- 1Chongqing Key Laboratory of Childhood Nutrition and Health, Department of Nephrology, Children's Hospital of Chongqing Medical University, National Clinical Research Center for Child Health and Disorders, Ministry of Education Key Laboratory of Child Development and Disorders, China International Science and Technology Cooperation Base of Child Development and Critical Disorders, Chongqing, China
- 2Big Data Center for Children's Medical Care, Children's Hospital of Chongqing Medical University, Chongqing, China
Background: Hand, foot, and mouth disease (HFMD) is a pediatric infectious disease prevalent in the Asia-Pacific region, requiring accurate forecasting for effective public health interventions. This study aims to compare the performance of time series foundation models (TimesFM and Moirai) with traditional methods (ARIMA and LSTM) in predicting HFMD outbreaks across various datasets and forecasting horizons.
Methods: The study analyzed weekly HFMD incidence data from Korea (2015–2024), Singapore (2012–2018), and Chongqing, China (2015–2024). Zero-shot versions of TimesFM (200 M and 500 M) and Moirai models were assessed against ARIMA and LSTM using forecasting horizons of 1 week, 5 weeks, and 10 weeks. Lookback windows of 50 and 100 weeks were used across experiments. Performance was evaluated based on forecasting accuracy across all datasets. Computational resource requirements were also analyzed.
Results: For 1-step predictions, ARIMA and Moirai delivered comparable results. TimesFM-500 M achieved the best performance for 5-step predictions with 100-week lookback windows across all datasets. For 10-step predictions, TimesFM-200 M performed well with 50-week lookback windows but showed weaker results with longer historical data. Foundation models demonstrated the potential for robust HFMD forecasting but required greater computational resources.
Conclusion: Time series foundation models can effectively predict HFMD outbreaks. While these models require more computational resources, their zero-shot capabilities simplify the forecasting process by eliminating the need for retraining.
1 Introduction
Hand, foot, and mouth disease (HFMD) is a common infectious disease that primarily affects children under 5 years of age and is caused by human enterovirus infections (1). Typical symptoms include fever, oral sores, and rashes on hands and feet. While most patients present mild symptoms and recover naturally within a week, some severe cases can lead to life-threatening complications (2). Over the past two decades, HFMD has caused multiple outbreaks worldwide, particularly in the Asia–Pacific region (3, 4). Accurate forecasting models are critical for improving disease surveillance, guiding medical resource allocation, and supporting targeted prevention strategies to mitigate public health risks (5, 6).
Autoregressive integrated moving average (ARIMA) and long short-term memory (LSTM) are traditional models widely used in epidemiological forecasting, including HFMD prediction in many countries and regions (7–10). However, ARIMA has basic limitations in capturing complex non-linear dynamics and long-term dependencies (11). LSTM networks overcome these limitations through their gated architecture, selectively retaining or forgetting information over long time sequences (12). This makes them excel at handling long sequence data and capturing long-term dependencies, and they have also shown good performance in epidemiological applications (13, 14). Nevertheless, LSTM models require extensive training data and computational resources for training (15, 16).
Recently, researchers have developed multiple time series foundation models (TSFMs) for time series analysis. These models use large-scale cross-domain pretraining to extract universal features for complex and heterogeneous prediction problems. TSFMs function as foundational building blocks for forecasting, classification, anomaly detection, and imputation. They offer effective out-of-the-box performance with minimal data requirements and can be fine-tuned for enhanced performance (17). Among these models, the masked encoder-based universal time series forecasting transformer (Moirai) from Salesforce and the time series foundation model (TimesFM) from Google are two representative models in terms of their architectural design, open-source availability, and flexibility of use. Moirai, which is based on a masked encoder architecture, learns universal time series features through pretraining on the LOTSA dataset. Its training objective is to reconstruct randomly masked segments of time series, enabling it to capture both the global context and local temporal patterns. The LOTSA dataset includes 27 billion observations across nine domains, such as healthcare, meteorology, economics, and transportation, including the COVID-19 time series. The pretraining objective of Moirai is to reconstruct randomly masked time series segments, enabling it to capture both global context and local temporal features, which are particularly important when dealing with heterogeneous and noisy data distributions like HFMD outbreaks (18). TimesFM adopts an autoregressive decoding structure with longer output patches, learning temporal patterns and contextual relationships through future sequence generation during pre-training. It supports dynamic prediction and handles long sequence generation, particularly in zero-shot and few-shot learning scenarios (19).
