- 1Bio-Manguinhos, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil
- 2Aggeu Magalhães Institute, Oswaldo Cruz Foundation, Recife, Brazil
- 3Department of Infectious Diseases and Microbiology, University of Pittsburgh, Pittsburgh, PA, United States
- 4Chemical Industry Information System – SIQUIM, School of Chemistry, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
- 5Laboratory of Respiratory Viruses, Exanthematous Viruses, Enteroviruses, and Viral Emergencies, Oswaldo Cruz Foundation, Rio de Janeiro, Brazil
- 6Worldwide Influenza Centre, The Francis Crick Institute, London, United Kingdom
- 7Academy of Intellectual Property and Innovation, National Institute of Industrial Property, Rio de Janeiro, Brazil
Highly Pathogenic Avian Influenza (HPAI) viruses, particularly H5N1 and H7N9, have long been considered potential pandemic threats, despite the absence of sustained human-to-human transmission. However, recent outbreaks in previously unaffected regions, such as Antarctica, suggest we may be shifting from theoretical risk to a more imminent threat. These viruses are no longer limited to avian populations. Their increasing appearance in mammals, including dairy cattle and domestic animals, raises the likelihood of viral reassortment and mutations that could trigger a human pandemic. If such a scenario unfolds, the world may face a crisis marked by high transmissibility and lethality, without effective vaccines readily available. Unlike the COVID-19 pandemic, when vaccines were rapidly developed despite inequities in access, the current influenza vaccine production model, largely reliant on slow, egg-based technologies, is insufficient for a fast-moving outbreak. While newer platforms show promise, they remain in early stages and cannot yet meet global demand, which alerts to the urgent need for accelerating vaccine and drug development, especially universal vaccines, next-generation vaccine platforms designed to provide broad, long-lasting protection against a wide spectrum of HPAI virus subtypes and strains. Here we propose a paradigmatic shift toward a more integrated, digitalized One Health surveillance system that links human, animal, and environmental data, especially in high-risk spillover regions. We underscore that Artificial Intelligence can revolutionize pandemic preparedness strategies, from improving early detection to speeding up vaccine and drug development and access to medical care, but should not be considered a stand-alone solution.
Introduction
Infections with zoonotic influenza viruses, including Highly Pathogenic Avian Influenza (HPAI) viruses pose a significant threat to global public health in scenarios where viruses could acquire the ability to transmit efficiently among humans and cause a pandemic. Particularly concerning are the avian influenza strains, such as H5N1 and H7N9, which can be, transmitted from birds to humans, with high case fatality rates (1), even considering that most of H7N9 zoonotic infections result from LPAI (Low Pathogenic Avian Influenza) viruses and not from HPAI viruses (2, 3). In addition, avian influenza viruses can mutate or reassort, potentially leading to more transmissible or lethal variants. Although to date there is no evidence of sustained human-to-human transmission of avian influenza viruses, this possibility of mutation or reassortment is concerning and requires urgent global strategies for preparedness (4, 5).
Understanding the epidemiological behavior of avian influenza viruses, the mechanisms underlying zoonotic spillover, and the potential public health ramifications of a highly lethal pandemic scenario is imperative for global preparedness. From a knowledge governance perspective, this calls for the urgent implementation of robust, integrated surveillance systems that encompass poultry farm environments, sylvatic animals, and human populations with high exposure risk. These systems must be complemented by the development and equitable distribution of rapid, point-of-care diagnostic tools and by the strategic deployment of non-pharmaceutical interventions aimed at slowing viral transmission in the absence of immediate pharmacological solutions.
In addition, we highlight the importance of international collaboration, risk communication, and equity considerations in resource allocation during vaccine and drug shortages (6). By addressing the unique challenges of this worst-case scenario, the article aims to contribute to a more resilient global preparedness framework, supported by quality data and Artificial Intelligence, capable of managing unprecedented public health crises. In a previous publication we emphasized the critical need for urgent and sustainable investments in vaccine innovation and global preparedness. The results of our publication warn that a future pandemic caused by avian influenza viruses could unfold in the absence of effective vaccines, given current technological and logistical limitations (7).
Key barriers include restricted access to vaccine patents, reliance on slow and labor-intensive egg-based production methods, and the insufficient advancement and technological limitation of the current mRNA platforms and universal influenza vaccine technologies. These challenges are particularly alarming in the context of viral evolution and adaptation within farmed animals and their human handlers, which heightens the risk of zoonotic spillover and widespread outbreaks. This potential vaccine gap underscores the urgent need for a comprehensive global pandemic preparedness framework. Such a model must prioritize the integration of genomic and antigenic surveillance within a unified One Health approach, while also fostering robust public-private partnerships and scaling up investment in innovative vaccine platforms. These efforts are essential not only for controlling highly pathogenic and low pathogenicity avian influenza viruses but also for anticipating and mitigating other emerging zoonotic threats (8).
Our article underscores the need for developing a universal influenza vaccine to reduce the risk of future pandemics, advocating for stronger international coordination led by organizations like World Health Organization (WHO) and Pan American Health Organization (PAHO) to improve vaccine accessibility and efficacy. Universal highly pathogenic avian influenza (HPAI) vaccines refer to next-generation vaccine platforms specifically designed to provide broad, long-lasting protection against a wide spectrum of HPAI virus subtypes and strains. Unlike conventional influenza vaccines that require frequent updates to match circulating strains, universal HPAI vaccines aim to target highly conserved viral regions—such as the hemagglutinin (HA) stalk domain, internal proteins (like NP and M1), or T-cell epitopes—that are less prone to antigenic drift and shift. By focusing on these conserved viral components, universal vaccines have the potential to induce cross-protective immune responses, reducing the need for strain-specific reformulation and offering a more effective tool for pandemic preparedness and control of both known and emerging HPAI variants (9, 10). Such vaccine candidates may utilize diverse platforms, including recombinant proteins, viral vectors, or mRNA technologies, and are under active investigation in both animal and early-phase human studies. Universal HPAI vaccines represent a critical innovation pathway toward overcoming the logistical and scientific limitations of current egg-based or strain-specific vaccines, especially in rapidly evolving outbreak scenarios (11, 12).
