the analysis of different compounds. The choice of the method largely depends on the chemical structure of the target compounds as well as their concentrations. Table 1 provides a summary of selected detection methods. More information regarding the governing principles together with application of these methods will be discussed in the following sections.
2 Polyphenols
2.1 Microalgae as a Source of Polyphenols and Recovery of Polyphenols From Biomass
Polyphenols are a structurally diverse group of organic compounds that contain at least one aromatic ring with one or more hydroxyl groups attached (Rein et al., 2013). As described in Figure 2, polyphenols can be divided into several classes depending on their structure: phenolic acids, flavonoids and non-flavonoids. Phenolic acids are carboxylic acids derived from either benzoic or cinnamic acids (Rentzsch et al., 2009). Flavonoids can be subdivided into different subgroups depending on the degree of unsaturation and oxidation of the C ring, currently there are about 6000 flavonoids (Panche et al., 2016). Non-flavonoids, which include stilbenes, tannins, lignans, among others.
FIGURE 2
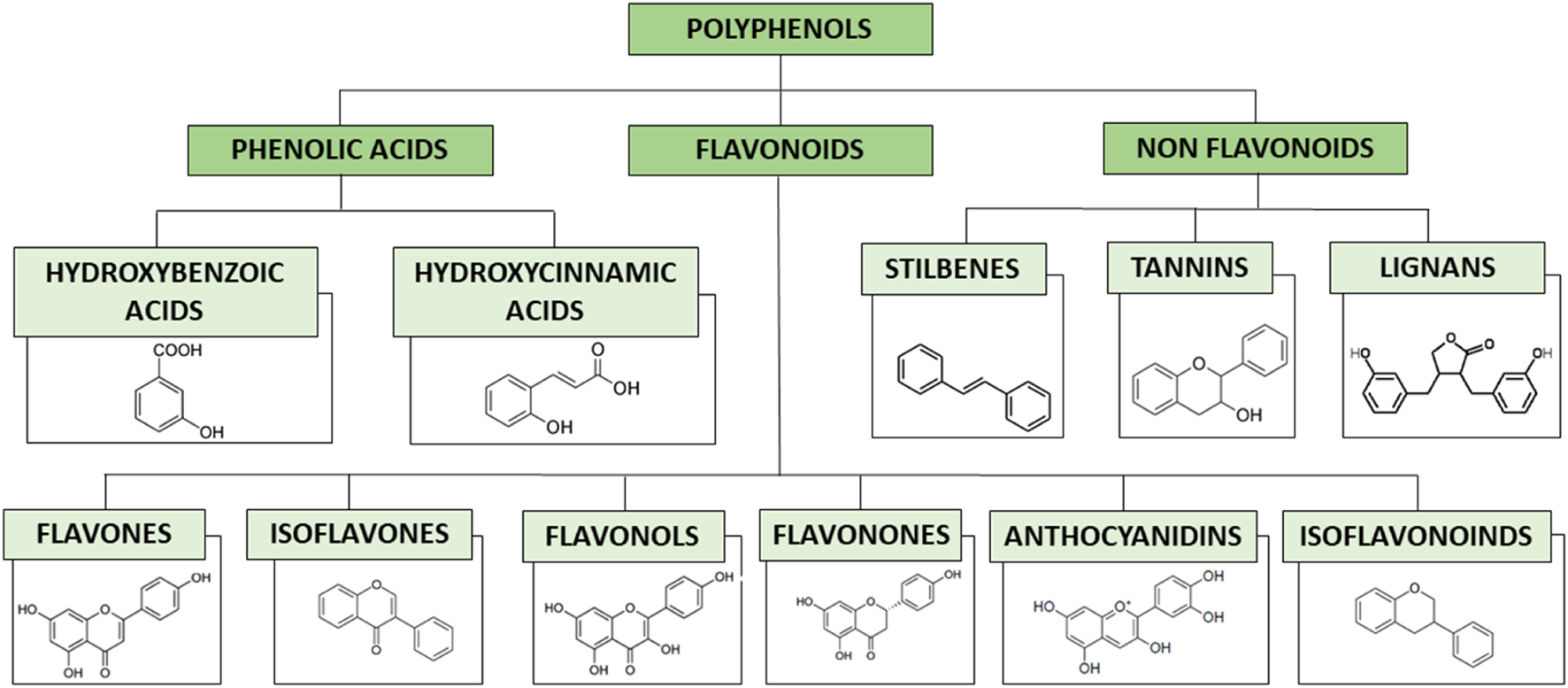
Classification of polyphenols with characteristic structures of each group.
Microalgae contain different types of polyphenols (Jerez-Martel et al., 2017). This diversity can be partly explained due to the different mechanism of adaptation that microalgae have been through, as they can be found in very extreme conditions (soils, ice, lakes, rivers, hot springs, and oceans, anywhere sunlight and water co-occur). The polyphenolic profile of Euglena cantabrica, for example, is comprised of gallic acid, protocatechuic acid, catechin, chlorogenic acid, and epicatechin (Jerez-Martel et al., 2017; Anwer et al., 2022; Sonkamble and Wagh, 2022). The main function of polyphenols in microalgae is to protect the cells from biotic and abiotic stresses and act as UV filters (Panche et al., 2016).
In humans, polyphenols serve as strong antioxidants that complement the functions of vitamins and enzymes as a defence against oxidative stress caused by excess reactive oxygen species (ROS). They can offer preventative measures against diseases, such as coronary heart disease, stroke, and cancers, and work as health-promoters with pharmacological properties, especially anti-inflammatory, antioxidant, antimutagenic and anticarcinogenic activities (Goleniowski et al., 2013; Panche et al., 2016). Although most of the evidence of the antioxidant activity of polyphenols is based on in vitro studies, increasing number of studies have indicated that they may have similar antioxidant functions in vivo (Tsao, 2010; Ajila et al., 2011; Mousavi et al., 2011; Goleniowski et al., 2013; Panche et al., 2016; Gu et al., 2019; Yu et al., 2020; Shen et al., 2022).
Being polar compounds, phenolic acids are generally extracted using hot or cold water while flavonoids are extracted using a mixture of water and organic solvents. However, for quantitative extraction of polyphenols, studies have shown water/methanol, water/ethanol or water/acetone solvent combinations with a trace of acid (acetic acid, formic acid or hydrochloric acids) to be the most effective solvents (Barkia et al., 2019). A recent study comparing the antioxidant capacities of different extracts (hexane, ethyl acetate and water) from 23 different microalgae showed that hexane (a highly non-polar solvent) obtained the highest extraction yield of antioxidant compounds from Chlamydomonas nivalis, Anabaena flos-aquae FACHB 245, Chlorella pyrenoidosa (Li et al., 2006). Such findings highlighted the need to select an optimized extraction solvent mixture based on solvent affinity with not only the target compounds but also the individual biomass matrices (Stalikas, 2007).
Due to increasing demand and future perspective uses for polyphenol extracts in the food and pharmaceutical sectors, green extraction methods such as ultrasound-assisted extraction (UAE), pressurized-liquid extraction (PLE), pulsed-electric field-assisted extraction, and supercritical fluid extraction using different green solvents (e.g., water) (Ojha et al., 2020) are fast gaining importance. A review on the best techniques to extract polyphenols from several complex matrices (including microalgae) found matrix solid phase dispersion (MSPD), microwave-assisted extraction (MAE) and supercritical fluid extraction (SFE) to be the most suitable extraction methods (Stalikas, 2007).
Since polyphenols are stable at low pH values, extraction processes are often carried out under mild acidic conditions to enhance solubilisation and increase yields (Friedman and Jürgens, 2000). Most polyphenols exist naturally in the free form. On the other hand, phenolic acids such as ferulic acid and lignans are often bound to structural materials with complex glycosylation patterns. Excessive use of acid, however, may cause hydrolysis of glycosides giving a different polyphenol profile than the original profile.
2.2 Identification and Quantification of Polyphenols
It is important to identify and quantify different polyphenol compounds found in microalgae for different purposes. The exact bioactive role played by each phytochemical in microalgae is not understood yet, mainly due to the lack of the use of advanced analytical techniques (Caporgno and Mathys, 2018; Cardoso et al., 2020). Moreover, little is known about the nature of polyphenolic compounds that are present in microalgae, since only a few articles have assessed the complete profile of phenolic compounds using chromatography methods (Saadaoui et al., 2020).
2.2.1 Spectrophotometry
Spectrophotometric methods are commonly used for the estimation of Total Phenolic Content (TPC) and Total Flavonoid Content (TFC) in microalgal polyphenol extracts (Haoujar et al., 2019). While spectrophotometric methods are rapid and simple, they lack the specificity for individual phenolic compounds. Interferences from non-polyphenolic components can also cause false readings and thus lead to erroneous results. For instance, some amino acids, metal ions and other reducing substances present in the sample have reducing powers and therefore can reduce chemical indicators such as DPPH (Tsao, 2010; Shahidi and Yeo, 2020). These methods should therefore only be used as a preliminary assessment of phenolic contents prior to more thorough chromatographic evaluation (Panche et al., 2016; Shahidi and Yeo, 2020).
2.2.2 High-Performance Liquid-Chromatography: Applications and Optimization
High-Performance Liquid-Chromatography (HPLC) can be used for the separation, identification and quantification of phenolic compounds recovered from complex matrices such as microalgae. As can be seen in Table 2, several studies have used HPLC to analyse the phenolic composition of microalgal biomass. Considering the relatively non-polar nature of phenolic compounds, reversed-phase columns (C-18) are the most used stationary phase for polyphenol chromatography. Separation of polyphenols is usually carried out in a gradient mode using a multi-component mobile phase (or eluant) constituting an aqueous buffer and an organic solvent (Matuszewski et al., 2003; Pitt, 2009; Machu et al., 2015; Safafar et al., 2015; Jerez-Martel et al., 2017; Papalia et al., 2019; Cardoso et al., 2020; Singh et al., 2020; Dahmen-Ben Moussa et al., 2022). The most commonly used organic solvent is methanol or acetonitrile. The mobile phase is often blended with a weak acid like formic acid, acetic acid, or trifluoroacetic acid to improve the retention, separation and peak shape by suppressing the ionization of surface silinol groups present in the stationary phase (Table 2) (Onofrejová et al., 2010; Safafar et al., 2015; Jerez-Martel et al., 2017; Papalia et al., 2019; Cardoso et al., 2020; Dahmen-Ben Moussa et al., 2022). Special attention must be paid to the pH of the mobile phase as extremely low pH values can lead to the hydrolysis of glycosidic polyphenols. Recently, researchers have also investigated Ultra-High Performance Liquid Chromatography (UHPLC) systems as it offers better resolution and sensitivity, while dramatically reducing solvent consumption and analysis time (Matuszewski et al., 2003; Pitt, 2009; Cutignano et al., 2016; Kukula-Koch et al., 2016; Castillo et al., 2022; Cinq-Mars et al., 2022).
TABLE 2
| Microalgae species | Stationary phase/mobile phase/detection unit | HPLC parameters | Compounds | Ref |
|---|---|---|---|---|
| Arthrospira platensis | RP Acclaim TM 120 C18 column (4.6 × 250 mm, 5 µm)/1% acetic acid solution (eluant A) and acetonitrile (eluant B)/diode array as the detector (DAD) | Flow rate: 0.7 ml/min, column T: 28°C, injection volume: 20 µL. Elution gradient:10–40% B in a linear fashion for 28 min, 40–60% B for 39 min, 60–90% B for 50 min | Gallic acid, catechin, caffeic acid, p-hydroxybenzoic acid, p-coumaric acid, ferulic acid, quercetin, genistein and kaempferol | Papalia et al. (2019) |
| Ankistrodesmus sp., Spirogyra sp., Euglena cantabrica, and Caespitella pascheri | RP Pursuit XRs C18 column (250 mm × 4.6 mm, 5 μm) and/Water enriched with 0.1% formic acid (eluant A) and methanol (eluant B)/DAD | Flow rate: 1.0 ml/min, injection volume: 20 μL, Elution gradient: 15–40% B for 13 min, 40% B kept isocratically for 5 min | Gallic, syringic, protocatechuic, and chlorogenic acids and the flavonoids (+) catechin and (-) epicatechin | Jerez-Martel et al. (2017) |
| Chlorella pyrenoidosa and Spirulina platensis | C18 Kinetex column (150 mm × 4.6 mm, i.d. 2.6 µm)/water–acetic acid (99:1, v/v) (eluant A) and water-acetonitrile-acetic acid (67:32:1, v/v/v) (eluant B)/DAD | Flow rate: 1 ml/min, injection volume: 10 μL at 23°C, elution gradient: 0–10 min: 90% A + 10% B, 10–16 min: 80% A + 20% B, 16–20 min: 60% A + 40% B, 20–25 min: 50% A + 50% B, 25–27 min: 60% A + 40% B, 27–35 min: 90% A + 10% B | Gallic acid; 4-hydroxybenzoic acid; catechin hydrate; epicatechin; catechin gallate; epigallocatechin; epigallocatechin gallate and pyrocatechol | Machu et al. (2015) |
| Phaeodactylum sp., Nannochloropsis sp., Chlorella sp., Dunaniella sp., and Desmodesmus sp | Prodigy ODS-3 column (250 mm × 46 mm, 5 µm particle)/phosphoric acid in de-ionized water, pH = 3 (eluant A) and acetonitrile (eluant B)/DAD | Flow rate: 0.9 ml/min, injection volume: 20 µL,. Elution gradient: 5% of B for 2 min, 5—40% B for 20 min, 40—100% B for 15 min and finally kept constant for 25 min | Gallic acid, 2,5 dihydroxy benzoic acid, 3,4 dihydroxy benzoic acid, caffeic, ferulic p-coumaric, salicylic and cinnamic acid | Safafar et al. (2015) |
| Spongiochloris spongiosa,; Spirulina platensis,; Anabaena doliolum,; Chlorella vulgaris, Nostoc 16 | Zorbax SB—C 18 column (75 mm × 4.6 mm, 3.5 μm)/0.2% acetic acid (eluant A) and acetonitrile (eluant B)/DAD | Flow rate: 1.1 ml/min, column T: 30°C, elution gradient: a linear gradient was applied, 0–4% B for 4 min, 4–17% B for 2 min, 17–30% B for 4 min, a decrease 30 to 4% B for 5 min | Gallic acid, protocatechuic acid, p-OH-benzoic acid, 3,4-diOHbenzaldehyde, chlorogenic acid, vanillic, caffeic acid, syringic acid, p-OH benzaldehyde, p-coumaric acid and ferulic acid | Klejdus et al. (2013) |
| Spongiochloris spongiosa | Zorbax SB-C18 column (150 mm × 4.6 mm, 3.5 µm)/0.2% acetic acidaq (eluant A) and acetonitrile (eluant B)/DAD and MS detector operated in a negative ESI mode | Flow rate: 1.1 ml/min, column T: 30°C, injection volume: 25µL, elution gradient: 0—4 min (4% B), 4—6 min (17% B), 6—10 min (30% B), and 10—15 min (4% B) | Protocatechuic acid, p-hydroxybenzoic acid, 2,3-dihydroxybenzoic acid, chlorogenic acid, vanillic acid, caffeic acid, p-coumaric acid, salicylic acid, cinnamic acid, p-hydroxybenzaldehyde, 3,4-dihydroxybenzaldehyde acid and vanillin | Onofrejová et al. (2010) |
| Nannochloris sp, T. suecica, P. tricornutum, and N. gaditana | Ascentis Express RP C18 column (150 mm × 4.6 mm, 2.7 µm,)/water/acetic acid (99.85/0.15v/v) (eluant A) and acetonitrile (eluant B)/PDA–(ESI)–MS | Flow rate: 1 ml/min, elution gradient: 0–5 min (5% B), 5–15 min (10% B), 15–30 min (20% B), 30–60 min (50% B), 60 min (100% B) | Protocatechuic acid, caffeic acid, quinic acid, catechin, kaempferol, quercetinetc. | Haoujar et al. (2019) |
| Spongiochloris spongiosa and Spirulina platensis | Zorbax SB–C18 column (75 mm × 4.6 mm, 3.5 µm)/0.2% acetic acid (eluant A) and acetonitrile (eluant B)/MS | Flow rate: 1.1 ml/min, column T: 30°C, elution gradient: linear, 0—4 min (4% B), 4—6 min (17% B), 6—10 min (30% B), and 10—15 min (4% B) | Benzoic acid derivatives, hydroxybenzaldehydes and cinnamic acid derivatives | Klejdus et al. (2009) |
| Cyanobacteria, Rhodophyta, Chlorophyta, Haptophyta and Ochrophyta | BEH-shield RP18 (3.0 × 150 mm, 1.7 µm)/water + 0.1% formic acid (eluant A) and acetonitrile + 0.1% formic acid (eluant B)/MS/MS | Flow rate: 0.5 ml/min, column T: 40°C, elution gradient: 0–26% B for 9.9 min, 26–65% B from 9.9 to 18.5 min, 65–100% B from 18.5 to 18.8 min and held constant at 100% B from 18.8 to 20.8 min | Several flavonoids such us quercetin and Kaempferol | Goiris et al. (2014) |
| Tetraselmis sp. IMP3, Tetraselmis sp. CTP4, Skeletonema sp | Mediterranea Sea18 column (15 × 0.21 cm, 5 μm)/methanol (eluant A) and 2.5% acetic acid aqueous solution (eluant B)/HPLC-DAD | Flow rate: 0.35 ml/min, injection volume: 20 μL, Elution gradient: 0–5 min (10% A), 5–10 min (10–30% A), 10–40 min (30–90% A), 40–45 min (90% A), 45–55 min (90–10% A), and 55–60 min (10% A) | Gallic acid, gentisic acid, catechin hydrate, caffeic acid, epicatechin, salicylic acid, luteolin-7-o-glucoside, and quercetin were the most abundant compounds studied | Cardoso et al. (2020) |
HPLC instrumentation and parameters used in recent microalgal studies for polyphenol analysis.
