- 1Department of Agronomy, Purdue University, West Lafayette, IN, United States
- 2U.S. Geological Survey, Great Lakes Science Center, Lake Michigan Ecological Research Station, Chesterton, IN, United States
Invasive plants such as Asiatic bittersweet (Celastrus orbiculatus Thunb.) are a significant problem for land managers as they impact plant species composition, disrupt nutrient dynamics and structure of native ecosystems, and are difficult to eradicate. As a result of the increasing abundance of Asiatic bittersweet across the eastern U.S., we have been investigating underlying factors potentially contributing to the success of this plant. Recently, ecologists have been investigating the role of plant-soil-microbe interactions contributing to plant invasion. This work has led to question: are there certain microbes (e.g., bacteria, fungi) contributing to the success of some invasive plants? We hypothesize that despite differences in geologic age of soils where Asiatic bittersweet has established in the Indiana Dunes National Park there are sufficient common factors that led to common bacterial taxa in their rhizosphere. The objectives were to determine differences and commonalities in the soil chemistry, plant community and bacterial communities of Asiatic bittersweet plants. To achieve these objectives, bittersweet plants were collected at thirteen locations in the national park from soils ranging in geologic age from 150 to over 14,500 years. Surrounding soil chemistry, plant cover and the 16S rRNA gene amplicon sequences of rhizosphere soil bacterial communities of these Asiatic bittersweet were compared. Asiatic bittersweet coverage of sampling sites ranged from 2 to 77% averaging 52 ± 2%. There were statistically significant differences (p < 0.05) in alpha diversity (Shannon, Faith’s PD and Pielou’s evenness) and beta diversity (Bray Curtis, Jaccard, unweighted Unifrac, weighted Unifrac) among the samples when grouped by soil age or habitat. Despite these differences in the bacterial communities from different soil ages and habitats, some bacterial taxa (e.g., Bacillus, Streptomyces, Sphingomonas and Rhizobiales) previously found in other studies to be beneficial to plant growth were found in every rhizosphere community sampled. These microbes provide insight into a possible contributing factor to the success of this invasive plant at the Indiana Dunes National Park, and a strategy for future work to reduce the impact of Asiatic bittersweet establishment and offer some new strategies to manage this nuisance species.
Introduction
Various microbiota-related strategies are used by invasive plants to outcompete other plants. Often through chemical secretions from the roots, plants can discourage the growth of adjacent native species through competition or kill them outright (Callaway and Ridenour, 2004). In addition, these secretions may discourage or encourage growth of certain microbes (bacteria, fungi) in the adjacent soil, giving the aggressive invader a growth advantage over other native plant species (Pascale et al., 2020). Another strategy is that bacteria or fungi can provide nutrients to the plant that it could not acquire otherwise. For example, the interactions between beneficial microbes and plants (e.g., nitrogen-fixing bacteria, mycorrhizal fungi) have been greatly exploited in crop production (Macik et al., 2020) and agroforestry for a long time. Recently, ecologists have begun to investigate the role of plant-soil-microbe interactions in facilitating plant invasion (Coats and Rumpho, 2014; Zhang et al., 2019; Shahrtash and Brown, 2021). But these studies are limited in number leading to a dearth of knowledge on the impacts of different invasive species on the soil microbial communities.
Throughout the eastern US, the highly invasive liana, Asiatic bittersweet (Celastrus orbiculatus Thunb.) (hereafter bittersweet) is dominating many landscapes (Pavlovic and Leicht Young, 2011; Leicht-Young and Pavlovic, 2012; Leicht-Young et al., 2013). Asiatic bittersweet is especially problematic due to its rapid growth, woody vine growth, ability to resprout from the roots when burned or damaged, hybridization with native American bittersweet (Celastrus scandens L.), and strangling trees, making them more susceptible to falling over. Rapid expansion of Asiatic bittersweet is a challenging problem for land managers due to their impacts on composition, nutrient dynamics, and structure of native ecosystems and because they are often difficult to eradicate. While confined to the boundary between the Dunes State Park and the town of Dune Acres in the 1980s within the Porter County in Indiana, monitoring of this species at the Indiana Dunes National Park (INDU) has shown a tenfold increase in frequency from 1998–2000 to 2010–2012 (Pavlovic et al., 2022). As a result of the increasing abundance of Asiatic bittersweet at INDU and across the eastern U.S., it is imperative that we gain a greater understanding of the underlying reasons for the success of this plant to develop strategies for its remediation before vulnerable ecosystems are lost. The long-term objective of this work is to identify methods to reduce and possibly minimize or eliminate this invasive plant from the park using existing (e.g., prescribed burning, mechanical removal) and emerging technologies such as microbial control of nuisance invasive plants (Kowalski et al., 2015; Trognitz et al., 2016; Shahrtash and Brown, 2021).
Asiatic bittersweet is known to alter soil chemistry by increasing pH and soil concentrations of certain nutrients, such as potassium, calcium, and magnesium (Leicht-Young et al., 2009), likely influencing nutrient dynamics, successional pathways, and plant community structure. Nitrogen mineralization and litter decomposition rates were higher in soils impacted by the bittersweet, indicating elevated microbial activities. However, microbial community structure and its role in nutrient cycling was not directly assessed in that study. We hypothesize that despite differences in the soil ecology of Asiatic bittersweet establishment in the Indiana Dunes region, there are certain bacterial taxa common in their rhizosphere potentially enabling this invasive plant to outcompete adjacent vegetation. The objectives of this study were to (1) determine the range of different soil chemistries and plant vegetation where Asiatic bittersweet had established at INDU, (2) identify the differences in bacterial community composition and structure in the rhizosphere of these plants, and 3) determine if there were rhizosphere bacteria common to these invasive plants. Asiatic bittersweet plants were collected at thirteen locations in INDU from soils ranging in age from 150 to over 14,500 years. Surrounding soil chemistry, plant cover and the 16S rRNA gene amplicon sequences of rhizosphere soil bacterial communities of these Asiatic bittersweet impacted locations were compared to determine differences and commonalities among locations with Asiatic bittersweet establishment. This information will provide insight into potential underlying factors contributing to the success of this invasive plant and provide strategies for future work to control the spread of Asiatic bittersweet.
Materials and methods
Field sites
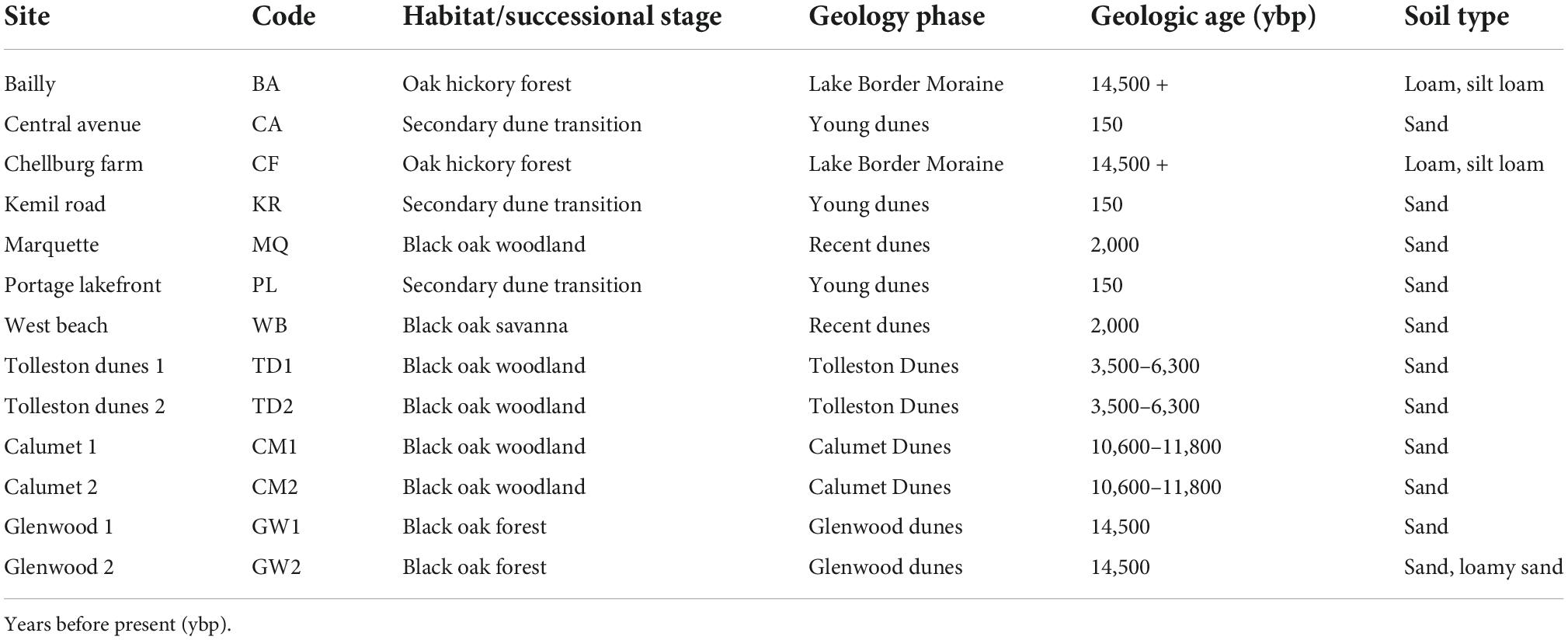
Asiatic bittersweet roots, bulk soil and adjacent vegetation were sampled on the lands of the Indiana Dunes National Park (41.63381°N, –87.05434°E), at the southern tip of Lake Michigan in Lake and Porter Counties, Indiana. The first herbarium collection of Asiatic bittersweet was collected in the Indiana Dunes in 1975 by Floyd Swink (Consortium of Midwestern Herbaria, 2022) and from 2000 to 2010 showed a tenfold increase in frequency across INDU (Pavlovic et al., 2022). Thirteen sites were selected across the Indiana Dunes incorporating sand dune and moraine habitats (Table 1). Sites encompassed a successional gradient from secondary dune-woodland transition to 14,500-year-old Glenwood dunes and two sites (Bailly and Chellburg Farm) on the older Lake Border Moraine [14.500 + years before present (ybp)] (Hansel et al., 1985; Argyilan et al., 2018). Habitats included secondary dunes transition, black oak savanna (recent and Tolleston dunes), woodland (Calumet dunes) and forest (Glenwood Dunes), and oak hickory forest. While the secondary dunes transition is sandwiched between grass dominated secondary dunes and oak savanna, these sites were on the edges of blowouts that formed approximately 150–200 years ago. However, blowout systems are undergoing a mosaic of disturbance occurring in the present time due to wind modifying sand movement across topographically complex landscape that produces patches of different successional stages. Consequently, we assigned woodland transitions to an age of 150 years rather than a younger age reflective of local blowout dynamics. Irrespective of the age assigned to the secondary dunes transition, they are younger than the other dunes/moraines and habitats. We sampled Asiatic bittersweet only and not American bittersweet (Celastrus scandens) because of concerns about its declining numbers and from the knowledge that sampling would kill the plants (Zaya et al., 2015).
