- 1Crop, Livestock and Environment Division, Japan International Research Center for Agricultural Sciences, Tsukuba, Japan
- 2Laboratory of Virology and Plant Biotechnology, Environmental Institute for Agricultural Research (INERA), Ouagadougou, Burkina Faso
- 3Central Region Agricultural Research Center, National Agriculture and Food Research Organization, Tsukuba, Japan
- 4Department of Natural Resources Management and Production Systems, Environmental Institute for Agricultural Research (INERA), Ouagadougou, Burkina Faso
Phosphate rocks (PR), the primary source of phosphorus (P), are often co-composted with organic materials to enhance P availability. However, the mechanisms of P solubilization in PR-enriched composts are not well elucidated. This study investigated such mechanisms by monitoring the changes in P fractions during composting and by determining the relationships between the physicochemical and biological parameters. Sorghum straw residues were composted alone (Comp), or with 10% PR (P-Comp), or with 10% PR and 10% rhizosphere soil (P-Comp-Soil), and samples were collected at 45, 60, and 180 days for analysis. The labile-P composed of H2O- and NaHCO3-extractable inorganic P (Pi) and organic P (Po), the moderately labile-P extracted by NaOH (Pi + Po), and the unavailable P formed of the HCl-P and residual-P, increased with the progress of the composting. At 180 days, P-Comp-Soil contained the highest amount of labile-P. There were strong and positive correlations between labile-P and the abundance of total fungi, phosphate-solubilizing fungi (PSF), alkaline phosphatase phoD, phosphonatase phnX, acid phosphatase aphA. Although total fungi were much fewer than total bacteria, the PSF mainly triggered the mineral P solubilization. The alkaline phosphatase phoD was the main enzyme leading the organic P mineralization, while the contribution of phosphonatase phnX, acid phosphatase aphA, and siderophore entA to the organic P solubilization was minor. Besides, the bacterial specific-transporter (pstS) gene increased with the increase of labile-P, allowing for immobilization of little fractions of P in microbial cells. This study highlighted the significant role of PSF and alkaline phosphatase in the P solubilization of PR-enriched composts. Furthermore, it showed the benefit of supplementing the PR-enriched composts with rhizosphere soil, a niche of ecologically important source of beneficial microbes.
Introduction
The rapid increase in the world’s human population expected to reach 9.1 billion by 2050 requires to increase food production by 70% (Krishnaraj and Dahale, 2014). Among the efforts to increase agricultural production, sustainable management of soil fertility will play a significant role. Phosphorus (P), the second most essential nutrient after nitrogen for plant growth, is, however, present at deficient low levels of available fractions in many ecosystems (Barber et al., 1963). This P deficiency has become a threat to soil fertility and crop production throughout the world (Ochwoh et al., 2005), and particularly in the humid tropics and subtropics, and sandy soils of the semi-arid tropics (Sanchez et al., 1997). P availability in soils depends on the P fractions, which influence the primary productivity of agricultural systems (Williams et al., 2013). Organic P (Po) fractions are found in plant remains, in composts, and microbial tissue. In contrast, inorganic P (Pi), which exists in various forms such as H2PO4 can form complexes with iron, aluminum, and calcium in soils and becomes unavailable for plant uptake (Theodorou and Plaxton, 1993; Khan and Joergensen, 2009). On the other hand, Sharma et al. (2013), supported by Pii et al. (2015), reported that organic P and inorganic P are present in enough concentrations. Still, their solubility and thus their availability is deficient. The low P availability in soils is corrected by applying phosphatic fertilizers with high levels of available P. These phosphatic fertilizers use phosphate rock as the primary source of P2O5. However, the supply of phosphate rock (PR) is limited in the whole world. Moreover, the high price of chemical fertilizers and phosphate rock (Sharma et al., 2013) added to the severe environmental threat due to the exploitation of phosphate rock reserves require to consider alternative strategies to sustain soil fertility and crop production. In this perspective, animal manures, straw, and other solid wastes are used as substrates for PR-enriched composts.
There have been suggestions to increase the nutrient bioavailability of PR-enriched composts by inoculations with phosphate-solubilizing microorganisms (Biswas and Narayanasamy, 2006) or phosphate-mineralizing microorganisms (Sunita, 2014). Their higher nutrient content makes these PR-enriched composts more efficient for plant growth, and they are more environmentally safe (Wei et al., 2015). However, the use of microorganisms as bio-fertilizers remains a hard-to-adopt technology, especially for smallholder farmers in developing countries. It requires access to inocula, appropriate storage, and proper handling ability during inoculation, which is not easily applicable by many farmers. Fortunately, it is well known that rhizosphere soils are reservoirs of numerous beneficial microbes (Barea et al., 2005; Vassilev et al., 2006), including P-solubilizing microorganisms. The addition of rhizosphere soil instead of pure microbial inocula may be an alternative way to supply PR-enriched composts with substantial P-solubilizing microbes and help improve P solubilization. However, this remains a hypothesis and should be further studied to clearly understand the magnitude of P solubilization in such PR-enriched composts with rhizosphere soil and the mechanisms that sustain it. Elucidating the P solubilization mechanisms is an essential step forward in the fabrication of better-quality composts with higher available P concentrations. In soil, microbial mineral P solubilization has long been associated with the production of low molecular weight organic acids (Bolan et al., 1997). Gluconic acid has been identified as the most frequently produced organic acid by phosphate-solubilizing bacteria (PSB) (Goldstein, 1995; Richardson and Simpson, 2011). It chelates divalent cations (e.g., Ca2+) in poorly soluble mineral phosphate forms, such as hydroxyapatite or tricalcium phosphate, thus releasing phosphate forms such as HPO42– and H2PO– available for plant uptake (Khan et al., 2010; Castagno et al., 2011). Gluconic acid produced from glucose oxidation requires the glucose dehydrogenase (GDH) enzyme (Deppenmeier et al., 2002) released by the activation of quinoprotein glucose dehydrogenase genes (gcd) (Chen et al., 2015). The activity of GDH, however, requires the redox cofactor pyrroloquinoline quinone (PQQ), whose biosynthesis involves a PQQ operon consisting of about 5–11 genes (Choi et al., 2008). Therefore, we targeted the pqq genes to study the abundance of PSB in this study. Since the mineral P solubilization is also carried out by fungi, we included the characterization of phosphate-solubilizing fungi (PSF). In soil, the solubilization of organic P depends much on the release of several enzymes such as acid and alkaline phosphatases, phosphonatases, phytases, and siderophores (Bergkemper et al., 2016). The determination of these enzymes by qPCR-based molecular techniques will give insights about their relative contribution to P solubilization of PR-enriched composts.
We designed this work to determine the changes in phosphorus fractions during the co-composting of sorghum straw residues with phosphate rock in the presence or absence of rhizosphere soil addition. We aimed at understanding the mechanisms that trigger such changes, focusing on the relationships between the physicochemical and biological properties.
Materials and Methods
Preparation and Monitoring of Composts
In this study, we used sorghum straw as an organic material substrate for the fabrication of three compost types. One of them was a sole compost (Comp) mainly composed of sorghum straw residues. The two other composts were enriched with the low-grade Burkina Faso phosphate rock (BPR) obtained from the Kodjari phosphate rock deposit and which contains 27.59% P2O5, 23.47% SiO2, 4.23% Al2O3, 2.98% Fe2O3, 34.39% CaO, 0.18% MgO, 0.53% K2O, 27.59% P2O5 (Nakamura et al., 2015). One BPR-enriched compost was without rhizosphere soil (P-Comp) while the other (P-Comp-Soil) received rhizosphere soil collected from farmers’ sorghum fields as a microbial source supplement. Just after harvesting the grains, several sorghum stands were uprooted. The loose soil was removed after shaking the root systems, and the soil adhering to the roots, defined as rhizosphere soil, was collected with care and used as inoculum during the composting. In detail, Comp comprised five layers of 20 kg oven-dried sorghum straw each and 460 g urea each. P-Comp was the same as Comp except that we added 10% BPR above each layer. P-Comp-Soil had the same composition as P-Comp, with the only difference that there was an addition of 10% of rhizosphere above each layer. Before making the compost piles, we chopped the sorghum straw to approximately 10 cm segments. An appropriate amount of urea was calculated and added to drop down the initial C/N ratio of sorghum straw from 55/1 to 25/1, which is crucial to initiate organic matter decomposition. There were three replications per compost type, giving a total of nine piles of 1.5 m × 1.5 m × 0.6 m size. The compost piles set on plastic sheets had their moisture levels adjusted to 65% before they were covered with the same plastic sheets. Compost piles were made on 22 March 2018 in INERA (National Institute for Environmental and Agricultural Research) Kamboinse (Burkina Faso) and monitored until 22 September 2018. Every 2 weeks, the piles were uncovered, and temperatures at the 20 cm deep layer were recorded in triplicates, using a Fluorescent portable dissolved oxygen meter DOP-01 (Automatic System Research Co., Ltd., Saitama, Japan). The moisture level was readjusted to 65% after the measurement. When returning the piles, we took necessary care not to contaminate a pile with the content of the others.
Compost Sampling
On days 45, 60, and 180 of composting, we collected a composite compost sample from each pile by combining aliquots of 3 sub-samples taken after turning the pile. Immediately after collection, each fresh sample (~500 g) was immediately put in tight plastic bags and stored at −20°C until chemical and molecular analysis. Just after the last sampling at 180 days, the piles were uncovered and left to air-dry entirely. The air-dried composts were put in nylon bags and stored at room temperature until further use.
Extraction of DNA From Compost, Soil, and Sorghum Straw Samples
Before DNA extraction, compost samples stored at −20°C were left at room temperature for 1 h to melt the attached ice. Following that, 200 mg of fresh compost per sample, 250 mg of the rhizosphere soil, and 100 mg sorghum straw were used for DNA extraction using DNeasy Soil Kit (Qiagen). We confirmed the presence of DNA bands by performing a 2% TAE agarose gel electrophoresis and assessed the DNA concentration by Qubit HS DNA Assay (Invitrogen, Carlsbad, CA). The moisture content of compost, soil, and sorghum straw at the time of DNA extraction was calculated after drying in a heat block an aliquot of known water content at 100°C for 3 h. The% moisture intervenes in subsequent calculations, especially during the processing of qPCR data. DNA was extracted at INERA (Burkina Faso) and transported to Japan in cold conditions together with air-dried composts, sorghum straw, and cold-stored soil used for chemical analysis. The transportation of samples from Burkina Faso to Japan was possible after obtaining transportation permits issued by the indicated authorities of both countries, accompanied by a Material Transfer Agreement.
Chemical Analysis
Nitrate (NO3–), Ammonia (NH4+), pH, Total Carbon, and Total Nitrogen
Air-dried compost and sorghum straw materials were ground into fine particles in a Vibrating Sample Mill TI-100 model (CMT Co., Ltd., Tokyo, Japan) for 1 min. Following that, 1 g ground air-dried material of each compost type or sorghum straw of known moisture content (determined after oven-drying at 70°C for 48 h) was added to 10 ml of 1M KCl in a 50 ml falcon tube, and incubated at 150 rpm, at 25°C, for 30 min to determine NH4-N and NO3-N according to Sarr et al. (2019). The soil’s NH4-N and NO3-N were assessed in the same way, except that the soil stored at −20°C was used. The pH and EC from air-dried compost samples were measured at a 10:1 distilled water-to-compost slurry by the glass electrode method (Thompson et al., 2001). However, soil EC was measured from a saturated paste of the soil (1:1 distilled water:soil). Total nitrogen (N) and total carbon (C) of all materials were determined by the dry combution method using an NC analyzer (Sumigraph NC-220: Sumika Chemical Analysis Service, Osaka, Japan).
