Abstract
New Approach Methodologies (NAMs) hold great potential to fill data gaps for chemicals and modernisation of chemical risk assessment practices. Current toxicity testing is based on conventional approaches with high reliability on in-vivo studies, but with time, regulators are trying to move towards in-vitro and in silico tools enabling efficient risk assessment strategies. Herein, we discuss about different emerging techniques which are or can become a NAM including both in-vitro and in silico models with particular focus on reducing animal studies and improving decision-making for hazard and exposure assessment. We also discussed about the way to strengthen the regulatory and public confidence in different NAMs and automation of these approaches. Some of these NAMs can help in identifying biochemical mechanisms for toxicity, calculate the point of departure (PoD), develop adverse outcome pathways (AOP), translate risk to multiple species and quantify uncertainty from predictions for multiple chemicals. Scientists and regulators can work together to frame robust guidelines for the practical application of these tools and ensure reproducible results.
Highlights
• Contribution of in-vitro and in silico approaches towards building NAM.
• Automation of different approaches for risk assessment.
• Current limitations in data reporting and steps to improve data fairification.
• Integrating high throughput approaches for risk assessment.
1 Introduction
Development of alternative approaches based on the 3Rs principle; reduce, replace, and refine which provides the framework for minimizing animal use in scientific testing has led to different visions for improving toxicological risk assessment, like “toxicology for 21st century”, and “new approach methodologies (NAMs)” for chemical safety risk assessment (Bearth et al., 2025; Sewell et al., 2024; van der Zalm et al., 2022). Currently, there are more than ten thousand chemicals present in the market with thousands of them lacking relevant data for meaningful risk assessment. Testing that vast number of chemicals will take decades to generate enough data, and additionally conducting animal tests are too expensive and ethically difficult, posing challenges to solve the issue of assessing risk from these chemicals to humans and the environment (Sewell et al., 2024). Hence, there is a clear need for NAMs, which can help in filling the existing gaps and accelerate the process of risk assessment.
NAMs in simple words can be defined as emerging technology, methodology, approach, or combination thereof, having the potential to improve risk assessment for fulfilling critical information gaps and avoid or reduce the reliance on animal studies (Kavlock et al., 2018). Regulators like USEPA (United States Environmental Protection Agency), EFSA (European Food Safety Authority), ECHA (European Chemical Agency), and other related agencies have been developing frameworks to implement and use NAMs for regulatory applications (Di Nicola et al., 2023). Integrated approaches for testing and assessment (IATA) combine different data sources, to conclude on the toxicity of chemicals and often include data from NAMs. According to the OECD, an IATA combines multiple sources of information for hazard identification, hazard characterization, and chemical safety assessment. IATA frameworks integrate and weigh all relevant existing evidence, while also guiding the generation of targeted new data when needed, to support regulatory decision-making on potential hazards and risks (OECD, 2016). For simple endpoints or local effects, animal studies can be replaced with in-vitro, in-chemico, or in silico approaches, however, for complex endpoints, in the current scenario the need for animal studies can be reduced, not eliminated. These methods as NAMs provide data to be used within IATA frameworks. However, by using a comprehensive “weight of evidence” approach and leveraging emerging NAMs there is fair possibility that by weighing and integrating different types of available information decision making on complex endpoints can also be taken without conducting additional animal studies. Further, NAMs can be used to inform population variability, by identifying susceptible populations especially pregnant females, infants, and occupational exposure workers by considering individual health risk and developing regulatory limits for narrow toxicological index chemicals (Di Nicola et al., 2023; Kavlock et al., 2018).
Apart from providing data for risk assessment, NAMs can also help in prioritization, especially supporting grouping approach or read-across. For instance, Point of Departure (PoD) refers to the dose at which a biological response is first observed and serves as the basis for extrapolations in risk assessment (Sturla, 2018). A PoD can be derived from different metrics, including the no-observed-adverse-effect level (NOAEL), lowest-observed-adverse-effect level (LOAEL), benchmark dose (BMD) (Izadi et al., 2012). PoDs can also be estimated using NAMs, which can help in setting safety values when data are limited. This approach can reduce the need for high-tier testing that is currently required to reach conclusions in risk assessment (Levorato et al., 2019). Another approach can be the utilization of publicly available data from ToxCast and toxicogenomic to prioritize chemicals based on bioactivity calculated through in silico models (Farmahin et al., 2017). These data can also be used to enrich Adverse Outcome Pathways (AOPs), which describe a sequence of causally linked biological events leading to an adverse health or ecotoxicological effect, and serve as a central framework for mechanistic risk assessment (Ankley et al., 2023; Villeneuve et al., 2014). Computational methods are therefore an integral component of NAMs, as they enable the integration and interpretation of large datasets for hazard and risk assessment. Importantly, computationally approaches have been demonstrated to have clear cost-effectiveness by enabling rapid, large-scale screening compared to resource-intensive animal bioassays and have already been adopted in regulatory contexts such as the U.S. EPA’s Endocrine Disruptor Screening Program and Toxic Substance Control Act prioritization efforts. International agencies including ECHA, Health Canada, and the European Food Safety authority (EFSA) have also used these data, reflecting the growing regulatory acceptance of computational toxicology approaches for chemical risk assessment (Thomas et al., 2019).
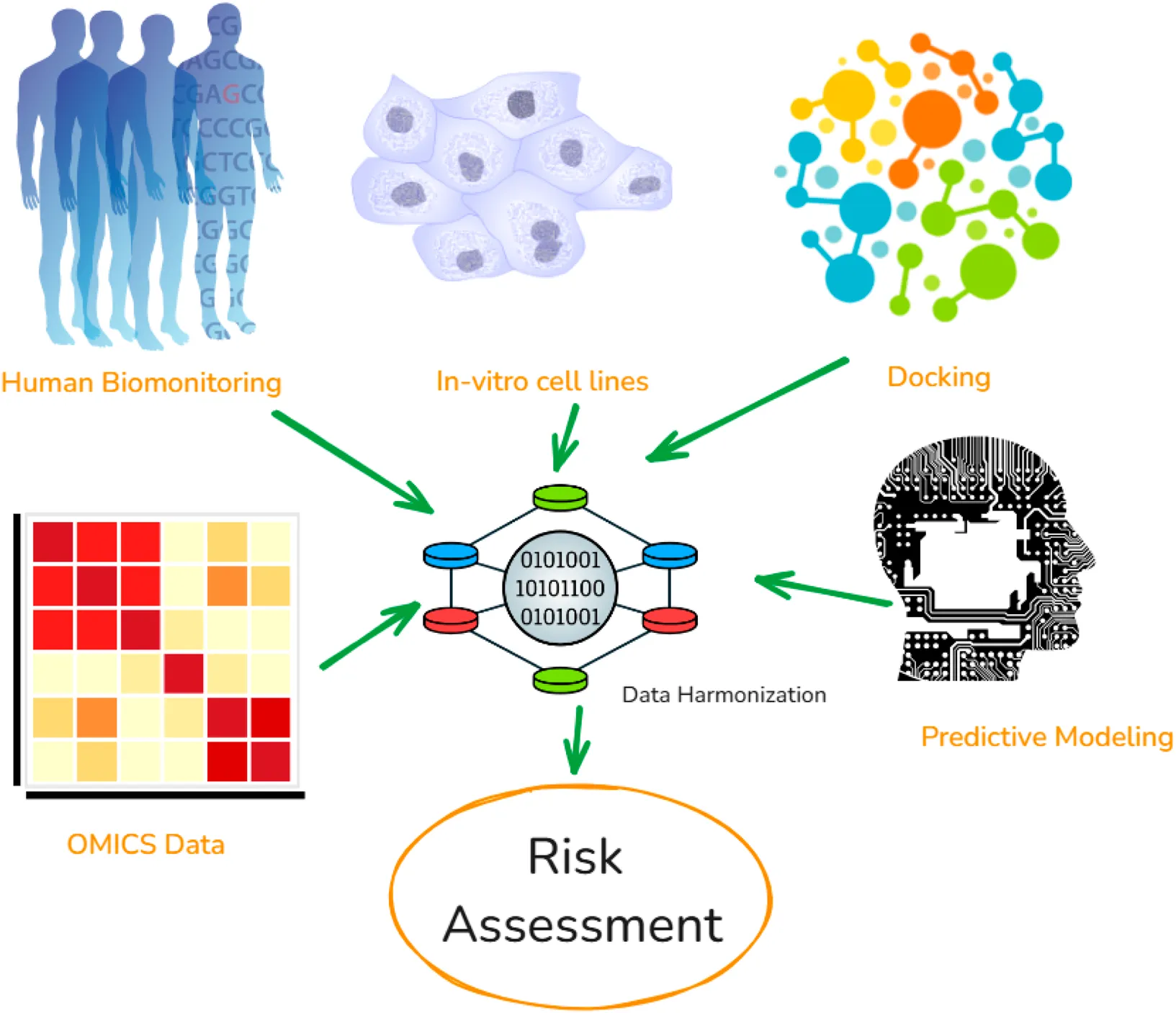
The objective of this review is to provide an overview on NAMs being used by risk assessors all over the world, highlight their limitations and challenges, discuss newly emerging NAMs which can help in environment and human health risk assessment in field like, high-throughput in-vitro, OMICS analysis, physics-based (molecular docking, molecular dynamic simulations, and density function theory), and data-driven methods (quantitative structure activity relationship: QSAR, physiologically based pharmacokinetic model: PBPK, systems biology models) (Figure 1). In addition, the review addresses approaches to promote the acceptance of these NAMs, including the role of AI/ML in their automation, and discusses the harmonization and FAIRification of data generated by NAMs, with emphasis on integrating evidence from in vitro and in silico studies to support AOPs.
FIGURE 1
Different approaches which can be utilized for developing New Approach Methodologies (NAMs), along with automation for supporting chemical risk assessment.
2 NAMs in chemical risk assessment
Integration of in-vitro and in silico approaches can be used to assess risk and identify the endpoints for regulatory purposes. In vitro approaches like 3D cell lines, organoids, spheroids, and microphysiological (MPS) systems are increasingly becoming famous and gaining acceptance by the researchers and toxicologists worldwide. But the OECD framework for the generation of data using in-vitro pipeline is still quite blurry, often limiting its usage for regulatory submissions. However, with increased focus on non-animal-based approaches by regulators, the translation of risk through in-vitro and in silico will become the standard framework in near future (Madden et al., 2020). In-silico based approaches can be divided into two categories, one being physics-based cheminformatic with the potential to use at early stages for extracting valuable insight about the chemical and the other being data driven models which can be used at later stages to translate the risk for humans. Techniques like molecular docking can be used to predict binding affinities and can help in prioritizing compounds with toxicity properties. Combining the initial data from cheminformatics to in-vitro or integrating multiple in silico models can help in building weight-of-evidence for newer and existing molecules.
Novel approaches like the OMICS pipeline, along with benchmark dose modelling (BMD) have the potential to reveal common biochemical response pathways and calculate different limit values like IC50 (Beale et al., 2022). However, due to different types of data being generated, standardisation, data quality, and interpretation are still the bottleneck for acceptance by regulators. The OECD OMICS reporting framework (OORF) has been developed to ensure the reproducibility and quality of data. It can be further used for calculating the PoD and as well combined with other in silico tools like PBK model for translation of risk (Farmahin et al., 2017).
Currently, PBK models are being used for risk translation, exposure reconstruction, chemical-chemical interaction, and much more to predict the fate of chemicals in living organisms (8,9). Toxicokinetic tools like httk, TK-plate, and others are gaining popularity and are continuously being improved to increase the incorporation into risk assessment. EFSA used a PBK model to calculate the tolerable weekly intake (TWI) for 4 PFAS considering immunotoxicity as the endpoint (Schrenk et al., 2020). Similarly, other in silico tools like QSAR for identifying chemical structural features with relevance for human and environmental hazard, systems biology models for evaluating toxicodynamics can support risk assessment and NAMs approaches. OECD toolbox currently uses a combination of multiple NAMs approaches like in-vitro, OMICS, PBK, QSAR, etc. to build weight of evidence for different chemicals and endpoints (Figure 1). Guidance for read-across and structurally similar compounds has also been published to support genotoxicity hazard by EFSA and ECHA (Henriquez et al., 2024). Read-across is an approach in which data from well-studied chemicals are used to predict the properties or toxicity of structurally or mechanistically similar chemicals, thereby filling data gaps while reducing reliance on animal testing. Recently, ECHA has recommended various in silico methods like nanoQSAR, grouping and read-across, PBK modelling, molecular dynamics simulations, and adverse outcome pathways for the prediction of risk posed by nanomaterials to humans and the environment (ECHA, 2022). Additionally, techniques like text mining and natural language processing support the curation of information from knowledge sources and facilitate hypothesis development for risk assessment. Implementations and advancement in QSAR, read-across, adverse outcome pathway (AOP), and integrated approach to testing and assessment (IATA) approaches, are always recommended by regulators for toxicological risk assessment (Table 1). Many of these approaches are already being accepted and used by pharmaceutical industries, but the regulators need to develop proper guidelines about using these tools considering their strengths and limitations for improving risk assessment (Table 1).
