- 1College of Desert Control Science and Engineering, Inner Mongolia Agricultural University, Hohhot, Inner Mongolia, China
- 2Key Laboratory of National Forestry and Grassland Administration for Desert Ecosystem Protection and Restoration, Hohhot, Inner Mongolia, China
Ulmus pumila L: is the primary tree species in the Hunshandake Sandy Land and plays a crucial role in controlling wind and sand movement and stabilizing the soil. Currently, Ulmus pumila L. is aging severely, and the inter-root bacterial community structure directly affects its resistance and nutrient uptake. Therefore, it is crucial to investigate the composition of the inter-root soil bacterial community and its driving factors during elm growth. In this study, high-throughput sequencing technology was utilized to determine the bacterial community composition and reveal the relationship between the inter-root bacterial community composition of U. pumila L. of different ages and microenvironmental factors based on basic soil properties and enzyme activities. The results showed that alkaline phosphatase and soil invertase levels increased with the increasing stand age. Compared with young forests (YF), soil total nitrogen, total phosphorus, and organic carbon in mature forests (MF) increased by 32.20%, 33.73%, and 17.65%, respectively. The contents of soil available nitrogen (AN) and available phosphorus showed the same trend in different forest ages. The variation trends of the total number of species and specific species of inter-root soil bacteria in U. pumila L. were consistent, and both showed a downward trend. Actinobacteria were significantly more abundant in young forests than in the others (P < 0.05). It showed a decreasing trend, and mature forests decreased by 29.23% compared to YF, while the abundance of Firmicutes increased. The results of the RDA analysis suggested that available nitrogen explained the most changes in soil bacterial community structure. AK and AN were strongly negatively correlated with Firmicutes but positively correlated with all other bacteria. This study provides useful information for the conservation and utilization of sparse elm forests and also helps provide a vital theoretical basis for understanding the ecological environment construction in the Hunshandake Sandy Land.
1 Introduction
The Hunshandake Sandy land is a typical semi-arid region, located in the farming-pastoral ecotone of northern China, where wind erosion and desertification are more serious (Liang et al., 2019). The area is also the nearest source of sand to the capital, Beijing (State Environmental Protection Administration, 2001), so it is important to control wind-sand flow, improve vegetation cover, and stabilize the environment. Ulmus pumila L. has a well-developed root system and is more resistant to drought; it is the major species with >95% proportion in the Hunshandake Sandy land (Li et al., 2003), and it dominates the sandy vegetation succession (Zhao et al., 2016). Thus, U. pumila L. plays an important role in controlling wind-sand movement and fixing sandy areas in the region. However, in recent years, the area of U. pumila L. has begun to decrease, and seedlings are slow to renew and age more severely (Su et al., 2014). This is mainly related to the role of inter-root soil bacteria (Liang et al., 2019). Inter-root soil bacteria play a crucial role in influencing nutrient cycling, soil structure, and enhancing plant resistance to environmental stresses, all of which are indispensable for the growth and survival of U. pumila L. seedlings. Consequently, comprehending the dynamics of the inter-root soil bacterial community and its correlation with the age of Ulmus pumila L. is of paramount importance for devising effective conservation strategies (Sun et al., 2020). Currently, the researchers primarily focus on the above-ground distribution pattern, community structure, seedling regeneration rate, and underground root distribution of sparse U. pumila L. (Tang et al., 2014), and little research has been conducted on inter-root soil bacteria. Therefore, it is extremely important to study the composition and structural characteristics of the inter-root soil bacterial community and its relationship with soil environmental factors during the growth of U. pumila L.
Soil microbes are an important constituent of the soil ecosystem, which are greatly meaningful in maintaining ecological service functions and promoting sustainable soil utilization (Bardgett and Putten, 2014; Heijden and Wagg, 2013). The inter-root microorganisms have an important influence on the growth and development of plants, whereas plants can change the soil environment by secreting bioactive molecules into the rhizosphere to regulate the local growth conditions (Chen et al., 2019). Soil bacteria account for the majority of the three major microbial groups, which function crucially in the conversion of matter and energy in the plant root eco-environment (Pang et al., 2000). Although the relative abundance distributions of major bacterial groups in plant rhizosphere are identical, the relative abundance of individual phyla varies to different degrees under different environments (Hamonts et al., 2018). Soil bacteria, as the most widely distributed group, prioritize decomposing the low-molecular-weight organic compounds (Tedersoo et al., 2008), and participate actively in the accumulation of soil organic matter and the cycling of nutrient elements (Wu and Lin, 2003; Sui et al., 2016). The rhizosphere bacteria of desert plants, for example, can help plants resist various environmental stresses, playing an important role in plant adaptation to different desertification environments (Hao et al., 2012). According to other reports, bacteria occupy the living space of plant roots by competing with other soil microbes for nutrients (Qin et al., 2017). The above studies further suggest the potentially vital associations between soil microbes and plants.
In this study, we analyzed the community composition and diversity of inter-root soil bacteria of elm trees of different ages by 16S rRNA gene sequencing. Combined with basic soil properties and enzyme activities, the relationship between the bacterial community composition and microenvironmental factors in the inter-root soil of U. pumila L. of different ages was revealed. This study aimed to investigate the response of U. pumila L. inter-root soil microbial community composition to age changes. This study can provide useful information for the conservation and utilization of sparse elm forests and also help to provide an important theoretical basis for understanding the ecological environment construction in the Hunshandake Sandy Land.