P Fractionation of Composts
P in composts was sequentially extracted using a modified procedure of Hedley et al. (1982) and Tiessen and Moir (2008). A 0.5 g dried sample in duplicate was extracted sequentially using 30 ml deionized water (H2O), 0.5 M NaHCO3 (pH 8.5), 0.1 M NaOH, and 1 M HCl. A portion of NaHCO3 and NaOH extracts was acidified to precipitate the suspended organic matter, and then the pH was adjusted. These supernatants were used to analyze the inorganic P (Pi). Total P content of NaHCO3, and NaOH extracts were determined after digestion by an acidified ammonium persulfate in an autoclave (103.4 kPa, 121°C). The difference between the total P and Pi in extracts was the organic P (Po). Residual P was extracted by HNO3-H2SO4 acid using high-performance microwave digestion unit (Microwave Laboratory System; Milestone mls 1,200 mega). As a check, we also determined separately the total in each compost type the ignition of the sample at 550°C for 4 h and extracted by 0.5 M HCl. The Pi in all extracts and digested solutions was measured colorimetrically according to the molybdenum-blue method of Murphy and Riley (1962), using UV-1800 (UV-VIS spectrophotometer, Hitachi). Since P-Com and P-Comp-Soil received BPR or BPR plus soil during composting, the amount of organic matter in a 1 g material would differ. Therefore, we determined the decomposition rate of organic matter by estimating the crude ash content based on TMECC protocol (Thompson et al., 2001).
Determination of Microbial Abundance and Genes Involved in P Solubilization by qPCR
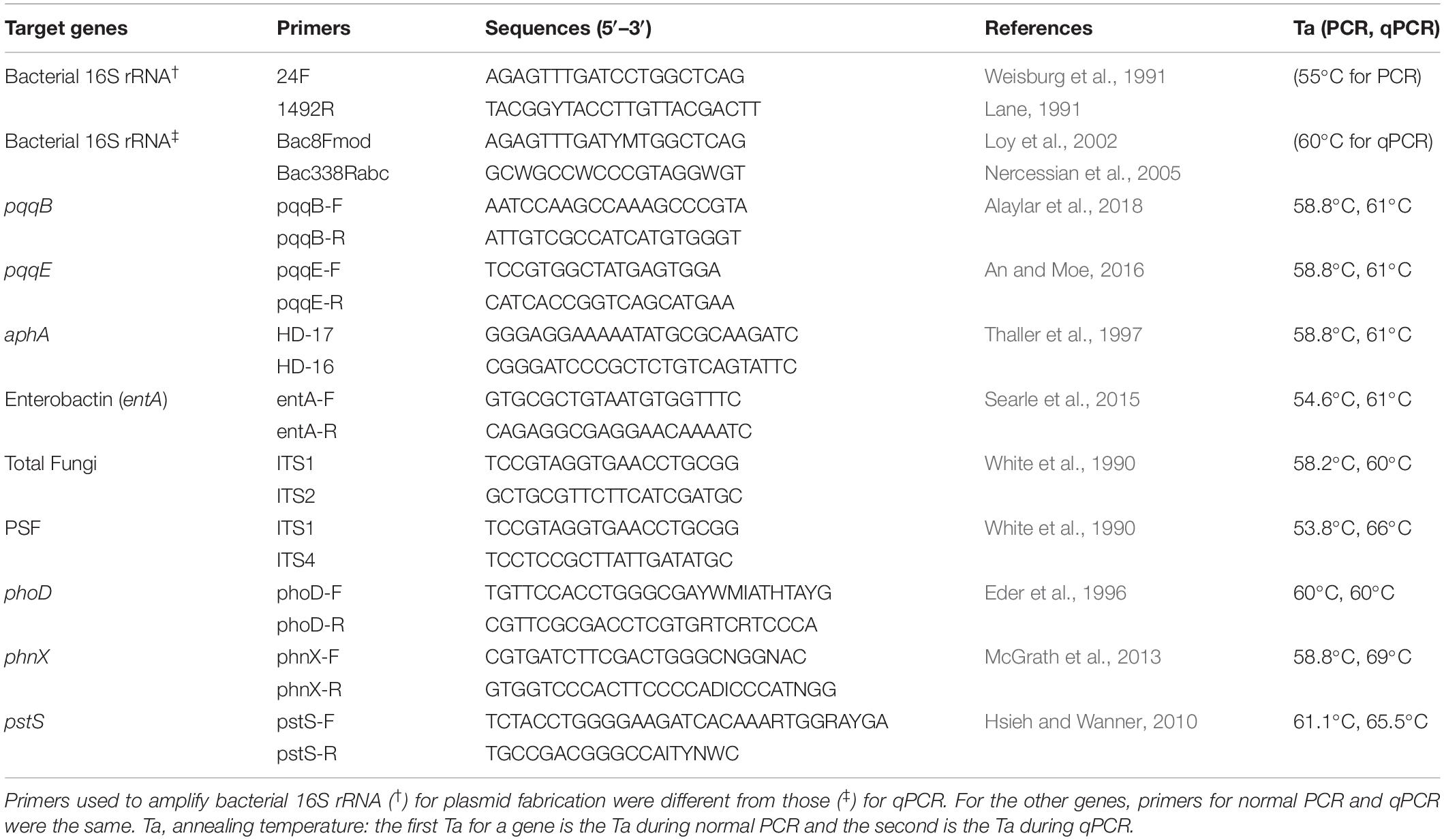
To perform qPCR, we first prepared plasmid standards. The target genes corresponded to 16S rRNA bacteria, ITS fungi, phosphate-solubilizing bacteria (Pyrroloquinoline Quinones pqqB and pqqE), PSF, alkaline phosphatase (phoD), acid phosphatase (aphA), phosphonatase (phnX), siderophore- mediated enterobactin (entA), phosphate specific transporter (pstS). At first, a soil sample was collected from the experimental field of the Japan International Research Center for Agricultural Sciences (JIRCAS) in Tsukuba, Japan (36.0535°N and 140.0792°E). The soil DNA was extracted by the Fast DNA Spin Kit for Soil (Mo-Bio, Carlsbad, CA) and used to amplify the target genes. Amplifications were carried out in triplicates for each gene in 20 μl reaction mixtures composed of 10 μl of 2 × Ampdirect Plus, 0.1 μl Biotaq HS, 10 μM of each primer, 7.7 μl sterile water and 2 μl DNA (~30 ng) purified using Nucleo Spin gDNA Clean-up (Macherey-Nagel). The PCR conditions consisted of an initial denaturation of 95°C for 10 min, followed by 35 cycles of 94°C for 30s, annealing temperature for 1 min, 72°C for 1 min, and a final elongation of 72°C for 7 min. The best annealing temperature (Table 1) for each primer set was determined by first performing a gradient PCR. PCR products were purified using Wizard SV Gel and PCR Clean-Up System (Promega, CA, United States) and quantified by NanoDrop. A 2% gel agarose electrophoresis was performed, and PCR products showing bands with the same size as the size of the target gene were ligated to pGEM-T-Easy Vector system 2 following the indicated instructions and cloned to JM109 competent cells. About 6–10 positive clones per gene were incubated for 16 h in LB Amp + liquid medium, and their plasmids were extracted by the QIAprep Spin Miniprep Kit (Qiagen). The integrity of plasmids was checked on 2% agarose gel, and concentrations were determined by NanoDrop method. Plasmids were sequenced by Fasmac Ltd., sequencing company (Kanagawa, Japan). Raw sequences were processed using MEGA version X (Kumar et al., 2018) to generate the exact nucleotide sequences of each gene after deleting the included sequences of the vector. The obtained edited sequences were subjected to a homology search via BLAST (Altschul et al., 1997) at DDBJ (the DNA Data Bank of Japan)1. For each gene, the plasmids showing sequences closer to that of the expected reference were used as standards for qPCR. The initial copy number of plasmid standards is necessary for downstream calculations. It was calculated as follow:
The length of the used vector (pGEM-T Easy system 2) is 3,015 bp. The insert’s length was determined after processing the sequences data in MEGA X as indicated above. The Avogadro’s number unit is in “number/mole.” The Dalton representing the value for dsDNA is in “g/mole of bp.” 10E + 9 is the conversion factor of g to ng. 2 nt refers to the two DNA brins. These calculations can be also be summarized into the simplified equation:
During qPCRs following Sarr et al. (2019), we used the prepared plasmid standards with known copy numbers and performed 3 biological replicates per sample. The conditions consisted of an initial denaturation of 98°C for 2 min, followed by 40 cycles of 98°C for 10 s, annealing temperature for 30 s, a melt curve at 65–90°C, and an increment at 0.5°C for 5 s. The amplification efficiencies-r2 values were 92.3%–0.993, 89.6%–0.999, 76.1%–0.998, 67.3%–1.000, 92.2%–0.999, 108.4%–0.999, 104%–0.998, 77.4%–0.999, 93.%–0.999, and 100.1%–0.999 for bacteria, fungi, PSF, phoD, phnX, aphA, entA, pstS, pqqB, and pqqE, respectively. Table 1 contains the primers used and the best qPCR annealing temperatures for each gene determined from a gradient qPCR.
Statistical Analyses
All physicochemical and biological properties obtained in the three sampling periods were subjected to a two-way ANOVA analysis to estimate the impact of treatments and composting days on these parameters, using CropStat ver. 7.2 (IRRI, Philippines). When no significant interaction was found between treatments and sampling days, the average value of the treatment at the three sampling dates was considered (n = 9). In such a condition, the average of the three treatments at each sampling period was included. However, in the presence of a significant interaction between treatments and the sampling days, a one-way ANOVA was applied to each sampling day separately to estimate the difference level between treatments. When a significant effect was detected (p < 0.05) in both two-way and one-way ANOVA, Fisher’s LSD was used to test the differences between treatments. Furthermore, all physicochemical and biological parameters were subjected to a principal component analysis (PCA) method using Kyplot version 5.0 (Kyence Co., Ltd., Japan). The results gathered in the significant principal components (PC1 and PC2) were discussed. The key parameters relating to P fractions and genes’ abundances were extracted and analyzed for Spearman’s rank correlations, PAST v.2.17 (Hammer et al., 2001). For the Spearman’s rank correlation analysis, we considered all the retrieved parameter data collected at 40, 60, and 180 days.