HPLC can be coupled to different detectors to identify and quantify polyphenols. The most commonly used detectors for polyphenols analysis are ultraviolet detector (UV/Vis) and diode array detectors (DAD) as can be seen in Table 1. Processing of chromatographic data obtained by HPLC-UV/Vis or HPLC-DAD is rather simple. It should, however, be noted that the identification of polyphenols using a fixed wavelength UV/Vis detector is prone to uncertainties even in the presence of standards, as there is always a risk of co-elution of other absorbing species which cannot be resolved by the detector (Matuszewski et al., 2003; Pitt, 2009; Do Nguyen et al., 2017; Soares et al., 2019; Fischer et al., 2021; Dahmen-Ben Moussa et al., 2022; Maalej et al., 2022). On the other hand, the DAD is capable of acquiring UV-vis spectra (190—800 nm) of analytes simultaneously, which aid in subsequent identification of the analytes (Ainsworth and Gillespie, 2007; Dahmen-Ben Moussa et al., 2022; Maalej et al., 2022). Overall, it is important to compare the retention time, peak shape and the UV spectra of a polyphenol by HPLC-DAD with those of a standard to confirm identity.
HPLC coupled with mass spectrometer (HPLC-MS) is also widely used for the identification and characterization of polyphenols due to its capability in determining the molecular mass and structural information of unknown compounds (Carrasco-Pancorbo et al., 2007; Zhu et al., 2008; Pitt, 2009; Gu et al., 2019; Hu et al., 2019; Peng et al., 2019). HPLC-MS analysis is often several hundred-fold more sensitive than HPLC-UV/Vis methods, thereby facilitating trace-level identification and quantification of phenolic compounds present in complex samples (Matuszewski et al., 2003; Pitt, 2009; Peng et al., 2019). HPLC system is connected to MS through different interfaces, where analytes are ionized prior to reaching the mass analyzer. The two well-known ionization techniques include electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI). Amongst several MS systems available on the market, ESI-MS is regarded as the most versatile ionization technique and is mainly preferred for the detection of ionic and polar compounds (Ajila et al., 2011). Therefore, polyphenols are commonly detected and identified using HPLC-ESI-MS systems. In this regard, several studies have been published using a ESI-MS detector to quantify polyphenols in microalgae (Guillaumie et al., 2006; Carrasco-Pancorbo et al., 2007; Zhu et al., 2008; Klejdus et al., 2009; Onofrejová et al., 2010; Wybraniec et al., 2013; Haoujar et al., 2019; Peng et al., 2019).
UHPLC coupled to tandem mass spectrometry (MS/MS) detector has recently been used to detect flavonoids in microalgae from different evolutionary lineages (Goiris et al., 2014; Cutignano et al., 2016). UHPLC often operates at low flow rates (0.2–0.6 ml/min) and can be directly connected to MS systems. The combination of UHPLC and MS makes a powerful system, offering extraordinary selectivity and sensitivity.
Today, there are different types of MS analyzers available, with quadrupole, ion-trap, and time-of-flight (TOF) mass analyzers being the most commonly used. Quadrupole mass analyzers are relatively cheap and provide acceptable selectivity, but have low resolution power (Matuszewski et al., 2003; Pitt, 2009; Cutignano et al., 2016). On the other hand, TOF and Orbitrap (a type of ion-trap mass analyzer) offer improved mass accuracy over a wide range of molecular weights, provide excellent peak resolution and allow for the measurements of isotopic patterns, all of which will help to elucidate the elemental composition of polyphenols (Matuszewski et al., 2003; Pitt, 2009). Given these advantages, more studies are expected to be carried out using Q-TOF for the analysis of microalgal polyphenols in the near future. HPLC-Q-TOF has recently been used for proteomics profiling on Dunaliella strain (Emami et al., 2015), and the identification of pigments in Tetraselmis suecica (Sansone et al., 2017). Considering the complexity of microalgal biomass, extracts should be purified by solid-phase extraction (SPE) or liquid-liquid extraction prior to HPLC analysis (Jerez-Martel et al., 2017) to aid separation, improve sensitivity and reduce interference (Stalikas, 2007).
Matrix-assisted laser desorption ionization time of-flight mass spectrometry (MALDI-TOF-MS) is another detection system that has been widely employed for characterizing polyphenols in biological matrices such as herbs, vegetables, fruits and by-products (Tzima et al., 2018; Gu et al., 2019; Peng et al., 2019). Its use for microalgal polyphenol analysis, however, has not yet been reported.
Even after obtaining UV/Vis spectra and MS fragmentation patterns, it is important to use authentic standards to confirm the identity of individual polyphenols. However, this approach is seldom followed, as polyphenol standards are either expensive or commercially unavailable (Matuszewski et al., 2003). Individual polyphenols can first be separated using thin layer chromatography (Stalikas, 2007) or preparative chromatography prior to HPLC to aid identification and then can be further characterized by Nuclear Magnetic Resonance (NMR).
2.2.3 Gas Chromatography: Applications and Optimization
Gas Chromatography (GC) has been widely used in microalgae analysis to characterize volatile compounds such as fatty acid methyl esters (
Yu et al., 2020) (described in section 3.3) and assess the presence of contaminants in
Chlorellaand
Spirulina(
Martín-Girela et al., 2020). GC is generally considered to be a more robust and sensitive technique compared to HPLC. GC can be commonly coupled with detection units such as flame ionization detectors (FID), electron capture detectors (ECD), or mass spectrometry (MS). Despite these advantages, GC is not widely used to separate and identify microalgal polyphenols and we have only managed to find a few studies that used GC for microalgae phenolic separation (
Table 3). The lack of widespread GC use could be attributed to the challenges that arise due to the physicochemical properties of polyphenols compounds:
1) High temperatures are needed to separate polyphenols using GC. This may lead to undesirable consequences such as thermal degradation of thermolabile phenolic compounds (Ameer et al., 2017).
2) Extra sample preparation steps to remove lipids from the extract (defatting) and release polyphenols through the cleavage of ester and glycosidic bonds (acid, alkali or enzymatic hydrolysis) are needed (Ajila et al., 2011).
3) Polyphenols are not volatile and require a time consuming derivatization step (silylation, methylation, or acetylation) to increase their volatility (Viñas and Campillo, 2014).
TABLE 3
| Microalgae species | Stationary phase/mobile phase/detection unit | GC parameters | Compounds | Ref |
|---|---|---|---|---|
| Spirulina maxima | A capillary polymethylphenylsiloxan-FI 95 column/no information/flame ionization detector (FID) | Oven temperature: 150°C for 3 min, 150–300°C at 5°C/min | Salicylic, trans-cinnamic, synaptic, chlorogenic, quimic and caffeic acids | Miranda et al. (1998) |
| Chlorella vulgaris | Capillary column/no information/FID | No information | Phenolic compounds, salicylic, trans cinnamic, synaptic, chlorogenic, chimic and caffeic acids | Miranda et al. (2001) |
| Chlorella vulgaris | DB5, (0.53 mm IDX-5µm film)/carrier gas: He (1 ml/min)/Mass Spectrometry (MS) | Oven temperature: 60°C for 0.5 min, 60–150°C at 10°C/min, 150–250°C at 10°C/min | Quercetin | Ahmed, (2016) |
| Chlorococcum minutum | HP-5MS column (30 m × 0.25 mm × 0.25 µm)/carrier gas: He (0.9 ml/min)/MS | Oven temperature: 60–250°C at 2°C/min | Flavonoid Lucenin-2 | Elshobary et al. (2020) |
| Chlorella sorokiniana and Chlorella minutissima | Column DB-5 (30 m × 0.25 mm × 0.25 μm)/carrier gas: He (1 ml/min)/MS | Oven temperature: 40°C for 1 min, 40 to 280°C at 3°C/min | Polyphenols were detected by TPC test and FTIR characterization. However, no polyphenols were detected through GC-MS screening | Olasehinde et al. (2019) |
GC instrumentation and parameters used in recent microalgal studies for polyphenol analysis.
Overall, it is evident that HPLC technique is the method of choice to separate and identify polyphenols compared to GC. However, GC can still be effectively used to identify and quantify selected polyphenols if the method is optimized. Table 3 summarises the instruments and operating parameters used for GC analysis of microalgal polyphenols.
To conclude, phenolic compounds are very diverse in microalgae. Spectrophotometric methods are not accurate enough for the determination of these compounds while HPLC coupled with different detectors seemed to be very useful. Especially, when unknown compounds are present, HPLC-MS is the choice. However, GC has not been widely used due to major obstacles such as high temperature, sample preparation.
3 Polyunsaturated Fatty Acids (PUFAs)
Fatty acids are the building blocks of lipids, one of the major constituents in living organisms. According to their carbon chain length and the number of double bonds present in the carbon chain, fatty acids are classified into different groups including short chain (less than 12 carbon atoms) and long chain fatty acids (more than 12 carbon atoms). Long chain fatty acids are further classified into saturated (no double bonds, SFA), monounsaturated (a single double bond, MUFA), and polyunsaturated fatty acids (more than double bonds, PUFA). Major natural SFAs are stearic acid (C18:0) and palmitic acid (C16:0) while oleic acid (C18:1) is the main naturally occurring MUFA. PUFAs include a broader range of fatty acids such as linoleic acid (C18:2), linolenic acid (C18:3), EPA (C20:5) and DHA (C22:6). PUFAs are normally classified into omega-3 and omega-6 fatty acids depending on the position of the initial double bond counted from the methyl end on the chain (Cretton et al., 2022; Patel et al., 2022). Omega-3 fatty acids are essential ingredients as they cannot the synthesized in human body and their uptake is through food. Although fish oil is considered as the main food source for omega-3 fatty acids, fish cannot also synthesize omega-3 oil. Microalgae are the main producers of omega-3 and their consumption by aquatic animals can enrich their fatty acid content in terms of EPA and DHA (Halim et al., 2012; Ryckebosch et al., 2012).
3.1 Microalgae as a Source of Long-Chain ω-3 Polyunsaturated Fatty Acids: Recovery and Transesterification
Long-chain ω-3 PUFAs in microalgae cells are principally accumulated in the polar lipid fractions that make up membrane lipids (phospholipids and glycolipids) though a smaller amount exists as part of biofuel-convertible storage neutral lipids (triacylglycerols) (Halim et al., 2012; Ryckebosch et al., 2014). A triacylglycerol (TAG) is a hydrophobic lipid molecule consisting of three fatty acids ester-bonded to a glycerol backbone, while phospholipid and glycolipid are polar lipid molecules that consist of two fatty acids bound to either a glycerol-phosphate group complex or a carbohydrate molecule (Halim et al., 2012). In microalgae cells, TAG is synthesized in the endoplasmic reticulum from the acylation of diacylglycerol (DAG), an intermediate by-product of membrane polar lipids. For this reason, membrane lipid and storage lipid are inherently interconnected and their levels fluctuate depending on the physiological state of the biomass (Jouhet et al., 2017).
The intracellular levels of microalgal ω-3 PUFAs vary significantly between taxonomic groups. Furthermore, intracellular fat content depends also on microalgal growth phase and culture conditions. For example, at early stages phospholipid content is higher since they are found in the cell membrane. On the other hand, at late stages of cultivation, neutral lipids are accumulated due to nitrogen or phosphorus starvation (Yan et al., 2010; Law et al., 2018; Halim et al., 2019; Cardoso et al., 2020). Promising ω-3 PUFA-rich microalgae strains include Nannochloropsis sp (EPA content = 9–38 wt% of total lipid, total lipid content = 16–68 wt% of biomass) and Phaeodactylum sp (EPA content = 2–36 wt% of total lipid, total lipid content = 15–40 wt% of biomass) for EPA commercial production and Isochrysis sp (DHA content = 1–40 wt% of total lipid, total lipid content = 7–33 wt% of biomass) and Schizochytrium (DHA content = 25–52 wt% of total lipid, total lipid content = 13–60 wt% of biomass) for DHA commercial production (Patil et al., 2007; Jakobsen et al., 2008; Ryckebosch et al., 2012; Xue et al., 2013; Singh et al., 2020; Kujawska et al., 2021).
The standard laboratory procedure for recovering ω-3 PUFAs from microalgal biomass involves 1) harvesting the cells from their suspension with a solid-liquid separation method (such as centrifugation, filtration, flocculation/filtration), 2) drying the resulting slurry with a non-thermal process (such as spray drying or freeze drying) to prevent fatty acid oxidation while preserving the functionality of ω-3 PUFAs, 3) extracting ω-3 PUFA lipids from the biomass powder with organic solvents (such as ethanol, acetone, chloroform/methanol), supercritical carbon dioxide or a mixture of supercritical carbon dioxide and organic solvents (Halim and Danquah, 2012). Cell disruption technologies (such as ultrasonication, microwave and high-pressure) are often integrated in the extraction system in order to accelerate mass transfer kinetics and enhance ω-3 PUFA yields. Other bioactive components, such as pigments (carotenoids and chlorophylls), vitamins, and polyphenols, are often co-extracted during the recovery process, contributing to the nutritional properties and health benefits of the ω-3 PUFA lipid extracts (Halim et al., 2012; Ryckebosch et al., 2012). Since long-chain ω-3 PUFAs tend to be more abundant in the polar lipid fractions, they are less effectively extracted by organic solvent extraction mixtures with low polarity (such as chloroform or hexane) and more readily extracted by polar organic solvents (such as acetone, ethanol and isopropanol) (Halim et al., 2012).
Transesterification reaction is needed to convert lipids recovered from microalgae biomass into volatile Fatty Acid Methyl Esters (FAMEs) suitable for GC vaporisation and analysis (Halim et al., 2012). During transesterification, the lipid is blended with methanol in the presence of a catalyst. The reaction cleaves fatty acids from triacylglycerol, phospholipid and glycolipid structures and methylate the free fatty acids to form FAMEs (Halim et al., 2012). Catalyst selection during transesterification is critical for accurate FAME analysis. The choice of using either a base (OH− ions from sodium hydroxide NaOH, potassium hydroxide KOH, calcium oxide CaO, lithium hydroxide LiOH, sodium methoxide NaOMe, potassium methoxide KOMe) or an acid catalyst (H+ ions from hydrochloric acid HCl, sulphuric acid H2SO4) will depend on the composition of the lipid fractions (Halim et al., 2012; Goh et al., 2019). A poor catalyst selection will result in partial or incomplete conversion of available ω-3 lipids into FAMEs, reducing the overall yield and reliability of the analysis. Recent studies have also shown that enzymes can also be used to successfully catalyse microalgal lipid transesterification (Goh et al., 2019). Lipase has a relatively low operational temperature range (25–50°C) which minimises the probability of lipid degradation, and has been proven effective in transesterifiying all lipid fractions (including free fatty acids) (Navarro López et al., 2016).
Table 4 provides a summary of the catalysts and operating conditions used in recent studies investigating transesterification of microalgal lipids. In Figure 3, the transesterification process steps are shown.
TABLE 4
| Microalgae species | Transesterification catalyst and conditions | FAME yield from available lipid (wt%) | Reference |
|---|---|---|---|
| Acutodesmus obliquus | calcium oxide at 75°C for 3.6 h | 86 | Pandit and Fulekar, (2017) |
| Auxenochlorella protothecoides | sulphuric acid at 70°C for 3 h | >90 | Silve et al. (2018) |
| Botryococcus sp | lipase B (Novozyme CAL-B) at 40°C for 6 h with ultrasonication | 88 | Sivaramakrishnan and Incharoensakdi, (2017) |
| Nannochloropsis oceanica | Hydrochloric acid at 85°C for 1 h | n/a | Jia et al. (2015) |
| Nannochloropsis gaditana | lipase at 40°C for 56 h | 99.5 | Navarro López et al. (2016) |
| Nannochloropsis sp. and Nannochloropsis gaditana | 10 wt% sulphuric acid at 55°C followed by 25 wt% potassium methoxide at 55°C | 47 | Halim et al. (2019) |
Lipid transesterification catalyst and conditions used in recent studies investigating the conversion of microalgal lipids to Fatty Acid Methyl Esters.
FIGURE 3
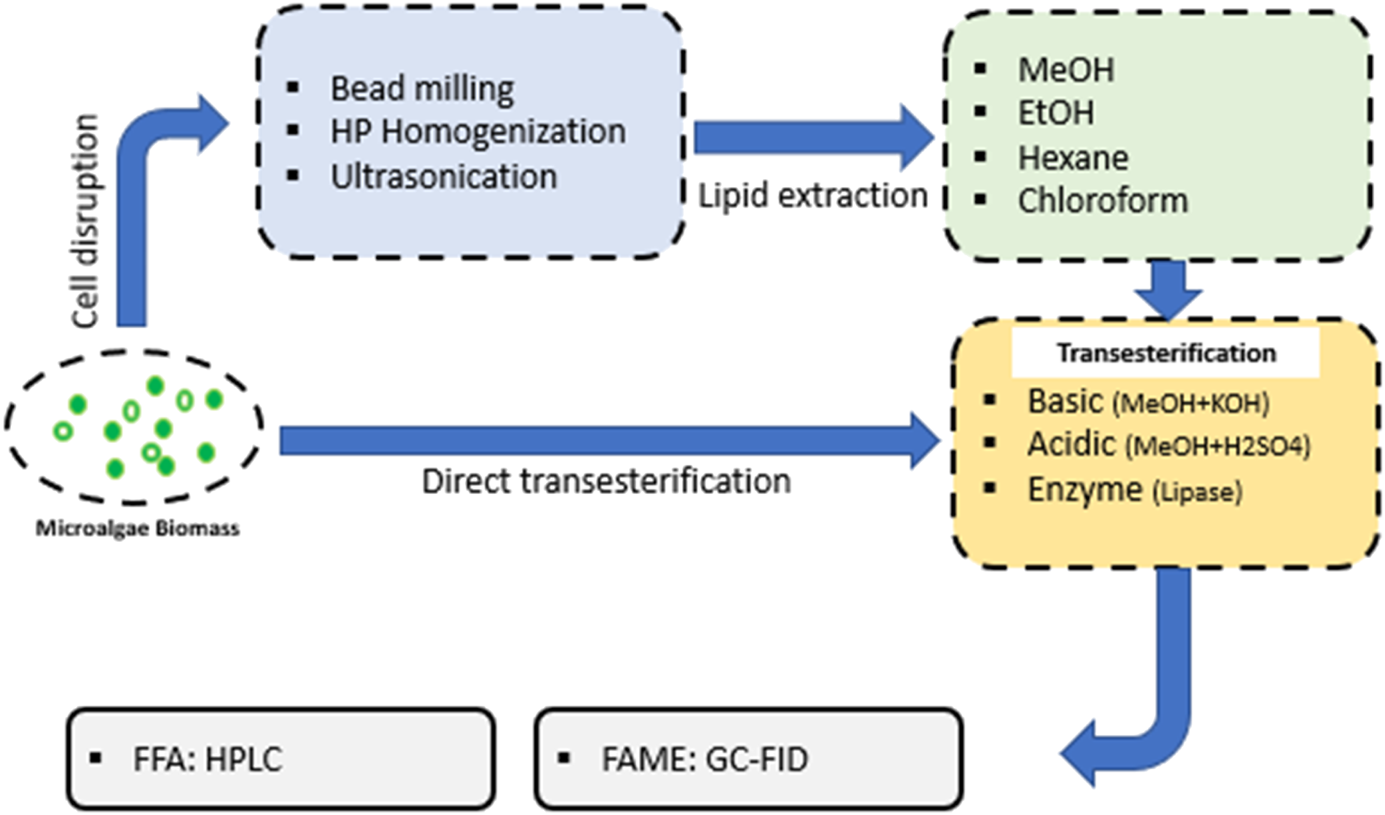
Illustration of the steps for the transesterification processes of lipids in order to form fatty acid methyl-esters (FAMEs) or free fatty acids (FFAs) before chromatographic analysis.