However, these TSFMs have not been applied to HFMD prediction. In this study, we evaluate the performance of these time series foundation models in HFMD forecasting and compare them with ARIMA and LSTM.
2 Methods
2.1 Data collection and study design
This study analyzes HFMD data collected from three regions in Asia. Data from Korea were obtained from the Korea Disease Control and Prevention Agency (KDCA) as publicly available weekly case counts (20). These data were collected by the KDCA through a well-established national reporting system in collaboration with designated surveillance institutions. Singaporean data were obtained from weekly reports published on the Singapore government's open data platform and were collected through healthcare reports, laboratory confirmations, and community-based monitoring programs. We excluded Singapore data after December 2018 because negligible HFMD cases were reported between January 2019 and December 2022 (21). For China, we obtained data from the Children's Hospital of Chongqing Medical University, covering the period from January 2015 to December 2024 (22). These data were collected following standardized hospital reporting protocols, and the original daily case counts from the hospital were aggregated into weekly totals for analysis. All datasets, accessed on January 10, 2025, consisted exclusively of aggregate case counts without any demographic or personally identifiable information.
The last 100 time points of each dataset were designated the test set to evaluate model performance. Comparative experiments were conducted across forecasting horizons of 1, 5, and 10 steps. For the LSTM, TimesFM, and Moirai models, performance comparisons were made under different lookback window lengths of 50 and 100.
2.2 ARIMA
In the ARIMA (p, d, q) model, parameter p denotes the order of autoregression, d represents the degree of differencing, and q indicates the order of the moving average. The appropriate (p, d, q) parameters are determined via the Akaike information criterion (AIC). For multistep forecasting, the model employs an iterative approach where single-step predictions are recursively fed as inputs until they reach the target forecast horizons. For all tested horizons (1-step, 5-step, and 10-step), the identified ARIMA parameters are 5,0,0 for the Korean dataset 5,0,2 for the Singapore dataset, and 1,1,0 for the CHCMU dataset.
2.3 LSTM
The LSTM model constructed in this study adopts a two-layer hidden layer structure, with each layer configured with 100-dimensional hidden units, and uses the GELU as the activation function. In terms of the training configuration, the model's batch size is 32, and the Adam optimizer with a learning rate of 0.001 is used. Unlike ARIMA models, which require iterative prediction, this LSTM architecture can directly output prediction sequences for multiple time steps (h=5 and h=10) through a single forward computation.
2.4 TimesFM and Moirai
This study used two sizes of TimesFM (TimesFM-1.0–200 M and TimesFM-2.0–500 M) (19) and three sizes of Moirai (Moirai-Small, Moirai-Base, and Moirai-Large with 14 M, 91 M, and 311 M parameters, respectively) (18). The input sequences were processed by sliding windows of 50 and 100 weeks. All the models were operated in zero-shot mode without additional fine-tuning on our datasets.
2.5 Evaluation of model performance
To assess the model's performance thoroughly, this study uses the root mean square error (RMSE) and mean absolute error (MAE). These metrics evaluate the model's performance from different perspectives. The formulas for calculating these metrics are provided.
where yi is the actual value at the i-th time point, ŷi is the predicted value at the i-th time point, and is the mean value of the actual values.
2.6 Model training setups
The training hardware configuration includes an NVIDIA RTX 4090 GPU and 24 GB of memory, with all the models trained on the same server environment. The software environment includes Python 3.10, which uses main libraries such as Sklearn, Statsmodels for ARIMA, and PyTorch for the LSTM, TimesFM and Moirai models.
3 Results
3.1 The datasets
The Korean dataset contains 524 data points, with an incidence of 12.41 ± 23.71 cases per week. The Singapore dataset consists of 365 data points, with an incidence of 644.96 ± 273.29 cases per week. The Chinese CHCMU dataset includes 518 data points, with an incidence of 282.43 ± 324.67 cases per week (Figure 1).
3.2 Single-step forecasting
For single-step prediction tasks, when the lookback window is set to 50 weeks, both ARIMA and Moirai achieve excellent performance across all three datasets with comparable prediction accuracies. Specifically, on the Korean dataset, the ARIMA model achieves an MAE of 4.286 and an RMSE of 6.193, whereas Moirai-Base shows similar performance, with an MAE of 4.303 and an RMSE of 6.302. For the CHCMU dataset, Moirai-Base attains an MAE of 48.763 and an RMSE of 129.987, approaching the performance of the ARIMA model, with an MAE of 50.068 and an RMSE of 124.427.