Results
Chemical Properties of Sorghum Straw, Soil, and Composts
Changes of P Fractions During Composting
The two-way ANOVA results showed non-significant interactions between treatments and sampling days for the three organic P fractions H2O-Po, NaHCO3-Po, and NaOH-Po. Therefore, each treatment’s combined data over the three sampling periods were considered, and they showed no significant differences among treatments, nor sampling days (Supplementary Table S1). For the remaining P fractions, significant interactions were found between treatments and sampling days, following two-way ANOVA. Thus, we applied a one-way ANOVA to treatments at each sampling day, and the results are reported in Table 2. At each sampling period (45, 60, and 180 days), significant p-values (p < 0.05) were obtained for all reported parameters, where the two BPR-enriched composts (P-Comp, P-Comp-Soil) showed significantly higher concentrations than the compost (Comp). Although not significant, the magnitude of the differences between P-Comp and P-Comp-Soil for these parameters increased by the composting age, to be relatively higher at 180 days than at 60 days. Along the composting period, we observed an increase of each P fraction’s concentration from 45 to 180 days (Table 2), which was more pronounced in BPR-enriched composts. For instance, total P at 180 days in P-Com and P-Comp-Soil was 2- and 2.3-fold, respectively, than their corresponding values at 45 days. These two BPR-enriched composts showed similar amounts of total P at 45 days. Their total P levels increased by 18.5 and 29% in P-Comp and P-Comp-Soil, respectively, at 60 days. At 180 days, the increase further reached 104 and 128% that at 45 days for P-Comp and P-Comp-Soil, respectively. As illustrated in Figure 1A, 86–88% of total P In BPR-enriched composts corresponded to the HCl-P fraction regardless of the composting period. The compost treatment (Comp) contained only 17–23% of HCl-P but had higher proportions of the other P fractions. Figure 1B shows the cumulative concentrations of labile-P fractions. While at 60 days, P-Comp and P-Comp-Soil showed similar total labile-P levels, the labile-P at 180 days was 13.05% higher in P-Comp-Soil than in P-Comp. Total labile-P in BPR-enriched composts at 180 days was twofold that at 45 days. Moreover, 60–67% of the labile-P in BPR-enriched composts was H2O_Pi.
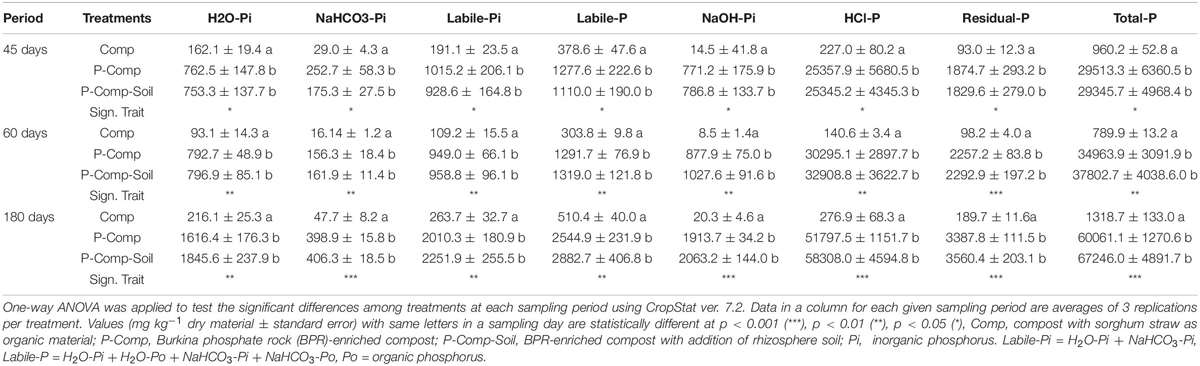
Table 2. LSD all-pairwise comparisons of P fractions following a one-way ANOVA applied to each compost sampling day.

Figure 1. Distribution of all P fractions in composts (A), and concentrations of labile-P fraction during the composting (B). Sampling was carried out at 45, 60, and 180 days. The data of each treatment at a given sampling period are averages of 3 replications per treatment. Comp, sole compost with sorghum straw as organic material; P-Comp, Burkina phosphate rock (BPR)-enriched compost; P-Comp-Soil, BPR-enriched composted with addition of rhizosphere soil.
Total and Inorganic Nitrogen, Total Carbon, C/N, pH, and EC During Composting
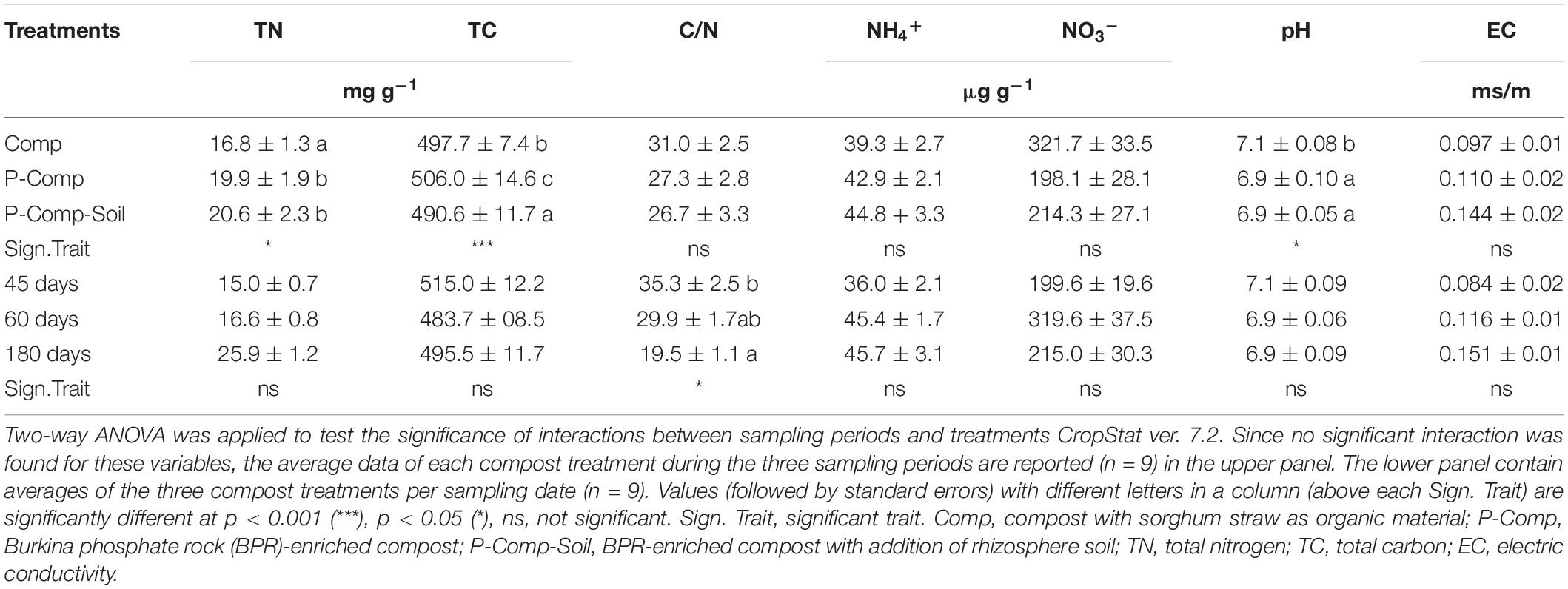
The results of the two-way ANOVA showed significant interactions between treatments and sampling days. The average data for each treatment over the three sampling periods are thus reported in Table 3. It revealed significant differences among treatments for total nitrogen (p < 0.05), total carbon (p < 0.001), and pH (p < 0.05), and among sampling periods for C/N ratio (p < 0.05). P-Comp and P-Comp-Soil contained significantly higher total nitrogen levels than Comp, and significantly lower pH than Comp. However, P-Comp-Soil had significantly less organic carbon contents, followed by Comp, then P-Comp-Soil. Considering the averages of parameters in the three compost treatments at each sampling date, we found an overall significantly less C/N ratio at 180 days than at 45 days. Still the C/N ratio between 60 days was like that at 45 days and that at 180 days.
Abundance of Microbes and Genes Involved in P Solubilization
The two-way ANOVA revealed non-significant interactions between treatments and sampling days for bacteria (16S rRNA) and PSF. The average data over the three sampling periods (n = 9) are thus reported in Supplementary Table S2. While there was no significant difference among treatments for the abundance of total bacteria, P-Comp-Soil contained significantly higher PSF abundance than Comp. However, the differences in PSF abundance between P-Comp and Comp, and P-Comp and P-Comp-Soil, was not significant, respectively. Table 5 shows that the overall abundance of bacteria and PSF was the same among sampling days.
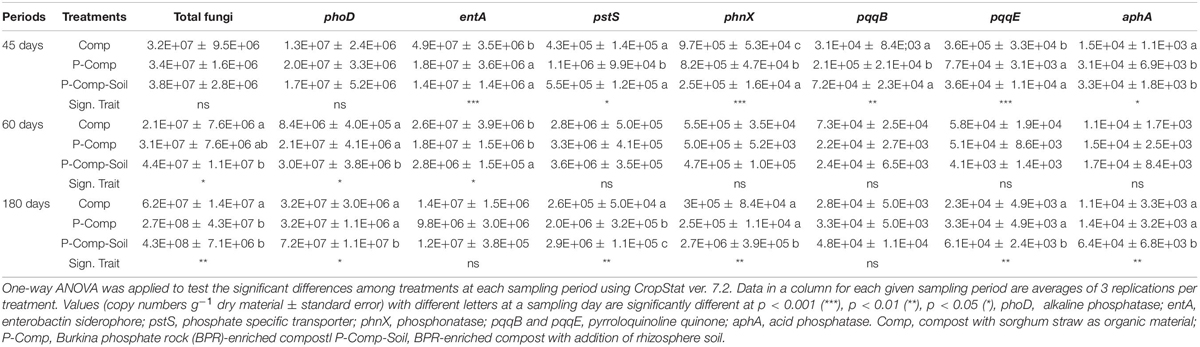
Table 4. LSD all-pairwise comparisons of microbial gene’s abundance following a one-way ANOVA applied to each compost sampling day.
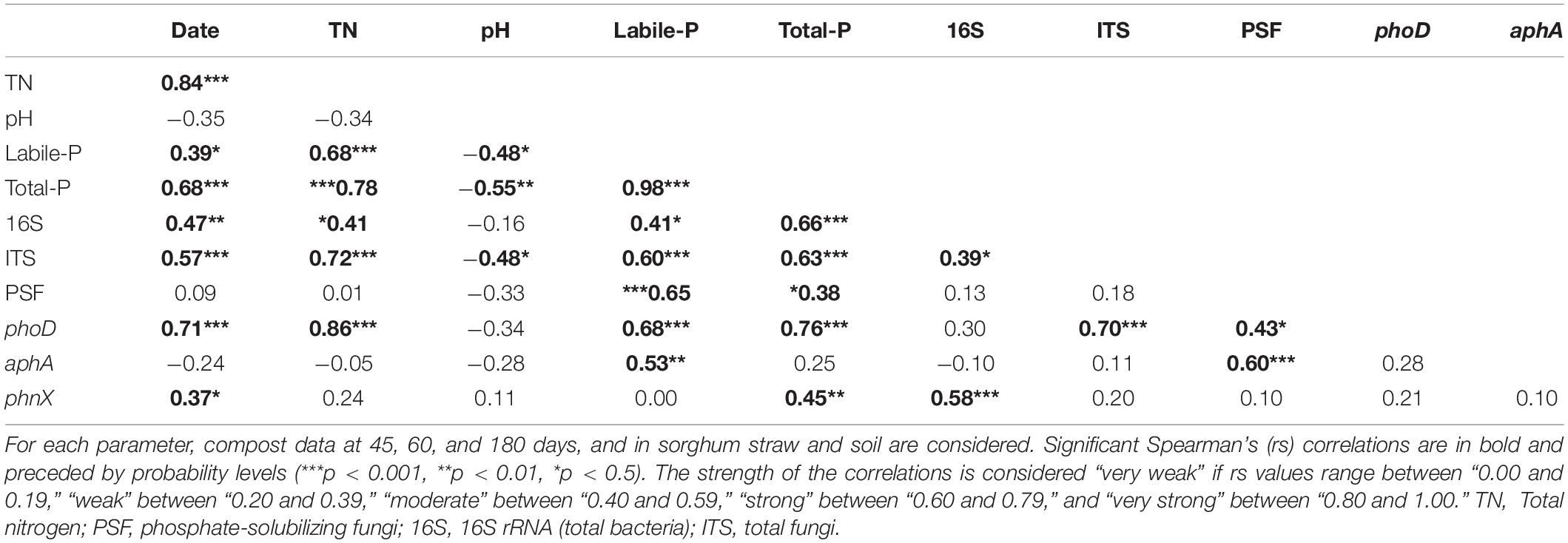
Table 5. Spearman’s correlations (rs) between theoretically selected significant chemical and microbiological compost parameters.