3.2 Chromatographic Analysis of ω-3 PUFAs: Governing Principles
The separation and analysis of ω-3 PUFAs recovered from microalgae biomass can be carried out with either 1) lipid transesterification and GC analysis or 2) HPLC-MS analysis. GC analysis is currently the universal method used for microalgal ω-3 PUFA determination because of its reliability, ease of sample preparation and results analysis (Goh et al., 2019). GC method enables the identification and absolute quantification of fatty acids (including ω-3 PUFAs) present in the microalgal lipid extract. The method is linear for a wide range of fatty acid concentrations and has a broad range of FA chain lengths. On the other hand, HPLC-MS fractionates lipids into their constituent classes prior to species and fatty acid determination. Because of this, the method provides additional layers of analysis compared to the standard fatty acid identification and quantification carried out by GC (Carrasco-Pancorbo et al., 2007; Goh et al., 2019; Rontani, 2022). HPLC-MS has the capacity to identify lipid classes and species present in the microalgal lipid, measure their relative abundance while at the same time evaluate the composition and distribution of fatty acids (including ω-3 PUFAs) in the different lipid classs. The multilayered nature of HPLC-MS analysis makes it is a very attractive lipid profiling tool to be used for microalgal studies investigating lipid metabolism, fatty acid biosynthesis and phylogenetics. The use of the method for microalgal analysis, however, is currently still limited given the high level of technical expertise needed for the operation of the equipment and the interpretation of the results. Section 3.3 presents a review of the basic operating principles associated with GC analysis of ω-3 PUFAs, while Section 3.3.1. provides an overview of the development of HPLC-MS for the analysis of microalgal lipid and ω-3 PUFAs.
3.3 Gas Chromatography: Applications and Optimization
Gas chromatography is currently the universal analytical technique used to determine the fatty acid compositions of microalgae (Tang and Row, 2013; Dębowski et al., 2020; Kujawska et al., 2021; Barati et al., 2022). During GC analysis, FAMEs are separated from each other based on their interaction with the stationary phase of the GC column and boiling points. FAMEs with stronger interaction to the stationary phase will stay in the column longer and thus have higher retention times compared to FAMEs with weaker interaction to the stationary phase. FAMEs with low boiling points will be vapourised more rapidly during temperature ramp and thus have shorter retention times compared to FAMEs with high boiling points. This combination of phase interaction and boiling point separation as a separation mechanism allows GC to resolve FAME molecules according to their chain length (carbon number excluding the carbon in the head methyl group), degree of unsaturation (number of double bonds), position of unsaturation (location of double bonds) and cis/trans geometric configuration of the double bonds (David and Vickers, 2005). The stationary phase has a substantial impact on the efficiency and success of fatty acid separation and determination.
Frequently used stationary phases for FAME separation are made of fused silica, polyethylene glycol (PEG) or cyanopropyl polysiloxane. PEG columns (also known as Wax columns) are commercially available under the tradename Omegawax, Carbowax-20M, CP-Wax 52CB, Supelcowax-10 and DB-Wax. Commercial variants of cyanopropyl polysiloxane column include DB-23, HP-88, CP-Sil88, BPX70, SP-2340, SP-2560 and J&W DB-FastFAME (Dobowy and Pyka, 2015; Zou and Wu, 2018). The polarity of the stationary phase determines the column’s ability to separate FAME molecules. PEG columns give relatively good separation of FAMEs from a lipid mixture according to their chain length but are generally considered to lack the ability to completely separate cis-trans isomers and different ω-3 PUFAs (Tang and Row, 2013). Cyanopropyl columns can be classified as either moderate polarity or high polarity. Cyanopropyl columns with moderate polarity (e.g., DB-23, DB-FasteFAME) provide excellent separation of ω-3 PUFAs from a complex FAME mixture. Detailed and successful separation of geometric (cis- and trans-) isomers of polyunsaturated FAMEs can, however, only be achieved using cyanopropyl columns with high polarity (e.g., HP-88, CP-Sil88) (Tang and Row, 2013). The thickness of stationary phase inside the column generally ranges from 0.15 to 0.25 μm.
Cyanopropyl columns w are generally recommended for GC analysis of microalgae lipids to ensure complete peak separation and accurate quantification of resolved ω-3 PUFAs. In their work evaluating the effect of stationary phase interaction on the analysis of microalgal FAME mixtures, David and Vickers (2005) determined that cyanopropyl columns with moderate polarity, such as DB-23, were able to completely resolve EPA (cis-20:5 n-3) and DHA (cis-22:6 n-3) from other FAMEs in the lipid mixture. Separation of ω-3 PUFAs using PEG columns (such as DB-Wax) was limited and co-elution of DHA (cis-22:6 n-3) and tetracosanoic acid (cis-24:1 n-9) was observed. Even though the cyanopropyl columns with high polarity (such as HP-88) had an excellent capacity for complete separation of cis- and trans- isomers, they lacked the ability to fully resolve ω-3 PUFAs from other FAMEs, with EPA (cis-20:5 n-3) co-eluting at the same time with docosadienoic acid (cis-22:2 n-6).
Findings from Woo et al. (2012) in their investigation of GC parameters on PUFA analysis from Chlorella vulgaris, Ankistrodesmus gracilis, and Scenedesmus quadricauda echoed those obtained from David and Vickers (2005). Columns with moderate polarity were found to be able to effectively resolve individual C16 - C18 PUFAs from their corresponding C16–C18 saturated and monounsaturated PUFAs in the microalgal FAME mixture. The authors noted the importance of choosing appropriate GC methods as artefact differences in fatty acids resulting from poorly conditioned GC operations can be significant and have adverse effects on the biomass fatty acid profiles.
Column dimension and carrier gas also play an important role in determining the efficiency of ω-3 PUFA separation during GC analysis. Zou and Wu (2018) investigated the effect of column length and carrier gas on the ability of cyanopropyl DB-FastFAME column in separating and resolving a 37-FAME standard mixture. Reducing column diameter was found to increase separation efficiency per unit length of the column, while the use of gas with a higher diffusivity (H2 instead of He) was shown to provide a faster analysis without compromising resolutions of the separated ω-3 PUFA peaks (Zou and Wu, 2018; Zotov et al., 2022).
Recent advancement in GC stationary phase includes the development of ionic liquid (IL) columns. In comparison with polyethylene glycol and cyanopropyl stationary phases, ionic liquid stationary phases showed comparable resolution, but lower column bleeding, resulting in more sensitive quantification. IL columns are also known to be able to separate geometric and positional fatty acid isomers. Gu and coworkers compared the performance of ionic liquid columns to PEG and cyanopropyl columns for the GC-MS analyses of algal fatty acids (Gu et al., 2011). The reduction in column bleeding of the ionic liquid columns (SLB-IL 82 and SLB-IL 100) meant that they were able to detect and quantify trace fatty acids in the samples more accurately than the conventional columns.
Identification and quantification individual microalgal FAMEs (including ω-3 PUFAs) after chromatographic separation is normally carried out with either a Flame Ionisation Detector (FID) or a Mass Spectrometer Detector (MSD). The purpose of the detector unit is to identify and measure the components that have been separated by the GC column. FID and MSD are the most commonly detectors used for algal FAMEs because of their low detection limits, being able to quantify fatty acids at nanogram (10−9) to femtogram (10−15) concentration levels (Tang and Row, 2013; Dobowy and Pyka, 2015). FID performs detection by burning organic compounds in the GC column effluent with a hydrogen flame (280°C) and measuring the total ions formed during the combustion. The level of ion signals is proportional to the concentration of organic species at the specific elution time of the column gas stream. Such indiscriminate ionisation of eluded FAMEs means that the detector is unable to identify the molecular signatures of the combusted components (Tang and Row, 2013; Dobowy and Pyka, 2015). FAME identification with GC-FID therefore solely relies on component separation by the GC column and retention time matching with known FAME standards. Limited availability of standards for certain FAME species can restrict the scope of fatty acid identification (and thus quantification) with GC-FID.
An MSD is able to identify and quantify different FAME species by ionising the molecules in the GC column effluent through electron bombardment and measuring the charged fragments formed from the ionisation process in accordance to distinct specific mass-to-charge ratio. Each ion species has its own unique isotopic mass-to-charge (M/Z) signature that can be directly compared to the MS database library for identification (Tang and Row, 2013; Dobowy and Pyka, 2015). Such operating principles provide the MS system with the capacity to facilitate both identification and measurement of FAME species eluting at a particular retention time from the column. FAME identification with GC-MSD is achieved through both the comparison of signature M/Z profile with database library and the confirmation of retention time with a known FAME standard. Table 5 provides a summary of the GC instrumentation and parameters used in recent microalgal studies for their ω-3 PUFA analysis.
TABLE 5
| Microalgae species | Stationary phase/mobile phase/detection unit | GC Parameters | Compounds | Ref |
|---|---|---|---|---|
| Nannochloropsis sp., Nannochloropsis gaditana | DB-23 (cyanopropyl polysiloxane)/Helium/FID | Column: 0.32 mm id x 30 m × 0.25 μm; Oven: 50–210 °C; FID: 300 °C | ω-3 FAMEs: C20:5 (n-3); other FAMEs: C14:0, C16:0, C16:1, C18:1, C18:2, C20:3 | Halim et al. (2019) |
| Nine different species, including Nannochloropsis gaditana, Nannochloropsis oculata, Pavlova lutheri, Phaeodactylum tricornutum and Rhodomonas salina | EC-Wax (PEG)/no information on mobile phase/FID | Column: 0.32 mm id x 30 m × 0.25 μm; Oven: 70–235°C | Only ω-3 FAMEs: C18:3 (n-3), C18:4 (n-3), C20:5 (n-3), C22:5 (n-3), C22:6 (n-3) | Ryckebosch et al. (2014) |
| Chlorella sp., Nannochloropsis sp | VF-5 MS (phenylmethyl polysiloxane)/Helium/MSD | Column: 10 m × 0.25 μm Oven: 50–230°C; Injection: 250°C | ω-3 FAMEs: C20:3 (n-3), C20:5 (n-3); other FAMEs: numerous saturated and monounsaturated fatty acids | Olmstead et al. (2013) |
| Twelve different species, including Synechococcus sp., Isochrysis galbana, Phaeodactylum tricornutum, Tetraselmis suecica, Nannochloropsis oceanica | DB-Wax (PEG)/no information/no information | Column: 0.1 mm id x 10 m × 0.1 μm; Oven: 70–250°C | ω-3 FAMEs: C18:3 (n-3), C18:4 (n-3), C20:5 (n-3), C22:6 (n-3); other FAMEs: numerous saturated and monounsaturated fatty acids | Patil et al. (2007) |
| Chlorella vulgaris, Ankistrodesmus gracilis, Scenedesmus quadricauda | Three different stationary phases: HP-5 (methylpolysiloxane), DB-Wax (PEG), Supelcowax (PEG)/Helium/FID | Column 1: 0.32 mm id x 30 m × 0.25 μm. Column 2: 0.25 mm id x 30 m × 0.25 μm. Column 3: 0.32 mm id x 60 m × 0.5 μm; Oven: 50–850°C | ω-3 FAMEs: C18:3 (n-3); other FAMEs: C16:0, C16:1, C18:0, C18:1, C18:2 | Woo et al. (2012) |
| 16 different microalgal species | Condition 1: DB-5MS (methylpolysiloxane)/He/MSD; Condition 2: DB Wax (PEG)/He/FID | For condition 1: column: 0.25 mm id x 30 m × 0.25 μm, oven: 45–320°C, injection: 240°C,; MSD: 230°C. For condition 2: column: 0.25 mm id x 30 m × 0.25μm, oven: 25–250°C, injection: 100°C,; FID: 280°C | ω-3 FAMEs: C18:3 (n-3); other FAMEs: numerous saturated and monounsaturated fatty acids | Kchech, (2017) |
| Chlorella vulgaris | EZ-Guard VF 5 ms (dimethylpolysiloxane)/no information/TOF-MS | Column: 0.25 mm id x 30 m × 0.25 μm | ω-3 FAMEs: C16:3 Δ7,10,13, C18:2 Δ9,12,15; other FAMEs: numerous saturated and monounsaturated fatty acids | Morschett et al. (2019) |
GC instrumentation and parameters used in recent microalgal studies involving ω-3 PUFA analysis.
Despite the routine use of FID for GC analysis of microalgal fatty acids, MSD is regarded to be the more superior detection unit for the following reasons: 1) the dual identification system (library comparison and peak matching) used in GC-MSD allows for a more accurate and reliable peak identification that is not limited by the availability of specific FAME standards, 2) GC-MSD has a broader range of detectable fatty acids than GC-FID, 3) GC-MSD is more sensitive to change in FAME concentration (higher ratio of Δ GC area signal/ΔFAME concentration) than GC-FID (Woo et al., 2012; Kchech, 2017). In their study comparing the performance of GC-FID and GC-MSD for the separation and quantification of FAME mixtures recovered from 16 different microalgae biomass, Kchech found that GC-MSD was able to detect a number of short-chained fatty acids (e.g. C8:0) and long-chained fatty acids (e.g. C24:1) in the microalgal lipid extracts that were not picked up by GC-FID (Kchech, 2017). They also found FAME quantification with GC-MSD to be more reproducible compared to that with GC-FID (For the same set of samples, relative standard deviation for FAME analysis by GC-MSD ranged between 0.51–24.56%, while that for GC-FID ranged between 6.61–41.91%).
The identification and quantification accuracy of GC analysis of microalgal ω-3 PUFAs can be improved by using Time-of-Flight Mass Spectrometry (TOF-MS) detection. This detection unit supplemented FAME identification with structural reconstruction of the respective FAME fragments and facilitated the determination of the specific stereochemistry of the fatty acids in the lipid extracts. Morschett and coworkers used this method to analyse the fatty acid composition of lipid extracted from C. vulgaris (Morschett et al., 2019). They successfully identified the stereoisomers of C16:1, C16:2, C16:3 and C18:3 in the lipid extracts as C16:1 Δ7, C16:2 Δ7, 10, C16:3 Δ7, 10, 13 and C18:3 Δ9, 12, 15. This detailed fingeprinting of C. vulgaris lipids allowed the authors to more accurately track the change in the biomass fatty acid composition and the transference of fatty acids between different glycerolipid classes as a function of physiological state.
One of the main limitations of using GC for fatty acid analysis (as opposed to the more targeted HPLC methods) is its inability to distinguish the lipid classes from which the ω-3 fatty acids were recovered/derivatised from (e.g,. free fatty acids, triacylglycerols, phospholipids and glycolipids) (Olmstead et al., 2013; Morschett et al., 2019). Obtaining detailed information on ω-3 fatty acid distribution amongst different lipid classes in microalgal biomass is critical to species screening, metabolic studies on ω-3 PUFA biosynthesis and development of optimal cultivation conditions aimed at maximum ω-3 production. Such analytical limitations can, however, be overcome by performing Thin Layer Chromatography (TLC) or Solid Phase Extraction (SPE) to fractionate the total biomass lipid extracts into different classes prior to transesterification and GC analysis.
Olmstead and coworkers used a simple and commercially available silica-based SPE system to successfully fractionate total lipid extracts from Chlorella sp. and Nannochloropsis sp. into their constituent neutral lipids (triacylglycerols and free fatty acids), phospholipids and glycolipids before subjecting the individual lipid fractions to transesterification and GC separation. This approach enabled them to demonstrate that the majority of ω-3 PUFAs (between 65–94 wt% of available ω-3 PUFAs) in Nannochloropsis biomass partitioned in the phospholipid and glycolipid fractions rather than the neutral lipid (or triacylglycerol) fraction (Olmstead et al., 2013). These findings have immediate implications to microalgal downstream processing as it highlighted the requirements for extraction solvents with high affinity to polar lipids for effective ω-3 PUFA recovery.
3.3.1 High-Performance Liquid Chromatography
HPLC-MS is a powerful method that can be used to not only identify lipid classes and species and measure their relative abundances but also to determine the composition of fatty acids (including ω-3 PUFAs) in each lipid class/species (Cajka and Fiehn, 2014; Dobowy and Pyka, 2015). This is in contrast to GC-based analysis which lacks the ability to separate out lipid classes and can thus only give global fatty acid information of the total biomass lipids (unless additional lipid fractionation techniques, such as TLC or SPE, is introduced to separate the lipid into individual classes prior to GC analysis). LC-MS lipidomic analysis can be untargeted (acquisition of full spectra of all lipid classes) or targeted for specific lipid classes. In addition to superior separation capacity, the method is sensitive at trace levels, has minimal background interference and does not require a separate transesterification step that can lead to artefact generation (Cajka and Fiehn, 2014). Absolute quantification of the amount of each lipid class with the HPLC -MS method, however, can be quite challenging given the varying degrees of ionization efficiencies for the different lipid classes and the large number of internal standards (at least one representative standard for each lipid class) required for such a calculation (Jouhet et al., 2017). Table 6 provides a summary of the HPLC instrumentation and parameters used in recent microalgal studies for ω-3 PUFA analysis.