In this article we examine the epidemiological dynamics of HPAI and LPAI viruses, zoonotic spillover pathways, and societal and healthcare implications of a highly lethal pandemic. We emphasize the necessity of robust One Health surveillance systems, innovative vaccine technologies, international collaboration, and the role of Artificial Intelligence (AI) in bolstering preparedness and response mechanisms.
Epidemiological scenario: potential routes of zoonotic spillover
Avian influenza viruses, including HPAI viruses, are characterized by their ability to mutate rapidly, spreading and enabling them to adapt to new hosts. The primary zoonotic transmission routes include direct contact with infected birds and animals, exposure to contaminated environments, like faeces, etc., and the preparation of, and consumption of undercooked poultry (meat or other animal products). Further human activities, such as intensive farming and wildlife trade, amplify the risk of spillover events, events (by increasing the instances of human-animal interactions).
Avian influenza presents a critical global health threat, as indicated in Table 1. According to WHO, from January 1 2003 to December 12, 2024, 954 confirmed human cases of HPAI influenza A (H5N1) virus infection were reported across 24 countries, with 464 fatalities. Cases and fatalities related to other HPAI viruses are indicated in Table 1. While the global trend has been a decline in H5N1 human cases since 2015, recent reports (13, 14) indicate an increasing number of new human infections. For instance, as of January 6, 2025, the United States had reported 66 confirmed human cases of H5N1 since 2024, with one fatality. Historically, cattle were not considered natural hosts for H5N1, however recent cases (such as in the US in 2024) indicate that the virus can infect dairy cows, particularly spreading their mammary glands. In addition, in January 2025, a human case of H5N1 was detected in England, the second symptomatic case in the UK.
It is important to note that although human cases have been relatively low, H5N1 has been spreading extensively among bird populations and has recently been increasingly detected in several mammalian species, including dairy cows and wild animals. For example, in the United States, since 2022, the United States Department of Agriculture Animal and Plant Health Inspection Service has reported HPAI A (H5N1) virus detections in more than 200 mammals (15).
Emerging evidence suggests that certain HPAI strains, particularly from clade 2.3.4.4b, have acquired mutations that may enhance their capacity to infect mammals. Among these, changes in the PB2 gene—such as E627K and D701N—have been identified as key molecular markers that improve viral replication efficiency in mammalian hosts. Although these mutations have been detected in sporadic cases, particularly in mammals exposed to infected birds, there is no conclusive evidence of sustained mammal-to-mammal transmission to date. Nonetheless, the detection of such adaptations underscores the need for heightened genomic surveillance at the human–animal interface, where the potential for zoonotic spillover is greatest.
Another critical genetic determinant in the adaptation of avian influenza viruses to mammalian hosts is the PB2 gene, which encodes one of the three polymerase subunits essential for viral replication. Specific mutations in PB2, notably E627K and D701N, have been repeatedly associated with enhanced viral replication efficiency at the lower temperatures found in the mammalian respiratory tract (16). These mutations can significantly increase the virulence and transmissibility of HPAI viruses in mammals, including humans. Studies following experimental infections and epidemiological investigations of zoonotic cases have consistently highlighted PB2 mutations as key markers for assessing pandemic potential. The capacity of these mutations to facilitate cross-species transmission underlines the importance of incorporating PB2 surveillance into global risk assessment frameworks for avian influenza viruses.
The transition of H7N9 from low pathogenic avian influenza (LPAI) to highly pathogenic avian influenza (HPAI) is marked by the acquisition of a polybasic cleavage site in the hemagglutinin (HA) protein, leading to increased virulence in poultry (17). However, this change does not necessarily enhance transmissibility or severity in humans. Notably, both LPAI and HPAI H7N9 strains have been linked to severe human infections, with case fatality rates around 40% (18).
HA cleavage site and receptor binding specificity
While the polybasic cleavage site facilitates systemic spread in avian hosts by enabling HA cleavage by ubiquitous proteases, human pathogenicity is more influenced by receptor-binding specificity and host factors than by the cleavage site alone.
Residue 226 mutation and receptor affinity
The Q226L amino acid substitution in the hemagglutinin (HA) protein shifts receptor-binding preference from avian-type (α-2,3-linked sialic acids) to human-type (α-2,6-linked sialic acids), potentially increasing the risk of zoonotic transmission (19–21). Importantly, this substitution in the H7 HA does not eliminate affinity for avian receptors, allowing the virus to infect both avian and human hosts.
However, it is important to note that H1N1 viruses, including the 1918 pandemic strain and the 2009 H1N1 strain, do not follow this receptor-binding model. Structural and functional assessments of the 1918 virus indicate that its HA adapted for human transmission through distinct mechanisms rather than solely relying on the Q226L mutation (22). Previous studies, such as those by Gamblin et al. (23) have analyzed receptor binding affinity for the 1918 virus, providing insights into its human adaptation. Additionally, research on ferret-transmissible H5N1 viruses has examined similar receptor-binding changes, further informing our understanding of zoonotic transmission (24, 25).
Implications for zoonotic transmission
While the Q226L mutation may facilitate initial cross-species transmission, additional mutations are likely required for sustained human-to-human transmission. While the acquisition of a polybasic cleavage site is a defining feature of HPAI viruses and enhances their pathogenicity in birds, it does not necessarily correlate with increased or decreased zoonotic risk. Both LPAI and HPAI strains have demonstrated the capacity to cause severe disease in humans under certain exposure conditions. Continuous surveillance of these mutations is crucial to assess their impact on transmissibility and pathogenicity.
Zoonotic risks and mortality rates
In Figure 1, we compare the main high-risk HPAI subtypes considering zoonotic risks and mortality rates. The classification of zoonotic risk for HPAI viruses is based on factors such as the frequency and severity of human infections, potential for human-to-human transmission, and genetic characteristics of the virus. While there is not a universally standardized 1-to-5 scale, various health organizations assess these risks to inform public health responses.