2 Materials and methods
2.1 Study area
As shown in Figure 1, this study area is located in Sanggen Dalai in the southern part of Xinlin Gol League, Inner Mongolia Autonomous Region (42°42′N, 115°55′E). The prevailing climate is of temperate semi-arid continental climate where winter is long, with an annual average temperature of 1°C–4°C, an average frost-free period of 105 days, an annual sunshine time of 3,200 h, an annual average wind speed of 4.2 m/s, and an annual precipitation of 320 mm. Affected by the Mongolian high-pressure air masses, the climate is dry and cold. summer is mild and humid due to the influence of the southeast monsoon, while there are often gales during spring. 90% of the region’s dunes are fixed and semi-fixed, with only a small percentage of mobile dunes. However, due to drought and overgrazing, an increasing number of semi-fixed dunes have been converted to mobile dunes. Zonal vegetation is the vegetation of the steppe desert. Vegetation on fixed and semi-fixed dunes is more productive and is therefore used as sandy pastures. Ulmus pumila L. is found on the windward and leeward slopes and lowlands of dunes. Ulmus pumila L. of different growth stages in this study were selected from the depression sampling sites.
2.2 Experimental design and sample collection
Soil samples were collected in August 2021. Ulmus pumila L. of the same altitude and different ages were selected as the study area, each with an area of 500 × 500 m. The age of the trees is classified according to the following categories: young forests aged ≤30 years (YF), middle-aged forests aged 31–50 years (MaF), and mature forests aged ≥51 years (MF). The distances between various sites were >50 m for each age class. Within each sampling site, three elms were randomly selected using the “S”-shaped sampling method, and a total of 12 inter-root soil samples were collected. Inter-root soil was collected at 1 m from the trunk of each tree in four directions, east, south, west, and north, at a depth of 5–30 cm. Initially, the first-order lateral roots were found with a shovel, and then the fine roots were traced along the direction of lateral root growth. To better ensure that the samples collected were rhizosphere soils of U. pumila L., about 150 g of soil samples were collected near the fine roots. After removing impurities such as stones, plant and animal debris, and roots by sieving, the soil samples from the four directions were evenly mixed in sealed bags and set aside. Record the sample number, sampling time, location, latitude, and longitude. There were three replicates per sample plot. Each soil sample was divided into three parts: one was stored at −80°C for the determination of soil bacterial community composition and diversity, another was stored at 4°C for the determination of soil enzyme activities, and the third was shade-dried under natural conditions for the determination of basic soil properties.
2.3 Determination of basic soil properties
Soil organic carbon (SOC, g·kg−1) was measured via the combustion method using an automated carbon analyzer (Vario MAX, Elementar, Germany). Total soil N (TN, g·kg−1) was determined using an automated C/N analyzer. The total soil P (TP, g·kg−1) was determined via a colorimetric method using an alkaline oxidation digestion procedure (Dick and Tabatabai, 1977). The continuous alkali hydrolyzed reduction diffusing method obtained the soil available nitrogen (AN) (Cornfield, 1960). The available potassium (AK) content in soil was obtained by NH4OAc extraction and flame photometry. The soil available phosphorus content (AP) was measured by the Olsen method (Olsen and Sommers, 1982).
The activity of alkaline phosphatase (ALP) was determined using benzene disodium phosphate as a substrate, and the amount of released phenol in 1 g of soil was quantified colorimetrically at 510 nm. Soil urease (SUE) activity was measured by sodium phenol–sodium hypochlorite colorimetry (Wang et al., 2019). Methods for the determination of soil invertase (SI) involve the colorimetric determination of reducing sugars that react with 3,5-dinitrosalicylic acid when soil is incubated with a buffered sucrose solution and toluene at 37°C for 24 h (Frankeberger and Johanson, 1983).
2.4 DNA extraction, PCR amplification, and bioinformatics analysis
An SDS (sodium dodecyl sulfate)- based method was used to extract total DNA from the Inter-root soil samples (Zhou, 1996). Use the OMEGA Soil DNA Kit (D5625-01) (Omega Bio-Tek, Norcross, GA, United States) reagent kit. The crude DNA extract was washed with 70% cold ethanol and placed in a fume hood to allow ethanol volatilization, and 100 µL of sterile deionized water was finally added to dissolve DNA in water. PCR products were then mixed in equimolar ratios, and the resultant pooled PCR products were purified using a GeneJETTM Gel Extraction Kit (Thermo Scientific, Waltham, MA, United States). After purification, the PCR product pools were sequenced on the Illumina HiSeq PE250 platform.
The V4 hypervariable regions of bacterial 16S rRNA genes were amplified using the forward primer 515F (50-GTGCCAGCMGCCGCGGTAA-30) and reverse primer 806R (50-GTGCCAGCMGCCGCGGTAA-30) to construct bacterial community libraries for HiSeq Illumina sequencing (Evans et al., 2014). Samples were demultiplexed using unique barcode sequences for samples, followed by cleaving of barcode and PCR primer sequences. Paired-end reads from PE250 two-terminal sequencing on the HiSeq platform were then joined using the FLASH software (V1.2.7, http://ccb.jhu.edu/software/FLASH/).
After obtaining the raw sequence data, quality control was performed using Trimmomatic (V0.36) with the parameters LEADING:20, TRAILING:20, SLIDINGWINDOW:4:20, and MINLEN:50 to remove low-quality reads and trim adapters. The filtered reads were then merged using FLASH (V1.2.7) with the parameters -M 100 and -m 200. The representative sequences of each ASV were annotated using the RDP Classifier (V2.14) against the Silva database (V138) with a confidence threshold of 0.8. Alpha diversity indices, including Shannon, Simpson, Chao1, and Pielou indices, were calculated using Mothur (V1.44.0). Beta diversity analysis was performed using QIIME (V1.9.1) to calculate the weighted UniFrac distance matrix, and Principal Coordinate Analysis (PCoA) was conducted to visualize the differences in bacterial community structures.
2.5 Statistical analysis
One-way ANOVA was used to statistically analyze the differences in soil properties and enzyme activities among the different groups. Fisher’s least significant difference (LSD) was used for significance testing, with a significance level of α = 0.05. We used SPSS 22.0 (IBM Corp., Armonk, NY, United States) statistical analysis software for data analysis and GraphPad Prism 9.0 for graphic plotting. The alpha diversity index of the bacterial community at different sequencing depths was calculated using Mothur software. Redundancy analysis (RDA) was performed using CANOCO software 5.0 to determine the relationship between inter-root soil bacterial species composition and soil properties of elm trees.
