- 1School of Public Policy and Management, China University of Mining and Technology, Xuzhou, China
- 2Low Carbon Energy Institute, China University of Mining and Technology, Xuzhou, China
- 3School of Environment Science and Spatial Informatics, China University of Mining and Technology, Xuzhou, China
- 4College of Resources and Environment, Northwest A&F University, Yangling, China
The microtopographic changes induced by coal mining subsidence caused a series of environmental problems such as soil erosion, and vegetation degradation in the mining area. However, the corresponding influence on surface vegetation and soil characteristic in different parts of the slope was completely different. To understand soil and vegetation degradation in coal mines and their future ecological restoration, it was crucial to investigate the origin. The relationship between soil microbial community diversity, structure, and taxa in the slope of subsidence area of different topographic locations in Daliuta coal mine, Shannxi, China, was determined by high throughput sequencing and molecular ecological network analysis. The relationship between the bacterial communities, environmental factors, and soil physicochemical properties was also investigated. We found a new topographic trait formed by surface subsidence to deteriorate the living environment of vegetation and the bacterial community. The vegetation coverage, soil water content, organic matter, and urease and dehydrogenase activities decreased significantly (p < 0.05). Although soil bacterial community diversity in the subsidence area did not differ significantly, the dominant taxa in different topographic locations varied. The molecular ecological networks representing bacterial community structure and function were also totally different. The networks in the middle and the top of the slope tend to be more complicated, and the interaction between species is obviously stronger than that of the bottom. However, the network in the bottom slope approached simplicity, and weak interaction, predominantly cooperative, was observed within and between modules. Meanwhile, the double stress of aridity and the lack of carbon source induced by subsidence also enhanced the capacity of the soil bacterial community to metabolize complex carbon sources at the bottom of the slope.
Introduction
Fossil fuels are used globally and are the dominant source of energy. However, the exploitation of fossil energy, especially that of coal, usually introduces super-incumbent topographic changes such as surface subsidence, fracture, and land sliding, which aggravate soil erosion and degradation. In addition, the regional biomass and diversity also decrease dramatically (Bian et al., 2012; Lei et al., 2016; Qi et al., 2016), causing huge damage to the local ecological system. The productivity of coal mines in China reached 3.6 billion tons in 2018 (Wang et al., 2019), amounting to half of the global production. The cumulative land area in China, damaged by mining has already reached 8 million hm2 with an annual increase at the rate of 48.8 thousand hm2 (Li, 2006). A few years ago, the damaged mine land scale was so huge that its artificial ecological restoration was nearly impossible. Thus, the self-recovering ability of the ecosystem was stimulated (Hu et al., 2013; Collier, 2015), which is influenced by multiple factors including soil physicochemical characteristics, the seed bank, and microbes, etc. (Li et al., 2018), wherein microbes have an important catalytic function (Harris, 2009; Ayala-Orozco et al., 2018). Many large-scale coal production bases of China are distributed in the semiarid and arid sandy grassland, whose ecological environment, once destroyed by mining, was easily degraded. Thus, it is essential to understand the influence of mining-induced change in topographic traits on the soil microbial community succession and interaction, and the potential of sandy grassland to restore the damaged ecological system of mine land.
Microbes are the key component of the ecosystem and are significant for global ecological restoration, environmental monitoring, pollution treatment, and biological protection, etc. (Alivisatos et al., 2015; Fierer, 2017). Recently, several studies were conducted on the microbial community of mining soil (Allison and Martiny, 2008), micro-influence of mining on the ecosystem (Ding et al., 2015), the relationship between soil-vegetation-microbe (Kardol and Wardle, 2010; Lozano et al., 2014), and the application of microbe in ecological restoration of mine land (Li et al., 2013). Soil microbial community composition and diversity are sensitive to disturbance, hence it can indicate changes in the soil environment (Zhang C. et al., 2016). Besides, species abundance and redundancy could increase the scale of adaptation of soil microbial community to environmental change and can possibly aid in resisting severe environmental deterioration (Allison and Martiny, 2008). However, soil stability (resistance and resilience) is determined by the structure or diversity of the microbial community and is also related to plant and soil properties, including the quantity and quality of coacervate and organic matter (Ding et al., 2015), etc. Thus, the main ecological processes could be characterized by soil-vegetation-microbe system. Mining disturbance changes the site condition of the soil environment and inevitably decreases the microbe adaptability. A decreasing trend is also observed in the microbial biomass (Jing et al., 2018). A decrease in soil microbial diversity due to mining was observed in Xilingol, Inner Mongolia (Shen et al., 2019). Soil damage due to coal mine exploitation negatively affects the microbial community and the corresponding ecosystem (Brown and Jumpponen, 2015). A decrease in soil microbe population was also reported after the subsidence of the mining area (Xiao et al., 2019). Likewise, the arbuscular mycorrhizal fungal community changed dramatically with the increasing reclamation time (Mao et al., 2019), while the reclamation shows a positive effect on the fungal community. Some researchers focused on the relationship between change in soil microbial community and environmental factors. The bacterial community structure and diversity were found to change after the mining disturbance (Dimitriu et al., 2010). Usually, higher plant diversity is favorable for enhanced preservation of soil carbon which is an important component of soil organic matter (SOM), as well as microbial activity (Prommer et al., 2020). However, the quantity and quality of SOM determine the composition of the microbial community (Ding et al., 2015). The composition and diversity of the soil microbial community are closely related to its function (Mendes et al., 2015). Meanwhile, soil matrices, microbial diversity as well as community composition influence the speed and direction of the vegetation community succession (Frouz et al., 2016). Interaction between soil, vegetation, and microbes drive the succession of the whole environment, although the microbial succession lags behind the vegetative succession. In turn, the ecological environment changes the taxonomic and functional diversity of the soil microbial community (Guo et al., 2018). The relationship between different species usually forms a complex ecological network, which controls the self-assembly of the bacterium community and thus influences the structure and function of the community (Zhou et al., 2010). Interaction between microbial genera is important for understanding the dynamics between microbial community assembly and environmental changes. Worldwide, specific microbial ecological networks have received great attention. However, previous studies seldom focused on the spatial heterogeneity of topographic traits due to mining subsidence which further influences the interaction between soil microbial taxa in different topographic locations (Delgado-Baquerizo et al., 2018b).
In the recent decade, high throughput sequencing has enabled the acquisition of significant information about the microbial world. Notwithstanding, the extraction, analysis, and summary of the massive information are always difficult (Wang and Brose, 2018). The molecular ecological network method uses visualization of the sequencing data to demonstrate the complicated and potential interaction between microbial taxa, investigate different ecological processes, ecological functional stability, complexity, and the function of key taxa (Deng et al., 2016). This method was used to study the relationship and the difference in network structure, interaction, and relationship of the key taxa of soil bacteria in the potato fields and their interaction with the environment with the increase in CO2 level (Zhou et al., 2010; Luo et al., 2014). Mining is a high-intensity disturbance and influences the soil nutrient cycle. Assessment of the interaction and relationship between microbial taxa, the search for key taxa that are adaptable to the subsidence environment, and the clarification of the main controlling factor of microbial community distribution in a coal mine were found to be important for reestablishing the self-recovering ability of mining land ecological system (Bi et al., 2019). This research hypothesizes that under the combined effect of soil and vegetation, topographical changes caused by mining differentially impact the structure of soil bacterial communities and their interspecific relationships. In this work, the Daliuta coal mine subsidence area in the Mu Us sandy grassland of Inner Mongolia was chosen as the study target. The development of soil bacterial community, their interaction, and the adaptability between the bacterial taxa and environment with different topographic traits were investigated by the high throughput sequencing technology and ecological network analysis method, and provide the possibility for further finer manipulation and management of microbial communities to serve the differentiated bio-remediation. The study is also important for the remediation sequence, direction, and procedures of differentiated sandy grassland.
Materials and Methods
Study Area
Daliuta coal mine has an area of 189.9 km2 and is situated between Yulin City, Shannxi Province and Inner Mongolia (110°12’–110°23’E, 39°13’–39°22’N). It belongs to the transition area between the Loess hilly landform and the sandstorm landform and has a semiarid climate in the temperate zone. It is situated at an altitude of 1,057–1,334 m, with an annual average temperature of 8.6°C. The area has an annual average precipitation of 340 mm, which is mainly stormy and occurs primarily from June to September. The annual evaporation amount was estimated to be 1,754 mm (Luo et al., 2020). Surface vegetation in this mine land is mainly drought-tolerant shrubs and herbs, for example, Artemisia halodendron, A. ordosica, Salix psammophila, Caragana intermedia, Lespedeza davurica, A. capillaris, Alfalfa, etc. Daliuta coal mine was officially exploited in 1996. Since then, the underground goaf area constitutes 70% of the total area of mine land (He et al., 2017). Long-term exploitation led to a decrease in the underground water level and soil water content, increased the depth of the surface dry sand layer, and caused severe damage to the vegetation. These were typical exploitation subsidence areas of degraded grassland. In this work, the sampling was done in the northeastern part of the Daliuta coal mine, at the 52302-working slope of the well-field III. The mining activities in the sampling area have been stopped since 2013. According to the monitoring of terrain and ground vegetation by remote sensing, the topography of the sampling area is stable at present (Huang et al., 2020), and the surface vegetation coverage has a high consistency in recent years (He et al., 2017). The sampling area are typical representative.
Soil Sample Collection and Analysis of Soil Physicochemical Properties
Eight samples each were collected from the top, middle, and bottom of the slope of a collapsing pit in the Daliuta coal mine on August 15–16, 2018, and were labeled as B (bottom), M (middle), and T (top) (Figure 1). The coverage degree of surface vegetation (PC) in the sampling site was determined through the customized grid plate (1 m × 1 m) with the grid area of 100 cm2 (10 cm × 10 cm). The sampling site was kept in the center of the grid, then the proportion between the grid number covered by the vertical projection of vegetation and the total grid number in the grid-scale was calculated (Chmura et al., 2016). Soil water content (SWC) and temperature (ST) were determined using the on-site quick measurement method with a temperature hydrometer (TR-6, Shunkeda, Guangdong). Five hundred grams of mixed surface soil sample was evenly collected from the depth of 0 to 10 cm near the contour in the three sites of the top, middle, and bottom of the slope, along the subsidence slope. Samples were sealed in the special sterile bag, kept in the car refrigerator, and sent to the lab. A portion of fresh soil was sealed with ice in a bubble chamber and was mailed directly to Shanghai Personal Biotechnology Co., Ltd. for sequencing. The remaining soil sample was naturally air-dried, and stones and animal and plant residues were removed. After grinding and filtering the soil by 100 mesh, physicochemical properties and enzyme activities of soil samples were determined.
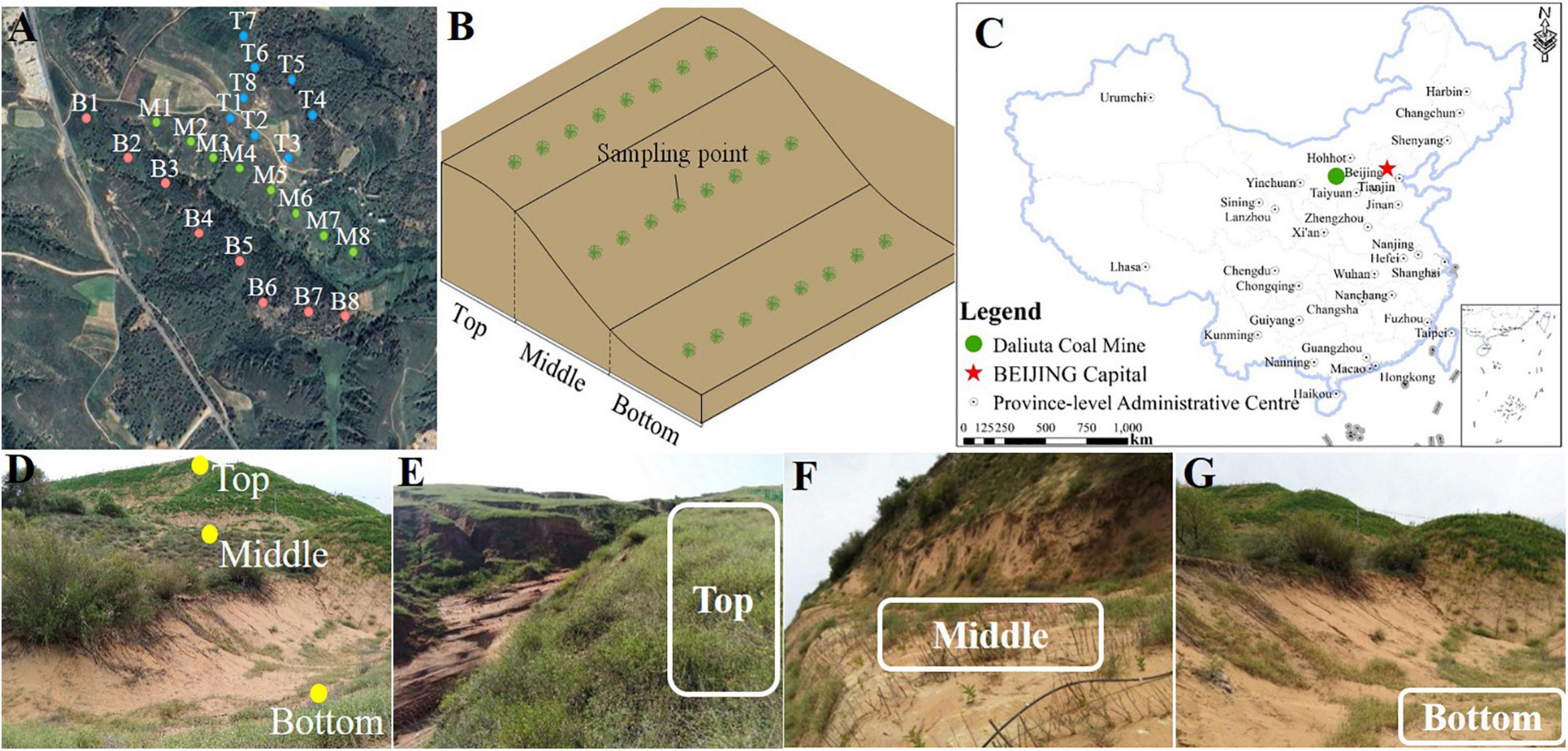
Figure 1. The study area and sampling sites of coal mining subsidence. (A) Sampling points of land subsidence in Daliuta coal mine, (B) Schematic diagram of the sampling point location, (C) Location of Daliuta coal mine, (D) Relative position of the sampling points. The photographs of the (E) Top, (F) Middle, and (G) Bottom of the slope.
