- Cloudera, Inc., Palo Alto, CA, United States
Precision public health is an emerging practice to more granularly predict and understand public health risks and customize treatments for more specific and homogeneous subpopulations, often using new data, technologies, and methods. Big data is one element that has consistently helped to achieve these goals, through its ability to deliver to practitioners a volume and variety of structured or unstructured data not previously possible. Big data has enabled more widespread and specific research and trials of stratifying and segmenting populations at risk for a variety of health problems. Examples of success using big data are surveyed in surveillance and signal detection, predicting future risk, targeted interventions, and understanding disease. Using novel big data or big data approaches has risks that remain to be resolved. The continued growth in volume and variety of available data, decreased costs of data capture, and emerging computational methods mean big data success will likely be a required pillar of precision public health into the future. This review article aims to identify the precision public health use cases where big data has added value, identify classes of value that big data may bring, and outline the risks inherent in using big data in precision public health efforts.
Introduction
This review article aims to identify the precision public health use cases where big data has added value, identify classes of value that big data may bring, and outline the risks inherent in using big data in precision public health efforts. This article focuses on surveying current practice, with a breadth of examples. The article does not include a critical review of the methods included in the big data and precision public health published research. It is hoped this article may pave the way for future researchers to measure the strengths and weaknesses, robustness, and validity of individual studies, interventions and outcomes. With the breadth of practice defined here, such follow-on in-depth critical review could identify precision public health best practices in design, methods, implementation, and analysis.
Methods
The terms “big data” and “precision public health”—two relatively new disciplines—often do not appear in the nomenclature of contemporary public health interventions and studies. Searching for the terms “big data” or “precision public health” returns a small fraction of the actual activity. Based on the lack of existing reviews and the complexity in identifying the intersection of precision public health and big data, the rationale of this narrative review article is to find examples of the use of big data in implementations of precision public health published in peer-reviewed academic journals. The author (a) reviewed a large number of public health studies to look for precision and big data, as well as related and follow-on studies, (b) identified and searched for specific types of big data being applied to public health, and (c) searched for uses of data in precision public health to identify big vs. small data—always using the definition of these terms rather than relying on the presence of the terms “big data” or “precision public health.”
Searches were performed using Google Scholar and Google. Examples of public health implementations—with and without big data—and precision public health implementations—with and without big data—only qualified for this article if they were published in peer-reviewed journals. In the presence of multiple qualifying examples, best attempts were made to limit examples to a single citation. In the presence of multiple examples, to reduce risk of bias and attempt to identify the most robust examples, the examples selected were those with the (a) most clearly identifiable public health use case, (b) clearest use of big data, (c) most “precision,” (d) in journals with the highest impact factor, that were (e) the most recent—and in that order of priority. Searches were concluded by July 20, 2017.
Search terms used were as follows:
1. For identifying implementations using big data volume, the term “public health” and each of the following: “big data,” “gene-wide,” “genome,” “genomic,” “germline,” “GWAS,” “imaging,” “molecular,” “multi-omic,” “pan-omic,” “phenome,” “PWAS,” “translational,” “video,” “whole exome,” and “whole genome.”
2. For identifying implementations using big data variety, the term “public health” and each of the following: “big data,” “drone,” “Facebook,” “Instagram,” “IoT,” “internet of things,” “linked,” “linked data,” “patient-centered,” “patient generated,” “mobile,” “mobile phone,” “registry,” “registries,” “secondary use,” “semantic,” “sensors,” “social media,” “surveys,” “Twitter,” “UAV,” “unmanned aerial vehicle,” “variety,” and “wearable.”
3. For identifying implementations using big data velocity, the term “public health” and each of the following: “big data,” “continuous,” “monitor,” “real-time,” “sensor,” “streams,” “streaming,” “velocity,” and “video.”
4. For identifying public health implementations—including programs, trials, innovations and experiments—using big data, the term “big data” and each of the following: “adverse drug event,” “ADE,” “adverse event,” “cohort,” “epidemic,” “epidemiology,” “health intervention,” “health risk,” “heterogeneous,” “homogeneous,” “human movement,” “outcomes,” “pandemic,” “pharmaco-epidemiology,” “population health,” “precision public health,” “prevention,” “public health,” “signal detection,” “surveillance,” “targeted intervention,” “tracking,” “vaccine,” “vector,” and “virus.”
Google Scholar also provides lists of more recent studies which have cited the current study. These lists were reviewed to identify if more recent studies existed that provided better examples of pertinent characteristics.
This method has a number of limitations. Google Scholar has limitations, including relying on the end user to discriminate which studies returned are from peer-reviewed journals. No review protocol exists independent of this review article. No study selection or summary measures were collected, and no meta-analysis was performed. No study characteristics were collected. No assessment of the validity of included studies was performed beyond their inclusion in peer-reviewed academic journals. No assessment of cumulative level bias risk was performed. No additional analysis methods were used. The selection of studies included was not independently reviewed. The scope of this narrative review precludes enumerating additional limitations. Limitations aside, the result of these methods is a collection of studies or programs where big data and precision public health—as these terms are defined in this article—are being used together. Through implementing these methods, this review article is the first to identify the scope and scale of big data’s role in precision public health, highlight classes of innovation, and identify the risks of using big data in this field.
Precision Public Health
“Precision public health is a new field driven by technological advances that enable more precise descriptions and analyzes of individuals and population groups, with a view to improving the overall health of populations” (1). The term was coined in Australia by Dr. Tarun Weeramanthri in 2013, and first found in print in 2014 (2). Dr. Muin Khoury and Dr. Sandro Galea describe precision public health as “improving the ability to prevent disease, promote health, and reduce health disparities in populations by applying emerging methods and technologies for measuring disease, pathogens, exposures, behaviors, and susceptibility in populations; and developing policies and targeted implementation programs to improve health” (3). Precision public health leverages big data and its enabling technologies to achieve a previously impossible level of targeting or speed (4). The Bill & Melinda Gates Foundation adds that precision public health “requires robust primary surveillance data, rapid application of sophisticated analytics to track the geographical distribution of disease, and the capacity to act on such information” (5). Precision public health works because “more-accurate methods for measuring disease, pathogens, exposures, behaviors, and susceptibility could allow better assessment of population health and development of policies and targeted programs for preventing disease” (4). Arnett & Claas add “Precision public health is characterized by discovering, validating, and optimizing care strategies for well-characterized population strata” (6). As for the size of the strata, Colijn et al. state “precision approaches must act at the right scale, which will often be intermediate—between “one size fits all” medicine and fully individualized therapies” (7).
The prominence of the term “precision” in the new practices of precision medicine and precision public health will invariably raise questions about their similarity. While precision medicine requires genetic, lifestyle, and environmental data to meet goals of more customized and potentially individualized clinical treatments, precision public health is about increased accuracy and granularity in defining public cohorts and delivering target interventions of many types (4–6). Precision medicine and precision public health are independent.
Big Data in Healthcare and Public Health
Big data has recently become a ubiquitous approach to driving insights, innovation and new interventions across economic sectors (8, 9). The United States National Institute of Standards and Technology defines big data as follows: “Big Data consists of extensive datasets—primarily in the characteristics of volume, variety, velocity, and/or variability—that require a scalable architecture for efficient storage, manipulation, and analysis,” (10). Decreases in costs of technology enabled the big data phenomenon to emerge (11). Data of “such a high volume, velocity and variety to require specific technology and analytical methods for its transformation into value” has a symbiotic relationship with the technology innovation on which it relies; the term big data often conflates the actual physical data with the unique technologies required to use it (12, 13).
In patient-specific healthcare, big data technology has helped enable greater scales of volume, variety and velocity (14, 15). Usable data volume has significantly increased in areas such as genomics (16, 17), molecular research (18, 19), medical image mining (20), and population health (21, 22). Enabling a variety of data to be integrated, for a more complete view of patient or population, has occurred in areas including air quality (23, 24), wearables (25, 26), patient generated content via the web (27), patient or physician movement (28, 29), medical studies (30), and critical care (31). Big data enabling increased velocity in healthcare was one of the earliest uses, in areas such as clinical prediction (32, 33), and diagnostics (15, 33). Current examples and future vision for use of big data exists in multiple and varying pathologies, including cancer (34), cardiology (35), epilepsy (36), family medicine (37), gastroenterology (38), nursing (39), pediatric ophthalmology (40), psychiatry (41, 42), and women’s health (43) as examples.
Barrett et al. state succinctly: “Big data can play a key role in both research and intervention activities and accelerate progress in disease prevention and population health” (44). Big data shows utility across the entire spectrum of public health disciplines. This capability ranges from “monitoring population health in real-time” to building “definitive extents and databases on the occurrence of many diseases” (45). Public health subject areas that include examples of the use of big data include community health (46), environmental health science (24, 47), epidemiology (48), infectious disease (45), maternal and child health (49), occupational health and safety (50), and nutrition (51). There is optimism and evidence for big data’s value in public health, both in research and in intervention (52).