TABLE 1
| Models | Methods | Strength | Limitation for regulatory acceptance | Strategies for mitigation |
|---|---|---|---|---|
| In vitro | 2D and 3D cells | Study of specific molecular mechanisms Mimics tissue architecture and cell-cell interaction Mimics the microarchitecture and physiological responses of human organs |
Under-representation of the target organ Lesser cell density than the actual tissue Monoclonal origin of cells does, impairs intracellular signaling |
Production of reproducible and high-quality scientific data by following the six principles of Good Cell and Tissue Culture Practice (GCCP) |
| OMICS | Identifying significant genes, proteins and metabolites involved in interfering with processes Significant pathways being altered at molecular level |
Lack of standardization Lack of transparency in data processing from raw data to an interpretable result Broad in scope and generate data that may be applicable to a wide range of toxicological endpoints |
Use of OORF for reporting to ensure critical details on study design, data quality, and regulatory relevance to enhance reproducibility | |
| In silico | Molecular Docking | Calculating binding energy of chemicals with receptors Stability and reactivity of toxic compounds Determination of LD50 by integrating it with deep learning |
Inadequacy in scoring function and algorithms can compromise the results | Post-processing of docking results can provide more accuracy in the docking results |
| MDS | Can simulate the behaviour of macromolecules | Every change in system due to quantum mechanics cannot be simulated due to approximations Understanding of the complete interaction is not possible because of the very short scale of time step |
Longer simulation time can provide better outcome Use of different optimized/designed force fields on the basis of specific contaminants A OECD reporting guideline can be helpful during regulatory filling where MDS has been used |
|
| DFT | Predict the reactivity and stability of compound | Empirical force fields cannot capture bond breaking and are limited by parametrization, advances such as reactive force fields | QM/MM hybrid methods, and polarizable force fields are making MD both more accurate and more broadly applicable for toxicological studies | |
| QSAR PBK |
Enables prediction of chemical properties or bioactivities by linking molecular structure to experimental outcomes, allowing efficient design of new compounds without extensive testing Estimating human exposure using IVIVE-based mechanistic PBK models, with the potential to integrate toxicokinetic with AOPs by utilizing in vitro assay results to quantify AOPs |
Lack of acceptance threshold in the assessment element such as applicability domain fit, reliability scoring, or goodness-of-fit criteria Limited validation datasets due to unavailability of PK data of many chemicals |
Ensuring predictive models are built on high-quality, well-characterized experimental data, including details on test variability, potential confounding factors, and chemical purity to enhance model reliability and structure-effect correlation Following OECD PBK and other reporting template while building models |
Different NAM approaches with strengths and limitations for acceptance in risk assessment.
2.1 In vitro methodologies
The field of in-vitro models is evolving rapidly and has become crucial for toxicology and chemical assessment (Wang et al., 2024). These models have gained popularity, especially after the implementation of the 3Rs (replacement, reduction and refinement) principle, as they offer a robust alternative to animal testing. In the section below, we will be discussing about advanced biological system which carries the potential to become NAM.
2.1.1 2D to 3D cell culture and organoids
Traditional in-vitro models mostly rely on 2D configurations utilizing animal and human cell lines. These models have been widely used because of their simplicity and cost-effectiveness, but they fail to capture the complexity of human physiology, including tissue architecture, cell-cell interactions and metabolic capacity (Wang et al., 2024). This limits their predictive value for human health outcomes. To address these limitations, a wide range of more advanced models has emerged.
The 3D cultures, organoids derived from stem cells, and microphysiological systems (MPS) such as organ-on-a-chip platforms better mimics tissue architecture (Cacciamali et al., 2022). These systems, more accurately, stimulate the complexity of human tissue and its interactions, allowing for investigations of organ-specific toxicity, barrier integrity, and multicellular responses to chemical exposures (Duval et al., 2017; Ravi et al., 2015). Since responses to substances can differ significantly across species, these models also aid in identifying diseases that might not be evident in animal models (Loewa et al., 2023). To provide a clearer understanding of in-vitro models applicability in chemical risk assessment, examples of these emerging models are presented, highlighting both their potential and limitations.
3D culture particularly organoids, have been used to study the toxic effect of different substances, such as pesticides, microplastics, or particulate matter, on different organs. Although being simpler than the whole organ, they exhibit fundamental features such as cellular organization, differentiation and interaction (Wan et al., 2024). For example, liver organoids have been applied to study metabolism-mediated hepatotoxicity, while brain organoids have been used to investigate neurodevelopmental toxicity. A review conducted by Cong et al. summarized the recent advances in relation to chemical risk assessment, concretely they recap the toxicity of microplastics and other environmental compounds in a wide variety of organoids (Cong et al., 2024).
In accordance with the examples mentioned above, Cheng et al. (2022), Cheng et al. (2023) demonstrated that liver organoids recapitulate hepatotoxic and lipotoxic effects induced by microplastics, alone or in combination with BPA. Through evaluations of cellular toxicity, lipid metabolism alterations, oxidative stress or inflammatory pathways, they predicted the potential risk of microplastics in liver fibrosis and cancer, highlighting the utility of organoids to assess chemical interactions. Similarly, brain organoids treated with organophosphate pesticides or particulate matter have been observed to produce autism-related disorders by reducing the CDH8 protein and affecting neuronal growth. In addition, exposure to diesel particulate matter was associated with altered mitochondrial function, while exposure to a common additive in plastic products reduced neuronal proliferation and migration, and induced apoptosis in brain organoids, further demonstrating the value of organoids in revealing specific mechanism of chemical toxicity (Albanese et al., 2020). Although organoids show great potential in toxicity assessment, their ability to stimulate complex pharmacokinetic processes is still limited. For example, they cannot fully replicate the blood-brain-barrier, which is crucial for evaluating drugs intended for central nervous system (CNS) disease (Wan et al., 2024).
In the same line, to better capture organ complexity MPS, such as organ-on-a-chip systems, have been developed. These microfluidic devices mimics the mechanical and biochemical environment of human organs, allowing multi-organ systems with diverse cell co-cultures and 3D tissue structures connected by microfluidic channels that replicate blood vessels (Bhatia and Ingber, 2014). Among the different application of these models, they have revolutionized respiratory disease research and have opened new possibilities for studying kidney function (Ashammakhi et al., 2018; Shrestha et al., 2020). A recent study, which evaluate nanoparticles toxicity using a lung-on-a-chip model demonstrated that organ-on-a-chip systems have a greater accuracy in assessing adverse effects compared to animal models (Shrestha et al., 2020). Specifically, Shrestha et al., found that nanoparticles exposure increased the expression of the intercellular adhesion molecule 1 and the production of reactive oxygen species. In addition, lung-on-a-chip models allowed the study of nanoparticles transport, demonstrating an acceleration of the toxic effect on lung, underscoring the importance of mechanical movements on toxicity assessment (Shrestha et al., 2020). Similarly, kidney-on-a-chip models are potentially relevant for chemical risk assessment, due to this organ is the main way of excretion of environmental compounds (Lepist and Ray, 2016). Li et al. found that cadmium exposure in a kidney-on-a-chip device with glomerular endothelial cells declined cell-viability and increased endothelial injury by increasing LDH in a dose-dependent manner. Consequently, a reduction in the tight junction protein was also observed, indicating that cadmium disrupt barrier integrity and increase permeability depending on the dose administered (Li et al., 2017). These studies help to understand the alterations produced by environmental chemicals that modify their absorption or excretion, making them more aggressive.
Other new advanced in-vitro approaches, particularly tissue engineering, have shown promising applications for chemical risk assessment (Cui et al., 2018). This strategy enables the creation of functional tissue models such as skin, bone, skeletal muscle and cardiovascular systems, which can be used for drug screening, toxicity testing and disease modeling (Mirshafiei et al., 2024). For instance, mesenchymal stem cells (MSCs) have been applied to regenerate bone tissue, differentiating it into osteoblasts with specific growth factors such as TGF-β, TGF-α or HGF, providing platform to assess compound-induced effect on bond formation and scaffold integration (Stamnitz and Klimczak, 2021). Similarly, human PSCs-derived cardiomyocytes, when combined with extracellular matrix scaffolds, can mimic myocardium contraction and electrical function, offering a model to evaluate cardiotoxicity of chemicals (Jackman et al., 2015). Despite these advances and their high potential for risk assessment, tissue-engineered systems remain in development, and challenges such as vascularization, scalability, and standardization of bio-ink still need to be addressed to facilitate its broader applications in chemical risk assessment (Cui et al., 2018).
Despite these advances, regulatory uptake of these methods has been limited due to challenges in validation, reproducibility and harmonization. Several OECD guidelines already incorporate in-vitro approaches which have been approved such as OECD TG 439, 487 and 496 used for the hazard identification of irritant chemicals or those with the potential to induce serious eye damage by the reconstruction of human epidermis or cornea, as well as genotoxicity test (OECD, 2021b; OECD, 2023; OECD, 2024). However, addressing complex endpoints such as repeated-dose toxicity, carcinogenicity or reproductive toxicity remains a significant challenge in the validation and implementation of in-vitro models (Miccoli et al., 2022; Schmeisser et al., 2023). Further progress will depend on standardized protocols, international validation efforts, and the integration of different methodological domains (ECHA, 2022; Schmeisser et al., 2023; Perkins et al., 2022).
2.1.2 In vitro data for hazard assessment
Many laboratories around the world are working to develop and optimize alternative in-vitro test methods for hazard assessment of compounds for either pre-screening of compounds or to identify toxicity and mechanism of action. Many developed in-vitro models as well as high throughput techniques like imaging, high content screening etc. have the potential to reduce cost and animal usage but the validation of such models and techniques is complex and challenging and it takes years for such approaches to get official regulatory acceptance. Nonetheless, there have been a lot of examples in recent years with pre-validated and validated methods related to genotoxicity, developmental neurotoxicity, safety testing in cosmetic products and much more to partially or fully replace animal testing for hazard assessment.
One good example is OECD guideline 493 which is well characterized, validated and accepted providing in-vitro procedure for identification of skin irritants. Reconstructed human epidermis composed of epithelial cell layers is recommended along with validated test method for cell viability. As per the regulation 1907/2006 for REACH, this alternative testing can be used for skin irritation and corrosion for substances allowing complete replacement of animal experimentation (Bas et al., 2021). Data shows that non-animal test methods have increased three times for skin corrosion/irritation, four times for eye irritation/damage and twenty times for skin sensitization for period 2017–2019 under REACH (Burden et al., 2021). There is also a lot of effort ongoing for developmental neurotoxicity NAM incorporating high content imaging (HCI) for apoptosis, proliferation, synaptogenesis and neurite outgrowth. Carsten et al. in his work evaluated a dataset of 92 chemicals for 57 assays including both microelectrode array, network formation assay (NFA) and HCI assay relevant to DNT. They found that NFA along with HCI can help in assessing key functional processes in neuronal development which is a good starting point for classifying DNT positive and negative chemicals. To improve classification of DNT NAM evaluation chemicals, addition of DNT NAM assay to integrated screening paradigm should include both neuronal and non-neuronal cell types which should represent important neurodevelopmental processes like neuronal crest cell migration and myelination that are not included currently (Carstens et al., 2022). Genotoxicity is another area where in-vitro test methods are advanced and accepted by regulators (OECD TG 471, 473, 476, 479, 481, 482, and 487). But, still many of alternative in-vitro test methods are not fully accepted by regulators due to lack of validation. Test method validation refers to a process based on scientifically sound principle by which relevance and reliability of a particular approach, method, process or test are established for a specific purpose (Kandárová and Letašiová, 2011). In brief, any in-vitro test method, whether new or updated, needs to be relevant and reliable, i.e., validated to be accepted by regulators. Establishing this aspect is a critical step in validation for NAM. Often, comparison of data generated using in-vitro test methods between laboratories and within labs is challenging as most researchers do not work with standardized and detailed protocols. Implementing standard operating procedures (SOP) in the laboratory for tests can help to achieve harmonization and also help with validation and regulatory acceptance of the test methods. Also, the concept of IATA in which different test methods are combined to predict one end point is challenging as there are multiple test guidelines. A successful example of such approach is endpoint for human skin sensitization where IATA is based on AOP describing the linkage between chemical interaction with a biological system at molecular, subcellular, cellular, tissue and organ level (Bas et al., 2021).
