- 1Department of Civil and Environmental Engineering, University of Tennessee, Knoxville, TN, United States
- 2Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN, United States
- 3Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
- 4Department of Biological Sciences, Michigan Technological University, Houghton, MI, United States
- 5Department of Microbiology, University of Tennessee, Knoxville, TN, United States
- 6Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 7Department of Earth and Planetary Sciences, University of Tennessee, Knoxville, TN, United States
- 8Institute for a Secure and Sustainable Environment, University of Tennessee, Knoxville, TN, United States
Clay-based flocculation techniques have been developed to mitigate harmful algal blooms; however, the potential ecological impacts on the microbial community are poorly understood. In this study, chemical measurements were combined with 16S rRNA sequencing to characterize the microbial community response to different flocculation techniques, including controls, clay flocculation, clay flocculation with zeolite, and clay flocculation with O2 added zeolite capping. Sediment bacterial biomass measured by PLFA were not significantly altered by the various flocculation techniques used. However, 16S rRNA sequencing revealed differences in water microbial community structure between treatments with and without zeolite capping. The differences were related to significant reductions of total nitrogen (TN), total phosphorus (TP) and ammonia (NH4+) concentration and increase of nitrate (NO3-) concentration in zeolite and O2 loaded zeolite capping. The relative abundance of ammonia oxidizing bacteria increased four-fold in zeolite capping microcosms, suggesting zeolite promoted absorbed ammonia removal in the benthic zone. Zeolite-capping promoted bacteria nitrogen cycling activities at the water-sediment interface. Potential pathogens that are usually adapted to eutrophic water bodies were reduced after clay flocculation. This study demonstrated clay flocculation did not decrease bacterial populations overall and may reduce regulatory indicators and pathogenic contaminants in water. Zeolite capping may also help prevent nutrients from being released back into the water thus preventing additional algal blooms.
IMPORTANCE
Despite the effectiveness of clay flocculation for removing harmful algal blooms as highlighted in numerous studies, the potential ecological impacts on the microbial community have rarely been investigated. Characterization of clay flocculation treated algal bloom water and sediment microbial community provides new insights into the ecological impacts of this algal bloom controlling technology.
Introduction
Clay based flocculation techniques have recently been developed to mitigate harmful algal blooms by algal sedimentation and have been used as a mitigation strategy in numerous places worldwide (Nam et al., 2004; Sengco and Anderson, 2004; Chen and Pan, 2012; Li and Pan, 2013). Initial research studies of clay flocculation mainly focused on the effect on internal phosphorus (P) and nitrogen (N) load management and toxicity reduction in the impacted water body (Li and Pan, 2015). Studies reported that P can be trapped by the flocculant and buried in sediment as long as 15 years (Cooke, 1999). Supplemental control measures often used, include zeolite, which can absorb ammonia via ion exchange (Wen et al., 2006). Addition of zeolite allows for the permanent removal of excess N by coupled nitrification and denitrification (Van et al., 1998; Gribsholt et al., 2006; Kuenen, 2008).
Few, if any, investigations have been published that examine the microbial community structure response to clay flocculation. In the current investigation, we performed high throughput sequencing of the 16S rRNA genes to analyze water and sediment microbial community responses to three clay flocculation approaches. We aimed to assess the ecological impact of clay flocculation on the indigenous microbial community: (1) to compare bacteria species diversity and microbial community composition among various treatments; (2) to elucidate latent associations of the microbial community with different environmental parameters. This study provided insight into how microbial communities and their activities will shift in a changing environmental engineering operation using clay flocculation.
Materials and Methods
Site Description and Experiment Setup
The field study site was located near CeTian reservoir in Datong, Shanxi province (China) (He et al., 2011). The reservoir has experienced extreme eutrophication because of the nutrients carried into it from intense agricultural in the associated watershed. The study site includes eight 800 m2 ponds, which were filled with algal contaminated water from the CeTian reservoir (Figure 1). The dominate algae were Scenedesmus quadricauda and Cyanobacteria. The most common Cyanobacteria OTU identification for the 16S rRNA from the water was Synechococcus.

Figure 1. Control (with algal bloom) and Clay treated (no algae in water only seaweed) ponds at China field site.
For this study, sediments were collected from one of the eight ponds and placed in 72 columns to a height of 20 cm. Algal contaminated water from the pond was used to fill the columns to 1 cm below the top. The treatments were clay flocculation (F), clay flocculation with added zeolite capping (Z), clay flocculation with O2 loaded zeolite capping (OZ), and control/no flocculation (C) (SI Figure 1). There were triplicate columns for each of the four treatments.
The F columns were treated with chitosan modified local soil (2 mg/L chitosan and 75 mg/L soil). In addition to chitosan modified local soil, the Z columns received a 1 cm layer of zeolite (sieved through 100 mesh), in order to act as a capping layer to prevent algae from getting back into the water. The OZ columns were treated with chitosan modified local soil and 1 cm O2 loaded zeolite-capping layer. The O2 loaded zeolite was prepared by putting sieved zeolite in a high-pressure cylinder and purging with pure O2 overnight. Columns were destructively sampled on the day of treatment (day 1 and on days 2, 4, 7, and 10). Samples were also collected before each treatment (day 0).
Water samples (50 mL) in the columns were collected from 10 cm above the sediment-water interface using a syringe and filtered through a 0.2 μm pore size corning™ RC syringe filter. The filters were immediately stored at −20°C for nutrient analysis. The remaining water in the column was filtered through a 47 mm diameter, 0.22 μm pore size, polyethylsulfone membrane filter (MO BIO Laboratories, Inc., Carlsbad, CA). The filters were immediately stored at −80°C for DNA extraction. Sediment cores were collected from columns using auger and thin walled tube samplers. The cores were sectioned at 3 cm depth intervals. Sub-samples were frozen at −80°C for both DNA and lipid analyses.