When the lookback window is extended to 100 weeks, for the Korean dataset, Moirai-Base records the lowest MAE (4.266), whereas TimesFM-500 M obtains the lowest RMSE (5.980). For the Singapore dataset, ARIMA performs best, with both the lowest MAE (79.298) and RMSE (99.901). For the CHCMU dataset, TimesFM-500 M records the lowest MAE (47.866), whereas ARIMA maintains the lowest RMSE (124.427). The prediction capability of the TimesFM models tends to improve as the lookback window length increases (Table 1). Visual comparisons of performance across all prediction steps and models are plotted in Supplementary Figure S1.
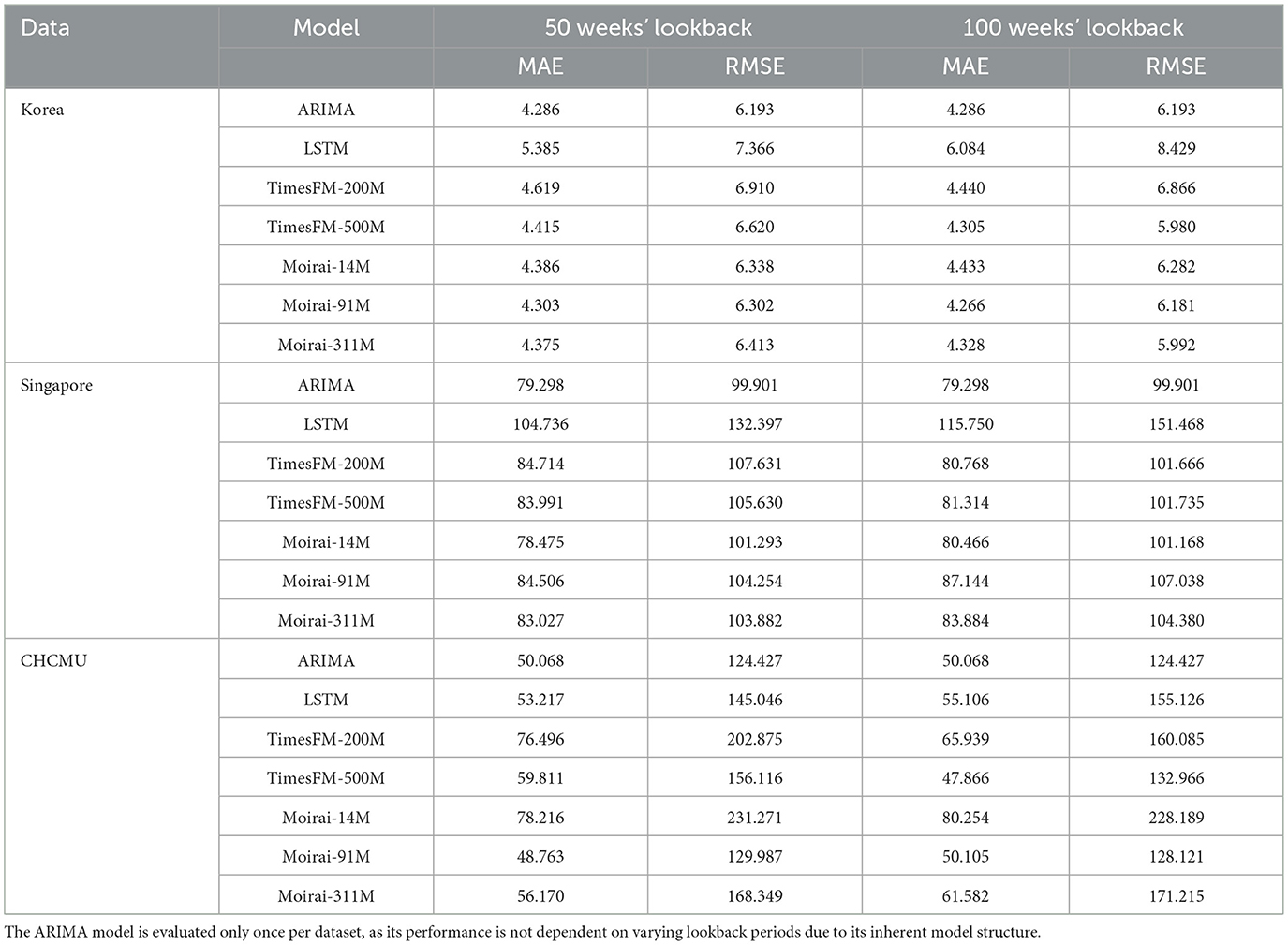
Table 1. Comparison of model performance in HFMD forecasting across Korea, Singapore, and CHCMU (1 week step).
