- Forest Research Institute, University of Sunshine Coast, Sippy Downs, QLD, Australia
Forest restoration influences several complementary ecosystem features and their trajectories, which ultimately can lead to diverse outcomes. However, integrated assessments of above- and below-ground responses to restoration efforts are lacking. For this reason, we conducted a review assessing advances in monitoring forest restoration, including the role of soil microbes in forest development. We reviewed advances in integrated forest restoration assessments measuring plant diversity, vegetation structure, ecosystem functions, and soil microbial communities based on environmental DNA. We also explored how considering soil microbial dynamics can improve restoration practices. Using the Scopus database, we searched for articles on forest restoration assessment that included the ecosystem features listed above. We extracted the location, biome, methodology and findings of each study. The novelty of the integrated approach was highlighted by the presence of a scattered and biased number of studies. Only 17 articles (out of 30) evaluated all the searched parameters together, with 14 of these studies being based in China. Microbial taxonomic diversity was affected by soil chemistry, plant diversity, and vegetation structure. Bacteria taxa richness fluctuated over time, whereas fungal taxa richness demonstrated hump-shaped and site-specific responses to forest restoration and tree community succession. Over time, bacteria functional groups’ relative abundance shifted from oligotrophic to copiotrophic taxa dominance, while for the fungal groups, the dominance changed from saprophytic to ectomycorrhizal in different forest biomes. This review provides general insights into the advances of restoration assessments considering microbial communities and the utility of analyzing multiple parameters to understand microbial community responses which, in turn, affect restoration outcomes.
1 Introduction
Forest restoration projects are important nature-based solutions promoted by agencies and governments to mitigate climate change effects (Di Sacco et al., 2021), ensure water quality, assist the provision of food and fiber, and for nature conservation. Specifically, forest restoration involves actions aimed to restore the historical or natural conditions of the previous forested state in a cleared and degraded area, and can include activities like tree planting, land-use exclusion, or other environment modifications to set the ecosystem on a desired restoration trajectory (Stanturf, 2005). Monitoring is essential to ensure the achievement of such restoration goals, hence, restoration must not be viewed as a “one-step” accomplishment determined by the total planted area. Specifically, the analysis of multiple aspects of the environment enables the recognition of challenges and trade-offs (e.g., higher biomass but lower biodiversity of trees) that influence the long-term effectiveness of restoration projects (Zhao, 2023).
Regardless of the value of estimating restoration outcomes (Abella and Covington, 2004; Herrick et al., 2006; Pastorok et al., 1997), integrated assessments of above- and below-ground responses are lacking (Bash and Ryan, 2002). In general, restoration evaluations were rare before the rise of contemporary, cost-effective monitoring techniques (e.g., remote sensing techniques) (Knoth et al., 2013; Stanturf et al., 2014), and comprehensive evaluations were seldom undertaken, despite early efforts for a broad inclusion of ecosystem characteristics measures, including biodiversity metrics. For example, Hobbs and Norton (1996) recommended the “Vital Ecosystem Attributes” introduced by Aronson et al. (1993), as parameters to evaluate the state of forest restoration, which included biodiversity, structure, and estimates of successional processes. Other authors suggested ecosystem functions’ central role in restoration practices (Falk, 2006; Herrick et al., 2006), while Stanturf et al. (2014) advised that species composition alone is an insufficient indicator for assessing long-term outcomes. The incorporation of different metrics (e.g., vegetation structure, litter production, species diversity) appeared only about 20 years ago, with applications to evaluate the long-term persistence of a restored ecosystem (Ruiz-Jaén and Aide, 2005b).
In restoration ecology, comprehensive assessments are fundamental to disentangle each compartment’s constituent influences (i.e., biotic and abiotic components, such as living organisms of different groups and chemical features respectively), enhancing the outcomes of restoration efforts (Adamo et al., 2021; He et al., 2023). The entire forest ecosystem and its functions are affected by restoration actions both above- (e.g., vegetation, invertebrates) and below-ground (e.g., soil nutrient cycling, soil microbial community composition). Multiple indicators demonstrate the presence of balanced and combined (or additive/synergistic) effects among structural, functional, and compositional characteristics of the environment, and organisms living in it (Zhao, 2023). Therefore, considering several attributes allows the comparison between places and projects, filling knowledge gaps while providing insights on ecosystem recovery dynamics.
1.1 Parameters considered
In this review, we focused on the description of plant-vegetation-ecosystem structure and soil microbial diversity (Figure. 1). We avoided the description of the reasons for which vegetation diversity and ecosystem functioning are assessed since these metrics are commonly measured in forest restoration. Then, we reported evidence of implementing the next-generation sequencing technique on environmental DNA to evaluate soil microbes, highlighting its value in restoration contexts.
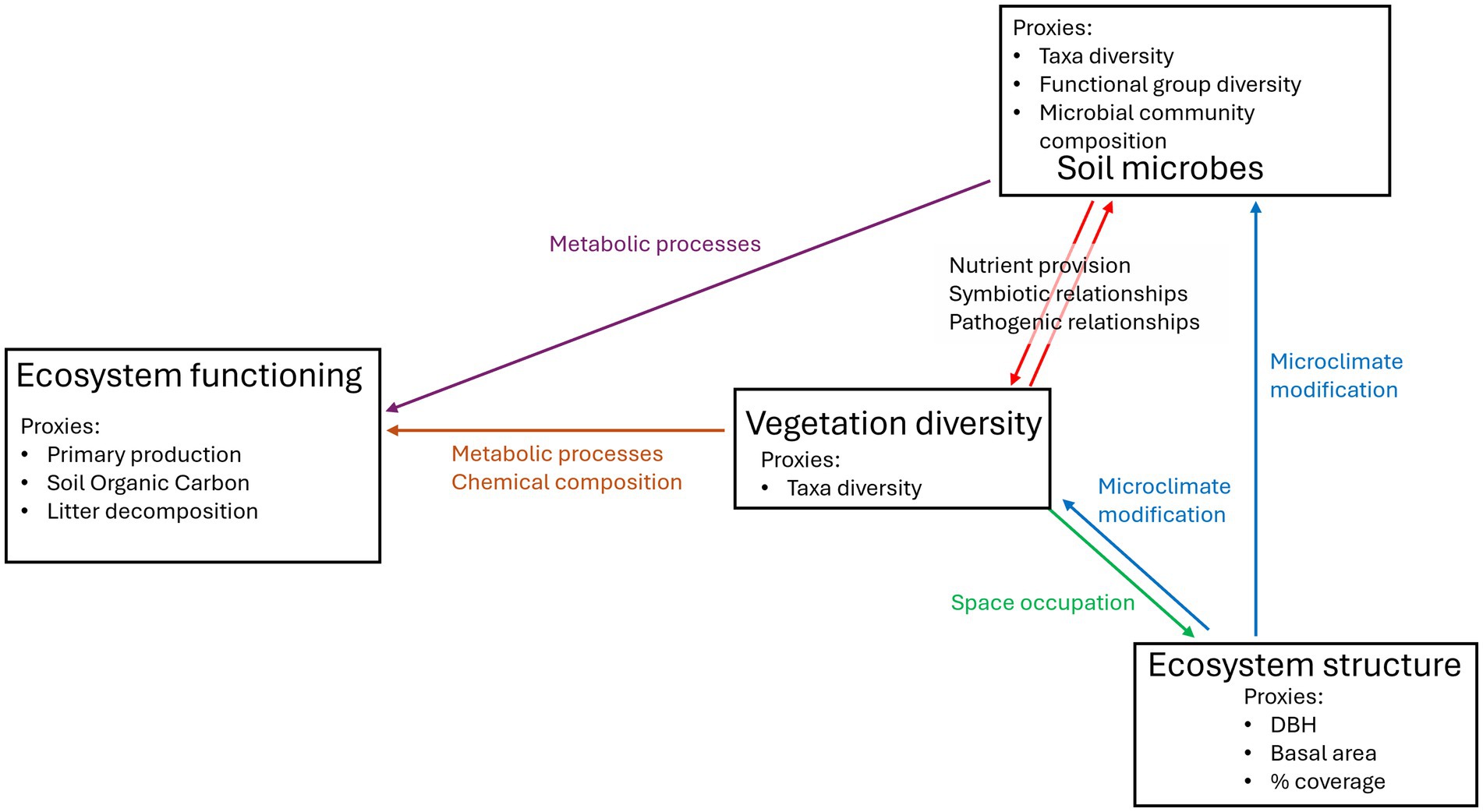
Figure 1. Interactions between vegetation diversity, ecosystem structure, ecosystem functioning, and soil microbes. In each box are reported the metrics commonly used to measure the environmental parameters. The colored arrows represent the links between environmental parameters given the processes reported.
1.1.1 Plant, vegetation, and ecosystem structure
The physical disposition of biotic and abiotic components in the environment (hereafter referred to as “ecosystem structure”) represents the ecosystem’s physical structure. As occurs for soil chemistry and vegetation composition, ecosystem structure is affected by degradation or restoration of the area (Cortina et al., 2006), and influences the establishment of microbial communities, plants, and animals.
Configuration of the biotic and abiotic elements in forest ecosystems can be perceived as a grade of structural complexity. For example, the spatial configuration creates different micro-climates, affecting the colonization of shade-tolerant vegetation and associated soil microbes (Lu et al., 2023). Common measures of forest structure include above-ground biomass (AGB), diameter at breast height (DBH), tree basal area, per cent cover, and tree density (stems/ha).
1.1.2 Soil microbes
Soil microbial community diversity has a central role in ecosystem processes linking biotic and abiotic conditions (Lemke and DeSalle, 2023). Examples of these are nutrient cycling, organic matter decomposition, and soil organic carbon (SOC) turnover (Bardgett and Van Der Putten, 2014; Harris, 2009; O’Rourke et al., 2015). These processes are affected by plant community composition and primary productivity, along with soil structure and microclimate characteristics (Feeney et al., 2006; Van Der Heijden et al., 2008; Wardle et al., 2004). Small variations in environmental resources and conditions also influence microbial composition and activity (e.g., water, temperature) (Fernandez Nuñez et al., 2021; Graham and Knelman, 2023; Lemke and DeSalle, 2023). In turn, while being altered by all these factors, soil microscopic organisms directly influence vegetation growth and ecosystem stability (Bardgett and Van Der Putten, 2014).
Considering soil microbial community characteristics (taxonomic and functional diversity and relative abundances) is fundamental to evaluating the consequences of forest restoration actions (Lemke and DeSalle, 2023). Given the physical attachment of microorganisms to their substrate, they can respond directly to variations in local conditions, in contrast to other living forms that easily move between places (e.g., birds, mammals) (Vasconcellos et al., 2014). Furthermore, microbial communities are composed of species that respond steadily and slowly to environmental variations (Graham and Knelman, 2023; Lemke and DeSalle, 2023), making their study effective for understanding both short- and long-term ecosystem dynamics (Bardgett and Van Der Putten, 2014; Li J. et al., 2020). Knowledge of soil microbes’ metabolism and ecology can improve the management of tree plantations or natural forest ecosystems, informing about the most suitable microbial symbionts to inoculate the plants with for their growth and health optimisation (Fernandez Nuñez et al., 2021).
The tight relationship of microorganisms with the local area, coupled with their high sensitivity to environmental variations (Yan et al., 2018), has contributed to their use as bioindicators in restoration practices (Graham and Knelman, 2023; Harris, 2003; Ma et al., 2022). Despite this evidence, soil microbe evaluation is not routinely included in forest restoration assessments (Pedrinho et al., 2024; Yan et al., 2018).