The mechanical composition of the soil was determined by the hydrometer method, and pH and electrical conductivity (EC) were determined by the potentiometry method (water: soil = 1:2.5) (Lei-ci, Shanghai, DDS-307A). The SOM was determined by potassium dichromate colorimetry (Jena, Germany, Specord 210 plus). Available phosphorus (AP), available potassium (AK), ammonium nitrogen (AN), and nitrate nitrogen (NN) were determined, respectively, by the Olsen method, ammonium acetate extraction flame photometer method (TAS-990, Puxi, Beijing), potassium chloride extraction ultraviolet spectrophotometry method, and calcium chloride extraction-ultraviolet spectrophotometry method. The activities of dehydrogenase (DHG), urease (URA), and protease (PRO) enzymes in soil were determined by the triphenyl tetrazolium chloride method (TTC), the sodium phenoxide-sodium hypochlorite colorimetry method, and the ninhydrin colorimetry method, respectively (Barahona and Iriarte, 2001; Malachowska-Jutsz and Matyja, 2019).
DNA Extraction, Purification, PCR Amplification, and 16S rRNA Sequencing
DNA from 24 soil samples was extracted according to the specification of the FastDNATM SPIN Kit for Soil (MP Biomedicals Corp., United States). The V4 and V5 zones of 16S rRNA in soil bacteria were PCR amplified from the three topographic locations. The primers used were 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). PCR conditions were: initial denaturation at 98°C for 2 min, 25 cycles of denaturation for 15 s at 98°C, annealing for 30 s at 55°C and elongation for 30 s at 72°C, followed by 5 min of terminal elongation at 72°C and then at 10°C. The size and quality of PCR amplified products from the three soil samples were determined by agarose gel (2%) electrophoresis. The gel elution kit (Axygen, United States) was used for the elution of the target fragments. Based on the above electrophoretic preliminary quantitation result, the PCR amplified elution product was quantified using the fluorescence reagent of Quant-iT picoGreen ds DNA Assay Kit and the microplate reader. Sequencing libraries of the soil bacteria from the top, middle, and bottom slopes were prepared by TruSeq nano DNA LT Library Prep Kit developed by Illumina Corp. The constructed library was quantitatively determined by Qubit and Q-PCR. The qualified library (Shanghai Personal Biotechnology Co., Ltd.) was sequenced by HiSeq2500 PE 2500 (Illumina, United States).
Construction of a Molecular Ecological Network of Soil Bacteria
Based on 16S rRNA sequencing data, the operational taxonomic unit (OTU) was classified adopting the principle of 97% similarity. The number of sample OTUs in each group was added. The frequency value of the top 80% of the OTUs was used for the construction of a molecular ecological network (Zhou et al., 2010; Deng et al., 2012). In this work, the molecular ecological network analysis pipeline was used for the construction and statistical analysis of the microbial molecular ecological network. Briefly, the data of the obtained OTUs were transformed by log10. The related matrix was calculated and transformed into a similarity matrix based on the Pearson correlation coefficient. According to the random matrix theory, the best similarity threshold was chosen from the similarity matrix and transformed into an adjacency matrix through reciprocity. The connection strength between each pair of nodes was coded by the adjacency matrix. The ecological community was predicted by analyzing the nearest adjacent distance of the related matrix eigenvalue. The basic properties of the network were analyzed by topological parameters of the network (Table 1 and Supplementary Table 3 in section 2 of Supplementary Material; Deng et al., 2012; Luo et al., 2014; Delgado-Baquerizo et al., 2018b; Doncheva et al., 2019). Each type of OTU was regarded as a network node. Total nodes represented the node number of the entire network, and the total links represented the connection number of all nodes. The larger the connection number, the closer the connection between nodes in the network was. The average connection number of each node was represented by average connectivity (Avg K). The more the average connection was, the more complex the interior of the network was. When the connection within one group of OTUs in the network was more and the connection outside the group was only a few, this group of OTUs could be regarded as a module. Module number represented the module quantity with more than five nodes which could be classified in the network. Modularity represented the proportion of OTUs which could be classified into modules in the network. The higher the extent of modularity was, the less the network redundancy was.
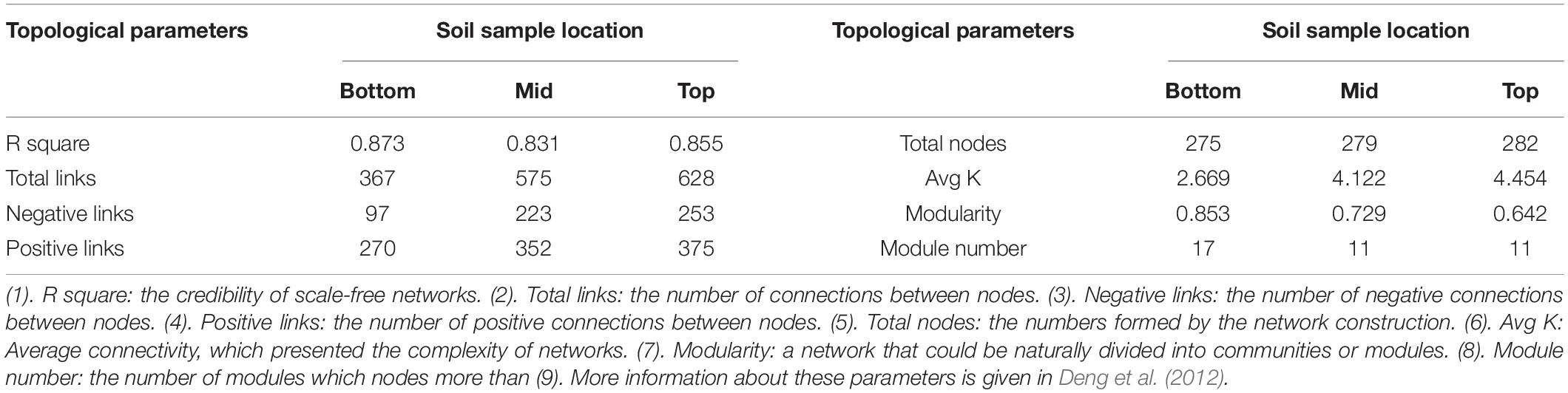
Table 1. Topological property of the empirical molecular ecological networks of microbial communities and their random networks at different locations of the subsidence area.
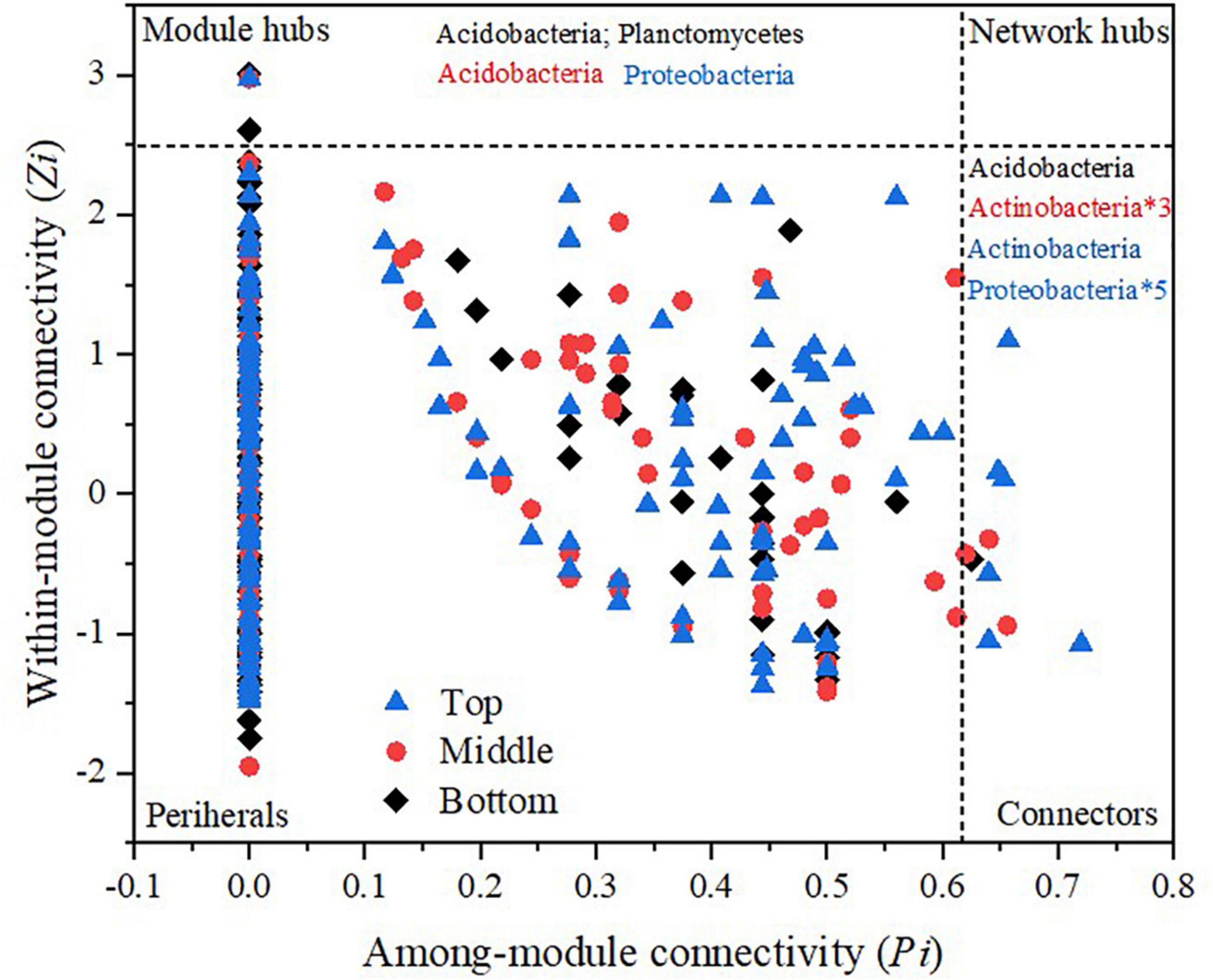
The distribution status of OTUs was demonstrated by the Z-P diagram which acted as the topological character in the module (Luo et al., 2014). Each dot represented one OTU in the data set. According to the scatter diagram of the connectivity within (Zi) and between (Pi) the module, the topological function of each OTU was confirmed. Module hubs, connectors, and network hubs were recognized (Olesen et al., 2007; Clauset et al., 2008), wherein OTUs with Zi ≥ 2.5 were recognized as module hubs, OTUs with Pi ≥ 0.62 were recognized as the connectors in the network, and OTUs which satisfied both the above conditions were recognized as the network hubs (Zhou et al., 2010). The network was visualized using the Cytoscape 3.8.0 software. One ecological network was divided into many modules and each module was a functional unit of the microbial system. In the network structure diagram, the node represented the taxa in the microbial community. The connection between nodes represented the interaction relationship between taxa. Blue and red colors represented positive and negative interactions, respectively (Zhou et al., 2010).
Statistical Analysis and Processing
Alpha diversity index and beta diversity index adopted the non-metric multidimensional scaling (NMDS) analysis, which was calculated and analyzed based on the galaxy platform1. One-way analysis of variance (ANOVA) of the physicochemical properties of different topographic locations was calculated using SPSS 21.0 software (IBM, United States). The construction and the calculation of characteristic parameters of molecular ecological network structure were based on the high throughput sequencing results of bacteria and used the MENA platform2. The structural diagram of the molecular ecological network was drawn using the Cytoscape 3.8.0 software. Soil microbial abundance diagrams were drawn by Origin Pro 2020 software (Origin Lab, United States). The gene functional prediction analysis predicted the function of soil bacteria using the PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) software3. For data treatment and drawing, Excel 2019 (Microsoft, United States) and R-project software were used (MathSoft, United States).