Geochemistry Measurements
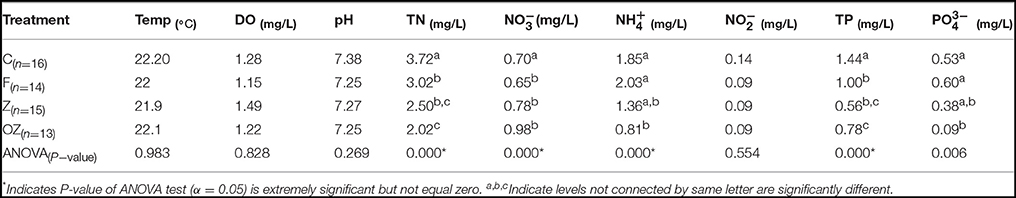
Temperature (Temp), pH, dissolved oxygen (DO), and oxidation-reduction potential (ORP) were measured by YSI 556 Handheld Multiparameter Instrument (Xylem Inc., OH, USA) before sampling. Total nitrogen (TN), ammonium (NH4-N), nitrate (NO3-N), total phosphorus (TP), and soluble reactive phosphate (PO4-P) were measured by the Research Center for Eco-Environmental Research Sciences, Chinese Academy of Sciences, Beijing, China using standard methods for water analysis in China (Hui, 2002).
Genomic DNA Extraction and PCR Amplification
Genomic DNA was extracted using PowerWater DNA isolation kit for water samples and PowerSoil DNA Isolation kit for sediment samples (MO BIO Laboratories, QIAGEN, San Diego, CA, USA) following the manufacturer's protocol. DNA extracts were purified using genomic DNA Clean and Concentrator kit (Zymo Research, Irvine, CA, USA). 16S rRNA genes were amplified using primer Phusion DNA polymerase (Thermo Scientific, Waltham, MA, USA) with primer pair 515f and barcoded 806r. DNA quality was determined using 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA) and DNA concentration was determined by KAPA SYBR® FAST qPCR Kits. Samples were diluted and pooled to a final concentration of 4 nM for sequencing.
Barcoded Amplicon 16S rRNA Sequencing
16S DNA library were prepared according to the protocol published by Caporaso et al. (2012), and sequenced in Illumina MiSeq platform at the University of Tennessee. Due to the poor quality of the reverse reads, only the forward reads were analyzed. Sequences were trimmed at a length of 250 bp and analyzed using QIIME v1.7.0 software package (Caporaso et al., 2012). Raw reads were assembled using join_paired_ends.py and then demultiplexed using split_libraries_fastq.py to and remove low quality scores. Chimeric sequences were identified and a total of 11 million sequences were retrieved after quality filtering and chimera checking. Open reference operational taxonomic units (OTUs) were picked via the UPARSE pipeline (Edgar, 2013) and taxonomy assignment was performed using rdp classifier trained against the Greengenes 16S rRNA gene database (http://greengenes.lbl.gov/ May 2013 release, 97%). OTUs with less than 0.005% relative abundance were removed. Alpha diversity (within sample diversity) was calculated for each sample using a variety of alpha diversity metrics in QIIME. Beta diversity (pairwise sample dissimilarity) was also calculated in QIIME using metrics including unweighted and weighted Unifrac, and Bray-Curtis. The OTU table for all samples was constructed from a biom file which is available in the supplemental info as a spreadsheet (see supplemental Hazen Chen otu_taxa.xlsx).
Phospholipid Fatty Acid Analysis
For each lyophilized sediment sample 10 g was extracted for total lipids using a two-step extraction methods where the final solvent ratio as 1:1:0.9 (v/v/v) for methanol:chloroform:buffer and water (White et al., 1979). Total lipid was fractionated on a silicic acid column into neutral, glycolipids and polar lipids by chloroform, acetone and methanol elution, respectively. Intact polar lipids were then methylated to recover fatty acid methyl esters (FAMEs). FAMEs were identified and quantified using an Agilent 6890 gas chromatograph coupled to an Agilent 5973 mass spectrometer. Microbial biomass in sediment was measured by converting picomoles of lipids per gram to viable cells per gram by multiplying bacterial size based conversion factor of 5.9 × 104 cells/pmol (Mills et al., 2006).
Statistical Analyses
Water, pH, ORP, Temp, nutrients, and biomass results were analyzed using ANOVA and Kruskal-Wallis tests in IBM SPSS. The 16S sequencing data were further analyzed with various methods in Prime 6 and PERMANOVA+ (Primer Inc. UK) (Clarke and Ainsworth, 1993). This included (1) tailed ANOVA test for microbial diversity alpha index, (2) hierarchical clustering for microbial community structure and composition, (3) non-metric multidimensional scaling (NMDS) based on weighted Unifrac distance matrix and environmental variables were fitted in NMDS, and (4) permutational multivariate analysis of variance using distance matrices to test whether the four treatments were different from each other. All tests were considered significant at α ≤ 0.05. Graphs were generated in R using “vegan” and “ggplot” packages (McMurdie and Holmes, 2013).
