- 1Neuroinformatics Lab, Institute of Cognitive Science, University of Osnabrück, Osnabrück, Germany
- 2Institute of Bio- and Geosciences, Plant Sciences (IBG-2), Jülich Research Centre, Jülich, Germany
- 3Working Group Remote Sensing and Digital Image Analysis, Institute of Computer Science, University of Osnabrück, Osnabrück, Germany
Environmental scientists often face the challenge of predicting a complex phenomenon from a heterogeneous collection of datasets that exhibit systematic differences. Accounting for these differences usually requires including additional parameters in the predictive models, which increases the probability of overfitting, particularly on small datasets. We investigate how Bayesian hierarchical models can help mitigate this problem by allowing the practitioner to incorporate information about the structure of the dataset explicitly. To this end, we look at a typical application in remote sensing: the estimation of leaf area index of white winter wheat, an important indicator for agronomical modeling, using measurements of reflectance spectra collected at different locations and growth stages. Since the insights gained from such a model could be used to inform policy or business decisions, the interpretability of the model is a primary concern. We, therefore, focus on models that capture the association between leaf area index and the spectral reflectance at various wavelengths by spline-based kernel functions, which can be visually inspected and analyzed. We compare models with three different levels of hierarchy: a non-hierarchical baseline model, a model with hierarchical bias parameter, and a model in which bias and kernel parameters are hierarchically structured. We analyze them using Markov Chain Monte Carlo sampling diagnostics and an intervention-based measure of feature importance. The improved robustness and interpretability of this approach show that Bayesian hierarchical models are a versatile tool for the prediction of leaf area index, particularly in scenarios where the available data sources are heterogeneous.
1 Introduction
The leaf area index (LAI) is a unitless measure (m2 m−2) of the one-sided leaf surface area of a plant relative to the soil surface area (Watson, 1947). It characterizes, among other variables, the photosynthetic performance of plants (Watson, 1947; Chen and Black, 1992; Montheith and Unsworth, 2007), the size and density of the crop’s canopy and thus serves as an important indicator for the plant’s growth stage and stand productivity (Schueller, 1992; Cox, 2002; Bréda, 2003; Mulla, 2013; Yan et al., 2019). It plays a major role in meteorological, ecological, and agronomical modeling (Moran et al., 1995; Broge and Mortensen, 2002; Kergoat et al., 2002; Asner et al., 2003; Yan et al., 2012; Anita et al., 2014), as well as for studying the influence of climate change on crop growth (Gao et al., 2005; Manea and Leishman, 2014).
Various non-destructive methods exist to measure or estimate LAI directly (Jonckheere et al., 2004), but they typically require taking a large number of manual measurements in the field. Since this is a laborious process and it can be difficult to control for confounding variables such as weather, alternative faster approaches to infer LAI from indirect measures, e.g., spectroscopy and (hyper-)spectral imaging, have been investigated (Liu et al., 2016; Din et al., 2017; Yan et al., 2019; He et al., 2020). A common approach makes use of vegetation indices (VIs), which can be computed from distinct wavebands of spectral measurements, to estimate LAI (Huete et al., 2002). These measures, while well understood and easy to calculate, have several limitations. For example, most of them are sensitive to more than one plant parameter (e.g., LAI and chlorophyll content) (Govaerts et al., 1999; Dorigo et al., 2007), and especially for wheat crops, the non-linear relationship between numerous VIs and the LAI can lead to saturation for moderate to high LAI values (LAI
However, the relationship between LAI and spectral reflectance is also affected by other factors, such as the crop type, phenology, Sun illumination, local micro-climate, the type of soil, or the amount of precipitation (Kjaer et al., 2016; Din et al., 2017), and it may vary throughout the life cycle of the crop. These effects can be included in spatio-temporal models of LAI (Mustafa et al., 2014; Seyednasrollah et al., 2018; Jin et al., 2019; Qiu et al., 2020; Ji et al., 2021), which can be applied to data from aerial or satellite surveillance. This has the potential to greatly simplify monitoring crop growth across large or remote areas (Sun et al., 2021; Wan et al., 2021; Zhang et al., 2021).
But the annotated training data required for such spatio-temporal models, i.e., matched measurements of reflectance spectra, ground-truth LAI, and potential confounding variables, is not available for every location and crop. For example, the data used in this study consists of distinct datasets of corresponding LAI measurements and reflectance spectra, each set acquired on a single field over a period of one or 2 days. In this situation, the amount of labeled training data that can be acquired is too limited to fit either a full spatio-temporal model or a separate model for each field and/or growth stage. It is possible, in principle, to train a single model that generalizes across these conditions by simply pooling multiple datasets that were acquired under different conditions (see (Siegmann and Jarmer, 2015), for example). However, since the association between each data point and the specific dataset it belongs to (and thus its location and time) is lost in the pooling process, such a model is likely to perform worse than a spatio-temporal model or a field- and growth-stage-specific model, given sufficient training data. Ideally, we would like to find a compromise between these two extremes, i.e., between a single model trained on the pooled data on the one hand and an independent model for each dataset, on the other hand, that allows us to generalize over all the available datasets yet makes specifically adjusted predictions for each dataset. To this end, we propose a hierarchical, parameter-efficient Bayesian model, which implicitly accounts for the influence of location and time by allowing the model parameters to vary across different datasets.
Bayesian hierarchical models similar to the one suggested here are especially appealing for environmental sciences (Britten et al., 2021), where they have seen increasingly widespread use. For example, several recent studies applied Bayesian hierarchical models to time series of multispectral satellite images in order to assess the effects of climate change through land surface phenology (Senf et al., 2017; Seyednasrollah et al., 2018; Qiu et al., 2020; Babcock et al., 2021) or other indicators such as Normalized Difference Vegetation Index (Wilson et al., 2011). A similar remote sensing approach has been used to predict LAI and its spatio-temporal evolution for bamboo (Xing et al., 2019) and other forests (Naithani et al., 2013; Schraik et al., 2019). For agronomical models of the LAI of food crops such as rice (Xu et al., 2019), Brazilian Cowpea (Soratto et al., 2020) or white winter wheat (Qu et al., 2008), local multispectral measurements are often used instead of–or in addition to–satellite images. In most of these studies, Bayesian hierarchical models are used to impose prior domain knowledge, combine multimodal data sources, and integrate data collected at multiple resolutions of space and/or time, all of which ultimately improve prediction performance. By contrast, our primary goal is to show how Bayesian hierarchical models and associated tools can be used to construct and diagnose simple and interpretable models for heterogeneous datasets, which commonly occur in environmental sciences.
Based on these considerations, we develop a Bayesian hierarchical model according to the following steps: 1) we filter the spectral measurements to remove noise, 2) apply basis splines with adaptively placed knots to extract features from the spectra, 3) train a Bayesian hierarchical model to predict LAI from these features on labeled data, 4) select and validate the best performing model, 5) and estimate the importance of the individual features for prediction. Our model learns an easily interpretable general relationship between reflectance spectra and LAI, as well as the dataset-specific deviations from that baseline. By using a variant of Markov Chain Monte Carlo (MCMC) sampling, we can incorporate domain knowledge or regularization through prior distributions of the parameters and provide a full posterior probability distribution over these parameters, which allows the quantification of uncertainty. We compare two variants of this hierarchical approach with a non-hierarchical alternative and find that it indeed offers a favorable trade-off between prediction accuracy and model complexity.
2 Methods
2.1 The Dataset
We evaluate our proposed model on a combination of four datasets, totaling 191 pairs of measured reflectance spectra (see also Supplementary Figure S1 for examples) and corresponding measurements of the LAI on fields covered by white winter wheat (lat. Triticum aestivum).