Big Data in Precision Public Health
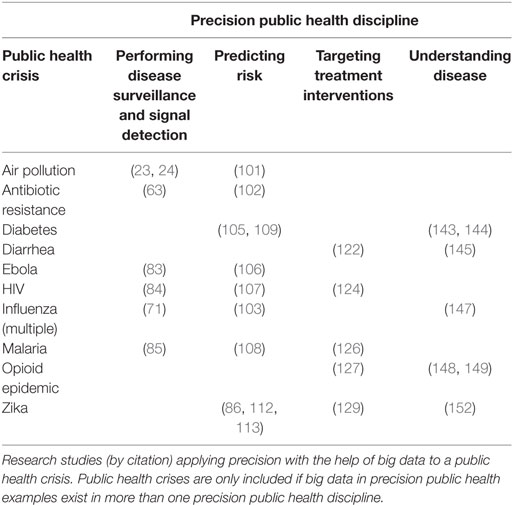
Today, use of big data has been shown to improve precision in select disciplines of public health. These areas include performing disease surveillance and signal detection (53, 54), predicting risk (55, 56), targeting interventions (6), and understanding disease (57). Research and proofs-of-concept with this data for these applications have been performed around the world. With the pace of technology innovation, and the speed at which precision health practitioners have embraced big data, there will likely be more public health disciplines, practices, approaches, and interventions implemented in the future or that are beyond the scope of this article (58, 59).
Performing Disease Surveillance and Signal Detection
Disease surveillance and signal detection are among the most commonly cited and revolutionary of the big data use cases in precision public health (45, 60–62). Precision signal detection or disease surveillance using big data has shown efficacy in air pollution (23, 24), antibiotic resistance (63), cholera (64), dengue (65, 66), drowning (67), drug safety (68, 69), electromagnetic field exposure (70), Influenza A H1N1 (71), Lyme disease (72), monitoring food intake (73), and whooping cough (74).
Disease surveillance often includes tracking affected individuals, i.e., human carriers, patients, or victims (75). Stoddard et al. stated in 2009: “Human movement is a critical, understudied behavioral component underlying the transmission dynamics of many vector-borne pathogens” (76). In the effort to track disease spread by human vectors, a premium is placed on information that is more recent and granular (77, 78). Thus, access to huge volumes of streaming real-time data generated by humans seems at once an ideal signal repository for identifying and tracking affected individuals, and definitionally big data (78).
Indeed, big data supports alternate and in some ways superior methods to track affected individuals (45, 62). Because affected individuals move so quickly and at such a wide range, the real-time capabilities of big data and big data technology are now critical in this discipline (79, 80). Studies have shown efficacy using mobile phone data in tracking movement in cholera (81), dengue (82), Ebola (83), human immunodeficiency virus (HIV) (84), malaria (85), rubella (85), and schistosomiasis (86). Other mechanisms that have shown efficacy or promise in tracking movement of affected individuals include air travel data (87), GPS data-loggers (88), magnetometers (89), Twitter (71), and web searches (65).
Predicting Risk
Effective signal detection often leads to attempts to predict future signals (90, 91). Predicting public health risk leads to a chance to implement preventive interventions (56, 92). Models predicting either disease spread or outcomes, using traditional or non-big data sources, have been developed across the spectrum of public health crises, including dengue (93), HIV (94), influenza (95), malaria (96), Rift Valley Fever (97), and tuberculosis (98).
One early example of using big data for public health prediction, Google Flu Trends, was a well-publicized failure (99). Since that episode, approaches to predicting risk using the internet and social media have shown special care to include merging big data with non-social media data sources, avoid overfitting models with relatively few cases, and being conscious of the risks of big data (56, 100).
Big data has been used for risk prediction of spread or outcomes in public health topics such as air pollution (101), antibiotic resistance (102), avian influenza A (103), blood lead levels (104), child abuse (49), diabetes (105), Ebola (106), HIV (107), malaria (108), gestational diabetes (109), smoking progression (110), West Nile (111), and Zika (86, 112, 113).
Targeting Treatment Interventions
Applying treatment interventions to homogeneous cohorts within a larger heterogeneous population has been advocated since Lalonde’s seminal report “A New Perspective on the Health of Canadians” in 1974 (114). Historical examples of adding precision to public health treatment populations include gonorrhea in the 1980s (115), HIV in the 1990s (116), breast cancer in the 2000s (117), and malaria in the 2010s (118). In 2010, the US Department of Health and Human Services said of those citizens with multiple chronic conditions: “Indeed, developing means for determining homogeneous subgroups among this heterogeneous population is viewed as an important step in the effort to improve the health status of the total population” (119).
Big data was leveraged in public health research identifying finer-grain treatment interventions in childhood asthma (120), childhood obesity (121), diarrhea (122), Hepatitis C (123), HIV (124), injectable drug use (125), malaria (126), opioid medication misuse (127), use of smokeless tobacco (128), and the Zika virus (129).
One clinical example at the intersection of identifying subpopulations for effective interventions and big data is personalized vaccinology or “vaccinomics” (130). Most vaccines today are applied in a one-size fits all model: the typical implementation assumes a homogenous population, uses the same vaccine and dosages for all patients, ignores replicated, empirical realities of a heterogeneous population, and does not use sophisticated genomic capabilities at hand (131, 132). While today’s vaccines are applied homogeneously, the results are individual: “The response to a vaccine is the cumulative result of non-random interactions with host genes, epigenetic phenomena, metagenomics and the microbiome, gene dominance, complementarity, epistasis, coinfections, and other factors” (133). Vaccinomics would focus on homogeneous subpopulations treated with vaccines, dosages and approaches that would “hold the promise of moving away from one standard vaccine against all human populations…to one where vaccines can be relatively easily tailor-fitted to individual, community and population specificity” (134).
Understanding Disease
Data volume and variety in epidemiology have grown consistently over time well before the age of big data (135–137). Contemporary exponential increases in data sizes, and perhaps more importantly increases in variety of data sources, make big data a valuable addition to the epidemiologist’s toolkit (64, 138). Glymour states “We recommend that social epidemiologists take advantage of recent revolutionary improvements in data availability and computing power to examine new hypotheses and expand our repertoire of study designs” (139). Big data may have added relevance in study designs that are patient-centric and precision-oriented (140).
“Person-oriented approaches, in contrast, focus on differences between individuals as characterized by configurations and patterns of variables. This is well in line with a precision-medicine approach to understanding disease risk, resilience, and treatment response in subpopulations of individuals” (140).
Big data is a component in studies that have shown new precision characteristics of such public health concerns as cholera (141), chikungunya (142), diabetes (143, 144), diarrhea (145), heatwave (146), influenza (147), opioid epidemic (148, 149), preterm birth (150), stunting (151), and Zika (152).
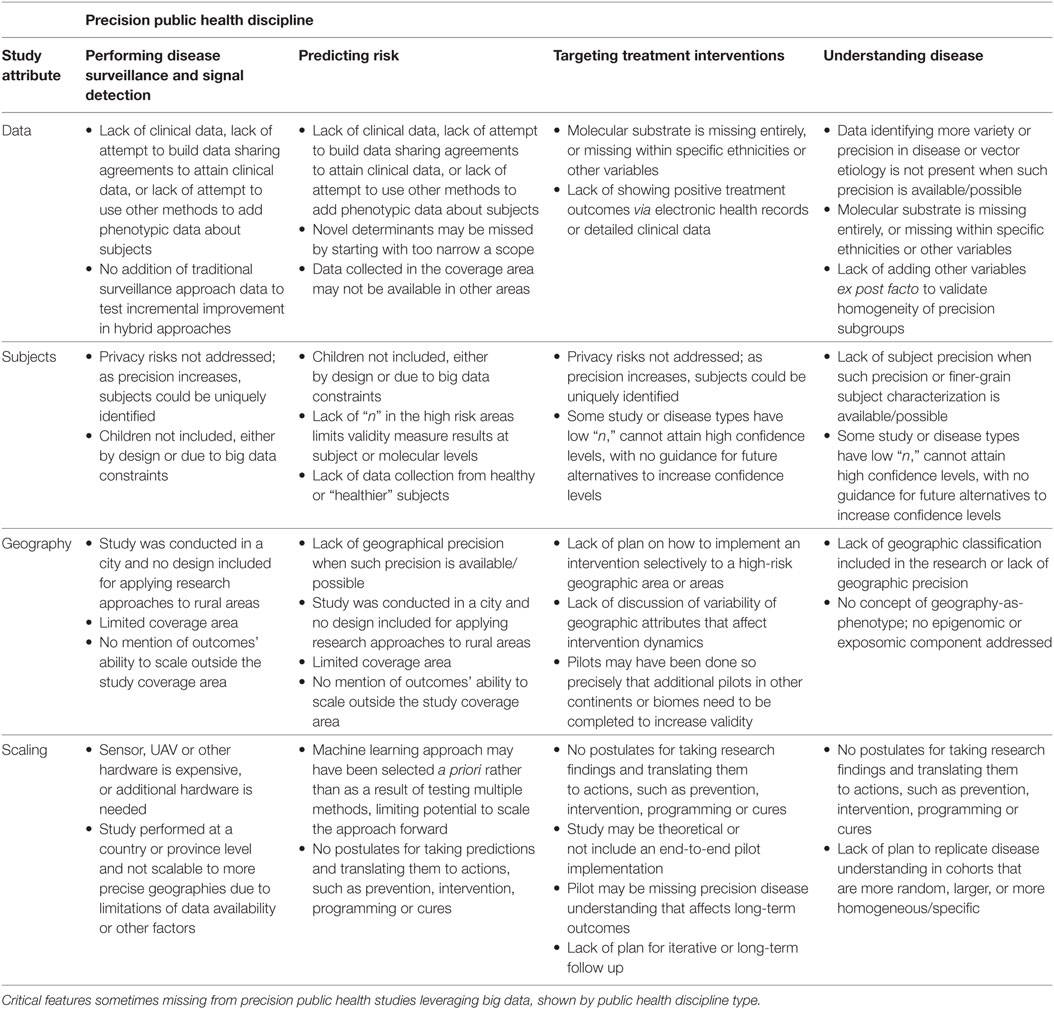
Table 1 summarizes the public health crises cited previously for which exists peer-reviewed research in at least two of the four precision public health disciplines. While the precision health research in Table 1 and in this article has peer-reviewed and exhaustive methods, there are some opportunity gaps that future research should consider and include. Table 2 lists critical gaps that occasionally exist in the research, grouped by precision public health discipline.