2.1.3 OMICS analysis
It is important to note that, in addition to demonstrating toxic effects, in-vitro models allow the acquisition of mechanistic information like the interaction between molecules, alterations in cell metabolism or the specific functions of each organ, understanding how chemical perturb biological pathways. This is essential for establishing safety thresholds and relevant human PoD (Zhang et al., 2018). Therefore, by integrating these advanced in-vitro systems with other approaches such as OMICS, it becomes possible to link cellular responses to AOPs and derive quantitative safety information, enhancing their relevance for chemical risk assessment (Perkins et al., 2022). In addition, integrating these innovative technologies with in silico approaches like IVIVE based PBK modelling can help in improving risk assessment (Kreutz et al., 2024; Zhang et al., 2018). This strengthens their regulatory relevance and highlights how in-vitro models, when combined with complementary techniques enhances our understanding of the biological processes underlying human exposure to chemicals.
The use of NAMs in the context of 3D cell cultures, MPS, and organoids presents a promising future for chemical risk assessment, providing more accurate, ethical, and human-relevant alternatives to traditional animal models. Although these in-vitro models have potential, they remain limited in their ability to reproduce human physiology and predict systemic toxicity (Schmeisser et al., 2023). To maximize their usefulness, NAMs must be standardized and validated for their regulatory acceptance and complemented with analytical approaches that provide specific information at the molecular level. To this end, OMICS is a potential new tool that allow us to capture cellular and molecular responses in our models. Combining in-vitro models with these techniques improves the interpretability, reproducibility and regulatory relevance of in-vitro models.
OMICS technologies provide the essential molecular and cellular readings needed to turn in-vitro systems into robust tools for chemical risk assessment. Toxicogenomics approaches, in particular, can help in characterizing human risk and have the potential to become NAMs by analyzing the toxicity of the chemicals at the molecular and cellular level. In the broader aspect, OMICS encompasses genomics, transcriptomics, metabolomics and proteomics study which can identify changes in the genotype and phenotype of the organism and relate them to the AOPs (Addicks et al., 2023).
Transcriptomics data obtained from cell lines can be used to identify activation of specific targets and also to calculate PoD based on changes in gene expression as an approach to screen large number of chemicals. This can identify molecular initiating events (MIE) and key events (KE) within the AOP to understand the chemical’s mode of action based on targeted differential gene expression identification (Harrill et al., 2024). Multiple studies used transcriptomics data for risk assessment. For example, Matteo et al. analyzed BPA and 15 data poor alternatives and their effects on human breast cancer cell lines to analyze the toxicity using the BMD and PoD analysis (Matteo et al., 2023). Similarly, Addicks et al., evaluated the effects of Per- and polyfluoroalkyl substances (PFAS) on the human liver spheroids using the transcriptomic PoD (t-PoD) (Addicks et al., 2023). This, high-throughput transcriptomics data allows us to look for responses across multiple molecular pathways compared to traditional in-vitro data. Although, multiple approaches have been applied to calculate molecular level PoD, there is no consensus on t-POD, primarily due to its variability across cell types and experimental conditions.
Metabolomics includes identifying and quantifying the complete set of metabolites whereas proteomics identifies and characterizes the entire set of proteins providing a holistic understanding of cellular response. Malinowska et al., illustrated integration of in-vitro metabolomics with high-throughput screening to support decision-making in risk assessment of chemicals using HepaRG cells and CdCl2 as a case study (Malinowska et al., 2022). Viant et al., demonstrated the usability and reliability of in-vivo metabolomics study of 8 chemicals in rats to group them based on the activity of the metabolites ensuring to make the analysis FAIR and more reproducible, following guidelines from OECD and ECHA, and reflect the need to establish standard quality control steps for processing the data and improving risk assessment (Viant et al., 2024).
To improve accuracy and overcome the limitation of averaging signals across heterogeneous cell population leading to false discoveries, single-cell omics techniques are gaining attention to provide better insights into the complexity of the tissue of interest. This can provide information about how chemicals affect specific cell types or pathways, identify rare toxicologically relevant cell populations, and enable molecular phenotyping. However, one of the challenges in this study is isolation of cells from whole tissue before analysis, which employs more complex and costly techniques like Magnetic-activated cell sorting (MACS), Fluorescence-activated cell sorting (FACS), cell barcoding and various microfluidic techniques. To address this limitation, several platforms have been developed (Gierahn et al., 2017). Single-cell omics have a wide range of applications in cancer-research, immunology, and developmental biology. Wu et al., used single-cell RNA sequencing and multi-omics analyses to characterize a novel signature comprising 33 genes related to tobacco carcinogens to elucidate the progression of bladder cancer through fibroblasts-induced immune invasion and epithelial mesenchymal transition (Wu et al., 2024).
In spite of obtaining accurate information with the single-cell omics technique, in response to toxicants, multimodal single-cell omics should be used effectively to understand cellular behavior and regulation, as well as to correlate findings with toxicity pathways and AOPs. An advanced approach like spatial transcriptomics provides insights into the spatial distribution of changes in gene expression in the tissue and captures the specific position of changes in the transcriptome of the tissue sample. The combination of single-cell omics strategies and spatial transcriptomics can be very useful in identifying the organ-specific toxicity, which can help in AOP identification and related biomarkers discovery with more precision. Chen et al., analyzed the progression of cerebral malaria in the murine brain and its treatment with artemisinin using spatiotemporal profiling, which demonstrates the potential of the technique to be used in risk assessment of the chemical (Chen et al., 2025).
Currently, the major challenge in omics analysis is data analysis and integration of data generated from multi-omics techniques, Since the dimension of data generated is different, Multi Omics Factor Analysis (MOFA) (Argelaguet et al., 2018) and Regularized Generalized Canonical Correlation Analysis (RGCCA) (Tenenhaus and Tenenhaus, 2011) are two of the promising algorithms that can be used for the integration of multi-omics datasets. A combination of multimodal single-cell omics analysis and one of these algorithms may help toxicologists with better risk assessment of the chemicals. For the data analysis of omics data, preprocessing is a crucial step for the quality of the analysis, which should be uniform and robust for better reliability of the results for the regulatory bodies. OECD provides a framework called OECD Omics Reporting Framework (OORF) for reporting elements for the regulatory use of omics data from laboratory-based toxicology studies. The main aim for developing this framework is to facilitate data sharing and promote reproducibility in omics toxicology to make it more harmonized and standardized. It provides guidelines for reporting different microarray data: RNA-seq, qRT-PCR, NMR metabolomics, and proteomics along with data analysis and experimental designs. It also provides standard preprocessing steps for the analysts to be followed for improved analysis and minimum false discovery rate (OECD Omics Reporting Framework, 2023).
2.2 In-silico methodologies
2.2.1 Physics based methods
These methods are frequently used in drug discovery but is less utilized in NAMs for human health risk assessment. In recent years, computational chemistry approaches such as molecular docking, density functional theory (DFT) calculations, molecular dynamics simulations, and binding energy calculations using Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA) and Molecular Mechanics Generalized Born Surface Area (MM/GBSA) have gained significant attention as several studies have been published in environmental science journals (Trisciuzzi et al., 2018; Zheng et al., 2025). These approaches help predict the interactions between hazardous chemicals and human receptors, providing insights into their binding mechanisms.
Molecular docking is a technique in computational chemistry which elucidate the interaction between a ligand and a receptor complex by simulating the spatial and energy matching between ligands and receptors. Through this the mechanistic of the toxicological effect due to any chemical can be understood. Through this technique large scale screening of chemicals based on their affinity to a specific target can be done. By providing information on binding energies, interaction residues and interaction patterns, Ortega-Vallbanaet et al. demonstrated that tools like DockTox can facilitate the virtual screening of small compounds that target MIE-associated proteins (Ortega-Vallbona et al., 2025). This approach hence has potential to inform AOPs and mechanistic understanding in risk assessment. Advances such as semi-empirical quantum charge calculations like PM6 have improved accuracy in pose prediction, particularly for charged systems and metalloproteins (Bikadi and Hazai, 2009). Importantly, docking can generate hypotheses about potential toxicological targets when experimental data are lacking, supporting the reduction of animal testing and aligning with the 3Rs principle. The potential of the method can be validated in the fact that EFSA in its scientific opinion document also suggest the use of docking for grouping of multiple chemicals based on structural similarity considering multiple features like class of chemicals, presence of functional groups, breakdown products or similar precursor to increase the confidence in the assessment (More et al., 2021). However, the accuracy of results depends heavily on the quality of input structures, and docking programs/algorithms used for the purpose. They generate binding poses that resemble experimentally determined structures; however, challenges remain in accurately reproducing ligand conformations and in accounting for protein flexibility, since most docking approaches still treat the receptor as rigid proteins, which reduces reliability when conformational flexibility is essential. Apart from this no single program deems fit for all the targets (Warren et al., 2006). False positives are common and docking predictions are rarely sufficient as standalone evidence. A critical limitation arises from the reliance on scoring functions as they rely on large approximations to simplify complex molecular recognition processes, which introduces inaccuracies and reduces predictive performance (Cross et al., 2009).
On the other hand, molecular dynamic simulations (MDS) apply Newtonian mechanics to model the movements and interactions of atoms over time. By solving the equations of motion, MDS enables investigation of protein flexibility, conformational changes, and stability during ligand binding. These simulations provide valuable insights into the dynamic behaviour of proteins and their complexes, offering a mechanistic understanding that is critical for both elucidating biological function and supporting chemical risk assessment. Beyond structural dynamics, MDS can quantify conformational stability, residual mobility, compactness, solvent-accessible surface area (SASA), essential dynamics, and Gibbs free energy (FEL) analysis. FEL analysis helps identify high- and low-energy transition states and visualizes the dynamic behaviour of the system. Furthermore, MM/PBSA and MM/GBSA calculations use molecular dynamics simulation data to estimate binding energy components such as van der Waals, electrostatic, polar solvation, SASA, and total binding energy (ΔG) of toxic chemicals with estrogen related receptor gamma (Pathak et al., 2024).
Density Functional Theory (DFT) can be used to calculate the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energies. The HOMO-LUMO gap is an important parameter for determining the stability and reactivity of toxic chemicals (Pathak and Kim, 2024). Apart from these characteristics DFT can also provide quantum-chemical descriptor such as atomic charge distributions, which when combined with energy gaps and stability profile captured the mechanistic aspects of metabolic activation and have been successfully applied to predict Ames mutagenicity of primary aromatic amines (Kuhnke et al., 2019). Conceptual DFT has also been successfully applied in screening of emerging pollutants where a rule of thumb was provided for differentiation between pollutants. The study gave evidence about the bioaccumulation of pollutant to be “too hard” which binds with protein e.g., PFAS compounds and “too soft” which binds with lipids. This approach overcame the barrier of the non-availability of experimental data where computational method can be applied to decipher the properties of any chemical (Li X. et al., 2024). These approaches can find their value as NAM since they provide atomic-level insights into the binding strength of chemicals with their target receptors in humans. They bridge the gap between hazardous chemicals and their receptors by revealing key interactions, facilitating the identification of hazardous chemicals with varying affinities for specific targets. This information can support risk assessment and aid in developing strategies to mitigate the effects of harmful chemicals by designing competitive inhibitors that disrupt their interaction with human receptors (Jaeger-Honz et al., 2025). Together, they can provide a tiered approach that balances computational cost with predictive power for chemical risk assessment.
2.2.2 Data driven methods
Computational modeling techniques like QSAR, read-across, PBPK, and IVIVE play a pivotal role in toxicology prediction and chemical safety assessment and can be used as an alternative to animal testing for REACH regulation (European Chemicals Agency, 2020).
QSAR/quantitative structure property relationship (QSPR) is one of the methods recommended by REACH for supporting the substance registration process which basically relates the set of descriptors (X) with the response (Y). These models can be used to predict physico-chemical properties of compounds as well as biological activity or toxicity based on molecular structure for newer and existing chemicals. However, to facilitate the consideration of QSAR models for regulatory purposes, it should follow the QSAR modeling reporting framework (QMRF) which can ensure reproducibility and confidence in model predictions. Apart from individual models, platforms like OECD QSAR toolbox is being used by regulators to perform initial screening of chemicals as well as to predict ADMET properties (Yordanova et al., 2019). However, still the data for environmental chemicals is limited compared to the drugs, as a result most QSAR models are trained on drug data, leading to biasness for the prediction of environmental chemicals (Moreau et al., 2022). Additionally, poor performance of QSAR model could be due to multiple factors, including data quality and relevance, choice of descriptors, model complexity, quantification of uncertainties, and ensuring predictions fall within the model’s applicability domain. These aspects should be carefully evaluated by model developers, users, and third-party assessors so that the poor performance of the model can be avoided (Cronin et al., 2025). Importantly, QSAR models not only support hazard prediction but can also generate input parameters that, when integrated with PBK framework, enhance toxicokinetic modelling and translation of in vitro data to in vivo exposure.