Each pair of measurements was taken on a different square plot of size 50 cm×50 cm. The LAI values of each plot were measured multiple times in a non-destructive way and averaged to a single value per plot. Five reflectance spectra were acquired and averaged for each plot using a spectroradiometer from a height of 1.4 m above ground with a nadir view and converted to absolute reflectance values using a reflectance standard of known reflectivity (Spectralon, Labsphere Inc., United States). The data were collected on four different fields in different years, corresponding to different stages in the plants’ growth cycle, and there are minor differences in the data collection procedure.
The first two sampling areas, which we call Field A and Field B in the following, are located near Köthen, Germany, which is a part of one of the most important agricultural regions in Germany. The region is distinctly dry, with 430 mm mean annual precipitation due to its location in the Harz mountains. The mean annual temperature varies between 8◦C to 9 ◦C. The study area has an altitude of 70 m above sea level and is characterized by a Loess layer up to 1.2 m deep that covers a slightly undulated tertiary plain. The predominant soil types of the region are Chernozems, in conjunction with Cambisols and Luvisols. At two locations in this region, 57 spectral measurements were recorded on seventh to eighth May 2011 using two ASD FieldSpec III spectroradiometers (ASD Inc., United States) with 25° field of view optics. Another 74 measurements were taken on 24th to May 25, 2012, using one ASD FieldSpec III (ASD Inc., United States) and one SVC HR-1024 spectroradiometer (Spectra Vista Corporation, United States) with 14° field of view. For each location, the corresponding LAI was measured non-destructively with a SunScan device (Delta-T Devices Ltd., United States) in 2011 and an LAI 2000 (LI-COR Inc., United States) in 2012, and, respectively, five and four LAI measurements were averaged per plot. Data from this study area was also used and described in more detail in (Siegmann and Jarmer, 2015).
The other two sampling areas, called Field C and Field D in the following, are located near Demmin, Germany. The region has a mean annual precipitation of 550 mm, and a mean annual temperature of 8◦C. Albeluvisols interspersed by Haplic Luvisols dominate the sand-rich area. The observed area is south of the river Tollense, where the ground elevation drops from 70 m to 7 m due to glacial moraines causing high variability in soil conditions. At two locations in this region, 26 spectral measurements were recorded on, and another 34 on using SVC HR-1024i spectroradiometer (Spectra Vista Corporation, United States) in nadir view 1.4 m above the ground using 14° field of view optics. In this case, six measurements of LAI [taken with LAI 2000 (LI-COR Inc., United States)] were averaged for each plot.
The recorded spectra cover wavelengths in the range from 350 to 2,500 nm, of which we use the range from 400 to 1,350 nm (for details see Table 1). We preprocess these spectra by smoothing with a first-order Gaussian filter with width σ = 10 nm.
For a summary of these parameters, see Table 1.
In the following, we reference specific subsets of this data. We introduce the following notation: all spectra-LAI-pairs for field j ∈ {1, …, 4} are numerically indexed by the set Jj, where J1, J2, J3, J4 correspond to the measurements from Field A, Field B, Field C, and Field D, respectively. We denote the ith ∈ Jj reflectance spectrum from dataset j by the function
2.2 Feature Extraction From Reflectance Spectra With a Spline Basis
The data collection and preprocessing steps outlined above result in reflectance spectra of wavelengths 400–1,350 nm at a resolution of 1 nm. Since this representation is much too high dimensional for direct use, we extract the most important information into a low dimensional representation by computing the inner product between the preprocessed reflectance spectra and a set of eleven cubic basis splines (B-splines) with adaptively placed knots (see Figure 1).
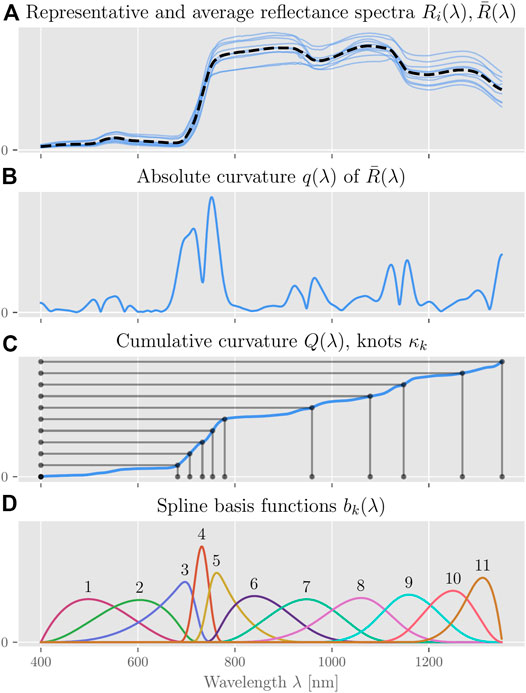
FIGURE 1. Adaptive knot-placement for B-Splines. (A) For the measured reflectance spectra Ri(λ), (B) we calculate the mean absolute curvature q(λ). (C) We then find knot positions such that the integral Q(λ) of this measure between any two successive knots κi, κi+1 is identical. (D) The result are 11 cubic spline basis functions bk(λ) with non-uniformly spaced knots.
The positions κi, i ∈ {1, …, 11} of the inner knots, which determine the shape of the individual basis splines, are chosen such that the cumulative absolute curvature Q (κi+1) − Q (κi) of the average reflectance spectrum is equal between any two successive knots i and i + 1. We compute the absolute curvature q(λ) by convolving the average reflectance spectrum
The eleven basis functions bk, k ∈ {1, …, 11} are generated from this knot vector κ using the standard Cox-DeBoor algorithm (Boor, 1978), where the multiplicity of the first and last knot is three, i.e., all basis functions go to zero at their respective start and end knots. The last basis function b12, which originates at knot κ10, is not used. This heuristic algorithm results in a proportionally larger number of knots, and thus higher spatial resolution, where the reflectance spectra have the largest absolute curvature and hence “have most structure”; see also (Pipa et al., 2012; Ferrer Arnau et al., 2013; Tjahjowidodo et al., 2015). For each of the data-subsets j ∈ {1, …, 4}, we can then compute our model’s feature or design matrix Xj using these basis functions bk(λ):
2.3 Bayesian Markov Chain Monte Carlo Regression for Predicting LAI
Our primary objective is to construct a simple, interpretable model that can reliably predict the LAI value of a wheat plot directly from a corresponding reflectance spectrum. We are additionally interested in analyzing the model’s confidence, how well it generalizes, and which features it relies on most to make a prediction. Since the total available data is limited and stems from four heterogeneous datasets, prior constraints are required to prevent overfitting.
In order to meet all of these requirements, we design three different (non-)hierarchical Bayesian generalized linear models (GLM) (McCullagh and Nelder, 1989; Gelman et al., 2013) of different complexity. For each of these, we infer a full posterior distribution over model parameters from training data and use this to provide a full posterior predictive distribution over LAI scores on testing data. To generate representative samples from these probability distributions, we use a specific type of Hamiltonian Monte Carlo sampling, namely No-U-Turn-Sampling (Hoffman and Gelman, 2014) (NUTS), as implemented by the probabilistic programming package pyMC3 (Salvatier et al., 2016).