Contributions of Big Data
Big data offers special contributions to precision public health in enabling a wider view of health variables through linking disparate or novel data (44, 153, 154) and enabling large study populations with volumes of multiomic data to identify “molecular cohorts” (155).
The technologies behind big data make it much easier to integrate a variety of data within a study (156). For example, because big data does not require investment in an a priori data schema, users can bring together a variety of different data and link it when the analytics are created (157). This enables researchers to link a mélange of unstructured disease and outcome data (158, 159). In their 2017 study, Harry Hemingway, in their completion of 33 studies using linked data with a total population of two million patients, said “Our findings clearly show that research using one of the NHS greatest assets—its data—is vital to innovate improvements in disease prevention, to make earlier diagnoses and to give the best treatments” (160). The inclusion of data variety increases the number of independent variables; one novel variable—or a combination of as yet uncompared variables—could end up being significant in defining relevant precision subpopulations (161, 162).
Examples of data that has been linked to help identify more precise cohorts of populations include: longitudinal health claims data (163, 164); secondary use anonymized electronic health records (159, 165); cohort studies, health surveys, and registries (166–168); environmental variables (104); molecular data such as from the genome, exposome, microbiome, or transcriptome (169–172); “mhealth” wearable and sensor data (173); mobile phone sensing data and self-reports (174); online patient generated content (175); and the semantic web (176).
The explosion of new volumes of genomic “big data” helped make possible the precision medicine movement (177). One of precision medicine’s promises was to lead to development of new treatments for subpopulations defined by their similarities at the molecular level (178, 179). Currently, translational efforts in precision medicine often work by identifying cohorts of patients who have or lack specific genomic or molecular biomarkers (132, 180). Since today’s precision medicine works at the granularity of disease subtypes and population strata and not at the “n of one” level, contemporary precision medicine really is—when applied to community crises—an example of precision public health (2).
Researchers agree that only by using very large sample sizes will genomic studies have the proper statistical power (181, 182). “These large case–control studies are essential for boosting the statistical power needed to detect the genetic variants responsible for rare diseases and can provide the necessary knowledge for use in the clinical setting,” (183). Big data has been a necessary component in the scale-up of genomic sample sizes, enabled by the decrease in cost of gene sequencing (183). Future versions of sovereign genomics programs in over ten countries have the potential to create data sets with millions of samples (184–186). These databases should be ideal platforms for research such as genome wide association studies, which have been used with over ten thousand cases per study in public health diseases such as Alzheimer’s disease (25,000+ cases), autism (16,000 cases), high blood pressure (200,000+ cases), posttraumatic stress disorder (10,000+ cases), and smoking (50,000+ cases) (187–191).
The most sophisticated precision approaches to public health today at once include data from multiple omic disciplines, can make use of linked phenotype data, and leverage novel or recent types of computation (7, 132, 192, 193). In targeting interventions, de novo or improved computational methods like geospatial risk modeling, latent class modeling, social molecular pathological epidemiology, and agent-based modeling simulation all benefit from big data to better identify these “intermediate” subpopulations (49, 122, 126, 193–196).
Risks
More work needs to be done both enumerating and evaluating the risks and challenges of using big data in precision public health.
1. Individuals could be stigmatized, even when not singularly identified, when they are stratified into small, observable cohorts, where they cannot maintain a “concealable stigmatized identity” (197).
2. Big data could enable non-consented individuals to identify patients’ or citizens’ identities either due to small cohorts or by “drilling through” the deeper and wider set of population data (198–200).
3. There are known drawbacks in increased reliance on a “high-risk” strategy, as originated by Rose, including ignoring population level determinants of health; taking focus away from a radical campaign that could have more sustainable positive effect for a larger population; risking missed interventions to borderline cases; or encouraging behaviors that continue to exist outside of social norms (201).
4. Big data risks targeting only relatively wealthier communities where data can be collected, or where big data expertise or distribution technologies are endemic (72, 202, 203).
5. For data collected through social media, crowdsourcing or similar channels, there may be more data about, in or from urban centers or areas of dense population, which will require additional computational governance (64).
6. Prevalence of large volumes of new types of individual health information available digitally risks that it could fall into the hands of unregulated commercial enterprises, or of insurance companies (204).
7. Experiencing governance gaps due to default use of existing governing legislation, rules or principles designed for data and technologies “that have now been superseded” by big data calls for more regulation (16, 205).
8. Applying novel big data without the appropriate controls, clinical interpretation, or statistical governance could lead to model overfitting, lack of accuracy, or results like Google Flu Trends, and could damage public faith in big data’s ability to add precision to public health or trust in contributing their own data (99, 206–208).
9. Big data brings unique challenges in data quality. Cai and Zhu created a big data quality framework with no less than 14 attributes by which any big data’s robustness should be assessed. Ignoring qualities like timeliness, accuracy, completeness or reliability leads to research weakness (209).
10. Performing healthcare research that includes big data is marked by, and needs, larger teams of diverse practitioners, often including informaticians, data scientists, computer scientists, physicians, researchers, and more—potentially leading to fewer studies and the challenges inherent in collaborating in large teams (59, 173).
11. Research that includes big data with high “variety” or linked data is likely to include a higher median number of data sources, which could require increased investment in cleaning and curating the data—resulting in slower scientific progress—or could compel the challenges of analyzing high dimensional data (210). For example, the high dimensionality of data found in both molecular and linked data incurs specific risk. Alyass et al. believe this data is “prone to high rates of false-positives due to chance alone…this requires researchers to adjust for multiple testing to control for type 1 error rates…or reduce dimensionality via sparse methods” (211).
Conclusion
Precision public health is exciting. Today’s public health programs can achieve new levels of speed and accuracy not plausible a decade ago. Adding precision to many parts of public health engagement has led and will lead to tangible benefits. Precision can enable public health programs to maintain the same efficacy while decreasing costs, or hold costs constant while delivering better, smarter, faster, and different education, cures and interventions, saving lives.
Precision public health does not require big data. That said, the future of big data in precision public health is assured, based on its successes and acceleration of use to date. Big data and the methods created to make it useful allow precision public health practitioners to operate at the top of their license and can bring more insight to cohort membership, disease pathways and treatments. Big data enables lower costs and more precision to find, educate, track, and help each high-risk citizen. In the future, precision public health needs, imperatives, mandates and techniques will drive new capabilities into big data.
Using big data in precision public health has risks. A number of risks were identified here and future study will expand these or identify more. Protecting the dignity, privacy, security of citizens and patients, while finding truly meaningful significant outcomes in a reasonable timeframe will take effort on the part of each and every researcher in this space.
What are the calls to action? Investment has increased, but additional investment and research are needed in many areas. First, more experimentation is needed to understand how to best create and mobilize open data, open science, open source communities, and open collaboration platforms. For context, the Observational Health Data Sciences and Informatics collaborative is a thriving global open science community focused on large scale population health outcomes and prediction. If such a collaborative existed for precision public health, one imagines practitioners could leverage shared best practices, data, open software, and opportunities. Second, there are opportunity gaps in training precision public health workers in countries with a dearth of data scientists, on-premise data storage and computational assets, or access to big data. For example, communities suffering public health crises increasingly desire to “learn how to use the information and improve their ability to respond to future outbreaks in the region,” rather than having their data removed for analysis by better funded nations (212). Third, follow-on research is needed in the area of big data in precision public health. Specifically, (a) best practices in performing data quality assessment along a broad range of attributes should be enumerated, (b) existing research should be scored along these attributes as well as those studies’ compliance with statistical best practices specific to big data and high dimensionality, (c) each area of value delivery—disease surveillance, predicting risk, targeting intervention and understanding disease—needs their own full treatment with regard to methods, data sources, data management, and more, (d) some critical framework ought to be created and proposed to systematically measure precision public health studies and programs, specific to and beyond big data, and (e) as precision public health becomes more mature, emerging trends should be noticed and evaluated. Fourth, more work is needed in areas of ethics, risk, and governance. The community should be watching for overreliance on big data-driven approaches that lead to decreases in radical whole-population solutions that increase baseline health norms. Fifth, the global economic opportunity of using big data prescriptively in public health has not been systematically measured, beyond specific country or disease successes. For context, organizations such as the United Nations, the World Bank, and the United States Agency for International Development have estimated economic impacts of individual epidemics. These or other institutions could convene a task force to estimate the economic benefit of applying precision to public health responses, as well as the relative contribution of big data. Sixth, precision public health centers of excellence in universities can help. Today, leaders in schools of public health are speaking and writing about precision public health; presumably academic courses, concentrations and centers will follow in stepwise progression. Seventh, new technical innovation must continue and needs investment. For example, this could include applying deep learning to precision public health use cases, or creating a novel free and open source data science software “pipeline” for geospatial event prediction.
Future precision public health will be transformative. It will include new applications, modifications, and uses of today’s assets, including social media and communication platforms, unmanned aerial vehicles, mobile applications, mobile sequencing, self-screening, sensors, vaccine or drug internet-of-things inventions, and more. Tomorrow, we could be looking up, wondering if a high-resolution satellite is mapping our neighborhood to predict the path of an infectious disease, or if a drone is approaching with a targeted intervention. With future applications of precision public health and the speed of big data adoption, tomorrow’s new public health students and young practitioners soon won’t think of the discipline as precision public health. They will only think of it as public health.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author is employed by Cloudera, Inc., a provider of big data technology.