Results
Geochemistry Characteristics of Treatments
Temperature (Temp), DO, pH, and -N in the column water samples were similar among the four treatments. The average geochemical characteristics and pairwise comparisons are shown in Table 1. In the four treatments the average DO concentration in the water column was 1.5 mg/L. The column was hypoxic because the freshly set anaerobic sediment consumed O2 at the bottom. Geochemical profile of the four treatments showed that TN and TP concentrations decreased dramatically in F, Z, and OZ. and concentrations remained constant in Z and OZ throughout the rest of the time points but increased gradually in C and F. Nitrate concentrations, however, increased gradually in Z and OZ after treatment was added and decreased in C and Z. The water columns treated by clay flocculation with capping and O2 loaded capping had significantly lower TN, TP, and concentration than samples from control. pH decreased to normal in later time points because of the removal of algal cells from the water column.
Microbial Biomass in Sediment
Due to small sample size for each treatment, we conducted non-parametric Kruskal-Wallis to test the differences of water biomass among treatments. The average PLFA concentration in the sediments of the four treatments did not differ significantly (ANOVA P = 0.9078) and corresponded to cell densities of 9.75 × 107 cells/g to 1.28 × 108 cells/g (SI Figure 2).
Alpha diversity was determined using Chao1 and Shannon metrics. The Chao1 estimator was calculated to predict the total number of OTUs (richness) in each water and sediment sample (Mills and Wassel, 1980). The Shannon index was used to evaluate how equally abundant the dominant species were (Lozupone and Knight, 2008). Chao 1 index was much higher in sediment samples, ranging from 350 to 1,600. Based on Chao1 alpha diversity metrics, sediment samples possessed twice the species richness as water samples. The evenness index, was similar for water and sediment samples, both ranging from 5 to 9. The median of Chao1 index was 320 in water. Although a wider spread of estimated OTUs, richness was not significantly different among the four treatments (ANOVA, P = 0.39). Species evenness measurements in the water samples indicated a non-significant variation among C, F, Z, and OZ (ANOVA, P = 0.186). When comparing Chao1 index within sediment samples, OZ treatment had a significantly lower Chao1 than Control (Tukey's HSD, P = 0.02).
Zeolite Capping and Sediment Samples
The dominant phyla did not change in the upper sediment (1–3 cm) samples among treatments (SI Figure 3). However, the percentage of Chloroflexi and Acidobacteria were found fewer in samples with zeolite/O2 loaded zeolite-capping treatments than that in control and flocculation only treatment. Firmicutes was enriched after flocculation with zeolite/O2 loaded zeolite capping, 10 times higher, than the percentage of control and flocculation only treatments. Proteobacteria was increased by only 5%. Some Proteobacteria play essential roles in nitrogen cycling. For example, Nitrosomonas nadales (Figure 2A) bacterium related to the ammonia oxidizing bacteria was enriched significantly in Z and OZ (ANOVA, P < 0.0001). The increased Methylophilales in Z and OZ were involved in denitrification in anoxic situations [(Ginige et al., 2004); Figure 2C]. Nitrososphaerales, a cosmopolitan ammonia oxidizing Archaea in soil, were enriched in Z and OZ (Tourna et al., 2011), even though they only comprised a small percentage of the microbial community (Figure 2A). Nitrospirales was also increased in Z and OZ (Figure 2B); it is a chemolithoautotrophic nitrite-oxidizing bacteria involved with the second step of nitrification (Cebron and Garnier, 2005; Lückera et al., 2010).
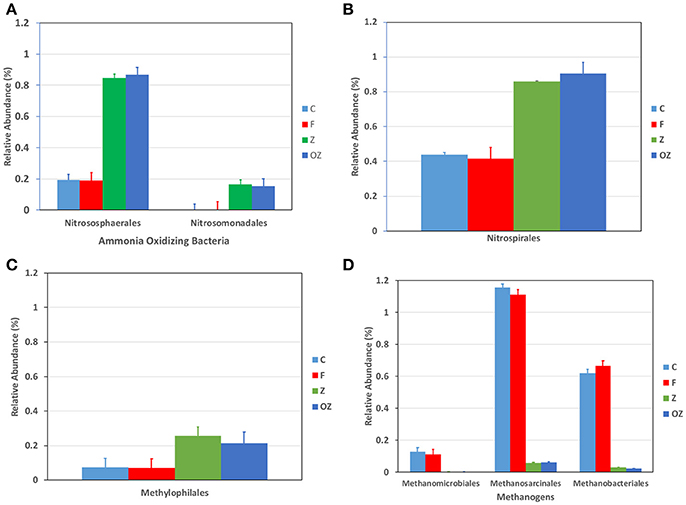
Figure 2. Changes in relative abundance of taxa that were involved with ammonia oxidization (A), nitrite oxidization (B), denitrification (C), and methane producing (D). C represents control treatments. F represents Clay flocculation only treatments; Z represents Clay flocculation with zeolite capping; OZ represents Clay flocculation with O2 added zeolite capping. All measurements were done in triplicate.
Community Structure in Water Samples
Obvious shifts in community structure at the phylum level were observed in flocculation-treated water samples (Figure 3). Proteobacteria remained at 40% through 10 days in the control but increased after the first 2 days in the treatments and then decreased at later time points. Actinobacteria varied with different treatments and kept increasing to the highest percentage in Z. A gradually decreasing trend was observed in the relative abundance of Planctomycetes. Verrucomicrobia was stable during 10 days. Deltaproteobacteria, including sulfate-reducing bacteria, were significantly enriched after flocculation (Tukey's HSD, P = 0.006). Elevated levels of Verrucomicrobiae were also characteristic of flocculation-treated water. The abundance of this cluster was negatively correlated with pH, and positively correlated with hydraulic retention time and temperature (Lindstrom et al., 2005). Members of the Verrucomicrobia have also been observed in both surface and hypolimnetic waters, suggesting a variety of metabolic strategies were going on within the treatment (Lindstrom et al., 2004). Cytophagia affiliated with Bacteroidetes were found significantly reduced in water samples after clay flocculation (Tukey's HSD, P = 0.03).