Results
Physicochemical Properties of Soil in Different Topographic Locations
Uneven topographic change in mine land caused different extents of disturbance, leading to variation in physicochemical properties of soil in different topographic locations of the mining land (Figure 2). In addition, soil physicochemical properties show some regularity with the change in the topographic altitude. First, with the intense topographic change, sand proportion (SP), PC, and SOM in different topographic locations demonstrated a dramatic variation. From the top to the bottom slope, PC decreased sharply by 39.62% (p < 0.05), leading to a reduction in SOM by as much as 87.62% (p < 0.05). A desertification phenomenon was observed in the soil of the middle and the bottom slopes, with a corresponding increase in SPs by 7.0 and 28.62% (p < 0.05), respectively. The EC also demonstrated a gradual but not significant decreasing trend from the top to the bottom of the slope. With the change in the soil layer structure, there was a dramatic difference in ST and SWC of the bottom, the top, and the middle slopes. SWC in the bottom slope decreased by more than one-third (p < 0.05). However, ST in the bottom slope was nearly 9°C higher than that in the middle and the top slope (p < 0.05). The AK, DHG, and URA were consistent between the bottom and the middle slope, and these were all significantly lower than those in the top slope. Compared with those of the top slope, AK, DHG, and URA decreased on an average, by 42.5% (0.23 mg kg–1), 38.3% (0.835 mg kg–1), and 65.8% (0.125 mg kg–1), respectively. There was no significant influence of topographic change on pH, AP, AN, NN, and PRO, wherein, NN content in the top slope was higher than that in the bottom and the top slopes. However, the contents of the other four indexes were lower in the middle slope compared with those in the bottom and top slopes. On the whole, with the increase in the intensity of topographic change, although there was no significant loss of main nutritional elements such as N, P, and K, the soil environment in the middle and bottom slope deteriorated dramatically.
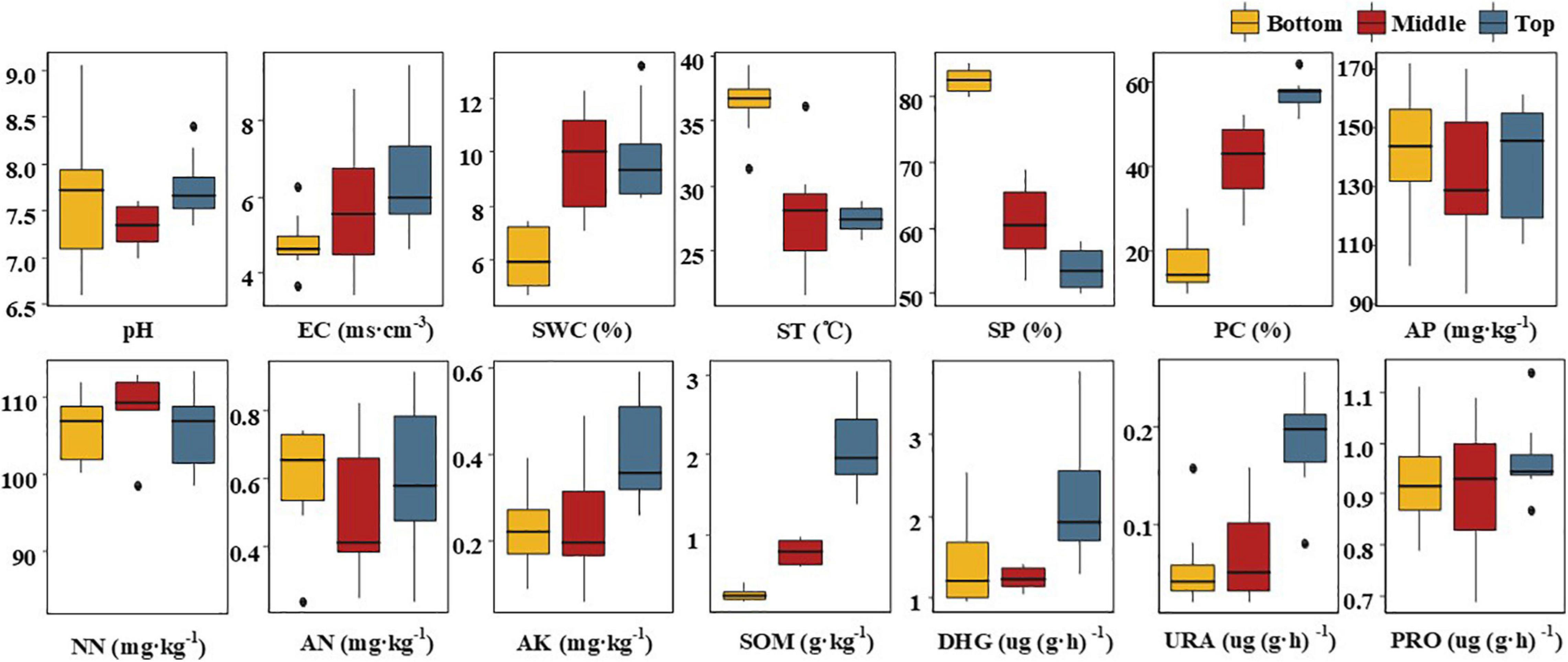
Figure 2. Box-plots of soil physicochemical properties at different locations of the subsidence area. SWC, soil water content; ST, soil temperature; SP, sand proportion; PC, plant cover ratio; AP, available P; NN, nitrate-nitrogen; AN, ammonium nitrogen; AK, available K; SOM, soil organic matter; DHG, dehydrogenase; URA, urease; PRO, protease. The Bottom, Middle, and Top are the positions of the subsidence area.
Structural Characteristics of Soil Bacterial Community in Different Topographic Locations
Soil Bacterial Community Diversity in Different Topographic Locations
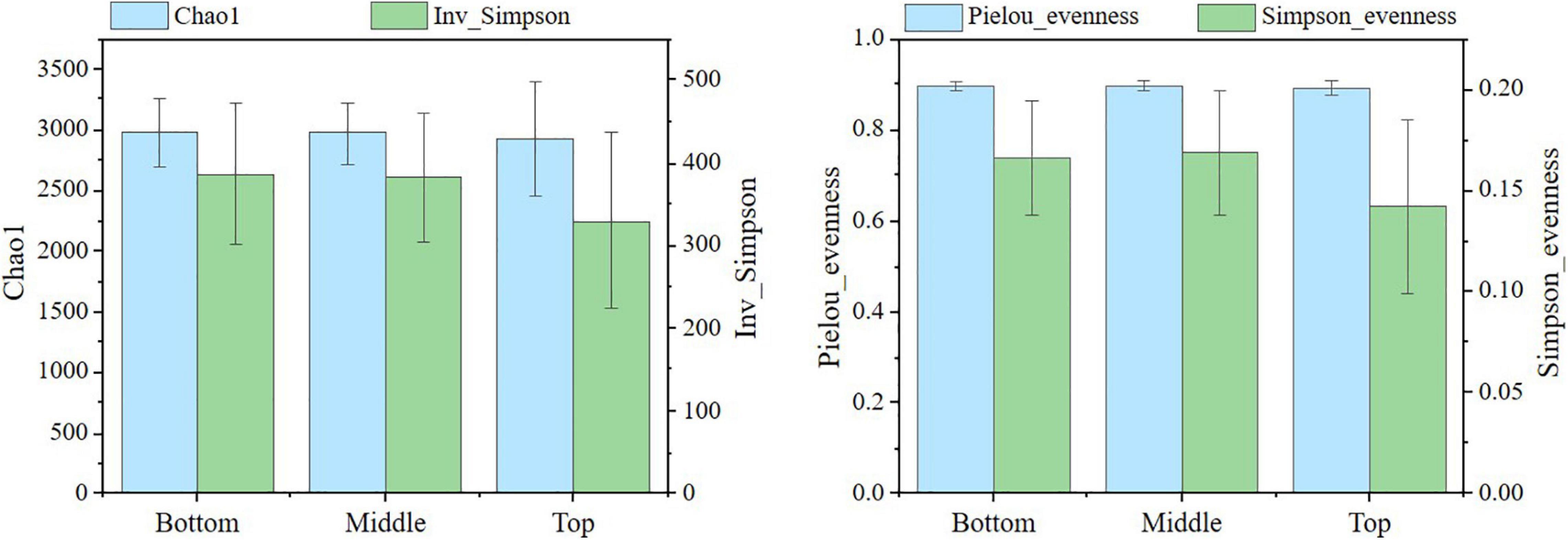
In this study, 924,281 effective sequences from 24 samples of three topographic locations were obtained. Nearly 33,194–44,787 sequences had almost even distribution, wherein the number of effective sequences determined in the middle and bottom slopes accounted for 92 and 96% of the effective sequences in the top slope, respectively. Overall, the number of bacteria in the middle and bottom slopes was slightly lower than that in the top slope. The estimation of soil bacterial alpha diversity is shown in Figure 3, wherein the Chao1 index and the Simpson index demonstrated the abundance of the bacterial community, and the Pielou-evenness index and the Simpson-evenness index represented the evenness of the microbial community. Analysis results of soil microbial beta diversity are shown in Figure 4.
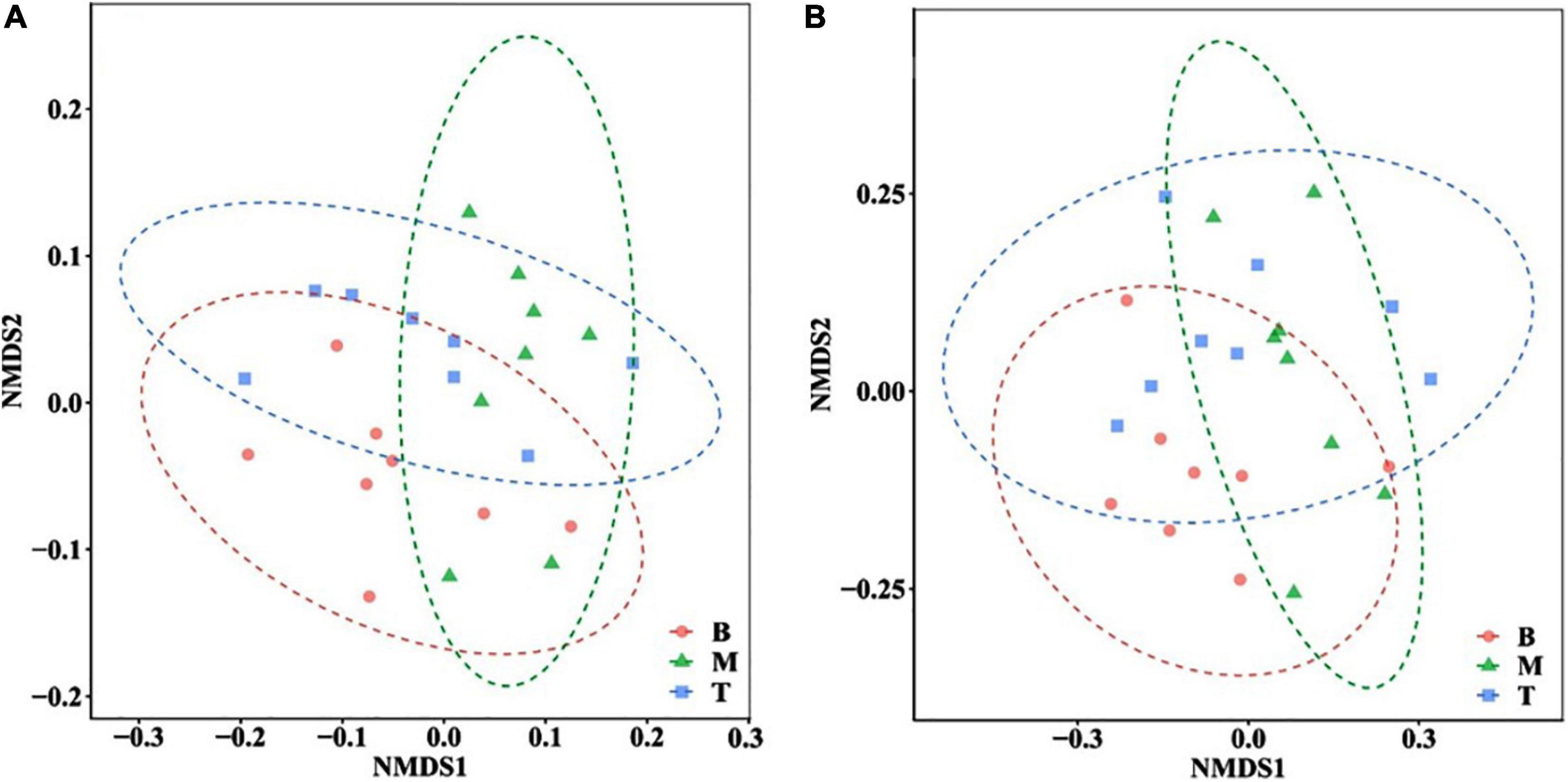
Figure 4. Non-metric multidimensional scaling (NMDS) analysis at (A) phylum and (B) genus level in different locations of the subsidence area.