References
1. Baynam G, Bauskis A, Pachter N, Schofield L, Verhoef H, Palmer RL, et al. 3-Dimensional facial analysis—facing precision public health. Front Public Health (2017) 5:31. doi:10.3389/fpubh.2017.00031
2. Severi G, Southey MC, English DR, Jung CH, Lonie A, McLean C, et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat (2014) 148(3):665–73. doi:10.1007/s10549-014-3209-y
3. Khoury MJ, Galea S. Will precision medicine improve population health? JAMA (2016) 316(13):1357–8. doi:10.1001/jama.2016.12260
4. Khoury MJ, Iademarco MF, Riley WT. Precision public health for the era of precision medicine. Am J Prev Med (2016) 50(3):398. doi:10.1016/j.amepre.2015.08.031
5. Dowell SF, Blazes D, Desmond-Hellmann S. Four steps to precision public health. Nat News (2016) 540(7632):189. doi:10.1038/540189a
6. Arnett DK, Claas SA. Precision medicine, genomics, and public health. Diabetes Care (2016) 39(11):1870–3. doi:10.2337/dc16-1763
7. Colijn C, Jones N, Johnston IG, Yaliraki S, Barahona M. Toward precision healthcare: context and mathematical challenges. Front Physiol (2017) 8:136. doi:10.3389/fphys.2017.00136
8. LaValle S, Lesser E, Shockley R, Hopkins MS, Kruschwitz N. Big data, analytics and the path from insights to value. MIT Sloan Manage Rev (2011) 52(2):21.
9. Lohr S. The age of big data. N Y Times (2012) 11(2012):SR1. Available from: http://www.nytimes.com/2012/02/12/sunday-review/big-datas-impact-in-the-world.html (Accessed on February 26, 2017).
10. National Institute of Standards and Technology. NIST Big Data Interoperability Framework: Volume 1, Definitions (NIST Special Publication 1500-1). (2015). Available from: http://nvlpubs.nist.gov/nistpubs/SpecialPublications/NIST.SP.1500-1.pdf
11. Cukier K, Mayer-Schoenberger V. The rise of big data: how it’s changing the way we think about the world. Foreign Aff (2013) 92:28. doi:10.2469/dig.v43.n4.65
12. De Mauro A, Greco M, Grimaldi M. What is big data? A consensual definition and a review of key research topics. In: Giannakopoulos G, Sakas DP, Kyriaki-Manessi D, editors. AIP Conference Proceedings, Vol. 1644. Madrid: AIP (2015). p. 97–104. doi:10.1063/1.4907823
13. Hu H, Wen Y, Chua TS, Li X. Toward scalable systems for big data analytics: a technology tutorial. IEEE Access (2014) 2:652–87. doi:10.1109/ACCESS.2014.2332453
14. Andreu-Perez J, Poon CC, Merrifield RD, Wong ST, Yang GZ. Big data for health. IEEE J Biomed Health Inform (2015) 19(4):1193–208. doi:10.1109/JBHI.2015.2450362
15. Belle A, Thiagarajan R, Soroushmehr SM, Navidi F, Beard DA, Najarian K. Big data analytics in healthcare. Biomed Res Int (2015) 2015:370194. doi:10.1155/2015/370194
16. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature (2015) 518(7538):197–206. doi:10.1038/nature14177
17. Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet (2012) 90(1):7–24. doi:10.1016/j.ajhg.2011.11.029
18. Altaf-Ul-Amin M, Afendi FM, Kiboi SK, Kanaya S. Systems biology in the context of big data and networks. Biomed Res Int (2014) 2014:11. doi:10.1155/2014/428570
19. Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature (2014) 509(7502):582–7. doi:10.1038/nature13319
20. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology (2015) 278(2):563–77. doi:10.1148/radiol.2015151169
21. Hripcsak G, Ryan PB, Duke JD, Shah NH, Park RW, Huser V, et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A (2016) 113(27):7329–36. doi:10.1073/pnas.1510502113
22. Slobogean GP, Giannoudis PV, Frihagen F, Forte ML, Morshed S, Bhandari M. Bigger data, bigger problems. J Orthop Trauma (2015) 29:S43–6. doi:10.1097/BOT.0000000000000463
23. Predić B, Yan Z, Eberle J, Stojanovic D, Aberer K. Exposuresense: integrating daily activities with air quality using mobile participatory sensing. Pervasive Computing and Communications Workshops (PERCOM Workshops), 2013 IEEE International Conference. IEEE (2013). p. 303–5.
24. Zheng Y, Liu F, Hsieh HP. U-air: When urban air quality inference meets big data. Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM (2013). p. 1436–44.
25. Chen M, Zhang Y, Li Y, Hassan MM, Alamri A. AIWAC: Affective interaction through wearable computing and cloud technology. IEEE Wireless Commun (2015) 22(1):20–7. doi:10.1109/MWC.2015.7054715
26. Jiang P, Winkley J, Zhao C, Munnoch R, Min G, Yang LT. An intelligent information forwarder for healthcare big data systems with distributed wearable sensors. IEEE Syst J (2016) 10(3):1147–59. doi:10.1109/JSYST.2014.2308324
27. Martínez P, Martínez JL, Segura-Bedmar I, Moreno-Schneider J, Luna A, Revert R. Turning user generated health-related content into actionable knowledge through text analytics services. Comput Industry (2016) 78:43–56. doi:10.1016/j.compind.2015.10.006
28. Frisby J, Smith V, Traub S, Patel VL. Contextual computing: a Bluetooth based approach for tracking healthcare providers in the emergency room. J Biomed Inform (2017) 65:97–104. doi:10.1016/j.jbi.2016.11.008
29. Qi B, Miao H, Yuan X, Xiao X. A patient tracking and positioning system based on improved DV-Hop algorithm. Information and Communication Technology Convergence (ICTC), 2015 International Conference on. IEEE (2015). p. 1297–9.
30. Gorenshteyn D, Zaslavsky E, Fribourg M, Park CY, Wong AK, Tadych A, et al. Interactive big data resource to elucidate human immune pathways and diseases. Immunity (2015) 43(3):605–14. doi:10.1016/j.immuni.2015.08.014
31. Celi LA, Mark RG, Stone DJ, Montgomery RA. “Big data” in the intensive care unit. Closing the data loop. Am J Respir Crit Care Med (2013) 187(11):1157–66. doi:10.1164/rccm.201212-2311ED
32. Bar-Or A, Healey J, Kontothanassis L, Van Thong JM. BioStream: a system architecture for real-time processing of physiological signals. Engineering in Medicine and Biology Society, 2004. IEMBS’04. 26th Annual International Conference of the IEEE. (Vol. 2), IEEE (2004). p. 3101–4.
33. Ahmad S, Ramsay T, Huebsch L, Flanagan S, McDiarmid S, Batkin I, et al. Continuous multi-parameter heart rate variability analysis heralds onset of sepsis in adults. PLoS One (2009) 4(8):e6642. doi:10.1371/journal.pone.0006642
34. Shaikh AR, Butte AJ, Schully SD, Dalton WS, Khoury MJ, Hesse BW. Collaborative biomedicine in the age of big data: the case of cancer. J Med Internet Res (2014) 16(4):e101. doi:10.2196/jmir.2496
35. Dilsizian SE, Siegel EL. Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep (2014) 16(1):1–8. doi:10.1007/s11886-013-0441-8
36. Ben-Menachem E. Epilepsy in 2015: the year of collaborations for big data. Lancet Neurol (2016) 15(1):6. doi:10.1016/S1474-4422(15)00356-7
37. Phillips RL Jr, Bazemore AW, DeVoe JE, Weida TJ, Krist AH, Dulin MF, et al. A family medicine health technology strategy for achieving the triple aim for US health care. Fam Med (2015) 47(8):628.
38. Wooden B, Goossens N, Hoshida Y, Friedman SL. Using big data to discover diagnostics and therapeutics for gastrointestinal and liver diseases. Gastroenterology (2017) 152(1):53–67. doi:10.1053/j.gastro.2016.09.065
39. Westra BL, Clancy TR, Sensmeier J, Warren JJ, Weaver C, Delaney CW. Nursing knowledge: big data science—implications for nurse leaders. Nurs Adm Q (2015) 39(4):304–10. doi:10.1097/NAQ.0000000000000130
40. Clark A, Ng JQ, Morlet N, Semmens JB. Big data and ophthalmic research. Surv Ophthalmol (2016) 61(4):443–65. doi:10.1016/j.survophthal.2016.01.003
41. McIntyre RS, Cha DS, Jerrell JM, Swardfager W, Kim RD, Costa LG, et al. Advancing biomarker research: utilizing ‘Big Data’ approaches for the characterization and prevention of bipolar disorder. Bipolar Disord (2014) 16(5):531–47. doi:10.1111/bdi.12162
42. Passos IC, Mwangi B, Kapczinski F. Big data analytics and machine learning: 2015 and beyond. Lancet Psychiatry (2016) 3(1):13–5. doi:10.1016/S2215-0366(15)00549-0
43. Stein P, Falco L, Kuebler F, Annaheim S, Lemkaddem A, Delgado-Gonzalo R, et al. Digital womens health based on wearables and big data. Fertil Steril (2016) 106(3):e113. doi:10.1016/j.fertnstert.2016.07.339
44. Barrett MA, Humblet O, Hiatt RA, Adler NE. Big data and disease prevention: from quantified self to quantified communities. Big data (2013) 1(3):168–75. doi:10.1089/big.2013.0027
45. Hay SI, George DB, Moyes CL, Brownstein JS. Big data opportunities for global infectious disease surveillance. PLoS Med (2013) 10(4):e1001413. doi:10.1371/journal.pmed.1001413
46. Consolvo S, McDonald DW, Toscos T, Chen MY, Froehlich J, Harrison B, et al. Activity sensing in the wild: a field trial of UbiFit garden In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM (2008). p. 1797–806. doi:10.1145/1357054.1357335
47. Braem B, Latre S, Leroux P, Demeester P, Coenen T, Ballon P. Designing a smart city playground: real-time air quality measurements and visualization in the city of things testbed. Smart Cities Conference (ISC2), 2016 IEEE International. IEEE (2016). p. 1–2.
