- 1The First Affiliated Hospital, College of Clinical Medicine of Henan University of Science and Technology, Luoyang, China
- 2School of Medical Technology and Engineering, Henan University of Science and Technology, Luoyang, China
- 3Clinical Laboratory Management Center, Luoyang Center for Disease Control and Prevention, Luoyang, Henan, China
- 4College of Animal Science and Technology, Henan University of Science and Technology, Luoyang, China
Introduction: This study aimed to investigate the prevalence of resistance to first-line anti-tuberculosis (TB) drugs and the molecular mechanisms underlying resistance mutations in patients with culture-positive Mycobacterium tuberculosis complex (MTBC). The findings provide a data basis for developing more precise and regionally tailored anti-TB treatment regimens.
Methods: From 2015 to 2022, a total of 3,605 strains isolated from 10 designated TB medical institutions in the main urban and county/township areas of Luoyang City, China, were confirmed as MTBC members through polymerase chain reaction (PCR) targeting a specific insertion sequence IS6110. Drug susceptibility testing using the proportional method was performed to analyze resistance patterns to first-line anti-TB drugs, namely, isoniazid (INH), rifampin (RFP), streptomycin (SM), and ethambutol (EMB). Molecular drug susceptibility testing was conducted on resistant strains using multicolor melting curve analysis (MMCA) to determine the mutation mechanisms associated with phenotypic resistance.
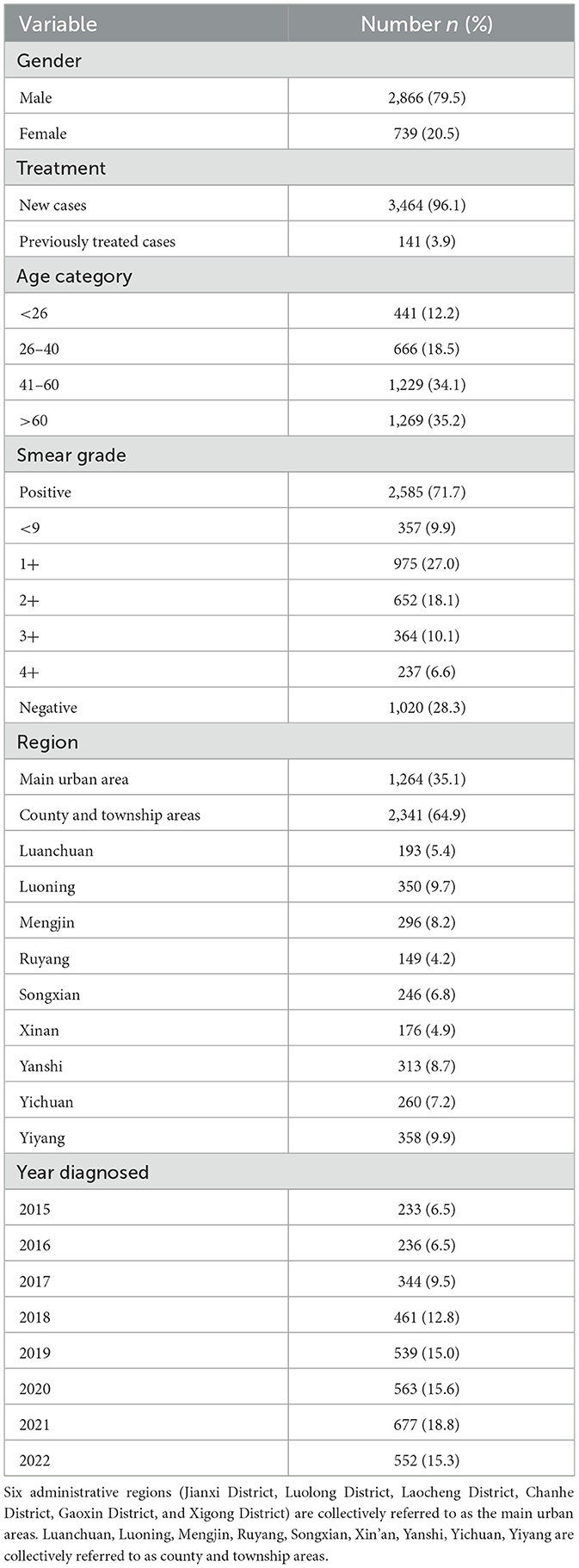
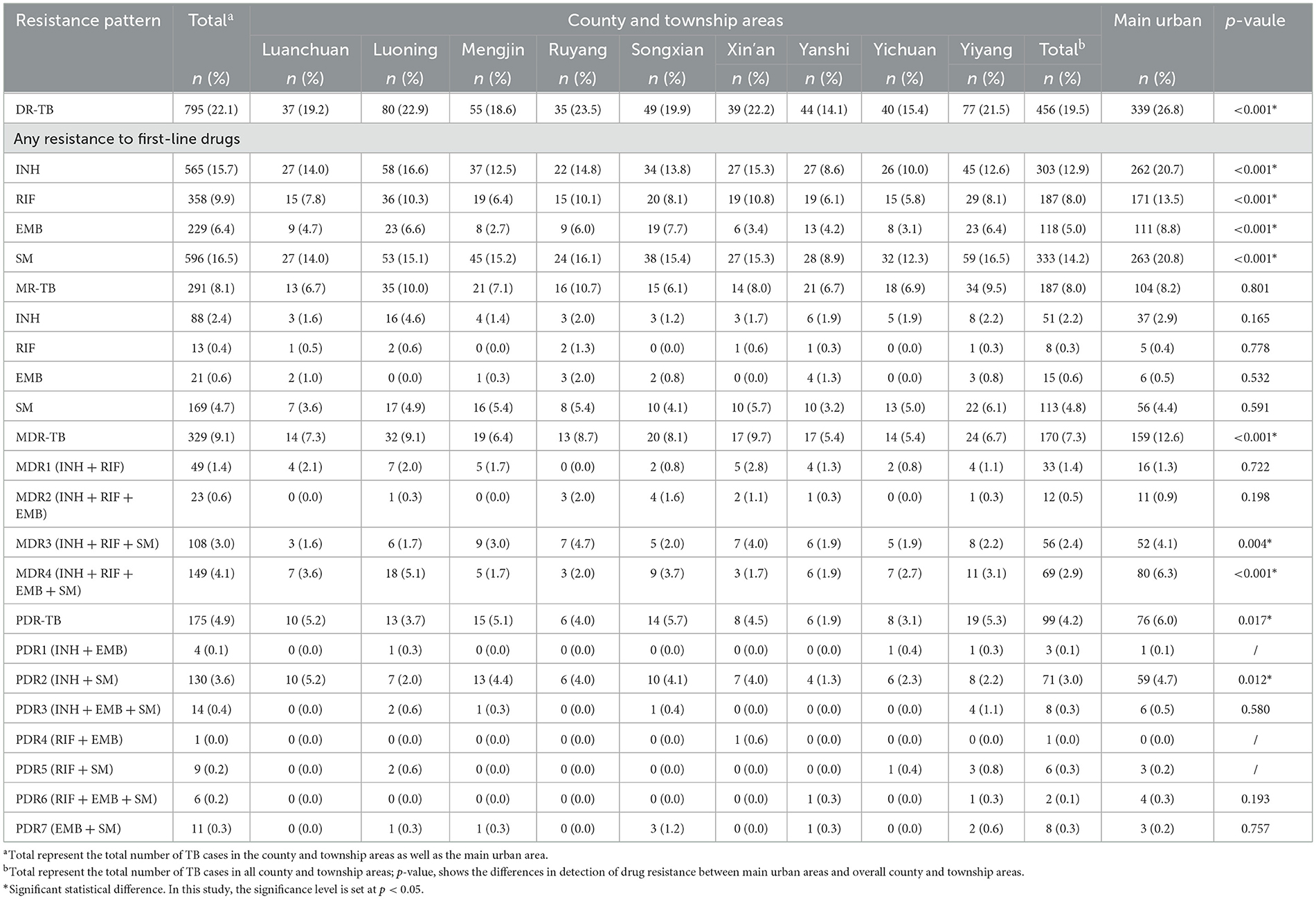
Results: Among the 3,605 culture-positive MTBC cases, 79.5% (2,866 cases) were male, 64.9% (2,341 cases) resided in county and township areas, and 64.8% (2,336 cases) were younger than 60 years. The resistance rates for first-line anti-TB drugs, from highest to lowest, were SM (16.5%), INH (15.7%), RFP (9.9%), and EMB (6.4%). The overall TB resistance rates were significantly higher in the main urban areas. During the study period, the proportion of mono-resistance tuberculosis (MR-TB), multidrug-resistant tuberculosis (MDR-TB) and polydrug-resistant tuberculosis (PDR-TB) decreased by 59.2% (12.9–5.3%), 40.3% (12.4–7.4%), and 68.3% (6.9–2.2%), respectively. The predominant resistance patterns for MDR-TB and PDR-TB were MDR4 (INH + RIF + EMB + SM) and PDR2 (INH + SM). The significant molecular mutations observed were rpsL43 for SM resistance (66.2%, 344 cases), katG315 for INH resistance (70.6%, 361 cases), rpoB529-533 for RFP resistance (54.0%, 183 cases), and embB306 for EMB resistance (56.5%, 108 cases). Resistance in MDR-TB and PDR-TB cases frequently involved combinations of hotspot mutations but was not strictly confined to these sites.