Read-across is an important data-driven approach widely applied in regulatory toxicology, particularly for data-gap filling in chemical risk assessment. It uses information from a source (data-rich) chemical to predict the properties of a target (data-poor) chemical, thereby reducing the need for experimental testing (Read-Across Assessment Framework, 2017). The approach typically relies on showing the similarity of an analogue to a target substance by comparing chemical structure, physicochemical properties, metabolism and toxicokinetics, toxicodynamics, and structural alerts identified through predictive QSAR approaches. Such similarities and differences should be clearly documented, ideally within a data matrix that highlights consistent trends across the category. The robustness of read-across can be strengthened by applying the ECHA assessment framework, which requires a hypothesis-based justification, evaluation of potential contradictions, credible extrapolations, and clear specification of substance identity and composition, including impurities. Collectively, these practices increase transparency and regulatory confidence in read-across predictions (Alexander-White et al., 2022; Read-Across Assessment Framework, 2017).
For toxicokinetics of chemicals, a generic PBK model can be used to predict the concentration-time profile or toxicokinetics of chemicals (Deepika & Kumar, 2023). Currently, QSAR along with PBK model can help in generating parameterization data required for building the model. Open access models like HTTK generate data for parameters like absorption rate constant based on Caco-2 cell lines, metabolic data, protein binding, etc., which helps in reducing dependence on animal studies. OECD PBK model reporting framework includes the guideline for developing confidence in the model with integration of in-vitro through IVIVE, QSAR, along with PBK for toxicokinetic prediction, which is a step toward next-generation PBK modeling (NG-PBK) (Paini et al., 2019). Next-generation models can utilize “big data” such as information from high-throughput or high-content in-vitro screening assays or omics technologies along with PBK for predictive toxicology and safety assessment (Ram et al., 2022). Quantitative IVIVE (QIVIVE) which combines in vitro data with in silico methods such as PBK modelling together with information on metabolism, transport, and binding to estimate the likelihood of harmful effects from environmental exposures also plays a key role in NAMs-based risk assessment by translating in vitro toxicity data to in vivo exposure, aiding chemical prioritization (Moreau et al., 2022). The IVIVE uses in-vitro high-throughput biological responses for the prediction of in-vivo exposure to estimate the safety threshold for humans. The utilization of PBK enables the consideration of factors like redistribution and metabolism in the results obtained from in-vitro test that ultimately translates it into exposure relevant at the organism level (Hines et al., 2022; Najjar et al., 2022). The reliability of QIVIVE-PBK is limited by constraints in clearance predictions, metabolic kinetics, and focus on parent chemical toxicity, necessitating improvements for broader application (Moreau et al., 2022). For instance, in a QIVIVE study predicting nicotine delivery product exposure, the model successfully estimated toxic effects but faced challenges in addressing the potential interaction occurring between the chemicals since modelling and parameterizing could be done only for nicotine (Moreau et al., 2024). In a similar manner another study highlights the challenge of building the QIVIVE model for chlorpyrifos exposure to predict developmental neurotoxicity since the model was limited to multiple oral exposures rather than the real-world exposure scenario, which occurs simultaneously from different routes like dermal and inhalation (Algharably et al., 2023).
Nonetheless, these in silico models have potential applications for different regulatory purposes and the potential to become NAMs by facilitating efficient translation of risk. For instance, USFDA supports the use of IVIVE and PBK for filling data gaps to support drug development (Hines et al., 2022). Similarly, these approaches can be used for risk assessment but the clear guidelines about its usage for regulatory purposes are still lacking. HTTK was used to calculate the PoD for 448 chemicals using in-vitro bioactivity data (Paul Friedman et al., 2020) and to prioritize chemicals for further screening, supporting the potential of these tools for facilitating interpretation of experimental data.
2.3 Approaches for regulatory acceptance of NAMs
NAMs despite having the potential to replace animal experiments usually suffer from the scrutiny of the risk assessors due to either lack of proper validation or huge uncertainty or lack of harmonization. In case of in-vitro studies, OECD has published a guidance document (OECD guidance document 211) with the intent to assess the relevance of the large amount of data generated by the alternative in-vitro tests which might be of practical value. To check the scientific robustness and context appropriateness, OECD suggest these methods to be reported in a definite framework which should be helpful in evaluating uncertainties and scientific confidence (OECD, 2014). In line to this, to address the problem of overlooking to the test method details, Krebs et al. developed a template called as ToxTemp to follow the requirements of the OECD guidance document 211 which can guide the users to get the information details along with inclusion of acceptance criteria for test elements with sufficient description of cells employed in the study (Krebs et al., 2019). To operationalize this, OECD introduced Harmonised Template 201 (OHT 201) in 2016, a standard format for reporting mechanistic and intermediate effects across NAMs, including in vitro, in silico, and ex vivo methods (Carnesecchi et al., 2023).
According to EFSA, to enhance regulatory acceptance, docking outcomes should be interpreted within a defined applicability domain, integrated with complementary in silico approaches (QSAR, ML) under a weight-of-evidence framework, and linked to mechanistic anchors such as MoA or AOPs. Use of standardized open-source tools (e.g., OECD QSAR Toolbox, VEGA) can further increase transparency and reproducibility, thereby strengthening regulatory confidence (More et al., 2021).
In a similar manner OECD guidance document for PBK modeling guides the development of PBK models with their validation to increase their scientific validity. The main objective of this guidance document is to offer a transparent and uniform structure for model assessment in order to facilitate communication between PBK model developers and regulators who evaluate and approve their use (OECD, 2021a). To further streamline the data-driven NAMs OECD has developed its toolbox. The primary advantages of the QSAR Toolbox are its ability to screen for environmental fate endpoints, acute ecotoxicity endpoints, and toxicity endpoints like mutagenicity, skin/eye irritation, and sensitization. Early in the R&D process, the toolbox can also be used to screen possible novel chemicals to find those that are most likely to have a favourable hazard profile. It also has the capacity for clustering chemicals for read-across study as the toolbox incorporates the information and tool from diverse sources into a logical workflow (OECD). A list of discussed NAMs with their strengths, limitations and mitigation strategies has been provided in Table 1.
3 Combining in-vitro and in silico methods with AOP
AOP represents the shift from conventional toxicity testing to more mechanistic framework for establishing disease map or moving to adverse outcome. In general, in-vitro test methods by informing the biological pathways, in silico models for quantification and prediction of events, and AI methods for data-gap filling along with other experimental data can help in enriching the information for an AOP (Corradi et al., 2022). AOPs have seen a significant growth in last few years with many in silico approaches being developed to make them suitable for human risk assessment by considering real-exposure scenarios. In short, an AOP can help in addressing specific endpoints but currently they are more qualitative rather than quantitative which limits their usage for regulatory purpose.
There is already a lot of ongoing work related to quantification of AOPs using in-vitro and in silico approaches where in-vitro assays can provide data on mechanistic end points and in silico approaches like Bayesian network, generalized linear or regression model or mechanistic modelling can incorporate the data into quantitative framework to predict adverse outcome. Sewer et al. used data from advanced organotypic airway model exposed to tobacco heating system aerosol or combustible cigarette smoke for AOP 411 which is on decreased lung function due to oxidative stress. They used regression model for key event relationship (KER) using gene expression and other data from in-vitro model. Two mathematical modelling-based approach: 1) empirical data based for KER related to oxidative stress, and 2) mechanistic systems biology-based approach for other key events related to AOP 411 was used. Overall, this is a good example of AOP-based approach aligning with IATA principles by integrating in-vitro data with computational modelling to predict decreased lung function with aerosol exposure (Sewer et al., 2024).
The huge mechanistic knowledge captured in AOP network (AOPN) can help in the shift towards non-animal and human-based NAM. Vinken et al. showed a framework for NAM to predict hepatotoxicity composed of in-vitro systems which can allow tiered testing at transcriptional, translational and functional level for enriching AOP network. In vitro systems and a battery of assays mechanistically anchored in an AOP network can be helpful towards NAM development. Some chemical-induced liver toxicity like cholestatic and steatotic drug induced liver injury (DILI) can be predicted by simple in-vitro models like monolayer culture of hepatocytes while some liver endpoint needs complex models like MPS (Vinken, 2024).
Another successful example of NAM is where the health-based guidance value was calculated for PFOA using in-vitro transcriptomics data (Figure 2). An in silico workflow was developed with PFOA as lead molecule comprising of calculating a) PoD for significant genes, b) calculating freely available PFOA in-vitro, and c) using QIVIVE for estimating in-vivo dose and later tolerable daily intake (Silva et al., 2024). This framework can provide vital information for risk assessment and, by including the uncertainty factor, can further help in reducing the risk and choosing a conservative value for a sensitive population. There are some limitations to this approach. For instance, the relevance of molecular pathway perturbations in predicting apical toxic endpoints, the linkage between molecular and apical changes and guidelines for the uncertainty factor for PoD need to be established (Cattaneo et al., 2023).
FIGURE 2
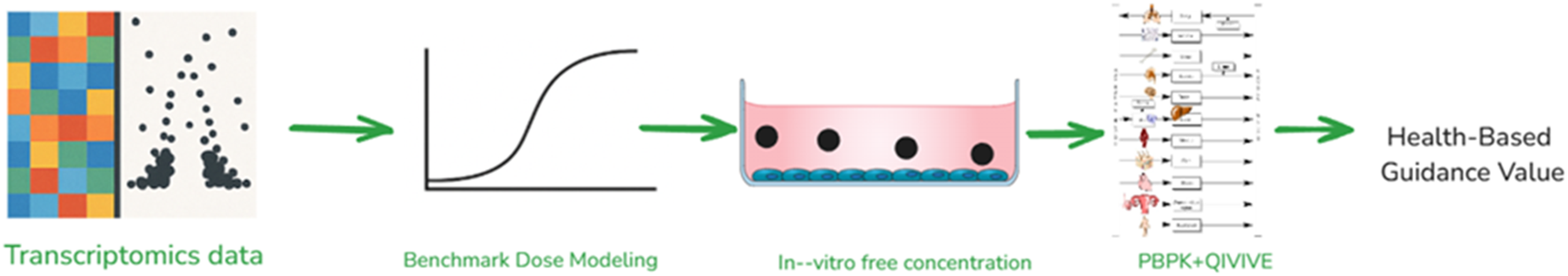
NAM based framework integrating in-vitro and in silico models for calculating human health-based guidance value.
NAMs carry the potential to transform risk assessment, especially with the improvement in computational modelling and high-throughput in-vitro data. NAM for specific endpoints like skin sensitization, skin and eye irritation, genotoxicity and endocrine activity are sufficient stand-alone with potential to inform about risk assessment. However, there is instance where a battery of NAMs has been used to conclude for the estimation of safety margin for pthalates; di(2-ethylhexyl) phthalate (DEHP) and di-n-butyl phthalate (DnBP) highlighting the value of integrating diverse NAMs within IATA. In this study, the AOP framework was applied to guide the selection of an in vitro assay for bioactivity testing, while Aggregate Exposure Pathways (AEPs) were developed to capture critical exposure information. These exposure data were then incorporated into a PBK model to predict internal metabolite concentrations. Finally, the model outputs were combined with in vitro-derived relative potency factors to calculate an internal margin of safety of ∼13,000 (Clewell et al., 2020). In a similar manner, a read-across case study on thirteen branched aliphatic carboxylic acids demonstrated how multiple NAMs can be integrated to strengthen regulatory confidence in data-gap filling. Structural similarity served as the starting point, but biological similarity was established using omics approaches—specifically, transcriptomic analysis of differentially expressed genes in human HepG2 liver cells. An AOP network for hepatic steatosis guided targeted in vitro testing of MIEs and KEs across multiple human liver models (HepG2, HepaRG spheroids, primary human hepatocyte), complemented by functional assays for mitochondrial dysfunction and lipid accumulation. In parallel, zebrafish embryo assays provided organism-level supporting evidence. To integrate the diverse mechanistic evidence, Dempster–Shafer decision theory (DST) was applied, allowing uncertainties to be quantified and to combine different evidence collected from in vitro assays. Collectively, these data supported a hypothesis-driven read-across, illustrating how in vitro NAMs, omics profiling, in silico integration, and AOP-guided testing can be combined within IATA to enhance confidence in predictions for chronic toxicity and reduce reliance on animal studies (Escher et al., 2022).
However, still NAM has some of the limitations especially to predict systemic toxicity, however efforts are ongoing in different project clusters like ASPIS which is a joint collaboration of three European projects for animal-free safety assessment of chemicals. A recent survey indicated that NAMs are being used by manufacturers at the internal level, but currently they are not considered sufficiently validated to be included in the REACH regulation. Also, the time needed for acceptance of NAM is quite high with limited NAM-based test guidelines adopted per year (Bearth et al., 2025). As a general perception, NAMs are considered less safe and traditional idea about animal testing being the “gold standard” needs to be changed to improve the acceptance of NAM in the regulatory framework.