48. Lardon J, Abdellaoui R, Bellet F, Asfari H, Souvignet J, Texier N, et al. Adverse drug reaction identification and extraction in social media: a scoping review. J Med Internet Res (2015) 17(7):e171. doi:10.2196/jmir.4304
49. Daley D, Bachmann M, Bachmann BA, Pedigo C, Bui MT, Coffman J. Risk terrain modeling predicts child maltreatment. Child Abuse Negl (2016) 62:29–38. doi:10.1016/j.chiabu.2016.09.014
50. Bragazzi NL, Dini G, Toletone A, Brigo F, Durando P. Leveraging big data for exploring occupational diseases-related interest at the level of scientific community, media coverage and novel data streams: the example of silicosis as a pilot study. PLoS One (2016) 11(11):e0166051. doi:10.1371/journal.pone.0166051
51. Hood L, Lovejoy JC, Price ND. Integrating big data and actionable health coaching to optimize wellness. BMC Med (2015) 13(1):4. doi:10.1186/s12916-014-0238-7
52. Burke-Garcia A, Scally G. Trending now: future directions in digital media for the public health sector. J Public Health (2014) 36(4):527–34. doi:10.1093/pubmed/fdt125
53. Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. Big data for infectious disease surveillance and modeling. J Infect Dis (2016) 214(Suppl 4):S375–9. doi:10.1093/infdis/jiw400
54. O’Shea J. Digital disease detection: a systematic review of event-based internet biosurveillance systems. Int J Med Inform (2017) 101:15–22. doi:10.1016/j.ijmedinf.2017.01.019
55. Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet (2016) 17(7):392–406. doi:10.1038/nrg.2016.27
56. Gandon S, Day T, Metcalf CJE, Grenfell BT. Forecasting epidemiological and evolutionary dynamics of infectious diseases. Trends Ecol Evol (2016) 31(10):776–88. doi:10.1016/j.tree.2016.07.010
57. Schneeweiss S. Improving therapeutic effectiveness and safety through big healthcare data. Clin Pharmacol Ther (2016) 99(3):262–5. doi:10.1002/cpt.316
58. Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med (2016) 375(13):1216. doi:10.1056/NEJMp1606181
59. Gu D, Li J, Li X, Liang C. Visualizing the knowledge structure and evolution of big data research in healthcare informatics. Int J Med Inform (2017) 98:22–32. doi:10.1016/j.ijmedinf.2016.11.006
60. Eysenbach G. Infodemiology and infoveillance: framework for an emerging set of public health informatics methods to analyze search, communication and publication behavior on the Internet. J Med Internet Res (2009) 11(1):e11. doi:10.2196/jmir.1157
61. Nsoesie EO, Brownstein JS. Computational approaches to influenza surveillance: beyond timeliness. Cell Host Microbe (2015) 17(3):275–8. doi:10.1016/j.chom.2015.02.004
62. Salathé M. Digital pharmacovigilance and disease surveillance: combining traditional and big-data systems for better public health. J Infect Dis (2016) 214(Suppl_4):S399–403. doi:10.1093/infdis/jiw281
63. MacFadden DR, Fisman D, Andre J, Ara Y, Majumder MS, Bogoch II, et al. A platform for monitoring regional antimicrobial resistance, using online data sources: resistanceopen. J Infect Dis (2016) 214(Suppl 4):S393–8. doi:10.1093/infdis/jiw343
64. Chunara R, Andrews JR, Brownstein JS. Social and news media enable estimation of epidemiological patterns early in the 2010 Haitian cholera outbreak. Am J Trop Med Hyg (2012) 86(1):39–45. doi:10.4269/ajtmh.2012.11-0597
65. Chan EH, Sahai V, Conrad C, Brownstein JS. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Negl Trop Dis (2011) 5(5):e1206. doi:10.1371/journal.pntd.0001206
66. Gomide J, Veloso A, Meira W Jr, Almeida V, Benevenuto F, Ferraz F, et al. Dengue surveillance based on a computational model of spatio-temporal locality of Twitter. Proceedings of the 3rd International Web Science Conference. ACM (2011). 3 p.
67. Claesson A, Svensson L, Nordberg P, Ringh M, Rosenqvist M, Djarv T, et al. Drones may be used to save lives in out of hospital cardiac arrest due to drowning. Resuscitation (2017) 114:152–6. doi:10.1016/j.resuscitation.2017.01.003
68. Correia RB, Li L, Rocha LM. Monitoring potential drug interactions and reactions via network analysis of instagram user timelines. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. (Vol. 21), NIH Public Access (2016). 492 p.
69. Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T, et al. Digital drug safety surveillance: monitoring pharmaceutical products in twitter. Drug Safety (2014) 37(5):343–50. doi:10.1007/s40264-014-0155-x
70. Joseph W, Aerts S, Vandenbossche M, Thielens A, Martens L. Drone based measurement system for radiofrequency exposure assessment. Bioelectromagnetics (2016) 37:195–9. doi:10.1002/bem.21964
71. Signorini A, Segre AM, Polgreen PM. The use of Twitter to track levels of disease activity and public concern in the US during the influenza A H1N1 pandemic. PLoS One (2011) 6(5):e19467. doi:10.1371/journal.pone.0019467
72. Pesälä S, Virtanen MJ, Sane J, Jousimaa J, Lyytikäinen O, Murtopuro S, et al. Health care professionals’ evidence-based medicine internet searches closely mimic the known seasonal variation of lyme borreliosis: a register-based study. JMIR Public Health Surveill (2017) 3(2):e19. doi:10.2196/publichealth.6764
73. Alajajian SE, Williams JR, Reagan AJ, Alajajian SC, Frank MR, Mitchell L, et al. The lexicocalorimeter: gauging public health through caloric input and output on social media. PLoS One (2017) 12(2):e0168893. doi:10.1371/journal.pone.0168893
74. Ghosh S, Chakraborty P, Nsoesie EO, Cohn E, Mekaru SR, Brownstein JS, et al. Temporal topic modeling to assess associations between news trends and infectious disease outbreaks. Sci Rep (2017) 7:40841. doi:10.1038/srep40841
75. Wilson ME. Travel and the emergence of infectious diseases. Emerg Infect Dis (1995) 1(2):39. doi:10.3201/eid0102.950201
76. Stoddard ST, Morrison AC, Vazquez-Prokopec GM, Soldan VP, Kochel TJ, Kitron U, et al. The role of human movement in the transmission of vector-borne pathogens. PLoS Negl Trop Dis (2009) 3(7):e481. doi:10.1371/journal.pntd.0000481
77. Aarestrup FM, Koopmans MG. Sharing data for global infectious disease surveillance and outbreak detection. Trends Microbiol (2016) 24(4):241–5. doi:10.1016/j.tim.2016.01.009
78. Simonsen L, Gog JR, Olson D, Viboud C. Infectious disease surveillance in the big data era: towards faster and locally relevant systems. J Infect Dis (2016) 214(Suppl 4):S380–5. doi:10.1093/infdis/jiw376
79. Kraemer MU, Hay SI, Pigott DM, Smith DL, Wint GW, Golding N. Progress and challenges in infectious disease cartography. Trends Parasitol (2016) 32(1):19–29. doi:10.1016/j.pt.2015.09.006
80. Tatem AJ. Mapping population and pathogen movements. Int Health (2014) 6(1):5–11. doi:10.1093/inthealth/ihu006
81. Finger F, Genolet T, Mari L, de Magny GC, Manga NM, Rinaldo A, et al. Mobile phone data highlights the role of mass gatherings in the spreading of cholera outbreaks. Proc Natl Acad Sci U S A (2016) 113(23):6421–6. doi:10.1073/pnas.1522305113
82. Wesolowski A, Qureshi T, Boni MF, Sundsøy PR, Johansson MA, Rasheed SB, et al. Impact of human mobility on the emergence of dengue epidemics in Pakistan. Proc Natl Acad Sci U S A (2015) 112:11887–92. doi:10.1073/pnas.1504964112
83. Wesolowski A, Buckee CO, Bengtsson L, Wetter E, Lu X, Tatem AJ. Commentary: containing the ebola outbreak - the potential and challenge of mobile network data. PLoS Curr (2014) 6. doi:10.1371/currents.outbreaks.0177e7fcf52217b8b634376e2f3efc5e
84. Isdory A, Mureithi EW, Sumpter DJ. The impact of human mobility on HIV transmission in Kenya. PLoS One (2015) 10:e0142805. doi:10.1371/journal.pone.0142805
85. Wesolowski A, Buckee CO, Engø-Monsen K, Metcalf CJE. Connecting mobility to infectious diseases: the promise and limits of mobile phone data. J Infect Dis (2016) 214(Suppl 4):S414–20. doi:10.1093/infdis/jiw273
86. Mari L, Gatto M, Ciddio M, Dia ED, Sokolow SH, De Leo GA, et al. Big-data-driven modeling unveils country-wide drivers of endemic schistosomiasis. Sci Rep (2017) 7:489. doi:10.1038/s41598-017-00493-1
87. Huff A, Allen T, Whiting K, Breit N, Arnold B. FLIRT-ing with Zika: a web application to predict the movement of infected travelers validated against the current Zika virus epidemic. PLoS Curr (2016) 8. doi:10.1371/currents.outbreaks.711379ace737b7c04c89765342a9a8c9
88. Vazquez-Prokopec GM, Bisanzio D, Stoddard ST, Paz-Soldan V, Morrison AC, Elder JP, et al. Using GPS technology to quantify human mobility, dynamic contacts and infectious disease dynamics in a resource-poor urban environment. PLoS One (2013) 8(4):e58802. doi:10.1371/journal.pone.0058802
89. Nguyen KA, Watkins C, Luo Z. Co-location epidemic tracking on London public transports using low power mobile magnetometer. arXiv preprint arXiv (2017):1704.00148. doi:10.1109/IPIN.2017.8115963