3.3 Five-step forecasting
For five-step prediction tasks, with a lookback window of 50 weeks, TimesFM-500 M achieves the best results on the Singapore dataset, with an MAE of 121.498 and an RMSE of 165.370, closely followed by ARIMA, with an MAE of 122.199 and an RMSE of 166.696. For the CHCMU dataset, TimesFM-200 M achieves the best performance, with an MAE of 121.026 and an RMSE of 349.123.
When the lookback window extends to 100 weeks, TimesFM-500 M shows significant improvement, achieving the highest prediction accuracy across all three datasets. For the Korean dataset, the MAE decreases from 7.201 to 6.252, a reduction of 13.18%, and the RMSE decreases from 67.01 to 60.53, a decrease of 9.69%. For the CHCMU dataset, the MAE decreases from 121.026 to 101.153, a decrease of 16.22%, whereas the RMSE improves from 349.123 to 269.97, a decrease of 22.62%. These results indicate that the prediction capability of TimesFM-500 M substantially improves with increasing lookback window length (Table 2).
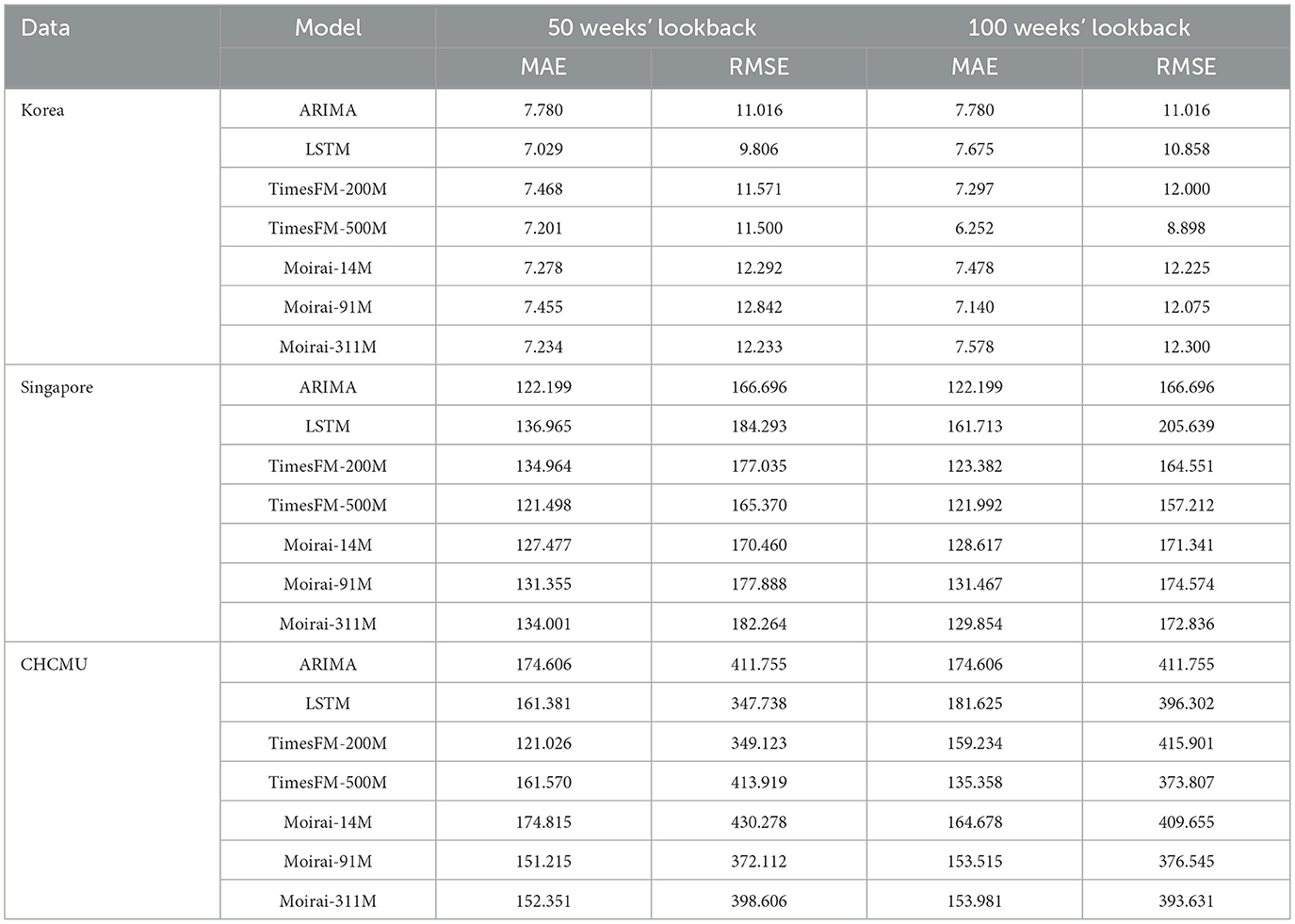
Table 2. Comparison of model performance in HFMD forecasting across Korea, Singapore, and the CHCMU (5-week step).
