- Collaborative Innovation Center for Vessel Pollution Monitoring and Control, College of Environmental Sciences and Engineering, Dalian Maritime University, Dalian, China
Antibiotic resistance genes (ARGs) in sludge propagate via horizontal gene transfer (HGT), necessitating advanced mitigation strategies. This study demonstrates TiO2 nanotube UV photocatalysis effectively degrades ARGs (70.6–82.5% reduction) and suppresses HGT by targeting mobile genetic elements (MGEs; 93.4–97.1% removal). Hierarchical TiO2 nanotubes (anatase phase) generated reactive oxygen species (ROS) inducing oxidative DNA damage and cell lysis, preferentially eliminating intracellular ARGs (33.5–46.6% decline) while converting them to extracellular forms. Mobile genetic elements (MGEs) (tnpA-04/intI1) were selectively fragmented via ROS, outperforming HOCl-based systems. Microbial analysis revealed Proteobacteria (e.g., Kofleria) as key ARG hosts, whose decline correlated with resistance reduction (p<0.05). Radiation-resistant Deinococcus dominated post-treatment communities but lacked ARG associations, indicating non-transmissible residual risks. Spatial-specific degradation mechanisms emerged: UV directly fragmented chromosomal ARGs, while ROS oxidized plasmid-borne MGEs, achieving dual ARG elimination and HGT blockade. The intracellular-to-extracellular ARG shift and host-MGE decoupling confirmed transmission disruption. This work establishes TiO2 photocatalysis as a paradigm for sludge treatment, synchronizing ARG removal and environmental risk mitigation through ROS-microbe-DNA interplay.
1 Introduction
The widespread use of antibiotics has greatly increased the risk of antimicrobial resistance (AMR) and the difficulty of wastewater monitoring (Islam et al., 2024a; Islam et al., 2024b). This is related to antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB), which poses a challenge to global health (Werkneh and Islam, 2023). Urban wastewater is a critical reservoir for resistance genes, as it accumulates wastewater from diverse sources containing ARGs, ARB, and antibiotic residues (La Rosa et al., 2025). During wastewater treatment, a significant proportion of resistance genes in the water phase accumulate in sludge, posing challenges for its safe disposal (Zhao et al., 2025). For instance, tetracycline - class ARGs have a high detection rate, being commonly detected in aquaculture wastewater and residual sludge (Chen et al., 2015). Effective disinfection and removal of ARB and ARGs are essential to mitigate secondary risks associated with ARG dissemination. However, traditional disinfection technologies like chlorine and ozone present certain issues. The chlorine-derived by-products in chlorine disinfection exhibit poor efficacy in eliminating ARGs. Some studies demonstrated that the abundance of ARGs in wastewater even increased after chlorine treatment (Karaolia et al., 2018; Liu et al., 2018). Regarding ozone, it might facilitate the propagation of ARGs in conventional treatment plants due to cell damage repair mechanisms and mutation processes (Zhuang et al., 2015). Moreover, the dense extracellular polymeric substances (EPS) in sludge adsorb ARGs via ion bridging, hydrophobic interactions, and polymer entanglement (Ding et al., 2015; Zhu et al., 2018). ARGs exist in both intracellular (iARG) and extracellular (eARG) forms, facilitating horizontal gene transfer (HGT) through mobile genetic elements (MGEs) such as plasmids, integrons, and transposons (Cai et al., 2022). Although eARGs occur at low concentrations, they can persist in soil or sediment for extended periods, posing long-term environmental risks (Deshpande and Fahrenfeld, 2022). Thus, effective elimination of both iARGs and eARGs is imperative during sludge treatment.
Advanced oxidation processes, especially photocatalytic technology, are effective means to reduce ARB and ARGs (Kalimuthu et al., 2025; Yuan et al., 2020). Chlorination, ozonation, and photocatalysis are commonly used methods. However, chlorination and ozonation often produce toxic by-products such as organic halides and have low degradation efficiency for ARGs.(Mazhar et al., 2020; Yuan et al., 2015). UV radiation induces DNA-protein crosslinking, DNA strand breaks, and pyrimidine dimer formation, significantly reducing resistance genes. The UV/peracetic acid system not only suppressed potential ARG host growth in antibiotic-contaminated wastewater but also curbed ARG dissemination during subsequent biological treatment (Yin et al., 2024). For sludge with dense EPS, UV photocatalysis effectively oxidizes organic matter, yet its ARG reduction potential remains underexplored. UV irradiation of nitrate during sludge fermentation pretreatment enhanced organic matter metabolism and short-chain fatty acid production, increasing decomposition rates by 2.3-fold and short-chain fatty acid yield by 12.7-fold (Lu et al., 2024). Municipal sludge biodrying combined with photocatalysis facilitated EPS degradation and improves dewaterability (Cai et al., 2023). Studies demonstrated that 10 μg/L TiO2 nanoparticles under UV irradiation decompose approximately 37% of organic content in Anzali Wetland sludge within 5 days (Marefat et al., 2023), highlighting TiO2 UV photocatalysis as a promising approach for ARG reduction in residual sludge. However, the complex EPS structure and ARG distribution in sludge pose challenges, and the efficiency and mechanisms of UV photocatalysis for ARG reduction in layered residual sludge remain unexplored.
This study first focused on UV-photocatalytic technology using TiO2 nanotubes to targeted eliminate ARGs in layered residual sludge, aiming to elucidate their degradation efficiency and HGT blocking mechanisms for ARGs in sludge. We successfully fabricated TiO2 nanotube catalysts that generate reactive oxygen species (ROS) under UV irradiation, effectively damaging microbial cells and DNA. The findings demonstrated that this technology effectively inhibits ARG dissemination through three synergistic mechanisms: degradation of MGEs, inactivation of host bacteria, and direct oxidative damage to DNA. These results provide a novel approach for controlling ARG contamination in wastewater treatment plants. Moreover, we identified practical implementation challenges, particularly the potential secondary enrichment of eARGs, which establishes a critical foundation for subsequent research in real-world applications.
