- 1School of Food Science and Engineering, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
- 2Shandong Aojing Biotechnology Co., Ltd., Zoucheng, China
Sweet-tasting protein is a kind of biomacromolecule that has remarkable sweetening power and is regarded as the promising sugar replacer in the future. Some sweet-tasting proteins has been used in foods and beverages. However, the structure and function relationship of these proteins is still elusive, and guidelines for their protein engineering is limited. It is well-known that the sweet-tasting proteins bind to and activate the sweet taste receptor T1R2/T1R3, thus eliciting their sweetness. The “wedge-model” for describing the interaction between sweet-tasting proteins and sweet taste receptor to elucidate their sweetness has been reported. In this perspective article, we revealed that the intramolecular interaction forces in sweet-tasting proteins is directly correlated to their properties (sweetness and stability). This intramolecular interaction pattern, named as “protein sector,” refers to a small subset of residues forming physically connections, which cooperatively affect the function of the proteins. Based on the analysis of previous experimental data, we suggest that “protein sector” of sweet-tasting proteins is pivotal for their sweet properties, which are meaningful guidelines for the future protein engineering.
Introduction
Sweet-tasting proteins are originated from the natural plants and exhibit extraordinary sweetening power, which are regarded as suitable replacers of sugars and artificial sweeteners in the future to improve the health of human beings, such as the control of obesity, diabetes, and hyperlipemia (1). Eight sweet proteins have been characterized so far (miraculin, monellin, thaumatin, mabinlin, pentadin, curculin, brazzein, and neoculin), with three proteins monellin, brazzein, and thaumatin being well-studied. However, some sweet-tasting proteins have intrinsic shortcomings (e.g., low sweetness or thermostability) that limit their extensive applications. In recent years, protein engineering of sweet-tasting proteins to improve their performance has drawn more and more attention of researchers and entrepreneurs, and many variants of these proteins with modified properties have been constructed (2, 3).
Structural Features of Sweet-Tasting Proteins
The sweet-tasting proteins consist of about 50–200 amino acids, with approximate molecular weight range from 6,500 to 30,000 Da. Interestingly, although these proteins display same properties (eliciting sweet sensation), they have no sequence identity and structural similarity. Indeed, the dimensional structures of many sweet-tasting proteins have been solved with X-ray diffraction or nuclear magnetic resonance (NMR), which show diversified spatial folding architectures including α-helix, β-sheet and random coils (loops) (Figures 1A–F).
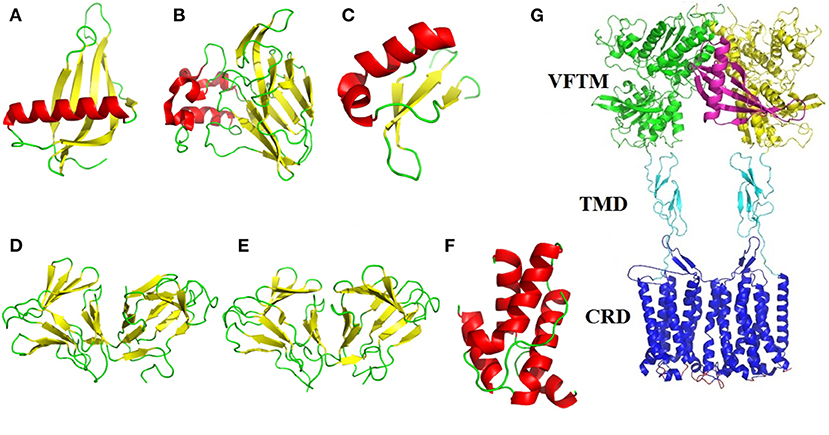
Figure 1. Structural illustration of sweet-tasting proteins and the sweet taste receptor. The three-dimensional structures of sweet-tasting proteins: (A) monellin (PDB: 2O9U); (B) thaumatin (PDB: 1RQW); (C) brazzein (PDB: 2LY5); (D) curculin (PDB: 2DPF); (E) neoculin (PDB: 2D04); (F) mabinlin II (PDB: 2DS2). The α-helix, β-sheet and loops in the structures were colored in red, yellow, and green, respectively. (G) Cartoon representation of the interaction between the sweet-tasting protein monellin (colored in purple) and the sweet taste receptor T1R2/T1R3. The VFTM (Venus flytrap module), CRD (cysteine-rich domain) and TMD (transmembrane domain) of the receptor are colored in green (T1R2) and yellow (T1R3), cyan and blue, respectively.