3 Results
3.1 Changes in the basic properties of inter-root soils of Ulmus pumila L. of different forest ages
The basic properties of inter-root soils of U. pumila L. of different stand ages showed distinct variation patterns. Compared with YF, TN, TP, and SOC levels in MF increased by 32.20%, 33.73%, and 17.65%, respectively (Figures 2a–c), but there was no significant difference (P > 0.05). The contents of all three were reduced in MaF compared with YF. As shown in Figures 2d, e, the contents of AN and AP displayed the same trend in different forest ages, and their contents in MF were significantly decreased compared with YF by 30.60% and 60.08%, respectively (P < 0.05). With the increase in forest age, the content of AK showed a significant downward trend (P < 0.05). MaF and MF decreased significantly by 14.99% and 70.56% compared with YF, respectively (Figure 2f). Among the soil enzyme activities, ALP and SI followed the same trend, both of which exhibited an increasing trend with the increasing forest age. As shown in Figure 2g, the activity of ALP was 2.06 and 1.52 times higher in MF than in YF and MaF, respectively. The activity of SUE showed a trend of decreasing and then increasing with the forest age (Figure 2h). The activities of SI were 102.83 U·L-1, 147.36 U·L-1, and 162.98 U·L-1 in YF, MaF, and MF, separately (Figure 2i). With the increasing forest age, MF significantly increased by 58.49% compared with YF (P < 0.05).
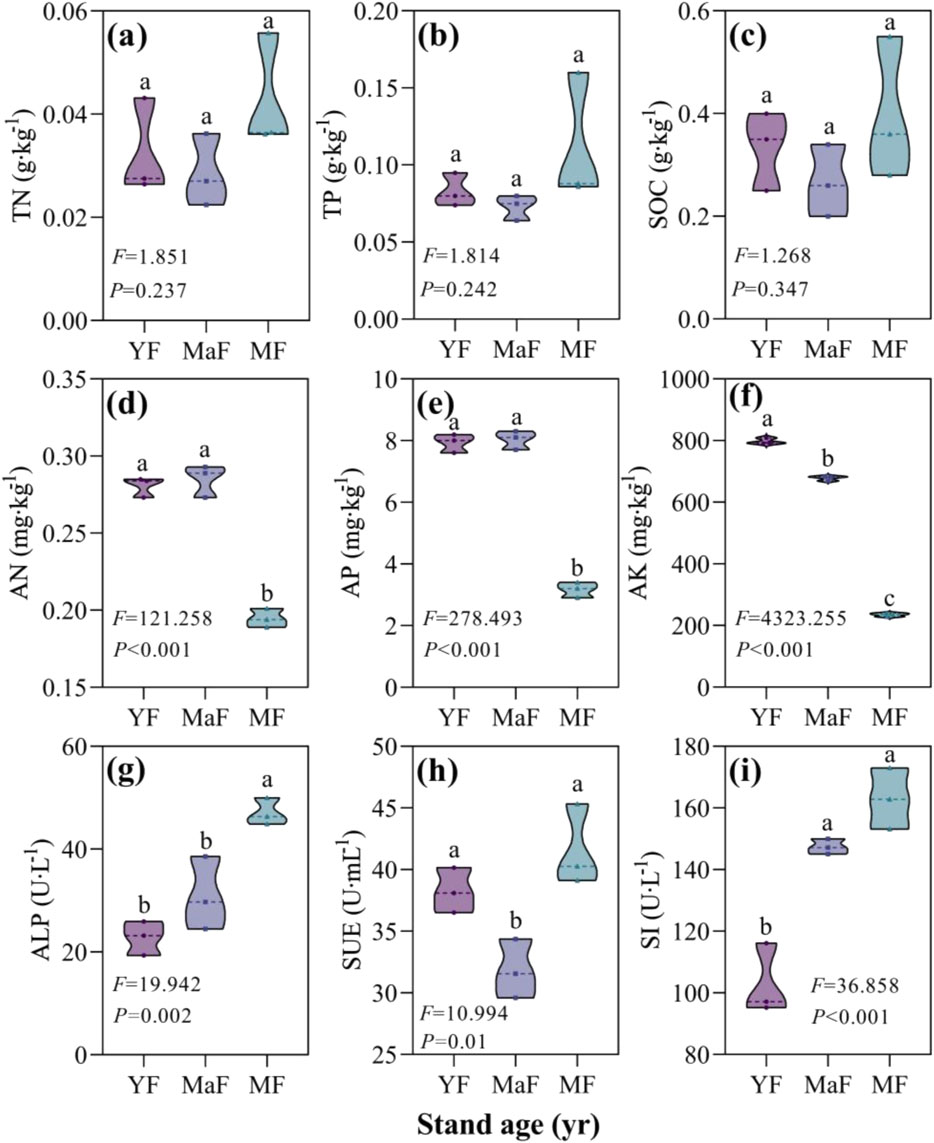
Figure 2. Basic properties of Ulmus pumila L. inter-root soils. Notes: YF: Young Forests; MaF: Middle-aged forests; MF: Mature Forests. Different letters indicate significant differences between different setting years (p < 0.05).
3.2 Characterization of inter-root soil bacterial community composition and structure in Ulmus pumila L. of different forest ages
With the increasing age of Ulmus pumila L., the number of soil bacteria ASVs decreased, which were 12,370, 11,265, and 8,857 in YF, MaF, and MF, respectively, and the number of ASVs common to the three soils was 1,949 (Figure 3). MaF and MF decreased by 8.93% and 28.40% compared with YF, respectively. In the three groups, the order of magnitude of the number of specific ASVs was YF (8,194) > MaF (7,106) > MF (5,279). The number of common ASVs in YF to MaF and MF were 3,353 and 2,772, respectively, while those of MaF and MF were 2,755. It can be observed that with the increase in tree age, the variation trends of the total number of species and specific species of inter-root soil bacteria in the U. pumila L. were consistent, and both showed a downward trend.