2.3.1 Model 1: A Baseline Model With Pooled Data
As a baseline (see Figure 2), we construct a simple generalized linear model, which we apply to all of the datasets j ∈ {1, …4} together. This model merely pools all available data but does not account for any systematic differences that might exist between the individual datasets. We assume the logarithm of the observed LAI scores to be normally distributed around an affine linear predictor μj with deviation σ, which is a model parameter with log-normal prior. The predictor μj is computed by the matrix-vector product between the dataset’s feature matrix Xj and the model’s weight vector w = (w1, …, w11), plus an additional bias parameter b. Including the unknown deviation parameter σ, the model thus has a total of 13 free parameters to be inferred from data. The individual parameters wk and b have normal priors with standard deviation 1 and 11, respectively, to allow the individual bias term to counteract the effect of all 11 weights, if necessary. The baseline model is described by Eq. 2.
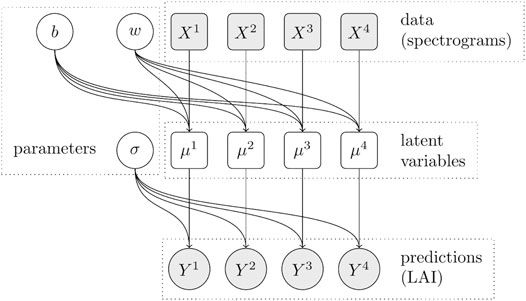
FIGURE 2. Dependency graph of the baseline model. For each dataset j (encoded in the feature matrix Xj and corresponding labels Yj), the prediction depends on the same three shared parameters b, w and σ. Circles represent random variables, rectangles represent deterministic variables, filled shapes represent observed variables.
2.3.2 Model 2: A Model With Hierarchical Bias
Our second model (see Figure 3) extends the baseline model by an additional bias parameter bj for each dataset and thus has a total of 17 free parameters. Due to the logarithmic link function, this additional parameter per dataset allows accounting for the overall variation in scale between the four different datasets. But rather than setting each parameter bj independently (and thus adding three full degrees of freedom), we constrain them to be clustered around a common bias value b*, which replaces the bias term b in the baseline model. Therefore, the prior for the new variables bj is a Normal distribution centered at b* with an order of magnitude smaller standard deviation of 11/10 = 1.1. The affine linear predictor μj then depends on the dataset-specific bias term bj. The hierarchical bias model is described by Eq. 3.
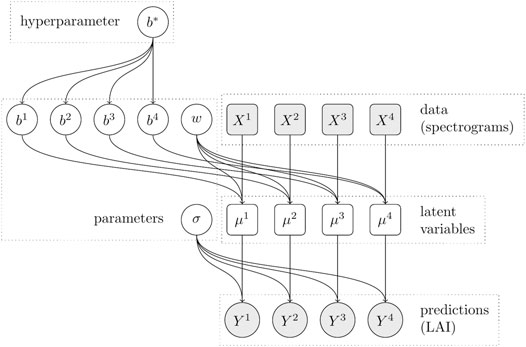
FIGURE 3. Dependency graph of the hierarchical model with dataset-specific bias terms. The predictions for each dataset j depend on an individual bias parameter bj, which in turn depends on the shared mean bias parameter b*.
2.3.3 Model 3: Full Hierarchical Model
Our third model (see Figure 4) extends the second model even further by also allowing the model weight vector w to vary for each dataset. Just like we did for the bias terms, we introduce the new parameter vectors wj, and we constrain the individual parameters
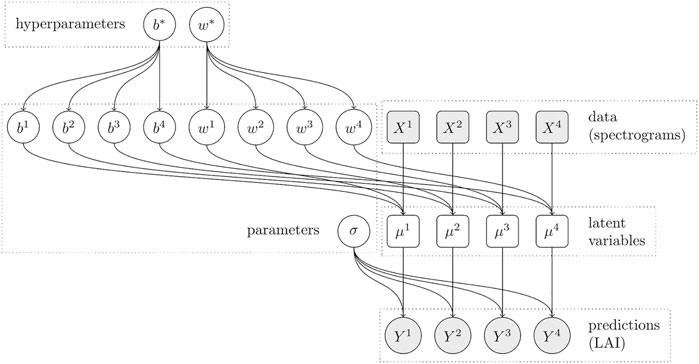
FIGURE 4. Dependency graph of the hierarchical model with dataset-specific bias and weight terms. The predictions for each dataset j now depend on an individual bias parameter bj and weight vector wj, which in turn depend on the shared bias parameter b* and the shared weight vector w*, respectively.
2.4 Model Selection Using Pareto-Smoothed Importance Sampling
To get an unbiased estimate of our model’s generalization error from the very limited available data, we would like to perform leave-one-out cross-validation (LOO-CV) and compute the expected log posterior predictive density (ELPD) for new data (Vehtari et al., 2019). Unfortunately, this is a prohibitively expensive computation when combined with MCMC sampling. However, the generated samples and their associated log-likelihood values contain sufficient information to estimate the LOO-CV ELPD by directly weighing the samples. This procedure is called Pareto-smoothed importance sampling (PSIS) (Vehtari et al., 2019). Combining these two methods, PSIS and LOO-CV, yields a validation method called PSIS-LOO-CV (Vehtari et al., 2019), which is beneficial in situations like this, where an MCMC sampling-based model is trained on a small dataset. As a result, we get for each model the ELPD score, which we use to compare the three proposed models, a parameter η, which can be interpreted as the effective number of degrees of freedom in the model, and the so-called Pareto shape parameters ki, which assess for each data point i in the dataset, how much it affects the ELPD estimation. For data points where ki exceeds 0.7, the PSIS-LOO-CV estimate becomes unreliable, which can also indicate an under-constrained model or an outlier in the data (Vehtari et al., 2019).
2.5 Evaluation of Feature Importance
To estimate the importance that our model assigns to each feature of the reflectance spectra, we calculate a model-agnostic measure of feature importance (Fisher et al., 2019) called model reliance (MR). Here, the importance of an individual feature is calculated as the relative change in the model’s error when the individual observations of only that feature are shuffled, compared to the error on non-shuffled data. This causal intervention intentionally breaks the dependence between different correlated features. Therefore, MR, unlike correlation analysis, is a causal tool to diagnose the model rather than the data. This is relevant here because the different features of our model are computed by taking the inner product between the reflectance spectra and a set of overlapping not independent basis functions and are hence certainly correlated. We use the same loss function as for the model selection, namely ELPD. Since this measure already estimates the logarithm of a quantity of interest (the posterior density), we use the difference between shuffled and non-shuffled ELPD instead of their ratio to estimate the logarithm of the MR for the posterior density. Because we are only interested in qualitatively ranking features by their importance, we normalize the resulting importance value of each feature by their average. To improve the robustness of this measure, the shuffling is repeated multiple times (here, ten times), and the results are averaged. Repeating this procedure for each feature of a model yields positive scores for ranking all features by their importance.
3 Results
In this section, we evaluate each of the three models presented above, namely the non-hierarchical model, the model with hierarchical bias term, and the full hierarchical model.