Monellin
Monellin is a sweet-tasting protein (molecular weight MW: 13,000 Da) about 3,000 times sweeter than sucrose on a weight basis, which was originally isolated from the fruit of the West African plant Dioscoreophyllum cumminsii Diels. The protein consists of two non-covalently associated polypeptide chains: an A chain of 44 amino acid residues and a B chain of 50 residues (4). A single-chain monellin (MNEI) was created by protein engineering in which the two natural chains are joined via a Gly-Phe dipeptide linker to improve its thermal stability (5). Interestingly, the spatial structures of native and single-chain monellin are very similar, which identically consist of a five-strand β-sheet partially “wrapped” around an α-helix (PDB: 4MON and 2O9U). Although the overall structure of monellin displays lower flexibility, the loops region in these structures show a high degree disorder, providing the structural plasticity that enables the protein to interact and optimize its large surface complementarity with the sweet taste receptor (6).
Brazzein
Brazzein is the smallest, heat-stable and intensely sweet protein (MW: 6,473 Da) derived from the ripe fruit of the West African plant Pentadiplandra brazzeana Baillon, which consists of 54 amino acid residues (7). The structure of brazzein was determined by NMR (PDB ID: 1BRZ and 2BRZ), which shows one short α-helix (residues 21–29) and three strands of antiparallel β-sheet held together by four disulfide bonds. The protein adopts a cysteine-stabilized αβ (CSαβ) fold stabilized by 17 interstrand α-helical hydrogen bonds and four disulfide bridges (8). The structure of brazzein (PDB: 4HE7) solved by X-ray diffraction is alike to its overall solution structure solved by NMR but with essential difference (2.0-2.2 Å rmsd for the Cα atoms) found in the loop and terminal regions (9). Furthermore, the brazzein fold exhibits similarity to a family of serine proteinase inhibitors based on the sequence comparisons, suggesting that brazzein could evolved from a serine proteinase inhibitor through a deletion mutation (8).
Thaumatin
Thaumatin is a 207 amino acids sweet-tasting protein (MW: 22,200 Da) with two major forms (I and II) and three minor forms (a, b, c), which is naturally derived from the fruit arils of a tropically grown plant Thaumatococcus daniellii Benth belonging to the family Marantaceae (10). The overall crystal structures of both natural and recombinant thaumatin I (PDB: 3AL7 and 2VHK) display three domains: a 11-strand flattened β-sandwich, a large disulfide-rich region and a small disulfide-rich region. The intramolecular eight disulfide bonds formed by sixteen cysteines are responsible for the stability of the protein (11). The quality of the 1.1 Å resolution of recombinant thaumatin I allowed the side chains of 20 residues to be modeled in two conformations and that of one residue (R76) to be modeled in three conformations. Study on the crystal structures of thaumation at different pHs has revealed that the increase in mobility of lysine residues as well as a loop region in domain II account for the pH-dependent sweetness of this sweet-tasting protein (12). Moreover, thaumatin has been approved and applied as both a sweetener and a favor-enhancer in many countries.
Mabinlin
Mabinlin is isolated from the seeds of Capparis masaikai Levl, which grows in subtropical regions within China. Based on its sequence, this sweet-tasting protein can be categorized into four members I, II, III, and IV (13). The mabinlin II (MW: 12,400 Da) consists of an A-chain of 33 amino acids and a B-chain of 72 amino acids, which are linked through two interchain disulfide bridges. The crystal structure of mabinlin II (PDB: 2DS2) was determined in 2008, which belongs to the “all alpha protein” in SCOP (Structural Classification of Proteins) classification. Specifically, the A-chain has two α-helixs and B-chain has three α-helixs (no β-sheet), and four disulfide bridges exist in the protein molecule (14). Interestingly, the separated B-chain can elicit the sweetness, whereas the A-chain can not, which is in accordance to the further findings that the B-chain with a unique (NL/I) tetralet motif is the sweetness determinant site, while the A-chain may play a role for the long aftertaste of this sweet-tasting protein.
Miraculin
Miraculin is a homodimeric sweet taste-modifying protein (191 amino acids, MW: 24,600 Da) which is isolated from the red berries of Richadella dulcifera, a shrub native to West Africa. Miraculin shows high amino acid sequence homology with the soybean trypsin inhibitor. The protein has no obvious taste at neutral pH. However, it has taste-modifying activity to convert sour stimuli to sweetness (15). Using human/mouse chimeric sweet taste receptors and molecular simulations, it has been revealed that miraculin binds with the human T1R2/T1R3 as an antagonist at neutral pH but functionally changes into an agonist at acidic pH, thus eliciting its sweet taste-modifying activity (16). Although the structure of miraculin is still not available, several crystal structures of miraculin-like proteins have been resolved, which show as a β-trefoil fold (PDB: 5YH4, IIR, etc.) (17).