Other than bacteria and PSF, the two-way ANOVA applied to all other biological parameters showed significant interactions between treatments and sampling days. Therefore, a one-way ANOVA was performed for each sampling period’s data, and the results are reported in Table 5. At 45 days, the significant LSD differences were obtained for entA, pstS, phnX, pqqB, and aphA. The siderophore entA gene’s abundance was substantially higher in Comp than in BRR-enriched composts. The phosphate specific transporter pstS gene was more abundant in P-Comp than in the two other treatments. The abundance of the phosphonatase phnX gene was significantly higher in Comp than in P-Comp, which contained more phnX abundance than the P-Comp-Soil. The PSB pqqB gene was more abundant in P-Comp, while pqqE abundance was higher in Comp. The acid phosphatase aphA gene was significantly higher in BPR-enriched composts (P-Comp and P-Comp-Soil).
At 60 days, the pattern of the significance levels differed from that at 45 days. The significant probability levels (p < 0.05) were found in total fungi, phoD, and entA genes. The total fungal abundance in P-Comp was the same as in Comp and P-Comp-Soil, but P-Comp-Soil contained a significantly higher abundance of fungi than Comp. The phoD gene’s abundance was markedly higher in P-Comp-Soil than in P-Comp and Comp. In contrast, P-Comp and Comp held significantly more copy numbers of the entA gene than P-Comp-Soil. At 180 days, except for entA and pqqB genes, all statistical analyses were significant (p < 0.05). The abundance of total fungi was significantly higher in the two BPR-enriched composts than in Comp. The phoD copy number in P-Comp-Soil was considerably higher than that in P-Comp and Comp. The pstS gene was substantially more abundant in P-Comp-Soil than in P-Comp, which had more pstS than Comp. Moreover, P-Comp-Soil contained significantly higher abundances of phnX, pqqE, and aphA genes than P-Comp and Comp.
Figure 2A showed the predominance of bacteria over fungi in all compost treatments regardless of the composting stage. At 45 days, the bacteria themselves accounted for 97–98.2% of the total bacteria and total fungi combined. Total bacteria and total fungi were in the same range at 60 and 45 days. However, the proportion of fungi increased in the three composts at 180 days. While at 45 days, the fungi accounted for only 1.8–3% of total bacteria total fungi combined, they reached 11–17.5% in the three compost treatments at 180 days. Although total bacteria dominated total fungi in composts, PSF significantly outnumbered the PSB genes pqqB and pqqE (Figure 2B). The Figure 2B revealed that 98.25–99.75% of P-solubilizing microbes in composts, regardless of the sampling day, were the PSF. The PSF’s proportion increased toward the compost maturation phase, where the ratio of pqqE and pqqB genes decreased. Figure 3 illustrates the distribution of phoD, phnX, and aphA genes, involved in the mineralization of organic P. It shows that the abundance of the alkaline phosphatase phoD was significantly higher than that of the phosphonatase phnX and acid phosphatase aphA. The phoD gene represented at least 93% of the total of these three genes. The proportion of phnX in composts was significantly higher than that of aphA which showed a very little abundances during the composting period. The Figure 3 shows that the percentage of phoD gene in Comp and P-Comp slightly increases over the composting period, to be higher at 180 days. The proportion of phnX followed an opposite trend since it was less at 180 days in these two compost types. In contrast, the percentage of phnX gene increased in P-Comp-Soil at 180 days compared to 45 and 60 days. In sorghum straw, the phnX (41%) and aphA (59%) were the only amplified enzymes, while the soil contained mostly the phoD gene (data not shown).
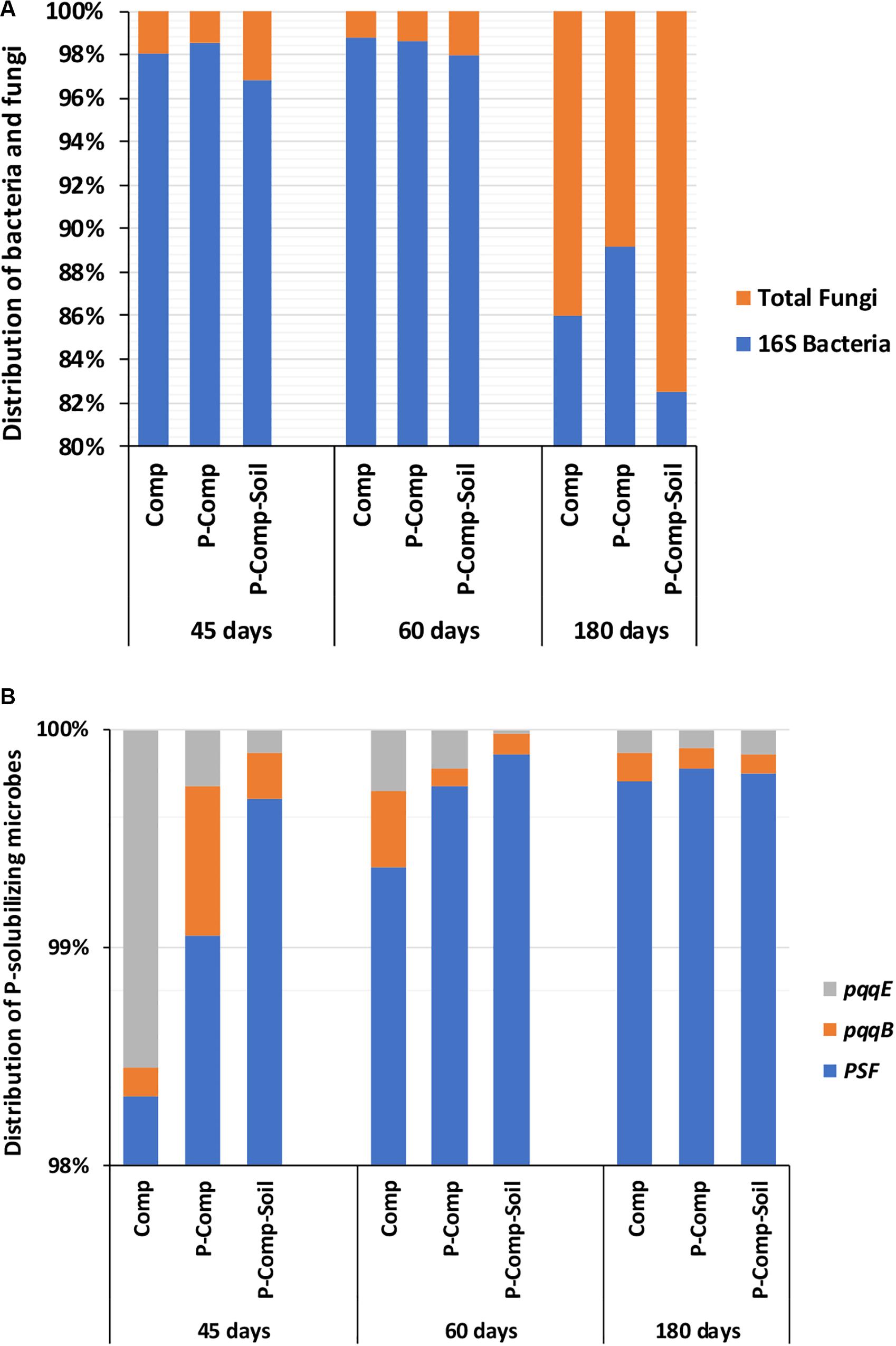
Figure 2. Distribution of bacteria and fungi (A), and P-solubilizing microbes in composts (B). Sampling was carried out at 45, 60, and 180 days. The data of each treatment at a given sampling period are averages of 3 replications per treatment. Comp, sole compost with sorghum straw as organic material; P-Comp, Burkina phosphate rock (BPR)-enriched compost; P-Comp-Soil, BPR-enriched composted with addition of rhizosphere soi; PSF, P-solubilizing fungi; pqq, pyrroloquinoline quinone.
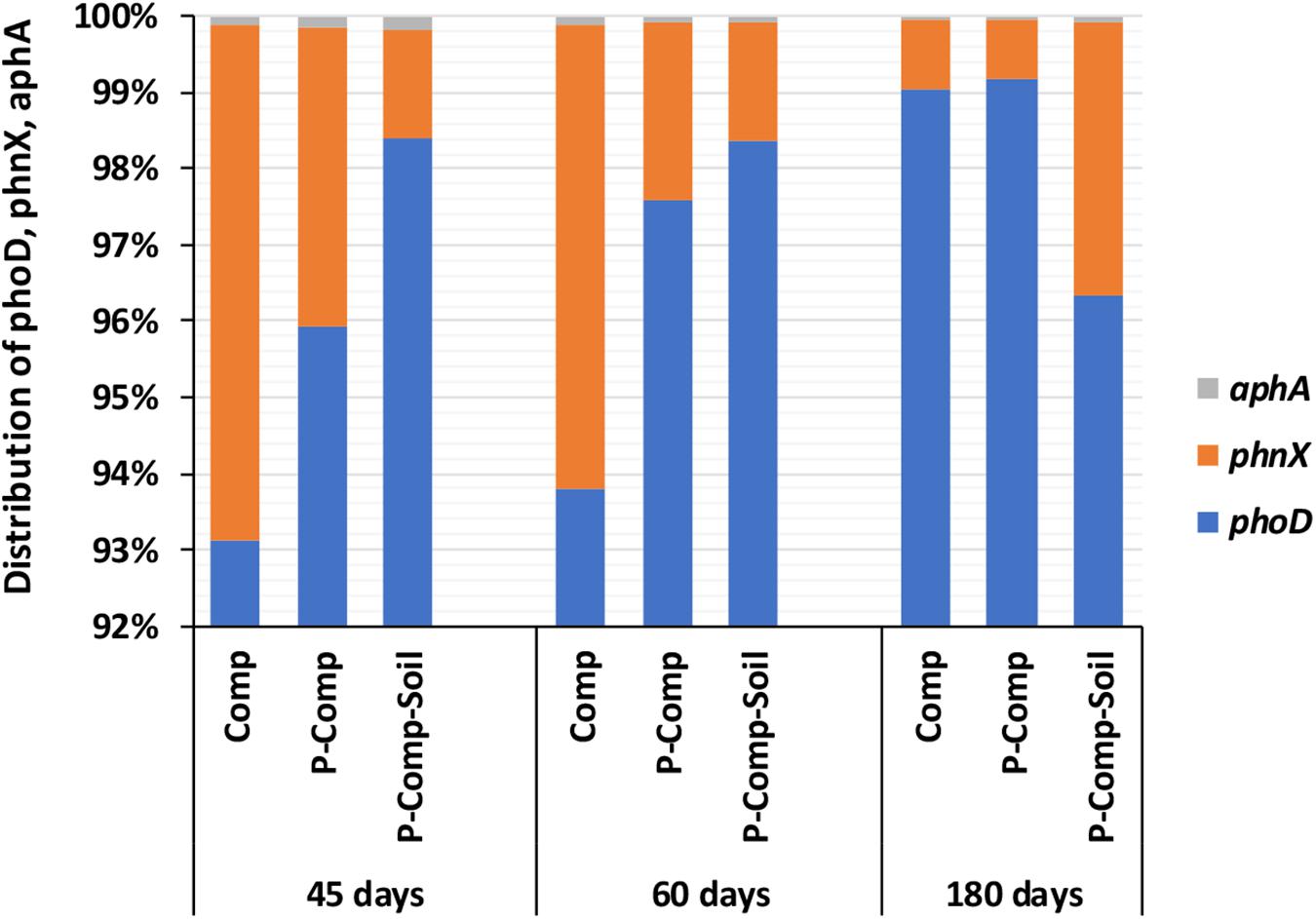
Figure 3. Distribution of phoD, phnX, and aphA genes involved in P mineralization. Sampling was carried out at 45, 60, and 180 days. The data of each treatment at a given sampling period are averages of 3 replications per treatment. Comp, sole compost with sorghum straw as organic material; P-Comp, Burkina phosphate rock (BPR)-enriched compost; P-Comp-Soil, BPR-enriched composted with addition of rhizosphere soil; phoD, alkaline phosphatase; phnX, phosphonatase; aphA, acid phosphatase.
