- Viral and Rickettsial Disease Laboratory, Center for Laboratory Sciences, California Department of Public Health, Richmond, CA, United States
The increasing incidence and global spread of mpox have prompted the World Health Organization to twice declare a Public Health Emergency of International Concern. Sustained human-to-human transmission, largely through sexual contact, and waning population immunity to smallpox have accelerated monkeypox virus evolution and driven the emergence of variants that can adversely affect the performance of existing molecular diagnostic tests. To minimize the risk of PCR target drop out and better detect and monitor emerging monkeypox virus variants, we have developed and validated a multiplex real-time PCR (MpoxEG4-plex rPCR) targeting highly conserved and essential orthopoxvirus genes for the detection of four analytes: orthopoxviruses, monkeypox virus, clade I monkeypox virus, and clade II monkeypox virus. The assay limit of detection was ≤ 9 genome copies per reaction, and the clinical accuracy, sensitivity, and specificity were > 96% for each analyte. The new assay was implemented to help confirm the first case of clade I mpox in the United States. The MpoxEG4-plex rPCR offers an accurate, informative, and reliable molecular diagnostic test for identifying cases and tracking case contacts in support of public health efforts to prevent and control the spread of mpox.
Introduction
The global emergence of mpox has prompted the World Health Organization to issue two declarations of Public Health Emergency of International Concern in response to outbreaks originating in African countries where mpox is endemic (1, 2). From 2022–2024, over 124,000 mpox cases and 272 deaths were reported by 128 countries (3). The initial outbreak, caused by a new monkeypox virus (MPXV) variant, clade IIb lineage B.1, was recognized in May 2022 with over 100,000 cases recorded worldwide and remains ongoing. In September 2023, a second outbreak caused by a new MPXV subclade, designated Ib, was detected in the Democratic Republic of Congo before spreading to neighboring African countries. Travel associated clade Ib cases have been reported by several countries outside of Africa, highlighting the potential for global spread and the need for heightened laboratory surveillance and updated molecular diagnostic tests for mpox.
MPXV is an enveloped, double-stranded DNA virus belonging to the genus Orthopoxvirus within the family Poxviridae. MPXV, along with other orthopoxviruses (OPXV) including variola (smallpox), vaccinia, camelpox, cowpox, Akhmeta, and borealpox, cause human disease characterized by the presence of localized or generalized vesiculopustular rash or lesions (4). Mpox is endemic to parts of Africa and is caused by two genetically and epidemiologically distinct clades, I and II, which are further divided into subclades a and b. Clade I MPXV is endemic to some central African countries including the Democratic Republic of Congo, Central African Republic, Republic of the Congo, and Gabon, whereas clade II is indigenous to some Western African countries including Nigeria, Ghana, Côte d’Ivoire, Liberia, and Sierra Leone. Both clades are endemic to Cameroon. Mpox caused by clade I has a higher case fatality rate (10.6%) than clade II (3.6%), however, more recent estimates indicate rates of 1–2% for both clades (5, 6). Historically, cases and outbreaks of mpox were due to zoonotic spillover events, with limited secondary transmission. More recently, larger outbreaks of mpox have been fueled by community transmission, particularly through sexual contact with sex workers and among men who have sex with men (7, 8). Selective pressures from sustained human-to-human transmission are believed to be driving accelerated MPXV evolution, leading to more transmissible strains such as clade IIb lineage B.1 and clade Ib (9, 10).
The MPXV genome is approximately 197 kbp in length and contains 193 open reading frames (ORFs) (11). The central region (approximately nucleotides 33,000 to 135,000) is highly conserved among poxviruses and contains essential genes that encode proteins involved in virus replication, gene transcription, and virion morphogenesis, whereas the terminal regions are more variable and contain mostly nonessential genes coding for proteins having roles in host adaptation, pathogenesis, and immune response modulation. At the left and right ends of the genome are identical inverted terminal repetitions (ITRs) of approximately 6.4 kbp. The terminal regions of poxvirus genomes, particularly the ITRs, are susceptible to genetic rearrangement, which can contribute to virus evolution and host adaptation through gene expansion and subsequent gene loss (12, 13). Similar gene duplication and gene deletion events have been described for MPXV, including the 2022 clade IIb and 2023 clade Ib outbreak strains (14–20). Nucleotide substitutions also contribute to MPXV evolution. Although MPXV has a slow rate of mutation relative to RNA viruses, the observation that recent outbreak strains have a higher-than-expected number of nucleotide substitutions supports sustained communicable spread and ongoing host adaptation (10, 16). Indeed, many of the observed nucleotide substitutions were indicative of host-mediated, antiviral apolipoprotein B mRNA editing catalytic polypeptide-like 3 (APOBEC3) cytosine deaminase activity (9, 16).
PCR is the gold standard for laboratory confirmation of mpox (21). Several real-time PCR (rPCR) assays have been developed for detection of mpox and have been designed to provide genus-, species-, clade-, or even subclade-specific information (22–27). However, the emergence of new variants can adversely impact the performance of PCR assays through genetic alterations in sequences targeted for primer or probe binding. This prospect is particularly concerning for PCR assays that target sequences within variable terminal regions and ITRs of the MPXV genome. PCR failures caused by deletions of MPXV species- and clade-specific targets located in the ITRs have been reported for clade Ib strains and a small subset of clade IIb lineage B.1 strains (20, 28). To circumvent this problem and improve the accuracy of PCR for mpox detection, we have developed and validated a quadplex rPCR assay (MpoxEG4-plex rPCR) targeting essential genes in the central conserved region of the MPXV genome to provide genus-, species-, and clade-specific information. This new assay enabled the rapid detection and confirmation of the first case of travel-associated clade I mpox in the United States (29).
Materials and methods
DNA and monkeypox viruses
DNA used in this study are listed in Supplementary Table 1. Inactivated MPXV clade Ia strain 2003-ROC-358 and clade IIa strain US_2003 were kindly provided by the Poxvirus & Rabies Branch of the Centers for Disease Control and Prevention (CDC). OPXV genome copies were determined by qPCR using the CDC’s Food and Drug Administration (FDA) 510(k) cleared non-variola Orthopoxvirus (NVO) rPCR assay and MPXV synthetic DNA PCR standard (National Institute of Standards and Technology, Gaithersburg, MD) (30).
Clinical and contrived specimens
Remnant nucleic acids from 61 lesion swab specimens collected in viral or universal transport media were used in this study. The panel included 30 MPXV-negative, 30 clade IIb MPXV-positive, and 1 clade Ib MPXV-positive specimens. Additionally, 30 clade Ia MPXV specimens were contrived by spiking 30 MPXV-negative lesion swab specimens with inactivated MPXV strain 2003-ROC-358 at concentrations ranging from 20,000 to 200,000,000 genome copies per 200 μL. The clinical specimens were submitted to the California Department of Public Health (CDPH) for diagnostic testing and surveillance purposes and were considered exempt from human subject regulations by the California Health and Human Services Agency Committee for the Protection of Human Subjects (Project #2025–058).
Nucleic acid extraction
Specimens were inactivated by combining 200 μL of each specimen with 200 μL of AL buffer (QIAGEN, Germantown, MD) and 20 μL of protease (QIAGEN) and incubating the mixture at 56°C for 15 min. Nucleic acids were extracted and purified using the NucliSENS easyMAG instrument (bioMérieux, Durham, NC) with an output volume of 100 μL. This procedure has been approved for use with the FDA 510(k) cleared NVO rPCR assay. The nucleic acids were tested immediately or stored at −80°C for retrospective testing.