2 Materials and methods
2.1 Sampling sites
The sewage sludge used in this experiment was collected from the secondary sedimentation tank of a sewage treatment plant in Dalian (121.6°E,38.9°N), which employs the anaerobic-anoxic-oxic process. After collection, the sludge was subjected to gravity sedimentation to discard the supernatant, and the resulting concentrated sludge was stored sealed at 4°C. The pH of the remaining sludge was 6.89 ± 0.1, with a moisture content of 96.50% ± 0.3%, a volatile solids/total solids (VS/TS) ratio of 47.65% ± 0.1%, a total chemical oxygen demand (TCOD) of 13,286 ± 328 mg/L, a soluble chemical oxygen demand (SCOD) of 564.3 ± 28.1 mg/L, dissolved carbohydrates of 72.4 ± 5.2 mg/L, dissolved proteins of 182.7 ± 9.5 mg/L, and an ammonia nitrogen of 296.3 ± 12.2 mg/L.
2.2 Preparation of TiO2 nanotubes network
The Ti foil was sequentially cleaned in acetone, anhydrous ethanol, and deionized water using ultrasonic treatment to remove surface oxides and residual oil (Supplementary Figure S1). Subsequently, NaOH solution (60 mL, 1–10 mol/L) and the Ti foil (50 × 30 × 3 mm) were added to a polytetrafluoroethylene liner. It is important to position the Ti foil vertically along the edge of the liner to maximize the contact area between the foil and the NaOH solution. The liner, containing the NaOH solution and Ti foil, was then placed into a high-pressure reactor for a reaction lasting 4 h at a temperature of 130°C–170°C.
Once the reactor cooled to room temperature, the Ti foil was carefully removed from the liner and immediately immersed in a 0.1 mol/L hydrochloric acid (HCl) solution for pickling, which lasted 12 h. The foil was then dried in a vacuum oven at 50°C and subsequently annealed in a muffle furnace at 500°C for 2 h. This series of steps resulted in the formation of a TiO2 nanotube network.
2.3 Photocatalytic experiment
The sewage sludge was subjected to photocatalytic experiments (Supplementary Figure S1), which were divided into three groups: original sludge (RS), TiO2 nanotube UV photocatalysis (UV-TiO2), and TiO2 nanotube treatment under dark conditions (Dark-TiO2). Each reaction took place in a 250 mL cylindrical glass reactor (φ = 8 cm) that was cleaned with alcohol and dried before use. The TiO2 nanotubes used in the photocatalytic reaction had an effective surface area of 30 cm2, which were dispersed in deionized water by ultrasonic treatment for 15 min before use. The photocatalytic reactor was set on a magnetic stirrer (450 rpm) inside a lightbox (CEL - LB70, Zhongjiaojinyuan). A 300 W xenon lamp in the lightbox, whose main wavelength was concentrated in the ultraviolet range (200–400 nm), served as the irradiation light source. To maintain a stable reaction temperature, a low - temperature thermostatic bath circulated 15°C cooling water around the reactor. The control and experimental sludge samples, collected from a local sewage treatment plant and filtered through a 0.2 mm sieve, were stirred (450 rpm) during the experiment. Each experiment was repeated three times to ensure accuracy.
2.4 TiO2 characterization
The phase structure of the samples was characterized using an X-ray diffractometer (XRD, Bede, Dur ham, UK), operating with CuKα radiation (λ = 0.15406 nm) at 40 kV and 35 mA. Diffraction patterns were collected in the 10°–80° range with a scan speed of 5°/min. The samples were Ti foil and nanoplates loaded with TiO2 nanotubes, with dimensions of 10 mm × 10 mm. The morphology of the samples was examined using a scanning electron microscope (SEM, Eva Ma 10, Zeiss, USA), which required high vacuum conditions for the measurements. The samples were cut into square pieces with a side length of 10 mm and adhered to the sample holder using conductive adhesive tape, ensuring close contact between the Ti foil and the TiO2 nanotube-loaded nanoplates. The samples were then analyzed at various magnifications.
2.5 Microbial and metagenomics analyses
2.5.1 DNA extraction
DNA in the sludge was extracted using the TIANamp Microbial DNA Kit (IANGEN Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. The quality of the extracted DNA was evaluated using a NanoDrop2000 spectrophotometer (Thermo Scientific, USA). The qualified DNA was then used to assess the abundance of ARGs, MGEs, 16S rRNA, and bacterial communities. A three-tier quality control system was implemented for all molecular experiments: Plasmid standards containing target ARGs (concentration gradient: 102–106 copies/μL) were used to validate qPCR sensitivity; 0.22 μm-filtered and sterilized wastewater samples were processed in parallel during DNA extraction to monitor reagent contamination; Duplicate no-template controls were included per 96-well plate, with abnormal amplifications automatically flagged by StepOnePlus software.
2.5.2 Detection of ARGs, MGEs, and 16S rRNA abundance via HT-qPCR
HT-qPCR was conducted using the SmartChip Real-time PCR system (Wafergen SmartChip, Fremont, CA). The reaction sites for HT-qPCR were provided by a gene chip containing 5184 nanopores, with amplification carried out using a 100 nL reaction volume. The primer sequences used in this study are illustrated in the Supplementary Table S1. All HT-qPCR reactions were performed in triplicate, with a minimum of two replicates required for a positive result. Data analysis was carried out using SmartChip qPCR software. The formulas for calculating the relative abundance (Equation 1) and absolute abundance (Equation 2) of each gene were based on the methods described by (Looft et al., 2012; Chen and Zhang, 2013). The absolute copy number of the 16S rRNA gene was determined using real-time fluorescence quantitative assays.
Where CT values refers to the cycle number at which the fluorescence signal of the amplification product reaches a preset threshold during the qPCR amplification process.
2.5.3 Detection of microbial communities by high-throughput sequencing
High-throughput sequencing of 16S rRNA was used to characterize bacterial communities using the universal primers 515-F(5′-GTGCCAGCMGCCGCGGTAA-3′) and 909-R (5′-CCCCGYCAATTCMTTTRAGT-3′). The original DNA fragment reads were merged using FLASH and then filtered using a QIIME quality filter. PCR chimeras were filtered in UCHIME. Operational Taxonomic Units (OTUs) were clustered at 97% similarity level to analyze species abundance and annotations (Tong et al., 2019).