Principal Component Analysis and Correlation Analysis of Significant Parameters
To determine which factor influences P fractions in composts, we performed a PCA using a total of 29 parameters, comprising the sampling dates, seventeen physicochemical parameters (Supplementary Table S1 and Tables 2, 3), and ten biological parameters (Supplementary Table S2 and Table 4). After the PCA, the principal components PC1 and PC2, which showed significant p levels (p < 0.001), were selected. Supplementary Table S3 shows the factor-loading matrix of the two chosen PCs. PC1 explained a positive very high factor loading for the 16S bacteria (0.997), and moderate factor loading for NaOH-Pi, HCl-Pi, Residual-P, total P, and pstS. This PC1 can be defined as the “bacterial-mediated changes in total P and recalcitrant P forms” even if the bacterial influence in these P forms was weak (Supplementary Figures S1G,H). Furthermore, Supplementary Figures S1A–F shows that PC1 explained by bacterial changes is weakly correlated to labile-P, or not at all related to ITS fungi, PSF, phoD, and aphA. The PC2 had higher factor loadings for most physicochemical properties, and biological factors such as fungi with a factor loading as high as 0.955, PSF, phosphatases (phoD, aphA), and phosphonatase (phnX). This PC2 can be defined as “compost properties change as influenced by fungi and enzymatic activities.” The major difference between the two PCs is that while bacteria lead the changes in PC1, fungi and the analyzed enzymes explain the properties changes in PC2. In the PC2, all P fractions, total P, total nitrogen, NH4+, EC, and sampling date, provided significant positive values, like the fungi, PSF, and phosphatases (phoD, aphA), and phosphonatase (phnX). In contrast, the C/N ratio, NO3–, and pH values were significantly negative. Although PC1 showed significant loads for NaOH-Pi, HCl-Pi, Residual-P, and total P, the loads of these parameters in the PC2 were much higher. Moreover, the PC2 had a more substantial load for the labile-P than PC1. Since PC2 explains powerfully the P fractions changes, including the labile-P, we plotted the PC2 scores obtained during PCA to some selected significant parameters such as labile-P, total fungi (ITS), PSF, phoD, aphA and phnX gene’s abundances (Figure 4) to assess how strong these properties influence individually the P fraction changes. Similarly to the results in Supplementary Table S3, Figure 4A confirms that labile-P positively, significantly correlates with PC2 scores. The same significant positive relationships existed between PC2 scores and total fungi (Figure 4B), PC2 scores and PSF (Figure 4C), PC2 scores and phoD (Figure 4D), PC2 scores and aphA (Figures 4E,F), and PC2 scores and phnX (Figures 4G,H). However, the significance of the relationships between PC2 scores and phoD, aphA, and phnX is higher when only data obtained at 180 days are considered (red dots of Figures 4D,F,H).
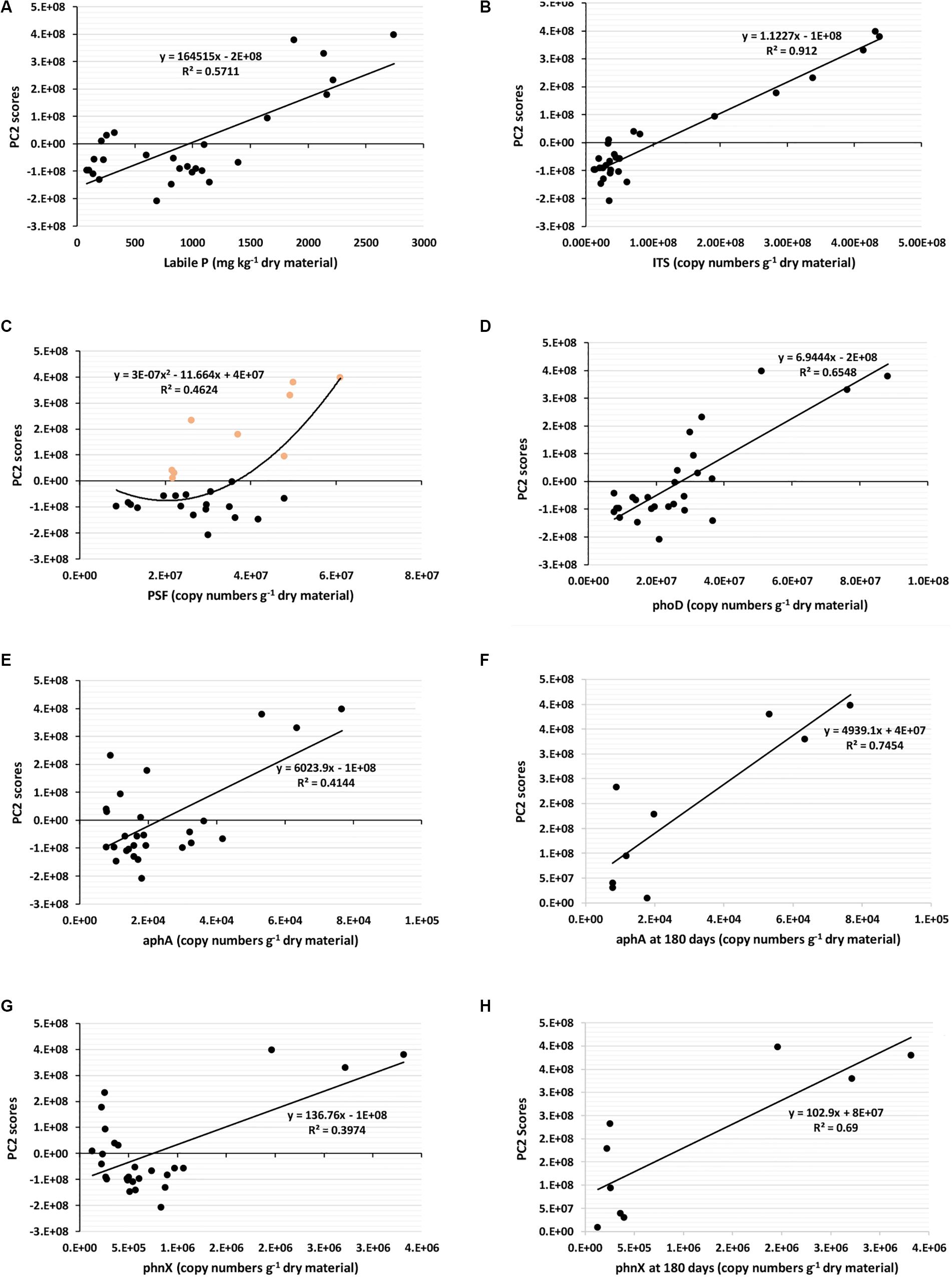
Figure 4. Relationships between PC2 scores and labile P, and gene’s abundances. All compost data at 45 days, 60 days, and 180 days are considered in (A–E,G). The orange dots at (C) are PSF abundances at 180 days. In (F,H), gene’s abundance obtained at 180 days were selected and plotted. ITS, Total fungi; PSF, phosphate-solubilizing fungi; phoD, alkaline phosphatase; aphA, acid phosphatase; phnX, phosphonatase.
The parameters for which we did not find significant differences among compost treatments after the two-way ANOVA, and those with non-significant loads in the PCA, all corresponding to the properties that do not directly influence the P fractions, were excluded from the Spearman’s correlation analysis. Spearman’s correlation test was performed to assess the details relationships between the significant parameters. The correlation’s results of these theoretically selected parameters are reported in Table 5. We found meaningful positive relationships between the sampling date and total nitrogen, labile-P, total P, microbial abundances, phoD, and phnX. The labile-P, which is one of the essential fractions in this study, was positively, strongly correlated to total nitrogen, and moderately negatively related to the pH. Furthermore, it was positively, very strongly interrelated to the concentration of total P. Moreover, Spearman’s correlations between labile-P and total fungi, PSF, alkaline phosphatase phoD, and acid phosphatase aphA were strongly positive. However, its positive relationship with total bacteria was moderate. Table 5 shows that the pH, other than with the labile-P, was also negatively correlated with total P (strongly) and fungi (moderately). The total nitrogen was positively, strongly to very strongly related to P, fungi, phoD, and moderately to total bacteria. There were positive and strong relationships between total P and total bacteria, total fungi, and phoD, while total P was moderately and weakly related with phnX, and PSF, respectively. We found that bacteria were positively strongly correlated with phnX, and weekly with fungi. Finally, the correlation was positive and strong between phoD and fungi, and moderate between phoD and PSF. These PSFs were positively strongly related to the acid phosphatase aphA. Such positive relationships were missing between total bacteria and phoD and aphA.
Discussion
In this study, we established three compost types using sorghum straw as organic material, supplemented with Burkina Faso phosphate rock (BPR) and a rhizosphere soil collected from farmer’s sorghum fields. Based on qPCR analysis, we compared the change evolution during composting of several microbial genes including 16S rRNA total bacteria, ITS fungi, PSF, phosphate solubilizing bacteria (pqqB and pqqE genes), alkaline phosphatase (phoD gene), acid phosphatase (aphA), phosphonatase (phnX gene), siderophore (enterobactin entA gene), and phosphate specific transporter (pstS gene). Also, we determined the different P fractions in composts, the total and available nitrogen levels, the pH, and the electric conductivity from samples collected at 45, 60, and 180 days incubation. Multivariate analyses were then performed to assess the relationships between compost P availability and functional genes.