The richness indexes and evenness indexes in Figure 3 indicate that the bacterial community richness and evenness were similar in the bottom and the middle slopes. Both of them were a little higher than that on the top slope. However, the standard deviations of the diversity indexes in the top slope were dramatically larger than those of the middle and bottom slopes, indicating that the bacterial communities in areas with greater changes (the middle and the bottom slopes) show an evolutionary trend of increase in diversity and decrease in spatial heterogeneity after the reshaping of the original topography. On the whole, considering the time scale of this study, the topographic change did not have a dramatic influence on the diversity of the soil bacterial community. Then, the highest 70% OTUs at the phylum level and the genus level were selected for NMDS analysis based on Bray-Curtis distance (Figure 4). Indeed, there was some difference in the microbial community structure between the bottom slope and other topographic locations on the level of phylum and genus. However, the group division was not dramatic.
Soil Bacterial Community Composition in Different Topographic Locations
Topographic change influenced the soil bacterial community composition, which varied among different topographic locations. In all samples, the highest 11 phyla accounted for more than 90% relative abundance (Figure 5). Among the dominant phyla (bacteria phyla with relative abundance > 1%) with relatively large abundance, Actinobacteria had a 13.3% decrease in relative abundance in the middle slope compared with that in the top slope. However, Proteobacteria increased by 15.9%. On the contrary, Actinobacteria increased by 6.8% in the bottom slope compared with that of the top slope, whereas the decrease in Proteobacteria was not significant. In addition, compared with those in the top slope, the relative abundance of Acidobacteria and Chloroflexi in the middle and the bottom slopes demonstrated a consistently decreasing trend. Among dominant phyla with small relative abundance, Planctomycetes, Gemmatimonadetes, Bacteroidetes, Cyanobacteria, and Euryarchaeota demonstrated a consistently increasing trend in the middle and the bottom slopes, compared with those in the top slope. However, only the relative abundance of Thaumarchaeota decreased by 24.7 and 50.6%, respectively, in the middle and bottom slopes. Among the above-mentioned phyla, the variation of Cyanobacteria and Euryarchaeota drew our attention, because the relative abundance of Cyanobacteria in the middle and top slopes increased by 1.2 and 1.9-fold (the non-representative sample M3 was excluded), respectively, and that of Euryarchaeota increased by 64% and 1.8-fold, respectively (Figure 5).
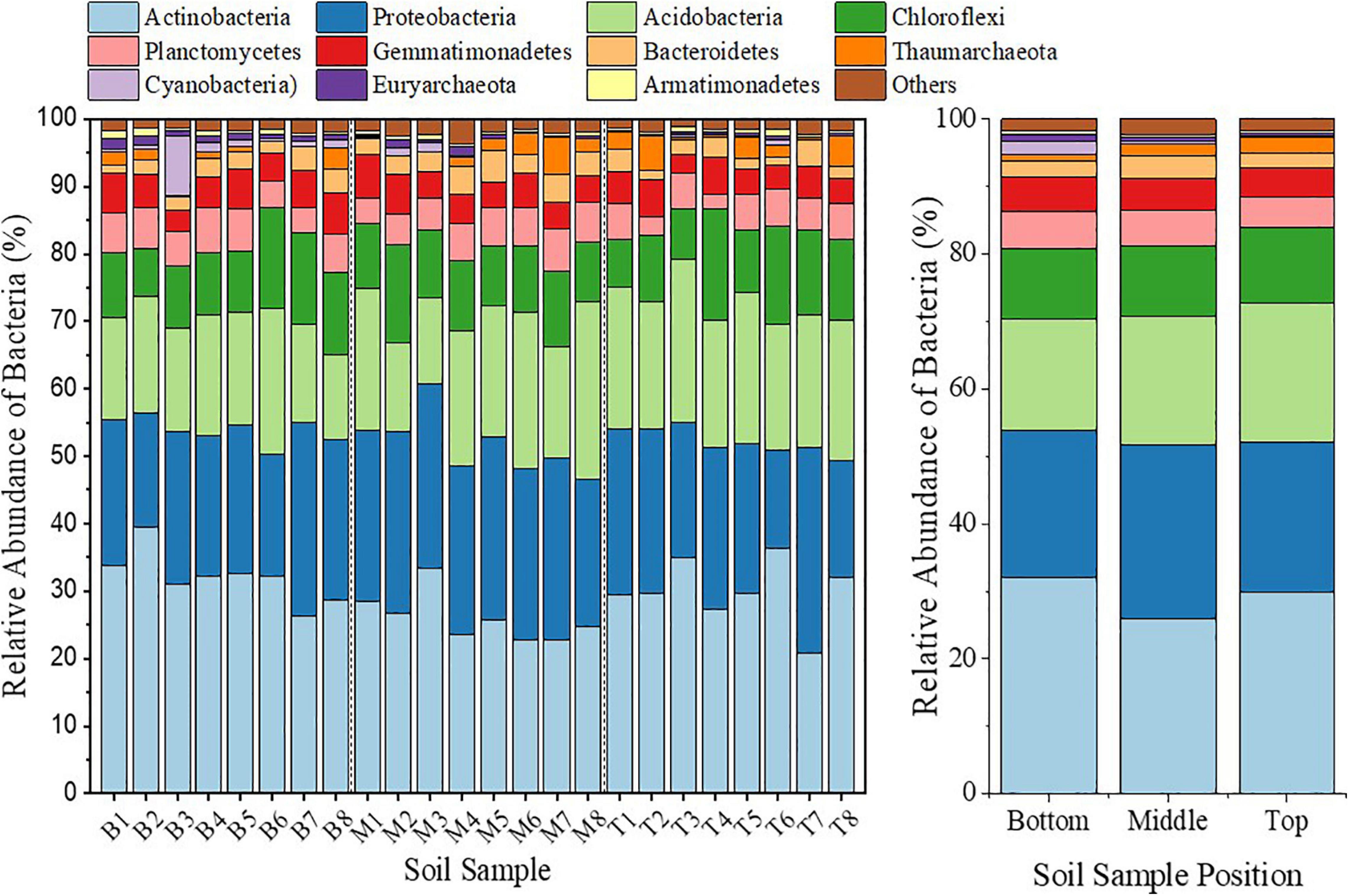
Figure 5. Bar diagram to represent the bacterial community analysis on the phylum level at different locations of the subsidence area.
The soil bacterial community composition on the genus level was analyzed to further verify the micro-differentiation of soil bacterial communities in different topographic locations. We could identify and classify 945 genera in 24 samples, of which 15 genera had relative abundance > 1%. Compared with that in the top slope, the relative abundance of Acinetobacter decreased greatly with the corresponding decrease in amplitude to 96.2% in the middle slope and 92.6% in the bottom slope, respectively. The decrease in the relative abundance of Solirubrobacter and Roseiflexus exceeded 35%. However, the relative abundance of RB41 which was the highest, decreased by 27.9 and 28.6% in the middle and the bottom slopes, respectively. Meanwhile, the relative abundance of Streptomyces and Gemmatimonas in the bottom slope increased greatly with the increase in amplitude to 103 and 111.4%, respectively. On the whole, when facing topographic change, the structure and composition of the soil bacterial community underwent a dramatic directional succession, in principle. However, there was a dramatic differentiation between different topographic locations.
The Correlation Between Soil Microbes and Environmental Factors
To clarify the relationship between the bacterial community succession and environmental factors, RDA analysis was conducted for soil microbial taxa in different topographic locations and environmental factors at the phylum level (Figure 6). On the whole, Axis 1 and Axis 2 could explain 47.29 and 13.04% of the sample differentiation, respectively, with relatively higher explanation reliability. Some clustering effect of soil samples was observed in coordinates of different topographic locations, wherein, the bottom clustering was dramatically separated from that at the top. Besides, there was a certain degree of overlapping in the coordinate system between the middle slope and those two axes.
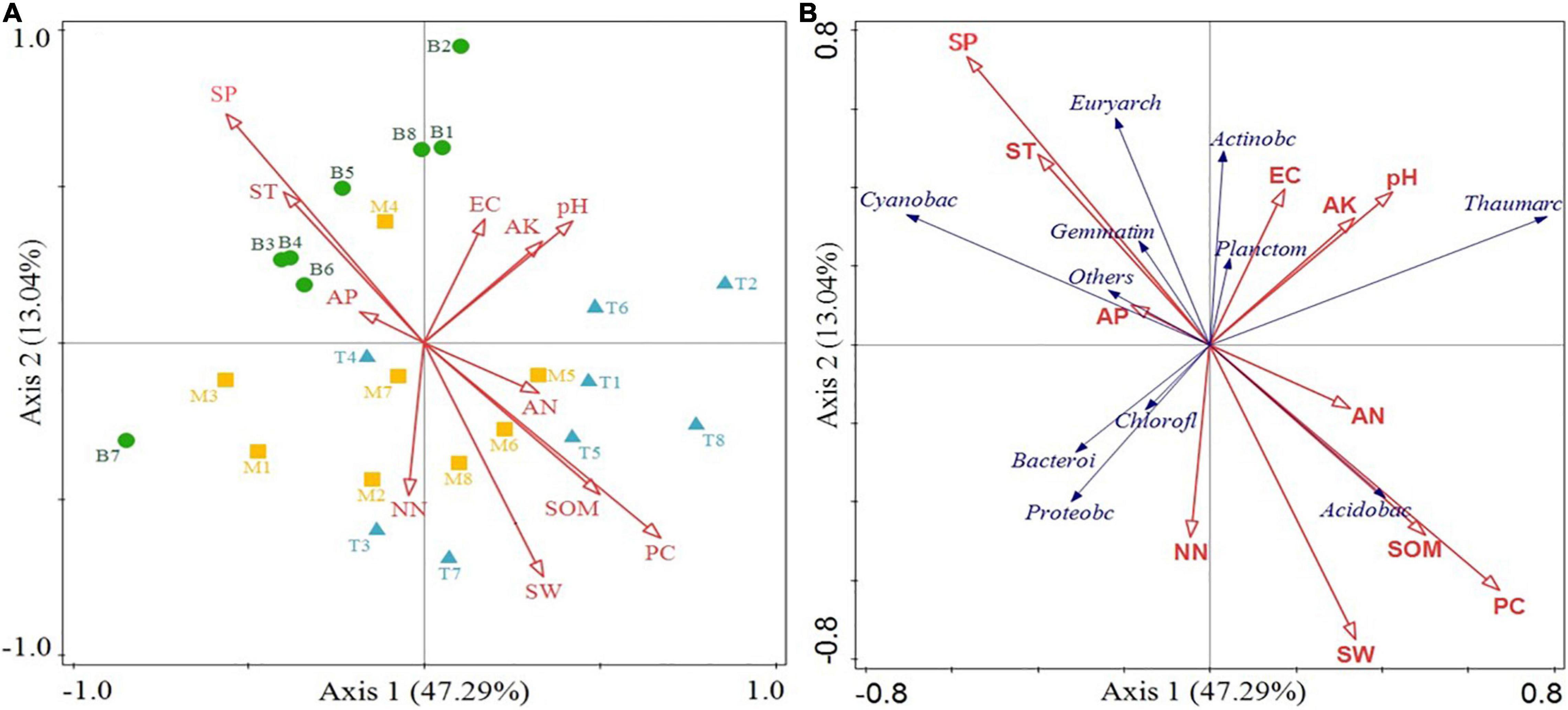
Figure 6. Redundancy analysis (RDA) of samples and environmental factors (A), environmental factors, and major phyla (B).
In the transition from the top to the bottom slope, there was a dramatic change in correlation between SP, SW, SOM, PC, and the sampling site (Figure 6A). The relatively long rays indicate that these factors were important and resulted in soil physicochemical property differentiation in different topographies. Figure 6B illustrates the correlation between the bacterial phyla and the environmental factors, represented by the cosine value of the intersection angle between species and environmental factors. In detail, Acidobacteria was correlated significantly positively with PC, SOM, SW, and AN but significantly negatively with SP and ST. However, correlations of Euryarchaeota, Gemmatimonadetes, and Cyanobacteria were the contrary, illustrating that Acidobacteria in the soil of the study area are more inclined to a living environment with sufficient water and SOM and have a certain preference for N. However, Euryarchaeota, Gemmatimonadetes, and Cyanobacteria are more tolerant to arid and barren environments. Actinobacteria and Planctomycetes dramatically and negatively correlated with NN and positively correlated with EC, AK, and pH. The correlations of these factors with Proteobacteria, Bacteroidetes, and Chloroflexi were the contrary, illustrating that Actinobacteria and Planctomycetes were more adaptable to the loss of N in the environment.
Soil Bacteria Molecular Ecological Network Characteristics in Different Topographic Locations
The bacterial molecular ecological network was constructed based on topographic locations. Network topological properties are mentioned in Table 1 and Supplementary Table 3 in section 2 of Supplementary Material. According to the construction method of molecular ecological network, OTUs with the highest 70% of the frequency in 24 sample sequences were selected. The network similarity threshold was set at 0.91. Considering the topological parameters (Table 1), the network connectivity distribution curve could satisfactorily simulate the power-law model (all R2 > 0.8), illustrating that the network was scale-free and reliable (Deng et al., 2012). The network topological properties of total nodes, total links, and Avg K demonstrated a consistent decreasing principle from the top to the bottom slope, indicating that the structural complexity of soil microbial community also decreases accordingly. Meanwhile, modularity and module numbers in the bottom slope network were dramatically higher than those in the middle and the top slopes, indicating the involvement of a higher proportion of bacteria in the material and energy cycle in the bottom slope. Besides, there were fewer redundant taxa in the network.