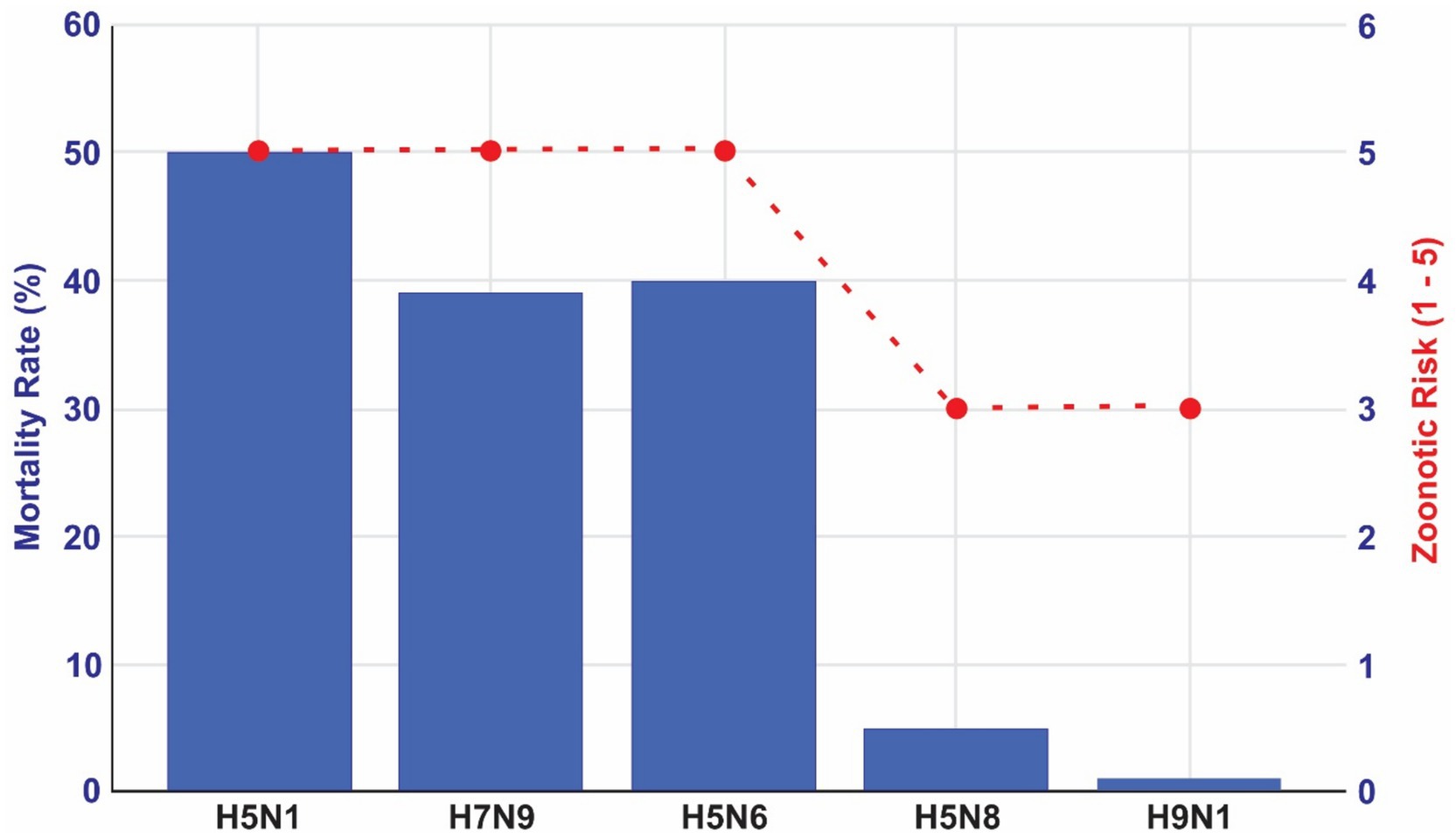
Figure 1. Zoonotic risks and mortality rates for the main HPAI viral subtypes. Source: elaborated by the authors based on World Health Organization (26) and Public Health Agency of Canada. Government of Canada (27).
The scores calculated in Figure 1 are scientifically based. We supported our classification of zoonotic risks based on two sources (26, 27). We supported our classification of zoonotic risks based on two sources, The Public Health Agency of Canada and WHO. The first one classifies certain influenza A virus subtypes, including H5N1, H5N6, and H7N9, as Risk Group 3 Human Pathogens due to their significant potential to cause serious human or animal disease (27). Similarly, WHO conducts risk assessments for specific HPAI and LPAI strains. In a 2022 assessment, the WHO evaluated the zoonotic risk of the A(H5N1) clade 2.3.4.4b viruses (28–30), considering factors like human cases, virus spread in wild and domestic animals, and genetic mutations. In Figure 1 we provide an indication of these zoonotic risks according to mortality rates, considering the possibility of overestimate of mortality rate due to underestimation of the number of infections especially.
These assessments, while not always presented on a numerical scale, provide a framework for scoring the relative zoonotic risks of different HPAI subtypes.
Accelerating HPAI vaccine innovation and development: technological gaps
Innovative approaches to vaccine development are urgently needed and key points include addressing technological development and production challenges to overcome technological gaps. Current egg-based vaccine production is slow and difficult to scale in an emergency, especially during an HPAI virus outbreak that could lead to massive animal culling. While mRNA technology shows promise, questions remain about the duration, breadth of protection and scalability (31). Due to these constraints, our previous study (7) provided evidence indicating that vaccine development for HPAI is lagging, with very few active patents and limited advancements in universal vaccines. This scenario hampers the global capacity to respond effectively to a potential pandemic. Substantial investment in universal influenza vaccines is crucial to address the limitations of current technologies.
In addition to the technological barriers previously discussed, a critical challenge in HPAI vaccine preparedness is the antigenic mismatch between stockpiled vaccines and the strain that causes a future outbreak. This scenario is increasingly likely given the accelerated antigenic drift and shift observed in H5Nx viruses. For example, while clade 2.3.4.4b viruses currently dominate outbreaks in Europe and North America, regions of Asia are still reporting circulation of other clades such as 2.3.2 and 2.3.3. This geographic heterogeneity in dominant clades increases the risk that existing vaccine stockpiles, often produced against earlier strains, will have reduced efficacy if deployed during a novel outbreak elsewhere. Moreover, regulatory and manufacturing timelines for updating avian influenza vaccines lag behind the pace of viral evolution, further compounding this gap. This underscores the urgency for next-generation HPAI vaccines that offer broader cross-clade protection, such as those targeting conserved viral epitopes or using novel platforms like mRNA and recombinant technologies (32, 33).