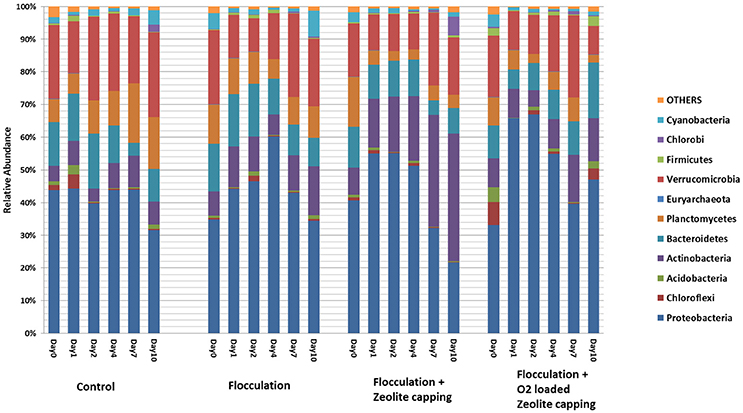
Figure 3. Relative abundance of dominant taxa at phylum level in water samples. The legend shows the most abundant phyla in these samples. The height of color bar represents the percentage of each taxon composed in the microbial community. X axis show each treatment at days 0, 1, 2, 4, 7, and 10.
Beta diversity was analyzed based on weighted Unifrac distance matrix by NMDS. Samples from C and F were clustered together, while samples from Z and OZ were clustered closely (Figure 4). The PERMANOVA analysis indicated distinct water microbial community structure between C and Z (Unique perms = 998, P = 0.001), as well as C and OZ (Unique perms = 997, P = 0.005). In addition, PERMANOVA test also indicated both treatment (Unique perms = 997, P = 0.001) and point-in-time (Unique perms = 998, P = 0.001) were factors contributing to variability in microbial community structure. Environmental parameters were fitted in NMDS (SI Table 1).
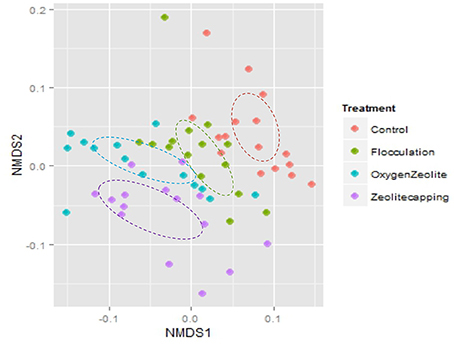
Figure 4. Non-metric Multidimensional scaling (NMDS) plot of weighted unifrac distance. Color of symbols represents different treatments. Analysis of Similarities test (ANOSIM) confirmed that C and F were similar (R = 0.018, P = 0.25) as well as that Z and OZ were similar (R = 0.012, P = 0.222), but Control and Flocculation treatments significantly differed from Zeolite-capping and O2 loaded Zeolite-capping treatments.
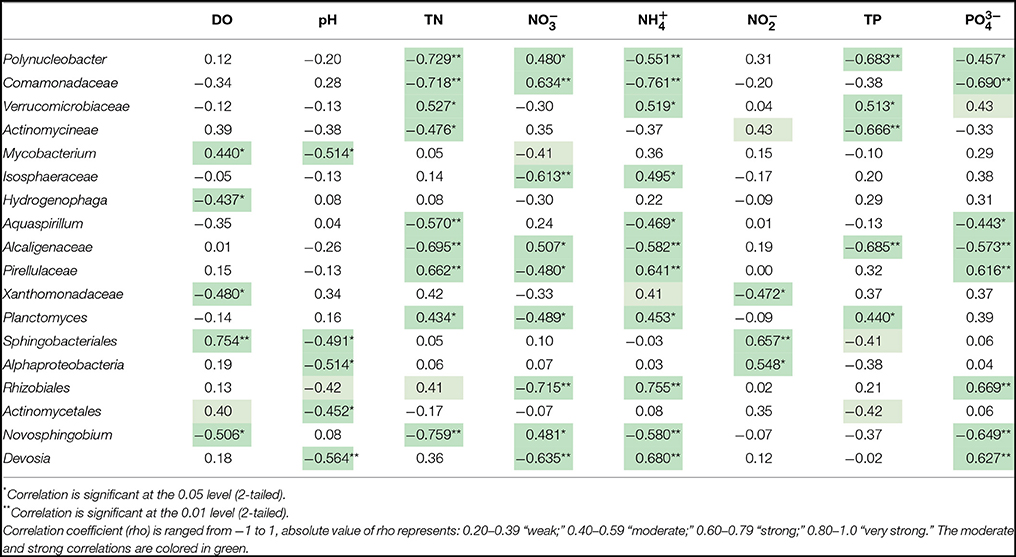
To identify which taxa were abundant in each sample, heatmap combined with hierarchical clustering was plotted (Figure 5). Relative abundance at the genus level of triplicates in each sample were averaged and then the most dominant 25 genera were depicted in the heatmap. The dendogram on the top of the heatmap shows the clusters of water samples, and samples in Z and OZ were separated from samples in C and F. Polynucleobacter, unclassified Comamonadaceae, unclassified Actinomycineae were present at a higher proportion in Z and OZ; while Planctomyces, unclassified Pirellulaceae, unclassified Rhizobiale, Devosia, and Hydrogenophage were not detected in Z and OZ.
