- 1Department of Microbiology, University of Georgia, Athens, GA, United States
- 2Department of Biology, Georgia Southern University, Statesboro, GA, United States
- 3Department of Geology and Geography, Georgia Southern University, Statesboro, GA, United States
Introduction
Aboveground plant surfaces (the phyllosphere) arguably represent the largest terrestrial habitat at ~6.4 × 108 km2 (Morris and Kinkel, 2002), which is double the global land surface (Vorholt, 2012). Microbes flourish in the phyllosphere, including bacteria (Redford et al., 2010), fungi (Kembel and Mueller, 2014), and archaea (Finkel et al., 2011), although bacteria generally dominate (Andrews and Harris, 2000). The phyllosphere bacterial community plays major roles in shaping plant ecophysiology, including: phytopathogeny (Saleem et al., 2017), leaf litter decomposition (Voríškova and Baldrian, 2013), and trace gas emissions (Bringel and Couée, 2015). While much attention has focused on how phyllosphere bacterial communities affect plant-atmosphere interactions (Remus-Emsermann and Schlechter, 2018), a current knowledge gap exists regarding how bacterial community composition in precipitation is altered by contact with the phyllosphere.
Vegetation significantly alters the amount and pattern of mass and energy inputs to the land surface. The phyllosphere is washed with tens to hundreds of liters over a single storm (Levia et al., 2011), sometimes in <0.25 h (Keim and Skaugset, 2004). When rainfall contacts forest canopies, it reaches the surface via two hydrologic fluxes: throughfall (TF) and stemflow (SF). TF is the portion of rain that drips from the canopy or passes through gaps, while SF is that which contacts and subsequently flows down stems, branches, and trunks. Recent work has shown that TF and SF contain up to 2 orders of magnitude higher concentrations of bacterial cells than open rainfall, resulting in >1016 cells ha−1 being delivered to the forest floor annually (Bittar et al., 2018). No work has assessed the taxonomic identities of bacteria transported by TF and SF, leaving open questions with strong ecohydrological implications: (1) Which bacterial taxa are TF and SF transporting to the surface? and, (2) Does the bacterial composition of TF, SF, and rainfall differ? Thus, this report examines how rainfall bacterial community composition is altered by interaction with Quercus virginiana (Mill., southern live oak) hosting the epiphyte, Tillandsia usneoides (L., Spanish moss) for 10 storms between March-September 2016 (Table S1). Sampled storms were evenly spread throughout the season (Figure S1). Bacterial community composition between rainfall, TF, and SF is compared, the potential sources of bacterial taxa are discussed, and additional questions generated by these measurements are raised.
Methods
Water samples were collected at the Skidaway Institute of Oceanography, Savannah, GA, USA (31.99°N, 81.02°W). Rain and throughfall samplers (0.18 m2, 0.5 m height, HDPE bins) were deployed immediately before each storm. Throughfall samplers were distributed beneath epiphyte-covered and bare canopy areas. Stemflow was collected by polyethylene tubing (2.5 cm diameter), cut lengthwise to function as gutters, and installed at 1.4 m height around the stems. All samplers were pre-cleaned with pH 2 ultrapure water (Milli-Q, Millipore) and triple-rinsed. Water samples were collected within 8 h of storm end, then 10–60 mL of each sample was filtered through 0.2 μm hydrophilic polypropylene syringe filters. Filters were refrigerated (4°C) in the dark for no more than 24 h until DNA extraction using PowerWater DNA Isolation Kits (Mo-BIO Laboratories) following manufacturer protocols.