OTUs which were vital within and between modules were identified by the Z-P diagram. Generally, the node with Zi ≥ 2.5 or Pi ≥ 0.62 was regarded as the key species (Zhou et al., 2010). There were more connection nodes in the top slope ecological network, wherein, five belonged to Proteobacteria and one belonged to Actinobacteria. In the middle network, there were three network nodes belonging to Actinobacteria. However, there was only 1 network connection node in the bottom slope which belonged to Acidobacteria. Thus, the key species with the highest connectivity in soil bacterial molecular ecological network at the top slope was Proteobacteria. Likewise, the key species in the middle and bottom slopes were Actinobacteria and Acidobacteria, respectively. There was a significant distinction between key species in different topographic locations, indicating that topographic change reshaped the structure and interspecies relationship of soil bacteria.
Based on the sequencing results of soil bacteria, bacterial ecological network diagrams were constructed. The relationship between different species in soil microbes is demonstrated in Figure 7. In detail, there were 34 key OTUs whose connectivity was larger than 10 in the top molecular ecological network (Figure 7C), wherein, the primary 33 were from modules 1 and 3, and were thus regarded as key modules. The structure of the middle network was also close (Figure 7B). There were 27 key OTUs, among these, 26 were from key modules 2, 3, and 9. Key OTUs in the middle and top slope also belonged mainly to Proteobacteria, Actinobacteria, Acidobacteria, and Chloroflexi. However, there was only one OTU node in the bottom slope that belonged to Acidobacteria with connectivity greater than 10 (Figure 7A). In the middle and top network, the connections within and between modules were stronger than those in the bottom, irrespective of being a positive or negative interaction. Although the structure of the bottom network was loose and the connection between nodes was weak, the interactions between and within modules were mainly positive.
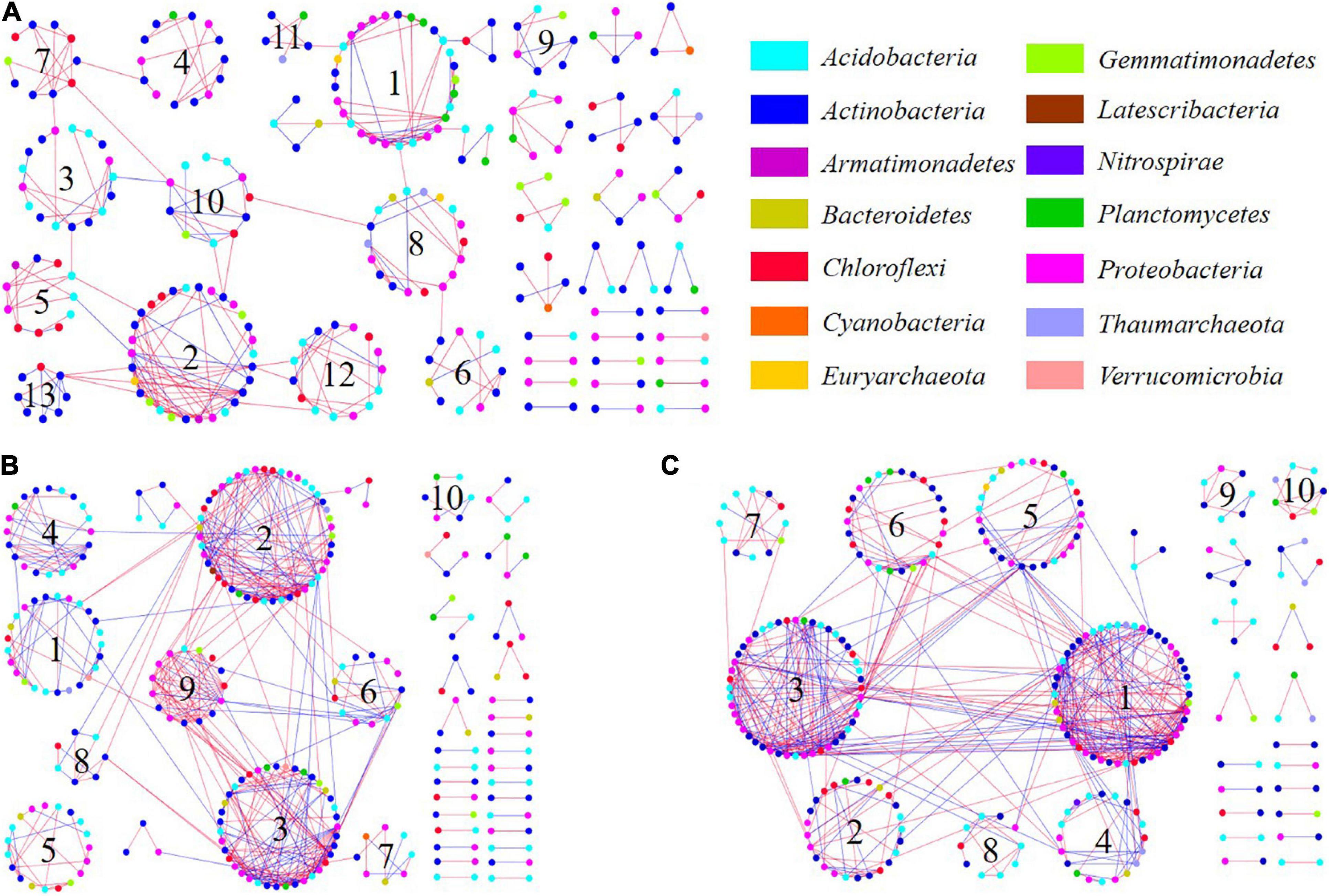
Figure 7. Soil bacterial molecular ecological networks and module structure of (A) the bottom of the slope, (B) the middle of the slope, and (C) the top of the slope. The connecting line represents the interaction of the target level on phylum; colors blue and red represent positive and negative interactions, respectively.
Soil Bacterial Functional Gene Prediction
In this work, OTUs of samples were selected and matched with the KEGG data library (Aoki-Kinoshita and Kanehisa, 2007) for the prediction of PICRUSt gene function (Douglas et al., 2020). The environment with drought and carbon stress clearly selected the bacteria in the middle and the bottom slopes (Figure 8) which responded to four functional categories of genes involved in biological metabolic pathway including cellular processes, environmental information processing, genetic information processing, and metabolism, wherein compared with that in the top slope, the secondary function genes for cell communication consistently and significantly increased in the middle and bottom slopes, and their proportions in all genes have increased by 7.3 and 38.8%, respectively. Meanwhile, the predicted differentiation analysis of secondary metabolism pathway by KEGG also illustrated that the genes for biodegradation and metabolism of xenobiotics, metabolism of terpenoids and polyketides, and lipid metabolism of bacterial communities in the bottom slope were strengthened in terms of function. However, the functional genes which metabolized complex carbon source in the middle slope did not increase, but in fact, decreased a little.
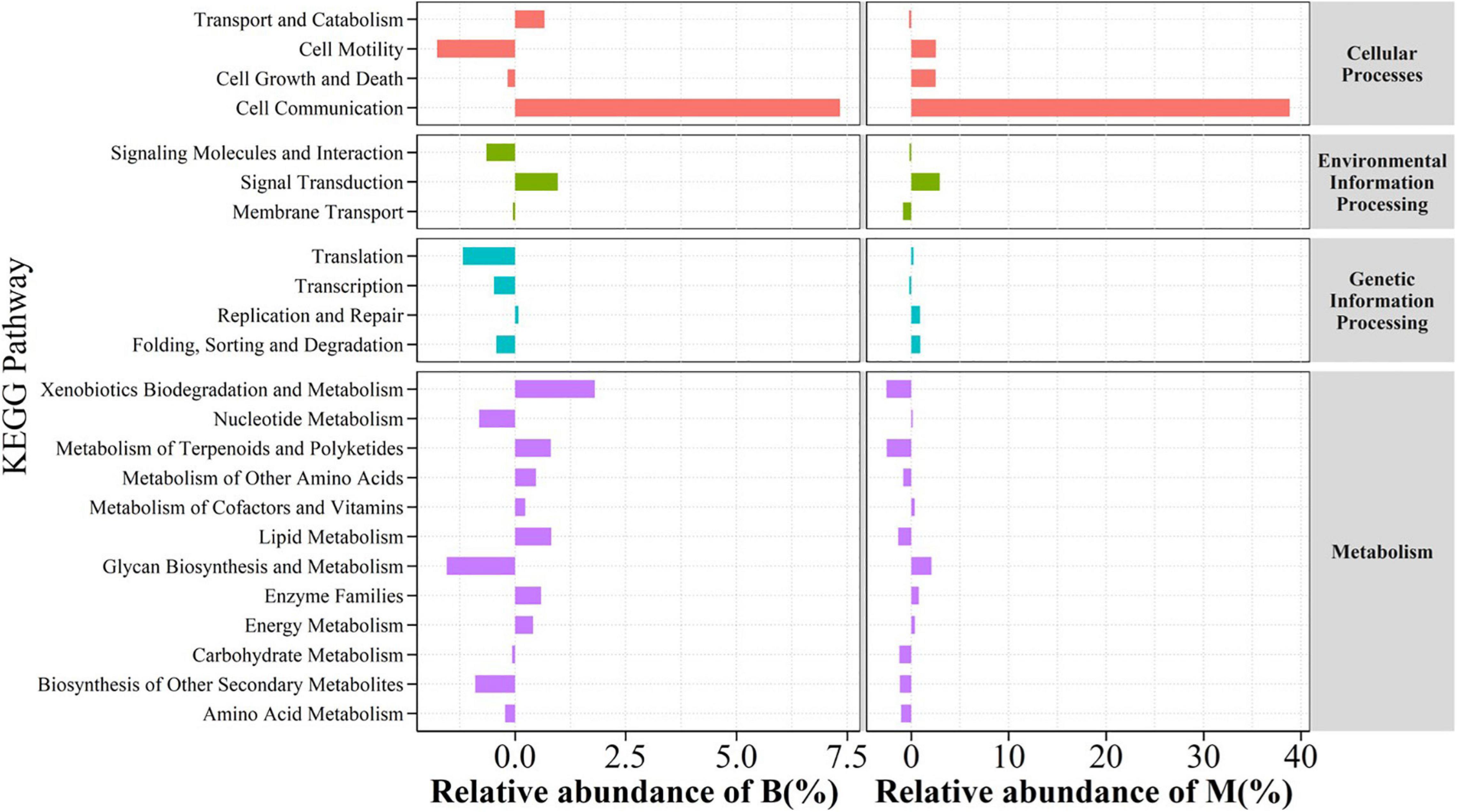
Figure 8. Functional genes analysis of relative abundance changes on Bottom compared to Top and relative abundance changes on Middle compared to Top of the slope.
Discussion
The Influence of the Topographic Change in Mine Land on Soil Physicochemical Properties
In an arid area, water is the primary factor influencing the site quality (Deng et al., 2020). Water in soil is the carrier of material and energy cycle between soil, plant, and microbes, and is responsible for important functions such as nutrient transfer, providing a reaction environment, and osmosis balance, etc. (Chen et al., 2016) In this study, the reshaping of mine land topography changed to originally similar site condition, especially the soil water content (Figure 2 and Supplementary Table 1 in section 2 of Supplementary Material), making the topographic condition the most basic factor to driving the variation in soil physicochemical properties in the interior of mining land. The mechanism of differentiation of physicochemical properties of mining soil due to topographic changes could be illustrated through three aspects such as soil, vegetation, and microbes.
The topographic change occurred because the overlaying soil of mine land subsided after losing support. The subsided top and the bottom slopes were usually accompanied by circular and curved fractures around the collapsing pit (Bian et al., 2012). The physical performance of soil is determined by its mechanical composition (Davidson et al., 2000). Due to the change in original topography, there is a change in structure, water conservation ability, and infiltration characteristic of local soil (Li et al., 2010; Ontl et al., 2013). Small particles of soil are lost along the fracture due to surface runoff and surface wind, causing a decrease in the clay particles in the middle and bottom slope and an increase in the sand proportion (Zu et al., 2004; Zhuang et al., 2007). However, topographic change usually results in plant mechanical damage, leading to a decrease in the surface water and soil conservation ability, which further accelerates the soil desertification process (Figure 2 and Supplementary Table 1 in section 2 of Supplementary Material). Meanwhile, topographic change directly altered the runoff direction of soil water (Michalet et al., 2002). Accordingly, the direction of transfer of the available nutrient in the soil which was easily soluble also converged to the bottom slope. However, a major portion of water in the soil and water-soluble nutrients were retained in the fracture between the middle and the bottom slope. In addition, since the bottom of the slope is closest to the breaking point of the overlying rock (Tiwari et al., 2019), the evaporation and infiltration of the cracks aggravate the loss of soil water, which is particularly obvious at the bottom slope. The dramatically lower water content in the bottom slope compared with that in the top slope in this study was consistent with that of a previous report (Doncheva et al., 2019). During the time of this investigation, the surface fracture was already filled with sand soil. In the area with abundant vegetation, the biological crust (function of Cyanobacteria) was formed, which prevented the evaporation of water in the soil and the loss of the soil nutrient (Warren et al., 2019). Meanwhile, with the increase of nitrogen-fixing bacteria (such as Thaumarchaeota and Rhizobiales), the available N could recover to its original level (Liang et al., 2011; Chen et al., 2018). In this work, we investigated the area in the rainy season. P could be massively accumulated due to the drying and wetting function, which also led to a higher AP content in the middle and the bottom slopes (Figure 2 and Supplementary Table 1 in section 2 of Supplementary Material).