2.6 Statistical analysis
The differences in gene removal across different treatments and the analysis of gene abundance among various culture systems were assessed using the Tukey test with SPSS 27 software (IBM, USA), where significance was set at p < 0.05. Pearson correlation analysis among ARGs, MGEs, and bacterial communities was conducted using SPSS 27 software (IBM, USA), with significance defined as p < 0.05. Heat maps were generated using TBtools. Bar charts and line charts were created with Origin 2018 (Origin Lab, USA). The network analysis chart, based on Pearson correlation results, was produced and modified using Gephi (v 0.9.2).
3 Results and analysis
3.1 Characterizations of TiO2 nanotubes network and ARG removal by TiO2 nanotubes UV photocatalysis
Figures 1a–c present SEM images of Ti foil and TiO2 nanotube composites. The untreated Ti foil (Figure 1a) shows a heterogeneous surface with steps and microcracks, lacking ordered structures. After nanotube growth (Figure 1b), a hierarchical TiO2 nanotube array forms, exhibiting a layered mesh-like architecture with increased surface area, enhancing reactive sites. Post sludge treatment (Figure 1c), the mesh structure remains stable despite partial degradation. XRD analysis (Figure 1d) revealed characteristic peaks of metallic titanium at 2θ = 38.42°, 40.17°, 53.0°, 62.94°, and 70.66. TiO2-modified samples show additional peaks at 25.3° and 48.04° (2θ), indicating high crystallinity, and anatase phase reflections at 53.88° and 55.06° (2θ), confirming crystalline TiO2 formation. These features were crucial for ARG photocatalytic degradation.
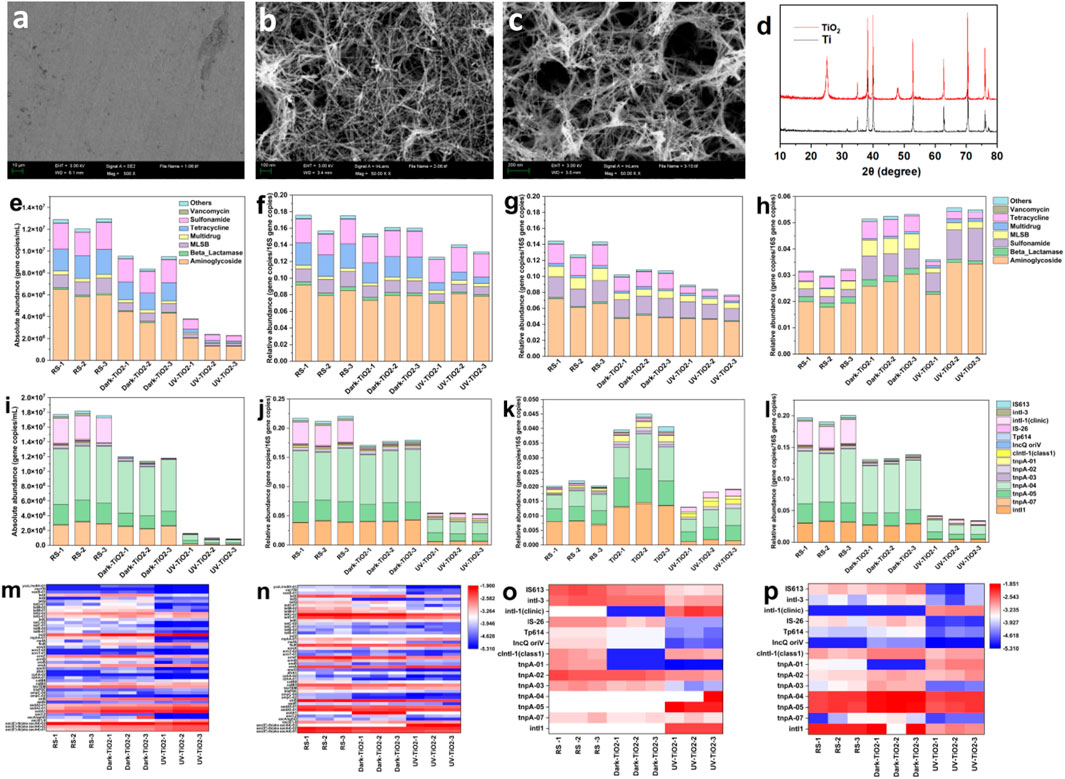
Figure 1. (a) SEM images of Ti foil; (b) SEM images of TiO2 nanotube; (c) XRD pattern of TiO2 nanotube after UV photocatalysis treatment; (d) XRD pattern of Ti foil and TiO2 nanotube; (e) absolute abundances of ARGs in different samples; (f) relative abundances of ARGs in different samples; (g) relative abundances of eARGs in different samples; (h) relative abundances of iARGs in different samples; (i) absolute abundances of MGEs in different samples; (j) relative abundances of MGEs in different samples; (k) relative abundances of eMGEs in different samples; (l) relative abundances of iMGEs in different samples; (m) relative abundance heatmap of eARGs in different samples; (n) relative abundance heatmap of iARGs in different samples; (o) relative abundance heatmap of eARGs in different samples; (p) relative abundance heatmap of iARGs in different samples.
Quantification of over 140 resistance genes revealed 48 ARG subtypes and 14 MGE subtypes. As illustrated in Figures 1e,f, the absolute abundance of ARGs in raw sludge (RS) ranged from 1.20 × 107 to 1.29 × 107 copies/mL, with aminoglycoside resistance genes predominating (5.81 × 106–6.51 × 106 copies/mL), followed by sulfonamide and tetracycline resistance genes. Consistent with findings by Su et al. (2018), Gao et al. (2012), aminoglycoside, tetracycline, and β-lactam resistance genes were identified as the most prevalent in urban wastewater in China, corroborating the presence of multidrug resistance genes in the RS observed in this study.