PCR inhibitors were removed from extracted DNA samples using Omega Bio-Tek MagBind® Total Pure NGS Kits (Norcross, GA, USA). A 1:1.2 sample to beads volume ratio was used to isolate bacterial genomic DNA. The V4 region of the 16S rRNA gene was amplified in each sample following methods in Tinker and Ottesen (2016), and submitted to the Georgia Genomics Facility for sequencing (Illumina MiSeq 250 × 250 bp, San Diego, CA). Sequences were analyzed using the mothur v.1.35.1 (Schloss et al., 2009) and MiSeq standard operating protocol with the following changes: 1 ambiguous base per read was allowed. SILVA release 123 was used for sequence alignment (Pruesse et al., 2007; Quast et al., 2012; Yilmaz et al., 2013) and the August 2013 Greengenes release was used for taxonomic classification with a bootstrap value of 70 (DeSantis et al., 2006; Wang et al., 2007; McDonald et al., 2012). Sequences were clustered into 97% identity operational taxonomic units (OTUs), and all samples were rarified to a constant depth of 5,564 sequences. Nonmetric MultiDimensional Scaling (NMDS) was calculated using weighted Bray-Curtis dissimilarities based on 97% OTUs after rarefication. Synoptic variables for each storm were determined using the US-NOAA's HYSPLIT atmospheric transport and dispersion model (Stein et al., 2015) under “normal” trajectory settings with GDAS 0.5 meteorology.
Results and Discussion
Bacterial Composition of Rainfall, Throughfall, and Stemflow
8,501,343 16S rRNA gene sequences from 174 samples passed quality filters, averaging 48,858 reads and 605 OTUs per sample (Table S2). The predominant phyla in all fluxes were Proteobacteria, Bacteriodetes, Firmicutes, Actiniobacteria, and OD1 (Figure 1). Across samples, Proteobacteria was the most abundant phylum, in some cases exceeding 50% of sequences (Figure 1). Bacteroidetes accounted for the next largest group of identifiable bacterial families at 15%, followed by Actinobacteria at 5–10% (Figure 1).
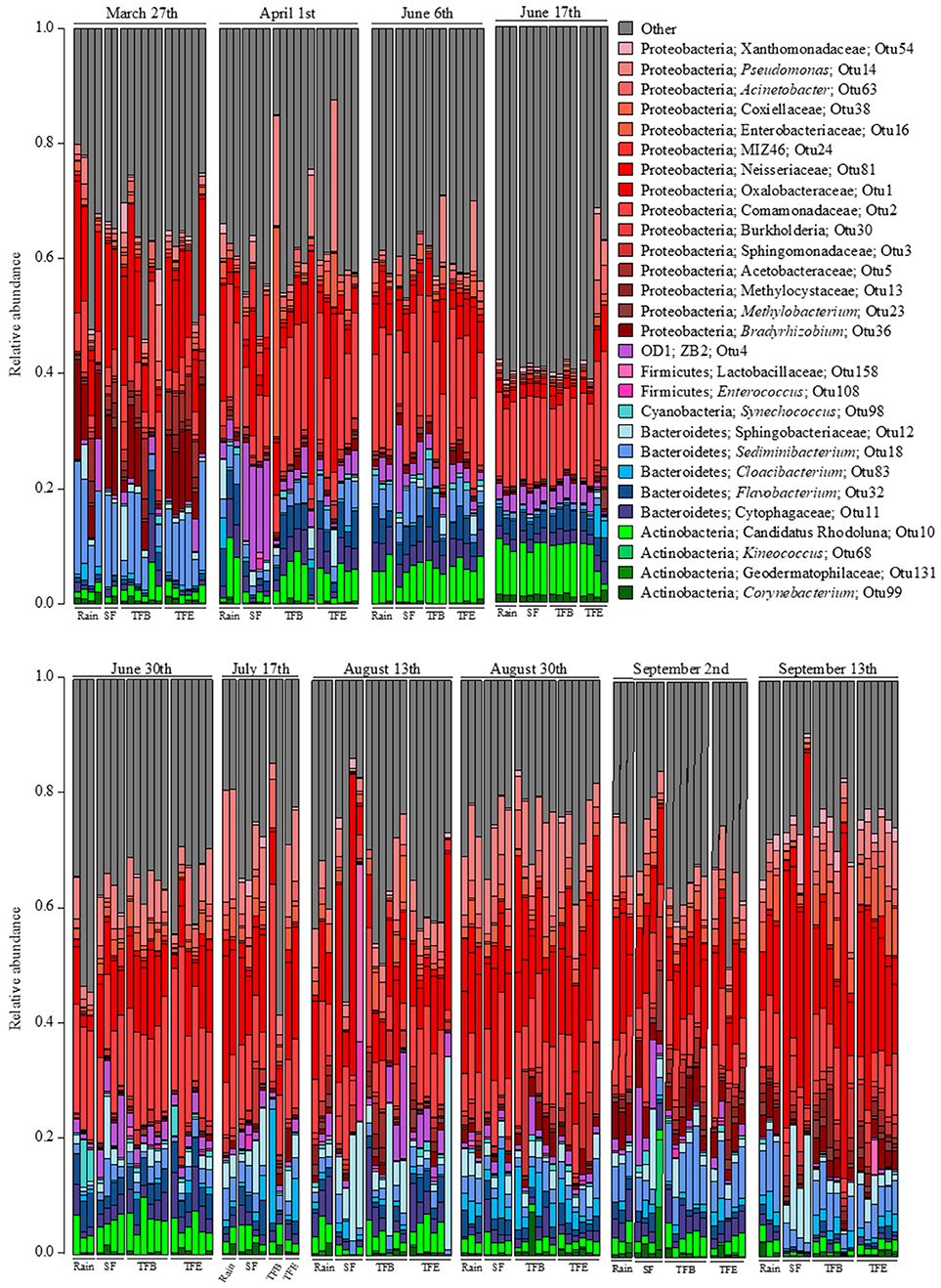
Figure 1. Relative abundances of taxa at the family level for each flux in each sampled storm. Families observed at <5% relative abundance in all samples were combined into the “other” category.
Many of these bacterial taxa are found as aerosols that catalyze ice formation, cloud growth and subsequent “bioprecipitation,” as well as on the phyllosphere. In particular, Proteobacteria are abundant in the phyllosphere (Vorholt, 2012) and in aerosols (Radosevich et al., 2002). Among the Proteobacteria, Pseudomonadaceae generally accounted for a larger proportion in bare and epiphyte throughfall (TFB, TFE, respectively), 