90. Gubler DJ. Surveillance for dengue and dengue hemorrhagic fever. Bull Pan Am Health Organ (1989) 23(4):397–404.
91. Langmuir AD. William Farr: founder of modern concepts of surveillance. Int J Epidemiol (1976) 5(1):13–8. doi:10.1093/ije/5.1.13
92. Godman B, Finlayson AE, Cheema PK, Zebedin-Brandl E, Gutiérrez-Ibarluzea I, Jones J, et al. Personalizing health care: feasibility and future implications. BMC Med (2013) 11(1):179. doi:10.1186/1741-7015-11-179
93. Naish S, Dale P, Mackenzie JS, McBride J, Mengersen K, Tong S. Climate change and dengue: a critical and systematic review of quantitative modelling approaches. BMC Infect Dis (2014) 14(1):167. doi:10.1186/1471-2334-14-167
94. Isham V. Mathematical modelling of the transmission dynamics of HIV infection and AIDS: a review. J Royal Stat Soc Ser A (Stat Soc) (1988) 151(1):5–30. doi:10.2307/2982179
95. Nsoesie EO, Brownstein JS, Ramakrishnan N, Marathe MV. A systematic review of studies on forecasting the dynamics of influenza outbreaks. Influenza Other Respi Viruses (2014) 8(3):309–16. doi:10.1111/irv.12226
96. Zinszer K, Verma AD, Charland K, Brewer TF, Brownstein JS, Sun Z, et al. A scoping review of malaria forecasting: past work and future directions. BMJ Open (2012) 2(6):e001992. doi:10.1136/bmjopen-2012-001992
97. Linthicum KJ, Anyamba A, Tucker CJ, Kelley PW, Myers MF, Peters CJ. Climate and satellite indicators to forecast rift valley fever epidemics in Kenya. Science (1999) 285(5426):397–400. doi:10.1126/science.285.5426.397
98. Ozcaglar C, Shabbeer A, Vandenberg SL, Yener B, Bennett KP. Epidemiological models of Mycobacterium tuberculosis complex infections. Math Biosci (2012) 236(2):77–96. doi:10.1016/j.mbs.2012.02.003
99. Lazer D, Kennedy R, King G, Vespignani A. The parable of Google flu: traps in big data analysis. Science (2014) 343(6176):1203–5. doi:10.1126/science.1248506
100. Yang S, Santillana M, Brownstein JS, Gray J, Richardson S, Kou SC. Using electronic health records and Internet search information for accurate influenza forecasting. BMC Infect Dis (2017) 17(1):332. doi:10.1186/s12879-017-2424-7
101. Chen J, Chen H, Wu Z, Hu D, Pan JZ. Forecasting smog-related health hazard based on social media and physical sensor. Inf Syst (2017) 64:281–91. doi:10.1016/j.is.2016.03.011
102. Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, et al. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep (2016) 6:27930. doi:10.1038/srep27930
103. Gilbert M, Golding N, Zhou H, Wint GW, Robinson TP, Tatem AJ, et al. Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia. Nat Commun (2014) 5:4116. doi:10.1038/ncomms5116
104. Sadler RC, LaChance J, Hanna-Attisha M. Social and built environmental correlates of predicted blood lead levels in the flint water crisis. Am J Public Health (2017) 107(5):763–9. doi:10.2105/AJPH.2017.303692
105. Phan TP, Alkema L, Tai ES, Tan KH, Yang Q, Lim WY, et al. Forecasting the burden of type 2 diabetes in Singapore using a demographic epidemiological model of Singapore. BMJ Open Diabetes Res Care (2014) 2(1):e000012. doi:10.1136/bmjdrc-2013-000012
106. Liu X, Speranza E, Muñoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, et al. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol (2017) 18(1):4. doi:10.1186/s13059-016-1137-3
107. Ireland ME, Schwartz HA, Chen Q, Ungar LH, Albarracín D. Future-oriented tweets predict lower county-level HIV prevalence in the United States. Health Psychol (2015) 34S:1252–60. doi:10.1037/hea0000279
108. Franke J, Gebreslasie M, Bauwens I, Deleu J, Siegert F. Earth observation in support of malaria control and epidemiology: MALAREO monitoring approaches. Geospat Health (2015) 10(1):335. doi:10.4081/gh.2015.335
109. White SL, Lawlor DA, Briley AL, Godfrey KM, Nelson SM, Oteng-Ntim E, et al. Early antenatal prediction of gestational diabetes in obese women: development of prediction tools for targeted intervention. PLoS One (2016) 11(12):e0167846. doi:10.1371/journal.pone.0167846
110. Pugach O, Cannon DS, Weiss RB, Hedeker D, Mermelstein RJ. Classification tree analysis as a method for uncovering relations between CHRNA5A3B4 and CHRNB3A6 in predicting smoking progression in adolescent smokers. Nicotine Tob Res (2017) 19(4):410–6. doi:10.1093/ntr/ntw197
111. Chuang TW, Wimberly MC. Remote sensing of climatic anom alies and West Nile virus incidence in the northern Great Plains of the United States. PLoS One (2012) 7(10):e46882. doi:10.1371/journal.pone.0046882
112. Bogoch II, Brady OJ, Kraemer MU, German M, Creatore MI, Brent S, et al. Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis (2016) 16(11):1237–45. doi:10.1016/S1473-3099(16)30270-5
113. McGough SF, Brownstein JS, Hawkins JB, Santillana M. Forecasting Zika incidence in the 2016 Latin America outbreak combining traditional disease surveillance with search, social media, and news report data. PLoS Negl Trop Dis (2017) 11(1):e0005295. doi:10.1371/journal.pntd.0005295
114. Lalonde M. A New Perspective on the Health of Canadians (The Lalonde Report). Ottawa: Minister of Supply and Services Canada (1974).
115. Hethcote HW, Yorke JA, Nold A. Gonorrhea modeling: a comparison of control methods. Math Biosci (1982) 58(1):93–109. doi:10.1016/0025-5564(82)90053-0
116. Richert CA, Peterman TA, Zaidi AA, Ransom RL, Wroten JE, Witte JJ. A method for identifying persons at high risk for sexually transmitted infections: opportunity for targeting intervention. Am J Public Health (1993) 83(4):520–4. doi:10.2105/AJPH.83.4.520
117. Gomez SL, Tan S, Keegan TH, Clarke CA. Disparities in mammographic screening for Asian women in California: a cross-sectional analysis to identify meaningful groups for targeted intervention. BMC Cancer (2007) 7(1):201. doi:10.1186/1471-2407-7-201
118. Bousema T, Griffin JT, Sauerwein RW, Smith DL, Churcher TS, Takken W, et al. Hitting hotspots: spatial targeting of malaria for control and elimination. PLoS Med (2012) 9(1):e1001165. doi:10.1371/journal.pmed.1001165
119. U.S. Department of Health and Human Services. Multiple Chronic Conditions—A Strategic Framework: Optimum Health and Quality of Life for Individuals with Multiple Chronic Conditions. Washington, DC: U.S. Department of Health and Human Services (2010).
120. Hose AJ, Depner M, Illi S, Lau S, Keil T, Wahn U, et al. Latent class analysis reveals clinically relevant atopy phenotypes in 2 birth cohorts. J Allergy ClinImmunol (2017) 139(6):1935–45. doi:10.1016/j.jaci.2016.08.046
121. Koning M, Hoekstra T, de Jong E, Visscher TL, Seidell JC, Renders CM. Identifying developmental trajectories of body mass index in childhood using latent class growth (mixture) modelling: associations with dietary, sedentary and physical activity behaviors: a longitudinal study. BMC Public Health (2016) 16(1):1128. doi:10.1186/s12889-016-3757-7
122. Lal A. Spatial modelling tools to integrate public health and environmental science, illustrated with infectious cryptosporidiosis. Int J Environ Res Public Health (2016) 13(2):186. doi:10.3390/ijerph13020186
123. Fraenkel L, Lim J, Garcia-Tsao G, Reyna V, Monto A, Bridges JF. Variation in treatment priorities for chronic hepatitis C: a latent class analysis. Patient (2016) 9(3):241. doi:10.1007/s40271-015-0147-7
124. Barral MF, Sousa AK, Santos AF, Abreu CM, Tanuri A, Soares MA. Identification of novel resistance-related polymorphisms in HIV-1 subtype C RT connection and RNAse H domains from patients under virological failure in Brazil. AIDS Res Hum Retroviruses (2017) 33(5):465–71. doi:10.1089/AID.2015.0376
125. Roth AM, Armenta RA, Wagner KD, Roesch SC, Bluthenthal RN, Cuevas-Mota J, et al. Patterns of drug use, risky behavior, and health status among persons who inject drugs living in San Diego, California: a latent class analysis. Subst Use Misuse (2015) 50(2):205–14. doi:10.3109/10826084.2014.962661
126. Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat Rev Microbiol (2014) 12(12):833–40. doi:10.1038/nrmicro3364
127. Cochran G, Hruschak V, Bacci JL, Hohmeier KC, Tarter R. Behavioral, mental, and physical health characteristics and opioid medication misuse among community pharmacy patients: a latent class analysis. Res Soc Admin Pharm (2016) 13(6):1055–61. doi:10.1016/j.sapharm.2016.11.005
128. Fu Q, Vaughn MG. A latent class analysis of smokeless tobacco use in the United States. J Community Health (2016) 41(4):850–7. doi:10.1007/s10900-016-0163-0
129. Castro LA, Fox SJ, Chen X, Liu K, Bellan SE, Dimitrov NB, et al. Assessing real-time Zika risk in the United States. BMC Infect Dis (2017) 17(1):284. doi:10.1186/s12879-017-2394-9
130. Poland GA, Ovsyannikova IG, Jacobson RM. Application of pharmacogenomics to vaccines. Pharmacogenomics(2009) 10(5):837–52. doi:10.2217/PGS.09.25
131. Pellegrino P, Falvella FS, Cheli S, Perrotta C, Clementi E, Radice S. The role of toll-like receptor 4 polymorphisms in vaccine immune response. Pharmacogenomics J (2016) 16(1):96–101. doi:10.1038/tpj.2015.21
132. Poland GA. The case for personalized vaccinology in the 21st century. Presented at the National Vaccine Advisory Committee Meeting on February 7th, 2017. (2017) Available from: https://www.hhs.gov/sites/default/files/poland_presentation.pdf
133. Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther (2007) 82(6):653–64. doi:10.1038/sj.clpt.6100415