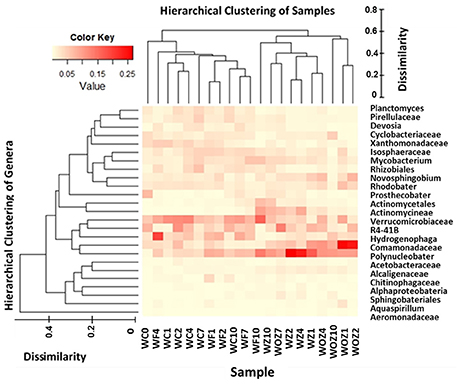
Figure 5. Heatmap combined hierarch clustering of sample types and top 25 genera. Relative abundance of most dominant 25 genera was depicted in heatmap. The color key indicated the relative abundance percentage scale. Hierarchical cluster analysis separated Control water (C) and Flocculation only water (F) samples from Flocculation with zeolite capping (Z) and with O2 added zeolite capping (OZ), which is shown on the top of heatmap. Hierarchical cluster analysis of genus level abundance is depicted on the left of heatmap. Dissimilarity score legend indicates the distance of clusters.
Flocculation with Zeolite/O2 Loaded Capping
In-depth 16S sequencing information was used to visualize the differences of water microbial community structure. The top 23 abundant genera and environmental parameters were subject to spearman correlation analysis. Spearman correlation (rho value) shown in Table 2 indicated that 18 genera were moderately or strongly correlated with physical and geochemical factors. Some of the important taxa were correlated with treatment type as well as nutrient condition. Polyuncleobacter, Comamonadaceae, Actinomycineae, Alcaligenaceae, and Novosphingobium were increased in flocculation when the capping layer was added. Addition of the capping layer delayed formation of eutrophic conditions.
Discussion
Geochemistry Characteristics of Treatments
Although DO was not a control factor in this study, it is critical for shaping water microbial community in natural systems (Yadav et al., 2014). The reduction of TN in the Z and OZ contributed to the decrease. in the water may be trapped by zeolite due to its ability to reversibly bind alkaline-earth cations inside the framework structure of zeolite and can easily be exchanged by surrounding positive ions (Wen et al., 2006). in the sediment can also be removed via biological pathways coupling nitrification and denitrification (Sebilo et al., 2006; Avrahami et al., 2011; Godos et al., 2014). The increased nitrate concentration in Z and OZ treatments implied a possibility of nitrification on the zeolite biofilm. Compared with nitrogen, not all the phosphorus in freshwater is bioavailable (Spears et al., 2007; Elser and Bennett, 2011). Autochthonous phosphorus reduction resulted from an inactivation agent binding the P in the water column and sealing it in the sediments (Gibbs and Ozkundakci, 2011; Van Oosterhout and Lurling, 2011; Pan et al., 2012). The rebound of TN and TP in control and Clay flocculation only treatments demonstrated that the zeolite capping layer prevented TN and TP from being released back into water.
Microbial Biomass was not Affected in Sediment
There was no significant difference in the means of universal bacteria gene copy number, PLFA, or cell density in the sediments of the four treatments. These results were consistent with reported research on sediment biomass (Polyanskaya et al., 2015). Bacteria biomass remained at a similar level as control before and after clay flocculation and capping with zeolite.
The evenness of the water microbial community structure was slightly decreased after clay flocculation, indicating potential interaction between some bacterial cells and flocculent particles. Clay was modified by chitosan, which tends to attract negative charged algal cells as well as specific bacteria cells (Zeng et al., 2008). Overall, there was no significant difference in evenness among the four treatments, suggesting that evenness was not significantly impacted despite a small decline after clay flocculation. Related research in the study of composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes found that richness of the bacterial communities is not significantly affected by the conditions created by cyanobacterial blooms (Eiler and Bertilsson, 2007). Thus, as the current study suggests microbial populations and species richness were not influenced by clay flocculation techniques.
Zeolite Capping Suggested Ammonia Oxidation in Sediment Samples
The zeolite capping layer absorbed ammonia from the water column and promoted sequential oxidation of ammonia via nitrite to nitrate at the water/sediment interface. Another Archaea member Euryarchaeota were significantly decreased in Z and OZ (ANOVA, P = 0.03), including Methanosarcinales, Methanobacteriales, and Methanomicrobiales (Figure 2D). They are methanogens in an anoxic environment, and respectively produce methane generally by degradation of organic matter, using hydrogen to reduce carbon dioxide, and using only carbon dioxide (Bonin and Boone, 2006; Garcia et al., 2006; Kendall and Boone, 2006). A possible explanation of methanogen reduction is that zeolite and O2 loaded zeolite used as the capping layer constantly brought the sediment in contact with oxygen, thus elevating ORP levels (Vera et al., 2014).
Distinct Community Structure in Water Samples
It has been reported that representatives of the classes Cytophagia and Flavobacteria play a central role in the degradation of biopolymers in marine and freshwater environments [(Sack et al., 2011); SI Figure 4]. In this study, limited nutrients, such as lower levels of ammonia and absence of organic compounds, may have inhibited growth of these bacteria after flocculation. The microbial community in C and F were strongly affected by total nitrogen, ammonia, and phosphate concentration, and Z and OZ were more shaped by both nitrate concentration and pH. Nutrients may shape bacterial community structure directly and/or indirectly. Previous studies reported TP concentration influenced the number of phosphate-decomposing bacteria (Li et al., 2005). The microbial community composition can be influenced by the exudates released by phytoplankton, and nitrogen and phosphorus, to regulate the biomass of phytoplankton (Haukka et al., 2006). The microbial community structures in control and clay flocculation only treatments were different from flocculation plus capping treatments. Nutrient concentrations are the main factors causing community structures and zeolite capping prevented nutrient release back into the water column.