4 Automation of different NAMs approaches
Over past few years, machine learning (ML) and artificial intelligence (AI) has played significant role in generating high-throughput data by in-vitro models, toxicogenomic and in silico modeling. AI approaches are becoming increasingly famous for toxicity prediction as NAM for next-generation risk assessment (NGRA). Integration of AI with high-throughput data can help in identification of bioactivity pattern across thousands of chemicals supporting prioritization of compounds and grouping them for reducing the experiments. Kim et al. used ToxCast database to develop mechanism-based toxicity-prediction models. Almost 1,485 bioassay dataset was collected and used for model training with 5 ML algorithms: logistic regression, decision tree, random forest, gradient boosting tree and XGBoost providing 24,500 models for 980 assays. 311 models were selected related to endpoints like acute toxicity, carcinogenicity, endocrine disruption and developmental and reproductive toxicity providing a starting point to incorporate AI based models for NGRA using in-vitro data (Kim et al., 2025).
New risk assessment methods can be developed by training ML models with large biological and chemical databases which can predict molecular interaction between chemicals and how the chemical is interacting with different receptors. ML models are being developed to predict ADME properties like caco-2 cell permeability, plasma unbound fraction, clearance to later incorporate in PBK models for predicting toxicokinetic of compounds (Li Y. et al., 2024; Vallance, 2016). Different type of ML based QSAR models are being developed by researchers to prioritize compounds and reduce the experimental burden (Pore and Roy, 2024).
With increasing computational power, large language models (LLMs) with contextual learning capabilities are emerging to improve automation, and with larger datasets they can be trained and fine-tuned to reduce the experimental burden for both existing and emerging chemicals (Klambauer et al., 2023). Recent advancements demonstrate the potential of LLMs like GPT-4 in the complex process of AOP construction, showing high accuracy in identifying MIEs, KEs, and linking them to adverse outcomes, thus accelerating data identification and synthesis from vast scientific literature (Shi and Zhao, 2024). LLMs can also help bridge multidisciplinary knowledge gaps and enhance standardization in AOP development (Shi and Zhao, 2024). Similarly, the GPT-3 model has been explored for data extraction and validation in the context of NAMs and AOPs, proving effective in recognizing semantic differences and extracting specific, often challenging, parameter-related entities from full-text articles, outperforming some traditional NLP tools in these areas (Viganò et al., 2024). This capability of LLMs to rapidly learn from limited examples and generate structured output can save significant manual effort in data curation and labelling, moving towards more automated and quantitative AOP mapping (Viganò et al., 2024).
Currently, efforts are ongoing toward automation of different approached for human safety assessment. KNIME analytics platform was used to develop automated workflow by integrating PBK, dynamics model and virtual cell-based assay (VCBA) model to evaluate chemical fate, kinetics and toxicity. VCBA can be used to guide in-vitro experiments especially test concentrations and time point for specific endpoint which can be later combined with PBK to calculate safe exposure levels for human (Sala Benito et al., 2017). Automated ML approaches are also coming up in recent years where autoML platforms like Vertex AI, dataiku and azure were employed by the researcher to develop a model for nanotoxicity prediction. However, none of three autoML platform outperform the conventional ML models, but it is beneficial for researchers with limited knowledge on ML based predictive model development (Xiao et al., 2024). Automation is not limited to computational models, but some work is ongoing for lab automation i.e., automation for compound dilution, cell seeding, media exchange, viability assessment and so on. Lab automation was achieved for embryonic stem cells (EST) for routine applications in 96- well format, enabling evaluation of several compounds in parallel (Witt et al., 2021). Automation in cell culturing and other experimental methods can help in improving reproducibility, achieving high standardization, low material consumption and increasing the confidence by reducing manual errors. Some work is also ongoing in European project like PARC where researchers are working on automated approach combining different in silico tools to develop an IATA for endocrine disruption. In a recent study, effort was made to develop a pipeline combining molecular docking simulation with chatGPT to simulate pharmacokinetic prediction of Oritavancin (Fatoki et al., 2024). Overall, it helped in deciphering pharmacological profile of drug along with clinical application. Similar work can be adopted for chemical risk assessment and improved by combining lab automation with molecular docking, QSAR, PBK and pharmacodynamic (PD) which can help in enriching biological knowledge about newer and existing compounds. Combining these approaches together through LLM and making them automatized can be a step towards IATA using NAMs data, however, there remain multiple challenges, especially the quality of data and format of output parameters and completeness of training data, addressing the “black box” nature of some LLMs, managing computational costs, and overcoming token limitations for full-text processing (Shi and Zhao, 2024; Viganò et al., 2024).
5 Harmonization and data fairification
Harmonization and data fairification are important pillars since implementing these steps can lead to the creation of machine-readable models, improve data reporting, and hence facilitate the application of NAMs. Experimental and measurement data are necessary and essential resources for modelling, analysis, drawing conclusions, and providing recommendations or guidelines. However, it is common that the background information needed in areas such as hazard, health, environment, epidemiology or toxicogenomics, among others, is found in different sources or databases, requiring their integration, which presents great difficulties (Mattes et al., 2004; Ball et al., 2022; Rozony, 2024; Samsa et al., 2005). These databases usually present heterogeneity in terms of format differences, lacking semantic integrity of form, structure, and terminologies or definitions, and may exist in structured, semi-structured or unstructured form (Samsa et al., 2005; Sindhu and Hegde, 2017). An example of this is presented in environmental risk studies, where the influence of the short- and long-term exposure to a pollutant on human health requires the unification of data from air quality, climate, meteorology and orography databases, together with public health data reflecting the outcomes or injuries. In addition, it is worth mentioning the quality of the data, which are shown to be redundant, with noise in the measurements, lack of quantity or even missing data that it is not always clear how to fill in, leading to the use of data from other sources that are sometimes used inappropriately (Ball et al., 2022). The difficulties increase in the current generation, where data grows exponentially and dynamically, which on the one hand, makes it difficult to analyze in real time and on the other hand, complicates its storage, availability, and use due to its large size.
Given the variability of the base data sources, there is a notorious need for harmonization in order to obtain standardized and quality data (Mattes et al., 2004; Ball et al., 2022). The difficulties present in this step in terms of low data quality, lack of standardization, and heterogeneity imply the need to involve experts who, on many occasions, must possess extensive knowledge in disparate fields such as database programming and manipulation, data analysis, and model development and simulation. This is where data science takes importance, which can notoriously help to reduce the amount of data and combine information from different domains into a single, reliable base from which to perform subsequent analyses, especially when manual manipulation is becoming impossible, given the large volume of real-time data. This requires novel methods for information manipulation and cleaning, and for this purpose NAMs present many advantages in extrapolation, data analysis and decision making. However, in the field of human health assessment, among others, data generated with NAMs are not performed or reported in a standardized way. Contrary to this, animal research benefits from established guidelines that simplify these aspects, as indicated in some reviews, which is a prerequisite for the implementation of alternative methodologies for risk assessment (Schmeisser et al., 2023). The use of these guidelines in human health would facilitate the extraction and interpretation of results and conclusions on the quality and usefulness of the information, as well as replicability. In this area, harmonization is even more relevant given the possibility of obtaining databases of great value and size, with information of great interest and quality, increasing the statistical power of future analyses (Cheng et al., 2024).
In the field of data processing, NAMs have strengths and limitations depending on their specific application. In terms of heterogeneity, ontology-based frameworks and tools such as schema-matching are a good option for data transformation and integration, due to their ability to standardize and structure semantics, defining terminological relations. However, it presents difficulties in terms of scalability, due to the need for manual intervention. In addition, schema-matching is limited in its use in large unstructured databases (Rozony, 2024). In this sense, tools such as Machine Learning (ML) or Artificial Intelligence (AI) allow reducing the manual cost by their automation capacity, thus dealing with scalability, as well as increasing data quality in their task of filtering, cleaning and searching for errors, inconsistencies or missing values, and in harmonization by the identification of patterns. However, both require a large amount of real and reliable data for the initial learning (Dinov, 2016). Moreover, another drawback is that these tools compromise data security (Rozony, 2024) and therefore present a significant limitation in the health field, given the sensitivity of these data and confidentiality, marked by the regulation of the competent institutions, which also affects their storage and use (Pezoulas and Fotiadis, 2024). Big healthcare is complex due to its lack of structure and heterogeneity, requiring extensive pre-processing. In these fields, blockchain technologies are decentralized and secure, accessible in real time, and therefore of great utility in their use on sensitive data, despite the fact that scalability shows some difficulty in certain studies (Rozony, 2024).
In order to deal with all the aspects mentioned above, there are studies that propose systems based on standardized models in terms of syntax and semantics, facilitating the use by other researchers (Horsburgh et al., 2009). Others have focused on the development of technological frameworks and platforms, such as standardized ecosystems for environmental sensor data manipulation that integrate data visualization, manipulation, collection, storage and modelling, in web-friendly formats (Borges et al., 2018; Bumberger et al., 2025). Thus, it can be seen that the multiple new methodologies present advantages and limitations. A key requirement is the uniformity in syntax, vocabulary, and format to ensure consistency. Additionally, the ability to process large datasets rapidly, and with confidentiality is essential to adhere to the FAIR (Findable, Accessible, Interoperable, Reusable) principles and use standardized interfaces (Ali et al., 2023; Kush et al., 2020; Pezoulas and Fotiadis, 2024). Open data sharing through platforms such as GitHub and Open Science Framework (Dinov, 2016) enables broader access for researchers. This is especially important in those domains where up to 80% of the total time is spent on data preprocessing (Huber et al., 2021).
To make the data and model machine-readable, ontology can be used, which integrates symbolic knowledge representations with text mining techniques to enhance the extraction and categorization of information from unstructured text data. Unlike traditional text mining, which relies heavily on statistical methods, it utilizes predefined ontologies to provide a structured framework for interpreting the semantic content of text.
Currently, there are multiple life-science related ontologies that can help in providing harmonized vocabulary and construct models by embedding ontology, especially to learn features for machine learning models (Kulmanov et al., 2021). Currently, there are more than 800 ontologies on the BioPortal to describe biomedical and biological entities. For instance, while building machine learning models, ontology is playing a role in predictions like drug-target protein function, protein-protein, gene-disease association, and other biologically relevant relationships. Gene ontology is one of the biggest ontologies which includes findings from more than 180,000 papers representing almost 1,000,000 annotations, providing a comprehensive model of biological systems (Gene Ontology, 2023). Recognizing the current lack of ontologies for many NAMs like PBK modeling and AOPs, multiple partners in the European Partnership for the Assessment of Risks from Chemicals (PARC) are actively developing solutions. For instance, partners within PARC’s Work Package 7 (WP7) recently developed PBPKO (pbpko). This new ontology and supporting tools are tailored for annotating PBK models to enhance their FAIRification.
AOP ontology was initially used in 2017 by Lyle Burgoon, with the motivation of developing of artificial intelligence approach to predict the adverse outcome of chemicals based on chemical screening data from high-throughput assays. This approach utilizes computational reasoners using first order logic to infer potential adverse outcome from specific chemical. The AOP Ontology organizes AOPs as a parent class with child groupings, including Developmental Toxicology AOP, Disease AOP, Liver Toxicity AOP, and Reproductive Toxicology AOP. Each grouping contains specific AOPs, such as NeuralTubeDefect and DiabetesMellitusType2. Specific instances, like “AOP Neural Tube Defect via Hoxb1,” are developed from these AOPs.
Within Work Package 7 of the PARC initiative, partners are collaborating with international experts to further refine the AOP ontology. The goal is to create a comprehensive AOP-ontology covering the entire domain, which will significantly simplify annotation processes and improve machine-readability. Overall advancing domain ontology represents a crucial step towards data harmonisation and developing more reproducible and robust predictive models. Ultimately, this refined biological domain ontology will be instrumental in advancing NAMs that support IATA.
6 Conclusion
Overall, this paper discusses about the different approaches which are or can become a part of NAMs. Integration of multiple techniques to extract useful information is one of the goals of IATA and other regulatory frameworks for risk assessment. This paper illustrates the scope and challenges in the existing frameworks and the level of complexity required to develop them including the approaches to increase their regulatory acceptance. Complete transition from in-vivo to in-vitro is still not possible nor is intended, since there could be challenges in translation of different output data into meaningful human or environmental risk assessment. Instead, a multifaceted integration of in vitro, in silico, omics-based, and data-driven approaches is required, with ongoing challenges in translating diverse outputs into meaningful human and environmental risk assessment. For some frameworks to convert into NAMs, data harmonization and processing steps are required. Inclusion of ontology and training large language models can help in accelerating the process of risk assessment; however, a major bottleneck remains the quality and amount of data required to perform this task.