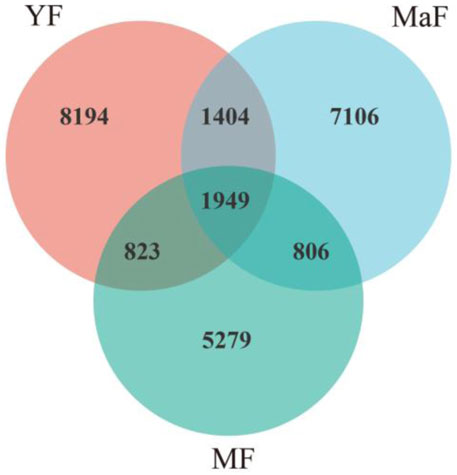
Figure 3. Venn diagram of the number of inter-root soil bacterial ASVs in Ulmus pumila L. of different forest ages.
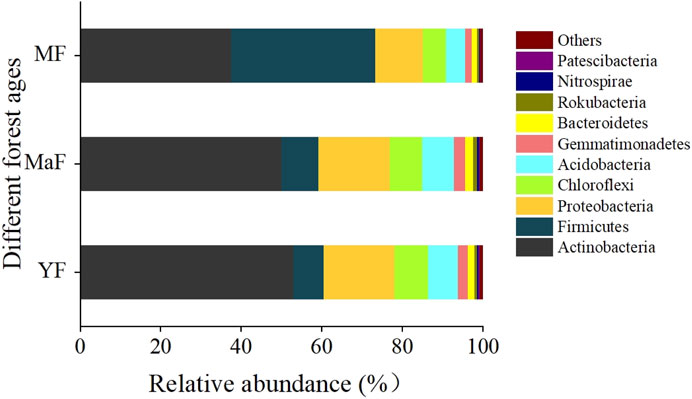
As shown in Figure 4, the bacterial community composition was similar across all sites at the phylum level. Still, there were differences in relative abundance. Bacteria of the same phylum in the inter-root soil of elm trees of different ages were, in order of mean value, Actinobacteria (46.71%), Firmicutes (17.49%), Proteobacteria (15.71%), Chloroflexi (7.39%), Acidobacteria (6.69%), Gemmatimonadetes (2.26%), Bacteroidetes (1.69%), Rokubacteria (0.63%), Nitrospirae (0.38%) and Patescibacteria (0.22%), accounting for over 99% of the total ASV abundance of all bacteria. Actinobacteria were significantly more abundant in YF than others (P < 0.05). It exhibited a decreasing trend with the increasing forest age, and MF decreased by 29.23% relative to YF, but the abundance of Firmicutes increased. Proteobacteria, Chloroflexi, and Acidobacteria demonstrated the same trend, and compared with YF, MF decreased by 32.49%, 31.32%, and 35.42%, respectively. The relative abundances of Gemmatimonadetes in YF, MaF, and MF were 2.41%, 2.80%, and 1.58%, respectively, whereas the abundances of Bacteroidetes and Rokubacteria could be ranked as MaF > YF > MF.
Based on the Weighted Unifrac distance, the inter-root soil bacterial communities of Ulmus pumila L. of different ages were analyzed by PCoA, and the combination of principal coordinates with the largest contribution rate was used for graphing. Based on the analysis results (Figure 5) PCoA1 was the principal coordinate component that explained as much as possible the variation of data, with an explanation rate of 30%; PCoA2 was the second highest, with an explanation rate of 13.7%, and the two principal components explained 43.7% of the bacterial community. Compared with MF, YF and MaF were closer in distance, and the composition of soil bacterial communities was more similar. The inter-root soil bacterial community composition became more variable with the increasing age of U. pumila L.
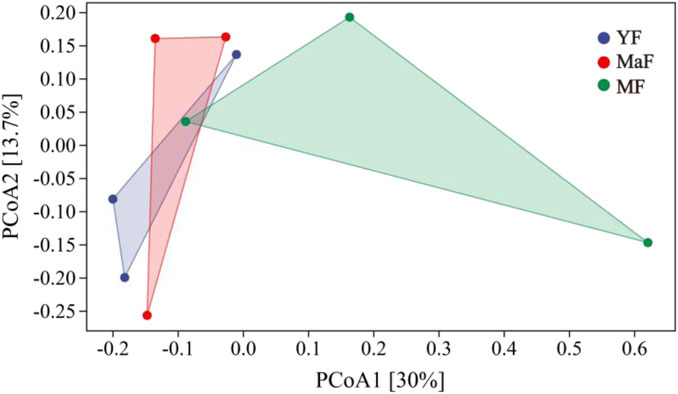
Figure 5. PCoA analysis of inter-root soil bacterial communities in Ulmus pumila L. of different forest ages.
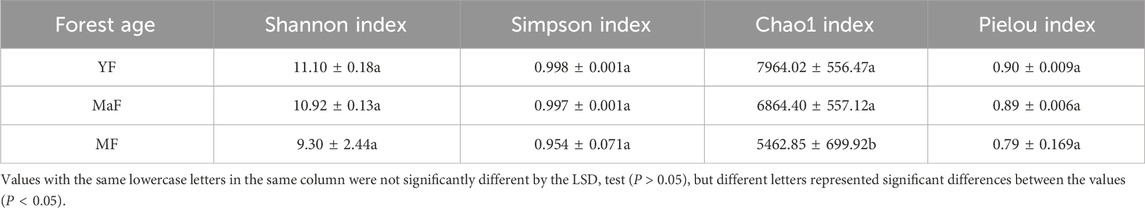
We compared the diversity calculated by the Shannon estimator, which ranged from 9.30 to 11.10 per sample (Table 1). The index was ranked in the order of magnitude as YF > MaF > MF. Simpson index did not show any significant difference in the rhizosphere soil of different ages (P > 0.05). The Chao1 index of YF, MaF, and MF shows a downward trend with increasing forest age, but the difference between YF and MaF is not significant (P > 0.05). Pielou index ranged from 0.79 to 0.90 in the rhizosphere soil of U. pumila L. of different ages. Both the rhizosphere soil bacterial diversity and richness indices of U. pumila L. showed a decreasing trend with the increasing forest age.