Evolution of Compost’s Physicochemical Parameters
The solid negative Spearman’s correlation between sampling dates and C/N ratio (Table 5) indicates a progress of the compost’s decomposition through composting time. At 180 days, the lower C/N ratio in the compost treatments enriched with Burkina Faso rock phosphate (BPR) revealed a better compost decomposition in these treatments, which led to higher N concentrations, inorganic P fractions, residual P, HCl-P, and total P as shown in Table 2. Wei et al. (2015) attributed the increase in total P to the so-called “concentration effect” occurring when C, hydrogen, and N are lost with the exit gas as CO2, H2O, and NH3, respectively, but P is retained in the sample. In this study, the addition of rhizosphere soil to the compost enriched with BPR led to some increases in total P and available P at 180 days. Still the increases were not significant compared to the BPR-enriched compost without soil. At this late composting phase, the labile-P in P-Comp-Soil was 13% higher than that in P-Comp, although these two treatments had the same concentration of available P at 60 days. Although the difference in available P was not significant between the two treatments, this result further supports that the release of available P in BPR-enriched composts is sustained better by the addition of rhizosphere soil during composting. The rhizosphere which is the narrow zone in the soil near roots (Kumar and Dubey, 2020) provides various ecosystem services, including the cycling of nutrients (Adle, 2016), and it is an enormous reservoir of microbial community and small soil-inhabiting animals (Malla et al., 2018; Dubey et al., 2019). However, the functionality of this rhizosphere microbiome depends on several factors such as the intrinsic natural abundance of microbes in the environment which also relates to the soil physicochemical properties, the type of plant since the nature of the released secondary molecules triggering microbial activities differ from plant to plant, the period of the season owing that microbes may function more actively during wet seasons than dry seasons (Soni et al., 2017; Vyas et al., 2018). In the present study, the sorghum rhizosphere soil used as inoculum was collected toward the end of the growing phase. It is plausible that at this period, the microbiome was less active than if it was collected during the vegetative stage, justifying why there was an increase of labile-P in BPR-enriched compost with soil, but not significant compared to BPR-enriched compost alone. Therefore, it may be necessary to optimize the soil addition rates to obtain a more substantial increase of available P in BPR-enriched composts. The labile inorganic P (Pi) corresponding to the sum of H2O-Pi and NaHCO3-Pi, and which led the variations of total labile-P in treatments compared to the labile organic P (Po) (Supplementary Table S1 and Table 2) amounted half of the available P in the sole compost and over 80% of the labile-P in BPR-enriched composts. We found an increase in the levels of labile Pi in treatments with the progress of the composting, supporting Sunita (2014), who also reported a rise of Pi during composting.
NaOH-extracted P, considered as the moderately soluble phosphorus pool (Ahmed et al., 2019), was not included in the readily available P in our study. In compost, NaOH extracted mostly the organic P fraction, while in BPR-enriched composts, it extracted 4–5 times more Pi than Po. The NaOH-extractable P fractions, including Fe-, Al-bound P, humic P, Ca-, Mg-bound P, and moderately labile-P (Lu et al., 2013), increased with the composting age (Table 2). The last two P fractions HCl-P and residual P are mainly composed of insoluble and stable forms of phosphorus, such as Ca-, Fe-, Al-bound P (Aulakh et al., 2003; Zicker et al., 2018). Our data showed that this non-labile-P pool followed the same increasing trend as the labile-P and the moderately labile-P (Table 2 and Figure 1A) and was higher in the BPR-enriched composts. The non-labile-P accounted for 30–35% of the total P in compost and 92–93% of that in BPR-enriched composts. Therefore, the addition of BPR during composting also increases the chance of supplying higher amounts of insoluble phosphorus. However, Ahmed et al. (2019) stated that HCl-P and residual P fractions of compost might be involved in long-term phosphorus cycling in soil. The higher NaOH-P and HCl-P levels at the end of the composting could be related to P immobilization by microorganisms, which can cause a slower buildup of residual P (Hedley et al., 1982). The mineral solubilization of the highly present CaO (34.39%) in BPR may release free Ca2+, which bounds to free Pi and increase the level of residual P when the composting proceeds.
Relationships Between Compost’s P Fractions and Microbiological Properties
The primary goal of this work was to determine the changes in P fractions during composting and to relate such changes with microbial functional genes in composts. Identifying the key factors contributing to increase the availability of P in natural ecosystems is crucial, as P is thought to override all other elements, including C and N (Elser, 2012). Adequate P availability positively influences the net productivity in agricultural systems (Williams et al., 2013), and it lays down the primordia of plant reproductive parts during the early phase of plant development (Satyaprakash et al., 2017). Although many soils can contain extensive total P stocks, it is widely accepted that the bioavailability of soluble orthophosphate, which can be used by most biota, is low (Rodriguez and Fraga, 1999). Fortunately, microorganisms have proven their ability to increase P availability by mineralization of organic-P and solubilization of inorganic-P (Richardson and Simpson, 2011). Recently, the study of microbes and genomes encoding enzymes responsible for P-cycling has been facilitated by the advance of high-throughput sequencing methods (Yao et al., 2018), eliminating the bias encountered in previous microbial isolation techniques. The qPCR-based molecular analyses applied in the present study provided substantial information on the evolution of P-cycling-related genes during the composting period.
We observed that as early as 45 days incubation, the total bacterial abundance in the three composts was already 4.85- to 9.85-fold than that of the used sorghum straw and soil (data not shown). This result indicates that bacteria may have increased tremendously during the thermophilic (55–60°C in average) phase in the first 2 weeks (microbial abundance not quantified during this period) to reach a pick as observed at 45 days. After that, they maintained a more a less stable number up to the end of the composting. Partanen et al. (2010) reported high concentrations of Actinobacteria and Bacillus spp. during the thermophilic phase of composting, which decompose hemicellulose, cellulose, and lignin, as also reported by Yu et al. (2007). The total fungi, which did not show much change during the first 2 months, increased significantly at 180 days in each compost treatment. This result reveals that fungi express well in composts even at lower temperatures. While the average daily temperature was 32°C, the temperature in composts dropped to 45°C at 45 days, and 35°C at 110 days (data not shown). The higher nutrient content in the BPR-enriched composts at 180 days could also partly explain the higher abundance of fungi in these treatments. This relationship between compost’s nutrients richness and fungal abundance was further supported by the highly significant correlation (r2 = 0.91) between PC2 scores, which explained most P fractions changes, including the labile-P, and the abundance of ITS fungi (Figure 4B). This observation, in addition to the positive strong Spearman’s correlations between the abundance of fungi and total N, labile-P, and total P (Table 5), justifies the positive interdependence between nutrient availability and fungal abundance. Furthermore, the study showed the significant contribution of PSF in the increase of labile-P (Table 5), particularly at the late stage of the composting (Figure 4C). The ability of PSFs to transform insoluble organic and inorganic phosphorus into soluble forms has been reported in many studies (Sharma et al., 2013; Kaur and Reddy, 2017; Wang et al., 2018).
In this study, although we found moderate but positive relationships between the abundance of total bacteria and the total nitrogen and labile-P (Table 5), the PCA (Supplementary Table S3) failed to relate the changes in P fractions to any of the analyzed phosphate solubilizing bacterial (PSB) genes pqqB and pqqE. This absence of correlation may arise from non-adequate PSB gene targets or the types of primers used, which might justify why we barely obtained noticeable differences in the abundance of these genes among the compost treatments. Zheng et al. (2017) previously indicated the difficulty of finding homologous genes among all mineral PSB (mPSB) regulating inorganic phosphate solubilization. However, since pqqA, pqqB, pqqC, pqqD, and pqqE genes are conserved and arranged in that order in the pqq operon (pqqABCDE) (Shen et al., 2012), we selected the pqqB and pqqE genes to study the mPSB. Given the substantial wide distribution of PSB in natural ecosystems including soils, freshwater, seawater, and sediments (Zhang et al., 2015; Liang et al., 2020; Wan et al., 2020), we cannot completely rule out a possible contribution of phosphate solubilizing bacteria in the increase of P fractions in our compost treatments. The moderate but positive correlation of 16S bacteria with the labile-P supports the possibility of bacteria to intervene in the P solubilization in the composts. As stated earlier, a lot of PSBs are responsible for the release of organic acids such as gluconic acid (Richardson and Simpson, 2011), which highly contribute to releasing available P for plant uptake from recalcitrant phosphorus forms (Castagno et al., 2011). Since the production of gluconic acid from glucose requires the oxidation of the glucose dehydrogenase (GDH) enzyme released by the activation of the gcd gene (Chen et al., 2015), and that the pyrroloquinoline quinone (PQQ) synthesized by the activation of the PQQ operon with 5–11 genes (Choi et al., 2008) acts as a cofactor, the obtained little concentrations of pqqB and pqqE genes are likely sufficient to trigger the activation of gcd genes and the production of gluconic acid by PSBs. Unfortunately, our attempt to estimate the abundance of gcd genes in our compost treatments was unsuccessful, probably due to non-adequate selected gcd primers or PCR conditions (data not shown). Moreover, we did not determine the levels and identity of organic acids, which might have indicated the presence or absence of PSB in the composts. After all, it is also possible that mPSB via the organic acids production pathway may be minor in this study. In this respect, Illmer and Schinner (1992, 1995), reported a solubilization of calcium phosphate by Pseudomonas even in the absence of organic acid production. Parks et al. (1990) also postulated that H+ excretion originating from NH4+ assimilation and or respiratory H2CO3 production could be the alternative mechanism of mineral phosphate solubilization. Furthermore, Kim et al. (1997) suggested that the release of organic acids may play an essential role in phosphate solubilization but is not the sole reason for the increase in Pi concentration. On the other hand, some reports (Kucey, 1983; Gyaneshwar et al., 1998; Sharma et al., 2013) pointed out that PSFs possess higher abilities to release P from recalcitrant inorganic P when compared to bacteria. These reports support our finding on the substantial role PSFs play in the mineralization of P in the compost treatments.
In general, the ability of phosphate solubilizing microbes to enhance the availability of P during P-cycling depends not only on the microbial abundance, but also on the nutritional richness of the substrate, the physiological and growth status of the organism (Alori et al., 2017), and the environmental conditions. Seshachala and Tallapragada (2012) revealed that among other factors, the interactions with other microorganisms in the substrate, and the physicochemical properties such as organic matter and pH, also influence microbial phosphate solubilization. In the present study, the clustering of the moderately negative pH load to the PC2 (Supplementary Table S3), indicated that a lower pH may also enhance the dissolution of P in BPR-enriched composts. Indeed, the two-way ANOVA had shown significantly lower pH values in the two BPR-enriched composts (Table 3). However, they were close to neutrality (6.86 and 6.91 in P-Comp-Soil and P-Comp, respectively). Even though lower pH values (acidic-oriented pH status) seemed to trigger P mineralization in composts, the abundance of the acid phosphatase aphA gene was very low compared to that of the alkaline phosphatase phoD gene. Alori et al. (2017) mentioned that in most soils, the pH where phosphate activities were reported ranges from neutral to acidic. Our investigation, which focused on composts and not on soil, also finds a similar range of pH from neutral to acidic, suitable to initiate acid phosphatase activities, even if the acid phosphatase gene abundance was low in this study. In contrast, the high abundance of the phoD gene in composts, and more markedly in the BPR-enriched compost with rhizosphere soil at 180 days, highlights the significant contribution of the alkaline phosphatase gene in P mineralization. Since, the primers of phoD used to encode the alkaline phosphatase covers a high diversity of bacteria and fungi, with a stronger incidence on bacteria (Bergkemper et al., 2016), our results on phoD abundance and its relationships with the compost’s physicochemical properties (Supplementary Table S3 and Table 5), indicate that not only the PSF but also phosphate-solubilizing bacteria may take parts in the mineralization of P, as we already stated above. However, Kutu et al. (2018) also reported that the release of alkaline phosphatase was tightly linked to the activity of fungi.