1.1.3 eDNA technology
Deoxyribose nucleic acid (DNA) fragments collected from any environmental substrate is called “environmental DNA” (eDNA), representing a mixture of genetic material from multiple organisms (Thomsen and Willerslev, 2015). Environmental DNA sampling is a useful technique to assess a high number of species and to infer their role in the ecosystem, especially when combined with other indicators (e.g., plant diversity, and soil carbon). The efficiency of this technique stems from the ability of DNA sequencing to rapidly and cost-effectively generate large amounts of information, enabling restoration efforts to leverage microbial data for improved management, such as identifying pathogenic taxa (Fernandez Nuñez et al., 2021). Before this, the isolation and growth of cultured microorganisms required significantly more effort and expense, preventing the implementation of their study in the forest restoration context.
Environmental DNA analysis has already proved to be an efficient method to assess ecosystem modification during ecological restoration. In tropical areas, this methodology provided information on microbial functional groups for future inoculations and management actions (Fernandez Nuñez et al., 2021), and recently eDNA sequencing was recommended as a method to monitor forest restoration outcomes at a landscape level, in combination with other ecosystem attributes (Mansourian and Stephenson, 2023). Nevertheless, eDNA evaluations in restoration programs are still rare (Yan et al., 2018) and poorly integrated with other ecosystem characteristics measures.
1.2 Objectives
This literature review aimed to assess the state of knowledge on the integrated evaluations of forest restoration actions, focusing on the relationships between the ecosystem parameters and soil microbes. We searched articles including valuations of vegetation diversity, ecosystem structure and functioning, and soil microbes, to understand if these parameters have been collectively assessed in forest restoration and how microbes change according to restoration development. Specifically, we investigated the presence of common patterns of variations of soil microbial diversity across the forest restoration chronosequence, considering the location and biome in which the research was conducted, using the biomes definition of Dinerstein et al. (2017), and disentangling general yet useful dynamics in restoration trajectory evaluation.
Our examination sought to answer the following questions: (i) What are the advances from using multiple parameters in restoration assessment (vegetation diversity, ecosystem structure and functioning, and soil microbes with eDNA analysis)? (ii) Are there general temporal and spatial patterns in microbial community characteristics (taxonomic diversity, functional diversity, and relative abundances) compared to other ecological parameters? Are these patterns consistent across different biomes or conditions? (iii) What are the gaps in integrated forest restoration assessments?
Finally, guided by the results we suggested future actions to improve the integrated evaluation of forest restoration, acknowledging the obstacles and opportunities for its implementation.
1.3 Challenges synthesizing different metrics and scales of assessment
Assessments of restoration outcomes currently lack an integral and standardized methodology, limiting their comparability and robustness. Several indexes and protocols have been implemented to measure vegetation diversity, its structure, soil fertility, and microbial community characteristics (Ruiz-Jaén and Aide, 2005b; Bartholomew et al., 2024). Each metric emphasizes different aspects of the parameter assessed, making direct comparisons impossible. For instance, both Shannon and Chao1 indices measure diversity, but the former calculates richness and diversity considering the observed relative abundance of taxa, while the latter evaluates estimated species richness (Chao, 1984; Shannon, 1949). Furthermore, multiple techniques are employed to perform microbial DNA extraction and sequencing (Hakimzadeh et al., 2024), limiting the comparison of results across studies and locations. In addition, the lack of standardized and generally adopted measures of dead wood presence imposes a significant limit on understanding the presence and dynamics of identified microbial taxa. Some taxa rely entirely on dead wood material for their survival (Baldrian et al., 2016), making this a fundamental information to reliably compare microbial communities across locations and stages of restoration.
Analyzing distinct restoration metrics implies trade-offs. The diverse number and type of metrics among studies impose simplifications, which can lower the quality of the resulting analysis, but ensure a shallow yet sufficient contrast, assuming the same robustness and reliability of the results for each study. General comparison between studies can still be accomplished because, despite the divergent metrics adopted, the overall aim of assessing restoration outcomes is a shared goal. For example, advances in the assessment of restoration dynamics from microbial analysis using the Shannon diversity index, and canopy coverage for vegetational structure, may seem comparable only to studies utilizing the same metrics. Nevertheless, comparing research exploiting a different diversity index (e.g., Pielou index), and average DBH as a structure metric is possible since the overall aim is to estimate the advances in integrated restoration evaluation (as in this review) and not the effect of the techniques and indexes adopted.
2 Methods
Articles were identified using keywords (present in the Abstract, Title, and Keywords sections) for eDNA, restoration, functions, structure, soil, and monitoring using the scientific database Scopus. In detail, we used the combinations of words, which after different trials, seemed to represent the best options (since the result of this combination included known articles related to the topic): “restor*” OR “reforest*” OR “reveget*” AND “high-throughput-sequencing” OR “metabarcoding” OR “edna” OR “environmental-dna” AND “soil” AND “function*” AND “structur*.” The words “reforestation” and “revegetation” were included to broaden the research to similar practices. The reason behind the inclusion of “high-throughput-sequencing,” and “metabarcoding” besides “edna” and “environmental-dna” is linked to the experienced occasions in which articles related to the topic were adopting these other words as terminology for the genetic analysis of microbes from soil samples. Finally, the words “soil,” “function*,” and “structur*” were included in the attempt to find studies explicitly investigating the relationships between the ecological parameters analyzed. The “eDNA” word was selected to focus the research on the cut-edge technique of eDNA, enhancing the likelihood of finding recent studies (i.e., given the usage of “eDNA”). The consideration of the selected enabled the identification of microbial taxonomic and functional variation while being aware of the other ecosystem characteristics. While focusing the research on the forest context, we did not include words for each biome or climatic region, to avoid the exclusion of articles not explicitly identifying the biogeographic context, or recognizing it through different words. This was done to improve the probability of including studies from different geographical regions, and therefore allowing for the comparison of microbial variations in different biomes while limiting the selection of spatially biased studies (e.g., deriving from one biome). After careful content analysis, the articles were filtered to converge to the review’s objectives. A total of 158 articles resulted from the research (on the 31st of January 2024), 132 of which were discarded because they related to topics outside of the scope of this review (post-mining restoration, chemical contamination, marine environment, non-forest biomes, and laboratory experiments among others). Including the word “OR monitor*” to assess the consideration of multiple parameters in restoration monitoring actions at the end of the searched words (thus selecting articles having either the word “structur*” and “monitor*”) yielded an additional 15 documents, 13 of which were discarded because the themes were not associated to this review objective, as per the articles rejected from the initial list of 158 articles. We also incorporated 2 relevant studies that were not found in the search but that the authors were aware of from experience. Further information on the usage of keywords is provided in Supplementary material S1.
We divided the studies according to biome type according to the definition of Dinerstein et al. (2017) to evaluate whether different patterns of soil microbial variation were observed over large-scale climatic gradients. Based on the biome categorization, and the study on bacteria or fungi separately or altogether, we evaluated the relationships underlying the variation of microbes’ diversity and functional groups in ecological restoration, synthesizing the reasons for their difference considering the authors’ explanations. For each article, we also assessed the species of trees present, soil type, land history, and the utilization of fertilizers or ploughing during site preparation which are known key factors influencing restoration outcomes (Pedrinho et al., 2024).
3 Results
3.1 Advances in integrated assessment of forest restoration
From a total of 175 articles, we retained 30 for this review (Supplementary Table S1). Approximately half (57%, 17 studies) of the retained articles evaluated multiple parameters: vegetation diversity, soil microbe characteristics, ecosystem functions such as soil organic carbon (SOC), and forest structure. Forty percent (12 studies) considered vegetation diversity, soil microbe characteristics, and ecosystem functions, and the remaining 7% (2 studies) examined the vegetation diversity, soil microbe characteristics. Seven of the articles (23%) used a list of dominant species as a metric for vegetational diversity, while the others included a more comprehensive list of species. Of the 30 studies, 30% (9) considered both bacteria and fungi, 30% (9) analyzed only bacteria, and 30% (9) solely studied fungi. For the last three studies, one considered the kingdom of Archaea in combination with bacteria and fungi, while in the remaining 2 articles, the analysis regarded only diazotroph bacteria and Archaea, respectively.
Ecosystem functions were more commonly assessed than ecosystem structure. Ninety-seven per cent of the articles (29 studies) evaluated at least one ecosystem function such as SOC, total carbon, and soil organic matter, and 76% (23) calculated SOC in combination with other soil parameters (e.g., pH, total nitrogen), with two studies also determining the decomposition rate of organic matter in the soil. Fifty-six per cent (17) of the retained articles included the per cent cover, crown, and canopy density. Tree DBH data was collected in 36% (11) of the articles studying forest structure. Field data collection for all the articles occurred between 2010 and 2021, with 23% (7) of the studies conducting fieldwork in 2019, and 20% (6) in 2015. The average age of the restored, reforested, or revegetated plot was 29.5 years, with three studies surveying variations within 5 years of project implementation.
Metrics used to assess taxa richness included Operational Taxonomic Units (OTUs, 28 studies), and Amplicon Sequence Variants (ASVs, 2 studies) (Fernandez Nuñez et al., 2021; Gómez-Aparicio et al., 2022). Calculation of ASVs is considered to be more accurate than OTUs for separating and clustering microbial groups based on phylogeny, but the methodology is congruent at the beta-diversity level, allowing the comparison of studies in this review (Chiarello et al., 2022). Indexes used to evaluate microbial richness and diversity were Shannon, Simpson, Pielou, ACE, Faith (PD), and Chao1. See Supplementary Table S2 and discussion by Kim et al. (2017) for a comparison between these diversity indexes.
Functional groups of bacteria and fungi were assessed in 22 of the 30 articles analyzed. The assessment of functional groups in these studies was based on the association of the evaluated taxa to the presence of genes encoding for specific functions. Soil enzyme activities were explored in one article (Huang et al., 2023). Sequencing of functional genes was performed in one study (Mirza et al., 2020), and a MicroResp system was used in two articles to characterize the metabolic profiling of microbes (Bonner et al., 2019; Smenderovac et al., 2017). Metabolic processes were deducted in two studies for bacteria soil communities (Deng et al., 2019; Li et al., 2023). The remaining articles assessed microbial functional groups using the following databases: FAPROTAX, FUNGuild, Fungal-Traits, PICRUSt2, and Index Fungorum.