Broadly neutralizing antibodies: recent findings for potential universal flu vaccine
A recent breakthrough study by researchers from the University of Pittsburgh, in collaboration with the NIH Vaccine Research Center, demonstrated that monkeys pretreated with a moderate dose of the broadly neutralizing antibody MEDI88521 were universally protected against HPAI viruses. In addition to confirming the antibody’s efficacy in preventing serious adverse health outcomes, the scientists established the minimum serum concentration required for protection in primate models (34) for further development of universal HPAI vaccines. For instance, deep learning approaches have been used to predict antigenic drift in H5N1 hemagglutinin variants, helping researchers anticipate viral escape mutations (35). Similarly, support vector machines and random forest algorithms have been applied to forecast epitope binding affinity and immunogenicity, enabling more targeted vaccine design (36). Recent studies have also demonstrated the use of neural network-based models to optimize mRNA vaccine sequences for enhanced expression and immunogenicity, which could be adapted for influenza vaccines in the future (37).
Pandemic preparedness and AI: enhancing genomic surveillance, knowledge governance and sustainability
Artificial Intelligence (AI) has begun to reshape the landscape of pandemic preparedness, particularly by enhancing the speed and precision of epidemiological surveillance and knowledge governance. In vaccine development, machine learning models can predict antigenic properties, simulate immune responses, and optimize candidate selection, accelerating preclinical pipelines (38, 39). In parallel, AI-powered epidemic intelligence systems can process diverse unstructured data—from genomic databases to wildlife surveillance—to detect abnormal patterns and emerging threats before they affect human populations.
These advances are especially relevant for HPAI, where rapid viral evolution and zoonotic spillover require integrated early warning systems. AI algorithms can assimilate real-time inputs from genomic sequencing, environmental monitoring, animal health records, and social behavior to anticipate outbreak hotspots. This predictive capacity enables health authorities to issue early alerts and prioritize surveillance resources more effectively.
Artificial Intelligence (AI) is emerging as a revolutionary tool, reshaping public health with advancements in analyzing and preventing future pandemic scenarios. Additionally, AI supports robust genomic and antigenic surveillance. Genomic analysis allows tracking of mutations and reassortments that enhance virulence or transmissibility, while antigenic assays assess how well existing immune responses recognize evolving HPAI strains. Integrating both approaches is critical to guide vaccine updates and antiviral strategies.
Beyond surveillance, AI offers transformative applications across operational domains of pandemic preparedness. When adequately designed and integrated within resilient health systems, AI can significantly enhance outbreak forecasting, optimize allocation of medical resources, and support rapid diagnostics. For example, deep learning models have been deployed to predict regional outbreak hotspots based on climatic and migratory bird data, as demonstrated during H5N1 outbreaks in Southeast Asia (40). AI-driven decision support systems have also been used to optimize stockpiling and distribution of antiviral medications and personal protective equipment in real-time emergency settings (41). Additionally, AI-powered diagnostic tools using image recognition and molecular data processing have accelerated point-of-care detection of avian influenza strains in field conditions (42). These efforts must also be grounded in the One Health framework, requiring international coordination and data sharing across human, animal, and environmental health domains to ensure comprehensive risk assessment (7, 26).
Designing AI to revolutionize pandemic preparedness
Artificial Intelligence (AI) is emerging as a revolutionary tool in public health, with its potential to analyze vast amounts of data, identify trends, and enable informed decision-making. For HPAI, AI could help identify conserved viral epitopes across multiple subtypes, guiding the rational design of universal or broadly protective vaccines. Recent studies have used machine learning models to predict conserved B-cell and T-cell epitopes in H5N1 and H7N9 hemagglutinin and neuraminidase proteins, accelerating preclinical evaluation of cross-protective candidates (35, 43). In pharmacological pipelines, AI-based virtual screening platforms have been applied to search large chemical libraries for molecules with predicted binding affinity to influenza polymerase and neuraminidase targets, significantly reducing lead compound identification time (6). Notably, deep learning frameworks have also been used to repurpose existing antiviral drugs against emerging HPAI strains by predicting off-target antiviral activities (44).
One of the most critical roles of AI is accelerating vaccine and drug discovery. For HPAI, AI can help identify conserved viral epitopes across subtypes, guiding development of universal or broadly protective vaccines. In pharmacological pipelines, it can screen large chemical libraries to identify potential antiviral compounds, reducing the time from discovery to clinical testing. Inclusive governance, characterized by equitable decision-making and transparent data sharing across countries and regions, is fundamental to effective global pandemic preparedness. This includes open access to viral genomic sequences, real-time epidemiological reporting, and collaborative use of AI-driven surveillance platforms to ensure timely detection and response to HPAI threats (45).
AI also facilitates strategic decision-making in resource-constrained scenarios. Algorithms can integrate epidemiological data, health system capacity, and demographic variables to prioritize vaccine allocation, deploy health workers, and anticipate regional surges in infection. Logistics systems enhanced by AI can ensure timely distribution of critical supplies such as PPE, antivirals, and ventilators, especially in underserved areas.
Moreover, AI can support integration of epizootic surveillance with immunization efforts. By linking real-time data from wildlife and livestock with mutation tracking, AI enables targeted containment and adaptive vaccination strategies. This is crucial to prevent the emergence of vaccine-resistant strains or hidden transmission pathways.
However, the success of AI depends on high-quality data inputs, inclusive governance, and ethical frameworks. It must be implemented as part of a broader transdisciplinary preparedness strategy, not as a stand-alone solution. AI’s greatest value lies in its ability to support rapid, data-driven action within a collaborative, globally coordinated response.
Beyond early warning and surveillance, AI also provides valuable tools for accelerating vaccine and drug development, optimizing resource-allocation, and integrating epizootic surveillance systems.