Statements
Author contributions
DD: Conceptualization, Writing – review and editing, Methodology, Investigation, Writing – original draft, Visualization. KB: Writing – original draft. SS: Writing – original draft. SK: Writing – original draft. RP: Writing – original draft. JB: Writing – original draft. OS: Writing – original draft. SV: Writing – original draft. VK: Writing – review and editing, Writing – original draft, Methodology, Resources, Visualization, Investigation, Supervision, Conceptualization, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. European Partnership for the Risk Assessment of Chemicals (PARC) and The Environmental Health and Exposome (ENVESOME). This study was financially supported by the European Union co-funded project European Partnership for the Risk Assessment of Chemicals (PARC) under grant agreement number 101057014. It was also financially supported by European Union co-funded project under Horizon 2020 program with grant agreement number 101157269. This publication only reflects author views, and they have no competing financial interest.
Acknowledgments
Authors would like to acknowledge reviewers for their valuable feedback which led to substantially improvement of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- ADMET
Absorption, Distribution, Metabolism, Excretion, and Toxicity; pharmacokinetic/toxicokinetic properties of chemicals
- AEP
Aggregate exposure pathway
- AI
Artificial intelligence
- AN
Artificial neural network
- AOP
Adverse outcome pathway; sequence of linked biological events leading to an adverse health or ecotoxicological effect
- BMD
Benchmark dose; dose–response modeling approach used to derive PoD
- BPA
Bisphenol A; chemical known for its endocrine disrupter property
- CNN
Convolutional neural networks
- CNS
Central nervous system
- DFT
Density functional theory; quantum chemistry method for predicting electronic structure of molecules
- DILI
Drug induced liver injury
- ECHA
European chemicals agency; EU regulatory authority managing chemical safety under REACH
- EFSA
European food safety authority; EU regulatory agency for food and chemical safety
- FEL
Free energy landscape; representation of conformational states in molecular simulations
- LD50
Lethal dose 50
- LLM
Large language model
- ΔG
Total binding energy
- GCCP
Good cell and tissue culture practices
- HOMO/LUMO
Highest occupied molecular orbital/Lowest unoccupied molecular orbital; descriptors of molecular stability and reactivity
- HepG2
A cell line exhibiting epithelial-like morphology that was isolated from a hepatocellular carcinoma of a 15-year-old, White, male youth with liver cancer
- IATA
Integrated approaches to testing and assessment; framework combining multiple data sources including NAMs
- IC50
Inhibitory concentration 50
- IVIVE
In vitro to in vivo extrapolation; translation of in vitro test data into in vivo exposure estimates
- KE
Key events
- LOAEL
lowest-observed-adverse-effect level; metric to derive PoD
- MACS/FACS
Magnetic-activated cell sorting/Fluorescence-activated cell sorting
- MIE
Molecular initiating events
- ML
Machine learning
- MM/PBSA and MM/GBSA
Molecular mechanics methods (Poisson–Boltzmann and Generalized Born surface area) to estimate binding free energies in molecular dynamics
- MOFA
Multi omics factor analysis
- MPS
Microphysiological systems (e.g., organ-on-a-chip); advanced in vitro models mimicking organ-level functions
- MSC
Mesenchymal stem cells
- NAM
New approach methodologies; emerging non-animal tools and approaches for chemical risk assessment
- NG-PBPK
New generation physiologically based pharmacokinetic modeling
- NGRA
New generation risk assessmen
- NLP
Natural language processing; AI techniques for extracting knowledge from scientific text
- NMR
Nuclear magnetic resonance; technique for structural and metabolomics analysis
- NOAEL
No-observed-adverse-effect level; metric to derive PoD
- OECD
Organization for economic co-operation and development; develops international testing guidelines for chemical safety
- OMICS
High-throughput biological data approaches such as genomics, transcriptomics, proteomics, and metabolomics
- OORF
OECD Omics reporting framework; guidelines for reporting and regulatory use of omics data
- PARC
Partnership for the assessment of risks from chemicals; EU-funded project supporting NAM development
- PBK
Physiologically based kinetic model; similar to PBPK, often used interchangeably
- PBPK
Physiologically based pharmacokinetic model; computational model simulating chemical ADME in organisms
- PFAS
Per- and Polyfluoroalkyl substances; a group of persistent environmental chemicals of regulatory concern
- PoD
Point of departure
- POS
Part-of-speech
- PPP1
Phosphoprotein phosphatases1
- QMRF
QSAR modeling reporting framework
- QSAR/QSPR
Quantitative structure–activity relationship/Quantitative structure property relationship; predictive models linking chemical structure to biological activity or toxicity
- REACH
Registration, evaluation, authorization and restriction of chemicals
- RF
Random forest
- RNN
Recurrent neural network
- SASA
Solvent accessible surface area; measure of protein surface exposed to solvent in molecular dynamics
- t-PoD
Transcriptomic PoD
- TWI
Tolerable weekly intake
- USEPA
United States environmental protection agency.
References
1
Addicks G. C. Rowan-Carroll A. Reardon A. J. F. Leingartner K. Williams A. Meier M. J. et al (2023). Per- and polyfluoroalkyl substances (PFAS) in mixtures show additive effects on transcriptomic points of departure in human liver spheroids. Toxicol. Sci.194 (1), 38–52. 10.1093/toxsci/kfad044
2
Albanese A. Swaney J. M. Yun D. H. Evans N. B. Antonucci J. M. Velasco S. et al (2020). Multiscale 3D phenotyping of human cerebral organoids. Sci. Rep.10 (1), 21487. 10.1038/s41598-020-78130-7
3
Alexander-White C. Bury D. Cronin M. Dent M. Hack E. Hewitt N. J. et al (2022). A 10-step framework for use of read-across (RAX) in next generation risk assessment (NGRA) for cosmetics safety assessment. Regul. Toxicol. Pharmacol.129, 105094. 10.1016/j.yrtph.2021.105094
4
Algharably E. A. Di Consiglio E. Testai E. Pistollato F. Bal-Price A. Najjar A. et al (2023). Prediction of in vivo prenatal chlorpyrifos exposure leading to developmental neurotoxicity in humans based on in vitro toxicity data by quantitative in vitro–in vivo extrapolation. Front. Pharmacol.14, 1136174. 10.3389/fphar.2023.1136174
5
Ali Z. Saddik A. Essifi B. Erraha B. Labbaci A. Ouessar M. (2023). ClimInonda: a web application for climate data management: a case study of the Bayech basin (Southwestern Tunisia). Sustainability15 (16), 12382. 10.3390/su151612382
6
Ankley G. T. Santana-Rodriguez K. Jensen K. M. Miller D. H. Villeneuve D. L. (2023). AOP Report: adverse outcome pathways for aromatase inhibition or androgen receptor agonism leading to male-biased sex ratio and population decline in fish. Environ. Toxicol. Chem.42 (4), 747–756. 10.1002/etc.5581
7
Argelaguet R. Velten B. Arnol D. Dietrich S. Zenz T. Marioni J. C. et al (2018). Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets. Mol. Syst. Biol.14 (6), e8124. 10.15252/msb.20178124
8
Ashammakhi N. Wesseling-Perry K. Hasan A. Elkhammas E. Zhang Y. S. (2018). Kidney-on-a-chip: untapped opportunities. Kidney Int.94 (6), 1073–1086. 10.1016/j.kint.2018.06.034
9
Ball N. Bars R. Botham P. A. Cuciureanu A. Cronin M. T. D. Doe J. E. et al (2022). A framework for chemical safety assessment incorporating new approach methodologies within REACH. Archives Toxicol.96 (3), 743–766. 10.1007/s00204-021-03215-9
10
Bas A. Burns N. Gulotta A. Junker J. Drasler B. Lehner R. et al (2021). Understanding the development, standardization, and validation process of alternative in vitro test methods for regulatory approval from a researcher perspective. Small17 (15), 2006027. 10.1002/smll.202006027
11
Beale D. J. Sinclair G. M. Shah R. Paten A. M. Kumar A. Long S. M. et al (2022). A review of omics-based PFAS exposure studies reveals common biochemical response pathways. Sci. Total Environ.845, 157255. 10.1016/j.scitotenv.2022.157255
12
Bearth A. Roth N. Jansen T. Holden L. Čavoški A. Di Consiglio E. et al (2025). New approach methodologies in human health risk assessment across European regulatory frameworks: status quo, barriers and drivers for regulatory acceptance and use. Environ. Int.196, 109279. 10.1016/j.envint.2025.109279
13
Bhatia S. N. Ingber D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol.32 (8), 760–772. 10.1038/nbt.2989
14
Bikadi Z. Hazai E. (2009). Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminformatics1 (1), 15. 10.1186/1758-2946-1-15
15
Borges M. C. Pallas F. Peise M. (2018). “Providing open environmental data—the scalable and web-friendly way,” in Advances and new trends in environmental informatics. Editors BungartzH.-J.KranzlmüllerD.WeinbergV.WeismüllerJ.WohlgemuthV. (Springer International Publishing), 21–37.
16
Bumberger J. Abbrent M. Brinckmann N. Hemmen J. Kunkel R. Lorenz C. et al (2025). Digital ecosystem for FAIR time series data management in environmental system science. SoftwareX29, 102038. 10.1016/j.softx.2025.102038
17
Burden N. Clift M. J. D. Jenkins G. J. S. Labram B. Sewell F. (2021). Opportunities and challenges for integrating new in vitro methodologies in hazard testing and risk assessment. Small17 (15), 2006298. 10.1002/smll.202006298
18
Cacciamali A. Villa R. Dotti S. (2022). 3D cell cultures: evolution of an ancient tool for new applications. Front. Physiology13, 836480. 10.3389/fphys.2022.836480
19
Carnesecchi E. Langezaal I. Browne P. Batista-Leite S. Campia I. Coecke S. et al (2023). OECD harmonised template 201: structuring and reporting mechanistic information to foster the integration of new approach methodologies for hazard and risk assessment of chemicals. Regul. Toxicol. Pharmacol.142, 105426. 10.1016/j.yrtph.2023.105426
20
Carstens K. E. Carpenter A. F. Martin M. M. Harrill J. A. Shafer T. J. Paul Friedman K. (2022). Integrating data from in vitro new approach methodologies for developmental neurotoxicity. Toxicol. Sci.187 (1), 62–79. 10.1093/toxsci/kfac018
21
Cattaneo I. Astuto M. C. Binaglia M. Devos Y. Dorne J. L. C. M. Fernandez Agudo A. et al (2023). Implementing new approach methodologies (NAMs) in food safety assessments: strategic objectives and actions taken by the European food safety authority. Trends Food Sci. Technol.133, 277–290. 10.1016/j.tifs.2023.02.006
22
Chen J. Bai Y. He X. Xiao W. Chen L. Wong Y. K. et al (2025). The spatiotemporal transcriptional profiling of murine brain during cerebral malaria progression and after artemisinin treatment. Nat. Commun.16 (1), 1540. 10.1038/s41467-024-52223-7
23
Cheng W. Li X. Zhou Y. Yu H. Xie Y. Guo H. et al (2022). Polystyrene microplastics induce hepatotoxicity and disrupt lipid metabolism in the liver organoids. Sci. Total Environ.806, 150328. 10.1016/j.scitotenv.2021.150328
24
Cheng W. Zhou Y. Xie Y. Li Y. Zhou R. Wang H. et al (2023). Combined effect of polystyrene microplastics and bisphenol A on the human embryonic stem cells-derived liver organoids: the hepatotoxicity and lipid accumulation. Sci. Total Environ.854, 158585. 10.1016/j.scitotenv.2022.158585
25
Cheng C. Messerschmidt L. Bravo I. Waldbauer M. Bhavikatti R. Schenk C. et al (2024). A general primer for data harmonization. Sci. Data11 (1), 152. 10.1038/s41597-024-02956-3
26
Clewell R. A. Leonard J. A. Nicolas C. I. Campbell J. L. Yoon M. Efremenko A. Y. et al (2020). Application of a combined aggregate exposure pathway and adverse outcome pathway (AEP-AOP) approach to inform a cumulative risk assessment: a case study with phthalates. Toxicol. In Vitro66, 104855. 10.1016/j.tiv.2020.104855
27
Cong J. Wu J. Fang Y. Wang J. Kong X. Wang L. et al (2024). Application of organoid technology in the human health risk assessment of microplastics: a review of progresses and challenges. Environ. Int.188, 108744. 10.1016/j.envint.2024.108744
28
Corradi M. P. F. de Haan A. M. Staumont B. Piersma A. H. Geris L. Pieters R. H. H. et al (2022). Natural language processing in toxicology: delineating adverse outcome pathways and guiding the application of new approach methodologies. Biomaterials Biosyst.7, 100061. 10.1016/j.bbiosy.2022.100061
29
Cronin M. T. D. Basiri H. Chrysochoou G. Enoch S. J. Firman J. W. Spînu N. et al (2025). The predictivity of QSARs for toxicity: recommendations for improving model performance. Comput. Toxicol.33, 100338. 10.1016/j.comtox.2024.100338
30
Cross J. B. Thompson D. C. Rai B. K. Baber J. C. Fan K. Y. Hu Y. et al (2009). Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J. Chem. Inf. Model.49 (6), 1455–1474. 10.1021/ci900056c
31
Cui H. Miao S. Esworthy T. Zhou X. Lee S. Liu C. et al (2018). 3D bioprinting for cardiovascular regeneration and pharmacology. Adv. Drug Deliv. Rev.132, 252–269. 10.1016/j.addr.2018.07.014
32
Deepika D. Kumar V. (2023). The role of “physiologically based pharmacokinetic model (PBPK)” new approach methodology (NAM) in pharmaceuticals and environmental chemical risk assessment. Int. J. Environ. Res. Public Health20 (Issue 4), 3473. 10.3390/ijerph20043473
33
Di Nicola M. R. Cattaneo I. Nathanail A. V. Carnesecchi E. Astuto M. C. Steinbach M. et al (2023). The use of new approach methodologies for the environmental risk assessment of food and feed chemicals. Curr. Opin. Environ. Sci. Health31, 1–8. 10.1016/j.coesh.2022.100416
34
Dinov I. D. (2016). Methodological challenges and analytic opportunities for modeling and interpreting Big Healthcare Data. GigaScience5 (1), 12. 10.1186/s13742-016-0117-6
35
Duval K. Grover H. Han L.-H. Mou Y. Pegoraro A. F. Fredberg J. et al (2017). Modeling physiological events in 2D vs. 3D cell culture. Physiology32 (4), 266–277. 10.1152/physiol.00036.2016
36
ECHA (2022). The use of alternative methods to predict the toxicity of chemicals. Available online at: https://www.qsartoolbox.org.