3.1 Model Predictions
First, we visualize the models’ accuracy and ability to generalize in a model-agnostic way by directly plotting predictions against the corresponding measured “ground-truth” values. For this purpose, we randomly select 80% of all available data (the training set, shown in blue) to infer model parameters, which we then use to predict the LAI for the remaining 20% of the data (the test set, shown in green). Due to the probabilistic nature of our models, a full posterior predictive distribution is given for each data point, which we summarize in Figure 5 (A),(C), and (E). We can observe that all three models make reasonable predictions, i.e., that the predicted LAI grows in proportion to the measured LAI. Because all our generalized linear models assume that the logarithms of the LAI scores are homoscedastic, the standard deviation of the predictive distribution increases with the measured LAI, as well. Rather than the raw residuals
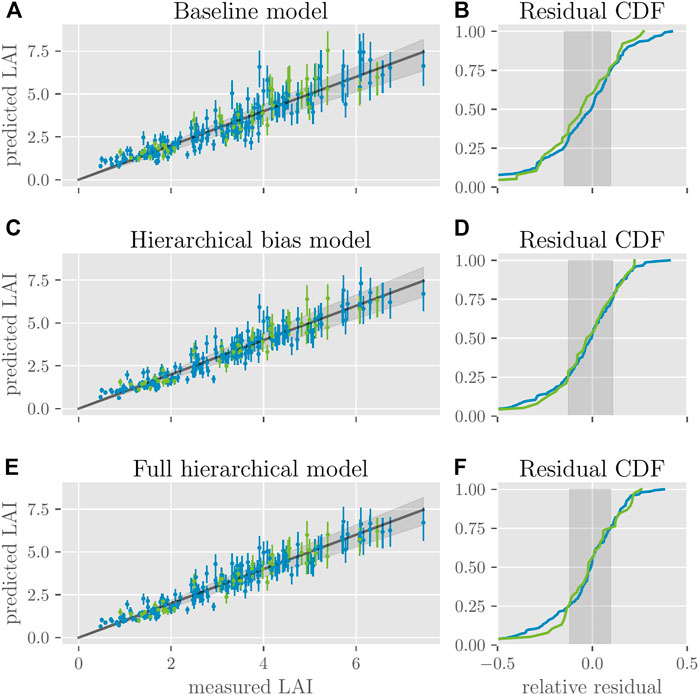
FIGURE 5. Model predictions of LAI. For the three models, (A), (C) and (E) plot for each data point (training data in blue and testing data in green) the predicted LAI values against the measured LAI values. Error bars indicate the interquartile range of the predictive distributions. Dots represent the expected value. The gray line shows the optimal predictions; the best 50% of model predictions lie within the gray cone around it. (B), (D), and (F) show the cumulative distribution function of the residuals, each normalized by the corresponding measured LAI value for training and testing data (blue and green lines). The gray areas show the same interquartile range as the cones in (A), (C), and (E).
3.2 Model Comparison
To quantify the generalization error of all three models more accurately, we use the PSIS-LOO-CV method on all available data to estimate the ELPD on novel data. This procedure yields several highly informative measures, which are summarized in Table 2. To verify the convergence of the sampling procedure for each model, we compare the marginal posterior distribution of each parameter across multiple chains and find no discrepancies or divergences (see also Supplementary Figures S2–S4). We can see that the highest ELPD (indicating the lowest generalization error) is achieved for the two hierarchical models, with little difference between them (−157.8 and −157.0, respectively, with a standard deviation of

TABLE 2. Comparison of the three models using PSIS-LOO-CV. The ELPD ± one standard deviation are listed for each model. #params denotes the number of parameters, η denotes the effective number of parameters. For each model, we show the number of data-points for which the Pareto shape parameter k falls into either of four different intervals.
However, due to the limited amount of training data and considering that the number of parameters ranges from 13 for the non-hierarchical baseline model to 17 for the model with hierarchical bias term to 61 for the full hierarchical model, we are also concerned with model complexity and the risk of overfitting. Since LOO-CV estimates generalization error directly, it does not need to explicitly penalize a large number of parameters, which is a significant advantage when comparing Bayesian hierarchical models. Instead, it allows us to estimate the model complexity of the three models by the so-called effective number of parameters η, which provides some intuition about how many degrees of freedom the model has to approximate the available data. As we see in Table 2, η = 13.3 is quite close to the parameter count of the non-hierarchical model on the pooled dataset. This only increases slightly to η = 15.0 for the hierarchical bias model, even though it has four additional parameters. However, adding another 44 parameters for the full hierarchical model increases η substantially to 24.9.
Since PSIS-LOO-CV emulates conventional LOO-CV, it provides additional information that can help us understand how prone each model is to overfitting: For each data point, the procedure yields the shape parameter k of a Pareto distribution, which indicates whether estimating the generalization error for that data point is reliable (k ∈ [ − ∞, 0.7], ideally k ≤ 0.5), potentially unreliable (k ∈ [0.7, 1]) or entirely unreliable (k ∈ [1, ∞])(Vehtari et al., 2019). As Table 2 shows, the full hierarchical model struggles with PSIS-LOO-CV for three data points, which may indicate that the model is more prone to overfitting to these potential “outliers” (see also Supplementary Figure S5).
As these numbers suggest, the model with a hierarchical bias term is the best choice because it is barely more complex than the non-hierarchical model, yet it performs at least as well as the full hierarchical model.
3.3 An Interpretable Kernel Function
As outlined above, all three models derive their predictions of LAI from a weighted linear combination of features, which we compute by taking inner products between the measured reflectance spectra and a set of B-spline basis functions. These linear operations can be equivalently expressed as taking the inner product between each reflectance spectrum and an inferred kernel function κj(λ), which provides a different, more interpretable perspective on the model.
To motivate this equivalent perspective, we look at how the reflectance spectra affect the linear predictors
Since the parameters
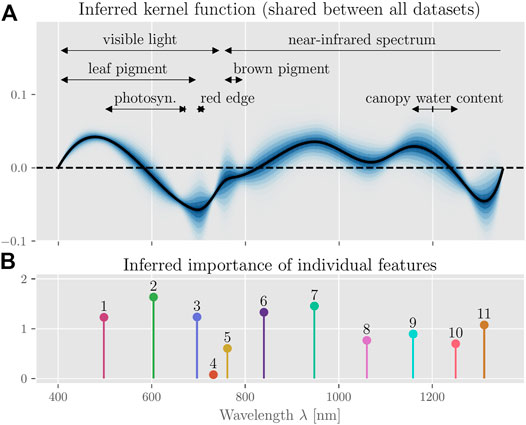
FIGURE 6. Inferred kernel function and feature importance. (A) shows the posterior distribution of the inferred kernel function. The black line represents the expected kernel. We can relate several ranges of the reflectance spectrum to physical phenomena, namely effects due to green leaf pigment [400–700 nm (Cen and Borman, 1990; Aparicio et al., 2000)] and photosynthetic capacity [495–680 nm, peak at 670 nm (Cen and Borman, 1990; Aparicio et al., 2000; Weber et al., 2012)] and the red edge region [690–720 nm (Zhang et al., 2016)] in the visible light range, as well as the canopy’s water content [1,150 nm–1,260 nm (Sims and Gamon, 2003), peak absorption at around 1,200 nm (Weber et al., 2012; Wang C et al., 2017)] in the near-infrared range. (B) shows a stem-plot of the relative importance of each feature (enumerated; normalized by the average feature importance) as well as the resulting estimated importance of each wavelength.
3.4 Feature Importance
In addition to the sign and magnitude of each feature’s contribution (which are determined by the inferred weights; c. f. Supplementary Figures S2–S4), we are also interested in how important each individual feature is for the model’s prediction. We quantify this via MR as described above. Figure 6B shows that, except feature four, all features are indeed important for the prediction accuracy of the model. The low importance of feature four centered around 730 nm is likely due to the narrow domain of basis function b4 (see Figure 1D), which indicates that this feature could be removed or an alternative knot-placement procedure could be chosen to reduce model complexity.
4 Discussion
Our results confirm that using a Bayesian hierarchical model not only leads to an improvement in the prediction accuracy over a non-hierarchical approach, but more importantly, it yields several qualitative benefits regarding interpretability, model complexity, and robustness.