TiO2 nanotube-based dark treatment reduced ARG abundance to 8.37–9.54 × 106 copies/mL (26.2%–30.4% reduction), potentially through nanotube-microbial membrane interactions (Thill et al., 2006). UV photocatalysis exhibited superior performance, achieving 70.6%–82.5% degradation efficiency (2.27–3.79 × 106 copies/mL), with 68.9%–93.2% elimination of aminoglycoside-, sulfonamide- and tetracycline-associated ARGs. Notably, TiO2 treatment decreased relative ARG abundance from 0.1566 to 0.1759 to 0.1251–0.1401 in raw sludge, demonstrating effective ARG amplification suppression. The enhanced efficacy arises from a dual mechanism: UV-activated TiO2 nanotubes generated ROS via photoexcitation, inducing oxidative stress-mediated lipid peroxidation, protein denaturation, and DNA damage (Bono et al., 2021; Das et al., 2022), while UV directly penetrated cellular structures to fragment resistance gene-carrying DNA (Barnes et al., 2013). This synergistic action disrupted cellular integrity, achieving significant ARG reduction (70%+ degradation efficiency) through combined physicochemical pathways.
3.2 Distribution and abundance of eARGs and iARGs
ARGs in sludge exist in both intracellular (iARGs) and extracellular (eARGs) forms, disseminating through distinct mechanisms. Intracellular 16S rRNA gene copies were less abundant in raw sludge (RS) compared to treated samples (Figures 1g,h). TiO2 nanotube dark treatment showed no significant change in gene copies, whereas photocatalytic treatment reduced extracellular 16S rRNA, indicating diminished DNA retention. The observed increase in eARGs post-photocatalysis likely resulted from cell lysis and subsequent intracellular DNA release.
Following TiO2 nanotube photocatalysis, iARGs decreased while eARGs increased, demonstrating the conversion of iARGs to eARGs. This shift highlighted the efficacy of TiO2 nanotube photocatalysis in redistributing i/eARGs within sludge. The decrease in the ratio of intracellular antibiotic resistance genes (iARGs) to extracellular antibiotic resistance genes (eARGs) indicates an accelerated accumulation of eARGs. This can potentially be attributed to bacterial cell death and the release of iARGs. Additionally, the adsorption of eARGs by sludge, which enables them to withstand nuclease attacks and persist in the environment, is also considered a potential contributing factor (Liu et al., 2018). As shown in Figures 1g,h, aminoglycoside, tetracycline, sulfonamide, β-lactam, and MLSB ARGs were predominant. Post-treatment, aminoglycoside and sulfonamide ARGs exhibited higher extracellular relative abundance, while tetracycline and MLSB ARGs were significantly reduced, underscoring the effectiveness of TiO2 nanotube photocatalysis in removing tetracycline, MLSB, and multidrug ARGs. Wang X. et al. (2024) observed a 7.8-fold iARG increase post-ClO2 disinfection in wastewater, while TiO2 nanotube photocatalysis conversely reduced iARGs but elevated eARGs, revealing the unique sludge ARG redistribution capability of nanotube photocatalysis. While ozonation typically induces iARG rebound, this study achieved stable ARG removal through enhanced oxidative targeting (Fu et al., 2023).
ARG resistance mechanisms are classified into antibiotic inactivation, cellular protection, and efflux pump categories. After TiO2 nanotube photocatalysis, eARGs with antibiotic inactivation mechanisms increased in relative abundance, while iARGs decreased. ARGs such as strB, prevalent in wastewater and soils, persisted extracellularly and were less impacted by photocatalysis (Supplementary Figure S2). Conversely, both eARGs and iARGs associated with cellular protection and efflux pump mechanisms declined, indicating greater susceptibility to TiO2 nanotube treatment (Supplementary Figure S2). This intracellular-to-extracellular ARG transition suggested that TiO2 nanotube photocatalysis preferentially degrades ARGs with cellular protection and efflux pump mechanisms, thereby reshaping the resistance mechanism profile in sludge.
3.3 Inhibition of ARG horizontal transfer via TiO2 nanotube UV photocatalysis
TiO2 nanotube UV photocatalysis demonstrated superior efficacy in suppressing MGEs, critical mediators of HGT and ARG dissemination. Analysis of 14 MGE subtypes revealed that UV photocatalysis reduced MGE abundance from 1.76 to 1.82 × 107 copies/mL in raw sludge to 8.73 × 105–1.61 × 106 copies/mL (93.4%–97.1% removal), outperforming Fe2+/Ca(ClO)2 systems (2.68-log intI1 removal) (Wang X. et al., 2024). This disparity stems from mechanistic differences: UV-activated TiO2 generates reactive oxygen species (ROS) preferentially targeting short DNA fragments (<5 kb MGEs), while HOCl oxidation in Fe2+/Ca(ClO)2 systems inadequately degraded chromosomal ARGs (>10 kb). Nanomaterial-enhanced oxidation studies (Wang et al., 2022; Wang X. et al., 2024) corroborated this mechanism, showing radical-dominated systems (e.g., TiO2/multi-walled carbon nanotubes composites) selectively degrade fragmented genetic elements via ·OH/ClO· generation.
Notably, intracellular MGEs (iMGEs; tnpA-04/intI1/tnpA-05-dominated) showed higher susceptibility to photocatalysis than extracellular counterparts (eMGEs; intI1-dominated). Although eMGEs exhibited selective enrichment (e.g., tnpA-01, clinical intI1), UV photocatalysis achieved pronounced suppression of intI1 (p < 0.01), directly impeding HGT-mediated ARG propagation. These findings established TiO2 photocatalysis as a dual-action strategy, simultaneously targeting iMGEs and eMGEs to block environmental ARG transmission.
3.4 Analysis of potential hosts of ARGs
Network analysis (Figure 2a) extended these findings by revealing taxon-specific co-occurrence patterns between bacterial communities and both eARGs/iARGs. This topological evidence suggests distinct horizontal transmission modalities for eARGs and iARGs at the genus level, with extracellular variants showing higher network centrality (ρ = 0.72, p < 0.01) indicative of cross-species transfer dominance. Significant Pearson correlations (p < 0.05, r > 0.8) between bacterial genera and ARGs/MGEs (e.g., aac(6′)-Ib-03, intI1) confirmed ARG mobility dependence on microbial consortia (Supplementary Figure S4), with Proteobacteria reduction (chlorine-sensitive bacteria, ClSBs) strongly correlating with ARG removal efficiency (Wang Y. et al., 2024). Multi-walled carbon nanotubes composites-enhanced systems selectively enriched chlorine-resistant taxa (e.g., Dechloromonas), validating microbial engineering strategies for ARG control (Wang et al., 2022).