Flocculation With Zeolite/O2 Loaded Capping Resulted in a Healthier Water Body
Polyuncleobacter is a lineage of Betaproteobacteria and it is able to grow in the absence of oxygen (Hahn et al., 2010). Positive correlation of minor freshwater taxa (such as Planctomycetes, Acidobacteria, and Chloroflexi) with nutrient concentrations implied that members of minor freshwater taxa might be warning signs of unhealthy water environments (Newton et al., 2011). For example, Xanthomonadaceae was decreased in the water (Tukey's HSD, P = 0.02). This family includes two plant-pathogenic genera, Xanthomonas and Xylella (Mhedbi-Hajri et al., 2011). Planctomyces and Pirellulaceae are members of the Planctomycetes, they are related to microbes capable of anaerobic ammonia oxidation, and are commonly found in eutrophic and polluted environments (Schlesner and Stackebrandt, 1986). Previous studies reported that the number of Planctomycetes increase with pH values ranging from 6.8 to 9.4, and are usually associated with algal blooms (Ward et al., 2006).
This study was one of the first to investigate the ecological impact of algal bloom water control techniques on microbial communities in microcosm experiments. High throughput 16S rRNA gene sequencing provided insight into this understudied topic. Our results revealed that water community structures were distinct between samples with and without zeolite capping treatments. The differences resulted in significant reduction of TN, TP, and concentrations and an increase of concentrations in zeolite/O2 loaded and zeolite capping. The enriched Nitrososphaerales, Methylophilale, Nitrososphaerales, and Nitrospirales in the sediments indicated ammonia oxidization via nitrite to nitrate during flocculation with zeolite/O2 capping. Bacteria biomass, richness, and evenness were not altered by the clay flocculation. Planctomyces, Pirellulaceae, and Xanthomonadaceae known to be associated with pathogens found in eutrophic water bodies were reduced. Future functional metagenomics studies should focus on describing the response of nitrification and denitrification genes to algal bloom control techniques. In addition, field pilot-scale testing will help describe this issue in a more practical and meaningful manner for monitoring and control.
Conclusions
In this study, chemical measurements were combined with 16S rRNA sequencing to characterize the microbial community response to different flocculation techniques, including controls, clay flocculation, clay flocculation with zeolite, and clay flocculation with O2 added zeolite capping. Sediment bacterial biomasses measured by PLFA were not significantly altered by the various flocculation techniques used. However, 16S rRNA sequencing revealed differences in water microbial community structure between treatments with and without zeolite capping. The differences were in the significant reductions of total nitrogen (TN), total phosphorus (TP) and ammonia () and increases in nitrate () in zeolite and zeolite/O2 capping. The relative abundance of ammonia oxidizing bacteria increased four-fold in zeolite capping microcosms, suggesting zeolite promoted absorbed ammonia removal in the benthic zone. This further suggested that zeolite-capping promoted the bacterial nitrogen cycling activities at the water-sediment interface. Potential pathogens that are usually found in eutrophic water bodies were reduced after clay flocculation. This study demonstrated clay flocculation may reduce regulatory indicators and pathogenic contaminants in water. Zeolite capping may also help prevent nutrients from being released back into the water. This work suggests that zeolite clay flocculation is an important in situ method to improve water quality when harmful algae blooms contaminate waterways.
Author Contributions
CC, TH, GP conceived experiments. CC, TH, ST, SP wrote paper. CC, ST, SP, FX, WS did analysis. CC, FX, WS, GP did field experiments.
Funding
Funded internally by the University of Tennessee and Chinese Academy of Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the support given by Dr. Gang Pan's research group in the Eco-Environmental Sciences Research Center, Chinese Academy of Sciences, Beijing. Parts of this manuscript were first reported in a conference: CC, GP, WS, FX, ST, SP, and TH contributed. How does clay flocculation of harmful algal blooms affect microbial community composition in water and sediment. October 22-24, 2015, Lafayette, In. Critical Zone Science, Sustainability, and Services in a Changing World, Purdue University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2018.00060/full#supplementary-material
The Supporting Information includes microcosm design, copy number of universal bacteria 16S rRNA gene from qPCR, biomass of sediment sample measured by PLFA, alpha diversity plots, microbial community composition in sediment, individual taxa percentage variation at class, order, family, and genus level.