The status of vegetation growth on the ground surface is a comprehensive macroscopic reflection of the influence of topographic change. The dynamic performance of vegetation change could characterize soil status with time (Wang et al., 2017). Surface subsidence caused mechanical damage to plant roots, and soil water and fertilizer loss also intensified competition among plants, resulting in a sharp decrease in vegetation coverage, which is consistent with the trend of vegetation change from the top to the bottom slope in this work (Figure 2). Meanwhile, drought caused by topographical changes can enhance the ability of bacterial communities to degrade soil organic carbon (Kuang et al., 2020), thereby accelerating the loss of SOM. This may be the reason why the SOM content shows greater differences than vegetation coverage.
The biological enzyme activity in the soil can indicate the soil nutrition status and the activity status of soil microbes (Wolinska et al., 2015), wherein, DHG can comprehensively indicate soil microbial characteristics (Zeng et al., 2017), and URA in the soil could be the validity index for the evaluation of soil fertility (Fu et al., 2020). Under the dual effects of soil and surface vegetation, the development and metabolism of bacteria in the middle and bottom slopes faced more serious water stress and carbon stress, so the DHG and URA enzyme activities of the top slope were significantly higher than those in the middle and bottom slopes (Figure 1 and Supplementary Table 1 in section 2 of Supplementary Material), which was also reported earlier (Doncheva et al., 2019). The PRO activity of the soil in the middle and the top slopes did not decrease and was possibly related to a significant increase in Gemmatimonas in the middle and the top slopes. Gemmatimonas has been reported to display strong PRO activity for the decomposition of complex carbonate compounds (Jiang et al., 2017).
The Adaptive Changes in Soil Bacterial Community Structure
Soil bacteria are sensitive to environmental change. Change in soil environment due to topographic change influences the composition, activity strength of bacterial community, and the material energy transformation process (Liu et al., 2016; Delgado-Baquerizo et al., 2018a), which further drives the directional succession of soil bacterial community. The proportions of Actinobacteria, Proteobacteria, Acidobacteria, and Chloroflexi in the top, middle and bottom slopes exceeded 80% (Figure 5). They comprised the primary functional modules of molecular ecological networks and dominated the development of the community taxa. Besides, there was no large difference in the number between those three levels. After surface subsidence of desertification mining land, the overlying soil was found to be generally dry and could not form nutrition pools (Yang et al., 2016). Fracture and land sliding also increased the soil porosity, making it favorable for the development of aerobic bacteria (Actinobacteria, Proteobacteria) (Frerichs et al., 2013; Banerjee et al., 2019) which became the dominant phyla. Meanwhile, the lack of carbon source in the middle and the bottom slopes (Figure 2) with relatively large topographic change granted developmental advantage to autotrophic bacteria (Acidobacteria, Chloroflexi) (Liu et al., 2016; Chen et al., 2017). Therefore, these four types of bacteria can adapt to harsh environments such as poor soil, lack of water, fracture, and squeezing after the topography of the mine changes. They constituted the main functional modules of the molecular ecological networks (Figure 7), and dominated the development of the community.
Although they are all in the context of environmental degradation, different topographic locations have different performances in soil-water environments and surface vegetation conditions (Figure 2). Thus, except for the succession trend, on the whole, bacterial communities demonstrated different directions of succession due to the topographic differentiation (Figure 7) as a response to different environmental stress. Cyanobacteria can adapt to hypoxic and oligotrophic environments (Figure 9), most of which can fix nitrogen (Percival and Williams, 2014), and can form biological soil crusts with algae and mosses (Ferrenberg et al., 2017), significantly improving surface soil nutrients and soil water holding capacity (Yang et al., 2012), etc. The biological crusts have strong physiological tolerance to adversity (Warren et al., 2019), and play an important role in improving the soil structure of the slope and the bottom of the slope, preventing soil erosion (Machado-de-Lima et al., 2019), and regulating the balance of precipitation infiltration and evapotranspiration (Moreira-Grez et al., 2019). Euryarchaeota is characterized by living in extreme environments (such as high salinity, high temperature and hypoxia) (Barberan et al., 2011), and a large number of ammonia oxidation functions are used to cope with the poor soil nutrients caused by mining subsidence (Schleper and Nicol, 2010). Both of them are negatively correlated with SOM and PC (Figure 6), and there is a significant increase in the middle and bottom of the slopes where the living conditions are relatively poor (Figure 5), which proves that Cyanobacteria and Euryarchaeota have higher tolerance to oligotrophic environment. Moreover, the bacterial species diversity in the middle and bottom slopes showed smaller spatial heterogeneity (Figure 3). These evidences all demonstrated the adaptive succession of bacterial communities in response to environmental degradation. Thaumarchaeota is able to dominate the nitrification process of the soil under low pH conditions due to its high affinity for ammonia (Zhang et al., 2012), and it also has two nutrient modes: autotrophic fixed CO2 and organic carbon metabolism (Walker et al., 2010; Konneke et al., 2014). However, in this study, Thaumarchaeota in the middle and bottom slopes showed a large decline (Figure 10) contrary to expectations. Considering that there is no significant difference in the overall pH of the collapsed area (Figure 2), and the phylum Thaumarchaeota has a wide range of adaptations to temperature, oxygen and moisture, we believe that this may be caused by the obstruction of the organic carbon metabolism pathway due to the sharp decrease in SOM. It can be seen that the deteriorated soil environment in the middle and bottom of the slopes also has a selective effect on the bacterial community.
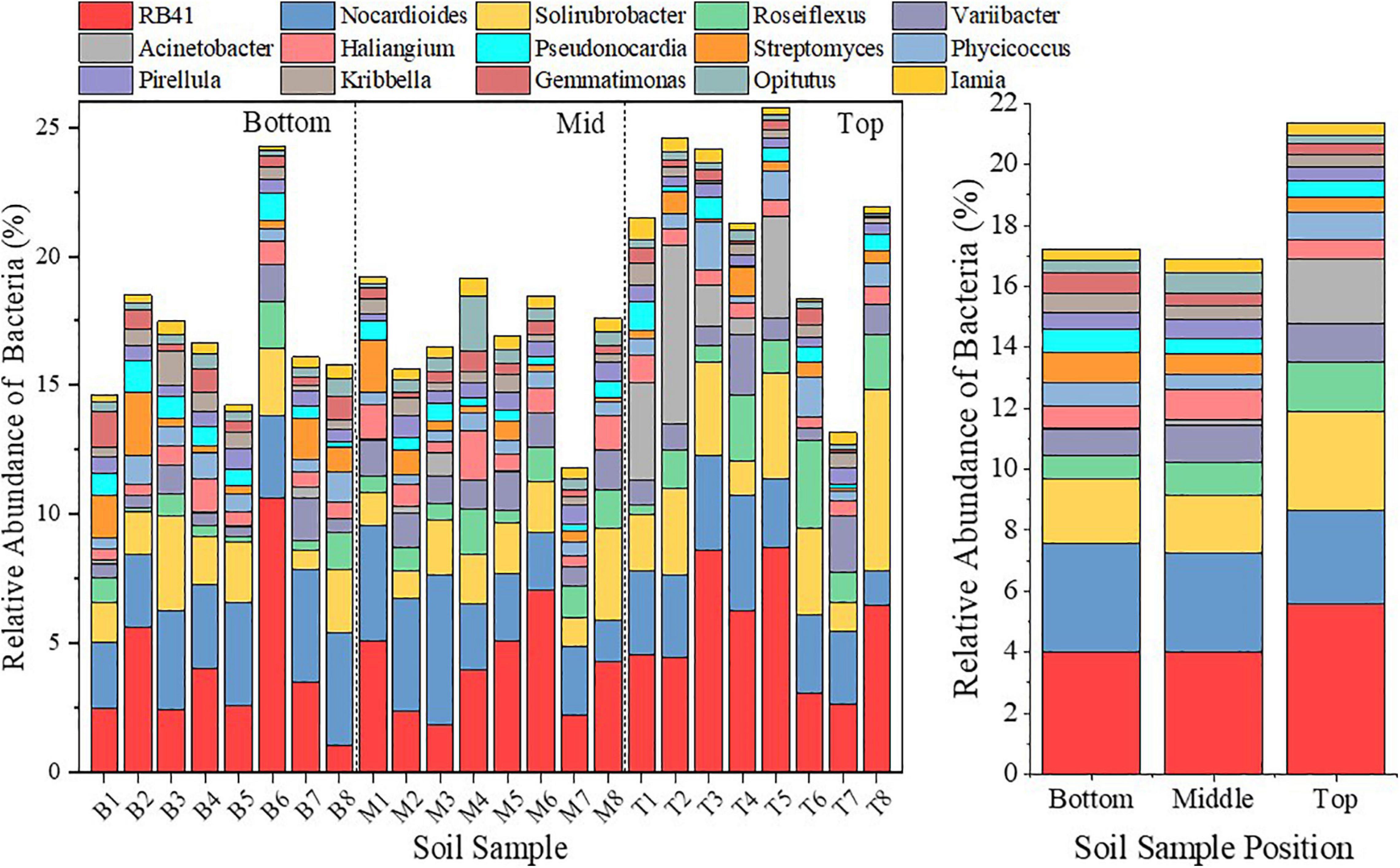
Figure 10. Bar diagram to represent the bacterial community analysis on the genus level at different locations of the subsidence area.
The bacterial community could better reflect the difference in directions of succession in terms of the bacteria at genus levels, and the effect of environmental selection is more obvious at the genus level. Among those, Gemmatimonas could secrete highly active PRO for the decomposition of complex carbohydrates (Jiang et al., 2017). As the most advanced Actinomycetes, the developed mycelial morphology of Streptomyces endows it with strong drought tolerance and barren resistance (Sun et al., 2017). Therefore, the numbers of both increased with a larger amplitude in the middle and bottom slopes (Figure 10). However, Acinetobacter, which lacks mobility and cannot circumvent (Barbe et al., 2004), almost disappeared in the middle and bottom slopes (Figure 10) due to its inability to respond to the sudden change of water and nutrition conditions in the earlier phase of topographic change. Solirubrobacter (Actinobacteria), Roseiflexus (Chloroflexi), and RB41 (Acidobacteria) are heterotrophic bacteria (Hanada et al., 2002; Singleton et al., 2003; Pascual et al., 2015), so the decrease of PC and SOM (Figure 10) concentration restricted their reproduction and development in the middle and bottom slopes from the carbon source. These evidences indicate that the direction of bacterial community succession is closely related to species composition of the community, and the environmental stress that exceeds the resistance and self-recovering ability of bacterial species will lead to the decline of the original community and the generation of new communities.
The Interaction Between Soil Bacterial Species
The interaction between species demonstrates the survival strategy of species with a series of external factors including resource availability and the extent of environment stress (Li et al., 2019). In our study, under the combined interaction of soil properties and surface vegetation, the key modules, key species (OTUs), and even the interspecific relationships within the bacterial community also changed (Figure 7). The Module hubs and Connectors in the molecular ecological network are critical in the soil nutrient cycle, and they often play a decisive role in the metabolic pathway of organic matter (Lynch and Neufeld, 2015; Jiang et al., 2016), thus becoming the core of the bacterial community. Changes in key species of bacterial communities also reflect changes in the relationship between bacterial species to a certain extent. From the top to the bottom slopes, the key species were transformed from Proteobacteria to Actinobacteria and then to Acidobacteria (Figure 9).
The key species of the top slope molecular ecological network are mainly composed of the genus Rhizobiales (Proteobacteria), which can symbiotically with plants (Erlacher et al., 2015), and the inorganic chemical energy class β-Proteobacteria (Proteobacteria), which can oxidize ammonia (Ferro et al., 2019). The abundant Alfalfa on the top of the slope provides favorable conditions for the development of the genus Rhizobiales (Song et al., 2017), and the growth-promoting effect of Rhizobiales on plants provides more nutrients for other bacteria (Zhang Z.Q. et al., 2016). As a result, they dominate the carbon and nitrogen cycle process of the top slope as the key species (Figure 9). Bacteria in the middle of the slope face fewer plants and the most complex topographical conditions, so the genus Nocardioides (Actinobacteria), which can use a variety of organic matter as carbon source, developed into the main key species in the metabolic process of the bacterial community by advantage of its developed hyphae morphology that can better absorb nutrients (Wu et al., 2019). The key species of the bottom slope network were mainly composed of anoxygenic chlorophototrophic Acidobacteria (Bryant, 2013). Meanwhile, the genus Planctomycetaceae (Planctomycetes) is also regarded as a key species of the bottom slope, and it has been reported to be highly correlated with the production of a large number of secondary metabolites (lantipeptide, terpene and type 1 polyketide synthases) (Jeske et al., 2013). Interestingly, in the prediction of functional genes, it was found that the genes related to metabolism of terpenoids and polyketides were significantly improved as well in the bottom slope bacteria (Figure 8). These findings suggest that in response to the extreme lack of carbon sources, the metabolic relationship of the bacterial community in the bottom of the slope was reshaped by the availability of carbon sources, so autotrophic bacteria and bacteria with strong secondary metabolism became the key species of community. Overall, the interaction within the bacterial community is mainly driven by the metabolism of nutrients.