Mpox essential genes 4-plex real-time PCR assay (MpoxEG4-plex rPCR)
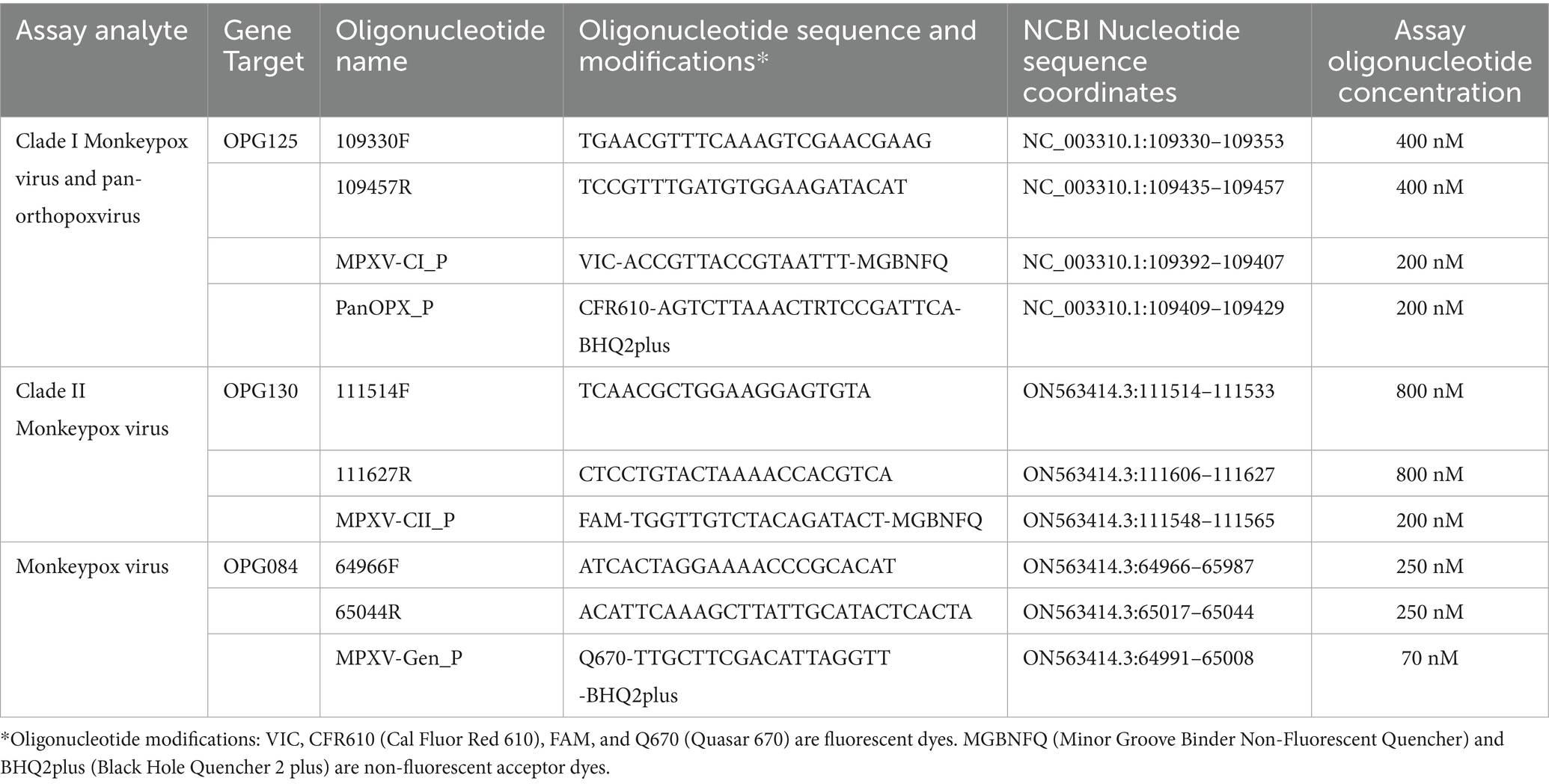
We assessed the suitability of 90 essential MPXV genes as targets for rPCR assay design (31). Essential gene ORFs and their genomic location were identified and mapped for clade I MPXV strain Zaire-96-I-16 (GenBank reference sequence NC_003310.1) using the coordinates provided by Shchelkunov et al. (11). ORF-by-ORF megablast searches and multiple sequence alignments of representative Orthopoxvirus complete genomes were performed using the National Center for Biotechnology Information’s (NCBI) nucleotide collection (nr/nt) database and bioinformatics tools. Regions of ORFs displaying significant sequence divergence at the MPXV species and clade levels were identified visually and recorded as possible candidates for rPCR design. Candidate regions were further refined by filtered megablast searches of the NCBI nr/nt database with only MPXV complete genomes selected as the search set to evaluate target region inclusivity or with MPXV sequences excluded from the search to assess target region exclusivity. Only candidate regions returning search results with 100% target inclusivity and off-target exclusivity were considered for rPCR design. The limited availability of suitable clade I MPXV candidate regions necessitated the selection of a single nucleotide polymorphism (SNP) for the clade I analyte target. The NCBI Primer-Blast tool was used for the design of OPXV-, MPXV- and clade-specific rPCR assays.
Three sets of primers and four differentially labeled fluorogenic probes were used in the MpoxEG4-plex rPCR. Table 1 provides the oligonucleotide sequences, modifications, concentrations, and MPXV genomic coordinates for the primer and probes. The fluorogenic probes were synthesized with either a minor groove binder (Life Technologies Corporation, Carlsbad, CA) or BHQplus (Biosearch Technologies, Novato, CA) modification. The 25 μL reaction consisted of the oligonucleotide primer and probes, 1X PerfeCTa Multiplex qPCR Supermix (Quantabio, Beverly, MA), and 5 μL of nucleic acid extract. Amplification and fluorescence detection were performed on the QuantStudio 5 DX real-time PCR system (Thermo Fisher Scientific, Waltham, MA) using the following parameters: 95°C for 8 min, 40 cycles of 95°C for 3 s and 63°C for 30 s.
A rPCR assay targeting human RNaseP was performed concomitantly as an endogenous control to ensure proper specimen collection and sample integrity.
Comparator rPCR assays
The FDA-cleared NVO real-time PCR assay targeting E9L was used as the comparator rPCR test for the detection of OPXV (30). For the detection of MPXV and clade Ia or clade II MPXV, a laboratory developed triplex rPCR validated by the CDPH for diagnostic use served as the comparator test. The MPXV triplex rPCR assay targets the G2R and C3L regions described by Li et al. (32) with minor modifications to improve inclusivity and enable multiplexing (Supplementary material). Only the MPXV generic analyte will be detected for clade Ib strains (C3L deletion) with this assay (27).
Determination of MpoxEG4-plex rPCR performance characteristics
To assess MPXV and OPXV primer and probe sequence inclusivity, 6,904 MPXV complete genome sequences (> 196,000 bp) with < 5% undetermined nucleotides and parsed by analyte (MPXV, clade I, or clade II) were downloaded from the GISAID Epipox database (accessed February 26, 2025) and interrogated using the python script PCR_strainer, and the PCR simulation program Thermonucleotide BLAST (33, 34). Sequences identified by PCR_strainer as having undetermined nucleotides in a target sequence were excluded, providing 6,886 MPXV, 260 clade I, 6,607 clade II, and 6,879 OPXV sequences for inclusivity analyses. Among the clade I sequences, 245 were assigned to subclade Ia and 15 to subclade Ib, and for the clade II sequences, 6 were assigned to subclade IIa and 6,601 to subclade IIb. Inclusivity results were documented as the number of sequences with 0, 1, 2, or ≥ 3 mismatches for each primer-probe set. In addition, 95 MPXV clade IIb A.2.2 complete genome sequences representing the proposed G.1 lineage were downloaded from the NCBI MPXV data hub (accessed July 5, 2025), and the MpoxEG4-plex rPCR primer and probe sequences were mapped to these genomes to assess inclusivity using Geneious Prime 2022.0.2 (Biomatters, Auckland, New Zealand). Finally, 292 non-MPXV orthopoxvirus sequences were available through the NCBI core nucleotide database and assessed for OPXV primer and probe sequence inclusivity using the NCBI multiple sequence alignment tool (accessed February 27, 2025). This cohort consisted of 1 Abatino, 6 Akhmeta, 1 borealpox, 6 buffalopox, 10 camelpox, 97 cowpox, 5 ectromelia, 3 horsepox, 1 rabbitpox, 1 raccoonpox, 1 skunkpox, 1 taterapox, 81 vaccinia, 77 variola, and 1 volepox complete genome sequences.