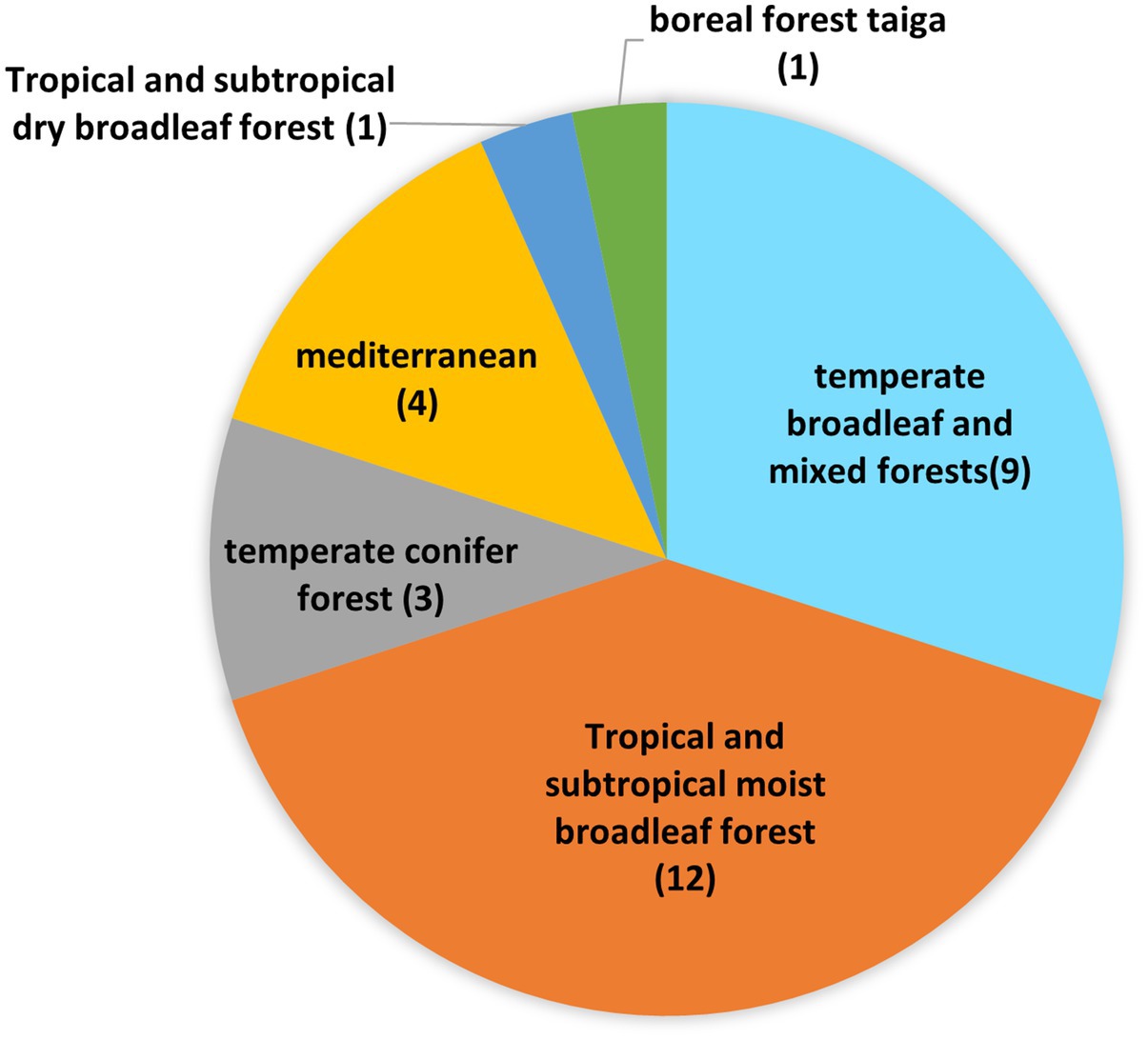
3.2 Geographic location and biomes assessed
Of the 30 articles evaluated, 17 (56%) collectively considered the vegetation diversity, ecosystem structure and functioning, and soil microbes studied through eDNA. Fourteen of these studies (82%) were conducted in China. Analyzed biomes, as defined by Dinerstein et al. (2017), included boreal forests/taiga, Mediterranean forests, woodlands and scrub, temperate broadleaf and mixed forests, temperate conifer forests, tropical and subtropical dry broadleaf forests, and tropical and subtropical moist broadleaf forests (Figure 2). Respectively, 100% (9) and 75% (9) of the studies conducted in the temperate broadleaf and mixed forests, and in tropical and subtropical moist broadleaf forests occurred in China.
3.3 Variation of bacteria taxonomic richness and functional groups considering ecological parameters in different biomes
Bacteria community composition became increasingly similar to remnant forest over vegetation succession in the Mediterranean and tropical biome. Bacterial taxonomic diversity and soil chemical characteristics remained the same in a Mediterranean biome over a successional chronosequence of 10 years, while bacterial taxa relative abundances over time resembled those in remnant reference sites (Gellie et al., 2017). The peak of diversity at this site occurred in the 7th year, whereas the community composition (taxa relative abundance) was increasingly similar to that of remnant sites over time (Yan et al., 2020). In this case, the soil physicochemistry was correlated with the age of the restoration. For example, nitrate and phosphorus were negatively correlated with time since restoration and ammonium and organic carbon significantly increased over the 10 years (Yan et al., 2020). In tropical and subtropical moist broadleaf forests, soil diazotrophs bacteria community composition increasingly converged with a primary forest over 40 years. Bacterial community composition also varied along with soil fertility (e.g., higher C content occurred in the pasture soil compared to primary forest sites) and was closely associated with plant species variation (Mirza et al., 2020).
Bacteria diversity showed discordant development patterns over the forest succession in temperate areas. In a temperate broadleaf and mixed forest biome in the Loess Plateau (China), bacteria showed higher diversity accompanied by higher soil C values in natural and artificial restoration plots (78 and 42 years old respectively) compared to plots of crops (Yang et al., 2018). In an additional study conducted in China, bacteria demonstrated an increase in diversity with vegetation restoration, and the soil chemical properties became comparable to the ones present in the reference site (Li et al., 2023). In 40-year-old mixed coniferous—broad-leaved plantations, coniferous plantations, and broad-leaved only plantations in North China, bacteria diversity and vegetation community structure were higher as the plantations got older compared to the early successional stage where shrubs dominated the landscape. Soil chemical properties also showed a differentiation between the early phases of succession and the forested stages (Qiu et al., 2022). Bacteria diversity decreased over a restoration period of 30 years of Pinus sylvestris and P. tabuliformis plantations in China, and soil carbon content increased during forest restoration (Deng et al., 2019). Similar results were found by Zhang et al. (2019), with decreasing bacterial diversity over a restoration period of 150 years with Querqus wutaishanica trees (in the Shandong Province, China) (Deng et al., 2019).
3.4 Fungi taxonomic richness and functional groups variation considering ecological parameters in different biomes
Our search revealed predominantly hump-back responses of fungal taxonomic diversity variations across biomes and time scales (Table 1). Research conducted in Mediterranean and tropical biome indicated that fungal diversity shows a hump-back pattern, and that saprophytic taxa are replaced by ectomycorrhizal ones over the forest development. In the Mediterranean biome, the fungal diversity reached a peak of diversity at the 8th year, and the early dominance of saprotrophic taxa shifted toward ectomycorrhizal taxa over a chronosequence (Yan et al., 2018). In the same biome, soil chemical properties showed a directional change toward the reference state, with increasing SOC and decreasing nitrate and phosphorus. In a tropical rain forest of Costa Rica, naturally regenerating plots of mixed species of different ages (from 5 to 30 years old and mature stage forest), displayed a fungal richness peak in the mid-successional stages (15–30 years), due to the presence of both early- and late-successional species (Adamo et al., 2021). In this study, researchers found that soil Mn concentration increased over the chronosequence, while pH, organic matter, N, and P showed no changes. In the above-ground sphere, tree richness significantly increased over the successional chronosequence (Adamo et al., 2021). Taxonomic diversity of plant pathogens, wood decomposers, and root-associated fungi did not differ over the sequence, while saprophytic fungi reached a peak in the mid-successional stage. Parasitic fungal taxonomic diversity increased over succession. In another moist tropical biome study of P. yunnanensis plantations up to 80 years, fungal diversity also assumed a “hump-back shape,” reaching the highest value at 60 years (Li S. et al., 2020). The hump-back-shaped function was also observed in woody species richness. Soil chemical variables (SOC, N, P) increased over the reforestation. Ectomycorrhizal fungi abundance initially decreased and then increased with the reforestation progression.
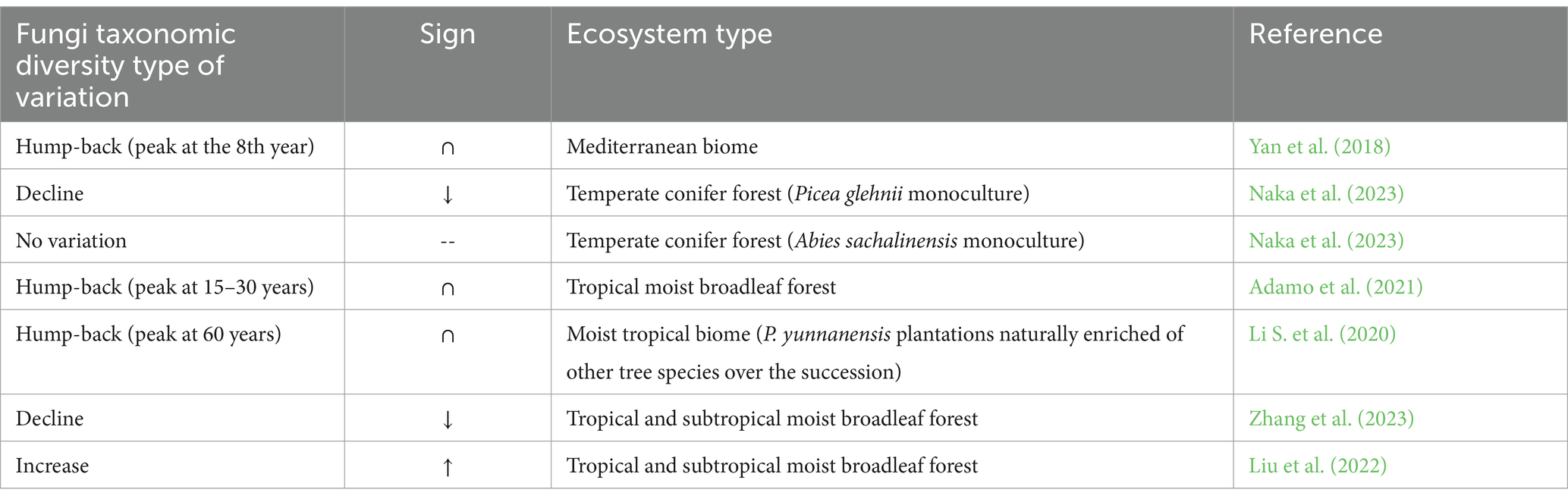
Table 1. Fungi taxonomic diversity variations across different time scales, and biomes in plantations and restoration forests.
The relative abundance of endophytic fungi also improved with reforestation, and the opposite occurred for the relative abundance of saprotrophic fungi. In 40–50-year-old plantations in tropical and subtropical moist biome Zhang et al. (2023) observed that fungal diversity declined with age. The transition from the grassland-shrub stage to the forested phase was associated with an increase in symbiotrophic fungi dominated by ectomycorrhiza and a decrease in saprotrophs. In another study, fungal diversity increased over the chronosequence in the moist tropical biome. The study, established in 1989, was undertaken in shrublands revegetated with different tree species: P. massoniana, Betula luminifera, and these two species as a mixture. The P. massoniana and mixed species forest (B. luminifera and P. massoniana), had the highest value of SOC, soil water content, total nitrogen, and total phosphorus, while pH value was comparable across stages of reforestation. The highest fungal taxonomic diversity was observed in mixed forest plots, and it was lower, but similar for the two monospecific forests. In both mixed and monospecific forests, fungal diversity was augmented with the increased age of reforestation (Liu et al., 2022). Still, the fungal taxonomic diversity and ectomycorrhizal relative abundance increased across a 30-year chronosequence, while bacteria taxonomic diversity and saprotrophic fungi relative abundance decreased in the New Caledonia tropical biome (Fernandez Nuñez et al., 2021). The decrease in saprotrophic: ectomycorrhizal fungi ratio indicated the restoration successional trajectory. SOC did not vary across the chronosequence, whereas total nitrogen decreased, and ammonium showed a peak in the mid-stage of forest restoration (Fernandez Nuñez et al., 2021).
Fungi in temperate biome plantations can display different patterns of variation. In the temperate conifer forest in Hokkaido Island Japan, fungal richness declined in monoculture plantations of Picea glehnii (a native species) and did not vary significantly over 49 years in Abies sachalinensis (also a native species) plantations or naturally regenerated forests (Naka et al., 2023). Ectomycorrhizal and saprophytic fungi recovered to the reference state of the natural forest in unmanaged regenerated forests, due to an increase of ectomycorrhizal fungi and a decrease of saprotrophic ones over the chronosequence (Naka et al., 2023).