3.4 Ten-step forecasting
For ten-step prediction tasks with a 50-week lookback window, TimesFM-200 M achieves the best overall performance. For the Korean dataset, it achieves an MAE of 7.344 and an RMSE of 11.503. For the CHCMU dataset, the lowest MAE of 154.640 is recorded. For the Singapore dataset, TimesFM-200 M has an MAE of 160.263 and an RMSE of 209.113, which are slightly higher than the performance of LSTM, with an MAE of 156.755 and an RMSE of 200.940. Unlike single-step and five-step predictions, when the lookback window increases to 100 weeks, all the TimesFM models show a decline in performance (Table 3).
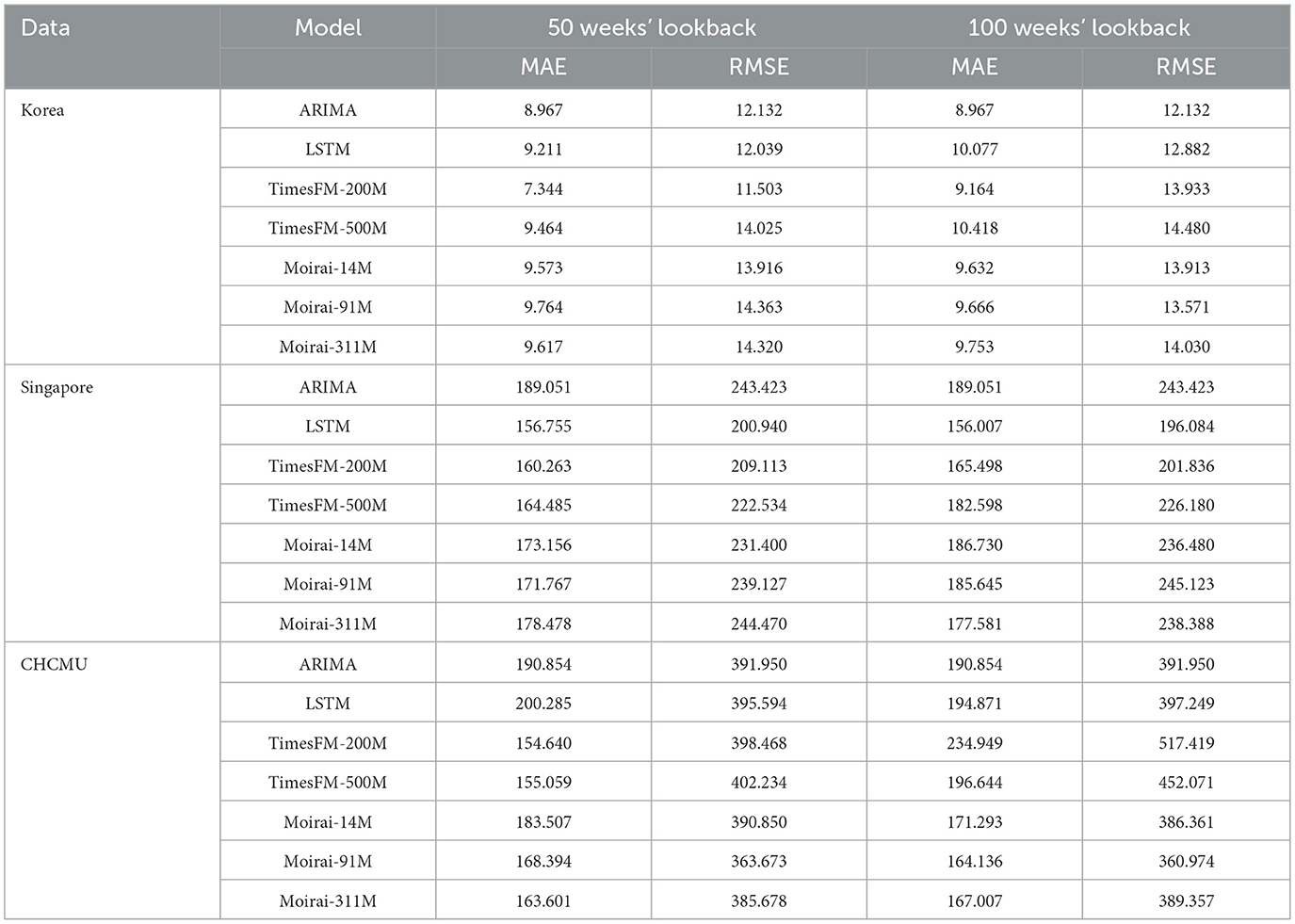
Table 3. Comparison of model performance in HFMD forecasting across Korea, Singapore, and the CHCMU (10-week step).