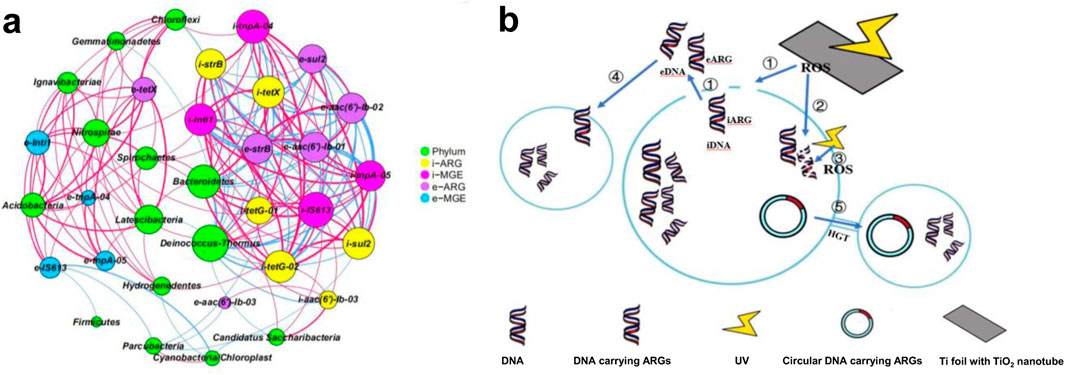
Figure 2. (a) Network analysis based on the co-occurrence of ARGs and their potential host bacteria after photocatalytic treatment of TiO2 nanotubes; (b) Schematic diagram of TiO2 nanotube photocatalytic degradation and blocking ARGs transfer mechanism.
TiO2 photocatalysis induced Deinococcus dominance (Supplementary Figure S3), exhibiting radiation resistance but no iARG association, suggesting its non-participation in ARG transmission. Its eARG correlation implied sludge antibiotic pollutant degradation. Concurrently, iARGs (i-aac(6′)-Ib-03, i-strB) and MGEs co-occurred with Kofleria (p < 0.01), whose post-treatment declined directly linked to iARG reduction. Strong iARG-MGE covariation (r > 0.85) confirmed HGT-driven co-dissemination. UV preferentially degraded eARGs, yet residual eARGs correlated with Deinococcus/Acidovorax, indicating transformation potential. Significant eARG-iARG negative correlations (e.g., e-strB vs. i-strB, p < 0.01) revealed iARG release via cell lysis. Chlorination preferentially inactivated chlorine-sensitive bacteria (ClSBs, e.g., Proteobacteria), while promoting the enrichment of resistant taxa such as Dechloromonas–a phenomenon that sustained iARG reservoirs through microbial community restructuring (Wang et al., 2022). Chlorination and TiO2 nanotube photocatalysis employed in this study shared almost no common host bacteria, as chlorination exhibits limited efficiency in removing iARGs (Liu and Kasuga, 2024).While, the TiO2 nanotube photocatalysis operated through a dual mechanism: (1) Reactive oxygen species (ROS)-mediated oxidative attack synergized with UV-induced DNA damage selectively eliminates iARG-hosting genera (e.g., Kofleria), and (2) radiation-resistant but non-ARG-associated taxa (e.g., Deinococcus) become ecologically dominant post-treatment. This strategic microbial succession simultaneously depleted iARG vectors and established biological barriers against horizontal gene transfer, thereby effectively disrupting the ARG transmission chain.
3.5 ARG degradation and transfer inhibition via TiO2 nanotube UV photocatalysis
The degradation of ARGs and MGEs by TiO2 nanotube UV photocatalysis, as demonstrated in Sections 3.3 and 3.4, revealed significant reductions in both absolute and relative abundances. ARG absolute abundance decreased by 70.6%–82.5%, with a relative abundance decline of 10.58%–28.83%. In contrast, MGEs exhibited higher degradation rates, with absolute and relative abundances reduced by 93.4%–97.1% and 74.27%–75.86%, respectively, indicating superior efficacy of TiO2 nanotube UV photocatalysis in degrading MGEs compared to ARGs. Distinct degradation patterns were observed in intracellular and extracellular environments. iARGs decreased by 33.53%–46.62%, while eARGs increased by 13.57%–87.41%. Similarly, iMGEs declined by 78.79%–82.96%, whereas eMGEs rose by 5.65%–36.18%. These trends highlighted the stronger degradation capability of TiO2 nanotube photocatalysis for MGEs, leading to significant compositional changes in ARGs and MGEs.
Horizontal transfer of ARGs via MGEs is evidenced by statistical correlations: significant ARG-MGE associations (Pearson r > 0.7) indicate transfer potential, while co-localized ARGs on mobile elements (e.g., plasmids) demonstrate MGE-driven co-dissemination (Sommer et al., 2017). Pearson analysis of intra/extracellular co-occurrence patterns further validated their functional linkage in horizontal transfer dynamics. Pearson correlation analysis revealed co-occurrence of ARGs and MGEs both inside and outside cells (Supplementary Figure S4), such as strB, sul2, tetX, tnpA-04, tnpA-05, IS613, and intI1, supporting the transferability of ARGs via MGEs. After TiO2 nanotube UV photocatalytic treatment, both iARGs and iMGEs showed significant decrease, indicating that TiO2 nanotube UV photocatalysis blocked HGT by degrading MGEs, thereby curbing the spread of ARGs. Chlorination might trigger a rebound effect of ARGs and MGEs due to its insufficient oxidative capacity to completely eliminate both iARGs and eARGs (Liu and Kasuga, 2024; Yoon et al., 2017). This limitation allows residual ARGs to proliferate rapidly through stress response mechanisms activated during the disinfection process. Co-occurrence of ARGs (e.g., strB, sul2, tetX) and MGEs (e.g., tnpA-04, tnpA-05, IS613, intI1) both intracellularly and extracellularly further corroborated ARG transferability Figure 2a. Post-treatment, significant reductions in both iARGs and iMGEs demonstrated that TiO2 nanotube UV photocatalysis inhibited HGT by degrading MGEs, thereby curbing ARG dissemination. Additionally, the significant negative correlation between iARGs and eARGs further supports ARG transfer disruption (Supplementary Figure S4). For example, intI-1(clinic), initially confined to RS cells, appeared both intracellularly and extracellularly after TiO2 nanotube UV photocatalysis. This suggests that reactive oxygen species (ROS) generated during photocatalysis damage bacterial cell structures, releasing iARGs and preventing their vertical transfer. The co-occurrence of intI1 with ARGs (sul1, aadA, tetA) suggested the potential dissemination of ARGs after chlorination, which corresponds to the prevalent rebound patterns observed in ARGs and MGEs (Liu and Kasuga, 2024).