TABLE 6
| Microalgae species | Stationary phase/mobile phase/detection unit | HPLC Parameters | Compounds | Ref |
|---|---|---|---|---|
| Botryococcus braunii | Luna Si column (5 μm, 4.6 × 250 mm)/Solvent A: acetone and Solvent B: hexane/detector: APCI/MS | Flow rate:1 ml/min, injection volume: 50 μL, eluction gradient: 98–93% B for 11 min, 93% B for 14 min, 93 to 74% B for 1 min, 74% B for 13 min, 74–98% B for 1 min, 98% B for 5 min. Mass spectrum recorded in + and—mode | Twenty nine lipid compounds, including 4 triacylglycerols (TAG), 2 diacylglycerols (DAG), 3 prenol lipids, 4 wax esters, 4 alkoxy ether lipids, 8 sterols and 3 sugar alcohols. Main FA in TAG: C16:0, C18:1, C18:2; main FA in DAG: C16:0, C18:1 | Donot et al. (2013) |
| Phaeodactylum tricornutum, Nannochloropsis gaditana | Diol column (5 μm, 3 × 150 mm)/Solvent A: hexane, isopropanol, water, 1 M ammonium acetate (625/350/24/1 v/v/v/v) and Solvent B: isopropanol, water, 1 M ammonium acetate (850/149/1 v/v/v)/detector: MS/MS | Flow rate: 200 μL/min, column T: 40°C, injection volume: 20 μL, elution gradient: 0–100% B for 30 min, 100% B for 15 min, 100–0% B for 5 min, 0% B for 20 min | Monogalactosyldiacylglycerol (MGDG), phosphatidylcholine (PC), triacylglycerol (TAG). Main FA in MGDG: C20:5 (n-3); main FA in PC: C20:5 (n-3), C16:1; main FA in TAG: C16:0, C16:1 | Jouhet et al. (2017) |
| Cyclotella cryptica, Nannochloropsis salina, Thalassiosira weissflogii, Alexandrium minutum, Alexandrium tamutum | Kinetex Biphenyl column (2.6 µm, 2.1 × 150 mm)/Solvent A: water and Solvent B: methanol/detector: MS | Flow rate: 0.3 ml/min, column T: 28°C, injection volume: 10 μL, elution gradient: 40–80% B for 2 min, 80–100% B for 13 min, 100% B for 7 min | Phospholipids (PL), sulfoquinovosyldiacylglycerols (SQDG), monogalactosyldiacylglycerols (MGDG), digalactosyldiacylglycerols (DGDG) and triacylglycerols (TAG). For Nannochloropsis salina, main FA in PL: C20:4 (n-3), C20:5 (n-3), C16:0, C16:1, main FA in SQDG: C16:0, main FA in MGDG: C20:5 (n-3), main FA in DGDG: C20:5 (n-3), main FA in TAG: C16:0, C16:1, C18:1 | Cutignano et al. (2016) |
HPLC instrumentation and parameters used in recent microalgal studies involving ω-3 PUFA analysis.
HPLC-MS lipidomic analysis of biological systems, such as microalgae biomass, typically involves the following steps (
Figure 4) (
Cajka and Fiehn, 2014;
Dobowy and Pyka, 2015;
Domingues and Calado, 2022;
Evans et al., 2022):
1) extraction of the lipids from the biomass using chloroform/methanol protocol with the addition of internal standards to cover the specific lipid classes,
2) LC separation of the lipid extract using short microbore columns with 2.6–2.8 μm particle size with C18 or C8 stationary phase,
3) electrospray ionisation of the separated lipid species in positve- and negative-ion modes followed by the capture, amplification and detection of ions with high-resolution MS. The detection can be can be carred out in an untargeted (full spectra acquisition) or targeted (multiple-reaction monitoring) mode.
4) data handling for the identification and (semi-) quantification of the detected lipid fragments based on the values of their mass-to-charge (M/Z) ratio (Cajka and Fiehn, 2014; Dobowy and Pyka, 2015). Phospholipids, glycolipids, di- and triacyglycerols are the most frequently targeted lipid classes for microalgal analysis (Cajka and Fiehn, 2014; Dobowy and Pyka, 2015).
FIGURE 4
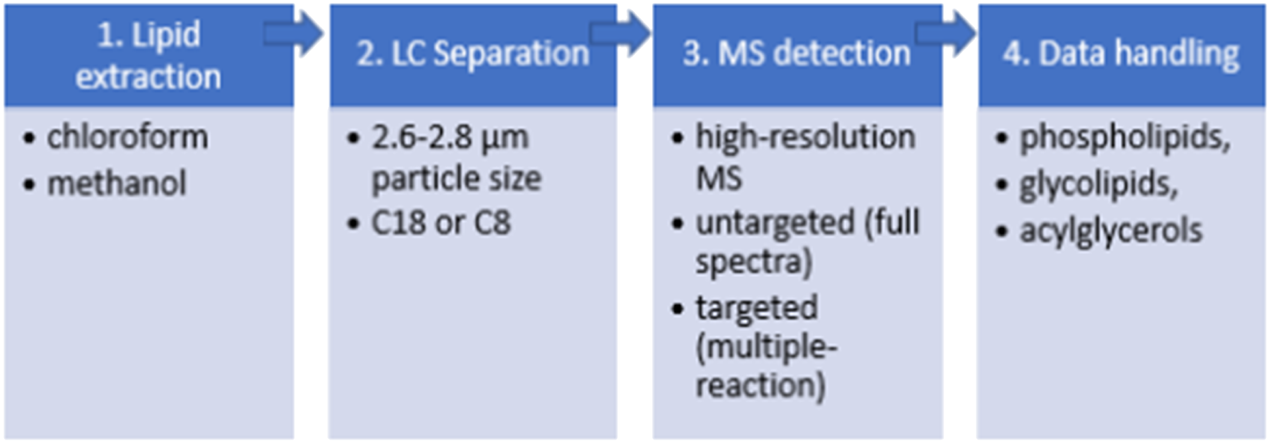
The pipeline of HPLC-MS lipidomic analysis for the analysis of microalgal lipids.
The comprehensive nature of HPLC-MS analysis makes it a very effective lipid profiling tool. It has the potential to significantly advance our insights into microalgal lipid metabolism and acclimation, uncovering novel lipid biosynthetic pathways and opening up new opportunities in strain screening and phylogenetic studies. The use of the method for microalgal lipid analysis, however, is currently still limited given the high level of technical expertise needed for the operation of the equipment and the processing and interpretation of the results. A number of studies have recently used hyphenated HPLC– MS platform with tandem Q-TOF (Yan et al., 2010, Yan et al., 2011 X.; Donot et al., 2013), QqQ (Jouhet et al., 2017) or Orbitrap (Cutignano et al., 2016) mass analyser to elucidate the detailed make-up of microalgal lipids in industrially relevant microalgae biomass (e.g. Botryococcus braunii, Phaeodactylum tricornutum, Nannochloropsis gaditana and Chlorella sp). The HPLC-MS method was used either for targeted analysis of the fluctuation of specific lipid classes in the investigated microalgae strains under certain cultivation/physiological states or untargeted analysis aimed at acquiring global lipidome profiles of the investigated microalgae strains.
The use of HPLC coupled with a tandem atmospheric pressure chemical ionization (APCI) –MS/MS detector enabled Donot and coworkers to determine the lipid class and fatty acid distribution of Botryococcus braunii’s lipid within a single analysis (Donot et al., 2013). Based on the m/z mass fragments of the lipid, twenty nine different componds were identified, including 4 triacylglycerols (TAG), 2 diacylglycerols (DAG), 3 prenol lipids, 4 wax esters, 4 alkoxy ether lipids, 8 sterols and 3 sugar alcohols. The authors were able to show that TAG and DAG molecules in Botryococcus braunii (race A) were composed of C16:0, C18:1 and C18:2 fatty acids. The absence of ω-3 PUFAs in B. braunii validated that research on the species should continue to focus on biofuel instead of antioxidant production.
Similarly, Jouhet and coworkers successfully used HPLC coupled with Electrospray Ionisation (ESI)-MS/MS detector to develop a protocol of determining the fatty acid distributions of two microalgal species Phaeodactylum tricornutum and Nannochloropsis gaditana (Jouhet et al., 2017). The accuracy of the HPLC- EIS-MS/MS method was demonstrated through successful production of corrected fatty acid distribution profiles that were very similar to those generated with TLC fractionation followed by transesterification and GC/MS. Unlike GC/MS that required additional TLC and transesterification steps, however, the HPLC-ESI-MS/MS method was able to establish this distribution without any pretreatment that may produce artefacts. The HPLC analysis verified that the majority of EPA (C20:5 n-3) in both P. tricornutum and N. gaditana biomass partitioned in the glycolipids fraction (∼25 and 60% of monogalactosyldiacylglycerol signal). The method was also used to track changes in N. gaditana’s fatty acid synthesis under nitrogen (N) deprivation. The accumulation of TAG during N-deplete cultivation was shown to be accompanied with de novo synthesis of C16:0 and C16:1 FAs and concomittant reduction in the global level of membrane lipids and C20:5 FA, suggestive of transference of ω-3 PUFAs to form saturated and monounsaturated fatty acids during nutrient depletion.
Cutignano and coworkers used UHPLC - MS/MS for comprehensive regiochemical assignments of fatty acids to different lipid classes (phospholipids PL, sulfoquinovosyldiacylglycerols SQDG, monogalactosyldiacylglycerols MGDG, digalactosyldiacylglycerols DGDG and triacylglycerols TAG) in the marine microalgae strains: Cyclotella cryptica, Nannochloropsis salina, Thalassiosira weissflogii, Alexandrium minutum and Alexandrium tamutum (Cutignano et al., 2016). The core-shell Biphenyl column coupled with a methanol/water elution gradient developed specifically for the chromatographic separation was found to yield good resolution between the lipid species and introduce minimal adduct formation, enabling the authors to assess unambiguously the fragmentation pattern of eluted lipid and carry out absolute quantitation of targeted neutral and polar individual lipid classes. The study found >350 individual lipid species between the five microalgal strains and confirmed that EPA was the principal component of MGDG and SQDG (both of which are sub-groups of glycolipids) in all of the investigated strains.
In conclusion, lipids are a major class of target compounds found in microalgae. Both HPLC and GC can be used for the determination of lipid composition. GC has shown advantages due to its robustness for fatty acid analysis and has gained a prominence while HPLC also offers benefits such as lipid classes analysis as well as fatty acid determination in their free form.
4 Pigments
Microalgae are associated with a large range of natural pigments. The major classes of pigments found in microalgae cells include three main groups of phycobiliproteins, chlorophylls and carotenoids (Figure 5). The pigments are not only crucial for the photosynthetic metabolism of microalgae but also provide antioxidant activities to protect the cells from damage. The produced and isolated pigments can be valuable products with several health benefits such as anticancer, anti-inflammatory, anti-obesity, neuroprotective, etc (Mimouni et al., 2012; Dewi et al., 2018; Ambati et al., 2019; Prabakaran et al., 2020). Microalgal pigments are exploited in a variety of industries ie. food dyes and health products. The most frequently used species for pigments production are Dunaliella salina, Haematococcus pluvialis, Chlorella spp., Muriellopsis spp., Scenedesmus spp., Spirulina (Arthrospira) spp., and Porphyridium spp (Hu et al., 2018; Silva et al., 2020).
FIGURE 5
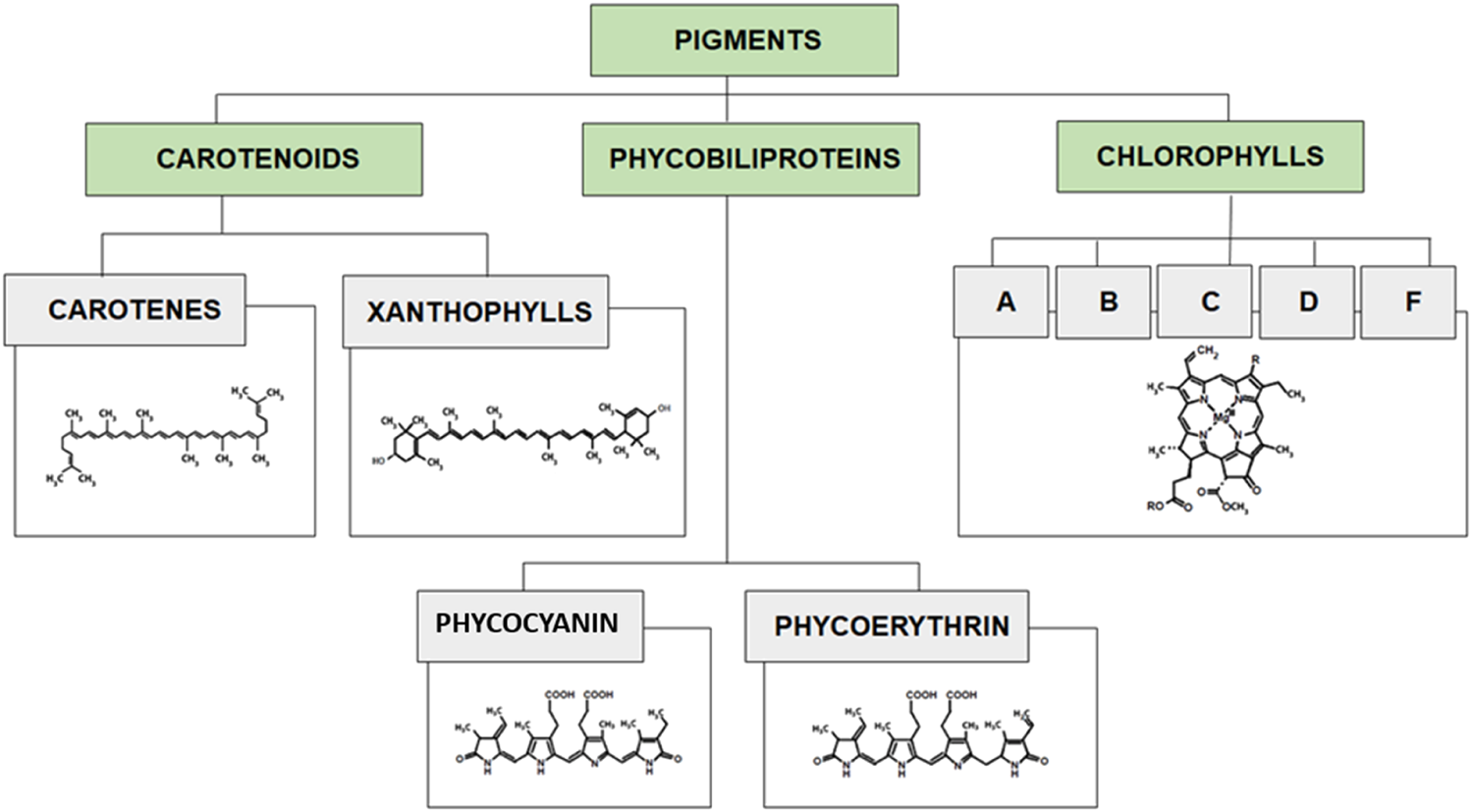
Classification of pigments with characteristic structures of each group.
4.1 Carotenoids
Carotenoids are isoprenoid molecules that can be chemically divided into carotenes (hydrocarbons; e.g., α-carotene, β-carotene, lycopene) and xanthophylls (oxygenated molecules; e.g., astaxanthin, lutein, canthaxanthin). Carotenoids light absorption range is between 400 and 550 nm. Carotenes are lipophilic molecules yellow to red in color required in the human diet. β-Carotene was the first high-value product commercially produced from microalgae Dunaliella salina, which is its richest source (14% DW) (Baker and Günther, 2004; Dufossé et al., 2005; Lou et al., 2020; Pourkarimi et al., 2020). Xanthophylls are polar molecules used in food and cosmetic additives. Main xanthophylls found in microalgae are astaxanthin and lutein. Astaxanthin found in Haematococcus pluvialis (0.2–5.0% DW) has powerful antioxidant capacity and high tinctorial properties (pinkish-red color) (Molino et al., 2018a). Lutein found in Scenedesmus, Chlorella, Muriellopsis spp, and many other microalgae species, is used as a natural yellow colorant and have been found to help in minimizing age-related blindness (macular degeneration and cataracts), and preventing cardiovascular diseases, atherosclerosis, and certain type of cancers (Low et al., 2020; Wang et al., 2022).
Due to their structure, carotenoids are only soluble in organic solvents, oils and fats. The conjugated double bonds, determining the unsaturated nature of carotenoids can cause oxidation and autoxidation reactions (if exposed to open air). Carotenoids are conventionally extracted using organic solvents. Non-polar carotenes and esterified xanthophyll are extracted using non-polar solvents (e.g., hexane, petroleum ether or tetrahydrofuran (THF). Polar carotenoids are extracted using polar solvents (e.g acetone, ethanol, and ethyl acetate). Current methods for the extraction of carotenoids include low pressure solvent extraction and supercritical fluid extraction. Recent review on the techniques studied to extract carotenoids from several matrices (including microalgae), highlights the importance of choosing the right solvent for an efficient extraction of carotenoids, and concluded that supercritical fluid extraction (SFE) was the superior green technique for extraction of carotenoids compared with other techniques such as Soxhlet, maceration, microwave-assisted extraction (MAE), ultrasound-assisted extraction (UAE), pressurized liquid extraction (PLE), pulsed electric field (PEF) assisted extraction and enzyme-assisted extraction (EAE) (Saini and Keum, 2018). SFE has been reported as a method to extract carotenoids in Botryococcus braunni, Chlorella vulgaris, Dunaliella salina and Spirulina maxima, Haematococcus pluvialis using supercritical CO2 as an extraction media (Molino et al., 2018b; Tirado and Calvo, 2019).
A summary of different methods used for the analysis of carotenoids is shown in Table 7. Different methods have been employed for the extraction of the pigments for analytical purposes including simple mixing, heating and ultrasound assisted extraction. The solvents used for the extraction of these pigments have been mostly based on non-aqueous systems as the pigments are fat soluble. The solvent systems included methanol, acetone, concentrated KOH solution, and diethyl ether. The chromatographic methods employed for the analysis of the pigments included TLC and HPLC. For HPLC analysis reversed phase C18, C8 or C30 columns have been used with different combination of solvents, as summarized in Table 7.