One important benefit of the Bayesian hierarchical approach is that an appropriate choice of priors and model structure allows us to integrate additional model parameters without excessively increasing model complexity. For example, the number of spectral features used in our model directly determines the scale of the respective spline basis functions, which determines the resolution of our kernel function. This can create a trade-off between a model with lower spectral resolution and a model with a larger number of parameters. In the Bayesian approach, we can choose the model with more parameters without the risk of overfitting if we formalize our uncertainty and prior assumptions about the parameters appropriately. This is particularly important for hierarchical models, where we might want to add a large number of parameters to account for the specific variations in each subset of the data. Compare, for example, our full hierarchical model with its 61 parameters to the non-hierarchical baseline model with 13 parameters. Here, the addition of 48 new parameters only increases the effective degrees of freedom of the model by 11 and appears to increase the risk of overfitting only moderately. In our hierarchical bias model, we directly incorporate the fact that each of the four data subsets was recorded at a different growth stage of the plants, which affects the expected LAI, and hence requires a separate bias parameter. However, by simultaneously inferring the shared prior distribution over these separate parameters, we can ensure that the prediction on any one subset of the data benefits from the information contained in all the others. Of course, a non-hierarchical model can also benefit from heterogeneous data (see e.g. (Siegmann and Jarmer, 2015)), but it may fail in subtle ways if systematic differences between the data subsets obscure the relevant associations within each dataset3. In general, Bayesian hierarchical models allow us to conveniently include additional information about the dataset, domain knowledge, and regularizing priors, all of which can help to reduce the model’s effective degrees of freedom. For the often small and heterogeneous datasets used in environmental sciences (Britten et al., 2021), this can be a major advantage over alternative machine learning approaches such as Random Forest Regression (Houborg and McCabe, 2018; Srinet et al., 2019) or Deep Learning (Wang T et al., 2017; Apolo-Apolo et al., 2020; Yamaguchi et al., 2021), which may require prohibitive amounts of training data due to their typically large number of parameters.
We employ MCMC sampling to generate unbiased samples of the full posterior distributions over parameters and predictions, which allow us to use additional diagnostic tools and error measures. For example, we can directly estimate posterior densities, credible intervals, and even generalization errors via sample-based methods such as PSIS-LOO-CV, which are more broadly applicable than information criteria such as the Akaike Information Criterion (AIC), the Widely Applicable Information Criterion (WAIC), or the Bayesian Information Criterion (BIC) (Vehtari et al., 2017). In particular, we saw that a hierarchical model might have a considerably larger number of parameters with a comparatively minor increase in model complexity, making any form of regularization based directly on the number of parameters difficult. Besides better diagnostics, sample-based measures can also provide insights about the data itself, e.g., indicating which data points are potential “outliers” that the model is susceptible to (see Supplementary Figure S5).
In addition to descriptive statistics, we also estimate feature importance using an intervention-based model-agnostic method that artificially breaks the dependence between naturally correlated features. Thus, we can infer exactly which features the model relies on for its prediction–independently from the magnitude of the respective parameters. Such information can help domain experts identify potential problems, e.g,. if supposedly relevant features are ignored, or irrelevant features are relied on. This simple example shows how methods from causal analysis (Pearl, 2009) can help explain or interpret the model in qualitatively different ways than descriptive statistics alone.
Because we use a generalized linear model, we can additionally analyze and interpret the model’s linear predictor directly in the measurement space. Since the individual features are extracted from the spectra using B-spline basis functions, this linear predictor is just an inner product between a reflectance spectrum and a kernel function plus an additive bias term. Due to the logarithmic link function, the bias term ultimately has a scaling effect on the LAI predictions. The kernel function directly shows which wavelengths are associated with higher LAI, e.g., short wavelengths of the visible spectrum and much of the near-infrared spectrum, or lower LAI, e.g., around 600–750 nm. These results can be directly linked to physical phenomena and examined with domain knowledge. For example, the positive association for short wavelengths in the range 400–550 nm may be attributable to the effect of green leaf pigment, which reflects light in the range 400–700 nm (Cen and Borman, 1990; Aparicio et al., 2000). Similarly, the pronounced drop around the red edge (690–720 nm (Zhang et al., 2016)), which is related to the plants’ chlorophyll content (Richardson et al., 2002; Liu et al., 2004), total nitrogen (Cheng et al., 2005; Clevers and Kooistra, 2009) and yield (Huang et al., 2004; Rasooli Sharabiani et al., 2014), may be attributed to the plants’ photosynthetic capacity (495–680 nm (Cen and Borman, 1990; Aparicio et al., 2000; Weber et al., 2012)) that peaks at around 670 nm.
Finally, we opted for a simplistic, interpretable model of LAI as a function of spectral power, but the hierarchical Bayesian modeling approach makes it easy to extend the proposed model further. For a larger dataset, the model complexity could be increased, either by choosing broader priors or by increasing the number of parameters to improve its accuracy, or the additional measurements could be used to reduce the uncertainty over the model’s parameters. The data used in this study consists of measurements taken at four distinct locations and points in time. By allowing the specific parameters for each of these datasets to vary independently around a shared set of global parameters, we thus indirectly account for the combined effect of location and time. If a much larger number of datasets is used, their spatio-temporal distribution could also be taken into account explicitly, for example, by forcing the specific parameters of the datasets to be correlated, depending on their proximity in time and space, through an appropriate choice of a joint prior distribution. This approach leads to a full spatio-temporal model, which could also predict LAI on crop sites for which no training data is available. One could also include additional levels of hierarchy (e.g., to extend the model to other related plant species or different geographical regions), or other factors such as soil moisture content (Rad et al., 2011), the influence of climate change and CO2 concentration on crop growth (Gao et al., 2005; Manea and Leishman, 2014), the effects of warming asymmetry due to climate change (Cox et al., 2020), effects of microclimate (Hardwick et al., 2015), the influence of the amount of soil conditioner on the crops (Su et al., 2015), and ammonium level in the soil.
Data Availability Statement
The datasets generated for this study and the corresponding source code can be found online at: https://github.com/ostojanovic/bayesian_lai.
Author Contributions
OS and JL—Conceptualization, Methodology, Project administration, Software, Visualization, Writing—original draft; GP—Supervision, Resources, Conceptualization, Validation, Writing—review and editing; BS and TJ—Data curation, Conceptualization, Validation, Writing—review and editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank our reviewers for their detailed and constructive feedback, which helped to improve this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.780814/full#supplementary-material
Footnotes
1This is equivalent to computing the absolute curvature of the smoothed average reflectance spectrum
2To simplify notation, we write wj and bj for the (possibly) dataset-specific weight and bias terms, and set wj = w or bj = b for models that don’t make these dataset-specific distinctions
3In Supplementary Figure S6, we show that this is indeed the case here, as wavelength around 1,200 nm lead to a pronounced dip in the kernel function when data from multiple datasets is pooled, but this association disappears if the model is instead fit to any individual dataset. Looking at the pooled dataset, we would therefore be led to conclude that lower spectral power around 1,200 nm is a strong predictor of higher LAI. While this is correct on the artificially pooled dataset, it appears to be incorrect on any individual datasets. This may be an instance of Simpson’s paradox (Pearl, 2014), which suggests that a hierarchical model is more appropriate here
References
Anita, S. M., Richard, F., and Wang, S. (2014). Assessing the Impact of Leaf Area index on Evapotranspiration and Groundwater Recharge across a Shallow Water Region for Diverse Land Cover and Soil Properties. J. Water Res. Hydraulic Eng. 3, 60–73.