3.6 Mechanism of photocatalytic degradation and transfer inhibition by TiO2 nanotubes under UV light
The differential degradation efficiencies of ARGs stem from ROS-mediated oxidative damage heterogeneity, where reactive oxygen species selectively target genomic loci based on gene copy numbers, deoxyribonucleotide composition, and sequence specificity (Li et al., 2022). This variability ultimately derives from TiO2 nanotube UV-photocatalyzed ARG mobilization-transformation processes in sludge, wherein ARG degradation is achieved through strategic suppression of horizontal transfer pathways. The degradation of ARGs and MGEs via TiO2 nanotube UV photocatalysis involved a complex mechanism, as illustrated in Figure 2b. TiO2 nanotube UV photocatalysis generated ROS, which damaged cellular components, including proteins, lipids, and DNA, leading to cell membrane rupture and the release of iARGs into the extracellular environment (Chen et al., 2021). This process reduced iARGs while increasing eARGs. Photocatalytic degradation operated through both direct and indirect pathways. Direct degradation occured when UV light penetrated the cell wall, interacting with nucleic acids to form dimers that inhibit DNA replication and degrade ARGs. Indirectly, UV light activated photosensitive substances within or outside bacteria, generating ROS that disrupt cellular structures, releasing iARGs and degrading eARGs. However, UV light can also promote eARG enrichment. This occurs because eDNA may bind to colloidal particles in sludge, reducing its exposure to UV light. Additionally, free eARGs can be assimilated by other bacteria through transformation, further facilitating their dissemination. The photocatalytic degradation of MGEs played a critical role in inhibiting HGT, as ROS oxidize MGEs, preventing ARG transfer between bacteria via conjugation. This mechanistic profile aligns with yet differs from the compartmentalized oxidation paradigm proposed in the study of (Wang Y. et al., 2024), where intracellular hypochlorous acid (HOCl) and extracellular hydroxyl radicals (·OH) exibit spatial-specific bactericidal actions. This fundamental commonality suggests that oxidant penetrability (cellular/envelope barriers) and DNA-targeting specificity collectively govern degradation efficiency across various advanced oxidation processes (AOPs).
The network analysis revealed correlations between ARGs, MGEs, and microbial communities under TiO2 nanotube UV treatment (Figure 2a). Specific genera, such as Kofleria, Nitrospira, and Ferruginibacter, were identified as potential hosts for ARGs like tetX and sul2. These genera also influenced ARG degradation and transformation in the environment, supporting the hypothesis that TiO2 nanotube treatment inhibits HGT by disrupting MGEs and reducing potential host bacteria. Conversely, the inefficiency of chlorination arises from two primary factors: cell membrane barriers that impede chlorine penetration, and preferential consumption of chlorine by cytoplasmic components rather than DNA molecules (Wang Y. et al., 2024; Liu and Kasuga, 2024).
3.7 Limitations
This study has several limitations that warrant consideration. Primarily, the laboratory-scale TiO2 photocatalytic systems investigated here have not been validated under real-world sludge treatment scenarios, creating a laboratory-to-reality knowledge gap. Three critical operational challenges remain unaddressed (1) complex matrix interference (e.g., competitive adsorption by humic acids): may substantially reduce ROS generation efficiency in heterogeneous sludge environments; (2) UV light penetration depth could be severely constrained in high-solid sludge (>2% total solids), potentially compromising treatment uniformity; and (3) the long-term stability of catalysts in continuous-flow operation requires verification, particularly regarding efficiency decay patterns after ≥10 reuse cycles. These scalability barriers must be systematically overcome before practical implementation.
4 Conclusion
This study explored the photocatalytic degradation of residual wastewater sludge using TiO2 nanotubes under UV light, with a focus on the removal of intracellular and extracellular ARGs and MGEs, as well as their effects on microbial communities.
(1) Following 3 h of UV treatment, the absolute abundance of ARGs decreased by 70.6%–82.5%, while relative abundance declined by 10.58%–28.83%. Aminoglycoside, sulfonamide, and tetracycline ARGs exhibited higher degradation efficiency. iARGs decreased by 33.53%–46.62%, whereas eARGs increased by 13.57%–87.41%. TiO2 nanotubes effectively degraded both ARGs and MGEs, inhibiting HGT.
(2) TiO2 nanotube UV photocatalysis reshaped microbial communities, reducing dominant genera such as Nitrospira and Terrimonas while promoting the growth of others like Deinococcus. Network analysis identified Kofleria and Nitrospira as key potential hosts for ARGs, highlighting the critical role of MGEs in gene transfer.
(3) UV-induced ROS damaged bacterial hosts, releasing iARGs and increasing eARGs. The degradation of MGEs and destruction of host bacteria effectively blocked HGT. Direct oxidation of intracellular and extracellular DNA further contributed to ARG degradation.