12%, compared to rainfall and SF, 5%, or less (Figure 1). Pseudomonas is common in aerosols above plant canopies and is active in ice nucleation and cloud microphysical processes leading to bioprecipitation (Morris et al., 2014). Other Proteobacteria families (Xanthomodaceae, Moraxellaceae, Enterobacteriaceae, etc.) are considered primary biological aerosols and have been found to be abundant down-wind from urban environments (Després et al., 2012)—like Savannah (Georgia, USA) near our site. Atmospheric transport of Acidobacteria to the tree canopy is also plausible as these have been observed in cloud water (Kourtev et al., 2011) and hailstones (Šantl-Temkiv et al., 2013); however, this phylum (and Actinobacteria) can be abundant on forest canopy surfaces (Kim et al., 2012).
Variability in Bacterial Community Structure Between Fluxes
Bacterial alpha and beta diversity was similar among samples of rainfall, SF, and TF from bare and epiphyte cover (Figure 2A). Dissimilarity, whether weighted or unweighted, had similar medians, interquartile ranges, non-outlier ranges, and even lower-level outliers across sample types (Figures 2B,C). Cross-flux variability was similar to intra-flux variability (see Figure S2). Thus, no trends in dissimilarity were observed in flux comparisons. These findings agree with an OTU-level permutational multivariate analysis of variance (PERMANOVA) that found no significant difference between fluxes (F = 1.29, p = 0.07). The path rainwater travels through the canopy does not appear to determine the community structure of bacteria delivered to the forest surface during storms. These findings are counterintuitive to previous findings; specifically, that (i) TF and SF bacterial concentrations are two orders of magnitude greater than in open rainfall at the same site (Bittar et al., 2018), (ii) SF and TF primarily flow along bark and leaf surfaces, respectively, and (iii) bark and leaf surfaces have been reported to host distinct bacterial communities (Lambais et al., 2014). It is seemingly paradoxical that rainwater entrained as TF and SF in the canopy becomes enriched in the same bacterial taxa carried by the rainwater itself. However, aerosolized bacteria that become cloud condensation nuclei (Bauer et al., 2003), and eventually rain, are also constantly deposited onto terrestrial surfaces during the dry period between storms (Tong and Lighthart, 2000). The ostensible paradox is, thus, resolved if the rainwater entrained as TF and SF is being enriched with similar bacterial taxa that were atmospherically deposited onto the canopy prior to the storm.