3.3 Correlation analysis between inter-root soil bacterial community composition and soil factors of Ulmus pumila L. of different forest ages
By using Pearson’s correlation analysis, the relationship between inter-root soil bacterial composition, basic soil properties, and enzyme activities of Ulmus pumila L. of different ages was explored (Figure 6). The results indicated that AP, AK, and AN were significantly positively correlated with Chloroflexi (P < 0.05). AP, AK, and AN were positively associated with Actinobacteria, Acidobacteria, Gemmatimonadetes, and Nitrospirae, but this relationship was not significant. AP and AK were also not significantly correlated with Proteobacteria (P > 0.05). AK and AN were significantly negatively correlated with SI (P < 0.05), but the negative correlation with SUE was not significant (P > 0.05). Among the bacterial community compositions, Actin. was significantly positively correlated with Proteobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes, and Nitrospirae, but significantly negatively correlated with Firmicutes (P < 0.05). The relative abundance of Proteobacteria showed a significant positive correlation with the other four species, including Chloroflexi, Acidobacteria, Gemmatimonadetes, and Bacteroidetes (P < 0.05). Rokubacteria revealed a significant positive correlation with Nitrospirae only (P < 0.05). Bacteroidetes were positively correlated with Acidobacteria, but negatively correlated with Firmicutes, and the relationship was not significant (P > 0.05).
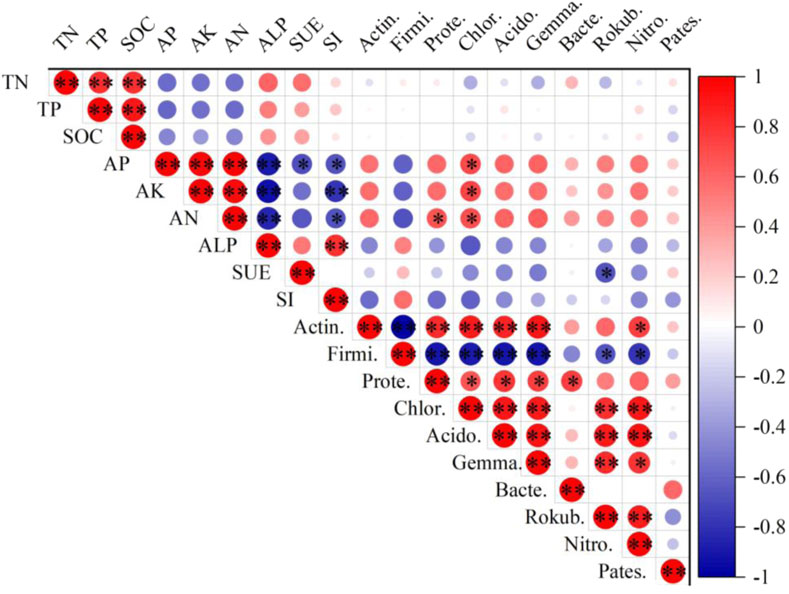
Figure 6. Correlations between soil basic properties and bacterial community composition at the phylum level. Notes: Actin, Actinobacteria; Firmi, Firmicutes; Prote, Proteobacteria; Chlor, Chloroflexi; Acido, Acidobacteria; Gemma, Gemmatimonadetes; Bacte, Bacteroidetes; Rokub, Rokubacteria; Nitro, Nitrospirae; Pates, Patescibacteria. * represents a significant correlation at the P < 0.05 level, and ** represents a highly significant correlation at the P < 0.01 level.
3.4 Redundancy analysis between inter-root soil bacterial community composition and soil factors for Ulmus pumila L. of different forest ages
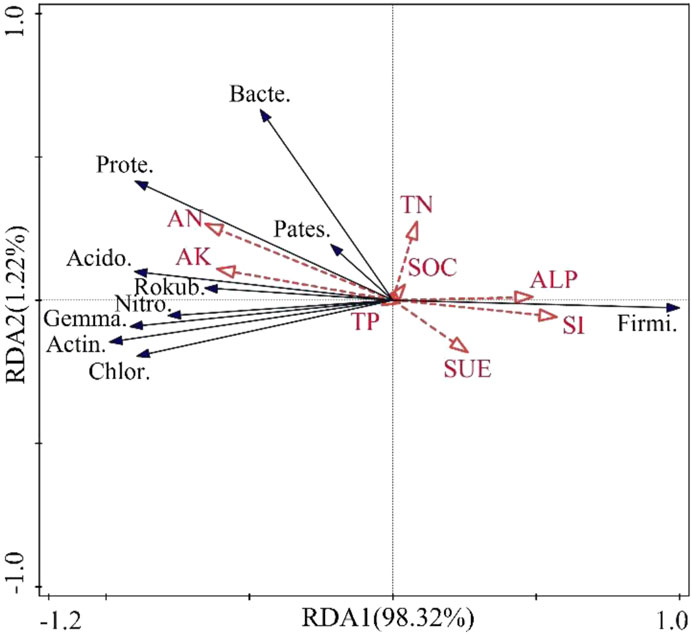
The relationship between soil basic properties and the bacterial community composition of Ulmus pumila L. of different ages was analyzed by redundancy analysis (RDA). The RDA results indicated that bacterial community compositions showed obvious correlations with soil environment factors (Figure 7). The total explanation of soil basic properties on the bacterial community was 99.54%. The explanatory rates for RDA1 and RDA2 were 98.32% and 1.22%, respectively. Among all the driving factors for the U. pumila L. growth processes, ALP, SI, SUE, SOC, and TN were positively correlated with the RDA1 axis. By contrast, AN, AK, and TP were negatively correlated with the RDA1 axis. AN, AK, TN, ALP, and SOC were positively correlated with the RDA2 axis, but SUE, SI, and TP were negatively related to the RDA2 axis. Among all the influencing factors, AN explained the most variations in soil bacterial community structure and had significant effects (P < 0.05). AK and AN were strongly negatively correlated for Firmicutes but positively correlated with all the other bacteria, e.g., Actinobacteria, Proteobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes, Bacteroidetes, and Rokubacteria. ALP and SI were positively correlated with Firmicutes but negatively correlated with Proteobacteria, Chloroflexi, Acidobacteria, and Gemmatimonadetes.