Aparicio, N., Villegas, D., Casadesus, J., Araus, J. L., and Royo, C. (2000). Spectral Vegetation Indices as Nondestructive Tools for Determining Durum Wheat Yield. Agron. J. 92, 83–91. doi:10.2134/agronj2000.92183x
Apolo-Apolo, O. E., Pérez-Ruiz, M., Martínez-Guanter, J., and Egea, G. (2020). A Mixed Data-Based Deep Neural Network to Estimate Leaf Area index in Wheat Breeding Trials. Agronomy 10, 175. doi:10.3390/agronomy10020175
Asner, G. P., Scurlock, J. M. O., and A. Hicke, J. (2003). Global Synthesis of Leaf Area index Observations: Implications for Ecological and Remote Sensing Studies. Glob. Ecol. Biogeogr. 12, 191–205. doi:10.1046/j.1466-822X.2003.00026.x
Babcock, C., Finley, A. O., and Looker, N. (2021). A Bayesian Model to Estimate Land Surface Phenology Parameters with Harmonized Landsat 8 and sentinel-2 Images. Remote Sensing Environ. 261, 112471. doi:10.1016/j.rse.2021.112471
Boor, C. d. (1978). A Practical Guide to Splines (Applied Mathematical Sciences). New York: Springer-Verlag.
Breda, N. J. J. (2003). Ground-based Measurements of Leaf Area index: a Review of Methods, Instruments and Current Controversies. J. Exp. Bot. 54, 2403–2417. doi:10.1093/jxb/erg263
Britten, G. L., Mohajerani, Y., Primeau, L., Aydin, M., Garcia, C., Wang, W.-L., et al. (2021). Evaluating the Benefits of Bayesian Hierarchical Methods for Analyzing Heterogeneous Environmental Datasets: A Case Study of marine Organic Carbon Fluxes. Front. Environ. Sci. 9, 28. doi:10.3389/fenvs.2021.491636
Broge, N. H., and Mortensen, J. V. (2002). Deriving green Crop Area index and Canopy Chlorophyll Density of winter Wheat from Spectral Reflectance Data. Remote Sensing Environ. 81, 45–57. doi:10.1016/S0034-4257(01)00332-7
Cen, Y.-P., and Bornman, J. F. (1990). The Response of Bean Plants to UV-B Radiation under Different Irradiances of Background Visible Light. J. Exp. Bot. 41, 1489–1495. doi:10.1093/jxb/41.11.1489
Chen, J. M., and Black, T. A. (1992). Defining Leaf Area index for Non-flat Leaves. Plant Cel Environ 15, 421–429. doi:10.1111/j.1365-3040.1992.tb00992.x
Cheng, Y., Hu, C., Dai, H., and Lei, Y. (2005). “Spectral Red Edge Parameters for winter Wheat under Different Nitrogen Support Levels,” in Proceedings of SPIE - The International Society for Optical Engineering, San Diego, CA, 2005. doi:10.1117/12.614759
Clevers, J. G. P. W., and Kooistra, L. (2009). “Using Hyperspectral Remote Sensing Data for Retrieving Canopy Water Content,” in WHISPERS ’09 - 1st Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Grenoble, France, 2009, 1–4. doi:10.1109/WHISPERS.2009.5289058
Cox, D. T. C., Maclean, I. M. D., Gardner, A. S., and Gaston, K. J. (2020). Global Variation in Diurnal Asymmetry in Temperature, Cloud Cover, Specific Humidity and Precipitation and its Association with Leaf Area index. Glob. Change Biol. 26, 7099–7111. doi:10.1111/gcb.15336
Cox, S. (2002). Information Technology: The Global Key to Precision Agriculture and Sustainability. Comput. Electron. Agric. 36, 93–111. doi:10.1016/S0168-1699(02)00095-9
Din, M., Zheng, W., Rashid, M., Wang, S., and Shi, Z. (2017). Evaluating Hyperspectral Vegetation Indices for Leaf Area index Estimation of Oryza Sativa L. At Diverse Phenological Stages. Front. Plant Sci. 8, 820. doi:10.3389/fpls.2017.00820
Dorigo, W. A., Zurita-Milla, R., de Wit, A. J. W., Brazile, J., Singh, R., and Schaepman, M. E. (2007). A Review on Reflective Remote Sensing and Data Assimilation Techniques for Enhanced Agroecosystem Modeling. Int. J. Appl. Earth Obs. Geoinf. 9, 165–193. doi:10.1016/j.jag.2006.05.003
Ferrer Arnau, L. J., Reig-Bolaño, R., Martí-Puig, P., Manjabacas, A., and Parisi-Baradad, V. (2013). Efficient Cubic Spline Interpolation Implemented with Fir Filters. Int. J. Comput. Inf. Syst. Ind. Manage. Appl. 105, 98–105.
Fisher, A., Rudin, C., and Dominici, F. (2019). All Models Are Wrong, but Many Are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously. J. Mach. Learn. Res. 20, 1–81.
Gao, Z., Gao, W., and Slusser, J. (2005). “The Response of Leaf Area index to Climate Change during 1981-2000 in China,” in Remote Sensing and Modeling of Ecosystems for Sustainability II (San Diego: International Society for Optics and Photonics), 5884, 58840S. doi:10.1117/12.612929
Gelman, A., Carlin, J. B., Stern, H. S., Dunson, D. B., Vehtari, A., and Rubin, D. B. (2013). Bayesian Data Analysis. Boca Raton: CRC Press.
Govaerts, Y. M., Verstraete, M. M., Pinty, B., and Gobron, N. (1999). Designing Optimal Spectral Indices: A Feasibility and Proof of Concept Study. Int. J. Remote Sensing 20, 1853–1873. doi:10.1080/014311699212524
Hardwick, S. R., Toumi, R., Pfeifer, M., Turner, E. C., Nilus, R., and Ewers, R. M. (2015). The Relationship between Leaf Area index and Microclimate in Tropical forest and Oil palm Plantation: Forest Disturbance Drives Changes in Microclimate. Agric. For. Meteorol. 201, 187–195. doi:10.1016/j.agrformet.2014.11.010
He, L., Ren, X., Wang, Y., Liu, B., Zhang, H., Liu, W., et al. (2020). Comparing Methods for Estimating Leaf Area index by Multi-Angular Remote Sensing in winter Wheat. Sci. Rep. 10, 13943. doi:10.1038/s41598-020-70951-w
Hoffman, M. D., and Gelman, A. (2014). The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res. 15, 1593–1623.