In terms of network structure, the bottom of the slope has the fewest nodes, but the highest degree of modularization (modularity = 85.3%, Table 1). High degree of modularity means that the community has lower species redundancy (Deng et al., 2012), and species redundancy tends to be positively correlated with the stability of the community (Coyte et al., 2015; Deng et al., 2016). Meanwhile, the number of positive links of the bottom slope network accounted for as high as 74.38% and the common cooperative relationship appeared in most modules, but the cooperation among species would reduce the stability of the community (Hernandez et al., 2021). Besides, the weak interconnection between the network modules at the bottom slope is also a manifestation of low community stability (Zhou et al., 2010). The network structures in the middle and the top slopes were similar, forming a metabolic functional group with a few modules as the core, and the relationship between species within the functional group is greatly strengthened. The higher redundancy and association strength indicate the more complex and stable community structures in the middle and bottom slopes (Coyte et al., 2015). Meanwhile, in the process of material metabolism, high transitivity (Transitivity) and short path distance (Avg PD, Supplementary Table 1) are also conducive to the stability of the community on the food webs (Meyer et al., 2020). In addition, the strong competition between the bacterial communities in the middle and top slopes is also a sign that the soil-plant ecosystem has regained a stable state (Bi et al., 2019). Overall, under severe environmental stress (bottom), soil bacteria tend to sacrifice community stability and choose to cooperate with each other to maintain the cyclic metabolism of substances, so as to ensure the energy requirements for growth, development and reproduction. Therefore, the changes in interspecific relationship may serve as evidence for the recovery of soil bacterial species and functional diversity (Zhao et al., 2019; Madi et al., 2020). Besides, the cell communication function genes also showed a similar change trend while the inter-species cooperation was greatly enhanced, which also confirmed the adaptive succession of inter-species interaction to environmental degradation at the genetic level.
Conclusion
Summarizing, these results reveals the impact of topographical changes in the mining area on the soil environment and bacterial communities. Based on the analysis of the results, we draw the following conclusions:
(1) The surface subsidence caused by coal mining reshaped the habitat of the desertification grassland. From the top to the bottom of the slope, the soil environment gradually deteriorates, and the lack of soil nutrients limits the development and reproduction of bacterial communities. Among them, PC and SOM are the biggest limiting factors in the study area.
(2) In order to deal with insufficient soil nutrients caused by terrain changes, bacterial communities tend to cooperate with each other. The interaction within the bacterial communities are mainly driven by the metabolism of nutrients.
(3) The degree of nutrition stress faced by different terrain parts is different, so the coping strategies adopted by soil bacterial communities are therefore different. Strong environmental pressure will weaken the relationship between soil bacteria and reduce the stability of the community.
The impact of topographical changes on soil bacteria is multifaceted. The carbon source may be the fundamental factor that governs the relationship between bacterial communities. The methods provided in this study are qualitative. Therefore, the specific relationship between soil carbon sources and bacterial communities is still needs further verification by quantitative research. The methods and perspectives this study provided may be able to attract the attention of researchers of grassland ecological restoration in subsided desertification mining areas, so that ecological restoration can be carried out according to local conditions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA694481).
Author Contributions
JM and ZL collected the samples. QZ, YY, and YW performed the experiments. QZ analyzed the data and along with JM and FC contributed in writing the manuscript. All authors were involved in the study design and contributed to the writing of the manuscript.
Funding
This work was supported by the National Natural Science Foundation (Nos. 41807515, 41907405, and 51974313) and the Natural Science Foundation of Jiangsu Province (No. BK20180641). We gratefully acknowledge these programs for financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.656708/full#supplementary-material
Footnotes
- ^ http://mem.rcees.ac.cn8080
- ^ http://ieg4.rccc.ou.edu/mena
- ^ http://github.com/picrust/picrust2/wiki
References
Alivisatos, A. P., Blaser, M. J., Brodie, E. L., Chun, M., Dangl, J. L., Donohue, T. J., et al. (2015). A unified initiative to harness Earth’s microbiomes. Science 350, 507–508. doi: 10.1126/science.aac8480
Allison, S. D., and Martiny, J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105, 11512–11519. doi: 10.1073/pnas.0801925105
Aoki-Kinoshita, K. F., and Kanehisa, M. (2007). Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 396, 71–91. doi: 10.1385/1-59745-515-6:71
Ayala-Orozco, B., Gavito, M. E., Mora, F., Siddique, I., Balvanera, P., Jaramillo, V. J., et al. (2018). Resilience of soil properties to land-use change in a tropical dry forest ecosystem. Land. Degrad. Dev. 29, 315–325. doi: 10.1002/ldr.2686
Banerjee, S., Walder, F., Buechi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Barahona, E., and Iriarte, A. (2001). An overview of the present state of standardization of soil sampling in Spain. Sci. Total Environ. 264, 169–174. doi: 10.1016/s0048-9697(00)00620-3
Barbe, V., Vallenet, D., Fonknechten, N., Kreimeyer, A., Oztas, S., Labarre, L., et al. (2004). Unique features revealed by the genome sequence of Acinetobacter sp ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32, 5766–5779. doi: 10.1093/nar/gkh910
Barberan, A., Fernandez-Guerra, A., Auguet, J.-C., Galand, P. E., and Casamayor, E. O. (2011). Phylogenetic ecology of widespread uncultured clades of the Kingdom Euryarchaeota. Mol. Ecol. 20, 1988–1996. doi: 10.1111/j.1365-294x.2011.05057.x
Bi, Y., Xie, L., Wang, J., Zhang, Y., and Wang, K. (2019). Impact of host plants, slope position and subsidence on arbuscular mycorrhizal fungal communities in the coal mining area of north-central China. J. Arid Environ. 163, 68–76. doi: 10.1016/j.jaridenv.2018.11.011
Bian, Z., Miao, X., Lei, S., Chen, S.-E., Wang, W., and Struthers, S. (2012). The challenges of reusing mining and mineral-processing wastes. Science 337, 702–703. doi: 10.1126/science.1224757
Brown, S. P., and Jumpponen, A. (2015). Phylogenetic diversity analyses reveal disparity between fungal and bacterial communities during microbial primary succession. Soil Biol. Biochem. 89, 52–60. doi: 10.1016/j.soilbio.2015.06.025
Bryant, D. A. (2013). “Green bacteria: chlorophyll biosynthesis, light-harvesting, reaction centers, and electron transport,” in Encyclopedia of Biological Chemistry, 2nd edn. eds W. J. Lennarz and M. D. Lane (Waltham: Academic Press), 501–509. doi: 10.1016/b978-0-12-378630-2.00159-6
Chen, F., Zhang, W., Ma, J., Yang, Y., Zhang, S., and Chen, R. (2017). Experimental study on the effects of underground CO2 leakage on soil microbial consortia. Int. J. Greenh. Gas Control 63, 241–248. doi: 10.1016/j.ijggc.2017.05.017
Chen, J., Mao, L., Mi, H., Lu, W., Ying, T., and Luo, Z. (2016). Involvement of abscisic acid in postharvest water-deficit stress associated with the accumulation of anthocyanins in strawberry fruit. Postharvest Biol. Technol. 111, 99–105. doi: 10.1016/j.postharvbio.2015.08.003
Chen, X., Zhang, L., Chen, M., Xing, Y., Hu, Z., and Liu, B. (2018). Aggregate distribution and organic carbon mineralization of reconstructed soils filled with coal gangue in a huainan coal mining area. China. Fresen. Environ. Bull. 27, 8882–8891.
Chmura, D., Salachna, A., and Sierka, E. (2016). Comparison of visual estimation of the canopy cover with the canopyscope assessment. Sylwan 160, 475–481.
Clauset, A., Moore, C., and Newman, M. E. J. (2008). Hierarchical structure and the prediction of missing links in networks. Nature 453, 98–101. doi: 10.1038/nature06830
Collier, M. J. (2015). Novel ecosystems and social-ecological resilience. Landsc. Ecol. 30, 1363–1369. doi: 10.1007/s10980-015-0243-z
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Davidson, E. A., Verchot, L. V., Cattanio, J. H., Ackerman, I. L., and Carvalho, J. E. M. (2000). Effects of soil water content on soil respiration in forests and cattle pastures of eastern Amazonia. Biogeochemistry 48, 53–69.
Delgado-Baquerizo, M., Eldridge, D. J., Hamonts, K., Reich, P. B., and Singh, B. K. (2018a). Experimentally testing the species-habitat size relationship on soil bacteria: a proof of concept. Soil Biol. Biochem. 123, 200–206. doi: 10.1016/j.soilbio.2018.05.016
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-Gonzalez, A., Eldridge, D. J., Bardgett, R. D., et al. (2018b). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Deng, Y., Jiang, Y.-H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinform. 13:113. doi: 10.1186/1471-2105-13-113
Deng, Y., Wang, S., Bai, X., Luo, G., Wu, L., Chen, F., et al. (2020). Vegetation greening intensified soil drying in some semi-arid and arid areas of the world. Agr. For. Meteorol. 292-293:108103 doi: 10.1016/j.agrformet.2020.108103
Deng, Y., Zhang, P., Qin, Y., Tu, Q., Yang, Y., He, Z., et al. (2016). Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environ. Microbiol. 18, 205–218. doi: 10.1111/1462-2920.12981
Dimitriu, P. A., Prescott, C. E., Quideau, S. A., and Grayston, S. J. (2010). Impact of reclamation of surface-mined boreal forest soils on microbial community composition and function. Soil Biol. Biochem. 42, 2289–2297. doi: 10.1016/j.soilbio.2010.09.001
Ding, J., Zhang, Y., Wang, M., Sun, X., Cong, J., Deng, Y., et al. (2015). Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol. Ecol. 24, 5175–5185. doi: 10.1111/mec.13384
Doncheva, N. T., Morris, J. H., Gorodkin, J., and Jensen, L. J. (2019). Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 18, 623–632. doi: 10.1021/acs.jproteome.8b00702
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Erlacher, A., Cernava, T., Cardinale, M., Soh, J., Sensen, C. W., Grube, M., et al. (2015). Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 6:53.
Ferrenberg, S., Tucker, C. L., and Reed, S. C. (2017). Biological soil crusts: diminutive communities of potential global importance. Front. Ecol. Environ. 15, 160–167. doi: 10.1002/fee.1469
Ferro, P., Vaz-Moreira, I., and Manaia, C. M. (2019). Betaproteobacteria are predominant in drinking water: are there reasons for concern? Crit. Rev. Microbiol. 45, 649–667.
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Frerichs, J., Oppermann, B. I., Gwosdz, S., Moeller, I., Herrmann, M., and Krueger, M. (2013). Microbial community changes at a terrestrial volcanic CO2 vent induced by soil acidification and anaerobic microhabitats within the soil column. FEMS Microbiol. Ecol. 84, 60–74. doi: 10.1111/1574-6941.12040
Frouz, J., Toyota, A., Mudrak, O., Jilkova, V., Filipova, A., and Cajthaml, T. (2016). Effects of soil substrate quality, microbial diversity and community composition on the plant community during primary succession. Soil Biol. Biochem. 99, 75–84. doi: 10.1016/j.soilbio.2016.04.024
Fu, Q., Abadie, M., Blaud, A., Carswell, A., Misselbrook, T. H., Clark, I. M., et al. (2020). Effects of urease and nitrification inhibitors on soil N, nitrifier abundance and activity in a sandy loam soil. Biol. Fertil. Soils 56, 185–194. doi: 10.1007/s00374-019-01411-5
Guo, Y., Chen, X., Wu, Y., Zhang, L., Cheng, J., Wei, G., et al. (2018). Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 635, 598–606. doi: 10.1016/j.scitotenv.2018.04.171
Hanada, S., Takaichi, S., Matsuura, K., and Nakamura, K. (2002). Roseiflexus castenholzii gen. nov., sp nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52, 187–193. doi: 10.1099/00207713-52-1-187
Harris, J. (2009). Soil microbial communities and restoration ecology: facilitators or followers? Science 325, 573–574. doi: 10.1126/science.1172975
He, Y., He, X., Liu, Z., Zhao, S., Bao, L., Li, Q., et al. (2017). Coal mine subsidence has limited impact on plant assemblages in an arid and semi-arid region of northwestern China. Ecoscience 24, 91–103.
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). Environmental stress destabilizes microbial networks. ISME J. 2:16.