4 Discussion
4.1 Effects of Ulmus pumila L. growth processes on inter-root soil properties and enzyme activities
The growth processes of Ulmus pumila L. significantly impact soil properties and microbial community structures, which in turn impact soil enzymatic activity (Liang et al., 2019). According to our study, the contents of AN and AP showed a consistent trend across different forest ages, among which, MF had significantly lower levels than YF. This may be because young forests require more nutrients for their growth in the early stages, while mature forests have already utilized sufficient nutrients, leaving fewer nutrients in the soil. In contrast, the contents of TN, TP, and SOC did not show any significant differences among different forest ages. This suggests that the plant maintains a balance between the utilization and accumulation of carbon, nitrogen, and phosphorus during long-term growth.
Soil enzymes play a crucial role in promoting substance decomposition and energy transformation in the soil. They are also one of the most important indicators of the level of comprehensive soil fertility and the ecological environment quality (Han et al., 2022). Our results demonstrated that soil enzyme activities, specifically ALP and SI, were significantly higher in MF than in YF. This indicates that a significant amount of catalytic mineralization of organophosphorus compounds is still required in the maturation stage of U. pumila L., aiming to provide sufficient phosphorus for soil microbes and plants. Urease, which is associated with the transformation of nitrogen-containing organic matter in the soil, showed a decreasing and then increasing trend with forest age, with the lowest activity being detected in the inter-root soil of middle-aged elm trees. This reveals that there is a dynamic demand for nitrogen in the growth of U. pumila L.
4.2 Effects of Ulmus pumila L. growth processes on the composition of inter-root soil bacterial communities
Although the overall bacterial community structure remained stable, the relative abundance of specific bacterial taxa varied with the age of U. pumila L. (Yang et al., 2019). The abundance of Actinobacteria was significantly higher in YF soils and showed a decreasing trend with the forest age, which decreased by 29.23% in MF compared with YF. This change may be related to the decomposition of organic matter and nutrient cycling in the soil, as actinobacteria exert a vital role in these processes (Liu and Wei, 2021). The relative abundance of Actinobacteria accounted for the largest percentage (46.71%), followed by Firmicutes (17.49%) and Proteobacteria (15.71%) in the inter-root soils of U. pumila L. at different ages. This is mainly because Actinobacteria exhibit a unique increase in survival rate upon environmental stress (Leggett et al., 2012). Proteobacteria were also shown to be the dominant bacterial group in the inter-root soils of U. pumila L. var. sabulosa, Populus deltoids, and Pinus tabulaeformis in previous studies (Liang et al., 2019; Gottel et al., 2011; Yu et al., 2013). They are the most common bacterial community in soils globally and grow faster, where they respond quickly to unstable carbon sources (Spain et al., 2009).
We also observed an increase in the abundances of Firmicutes and a decrease in the abundance of Proteobacteria, Chloroflexi, and Acidobacteria as the elm trees aged. These changes may be related to the availability of nutrients including nitrogen and phosphorus in the soil, as these bacterial taxa play a key role in nutrient cycling. Proteobacteria are usually associated with the process of nitrogen transformation in the soil, while Acidobacteria are related to the decomposition of soil organic matter (Wu et al., 2024).
4.3 Relationship between inter-root soil properties and bacterial communities during the Ulmus pumila L. growth processes
The relationship between soil bacterial community composition and soil factors was investigated using Pearson’s correlation analysis and RDA. The results demonstrated that AP, AK, and AN were significantly and positively correlated (P < 0.05) with Chloroflexi, consistent with previous studies showing the effect of soil nutrients on bacterial communities (Huang et al., 2023). However, this relationship was not significant for Actinobacteria, Acidobacteria, Gemmatimonadetes, and Nitrospirae, possibly attributed to a lag in the response of these bacteria to soil nutrients or interference from other environmental factors (Wang et al., 2023). In addition, AK and AN were significantly (P < 0.05) negatively correlated with soil enzyme activities (SI), indicating that soil enzyme activities may be inhibited under high nutrient conditions, therefore affecting the structure and function of bacterial communities (Zheng et al., 2018). The significant positive correlation of Actinobacteria with other bacterial groups such as Proteobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes, and Nitrospirae suggests the synergistic relationships, while the significant negative correlation with Firmicutes indicates the competitive relationships (Zhang, 2024). The significant positive correlation between Proteobacteria and several other bacteria demonstrates that these bacteria may jointly participate in ecological functions such as organic matter decomposition and nutrient cycling in the soil ecosystem (Wei et al., 2018). RDA further revealed that AN had the highest explanatory rate and a significant effect on the variation of soil bacterial community structure, which may be because that nitrogen is a key element for plant growth, and its content and morphology directly affect the plant growth status and the soil microbial activities (Hermans et al., 2020). AK and AN were strongly negatively correlated with Firmicutes, and positively with other bacteria, including Actinobacteria, Proteobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes, Bacteroidetes, and Rokubacteria. This selective effect may be related to the physiological properties and ecological niche of the bacteria (Wang et al., 2023).