3.5 Parallel trends of bacteria and fungi taxonomic richness variations considering ecological parameters in different biomes
The decline in the number of trees sporadically affected the taxonomic richness of bacteria and fungi in the Mediterranean biome but not in the Boreal one. We retained two studies considering the decline of tree numbers to better understand the patterns of richness and diversity variation of fungi and bacteria. In a Jack pine plantation in Canada (boreal forest/taiga biome), the number of trees diminished due to harvesting, and this action did not impact bacteria, Archaea, and fungi taxonomic richness. However, the decline negatively affected the microbial metabolic activity (Smenderovac et al., 2017). In a study conducted in the Mediterranean biome, the ASVs variation displayed an increase in bacteria and fungi diversity with a decrease in the number of trees caused by a pathogen invasion 25 years earlier (Gómez-Aparicio et al., 2022). Diversity and abundance of ectomycorrhiza declined with the reduced tree numbers, and saprotrophic fungi increased, both in diversity and abundance (Gómez-Aparicio et al., 2022).
Bacterial and fungal taxonomic diversity can display different patterns of co-variation. An afforestation study conducted in grasslands of the Loess Plateau (China), demonstrated that after 15 years of P. tabuliformis plantation, bacterial taxonomic diversity did not vary significantly, whereas fungi taxonomic diversity decreased (Wang et al., 2019). At this location, SOC decreased, along with soil nitrogen content. The relative abundance of ectomycorrhizal fungi increased, and the relative abundance of biotrophic fungi decreased (Wang et al., 2019). In another afforestation plantation of P. tabuliformis in Shaanxi Province (China), bacterial taxonomic diversity decreased over 60 years, whilst fungi taxonomic richness and SOC increased. Fungal and bacterial taxonomical community composition varied according to the succession stage (Liu et al., 2019). In P. sylvestris plantations (species native to the study location), bacterial communities had the highest diversity in one of the latest assessed stages (at age 41 of 59 years), and fungal diversity decreased until the age of 41 (Bi et al., 2021). Community composition changed, SOC initially decreased and then returned to the young stand values (15 and 30 years old). Available nitrogen and total nitrogen increased and then diminished to reach a value lower than that observed in the earlier stages. Available phosphorus significantly decreased with age (Bi et al., 2021). In Robinia pseudoacacia plantations (a nitrogen-fixing tree species with a symbiotic relationship with Rhizobium bacteria) on the Loess Plateau, taxonomic diversity of bacteria and fungi, SOC, and ammonium (NH4+) were positively correlated with stand age over 45 years. Bacteria functional traits indicated a shift from oligotrophs to copiotrophs as soils transitioned from nutrient-poor to nutrient-rich (Zhou et al., 2022).
In tropical forests, the vegetation structure and composition affect the bacteria and fungi community. In a study focused on reforestation with P. massoniana (over 30 years), the presence of an understory layer was associated with higher bacteria and fungi taxonomic diversity while being positively correlated with nitrogen-fixing bacteria and negatively correlated with ectomycorrhizal fungi. Taxonomic diversity of bacteria and fungi was also correlated with SOC, nitrogen, and phosphorus, and the community composition varied according to ferns’ presence and the successional stage (Lu et al., 2023).
Bacteria and fungi diversity displayed different patterns and relationships with ecosystem function in native and exotic tropical plantations. In native plantations of P. massoniana, Cunninghamia lanceolata and Schima wallichii, Castanopsis hystrix, Michelia macclurei and Cinnamomum burmannii in Southern China, bacteria and fungi biomass and Shannon diversity were lower than in exotic monocultures of Acacia mangium, but their richness was higher. The authors highlight the possible role of the tree species attributes in shaping the microbial community diversity (e.g., lower substrate quality furnished by the native conifer species for determined microbial group). A significant relationship between soil chemical compounds was found. Above- and below-ground composition was correlated for bacteria in sites where restoration occurred with native trees. Nitrogen and SOC stocks were similar among exotic and native forests, while the total phosphorus was higher in native plantations (Wei et al., 2023).
In the Philippines, 20 years old Swietenia macrophylla plantations displayed variation in microbial community functions (substrate use, respiration, and enzyme activity) convergent to the remnant reference state (Bonner et al., 2019). Microbial community composition was further along the path of recovery in 20-year-old “rainforestation” plantations (i.e., plantations of highly diverse and mainly native species) than in monocultures of S. macrophylla. The SOC was significantly higher in the monoculture plantations and was better explained by microbial composition variation than the land-use.
4 Discussion
4.1 Bias and geographic gaps
Despite some advances in assessment using multiple parameters (vegetation diversity, ecosystem structure, ecosystem functioning, and soil microbes’ assessment with eDNA analysis), our review revealed a geographical gap in the application of these studies. We retrieved a list of less than 200 studies, with the first article published in 2018, substantially later than the publication of articles considering general aspects of forest restoration. Integrated evaluation in forest restoration outcomes is at the frontier stage, as is eDNA analysis. The first analysis based on environmental DNA was reported by Ogram et al. in the late 1980s (Ogram et al., 1987).
Our review highlights the lack of studies in different areas and the initial stages of forest succession. We confirmed the geographical gap found by other authors (i.e., lack of studies in less developed countries) (Ruiz-Jaén and Aide, 2005a; Wortley et al., 2013), extending it further to include North American locations that Ruiz-Jaén and Aide (2005a), and Wortley et al. (2013) evaluated as being properly assessed. Tropical and subtropical coniferous forests also lacked integrated evaluation, and a bias of studies was represented by much of the research being conducted in China, particularly on the Loess Plateau. While the scarcity of studies and biases toward geographical region may prevent the identification of other microbial variation patterns, the large occurrence of studies based in China did not hinder the research of a general microbial pattern of variation, since these analyses considered multiple biomes (temperate and tropical forests) present within the borders of this state and in other parts of the world. Restoration success is mostly evaluated after a long time (the average age of examined plots was 29 years) highlighting the absence of analyses for the early phases of forest restoration.
Bacteria and fungi were not always studied together, despite their reciprocal influences on their diversity and community composition. Study objectives, authors’ experience, and the costs of the primers and machines used influenced the selection of one of the two microbial groups. Archaea were the least investigated, despite their importance in ecosystem dynamics. While bacteria genetic libraries and ecology are becoming well-established, effort is required to build libraries for fungi and Archaea. Given this limitation, we suggest analysis to acquire baselines for these latest groups, concurrently improving bacteria’s available information, for which progress is expected for the exploitation of their metabolic activities in particular. In parallel to the enhancement of fungi and Archaea databases, bacteria may be used as a common metric to assess restoration success.
Soil characteristics were usually examined, while ecosystem structure was generally neglected. Most articles observed soil characteristics (93%), allowing possible comparison among different locations. Interestingly, one article measured the organic matter decomposition, which may lead to fruitful information about ecosystem functioning (Philpott et al., 2018). Ecosystem structure was mostly overlooked, despite its importance for microbial community dynamics in restoration contexts (Lu et al., 2023). Although succession stage and forest age may be directly related to vegetation structure, careful consideration is needed on the metrics used. For example, diameter at breast height (DBH) increases over a chronosequence but Deng et al. (2019), and Zhang et al. (2019) found that stand density decreases over this time. The explicit incorporation of measures of ecosystem structure in environmental dynamics analyses must be acknowledged, given the simple ways to acquire this data and the importance of it in restoration success and microbial variation. Evidence of the direct influence of ecosystem structure on soil microbes was provided by Fernandez Nuñez et al. (2021) and Lu et al. (2023), and it will be discussed in the following paragraphs.
4.1.1 Bacterial taxonomic richness and functional variation in different biomes: drivers, and hypotheses
Divergent trends of bacteria diversity were found within and among different biomes analyzed, suggesting that the classification of studies according to the biome in which they were conducted was not sufficient to reveal shared patterns of response. Yearly evaluations over 10 years may expose variation at the micro-scale, while an assessment every 5 years may be insufficient to reveal any temporal pattern. Macro-scale climatic features did not explain results variability, highlighting the importance of smaller-scale ecosystem characteristics. The effect of micro-habitat features was already reported in microbial community predictions, considering the deterministic processes that drive them in the late stages of succession when soil chemistry and depletion of resources exert strong selective effects (Dini-Andreote et al., 2015; Yan et al., 2023).
Across different types of ecosystems, spatial variation of bacteria taxonomic diversity (resembling the temporal variation in a space-for-time approach) was attributed to the modification of soil chemistry due to plant species changes (Gellie et al., 2017), strengthening the hypothesized connections between bacteria, tree species and soil nutrients (Li et al., 2023; Lu et al., 2023; Qiu et al., 2022; Yang et al., 2018; Zhou et al., 2022). Soil nutrient variation guided the transition from oligotroph to copiotroph bacteria in the temperate biome Zhou et al. (2022). Increase in the age of R. pseudoacacia plantations was accompanied by the shift from bacteria adapted to harsh soil conditions (oligotrophs) to bacteria communities thriving in nutrient-rich soils (copiotrophs). This dynamic may be ascribed to the dominant tree species (R. pseudoacacia), part of the Fabaceae family, which benefits from rhizobium symbiosis with soil nitrogen-fixing bacteria (Liu et al., 2018). The enrichment over time of soil of roots exudates and soil organic carbon (compounds closely related to microbial activity) is an established pattern (Allek et al., 2023), leading copiotrophic bacteria to replace oligotrophic ones. Normally, taxa thriving in unstable and nutrient-depleted environments would increase their abundance in more favorable conditions. However, copiotrophic bacteria can outcompete oligotrophic bacteria because of their ability to absorb nutrients faster (Liang et al., 2021), while having higher growth rates (Li et al., 2021). Copiotrophic bacteria have also been associated with lower affinity to the substrate than oligotrophs ones, and with the preferential utilization of unstable carbon, while oligotrophs are associated with recalcitrant soils (Liao et al., 2023). The increase of SOC and available nutrients (i.e., NH4+) in the study by Zhou et al. (2022) supports these findings since the presence of such compounds is indicative of copiotrophic bacteria occurrence.
The development of a more structurally complex environment during reforestation leads to an increase in nitrogen-fixers and a decrease in hydrocarbon-degrading bacteria in a tropical biome (Lu et al., 2023). This increase in the relative abundance of nitrogen-fixing bacteria with reforestation and fern understory occurrence has been linked to plant species favoring the establishment of such groups of bacteria, given the symbiotic relationships they can establish with microorganisms, promoting nutrient availability. For example, P. massoniana can develop symbiotic relationships with ectomycorrhizal fungi (Lu et al., 2023). Increases in nutrient concentration led bacterial communities to shift toward a major abundance of copiotrophic taxa such as Proteobacteria. Moreover, copiotrophs can supply the soil with the nutrients needed by both nitrogen-fixers and trees, thanks to the symbiotic relationship they are part of, further enhancing their relative abundance (Figure 3). This is supported by multiple studies across the tropics (Batterman et al., 2013; Gehring et al., 2005; Gei et al., 2018; Taylor et al., 2019).