Curculin and Neoculin
Curculin is isolated from Curculigo latifolia, a plant grown in Malaysia. The homodimeric form of this protein (114 amino acids, MW: 14,600 Da) exhibits sweet taste-modifying activity as miraculin (18). However, its heterodimeric isoform (named as neoculin) exhibits both sweet-tasting and taste-modifying activities (19). Both crystal structures of curculin (PDB: 2DPF) and neoculin (PDB: 2D04) have been determined, which adopt very similar backbone conformation and domain arrangement (20). It is revealed that curculin exhibits sweetness and taste-modifying activities through its partially overlapping but distinct molecular surfaces, which are suggested to be involved in the interaction with the sweet taste receptor.
Pentadin
Pentadin (500 times sweeter than sucrose on a weight basis, MW: 12,000 Da) has the same plant origin as brazzein (Pentadiplandra brazzeana Baillon), and its subunits are linked by an intramolecular disulfide bridge (21). The sweet properties of pentadin are more like that of monellin than that of thaumatin. However, there is no sequence and structural information reported about this sweet-tasting protein until now.
Eliciting the Sweetness: Interaction Between the Sweet-Tasting Proteins and Sweet Taste Receptor
How the sweet-tasting proteins elicit their sweetness has been an intriguing question for a long time. In 2001–2002, scientists revealed that the heterodimeric receptor T1R2/T1R3 located in the membrane of taste bud cells mediates the sweet taste sensation upon the stimulus of various sweeteners (22, 23). The sweeteners (including sweet-tasting proteins) bind to, interact with, activate the receptor, then trigger a series of signal cascades (G protein activation, phospholipase C-β2 motivation, Ca2+ release, cell depolarization, etc.), and ultimately elicit the sweet sensation (24). However, structural determination of this membrane protein G protein-coupled receptor (GPCR) is still a big challenge, and spatial information of sweetener-receptor complex is lacking now. Nevertheless, molecular modeling and docking have been extensively performed to investigate the sweetener-receptor interaction, which could be verified by further functional mutagenesis analysis(25–27).
The most popular model elucidating the interaction between sweet-tasting proteins and receptor is called as wedge-model, proposed by prof. Temussi, in which the surface charge complementarity between the sweet-tasting proteins (or its amino acids) and the sweet taste receptor mediates their interaction thus determining the sweetness of proteins (Figure 1G) (28). This model has been broadly accepted according to molecular simulation and experimental validation (29, 30).
Strategies for Optimization of the Properties of Sweet-Tasting Proteins
Guidance for protein engineering of the sweet-tasting proteins is primarily based on the above described wedge-model. Accordingly, mutated residues of the sweet-tasting protein variants were mainly focused on those on their protein surface, and removing negative charge or increasing positive charge is generally accompanied with improved sweetness, which is consistent with their charge complementarity with the interactive residues (negative charge) in sweet taste receptor. For instance, mutants E2N and Y65R of MNEI, E41K, D40K, and E53R of brazzein, and D21N of thaumatin with significantly improved sweetness have been constructed (31–35). Notably, structural calculations and quantitative structure-activity relationship (QSAR) investigations have been popularly performed in recent years to improve the properties of sweet-tasting proteins with some novel mutants having been identified (e.g., S76Y of MNEI) (36, 37).
“Proteins Sectors” is Correlated to the Sweetness of Sweet-Tasting Proteins
In 2009, Halabi et al. uncovered that biological properties of proteins arise from the cooperative action of their amino acid residues, and the pattern of residue cooperativity is generally called as “protein sector,” in which a small subset of residues forms an interactive architecture and physically connected networks, and each sector is physically connected in the tertiary structure and has a distinct functional role (38, 39). In recent years, other interdisciplinary methods and techniques have been adopted to identify the protein sectors (40, 41).