4 Discussion
This study presents the first evaluation of TSFMs for predicting the incidence of HFMD, comparing the performance of TimesFM and Moirai with that of traditional models (ARIMA and LSTM). Using weekly incidence data from Korea, Singapore, and Chongqing, China, the comparison examines different temporal scales for both prediction horizons and historical windows. The findings reveal that for single-step prediction, ARIMA and Moirai achieve comparable and excellent performance. For five-step prediction with a 100-week lookback window, TimesFM-500 M demonstrates superior performance across all three datasets. For ten-step prediction, TimesFM-200 M performs well but does not benefit from increased historical windows.
Foundation time series models outperform traditional approaches across multiple prediction settings without task-specific fine-tuning. This is attributed to their ability to leverage large-scale pretraining on diverse datasets and advanced architectures, such as Moirai's multi-scale projection and TimesFM's patch-based decoding, which enhance their capacity to model temporal patterns and adapt to geographic variability (23–25). Emerging research suggests that TSFM performance improves with increases in model scale, data diversity, and computational resources, highlighting these factors as key to their predictive success across varied time-series scenarios (26). However, traditional models such as ARIMA remain highly effective in specific use cases, indicating that model selection should be context dependent.
While time series foundation models require more computational resources for training and inference, they offer the advantage of zero-shot prediction, which eliminates the need for retraining on new datasets and reduces long-term costs (18). In contrast, traditional methods use less computational power but need expertise to determine the best parameters and architectures for each specific task and dataset. Thus, in cases where computational resources are available and quick deployment is needed, pre-trained models may be the better choice.
The performance differences between TimesFM and Moirai may be related to their architectural designs. TimesFM's decoder-only architecture appears to benefit long-term prediction tasks, possibly because of its ability to generate predictions incrementally and adjust them on the basis of previous predictions. This characteristic in handling temporal dynamics might contribute to improved long-term prediction accuracy (27), which matches our observations of relatively better performance in medium- to long-term predictions.
This study has several limitations. The geographical focus is narrow, and validation was limited to HFMD, which affects the generalizability of the findings. Uncertainty quantification was not explicitly addressed, and model performance under operational constraints, such as limited computational resources, was not tested. Further work should include fine-tuning for specific tasks to optimize pre-trained model performance.
In summary, this study explores the potential of time series foundation models in predicting the incidence of HFMD. The demonstrated zero-shot prediction capabilities and relatively better performance in certain settings offer new technical options for HFMD warning systems, with implications for enhancing disease surveillance and informing epidemic prevention strategies. However, its applicability to other infectious diseases remains to be validated. Future studies could improve prediction accuracy and broaden applicability by refining model architecture, validating against other diseases, and incorporating multisource data alongside epidemiological data to better understand the dynamics of disease transmission.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: The Korean HFMD data are publicly accessible from the Korea Disease Control and Prevention Agency (KDCA) Infectious Disease Portal (https://dportal.kdca.go.kr/pot/is/st/hfmd.do). Singapore HFMD data are available via Singapore's open data platform (https://data.gov.sg/datasets?agencies=Ministry+of+Health+(MOH)&page=1&query=infectious&resultId=d_ca168b2cb763640d72c4600a68f9909e). Data from Children's Hospital of Chongqing Medical University are available upon reasonable request by contacting the corresponding author.