Sample collection
Five independent Asiatic bittersweet plants were selected at each sampling site and each centered in 0.5 × 0.5 m quadrats for a total of 65 quadrats (13 sites × 5 plants). Samples were collected between August 24 and September 1 2017. The coverage and identity of herbaceous groundlayer vegetation surrounding the bittersweet plant was assessed within each quadrat. Representative bulk soil samples (multiple 10 cm × 1.0 cm soil cores) from each sampling location were collected from the quadrats for soil chemistry analysis. The soil core subsamples were pooled in the field and thoroughly mixed in the lab. From each of the five quadrats, one whole bittersweet plant was carefully excavated for rhizosphere analysis. Loose soil was shaken off the roots that were then placed into sterile bags for rhizosphere soil separation in the laboratory. All samples were placed on ice for transport to the lab. Bulk soils were stored at –20°C until processed. Root samples were processed after returning to the laboratory to collect closely adhered rhizosphere soils. The roots were placed in sterile centrifuge tubes containing phosphate buffer then vigorously shaken for 5 min then centrifuged. After centrifugation the buffer was poured off and the remaining rhizosphere soil was stored at –20°C until processed.
Soil chemistry
A portion of the pooled soil samples (for each of 65 locations) was sent to A&L Great Lakes in Fort Wayne, Indiana (N = 14, methods numbers follow (Soil Science Society of America, 2011) procedures based on (Page et al., 1982), for soil texture determination (hydrometer method) and a range of chemical parameters, including soil pH (Method 122.6), cation exchange capacity (CEC), base saturation of cations (% for Ca, Mg, Na, K Black, 1965), macronutrients (N, P (Method 24–5.1), K (Method 13–3.3.1), organic matter (Walkley-Black technique, Method 29–3.5.2), total and organic carbon, total N and micronutrients (S, Zn, Fe, Cu, B, Mn: various extractions and atomic absorption spectrometry).
Rhizosphere bacterial community analysis
Deoxyribonucleic acid extraction
A subsample (0.5 g) of rhizosphere soil was used for deoxyribonucleic acid (DNA) extraction using FastDNA™ kit (MP Biomedicals, LLC, Solon, OH, USA) following the manufacturer’s protocol. Agarose gels and spectrophotometry (Nanodrop 1,000 spectrophotometer; Thermo Scientific, Wilmington, DE) were used to ensure nucleic acid purity and quality. A NanoDrop 3,300 Fluorospectrometer (Thermo Fisher Scientific, Wilmington, DE) was used to quantify DNA that was stained with Hoechst dye.
Illumina 16S rRNA gene sequencing
Polymerase chain reaction (PCR) amplification of the 16S rRNA gene in soil samples was performed using V3-V4 region primers (343-forward TAC GGR AGG CAG CAG and 804-reverse CTA CCR GGG TAT CTA ATC C) (Liu et al., 2008; Nossa et al., 2010). PCR protocol and addition of primer tags were according to the manufacturer’s protocol (Illumina, San Diego, CA), with the exception that the first PCR reaction was performed for only 15 cycles and the second for 5 cycles to minimize PCR artifacts. PCR reactions were carried out using ∼10 ng of template DNA in Q5® High Fidelity DNA Polymerase 2 × master mix (New England Biolabs, Ipswich, MA, USA). PCR amplicons were purified with AxyPrepMag PCR clean-up kit (Axygen Scientific, Union City, CA, USA) and subsequently quantified using a NanoDrop 3,300 Fluorospectrometer, after staining with the QuantiFluor dsDNA System (Promega). Equimolar amounts of amplicons from each sample were combined and sequenced (2 × 250 paired-end) using a MiSeq Illumina system at the Purdue Genomics core facilities, West Lafayette, IN, USA. Nucleotide sequences are available in NCBI-SRA database under accession number PRJNA826510.
16S rRNA gene amplicon sequence analysis
After primer tag removal using Illumina software, sequences were analyzed using the QIIME2 pipeline (version 2021.1) (Bolyen et al., 2019). DADA2 (Callahan et al., 2016) was used to check sequence quality, denoise, merge paired end reads and select amplicon sequence variants (ASV). ASVs with less than 0.1% frequency of the mean sample depth or found in only one sample were then filtered from the dataset. Taxonomic assignments were made using the Silva data set (version 138_99) (Yilmaz et al., 2014) then chloroplast, mitochondria and non-assigned sequences were filtered from the dataset. Datasets were rarified to use equal number of reads (12,600 reads) for alpha and beta community analyses. Bacterial alpha-diversity measurements calculated were Shannon diversity (richness and evenness), Chao1 index (richness), observed-features (ASVs), Faith’s phylogenetic diversity (PD) and Pielou’s evenness. Beta diversity metrics tested included both phylogenetic Unifrac distances (weighted and un-weighted) (Lozupone et al., 2011) and non-phylogenetic distances (Jaccard and Bray Curtis). Completeness of ASV representation in each sample was estimated by Good’s coverage, which was > 99.999% in all samples indicating that almost all the taxa were included in the analysis. Relative distribution of β-diversity measures among samples were visualized using Principal Coordinate Analysis (PCoA).
Statistical analyses
Prior to Principal components analysis (PCA) and cluster analysis, correlations among soil variables were calculated and variable were removed that had high correlations with other soil variables. Selected soil variables, P ppm, K ppm, pH, micronutrients ppm (Cu, Zn, Mn, Fe, S, B), total C and total N were standardized and centered in PCORD since variable units of measurement were different. PCA and cluster analysis was conducted in PAST (PAleontological STatistics) (Hammer et al., 2001). PCA was used to determine whether distinct clusters of chemistry were found across successional stages and ages and then were tested using discriminant analysis (SYSTAT, 2004) and a multiple response permutation procedure (MRPP) (McCune and Mefford, 2006).
Ground layer vegetation presence/absence and cover by growth form (forbs, graminoids, shrubs, trees, and lianas) was ordinated using non-metric multidimensional scaling (NMS) and NMS results were compared to soil chemistry variables.
For the microbial data most of the statistical comparisons were calculated for the samples grouped by geologic age and habitat. But note that habitat agglomerated geologic age groups within the oak woodland category (Table 2). Significant differences among alpha-diversity measurements were determined using Kruskal Wallis for all groups as well as pairwise comparisons followed by Benjamini-Hochberg FDR correction. Beta diversity differences among communities were determined using 999 permutations of perMANOVA (Anderson, 2001) of all groups and pairwise comparisons with Benjamini-Hochberg FDR correction. Then, permDISP (permutational analysis of multivariate dispersions) (Anderson et al., 2006) determined if dispersion could be contributing to significant differences. Differences were considered significant if p < 0.05 or q < 0.05 for pairwise comparisons.
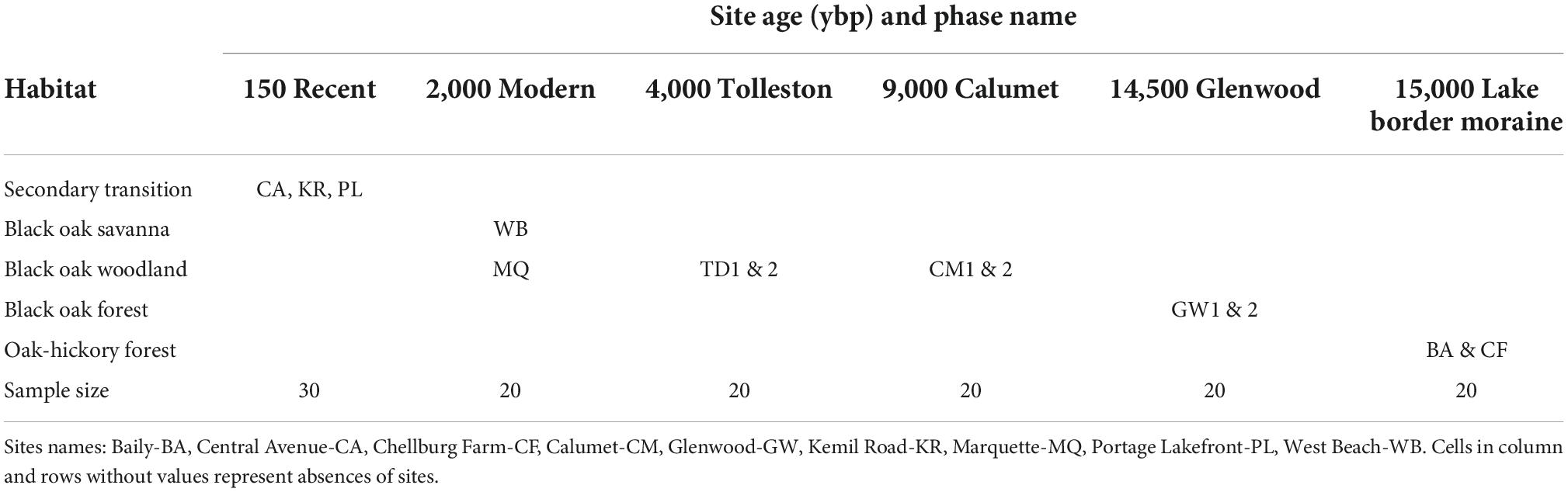
Table 2. Crosswalk between habitat and site age (ybp) for 13 sites sampled for rhizosphere bacteria, soil chemistry and vegetation.
Differential abundances in taxonomy across all habitats as well as geologic age were determined using Analysis of Composition of Microbiomes (ANCOM) (Mandal et al., 2015). ANCOM was used to identify differences in the log-transformed counts of taxa between habitat groups and geologic age groups. Bacterial taxa (genus and species level) common to all rhizospheres were determined using the “core feature” command in the feature tables plug-in in QIIME2. Taxa overlapping among the various habitats was illustrated using a Venn diagram program available from the University of Ghent Bioinformatics and Evolutionary Genomics group.1
Canonical correspondence analysis (CCA) was used to find associations among the bacterial genera and differentiating soil and vegetation matrix factors using PAST and PCORD (McCune and Mefford, 2006). After removal of co-correlating factors, explanatory variables included in the preliminary model were dune age, slope, aspect, soil variables (pH, CEC,% organic matter, and ppm of B, Cu, Fe, K, Mn, P, S, and Zn), and vegetation (total vegetation, Shannon diversity (D), litter cover, bare ground cover, bittersweet and graminoids). CCA analysis in Vegan using the function ordistep were used to identify which soil and vegetation variables significantly contributed to the relationship with the bacterial genera (R Core Team, 2022).