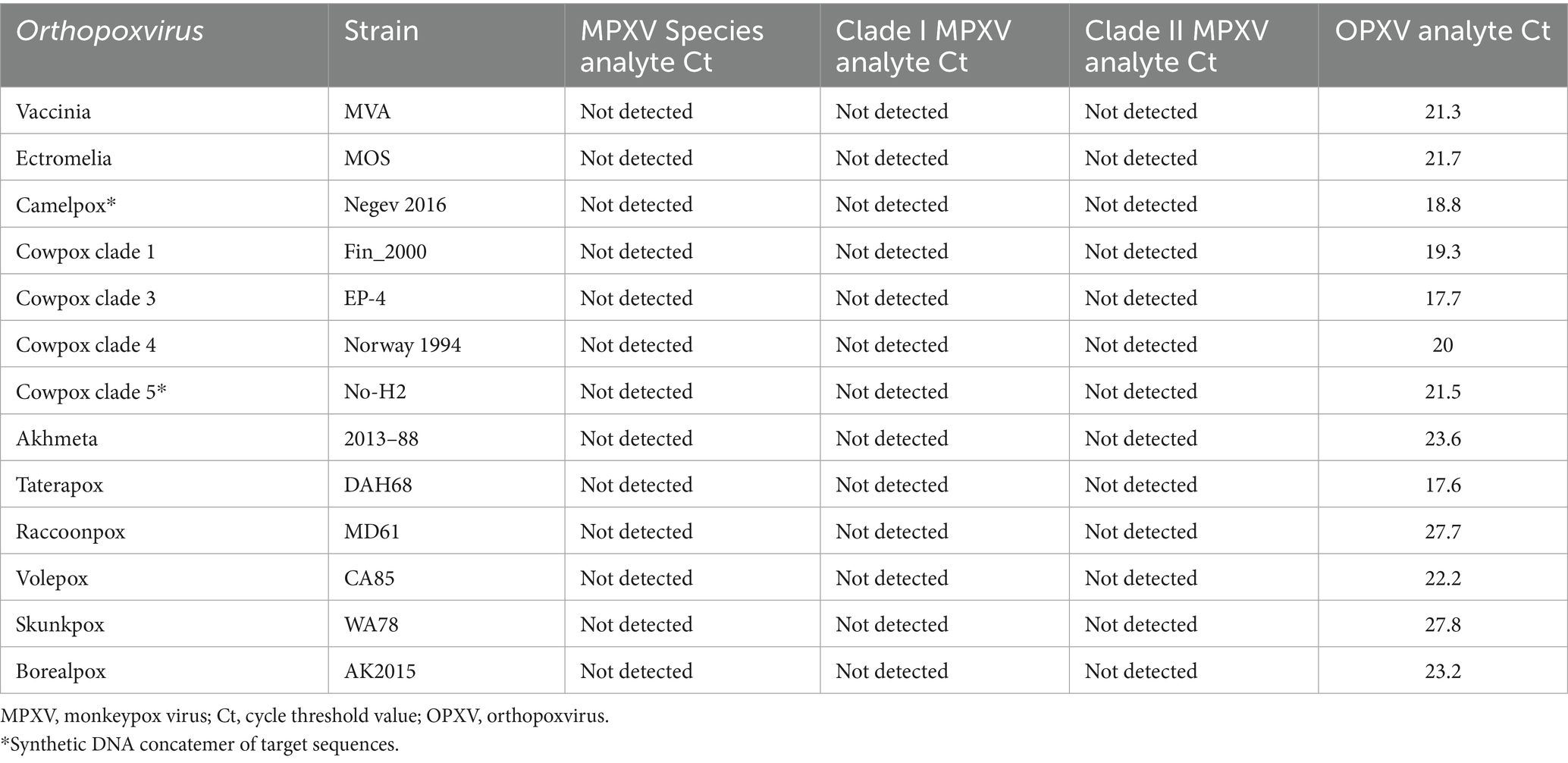
In silico exclusivity of the primer and probe sequences was assessed through individual blastn searches against the NCBI reference genome sequences for Homo sapiens and 33 microorganisms as recommended by the FDA (35). DNA from organisms sharing ≥ 80% sequence homology with a primer or probe sequence and from 10 additional OPXV were tested with the MpoxEG4-plex assay for cross-reactivity (Supplementary Table 1).
Assay analytical sensitivity or limit of detection (LOD) was determined for each analyte using inactivated and quantified MPXV strains 2003-ROC-358 (clade I) and US_2003 (clade II) spiked into pooled MPXV-negative lesion swab matrix and processed through the entire test workflow. A preliminary LOD was established for decreasing three-fold serial concentrations ranging from 243 to 0.33 genome copies per reaction tested in replicates of three. A second run was then performed at the preliminary LOD and two immediately higher concentrations in replicates of seven. The data from these two runs were combined and the LOD for each analyte defined as the concentration for which all 10 replicates were detected.
Accuracy, sensitivity, and specificity were determined for each analyte by comparison of test results from the MpoxEG4-plex rPCR with those from the comparator tests for the panel of 61 clinical and 30 contrived specimens.
Results
Design and development of the MpoxEG4-plex rPCR assay
Three gene targets, OPG084, OPG125, and OPG130, were selected for assay design based on systematic ORF-by-ORF alignment and comparison of 90 essential gene sequences from representative OPXV genomes (Table 1) (31). The OPG084 and OPG130 primer and probe sets detect MPXV species and clade II MPXV strains, respectively, whereas the OPG125 primer and duplex probe set differentially detects clade I MPXV and OPXV. In silico analyses of complete genome sequences revealed that the primer and probe sets are highly inclusive with perfect matches to 99.4, 100, 99.9, and 99.8% of the MPXV, clade I, clade II, and OPXV target sequences, respectively (Table 2). The OPG084 primer and probe sequences are highly conserved among MPXV and broadly divergent among other OPXV (Table 2; Figure 1A). Similarly, the OPG130 primer and probe sequences are highly conserved among clade II MPXV strains but display a significant number of nucleotide mismatches with other OPXV species (Table 2; Figure 1B). The ability of the OPG130 target to distinguish clade II MPXV strains from clade I strains, however, is largely dependent on two nucleotide mismatches between the probe sequence and clade I sequences (Figure 1B). The OPG125 primer sequences and OPXV probe sequence are widely conserved among both Old and New World OPXV (Table 2; Figure 1C). This target utilizes a second probe sequence that is highly conserved among clade I MPXV strains and allows differentiation from clade II MPXV and other OPXV based largely on a SNP (Table 2; Figure 1C). The differentiation of the clade I and clade II analytes were facilitated by the design and use of allelic discrimination probes.