The phosphonatase gene, phnX in the present study, encodes the phosphonatase enzyme, which cleaves the relatively stable carbon-phosphorus bonds occurring in natural and synthetic organophosphonates via C-P lyases and phosphonoacetaldehyde hydrolases (Hsieh and Wanner, 2010). Our PCA results (Supplementary Table S3) showed that apart from phosphatases, the gene encoding the phosphonatase enzyme also plays a significant role in the P mineralization in composts. The effect of phnX on P fractions may be more active toward compost maturity (Figure 4H). The siderophore entA gene and the phosphate specific transporter pstS were not clustered to the PC2. Thus, their involvement in the P fractions changes during the composting period was nil (entA) or minor (pstS). Instead, the pstS load in the PC1 was significant same as that of total bacteria. Because phosphate specific transporters are harbored by bacteria (Hudek et al., 2016), the significance of the 16S bacteria load in the PC1 may have pulled up that of pstS. Moreover, the labile-P load on the PC1 that also explains pstS, was significant but weak (0.382), indicating the existence relationship between labile-P and pstS. In this respect, we found that at 180 days, the abundance of pstS was significantly higher in P-Comp-Soil, followed by P-Comp, then Comp. It is known that available P is a useful nutrient for microbes. Therefore, the labile-P in composts can activate the expression of the bacterial phosphate-specific transporter (pstS) gene to import the nutrient in microbial cells. In this regard, Vuppada et al. (2018) reported that bacterial cells must maintain intracellular Pi pools for optimal growth and have developed strategies to sense Pi and control the expression of genes to fit best their environmental circumstances. Pst (phosphate specific transporter) and Pit (phosphate inorganic transporter) are the two forms of Pi import systems in bacteria. The Pit import system is activated when the external phosphate concentration is lower than 20 μM (Hudek et al., 2016), while the Pst import system is induced at this concentration (Rao and Torriani, 1990). Since the labile-Pi levels of our compost treatments (Table 2) were all above this concentration, the periplasmic substrate-binding protein (pstS) can be well activated for the transport and immobilization of Pi in microbes. The immobilized P in bacterial cells can later by solubilized and made available in soil upon compost application to agricultural fields.
Conclusion
This study evaluated the mechanisms of P solubilization of Burkina Faso phosphate rock-enriched composts, using sorghum straw as organic material. It demonstrated that the addition of rhizosphere as a microbial source to composts helps improve the solubility of the used low-grade phosphate rock. Quantitative PCR data revealed that although bacteria are substantially more abundant than fungi during the composting process, the PSF significantly increased during the late composting phase and were the leading players of the mineral P solubilization. Furthermore, the alkaline phosphatase (phoD), which dominated the genes encoding for the enzymes involved in organic P mineralization increased its abundance with the progress of the composting and played a substantial role in the organic P mineralization. The increase of inorganic P in compost solution favored the transport of Pi to microbial cells. However, such immobilized P can be later decomposed and released in soil upon application of composts in fields. Based on the observed increase of labile-P in phosphate rock-enriched compost following soil addition, but which was not very significant compared to the enriched compost without soil, we hypothesize that a soil collected just at crop harvest or collected during crop cultivation and stored at the appropriate temperature and light conditions, may provide a superior solubilization potential due to the existence of a more active microbial community.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
PSS, MF, EC, and SN: conceptualization. PSS, EBT, and MF: methodology. PSS and SN: validation. PSS and MF: formal analysis. PSS, EBT, MF, EC, and ANZ: investigation. PSS, MF, and ANZ: data curation. PSS: writing-original draft preparation. PSS and SN: writing-review and editing. PSS, EBT, MF, ANZ, EC, and SN: visualization. SN: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Japan Science and Technology Agency (JST) and the Japan International Cooperation Agency (JICA) through the Burkina SATREPS project No JPMJSA1609.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our sincere gratitude to Science and Technology Research Partnerships (SATREPS) program. We are very thankful to Dr. Fujio Nagumo of JIRCAS for the guidance and for facilitating the establishment of the compost trials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2020.559195/full#supplementary-material
FIGURE S1 | Relationships between PC1 scores and labile P, total P, residual P, and gene’s abundances.
Footnotes
References
Adle, S. (2016). titleRhzosphere, food security, and climate change: A critical role for plant-soil research. Rhizosphere 1, 1–3. doi: 10.1016/j.rhisph.2016.08.005
Ahmed, W., Jing, H., Kaillou, L., Qaswar, M., Khan, M. N., Jin, C., et al. (2019). Changes in phosphorus fractions associated with soil chemical properties under long-term organic and inorganic fertilization in paddy soils of southern China. PLoS One 14:e0216881. doi: 10.1371/journal.pone.0216881
Alaylar, B., Güllüce, M., Karadayi, G., and Karadayi, M. (2018). Isolation of PGPR strains with phosphate solubilizing activity from Erzurum and their molecular evaluation by using newly designed specific primer for pqqB gene. Int. J. Sci. Eng. Res. 9, 103–105.
Alori, E. T., Glick, B. R., and Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential use in sustainable agriculture. Front. Microbiol. 8:971. doi: 10.3389/fmicb.2017.00971
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
An, R., and Moe, L. A. (2016). Regulation of pyrroloquinoline quinone-dependent glucose dehydrogenase activity in the model rhizosphere-dwelling bacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 82, 4955–4964. doi: 10.1128/AEM.00813-16
Aulakh, M. S., Kabba, B. S., Baddesha, H. S., Bahi, G. S., and Mps, G. (2003). Crop yields and phosphorus fertilizer transformations after 25 years of applications to a subtropical soil under groundnut-based cropping systems. Field Crops Res. 83, 283–296. doi: 10.1016/S0378-4290(03)00078-9
Barber, S. A., Walker, J. M., and Vasey, E. H. (1963). Mechanisms for the movement of plant nutrients from the soil and fertilizer to the plant root. J. Agric. Food. Chem. 11, 204–207. doi: 10.1021/jf60127a017
Barea, J. M., Pozo, M. J., Azcon, R., and Azcon-Agular, C. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. doi: 10.1093/jxb/eri197
Bergkemper, F., Kublik, S., Lang, F., Krüger, J., Vestergaard, G., Schloter, M., et al. (2016). Novel ologonucleotide primers reveal a high diversity of microbes which drives phosphorus turnover in soil. J. Microbiol. Methods 125, 91–97. doi: 10.1016/j.mimet.2016.04.011
Biswas, D. R., and Narayanasamy, G. (2006). Rock phosphate enriched compost: an approach to improve low-grade Indian rock phosphate. Bioresour. Technol. 97, 2243–2251. doi: 10.1016/j.biortech.2006.02.004
Bolan, N. S., Elliot, J., Gregg, P. E. H., and Well, S. (1997). Enhanced dissolution of phosphate rocks in the rhizosphere. Biol. Fert. Soils 24, 169–174. doi: 10.1007/s003740050226
Castagno, L. N., Estrella, M. J., Sannazzaro, A. I., Grassano, A. E., and Ruiz, O. A. (2011). Phosphate-solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the Salado River Basin (Argentina). J. Appl. Microbiol. 110, 1151–1165. doi: 10.111/j.1365-2672.2011.04968.x
Chen, W., Yang, F., Zhang, L., and Wang, J. (2015). Organic acid secretion and phosphate solubilizing efficiency of Pseudomonas sp. PSB12: effects of phosphorus forms and carbon sources. Geomicrobiol. J. 33, 870–877. doi: 10.1080/01490451.2015.1123329
Choi, O., Kim, J., Kim, J. G., Jeong, Y., Moon, J. S., Park, C. S., et al. (2008). Pyrroloquinonline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiol. 146, 657–668. doi: 10.1104/pp.107.112748
Deppenmeier, U., Hoffmeister, M., and Prust, C. (2002). Biochemistry and biotechnical applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60, 233–242. doi: 10.1007/s00253-002-1114-5
Dubey, A., Kumar, A., Abd Allah, E. F., Hashem, A., and Khan, M. L. (2019). Growing more with less: breeding and developing drought resilient soybean to improve food security. Ecol. Ind. 105, 425–437. doi: 10.1016/j.ecolind.2018.03.003
Eder, S., Shi, L., Jensen, K., Yamane, K., and Hulett, F. M. (1996). A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of the Pho regulon gene, phoD. Microbiology 142, 2041–2047. doi: 10.1099/13500872-142-8-2041
Elser, J. J. (2012). Phosphorus: a limiting nutrient for humanity? Curr. Opin. Biotechnol. 23, 833–838. doi: 10.1016/j.copbio.2012.03.001
Goldstein, A. H. (1995). Recent progress in understanding the molecular and biochemistry of calcium phosphate solubilization by gram negative bacteria. Biol. Agric. Hortic. 12, 185–193. doi: 10.1080/01448765.1995.9754736
Gyaneshwar, P., Kumar, G. N., and Parekh, I. J. (1998). Effect of buffering on the phosphate-solubilizing ability of microorganisms. World J. Microb. Biotechnol. 13, 669–673. doi: 10.1023/A:1008852718733
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST. Paleontological statistics software Package for education and data analysis. Palaeontol. Electron. 4:9.
Hedley, M. J., Stewart, J. W. B., and Chauhan, B. S. (1982). Changes in inorganic and organic soil phosphorus fractions induced by cultivation practices and by laboratory incubations. Soil Sci. Soc. Am. J. 46, 970–976. doi: 10.2136/sssaj1982.03615995004600050017x
Hsieh, Y. J., and Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 15, 198–203. doi: 10.1016/j.mib.2010.01.014
Hudek, L., Premachandra, D., Webster, W. A. J., and Bräu, L. (2016). Role of phosphate transport system component PstB1 in phosphate internalization by Nostoc punctiforme. Appl. Environ. Microbiol. 82, 6344–6356. doi: 10.1128/AEM.01336-16
Illmer, P., and Schinner, F. (1992). Solubilization of inorganic phosphates by microorganisms isolated from forest soils. Soil Biol. Biochem. 24, 389–395. doi: 10.1016/0038-0717(92)90199-8
Illmer, P., and Schinner, F. (1995). Solubilization of organic calcium phosphates – solubilization mechanisms. Soil Biol. Biochem. 27, 257–263. doi: 10.1016/0038-0717(94)00190-C
Kaur, G., and Reddy, M. S. (2017). “Role of phosphate-solubilizing fungi in sustainable agriculture,” in Developments in Fungal Biology and Applied Mycology, eds T. Satyanarayaa, S. Deshmukh, and B. Johri (Singapore: Springer), 391–412. doi: 10.1007/978-981-10-4768-8_20
Khan, K. S., and Joergensen, R. G. (2009). Changes in microbial biomass and P fractions in biogenic household waste compost amended with inorganic P fertilizers. Bioresour. Technol. 100, 303–309. doi: 10.1016/j.biortech.2008.06.002
Khan, M. S., Zaidi, A., Ahemad, M., Oves, M., and Wani, P. A. (2010). Plant growth promotion by phosphate solubilizing fungi-current perspective. Arch. Agron. Soil Sci. 56, 73–98. doi: 10.1080/03650340902806469
Kim, K. Y., McDonald, G. A., and Jordan, D. (1997). Solubilization of hydroxyapatite by Enterobacter aglomerans and cloned Escherichia coli in culture medium. Biol. Fert. Soils 24, 347–352. doi: 10.1007/s003740050256
Krishnaraj, P. U., and Dahale, S. (2014). Mineral phosphate solubilization: concepts and prospects in sustainable agriculture. Proc. Indian Natn. Sci. Acad. 80, 389–405. doi: 10.16943/ptinsa/2014/v80i2/55116
Kucey, R. M. N. (1983). Phosphate-solubilizing bacteria and fungi in various cultivated and virgin Alberta soils. Can. J. Soil Sci. 63, 671–678. doi: 10.4141/cjss83-068
Kumar, A., and Dubey, A. (2020). Rhizosphere microbiome: engineering bacterial competitiveness for enhancing crop production. J. Ad. Res. 24, 337–352. doi: 10.1016/j.jare.2020.04.014
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGAX: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kutu, F. R., Mokase, T. J., Dada, O. A., and Rhode, O. H. J. (2018). Assessing microbial population dynamics, enzyme activities and phosphorus availability indices during phospho-compost production. Int. J. Recycl. Org. Waste Agric. 8, 87–97. doi: 10.1007/s40093-018-0231-9
Lane, D. J. (1991). “S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackbrandt and M. Goodfellow (New York, NY: John Wiley & Sons Ltd).