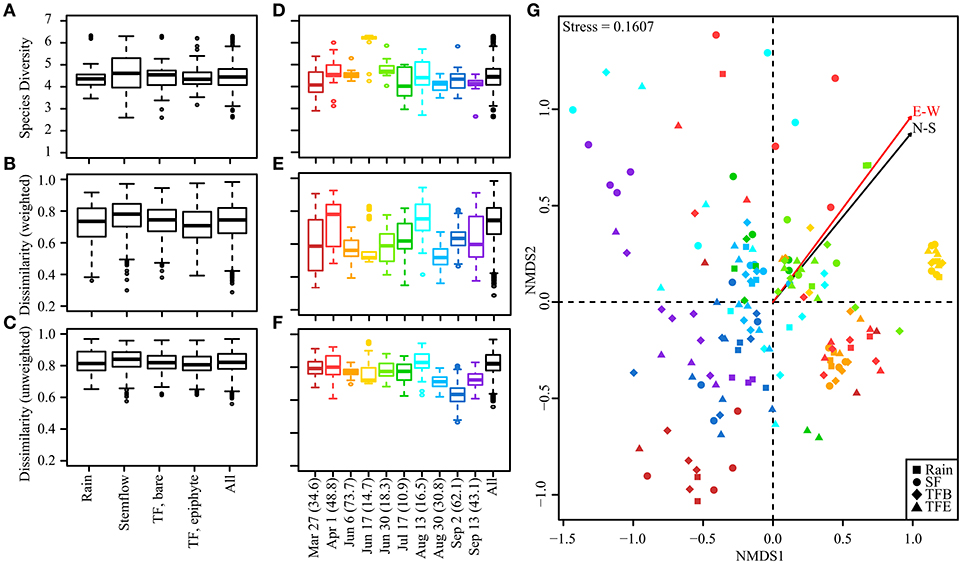
Figure 2. Boxplots presenting the median (bar), interquartile range (box), non-outlier range (whiskers) and outliers (circles) for Shannon diversity and Bray-Curtis dissimilarity indices. Alpha diversity (Shannon index) and beta diversity (abundance-weighted and unweighted Bray-Curtis dissimilarity) is plotted for (A–C) flux types across storms and (D–F) individual storms. Bacterial communities from samples collected from all storms represented on a 2-D NMDS plot (G). Points are colored according to storm (see D–F for color key) while shapes show flux/sample type. Rainfall amount (mm) for each storm is provided parenthetically in the x-axis of (D–F). Arrows represent fits for the N-S and E-W components of HYSPLIT back-trajectories of the storm source (Table S1).
This raises questions regarding the dynamics of the phyllosphere bacterial community. For instance, are there two community factions in the phyllosphere: weakly-attached and/or atmospherically-deposited bacteria vs. strongly-attached phyllosphere bacteria? What determines which of these groups a bacterium falls into? Do specific factions of phyllosphere bacteria form biofilms that protect them from being scoured away by rainwater, and why do these protective strategies not extend to atmospherically-deposited bacteria? Findings by Lymperopoulou et al. (2016) suggest that bacterial taxa located on plant leaf surfaces impact bacterial communities found in surrounding air, supporting the hypothesis that loosely-associated phyllosphere bacteria are shared with environments surrounding the plant surface. This report and other work at this site (Bittar et al., 2018) suggests that weakly-attached bacteria are numerically abundant. However, the level of activity of these bacteria on the leaf surface and how they shape the structure and function of the total phyllosphere community remains in question.