Conclusion: Tuberculosis resistance rates have declined over time, with distinct regional variations in resistance patterns. Significant molecular mutations responsible for drug resistance predominantly involve common hotspot mutations, but they are not limited to these.
Introduction
Despite ongoing efforts to control tuberculosis (TB), drug-resistant TB (DR-TB) remains a significant public health challenge. DR-TB is defined as resistance to one or more anti-TB drugs (1, 2), with common subtypes including mono-resistance tuberculosis (MR-TB) (resistance to a single first-line anti-TB drug), multidrug-resistant tuberculosis (MDR-TB; resistance to at least isoniazid [INH] and rifampin [RFP]), and polydrug-resistant tuberculosis (PDR-TB; resistance to more than one first-line drug, excluding concurrent resistance to both INH and RFP), extensively drug-resistant TB (XDR-TB, resistance to INH, RFP, fluoroquinolones, and at least one second-line injectable drug), posing additional challenges to global TB control efforts (3–5). Without effective interventions, achieving the Sustainable Development Goal of ending the TB epidemic will remain difficult (6). Surveillance of DR-TB prevalence and resistance mechanisms is essential to improving TB diagnosis and treatment strategies, thereby reducing the disease's social and economic burden (7, 8). Multicolor melting curve analysis (MMCA) is widely used in China as a rapid molecular diagnostic tool for TB, providing valuable clinical treatment guidance and insights into molecular resistance mechanisms (9). However, molecular testing alone cannot fully replace in vitro drug susceptibility testing. Traditional TB culture and drug susceptibility testing, performed under quality assurance protocols, remain critical components of DR-TB diagnosis (10). Integrating molecular diagnostics with conventional laboratory techniques is highly beneficial for optimizing TB control strategies.
Unfortunately, local data are scarce on in vitro drug susceptibility patterns and corresponding molecular resistance mechanisms, which impedes efforts to implement precise TB prevention and control measures (11–13). This study investigates the resistance of culture-positive TB patients to first-line drugs in designated TB medical institutions from 2015 to 2022, analyzing molecular mutation mechanisms in conjunction with rapid molecular diagnostic techniques. The findings fill a critical gap in research on molecular drug resistance in patients with persistent bacterial excretion and provide novel insights into TB prevention and treatment. These results are expected to assist local health authorities in formulating more precise and effective policies, facilitating the transition from traditional empirical treatment to precision therapy based on molecular resistance profiles. This, in turn, is anticipated to enhance clinical treatment outcomes and mitigate the transmission risk of DR-TB.
Methods
Study design and data collection
This study was conducted between January 2015 and December 2022. Sputum samples from TB patients attending 10 designated TB institutions in one main urban area and nine county and township areas of Luoyang City, China, were cultured using modified Lowenstein-Jensen medium (modified L-J medium; Henan Sainuote Biotechnology Co., Ltd., Henan, China) (Figure 1). Positive cultures underwent proportional drug susceptibility testing. MMCA was performed on phenotypically resistant MTBC isolates to assess resistance patterns to first-line anti-TB drugs and the corresponding molecular mechanisms in patients with persistent bacterial excretion. Patient demographics, including age, gender, and regional distribution, were obtained from the local TB reporting network and electronic medical record systems during diagnosis and treatment.
Figure 1. Map of the study area. One main urban area and nine county and township areas (Mengjin, Xin'an, Luanchuan, Songxian, Ruyang, Yiyang, Luoning, Yichuan, and Yanshi).
Mycobacteria isolation and preliminary strain identification
Following standard guidelines for routine TB diagnosis and treatment monitoring, three consecutive sputum samples (morning, evening, and random) were collected from each patient and sent to Luoyang Infectious Disease Hospital for further analysis. The samples were smeared, acid-fast stained, examined microscopically, and recorded. Strains were isolated by culturing on Lowenstein-Jensen egg-based medium, as the International Union Against Tuberculosis and Lung Disease recommended. Small cauliflower-like colonies on the slant surface of the medium were subsequently confirmed as Mycobacterium tuberculosis complex (MTBC) members using p-nitrobenzoic acid (PNB) and thiophene-2-carboxylic acid hydrazide (TCH) medium tests (Henan Sainuote Biotechnology Co., Ltd., Henan, China), as well as fluorescence PCR targeting the IS6110 insertion sequence (MTBC Nucleic Acid Detection Kit, Xiamen Zeesan Biotech Co., Ltd., Xiamen, China).
The strain identification was performed using a 30-μL PCR reaction system (Xiamen Zeesan Biotech Co., Ltd., Xiamen, China) with the following thermal cycling conditions: initial uracil-DNA glycosylase (UNG) treatment at 50°C for 2 min (1 cycle) to prevent carryover contamination; pre-denaturation at 95°C for 10 min (1 cycle); touchdown PCR consisting of 10 cycles of denaturation at 95°C for 10 s, annealing starting at 71°C (decreasing by 1°C per cycle) for 15 s, and extension at 78°C for 15 s; followed by 45 amplification cycles of denaturation at 95°C for 10 s, annealing at 61°C for 15 s [with fluorescein amidite monitoring (FAM) and hexachlorofluorescein (HEX) fluorescence signal acquisition], and extension at 78°C for 15 s.
Phenotypic susceptibility testing
Proportional drug susceptibility testing was conducted using MTBC susceptibility medium (modified L-J culture method, Henan Sainuote Biotechnology Co., Ltd., Henan, China) (14) and incubated at 36°C with 5–10% CO2 for 4 weeks. A colony count ratio exceeding 1% in drug-containing medium relative to drug-free control medium was considered indicative of drug resistance. The critical drug concentrations were as follows: 0.2-μg/mL isoniazid (INH), 40-μg/mL rifampicin (RFP), 2-μg/mL-ethambutol (EMB), and 4.0-μg/mL streptomycin (SM).