“Proteins sector” in sweet-tasting proteins has not been reported until now. By analyzing previous experimental data, we highlight herein that the intramolecular interaction forces in sweet-tasting proteins can significantly affect the sweetness of these proteins. For instance, a G16A mutation located in the core of sweet protein monellin could induce flexibility changes of protein surface via propagation effects mediated by hydrophobic interactions, which led to a 10-fold decrease of sweetness (42). Two mutants of MNEI Q28K/C41S/Y65R and E23Q/Q28K/C41S/Y65R were reported with around 1.5 to 2.5-fold enhancement of sweetness than the wild-type protein. We compared their solved structures (PDB: 5LC6 and 5LC7) with that of the wild-type (PDB: 1IV7) (43), and indicated that the mutated residue S41 in both mutants adopt different conformations relative to C41 in wild-type, which lead to formation of a hydrogen bond with a water molecule that connects mutated S41 to main chain atoms of P40, I38, and Y63 via hydrogen bonds (Figures 2A,B). Similar arrangement was also found for mutant Y65R that resulted in modified interactions in the protein (2). Therefore, it can be suggested that reorganizations of intramolecular interaction network (“protein sectors”) account for the conformations changes of the proteins and their orientation on the receptor, thus affecting their sweetness.
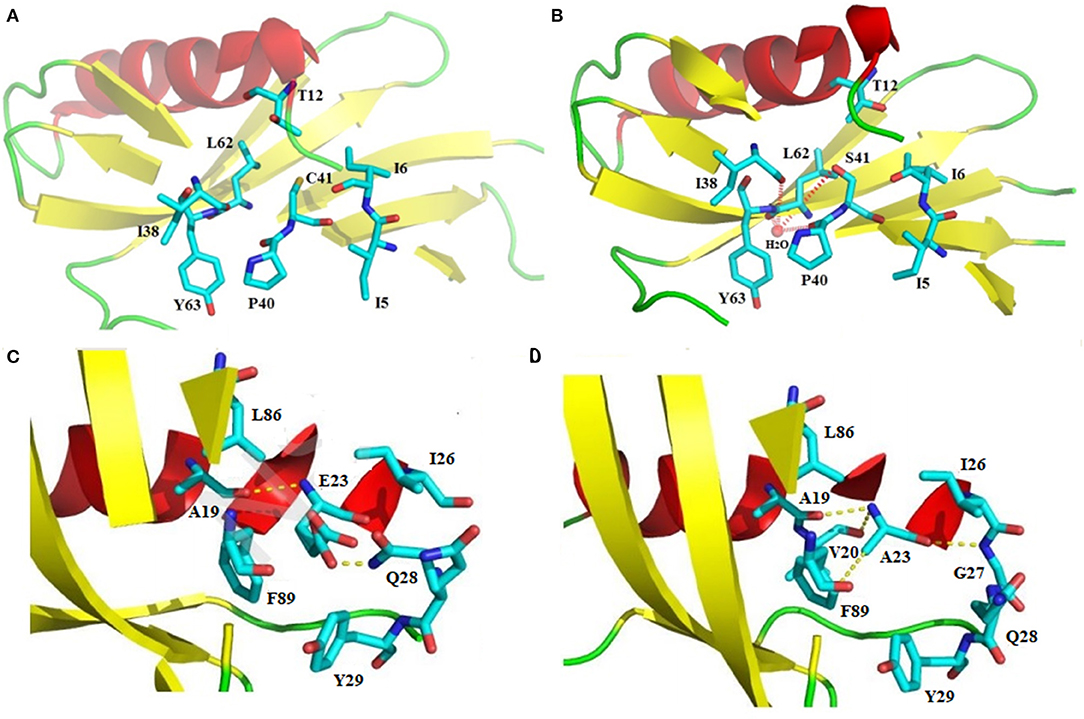
Figure 2. Structural illustration of the relationship between the intramolecular interaction patterns and the properties of sweet-tasting proteins. Formation of the intramolecular hydrogen bonds interaction network upon the C41S mutation of MNEI: (A) Spatial arrangement around C41 in the wild-type sweet-tasting protein monellin (PDB: 1IV7); (B) Formation of the intramolecular hydrogen bonds interaction network (red dashed lines) around the mutated S41, which accounts for the improved sweet potency of the mutants Q28K/C41S/Y65R and E23Q/Q28K/C41S/Y65R (PDB: 5LC6 and 5LC7). Comparison of the intramolecular interaction networks of the wild-type MNEI and its mutant E2N/E23A; (C) Intramolecualr interaction forces around the E23 site of the wild-type MNEI. The hydrogen bonds were indicated with yellow dashed lines (PDB: 2O9U); (D) Modified intramolecualr interaction network around the A23 site of E2N/E23A mutant (PDB: 5Z1P), which accounts for its significantly improved thermostability.