Results
PCA of soil chemistry produced three components explaining, respectively, 42, 22, and 17% of the variation for a total of 81% (Figure 1A). The first axis separates soils with higher percentage sand from moraine soils. The second axis separates the sandy soils along the temporal chronosequence. The first principal component is strongly related to concentrations (mg/kg) of K, Mn, B, Zn, and total C and N. The second axis is an increasing pH gradient with negative loadings on P (mg/kg) and Fe (ppm). The third axis has highest loadings on Zn and Cu (ppm), an axis that is largely determined by one Calumet Dune sample that has high values of copper and zinc (10 ppm Cu, 20 mg/kg Zn). Sites with high soil iron included Glenwood 1 and 2, West Beach (> 50 mg/kg) whereas Central Ave., Kemil Road, and Portage Lakefront had low values (< 5 mg/kg). Soils were classified into four classes (discriminant Wilkes Lambda 39 138 = 34.6, P < 0.001 with equal priors and 98% successful classification); i) Seventeen samples with high pH (5.1–7.9), sand content (94–96% and organic C/N ratio (11.3–82.1) representing young dune near Lake Michigan, ii) ten with high% silt,%clay, CEC, total C, total N, K ppm, Mn ppm and B ppm representing moraine soils (i.e., oak hickory), iii) 13 typical older sand dunes with low pH and high P, S mg/kg and Cu mg/kg, and lastly iv) 25 old dune samples with industrial impacts, low pH and high Zn and Fe (mg/kg). Soils were significantly different among geologic phase ages and habitats, but MRPP had a larger test statistic with dune age (MRPP dune age: T = –27.7, A = 0.67. P < 0.001, MRPP habitat: T = –25.1, A = 0.53, P < 0.001).
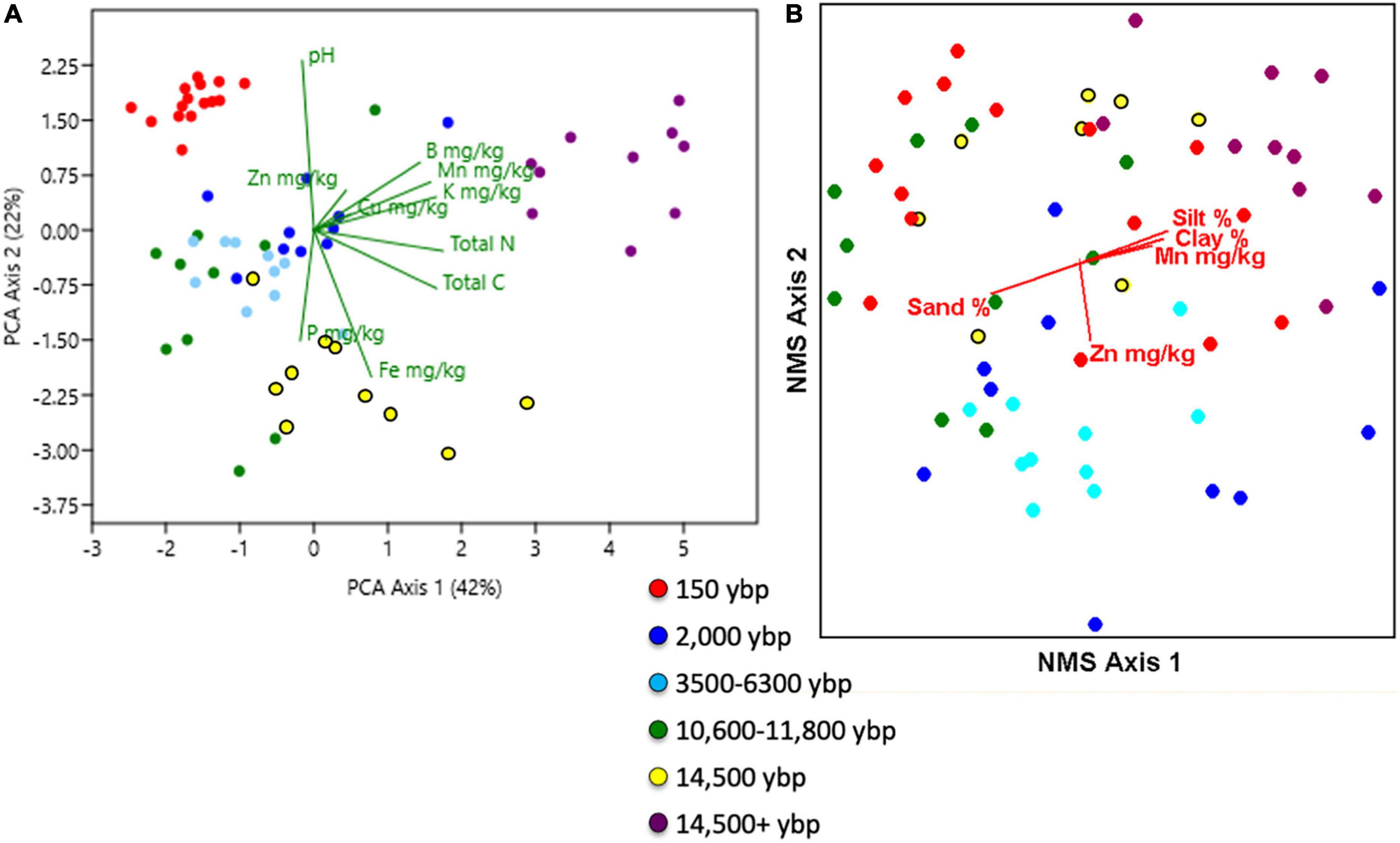
Figure 1. (A) Two dimensional principal components analysis (PCA) of Asian bittersweet bulk soil sample chemistry among the 13 sites. (B) Two dimensional non-metric multidimensional scaling ordination of vegetation presence/absence by species. Red biplots show the direction and magnitude of soil environmental variable corelates with vegetation composition. For both plots, each dot represents a sample and is colored by dune ages listed in the legend. Chemical abbreviations are B = boron, C = carbon, CU = copper, Fe - iron, K= potassium, Mn = manganese, N = nitrogen, P = phosphorous, and Zn = zinc. Samples were collected at Indiana Dunes National Park in 2017.
Vegetation composition
A total of 87 plant species were sampled including five exotic species: Asiatic bittersweet (Celastrus ortbiculatus), burning bush (Euonymus alatus), Asiatic bush honeysuckle (Lonicera japonica), and bouncing bet (Saponaria officinale), and 82 native species (nomenclature follows (Wilhelm and Rericha, 2017). All vegetation plots had Asiatic bittersweet ranging from 2 to 77% cover, averaging 52 ± 2% (± standard error (SE) and throughout) of the total cover of vegetation (7.5–100%). Mean cover of bittersweet was double in the black oak savanna (42 ± 7) and oak hickory forest (35 ± 7) compared to the secondary dune transition (15 ± 3) [F(5, 60) = 28, P < 0.001]. All other comparisons were not significant. Average plant species richness per plot was 5 ± 1 ranging from 0 to 15 species per plot. Most common native species were Virginia creeper (Parthenocissus quinquefolia), jack-in-the -pulpit (Ariseama triphyllum), blue-stemmed goldenrod (Solidago caesia), sassafras (Sassafras albidum), and poison ivy (Toxicodendron radicans). Cover of native species was 21 ± 2 representing 45 ± 3% of the vegetation cover (0–93%). NMS of the presence absence vegetation data produced three ordination axes explaining the variation in vegetation composition (Figure 1B). Axis one was highly negatively correlated with percent sand (–0.55) and positively correlated with percent silt (0.55) and clay (0.53) representing a parent material productivity gradient with concentrations of manganese increasing along the axis. Zinc ppm (r = –0.51) was negatively correlated with axis 2. Silt loam oak-hickory sites were clustered in the upper right hand of the ordination graph but were not wholly distinct from the rest of the plots that had sandier soils (Figure 1B). Plants such as sugar maple (Acer saccharum), multiflora rose (Rosa multiflora), bedstraw (Galium aparine), wild lettuce (Lactuca canadensis) and gray dogwood (Cornus canadensis) distinguished the moraine soil sites from the sandy sites. NMS of growth form data produced a two-dimensional ordination (not shown) with the first axis negatively correlated with total vegetation cover (r = –0.69) and manganese ppm (–0.49) and positively correlated with bare ground (r = 0.42). Five growth form vegetation clusters were identified but they were based on growth form structure and were not related to habitat or dune age. Groups represented relative dominance of growth forms and one group stood out because of low vegetation cover compared to most of the plots and these were distributed among habitats and ages.
16S rRNA gene sequence analyses
PCR of one sample from the Central Avenue site (secondary dune) was unsuccessful and was excluded from all sequence analyses. Analysis of the remaining 64 samples produced 3,434,286 paired-end reads, with read numbers in individual samples ranging from 12,618 to 204,216 (mean reads 53,660 ± 35,941 S.D) after trimming, merging, and filtering for quality and non-bacterial reads. The read cutoff used for alpha and beta diversity analyses was 12,600. Goods coverage had a mean of 99.9% ranging from 99.9 to 100%.
Taxonomic annotations
Taxonomic annotations of the 16S rRNA gene sequences indicated there were 25 bacterial phyla represented in the collected rhizosphere soil samples. The phyla Proteobacteria (42.3 ± 1.2%, mean ± standard error), Actinobacteria (31.9 ± 1.2%), Acidobacteriota (15.3 ± 0.7%), Bacteroidota (3.5 ± 0.1%), Chloroflexi (2.1 ± 0.2%), Firmicutes (1.4 ± 0.2%), Myxococcota (1.3 ± 0.1%), Gemmatimonadota (0.9 ± 0.1%), and Methylomirabilota (0.5 ± 0.1%) cumulatively represent over 98% of the sequences (Figure 2). Other phyla and candidate phyla included, Bdellovibrionota, Cyanobacteria, Dependentiae, Desulfobacterota, Elusimicrobiota, Entotheonellaeota, Latescibacterota, MBNT15, Myxococcota, NB1-j, Nitrospirota, Patescibacteria, Planctomycetota, RCP2-54, SAR324_clade (Marine_group_B), Verrucomicrobiota, and WPS-2. These phyla represent 58 classes, 144 orders, 245 families, and 523 genera.