Resistance gene analysis
MMCA technology enables the simultaneous detection of multiple mutation sites associated with resistance to various anti-TB drugs within a single PCR reaction system. This technique exhibits high specificity for known single-nucleotide polymorphism (SNP) loci and offers advantages such as high reproducibility, ease of operation, low cost, and minimal experimental and personnel requirements, making it particularly suitable for large-scale clinical testing. MMCA has been widely applied and validated in clinical practice (9, 15). In this study, MMCA was utilized to analyze molecular mutations associated with resistance to first-line anti-TB drugs (INH, RFP, streptomycin [SM], and EMB) in strains exhibiting resistance to at least one of these drugs in drug susceptibility testing (DST).
The genomic DNA was extracted using the boiling lysis method. First, a single loopful of MTBC colonies was scraped with a 22-standard wire gauge (22SWG) inoculation loop and suspended in 250 μL of TB DNA extraction buffer. Then, the suspension was heated at 99°C for 20 min. After that, it was centrifuged at 14,000 rpm for 10 min. Finally, the supernatant was transferred to a new 1.5-mL centrifuge tube, serving as the PCR amplification template.
The PCR protocol for molecular drug sensitivity is as follows: PCR amplification was performed in a 25-μL reaction volume (Xiamen Zeesan Biotech Co., Ltd., Xiamen, China) with the following protocol: initial UNG treatment at 50°C for 2 min (1 cycle); pre-denaturation at 95°C for 10 min (1 cycle); touchdown PCR for 10 cycles (denaturation at 95°C for 10 s, annealing starting at 71°C with 1°C decrease per cycle for 15 s; and extension at 78°C for 15 s), followed by 45 amplification cycles (denaturation at 95°C for 10 s, annealing at 61°C for 26 s with fluorescence signal acquisition and extension at 78°C for 15 s). Melting curve analysis was performed by incubation at 95°C for 2 min and 40°C for 2 min, followed by gradual heating from 40°C to 85°C at a rate of 0.04°C/s, with continuous fluorescence monitoring [FAM and Tetrachlorofluorescein (TET) channels] at 1°C intervals.
The molecular drug resistance detection kit (MMCA method), including amplification reagents, extraction reagents, and negative/positive control reagents, was obtained from Xiamen Zeesan Biotech Co., Ltd., Xiamen, China.
INH resistance (INH-R) was detected through analysis of the ahpC promoter region (−44 to −30 and −15 to 3 sites), inhA94, the inhA promoter region (−17 to −8 sites), and katG315 mutations. Specific resistance sites included mutations in the INH detection region: ahpC promoter −44A, −42C, −39T, −34C, −32A, −30T, −15T, −12T, −10A, −10G, −10T, −9A, −6A, −4G, and ahpC4T; inhA94GCG; inhA promoter −17T, −16G, −15T, −11T, −8A, −8C; katG315AGG, katG315CGC, katG315CTC, katG315ACC, katG315AAC, katG315ATC, and katG315ACA. RFP resistance (RFP-R) was assessed by detecting mutations in codons 507–533 of the rpoB gene, encompassing the 81-base pair rifampicin resistance-determining region (RRDR). SM resistance (SM-R) was identified through mutations in codon 43 and codon 88 of the rpsL gene, as well as codons 513–517 and codons 905–908 of the rrs gene. EMB resistance (EMB-R) was evaluated by detecting mutations at codons 306, 406, and 497, as well as codons 368, 378, and 380 of the embB gene.
Quality control
Two independent professionals conducted experimental procedures, data analysis, and quality control measures. The National Reference Laboratory of China regularly evaluated the laboratory. Positive control strain H37Rv (ATCC 27294) and negative control strain Escherichia coli (ATCC 25922) were provided by the Chinese Center for Disease Control and Prevention. Negative and positive controls were supplied with commercial kits. Standard operating procedures for drug resistance testing and instrument usage (commercial reagents and controls provided by Xiamen Zeesan Biotech Co., Ltd., Xiamen, China) were strictly followed, with intra-batch and inter-batch quality control conducted as required.
Inclusion and exclusion criteria
The study enrolled patients who sought medical treatment at designated TB medical institutions with sputum specimens that were positive for MTBC in solid culture. Patients with negative MTBC culture results and duplicate cases of patients identified through demographic information (e.g., gender, date of birth, TB treatment history, and regional distribution) were excluded.
Data analysis
Descriptive statistics were performed for both categorical and numerical variables. Line graphs and stacked bar charts were utilized to visualize temporal trends in the number and proportion of different resistance patterns. The Pearson chi-squared test was applied to examine associations between tuberculosis resistance patterns in the main urban area and those in county and township areas, with Fisher's exact test used as follows: when the sample size is 40 or more but 1 ≤ expected frequency < 5; when the sample size is smaller than 40; when there is any expected frequency < 1; and when the chi-squared test probability p-value is close to 0.05. Statistical significance was determined at a 95% confidence level, with a p-value of < 0.05 considered significant. All data analyses were conducted using STATA/SE 15.1 software (Stata Statistical Software: Release 15. College Station, TX, USA).
Results
Clinical and demographic characteristics of participants
From January 2015 to December 2022, a total of 3,605 sputum specimens from non-duplicative TB patients in 10 designated TB medical institutions in Luoyang City, Henan Province, China were successfully cultured for MTBC. Of these, 79.5% (2,866 cases) were male and 20.5% (739 cases) were female. Regarding geographic distribution, 64.9% (2,341 cases) of cases were from county and township areas, while 35.1% (1,264 cases) were from the main urban area. The age group above 60 years constituted the highest proportion at 35.2% (1,269), followed by the 41–60 years age group at 34.1% (1,229 cases). Sputum smear positive was observed in 71.7% (2,585 cases), with 61.8% (2,228 cases) graded at least 1+. The peak culture-positive rate was recorded in 2021 at 18.8% (677 cases), as shown in Table 1.
Resistance time patterns
During the study period, the overall DR-TB detection rate in this region was 22.1% (795 cases), with a reduction of 53.7% (from 32.2 to 14.9%). The resistance rate of first-line anti-TB drugs from high to low was SM (16.5%) > INH (15.7%) > RFP (9.9%) > EMB (6.4%). The resistance rates of RFP, INH, and SM declined steadily until 2019, followed by a more rapid decline from 2019 to 2022, with overall reductions of 48.0% (from 15.0 to 7.8%), 38.6% (from 21.0 to 12.9%), and 70.3% (from 23.2 to 6.9%), respectively. The EMB resistance rate increased from 4.3% in 2015, peaked at 10.0% in 2019, and then dropped to 4.2% in 2022, showing a marginal overall decline of 2.3%. Between 2015 and 2020, SM-R and INH-R were the predominant resistance patterns; however, from 2020 to 2022, the prevalence of SM-R declined, while INH-R and RFP-R increased, ultimately making INH-R and RFP-R the most common resistance patterns by the end of 2022 (Figure 2A). The detection rate of MR-TB was 8.1% (291 cases), with 59.2% reduction (from 12.9% in 2015 to 5.3% in 2022). Of which, the decline rate of MR-TB (SM) was the most remarkable, reaching 90.9% (from 7.7% in 2015 to 0.7% in 2022). Conversely, MR-TB (INH) showed an increasing trend, rising by 65.4% (from 2.6% in 2015 to 4.3% in 2022) (Figure 2B). The detection rate of MDR-TB was 9.1% (329 cases), and declined by 40.3% (from 12.4% in 2015 to 7.4% in 2022). In 2015, MDR3 (INH + RIF + SM) was the predominant pattern (6.9%), followed by a rapid decline. During 2016–2021, MDR4 (INH + RIF + EMB + SM) was predominant. However, after 2019, its prevalence decreased, and MDR1 (INH + RIF) became the most common pattern by 2022 (Figure 2C). The PDR-TB detection rate was 4.9% (179 cases), and seven PDR-TB patterns were observed, with PDR2 (INH + SM) being the most prevalent (3.2%), followed by PDR3 (INH + EMB + SM) at 0.4% and PDR7 (EMB + SM) at 0.3%. The overall detection rate of PDR-TB declined by 68.3% (from 6.9% in 2015 to 2.2% in 2022), with PDR2 (INH + SM) decreasing more substantially by 91.1% (from 5.6% in 2015 to 0.5% in 2022). By 2022, PDR7 (EMB + SM) was the dominant pattern, accounting for 0.9% (Figure 2D).