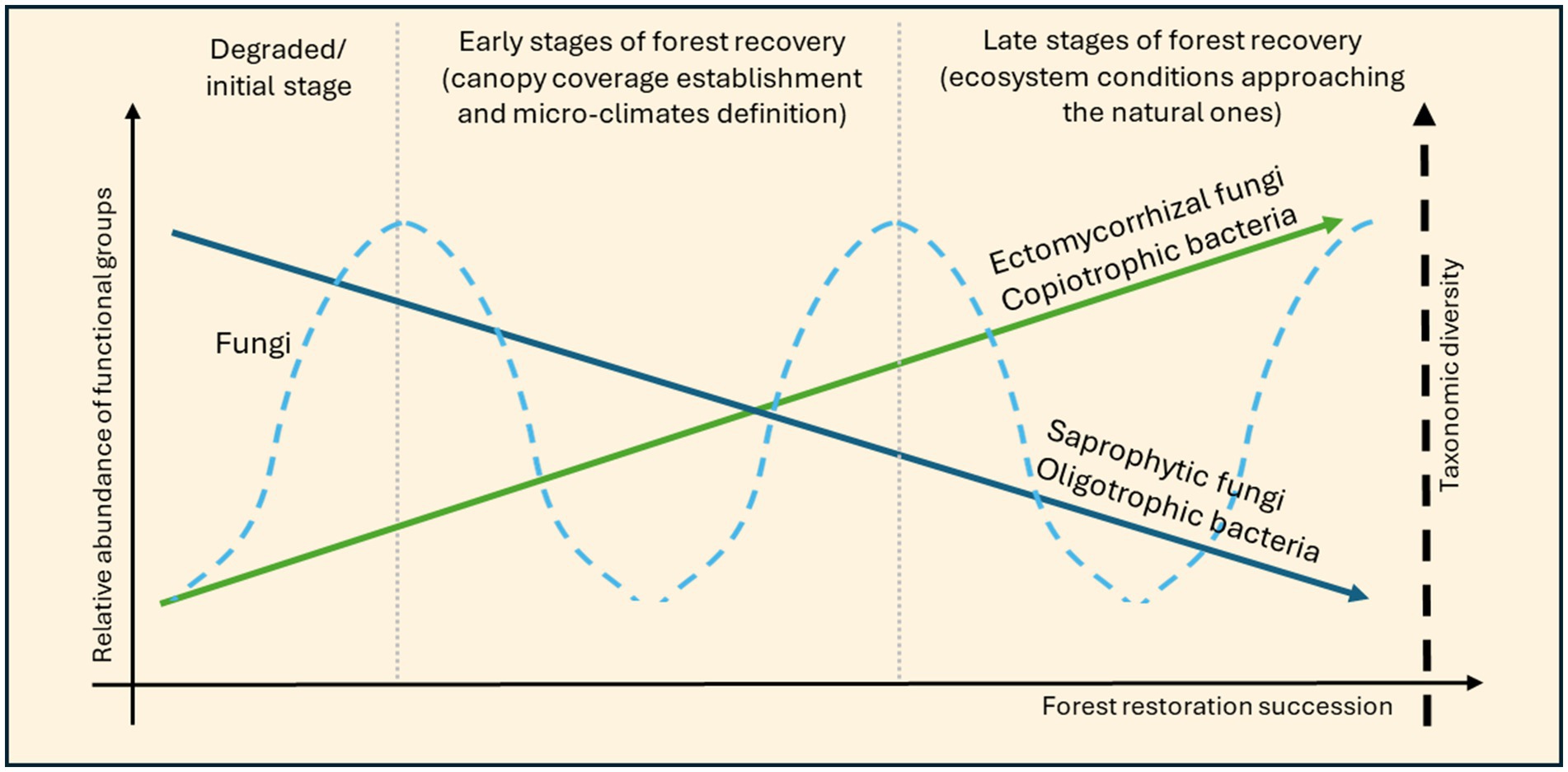
Figure 3. General representation of the hypothetical fluctuations of bacteria and fungi relative abundance of functional groups, and fungi taxonomic diversity, across the forest restoration development. The extent and the period to which these variations occur will depend on the starting conditions of each location and their particular plant community, ecosystem structure, and soil nutrient development. The blue dashed line represents fungi taxonomic diversity, while the solid blue and green lines represent hypothesized trajectories of fungi and bacteria relative abundance of specific functional groups.
Land-use history (i.e., the legacy effect) was demonstrated as an important factor in shaping the microbial community (Kielak et al., 2008; Wang et al., 2022). While it is reasonable to attribute the increase of bacteria taxonomic diversity to the ingression of opportunistic and fast-growing taxa following the vegetation transition, their lagged response or even lack of variation may be ascribed to the soil’s chemical properties. Specifically, bacteria response reflects: the agricultural history through chemical compound concentration from prior manipulation (Kielak et al., 2008); the specific “strength” of previous land uses; their resilience, given the numerous strategies and processes they are capable of (e.g., capacity to build protective biofilms) (Wang et al., 2019).
From the outcomes of each analyzed study, we argue that soil parental material and historical filters affect the plant species that colonize the area, while plant species influence soil chemistry and bacterial communities, which interact to shape forest succession and restoration achievement.
4.1.2 Fungi taxonomic richness variation in different biomes: drivers, and hypotheses
Fungi taxonomic richness varies according to a hump-back function across different biomes, reflecting vegetational changes (Figure 3) (Adamo et al., 2021; Fernandez Nuñez et al., 2021; Li S. et al., 2020; Yan et al., 2018). Over the chronosequence, plant species of later stages overlap with those already present. The intersection between vegetational species increases the vegetational richness, which seems to be mirrored by fungi taxonomic diversity (Adamo et al., 2021; Liu et al., 2020). An increase in vegetational diversity occurs each time a stage transits to another, and this dynamic is reproduced by the fungi community as observed in several studies (dashed line in Figure 3). For instance, the result of Zhang et al. (2023) might represent the final part of that “hump-back shape” (declining) diversity function, occurring in a phase of decrease in vegetational richness, since the study observed the later phases of the transition from a shrub stage to a forested one. The fluctuations in diversity values observed by Adamo et al. (2021) in naturally regenerating sites of 15–30 years may illustrate the full range of the response curve occurring through forest succession.
The hump-back-shaped function of fungi taxonomic diversity occurs at a different period according to biome and tree species traits over the restoration chronosequence (Yan et al., 2018; Adamo et al., 2021). Although soil chemistry is a common influencing factor, various limitations may be present in each biome. A slower increase in fungal taxonomic diversity in tropical areas compared to Mediterranean areas might reflect constraints in micro-elements or molecules tightly bound to organic compounds. We hypothesized that this dynamic may be due to the lack of humic acids and their nutrient-locking effect in the Mediterranean biome’s dry soil. The scarcity of humic acids makes the nutrients (when available) readily accessible to the organisms. Conversely, humic acids and nutrients are abundant in the tropical biome and tightly bound to each other, obstructing the acquisition of nutritional resources by fungi.
Tree species, or even tree taxonomic families, may influence the fungal taxonomic diversity variation patterns more than biome large-scale characteristics. In the temperate biome, fungal diversity in monoculture plantations of P. glehnii and Abies sachalinensis showed a decline over time (Naka et al., 2023), similar to that observed by Wang et al. (2019), in plantation of P. tabuliformis. This outcome may be ascribed to the characteristics of the trees adopted, being part of the same family (Pinaceae). Corroborating this hypothesis, are the results of Zhou et al. (2022), Liu et al. (2019), which showed a different variation of fungi diversity in forests of different tree families in the same temperate biome. Given these findings, we argue that tree species diversity may play a major role in shaping fungi communities, affecting the taxonomic richness variation of these microorganisms more than larger-scale constraints (like climate).
The vegetational transitional phase in which restoration evaluation is conducted must be considered to appreciate the whole fungi taxonomic diversity dynamic. The identified fungi taxonomic dynamic of Naka et al. (2023), Zhang et al. (2023), and Wang et al. (2019) contrasted with the ones of Lu et al. (2023) and Li S. et al. (2020). These two latest studies focused on the analysis of soil between 0 and 10 cm of depth, while the former considered a soil depth of 5 cm. We argue that this difference, along with the diverse timeframe of analyses, and provinces in which the sampling occurred may have caused the disagreement. In addition, while the trees’ taxonomic family may be enough to foresee fungal taxonomic diversity variations, the maturity of the forest (reflecting the establishment of symbiotic relationships and soil chemistry variation), the transition phase occurring in the ecosystem (e.g., from grasslands to forest), and the abiotic disturbances occurring in each location (e.g., destructive events which delay the fungal diversity) can influence the period over which the hump-back-shaped function is expressed.
A common pattern of variation of relative abundance of fungi functional groups is shared between biomes: ectomycorrhizal fungi increase while saprotrophic fungi decline over time (Figure 3) (Adamo et al., 2021; Fernandez Nuñez et al., 2021; Li S. et al., 2020; Naka et al., 2023; Wang et al., 2019; Yan et al., 2018; Zhang et al., 2023). This trend can be attributed to the Gadgil effect, for which ectomycorrhizal fungi outcompete saprotrophic fungi thanks to the symbiotic relationships they are capable of Gadgil and Gadgil (1971, 1975). Tree species of older successional stages could be considered causal in shaping this pattern. Symbiotic relationships between plants and fungi are also present in the early stages of succession, but they are largely expressed by bacteria, for example between Medicago sativa pastures and the Rhizobium genera (Liu et al., 2018). Conversely, conifers and broadleaved trees of later successional stages generally have symbiotic relationships with fungi. This variation of cooperative protagonists (from bacteria to fungi) may lead to the increase in relative abundance of symbiotic fungi, sustained by and supporting tree growth over the succession, while saprotrophic fungi struggle to retain their niche. Early-stage exploitation of symbiotic relationships by bacteria may not affect fungi niches, given the different resources and time of growth of the two, allowing saprophytic fungi to dominate the environment. In later successional stages, the development of symbiotic relationships between ectomycorrhizal fungi and plants can promote the success of this fungi functional group, restricting the available niche for saprophytic ones. Nutrients in the soil also increase over the restoration succession, but their exploitation may be difficult for saprotrophic fungi, which are not aided in their access to these compounds as ectomycorrhizal fungi are (given their symbioses with trees).
4.1.3 Parallel above- and below-ground parameters variation over restoration
All the ecosystem characteristics analyzed changed over the restoration stages with shared patterns across biomes. Vegetation diversity and richness are directly influenced by restoration practices (e.g., the choice of plantation species) and the natural progression of ecosystem succession, resulting in increased plant richness in all the articles reviewed. Plant richness affects soil chemistry, soil microbial taxonomic and functional diversity relative abundances, and ecosystem structure (e.g., Gellie et al., 2017; Li et al., 2023; Lu et al., 2023). Considering all the selected studies, we conclude that ecosystem functioning and soil microbial communities change abruptly, while ecosystem structure develops slowly, following tree species variation. We argue that it is better to couple all these parameters to understand the restoration success and make them indicators of this process, considering their different time of modification. One example of the possible coupling between slow- and fast-changing traits is the consideration of fungi functional group variation and tree species diversity.
Ecosystem structure evolves according to each context, while ecosystem functioning proxies increase over the forest succession. Ecosystem structure is influenced by the restoration stage (i.e., age since the action of restoration occurred) and natural events, both abiotic, like climatic events, and biotic, such as the ingression of plant pathogens (Gómez-Aparicio et al., 2022; Smenderovac et al., 2017). Controlled field experiments undertaken to isolate the effect of ecosystem structure revealed clear trends for microbial community variations. For instance, Lu et al. (2023) found that understory manipulation significantly affected bacteria and fungi taxonomic and functional diversity. Ecosystem functioning (e.g., soil fertility represented by SOC) generally increases over restoration chronosequence or remains at high values depending on the starting point (Bonner et al., 2019).
The literature indicated native trees may sustain higher microbial richness than exotic trees but have lower microbial biomass. In native plantations of P. massoniana and C. lanceolata, microbial biomass was found to be lower than in exotic monocultures of A. mangium, and this was ascribed to the features of the tree species (broadleaved, coniferous, or N-fixing species), rather than the tree origin (exotic or native) or planting configuration (Wei et al., 2023). The major influence of functional and morphological traits over the tree species origin may be caused by the planting of both exotic and native species, thus removing the “geographical filter” that would have obstructed the establishment of the exotic taxa. The presence of competitive traits in exotic species (e.g., the ability to create symbiotic relationships) enhances their potential to sustain higher microbial abundance.