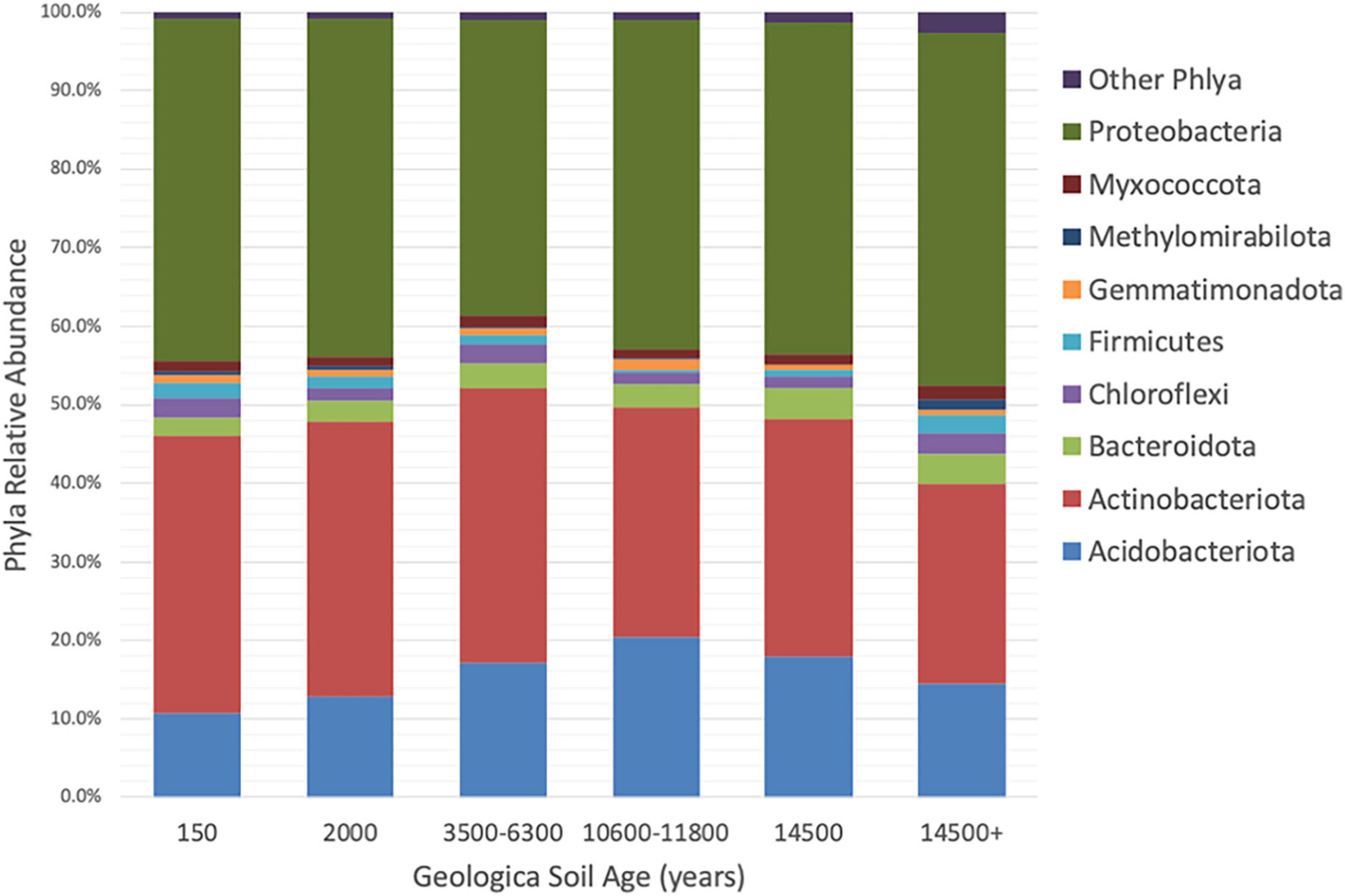
Figure 2. Comparison of relative abundances of phyla in Asiatic bittersweet rhizosphere soils collected from soils of increasing geologic ages (ybp). Only phyla with relative abundance > 1% in at least one soil age is included. Other phyla are represented by Bdellovibrionota, Cyanobacteria, Dependentiae, Desulfobacterota, Elusimicrobiota, Entotheonellaeota, Latescibacterota, MBNT15, Myxococcota, NB1-j, Nitrospirota, Patescibacteria, Planctomycetota, RCP2-54, SAR324_clade (Marine_group_B), Verrucomicrobiota, and WPS-2. Samples were collected at Indiana Dunes National Park in 2017.
Alpha diversity
Rarefaction curves reaching or approaching a plateau of all the alpha diversity metrics tested indicated 12,600 reads were adequate for making community comparisons (Supplementary Figure 1). Kruskal Wallis comparisons of alpha diversity metrics indicated there were significant differences for Shannon, Faith’s PD and Pielou’s evenness but not for the richness metrics Chao1 or observed features for samples grouped by geologic soil age or by habitat. Pairwise comparisons indicated there were some significant differences in the alpha diversity metrics for Faith’s PD and Pielou’s evenness between geologic age (Figures 3A,C) and habitat samples (Figures 3B,D). Faith’s PD and Pielou’s evenness values were significantly greater between the 14,000 + and 150 ybp samples. The Pielou’s evenness value was significantly greater for the oak-hickory forest habitat compared to secondary dune. Faith’s PD was significantly greater for both the oak-hickory forest and black-oak woodlands compared to secondary dune transition as well as to black oak forests.
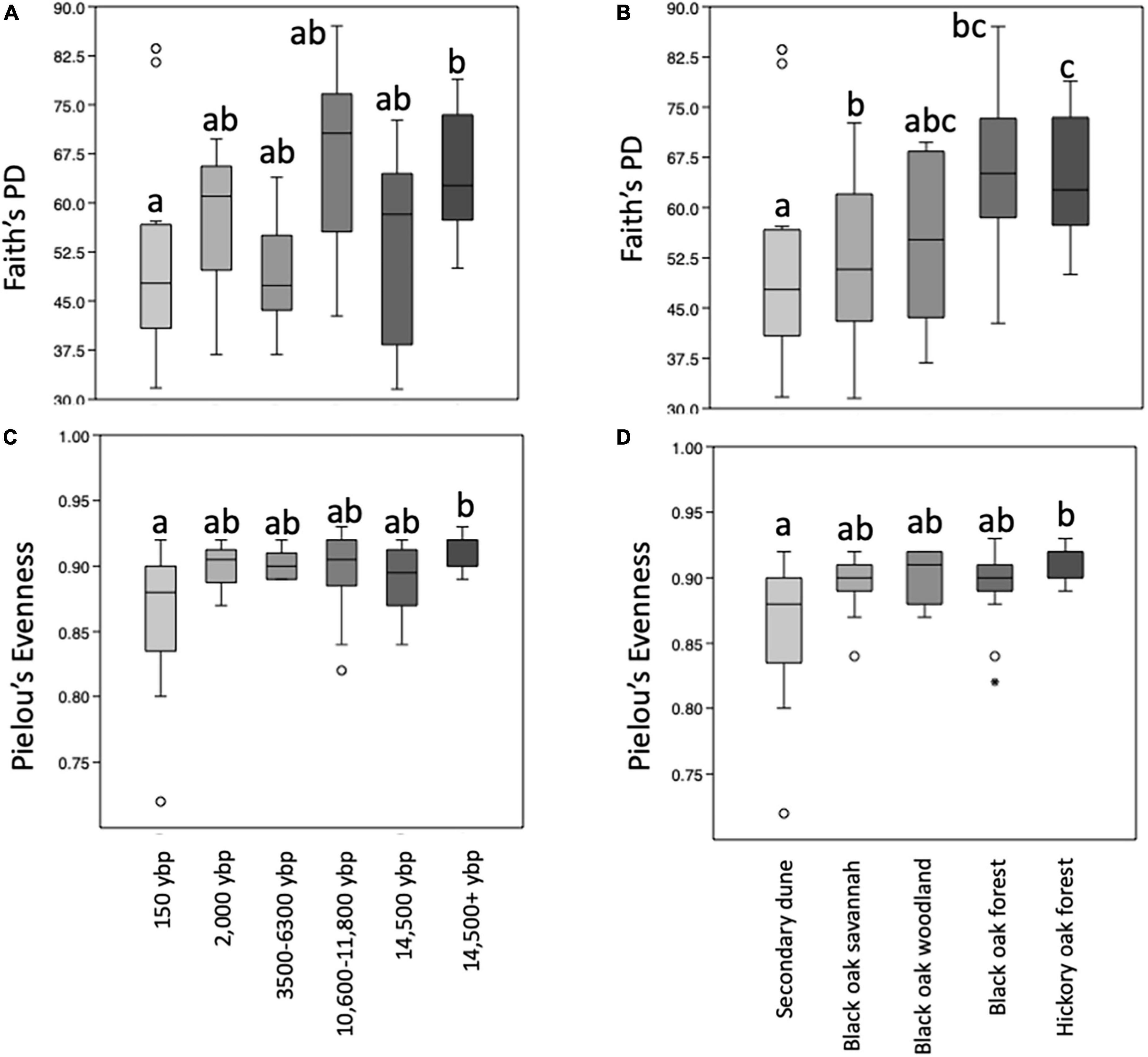
Figure 3. Boxplots representing the medians and upper and lower quartile values of within sample (alpha) diversity metrics (A,B) PD whole tree (Faith’s phylogenetic diversity (PD), richness) and (C,D) Pielou’s evenness of samples from different soil geologic ages (A,B) and habitats (C,D). Kruskal Wallis indicates overall differences for all metrics tested are significant (p < 0.02) and pairwise-tests with 999 permutations with significant differences at q < 0.05 illustrated by different letters above bars. Outliers are represented by open circles. Samples were collected at Indiana Dunes National Park in 2017.
Beta diversity
PCoA of all the beta diversity metrics showed clear separation of samples by geologic age (representative example using unweighted Unifrac, Figure 4A). The unweighted Unifrac figure shows age separated mainly along PCoA1 but 14,500 ybp and 14,500 + ybp separated along PCoA2. PerMANOVA indicated the overall difference was significant for all the beta diversity metrics (p = 0.001) and pairwise comparisons also differed except for 2,000 compared to the 3,500–6,300 ybp samples for weighted Unifrac (Supplementary Table 1). However, permDISP was also significant for overall comparisons of all metrics (p < 0.05). The PCoA plot illustrates that there are differences in dispersion of samples among the different geologic age. But the 150 ybp samples clearly separate from the 14,500 and 14,500 + ybp samples along the first axis. There is overlap between the 2,000, 3,500–6,300, and 10,600–11,800 samples but clusters can be differentiated along axis 2.