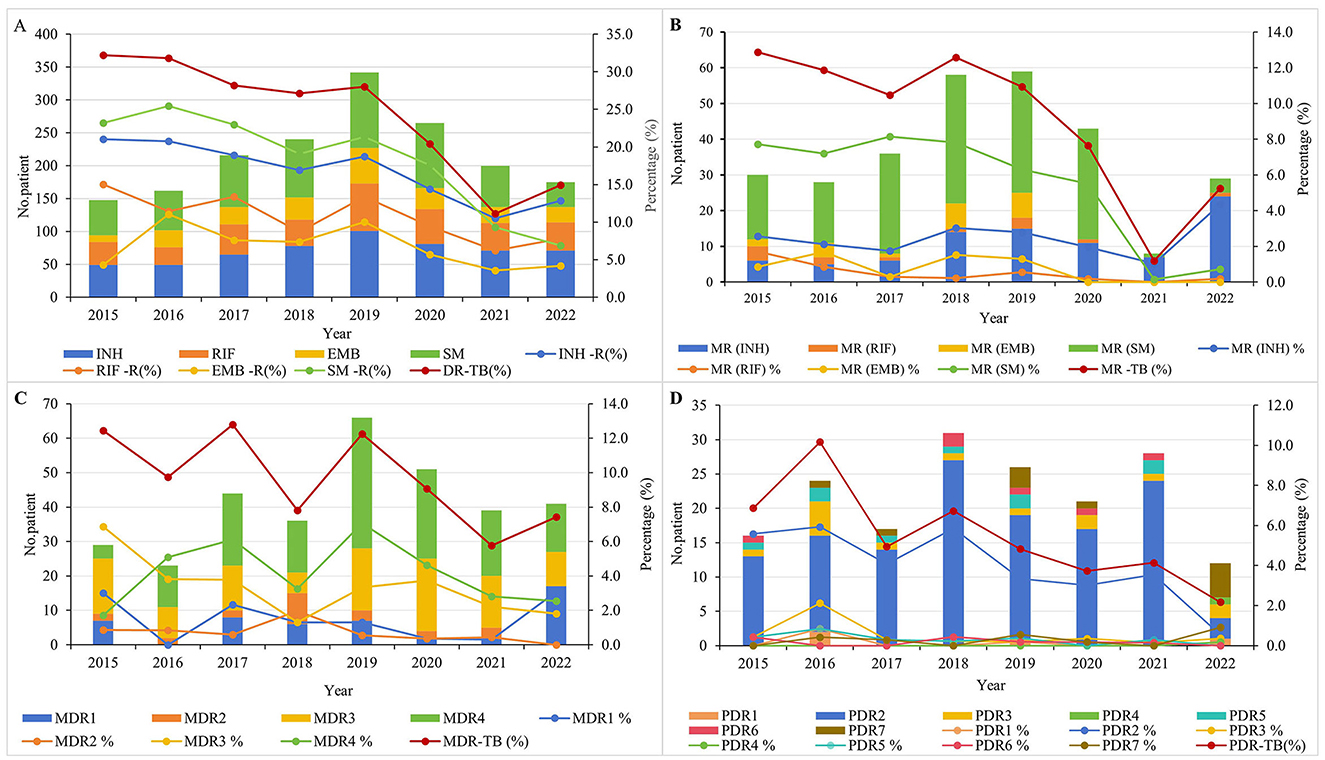
Figure 2. Time trend of different drug resistance patterns of tuberculosis in 2015–2022. (A) Time trends of first-line drug resistance for INH, RIF, EMB, and SM. (B) Time trends for different resistance patterns in MR-TB. (C) Time trends for different resistance patterns in MDR-TB. (D) Time trends for different resistance patterns in PDR-TB.
Spatial pattern of drug resistance
Significant differences in detection rates were observed between the main urban area and county/township areas for INH-R, RFP-R, EMB-R, SM-R, MDR-TB, and PDR-TB (p < 0.001 for all comparisons except PDR-TB, which was p = 0.017). No significant difference was found in the detection rates of MR-TB (p = 0.801). The main urban area had a drug-resistant TB detection rate of 26.8% (339 cases), whereas Ruyang, Luoning, Xin'an, and Yiyang exhibited relatively high rates of 23.5% (35 cases), 22.9% (80 cases), 22.2% (39 cases), and 21.5% (77 cases), respectively. Ruyang had the highest MR-TB rate (10.7%, 16 cases), followed by Luoning (10.0%, 35 cases), Yiyang (9.5%, 34 cases), the main urban area (8.2%, 104 cases), and Songxian with the lowest rate (6.1%, 15 cases). Xin'an (9.7%, 17 cases), Luoning (9.1%, 32 cases), Ruyang (8.7%, 13 cases), and Songxian (8.1%, 20 cases) had relatively high MDR-TB detection rates. Regarding PDR-TB, the highest detection rates were observed in Songxian (5.7%, 14 cases), Yiyang (5.3%, 19 cases), Luanchuan (5.2%, 10 cases), and Mengjin (5.1%, 15 cases), while Yanshi had the lowest (1.9%, 6 cases). The distribution of MDR patterns varied, with MDR4 (INH + RFP + EMB + SM) being predominant in the main urban area, Luoning, Songxian, Luanchuan, Yiyang, and Yichuan. However, in Mengjin, Ruyang, and Xin'an, MDR3 (INH + RFP + SM) was the predominant pattern. PDR patterns also varied regionally, with PDR2 (INH + SM) identified exclusively in Luanchuan and Ruyang, and a single case of PDR4 (RFP + EMB) detected in Xin'an (Table 2).
Molecular mutation mechanisms identified by multicolor melting curve analysis
The molecular resistance rates for INH, RFP, SM, and EMB were 90.4% (511/565), 94.7% (339/358), 87.2% (520/596), and 83.4% (191/229), respectively. INH-R was primarily attributed to KatG315 alterations, accounting for 70.6% (361 cases), including mutations at 62.8% (321 cases) and deletions at 7.8% (40 cases). Additional mutations were identified in the inhA promoter region (−17 to −8 sites) at 15.7% (80 cases), and the ahpC promoter region (−44 to −30 and −15 to 3 sites) at 4.9% (25 cases). RFP-R was predominantly due to rpoB529-533 mutations (54.0%, 183 cases), followed by mutations in rpoB521-528 (18.9%, 64 cases), rpoB513-520 (10.3%, 35 cases), and rpoB507-512 (7.1%, 24 cases). SM-R was primarily associated with rpsL43 mutations (66.2%, 344 cases), with additional mutations observed in rpsL88 (18.7%, 97 cases) and rrs513-517 (6.3%, 33 cases). EMB-R was linked to mutations in embB306 (56.5%, 108 cases), embB406 (24.1%, 46 cases), and embB497 (15.2%, 29 cases). MR-TB (SM) was predominantly caused by mutations in rpsL43 (37.9%, 99 cases), rpsL88 (9.2%, 24 cases), and rrs513-517 (6.1%, 16 cases). MR-TB (INH) was mainly attributed to mutations in KatG315 (18.0%, 47 cases) and the inhA promoter region (−17 to −8 sites) (4.6%, 12 cases). MR-TB (EMB) and MR-TB (RFP) were due to mutation in embB306 (3.8%, 10 cases) and rpoB529-533 (2.7%, 7 cases), respectively. The primary molecular mechanisms underlying MDR-TB involved combined mutations in KatG315 & rpoB529-533 & embB306 & rpsL43 (13.6%, 40 cases), KatG315 & rpoB529-533 & rpsL43 (10.5%, 31 cases), and KatG315 & rpoB529-533 & embB406 & rpsL88 (9.8%, 29 cases). The predominant mechanism for PDR-TB was the combined mutation of KatG315 & rpsL43 (60.3%, 94 cases) (Table 3).