Fungi taxonomic diversity seems to change faster than bacteria, but the influence of each ecosystem feature (e.g., parental material of the soil) makes the recognition of common patterns difficult. Nevertheless, soil chemistry and pH are generally related to a specific microbial community composition (Li et al., 2023). An acidic pH favors fungi, while alkaline or neutral conditions are beneficial for bacteria (Rousk et al., 2009). Bacteria diversity appears to increase over time, without extreme and fast responses demonstrated by fungi. Still, initial conditions and specific features of the ecosystem dictate the evolution of the trend. We hypothesized that fungi may change more quickly in comparison to bacteria given their specificity for plant species composition (Chen et al., 2022), and their greater sensitivity to highly variable parameters (e.g., humidity) (Yan et al., 2018), for which bacteria developed coping strategies (e.g., creating biofilms). The difference in temporal fluctuations of bacteria and fungi is corroborated by bacteria resilience that pushes their community variation later in time (Mirza et al., 2020; Wang et al., 2019), and the sensitivity of fungi, which allows for steady and measurable changes in their diversity (Yan et al., 2018).
The causes and consequences of above and below-ground community variations are intertwined. We argue that this interdependence further stresses the importance of holistically assessing ecosystems to demonstrate successful outcomes of restoration projects. The analysis of nutrient concentration, ecosystem structure, and plant community at the start of the project and throughout its development can help understand if additional interventions are needed to achieve the desired restoration trajectory (when the assessed values are compared to the ones of the aimed state). For instance, a modification in the bacterial community could help identify an environmental change that may be conducive to invasive plants that would be deleterious to the project goals.
4.2 Recommendations and future research directions
The infancy of integrated restoration evaluations hinders a comprehensive assessment of trends across restoration ages and biomes, but early results suggest the inclusion of different parameter analyses, the usage of innovative methods, and coordinated efforts between stakeholders to assess the restoration outcomes and plan future actions (Table 2). For instance, while data on vegetation and ecosystem properties are often collected through time as occurred in research projects conducted in Costa Rica (Aguilar-Arias et al., 2012; Morales-Salazar et al., 2013), we suggest the inclusion of soil microbial community monitoring to better understand restoration ecosystem dynamics. The monitoring of restoration project success requires technical improvements, additional studies to fill geographical gaps, and clarification of microbial community patterns. Furthermore, divergent trends regarding the extent and type of soil microbes’ taxonomic diversity variation along restoration gradients and locations corroborate the need for research in different geographies, while developing a standardized protocol that can be applied in dissimilar ecosystems, producing comparable data (thus overcoming the related challenges stated in section 1.3).
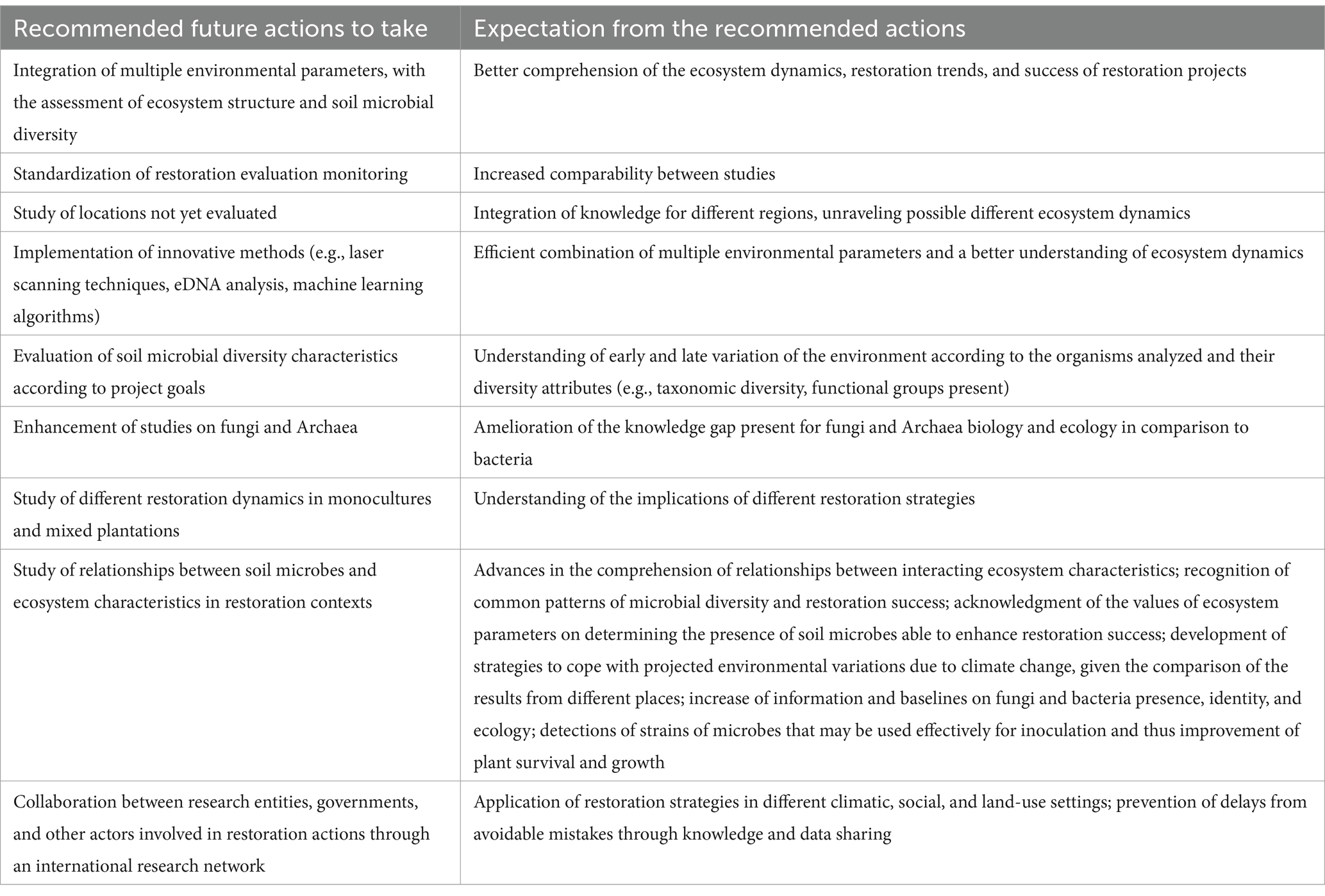
Table 2. Recommended actions for evaluating restoration and outcomes expected from their implementation.
As a standardized protocol, we highly recommend the assessment of restoration through the assessment of microbial community characteristics (taxonomic and functional diversity assessed through eDNA sequencing); evaluation of vegetation layers (coverage and taxonomy and/or functional diversity); characterization of the spatial disposition of biotic and abiotic components, tree density (stems/ha) and occurrence of abiotic features (e.g., rocks, streams, faults). For this latest evaluation, the advantage of remote sensing and laser scanning techniques to accurately monitor vegetation cover and topography (Buckley et al., 2023) has great potential.
Restoration assessment integrating plant diversity, ecosystem structure, soil microbial community, and ecosystem function metrics can significantly improve the outcome of restoration actions. Innovative methods enhance efficiency by streamlining the aggregation of multiple parameters. Remote sensing, and eDNA analysis, are great examples of innovative and cost-effective techniques that will bring environmental study to a new insightful level. Additionally, advanced practices to conduct ecosystem assessments are being produced (e.g., eDNA protocols for sample collection), as well as pioneering statistical analyses and increasing the processing power of computers (reaching the quantum field) allowing the use of cutting-edge machine learning algorithms. Examples of these are the structural equation models found in the analyses of Zhou et al. (2022), deep learning methods to investigate metabarcoding data (Lamperti et al., 2023), and revolutionary classification functions (Novello and Tonello, 2024). Plant diversity and SOC are metrics commonly implemented in restoration evaluation, but the inclusion of analyses on ecosystem structure and soil microbes’ diversity is yet to be established. Recognizing relationships and expected trajectories of all these environmental characteristics will allow stakeholders to understand what to aim for, the constraints and the best strategy to adopt to achieve the restoration aims and how they can measure the progress toward them.
The understanding of microbial community variation along with ecosystem structure yields accurate results and better hypotheses for the recognition of successional dynamics (Lu et al., 2023). The most used measure of ecosystem structure is ground cover percentage of herbaceous vegetation, and canopy cover for trees, not always in combination with DBH and other metrics. We suggest the holistic development and utilization of additional ecosystem structure parameters that can better inform restoration ecosystem dynamics.
Preliminary studies on microbial diversity may be useful to detect environmental variation and inform, further study on fungi or bacteria according to the time scale of restoration assessment. Assessing the microbial taxonomic diversity variations through time can demonstrate the successional “phase” of the ecosystem when it is not clear from the morphology or vegetational species composition. Meanwhile, the functional community composition may indicate the recovery toward the reference state (Fernandez Nuñez et al., 2021). In addition, by acknowledging the different times required by bacteria and fungi taxonomic diversity to significantly change, restoration assessments can be designed that consider trade-offs between costs and comprehensive analysis. For example, when the assessment is done in the early successional stages and the budget does not allow the investigation of different microbial groups, fungi should be the preferred focus. The opposite may apply to later-stage evaluations.
Constructive results for forest restoration management will arise through directed research on its ecological dynamics. The recent rise in the number of articles focused on integrated restoration measurements permits speculation on a future escalation of efforts to better understand ecosystem dynamics in restoration projects. The upsurge of integrated studies has been aided by the broadening of the eDNA methodology usage and the acknowledgement of the importance of soil and its microbial community.
Studies focused on different patterns of variations in monoculture and mixed plantations (e.g., if the number of ASVs reaches the same value between plantation types) are needed to understand the implications of plantation types on the ecosystem. For example, if after restoration the ASVs values are similar to the previous state, the success of the restoration could be questioned, enabling the understanding of the legacy effect of the pre-restoration biome.
Research unraveling the relationships between soil microbes and ecosystem characteristics in restoration contexts will lead to the following outcomes: (i) advances in the comprehension of relationships between ecosystem characteristics; (ii) recognition of common patterns of microbial diversity and restoration success; (iii) acknowledgement of ecosystem parameters values in determining the presence of soil microbes able to enhance restoration success; (iv) development of strategies to cope with projected environmental variations due to climate change, given the comparison of the results from different places (e.g., constraints leading to restoration success in dry locations may be applied to areas facing increased aridity due to climatic changes); (v) increase of information on fungi and bacteria presence, identity, and ecology; (vi) detections of strains of microbes that may be used for inoculation to improve plant survival and growth (Fernandez Nuñez et al., 2021).
We advocate the importance of collaboration and knowledge sharing between research entities, governments, and other actors involved in restoration programs. The establishment of an international research network would promote the application of restoration strategies in different climatic, social, and land use settings, learning from mistakes, and taking advantage of successful approaches. This should lead to a more coordinated effort toward restoration projects’ success, preventing delays from avoidable mistakes.
5 Conclusion
Plant diversity increases over the restoration chronosequence, while ecosystem structure and functioning develop according to each ecosystem condition. Despite the presence of a geographical bias and the lack of multiple parameters consideration in standardized methods, we were able to highlight general yet not exhaustive trends of microbial community characteristics variations over forest restoration. Bacteria taxonomic diversity fluctuates, and fungi taxa richness displays a hump-back variation across different biomes, suggesting underlying “common rules” for their changes. Bacterial taxa adapted to harsh conditions are outcompeted by the taxa able to better exploit the eutrophic conditions of the later stages of forest restoration, while symbiotic fungi dominate over saprophytic ones across the chronosequence. Soil microbial community variation is affected by large-scale modifications and fine-scale adjustments of each location, and research is needed to understand possible common microbial variations across biomes and over the restoration process. Bringing new insights into restoration results assessments, our review emphasizes the value of integrated assessments for significantly better comprehension and management of restoration processes.
Author contributions
JV: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. RC: Conceptualization, Investigation, Supervision, Validation, Writing – review & editing. AC: Conceptualization, Supervision, Validation, Writing – review & editing. DL: Conceptualization, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research received support from the University of the Sunshine Coast (UniSC) through the UniSC International Research Scholarship, Australia. Additionally, it forms a part of Project Tarsier at the Tropical Forests and People Research Centre at UniSC. The Forest Research Institute of UniSC provided funds to cover the open-access publishing costs for this paper.