This study provides new insights in understanding the interactions between the inter-root soil bacterial community of U. pumila L. and soil factors. It helps to reveal the ecological functions and regulatory mechanisms of soil microorganisms in desert ecosystems. However, there are some limitations of the study, such as the limitation of sampling time, and future studies may increase sampling in different seasons and different geographical areas to further validate and deepen the conclusions of this study. Moreover, this study provides a stronger scientific basis for managing and conserving soil ecosystems.
5 Conclusion
This study characterized the composition and diversity of soil bacterial communities in the inter-root soils of elm trees of different ages. The results revealed that ALP and SI increased with the increasing stand age. Compared with YF, TN, TP, and SOC contents in MF increased by 32.20%, 33.73%, and 17.65%, respectively, but there was no significant difference (P > 0.05). The contents of AN and AP showed the same trend in different forest ages. The variation trends of the total number of species and specific species of inter-root soil bacteria in U. pumila L. were consistent, and both showed a downward trend. Actinobacteria were significantly more abundant in YF than in the others (P < 0.05). Its abundance exhibited a decreasing trend, and that in MF decreased by 29.23% compared with YF, but the abundance of Firmicutes increased. AP, AK, and AN were significantly positively correlated with Chloroflexi. AK and AN were significantly negatively correlated with SI (P < 0.05), but the negative correlation with SUE was not significant (P > 0.05). The results of the RDA analysis indicated that AN explained the most changes in soil bacterial community structure and had substantial effects (P < 0.05). AK and AN were strongly negatively correlated with Firmicutes but positively correlated with all the other bacteria. This study provides important data to support the understanding of the response of inter-root bacterial communities of elm trees to age changes in the Hunsandak hinterland.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
BH: Data curation, Methodology, Writing – original draft. YM: Data curation, Funding acquisition, Investigation, Software, Writing – original draft. GL: Validation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the Inner Mongolia Autonomous Region “Listed and Commanded” Project (2024JBGS0023) and Inner Mongolia Autonomous Region Natural Science Foundation (2020ZD05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bardgett, R. D., and Putten, W. H. V. D. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi:10.1038/nature13855
Chen, S., Waghmode, T. R., Sun, R., Kuramae, E. E., Hu, C., and Liu, B. (2019). Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7, 136–148. doi:10.1186/s40168-019-0750-2
Cornfield, A. H. (1960). Ammonia released on treating soils with N sodium hydroxide as a possible means of predicting the nitrogen-supplying power of soils. Nature 187, 260–261. doi:10.1038/187260a0
Dick, W. A., and Tabatabai, M. A. (1977). An alkaline oxidation method for determination of total phosphorus in soils. Soil Sci. Soc. Am. J. 41, 511–514. doi:10.2136/sssaj1977.03615995004100030015x
Evans, C. C., LePard, K. J., Kwak, J. W., Stancukas, M. C., Laskowski, S., Dougherty, J., et al. (2014). Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. Plos one 9 (3), e92193. doi:10.1371/journal.pone.0092193
Frankeberger, W. T., and Johanson, J. B. (1983). Method of measuring invertase activity in soils. Plant Soil 74, 301–311. doi:10.1007/bf02181348
Gottel, N. R., Castro, H. F., Kerley, M., Yang, Z., Pelletier, D. A., Podar, M., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. doi:10.1128/aem.05255-11
Hamonts, K., Trivedi, P., Garg, A., Janitz, C., Grinyer, J., Holford, P., et al. (2018). Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 20, 124–140. doi:10.1111/1462-2920.14031
Hao, X. X. P., Johnstone, L., Miller, S. J., Rensing, C., and Wei, G. (2012). Genome sequence and mutational analysis of plant-growth-promoting bacterium Agrobacterium tumefaciens CCNWGS0286 isolated from a zinc-lead mine tailing. Appl. Environ. Microbiol. 78 (15), 5384–5394. doi:10.1128/aem.01200-12
Heijden, MGAVD, Wagg, C., Gordon, A. M., and Klironomos, J. N. (2013). Growth response of crops to soil microbial communities from conventional monocropping and tree-based intercropping systems. Plant Soil 363, 345–356. doi:10.1007/s11104-012-1321-5
Hermans, S. M., Buckley, H. L., Case, B. S., Curran-Cournane, F., Taylor, M., and Lear, G. (2020). Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome 8 (1), 79. doi:10.1186/s40168-020-00858-1
Hu, P. C., Yin, J., Wei, X. D., and Wang, C. (2022). Effects of different water and nitrogen treatments on potato quality and soil urease activity. Jiangsu Agric. Sci. 50 (6), 87–92. doi:10.15889/j.issn.1002-1302.2022.06.014
Huang, J., Liu, X., Liu, J., Zhang, Z., Zhang, W., Qi, Y., et al. (2023). Changes of soil bacterial community, network structure, and carbon, nitrogen and sulfur functional genes under different land use types. Catena 231, 107385. doi:10.1016/j.catena.2023.107385
Leggett, M. J., McDonnell, G., Denyer, S. P., Setlow, P., and Maillard, J. Y. (2012). Bacterial spore structures and their protective role in biocide resistance. J. Appl. Microbiol. 113, 485–498. doi:10.1111/j.1365-2672.2012.05336.x
Li, Y. G., Jiang, G. M., Niu, S. L., Liu, M., Peng, Y., Yu, S., et al. (2003). Gas exchange and water use efficiency of three native tree species in Hunshandak Sandland of China. Photosynthetica 41, 227–232. doi:10.1023/b:phot.0000011955.12025.dc
Liang, T., Yang, G., Ma, Y., Yao, Q., Ma, Y., Ma, H., et al. (2019). Seasonal dynamics of microbial diversity in the rhizosphere of Ulmus pumila L. var. sabulosa in a steppe desert area of Northern China. PeerJ 7, e7526. doi:10.7717/peerj.7526
Liu, X. H., and Wei, T. X. (2021). High-throughput Sequencing analysis of soil bacterial community in the grain for green project areas of the loess plateau. Environ. Sci. 42 (9), 4489–4499. doi:10.13227/j.hjkx.202012233
Olsen, S. R., and Sommers, L. E. (1982). “Phosphorous,” in Methods of soil analysis, Part 2, chemical and microbial properties. Agronomy monograph. Editors A. L. Page, R. H. Miller, and D. R. Keeney (Madison, Wisconsin: Agronomy Society of America), 9, 403–430.