Discussion
TB is a globally prevalent respiratory disease caused by MTBC and poses a significant threat to human health. In 2022, ~10.6 million new TB cases were reported worldwide, with 7.4% occurring in China (4). In the same year, TB resulted in 1.3 million deaths among HIV-negative individuals globally, including ~33,000 fatalities in China (4). To date, TB remains the leading cause of mortality from a single infectious agent. The influence of gender on TB susceptibility remains a subject of debate. Data demonstrate the male-to-female ratio (M/F ratio) of TB incidence ranges from 1.2:1 to 4.9:1, with consistently higher ratios observed in Asian populations compared to African cohorts (16). In Ethiopia, Nigeria, Sudan, Uganda, and other regions, TB prevalence is highest among individuals aged 35–54 years. In contrast, in some parts of China, the incidence among individuals aged 65 years is twice that observed in younger populations (17). Rural areas bear the most significant TB burden, mainly due to disparities in economic development (18, 19). In the present study, culture-positive TB incidence was 3.9 times higher in males than females, 1.9 times higher in county and township areas than in the main urban area, and 2.3 times higher in individuals over 40 years than in those under 40 years. These findings align with our previous reports on rapid molecular TB detection (12). The observed gender-related differences may be attributed to both biological factors (e.g., differences in lung physiology, immune system regulation by sex hormones, and genetic susceptibility) and sociobehavioral factors (e.g., lifestyle, occupation, and socioeconomic status) (20).
The overall detection rate of DR-TB exhibited a declining trend from 2015 to 2022. Prior to 2019, variations in TB resistance patterns remained relatively stable; however, a sharp decline occurred thereafter. This situation aligns closely with the overall reduction in TB incidence in China over the past decade (13). Overall, the observed decline in drug resistance rates results from sustained efforts in TB control, validating the effectiveness of anti-TB strategies and highlighting the impact of prevention and control policies. Indeed, the widespread adoption of rapid molecular diagnostic techniques, extensive public health education and awareness campaigns, expanded medical insurance coverage, and the global TB control agenda have all contributed to progress in local TB control efforts. However, the COVID-19 pandemic in 2019 led to a significant diversion of medical resources, hindering timely TB treatment and case reporting (21). Consequently, the observed decline in TB resistance may not accurately reflect the actual situation, necessitating our close attention and an objective approach (22). Some studies suggest that, to achieve a phased victory in TB control, the annual average decline in TB incidence in China from 2022 to 2025 must reach 14.5%, thereby contributing to the realization of the “End TB Strategy” by 2035 (4, 13). However, the average annual decline rate of TB resistance in this region is only 9.8%, presenting substantial challenges to local TB control efforts. Moreover, an in-depth analysis of local resistance patterns indicates their prevalence has evolved. So, the decline of phenotypic resistance rate cannot be used as the only indicator to measure the effectiveness of local anti-TB (23, 24). DR-TB management remains a formidable challenge, necessitating the formulation of more precise and effective policies, the implementation of targeted prevention and control measures tailored to specific resistance patterns, and the intensification of efforts to contain DR-TB transmission in the post-COVID-19 era.
MTBC continues to circulate locally, exhibiting significant regional variations in distribution and resistance patterns. Phenotypic drug resistance testing for SM and INH revealed relatively high resistance rates of 16.5 and 15.7%, respectively. This may be attributed to the prolonged use of these two drugs in first-line anti-TB regimens, underscoring the need to further strengthen Directly Observed Therapy Shortcourse (DOTS) regimens to better control TB resistance (11, 13). The local TB resistance rate (22.1%) is lower than the national drug resistance baseline survey estimate (39.12%) but higher than those reported in Shandong Province (18.7%) and Jilin City (12.24%), and comparable to that of Shanghai (21%) (8, 16, 25, 26). The local multi-drug resistance rate (9.1%) exceeds that of the aforementioned three regions (3.22%, 8.16%, and 4.98%) as well as the national average (4.5%) (26, 27). These findings suggest there remains substantial room for improvement in DR-TB management. The geographical distribution of the TB burden in China is heterogeneous, with a higher disease burden observed in the western regions (13). Variability in TB distribution across provinces is also evident; for example, TB cases in Xinjiang are predominantly concentrated in the southern region, whereas in Tibet, cases are primarily found in the southeastern areas (28). Locally, resistance rates were highest in main urban areas (see Table 2). In China, due to the nature of the medical insurance system, household registration status has a limited impact on the treatment outcomes of confirmed TB patients. TB patients from different residential areas often converge at specific hospitals for standardized treatment (29), leading to the concentration of complex and difficult-to-treat cases in urban areas, which may account for the higher TB resistance rates observed in these areas. Additionally, factors such as high mental stress, low BMI, caused by societal influences (e.g., aesthetic preferences in urban populations), are also associated with an increased risk of TB resistance (30). In recent years, the rapid development of tourism in Luoyang has significantly contributed to the local economy, attracting a substantial influx of migrants. This increased mobility facilitates the rapid spread of TB, and poor treatment adherence among migrants is a critical factor contributing to high TB resistance (31). The regional disparities in phenotypic drug resistance are potentially influenced by variations in treatment regimens, the intensity of control measures, the intrinsic resistance of MTBC, and exposure to drug-resistant infection sources. Luoyang, situated in western Henan Province, is an industrial city characterized by hilly terrain and an underdeveloped economy. The diverse industrial structures and demographic profiles in different areas also contribute to regional variations in drug resistance. Therefore, developing targeted control strategies, rationally allocating resources, strengthening regional cooperation and information sharing, and collaboratively addressing the challenge of TB drug resistance remain key priorities for TB control efforts.
Rapid detection based on molecular biology provides essential support for the early diagnosis and treatment of drug-resistant TB. Molecular resistance characteristics result from the interplay of multiple factors. This study identified 1,561 strains with molecular mutation among 1,748 phenotypically resistant MTBC strains (Tables 2, 3), indicating that the local TB resistance mechanism is complex. In addition to common molecular mutations, other resistance mechanisms or non-hotspot mutations contribute to the spread of drug-resistant tuberculosis in the region. Mutations in target genes or regulatory regions of antimicrobial agents are highly correlated with TB resistance. Monitoring these hotspot mutations remains clinically valuable for predicting TB resistance, even though molecular resistance does not fully represent phenotypic resistance (32, 33).