1. Accelerating vaccine and drug development
For HPAI, AI could help identify structurally conserved regions of viral proteins across multiple strains, supporting the development of vaccines that offer broad protection. In drug discovery, AI can analyze vast libraries of chemical compounds to identify potential antiviral candidates, drastically reducing the timeline from research to deployment.
2. Optimizing resource allocation
In a pandemic scenario marked by high lethality and scarce resources, AI supported by quality data could assist policymakers in making data-driven decisions about resource distribution. AI models could help a timely response to a broad range of indicators, such as population density, healthcare infrastructure, and disease transmission patterns to prioritize vaccine allocation, to deploy healthcare personnel, and to optimize hospital capacities.
AI-based logistics systems can predict areas likely to experience surges in cases, enabling timely delivery of critical supplies like personal protective equipment and ventilators. This proactive approach could ensure that even resource-limited regions are adequately supported.
3. Integrating epizootic surveillance and immunization
AI can play a critical role in supporting an integrated “big data” monitoring system that combines epizootic surveillance and immunization, if vaccines are not available. This integrated system can be a powerful tool to prevent and contain outbreaks, identifying viral circulation, monitoring mutations, and detecting early infections in domestic and wild birds. This information is essential for guiding targeted immunization programs and adjusting vaccine formulations to match emerging strains. Molecular diagnostics and genomic sequencing enhance the ability to track viral evolution, while international cooperation through organizations such as the World Organization for Animal Health (WOAH) and the Food and Agriculture Organization (FAO) facilitates data sharing and coordinated responses. Without comprehensive vaccine-oriented surveillance, immunization efforts may become ineffective due to the emergence of vaccine-resistant variants or undetected transmission routes. This AI strategy alongside stringent biosecurity measures and global cooperation, including geo-politically sensitive routes, is essential to mitigating the threat of HPAI and preventing future pandemics.
Conclusion
Global epidemiological reports indicate that we might be entering a new era of Avian Flu, with the H5N1 strain spreading more rapidly among mammals. Although cases have been linked to infected wild birds and livestock farms, the virus is now spreading not only among birds and domestic animals, but increasingly infecting mammals.
Indeed, H5N1 viruses have been found in both wild and captive mammals, and they can sometimes cause fatalities as well as severe illness. Additionally, H5N1 detections in domestic cats are gaining attention. The US Department of Agriculture’s Animal and Plant Health Inspection Service reports that the HPAI H5N1 strain was found in a domestic cat in Colorado State on 2025/01/31 (15). Notably the B3.13 strain of the Eurasian 2.3.4.4b clade H5N1 virus has been spreading in animals not historically attributed as reservoirs for the HPAI virus (46). In relation to human infection, the World Health Organization (WHO) reported from 24 countries that between 2003 (beginning) and 2024 (2024/12/12), there were 954 human cases of H5N1, resulting in 464 fatalities, or 48.6% of the total zoonotic cases from avian influenza viruses (26).
A possible extreme scenario, in which a mutated strain becomes highly transmissible among humans, would create a global health crisis marked by significant morbidity and mortality. Preparing for a potential HPAI pandemic requires a multifaceted transdisciplinary approach that addresses epidemiological, technological, and societal challenges.
The possible absence of an effective HPAI vaccine for human in a highly lethal pandemic scenario, contrasting with rapid vaccine development in the COVID-19 pandemic, highlights the urgency of accelerating investment in innovative solutions and equitable global strategies. By leveraging AI strategies, fostering international collaboration and strengthening innovation funding mechanisms, the global health community can build a more resilient and sustainable innovation governance system capable of responding to unprecedented crises.
What is needed is a shift toward faster action and a coordinated, inclusive strategy that prioritizes preparedness before a next pandemic begins. The time to act is now.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
CP: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. EM: Formal analysis, Writing – review & editing. AO: Formal analysis, Writing – review & editing. SS: Formal analysis, Writing – review & editing. MS: Formal analysis, Writing – review & editing. JM: Formal analysis, Writing – review & editing. AA: Formal analysis, Writing – review & editing. AH: Formal analysis, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors thank the financial support of Bio-Manguinhos, Oswaldo Cruz Foundation Brazil, of the Chemical School of the Federal University of Rio de Janeiro, of the Brazilian National Institute of Industrial Property -INPI and of the University of Pittsburgh, US.
Acknowledgments
The authors thank Jose Viña for the technical support in the design of the Table 1 and Figure 1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^MEDI8852 is a fully human monoclonal antibody that targets the highly conserved stem region of influenza A hemagglutinin (HA), offering robust protection in preclinical models of H5N1and H7N9, even when administered up to 72 h post-exposure, and outperforming oseltamivir in key measures of survival and disease severity. It neutralizes all 18 subtypes of influenza A, including both Group I (H5N1) and Group II (H7N9) strains (67).
References
1. World Health Organization. (2025). Avian influenza weekly update number 997. Available online at: https://cdn.who.int/media/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20250516.pdf?download=true&sfvrsn=cf54905c_1 (accessed June 9, 2025).
2. Gao, R, Cao, B, Hu, Y, Feng, Z, Wang, D, Hu, W, et al. Human infection with a novel avian-origin influenza a (H7N9) virus. N Engl J Med. (2013) 368:1888–97. doi: 10.1056/NEJMoa1304459
3. Hou, Y, Deng, G, Cui, P, Zeng, X, Li, B, Wang, D, et al. Evolution of H7N9 highly pathogenic avian influenza virus in the context of vaccination. Emerg Microbes Infect. (2024) 13:2343912. doi: 10.1080/22221751.2024.2343912
4. Peacock, TP, Moncla, L, Dudas, G, Vaninsberghe, D, Sukhova, K, Lloyd-Smith, JO, et al. The global H5N1 influenza panzootic in mammals. Nature. (2025) 637:304–13. doi: 10.1038/s41586-024-08054-z
5. Caserta, LC, Frye, EA, Butt, SL, Laverack, M, Nooruzzaman, M, Covaleda, LM, et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature. (2024) 634:669–76. doi: 10.1038/s41586-024-07849-4
6. Torreele, E. Tackling vaccine inequity in 2023: have we made progress? Expert Rev Vaccines. (2024) 23:1–4. doi: 10.1080/14760584.2023.2292771
7. Possas, C, Marques, ET, Oliveira, A, Schumacher, S, Mendes, FML, Siqueira, MM, et al. Vaccine preparedness for highly pathogenic avian influenza: innovation and intellectual property issues In: C Keswani and C Possas, editors. Intellectual property issues in life sciences disputes and controversies. Boca Raton, FL: CRC Press (2025). 100–42.