References
Avrahami, S., Jia, Z. J., Neufeld, J. D., Murrell, J. C., Conrad, R., and Kusel, K. (2011). Active autotrophic ammonia-oxidizing bacteria in biofilm enrichments from simulated creek ecosystems at two ammonium concentrations respond to temperature manipulation. Appl. Environ. Microb. 77, 7329–7338. doi: 10.1128/AEM.05864-11
Bonin, A. S., and Boone, D. R. (2006). The order Methanobacteriales 3, 231–243. doi: 10.1007/0-387-30743-5_11
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cebron, A., and Garnier, J. (2005). Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower Seine River (France). Water Res. 39, 4979–4992. doi: 10.1016/j.watres.2005.10.006
Chen, J., and Pan, G. (2012). Harmful algal blooms mitigation using clay/soil/sand modified with xanthan and calcium hydroxide. J. Appl. Phycol. 24, 1183–1189. doi: 10.1007/s10811-011-9751-7
Clarke, K. R., and Ainsworth, M. (1993). A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92, 205–219. doi: 10.3354/meps092205
Cooke, E. B. W. G. D. (1999). Effectiveness and longevity of phosphorus inactivation with alum. Lake Res. Manag. 15, 5–27. doi: 10.1080/07438149909353948
de Godos, I., Vargas, V. A., Guzman, H. O., Soto, R., Garcia, B., Garcia, P. A., et al. (2014). Assessing carbon and nitrogen removal in a novel anoxic-aerobic cyanobacterial-bacterial photobioreactor configuration with enhanced biomass sedimentation. Water Res. 61, 77–85. doi: 10.1016/j.watres.2014.04.050
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–1000. doi: 10.1038/nmeth.2604
Eiler, A., and Bertilsson, S. (2007). Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 9, 838–838. doi: 10.1111/j.1462-2920.2006.01231.x
Elser, J., and Bennett, E. (2011). Phosphorus cycle: a broken biogeochemical cycle. Nature 478, 29–31. doi: 10.1038/478029a
Garcia, J. L., Ollivier, B., and Wnitman, W. B. (2006). The order of methanomicrobiales. Prokaryotes 3, 208–230. doi: 10.1007/0-387-30743-5_10
Gibbs, M., and Ozkundakci, D. (2011). Effects of a modified zeolite on P and N processes and fluxes across the lake sediment-water interface using core incubations. Hydrobiologia 661, 21–35. doi: 10.1007/s10750-009-0071-8
Ginige, M. P., Hugenholtz, P., Daims, H., Wagner, M., Keller, J., and Blackall, L. L. (2004). Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microb. 70, 588–596. doi: 10.1128/AEM.70.1.588-596.2004
Gribsholt, B., Struyf, E., Tramper, A., Andersson, M. G. I., Brion, N., De Brabandere, L., et al. (2006). Ammonium transformation in a nitrogen-rich tidal freshwater marsh. Biogeochemistry 80, 289–298. doi: 10.1007/s10533-006-9024-8
Hahn, M. W., Lang, E., Brandt, U., Lunsdorf, H., Wu, Q. L., and Stackebrandt, E. (2010). Polynucleobacter cosmopolitanus sp nov., free-living planktonic bacteria inhabiting freshwater lakes and rivers. Int. J. Syst. Evol. Micr. 60, 166–173. doi: 10.1099/ijs.0.010595-0
Haukka, K., Kolmonen, E., Hyder, R., Hietala, J., Vakkilainen, K., Kairesalo, T., et al. (2006). Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb. Ecol. 51, 137–146. doi: 10.1007/s00248-005-0049-7
He, G. J., Fang, H. W., Bai, S., Liu, X. B., Chen, M. H., and Bai, J. (2011). Application of a three-dimensional eutrophication model for the Beijing Guanting Reservoir, China. Ecol. Model. 222, 1491–1501. doi: 10.1016/j.ecolmodel.2010.12.006
Hui, W. (2002). Monitoring and Analytical Method on Water and Wastewater, 4th Ed. Beijing: China Environmental Science. (In Chinese).
Kendall, M. M., and Boone, D. R. (2006). The order methanosarcinales. Prokaryotes 3, 244–256. doi: 10.1007/0-387-30743-5_12
Kuenen, J. G. (2008). Anammox bacteria: from discovery to application. Nat. Rev. Microbiol. 6, 320–326. doi: 10.1038/nrmicro1857
Li, C., Yuan, H. L., and Huang, H. Z. (2005). Vertical distribution of phosphorus and P-dissolving/decomposing bacteria in the sediment of Guanting reservoir. Sci. China Ser D 48, 285–294. doi: 10.1360/05yd0006
Li, H., and Pan, G. (2015). Simultaneous removal of harmful algal blooms and microcystins using microorganism- and chitosan-modified local soil. Environ. Sci. Technol. 49, 6249–6256. doi: 10.1021/acs.est.5b00840
Li, L., and Pan, G. (2013). A universal method for flocculating harmful algal blooms in marine and fresh waters using modified sand. Envirron. Sci. Technol. 47, 4555–4562. doi: 10.1021/es305234d
Lindström, E. S., Kamst-Van Agterveld, M. P., and Zwart, G. (2005). Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microb. 71, 8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005
Lindstrom, E. S., Vrede, K., and Leskinen, E. (2004). Response of a member of the verrucomicrobia, among the dominating bacteria in a hypolimnion, to increased phosphorus availability. J. Plankton Res. 26, 241–246. doi: 10.1093/plankt/fbh010
Lozupone, C. A., and Knight, R. (2008). Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32, 557–578. doi: 10.1111/j.1574-6976.2008.00111.x
Lücker, S., Wagnera, M., and Maixnera, F. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484. doi: 10.1073/pnas.1003860107
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Mhedbi-Hajri, N., Jacques, M. A., and Koebnik, R. (2011). Adhesion mechanisms of plant-pathogenic Xanthomonadaceae. Adv. Exp. Med. Biol. 715, 71–89. doi: 10.1007/978-94-007-0940-9_5
Mills, A. L., and Wassel, R. A. (1980). Aspects of diversity measurement for microbial communities. Appl. Environ. Microb. 40, 578–586.