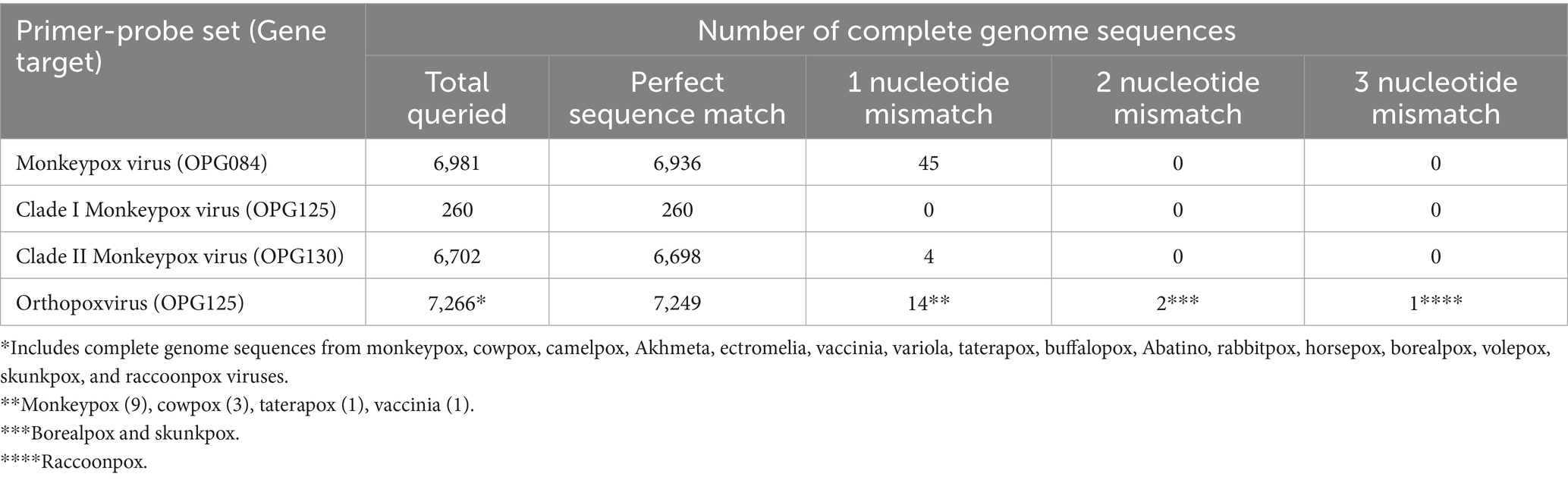
Table 2. In silico analyses of mpox essential genes 4-plex real-time PCR primer and probe sequences for inclusivity.
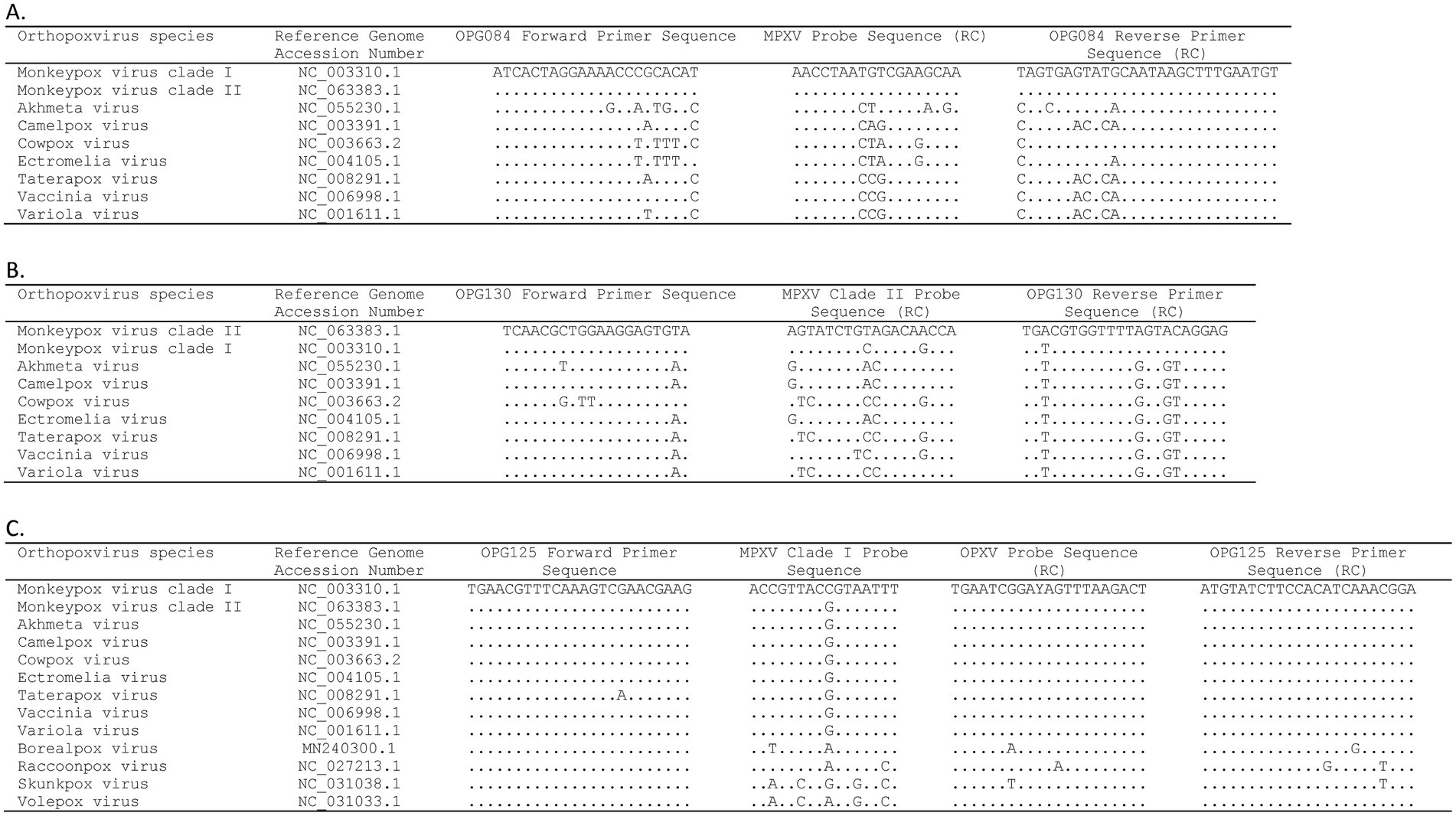
Figure 1. Alignment of mpox essential genes 4-plex real-time PCR (A) OPG084, (B) OPG130, and (C) OPG125 primer and probe sequences with orthopoxvirus reference sequences. Dots indicate identical nucleotide for that position. RC, reverse complement.
MpoxEG4-plex rPCR assay performance characteristics
Assay cross-reactivity was first assessed by in silico analyses of primer and probe sequences for ≥ 80% homology with the human genome sequence and the genome sequences of 33 microorganisms, as specified in the FDA’s Emergency Use Authorization Template for Mpox (35). Significant sequence homology was observed with 24 genome sequences. Assay cross-reactivity with representative DNA from these 23 organisms (variola virus excluded) and 9 additional OPXV strains was not detected (Supplementary Table 1). As expected, DNA representing 13 OPXV strains were detected for the OPXV analyte but not for the MPXV analytes (Table 3).
The assay LOD was determined for each analyte using quantified inactivated clade I and clade II viruses spiked into pooled MPXV-negative lesion swab matrix and processed through the entire assay workflow. LOD determinations of 1 copy and 3 copies per reaction were established for the MPXV analyte with the clade I and clade II viruses, respectively, 3 copies per reaction for the OPXV analyte with both viruses, 3 copies per reaction for the clade I MPXV analyte, and 9 copies per reaction for the clade II MPXV analyte (Tables 4, 5).