In summary, TiO2 nanotube UV photocatalysis represents a promising approach for degrading ARGs and preventing their dissemination in wastewater sludge.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
HY: Conceptualization, Methodology, Software, Writing – original draft, Writing – review and editing. XZ: Conceptualization, Methodology, Software, Writing – original draft, Writing – review and editing. JZ: Investigation, Visualization, Writing – review and editing. TS: Investigation, Visualization, Writing – original draft. YZ: Conceptualization, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 52370127), Young Elite Scientists Sponsorship Program by China Association for Science and Technology (No. 2022QNRC001), Project LJ222410151015 supported by Fundamental Research Funds of higher education Department of Liaoning Province (Program for Liaoning Innovative Research Team in University), Dalian Outstanding Youth Science and Technology Talent Program (2024RY020), and Fundamental Research Funds for the Central Universities (No. 3132023518).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2025.1590101/full#supplementary-material
References
Barnes, R. J., Molina, R., Xu, J., Dobson, P. J., and Thompson, I. P. (2013). Comparison of TiO2 and ZnO nanoparticles for photocatalytic degradation of methylene blue and the correlated inactivation of gram-positive and gram-negative bacteria. J. Nanoparticle Res. 15 (2), 1432. doi:10.1007/s11051-013-1432-9
Bono, N., Ponti, F., Punta, C., and Candiani, G. (2021). Effect of UV irradiation and TiO2-photocatalysis on airborne bacteria and viruses: an overview. Materials 14 (5), 1075. doi:10.3390/ma14051075
Cai, C., Huang, X., and Dai, X. (2022). Differential variations of intracellular and extracellular antibiotic resistance genes between treatment units in centralized sewage sludge treatment plants. Water Res. 222, 118893. doi:10.1016/j.watres.2022.118893
Cai, L., Cao, M.-K., Zheng, G.-D., Wang, X.-Y., Guo, H.-T., and Jiang, T. (2023). Sludge biodrying coupled with photocatalysis improves the degradation of extracellular polymeric substances. J. Environ. Manag. 345, 118590. doi:10.1016/j.jenvman.2023.118590
Chen, B., Hao, L., Guo, X., Wang, N., and Ye, B. (2015). Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ. Sci. Pollut. Res. 22, 13950–13959. doi:10.1007/s11356-015-4636-y
Chen, H., and Zhang, M. (2013). Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from hangzhou, China. Environ. Sci. & Technol. 47, 8157–8163. doi:10.1021/es401091y
Chen, L., Gao, S., Lou, L., and Zhou, Z. (2021). Removal of intracellular and extracellular antibiotic resistance genes from swine wastewater by sequential electrocoagulation and electro-fenton processes. Environ. Eng. Sci. 38 (2), 74–80. doi:10.1089/ees.2020.0203
Das, S., Singh, V., and Paul, S. (2022). Surface conjugation of titanium dioxide nanoparticles on nano-graphene oxide enhances photocatalytic degradation of azo dyes under sunlight. Environ. Sci. Pollut. Res. 29 (26), 40226–40240. doi:10.1007/s11356-022-18796-w
Deshpande, A. S., and Fahrenfeld, N. L. (2022). Abundance, diversity, and host assignment of total, intracellular, and extracellular antibiotic resistance genes in riverbed sediments. Water Res. 217, 118363. doi:10.1016/j.watres.2022.118363
Ding, Z., Bourven, I., Guibaud, G., van Hullebusch, E. D., Panico, A., Pirozzi, F., et al. (2015). Role of extracellular polymeric substances (EPS) production in bioaggregation: application to wastewater treatment. Appl. Microbiol. Biotechnol. 99 (23), 9883–9905. doi:10.1007/s00253-015-6964-8
Fu, Q., Chen, Z., Yu, Z., Wu, Y., Bao, H., Guo, J., et al. (2023). Ozonation enables to suppress horizontal transfer of antibiotic resistance genes in microbial communities during swine manure composting. Chem. Eng. J. 462, 142218. doi:10.1016/j.cej.2023.142218
Gao, P., Mao, D., Luo, Y., Wang, L., Xu, B., and Xu, L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46 (7), 2355–2364. doi:10.1016/j.watres.2012.02.004
Islam, Md. A., Kumar, R., Sharma, P., Zhang, S., Bhattacharya, P., and Tiwari, A. (2024a). Wastewater-based surveillance of mpox (monkeypox): an early surveillance tool for detecting hotspots. Curr. Pollut. Rep. 10 (2), 312–325. doi:10.1007/s40726-024-00299-6
Islam, Md. A., Rakib, S. H., Bhattacharya, P., Jakariya, Md., Haque, M. M., and Tiwari, A. (2024b). Integrated strategy: identifying SARS-CoV-2 strains in communities via wastewater monitoring and clinical diagnosis. Sci. Total Environ. 912, 168617. doi:10.1016/j.scitotenv.2023.168617
Kalimuthu, J., Sigdel, P., Ishii, S., Kolasinski, K. W., Saha, D., and Gadhamshetty, V. (2025). Visible-light-activated removal of antibiotic resistance genes using electrospun plasmonic photocatalyst of carbon nitrides and silver nanoparticles. Environ. Eng. Sci. 42 (4), 141–154. doi:10.1089/ees.2024.0309
Karaolia, P., Michael-Kordatou, I., Hapeshi, E., Drosou, C., Bertakis, Y., Christofilos, D., et al. (2018). Removal of antibiotics, antibiotic-resistant bacteria and their associated genes by graphene-based TiO2 composite photocatalysts under solar radiation in urban wastewaters. Appl. Catal. B Environ. 224, 810–824. doi:10.1016/j.apcatb.2017.11.020
La Rosa, M. C., Maugeri, A., Favara, G., La Mastra, C., Magnano San Lio, R., Barchitta, M., et al. (2025). The impact of wastewater on antimicrobial resistance: a scoping review of transmission pathways and contributing factors. Antibiotics 14 (2), 131. doi:10.3390/antibiotics14020131
Li, H., Song, R., Wang, Y., Zhong, R., Wang, T., Jia, H., et al. (2022). Environmental free radicals efficiently inhibit the conjugative transfer of antibiotic resistance by altering cellular metabolism and plasmid transfer. Water Res. 209, 117946. doi:10.1016/j.watres.2021.117946