8. Food and Agriculture Organization of the United Nations. World Organisation for Animal Health. (2024). Global strategy for the prevention and control of highly pathogenic avian influenza (2024–2033). Available online at: https://openknowledge.fao.org/server/api/core/bitstreams/6fff62da-80e1-43ab-94ee-3a5b69940b7c/content (accessed June 9, 2025).
9. Nachbagauer, R, and Krammer, F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect. (2017) 23:222–8. doi: 10.1016/j.cmi.2017.02.009
10. Sridhar, S, Brokstad, KA, and Cox, RJ. Influenza vaccination strategies: comparing inactivated and live attenuated influenza vaccines. Vaccines. (2015) 3:373–89. doi: 10.3390/vaccines3020373
11. Erbelding, EJ, Post, DJ, Stemmy, EJ, Roberts, PC, Augustine, AD, Ferguson, S, et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis. (2018) 218:347–54. doi: 10.1093/infdis/jiy103
12. Krammer, F. The human antibody response to influenza a virus infection and vaccination. Nat Rev Immunol. (2019) 19:383–97. doi: 10.1038/s41577-019-0143-6
13. Centers for Disease Control and Prevention. (2025). Avian influenza (bird flu). Past reported global human cases with highly pathogenic avian influenza a(H5N1) (HPAI H5N1) by country, 1997–2025. Available online at: https://www.cdc.gov/bird-flu/php/avian-flu-summary/chart-epi-curve-ah5n1.html (accessed June 9, 2025).
14. World Health Organization. (2025). Avian influenza weekly update number 983. Available online at: https://cdn.who.int/media/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20250131.pdf?sfvrsn=5f006f99_149 (accessed June 9, 2025).
15. Animal and Plant Health Inspection Service. U.S. department of agriculture. (2025). Detections of highly pathogenic influenza in mammals. Available online at: https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections/mammals (accessed June 9, 2025).
16. Mok, CK, Lee, HH, Lestra, M, Nicholls, JM, Chan, MC, Sia, SF, et al. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel a/H7N9 influenza virus in mammalian hosts. J Virol. (2014) 88:3568–76. doi: 10.1128/JVI.02740-13
17. Song, W, Huang, X, Guan, W, Chen, P, Wang, P, Zheng, M, et al. Multiple basic amino acids in the cleavage site of H7N9 hemagglutinin contribute to high virulence in mice. J Thorac Dis. (2021) 13:4650–60. doi: 10.21037/jtd-21-226
18. Chang, P, Sadeyen, JR, Bhat, S, Daines, R, Hussain, A, Yilmaz, H, et al. Risk assessment of the newly emerged H7N9 avian influenza viruses. Emerg Microbes Infect. (2023) 12:2172965. doi: 10.1080/22221751.2023.2172965
19. Lin, TH, Zhu, X, Wang, S, Zhang, D, McBride, R, Yu, W, et al. A single mutation in bovine influenza H5N1 hemagglutinin switches specificity to human receptors. Science. (2024) 386:1128–34. doi: 10.1126/science.adt0180
20. Vines, A, Wells, K, Matrosovich, M, Castrucci, MR, Ito, T, and Kawaoka, Y. The role of influenza a virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. (1998) 72:7626–31. doi: 10.1128/JVI.72.9.7626-7631.1998
21. Liu, WJ, Xiao, H, Dai, L, Liu, D, Chen, J, Qi, X, et al. Avian influenza a (H7N9) virus: from low pathogenic to highly pathogenic. Front Med. (2021) 15:507–27. doi: 10.1007/s11684-020-0814-5
22. Taubenberger, JK, Kash, JC, and Morens, DM. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci Transl Med. (2019) 11:502. doi: 10.1126/scitranslmed.aau5485
23. Gamblin, SJ, Haire, LF, Russell, RJ, Stevens, DJ, Xiao, B, Ha, Y, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. (2004) 303:1838–42. doi: 10.1126/science.1093155
24. Xiong, X, Coombs, PJ, Martin, SR, Liu, J, Xiao, H, McCauley, JW, et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. (2013) 497:392–6. doi: 10.1038/nature12144
25. Herfst, S, Schrauwen, EJ, Linster, M, Chutinimitkul, S, de Wit, E, Munster, VJ, et al. Airborne transmission of influenza a/H5N1 virus between ferrets. Science. (2012) 336:1534–41. doi: 10.1126/science.1213362
26. World Health Organization. (2025). Avian influenza weekly update #986. Available online at: https://www.who.int/westernpacific/publications/m/item/avian-influenza-weekly-update---986--21-february-2025 (accessed June 9, 2025).
27. Public Health Agency of Canada. Government of Canada. (2024). Statement from the Public Health Agency of Canada: Update on avian influenza and risk to Canadians Available online at: https://www.canada.ca/en/public-health/news/2024/11/update-on-avian-influenza-and-risk-to-canadians.html (accessed June 9, 2025).
28. Food and Agriculture Organization of the United Nations, World Health Organization, & 418 World Organization for Animal Health. (2025). Updated joint FAO/WHO/WOAH public health assessment of recent influenza a(H5) virus events in animals and people. Assessment based on data as of 1 march 2025. Available online at: https://cdn.who.int/media/docs/default-source/influenza/human-animal-interface-risk-assessments/2025_04_17_fao-woah-who_h5n1_assessment.pdf?download=true&sfvrsn=9bc6cc8e_1 (accessed June 9, 2025).
29. World Health Organization. (2020). Tool for influenza pandemic risk assessment (TIPRA). Available online at: https://www.who.int/publications/i/item/tool-for-influenza-pandemic-risk-assessment-(tipra)-2nd-edition (accessed June 9, 2025).