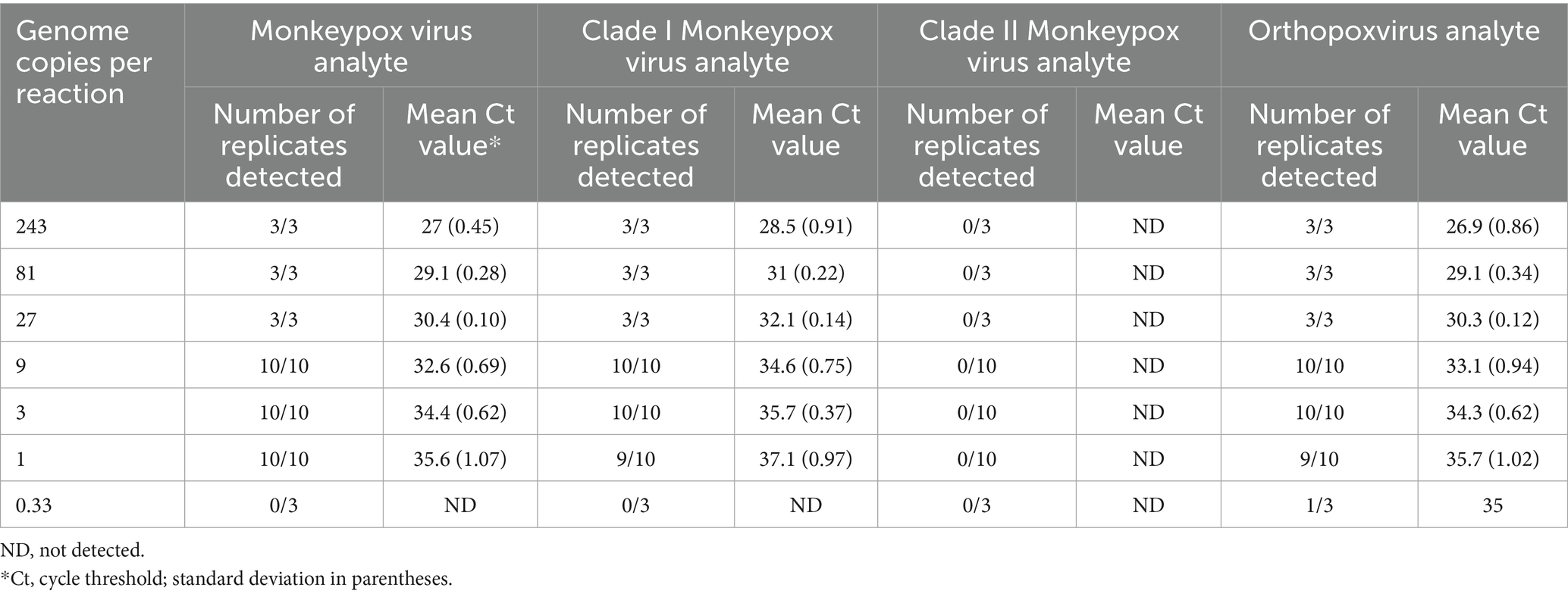
Table 4. Mpox essential genes 4-plex real-time PCR limit of detection analysis for clade I monkeypox virus.
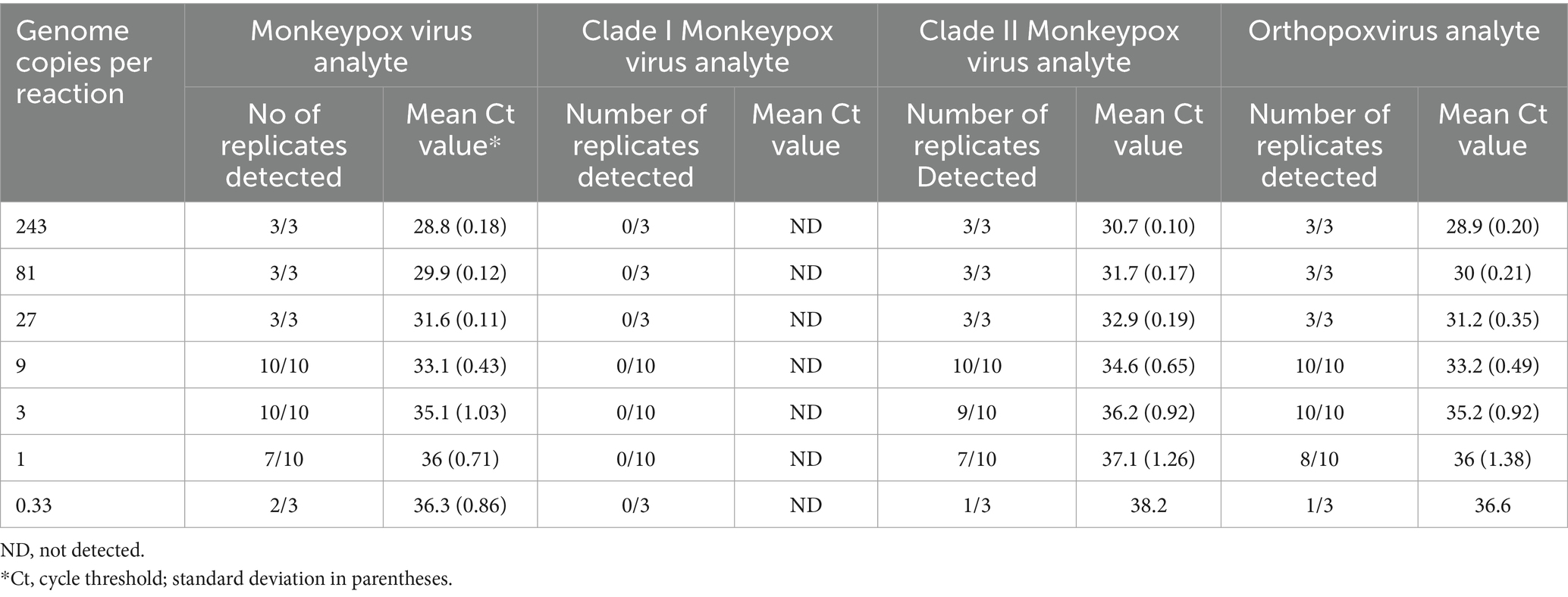
Table 5. Mpox essential genes 4-plex real-time PCR limit of detection analysis for clade II monkeypox virus.
Clinical agreement was determined by retrospectively testing 61 clinical specimens (1 clade Ib MPXV positive, 30 clade IIb MPXV positives, and 30 MPXV negatives) and 30 contrived clade Ia MPXV positive specimens and comparing test results with the FDA-cleared NVO assay or the laboratory developed triplex assay for the detection of MPXV species, clade Ia MPXV, and clade II MPXV (Supplementary Table 2). The accuracy, sensitivity, and specificity of the MpoxEG4-plex assay were all 100% for the OPXV, MPXV, and clade I MPXV analytes, and 98.9, 96.7, and 100% for the clade II MPXV analyte, respectively. Only a single discordant analyte result was observed; clinical specimen 54 was positive for the clade II MPXV analyte (Ct = 34) with the comparator test but negative for the MpoxEG4-plex rPCR clade II analyte. The OPXV (Ct = 37.1) and MPXV (Ct = 36.9) analytes were detected for this specimen with the MpoxEG4-plex rPCR assay. As expected, the clade Ib specimen (clinical specimen 61) was detected for the OPXV, MPXV, and clade I analytes with the MpoxEG4-plex rPCR assay and the non-variola OPXV and MPXV analytes with the comparator tests. With the triplex MPXV rPCR comparator test, this specimen was positive for the generic MPXV analyte but negative for the clade Ia (C3L target) and clade II analytes. Whole genome sequencing (WGS) confirmed this specimen as clade Ib (36).