Liu, M., and Kasuga, I. (2024). Impact of chlorine disinfection on intracellular and extracellular antimicrobial resistance genes in wastewater treatment and water reclamation. Sci. Total Environ. 949, 175046. doi:10.1016/j.scitotenv.2024.175046
Liu, S.-S., Qu, H.-M., Yang, D., Hu, H., Liu, W.-L., Qiu, Z.-G., et al. (2018). Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 136, 131–136. doi:10.1016/j.watres.2018.02.036
Looft, T., Johnson, T. A., Allen, H. K., Bayles, D. O., Alt, D. P., Stedtfeld, R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. 109 (5), 1691–1696. doi:10.1073/pnas.1120238109
Lu, Y., Liu, Z., Cui, Z., Li, D., Duan, Y., Chen, X., et al. (2024). Feasibility and mechanism exploration of enhancing short chain fatty acid production assisted by nitrate photolysis during sludge fermentation. Chem. Eng. J. 496, 154323. doi:10.1016/j.cej.2024.154323
Marefat, A., Karbassi, A., Aghabarari, B., and Rodríguez-Castellón, E. (2023). Photodegradation of organic-derived sludge in Anzali wetland using titanium dioxide (TiO2). Int. J. Environ. Sci. Technol. 20 (5), 4873–4882. doi:10.1007/s13762-023-04860-6
Mazhar, M. A., Khan, N. A., Ahmed, S., Khan, A. H., Hussain, A., Rahisuddin, , et al. (2020). Chlorination disinfection by-products in municipal drinking water – a review. J. Clean. Prod. 273, 123159. doi:10.1016/j.jclepro.2020.123159
Sommer, M. O. A., Munck, C., Toft-Kehler, R. V., and Andersson, D. I. (2017). Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat. Rev. Microbiol. 15 (11), 689–696. doi:10.1038/nrmicro.2017.75
Su, J.-Q., An, X.-L., Li, B., Chen, Q.-L., Gillings, M. R., Chen, H., et al. (2018). Correction to: metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 6 (1), 127. doi:10.1186/s40168-018-0504-6
Thill, A., Zeyons, O., Spalla, O., Chauvat, F., Rose, J., Auffan, M., et al. (2006). Cytotoxicity of CeO 2 nanoparticles for Escherichia coli. physico-chemical insight of the cytotoxicity mechanism. Environ. Sci. & Technol. 40 (19), 6151–6156. doi:10.1021/es060999b
Tong, J., Tang, A., Wang, H., Liu, X., Huang, Z., Wang, Z., et al. (2019). Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresour. Technol. 272, 489–500. doi:10.1016/j.biortech.2018.10.079
Wang, M., Wang, Y., Ni, X., Hou, X., Ma, D., Li, Q., et al. (2022). How multi-walled carbon nanotubes in wastewater influence the fate of coexisting antibiotic resistant genes in the subsequent disinfection process. Chemosphere 302, 134641. doi:10.1016/j.chemosphere.2022.134641
Wang, X., Lu, Y., Yan, Y., Wang, R., Wang, Y., Li, H., et al. (2024a). Pivotal role of intracellular oxidation by HOCl in simultaneously removing antibiotic resistance genes and enhancing dewaterability during conditioning of sewage sludge using Fe2+/Ca(ClO)2. Water Res. 254, 121414. doi:10.1016/j.watres.2024.121414
Wang, Y., Lu, Y.-W., and Liu, H. (2024b). Nanowire electroporation-induced cell pores on antibiotic-resistant bacteria to promote chlorine permeation for eliminating intracellular antibiotic resistance genes. Chem. Eng. J. 479, 147801. doi:10.1016/j.cej.2023.147801
Werkneh, A. A., and Islam, M. A. (2023). Post-treatment disinfection technologies for sustainable removal of antibiotic residues and antimicrobial resistance bacteria from hospital wastewater. Heliyon 9 (4), e15360. doi:10.1016/j.heliyon.2023.e15360
Yin, W., Liu, T., Chen, J., Zhang, L., Ji, R., Xu, Y., et al. (2024). Using UV/peracetic acid as pretreatment for subsequent bio-treatment of antibiotic-containing wastewater treatment: mitigating microbial inhibition and antibiotic resistance genes proliferation. J. Hazard. Mater. 470, 134166. doi:10.1016/j.jhazmat.2024.134166
Yoon, Y., Chung, H. J., Wen Di, D. Y., Dodd, M. C., Hur, H.-G., and Lee, Y. (2017). Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2. Water Res. 123, 783–793. doi:10.1016/j.watres.2017.06.056
Yuan, Q., Zhang, D., Yu, P., Sun, R., Javed, H., Wu, G., et al. (2020). Selective adsorption and photocatalytic degradation of extracellular antibiotic resistance genes by molecularly-imprinted graphitic carbon nitride. Environ. Sci. & Technol. 54 (7), 4621–4630. doi:10.1021/acs.est.9b06926
Yuan, Q.-B., Guo, M.-T., and Yang, J. (2015). Fate of antibiotic resistant bacteria and genes during wastewater chlorination: implication for antibiotic resistance control. PLOS ONE 10 (3), e0119403. doi:10.1371/journal.pone.0119403
Zhao, B., Zhang, R., Jin, B., Yu, Z., Wen, W., Zhao, T., et al. (2025). Sludge water: a potential pathway for the spread of antibiotic resistance and pathogenic bacteria from hospitals to the environment. Front. Microbiol. 16, 1492128. doi:10.3389/fmicb.2025.1492128
Zhu, Y., Wang, Y., Zhou, S., Jiang, X., Ma, X., and Liu, C. (2018). Robust performance of a membrane bioreactor for removing antibiotic resistance genes exposed to antibiotics: role of membrane foulants. Water Res. 130, 139–150. doi:10.1016/j.watres.2017.11.067
Keywords: antibiotic resistance gene, wastewater sludge, UV photocatalysis, TiO2 nanotube, extracellular antibiotic resistance gene, intracellular antibiotic resistance gene
Citation: Yu H, Zhang X, Zhao J, Sun T and Zhu Y (2025) Mechanism of TiO2 nanotube UV-photocatalytic degradation of antibiotic resistance genes in the wastewater sludge and blocking of the transfer. Front. Environ. Sci. 13:1590101. doi: 10.3389/fenvs.2025.1590101
Received: 08 March 2025; Accepted: 23 April 2025;
Published: 06 May 2025.
Edited by:
Yalçın Tepe, Giresun University, TürkiyeReviewed by:
Oscar Manuel Rodriguez Narvaez, Centro de Innovación Aplicada en Tecnologías Competitivas (CIATEC), MexicoMd. Aminul Islam, Tulane University, United States
Praveen Pratap Singh, United College of Engineering and Research, India
Copyright © 2025 Yu, Zhang, Zhao, Sun and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Yu, eXVoYW5nODkxQDEyNi5jb20=, eXVoYW5nQGRsbXUuZWR1LmNu
 Hang Yu
Hang Yu Xu Zhang
Xu Zhang