Acknowledgments
The authors acknowledge the valuable comments of Tomiwa Oluwajuwon.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2025.1540513/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Articles retained and parameters evaluated (order by newest to oldest). Symbols: ↔ variation of microbial groups not possible to define in terms of increase or decrease (e.g., different variation according to the taxa); ∩ hump-back variation of taxonomic diversity; ↑ increase of taxonomic diversity; ↓ decrease of taxonomic diversity. Functional variation is omitted because deriving from in-depth analysis.
SUPPLEMENTARY TABLE S2 | Diversity indexes commonly used in microbial analysis. Table adapted from the study of Kim et al. (2017).
References
Abella, S. R., and Covington, W. W. (2004). Monitoring an Arizona ponderosa pine restoration: sampling efficiency and multivariate analysis of understory vegetation. Restor. Ecol. 12, 359–367. doi: 10.1111/j.1061-2971.2004.00317.x
Adamo, I., Ortiz-Malavasi, E., Chazdon, R., Chaverri, P., Steege, H. T., and Geml, J. (2021). Soil fungal community composition correlates with site-specific abiotic factors, tree community structure, and forest age in regenerating tropical rainforests. Biology 10:1120. doi: 10.3390/biology10111120
Aguilar-Arias, H., Ortiz-Malavassi, E., and Vílchez-Alvarado, B. (2012). Páginas 22 aMzEta3VydUB0ZWMuYWMuY3I=-ISSN: 2215-2504. Revista Forestal Mesoamericana Kurú 22, 22–31.
Allek, A., Viany Prieto, P., Korys, K. A., Rodrigues, A. F., Latawiec, A. E., and Crouzeilles, R. (2023). How does forest restoration affect the recovery of soil quality? A global meta-analysis for tropical and temperate regions. Restor. Ecol. 31:e13747. doi: 10.1111/rec.13747
Aronson, J., Floret, C., Le Floch, E., Ovalle, C., and Pontanier, R. (1993). Restoration and rehabilitation of degraded ecosystems in arid and semi-arid lands. I. A view from the south. Restor. Ecol. 1, 8–17. doi: 10.1111/j.1526-100X.1993.tb00004.x
Baldrian, P., Zrůstová, P., Tláskal, V., Davidová, A., Merhautová, V., and Vrška, T. (2016). Fungi associated with decomposing deadwood in a natural beech-dominated forest. Fungal Ecol. 23, 109–122. doi: 10.1016/j.funeco.2016.07.001
Bardgett, R. D., and Van Der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Bartholomew, D. C., Mosyaftiani, A., Morgan, B., Shah, T., Shaw, K., Stillman, C., et al. (2024). The global biodiversity standard: manual for assessment and best practices authors. Richmond (GB), UK & SER Washington, DC: Botanic Gardens Conservation International & Society for Ecological Restoration.
Bash, J. S., and Ryan, C. M. (2002). Stream restoration and enhancement projects: is anyone monitoring? Environ. Manag. 29, 877–885. doi: 10.1007/s00267-001-0066-3
Batterman, S. A., Hedin, L. O., Van Breugel, M., Ransijn, J., Craven, D. J., and Hall, J. S. (2013). Key role of symbiotic dinitrogen fixation in tropical forest secondary succession. Nature 502, 224–227. doi: 10.1038/nature12525
Bi, B., Zhang, H., Yuan, Y., Wu, Z., Wang, Y., and Han, F. (2021). Dynamic changes of soil microbial community in Pinus sylvestris var. mongolica plantations in the Mu Us Sandy Land. J. Environ. Manag. 287:112306. doi: 10.1016/j.jenvman.2021.112306
Bonner, M. T. L., Herbohn, J., Gregorio, N., Pasa, A., Avela, M. S., Solano, C., et al. (2019). Soil organic carbon recovery in tropical tree plantations may depend on restoration of soil microbial composition and function. Geoderma 353, 70–80. doi: 10.1016/j.geoderma.2019.06.017
Buckley, H. L., Hall, D., Jarvis, R. M., Smith, V., Walker, L. A., Silby, J., et al. (2023). Using long-term experimental restoration of agroecosystems in Aotearoa New Zealand to improve implementation of nature-based solutions for climate change mitigation. Front. For. Glob. Change 5:950041. doi: 10.3389/ffgc.2022.950041
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chen, Y., Xi, J., Xiao, M., Wang, S., Chen, W., Liu, F., et al. (2022). Soil fungal communities show more specificity than bacteria for plant species composition in a temperate forest in China. BMC Microbiol. 22:208. doi: 10.1186/s12866-022-02591-1
Chiarello, M., McCauley, M., Villéger, S., and Jackson, C. R. (2022). Ranking the biases: the choice of OTUs vs. ASVs in 16S rRNA amplicon data analysis has stronger effects on diversity measures than rarefaction and OTU identity threshold. PLoS One 17:e0264443. doi: 10.1371/journal.pone.0264443
Cortina, J., Maestre, F. T., Vallejo, R., Baeza, M. J., Valdecantos, A., and Pérez-Devesa, M. (2006). Ecosystem structure, function, and restoration success: are they related? J. Nat. Conserv. 14, 152–160. doi: 10.1016/j.jnc.2006.04.004
Deng, J., Zhang, Y., Yin, Y., Zhu, X., Zhu, W., and Zhou, Y. (2019). Comparison of soil bacterial community and functional characteristics following afforestation in the semi-arid areas. PeerJ 2019:e7141. doi: 10.7717/peerj.7141
Di Sacco, A., Hardwick, K. A., Blakesley, D., Brancalion, P. H. S., Breman, E., Cecilio Rebola, L., et al. (2021). Ten golden rules for reforestation to optimize carbon sequestration, biodiversity recovery and livelihood benefits. Glob. Chang. Biol. 27, 1328–1348. doi: 10.1111/gcb.15498
Dinerstein, E., Olson, D., Joshi, A., Vynne, C., Burgess, N. D., Wikramanayake, E., et al. (2017). An ecoregion-based approach to protecting half the terrestrial realm. Bioscience 67, 534–545. doi: 10.1093/biosci/bix014
Dini-Andreote, F., Stegen, J. C., Van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 112, 1326–1332. doi: 10.1073/pnas.1414261112
Falk, D. A. (2006). Process-centred restoration in a fire-adapted ponderosa pine forest. J. Nat. Conserv. 14, 140–151. doi: 10.1016/j.jnc.2006.04.005
Feeney, D. S., Crawford, J. W., Daniell, T., Hallett, P. D., Nunan, N., Ritz, K., et al. (2006). Three-dimensional microorganization of the soil-root-microbe system. Microb. Ecol. 52, 151–158. doi: 10.1007/s00248-006-9062-8
Fernandez Nuñez, N., Maggia, L., Stenger, P. L., Lelievre, M., Letellier, K., Gigante, S., et al. (2021). Potential of high-throughput eDNA sequencing of soil fungi and bacteria for monitoring ecological restoration in ultramafic substrates: the case study of the new Caledonian biodiversity hotspot. Ecol. Eng. 173:106416. doi: 10.1016/j.ecoleng.2021.106416
Gadgil, R. L., and Gadgil, P. D. (1975). Suppression of litter decomposition by mycorrhizal roots of Pinus radiata. N. Z. J. For. Sci. 5, 33–41.
Gehring, C., Vlek, P. L. G., De Souza, L. A. G., and Denich, M. (2005). Biological nitrogen fixation in secondary regrowth and mature rainforest of Central Amazonia. Agric. Ecosyst. Environ. 111, 237–252. doi: 10.1016/j.agee.2005.06.009
Gei, M., Rozendaal, D. M. A., Poorter, L., Bongers, F., Sprent, J. I., Garner, M. D., et al. (2018). Legume abundance along successional and rainfall gradients in neotropical forests. Nat. Ecol. Evol. 2, 1104–1111. doi: 10.1038/s41559-018-0559-6
Gellie, N. J. C., Mills, J. G., Breed, M. F., and Lowe, A. J. (2017). Revegetation rewilds the soil bacterial microbiome of an old field. Mol. Ecol. 26, 2895–2904. doi: 10.1111/mec.14081
Gómez-Aparicio, L., Domínguez-Begines, J., Villa-Sanabria, E., García, L. V., and Muñoz-Pajares, A. J. (2022). Tree decline and mortality following pathogen invasion alters the diversity, composition and network structure of the soil microbiome. Soil Biol. Biochem. 166:108560. doi: 10.1016/j.soilbio.2022.108560
Graham, E. B., and Knelman, J. E. (2023). Implications of soil microbial community assembly for ecosystem restoration: patterns, process, and potential. Microb. Ecol. 85, 809–819. doi: 10.1007/s00248-022-02155-w
Hakimzadeh, A., Abdala Asbun, A., Albanese, D., Bernard, M., Buchner, D., Callahan, B., et al. (2024). A pile of pipelines: an overview of the bioinformatics software for metabarcoding data analyses. Mol. Ecol. Resour. 24:e13847. doi: 10.1111/1755-0998.13847
Harris, J. A. (2003). Measurements of the soil microbial community for estimating the success of restoration. Eur. J. Soil Sci. 54, 801–808. doi: 10.1046/j.1365-2389.2003.00559.x
Harris, J. (2009). Soil microbial communities and restoration ecology: facilitators or followers? Science 35, 573–574. doi: 10.1126/science.1176295
He, J., Zhang, J., Wang, J., Dong, Z., Meng, Z., Xu, R., et al. (2023). Natural restoration enhances soil multitrophic network complexity and ecosystem functions in the loess plateau. Catena 226:107059. doi: 10.1016/j.catena.2023.107059
Herrick, J. E., Schuman, G. E., and Rango, A. (2006). Monitoring ecological processes for restoration projects. J. Nat. Conserv. 14, 161–171. doi: 10.1016/j.jnc.2006.05.001
Hobbs, R. J., and Norton, D. A. (1996). Towards a conceptual framework for restoration ecology. Restorat. Ecol. 4, 93–110. doi: 10.1111/j.1526-100X.1996.tb00112.x
Huang, T., Wang, Y., Wang, X., Ma, L., and Yang, X. (2023). Discrepant diversity patterns and function of bacterial and fungal communities on an earthquake-prone mountain gradient in Northwest Sichuan, China. Front. Microbiol. 14:1217925. doi: 10.3389/fmicb.2023.1217925
Kielak, A., Pijl, A. S., Van Veen, J. A., and Kowalchuk, G. A. (2008). Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol. Ecol. 63, 372–382. doi: 10.1111/j.1574-6941.2007.00428.x
Kim, B. R., Shin, J., Guevarra, R. B., Lee, J. H., Kim, D. W., Seol, K. H., et al. (2017). Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Knoth, C., Klein, B., Prinz, T., and Kleinebecker, T. (2013). Unmanned aerial vehicles as innovative remote sensing platforms for high-resolution infrared imagery to support restoration monitoring in cut-over bogs. Appl. Veg. Sci. 16, 509–517. doi: 10.1111/avsc.12024
Lamperti, L., Sanchez, T., Si Moussi, S., Mouillot, D., Albouy, C., Flück, B., et al. (2023). New deep learning-based methods for visualizing ecosystem properties using environmental DNA metabarcoding data. Mol. Ecol. Resour. 23, 1946–1958. doi: 10.1111/1755-0998.13861
Lemke, M., and DeSalle, R. (2023). The next generation of microbial ecology and its importance in environmental sustainability. Microb. Ecol. 85, 781–795. doi: 10.1007/s00248-023-02185-y
Li, S., Huang, X., Shen, J., Xu, F., and Su, J. (2020). Effects of plant diversity and soil properties on soil fungal community structure with secondary succession in the Pinus yunnanensis forest. Geoderma 379:114646. doi: 10.1016/j.geoderma.2020.114646