Discussion
We have developed and validated a multiplex rPCR assay targeting three essential genes for the detection of OPXV genus, MPXV species, and MPXV clades I and II. All three gene targets, OPG125, OPG084, and OPG130, are located within the central conserved region of the MPXV genome and encode proteins having an essential role in either viral replication or morphogenesis (31). OPG125 (MPXV ORF E13L) encodes a scaffold protein that is highly conserved among poxviruses and considered essential for morphogenesis of vaccina virus (homolog D13L) (37). The OPG125 target is amplified using primer sequences broadly conserved among OPXV; the amplicon is detected with two differentially labeled probes: one recognizing an OPXV-specific sequence and the other targeting a MPXV clade I-specific SNP in a separate region of the amplicon. The MPXV species target, OPG084 (MPXV ORF I8R), encodes a DNA and RNA helicase, which is required for early gene transcription in vaccinia virus (38). For clade II MPXV detection, the assay targets OPG130 (MPXV ORF A5L), an immunodominant 39-kDa virion core protein that is required for vaccinia virus maturation (39).
Validation of the MpoxEG4-plex assay established an analytical sensitivity of ≤ 9 genome copies per reaction for all four analytes and confirmed that the targets were highly specific for the intended analyte by both in silico analyses and cross-reactivity testing. Assay accuracy, sensitivity, and specificity were > 96% for all analytes with only a single discordant analyte result observed relative to the comparator tests: a clade II analyte result was falsely negative for one specimen, likely due to the MPXV nucleic acid concentration being at the lower limit of analyte detection. Overall, the performance characteristics of the MpoxEG4-plex rPCR assay were equivalent to the comparator tests but with added inclusivity for clade Ib. Notably, implementation of the MpoxEG4-plex rPCR assay was instrumental in recognizing the first clade I infection in a traveler returning to the United States from Eastern Africa (36).
Numerous PCR targets have been explored for the detection of OPXV and MPXV (22–27). However, many of the MPXV species- and clade-specific assays target genes that are nonessential and located within the variable terminal regions of the genome. While sequence diversity within these regions may offer species- and clade-specific targets, they also may be subject to gene loss through genetic rearrangement. Two commonly used PCR targets for MPXV species and clade II identification are located within OPG002 (MPXV ORFs J2L and J2R; often referred to as G2R, the variola homolog), a gene present in both ITRs (32). False negative PCR results attributed to a 600-bp deletion within both copies of OPG002 and consequently, the loss of MPXV species and clade II targets have been recently reported (28). Fortunately, the appearance of strains with this deletion seems to have been geographically isolated and transient. In contrast, a more sustained and diagnostically consequential deletion emerged with subclade Ib (20). These strains possess a 1,114-bp deletion encompassing OPG032 (MPXV ORF D14L; vaccinia homolog C3L) that adversely impacts the performance of a commonly used clade I-specific PCR (27). These two examples of gene deletion illustrate the potential risk associated with targeting the terminal variable regions of the MPXV genome for the design of PCR assays, particularly given the propensity of poxviruses to evolve and adapt to new hosts through gene expansion and subsequent deletion.
Nucleotide substitutions also play an important role in poxvirus evolution and host adaptation. Poxviruses have an estimated substitution rate of 1 to 2 nucleotides per genome per year (40). The substitution rate for clade IIb MPXV, however, increased 6 to 12-fold in the last 10 years, likely because of sustained human-to-human transmission (16). Substitutions observed with the emergent 2022 outbreak clade IIb variant B.1 and 2023 outbreak clade Ib strains are predominantly driven by host-mediated, antiviral APOBEC3 cytosine deaminase activity (nucleotide replacements GA > AA and TC > TT) (16, 18). Remarkably, the 2022 lineage B.1 outbreak sequence differed by 46 nucleotide substitutions from a 2018 lineage A.1 reference sequence with nearly 90% of the replacements likely associated with APOBEC3 activity (16). This accelerated evolution can impact the performance of MPXV PCR assays. PCR primer sequence mismatches with clade IIb lineage B.1 sequences have been shown to reduce the analytical sensitivity of MPXV PCR assays (41, 42). Moreover, several recent studies using in silico analyses of MPXV genome sequence compilations have documented a significant occurrence of primer and probes sequence mismatches that may impact the performance of many published PCR assays (42–45). Conversely, the MpoxEG4-plex rPCR primer and probe sequences are highly inclusive with perfect matches of > 99.4% to the intended OPXV/MPXV genome sequences (Table 2).
The appearance of APOBEC3 mutations in clade IIb, lineage B.1 genomes follow a nonrandom genome distribution including several mutational hotspots, and a higher-than-expected frequency of synonymous mutations and mutations in noncoding regions and inverted repeats (9, 46). Sustained human-to-human transmission likely will continue to drive MPXV diversity through the accumulation of APOBEC3 nucleotide substitutions that do not significantly impact viral fitness. Future APOBEC3 mutations are predicted to manifest in TC dinucleotide hotspots present in the ITRs, large coding sequences (> 1.8 kbp), and transcriptionally active genes (47, 48). The selection of PCR targets located within the central conserved region of the MPXV genome, and the presumed functional and structural constraints of essential genes minimizes the risk of MpoxEG4-plex rPCR primer and probe sequence mismatches as a consequence of future APOBEC3 mutations. Moreover, the multianalyte design of the MpoxEG4-plex assay will assist in the rapid recognition of discordant results should a MPXV variant emerge with target sequence alterations. Nevertheless, assay inclusivity should be monitored for drift by periodic queries of MPXV genome databases for primer and probe sequence mismatches. For example, the MpoxEG4-plex rPCR primer and probes sequences were reassessed with the emergence of newly described clade IIb A.2.2.1 (proposed lineage G.1) strains associated with an outbreak in Sierra Leone and found to match this lineage perfectly (49).
A limitation of our study was the paucity of clade I positive specimens and strains available for testing. Clade I MPXV, but not clade II, is a select agent in the United States, and its transfer, possession, and use are subject to federal regulation. To address the lack of clade I clinical specimens, we contrived specimens using known concentrations of a single inactivated clade Ia strain. In addition, we received and tested a suspected clade I MPXV specimen during assay validation that was confirmed as clade Ib by WGS (36). While only two clade I strains were evaluated with the MpoxEG4-plex assay, in silico analyses supported excellent primer-probe binding inclusivity for clade I genome sequences. Nonetheless, assay robustness would benefit from additional testing using a diverse panel of MPXV strains. Another limitation was the evaluation of only a single specimen type, lesion swabs. Whereas lesion material is considered the optimal specimen for MPXV PCR, validation of additional specimen types such as rectal and oropharyngeal swabs would be useful for testing patients lacking an obvious rash or lesion and may contribute to case finding and public health intervention efforts (e.g., contact tracing) (21).
With sustained human-to-human transmission, MPXV will continue to evolve at an accelerated pace and new variants will likely emerge that may present diagnostic challenges for existing molecular tests (50). The multianalyte configuration of the MpoxEG4-plex assay and the selection of highly conserved essential gene targets will reduce the risk of false-negative test results due to emerging MPXV variants. Moreover, the ability of the MpoxEG4-plex assay to provide clade level information will facilitate tracking clade distributions and introductions into nonendemic regions. Finally, the new assay promises to provide rapid, accurate, and informative test results to help guide the public health response to prevent and control the spread of mpox.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the California Health and Human Services Agency for the Protection of Human Subjects. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
WP: Writing – review & editing, Conceptualization, Methodology, Investigation, Writing – original draft, Validation, Formal analysis. AE: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. JH: Conceptualization, Resources, Validation, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project was supported by funding from the U. S. Centers for Disease Control and Prevention (CDC), Mpox Crisis Response Cooperative Agreement [TP22-2201].