Interstorm Variability in Bacterial Community Composition
In contrast to the similarity of bacterial community structure among fluxes, large variability was observed in bacterial community composition between storms (Figures 2D–F). These differences were found to be significant by PERMANOVA (F = 7.48, p = 0.001). Similarity in bacterial community composition between storms was visualized using NMDS, where proximity of samples within a 2-dimensional space indicates community similarity (Figure 2G). The stress value (0.16) of this analysis is below the 0.2 threshold typically indicative of good representation in the selected space. The observation that bacterial communities from the same hydrologic flux (rainfall, TF, or SF) do not cluster together suggests low similarity within sample types (Figure 2G), indicating again that the path rain takes through the phyllosphere to the forest floor (TF vs. SF) does not strongly influence bacterial community composition. However, interstorm variability appears to influence sample clustering by NMDS. HYSPLIT back-trajectories for each storm is strongly predictive of the positioning of individual storms within the NMDS (N-S: r2 = 0.5534, p = 9.999e-05; E-W: r2 = 0.3955, p = 9.999e-05). The N-S and E-W components of the storm trajectories show similar relationships with community data, and PERMANOVA found both trajectory sets to be significant (p = 0.001), suggesting an overall trend in differential community structure for storms with NE vs. SW trajectories. One event (17-Jun-2016) exhibited a distinct cluster within the NMDS plots, perhaps due to its radically different back-trajectory (Figure 2G). However, this storm is not solely responsible for the observed relationships, as NMDS analyses that exclude this storm still show significant relationships between sample positions and storm HYSPLIT trajectories.
Biogeochemical Implications
Previous work at this site demonstrated that SF may supply a rapid and substantial bacterial flux (30 × 109 cells m−2 h−1) to soils during storms (Bittar et al., 2018). Members of the most abundant phylum in canopy-derived hydrologic fluxes, Proteobacteria, play key roles in carbon, sulfur, and nitrogen cycling (Kersters et al., 2006). Some examples found to be abundant in TF and SF include (i) members of the Oxalobacteriaceae, known to include bacteria that can fix atmospheric nitrogen, (ii) genera of Sphingomonadaceae, members of which can metabolize aromatic carbon compounds, and (iii) Enterobacteriaceae and Pseudomonadaceae, members of which can oxidize sulfur (Kersters et al., 2006). β- and γ-Proteobacteria (classes to which many of the taxa in our samples belong) are reported to be some of the most prominent genera in litter decomposition (Aneja et al., 2006), although Actinobacteria and Bacteroidetes (including genera identified in TF and SF) have been found to be abundant during litter decomposition as well (Tláskal et al., 2016). Although less abundant in TF and SF, Acidobacteria are capable of metabolizing a large range of organic carbon compounds, nitrate and nitrite reduction, forming biofilms, stabilizing soil structures, and producing antimicrobial compounds in soils (Ward et al., 2009). Together, these groups have the metabolic potential to shape litter degradation and biogeochemical cycles in the forest floor should they survive transport and become established in this environment.
Conclusions
We report the composition and structure of bacteria exchanged between the phyllosphere and forest floor during storm events. Bacterial communities, surprisingly, did not differ significantly in water sampled from various fluxes: rainfall (no canopy interaction), SF (bark-dominated flowpath), and TF (leaf- and epiphyte-dominated flowpaths). Significant differences in bacterial community composition were observed across all fluxes between storms. This suggests that storm characteristics may shape the bacterial community of sub-canopy rainfall. Findings indicate that, although SF and TF are enriched in bacteria compared to rainfall (Bittar et al., 2018), this enrichment may not represent a disruption in the local phyllosphere bacterial communities, along which TF and SF drain. Rather, TF and SF drainage during storms may primarily suspend and transport atmospherically-deposited biological aerosols. Future work should assess the influence of tree species and stand structure. Finally, should the bacteria transported by TF and SF be able to access or establish microbial communities at the forest floor, there may be broad biogeochemical implications for the litter layer, topsoil, and rhizosphere. Thus, we suggest future research assess the fate and potential function of microbes in TF and SF at the forest surface.
Author Contributions
JV and PP conceived and designed the sampling plan. PP collected samples and extracted DNA. MT performed the library preparation sequencing and related analyses. EO directed and trained MT and participated in the design and coordination of 16S rRNA gene sequencing library preparation and data analysis. All authors contributed to the manuscript writing.