TABLE 7
| Pigment | HPLC column type | Mobile phase | Source | Ref |
|---|---|---|---|---|
| Lutein | Reversed phase C18, reversed phase C8, reversed-phase C30 | Methanol:dichloromethane, Acetonitrile:distilled water, acetonitrile/methanol, methanol:ethyl acetate, methanol/acetone, ethyl acetate | Chlorella pyrenoidosa, Chlorella protothecoides, Chlorella vulgaris, Chlorella pyrenoidosa, Chlorella zofingiensis, Tetradedesmus sp., Coccomyxa acidophila, Dunaliella | (Shi et al., 1997, 2000; Wu et al. (2009); Cordero et al., 2010, 2011; Yen et al. (2011); Van Wagenen et al. (2015); Chen et al., 2016, 2017; Hamed et al. (2021) |
| Violaxanthin | Reversed phase C8, reversed phase C18 | Ethyl acetate, Acetonitrile/distilled water, dichloromethane/acetonitrile/methanol | Chlorella sorokiniana, Coccomyxa acidophila, Scenedesmus dimorphus | Casal et al. (2011); Van Wagenen et al. (2015); Ahmad et al. (2022) |
| Astaxanthin | Reversed phase C8 | Ethyl acetate, Acetonitrile/distilled water, dichloromethane/methanol/acetonitrile/water, acetonitrile/methanol/Tris–HCl, methanol/ethyl acetate, dichloromethane/acetonitrile/methanol | Chlorella sorokiniana, Haematococcus pluvialis, Chlorella zofingiensis, Scenedesmus dimorphus | Ip et al. (2004); Sun, (2006); Liu et al. (2013); Van Wagenen et al. (2015); Ahmad et al. (2022) |
| Fucoxanthin | Reversed phase C8, C30 | Water/methanol/acetonitrile, methanol/methyl tert-butyl ether/water | Chlorella sorokiniana, Phaeodactylum tricornutum, Nitzschia laevis | Van Wagenen et al. (2015); McClure et al. (2018); Lu et al. (2019); Jiang et al. (2022) |
| Zeaxanthin | Reversed phase C8, reversed phase C18 | Ethyl acetate, Acetonitrile/distilled water, dichloromethane/acetonitrile/methanol | Chlorella sorokiniana, Coccomyxa acidophila, Scenedesmus dimorphus | Casal et al. (2011); Van Wagenen et al. (2015); Ahmad et al. (2022) |
| Carotene | Reversed phase C8, reversed phase C18 | Methanol/dichloromethane,; Acetonitrile/distilled water, ethyl acetate, dichloromethane/acetonitrile/methanol | Chlorella sorokiniana, Coccomyxa acidophila, Scenedesmus dimorphus, Dunaliella | Casal et al. (2011); Van Wagenen et al. (2015); Hamed et al. (2021); Ahmad et al. (2022) |
A summary of chromatographic methods employed for the evaluation of different microalgal carotenoids.
4.2 Chlorophylls
Chlorophylls are tetrapyrroles with a centrally bound magnesium ion. Chemically, chlorophylls can be divided into chlorophylls a, b, c, d, e, and f. Chlorophylls show two absorption bands: 450–475 and 630–675 nm.
Chlorophylls are the green pigments responsible for the photosynthesis process in plants and algae. They have small differences in their absorption spectra (chlorophyll a is blue-green in color, chlorophyll b is brilliant green, chlorophyll c is yellow-green, chlorophyll d is brilliant/forest green, and chlorophyll f is emerald green). Microalgae Chlorella contains the two main types of chlorophylls (a, b) (up to 4.5% DW). Chlorophylls are chemically unstable molecules which make their analysis and characterization difficult in microalgae evidencing the need for specific tools such as chromatographic techniques for reliable results (Fernandes et al., 2020).
Most common methods for chlorophyll a and b extraction include solvent extraction using relatively polar organic solvents (e.g. dimethyl formamide, sec-butanol, methanol) that allows a rapid and efficient extraction from microalgae with a good stability of pigments (Schumann, Häubner, Klausch, & Karsten, 2005). A range of extraction techniques have been studied to maximize chlorophylls extraction, including MAE extraction for the extraction of chlorophyll a and b in Dunaliella tertiolecta and Cylindrotheca closterium (Ahmad et al., 2022).
After successful extraction, chlorophylls can be analysed by reversed phase HPLC using C18 or C30 columns. Mixtures of different polar aprotic solvents (dichloromethane, acetonitrile, acetone) and protic solvents (Diethyl ether, methyl tert-butyl ether, methanol, water, etc.) can be employed as mobile phase. Elution gradients (instead of isocratic operation) are preferred for more accurate results (Maroneze et al., 2019; Fernandes et al., 2020; Ahmad et al., 2022). In a recent research, Fernandes et al., revealed that HPLC-PDA with a C30 column can successfully separate different type of chlorophylls when a mixture of methanol:methyl tert-butyl ether:water is used as the mobile phase with a changing gradient (Fernandes et al., 2020).
4.3 Phycobiliproteins
Phycobiliproteins (PBPs) are unique pigments produced exclusively by cyanobacteria and microalgae. These pigments are composed of proteins that are covalently bound to chromophores called phycobilins, belonging to open-chain tetrapyrroles including phycocyanin (blue pigment), phycoerythrin (red pigment), and allophycocyanin (light-blue pigment).
Phycocyanin from Spirulina spp. and phycoerythrin from Porphyridium spp. are two of the most well-known phycobiliproteins produced commercially (Christaki et al., 2015). PBPs are auxiliary pigments, and highly fluorescent proteins with linear prosthetic groups (bilins) linked to specific cysteine residues. Depending on their composition and content of chromophores, PBPs as absorb at different wavelengths: phycocyanins (PC, λmax = 610–625 nm), phycoerythrins (PE, λmax = 490–570 nm), and allophycocyanins (APC, λmax = 650–660 nm) (Arashiro et al., 2020). Phycobiliproteins present antioxidant; anti-inflammatory, and anticancer activities (Pan-utai and Iamtham, 2019; Rodrigues et al., 2020).
A summary of different methods employed for the extraction and chromatographic analysis of phycobiliproteins is shown in Figure 6. Effective extraction of phycocyanin has always been a challenging issue, as the protein is bound to the cell structure of microalgae. Therefore, there is a need for cell disruption techniques such as high pressure homogenisation (Moraes et al., 2011), osmotic treatment (Doke, 2005), enzymatic treatment (Bhaskar et al., 2005), grinding, freeze-thawing, cold plasma, microwave treatment, high pressure processing, pulsed electric fields, and ultrasound assisted extraction (Soni et al., 2006; Chen et al., 2014). The pigments are water soluble and thus can be extracted directly using cold and hot aqueous system once they are liberated from cellular matrix by cell disruption. However, other novel solvents such as protic ionic liquids could be employed for simultaneous extraction and purification (Rodrigues et al., 2020).
FIGURE 6
Schematic representation of different methods for the extraction, purification, and analysis of phycocyanin from microalgae.
Chromatography plays a major role in both the purification and analysis of phycocyanin. The majority of the methods used in the literature rely on ammonium sulfate precipitation followed by a range of different chromatographic separation methods for the purification of phycocyanin. Application of ion chromatography, gel filtration chromatography and affinity chromatography alone or in combination with each other have been reported in the literature (Patel et al., 2005; Yan S.-G. et al., 2011; Song et al., 2013; Kissoudi et al., 2018).
Phycocyanin from S. platensis was purified using reversed phase HPLC equipped with PDA and fluorescence detector. Aqueous acetonitrile and trifluoroacetic acid were used in the mobile phase (Prabakaran et al., 2020). In a work by Rossano et al. (2003) purification of R-phycoerythrins from Corallina elongata was performed by a two-step chromatographic method using ion-exchange and gel filtration chromatography. Some researchers (Liu et al., 2005; Kissoudi et al., 2018; Zhao et al., 2020) coupled ion-exchange and affinity chromatography in a single run and introduced a one-step chromatography method to separate and purify phycocyanins, including initial precipitation of the target compound on the column followed by pH gradient elution. Some studies also employed a multistep procedure to purify phycocyanins from S. platensis. Initially ammonium ion was used to precipitate the proteins, then dialysis was performed to exclude small molecules followed by gel filtration chromatography by a Sephadex-G-100 column. The obtained fractions then were introduced to a reversed phase HPLC using a C5 column and eluted aqueous acetonitrile solution containing 0.1% (v/v) trifluoroacetic acid was used as the mobile phase.
Pigments are a diverse range of compounds with different properties found in microalgae. Reversed phase HPLC equipped with different column and detector types has been extensively used for the analysis of pigments. However, ion exchange HPLC and gel-filtration HPLC suggest several benefits for some pigment classes found in microalgae such as phycobiliproteins, as these compounds possess large molecules and proteins are found in their structure.
5 Concluding Remarks, Further Trends and Challenges
The results of this review paper indicated that, LC and GC methods coupled to different type of detectors provide powerful and reliable analytical tools for the identification and quantification of polyphenols, ω-3 PUFAs, and pigments present in different microalgae species. There could be limitations for each method depending on the nature of the compounds under investigation. The choice of the appropriate instrument and method is carried out based on different factors including volatility, solubility, sample preparation requirement, and accuracy.
Chromatographic techniques have been proved to be critical in not only building our understanding of microalgal metabolism, biosynthesis pathways and acclimation to adverse cultivation conditions but also in measuring the yields and effectiveness of downstream processes developed for the recovery of these high-value metabolites from the biomass. As such, chromatographic techniques play an indispensable analytical role in establishing both the technological viability and the economic feasibility of commercial-scale production of microalgal polyphenols, PUFAs and pigments for nutraceutical and food applications. In addition to LC and GC methods described in Section 3, 4, and 5, there are other chromatographic methods that can potentially be applied in the development of microalgal biotechnology.
In addition to sample analysis, chromatography can also be used for sample preparation and purification. Preparative chromatography is a powerful method that can be used to fractionate/purify microalgal lipid extract to constituent lipid, polyphenol and pigments classes prior to analytical chromatography. Preparative chromatography, however, has scarcely been applied for microalgae. We were only able to find a few studies demonstrating the use preparative chromatography for the isolation of peptides from Spirulina Plantensis (Hu et al., 2019), the separation of pigments from Nitzschia laevis (Sun et al., 2019) or polyphenol fractionation from Chlamydomonas sp. KSF108 (Khanh Tran et al., 2019).
Multi-dimensional preparative chromatographic techniques such as GC x GC or/and LC x LC coupled to MS system can be used to enable both the purification and identification of new phytochemicals in microalgae matrices and are particularly useful for sample screening with minimal clean-up. Countercurrent chromatography has been already used in microalgae matrices to separate pigments and polyphenols. Li and coworkers presented a study using preparative High-Speed Countercurrent Chromatography (HSCCC) with biphasic solvent system (n-hexane-ethanol-water in the ratio of 10:9:1 v/v) for the purification of crude extract of canthaxanthin from the microalga Chlorella zofingiensis to a purity level of about 98.7% (Li et al., 2006). Another emerging chromatographic technique that can be used in industrial scale for product purification is Expanded Bed Adsorption (EBA) chromatography that was recently described for isolation of phycoerythrin) from the microalga Porphyridium cruentum, using dimethylaminoethyl cellulose (DEAE-c) as a resin (Bermejo et al., 2003).
Supercritical fluid chromatography (SFC) is a relatively recent addition to analytical chromatographic technique. The difference of supercritical fluid chromatography from other chromatographic techniques is the use of a supercritical fluid as the mobile phase. This has the advantages of reducing solvent requirements and retention time, while also being able to analyze thermolabile compounds that GC cannot (Ajila et al., 2011). In this regard, SFC using a C18 column and a 2-ethyl pyridine column was recently used for the quantitative determination of 8 different carotenoids in the extracts of Scenedesmus sp. The method was able to validate and quantify the 8 carotenoids in Scenedesmus sp. extract. A recent study has also successfully coupled supercritical fluid extraction and supercritical fluid chromatography-tandem mass-spectrometry for the sequential extraction and characterization of 28 apocarotenoids from four different microalgae strains: Chlamydomonas sp, Tetraselmis chuii S, Nannochloropsis gaditana and Chlorella sorokiniana (Zoccali et al., 2019).
Statements
Author contributions
All authors have read and agreed to the published version of the manuscript. Conceptualization, RA; writing—original draft preparation, RA, RH, HK; writing—review, RA, MMP, HK, and RH; supervision, BT; manuscript proof reading, RH; editing, RA; project administration, RA.
Funding
Enterprise Ireland Project ID: MF 2020 0108.
Acknowledgments
We acknowledge Enterprise Ireland and the European Union’s Horizon 2020 Research and innovation Programme under the Marie Skłodowska-Curie Co-funding of regional, national and international programmes Grant agreement No: 847402.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abrahamsson V. Rodriguez-Meizoso I. Turner C. (2012). Determination of Carotenoids in Microalgae Using Supercritical Fluid Extraction and Chromatography. J. Chromatogr. A1250, 63–68. 10.1016/j.chroma.2012.05.069
2
Adetunji C. O. Olaniyan O. T. Anani O. A. Bodunrinde R. E. Osemwegie O. O. Ubi B. E. (2022). “Integrated Processes for Production of Pharmaceutical Products from Agro-Wastes,” in Biomass, Biofuels, Biochemicals. Editors VarjaniS.PandeyA.BhaskarT.MohanS. V.TsangD. C. W. (Elsevier), 439–461. 10.1016/b978-0-323-89855-3.00021-2
3
Ahmad N. Mounsef J. R. Lteif R. (2022). Pigment Production by Scenedesmus Dimorphus Using Different Low‐cost and Alternative Culture Media. J. Chem. Technol. Biotechnol.97, 287–294. 10.1002/jctb.6940
4
Ahmed E. A. (2016). Antimicrobial Activity of Microalgal Extracts Isolated from Baharia Oasis. Egypt.
5
Ainsworth E. A. Gillespie K. M. (2007). Estimation of Total Phenolic Content and Other Oxidation Substrates in Plant Tissues Using Folin-Ciocalteu Reagent. Nat. Protoc.2, 875–877. 10.1038/nprot.2007.102
6
Ajila C. M. Brar S. K. Verma M. Tyagi R. D. Godbout S. Valéro J. R. (2011). Extraction and Analysis of Polyphenols: Recent Trends. Crit. Rev. Biotechnol.31, 227–249. 10.3109/07388551.2010.513677
7
Akbarizareh M. Ofoghi H. Hadizadeh M. (2019). Assessment of Phenolic Components of the Microalgae Spirulina Platensis Using Two Methods of Chromatography, TLC and HPLC. J. Mar. Biol.11, 13–24.
8
Al-Zuhair S. Ashraf S. Hisaindee S. Darmaki N. A. Battah S. Svistunenko D. et al (2017). Enzymatic Pre-treatment of Microalgae Cells for Enhanced Extraction of Proteins. Eng. Life Sci.17, 175–185. 10.1002/elsc.201600127
9
Alalayah W. M. Al-Zahrani A. Edris G. Demirbas A. (2017). Kinetics of Biological Hydrogen Production from Green Microalgae Chlorella Vulgaris Using Glucose as Initial Substrate. Energy Sources, Part A Recovery, Util. Environ. Eff.39, 1210–1215. 10.1080/15567036.2017.1315755
10
Ambati R. R. Gogisetty D. Aswathanarayana R. G. Ravi S. Bikkina P. N. Bo L. et al (2019). Industrial Potential of Carotenoid Pigments from Microalgae: Current Trends and Future Prospects. Crit. Rev. Food Sci. Nutr.59, 1880–1902. 10.1080/10408398.2018.1432561
11
Ameer K. Shahbaz H. M. Kwon J.-H. (2017). Green Extraction Methods for Polyphenols from Plant Matrices and Their Byproducts: A Review. Compr. Rev. Food Sci. Food Saf.16, 295–315. 10.1111/1541-4337.12253
12
Anwer S. S. Sdiq K. H. Muhammad K. R. Aladdin L. M. (2022). Phenolic Compound and Fatty Acid Properties of Some Microalgae Species Isolated from Erbil City. Braz. J. Biol.82. 10.1590/1519-6984.256927
13
Archer S. Tarran G. Stephens J. Butcher L. Kimmance S. (2011). Combining Cell Sorting with Gas Chromatography to Determine Phytoplankton Group-specific Intracellular Dimethylsulphoniopropionate. Aquat. Microb. Ecol.62, 109–121. 10.3354/ame01464
14
Richmond A. Hu Q. (Editors) (2013). Handbook of Microalgal Culture (Oxford, UK: John Wiley & Sons). 10.1002/9781118567166
15
Ashaolu T. J. Samborska K. Lee C. C. Tomas M. Capanoglu E. Tarhan Ö. et al (2021). Phycocyanin, a Super Functional Ingredient from Algae; Properties, Purification Characterization, and Applications. Int. J. Biol. Macromol.193, 2320–2331. 10.1016/j.ijbiomac.2021.11.064
16
Baker R. Günther C. (2004). The Role of Carotenoids in Consumer Choice and the Likely Benefits from Their Inclusion into Products for Human Consumption. Trends Food Sci. Technol.15, 484–488. 10.1016/j.tifs.2004.04.0094
17
Barati B. Zafar F. F. Qian L. Wang S. El-Fatah Abomohra A. (2022). Bioenergy Characteristics of Microalgae under Elevated Carbon Dioxide. Fuel321, 123958. 10.1016/j.fuel.2022.123958
18
Barkia I. Saari N. Manning S. R. (2019). Microalgae for High-Value Products towards Human Health and Nutrition. Mar. Drugs17, 304. 10.3390/md17050304
19
Bermejo R. Gabriel Acién F. Ibáñez M. J. Fernández J. M. Molina E. Alvarez-Pez J. M. (2003). Preparative Purification of B-Phycoerythrin from the Microalga Porphyridium Cruentum by Expanded-Bed Adsorption Chromatography. J. Chromatogr. B790, 317–325. 10.1016/s1570-0232(03)00168-5
20
Bernal J. L. Martín M. T. Toribio L. (2013). Supercritical Fluid Chromatography in Food Analysis. J. Chromatogr. A1313, 24–36. 10.1016/j.chroma.2013.07.022
21
Bhaskar S. U. Gopalaswamy G. Raghu R. (2005). A Simple Method for Efficient Extraction and Purification of C-Phycocyanin from Spirulina Platensis Geitler.
22
Bielser J.-M. Aeby M. Caso S. Roulet A. Broly H. Souquet J. (2021). Continuous Bleed Recycling Significantly Increases Recombinant Protein Production Yield in Perfusion Cell Cultures. Biochem. Eng. J.169, 107966. 10.1016/j.bej.2021.107966
23
Birch A. J. Donovan F. W. Moewus F. (1953). Biogenesis of Flavonoids in Chlamydomonas Eugametos. Nature172, 902–904. 10.1038/172902a0
24
Buono S. Langellotti A. L. Martello A. Rinna F. Fogliano V. (2014). Functional Ingredients from Microalgae.