37
Escher S. E. Aguayo-Orozco A. Benfenati E. Bitsch A. Braunbeck T. Brotzmann K. et al (2022). Integrate mechanistic evidence from new approach methodologies (NAMs) into a read-across assessment to characterise trends in shared mode of action. Toxicol. In Vitro79, 105269. 10.1016/j.tiv.2021.105269
38
European Chemicals Agency (2020). The use of alternatives to testing on animals for the REACH regulation. European Chemicals Agency. 10.2823/092305
39
Farmahin R. Williams A. Kuo B. Chepelev N. L. Thomas R. S. Barton-Maclaren T. S. et al (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Archives Toxicol.91 (5), 2045–2065. 10.1007/s00204-016-1886-5
40
Fatoki T. H. Balogun T. C. Ojewuyi A. E. Omole A. C. Olukayode O. V. Adewumi A. P. et al (2024). In silico molecular targets, docking, dynamics simulation and physiologically based pharmacokinetics modeling of oritavancin. BMC Pharmacol. Toxicol.25 (1), 79. 10.1186/s40360-024-00804-z
41
Gene Ontology (2023). A study on valid in silico modelling tools and read-across approaches, including creation of case studies on read-across for specific (types of) nanomaterials. Available online at: https://www.geneontology.org/.
42
Gierahn T. M. Wadsworth M. H. Hughes T. K. Bryson B. D. Butler A. Satija R. et al (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods14 (4), 395–398. 10.1038/nmeth.4179
43
Harrill J. A. Everett L. J. Haggard D. E. Bundy J. L. Willis C. M. Shah I. et al (2024). Exploring the effects of experimental parameters and data modeling approaches on in vitro transcriptomic point-of-departure estimates. Toxicology501, 153694. 10.1016/j.tox.2023.153694
44
Henriquez J. E. Badwaik V. D. Bianchi E. Chen W. Corvaro M. LaRocca J. et al (2024). From pipeline to plant protection products: using new approach methodologies (NAMs) in agrochemical safety assessment. J. Agric. Food Chem.72 (19), 10710–10724. 10.1021/acs.jafc.4c00958
45
Hines D. E. Bell S. Chang X. Mansouri K. Allen D. Kleinstreuer N. (2022). Application of an accessible interface for pharmacokinetic modeling and in vitro to in vivo extrapolation. Front. Pharmacol.13, 864742. 10.3389/fphar.2022.864742
46
Horsburgh J. S. Tarboton D. G. Piasecki M. Maidment D. R. Zaslavsky I. Valentine D. et al (2009). An integrated system for publishing environmental observations data. Environ. Model. Softw.24 (8), 879–888. 10.1016/j.envsoft.2009.01.002
47
Huber R. D’Onofrio C. Devaraju A. Klump J. Loescher H. W. Kindermann S. et al (2021). Integrating data and analysis technologies within leading environmental research infrastructures: challenges and approaches. Ecol. Inf.61, 101245. 10.1016/j.ecoinf.2021.101245
48
Izadi H. Grundy J. E. Bose R. (2012). Evaluation of the benchmark dose for point of departure determination for a variety of chemical classes in applied regulatory settings. Risk Anal.32 (5), 830–835. 10.1111/j.1539-6924.2011.01732.x
49
Jackman C. P. Shadrin I. Y. Carlson A. L. Bursac N. (2015). Human cardiac tissue engineering: from pluripotent stem cells to heart repair. Curr. Opin. Chem. Eng.7, 57–64. 10.1016/j.coche.2014.11.004
50
Jaeger-Honz S. Hackett R. Fotler R. Dietrich D. R. Schreiber F. (2025). Conformation and binding of 12 Microcystin (MC) congeners to PPP1 using molecular dynamics simulations: a potential approach in support of an improved MC risk assessment. Chemico-Biological Interact.407, 111372. 10.1016/j.cbi.2025.111372
51
Kandárová H. Letašiová S. (2011). Alternative methods in toxicology: pre-validated and validated methods. Interdiscip. Toxicol.4 (3), 107–113. 10.2478/v10102-011-0018-6
52
Kavlock R. J. Bahadori T. Barton-Maclaren T. S. Gwinn M. R. Rasenberg M. Thomas R. S. (2018). Accelerating the pace of chemical risk assessment. Chem. Res. Toxicol.31 (5), 287–290. 10.1021/acs.chemrestox.7b00339
53
Kim D. Ahn S. Jeong J. Choi J. (2025). Part I. Systematic development of machine learning models for predicting mechanism-based toxicity from in vitro ToxCast bioassay data. Comput. Toxicol.35, 100371. 10.1016/j.comtox.2025.100371
54
Klambauer G. Clevert D. A. Shah I. Benfenati E. Tetko I. V. (2023). Introduction to the special issue: AI meets toxicology. Chem. Res. Toxicol.36 (8), 1163–1167. 10.1021/acs.chemrestox.3c00217
55
Krebs A. Waldmann T. Wilks M. F. van Vugt-Lussenburg B. M. A. van der Burg B. Terron A. et al (2019). Template for the description of cell-based toxicological test methods to allow evaluation and regulatory use of the data. ALTEX - Altern. Animal Exp.36 (4), 682–699. 10.14573/altex.1909271
56
Kreutz A. Chang X. Hogberg H. T. Wetmore B. A. (2024). Advancing understanding of human variability through toxicokinetic modeling, in vitro-in vivo extrapolation, and new approach methodologies. Hum. Genomics18 (1), 129. 10.1186/s40246-024-00691-9
57
Kuhnke L. ter Laak A. Göller A. H. (2019). Mechanistic reactivity descriptors for the prediction of ames mutagenicity of primary aromatic amines. J. Chem. Inf. Model.59 (2), 668–672. 10.1021/acs.jcim.8b00758
58
Kulmanov M. Smaili F. Z. Gao X. Hoehndorf R. (2021). Semantic similarity and machine learning with ontologies. Briefings Bioinforma.22 (4), bbaa199. 10.1093/bib/bbaa199
59
Kush R. D. Warzel D. Kush M. A. Sherman A. Navarro E. A. Fitzmartin R. et al (2020). FAIR data sharing: the roles of common data elements and harmonization. J. Biomed. Inf.107, 103421. 10.1016/j.jbi.2020.103421
60
Lepist E.-I. Ray A. S. (2016). Renal transporter-mediated drug-drug interactions: are they clinically relevant?J. Clin. Pharmacol.56 (S7), S73–S81. 10.1002/jcph.735
61
Levorato S. Rietjens I. M. C. M. Carmichael P. L. Hepburn P. A. (2019). Novel approaches to derive points of departure for food chemical risk assessment. Curr. Opin. Food Sci.27, 139–144. 10.1016/j.cofs.2019.02.016
62
Li Z. Jiang L. Tao T. Su W. Guo Y. Yu H. et al (2017). Assessment of cadmium-induced nephrotoxicity using a kidney-on-a-chip device. Toxicol. Res.6 (3), 372–380. 10.1039/C6TX00417B
63
Li X. Anderson J. S. M. Jobst K. J. (2024a). Bioaccumulative chemicals are either too hard or too soft: conceptual density functional theory as a screening tool for emerging pollutants. Environ. Int.183, 108388. 10.1016/j.envint.2023.108388
64
Li Y. Wang Z. Li Y. Du J. Gao X. Li Y. et al (2024b). A combination of machine learning and PBPK modeling approach for pharmacokinetics prediction of small molecules in humans. Pharm. Res.41 (7), 1369–1379. 10.1007/s11095-024-03725-y
65
Loewa A. Feng J. J. Hedtrich S. (2023). Human disease models in drug development. Nat. Rev. Bioeng.1 (8), 545–559. 10.1038/s44222-023-00063-3
66
Madden J. C. Enoch S. J. Paini A. Cronin M. T. D. (2020). A review of in silico tools as alternatives to animal testing: principles, resources and applications. Altern. Laboratory Animals48 (4), 146–172. 10.1177/0261192920965977
67
Malinowska J. M. Palosaari T. Sund J. Carpi D. Bouhifd M. Weber R. J. M. et al (2022). Integrating in vitro metabolomics with a 96-well high-throughput screening platform. Metabolomics18 (1), 11. 10.1007/s11306-021-01867-3
68
Matteo G. Leingartner K. Rowan-Carroll A. Meier M. Williams A. Beal M. A. et al (2023). In vitro transcriptomic analyses reveal pathway perturbations, estrogenic activities, and potencies of data-poor BPA alternative chemicals. Toxicol. Sci.191 (2), 266–275. 10.1093/toxsci/kfac127
69
Mattes W. B. Petttit S. D. Sansone S.-S. Bushel P. R. Waters M. D. (2004). Database development in toxicogenomics: issues and efforts. Environ. Health Perspect.112 (4), 495–505. 10.1289/ehp.6697
70
Miccoli A. Marx-Stoelting P. Braeuning A. (2022). The use of NAMs and omics data in risk assessment. EFSA J.20 (S2), e200908. 10.2903/j.efsa.2022.e200908
71
Mirshafiei M. Rashedi H. Yazdian F. Rahdar A. Baino F. (2024). Advancements in tissue and organ 3D bioprinting: current techniques, applications, and future perspectives. Mater. Des.240, 112853. 10.1016/j.matdes.2024.112853
72
More S. J. Bampidis V. Benford D. Bragard C. Hernandez-Jerez A. Bennekou S. H. et al (2021). Guidance Document on Scientific criteria for grouping chemicals into assessment groups for human risk assessment of combined exposure to multiple chemicals. EFSA J.19 (12), e07033. 10.2903/j.efsa.2021.7033
73
Moreau M. Mallick P. Smeltz M. Haider S. Nicolas C. I. Pendse S. N. et al (2022). Considerations for improving metabolism predictions for in vitro to in vivo extrapolation. Front. Toxicol.4, 894569. 10.3389/ftox.2022.894569
74
Moreau M. Simms L. Andersen M. E. Trelles Sticken E. Wieczorek R. Pour S. J. et al (2024). Use of quantitative in vitro to in vivo extrapolation (QIVIVE) for the assessment of non-combustible next-generation product aerosols. Front. Toxicol.6, 1373325. 10.3389/ftox.2024.1373325
75
Najjar A. Punt A. Wambaugh J. Paini A. Ellison C. Fragki S. et al (2022). Towards best use and regulatory acceptance of generic physiologically based kinetic (PBK) models for in vitro-to-in vivo extrapolation (IVIVE) in chemical risk assessment. Archives Toxicol.96 (12), 3407–3419. 10.1007/s00204-022-03356-5
76
OECD (2014). Joint meeting of the chemicals committee and the working party on chemicals, pesticides and biotechnology guidance document for describing non-guideline in vitro test methods Series on Testing and Assessment No. 211 JT03368605.
77
OECD (2016). Joint meeting of the chemicals committee and the working party on chemicals, pesticides and biotechnology guidance document on the reporting of defined approaches to be used within integrated approaches to testing and assessment Series on Testing and Assessment No. 255 JT03403921.