Funding
The study was supported by NSF-0841146.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was performed with the support of the Georgia Genomics and Bioinformatics Core at the University of Georgia. Thanks to Drs. J. Scott Harrison and Tiehang Wu at Georgia Southern University for assistance in the lab and helpful comments during writing the study, the staff at SkIO for maintaining and securing the research site, and students who assisted in site maintenance or sample processing (Elliott Lewis, Julius Lewis, L. Dean Moore, Ansley Whitetree). All sequences associated with this work have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession SRP133049.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2018.00007/full#supplementary-material
References
Andrews, J. H., and Harris, R. F. (2000). The ecology and biogeography of microorganisms on plant surfaces. Ann. Rev. Phytopathol. 38, 145–180. doi: 10.1146/annurev.phyto.38.1.145
Aneja, M. K., Sharma, S., Fleischmann, F., Stich, S., Heller, W., Bahnweg, G., et al. (2006). Microbial colonization of beech and spruce litter—Influence of decomposition site and plant litter species on the diversity of microbial community. Microb. Ecol. 52, 127–135. doi: 10.1007/s00248-006-9006-3
Bauer, H., Giebl, H., Hitzenberger, R., Kasper-Giebl, A., Reischl, G., Zibuschka, F., et al. (2003). Airborne bacteria as cloud condensation nuclei. J. Geophys. Res. 108:4658. doi: 10.1029/2003JD003545
Bittar, T., Pound, P., Whitetree, A., Moore, L. D., and Van Stan, J. T. (2018). Estimation of throughfall and stemflow bacterial flux in a subtropical oak-cedar forest. Geophys. Res. Lett. 45, 1410–1418. doi: 10.1002/2017GL075827
Bringel, F., and Couée, I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6:486. doi: 10.3389/fmicb.2015.00486
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Després, V. R., Huffman, J. A., Burrows, S. M., Hoose, C., Safatov, A. S., Buryak, G., et al. (2012). Primary biological aerosol particles in the atmosphere: a review. Tellus B 64:15598. doi: 10.3402/tellusb.v64i0.15598
Finkel, O. M., Burch, A. Y., Lindow, S. E., Post, A. F., and Belkin, S. (2011). Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Environ. Microbiol. 77, 7647–7655. doi: 10.1128/AEM.05565-11
Keim, R. F., and Skaugset, A. E. (2004). A linear system model of dynamic throughfall rates beneath forest canopies. Water Resour. Res. 40, W05208. doi: 10.1029/2003WR002875
Kembel, S. W., and Mueller, R. C. (2014). Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92, 303–311. doi: 10.1139/cjb-2013-0194
Kersters, K., De Vos, P., Gillis, M., Swings, J., Vandamme, P., and Stackebrandt, E. (2006). Introduction to the Proteobacteria. Prokaryotes 5, 3–37. doi: 10.1007/0-387-30745-1_1
Kim, M., Singh, D., Lai-Hoe, A., Go, R., Rahim, R. A., Ainuddin, A. N., et al. (2012). Distinctive phyllosphere bacterial communities in tropical trees. Microb. Ecol. 63, 674–681. doi: 10.1007/s00248-011-9953-1
Kourtev, P. S., Hill, K. A., Shepson, P. B., and Konopka, A. (2011). Atmospheric cloud water contains a diverse bacterial community. Atmos. Environ. 45, 5399–5405. doi: 10.1016/j.atmosenv.2011.06.041
Lambais, M. R., Lucheta, A. R., and Crowley, D. E. (2014). Bacterial community assemblages associated with the phyllosphere, dermosphere, and rhizosphere of tree species of the Atlantic forest are host taxon dependent. Microb. Ecol. 68, 567–574. doi: 10.1007/s00248-014-0433-2
Levia, D. F., Keim, R. F., Carlyle-Moses, D. E., and Frost, E. E. (2011). “Throughfall and stemflow in wooded ecosystems,” in Forest Hydrology and Biogeochemistry, eds D. F. Levia, D. E. Carlyle-Moses, and T. Tanaka (Heidelberg, Springer), 425–443. doi: 10.1007/978-94-007-1363-5_21