25
Cajka T. Fiehn O. (2014). Comprehensive Analysis of Lipids in Biological Systems by Liquid Chromatography-Mass Spectrometry. TrAC Trends Anal. Chem.61, 192–206. 10.1016/j.trac.2014.04.017
26
Caporgno M. P. Mathys A. (2018). Trends in Microalgae Incorporation into Innovative Food Products with Potential Health Benefits. Front. Nutr.5, 58. 10.3389/fnut.2018.00058
27
Cardoso C. Pereira H. Franca J. Matos J. Monteiro I. Pousão-Ferreira P. et al (2020). Lipid Composition and Some Bioactivities of 3 Newly Isolated Microalgae (Tetraselmis Sp. IMP3, Tetraselmis Sp. CTP4, and Skeletonema sp.). Aquacult Int.28, 711–727. 10.1007/s10499-019-00489-w
28
Carrasco-Pancorbo A. Neusüß C. Pelzing M. Segura-Carretero A. Fernández-Gutiérrez A. (2007). CE- and HPLC-TOF-MS for the Characterization of Phenolic Compounds in Olive Oil. Electrophoresis28, 806–821. 10.1002/elps.200600382
29
Casal C. Cuaresma M. Vega J. M. Vilchez C. (2011). Enhanced Productivity of a Lutein-Enriched Novel Acidophile Microalga Grown on Urea. Mar. Drugs9, 29–42. 10.3390/md9010029
30
Castillo A. Pereira S. Otero A. Garcia-Jares C. Lores M. (2022). Multicomponent Bioactive Extract from Red Stage Haematococcus pluvialis Wet Paste: Avoiding the Drying Step and Toxic Solvents. J. Appl. Phycol., 1–17. 10.1007/s10811-022-02712-3
31
Chen C.-Y. JesiscaHsieh C. Hsieh C. Lee D.-J. Chang C.-H. Chang J.-S. (2016). Production, Extraction and Stabilization of Lutein from Microalga Chlorella Sorokiniana MB-1. Bioresour. Technol.200, 500–505. 10.1016/j.biortech.2015.10.071
32
Chen H.-W. Yang T.-S. Chen M.-J. Chang Y.-C. Wang E. I.-C. Ho C.-L. et al (2014). Purification and Immunomodulating Activity of C-Phycocyanin from Spirulina Platensis Cultured Using Power Plant Flue Gas. Process Biochem.49, 1337–1344. 10.1016/j.procbio.2014.05.006
33
Chen J.-H. Chen C.-Y. Chang J.-S. (2017). Lutein Production with Wild-type and Mutant Strains of Chlorella Sorokiniana MB-1 under Mixotrophic Growth. J. Taiwan Inst. Chem. Eng.79, 66–73. 10.1016/j.jtice.2017.04.022
34
Chollet C. Boutet-Mercey S. Laboureur L. Rincon C. Méjean M. Jouhet J. et al (2019). Supercritical Fluid Chromatography Coupled to Mass Spectrometry for Lipidomics. J. Mass Spectrom.54, 791–801. 10.1002/jms.4445
35
Cinq-Mars M. Bourdeau N. Marchand P. Desgagné-Penix I. Barnabé S. (2022). Characterization of Two Microalgae Consortia Grown in Industrial Wastewater for Biomass Valorization. Algal Res.62, 102628. 10.1016/j.algal.2021.102628
36
Cordero B. F. Obraztsova I. Couso I. Leon R. Vargas M. A. Rodriguez H. (2011). Enhancement of Lutein Production in Chlorella Sorokiniana (Chorophyta) by Improvement of Culture Conditions and Random Mutagenesis. Mar. Drugs9, 1607–1624. 10.3390/md9091607
37
Cordero B. F. Obraztsova I. Martín L. Couso I. León R. Ángeles Vargas M. et al (2010). Isolation and Characterization of a Lycopene β-Cyclase gene From the Astaxanthin-Producing Green Alga Chlorella Zofingiensis (Chlorophyta)1. J. Phycol.46, 1229–1238. 10.1111/j.1529-8817.2010.00907.x
38
Cretton M. Malanga G. Mazzuca Sobczuk T. Mazzuca M. (2022). Marine Lipids as a Source of High-Quality Fatty Acids and Antioxidants. Food Rev. Int., 1–24. 10.1080/87559129.2022.2042555
39
Cutignano A. Luongo E. Nuzzo G. Pagano D. Manzo E. Sardo A. et al (2016). Profiling of Complex Lipids in Marine Microalgae by UHPLC/tandem Mass Spectrometry. Algal Res.17, 348–358. 10.1016/j.algal.2016.05.016
40
Dahmen-Ben Moussa I. Belhaj D. Ayadi H. (2022). Optimization of Extraction Process of Polyphenols from Wild Halotolerant Cyanobacteria, Phormidium Versicolor (NCC 466). Biomass Convers. Biorefinery, 1–11. 10.1007/s13399-022-02651-6
41
David F. Vickers A. K. (2005). Column Selection for the Analysis of Fatty Acid Methyl Esters Application. Agil. Appl. Note, 1–12.
42
Dębowski M. Zieliński M. Kazimierowicz J. Kujawska N. Talbierz S. (2020). Microalgae Cultivation Technologies as an Opportunity for Bioenergetic System Development—Advantages and Limitations. Sustainability12, 9980.
43
Derrien A. Coiffard L. J. M. Coiffard C. De Roeck-Holtzhauer Y. (1998). Free Amino Acid Analysis of Five Microalgae. J. Appl. Phycol.10, 131–134. 10.1023/a:1008003016458
44
Dewi I. C. Falaise C. Hellio C. Bourgougnon N. Mouget J.-L. (2018). “Anticancer, Antiviral, Antibacterial, and Antifungal Properties in Microalgae,” in Microalgae in Health and Disease Prevention (Elsevier), 235–261. 10.1016/b978-0-12-811405-6.00012-8
45
Dogra B. Amna S. Park Y. I. Park J. K. (2017). Biochemical Properties of Water Soluble Polysaccharides from Photosynthetic Marine Microalgae Tetraselmis Species. Macromol. Res.25, 172–179. 10.1007/s13233-017-5016-x
46
Doke J. M. (2005). An Improved and Efficient Method for the Extraction of Phycocyanin from Spirulina Sp. Int. J. Food Eng.1. 10.2202/1556-3758.1037
47
Dołowy M. Pyka A. (2015). Chromatographic Methods in the Separation of Long-Chain Mono-And Polyunsaturated Fatty Acids, 2015, 1–20. 10.1155/2015/120830Chromatographic Methods in the Separation of Long-Chain Mono- and Polyunsaturated Fatty AcidsJ. Chem.
48
Domingues M. R. Calado R. (2022). Lipids of Marine Algae-Biomolecules with High Nutritional Value and Important Bioactive Properties. Biomolecules12, 134. 10.3390/biom12010134
49
Donot F. Cazals G. Gunata Z. Egron D. Malinge J. Strub C. et al (2013). Analysis of Neutral Lipids from Microalgae by HPLC-ELSD and APCI-MS/MS. J. Chromatogr. B942-943, 98–106. 10.1016/j.jchromb.2013.10.016
50
Donot F. Strub C. Fontana A. Jouy N. Delbes C. Gunata Z. et al (2016). Rapid Analysis and Quantification of Major Neutral Lipid Species and Free Fatty Acids by HPLC‐ELSD from Microalgae. Eur. J. Lipid Sci. Technol.118, 1550–1556. 10.1002/ejlt.201500545
51
Dufossé L. Galaup P. Yaron A. Arad S. M. Blanc P. Chidambara Murthy K. N. et al (2005). Microorganisms and Microalgae as Sources of Pigments for Food Use: a Scientific Oddity or an Industrial Reality?Trends Food Sci. Technol.16, 389–406. 10.1016/j.tifs.2005.02.006
52
Sun D.-W. (Editor) (2006). “Physical-chemical Principles in Freezing,” Handbook of Frozen Food Packaging and Processing (Boca Raton, FL: Taylor & Francis).
53
Elshobary M. E. El-Shenody R. A. Ashour M. Zabed H. M. Qi X. (2020). Antimicrobial and Antioxidant Characterization of Bioactive Components from Chlorococcum Minutum. Food Biosci.35, 100567. 10.1016/j.fbio.2020.100567
54
Emami K. Hack E. Nelson A. Brain C. M. Lyne F. M. Mesbahi E. et al (2015). Proteomic-based Biotyping Reveals Hidden Diversity within a Microalgae Culture Collection: An Example Using Dunaliella. Sci. Rep.5, 1–15. 10.1038/srep10036
55
Evans T. W. Elling F. J. Li Y. Pearson A. Summons R. E. (2022). A New and Improved Protocol for Extraction of Intact Polar Membrane Lipids from Archaea. Org. Geochem.165, 104353. 10.1016/j.orggeochem.2021.104353
56
Fagundes M. B. Vendruscolo R. G. Wagner R. (2020). “Sterols from Microalgae,” in Handbook of Microalgae-Based Processes and Products (Elsevier), 573–596. 10.1016/b978-0-12-818536-0.00021-x
57
Fernandes A. S. Petry F. C. Mercadante A. Z. Jacob-Lopes E. Zepka L. Q. (2020). HPLC-PDA-MS/MS as a Strategy to Characterize and Quantify Natural Pigments from Microalgae. Curr. Res. Food Sci.3, 100–112. 10.1016/j.crfs.2020.03.009
58
Fernandes T. V. Trebuch L. M. Wijffels R. H. (2022). “Microalgae-based Technologies for Circular Wastewater Treatment,” in Integrated Wastewater Management and Valorization Using Algal Cultures (Elsevier), 81–112. 10.1016/b978-0-323-85859-5.00001-4
59
Fischer J. Treblin M. Sitz T. Rohn S. (2021). Development of a Targeted HPLC-ESI-QqQ-MS/MS Method for the Quantification of Sulfolipids from a Cyanobacterium, Selected Leafy Vegetables, and a Microalgae Species. Anal. Bioanal. Chem.413, 1941–1954. 10.1007/s00216-021-03164-3
60
Fredrick J. F. (1975). Affinity Chromatography Studies of the De Novo Glucan Synthesizing Phosphorylase Isozyme of Blue-Green Algae. Plant Sci. Lett.5, 131–135. 10.1016/0304-4211(75)90054-1
61
Friedman M. Jürgens H. S. (2000). Effect of pH on the Stability of Plant Phenolic Compounds. J. Agric. Food Chem.48, 2101–2110. 10.1021/jf990489j
62
Gill B. S. Navgeet, Qiu F. (2020). “Technologies for Extraction and Production of Bioactive Compounds,” in Biotechnological Production of Bioactive Compounds (Elsevier), 1–36. 10.1016/b978-0-444-64323-0.00001-1
63
Goh B. H. H. Ong H. C. Cheah M. Y. Chen W.-H. Yu K. L. Mahlia T. M. I. (2019). Sustainability of Direct Biodiesel Synthesis from Microalgae Biomass: A Critical Review. Renew. Sustain. Energy Rev.107, 59–74. 10.1016/j.rser.2019.02.012
64
Goiris K. Muylaert K. Voorspoels S. Noten B. De Paepe D. E Baart G. J. et al (2014). Detection of Flavonoids in Microalgae from Different Evolutionary Lineages. J. Phycol.50, 483–492. 10.1111/jpy.12180
65
Goleniowski M. Bonfill M. Cusido R. Palazón J. (2013). “Phenolic Acids,” in Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes (Springer Berlin Heidelberg), 1951–1973. 10.1007/978-3-642-22144-6_64
66
Gómez-Ordóñez E. Jiménez-Escrig A. Rupérez P. (2012). Molecular Weight Distribution of Polysaccharides from Edible Seaweeds by High-Performance Size-Exclusion Chromatography (HPSEC). Talanta93, 153–159.
67
González-Hourcade M. Del Campo E. M. Braga M. R. Salgado A. Casano L. M. (2020). Disentangling the Role of Extracellular Polysaccharides in Desiccation Tolerance in Lichen-Forming Microalgae. First Evidence of Sulfated Polysaccharides and Ancient Sulfotransferase Genes. Environ. Microbiol.22, 3096–3111. 10.1111/1462-2920.15043
68
Goswami R. K. Mehariya S. Karthikeyan O. P. Gupta V. K. Verma P. (2022). Multifaceted Application of Microalgal Biomass Integrated with Carbon Dioxide Reduction and Wastewater Remediation: A Flexible Concept for Sustainable Environment. J. Clean. Prod.339, 130654. 10.1016/j.jclepro.2022.130654
69
Gu C. Howell K. Dunshea F. R. Suleria H. A. R. (2019). LC-ESI-QTOF/MS Characterisation of Phenolic Acids and Flavonoids in Polyphenol-Rich Fruits and Vegetables and Their Potential Antioxidant Activities. Antioxidants8, 405. 10.3390/antiox8090405
70
Gu Q. David F. Lynen F. Vanormelingen P. Vyverman W. Rumpel K. et al (2011). Evaluation of Ionic Liquid Stationary Phases for One Dimensional Gas Chromatography-Mass Spectrometry and Comprehensive Two Dimensional Gas Chromatographic Analyses of Fatty Acids in Marine Biota. J. Chromatogr. A1218, 3056–3063. 10.1016/j.chroma.2011.03.011
71
Guidetti Vendruscolo R. Bittencourt Fagundes M. Jacob-Lopes E. Wagner R. (2019). Analytical Strategies for Using Gas Chromatography to Control and Optimize Microalgae Bioprocessing. Curr. Opin. Food Sci.25, 73–81. 10.1016/j.cofs.2019.02.008
72
Guillaumie F. Justesen S. F. L. Mutenda K. E. Roepstorff P. Jensen K. J. Thomas O. R. T. (2006). Fractionation, Solid-phase Immobilization and Chemical Degradation of Long Pectin Oligogalacturonides. Initial Steps towards Sequencing of Oligosaccharides. Carbohydr. Res.341, 118–129. 10.1016/j.carres.2005.10.011
73
Hahn T. Zayed A. Kovacheva M. Stadtmüller R. Lang S. Muffler K. et al (2016). Dye Affinity Chromatography for Fast and Simple Purification of Fucoidan from Marine Brown Algae. Eng. Life Sci.16, 78–87. 10.1002/elsc.201500044
74
Halim R. Danquah M. K. (2012). “Bioprocess Development for Chlorophyll Extraction from Microalgae,” in Advanced Biofuels and Bioproducts (New York: Springer), 807–832. 10.1007/978-1-4614-3348-4_34
75
Halim R. Danquah M. K. Webley P. A. (2012). Extraction of Oil from Microalgae for Biodiesel Production: A Review. Biotechnol. Adv.30, 709–732. 10.1016/j.biotechadv.2012.01.001
76
Halim R. Hill D. R. A. Hanssen E. Webley P. A. Blackburn S. Grossman A. R. et al (2019). Towards Sustainable Microalgal Biomass Processing: Anaerobic Induction of Autolytic Cell-Wall Self-Ingestion in Lipid-Rich Nannochloropsis Slurries. Green Chem.21, 2967–2982. 10.1039/c8gc03186j
77
Hamed I. Işık O. Ak Çimen B. Uslu L. Kafkas E. Zarifikhosroshahi M. (2021). Influence of Stress Factors on Growth and Pigment Production in Three Dunaliella Species Cultivated Outdoors in Flat-Plate Photobioreactors. Plant Biosyst. - Int. J. Deal. all Aspects Plant Biol.155, 179–187. 10.1080/11263504.2020.1727985
78
Haoujar I. Cacciola F. Abrini J. Mangraviti D. Giuffrida D. Oulad El Majdoub Y. et al (2019). The Contribution of Carotenoids, Phenolic Compounds, and Flavonoids to the Antioxidative Properties of Marine Microalgae Isolated from Mediterranean Morocco. Molecules24, 4037. 10.3390/molecules24224037
79
Herrero M. Cifuentes A. Ibanez E. (2006). Sub- and Supercritical Fluid Extraction of Functional Ingredients from Different Natural Sources: Plants, Food-By-Products, Algae and microalgaeA Review. Food Chem.98, 136–148. 10.1016/j.foodchem.2005.05.058
80
Hori K. Sato Y. Ito K. Fujiwara Y. Iwamoto Y. Makino H. et al (2007). Strict Specificity for High-Mannose Type N-Glycans and Primary Structure of a Red Alga Eucheuma Serra Lectin. Glycobiology17, 479–491. 10.1093/glycob/cwm007
81
Hrebien V. Deschaseaux E. Eickhoff W. Swan H. B. Eyre B. D. (2020). Quantification of Isoprene in Coastal Ecosystems by Gas Chromatography-Mass Spectrometry Using Cumulative Headspace Injections. Limnol. Oceanogr. Methods18, 374–382. 10.1002/lom3.10363
82
Hu J. Nagarajan D. Zhang Q. Chang J.-S. Lee D.-J. (2018). Heterotrophic Cultivation of Microalgae for Pigment Production: A Review. Biotechnol. Adv.36, 54–67. 10.1016/j.biotechadv.2017.09.009
83
Hu S. Fan X. Qi P. Zhang X. (2019). Identification of Anti-diabetes Peptides from Spirulina Platensis. J. Funct. Foods56, 333–341. 10.1016/j.jff.2019.03.024
84
Ip P.-F. Wong K.-H. Chen F. (2004). Enhanced Production of Astaxanthin by the Green Microalga Chlorella Zofingiensis in Mixotrophic Culture. Process Biochem.39, 1761–1766. 10.1016/j.procbio.2003.08.003
85
Jakobsen A. N. Aasen I. M. Josefsen K. D. Strøm A. R. (2008). Accumulation of Docosahexaenoic Acid-Rich Lipid in Thraustochytrid Aurantiochytrium Sp. Strain T66: Effects of N and P Starvation and O2 Limitation. Appl. Microbiol. Biotechnol.80, 297–306. 10.1007/s00253-008-1537-8
86
Jeffrey S. (1961). Paper-chromatographic Separation of Chlorophylls and Carotenoids from Marine Algae. Biochem. J.80, 336–342. 10.1042/bj0800336
87
Jerez-Martel I. García-Poza S. Rodríguez-Martel G. Rico M. Afonso-Olivares C. Gómez-Pinchetti J. L. (2017). Phenolic Profile and Antioxidant Activity of Crude Extracts from Microalgae and Cyanobacteria Strains. J. Food Qual.2017, 1–8. 10.1155/2017/2924508
88
Jia J. Han D. Gerken H. G. Li Y. Sommerfeld M. Hu Q. et al (2015). Molecular Mechanisms for Photosynthetic Carbon Partitioning into Storage Neutral Lipids in Nannochloropsis Oceanica under Nitrogen-Depletion Conditions. Algal Res.7, 66–77. 10.1016/j.algal.2014.11.005
89
Jiang J. Huang J. Zhang H. Zhang Z. Du Y. Cheng Z. et al (2022). Potential Integration of Wastewater Treatment and Natural Pigment Production by Phaeodactylum Tricornutum: Microalgal Growth, Nutrient Removal, and Fucoxanthin Accumulation. J. Appl. Phycol.10.1007/s10811-022-02700-7
90
Jiang X. Wang H. Hu E. Lei Z. Fan B. Wang Q. (2020). Efficient Adsorption of Uranium from Aqueous Solutions by Microalgae Based Aerogel. Microporous Mesoporous Mater.305, 110383. 10.1016/j.micromeso.2020.110383
91
Jouhet J. Lupette J. Clerc O. Magneschi L. Bedhomme M. Collin S. et al (2017). LC-MS/MS versus TLC Plus GC Methods: Consistency of Glycerolipid and Fatty Acid Profiles in Microalgae and Higher Plant Cells and Effect of a Nitrogen Starvation. PLoS One12, e0182423. 10.1371/journal.pone.0182423
92
Jumaah F. Plaza M. Abrahamsson V. Turner C. Sandahl M. (2016). A Fast and Sensitive Method for the Separation of Carotenoids Using Ultra-high Performance Supercritical Fluid Chromatography-Mass Spectrometry. Anal. Bioanal. Chem.408, 5883–5894. 10.1007/s00216-016-9707-5
93
Kchech A. (2017). The Quantification of Fatty Acids in Microalgae Using GC-fid and GS-ms.