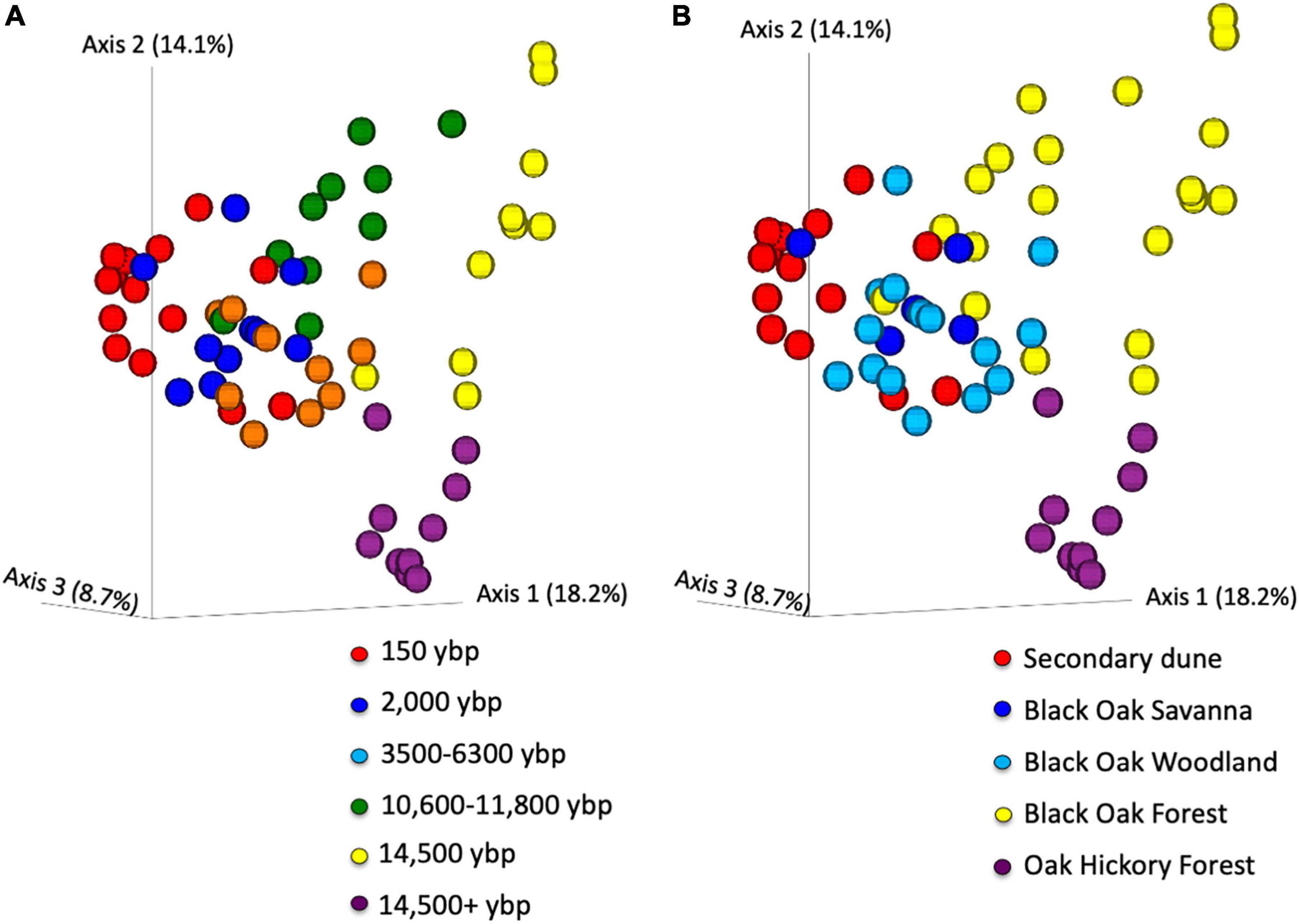
Figure 4. Principal coordinate analysis of unweighted Unifrac beta diversity metrics of Asiatic bittersweet rhizosphere bacterial communities. Data points colored by (A) geologic age of soils (ybp- years before present), and (B) habitat type. perMANOVA comparisons of samples by age or habitat, overall differences significant p = 0.001 and all pairwise comparisons q < 0.02. Overall permDISP comparison also significant p < 0.03 for age and habitat. Pairwise analysis indicates that dispersion differences are significant (p < 0.05) between over 14,500 + y samples and all other ages except 14,000 y. For habitats, pairwise permdisp differences significant (p < 0.04) for oak hickory forest compared to secondary dune and to black oak forests, also black oak woodland compared to secondary dune and black oak forests. Samples were collected at Indiana Dunes National Park in 2017.
The differences noted in the PCoA plots of the geologic ages was also reflected when plots were differentiated by habitat (Figure 4B and Supplementary Table 2). As expected, the PCoA plot shows clear separation of secondary dune samples from the black oak and oak-hickory forest samples along PCoA1 and PCoA2. The differences are significant for habitat pairwise comparisons for all metrics except weighted Unifrac where black oak savanna did not differ from black oak woodland and secondary dune (Supplementary Figure 2 and Supplementary Table 2). Similarly, overall permDISP differed significantly but not for all pairwise comparisons.
Differentially abundant and common bacterial taxa
ANCOM identified similar bacterial taxa differing among the geologic age (Supplementary Table 3) and the habitat samples (Supplementary Table 4). There were six taxa that were highly significant based on volcano plots of the data (Supplementary Figure 3). In the oldest soils (14,500 + ybp) and oak-hickory forests there were significantly more uncultured Bacteriovoracaceae, Comamonas, Geobacter, and a Latescibacterota (Figure 5). In the 14,500 + ybp soils there were more Aurantisolimonas whereas there were more Starkeya found in the oak-hickory forest habitat (Supplementary Figure 4). In the youngest soils (150 ybp) and secondary dune transition there were more Promicromonospora.
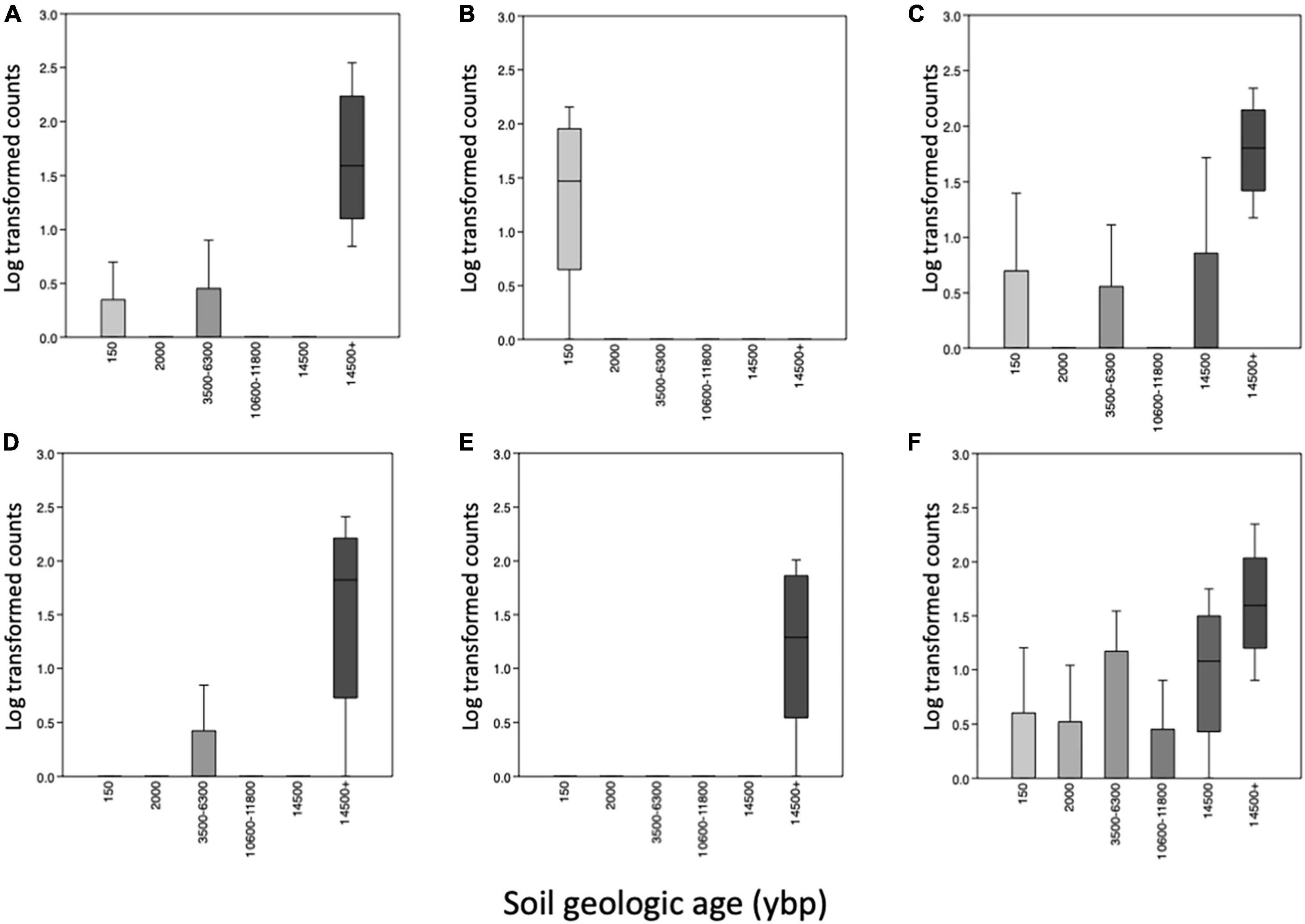
Figure 5. Boxplot of medians and SD (standard deviations) of differentially abundant taxa identified using Analysis of Composition of Microbiomes (ANCOM) of microbial communities comparing the various soil geologic ages (ybp, years before present) along the y-axis. Taxa with W-values > 439 and clr > 34, (A) an uncultured Bacteriovoraceae (p__Bdellovibrionota), (B) Promicromonospora (p_Actinobacteriota), (C) Comamonas (p_Proteobacteria), (D) Geobacter (p Desulfobacterota), (E) an unnamed Latescibacterota and (F) Aurantisolimonas (p_Bacteoidota). W is number of time null hypothesis that were rejected and clr is the center log ratio.
More importantly for the objective of this study a Venn diagram comparison of the taxa present in each habitat indicated 290 of the 523 taxa (55.4%) identified at the genus level were found in at least one sample from every habitat (Supplementary Figure 5). The QIIME2 “core feature” program identified 23 taxa at the genus level found in every rhizosphere sample (Table 3). Analyses at the species are not included because none of the common genera were identified at the species level. A number of these core taxa are thought to be plant growth promoting bacteria, including seven taxa in the order Rhizobiales (Devosia, Hyphomicrobium, Labrys, Mesorhizobium, Bradyrhizobium, and two un-named Xanthobacteraceae genera), Bacillus, Streptomyces and Sphingomonas.