30. Yamaji, R, Zhang, W, Kamata, A, Adlhoch, C, Swayne, DE, Pereyaslov, D, et al. Pandemic risk characterisation of zoonotic influenza a viruses using the tool for influenza pandemic risk assessment (TIPRA). Lancet Microbe. (2024) 6:100973. doi: 10.1016/j.lanmic.2024.100973
31. Possas, C, Marques, ETA, Oliveira, A, Schumacher, S, Mendes, F, Siqueira, MM, et al. Highly pathogenic avian influenza vaccines: challenges for innovation. MedRxiv. (2023). doi: 10.1101/2023.05.31.23290790
32. World Health Organization. (2025). Genetic and antigenic characteristics of zoonotic influenza a viruses and development of candidate vaccine viruses for pandemic preparedness. Available online at: https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-northern-hemisphere-recommendation-2025-2026/202502_zoonotic_vaccinvirusupdatev2.pdf?sfvrsn=b7ee9689_13 (accessed June 9, 2025).
33. Le, TH, and Nguyen, NT. Evolutionary dynamics of highly pathogenic avian influenza a/H5N1 HA clades and vaccine implementation in Vietnam. Clin Exp Vaccine Res. (2014) 3:117–27. doi: 10.7774/cevr.2014.3.2.117
34. Kanekiyo, M, Gillespie, RA, Cooper, K, Canedo, VG, Castanha, PMS, Pegu, A, et al. Pre-exposure antibody prophylaxis protects macaques from severe influenza. Science. (2025) 387:534–41. doi: 10.1126/science.ado6481
35. Yin, R, Tran, VH, Zhou, X, Zheng, J, and Kwoh, CK. Predicting antigenic variants of H1N1 influenza virus based on epidemics and pandemics using a stacking model. PLoS One. (2018) 13:e0207777. doi: 10.1371/journal.pone.0207777
36. Chen, J, Liu, H, Yang, J, and Chou, KC. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. (2007) 33:423–8. doi: 10.1007/s00726-006-0485-9
37. Rosa, SS, Nunes, D, Antunes, L, Prazeres, DMF, Marques, MPC, and Azevedo, AM. Maximizing mRNA vaccine production with Bayesian optimization. Biotechnol Bioeng. (2022) 119:3127–39. doi: 10.1002/bit.28216
38. Syrowatka, A, Kuznetsova, M, Alsubai, A, Beckman, AL, Bain, PA, Craig, KJT, et al. Leveraging artificial intelligence for pandemic preparedness and response: a scoping review to identify key use cases. NPJ Digit Med. (2021) 4:96. doi: 10.1038/s41746-021-00459-8
39. Possas, C, Marques, ETA, Kuchipudi, SV, Kumar, P, Kim, JH, and Homma, A. Disease X in the tropics, preventing the next pandemic: how to accelerate spillover prevention and vaccine preparedness? Front in Tropical Dis. (2024) 5:1417065. doi: 10.3389/fitd.2024.1417065
40. Kjær, LJ, Kirkeby, CT, Boklund, AE, Hjulsager, CK, Fox, AD, and Ward, MP. Prediction models show differences in highly pathogenic avian influenza outbreaks in Japan and South Korea compared to Europe. Sci Rep. (2025) 15:6783. doi: 10.1038/s41598-025-91384-3
41. Koyuncu, M, and Erol, R. Optimal resource allocation model to mitigate the impact of pandemic influenza: a case study for Turkey. J Med Syst. (2010) 34:61–70. doi: 10.1007/s10916-008-9216-y
42. Astill, J, Dara, R, Fraser, EDG, and Sharif, S. Detecting and predicting emerging disease in poultry with the implementation of new technologies and big data: a focus on avian influenza virus. Front Vet Sci. (2018) 5:263. doi: 10.3389/fvets.2018.00263
43. Ton, AT, Gentile, F, Hsing, M, Ban, F, and Cherkasov, A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease using deep docking of 1.3 billion compounds. Mol Inform. (2020) 39:28. doi: 10.1002/minf.202000028
44. Stokes, JM, Yang, K, Swanson, K, Jin, W, Cubillos-Ruiz, A, Donghia, NM, et al. A deep learning approach to antibiotic discovery. Cell. (2020) 180:688–702. doi: 10.1016/j.cell.2020.01.021
45. Jit, M, Ananthakrishnan, A, McKee, M, Wouters, OJ, Beutels, P, and Teerawattananon, Y. Multi-country collaboration in responding to global infectious disease threats: lessons for Europe from the COVID-19 pandemic. Lancet Reg Health Eur. (2021) 9:100221. doi: 10.1016/j.lanepe.2021.100221
46. Colorado Veterinary Medical Association. (2024). Influenza a (highly pathogenic avian influenza H5N1) in domestic cats. Available online at: https://colovma.org/influenza-a-highly-pathogenic-avian-influenza-h5n1-in-domestic-cats/ (accessed June 9, 2025).
47. Pan American Health Organization. World Health Organization. (2025). Epidemiological update avian influenza a (H5N1) in the American regions. Available online at: https://www.paho.org/en/documents/epidemiological-update-avian-influenza-ah5n1-americas-region-24-january-2025#:~:text=24%20January%202025-,Epidemiological%20Update%20%2D20Avian20Influenza%20A(H5N1)%20in%20the,Americas%20Region%20%2D202420January%202025&text=Between%202022%20and20as20of,H5N1%20avian%20influenza%20to%20WOAH (accessed June 9, 2025).
48. Chan, PKS. Outbreak of avian influenza a(H5N1) virus infection in Hong Kong in 1997. Clin Infect Dis. (2002) 34:S58–64. doi: 10.1086/338820
49. Centers for Disease Control and Prevention. (2023). Emergence and evolution of H5N1 bird flu. Available online at: https://archive.cdc.gov/www_cdc_gov/flu/avianflu/communication-resources/bird-flu-origin-infographic.html#:~:text=2003%2D2005%20H5N1%20Spreads,the%20Middle%20East%20and%20Europe (accessed June 9, 2025).