Houborg, R., and McCabe, M. F. (2018). A Hybrid Training Approach for Leaf Area index Estimation via Cubist and Random Forests Machine-Learning. ISPRS J. Photogram. Remote Sensing 135, 173–188. doi:10.1016/j.isprsjprs.2017.10.004
Huang, W., Wang, J., Wang, J., Liu, L., Zhao, C., and Wan, H. (2004). “Application of Red Edge Variables in winter Wheat Nutrition Diagnosis,” in IGARSS 2004-IEEE International Geoscience and Remote Sensing Symposium, Anchorage, AK, 2004, 4052–4055. doi:10.1109/IGARSS.2004.1370020
Huete, A., Didan, K., Miura, T., Rodriguez, E. P., Gao, X., and Ferreira, L. G. (2002). Overview of the Radiometric and Biophysical Performance of the Modis Vegetation Indices. Remote Sensing Environ. 83, 195–213. doi:10.1016/S0034-4257(02)00096-2
Ji, J., Li, X., Du, H., Mao, F., Fan, W., Xu, Y., et al. (2021). Multiscale Leaf Area index Assimilation for Moso Bamboo forest Based on sentinel-2 and Modis Data. Int. J. Appl. Earth Obs. Geoinf. 104, 102519. doi:10.1016/j.jag.2021.102519
Jin, H., Xu, W., Li, A., Xie, X., Zhang, Z., and Xia, H. (2019). Spatially and Temporally Continuous Leaf Area index Mapping for Crops through Assimilation of Multi-Resolution Satellite Data. Remote Sensing 11, 2517. doi:10.3390/rs11212517
Jonckheere, I., Fleck, S., Nackaerts, K., Muys, B., Coppin, P., Weiss, M., et al. (2004). Review of Methods for In Situ Leaf Area index Determination. Agric. For. Meteorol. 121, 19–35. doi:10.1016/j.agrformet.2003.08.027
Kergoat, L., Lafont, S., Douville, H., Berthelot, B., Dedieu, G., Planton, S., et al. (2002). Impact of Doubled CO2 on Global‐scale Leaf Area index and Evapotranspiration: Conflicting Stomatal Conductance and LAI Responses. J.‐Geophys. Res. 107, ACL 30–1–ACL 30–16. doi:10.1029/2001JD001245
Kjaer, K. H., Petersen, K. K., and Bertelsen, M. (2016). Protective Rain Shields Alter Leaf Microclimate and Photosynthesis in Organic Apple Production. Acta Hortic. 1134, 317–326. doi:10.17660/ActaHortic.2016.1134.42
Liu, K., Zhou, Q.-b., Wu, W.-b., Xia, T., and Tang, H.-j. (2016). Estimating the Crop Leaf Area index Using Hyperspectral Remote Sensing. J. Integr. Agric. 15, 475–491. doi:10.1016/S2095-3119(15)61073-5
Liu, L., Wang, J., Huang, W., Zhao, C., Zhang, B., and Tong, Q. (2004). Estimating winter Wheat Plant Water Content Using Red Edge Parameters. Int. J. Remote Sensing 25, 3331–3342. doi:10.1080/01431160310001654365
Manea, A., and Leishman, M. R. (2014). Leaf Area Index Drives Soil Water Availability and Extreme Drought-Related Mortality under Elevated CO2 in a Temperate Grassland Model System. PLoS ONE 9, e91046. doi:10.1371/journal.pone.0091046
Montheith, J., and Unsworth, M. (2007). Principles of Environmental Physics. San Diego, CA, USA: Academic Press.
Moran, M. S., Maas, S. J., and Pinter, P. J. (1995). Combining Remote Sensing and Modeling for Estimating Surface Evaporation and Biomass Production. Remote Sensing Rev. 12, 335–353. doi:10.1080/02757259509532290
Mulla, D. J. (2013). Twenty Five Years of Remote Sensing in Precision Agriculture: Key Advances and Remaining Knowledge Gaps. Biosyst. Eng. 114, 358–371. doi:10.1016/j.biosystemseng.2012.08.009
Mustafa, Y. T., Tolpekin, V. A., and Stein, A. (2014). Improvement of Spatio-Temporal Growth Estimates in Heterogeneous Forests Using Gaussian Bayesian Networks. IEEE Trans. Geosci. Remote Sensing 52, 4980–4991. doi:10.1109/TGRS.2013.2286219
Naithani, K. J., Baldwin, D. C., Gaines, K. P., Lin, H., and Eissenstat, D. M. (2013). Spatial Distribution of Tree Species Governs the Spatio-Temporal Interaction of Leaf Area index and Soil Moisture across a Forested Landscape. PLOS ONE 8, e58704. doi:10.1371/journal.pone.0058704
Nguy-Robertson, A. L., Peng, Y., Gitelson, A. A., Arkebauer, T. J., Pimstein, A., Herrmann, I., et al. (2014). Estimating green Lai in Four Crops: Potential of Determining Optimal Spectral Bands for a Universal Algorithm. Agric. For. Meteorol. 192-193, 140–148. doi:10.1016/j.agrformet.2014.03.004
Pearl, J. (2009). Causal Inference in Statistics: An Overview. Stat. Surv. 3, 96–146. doi:10.1214/09-ss057
Pearl, J. (2014). Comment: Understanding Simpson's Paradox. Am. Stat. 68, 8–13. doi:10.1080/00031305.2014.876829
Pipa, G., Chen, Z., Neuenschwander, S., Lima, B., and Brown, E. N. (2012). Mapping of Visual Receptive Fields by Tomographic Reconstruction. Neural Comput. 24, 2543–2578. doi:10.1162/NECO_a_00334
Qiu, T., Song, C., Clark, J. S., Seyednasrollah, B., Rathnayaka, N., and Li, J. (2020). Understanding the Continuous Phenological Development at Daily Time Step with a Bayesian Hierarchical Space-Time Model: Impacts of Climate Change and Extreme Weather Events. Remote Sensing Environ. 247, 111956. doi:10.1016/j.rse.2020.111956
Qu, Y., Wang, J., Wan, H., Li, X., and Zhou, G. (2008). A Bayesian Network Algorithm for Retrieving the Characterization of Land Surface Vegetation. Remote Sensing Environ. 112, 613–622. doi:10.1016/j.rse.2007.03.031
Rad, M. H., Assare, M. H., Banakar, M. H., and Soltani, M. (2011). Effects of Different Soil Moisture Regimes on Leaf Area Index, Specific Leaf Area and Water Use Efficiency in Eucalyptus (Eucalyptus camaldulensis Dehnh) under Dry Climatic Conditions. Asian J. Plant Sci. 10, 294–300. doi:10.3923/ajps.2011.294.300
Rasooli Sharabian, V., Noguchi, N., and Ishi, K. (2014). Significant Wavelengths for Prediction of winter Wheat Growth Status and Grain Yield Using Multivariate Analysis. Eng. Agric. Environ. Food 7, 14–21. doi:10.1016/j.eaef.2013.12.003
Richardson, A. D., Duigan, S. P. S., and Berlyn, G. P. (2002). An Evaluation of Noninvasive Methods to Estimate Foliar Chlorophyll Content. New Phytol. 153, 185–194. doi:10.1046/j.0028-646X.2001.00289.x
Salvatier, J., Wiecki, T. V., and Fonnesbeck, C. (2016). Probabilistic Programming in Python Using PyMC3. PeerJ Comput. Sci. 2, e55. doi:10.7717/peerj-cs.55
Schraik, D., Varvia, P., Korhonen, L., and Rautiainen, M. (2019). Bayesian Inversion of a forest Reflectance Model Using sentinel-2 and Landsat 8 Satellite Images. J. Quantitative Spectrosc. Radiative Transfer 233, 1–12. doi:10.1016/j.jqsrt.2019.05.013
Schueller, J. K. (1992). A Review and Integrating Analysis of Spatially-Variable Control of Crop Production. Fertilizer Res. 33, 1–34. doi:10.1007/BF01058007
Senf, C., Pflugmacher, D., Heurich, M., and Krueger, T. (2017). A Bayesian Hierarchical Model for Estimating Spatial and Temporal Variation in Vegetation Phenology from Landsat Time Series. Remote Sensing Environ. 194, 155–160. doi:10.1016/j.rse.2017.03.020