78
OECD (2021a). Guidance document on the characterisation, validation and reporting of pbk models for regulatory purposes Series on Testing and Assessment No Guidance document on the characterisation, validation and reporting of Physiologically Based Kinetic (PBK) models for regulatory purposes PUBE. Available online at: https://www.oecd.org/chemicalsafety/.
79
OECD (2021b). Test No. 439: in vitro skin irritation: reconstructed human epidermis test method, OECD guidelines for the testing of chemicals, section 4. Paris: OECD Publishing. 10.1787/9789264242845-en
80
OECD (2023). OECD Omics Reporting Framework (OORF): Guidance on reporting elements for the regulatory use of omics data from laboratory-based toxicology studies, OECD Series on Testing and Assessment, Paris: OECD Publishing. 10.1787/6bb2e6ce-en
81
OECD (2024). Test No. 496: in vitro macromolecular test method for identifying chemicals inducing serious eye damage and chemicals not requiring classification for eye irritation or serious eye damage, OECD guidelines for the testing of chemicals, section 4. Paris: OECD Publishing.
82
OECD Omics Reporting Framework (2023). Guidance on reporting elements for the regulatory use of omics data from laboratory-based toxicology studies. OECD. 10.1787/6bb2e6ce-en
83
Ortega-Vallbona R. Talavera-Cortés D. Carpio L. E. Coto Palacio J. Roncaglioni A. Garcia De Lomana M. et al (2025). DockTox: targeting molecular initiating events in organ toxicity through molecular docking. Toxicology515, 154155. 10.1016/j.tox.2025.154155
84
Paini A. Leonard J. A. Joossens E. Bessems J. G. M. Desalegn A. Dorne J. L. et al (2019). Next generation physiologically based kinetic (NG-PBK) models in support of regulatory decision making. Comput. Toxicol.9, 61–72. 10.1016/j.comtox.2018.11.002
85
Pathak R. K. Kim J.-M. (2024). A computational assay for identifying millet-derived compounds that antagonize the interaction between bisphenols and estrogen-related receptor gamma. Front. Pharmacol.15, 1435254. 10.3389/fphar.2024.1435254
86
Pathak R. K. Jung D.-W. Shin S.-H. Ryu B.-Y. Lee H.-S. Kim J.-M. (2024). Deciphering the mechanisms and interactions of the endocrine disruptor bisphenol A and its analogs with the androgen receptor. J. Hazard. Mater.469, 133935. 10.1016/j.jhazmat.2024.133935
87
Paul Friedman K. Gagne M. Loo L.-H. Karamertzanis P. Netzeva T. Sobanski T. et al (2020). Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol. Sci.173 (1), 202–225. 10.1093/toxsci/kfz201
88
Perkins E. J. Woolard E. A. Garcia-Reyero N. (2022). Integration of adverse outcome pathways, causal networks and omics to support chemical hazard assessment. Front. Toxicol.4, 786057. 10.3389/ftox.2022.786057
89
Pezoulas V. C. Fotiadis D. I. (2024). The pivotal role of data harmonization in revolutionizing global healthcare: a framework and a case study. Connected Health Telemedicine3 (2). 10.20517/chatmed.2023.37
90
Pore S. Roy K. (2024). Insights into pharmacokinetic properties for exposure chemicals: predictive modelling of human plasma fraction unbound (fu) and hepatocyte intrinsic clearance (Clint) data using machine learning. Digit. Discov.3 (9), 1852–1877. 10.1039/D4DD00082J
91
Ram R. N. Gadaleta D. Allen T. E. H. (2022). The role of “big data” and “in silico” New Approach Methodologies (NAMs) in ending animal use – a commentary on progress. Comput. Toxicol.23, 100232. 10.1016/j.comtox.2022.100232
92
Ravi M. Paramesh V. Kaviya S. R. Anuradha E. Solomon F. D. P. (2015). 3D cell culture systems: advantages and applications. J. Cell. Physiology230 (1), 16–26. 10.1002/jcp.24683
93
Read-Across Assessment Framework (2017). Considerations on multi-constituent substances and UVCBs. 10.2823/794394
94
Rozony F. Z. (2024). A systematic review of big data integration challenges and solutions for heterogeneous data sources. Available online at: https://aithor.com/paper-summary/a-systematic-review-of-big-data-integration-challenges-and-solutions-for-heterogeneous-data-sources.
95
Sala Benito J. V. Paini A. Richarz A.-N. Meinl T. Berthold M. R. Cronin M. T. D. et al (2017). Automated workflows for modelling chemical fate, kinetics and toxicity. Toxicol. In Vitro45, 249–257. 10.1016/j.tiv.2017.03.004
96
Samsa G. Hu G. Root M. (2005). Combining information from multiple data sources to create multivariable risk models: illustration and preliminary assessment of a new method. J. Biomed. Biotechnol.2005 (2), 113–123. 10.1155/jbb.2005.113
97
Schmeisser S. Miccoli A. von Bergen M. Berggren E. Braeuning A. Busch W. et al (2023). New approach methodologies in human regulatory toxicology – not if, but how and when. Environ. Int.178, 108082. 10.1016/j.envint.2023.108082
98
Schrenk D. Bignami M. Bodin L. Chipman J. K. del Mazo J. Grasl-Kraupp B. et al (2020). Risk to human health related to the presence of perfluoroalkyl substances in food. EFSA J.18 (9), e06223. 10.2903/j.efsa.2020.6223
99
Sewell F. Alexander-White C. Brescia S. Currie R. A. Roberts R. Roper C. et al (2024). New approach methodologies (NAMs): identifying and overcoming hurdles to accelerated adoption. Toxicol. Res.13 (2), tfae044. 10.1093/toxres/tfae044
100
Sewer A. Talikka M. Calvino-Martin F. Luettich K. Iskandar A. (2024). Quantitative modeling of in vitro data using an adverse outcome pathway for the risk assessment of decreased lung function in humans. Toxicol. Lett.393, 107–113. 10.1016/j.toxlet.2024.02.001
101
Shi H. Zhao Y. (2024). Integration of advanced large language models into the construction of adverse outcome pathways: opportunities and challenges. Environ. Sci. Technol.58 (35), 15355–15358. 10.1021/acs.est.4c07524
102
Shrestha J. Razavi Bazaz S. Aboulkheyr Es H. Yaghobian Azari D. Thierry B. Ebrahimi Warkiani M. et al (2020). Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol.40 (2), 213–230. 10.1080/07388551.2019.1710458
103
Silva A. C. E. Loizou G. D. McNally K. Osborne O. Potter C. Gott D. et al (2024). A novel method to derive a human safety limit for PFOA by gene expression profiling and modelling. Front. Toxicol.6, 1368320. 10.3389/ftox.2024.1368320
104
Sindhu C. S. Hegde N. P. (2017). Handling complex heterogeneous healthcare big data. Available online at: https://api.semanticscholar.org/CorpusID:113404995.
105
Stamnitz S. Klimczak A. (2021). Mesenchymal stem cells, bioactive factors, and scaffolds in bone repair: from research perspectives to clinical practice. Cells10 (8), 1925. 10.3390/cells10081925
106
Sturla S. J. (2018). Point of departure. Chem. Res. Toxicol.31 (1), 2–3. 10.1021/acs.chemrestox.7b00341
107
Tenenhaus A. Tenenhaus M. (2011). Regularized generalized canonical correlation analysis. Psychometrika76 (2), 257–284. 10.1007/s11336-011-9206-8
108
Thomas R. S. Bahadori T. Buckley T. J. Cowden J. Deisenroth C. Dionisio K. L. et al (2019). The next generation blueprint of computational toxicology at the U.S. Environmental protection agency. Toxicol. Sci.169 (2), 317–332. 10.1093/toxsci/kfz058
109
Trisciuzzi D. Alberga D. Leonetti F. Novellino E. Nicolotti O. Mangiatordi G. F. (2018). “Molecular docking for predictive toxicology,” in Computational toxicology: methods and protocols. Editor NicolottiO. (New York: Springer), 181–197. 10.1007/978-1-4939-7899-1_8
110
Vallance P. (2016). Industry–academic relationship in a new Era of drug discovery. J. Clin. Oncol.34, 3570–3575. 10.1200/JCO.2016.68.4217
111
van der Zalm A. J. Barroso J. Browne P. Casey W. Gordon J. Henry T. R. et al (2022). A framework for establishing scientific confidence in new approach methodologies. Archives Toxicol.96 (11), 2865–2879. 10.1007/s00204-022-03365-4
112
Viant M. R. Amstalden E. Athersuch T. Bouhifd M. Camuzeaux S. Crizer D. M. et al (2024). Demonstrating the reliability of in vivo metabolomics based chemical grouping: towards best practice. Archives Toxicol.98 (4), 1111–1123. 10.1007/s00204-024-03680-y
113
Viganò E. L. Ballabio D. Roncaglioni A. (2024). Artificial intelligence and machine learning methods to evaluate cardiotoxicity following the adverse outcome pathway frameworks. Toxics12 (1), 87. 10.3390/toxics12010087
114
Villeneuve D. L. Crump D. Garcia-Reyero N. Hecker M. Hutchinson T. H. LaLone C. A. et al (2014). Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci.142 (2), 312–320. 10.1093/toxsci/kfu199
115
Vinken M. (2024). Adverse outcome pathway networks as the basis for the development of new approach methodologies: liver toxicity as a case study. Curr. Opin. Toxicol.40, 100504. 10.1016/j.cotox.2024.100504
116
Wan Y. Ding J. Jia Z. Hong Y. Tian G. Zheng S. et al (2024). Current trends and research topics regarding organoids: a bibliometric analysis of global research from 2000 to 2023. Heliyon10 (12), e32965. 10.1016/j.heliyon.2024.e32965
117
Wang L. Hu D. Xu J. Hu J. Wang Y. (2024). Complex in vitro model: a transformative model in drug development and precision medicine. Clin. Transl. Sci.17 (2), e13695. 10.1111/cts.13695
118
Warren G. L. Andrews C. W. Capelli A. M. Clarke B. LaLonde J. Lambert M. H. et al (2006). A critical assessment of docking programs and scoring functions. J. Med. Chem.49 (20), 5912–5931. 10.1021/jm050362n
119
Witt G. Keminer O. Leu J. Tandon R. Meiser I. Willing A. et al (2021). An automated and high-throughput-screening compatible pluripotent stem cell-based test platform for developmental and reproductive toxicity assessment of small molecule compounds. Cell Biol. Toxicol.37 (2), 229–243. 10.1007/s10565-020-09538-0
120
Wu X. Yang X. Dai Y. Zhao Z. Zhu J. Guo H. et al (2024). Single-cell sequencing to multi-omics: technologies and applications. Biomark. Res.12 (1), 110. 10.1186/s40364-024-00643-4
121
Xiao X. Trinh T. X. Gerelkhuu Z. Ha E. Yoon T. H. (2024). Automated machine learning in nanotoxicity assessment: a comparative study of predictive model performance. Comput. Struct. Biotechnol. J.25, 9–19. 10.1016/j.csbj.2024.02.003
122
Yordanova D. Schultz T. W. Kuseva C. Tankova K. Ivanova H. Dermen I. et al (2019). Automated and standardized workflows in the OECD QSAR Toolbox. Comput. Toxicol.10, 89–104. 10.1016/j.comtox.2019.01.006
123
Zhang Q. Li J. Middleton A. Bhattacharya S. Conolly R. B. (2018). Bridging the data gap from in vitro toxicity testing to chemical safety assessment through computational modeling. Front. Public Health6, 261. 10.3389/fpubh.2018.00261
124
Zheng Z.-Y. Wei X.-P. Yang Y.-T. Ni H.-G. (2025). Prediction and mechanism of combined toxicity of surfactants and antibiotics in aquatic environment based on in silico method. J. Hazard. Mater.488, 137390. 10.1016/j.jhazmat.2025.137390
Summary
Keywords
new approach methodologies (NAM), point of departure (POD), in-vitro , omics, in-silico , machine learning, data harmonization
Citation
Deepika D, Bharti K, Sharma S, Kumar S, Pathak RK, Biosca Brull J, Sabuz O, García Vilana S and Kumar V (2025) Advancing human health risk assessment: the role of new approach methodologies. Front. Toxicol. 7:1632941. doi: 10.3389/ftox.2025.1632941
Received
22 May 2025
Accepted
18 September 2025
Published
07 October 2025
Volume
7 - 2025
Edited by
Iseult Lynch, University of Birmingham, United Kingdom
Reviewed by
Thomas Eckart Exner, Seven Past Nine, Slovenia
Rebecca Ram, Lush Prize, United Kingdom
Updates
Copyright
© 2025 Deepika, Bharti, Sharma, Kumar, Pathak, Biosca Brull, Sabuz, García Vilana and Kumar.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vikas Kumar, vikas.kumar@urv.cat
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.