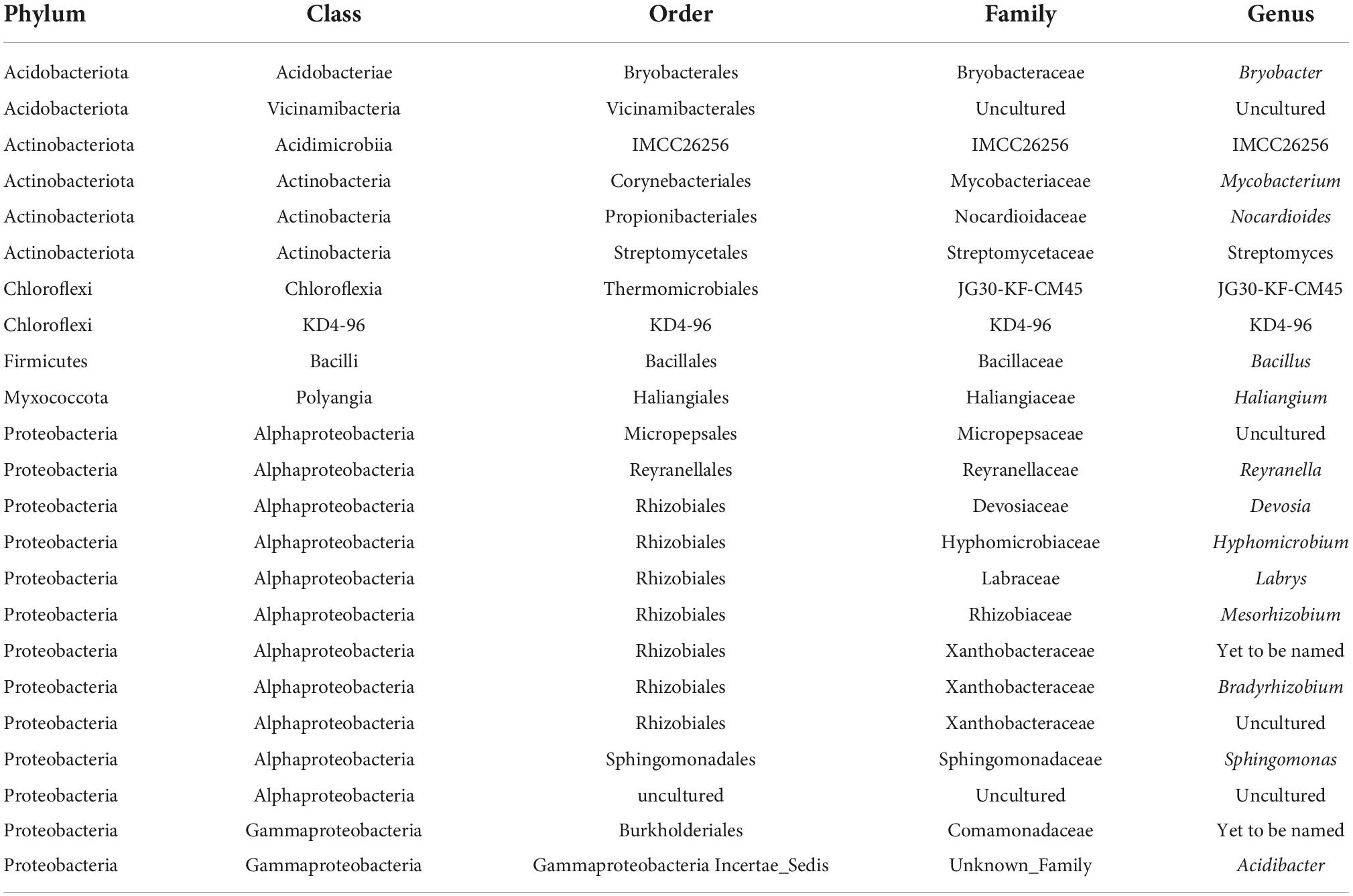
Table 3. Bacteria at genus level of classification present in 64 samples collected at Indiana Dunes National Park in 2017.
Bacterial community association with soil chemistry and ground layer vegetation
Significant relationships existed between the bacterial genera distribution and soil chemistry and associated plant communities based on CCA (species- environmental correlation by axes 0 0.977–0.90–0.88, p < 0.001). The first and second canonical axes explained, respectively, 12 and 8% of the variation with a cumulative 20% and adding the third axis only increased the cumulative variation explained to 24%. The first axis represented the soil geologic age and nutrient gradient with secondary dunes transition bacterial communities on the left, black oak forest and oak hickory forest bacterial communities clustered on the right (Figure 6 and Supplementary Table 5). Oak hickory forest bacterial communities also clustered toward the right side of the plot but separated from the other samples along the second axis. The significant chemical factors relating to bacterial genera communities were pH (P = 0.005), total N (P = 0.005), dune age (P = 0.005), graminoid cover (P = 0.025), Zn (P = 0.03), total C (P = 0.04) and with Fe and slope included but did not individually contribute to the model (Figure 6, biplots and Supplementary Table 5). Zn is absent in Figure 6 since axis 3 is not shown (see factor loadings (Supplementary Table 5).
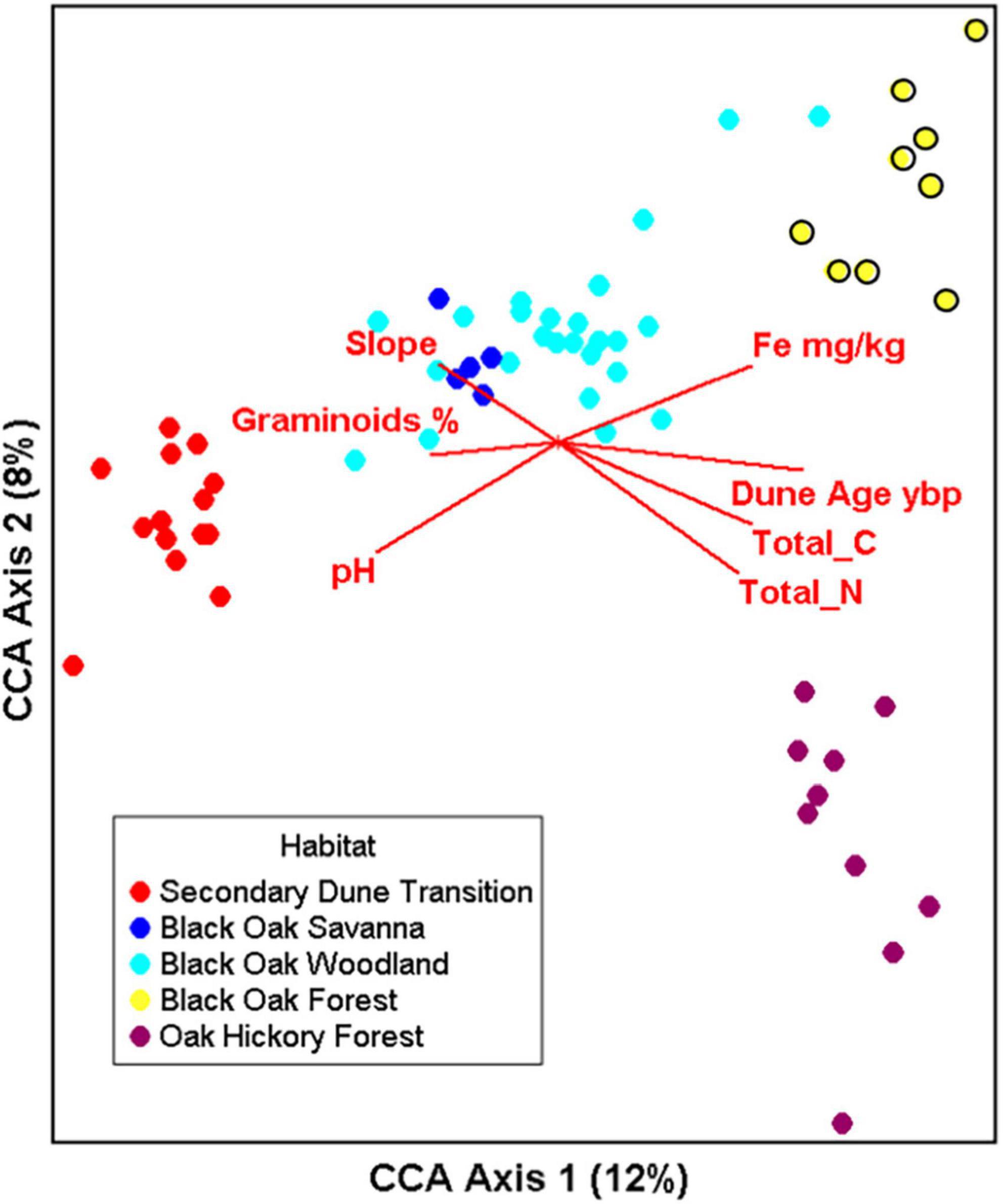
Figure 6. Canonical correspondence analysis (CCA) illustrating the relationship of bacterial genera (log transformed counts) to dune age, slope, soil chemical factors (pH, CEC, OM (% organic matter), concentrations of Fe, K, Mn, P, B ppm), % bare ground, litter cover, and graminoids. CCA1 and 2 explain 12.0 and 8.0% of the total constrained variation, respectively. Number of samples n = 64, overall p < 0.001 with 999 permutations. All variable tested in the model are listed in Supplementary Table 5.
Discussion
This study shows the broad range of soil conditions across a geologic age gradient in which the Asiatic bittersweet has successfully invaded in the Indiana Dunes region. These soils varying in geologic age differed in soil texture and productivity relative to substrate type, sand vs. clay. Within the sand dune habitats pH and total C and N varied in opposite directions, but otherwise the soils were fundamentally similar in physical structure and chemistry. Distinct microbiome communities are associated with the rhizospheres of Asiatic bittersweet. However, despite the differences there are also bacterial taxa common to all locations, indicating that these soils from different geologic ages each did not select for different bacterial communities. Indicating that these soils from different geologic ages did not differ substantially to select for entirely different microbial communities. Using alpha and beta diversity metrics to compare microbial communities in the rhizosphere of plants growing in soils of different ages and habitats, we show there are distinct differences in the composition and structure of rhizosphere microbiomes associated with Asiatic bittersweet along this soil chronosequence. Despite these microbiome differences, we found over half (290 of 523 total) of the bacterial genera were found in at least one sample from each soil age and 23 of them were found in every sample. These findings suggest that despite the recent entry of Asiatic bittersweet into these various habitats (since the 1970’s), there are certain bacterial communities, which have been shown by others to promote plant growth (e.g., Bacillus, Streptomyces), potentially associated with Asiatic bittersweet success in the park’s landscapes. But these taxa are very common soil bacteria (Delgado-Baquerizo et al., 2018) and unlikely newly introduced with the Asiatic bittersweet. Therefore, without further research we do not know why these taxa are common to these rhizospheres. Also, some groups, such as the Streptomyces, have both beneficial (Vurukonda et al., 2018) and pathogenic representatives (Loria et al., 2006).
The differences in soil chemical properties found in this study reflect trends reported over the last 50 years in the Indiana Dunes and Great Lakes sand dunes in general. Both Olson (1958) and Lichter (1998b) found that soil pH decreased from a nearshore peak between 7 and 8 to a low between 4 and 5 during the first 1,000 years of soil development. Other dune systems worldwide show a decrease in pH with time, but the magnitude of change and range varies (Nelson et al., 2020; Pollmann et al., 2020; Gaiero et al., 2021). Similarly, Lichter (1998a) at Wilderness State Park in lower peninsula of Michigan found that soil carbon and nitrogen asymptotically peaked between 500 and 1,000 years, respectively, at 40 and 2.3 mg/ha. The sometimes moderate to high concentrations of metals (Fe, Mn, Zn, Cu) are reflective of industrial steel production and human impacts on the Indiana Dunes soil (Cole et al., 1990; Esser et al., 1991; Lichter, 1998b). Despite this difference in soil chemical properties the Asian bittersweet has been able to rapidly establish across the Indiana dunes chronosequence (Pavlovic et al., 2022). Soil properties do not appear to influence invasion except for hydric soils where we have found that it has not been able to invade.