Pang, X., Zhang, F. S., and Wang, J. G. (2000). Effect of different nitrogen levels on SMB-N and microbial activity. Plant Nutr. Fertilizer Sci. 6 (4), 476–480.
Qin, Y., Fu, Y., Kang, W., Li, H., Gao, H., Vitalievitch, K. S., et al. (2017). Isolation and identification of a cold-adapted bacterium and its characterization for biocontrol and plant growth-promoting activity. Ecol. Eng. 105, 362–369. doi:10.1016/j.ecoleng.2017.04.045
Spain, A. M., Krumholz, L. R., and Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi:10.1038/ismej.2009.43
State Environmental Protection Administration (2001). State of the environment report in China. Beijing.
Su, H., Li, Y. G., Liu, W., Xu, H., and Sun, O. J. (2014). Changes in water use with growth in Ulmus pumila in semiarid sandy land of northern China. Trees 28, 41–52. doi:10.1007/s00468-013-0928-3
Sui, X., Zhang, R. T., Yang, L. B., Xu, N., Zhong, H. X., Wang, J. F., et al. (2016). Functional diversity of soil bacterial community in different types of Plaque chinensis wetlands in the Sanjiang Plain. Environ. Sci. Res. 29 (10), 1479–1486. doi:10.13198/j.issn.1001-6929.2016.10.11
Sun, J. P., Liu, Y. H., Zuo, Y. M., Han, M. L., Zhang, H. W., and Lv, J. J. (2020). The bacterial community structure and function of Suaeda salsa rhizosphere soil. Chin. J. Eco-Agriculture 28 (10), 1618–1629. doi:10.13930/j.cnki.cjea.200160
Tang, J., Jiang, D. M., and Wang, Y. C. (2014). Progress in study on seed seedling regeneration of elm in open forest grassland. Chin. J. Ecol. 33, 1114–1120. doi:10.13292/j.1000-4890.2014.0118
Tedersoo, J., Jairus, T., Horton, B. M., Abarenkov, K., Suvi, T., Saar, I., et al. (2008). Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 180 (2), 479–490. doi:10.1111/j.1469-8137.2008.02561.x
Wang, X., Huang, T., Li, Y., Zhao, G., and Zhao, J. (2023). Elevational characteristics of soil bacterial community and their responses to soil translocation at a mountainside in northwest Sichuan, China. Sci. Rep. 13 (1), 17906. doi:10.1038/s41598-023-44811-2
Wang, Y. G., Liu, J. J., Liu, Y. X., and Ma, L. (2019). Study on influencing factors of determination of soil urease activity by phenol-sodium hypochlorite colorimetric method. Soil Bull. 50 (5), 1166–1170. doi:10.19336/j.cnki.trtb.2019.05.22
Wei, H., Peng, C., Yang, B., Song, H., Li, Q., Jiang, L., et al. (2018). Contrasting soil bacterial community, diversity, and function in two forests in China. Front. Microbiol. 9, 1693. doi:10.3389/fmicb.2018.01693
Wu, J. F., and Lin, X. G. (2003). The role of soil microorganisms in promoting plant growth. Soil (1), 19–22.
Wu, W. W., Han, X., Wang, J. P., Sun, N. X., and Li, Y. (2024). Alterations with plantation years of fructus aurantii on rhizosphere microbiome and soil properties. Acta Pedol. Sin. 61 (6), 1729–1740.
Yang, G. R., Ma, Y., Jiang, B. Ma H., J., , Lü C., , et al. (2019). Analysis of the bacterial community and diversity in tea plantation soil via 16S rDNA sequencing. Acta Ecol. Sin. 39 (22), 8452–8461. doi:10.5846/stxb201808141733
Yu, H. X., Wang, C. Y., and Tang, M. (2013). Fungal and bacterial communities in the rhizosphere of Pinus tabulaeformis related to the restoration of plantations and natural secondary forests in the Loess Plateau, Northwest China. Sci. World J. 2013, 606480–13. doi:10.1155/2013/606480
Zhang, L. (2024). Effects of mixed biocrusts on soil nutrients and bacterial community structure: a case study from Hilly Loess Plateau, China. Sci. Rep. 14 (1), 21265. doi:10.1038/s41598-024-71927-w
Zhao, W., Hu, Z. M., Yang, H., Zhang, L. M., Guo, Q., Wu, Z. Y., et al. (2016). Carbon density characteristics of sparse Ulmus pumila forest and Populus simonii plantation in Onqin Daga Sandy Land and their relationships with stand age. Chin. J. Plant Ecol. 40, 318–326. doi:10.17521/cjpe.2015.1080
Zheng, H., Liu, Y., Zhang, J., Chen, Y., Yang, L., Li, H., et al. (2018). Factors influencing soil enzyme activity in China’s forest ecosystems. Plant Ecol. 219, 31–44. doi:10.1007/s11258-017-0775-1
Keywords: inter-root soil, bacterial community composition, Ulmus pumila L., different ages, bacterial diversity
Citation: Hu B, Ma Y and Li G (2025) Inter-root soil bacterial community composition and its driving factors in Ulmus pumila L. of different ages. Front. Environ. Sci. 13:1557315. doi: 10.3389/fenvs.2025.1557315
Received: 08 January 2025; Accepted: 11 June 2025;
Published: 01 July 2025.
Edited by:
Anoop Kumar Srivastava, Central Citrus Research Institute (ICAR), IndiaCopyright © 2025 Hu, Ma and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gangtie Li, MTM4NDg4MTcxODNAMTYzLmNvbQ==
 Bo Hu
Bo Hu Yunxia Ma1,2
Yunxia Ma1,2