Lymperopoulou, D. S., Adams, R. I., and Lindow, S. E. (2016). Contribution of vegetation to the microbial composition of nearby outdoor air. Appl. Environ. Microb. 82, 3822–3833. doi: 10.1128/AEM.00610-16
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Morris, C. E., Conen, F., Huffman, J. A., Phillips, V., Pöschl, U., and Sands, D. C. (2014). Bioprecipitation: a feedback cycle linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob. Change Biol. 20, 341–351. doi: 10.1111/gcb.12447
Morris, C. E., and Kinkel, L. L. (2002). “Fifty years of phyllosphere microbiology: significant contributions to research in related fields,” in Phyllosphere Microbiology,” eds S. E. Lindow, E. I. Hecht-Poinar, and V. Elliot (St. Paul, MN: APS Press), 365–375.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Radosevich, J. L., Wilson, W. J., Shinn, J. H., DeSantis, T. Z., and Andersen, G. L. (2002). Development of a high-volume aerosol collection system for the identification of air-borne micro-organisms. Lett. Appl. Microbiol. 34, 162–167. doi: 10.1046/j.1472-765x.2002.01048.x
Redford, A. J., Bowers, R. M., Knight, R., Linhart, Y., and Fierer, N. (2010). The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 12, 2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x
Remus-Emsermann, M. N. P., and Schlechter, R. O. (2018). Phyllosphere microbiology: at the interface between microbial individuals and the plant host. N. Phytol. 21, 1327–1333. doi: 10.1111/nph.15054
Saleem, M., Meckes, N., Pervaiz, Z. H., and Traw, M. B. (2017). Microbial interactions in the phyllosphere increase plant performance under herbivore biotic stress. Front. Microbiol. 8:41. doi: 10.3389/fmicb.2017.00041
Šantl-Temkiv, T., Finster, K., Dittmar, T., Hansen, B. M., Thyrhaug, R., Nielsen, N. W., et al. (2013). Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS ONE 8:e53550. doi: 10.1371/journal.pone.0053550
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Stein, A. F., Draxler, R. R., Rolph, G. D., Stunder, B. J. B., and Cohen, M. D. (2015). NOAA's HYSPLIT atmospheric transport and dispersion modelling system. Bull. Amer. Meteorol. Soc. 96, 2059–2077. doi: 10.1175/BAMS-D-14-00110.1
Tinker, K. A., and Ottesen, E. A. (2016). The core gut microbiome of the American cockroach, Periplaneta Americana, is stable and resilient to dietary shifts. Appl. Environ. Microbiol. 82, 6603–6610. doi: 10.1128/AEM.01837-16
Tláskal, V., Voríšková, J., and Baldrian, P. (2016). Bacterial succession on decomposing leaf litter exhibits a specific occurrence pattern of cellulolytic taxa and potential decomposers of fungal mycelia. FEMS Microbiol. Ecol. 92:fiw177. doi: 10.1093/femsec/fiw177
Tong, Y., and Lighthart, B. (2000). The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Sci. Technol. 32, 393–403. doi: 10.1080/027868200303533
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Voríškova, J., and Baldrian, P. (2013). Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486. doi: 10.1038/ismej.2012.116
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Keywords: throughfall, stemflow, plant-soil interactions, Quercus virginiana, epiphyte
Citation: Teachey ME, Pound P, Ottesen EA and Van Stan JT (2018) Bacterial Community Composition of Throughfall and Stemflow. Front. For. Glob. Change 1:7. doi: 10.3389/ffgc.2018.00007
Received: 14 September 2018; Accepted: 25 October 2018;
Published: 13 November 2018.
Edited by:
Steven George McNulty, USDA Southeast Climate Hubs, United StatesReviewed by:
James A. Coker, University of Maryland University College, United StatesSeyed Mohammad Moein Sadeghi, University of Tehran, Iran
Copyright © 2018 Teachey, Pound, Ottesen and Van Stan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John T. Van Stan, anZhbnN0YW5AZ2VvcmdpYXNvdXRoZXJuLmVkdQ==
†These authors have contributed equally to this work
 Morgan E. Teachey1†
Morgan E. Teachey1† John T. Van Stan
John T. Van Stan