Ethics statement
The studies involving humans were approved by Institutional Review Board of Children's Hospital of Chongqing Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YW: Investigation, Data curation, Writing – original draft, Methodology, Software, Visualization, Formal analysis. GH: Visualization, Investigation, Software, Data curation, Formal analysis, Writing – original draft, Methodology. CC: Methodology, Writing – original draft, Software, Validation. QL: Supervision, Project administration, Conceptualization, Resources, Writing – review & editing. XX: Funding acquisition, Resources, Conceptualization, Writing – review & editing, Supervision, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2022YFC2704900). The funders had no role in the design of the study, data collection, analysis, interpretation of the results, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2025.1634138/full#supplementary-material
References
1. Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, et al. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis. (2014) 14:308–18. doi: 10.1016/S1473-3099(13)70342-6
2. Wang Y, Zhao H, Ou R, Zhu H, Gan L, Zeng Z, et al. Epidemiological and clinical characteristics of severe hand-foot-and-mouth disease (HFMD) among children: a 6-year population-based study. BMC Public Health. (2020) 20:801. doi: 10.1186/s12889-020-08961-6
3. Bubba L, Broberg EK, Jasir A, Simmonds P, Harvala H. Circulation of non-polio enteroviruses in 24 EU and EEA countries between 2015 and 2017: a retrospective surveillance study. Lancet Infect Dis. (2020) 20:350–61. doi: 10.1016/S1473-3099(19)30566-3
4. Zhu P, Ji W, Li D, Li Z, Chen Y, Dai B, et al. Current status of hand-foot-and-mouth disease. J Biomed Sci. (2023) 30:15. doi: 10.1186/s12929-023-00908-4
5. Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. Big data for infectious disease surveillance and modeling. J Infect Dis. (2016) 214:S375–s9. doi: 10.1093/infdis/jiw400
6. Mizan T, Taghipour S. Medical resource allocation planning by integrating machine learning and optimization models. Artif Intell Med. (2022) 134:102430. doi: 10.1016/j.artmed.2022.102430
7. Peng Y, Yu B, Wang P, Kong DG, Chen BH, Yang XB. Application of seasonal auto-regressive integrated moving average model in forecasting the incidence of hand-foot-mouth disease in Wuhan, China. J Huazhong Univ Sci Technolog Med Sci. (2017) 37:842–8. doi: 10.1007/s11596-017-1815-8
8. Wang Y, Xu C, Zhang S, Yang L, Wang Z, Zhu Y, et al. Development and evaluation of a deep learning approach for modeling seasonality and trends in hand-foot-mouth disease incidence in mainland China. Sci Rep. (2019) 9:8046. doi: 10.1038/s41598-019-44469-9
9. Yoshida K, Fujimoto T, Muramatsu M, Shimizu H. Prediction of hand, foot, and mouth disease epidemics in Japan using a long short-term memory approach. PLoS ONE. (2022) 17:e0271820. doi: 10.1371/journal.pone.0271820
10. Zhu H, Chen S, Liang R, Feng Y, Joldosh A, Xie Z, et al. Study of the influence of meteorological factors on HFMD and prediction based on the LSTM algorithm in Fuzhou, China. BMC Infect Dis. (2023) 23:299. doi: 10.1186/s12879-023-08184-1
11. Siami-Namini S, Tavakoli N, Namin AS, editors. A comparison of ARIMA and LSTM in forecasting time series. In: 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA). Orlando, FL: IEEE (2018). doi: 10.1109/ICMLA.2018.00227
12. Graves A, Graves A. Long short-term memory. In; Supervised Sequence Labelling With Recurrent Neural Networks. Berlin: Springer (2012). p. 37–45. doi: 10.1007/978-3-642-24797-2_4
13. Lou HR, Wang X, Gao Y, Zeng Q. Comparison of ARIMA model, DNN model and LSTM model in predicting disease burden of occupational pneumoconiosis in Tianjin, China. BMC Public Health. (2022) 22:2167. doi: 10.1186/s12889-022-14642-3
14. Tsan YT, Chen DY, Liu PY, Kristiani E, Nguyen KLP, Yang CT. The prediction of influenza-like illness and respiratory disease using LSTM and ARIMA. Int J Environ Res Public Health. (2022) 19:1858. doi: 10.3390/ijerph19031858
15. Wang G, Wei W, Jiang J, Ning C, Chen H, Huang J, et al. Application of a long short-term memory neural network: a burgeoning method of deep learning in forecasting HIV incidence in Guangxi, China. Epidemiol Infect. (2019) 147:e194. doi: 10.1017/S095026881900075X
16. Zhang R, Song H, Chen Q, Wang Y, Wang S, Li Y. Comparison of ARIMA and LSTM for prediction of hemorrhagic fever at different time scales in China. PLoS ONE. (2022) 17:e0262009. doi: 10.1371/journal.pone.0262009
17. Liang Y, Wen H, Nie Y, Jiang Y, Jin M, Song D, et al. Foundation models for time series analysis: a tutorial and survey. In: Proceedings of the 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining; New York, NY: Association for Computing Machinery. (2024). p. 6555–65. doi: 10.1145/3637528.3671451
18. Woo G, Liu C, Kumar A, Xiong C, Savarese S, Sahoo D, et al. Unified training of universal time series forecasting transformers. In: Proceedings of the 41st International Conference on Machine Learning (ICML 2024). Vienna: ICML (2024).