94
Khanh Tran H. N. Kim J. A. Youn U. J. Kim S. Woo M. H. Min B. S. (2019). Investigation of Chemical Compounds from Chlamydomonas Sp. KSF108 (Chlamydomonadaceae). Biochem. Syst. Ecol.83, 4–6. 10.1016/j.bse.2018.12.009
95
Kissoudi M. Sarakatsianos I. Samanidou V. (2018). Isolation and Purification of Food-Grade C-Phycocyanin from Arthrospira Platensis and its Determination in Confectionery by HPLC with Diode Array Detection. J. Sep. Sci.41, 975–981. 10.1002/jssc.201701151
96
Klejdus B. Kopecký J. Benešová L. Vacek J. (2009). Solid-phase/supercritical-fluid Extraction for Liquid Chromatography of Phenolic Compounds in Freshwater Microalgae and Selected Cyanobacterial Species. J. Chromatogr. A1216, 763–771. 10.1016/j.chroma.2008.11.096
97
Klejdus B. Lojková L. Vlcek J. (2013). Hyphenated Solid Phase Extraction/Supercritical Fluid Extraction Methods for Extraction of Phenolic Compounds from Algae. Cac10, 86–98. 10.2174/1573411011410010008
98
Kroumova A. B. Wagner G. J. (1995). Methods for Separation of Free, Short, Medium, and Long Chain Fatty Acids and for Their Decarboxylation. Anal. Biochem.225, 270–276. 10.1006/abio.1995.1153
99
Kujawska N. Talbierz S. Dębowski M. Kazimierowicz J. Zieliński M. (2021). Optimizing Docosahexaenoic Acid (DHA) Production by Schizochytrium Sp. Grown on Waste Glycerol. Energies14, 1685. 10.3390/en14061685
100
Kukula-Koch W. Koch W. Angelis A. Halabalaki M. Aligiannis N. (2016). Application of pH-Zone Refining Hydrostatic Countercurrent Chromatography (hCCC) for the Recovery of Antioxidant Phenolics and the Isolation of Alkaloids from Siberian Barberry Herb. Food Chem.203, 394–401. 10.1016/j.foodchem.2016.02.096
101
Kumari V. B. C. Patil S. M. Ramu R. Shirahatti P. S. Kumar N. Sowmya B. P. et al (2022). “Chromatographic Techniques: Types, Principles, and Applications,” in Analytical Techniques in Biosciences (Elsevier), 73–101. 10.1016/b978-0-12-822654-4.00013-0
102
Laboureur L. Ollero M. Touboul D. (2015). Lipidomics by Supercritical Fluid Chromatography. Ijms16, 13868–13884. 10.3390/ijms160613868
103
Lage S. Gentili F. G. (2018). Quantification and Characterisation of Fatty Acid Methyl Esters in Microalgae: Comparison of Pretreatment and Purification Methods. Bioresour. Technol.257, 121–128. 10.1016/j.biortech.2018.01.153
104
Law S. Q. K. Halim R. Scales P. J. Martin G. J. O. (2018). Conversion and Recovery of Saponifiable Lipids from Microalgae Using a Nonpolar Solvent via Lipase-Assisted Extraction. Bioresour. Technol.260, 338–347. 10.1016/j.biortech.2018.03.129
105
Li H.-B. Fan K.-W. Chen F. (2006). Isolation and Purification of Canthaxanthin from the microalgaChlorella Zofingiensis by High-Speed Counter-current Chromatography. J. Sep. Sci.29, 699–703. 10.1002/jssc.200500365
106
Liu J. Sun Z. Zhong Y. Gerken H. Huang J. Chen F. (2013). Utilization of Cane Molasses towards Cost-Saving Astaxanthin Production by a Chlorella Zofingiensis Mutant. J. Appl. Phycol.25, 1447–1456. 10.1007/s10811-013-9974-x
107
Liu L.-N. Chen X.-L. Zhang X.-Y. Zhang Y.-Z. Zhou B.-C. (2005). One-step Chromatography Method for Efficient Separation and Purification of R-Phycoerythrin from Polysiphonia urceolata. J. Biotechnol.116, 91–100. 10.1016/j.jbiotec.2004.09.017
108
Lou S. Zhu X. Zeng Z. Wang H. Jia B. Li H. et al (2020). Identification of microRNAs Response to High Light and Salinity that Involved in Beta-Carotene Accumulation in Microalga Dunaliella Salina. Algal Res.48, 101925. 10.1016/j.algal.2020.101925
109
Low K. L. Idris A. Mohd Yusof N. (2020). Novel Protocol Optimized for Microalgae Lutein Used as Food Additives. Food Chem.307, 125631. 10.1016/j.foodchem.2019.125631
110
Lu X. Liu B. He Y. Guo B. Sun H. Chen F. (2019). Novel Insights into Mixotrophic Cultivation of Nitzschia Laevis for Co-production of Fucoxanthin and Eicosapentaenoic Acid. Bioresour. Technol.294, 122145. 10.1016/j.biortech.2019.122145
111
Maalej A. Dahmen-Ben Moussa I. Karray F. Chamkha M. Sayadi S. (2022). Olive Oil By-Product’s Contribution to the Recovery of Phenolic Compounds from Microalgal Biomass: Biochemical Characterization, Anti-melanogenesis Potential, and Neuroprotective Effect. Biomass Convers. Biorefinery, 1–13.
112
Machu L. Misurcova L. Vavra Ambrozova J. Orsavova J. Mlcek J. Sochor J. et al (2015). Phenolic Content and Antioxidant Capacity in Algal Food Products. Molecules20, 1118–1133. 10.3390/molecules20011118
113
Markham K. R. Porter L. J. (1969). Flavonoids in the Green Algae (Chlorophyta). Phytochemistry8, 1777–1781. 10.1016/s0031-9422(00)85968-3
114
Maroneze M. M. Zepka L. Q. Lopes E. J. Pérez-Gálvez A. Roca M. (2019). Chlorophyll Oxidative Metabolism during the Phototrophic and Heterotrophic Growth of Scenedesmus Obliquus. Antioxidants8, 600. 10.3390/antiox8120600
115
Martín-Girela I. Albero B. Tiwari B. K. Miguel E. Aznar R. (2020). Screening of Contaminants of Emerging Concern in Microalgae Food Supplements. Separations7, 28. 10.3390/separations7020028
116
Matsubara A. Bamba T. Ishida H. Fukusaki E. Hirata K. (2009). Highly Sensitive and Accurate Profiling of Carotenoids by Supercritical Fluid Chromatography Coupled with Mass Spectrometry. J. Sep. Sci.32, 1459–1464. 10.1002/jssc.200800699
117
Matuszewski B. K. Constanzer M. L. Chavez-Eng C. M. (2003). Strategies for the Assessment of Matrix Effect in Quantitative Bioanalytical Methods Based on HPLC−MS/MS. Anal. Chem.75, 3019–3030. 10.1021/ac020361s
118
McClure D. D. Luiz A. Gerber B. Barton G. W. Kavanagh J. M. (2018). An Investigation into the Effect of Culture Conditions on Fucoxanthin Production Using the Marine Microalgae Phaeodactylum Tricornutum. Algal Res.29, 41–48. 10.1016/j.algal.2017.11.015
119
McSheehy S. Szpunar J. (2000). Speciation of Arsenic in Edible Algae by Bi-dimensional Size-Exclusion Anion Exchange HPLC with Dual ICP-MS and Electrospray MS/MS Detection. J. Anal. At. Spectrom.15, 79–87. 10.1039/a906890b
120
Megawati D. S. Fasya A. G. Pratiwi R. A. Maghfiroh N. (2020). “Pharmacology Potency of Thin Layer Chromatography Steroid Isolates of Chlorella Sp. Chloroform Fraction,” in IOP Conference Series: Earth and Environmental Science (United Kingdom: IOP Publishing). 10.1088/1755-1315/456/1/012012
121
Mimouni V. Ulmann L. Pasquet V. Mathieu M. Picot L. Bougaran G. et al (2012). The Potential of Microalgae for the Production of Bioactive Molecules of Pharmaceutical Interest. Cpb13, 2733–2750. 10.2174/138920112804724828
122
Miranda M. S. Sato S. Mancini-Filho J. (2001). Antioxidant Activity of the Microalga Chlorella Vulgaris Cultered on Special Conditions. Boll. Chim. Farm.140, 165–168.
123
Miranda M. S. Cintra R. G. Barros S. B. M. Mancini-Filho J. (1998). Antioxidant Activity of the Microalga Spirulina Maxima. Braz J. Med. Biol. Res.31, 1075–1079. 10.1590/S0100-879X1998000800007
124
Moazami N. Ranjbar R. Ashori A. Tangestani M. Nejad A. S. (2011). Biomass and Lipid Productivities of Marine Microalgae Isolated from the Persian Gulf and the Qeshm Island. biomass bioenergy35, 1935–1939. 10.1016/j.biombioe.2011.01.039
125
Mohsenpour S. F. Hennige S. Willoughby N. Adeloye A. Gutierrez T. (2021). Integrating Micro-algae into Wastewater Treatment: A Review. Sci. Total Environ.752, 142168. 10.1016/j.scitotenv.2020.142168
126
Molino A. Mehariya S. Iovine A. Larocca V. Di Sanzo G. Martino M. et al (2018a). Extraction of Astaxanthin and Lutein from Microalga Haematococcus pluvialis in the Red Phase Using CO2 Supercritical Fluid Extraction Technology with Ethanol as Co-solvent. Mar. Drugs16, 432. 10.3390/md16110432
127
Molino A. Rimauro J. Casella P. Cerbone A. Larocca V. Chianese S. et al (2018b). Extraction of Astaxanthin from Microalga Haematococcus pluvialis in Red Phase by Using Generally Recognized as Safe Solvents and Accelerated Extraction. J. Biotechnol.283, 51–61. 10.1016/j.jbiotec.2018.07.010
128
Monteiro C. M. Castro P. M. L. Malcata F. X. (2012). Metal Uptake by Microalgae: Underlying Mechanisms and Practical Applications. Biotechnol. Prog.28, 299–311. 10.1002/btpr.1504
129
Moraes C. C. Sala L. Cerveira G. P. Kalil S. J. (2011). C-phycocyanin Extraction from Spirulina Platensis Wet Biomass. Braz. J. Chem. Eng.28, 45–49. 10.1590/s0104-66322011000100006
130
Moreau R. A. Nyström L. Whitaker B. D. Winkler-Moser J. K. Baer D. J. Gebauer S. K. et al (2018). Phytosterols and Their Derivatives: Structural Diversity, Distribution, Metabolism, Analysis, and Health-Promoting Uses. Prog. Lipid Res.70, 35–61. 10.1016/j.plipres.2018.04.001
131
Morelli E. Scarano G. (2004). Copper-induced Changes of Non-protein Thiols and Antioxidant Enzymes in the Marine Microalga Phaeodactylum Tricornutum. Plant Sci.167, 289–296. 10.1016/j.plantsci.2004.04.001
132
Morschett H. Gätgens J. Wiechert W. Oldiges M. (2019). Rapid and Comprehensive Evaluation of Microalgal Fatty Acids via Untargeted Gas Chromatography and Time‐of‐flight Mass Spectrometry. Eng. Life Sci.19, 1006–1011. 10.1002/elsc.201900092
133
Mousavi Z. E. Mousavi S. M. Razavi S. H. Emam-Djomeh Z. Kiani H. (2011). Fermentation of Pomegranate Juice by Probiotic Lactic Acid Bacteria. World J. Microbiol. Biotechnol.27, 123–128. 10.1007/s11274-010-0436-1
134
Navarro López E. Robles Medina A. González Moreno P. A. Esteban Cerdán L. Martín Valverde L. Molina Grima E. (2016). Biodiesel Production from Nannochloropsis Gaditana Lipids through Transesterification Catalyzed by Rhizopus Oryzae Lipase. Bioresour. Technol.203, 236–244. 10.1016/j.biortech.2015.12.036
135
Nguyen H. T. D. Ramli A. Kee L. M. (2017). A Review on Methods Used in Analysis of Microalgae Lipid Composition. J. Jpn. Inst. Energy96, 532–537. 10.3775/jie.96.532
136
Ojha K. S. Aznar R. O'Donnell C. Tiwari B. K. (2020). Ultrasound Technology for the Extraction of Biologically Active Molecules from Plant, Animal and Marine Sources. TrAC Trends Anal. Chem.122, 115663. 10.1016/j.trac.2019.115663
137
Olander A. Lawson C. A. Possell M. Raina J.-B. Ueland M. Suggett D. J. (2021). Comparative Volatilomics of Coral Endosymbionts from One-And Comprehensive Two-Dimensional Gas Chromatography Approaches. Mar. Biol.168, 1–13. 10.1007/s00227-021-03859-2
138
Olasehinde T. A. Odjadjare E. C. Mabinya L. V. Olaniran A. O. Okoh A. I. (2019). Chlorella Sorokiniana and Chlorella Minutissima Exhibit Antioxidant Potentials, Inhibit Cholinesterases and Modulate Disaggregation of β-amyloid Fibrils. Electron. J. Biotechnol.40, 1–9. 10.1016/j.ejbt.2019.03.008
139
Olmstead I. L. D. Hill D. R. A. Dias D. A. Jayasinghe N. S. Callahan D. L. Kentish S. E. et al (2013). A Quantitative Analysis of Microalgal Lipids for Optimization of Biodiesel and Omega-3 Production. Biotechnol. Bioeng.110, 2096–2104. 10.1002/bit.24844
140
Onofrejová L. Vašíčková J. Klejdus B. Stratil P. Mišurcová L. Kráčmar S. et al (2010). Bioactive Phenols in Algae: The Application of Pressurized-Liquid and Solid-phase Extraction Techniques. J. Pharm. Biomed. Analysis51, 464–470. 10.1016/j.jpba.2009.03.027
141
Pajic I. Kljajic Z. Dogovic N. Sladic D. Juranic Z. Gasic M. J. (2002). A Novel Lectin from the Sponge Haliclona Cratera: Isolation, Characterization and Biological Activity. Comp. Biochem. Physiology Part C Toxicol. Pharmacol.132, 213–221. 10.1016/S1532-0456(02)00068-6
142
Pan-utai W. Iamtham S. (2019). Extraction, Purification and Antioxidant Activity of Phycobiliprotein from Arthrospira Platensis. Process Biochem.82, 189–198. 10.1016/j.procbio.2019.04.014
143
Panche A. N. Diwan A. D. Chandra S. R. (2016). Flavonoids: An Overview. J. Nutr. Sci.5. 10.1017/jns.2016.41
144
Pandit P. R. Fulekar M. H. (2017). Egg Shell Waste as Heterogeneous Nanocatalyst for Biodiesel Production: Optimized by Response Surface Methodology. J. Environ. Manag.198, 319–329. 10.1016/j.jenvman.2017.04.100
145
Papalia T. Sidari R. Panuccio M. R. (2019). Impact of Different Storage Methods on Bioactive Compounds in Arthrospira Platensis Biomass. Molecules24, 2810. 10.3390/molecules24152810
146
Patel A. Desai S. S. Mane V. K. Enman J. Rova U. Christakopoulos P. et al (2022). Futuristic Food Fortification with a Balanced Ratio of Dietary ω-3/ω-6 Omega Fatty Acids for the Prevention of Lifestyle Diseases. Trends Food Sci. Technol. 10.1016/j.tifs.2022.01.006
147
Patel A. Mishra S. Pawar R. Ghosh P. K. (2005). Purification and Characterization of C-Phycocyanin from Cyanobacterial Species of Marine and Freshwater Habitat. Protein Expr. Purif.40, 248–255. 10.1016/j.pep.2004.10.028
148
Patil V. Källqvist T. Olsen E. Vogt G. Gislerød H. R. (2007). Fatty Acid Composition of 12 Microalgae for Possible Use in Aquaculture Feed. Aquacult Int.15, 1–9. 10.1007/s10499-006-9060-3
149
Peng D. Zahid H. F. Ajlouni S. Dunshea F. R. Suleria H. A. R. (2019). LC-ESI-QTOF/MS Profiling of Australian Mango Peel By-Product Polyphenols and Their Potential Antioxidant Activities. Processes7, 764. 10.3390/pr7100764
150
Peraman M. Nachimuthu S. (2019). Identification and Quantification of Fucoxanthin in Selected Carotenoid-Producing Marine Microalgae and Evaluation for Their Chemotherapeutic Potential. Phcog Mag.15, 243. 10.4103/pm.pm_64_19
151
Pitt J. J. (2009). Principles and Applications of Liquid Chromatography-Mass Spectrometry in Clinical Biochemistry. Clin. Biochem. Rev.30, 19–34.