Li, J., Shangguan, Z., and Deng, L. (2020). Dynamics of soil microbial metabolic activity during grassland succession after farmland abandonment. Geoderma 363:114167. doi: 10.1016/j.geoderma.2019.114167
Li, Y., Wang, Z. B., Zhang, X. Y., Dang, Y. R., Sun, L. L., Zhang, W. P., et al. (2021). Experimental evidence for long-term coexistence of copiotrophic and oligotrophic bacteria in pelagic surface seawater. Environ. Microbiol. 23, 1162–1173. doi: 10.1111/1462-2920.15321
Li, Q. Y., Zhu, C. L., Yu, J. B., Wu, X. Y., Huang, S. Q., Yang, F., et al. (2023). Response of soil bacteria of Dicranopteris dichotoma populations to vegetation restoration in red soil region of China. J. Soil Sci. Plant Nutr. 23, 456–468. doi: 10.1007/s42729-022-01058-6
Liang, Y., Pan, F., Ma, J., Yang, Z., and Yan, P. (2021). Long-term forest restoration influences succession patterns of soil bacterial communities. Environ. Sci. Pollut. Res. 28, 20598–20607. doi: 10.1007/s11356-020-11849-y
Liao, J., Dou, Y., Yang, X., and An, S. (2023). Soil microbial community and their functional genes during grassland restoration. J. Environ. Manag. 325:116488. doi: 10.1016/j.jenvman.2022.116488
Liu, G. Y., Chen, L. L., Shi, X. R., Yuan, Z. Y., Yuan, L. Y., Lock, T. R., et al. (2019). Changes in rhizosphere bacterial and fungal community composition with vegetation restoration in planted forests. Land Degrad. Dev. 30, 1147–1157. doi: 10.1002/ldr.3275
Liu, A., Contador, C. A., Fan, K., and Lam, H. M. (2018). Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 871:1860. doi: 10.3389/fpls.2018.01860
Liu, J., Le, T. H., Zhu, H., Yao, Y., Zhu, H., Cao, Y., et al. (2020). Afforestation of cropland fundamentally alters the soil fungal community. Plant Soil 457, 279–292. doi: 10.1007/s11104-020-04739-2
Liu, L., Zhou, G., Dang, P., Chen, J., Shang, H., Qiu, L., et al. (2022). Differences of soil fungal community structure under three afforestation modes in rocky desertification region of Western Hunan Province. Acta Ecol. Sin. 42, 4150–4159. doi: 10.3390/f15010089
Lu, Y., Lyu, M., Xiong, X., Deng, C., Jiang, Y., Zeng, M., et al. (2023). Understory ferns promote the restoration of soil microbial diversity and function in previously degraded lands. Sci. Total Environ. 870:161934. doi: 10.1016/j.scitotenv.2023.161934
Ma, F., Wang, C., Zhang, Y., Chen, J., Xie, R., and Sun, Z. (2022). Development of microbial indicators in ecological systems. Int. J. Environ. Res. Public Health 19:13888. doi: 10.3390/ijerph192113888
Mansourian, S., and Stephenson, P. J. (2023). Exploring challenges and lessons for monitoring Forest landscape restoration. Curr. Landsc. Ecol. Rep. 8, 159–170. doi: 10.1007/s40823-023-00092-z
Mirza, B. S., Mcglinn, D. J., Bohannan, B. J. M., Nüsslein, K., Tiedje, J. M., Rodrigues, J. L. M., et al. (2020). Diazotrophs show signs of restoration in Amazon rain forest soils with ecosystem rehabilitation. Appl. Environ. Microbiol. 86:e00195-20. doi: 10.1128/AEM.00195-20
Morales-Salazar, M. S., Vílchez-Alvarado, B., Chazdon, R. L., Ortiz-Malavasi, E., and Guevara-Bonilla, M. (2013). Corredor Biológico Osa, Costa Rica. Available online at: www.tec.ac.cr/revistaforestal (Accessed November 20, 2024).
Naka, M., Masumoto, S., Nishizawa, K., Matsuoka, S., Tatsumi, S., Kobayashi, Y., et al. (2023). Long-term consequences on soil fungal community structure: monoculture planting and natural regeneration. Environ. Manag. 73, 777–787. doi: 10.1007/s00267-023-01917-7
Novello, N., and Tonello, A. M. (2024). F-divergence based classification: beyond the use of cross-entropy, Proceedings of the 41st international conference on machine learning, 38448–38473.
O’Rourke, S. M., Angers, D. A., Holden, N. M., and Mcbratney, A. B. (2015). Soil organic carbon across scales. Glob. Chang. Biol. 21, 3561–3574. doi: 10.1111/gcb.12959
Ogram, A., Sayler, G. S., and Barkay, T. (1987). The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 7, 57–66. doi: 10.1016/0167-7012(87)90025-X
Pastorok, R. A., Macdonald, A., Sampson, J. R., Wilber, P., Yozzo, D. J., and Titre, J. P. (1997). An ecological decision framework for environmental restoration projects. Ecol. Eng. 9, 89–107. doi: 10.1016/S0925-8574(97)00036-0
Pedrinho, A., Mendes, L. W., de Araujo Pereira, A. P., Araujo, A. S. F., Vaishnav, A., Karpouzas, D. G., et al. (2024). Soil microbial diversity plays an important role in resisting and restoring degraded ecosystems. Plant Soil 500, 325–349. doi: 10.1007/s11104-024-06489-x
Philpott, T. J., Barker, J. S., Prescott, C. E., and Grayston, S. J. (2018). Limited effects of variable-retention harvesting on fungal communities decomposing fine roots in coastal temperate rainforests. Appl. Environ. Microbiol. 84:e02061-17. doi: 10.1128/AEM.02061-17
Qiu, Z., Shi, C., Zhao, M., Wang, K., Zhang, M., Wang, T., et al. (2022). Improving effects of afforestation with different forest types on soil nutrients and bacterial community in Barren Hills of North China. Sustainability (Switzerland) 14:1202. doi: 10.3390/su14031202
Rousk, J., Brookes, P. C., and Bååth, E. (2009). Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75, 1589–1596. doi: 10.1128/AEM.02775-08
Ruiz-Jaén, M. C., and Aide, T. M. (2005a). Restoration success: how is it being measured? Restor. Ecol. 13, 569–577. doi: 10.1111/j.1526-100X.2005.00072.x
Ruiz-Jaén, M. C., and Aide, T. M. (2005b). Vegetation structure, species diversity, and ecosystem processes as measures of restoration success. For. Ecol. Manag. 218, 159–173. doi: 10.1016/j.foreco.2005.07.008
Shannon, C. E. (1949). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Smenderovac, E. E., Webster, K., Caspersen, J., Morris, D., Hazlett, P., and Basiliko, N. (2017). Does intensified boreal forest harvesting impact soil microbial community structure and function? Can. J. For. Res. 47, 916–925. doi: 10.1139/cjfr-2016-0468
Stanturf, J. A. (2005). “What is forest restoration” in Restoration of boreal and temperate forests (Boca Raton: CRC Press), 3–11.
Stanturf, J. A., Palik, B. J., and Dumroese, R. K. (2014). Contemporary forest restoration: a review emphasizing function. For. Ecol. Manag. 331, 292–323. doi: 10.1016/j.foreco.2014.07.029
Taylor, B. N., Chazdon, R. L., and Menge, D. N. L. (2019). Successional dynamics of nitrogen fixation and forest growth in regenerating Costa Rican rainforests. Ecology 100:e02637. doi: 10.1002/ecy.2637
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA - an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Van Der Heijden, M. G. A., Bardgett, R. D., and Van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Vasconcellos, R. L. F., Zucchi, T. D., Taketani, R. G., Andreote, F. D., and Cardoso, E. J. B. N. (2014). Bacterial community characterization in the soils of native and restored rainforest fragments. Anton. Leeuw. Int. J. Gen. Mol. Microbiol. 106, 947–957. doi: 10.1007/s10482-014-0264-7
Wang, F., Li, Z., Fu, B., Lü, Y., Liu, G., Wang, D., et al. (2022). Short-term grazing exclusion alters soil bacterial co-occurrence patterns rather than community diversity or composition in temperate grasslands. Front. Microbiol. 13:824192. doi: 10.3389/fmicb.2022.824192
Wang, K., Zhang, Y., Tang, Z., Shangguan, Z., Chang, F., Jia, F., et al. (2019). Effects of grassland afforestation on structure and function of soil bacterial and fungal communities. Sci. Total Environ. 676, 396–406. doi: 10.1016/j.scitotenv.2019.04.259
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Heikki, S., van der Putten Wim, H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Wei, L., Bergeron, Y., De Frenne, P., Verheyen, K., Tian, L., Ren, H., et al. (2023). Above- and belowground composition and diversity of subtropical plantations and their relationships with soil nutrient stocks. Plant Soil 495, 235–252. doi: 10.1007/s11104-023-06317-8
Wortley, L., Hero, J. M., and Howes, M. (2013). Evaluating ecological restoration success: a review of the literature. Restor. Ecol. 21, 537–543. doi: 10.1111/rec.12028
Yan, D. F., Gellie, N. J. C., Mills, J. G., Connell, G., Bissett, A., Lowe, A. J., et al. (2020). A soil archaeal community responds to a decade of ecological restoration. Restor. Ecol. 28, 63–72. doi: 10.1111/rec.13033
Yan, G., Luo, X., Huang, B., Wang, H., Sun, X., Gao, H., et al. (2023). Assembly processes, driving factors, and shifts in soil microbial communities across secondary forest succession. Land Degrad. Dev. 34, 3130–3143. doi: 10.1002/ldr.4671
Yan, D. F., Mills, J. G., Gellie, N. J. C., Bissett, A., Lowe, A. J., and Breed, M. F. (2018). High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biol. Conserv. 217, 113–120. doi: 10.1016/j.biocon.2017.10.035
Yang, Y., Dou, Y., and An, S. (2018). Testing association between soil bacterial diversity and soil carbon storage on the loess plateau. Sci. Total Environ. 626, 48–58. doi: 10.1016/j.scitotenv.2018.01.081
Zhang, Z., Li, X., and Ji, X. (2023). Effects of afforestation on soil fungi in rocky mountain areas of North China. Land Degrad. Dev. 34, 5454–5467. doi: 10.1002/ldr.4857
Zhang, X., Liu, S., Huang, Y., and Fu, S. (2019). Changes on community structure and diversity of soil bacterial community during the succession of Quercus wutaishanica. Scientia Silvae Sinicae 55, 193–202.
Zhao, W. (2023). Progress and prospects in assessing the multidimensional environmental impacts of global vegetation restoration. Appl. Sci. 13:11426. doi: 10.3390/app132011426
Keywords: forest restoration, microorganisms, eDNA, soil, restoration evaluation, bacteria, fungi
Citation: Vivian J, Chazdon RL, Catling AA and Lee DJ (2025) Early evidences of links between soil microbes and forest restoration through multiple ecosystem metrics. Front. For. Glob. Change. 8:1540513. doi: 10.3389/ffgc.2025.1540513
Edited by:
Enrica Picariello, University of Sannio, ItalyReviewed by:
József Geml, Eszterházy Károly Catholic University, HungaryJesús Aguirre Gutiérrez, University of Oxford, United Kingdom
Copyright © 2025 Vivian, Chazdon, Catling and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenny Vivian, amVubnkudml2aWFuQHJlc2VhcmNoLnVzYy5lZHUuYXU=
†ORCID: Jenny Vivian, orcid.org/0000-0003-1518-3121
 Jenny Vivian
Jenny Vivian Robin L. Chazdon
Robin L. Chazdon Alexandra A. Catling
Alexandra A. Catling David J. Lee
David J. Lee