It was reported that the sweetness of a series of variants of the sweet-tasting protein brazzein is correlated to the patterns of hydrogen bonds detected by NMR spectroscopy. Specifically, three sweeter variants exhibited the same pattern of hydrogen bonds, whereas all three “non-sweet” variants lacked one hydrogen bond at the middle of the α-helix, where it is kinked, and one hydrogen bond in the middle of β-helixs II and III, where they are twisted (44). Similar structural rearrangements were also described in other variants of brazzein (45). These results highlight the significance of intramolecular interaction patterns (“protein sectors”) for the sweetness of sweet-tasting proteins.
Besides these findings, studies on multiple mutations indicated that there were combinatorial effects of mutated residues for the sweetness of sweet-tasting proteins. For example, the multiple-sites mutant H31R/E36D/E41A of sweet-tasting protein brazzein displayed significantly improved sweetness than those of three double-sites mutants (H31R/E36D, H31R/E41A, and E36D/E41A) and three single-site mutants (H31R, E36D, and E41A) (46). These results further underline the essential roles of intramolecular interaction organization (“protein sectors”) in the sweet-tasting proteins for their performance.
“Proteins Sectors” Determines the Stability of Sweet-Tasting Proteins
Stability is another important property of sweet-tasting proteins, and their intramolecular interaction forces are shown to be also critical for their stability. For example, the most thermostable sweet protein brazzein harbors four intramolecular disulfide bonds, which are essential for its thermostability (47). Moreover, intramolecular disulfide bonds are prevalent in other sweet-tasting proteins, such as thaumatin and mabinlin (11, 14). We have solved the crystal structure of E2N/E23A mutant of MNEI (the single-chain monellin) (PDB: 5Z1P), and indicated that compared to the wild-type protein, mutation of E23 to A could resulted in new hydrogen bonds with V20 and G27 as well as an enhanced C-H…π bond interaction with F89, which are responsible for its improved thermostability (Tm values 84.9 and 74.2°C for E2N/E23A and wild-type MNEI, respectively) (Figures 2C,D) (48).
In another mutant of MNEI (E23Q/Q28K/C41S/Y65R), E23Q mutation was reported to induce conformational arrangements of surrounding residues and establish new hydrogen bonds with Y29 and G30. The Q28K mutation plays a concerted role, which hydrogen bonds with N90. All these new interactions establish a stabilizing hydrogen bonds network that account for the improved stability of the mutated protein (43). These results together suggest the crucial roles of intramolecular interaction patterns (“protein sectors”) for the stability of sweet-tasting proteins.
Discussion and Future Outlook
Based on the above analysis, it is evident that “protein sector” in sweet-tasting proteins is significant for understanding their structure and function relationship, which is essential for the protein engineering of these biomacromolecules. However, our knowledge about the intramolecular interaction organization of sweet-tasting proteins, especially those determining their properties, is still limited. For instance, how connection patterns of each amino acid shape the full landscape of “protein sector” in the proteins and determine their properties? Is there an universal pattern among different sweet-tasting proteins (49)?
Because “protein sector” in sweet-tasting proteins is a global network composed by many different amino acids, thus in the future a large number of multiple mutations are needed to elucidate the function of each residue and their overall performances. Furthermore, because most findings related to the “protein sector” in sweet-tasting proteins are from monellin (or single chain monellin, MNEI), it is needed to perform more extensive studies toward other sweet-tasting proteins to illuminate the universal mechanism of intramolecular organization in these miraculous proteins. Moreover, more details of dynamic conformations of sweet-tasting proteins and their variants are promising to uncover the intrinsic assembling of “protein sectors” as well as their relationship with the properties of these proteins (50, 51). Prospectively, our new insight into the structure-activity relationship of sweet-tasting proteins-“protein sectors” would provide meaningful guidelines for their protein engineering, which could greatly accelerate the improvement of their properties and promote the application of sweet-tasting proteins in foods and beverages (52, 53).
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BL and XZ conceived and designed the data analysis. CW and BL wrote the manuscript. XZ and YZ helped to analyze the data. All authors agree to be accountable for the content of this work.
Funding
This research was funded by the Key Research & Development Project of Shandong Province, Major Science and Technology Innovation Project (2020CXGC010604), Natural Science Foundation of Shandong Province (ZR2020KC035) and the National Natural Science Foundation of China (31970935).