Liang, J., Liu, J., Jia, P., Yang, T., Zeng, Q., Zhang, S., et al. (2020). Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J. 14, 1600–1613. doi: 10.1038/s41396-020-0632-4
Loy, A., Lehner, A., Lee, N., Adamczyk, J., Meier, H., Ernst, J., et al. (2002). Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68, 5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002
Lu, D., Yan, B., Wang, L., Deng, Z., and Zhang, Y. (2013). Changes in phosphorus fractions and nitrogen forms during composting of pig manure with rice straw. J. Integr. Agric. 12, 1855–1864. doi: 10.1016/S2095-3119(13)60400-1
Malla, M. A., Dubey, A., Yadav, S., Kumar, A., Hashem, A., and Abd Allah, E. F. (2018). Understanding and designing the strategies for the microbes-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 9:1132. doi: 10.3389/fmicb.2018.01132
McGrath, J. W., Chin, J. P., and Quinn, J. P. (2013). Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat. Rev. Microbiol. 11, 412–419. doi: 10.1038/nrmicro3011
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Nakamura, S., Iwai, T., Toriyama, K., Tobita, S., Matsunaga, R., Fukuda, M., et al. (2015). Solubilization of Burkina Faso phosphate rock through calcination method. Jap. J. Soil Sci. Plant Nutr. 86, 535–539. doi: 10.20710/dojo.86.6_534
Nercessian, O., Fouquet, Y., Pierre, C., Prieur, D., and Jeanthon, C. (2005). Diversity of bacteria and archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ. Microbiol. 7, 698–714. doi: 10.1111/j.1462-2920.2005.00744.x
Ochwoh, V. A., Claassens, A. S., and de Jager, P. C. (2005). Chemical changes of applied and native phosphorus during incubation and distribution into different soil phosphorus pools. Comm. Soil Sci. Plant Anal. 36, 535–556. doi: 10.1081/CSS-200043281
Parks, E. S., Olson, G. J., Brickman, F. E., and Baldi, F. (1990). Characterization of high-performance liquid chromatography (HPLC) of the solubilization of phosphorus in iron ore by a fungus. Indian J. Microbiol. 5, 183–190. doi: 10.1007/BF01573868
Partanen, P., Hultman, J., Paulin, L., Auvinen, P., and Romantschuk, M. (2010). Bacterial diversity at different stages of the composting process. BMC Microbiol. 10:94. doi: 10.1186/1471-2180-10-94
Pii, Y., Mimmo, T., Tomasi, N., Terzano, R., Cesco, S., and Crecchio, C. (2015). Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. Biol. Fert. Soils 51, 403–415. doi: 10.1007/s00374-015-0996-1
Rao, N. N., and Torriani, A. (1990). Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4, 1083–1090. doi: 10.1111/j.1365-2958.1990.tb00682.x
Richardson, A. E., and Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability. Plant Physiol. 156, 989–996. doi: 10.1104/pp.111.175448
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/s0734-9750(99)00014-2
Sanchez, P. A., Shepherd, K. D., Soule, M. J., Place, F. M., Buresh, R. J., Izac, M. N., et al. (1997). “Soil fertility replenishment in Africa: an investment in natural resources capital,” in Replenishing Soil Fertility in Africa, eds R. J. Buresh, P. A. Sanchez, and F. Calhoun (Madison, WI, USA: SSSA Special Publication No 51), 1–46.
Sarr, P. S., Ando, Y., Nakamura, S., Deshpande, S., and Subbarao, G. V. (2019). Sorgoleone release from sorghum roots shapes the composition of nitrifying populations, total bacterial and archaea and determines the level of nitrification. Biol. Fert. Soils 56, 145–166. doi: 10.1007/s00374-019-01405-3
Satyaprakash, M., Nikitha, T., Reddi, E. U. B., Sadhana, B., and Vani, S. S. (2017). A review on phosphorus and phosphate solubilizing bacteria and their role in plant nutrition. Int. J. Curr. Microbiol. Appl. Sci. 6, 2133–2144. doi: 10.20546/ijcmas.2017.604.251
Searle, L. J., Méric, G., Porcelli, I., Sheppard, S. K., and Lucchini, S. (2015). Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of Escherichia coli. PLoS One 10:e0117906. doi: 10.1371/journal.pone.0117906
Seshachala, U., and Tallapragada, P. (2012). Phosphate solubilizers from the rhizosphere of Piper nigrum L. in Karnataka, India. Chil. J. Agric. Res. 72, 397–403. doi: 10.4067/S078-58392012000300014
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate-solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587. doi: 10.1186/2193-1801-2-587
Shen, Y. Q., Bonnot, F., Imsand, E. M., RoseFigura, J. M., Sjolander, K., and Klinman, J. P. (2012). Distribution and properties of genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51, 2265–2275. doi: 10.1021/bi201763d
Soni, R., Kumar, V., Suyal, D. C., Jain, L., and Goel, R. (2017). “Metagenomics of plant rhizosphere microbiome,” in Understanding Host-Microbiome Interactions – An Omics Approach, eds R. Singh, R. Kothari, P. Koringa, and S. Singh (Singapore: Springer), 193–205. doi: 10.1007/978-981-10-5050-3_12
Sunita, G. (2014). Effect of fungal consortium and animal manure amendments on phosphorus fractions of paddy-straw compost. Int. Biodeter. Biodegr. 94, 90–97. doi: 10.1016/j.ibiod.2014.06.023
Thaller, M. C., Schippa, S., Bonci, A., Cresti, S., and Rossolini, G. M. (1997). Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol. Let. 146, 191–198. doi: 10.1016/s0378-1097(96)00474-0
Theodorou, M. E., and Plaxton, W. C. (1993). Metabolic adaptations of plant respiration to nutritional phosphate deprivation. Plant Physiol. 101, 339–344. doi: 10.1104/pp.101.2.339
Thompson, W. H., Leege, P. B., Millner, P. D., and Watson, M. E. (2001). Test Methods for the Examination of Composting and Compost. Available online from: http://tmecc.org/tmecc/ (last accessed March 2019)
Tiessen, H., and Moir, J. O. (2008). “Characterization of available P by sequential extraction,” in Soil Sampling and Methods of Analysis, 2nd Edn, eds M. R. Carter and E. G. Gregorich (Boca Raton, FL: CRC Press), 321–334.
Vassilev, N., Vassileva, M., and Nikolaeva, I. (2006). Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl. Microbiol. Biotechnol. 71, 137–144. doi: 10.1007/s00253-006-0380-z
Vuppada, R. K., Hansen, C. R., Strickland, K. A. P., Kelly, K. M., and McCleary, W. R. (2018). Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol. 18:8. doi: 10.1186/s12866-017-1126-z
Vyas, P., Kumar, A., Kumar, D., and Dubey, A. (2018). Screening and characterization of Achromobacter xylosoxidans isolated from rhizosphere of Jatropha curcas L. (Energy crop) for plant-growth promoting traits. J. Adv. Res. Biotechnol. 3, 1–8. doi: 10.15226/2475-4714/3/1/00134
Wan, W., Qin, Y., Wu, H., Zuo, W., He, H., Tan, J., et al. (2020). Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capability and their potential for lead immobilization. Front. Microbiol. 11:752. doi: 10.3389/fmicb.2020.00752
Wang, X., Wang, C., Sui, J., Liu, Z., Li, Q., Ji, C., et al. (2018). Isolation and characterization of phosphofungi, and screening of their plant growth-promoting activities. AMB Expr. 8:63. doi: 10.1186/s13568-018-0593-4
Wei, Y., Zhao, Y., Xi, B., Wei, Z., Li, X., and Cao, Z. (2015). Changes in phosphorus fractions during organic wastes composting from different sources. Bioresour. Technol. 189, 349–356. doi: 10.1016/j.biortech.2015.04.031
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
White, T. J., Bruns, T. F., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Williams, A., Börjesson, G., and Hedlund, K. (2013). The effect of 55 years of different inorganic fertilizer regimes on soil properties and microbial community composition. Soil Biol. Biochem. 67, 41–46. doi: 10.1016/j.soilbio.2013.08.008
Yao, Q., Li, Z., Song, Y., Wright, S. J., Guo, X., Tringe, S. G., et al. (2018). Community proteogenomics reveals the systemic impact of phosphorus availability on microbial functions in tropical soil. Nat. Ecol. Evol. 2, 499–509. doi: 10.1038/s41559-017-0463-5
Yu, H., Zeng, G., Huang, H., Xi, X., Wang, R., Huang, D., et al. (2007). Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation 18, 793–802. doi: 10.1007/s10532-007-9108-8
Zhang, S., Liao, S. A., Yu, X., Lu, H., Xian, J. A., Guo, H., et al. (2015). Microbial diversity of mangrove sediment in Shenzhen Bay and gene cloning characterization of an isolated phytase-producing strains of SPC09 B. cereus. Appl. Microbiol. Biotechnol. 99, 5339–5350. doi: 10.1007/s00253-015-6405-8
Zheng, B. X., Hao, X. L., Ding, K., Zhou, G. W., Chen, Q. L., Zhang, J. B., et al. (2017). Long-term nitrogen fertilization decreased the abundance of inorganic phosphate solubilizing bacteria in an alkaline soil. Sci. Rep. 7:42284. doi: 10.1038/srep42284
Keywords: P solubilization, phosphate rock-enriched compost, alkaline phosphatase, sorghum straw, P fractions, phosphate-solubilizing fungi, Burkina phosphate rock
Citation: Sarr PS, Tibiri EB, Fukuda M, Zongo AN, Compaore E and Nakamura S (2020) Phosphate-Solubilizing Fungi and Alkaline Phosphatase Trigger the P Solubilization During the Co-composting of Sorghum Straw Residues With Burkina Faso Phosphate Rock. Front. Environ. Sci. 8:559195. doi: 10.3389/fenvs.2020.559195
Received: 05 May 2020; Accepted: 31 August 2020;
Published: 25 September 2020.
Edited by:
Hannes Schmidt, University of Vienna, AustriaReviewed by:
Carsten W. Mueller, University of Copenhagen, DenmarkXiaodong Chen, ZheJiang Academy of Agricultural Sciences, China
Copyright © 2020 Sarr, Tibiri, Fukuda, Zongo, Compaore and Nakamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Papa Saliou Sarr, c2FsaW91QGFmZnJjLmdvLmpw
 Papa Saliou Sarr
Papa Saliou Sarr Ezechiel Bionimian Tibiri
Ezechiel Bionimian Tibiri Monrawee Fukuda
Monrawee Fukuda Armel Nongma Zongo
Armel Nongma Zongo Emmanuel Compaore4
Emmanuel Compaore4