Acknowledgments
We are grateful to Todd Smith and Christina Hutson of the Poxvirus and Rabies Branch of the Centers for Disease Control and Prevention for providing inactivated monkeypox virus and orthopoxvirus nucleic acids, and our invaluable public health partners in affected local jurisdictions across California. We also thank Giorgio Cosentino, Monica Haw, Chantha Kath, Alexa Quintana, and Ricardo Berumen of the Vaccine Preventable Diseases section of VRDL for technical support, and Shiffen Getabecha of VRDL for specimen tracking. We gratefully acknowledge all data contributors, the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2025.1626030/full#supplementary-material
References
1. World Health Organization. WHO director-general declares ongoing monkeypox outbreak a public health emergency of international concern. (2022). Available online at: https://www.who.int/italy/news/item/23-07-2022-who-director-general-declares-the-ongoing-monkeypox-outbreak-a-public-health-event-of-international-concern (Accessed 23 July 2022).
2. World Health Organization. WHO director-general declares mpox outbreak a public health emergency of international concern. (2024). Available online at: https://www.who.int/news/item/14-08-2024-who-director-general-declares-mpox-outbreak-a-public-health-emergency-of-international-concern (Accessed 14 August 2024).
3. World Health Organization. 2022-24 Mpox (Monkeypox) outbreak: global trends. (2025). Available online at: https://worldhealthorg.shinyapps.io/mpx_global/#4_Global_situation_update (Accessed 5 February 2025).
4. Hata, DJ, Powell, EA, Starolis, MW, and Realegeno, SE. What the pox? Review of poxviruses affecting humans. J Clin Virol. (2024) 174:105719. doi: 10.1016/j.jcv.2024.105719
5. Bunge, EM, Hoet, B, Chen, L, Lienert, F, Weidenthaler, H, Baer, LR, et al. The changing epidemiology of human monkeypox-a potential threat? A systematic review. PLoS Negl Trop Dis. (2022) 16:e0010141. doi: 10.1371/journal.pntd.0010141
6. National Institutes of Health. The antiviral tecovirimat is safe but did not improve clade I mpox resolution in Democratic Republic of the Congo. (2024). Available online at: https://www.nih.gov/news-events/news-releases/antiviral-tecovirimat-safe-did-not-improve-clade-i-mpox-resolution-democratic-republic-congo (Accessed 15 August 2024).
7. Angelo, KM, Smith, T, Camprubí-Ferrer, D, Balerdi-Sarasola, L, Díaz Menéndez, M, Servera-Negre, G, et al. Epidemiological and clinical characteristics of patients with monkeypox in the GeoSentinel network: a cross-sectional study. Lancet Infect Dis. (2023) 23:196–206. doi: 10.1016/S1473-3099(22)00651-X
8. Brosius, I, Vakaniaki, EH, Mukari, G, Munganga, P, Tshomba, JC, De Vos, E, et al. Epidemiological and clinical features of mpox during the clade Ib outbreak in south Kivu, Democratic Republic of the Congo: a prospective cohort study. Lancet. (2025) 29, 405:547–59. doi: 10.1016/S0140-6736(25)00047-9
9. O'Toole, Á, Neher, RA, Ndodo, N, Borges, V, Gannon, B, Gomes, JP, et al. Apobec3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016. Science. (2023) 382:595–600. doi: 10.1126/science.adg8116
10. Vakaniaki, EH, Kacita, C, Kinganda-Lusamaki, E, O'Toole, Á, Wawina-Bokalanga, T, Mukadi-Bamuleka, D, et al. Sustained human outbreak of a new MPXV clade I lineage in eastern Democratic Republic of the Congo. Nat Med. (2024) 30:2791–5. doi: 10.1038/s41591-024-03130-3
11. Shchelkunov, SN, Totmenin, AV, Safronov, PF, Mikheev, MV, Gutorov, VV, Ryazankina, OI, et al. Analysis of the monkeypox virus genome. Virology. (2002) 297:172–94. doi: 10.1006/viro.2002.1446
12. Elde, NC, Child, SJ, Eickbush, MT, Kitzman, JO, Rogers, KS, Shendure, J, et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. (2012) 150:831–41. doi: 10.1016/j.cell.2012.05.049
13. Senkevich, TG, Yutin, N, Wolf, YI, Koonin, EV, and Moss, B. Ancient gene capture and recent gene loss shape the evolution of Orthopoxvirus-host interaction genes. mBio. (2021) 12:e0149521. doi: 10.1128/mBio.01495-21
14. Gigante, CM, Plumb, M, Ruprecht, A, Zhao, H, Wicker, V, Wilkens, K, et al. Genomic deletions and rearrangements in monkeypox virus from the 2022 outbreak, USA. bioRxiv. (2022). doi: 10.1101/2022.09.16.508251
15. Monzón, S, Varona, S, Negredo, A, Vidal-Freire, S, Patiño-Galindo, JA, Ferressini-Gerpe, N, et al. Monkeypox virus genomic accordion strategies. Nat Commun. (2024) 15:3059. doi: 10.1038/s41467-024-46949-7
16. Isidro, J, Borges, V, Pinto, M, Sobral, D, Santos, JD, Nunes, A, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. (2022) 28:1569–72. doi: 10.1038/s41591-022-01907-y
17. Sereewit, J, Lieberman, NAP, Xie, H, Bakhash, SAKM, Nunley, BE, Chung, B, et al. ORF-interrupting mutations in Monkeypox virus genomes from Washington and Ohio, 2022. Viruses. (2022) 14:2393. doi: 10.3390/v14112393
18. Brinkmann, A, Kohl, C, Pape, K, Bourquain, D, Thürmer, A, Michel, J, et al. Extensive ITR expansion of the 2022 Mpox virus genome through gene duplication and gene loss. Virus Genes. (2023) 59:532–40. doi: 10.1007/s11262-023-02002-1
19. Desingu, PA, Rubeni, TP, Nagarajan, K, and Sundaresan, NR. Molecular evolution of 2022 multi-country outbreak-causing monkeypox virus clade IIb. iScience. (2023) 27:108601. doi: 10.1016/j.isci.2023.108601
20. Masirika, LM, Udahemuka, JC, Schuele, L, Ndishimye, P, Otani, S, Mbiribindi, JB, et al. Ongoing mpox outbreak in Kamituga, south Kivu province, associated with monkeypox virus of a novel clade I sub-lineage, Democratic Republic of the Congo, 2024. Euro Surveill. (2024) 29:2400106. doi: 10.2807/1560-7917.ES.2024.29.11.2400106
21. World Health Organization. Diagnostic testing for the monkeypox virus (MPXV). Interim guidance. (2024). Available online at: https://www.who.int/publications/i/item/WHO-MPX-Laboratory-2024.1 (Accessed 10 May 2024).