50. Kamel, M, Aleya, S, Almagharbeh, WT, Aleya, L, and Abdel-Daim, MM. The emergence of highly pathogenic avian influenza H5N1 in dairy cattle: implications for public health, animal health, and pandemic preparedness. Eur J Clin Microbiol Infect Dis. (2025) 14:1–17. doi: 10.1007/s10096025-05147-z
51. Kim, S-H. Challenge for one health: co-circulation of zoonotic H5N1 and H9N2 avian influenza viruses in Egypt. Viruses. (2018) 10:121. doi: 10.3390/v10030121
52. Li, H, Li, Q, Li, B, Guo, Y, Xing, J, Xu, Q, et al. Continuous Reassortment of clade 2.3.4.4 H5N6 highly Pathogenetic avian influenza viruses demonstrating high risk to public health. Pathogens. (2020) 9:670. doi: 10.3390/pathogens9080670
53. Zhu, W, Li, X, Dong, J, Bo, H, Liu, J, Yang, J, et al. Epidemiologic, clinical, and genetic characteristics of human infections with influenza a(H5N6) viruses, China. Emerg Infect Dis. (2022) 28:1332–44. doi: 10.3201/eid2807.212482
54. Li, Y, Li, M, Li, Y, Tian, J, Bai, X, Yang, C, et al. Outbreaks of highly pathogenic avian influenza (H5N6) virus subclade 2.3.4.4h in swans, Xinjiang, Western China, 2020. Emerg Infect Dis. (2020) 26:2956–60. doi: 10.3201/eid2612.201201
55. Adlhoch, C, Brown, IH, Angelova, SG, Bálint, Á, Bouwstra, R, Buda, S, et al. Highly pathogenic avian influenza a(H5N8) outbreaks: protection and management of exposed people in Europe, 2014/15 and 2016. Euro Surveill. (2016) 21:30419. doi: 10.2807/1560-7917.ES.2016.21.49.30419
56. More, S, Bicout, D, Bøtner, A, Butterworth, A, Calistri, P, Depner, K, et al. Avian influenza. EFSA J. (2017) 15:e04991. doi: 10.2903/j.efsa.2017.4991
57. Graziosi, G, Lupini, C, Catelli, E, and Carnaccini, S. Highly pathogenic avian influenza (HPAI) H5 clade 2.3.4.4b virus infection in birds and mammals. Animals. (2024) 14:372. doi: 10.3390/ani14091372
58. Rafique, S, Rashid, F, Mushtaq, S, Ali, A, Li, M, Luo, S, et al. Global review of the H5N8 avian influenza virus subtype. Front Microbiol. (2023) 14:1200681. doi: 10.3389/fmicb.2023.1200681
59. Zhang, Z, and Lei, Z. The alarming situation of highly pathogenic avian influenza viruses in 2019–2023. Glob Med Genet. (2024) 11:200–13. doi: 10.1055/s-0044-1788039
60. Liu, Y, Chen, Y, Yang, Z, Lin, Y, Fu, S, Chen, J, et al. Evolution and antigenic differentiation of avian influenza a(H7N9) virus, China. Emerg Infect Dis. (2024) 30:1218–22. doi: 10.3201/eid3006.230530
61. Liu, K, Qi, X, Bao, C, Wang, X, and Liu, X. Novel H10N3 avian influenza viruses: a potential threat to public health. Lancet Microbe. (2024) 5:e417. doi: 10.1016/S26665247(23)00409-3
62. Ding, S, Zhou, J, Xiong, J, Du, X, Yang, W, Huang, J, et al. Continued evolution of H10N3 influenza virus with adaptive mutations poses an increased threat to mammals. Virol Sin. (2024) 39:546–55. doi: 10.1016/j.virs.2024.06.005
63. Alvarez, J, Boklund, A, Dippel, S, Dórea, F, Figuerola, J, Herskin, MS, et al. Preparedness, prevention and control related to zoonotic avian influenza. EFSA J. (2025) 23:e9191. doi: 10.2903/j.efsa.2025.9191
64. Zhu, W, Yang, L, Han, X, Tan, M, Zou, S, Li, X, et al. Origin, pathogenicity, and transmissibility of a human isolated influenza a (H10N3) virus from China. Emerg Microbes Infect. (2025) 14:2432364. doi: 10.1080/22221751.2024.2432364
65. Chrzastek, K, Lee, DH, Gharaibeh, S, Zsak, A, and Kapczynski, DR. Characterization of H9N2 avian influenza viruses from the Middle East demonstrates heterogeneity at amino acid position 226 in the hemagglutinin and potential for transmission to mammals. Virology. (2018) 518:195–201. doi: 10.1016/j.virol.2018.02.016
66. Gu, M, Xu, L, Wang, X, and Liu, X. Current situation of H9N2 subtype avian influenza in China. Vet Res. (2017) 48:49. doi: 10.1186/s13567-017-0453-2
Keywords: HPAI, pandemic preparedness, mutation, spillover, vaccine governance, artificial intelligence
Citation: Possas C, Marques ETA, Oliveira A, Schumacher S, Siqueira MM, McCauley J, Antunes A and Homma A (2025) Highly pathogenic avian influenza: pandemic preparedness for a scenario of high lethality with no vaccines. Front. Public Health. 13:1613869. doi: 10.3389/fpubh.2025.1613869
Edited by:
Sneha Vishwanath, University of Cambridge, United KingdomReviewed by:
Joanne Marie Montoya Del Rosario, Diosynvax Ltd., United KingdomCopyright © 2025 Possas, Marques, Oliveira, Schumacher, Siqueira, McCauley, Antunes and Homma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Possas, Y3Jpc3RpbmEucG9zc2FzQGJpby5maW9jcnV6LmJy; Ernesto T. A. Marques, bWFycXVlc0BwaXR0LmVkdQ==
 Cristina Possas
Cristina Possas Ernesto T. A. Marques
Ernesto T. A. Marques Alessandra Oliveira
Alessandra Oliveira Suzanne Schumacher
Suzanne Schumacher Marilda M. Siqueira
Marilda M. Siqueira John McCauley
John McCauley Adelaide Antunes
Adelaide Antunes Akira Homma
Akira Homma