Mutations in katG315, rpoB529-533, rpsL43, and embB306 were highly associated with INH-R, RFP-R, SM-R, and EMB-R, respectively, in this region, which represents the most common hotspot mutations of TB resistance. However, regional differences exist in the location and frequency of these gene mutations. The detection rate of INH-R caused by katG315 mutations varies globally from 42 to 90% (34). Locally, this rate was 75.1% (384 cases), with 70.6% (361 cases) attributable to independent mutations at the katG315 site. This proportion is significantly higher than that reported in Japan and Italy (28–38%), Western Africa and Spain (43–46%), and southern India and Australia (61–66%), but lower than that in Eastern Europe, Russia, and South Africa (86–97%), while being similar to that in Switzerland (71%) (35–37). A high detection rate of katG315 mutations suggests that molecular diagnostic techniques targeting this site can be effectively utilized for the rapid screening of INH-R strains and early intervention in patients. In general, mutations in the inhA promoter region occur in 12–42% of INH-R strains (38). The mutation proportion in the inhA promoter region (−17 to −8 sites) locally is 15.7% (80 cases), which is consistent with the national average in China (15.2%) (39) but lower than that in India (28.1%) (37) and Canada (35.56%) (40), while higher than that in Uganda, Poland and Kyrgyz Republic (3.7–7%) (37). Additionally, the presence of mutations in other genes and combined mutations suggests that addressing the complex challenge of INH-R requires consideration of heterogeneous resistance in local DR-TB management and control. Reports indicate that more than 96.0% of RFP resistance is due to mutations in the RRDR of the rpoB gene (41). Among these, single-site mutations make up 87.3%, two-site mutations account for 5.1%, and multisite combined mutations are 0.3% (42). These findings closely align with our data, which identified RRDR variations in 94.7% of RFP-R strains, with single-, two-, and multisite mutations occurring at 90.3%, 8.8%, and 0.9%, respectively. However, regional variations persist. For example, the mutation rate of the RRDR in Wuhan reaches 97.93% (41), 86.1% in Africa, and 76.4% in Iran (43). Locally, SM-R due to mutations in rpsL43 accounts for 66.2%, which is comparable to findings from Zhejiang (65.0%) (39) and Guangzhou (67.15%) (41, 44, 45), higher than in Yunnan (54.2%) (46) but lower than in Beijing (72.0%) (47) and Tianjin (81.13%) (48). Compared with foreign studies, it is higher than that in North India (48.9%) (49) and South Korea (52.8%) (50), and lower than that in Poland (71.9%) and Singapore (80.4%) (51). The embB gene, particularly the embB306 locus, is highly correlated with EMB-R (52). The local embB306 mutation rate (56.5%) exceeds that reported in Mexico (33.0%) and Russia (48.28%), but is lower than in Germany (68.0%) and Hunan (71.4%), and comparable to findings in Hebei (56.45%) and France (56.34%) (53–55). The emergence of local MDR-TB and PDR-TB primarily results from combinations of mutations in single anti-TB drug resistance hotspot genes, though additional mechanisms may be involved. Notably, MDR-TB presents a particularly complex drug resistance mechanism, highlighting its critical role in TB management. The resistance mechanisms of MTBC are intricate, with interactions among resistance sites following a linkage mechanism. Resistance outcomes are influenced by the gene environment in which mutations occur and by concurrent mutations in other resistance-associated loci. For example, the embB306 mutation significantly contributes to the development of RIF-R and INH-R, thereby promoting MDR-TB. Moreover, resistance-related mutations are also observed in susceptible strains, reflecting polymorphisms in evolutionary processes (56, 57).
These regional variations in resistance mutation sites underscore the necessity of formulating precise prevention and control strategies and personalized treatment plans. Furthermore, they provide direction for developing advanced drug resistance detection technologies, underscoring the global challenges in TB control. Sharing resistance gene data across countries is essential to enhance the global understanding of TB resistance trends and facilitate collaborative research on effective countermeasures.
This study provides insights into the prevalence of local TB resistance over the past 8 years. By integrating analyses of phenotypic and molecular resistance mechanisms, regional differences in resistance patterns across various administrative divisions of the city have been examined. It should be noted that, although in-depth sequencing was not performed to verify the specific locations of the drug-resistant sites, and due to certain limitations, we were unable to uncover other drug resistance mechanisms apart from mutations of standard hotspot drug-resistant genes, our findings contribute to the exploration of disparities in medical resource distribution, patient management, and public health awareness across regions, thereby identifying potential factors driving drug resistance variations and informing strategies for improving public health services. The study also enhances the understanding of local DR-TB characteristics, strengthens regional cooperation and exchanges, and promotes technological advancements in TB prevention and control. Additionally, the study supplements the epidemiological data on TB in China and, to some extent, contributes to the global dataset on this disease.
Conclusion
Regional variations and complex drug resistance patterns continue to pose significant challenges in the fight against TB. The temporal prevalence pattern of local DR-TB shares similarities with findings from domestic and international studies while exhibiting distinct regional characteristics. Resistance can arise through mechanisms beyond common genetic mutations, and MDR/PDR-TB resistance is no longer merely an accumulation of mutations at hotspot loci. This intricate resistance mechanism further complicates local TB control efforts. Continuous updates on the epidemiology of drug-resistant TB are essential to assess the scale of TB transmission in the region accurately and to develop more effective and scientifically informed anti-TB strategies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Ethics Review Committee of The First Affiliated Hospital and Clinical Medical College, Henan University of Science and Technology (Approval Registration Number−2022/03/B070), and the need for informed consent was waived by this Ethics Committee considering the retrospective nature of the study and the anonymous clinical data used for analysis. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
ZW: Writing – original draft, Writing – review & editing. LX: Writing – original draft, Writing – review & editing. TG: Writing – original draft. LL: Writing – original draft. QZ: Writing – review & editing. JL: Writing – review & editing. XZ: Writing – review & editing. ZZ: Writing – review & editing. YX: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Nature Science Foundation of China (32072899), Henan Science and Technology Development Plan (212102310741), and Joint Construction Project of Henan Province (LHGJ20200589).
Acknowledgments
We would like to thank all study participants, administrative staff, and medical staff at each designated TB health facility—special thanks to Luoyang Center for Disease Control and Prevention (CDC) for their exceptional support of the study. We also particularly thank all the experts from the College of Animal Science and the College of Medical Engineering Technology of Henan University of Science and Technology for their strong support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2025.1543647/full#supplementary-material
Abbreviations
TB, tuberculosis; MTBC, Mycobacterium tuberculosis complex; INH, isoniazid; RFP, rifampin; SM, streptomycin; EMB, ethambutol; DR-TB, drug-resistant tuberculosis; MR-TB, mono-resistant tuberculosis; MDR-TB, multidrug-resistant tuberculosis; PDR-TB, polydrug-resistant tuberculosis; INH-R, isoniazid resistance; RFP-R, rifampin resistance; SM-R, streptomycin resistance; EMB-R, ethambutol resistance; MMCA, multicolor melting curve analysis.
References
1. Kim HY, Heysell SK, Mpagama S, Marais BJ, Alffenaar JW. Challenging the management of drug-resistant tuberculosis. Lancet. (2020) 395:783. doi: 10.1016/S0140-6736(20)30049-0
2. Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR, Jr. Management of drug-resistant tuberculosis. Lancet. (2019) 394:953–66. doi: 10.1016/S0140-6736(19)31882-3
3. WHO. Definitions and Reporting Framework for Tuberculosis-2013 Revision. Geneva: World Health Organization (2013). Available online at: https://www.who.int/publications/i/item/9789241505345 (accessed July 20, 2024).
4. WHO. Global Tuberculosis Report. Geneva: World Health Organization (2023). Available online at: https://www.who.int/publications/i/item/9789240083851 (accessed July 20, 2024).
5. Dheda K, Gumbo T, Maartens G, Dooley KE, McNerney R, Murray M, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. (2017) 15:S2213-2600(17)30079-6.