22. Huggett, JF, French, D, O'Sullivan, DM, Moran-Gilad, J, and Zumla, A. Monkeypox: another test for PCR. Euro Surveill. (2022) 27:2200497. doi: 10.2807/1560-7917.ES.2022.27.32.2200497
23. Michel, J, Targosz, A, Rinner, T, Bourquain, D, Brinkmann, A, Sacks, JA, et al. Evaluation of 11 commercially available PCR kits for the detection of monkeypox virus DNA, Berlin, July to September 2022. Euro Surveill. (2022) 27:2200816. doi: 10.2807/1560-7917.ES.2022.27.45.2200816
24. Huo, S, Chen, Y, Lu, R, Zhang, Z, Zhang, G, Zhao, L, et al. Development of two multiplex real-time PCR assays for simultaneous detection and differentiation of monkeypox virus IIa, IIb, and I clades and the B.1 lineage. Biosaf Health. (2022) 4:392–8. doi: 10.1016/j.bsheal.2022.10.005
25. Mills, MG, Juergens, KB, Gov, JP, McCormick, CJ, Sampoleo, R, Kachikis, A, et al. Evaluation and clinical validation of monkeypox (mpox) virus real-time PCR assays. J Clin Virol. (2023) 159:105373. doi: 10.1016/j.jcv.2022.105373
26. Mostafa, HH, Wall, G, Su, S-C, Hysa, G, Gong, L, Dadjeu, UC, et al. Multi-center evaluation of the research use only NeuMoDx monkeypox virus (MPXV) fully automated real-time PCR assay. J Clin Microbiol. (2024) 62:e0002824. doi: 10.1128/jcm.00028-24
27. Schuele, L, Masirika, LM, Udahemuka, JC, Siangoli, FB, Mbiribindi, JB, Ndishimye, P, et al. Real-time PCR assay to detect the novel clade Ib monkeypox virus, September 2023 to may 2024. Euro Surveill. (2024) 29:2400486. doi: 10.2807/1560-7917.ES.2024.29.32.2400486
28. Garrigues, JM, Hemarajata, P, Lucero, B, Alarcón, J, Ransohoff, H, Marutani, AN, et al. Identification of human Monkeypox virus genome deletions that impact diagnostic assays. J Clin Microbiol. (2022) 60:e0165522. doi: 10.1128/jcm.01655-22
29. Centers for Disease Control and Prevention. Health alert network. First case of clade I Mpox diagnosed in the United States. (2024). https://www.cdc.gov/han/2024/han00519.html (Accessed 18 November 2024).
30. Aden, TA, Blevins, P, York, SW, Rager, S, Balachandran, D, Hutson, CL, et al. Rapid diagnostic testing for response to the Monkeypox outbreak - laboratory response network, United States, may 17-June 30, 2022. MMWR Morb Mortal Wkly Rep. (2022) 71:904–7. doi: 10.15585/mmwr.mm7128e1
31. Upton, C, Slack, S, Hunter, AL, Ehlers, A, and Roper, RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. (2003) 77:7590–600. doi: 10.1128/jvi.77.13.7590-7600.2003
32. Li, Y, Zhao, H, Wilkins, K, Hughes, C, and Damon, IK. Real-time PCR assays for the specific detection of monkeypox virus west African and Congo Basin strain DNA. J Virol Methods. (2010) 169:223–7. doi: 10.1016/j.jviromet.2010.07.012
33. Kuchinski, KS, Jassem, AN, and Prystajecky, NA. Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline. J Clin Virol. (2020) 131:104581. doi: 10.1016/j.jcv.2020.104581
34. Gans, JD, and Wolinsky, M. Improved assay-dependent searching of nucleic acid sequence databases. Nucleic Acids Res. (2008) 36:e74. doi: 10.1093/nar/gkn301
35. US Food and Drug Administration. (2022). Monkeypox (mpox) emergency use authorizations for medical devices. EUA template for developers of molecular diagnostic tests for Monkeypox [mpox]. Available online at: https://www.fda.gov/medical-devices/emergency-use-authorizations-medical-devices/monkeypox-mpox-emergency-use-authorizations-medical-devices#templates (Accessed 24 December 2024).
36. Levy, V, Branzuela, A, Hsieh, K, Getabecha, S, Berumen, R 3rd, Saadeh, K, et al. First clade Ib monkeypox virus infection reported in the Americas - California, November 2024. MMWR Morb Mortal Wkly Rep. (2025) 74:44–9. doi: 10.15585/mmwr.mm7404a1
37. Zhang, Y, and Moss, B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. (1992) 187:643–53. doi: 10.1016/0042-6822(92)90467-4
38. Bayliss, CD, and Smith, GL. Vaccinia virion protein I8R has both DNA and RNA helicase activities: implications for vaccinia virus transcription. J Virol. (1996) 70:794–800. doi: 10.1128/JVI.70.2.794-800.1996
39. Williams, O, Wolffe, EJ, Weisberg, AS, and Merchlinsky, M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J Virol. (1999) 73:4590–9. doi: 10.1128/JVI.73.6.4590-4599.1999
40. Firth, C, Kitchen, A, Shapiro, B, Suchard, MA, Holmes, EC, and Rambaut, A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. (2010) 27:2038–51. doi: 10.1093/molbev/msq088
41. Gigante, CM, Korber, B, Seabolt, MH, Wilkins, K, Davidson, W, Rao, AK, et al. Multiple lineages of monkeypox virus detected in the United States, 2021-2022. Science. (2022) 378:560–5. doi: 10.1126/science.add4153
42. Wu, F, Oghuan, J, Gitter, A, Mena, KD, and Brown, EL. Wide mismatches in the sequences of primers and probes for monkeypox virus diagnostic assays. J Med Virol. (2023) 95:e28395. doi: 10.1002/jmv.28395
43. Vatsyayan, A, Arvinden, VR, and Scaria, V. Systematic in-silico evaluation of the diagnostic impact of Mpox genome variants in the current outbreak. Mol Diagn Ther. (2023) 27:275–80. doi: 10.1007/s40291-022-00629-8
44. Ghate, SD, Suravajhala, P, Patil, P, Vangala, RK, Shetty, P, and Rao, RSP. Molecular detection of monkeypox and related viruses: challenges and opportunities. Virus Genes. (2023) 59:343–50. doi: 10.1007/s11262-023-01975-3
45. Song, K, Brochu, HN, Zhang, Q, Williams, JD, and Iyer, LK. An in silico analysis of PCR-based Monkeypox virus detection assays: a case study for ongoing clinical surveillance. Viruses. (2023) 15:2327. doi: 10.3390/v15122327
46. Dobrovolná, M, Brázda, V, Warner, EF, and Bidula, S. Inverted repeats in the monkeypox virus genome are hot spots for mutation. J Med Virol. (2023) 95:e28322. doi: 10.1002/jmv.28322
47. Forni, D, Cagliani, R, Pozzoli, U, and Sironi, M. An APOBEC3 mutational signature in the genomes of human-infecting orthopoxviruses. mSphere. (2023) 8:e0006223. doi: 10.1128/msphere.00062-23
48. Delamonica, B, Davalos, L, Larijani, M, Anthony, SJ, Liu, J, and MacCarthy, T. Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes. Virus Evol. (2023) 9:1–13. doi: 10.1093/ve/vead047
49. Sahr, F, Grant, DS, Harding, D, Sandi, JD, and Campbell, AKO. Genomic epidemiology of Mpox virus in Sierra Leone. (2025). Available online at: https://virological.org/t/genomic-epidemiology-of-mpox-virus-in-sierra-leone/995 (Accessed July 05, 2025).
Keywords: mpox, orthopoxvirus, multiplex polymerase chain reaction, essential genes, disease outbreaks, public health surveillance
Citation: Probert WS, Espinosa A and Hacker JK (2025) Clade I or clade II? Targeting essential viral genes to differentiate monkeypox virus clades by multiplex real-time PCR. Front. Public Health. 13:1626030. doi: 10.3389/fpubh.2025.1626030
Edited by:
David Sue, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Khemraj Budachetri, Eurofins BioPharma Testing Columbia, United StatesKuncheng Song, Laboratory Corporation of America Holdings (LabCorp), United States
Copyright © 2025 Probert, Espinosa and Hacker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William S. Probert, V2lsbC5Qcm9iZXJ0QGNkcGguY2EuZ292
 William S. Probert
William S. Probert Alex Espinosa
Alex Espinosa Jill K. Hacker
Jill K. Hacker