The ground layer vegetation composition alone did not separate into strong communities due to the dominance of invasive species and the small scale of the plots (Figure 1B. Soil ages and communities (not shown) are intermixed. Oak hickory understories on the moraine were distinctive compared to those on sandy soil that produce wide variation in composition with dune ages overlapping (upper right in Figure 1B). Bittersweet cover at the ground level was highest in the oak hickory forest and black oak savanna. The oak hickory samples were from secondary oak hickory forest at the edge of the older old growth oak hickory forest in the adjacent ravine, so canopy closure was less than in old growth forests. The black oak savanna was burned that resulted in higher densities of stems than otherwise would occur (Pavlovic et al., 2016 and pers. obs.). Graminoids were expected to be prevalent in the secondary dune transition, but their low abundance elsewhere may indicate how bittersweet shade impacts the native plant community in forested communities. In higher light levels (>70% PAR), bittersweet seedlings produced more leaves, thicker shoots, and longer and heavier roots compared to those grown in lower light levels (Greenberg et al., 2001).
The differences in microbiome composition along the soil chronosequence reported here have been also seen in other studies (Alfaro et al., 2017; Yeoh et al., 2017; Turner et al., 2019). Similar to other studies, pH appears to one of the main factors driving the differences in microbial community composition (Alfaro et al., 2017; Yeoh et al., 2017; Turner et al., 2019). The higher alpha diversity values in the oldest soils here was also reported in the Jurien Bay (Western Australia) dune chronosequence but this increase occurs up to soils in the 105 B.P. years and in much older soils the diversity decreases (Turner et al., 2019). Since these other studies were performed on dunal chronosequence soils in Australia and Peru these patterns of diversity differences are likely general to dunal chronosequences and not specific to specific geographic regions.
A model of the relationship among the soil factors, plant and microbial communities using CCA indicates that the main chemical factors associated with the different bacterial communities in the Indiana Dune soils of different geologic ages are pH, total N & C, and Zn content. Other studies have shown that soil chemical factors, especially pH, are major determinants of microbial communities (Lauber et al., 2009; Bahram et al., 2018). In soils of different ages plant succession and the members of the plant community (e.g., oak leaf litter) would contribute to the differences in soil pH (Olson, 1958) and as well as macronutrients such as carbon and nitrogen. Differences in bacterial communities potentially influenced by the plant communities are also evident in the CCA with grasses (Graminoids) more dominant in the younger soils making up the secondary dune transition with the shrubs (Liana) in the black-oak forest. These findings are similar to others along a soil chronosequence where soil pH declines with dune age (carbonate weathering, see Tombeur et al., 2020; Nelson et al., 2021), soil organic matter increases, and bacterial communities shift in composition as soil chemistry and vegetation changes (e.g., Cline and Zak, 2015; Alfaro et al., 2017; Yeoh et al., 2017). Leaf litter of Asiatic bittersweet has been shown to increase soil pH, and levels of potassium, calcium and magnesium (Leicht-Young et al., 2009). The increase in pH would counter the successional trend to decrease pH, but we do not have data to support that hypothesis from this study.
Despite the soil, vegetation and microbial differences expected along a soil chronosequence there were still many bacteria genera common among the soils. A set of core plant community associated rhizosphere bacteria was also found along a dunal chronosequence in the Great Sandy National Park on the Queensland coast of Australia (Yeoh et al., 2017). The authors thought that this core root microbiome had evolved over the 400-million-year history of the terrestrial plants in that park. While our dune system was less than 20 thousand years old, the native plant communities and bacterial communities might have coevolved for a longer time. But in our study the Asiatic bittersweet is a newly introduced species to these locations but were still able to out compete the native plant species. This is partially due to some of its growth strategies, such as, rapid growth with copious roots, and vine like habit enabling it to strangle trees (Fryer, 2011). But there is major lingering question. What factors do bacteria contribute to its rapid growth and successful invasion along this soil chronosequence?
Many of the bacteria genera common to the rhizosphere in all the samples are known to be involved in plant growth promotion and nitrogen acquisition. There are a number of taxa in the order Rhizobiales found in the rhizosphere samples. Although this group typically infects and forms nitrogen-fixing nodules in leguminous plants species, there is mounting evidence these bacteria are also beneficial to non-leguminous plants (Garrido-Oter et al., 2018; Fang et al., 2019). Laboratory growth experiments using a Rhizobium strain isolated from the non-leguminous invasive plant species Ageratina adenophora showed it was the best at improving plant growth (Fang et al., 2019). Unfortunately, we do not have direct evidence that any of the Rhizobiales associated with these bittersweet are fixing nitrogen or providing some other growth advantage to the plants. Rhizobia were also found as part of the core microbiome associated with the roots in the Australian dunal study (Yeoh et al., 2017). This suggests that although some bacterial taxa have possibly evolved along with the plants over geologic time, this invasive species potentially has mechanisms to out compete those plants to successfully invade diverse ecosystems. Since there is an apparent a core of microbes with plant growth promoting traits, we hypothesize that these Asiatic bittersweet plants do not need to enhance the growth of new microbes for invasion success but only some means of optimizing attraction of existing growth promoting bacteria to their rhizosphere. Since all the soil biota were not assessed in this study, it is possible that some other biota (e.g., fungi) are also contributing to the success of these plants. For instance, Asiatic bittersweet has been known to harbor endomychorrhizae in its roots possibly aiding its invasion ability (Leicht-Young et al., 2016). But it is likely that soil biota contributes to the success of these invasive plants to outcompete the indigenous plants.
The identification of some common bacteria from this study might provide a microbe-based means to control this plant species in diverse habitats. One possible means of reducing unwanted bacteria is to use taxa specific phages to reduce their numbers. Phage therapy is being used in medicine to treat human pathogens (Pirnay et al., 2022; Venturini et al., 2022) and more recently to control plant pathogens (Ye et al., 2019). Unfortunately, the bacteria associated here with Asiatic bittersweet can be beneficial to other plants in this ecosystem therefore it may not be reasonable to eliminate them. The Asiatic bittersweet is fairly widespread from Ontario and Quebec in Canada to mainly the states in the Atlantic seaboard and into Great Lakes region but its distribution is expanding (Fryer, 2011). There might be antagonistic microorganisms (bacteria, fungi) that can have parasitic relationships with plants rather than beneficial organisms that can be used to control these plants. Other possibilities would be application of nematodes to prey on the roots of the target plants (Wilschut and Geisen, 2021). Therefore, additional studies are needed to better understand the interactions between bittersweet and soil microbial communities, including fungi, to better understand the role of these communities in plant invasion and to identify potential microbiota for controlling this nuisance species. One weakness of this work is that funds were not available to determine the distribution of fungi in these samples. This study now paves the way for future studies to test microbial-plant interactions using controlled greenhouse experiments (Bickford et al., 2022). Elucidating the mechanisms used by invasive plants for a competitive advantage over native species also has broader implications for microbially influenced plant-plant interactions. For example, similar mechanisms may be used by weeds to provide them competitive advantage over crop plants. At this time, we do not know if there are common signals produced that give some plants a competitive advantage over others.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA826510. Soil and plant data are available in USGS data release https://doi.org/10.5066/P99W7Y45.
Author contributions
CN, NP, and MB conceived and designed the research. CN performed the bioinformatics and statistical analyses for the bacterial community data. NP collected the field data, plant identification, and statistical analyses of the plant and soil data. CN wrote the article with significant contribution from NP. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the US Geological Survey. The USDA National Institute of Food and Agricultural Hatch grant IND01825 provided partial funding to CN. Any use of trade, product, or firm names was for descriptive purposes only and did not imply endorsement by the U.S. Government.
Acknowledgments
We thank Erin Argylan and Todd Thompson for assistance with dune formation dates, Andrew Davies and Justin Chapelle for collecting the field samples, and Stephanie Grebinoski for laboratory assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.999099/full#supplementary-material
Footnotes
References
Alfaro, F. D., Manzano, M., Marquet, P. A., and Gaxiola, A. (2017). Microbial communities in soil chronosequences with distinct parent material: the effect of soil pH and litter quality. J. Ecol. 105, 1709–1722. doi: 10.1111/1365-2745.12766
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Anderson, M. J., Ellingsen, K. E., and Mcardle, B. H. (2006). Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693. doi: 10.1111/j.1461-0248.2006.00926.x
Argyilan, E. P., Johnston, J. W., Lepper, K., Monaghan, G. W., and Thompson, T. A. (2018). “Lake level, shoreline, and dune behavior along the Indiana southern shore of Lake Michigan,” in Ancient oceans, orogenic uplifts, and glacial ice: Geologic crossroads in America’s heartland: Geological society of america field guide, Vol. 51, ed. L. J. Florea. 181–203. doi: 10.1130/2018.0051(08)
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Bickford, W. A., Goldberg, D. E., Zak, D. R., Snow, D. S., and Kowalski, K. P. (2022). Plant effects on and response to soil microbes in native and non-native Phragmites australis. Ecol. Appl. 32:e2565. doi: 10.1002/eap.2565
Black, C. A. (1965). Methods of Soil Analysis. Madison, WIS: American Society of Agronomy. doi: 10.2134/agronmonogr9.1
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callaway, R. M., and Ridenour, W. M. (2004). Novel weapons: invasive success and the evolution of increased competitive ability. Front. Ecol. Environ. 2, 436–443. doi: 10.1890/1540-9295(2004)002[0436:NWISAT]2.0.CO;2
Cline, L. C., and Zak, D. R. (2015). Initial colonization, community assembly and ecosystem function: fungal colonist traits and litter biochemistry mediate decay rate. Mol. Ecol. 24, 5045–5058. doi: 10.1111/mec.13361
Coats, V. C., and Rumpho, M. E. (2014). The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front. Microbiol. 5:368. doi: 10.3389/fmicb.2014.00368
Cole, K. L., Engstrom, D. R., Futyma, R. P., and Stottlemyer, R. (1990). Past atmospheric deposition of metals in northern Indiana measured in a peat core from Cowles Bog. Environ. Sci. Technol. 24, 543–549. doi: 10.1021/es00074a013
Consortium of Midwestern Herbaria (2022). Available online at: https://midwestherbaria.org/portal/index.php (accessed March 10, 2022).