Mills, C. T., Dias, R. F., Graham, D., and Mandernack, K. W. (2006). Determination of phospholipid fatty acid structures and stable carbon isotope compositions of deep-sea sediments of the Northwest Pacific, ODP site 1179. Mar. Chem. 98, 197–209. doi: 10.1016/j.marchem.2005.10.001
Nam, P. H., Fujimori, A., and Masuko, T. (2004). Flocculation characteristics of organo-modified clay particles in poly(L-lactide)/montmorillonite hybrid systems. E-Polymers 4, 40–46. doi: 10.1515/epoly.2004.4.1.40
Newton, R. J., Jones, S. E., Eiler, A., McMahon, K. D., and Bertilsson, S. (2011). A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. R 75, 14–49. doi: 10.1128/MMBR.00028-10
Pan, G., Dai, L., Li, L., He, L., Li, H., Bi, L., et al. (2012). Reducing the recruitment of sedimented algae and nutrient release into the overlying water using modified soil/sand flocculation-capping in eutrophic lakes. Environ. Sci. Technol. 46, 5077–5084. doi: 10.1021/es3000307
Polyanskaya, L. M., Pinchuk, I. P., and Zvyagintsev, D. G. (2015). Assessment of the number, biomass, and cell size of bacteria in different soils using the “cascade” filtration method. Eurasian Soil Sci. 48, 288–293. doi: 10.1134/S1064229315030096
Sack, E. L. W., Wielen, P. W. J. J. V. D., and Kooij, D. V. D. (2011). Flavobacterium johnsoniae as a model organism for characterizing biopolymer utilization in oligotrophic freshwater environments. Appl. Environ. Microb. 77, 6931–6938. doi: 10.1128/AEM.00372-11
Schlesner, H., and Stackebrandt, E. (1986). Assignment of the genera Planctomyces and pirella to a new family planctomycetaceae fam-nov and description of the order planctomycetales ord-nov. Syst. Appl. Microbiol. 8, 174–176. doi: 10.1016/S0723-2020(86)80072-8
Sebilo, M., Billen, G., Mayer, B., Billiou, D., Grably, M., Garnier, J., et al. (2006). Assessing nitrification and denitrification in the seine river and estuary using chemical and isotopic techniques. Ecosystems 9, 564–577. doi: 10.1007/s10021-006-0151-9
Sengco, M. R., and Anderson, D. M. (2004). Controlling harmful algal blooms through clay Flocculation. J. Eukaryot. Microbiol. 51, 169–172. doi: 10.1111/j.1550-7408.2004.tb00541.x
Spears, B. M., Carvalho, L., and Paterson, D. M. (2007). Phosphorus partitioning in a shallow lake: implications for water quality management. Water Environ. J. 21, 47–53. doi: 10.1111/j.1747-6593.2006.00045.x
Tourna, M., Stieglmeier, M., Spang, A., Konneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U.S.A. 108, 8420–8425. doi: 10.1073/pnas.1013488108
Van, F., Boers, P. C. M., and Lijklema, L. (1998). Anoxic N-2 fluxes from freshwater sediments in the absence of oxidized nitrogen compounds. Water Res. 32, 407–409. doi: 10.1016/S0043-1354(97)00279-0
Van Oosterhout, F., and Lurling, M. (2011). Effects of the novel ‘Flock & Lock' lake restoration technique on Daphnia in Lake Rauwbraken (The Netherlands). J. Plankton Res. 33, 255–263. doi: 10.1093/plankt/fbq092
Vera, I., Araya, F., Andres, E., Saez, K., and Vidal, G. (2014). Enhanced phosphorus removal from sewage in mesocosm-scale constructed wetland using zeolite as medium and artificial aeration. Environ. Technol. 35, 1639–1649. doi: 10.1080/09593330.2013.877984
Ward, N., Staley, J. T., Fuerst, J. A., Giovannoni, S., Schlesner, H., and Stackebrandt, E. (2006). The order planctomycetales, including the genera Planctomyces, pirellula, gemmata and isosphaera and the candidatus genera brocadia, kuenenia and scalindua. Prokaryotes 7, 757–793. doi: 10.1007/0-387-30747-8_31
Wen, D., Ho, Y. S., and Tang, X. Y. (2006). Comparative sorption kinetic studies of ammonium onto zeolite. J. Hazard. Mater. 133, 252–256. doi: 10.1016/j.jhazmat.2005.10.020
White, D. C., Davis, W. M., Nickels, J. S., King, J. D., and Bobbie, R. J. (1979). Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40, 51–62. doi: 10.1007/BF00388810
Yadav, T. C., Khardenavis, A. A., and Kapley, A. (2014). Shifts in microbial community in response to dissolved oxygen levels in activated sludge. Bioresour. Technol. 165, 257–264. doi: 10.1016/j.biortech.2014.03.007
Keywords: algal blooms, microbial ecology, water quality, pathogens, clay flocculation, Phosphorus, Nitrogen
Citation: Chen C, Pan G, Shi W, Xu F, Techtmann SM, Pfiffner SM and Hazen TC (2018) Clay Flocculation Effect on Microbial Community Composition in Water and Sediment. Front. Environ. Sci. 6:60. doi: 10.3389/fenvs.2018.00060
Received: 14 December 2017; Accepted: 07 June 2018;
Published: 25 June 2018.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Pia Ines Scherer, Technische Universität München, GermanyWenjing Zhang, Xiamen University, China
Copyright © 2018 Chen, Pan, Shi, Xu, Techtmann, Pfiffner and Hazen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Terry C. Hazen, dGNoYXplbkB1dGsuZWR1
 Chunyi Chen
Chunyi Chen Gang Pan
Gang Pan Wenqing Shi3
Wenqing Shi3 Feng Xu
Feng Xu Stephen M. Techtmann
Stephen M. Techtmann Susan M. Pfiffner
Susan M. Pfiffner Terry C. Hazen
Terry C. Hazen