6. GBD Tuberculosis Collaborators. Global, regional, and national burden of tuberculosis, 1990-2016: results from the Global Burden of Diseases, Injuries, and Risk Factors 2016 Study. Lancet Infect Dis. (2018) 18:1329–49. doi: 10.1016/S1473-3099(18)30625-X
7. Mendis C, Thevanesam V, Kumara A, Wickramasinghe S, Madegedara D, Gamage C, et al. Insight into genetic diversity of Mycobacterium tuberculosis in Kandy, Sri Lanka reveals predominance of the Euro-American lineage. Int J Infect Dis. (2019) 87:84–91. doi: 10.1016/j.ijid.2019.07.001
8. Zhao Y, Xu S, Wang L, Chin DP, Wang S, Jiang G, et al. National survey of drug resistant tuberculosis in China. N Engl J Med. (2012) 366:2161–70. doi: 10.1056/NEJMoa1108789
9. Xu Y, Liang B, Du C, Tian XS, Cai XS, Hou YJ, et al. Rapid identification of clinically relevant mycobacterium species by multicolor melting curve analysis. J Clin Microbiol. (2019) 57:e01096–18. doi: 10.1128/JCM.01096-18
10. Bakuła Z, Napiórkowska A, Bielecki J, Augustynowicz-Kopeć E, Zwolska Z, Jagielski T. Mutations in the embB gene and their association with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates from Poland. Biomed Res Int. (2013) 2013:167954. doi: 10.1155/2013/167954
11. Wang SH, Chen RQ, Chang WJ, Su RY, Ma XG, Zheng DW, et al. Drug resistance of Mycobacterium tuberculosis in five monitoring sites of Henan Province, 2013-2018. Chin J Infect Control. (2023) 22:629–36. doi: 10.12138/j.issn.1671-9638.20233646
12. Wang Z, Hou Y, Guo TF, Jiang T, Xu LY, Hu HX et al. Epidemiological characteristics and risk factors of multidrug-resistant tuberculosis in Luoyang, China. Front in Public Health. (2023) 11:1117101. doi: 10.3389/fpubh.2023.1117101
13. Wang XY, Jiang ML, Pang YJ, Sun DJY, Yu CQ, Wang L, et al. Current status of tuberculosis burden in China. Chin J Epidemiol. (2024) 45:857–64. doi: 10.3760/cma.j.cn112338-20240311-00111
14. World Health Organization. Guidelines for Surveillance of Drug Resistance in Tuberculosis, 5th ed. Geneva: WHO (2015). Available online at: https://www.who.int/publications/i/item/9789241549134 (accessed July 20, 2024).
15. Yu M, Xu XX, Yan P, Zhang BQ, Liu CT, Wu JQ, et al. Clinical application of multicolor melting curve analysis in detection of drug resistance of Mycobacterium tuberculosis in sputum samples. J Tubercul Lung Health. (2018) 7:268–74. doi: 10.3969/j.issn.2095-3755.2018.04.009
16. An QQ, Song WM, Liu JY, Tao NN, Liu Y, Zhang QY, et al. Primary drug-resistance pattern and trend in elderly tuberculosis patients in Shandong, China, from 2004 to 2019. Infect Drug Resist. (2020) 13:4133–45. doi: 10.2147/IDR.S277203
17. Abuaku B, Tan H, Li X, Chen M, Huang X. Treatment default and death among tuberculosis patients in Hunan, China. Scand J Infect Dis. (2010) 42:281–7. doi: 10.3109/00365540903493723
18. Bele S, Jiang W, Lu H, You H, Fan H, Huang LF, et al. Population aging and migrant workers: bottlenecks in tuberculosis control in rural China. PLoS ONE. (2014) 9:e88290. doi: 10.1371/journal.pone.0088290
19. Wang LX, Zhang H, Ruan YZ, Chin DP, Xia YY, Cheng SM, et al. Tuberculosis prevalence in China, 1990-2010; a longitudinal analysis of national survey data. Lancet. (2014) 383:2057–64. doi: 10.1016/S0140-6736(13)62639-2
20. Dabitao D, Bishai WR. Sex and gender differences in tuberculosis pathogenesis and treatment outcomes. Curr Top Microbiol Immunol. (2023) 441:139–83. doi: 10.1007/978-3-031-35139-6_6
21. TB/COVID-19 Global Study Group. Tuberculosis and COVID-19 co-infection: description of the global cohort. Eur Respir J. (2022) 59:2102538. doi: 10.1183/13993003.02538-2021
22. Trajman A, Felker I, Alves LC, Coutinho I, Osman M, Meehan SA, et al. The COVID-19 and TB syndemic: the way forward. Int J Tuberc Lung Dis. (2022) 26:710–9. doi: 10.5588/ijtld.22.0006
23. Pei SJ, Song ZX, Yang W, He WC, Ou XC, Zhao B, et al. The catalogue of Mycobacterium tuberculosis mutations associated with drug resistance to 12 drugs in China from a nationwide survey: a genomic analysis. Lancet Microbe. (2024) 5:100899. doi: 10.1016/S2666-5247(24)00131-9
24. Yao L, Tang SJ. Annual progress of new drugs and new regimens for anti-tuberculosis treatment 2024. Chin J Tubercul Respir Dis. (2025) 48:170–5. doi: 10.3760/cma.j.cn112147-20241016-00615
25. Wang M, Li J, Zhang YY, Wang LL, Yun CL, Jiang Y, et al. The status and risk factors of drug resistance tuberculosis in Shanghai, 2013-2017. Chin J Antitubercul. (2019) 12:1269–76. doi: 10.3969/j.issn.1000-6621.2019.12.007
26. Song Y, Wan L, Chen SS, Xu YJ, Liu ZG, Zhao XQ, et al. Analysis on drug resistance of Mycobacterium tuberculosis and influencing factors in six provinces of China. Chin J Epidemiol. (2016) 37:945–8. doi: 10.3760/cma.j.issn.0254-6450.2016.07.008
27. Chen YM, Wen WP, Wu HZ, Xu LY, Peng KH, Yu ML. Analysis of monitoring results of tuberculosis drug-resistance in Guangdong Province from 2016 to 2020. Chin J Antitubercul. (2022) 44:685–9. doi: 10.19982/j.issn.1000-6621.20220028
28. Hu Mg, Feng YQ, Li T, Zhao YL, Wang JF, Xu CD, et al. Unbalanced risk of pulmonary tuberculosis in China at the subnational scale: spatiotemporal analysis. JMIR Public Health Surveill. (2022) 8:e36242. doi: 10.2196/36242
29. Zhan JH, Xiong GC, Luo D, Peng YP, Chen XW, Zeng LB, et al. Characteristics and treatment outcome of culture-positive tuberculosis patients among rural and urban residents in Jiangxi, China: a retrospective cross-sectional study. Asia-Pacific J Public Health. (2023) 35:291–4. doi: 10.1177/10105395231169083
30. Li XX, Lu W, Zu RQ, Zhu LM, Yang HT, Chen C, et al. Comparing risk factors for primary multidrug-resistant tuberculosis and primary drug-susceptible tuberculosis in Jiangsu province, China: a matched-pairs case-control study. Am J Trop Med Hyg. (2015) 92:280–5. doi: 10.4269/ajtmh.13-0717
31. Lin BC, Shao JP. Epidemiological characteristics of 2271 cases of pulmonary tuberculosis in floating population of Wenling, Zhejiang. Dis Surveill. (2012) 27: 592–4, 8. doi: 10.3784/j.issn.1003-9961.2012.8.003
32. Xu Y, Wu J, Liao S, Sun Z. Treating tuberculosis with high doses of anti-TB drugs: mechanisms and outcomes. Ann Clin Microbiol Antimicrob. (2017) 16:67. doi: 10.1186/s12941-017-0239-4
33. Cheng S, Cui Z, Li Y, Hu Z. Diagnostic accuracy of a molecular drug susceptibility testing method for the antituberculosis drug ethambutol: a systematic review and meta-analysis. J Clin Microbiol. (2014) 52:2913–24. doi: 10.1128/JCM.00560-14