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Esser, K. B., Helmke, P. A., and Bockheim, J. G. (1991). Trace element contamination of soils in the Indiana Dunes. J. Environ. Qual. 20, 492–496. doi: 10.2134/jeq1991.00472425002000020025x
Fang, K., Bao, Z.-S.-N., Chen, L., Zhou, J., Yang, Z.-P., Dong, X.-F., et al. (2019). Growth-promoting characteristics of potential nitrogen-fixing bacteria in the root of an invasive plant Ageratina adenophora. PeerJ 7:e7099. doi: 10.7717/peerj.7099
Fryer, J. L. (2011). Celastrus orbiculatus. Fire Effects Information System. Available online at: https://www.fs.usda.gov/database/feis/plants/vine/celorb/all.html (accessed March 10, 2022).
Gaiero, J. R., Tosi, M., Bent, E., Boitt, G., Khosla, K., Turner, B. L., et al. (2021). Soil microbial communities influencing organic phosphorus mineralization in a coastal dune chronosequence in New Zealand. FEMS Microbiol. Ecol. 97:fiab034. doi: 10.1093/femsec/fiab034
Garrido-Oter, R., Nakano, R. T., Dombrowski, N., Ma, K.-W., Mchardy, A. C., and Schulze-Lefert, P. (2018). Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24, 155–167.e15. doi: 10.1016/j.chom.2018.06.006
Greenberg, C. H., Smith, L. M., and Levey, D. J. (2001). Fruit fate, seed germination and growth of an invasive vine - an experimental test of ‘sit and wait’ strategy. Biol. Invasions 3, 363–372. doi: 10.1023/A:1015857721486
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9.
Hansel, A. K., Mickelson, D. M., Schneider, A. F., and Larsen, C. E. (1985). “Late wisconsinan and holocene history of the Lake Michigan Basin,” in Quaternary Evolution of the Great Lakes, eds P. F. Karrow and P. E. Calkin (Ottawa, ON: Geological Association of Canada).
Kowalski, K. P., Bacon, C., Bickford, W., Braun, H., Clay, K., Leduc-Lapierre, M., et al. (2015). Advancing the science of microbial symbiosis to support invasive species management: a case study on Phragmites in the Great Lakes. Front. Microbiol. 6:95. doi: 10.3389/fmicb.2015.00095
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-Based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Leicht-Young, S. A., Healy, R., and Del Tredici, P. (2016). Observation of mycorrhizal colonization in roots in natural populations of Celastrus orbiculatus Thunb. J. Torrey Bot. Soc. 143, 322–324.
Leicht-Young, S. A., O’donnell, H., Latimer, A. M., and Silander, J. A. Jr (2009). Effects of an invasive plant species, Celastrus orbiculatus, on soil composition and processes. Am. Midland Nat. 161, 219–231. doi: 10.1674/0003-0031-161.2.219
Leicht-Young, S. A., and Pavlovic, N. B. (2012). Encroachment of oriental bittersweet into Pitcher’s thistle habitat. Nat. Areas J. 32, 171–176. doi: 10.3375/043.032.0206
Leicht-Young, S. A., Pavlovic, N. B., and Grundel, R. (2013). Susceptibility of eastern US habitats to invasion of Celastrus orbiculatus (oriental bittersweet) following fire. Forest Ecol. Manag. 302, 85–96. doi: 10.1016/j.foreco.2013.03.019
Lichter, J. (1998a). Primary succession and forest development on coastal Lake Michigan sand dunes. Ecol. Monographs 68, 487–510. doi: 10.1890/0012-9615(1998)068[0487:PSAFDO]2.0.CO;2
Lichter, J. (1998b). Rates of weathering and chemical depletion in soils across a chronosequence of Lake Michigan sand dunes. Geoderma 85, 255–282. doi: 10.1016/S0016-7061(98)00026-3
Liu, Z., Desantis, T. Z., Andersen, G. L., and Knight, R. (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36:e120. doi: 10.1093/nar/gkn491
Loria, R., Kers, J., and Joshi, M. (2006). Evolution of plant pathogenicity in Streptomyces. Ann. Rev. Phytopathol. 44, 469–487. doi: 10.1146/annurev.phyto.44.032905.091147
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Macik, M., Gryta, A., and Frac, M. (2020). Biofertilizers in agriculture: an overview on concepts, strategies and effects on soil microorganisms. Adv. Agron. 162, 31–87. doi: 10.1016/bs.agron.2020.02.001
Mandal, S., Van Treuren, W., White, R., Eggesbø, M., Knight, R., and Peddada, S. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
McCune, B., and Mefford, M. J. (2006). PC-ORD. Multivariate Analysis of Ecological Data. MjM Software: Gleneden Beach, OR.
Nelson, L. A., Cade-Menun, B. J., Walker, I. J., and Sanborn, P. (2020). Soil hhosphorus dynamics across a holocene chronosequence of Aeolian sand dunes in a hypermaritime environment on Calvert Island, BC, Canada. Front. Forests Global Change 3:83. doi: 10.3389/ffgc.2020.00083
Nelson, L. A., Sanborn, P., Cade-Menun, B. J., Walker, I. J., and Lian, O. B. (2021). Pedological trends and implications for forest productivity in a Holocene soil chronosequence, Calvert Island, British Columbia, Canada. Canadian J. Soil Sci. 101, 654–672. doi: 10.1139/cjss-2021-0033
Nossa, C. W., Oberdorf, W. E., Yang, L., Aas, J. A., Paster, B. J., Desantis, T. Z., et al. (2010). Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 16, 4135–4144. doi: 10.3748/wjg.v16.i33.4135
Olson, J. S. (1958). Rates of succession and soil changes on Southern Lake Michigan sand dunes. Botanical Gazette 119, 125–170. doi: 10.1086/335973
Page, A. L., Miller, R. H., and Keeney, D. R. (1982). Methods of soil analysis: Part 2 -chemical and microbiological properties. Madison, WI: American Society of Agronomy, Soil Science Society of America.
Pascale, A., Proietti, S., Pantelides, I. S., and Stringlis, I. A. (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10:1741. doi: 10.3389/fpls.2019.01741
Pavlovic, N. B., Plampin, B., Tonkovich, G. S., and Hamilla, D. (2022). “Special flora and vegetation of the Indiana dunes national park”, in Natural Resource Reports. (in press).
Pavlovic, N. B., and Leicht Young, S. A. (2011). Are temperate mature forests buffered from invasive lianas? J. Torrey Botan. Soc. 138, 85–92. doi: 10.3159/10-RA-055.1
Pavlovic, N. B., Leicht-Young, S. A., and Grundel, R. (2016). Oriental bittersweet (Celastrus orbiculatus): Spreading by fire. For. Ecol. Manage. 364, 183–194. doi: 10.1016/j.foreco.2015.12.036
Pirnay, J. P., Ferry, T., and Resch, G. (2022). Recent progress toward the implementation of phage therapy in Western medicine. FEMS Microbiol. Rev. 46:fuab040. doi: 10.1093/femsre/fuab040
Pollmann, T., Tsukamoto, S., Frechen, M., and Giani, L. (2020). Rate of soil formation in Arenosols of dunes on Spiekeroog Island (Germany). Geoderma Regional 20:e00246. doi: 10.1016/j.geodrs.2019.e00246
R Core Team (2022). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Shahrtash, M., and Brown, S. P. (2021). A path forward: promoting microbial-based methods in the control of invasive plant species. Plants 10:943. doi: 10.3390/plants10050943
Soil Science Society of America (2011). Recommended Chemical Soil Test Procedures for the North Central Region (NCR-13). Columbia, MI: North Central Regional Research.
Tombeur, F., Turner, B. L., Laliberté, E., Lambers, H., and Cornelis J T. (2020). Silicon dynamics during 2 million years of soil development in a coastal dune chronosequence under a Mediterranean climate. Ecosystems 23, 1614–1630. doi: 10.1007/s10021-020-00493-9
Trognitz, F., Hackl, E., Widhalm, S., and Sessitsch, A. (2016). The role of plant-microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 92, fiw138. doi: 10.1093/femsec/fiw138
Turner, B. L., Zemunik, G., Laliberté, E., Drake, J. J., Jones, F. A., and Saltonstall, K. (2019). Contrasting patterns of plant and microbial diversity during long-term ecosystem development. J. Ecol. 107, 606–621. doi: 10.1111/1365-2745.13127
Venturini, C., Fabijan, A. P., Lubian, A. F., Barbirz, S., and Iredell, J. (2022). Biological foundations of successful bacteriophage therapy. EMBO Mol. Med. 14:e12435. doi: 10.15252/emmm.202012435
Vurukonda, S., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19:952. doi: 10.3390/ijms19040952
Wilhelm, G., and Rericha, L. (2017). Flora of the Chicago Region: A Floristic and Ecological Synthesis. Indianapolis, Ind: Indiana Academy of Science.
Wilschut, R. A., and Geisen, S. (2021). Nematodes as drivers of plant performance in natural systems. Trends Plant Sci. 26, 237–247. doi: 10.1016/j.tplants.2020.10.006
Ye, M., Sun, M. M., Huang, D., Zhang, Z. Y., Zhang, H., Zhang, S. T., et al. (2019). A review of bacteriophage therapy for pathogenic bacteria inactivation in the soil environment. Environ. Int. 129, 488–496. doi: 10.1016/j.envint.2019.05.062
Yeoh, Y. K., Dennis, P. G., Paungfoo-Lonhienne, C., Weber, L., Brackin, R., Ragan, M. A., et al. (2017). Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8:215. doi: 10.1038/s41467-017-00262-8
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and All-species Living Tree Project (LTP) taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zaya, D. N., Leicht-Young, S. A., Pavlovic, N. B., Feldheim, K. S., and Ashley, M. S. (2015). Genetic characterization of invasion, hybridization, and native species decline in Celastrus. Biol. Invasions 17, 2975–2988. doi: 10.1007/s10530-015-0926-z
Keywords: ecosystem, invasive species, soil chronosequence, integrated pest management, plant-microbe interactions, Celastrus orbiculatus
Citation: Nakatsu CH, Pavlovic NB and Byappanahalli MN (2022) Bacteria common to rhizosphere communities of Asiatic bittersweet across a post-glacial landscape. Front. Ecol. Evol. 10:999099. doi: 10.3389/fevo.2022.999099
Received: 20 July 2022; Accepted: 26 October 2022;
Published: 16 November 2022.
Edited by:
Arnaldo Marín, University of Murcia, SpainReviewed by:
Xingang Zhou, Northeast Agricultural University, ChinaTong Li, Zhengzhou University, China
At least a portion of this work is authored by Noel B. Pavlovic and Muruleedhara N. Byappanahalli on behalf of the U.S. Government and as regards Noel B. Pavlovic, Muruleedhara N. Byappanahalli and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cindy H. Nakatsu, Y25ha2F0c3VAcHVyZHVlLmVkdQ==
†These authors have contributed equally to this work
 Cindy H. Nakatsu
Cindy H. Nakatsu Noel B. Pavlovic
Noel B. Pavlovic Muruleedhara N. Byappanahalli
Muruleedhara N. Byappanahalli