134. Nandy A, Basak SC. Viral epidemics and vaccine preparedness. J Mol Pathol Epidemiol (2017) 2:S1.
135. Arzberger P, Schroeder P, Beaulieu A, Bowker G, Casey K, Laaksonen L, et al. Promoting access to public research data for scientific, economic, and social development. Data Sci J (2004) 3:135–52. doi:10.2481/dsj.3.135
136. Hammond EC, Irwin J, Garfinkel L. Data-processing and analysis in epidemiological research. Am J Public Health Nations Health (1967) 57(11):1979–84. doi:10.2105/AJPH.57.11.1979
137. Lopez AD. The evolution of the global burden of disease framework for disease, injury and risk factor quantification: developing the evidence base for national, regional and global public health action. Global Health (2005) 1(1):5. doi:10.1186/1744-8603-1-5
138. Kao RR, Haydon DT, Lycett SJ, Murcia PR. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol (2014) 22(5):282–91. doi:10.1016/j.tim.2014.02.011
139. Glymour MM, Osypuk TL, Rehkopf DH. Invited commentary: off-roading with social epidemiology—exploration, causation, translation. Am J Epidemiol (2013) 178(6):858–63. doi:10.1093/aje/kwt145
140. Johnson SB, Little TD, Masyn K, Mehta PD, Ghazarian SR. Multidisciplinary design and analytic approaches to advance prospective research on the multilevel determinants of child health. Ann Epidemiol (2017) 27(6):361–70. doi:10.1016/j.annepidem.2017.05.008
141. Geethanjali C, Bhanumathi S. Generating drug-gene association for Vibrio cholerae using ontological profile similarity. Indian J Sci Technol (2016) 9(33). doi:10.17485/ijst/2016/v9i33/99620
142. Rujirojindakul P, Chongsuvivatwong V, Limprasert P. Association of ABO blood group phenotype and allele frequency with chikungunya fever. Adv Hematol (2015) 2015:543027. doi:10.1155/2015/543027
143. Ross MC, Muzny DM, McCormick JB, Gibbs RA, Fisher-Hoch SP, Petrosino JF. 16S gut community of the Cameron County Hispanic cohort. Microbiome (2015) 3(1):7. doi:10.1186/s40168-015-0072-y
144. Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes (2017) 66(11):2888–902. doi:10.2337/db16-1253
145. Bustamante M, Standl M, Bassat Q, Vilor-Tejedor N, Medina-Gomez C, Bonilla C, et al. A genome-wide association meta-analysis of diarrhoeal disease in young children identifies FUT2 locus and provides plausible biological pathways. Hum Mol Genet (2016) 25(18):4127–42. doi:10.1093/hmg/ddw264
146. Xiao J, Spicer T, Jian L, Yun GY, Shao C, Nairn J, et al. Variation in population vulnerability to heat wave in Western Australia. Front Public Health (2017) 5:64. doi:10.3389/fpubh.2017.00064
147. Barber MF, Elde NC. Escape from bacterial iron piracy through rapid evolution of transferrin. Science (2014) 346(6215):1362–6. doi:10.1126/science.1259329
148. Kringel D, Ultsch A, Zimmermann M, Jansen JP, Ilias W, Freynhagen R, et al. Emergent biomarker derived from next-generation sequencing to identify pain patients requiring uncommonly high opioid doses. Pharmacogenomics J (2016) 17(5):419–26. doi:10.1038/tpj.2016.28
149. Smith AH, Jensen KP, Li J, Nunez Y, Farrer LA, Hakonarson H, et al. Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Mol Psychiatry (2017) 22(3):346–52. doi:10.1038/mp.2016.257
150. Newnham JP, Kemp MW, White SW, Arrese CA, Hart RJ, Keelan JA. Applying precision public health to prevent preterm birth. Front Public Health (2017) 5:66. doi:10.3389/fpubh.2017.00066
151. Danaei G, Andrews KG, Sudfeld CR, Fink G, McCoy DC, Peet E, et al. Risk factors for childhood stunting in 137 developing countries: a comparative risk assessment analysis at global, regional, and country levels. PLoS Med (2016) 13(11):e1002164. doi:10.1371/journal.pmed.1002164
152. Faria NR, da Silva Azevedo RDS, Kraemer MU, Souza R, Cunha MS, Hill SC, et al. Zika virus in the Americas: early epidemiological and genetic findings. Science (2016) 352(6283):345–9. doi:10.1126/science.aaf5036
153. Hansen M, de Klerk N, Stewart L, Bower C, Milne E. Linked data research: a valuable tool in the ART field. Hum Reprod (2015) 30(12):2956–7. doi:10.1093/humrep/dev247
154. Millett ER, Quint JK, De Stavola BL, Smeeth L, Thomas SL. Improved incidence estimates from linked vs. stand-alone electronic health records. J Clin Epidemiol (2016) 75:66–9. doi:10.1016/j.jclinepi.2016.01.005
155. Saleheen D, Zhao W, Young R, Nelson CP, Ho WK, Ferguson JF, et al. Loss of cardio-protective effects at the ADAMTS7 locus due to gene-smoking interactions. Circulation (2017) 135(24):2336–53. doi:10.1161/CIRCULATIONAHA.116.022069
156. Strohbach M, Daubert J, Ravkin H, Lischka M. Big data storage. In: New Horizons for a Data-Driven Economy. Cham: Springer International Publishing (2016). p. 119–41. doi:10.1007/978-3-319-21569-3_7
157. Brennan PF, Bakken S. Nursing needs big data and big data needs nursing. J Nurs Scholarsh (2015) 47(5):477–84. doi:10.1111/jnu.12159
158. Miani C, Robin E, Horvath V, Manville C, Cave J, Chataway J. Health and healthcare: assessing the real world data policy landscape in Europe. Rand Health Q (2014) 4(2):15.
159. Bonner S, McGough AS, Kureshi I, Brennan J, Theodoropoulos G, Moss L, et al. Data quality assessment and anomaly detection via map/reduce and linked data: a case study in the medical domain. Big Data (Big Data), 2015 IEEE International Conference. IEEE (2015). p. 737–46.
160. Hemingway H, Feder GS, Fitzpatrick NK, Denaxas S, Shah AD, Timmis AD. Using nationwide ‘big data’ from linked electronic health records to help improve outcomes in cardiovascular diseases: 33 studies using methods from epidemiology, informatics, economics and social science in the ClinicAl disease research using LInked Bespoke studies and Electronic health Records (CALIBER) programme. Programme Grants Appl Res (2017) 5(4): doi:10.3310/pgfar05040
161. Collyer ML, Sekora DJ, Adams DC. A method for analysis of phenotypic change for phenotypes described by high-dimensional data. Heredity (2015) 115(4):357–65. doi:10.1038/hdy.2014.75
162. Mooney SJ, Westreich DJ, El-Sayed AM. Epidemiology in the era of big data. Epidemiology (2015) 26(3):390. doi:10.1097/EDE.0000000000000274
163. Lin KJ, Schneeweiss S. Considerations for the analysis of longitudinal electronic health records linked to claims data to study the effectiveness and safety of drugs. Clin Pharmacol Ther (2016) 100(2):147–59. doi:10.1002/cpt.359
164. Setiawan VW, Virnig BA, Porcel J, Henderson BE, Le Marchand L, Wilkens LR, et al. Linking data from the multiethnic cohort study to medicare data: linkage results and application to chronic disease research. Am J Epidemiol (2015) 181(11):917–9. doi:10.1093/aje/kwv055
165. Finlayson SG, LePendu P, Shah NH. Building the graph of medicine from millions of clinical narratives. Sci Data (2014) 1:140032. doi:10.1038/sdata.2014.32
166. Hall ES, Goyal NK, Ammerman RT, Miller MM, Jones DE, Short JA, et al. Development of a linked perinatal data resource from state administrative and community-based program data. Matern Child Health J (2014) 18(1):316–25. doi:10.1007/s10995-013-1236-7
167. Kent EE, Malinoff R, Rozjabek HM, Ambs A, Clauser SB, Topor MA, et al. Revisiting the surveillance epidemiology and end results cancer registry and Medicare health outcomes survey (SEER-MHOS) linked data resource for patient-reported outcomes research in older adults with cancer. J Am Geriatr Soc (2016) 64(1):186–92. doi:10.1111/jgs.13888
168. Sanmartin C, Decady Y, Trudeau R, Dasylva A, Tjepkema M, Finès P, et al. Linking the Canadian community health survey and the Canadian mortality database: an enhanced data source for the study of mortality. Health Rep (2016) 27(12):10.