34. Jagielski T, Bakuła Z, Roeske K, Kamiński M, Napiórkowska A, Augustynowicz-Kopeć E, et al. Mutation profiling for detection of isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother. (2015) 70:3214–21. doi: 10.1093/jac/dkv253
35. Jagielski T, Grzeszczuk M, Kamiński M, Roeske K, Napiórkowska A, Stachowiak R, et al. Identification and analysis of mutations in the katG gene in multidrug-resistant Mycobacterium tuberculosis clinical isolates. Pneumonol Alergol Pol. (2013) 81:298–307. doi: 10.5603/ARM.34789
36. Haeili M, Fooladi AI, Bostanabad SZ, Sarokhalil DD, Siavoshi F, Feizabadi MM. Rapid screening of rpoB and katG mutations in Mycobacterium tuberculosis isolates by high-resolution melting curve analysis. Indian J Med Microbiol. (2014) 32:398–403. doi: 10.4103/0255-0857.142245
37. Charan AS, Gupta N, Dixit R, Arora P, Patni T, Antony K, et al. Pattern of InhA and KatG mutations in isoniazid monoresistant Mycobacterium tuberculosis isolates. Lung India. (2020) 37:227–31. doi: 10.4103/lungindia.lungindia_204_19
38. Yang Y, Zhang XR. Study on the correlation between isoniazid-resistant gene mutations of Mycobacterium tuberculosis and the level of drug resistance. J Nanjing Med Univ. (2023) 43:392–6. doi: 10.7655/NYDXBNS20230314
39. Wu KY, Lu HW, Zhang MW, Zhu YL, Li XC, Pan JH, et al. Analysis on characteristic of drug resistance-associated gene mutations and the correlation with genotypes among Mycobacterium tuberculosis isolates in Zhejiang Province. Chin J Antitubercul. (2022) 44:1126–34. doi: 10.19982/j.issn.1000-6621.20220224
40. Spinato J, Boivin É, Bélanger-Trudelle É, Fauchon H, Tremblay C, Soualhine H. Genotypic characterization of drug resistant Mycobacterium tuberculosis in Quebec, 2002-2012. BMC Microbiol. (2016) 16:164. doi: 10.1186/s12866-016-0786-4
41. Wei L, Chen J, Deng Z, Zhang Z, Zhang Z, Duan Q. Genetic diversity and transmission pattern of multidrug-resistant tuberculosis based on whole-genome sequencing in Wuhan, China. Front Public Health. (2025) 13:1442987. doi: 10.3389/fpubh.2025.1442987
42. Xiang M, Zhang H, Chen L. Mechanisms of rifampicin resistance in Mycobacterium tuberculosis and research advances. Hainan Med J. (2017) 28:2853–6. doi: 10.3969/j.issn.1003-6350.2017.17.031
43. Velayati AA, Farnia P, Mozafari M, Sheikholeslami M, Karahrudi MF, Tabarsi P, et al. High prevalence of rifampin-monoresistant tuberculosis: a retrospective analysis among Iranian pulmonary tuberculosis patients. Am J Trop Med Hyg. (2014) 90:99–105. doi: 10.4269/ajtmh.13-0057
44. Chen Y, Weng QL, Meng FR. Mutation characteristics of streptomycin resistance genes in Mycobacterium tuberculosis. China Trop Med. (2018) 18:534–7. doi: 10.13604/j.cnki.46-1064/r.2018.06.05
45. Almeida Da Silva P, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. (2011) 66:1417–30. doi: 10.1093/jac/dkr173
46. Li D, Song Y, Zhang CL, Li X, Xia X, Zhang AM. Screening mutations in drug-resistant Mycobacterium tuberculosis strains in Yunnan, China. J Infect Public Health. (2017) 10:630–6. doi: 10.1016/j.jiph.2017.04.008
47. Wan L, Liu HC, Li MC, Jiang Y, Zhao XQ, Liu ZG, et al. Genomic analysis identifies mutations concerning drug-resistance and beijing genotype in multidrug-resistant Mycobacterium tuberculosis isolated from China. Front Microbiol. (2020) 11:1444. doi: 10.3389/fmicb.2020.01444
48. Mu C, Sun R, Wang ZR, Zhao H, Wang CH. Molecular characterization of mutations associated with resistance among 72 multidrug-resistant strains of Mycobacterium tuberculosis by whole genome sequencing. China Tropical Medicine. (2023) 23:725–9. doi: 10.13604/j.cnki.46-1064/r.2023.07.09
49. Yadav R, Sethi S, Dhatwalia SK, Gupta D, Mewara A, Sharma M. Molecular characterisation of drug resistance in Mycobacterium tuberculosis isolates from North India. Int J Tuberc Lung Dis. (2013) 17:251–7. doi: 10.5588/ijtld.12.0319
50. Jnawali HN, Yoo H, Ryoo S, Lee KJ, Kim BJ, Koh WJ, et al. Molecular genetics of Mycobacterium tuberculosis resistant to aminoglycosides and cyclic peptide capreomycin antibiotics in Korea. World J Microbiol Biotechnol. (2013) 29:975–82. doi: 10.1007/s11274-013-1256-x
51. Jagielski T, Ignatowska H, Bakuła Z, Dziewit Ł, Napiórkowska A, Augustynowicz-Kopeć E, et al. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE. (2014) 9:e100078. doi: 10.1371/journal.pone.0100078
52. Plinke C, Walter K, Aly S, Ehlers S, Niemann S. Mycobacterium tuberculosis embB codon 306 mutations confer moderately increased resistance to ethambutol in vitro and in vivo. Antimicrob Agents Chemother. (2011) 55:2891–6. doi: 10.1128/AAC.00007-10
53. Li Y, Wang Y, Zhang Z, Gao H, Wang H, Cao J, et al. Association between embB codon 306 mutations, phenotypic resistance profiles, and genotypic characterization in clinical Mycobacterium tuberculosis isolates from Hebei, China. Antimicrob Agents Chemother. (2016) 60:7295–302. doi: 10.1128/AAC.00532-16
54. Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from Northwestern Russia: implications for genotypic resistance testing. J Clin Microbiol. (2002) 40:3810–3. doi: 10.1128/JCM.40.10.3810-3813.2002
55. Brossier F, Sougakoff W, Bernard C, Petrou M, Adeyema K, Pham A, et al. Molecular analysis of the embCAB locus and embR gene involved in ethambutol resistance in clinical isolates of Mycobacterium tuberculosis in France. Antimicrob Agents Chemother. (2015) 59:4800–8. doi: 10.1128/AAC.00150-15
56. Gupta A, Sinha P, Nema V, Gupta PK, Chakraborty P, Kulkarni S, et al. Detection of Beijing strains of MDR M. tuberculosis and their association with drug resistance mutations in katG, rpoB, and embB genes. BMC Infect Dis. (2020) 20:752. doi: 10.1186/s12879-020-05479-5
Keywords: resistance, culture-positive, tuberculosis, molecular epidemiology, multicolor melting curve analysis
Citation: Wang Z, Xu L, Guo T, Li L, Zhang Q, Liu J, Zu X, Zhao Z and Xue Y (2025) Resistance characteristics of culture-positive tuberculosis from 2015 to 2022. Front. Public Health 13:1543647. doi: 10.3389/fpubh.2025.1543647
Received: 11 December 2024; Accepted: 07 May 2025;
Published: 23 May 2025.
Edited by:
Sivakumar Shanmugam, National Institute of Research in Tuberculosis (ICMR), IndiaReviewed by:
Nathan Mugenyi, Victoria University, UgandaNagwan El Menofy, Al-Azhar University, Egypt
Copyright © 2025 Wang, Xu, Guo, Li, Zhang, Liu, Zu, Zhao and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Xue, eHVleXVuNjY4OEAxNjMuY29t; Zhanqin Zhao, emhhb3poYW5xaW5AMTI2LmNvbQ==
 Zhenzhen Wang
Zhenzhen Wang Liyang Xu2,3
Liyang Xu2,3 Xiangyang Zu
Xiangyang Zu Zhanqin Zhao
Zhanqin Zhao Yun Xue
Yun Xue