Hu, C. C., Chen, C., Yu, Y., Zhou, X. B., Lin, S., and Zhou, S. J. (2013). “Study on the direction of land reclamation and ecological reconstruction in jiaozuo coal mining area under the angle of central plains economic zone,” in Advances in Environmental Technologies, Pts 1-6, eds J. Zhao, R. Iranpour, X. Li, and B. Jin. (Amsterdam: Elsevier), 4991–4995. doi: 10.4028/www.scientific.net/amr.726-731.4991
Huang, J. L., Bai, Y., Lei, S. G., and Deng, K. Z. (2020). Time-Series SBAS Pixel offset tracking method for monitoring three-dimensional deformation in a mining area. IEEE Access 8, 118787–118798. doi: 10.1109/access.2020.3004460
Jeske, O., Jogler, M., Petersen, J., Sikorski, J., and Jogler, C. (2013). From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104, 551–567. doi: 10.1007/s10482-013-0007-1
Jiang, Y. J., Li, S. Z., Li, R. P., Zhang, J., Liu, Y. H., Lv, L. F., et al. (2017). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109, 145–155. doi: 10.1016/j.soilbio.2017.02.010
Jiang, Y., Liang, Y., Li, C., Wang, F., Sui, Y., Suvannang, N., et al. (2016). Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem. 95, 250–261. doi: 10.1016/j.soilbio.2016.01.007
Jing, Z. R., Wang, J. M., Zhu, Y. C., and Feng, Y. (2018). Effects of land subsidence resulted from coal mining on soil nutrient distributions in a loess area of China. J. Clean. Prod. 177, 350–361. doi: 10.1016/j.jclepro.2017.12.191
Kardol, P., and Wardle, D. A. (2010). How understanding aboveground-belowground linkages can assist restoration ecology. Trends Ecol. Evol. 25, 670–679. doi: 10.1016/j.tree.2010.09.001
Konneke, M., Schubert, D. M., Brown, P. C., Hugler, M., Standfest, S., Schwander, T., et al. (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. U. S. A. 111, 8239–8244. doi: 10.1073/pnas.1402028111
Kuang, E., Xu, M., Colinet, G., Chi, F., Su, Q., Zhu, B., et al. (2020). Degradation characteristics of maize straw under different buried depths in northeast black soil and their effects on soil carbon and nitrogen. Int. J. Agric. Biol. 24, 77–84.
Lei, K., Pan, H., and Lin, C. (2016). A landscape approach towards ecological restoration and sustainable development of mining areas. Ecol. Eng. 90, 320–325. doi: 10.1016/j.ecoleng.2016.01.080
Li, C., Li, Y., Ma, J., Fan, L., and Wang, Q. (2010). Spatial heterogeneity of soil chemical properties between Haloxylon persicum and Haloxylon ammodendron populations. J. Arid Land 2, 257–265.
Li, J., Xin, Z., Yan, J., Li, H., Chen, J., and Ding, G. (2018). Physicochemical and microbiological assessment of soil quality on a chronosequence of a mine reclamation site. Eur. J. Soil Sci. 69, 1056–1067.
Li, M. S. (2006). Ecological restoration of mineland with particular reference to the metalliferous mine wasteland in China: a review of research and practice. Sci. Total Environ. 357, 38–53.
Li, S.-P., Bi, Y.-L., Kong, W.-P., Wang, J., and Yu, H.-Y. (2013). Effects of the arbuscular mycorrhizal fungi on environmental phytoremediation in coal mine areas. Huan Jing Ke Xue 34, 4455–4459.
Li, X. X., Ma, J., Yang, Y. J., Hou, H. P., Liu, G. J., and Chen, F. (2019). Short-term response of soil microbial community to field conversion from dryland to paddy under the land consolidation process in North China. Agric. Open Access J. 9, 1–17.
Liang, Y., Van Nostrand, J. D., Deng, Y., He, Z., Wu, L., Zhang, X., et al. (2011). Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J. 5, 403–413.
Liu, J., Sui, Y., Yu, Z., Yao, Q., Shi, Y., Chu, H., et al. (2016). Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Energy Sci. Eng. 95, 212–222.
Lozano, Y. M., Hortal, S., Armas, C., and Pugnaire, F. I. (2014). Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol. Biochem. 78, 298–306.
Luo, C., Rodriguez-R, L. M., Johnston, E. R., Wu, L., Cheng, L., Xue, K., et al. (2014). Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl. Environ. Microb. 80, 1777–1786.
Luo, Z. B., Ma, J., Chen, F., Li, X. X., Zhang, Q., and Yang, Y. J. (2020). Adaptive development of soil bacterial communities to ecological processes caused by mining activities in the loess plateau. China. Microorganisms 8:477.
Lynch, M. D. J., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229.
Machado-de-Lima, N. M., Camara Fernandes, V. M., Roush, D., Ayuso, S. V., Rigonato, J., Garcia-Pichel, F., et al. (2019). The compositionally distinct cyanobacterial biocrusts from brazilian savanna and their environmental drivers of community diversity. Front. Microbiol. 10:2798.
Madi, N., Vos, M., Murall, C. L., Legendre, P., and Shapiro, B. J. (2020). Does diversity beget diversity in microbiomes? Elife 9:e5899.
Malachowska-Jutsz, A., and Matyja, K. (2019). Discussion on methods of soil dehydrogenase determination. Int. J. Environ. Sci. Technol. 16, 7777–7790.
Mao, L., Pan, J., Jiang, S., Shi, G., Qin, M., Zhao, Z., et al. (2019). Arbuscular mycorrhizal fungal community recovers faster than plant community in historically disturbed Tibetan grasslands. Soil Biol. Biochem. 134, 131–141.
Mendes, L. W., Tsai, S. M., Navarrete, A. A., de Hollander, M., van Veen, J. A., and Kuramae, E. E. (2015). Soil-borne microbiome: linking diversity to function. Microb. Ecol. 70, 255–265.
Meyer, J. M., Leempoel, K., Losapio, G., and Hadly, E. A. (2020). Molecular ecological network analyses: an effective conservation tool for the assessment of biodiversity, trophic interactions, and community structure. Front. Ecol. Evol. 8:588430.
Michalet, R., Gandoy, C., Joud, D., Pages, J. P., and Choler, P. (2002). Plant community composition and biomass on calcareous and siliceous substrates in the northern French Alps: comparative effects of soil chemistry and water status. Arct. Antarct. Alp. Res. 34, 102–113.
Moreira-Grez, B., Tam, K., Cross, A. T., Yong, J. W. H., Kumaresan, D., Nevill, P., et al. (2019). The bacterial microbiome associated with arid biocrusts and the biogeochemical influence of biocrusts upon the underlying soil. Front. Microb. 10:2143.
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. U. S. A. 104, 19891–19896.
Ontl, T. A., Hofmockel, K. S., Cambardella, C. A., Schulte, L. A., and Kolka, R. K. (2013). Topographic and soil influences on root productivity of three bioenergy cropping systems. New Phytol. 199, 727–737.
Pascual, J., Wuest, P. K., Geppert, A., Foesel, B. U., Huber, K. J., and Overmann, J. (2015). Novel isolates double the number of chemotrophic species and allow the first description of higher taxa in Acidobacteria subdivision 4. Syst. Appl. Microbiol. 38, 534–544.
Percival, S. L., and Williams, D. W. (2014). “Chapter five - cyanobacteria,” in Microbiology of Waterborne Diseases, 2nd edn. eds S. L. Percival, M. V. Yates, D. W. Williams, R. M. Chalmers, and N. F. Gray (London: Academic Press), 79–88.
Prommer, J., Walker, T. W. N., Wanek, W., Braun, J., Zezula, D., Hu, Y., et al. (2020). Increased microbial growth, biomass, and turnover drive soil organic carbon accumulation at higher plant diversity. Glob. Chang. Biol. 26, 669–681.
Qi, Y., Stern, N., Wu, T., Lu, J., and Green, F. (2016). China’s post-coal growth. Nat. Geosci. 9, 564–566.
Schleper, C., and Nicol, G. W. (2010). “∗Ammonia-oxidising archaea - physiology, ecology and evolution,” in Advances in Microbial Physiology, Vol 57, ed. R. K. Poole. (Cambridge, MA: Academic Press)
Shen, C., Ma, D., Sun, R., Zhang, B., Li, D., and Ge, Y. (2019). Long-term stacking coal promoted soil bacterial richness associated with increased soil organic matter in coal yards of power plants. J. Soil Sediment. 19, 3442–3452.
Singleton, D. R., Furlong, M. A., Peacock, A. D., White, D. C., Coleman, D. C., and Whitman, W. B. (2003). Solirubrobacter pauli gen. nov., sp nov., a mesophilic bacterium within the Rubrobacteridae related to common soil clones. Int. J. Syst. Evol. Microbiol. 53, 485–490.
Song, T., Xu, H., Sun, N., Jiang, L., Tian, P., Yong, Y., et al. (2017). Metabolomic Analysis of Alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Front. Plant Sci. 8:1208.
Sun, D., Liu, C., Zhu, J., and Liu, W. (2017). Connecting metabolic pathways: sigma factors in Streptomyces spp. Front. Microb. 8:2546.
Tiwari, S., Singh, C., Boudh, S., Rai, P. K., Gupta, V. K., and Singh, J. S. (2019). Land use change: a key ecological disturbance declines soil microbial biomass in dry tropical uplands. J. Environ. Manag. 242, 1–10.
Walker, C. B., de la Torre, J. R., Klotz, M. G., Urakawa, H., Pinel, N., Arp, D. J., et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A 107, 8818–8823.
Wang, J., Wang, P., Qin, Q., and Wang, H. (2017). The effects of land subsidence and rehabilitation on soil hydraulic properties in a mining area in the Loess Plateau of China. Catena 159, 51–59.
Wang, S., and Brose, U. (2018). Biodiversity and ecosystem functioning in food webs: the vertical diversity hypothesis. Ecol. Lett. 21, 9–20.
Wang, W. L., Li, Z., Lyu, J. F., Zhang, H., Yue, G. X., and Ni, W. D. (2019). An overview of the development history and technical progress of China’s coal-fired power industry. Front. Energy 13, 417–426.
Warren, S. D., St Clair, L. L., Stark, L. R., Lewis, L. A., Pombubpa, N., Kurbessoian, T., et al. (2019). Reproduction and dispersal of biological soil crust organisms. Front. Ecol. Evol. 7:344.
Wolinska, A., Rekosz-Burlaga, H., Goryluk-Salmonowicz, A., Blaszczyk, M., and Stepniewska, Z. (2015). Bacterial abundance and dehydrogenase activity in selected agricultural soils from lublin region. Poly. J. Environ. Stud. 24, 2677–2682.
Wu, S., Xia, X., Zhou, Z., Wang, D., and Wang, G. (2019). Nocardioides gansuensis sp. nov., isolated from geopark soil. Int. J. Syst. Evol. Microbiol. 69, 390–396.
Xiao, L., Bi, Y., Du, S., Wang, Y., and Guo, C. (2019). Effects of re-vegetation type and arbuscular mycorrhizal fungal inoculation on soil enzyme activities and microbial biomass in coal mining subsidence areas of Northern China. Catena 177, 202–209.
Yang, D., Bian, Z., and Lei, S. (2016). Impact on soil physical qualities by the subsidence of coal mining: a case study in Western China. Environ. Earth Sci. 75:652.
Yang, K., Zhao, Y.-G., and Ma, X.-X. (2012). Water stability of biological soil crusts in hilly regions of Loess Plateau. Northwest China. Ying Yong Sheng Tai Xue Bao 23, 173–177.
Zeng, Y. H., Selyanin, V., Lukes, M., Dean, J., Kaftan, D., Feng, F. Y., et al. (2017). Erratum: characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca. Int. J. Syst. Evol. Microbiol. 67, 521–522.
Zhang, C., Liu, G., Xue, S., and Wang, G. (2016). Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 97, 40–49.
Zhang, L.-M., Hu, H.-W., Shen, J.-P., and He, J.-Z. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045.
Zhang, Z. Q., Shao, L. H., Chang, L. Q., Cao, Y. M., Zhang, T., Wang, Y. F., et al. (2016). Effect of rhizobia symbiosis on lignin levels and forage quality in alfalfa (Medicago sativa L.). Agric. Ecosyst. Environ. 233, 55–59.
Zhao, K., Li, J., Huang, T., Yuan, Y., Lin, J., Yue, B., et al. (2019). Coexistence of microbial species in structured communities by forming a hawk-dove game like interactive relationship. Front. Microb. 10:807.
Zhou, J., Deng, Y., Luo, F., He, Z., Tu, Q., and Zhi, X. (2010). Functional Molecular Ecological Networks. Mbio 1:e00169-10.
Zhuang, P., Yang, Q. W., Wang, H. B., and Shu, W. S. (2007). Phytoextraction of heavy metals by eight plant species in the field. Water Air Soil Pollut. 184, 235–242.
Keywords: mining subsidence, soil bacterial, molecular ecological network, ecological restoration, microtopography, metabolic function
Citation: Zhang Q, Ma J, Yang Y, Luo Z, Wang Y and Chen F (2021) Mining Subsidence-Induced Microtopographic Effects Alter the Interaction of Soil Bacteria in the Sandy Pasture, China. Front. Environ. Sci. 9:656708. doi: 10.3389/fenvs.2021.656708
Received: 21 January 2021; Accepted: 25 March 2021;
Published: 16 April 2021.
Edited by:
Simone Priori, University of Tuscia, ItalyReviewed by:
Loredana Canfora, Council for Agricultural and Economics Research (CREA), ItalyFernando San José Martínez, Polytechnic University of Madrid, Spain
Copyright © 2021 Zhang, Ma, Yang, Luo, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Ma, amluZ21hMjAxM0BjdW10LmVkdS5jbg==; Fu Chen, Y2hlbmZ1QGN1bXQuZWR1LmNu
 Qi Zhang
Qi Zhang Jing Ma2*
Jing Ma2* Fu Chen
Fu Chen