169. Croes K, De Coster S, De Galan S, Morrens B, Loots I, Van de Mieroop E, et al. Health effects in the Flemish population in relation to low levels of mercury exposure: from organ to transcriptome level. Int J Hyg Environ Health (2014) 217(2):239–47. doi:10.1016/j.ijheh.2013.06.004
170. Findley K, Williams DR, Grice EA, Bonham VL. Health disparities and the microbiome. Trends Microbiol (2016) 24(11):847–50. doi:10.1016/j.tim.2016.08.001
171. Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr (2013) 98(1):111–20. doi:10.3945/ajcn.112.056689
172. Rozek LS, Dolinoy DC, Sartor MA, Omenn GS. Epigenetics: relevance and implications for public health. Annu Rev Public Health (2014) 35:105–22. doi:10.1146/annurev-publhealth-032013-182513
173. Carreiro S, Chai PR, Carey J, Chapman B, Boyer EW. Integrating personalized technology in toxicology: sensors, smart glass, and social media applications in toxicology research. J Med Toxicol (2017) 13(2):166–72. doi:10.1007/s13181-017-0611-y
174. Triantafyllidis AK, Velardo C, Salvi D, Shah SA, Koutkias VG, Tarassenko L. A survey of mobile phone sensing, self-reporting, and social sharing for pervasive healthcare. IEEE J Biomed Health Inform (2017) 21(1):218–27. doi:10.1109/JBHI.2015.2483902
175. Ji X, Chun SA, Cappellari P, Geller J. Linking and using social media data for enhancing public health analytics. J Inform Sci (2017) 43(2):221–45. doi:10.1177/0165551515625029
176. Xie L, Draizen EJ, Bourne PE. Harnessing big data for systems pharmacology. Annu Rev Pharmacol Toxicol (2017) 57:245–62. doi:10.1146/annurev-pharmtox-010716-104659
177. Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Eng J Med (2012) 366(6):489–91. doi:10.1056/NEJMp1114866
178. Fradkin JE, Hanlon MC, Rodgers GP. NIH Precision Medicine Initiative: implications for diabetes research. Diabetes Care (2016) 39(7):1080–4. doi:10.2337/dc16-0541
179. Gligorijević V, Malod-Dognin N, Pržulj N. Integrative methods for analyzing big data in precision medicine. Proteomics (2016) 16(5):741–58. doi:10.1002/pmic.201500396
180. Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer (2016) 16(8):525–37. doi:10.1038/nrc.2016.56
181. Burton PR, Hansell AL, Fortier I, Manolio TA, Khoury MJ, Little J, et al. Size matters: just how big is BIG? Quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol (2009) 38(1):263–73. doi:10.1093/ije/dyn147
182. Ma’n HZ, Junker A, Knoppers BM, Rahimzadeh V. Streamlining review of research involving humans: Canadian models. J Med Genet (2015) 52(8):566–9. doi:10.1136/jmedgenet-2014-102640
183. Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol (2013) 425(21):4047–63. doi:10.1016/j.jmb.2013.08.008
184. Althani A. Qatar biobank and Qatar genome programs road map. J Tissue Sci Eng (2015) 6:157. doi:10.4172/2157-7552.1000157
185. Nimmesgern E, Benediktsson I, Norstedt I. Personalized medicine in Europe. Clin Transl Sci (2017) 10(2):61–3. doi:10.1111/cts.12446
186. Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big data: astronomical or genomical? PLoS Biol (2015) 13(7):e1002195. doi:10.1371/journal.pbio.1002195
187. Escott-Price V, Bellenguez C, Wang L-S, Choi S-H, Harold D, Jones L, et al. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS One (2014) 9(6):e94661. doi:10.1371/journal.pone.0094661
188. Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet (2016) 48(10):1171–84. doi:10.1038/ng.3667
189. Justice AE, Winkler TW, Feitosa MF, Graff M, Fisher VA, Young K, et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat Commun (2017) 8:14977. doi:10.1038/ncomms14977
190. The Autism Spectrum Working Group of the Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24. 32 and a significant overlap with schizophrenia. Mol Autism (2017) 8:21. doi:10.1186/s13229-017-0137-9
191. Chen CY, Stein M, Ursano R, Cai T, Gelernter J, Heeringa S, et al. Genome-wide association study of posttraumatic stress disorder symptom domains in two cohorts of United States army soldiers. Biol Psychiatry (2017) 81(10):S91–2. doi:10.1016/j.biopsych.2017.02.236
192. Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol (2016) 52(3):265–75. doi:10.1007/s00535-016-1272-3
193. Nishi A, Milner DA Jr, Giovannucci EL, Nishihara R, Tan AS, Kawachi I, et al. Integration of molecular pathology, epidemiology and social science for global precision medicine. Expert Rev Mol Diagn (2016) 16(1):11–23. doi:10.1586/14737159.2016.1115346
194. Jung T, Wickrama KAS. An introduction to latent class growth analysis and growth mixture modeling. Soc Personality Psychol Compass (2008) 2(1):302–17. doi:10.1111/j.1751-9004.2007.00054.x
195. Speybroeck N, Van Malderen C, Harper S, Müller B, Devleesschauwer B. Simulation models for socioeconomic inequalities in health: a systematic review. Int J Environ Res Public Health (2013) 10(11):5750–80. doi:10.3390/ijerph10115750
196. Zhang X, Pérez-Stable EJ, Bourne PE, Peprah E, Duru OK, Breen N, et al. Big data science: opportunities and challenges to address minority health and health disparities in the 21st Century. Ethn Dis (2017) 27(2):95–106. doi:10.18865/ed.27.2.95
197. Quinn DM, Chaudoir SR. Living with a concealable stigmatized identity: the impact of anticipated stigma, centrality, salience, and cultural stigma on psychological distress and health. J Pers Soc Psychol (2009) 97(4):634. doi:10.1037/a0015815
198. Narayanan A, Shmatikov V. Myths and fallacies of personally identifiable information. Commun ACM (2010) 53(6):24–6. doi:10.1145/1743546.1743558
199. Ohm P. Broken promises of privacy: responding to the surprising failure of anonymization. Ucla L Rev (2009) 57:1701.
200. Sweeney L. Weaving technology and policy together to maintain confidentiality. The J Law Med Ethics (1997) 25(2-3):98–110. doi:10.1111/j.1748-720X.1997.tb01885.x
201. Rose G. Sick individuals and sick populations. Int J Epidemiol (2001) 30(3):427–32. doi:10.1093/ije/30.3.427
203. Lupton D. Health promotion in the digital era: a critical commentary. Health Promot Int (2015) 30(1):174–83. doi:10.1093/heapro/dau091
204. Kostkova P, Brewer H, de Lusignan S, Fottrell E, Goldacre B, Hart G, et al. Who owns the data? Open data for healthcare. Front Public Health (2016) 4:7. doi:10.3389/fpubh.2016.00007
205. Vayena E, Salathé M, Madoff LC, Brownstein JS. Ethical challenges of big data in public health. PLoS Comput Biol (2015) 11(2):e1003904. doi:10.1371/journal.pcbi.1003904
206. Belgrave D, Henderson J, Simpson A, Buchan I, Bishop C, Custovic A. Disaggregating asthma: big investigation versus big data. J Allergy ClinImmunol (2017) 139(2):400–7. doi:10.1016/j.jaci.2016.11.003
207. Mascalzoni D, Dove ES, Rubinstein Y, Dawkins HJ, Kole A, McCormack P, et al. International charter of principles for sharing bio-specimens and data. Eur J Hum Genet (2015) 23(6):721. doi:10.1038/ejhg.2014.197
208. Santillana M, Nguyen AT, Louie T, Zink A, Gray J, Sung I, et al. Cloud-based electronic health records for real-time, region-specific influenza surveillance. Sci Rep (2016) 6:25732. doi:10.1038/srep25732
209. Cai L, Zhu Y. The challenges of data quality and data quality assessment in the big data era. Data Sci J (2015) 14:2. doi:10.5334/dsj-2015-002
210. Johnstone IM, Titterington DM. Statistical challenges of high-dimensional data. Philos Trans A Math Phys Eng Sci (2009) 367:4237–53. doi:10.1098/rsta.2009.0159
211. Alyass A, Turcotte M, Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genomics (2015) 8(1):33. doi:10.1186/s12920-015-0108-y
Keywords: precision public health, big data, computational epidemiology, infectious disease surveillance, precision population health
Citation: Dolley S (2018) Big Data’s Role in Precision Public Health. Front. Public Health 6:68. doi: 10.3389/fpubh.2018.00068
Received: 25 July 2017; Accepted: 20 February 2018;
Published: 07 March 2018
Edited by:
Hugh J. S. Dawkins, Government of Western Australia Department of Health, AustraliaReviewed by:
Gareth Baynam, Genetic Services of Western Australia, AustraliaDavid Preen, University of Western Australia, Australia
Ori Gudes, University of New South Wales, Australia
Emmanuel D. Jadhav, Ferris State University, United States
Copyright: © 2018 Dolley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shawn Dolley, c2hhd24uZG9sbGV5QGdtYWlsLmNvbQ==
 Shawn Dolley
Shawn Dolley