19. Das A, Kong W, Sen R, Zhou Y, editors. A decoder-only foundation model for time-series forecasting. In: Proceedings of the 41st International Conference on Machine Learning (ICML 2024). Vienna: PMLR (2024).
20. Infectious Disease Portal. Korea Disease Control and Prevention Agency. (2025). Available online at: https://dportal.kdca.go.kr/pot/ (Accessed January 10, 2025).
21. Ministry of Health. Weekly Infectious Disease Bulletin Cases: data.gov.sg. (2024). Available online at: https://data.gov.sg/datasets/d_ca168b2cb763640d72c4600a68f9909e/view (Accessed January 10, 2025).
22. Wan Y, Song P, Liu J, Xu X, Lei X, A. hybrid model for hand-foot-mouth disease prediction based on ARIMA-EEMD-LSTM. BMC Infect Dis. (2023) 23:879. doi: 10.1186/s12879-023-08864-y
23. Ma Q, Liu Z, Zheng Z, Huang Z, Zhu S, Yu Z, et al. A survey on time-series pre-trained models. IEEE Trans Knowl Data Eng. (2024) 36:7536–55. doi: 10.1109/TKDE.2024.3475809
24. Huynh QT, Nguyen TH, Vu DT, Ngo MM, editors. anomaly detection for vietnamese financial market. In: 2024 18th International Conference on Advanced Computing and Analytics (ACOMPA). Ben Cat: IEEE (2024). doi: 10.1109/ACOMPA64883.2024.00016
25. Saravanan HK, Dwivedi S, Praveen P, Arjunan P, editors. Analyzing the performance of time series foundation models for short-term load forecasting. In: Proceedings of the 11th ACM International Conference on Systems for Energy-Efficient Buildings, Cities, and Transportation. New York, NY: Association for Computing Machinery (2024). doi: 10.1145/3671127.3699536
26. Yao Q, Yang C-HH, Jiang R, Liang Y, Jin M, Pan S. Towards neural scaling laws for time series foundation models. arXiv [Preprint]. (2024). doi: 10.48550/arXiv.2410.12360
Keywords: HFMD, time series analysis, foundation models, epidemic prediction, epidemiological modeling
Citation: Wang Y, Huang G, Chen C, Li Q and Xu X (2025) A comparative study of time series foundation models for hand, foot, and mouth disease forecasting: TimesFM, Moirai, and traditional approaches. Front. Public Health 13:1634138. doi: 10.3389/fpubh.2025.1634138
Received: 23 May 2025; Accepted: 05 September 2025;
Published: 25 September 2025.
Edited by:
Khalid Hattaf, Centre Régional des Métiers de l'Education et de la Formation (CRMEF), MoroccoReviewed by:
P. Kabbilawsh, NIT Calicut, IndiaImam Tahyudin, Amikom University Purwokerto, Indonesia
Copyright © 2025 Wang, Huang, Chen, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiu Li, bGlxaXU4MDlAaG9zcGl0YWwuY3FtdS5lZHUuY24=; Ximing Xu, eGltaW5nQGhvc3BpdGFsLmNxbXUuZWR1LmNu
†These authors share first authorship
 Yanqin Wang
Yanqin Wang Guoyu Huang
Guoyu Huang Ceran Chen2
Ceran Chen2 Qiu Li
Qiu Li Ximing Xu
Ximing Xu