152
Pourkarimi S. Hallajisani A. Alizadehdakhel A. Nouralishahi A. Golzary A. (2020). Factors Affecting Production of Beta-Carotene from Dunaliella Salina Microalgae. Biocatal. Agric. Biotechnol.29, 101771. 10.1016/j.bcab.2020.101771
153
Prabakaran G. Sampathkumar P. Kavisri M. Moovendhan M. (2020). Extraction and Characterization of Phycocyanin from Spirulina Platensis and Evaluation of its Anticancer, Antidiabetic and Antiinflammatory Effect. Int. J. Biol. Macromol.153, 256–263. 10.1016/j.ijbiomac.2020.03.009
154
Rea G. Antonacci A. Lambreva M. Pastorelli S. Tibuzzi A. Ferrari S. et al (2011). Integrated Plant Biotechnologies Applied to Safer and Healthier Food Production: The Nutra-Snack Manufacturing Chain. Trends Food Sci. Technol.22, 353–366. 10.1016/j.tifs.2011.04.005
155
Rein M. J. Renouf M. Cruz-Hernandez C. Actis-Goretta L. Thakkar S. K. da Silva Pinto M. (2013). Bioavailability of Bioactive Food Compounds: A Challenging Journey to Bioefficacy. Br. J. Clin. Pharmacol.75, 588–602. 10.1111/j.1365-2125.2012.04425.x
156
Rentzsch M. Wilkens A. Winterhalter P. (2009). “Non-flavonoid Phenolic Compounds,” in Wine Chemistry and Biochemistry (New York: Springer), 509–527. 10.1007/978-0-387-74118-5_23
157
Rodrigues R. D. P. Silva A. S. e. Carlos T. A. V. Bastos A. K. P. de Santiago-Aguiar R. S. Rocha M. V. P. (2020). Application of Protic Ionic Liquids in the Microwave-Assisted Extraction of Phycobiliproteins from Arthrospira Platensis with Antioxidant Activity. Sep. Purif. Technol.252, 117448. 10.1016/j.seppur.2020.117448
158
Rontani J.-F. (2022). Use of Gas Chromatography-Mass Spectrometry Techniques (GC-MS, GC-MS/MS and GC-QTOF) for the Characterization of Photooxidation and Autoxidation Products of Lipids of Autotrophic Organisms in Environmental Samples. Molecules27, 1629. 10.3390/molecules27051629
159
Ryckebosch E. Bruneel C. Muylaert K. Foubert I. (2012). Microalgae as an Alternative Source of Omega-3 Long Chain Polyunsaturated Fatty Acids. Lipid Technol.24, 128–130. 10.1002/lite.201200197
160
Ryckebosch E. Bruneel C. Termote-Verhalle R. Goiris K. Muylaert K. Foubert I. (2014). Nutritional Evaluation of Microalgae Oils Rich in Omega-3 Long Chain Polyunsaturated Fatty Acids as an Alternative for Fish Oil. Food Chem.160, 393–400. 10.1016/j.foodchem.2014.03.087
161
Saadaoui I. Rasheed R. Abdulrahman N. Bounnit T. Cherif M. Al Jabri H. et al (2020). Algae-Derived Bioactive Compounds with Anti-lung Cancer Potential. Mar. Drugs18, 197. 10.3390/md18040197
162
Safafar H. van Wagenen J. Møller P. Jacobsen C. (2015). Carotenoids, Phenolic Compounds and Tocopherols Contribute to the Antioxidative Properties of Some Microalgae Species Grown on Industrial Wastewater. Mar. Drugs13, 7339–7356. 10.3390/md13127069
163
Saini R. K. Keum Y.-S. (2018). Carotenoid Extraction Methods: A Review of Recent Developments. Food Chem.240, 90–103. 10.1016/j.foodchem.2017.07.099
164
Sandau E. Sandau P. Pulz O. (1996). Heavy Metal Sorption by Microalgae. Acta Biotechnol.16, 227–235. 10.1002/abio.370160402
165
Sansone C. Brunet C. (2019). Promises and Challenges of Microalgal Antioxidant Production, 8, 199. 10.3390/antiox8070199Promises and Challenges of Microalgal Antioxidant ProductionAntioxidants
166
Sansone C. Galasso C. Orefice I. Nuzzo G. Luongo E. Cutignano A. et al (2017). The Green Microalga Tetraselmis Suecica Reduces Oxidative Stress and Induces Repairing Mechanisms in Human Cells. Sci. Rep.7, 1–12. 10.1038/srep41215
167
Sathya S. (2017). Separation of Algal Pigments by Thin Layer Chromatography (TLC) and High Performance Liquid Chromatography (HPLC). World J. Pharm. Res.6, 1275–1284.
168
Scarano G. Morelli E. (2002). Characterization of Cadmium-And Lead-Phytochelatin Complexes Formed in a Marine Microalga in Response to Metal Exposure. Biometals15, 145–151. 10.1023/a:1015288000218
169
Schwenzfeier A. Wierenga P. A. Gruppen H. (2011). Isolation and Characterization of Soluble Protein from the Green Microalgae Tetraselmis Sp. Bioresour. Technol.102, 9121–9127. 10.1016/j.biortech.2011.07.046
170
Shahidi F. Yeo J. (2020). Should the In Vitro Colorimetric Assays in Antioxidant and Lipid Oxidation Evaluation Be Abandoned?: A Critical Review Focusing on Bioactive Molecule Screening Assays in In Vitro and In Vivo Models. Jfb9. 10.31665/jfb.2020.9215
171
Shen N. Wang T. Gan Q. Liu S. Wang L. Jin B. (2022). Plant Flavonoids: Classification, Distribution, Biosynthesis, and Antioxidant Activity. Food Chem, 383, 132531. 10.1016/j.foodchem.2022.132531
172
Shi X.-M. Chen F. Yuan J.-P. Chen H. (1997). Heterotrophic Production of Lutein by Selected Chlorella Strains. J. Appl. Phycol.9, 445–450. 10.1023/A:1007938215655
173
Shi X.-M. Zhang X.-W. Chen F. (2000). Heterotrophic Production of Biomass and Lutein by Chlorella Protothecoides on Various Nitrogen Sources. Enzyme Microb. Technol.27, 312–318. 10.1016/s0141-0229(00)00208-8
174
Silva A. J. Cavalcanti V. L. R. Porto A. L. F. Gama W. A. Brandão-Costa R. M. P. Bezerra R. P. (2019). The Green Microalgae Tetradesmus Obliquus (Scenedesmus Acutus) as Lectin Source in the Recognition of ABO Blood Type: Purification and Characterization. J. Appl. Phycol., 1–8. 10.1007/s10811-019-01923-5
175
Silva S. C. Ferreira I. C. F. R. Dias M. M. Barreiro M. F. (2020). Microalgae-derived Pigments: A 10-year Bibliometric Review and Industry and Market Trend Analysis. Molecules25, 3406. 10.3390/molecules25153406
176
Silve A. Papachristou I. Wüstner R. Sträßner R. Schirmer M. Leber K. et al (2018). Extraction of Lipids from Wet Microalga Auxenochlorella Protothecoides Using Pulsed Electric Field Treatment and Ethanol-Hexane Blends. Algal Res.29, 212–222. 10.1016/j.algal.2017.11.016
177
Singh S. K. Kaur R. Bansal A. Kapur S. Sundaram S. (2020). “Biotechnological Exploitation of Cyanobacteria and Microalgae for Bioactive Compounds,” in Biotechnological Production of Bioactive Compounds (Elsevier), 221–259. 10.1016/b978-0-444-64323-0.00008-4
178
Sivaramakrishnan R. Incharoensakdi A. (2017). Direct Transesterification of Botryococcus Sp. Catalysed by Immobilized Lipase: Ultrasound Treatment Can Reduce Reaction Time with High Yield of Methyl Ester. Fuel191, 363–370. 10.1016/j.fuel.2016.11.085
179
Soares A. T. da Costa D. C. Vieira A. A. H. Antoniosi Filho N. R. (2019). Analysis of Major Carotenoids and Fatty Acid Composition of Freshwater Microalgae. Heliyon5, e01529. 10.1016/j.heliyon.2019.e01529
180
Song W. Zhao C. Wang S. (2013). A Large-Scale Preparation Method of High Purity C-Phycocyanin. Int. J. Biosci. Biochem. Bioinforma.3, 293–297.
181
Soni B. Kalavadia B. Trivedi U. Madamwar D. (2006). Extraction, Purification and Characterization of Phycocyanin from Oscillatoria Quadripunctulata-Isolated from the Rocky Shores of Bet-Dwarka, Gujarat, India. Process Biochem.41, 2017–2023. 10.1016/j.procbio.2006.04.018
182
Sonkamble V. Wagh N. (2022). “An Innovative Role of Bioactive Compounds from Microalgae,” in An Integration of Phycoremediation Processes in Wastewater Treatment (Elsevier), 313–336. 10.1016/b978-0-12-823499-0.00023-7
183
Stalikas C. D. (2007). Extraction, Separation, and Detection Methods for Phenolic Acids and Flavonoids. J. Sep. Sci.30, 3268–3295. 10.1002/jssc.200700261
184
Stein J. Kulemeier J. Lembcke B. Caspary W. F. (1992). Simple and Rapid Method for Determination of Short-Chain Fatty Acids in Biological Materials by High-Performance Liquid Chromatography with Ultraviolet Detection. J. Chromatogr. B Biomed. Sci. Appl.576, 53–61. 10.1016/0378-4347(92)80174-o
185
Stirk W. A. Ördög V. Van Staden J. Jäger K. (2002). Cytokinin-and Auxin-like Activity in Cyanophyta and Microalgae. J. Appl. Phycol.14, 215–221. 10.1023/a:1019928425569
186
Sun P. Wong C.-C. Li Y. He Y. Mao X. Wu T. et al (2019). A Novel Strategy for Isolation and Purification of Fucoxanthinol and Fucoxanthin from the Diatom Nitzschia Laevis. Food Chem.277, 566–572. 10.1016/j.foodchem.2018.10.133
187
Tang B. Row K. H. (2013). Development of Gas Chromatography Analysis of Fatty Acids in Marine Organisms. J. Chromatogr. Sci.51, 599–607. 10.1093/chromsci/bmt005
188
Tarafder A. Miller L. (2021). Chiral Chromatography Method Screening Strategies: Past, Present and Future. J. Chromatogr. A1638, 461878. 10.1016/j.chroma.2021.461878
189
Tirado D. F. Calvo L. (2019). The Hansen Theory to Choose the Best Cosolvent for Supercritical CO2 Extraction of β-carotene from Dunaliella Salina. J. Supercrit. Fluids145, 211–218. 10.1016/j.supflu.2018.12.013
190
Tsao R. (2010). Chemistry and Biochemistry of Dietary Polyphenols. Nutrients2, 1231–1246. 10.3390/nu2121231
191
Tzima K. Brunton N. Rai D. (2018). Qualitative and Quantitative Analysis of Polyphenols in Lamiaceae Plants-A Review. Plants7, 25. 10.3390/plants7020025
192
Van Wagenen J. De Francisci D. Angelidaki I. (2015). Comparison of Mixotrophic to Cyclic Autotrophic/heterotrophic Growth Strategies to Optimize Productivity of Chlorella Sorokiniana. J. Appl. Phycol.27, 1775–1782. 10.1007/s10811-014-0485-1
193
Viegas C. V. Mendes L. B. B. João R. R. Díaz G. C. Aranda D. A. G. Cruz Y. R. (2020). Quantification of Lipid Content and Identification of the Main Lipid Classes Present in Microalgae Extracts Scenedesmus Sp. For Obtaining Fatty Compounds for Biofuel Production. Epe12, 273–287. 10.4236/epe.2020.126017
194
Viñas P. Campillo N. (2014). “Gas Chromatography-Mass Spectrometry Analysis of Polyphenols in Foods,” in Polyphenols in Plants: Isolation, Purification and Extract Preparation (Elsevier), 103–157. 10.1016/B978-0-12-397934-6.00007-3
195
Wang J. Chen R. Fan L. Cui L. Zhang Y. Cheng J. et al (2021). Construction of Fungi-Microalgae Symbiotic System and Adsorption Study of Heavy Metal Ions. Sep. Purif. Technol.268, 118689. 10.1016/j.seppur.2021.118689
196
Wang J. Hu X. Chen J. Wang T. Huang X. Chen G. (2022). The Extraction of β-Carotene from Microalgae for Testing Their Health Benefits. Foods11, 502. 10.3390/foods11040502
197
Wilde E. W. Benemann J. R. (1993). Bioremoval of Heavy Metals by the Use of Microalgae. Biotechnol. Adv.11, 781–812. 10.1016/0734-9750(93)90003-6
198
Woo S.-G. Yoo K. Lee J. Bang S. Lee M. On K. et al (2012). Comparison of Fatty Acid Analysis Methods for Assessing Biorefinery Applicability of Wastewater Cultivated Microalgae. Talanta97, 103–110. 10.1016/j.talanta.2012.04.002
199
Wu Z.-Y. Qu C.-B. Shi X.-M. (2009). Biochemical System Analysis of Lutein Production by Heterotrophic Chlorella Pyrenoidosa in a Fermentor. Food Technol. Biotechnol.31, 450.
200
Wybraniec S. Starzak K. Skopińska A. Nemzer B. Pietrzkowski Z. Michałowski T. (2013). Studies on Nonenzymatic Oxidation Mechanisms in Neobetanin, Betanin, and Decarboxylated Betanins. J. Agric. Food Chem.61, 6465–6476. 10.1021/jf400818s
201
Xie D. Ji X. Zhou Y. Dai J. He Y. Sun H. et al (2022). Chlorella Vulgaris Cultivation in Pilot-Scale to Treat Real Swine Wastewater and Mitigate Carbon Dioxide for Sustainable Biodiesel Production by Direct Enzymatic Transesterification. Bioresour. Technol.349, 126886. 10.1016/j.biortech.2022.126886
202
Xue Z. Sharpe P. L. Hong S.-P. Yadav N. S. Xie D. Short D. R. et al (2013). Production of Omega-3 Eicosapentaenoic Acid by Metabolic Engineering of Yarrowia Lipolytica. Nat. Biotechnol.31, 734–740. 10.1038/nbt.2622
203
Yan S.-G. Zhu L.-P. Su H.-N. Zhang X.-Y. Chen X.-L. Zhou B.-C. et al (2011a). Single-step Chromatography for Simultaneous Purification of C-Phycocyanin and Allophycocyanin with High Purity and Recovery from Spirulina (Arthrospira) Platensis. J. Appl. Phycol.23, 1–6. 10.1007/s10811-010-9525-7
204
Yan X. Chen D. Xu J. Zhou C. (2011b). Profiles of Photosynthetic Glycerolipids in Three Strains of Skeletonema Determined by UPLC-Q-TOF-MS. J. Appl. Phycol.23, 271–282. 10.1007/s10811-010-9553-3
205
Yan X. Li H. Xu J. Zhou C. (2010). Analysis of Phospholipids in Microalga Nitzschia Closterium by UPLC-Q-TOF-MS. Chin. J. Ocean. Limnol.28, 106–112. 10.1007/s00343-010-9263-3
206
Yen H.-W. Sun C.-H. Ma T.-W. (2011). The Comparison of Lutein Production by Scenesdesmus Sp. In the Autotrophic and the Mixotrophic Cultivation. Appl. Biochem. Biotechnol.164, 353–361. 10.1007/s12010-010-9139-1
207
Yu J. Ma D. Qu S. Liu Y. Xia H. Bian F. et al (2020). Effects of Different Probiotic Combinations on the Components and Bioactivity of Spirulina. J. Basic Microbiol.60, 543–557. 10.1002/jobm.201900699
208
Zhao P. Wang X. Niu J. He L. Gu W. Xie X. et al (2020). Agar Extraction and Purification of R-Phycoerythrin from Gracilaria Tenuistipitata, and Subsequent Wastewater Treatment by Ulva Prolifera. Algal Res.47, 101862. 10.1016/j.algal.2020.101862
209
Zhou X. Yi M. Ding L. He S. Yan X. (2019). Isolation and Purification of a Neuroprotective Phlorotannin from the Marine Algae Ecklonia Maxima by Size Exclusion and High-Speed Counter-current Chromatography. Mar. Drugs17, 212. 10.3390/md17040212
210
Zhu W.-x. Yang J.-z. Wei W. Liu Y.-f. Zhang S.-s. (2008). Simultaneous Determination of 13 Aminoglycoside Residues in Foods of Animal Origin by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry with Two Consecutive Solid-phase Extraction Steps. J. Chromatogr. A1207, 29–37. 10.1016/j.chroma.2008.08.033
211
Zoccali M. Giuffrida D. Salafia F. Socaciu C. Skjånes K. Dugo P. et al (2019). First Apocarotenoids Profiling of Four Microalgae Strains. Antioxidants8, 209. 10.3390/antiox8070209
212
Zotov V. A. Bessonov V. V. Risnik D. V. (2022). Methodological Aspects of the Analysis of Fatty Acids in Biological Samples. Appl. Biochem. Microbiol.58, 83–95. 10.1134/s0003683822010112
213
Zou Y. Wu H. (2018). Improving the Analysis of 37 Fatty Acid Methyl Esters. USA.
Summary
Keywords
Liquid Chromatography, gas chromatography, bioactive compounds, Microalgae, PUFA, Polyphenols, pigments
Citation
Kiani H, Aznar R, Poojary MM, Tiwari BK and Halim R (2022) Chromatographic Techniques to Separate and Identify Bioactive Compounds in Microalgae. Front. Energy Res. 10:904014. doi: 10.3389/fenrg.2022.904014
Received
25 March 2022
Accepted
20 May 2022
Published
20 June 2022
Volume
10 - 2022
Edited by
Marcin Debowski, University of Warmia and Mazury in Olsztyn, Poland
Reviewed by
Joanna Kazimierowicz, Bialystok University of Technology, Poland
Angela Sardo, Stazione Zoologica Anton Dohrn Napoli, Italy
Updates
Copyright
© 2022 Kiani, Aznar, Poojary, Tiwari and Halim.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronald Halim, ronald.halim@ucd.ie
This article was submitted to Bioenergy and Biofuels, a section of the journal Frontiers in Energy Research
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.