Serrano, L., Filella, I., and Peñuelas, J. (2000). Remote Sensing of Biomass and Yield of winter Wheat under Different Nitrogen Supplies. Crop Sci. 40, 723–731. doi:10.2135/cropsci2000.403723x
Seyednasrollah, B., Swenson, J. J., Domec, J.-C., and Clark, J. S. (2018). Leaf Phenology Paradox: Why Warming Matters Most where it Is Already Warm. Remote Sensing Environ. 209, 446–455. doi:10.1016/j.rse.2018.02.059
Siegmann, B., and Jarmer, T. (2015). Comparison of Different Regression Models and Validation Techniques for the Assessment of Wheat Leaf Area index from Hyperspectral Data. Int. J. Remote Sensing 36, 4519–4534. doi:10.1080/01431161.2015.1084438
Sims, D. A., and Gamon, J. A. (2003). Estimation of Vegetation Water Content and Photosynthetic Tissue Area from Spectral Reflectance: A Comparison of Indices Based on Liquid Water and Chlorophyll Absorption Features. Remote Sensing Environ. 84, 526–537. doi:10.1016/S0034-4257(02)00151-7
Soratto, R. P., Matoso, A. O., Gilabel, A. P., Fernandes, F. M., Schwalbert, R. A., and Ciampitti, I. A. (2020). Agronomic Optimal Plant Density for Semiupright Cowpea as a Second Crop in southeastern brazil. Crop Sci. 60, 2695–2708. doi:10.1002/csc2.20232
Srinet, R., Nandy, S., and Patel, N. R. (2019). Estimating Leaf Area index and Light Extinction Coefficient Using Random forest Regression Algorithm in a Tropical Moist Deciduous forest, india. Ecol. Inform. 52, 94–102. doi:10.1016/j.ecoinf.2019.05.008
Su, L., Wang, Q., Wang, C., and Shan, Y. (2015). Simulation Models of Leaf Area Index and Yield for Cotton Grown with Different Soil Conditioners. PLoS ONE 10, e0141835. doi:10.1371/journal.pone.0141835
Sun, B., Wang, C., Yang, C., Xu, B., Zhou, G., Li, X., et al. (2021). Retrieval of Rapeseed Leaf Area index Using the Prosail Model with Canopy Coverage Derived from Uav Images as a Correction Parameter. Int. J. Appl. Earth Obs. Geoinf. 102, 102373. doi:10.1016/j.jag.2021.102373
Tjahjowidodo, T., Dung, V., and Han, M. (2015). “A Fast Non-uniform Knots Placement Method for B-Spline Fitting,” in 2015 IEEE International Conference on Advanced Intelligent Mechatronics (AIM), Busan, South Korea, 2015 (IEEE), 1490–1495. doi:10.1109/aim.2015.7222752
Vehtari, A., Gelman, A., and Gabry, J. (2017). Practical Bayesian Model Evaluation Using Leave-One-Out Cross-Validation and WAIC. Stat. Comput. 27, 1413–1432. doi:10.1007/s11222-016-9696-4
Vehtari, A., Simpson, D., Gelman, A., Yao, Y., and Gabry, J. (2019). Pareto Smoothed Importance Sampling. arXiv. arXiv:1507.02646 [stat].
Wan, L., Zhang, J., Dong, X., Du, X., Zhu, J., Sun, D., et al. (2021). Unmanned Aerial Vehicle-Based Field Phenotyping of Crop Biomass Using Growth Traits Retrieved from Prosail Model. Comput. Electron. Agric. 187, 106304. doi:10.1016/j.compag.2021.106304
Wang C, C., Feng, M., Yang, W., Ding, G., Xiao, L., Li, G., et al. (2017). Extraction of Sensitive Bands for Monitoring the winter Wheat (triticum Aestivum) Growth Status and Yields Based on the Spectral Reflectance. PLOS ONE 12, e0167679. doi:10.1371/journal.pone.0167679
Wang T, T., Xiao, Z., and Liu, Z. (2017). Performance Evaluation of Machine Learning Methods for Leaf Area index Retrieval from Time-Series Modis Reflectance Data. Sensors 17, 81. doi:10.3390/s17010081
Watson, D. J. (1947). Comparative Physiological Studies on the Growth of Field Crops: I. Variation in Net Assimilation Rate and Leaf Area between Species and Varieties, and within and between Years. Ann. Bot. 11, 41–76. doi:10.1093/oxfordjournals.aob.a083148
Weber, V. S., Araus, J. L., Cairns, J. E., Sanchez, C., Melchinger, A. E., and Orsini, E. (2012). Prediction of Grain Yield Using Reflectance Spectra of Canopy and Leaves in maize Plants Grown under Different Water Regimes. Field Crops Res. 128, 82–90. doi:10.1016/j.fcr.2011.12.016
Wilson, A. M., Silander, J. A., Gelfand, A., and Glenn, J. H. (2011). Scaling up: Linking Field Data and Remote Sensing with a Hierarchical Model. Int. J. Geograph. Inf. Sci. 25, 509–521. doi:10.1080/13658816.2010.522779
Xing, L., Li, X., Du, H., Zhou, G., Mao, F., Liu, T., et al. (2019). Assimilating Multiresolution Leaf Area index of Moso Bamboo forest from Modis Time Series Data Based on a Hierarchical Bayesian Network Algorithm. Remote Sensing 11, 56. doi:10.3390/rs11010056
Xu, X. Q., Lu, J. S., Zhang, N., Yang, T. C., He, J. Y., Yao, X., et al. (2019). Inversion of rice Canopy Chlorophyll Content and Leaf Area index Based on Coupling of Radiative Transfer and Bayesian Network Models. ISPRS J. Photogramm. Remote Sensing 150, 185–196. doi:10.1016/j.isprsjprs.2019.02.013
Yamaguchi, T., Tanaka, Y., Imachi, Y., Yamashita, M., and Katsura, K. (2021). Feasibility of Combining Deep Learning and Rgb Images Obtained by Unmanned Aerial Vehicle for Leaf Area index Estimation in rice. Remote Sensing 13, 84. doi:10.3390/rs13010084
Yan, G., Hu, R., Luo, J., Weiss, M., Jiang, H., Mu, X., et al. (2019). Review of Indirect Optical Measurements of Leaf Area index: Recent Advances, Challenges, and Perspectives. Agric. For. Meteorol. 265, 390–411. doi:10.1016/j.agrformet.2018.11.033
Yan, H., Wang, S. Q., Billesbach, D., Oechel, W., Zhang, J. H., Meyers, T., et al. (2012). Global Estimation of Evapotranspiration Using a Leaf Area index-based Surface Energy and Water Balance Model. Remote Sensing Environ. 124, 581–595. doi:10.1016/j.rse.2012.06.004
Zhang, J., Cheng, T., Guo, W., Xu, X., Qiao, H., Xie, Y., et al. (2021). Leaf Area index Estimation Model for UAV Image Hyperspectral Data Based on Wavelength Variable Selection and Machine Learning Methods. Plant Methods 17, 49. doi:10.1186/s13007-021-00750-5
Keywords: bayesian hierarchical model, leaf area index, interpretability, markov chain monte carlo sampling, remote sensing, reflectance spectra
Citation: Stojanović O, Siegmann B, Jarmer T, Pipa G and Leugering J (2022) Bayesian Hierarchical Models can Infer Interpretable Predictions of Leaf Area Index From Heterogeneous Datasets. Front. Environ. Sci. 9:780814. doi: 10.3389/fenvs.2021.780814
Received: 21 September 2021; Accepted: 20 December 2021;
Published: 11 January 2022.
Edited by:
Christos H. Halios, Public Health England, United KingdomReviewed by:
Tong Qiu, Duke University, United StatesQiuyan Yu, New Mexico State University, United States
Katerina Trepekli, University of Copenhagen, Denmark
Copyright © 2022 Stojanović, Siegmann, Jarmer, Pipa and Leugering. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivera Stojanović, b3N0b2phbm92aWNAdW9zLmRl
 Olivera Stojanović
Olivera Stojanović Bastian Siegmann
Bastian Siegmann Thomas Jarmer
Thomas Jarmer Gordon Pipa
Gordon Pipa Johannes Leugering
Johannes Leugering