Conflict of Interest
YZ was employed by the company Shandong Aojing Biotechnology Co., Ltd., Zoucheng, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Faus I. Recent developments in the characterization biotechnological production of sweet-tasting proteins. Appl Microbiol Biotechnol. (2000) 53:145–51. doi: 10.1007/s002530050001
2. Rega MF, Di Monaco R, Leone S, Donnarumma F, Spadaccini R, Cavella S, et al. Design of sweet protein based sweeteners: hints from structure–function relationships. Food Chem. (2015) 173:1179–86. doi: 10.1016/j.foodchem.2014.10.151
3. Liu Q, Li L, Yang L, Liu T, Cai C, Liu B. Modification of the sweetness and stability of sweet-tasting protein monellin by gene mutation and protein engineering. Biomed Res Int. (2016) 2016:3647173. doi: 10.1155/2016/3647173
4. Kondo K, Miura Y, Sone H, Kobayashi K, Iijima H. High-level expression of a sweet protein, monellin, in the food yeast Candida utilis. Nat Biotechnol. (1997) 15:453–7. doi: 10.1038/nbt0597-453
5. Kim SH, Kang CH, Kim R, Cho JM, Lee YB, Lee TK. Redesigning a sweet protein: increased stability and renaturability. Protein Eng. (1989) 2:571–5. doi: 10.1093/protein/2.8.571
6. Hobbs JR, Munger SD, Conn GL. Monellin (MNEI) at 1.15 resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2007) 63:162–7. doi: 10.1107/S1744309107005271
7. Hellekant G, Danilova V. Brazzein a small, sweet protein: discovery and physiological overview. Chem Senses. (2005) 30:i88–9. doi: 10.1093/chemse/bjh127
8. Caldwell JE, Abildgaard F, Dzakula Z, Ming D, Hellekant G, Markley JL. Solution structure of the thermostable sweet-tasting protein brazzein. Nat Struct Biol. (1998) 5:427–31. doi: 10.1038/nsb0698-427
9. Nagata K, Hongo N, Kameda Y, Yamamura A, Sasaki H, Lee WC, et al. The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana. Acta Crystallogr D Biol Crystallogr. (2013) 69:642–7. doi: 10.1107/S0907444913001005
10. van der Wel H, Loeve K. Isolation and characterization of thaumatin I and II. the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur J Biochem. (1972) 31:221–5. doi: 10.1111/j.1432-1033.1972.tb02522.x
11. Masuda T, Ohta K, Mikami B, Kitabatake N. High-resolution structure of the recombinant sweet-tasting protein thaumatin. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2011) 67:652–8. doi: 10.1107/S174430911101373X
12. Masuda T, Ohta K, Mikami B, Kitabatake N, Tani F. Atomic structure of the sweet-tasting protein thaumatin I at pH 8.0 reveals the large disulfide-rich region in domain II to be sensitive to a pH change. Biochem Biophys Res Commun. (2012) 419:72–6. doi: 10.1016/j.bbrc.2012.01.129
13. Gu W, Xia Q, Yao J, Fu S, Guo J, Hu X. Recombinant expressions of sweet plant protein mabinlin II in Escherichia coli and food-grade Lactococcus lactis. World J Microbiol Biotechnol. (2015) 31:557–67. doi: 10.1007/s11274-015-1809-2
14. Li DF, Jiang P, Zhu DY, Hu Y, Max M, Wang DC. Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness. J Struct Biol. (2008) 162:50–62. doi: 10.1016/j.jsb.2007.12.007
15. Theerasilp S, Kurihara Y. Complete purification and characterization of the taste-modifying protein, miraculin, from miracle fruit. J Biol Chem. (1988) 263:11536–9. doi: 10.1016/S0021-9258(18)37991-2
16. Koizumi A, Tsuchiya A, Nakajima K, Ito K, Terada T, Shimizu-Ibuka A, et al. Human sweet taste receptor mediates acid-induced sweetness of miraculin. Proc Natl Acad Sci USA. (2011) 108:16819–24. doi: 10.1073/pnas.1016644108
17. Gahloth D, Selvakumar P, Shee C, Kumar P, Sharma AK. Crystal structure of Miraculin like protein from seeds of Murraya koenigii. Arch Biochem Biophys. (2010) 494:15–22. doi: 10.1016/j.abb.2009.11.008
18. Yamashita H, Akabane T, Kurihara Y. Activity and stability of a new sweet protein with taste-modifying action, curculin. Chem Senses. (1995) 20:239–43. doi: 10.1093/chemse/20.2.239
19. Shirasuka Y, Nakajima K, Asakura T, Yamashita H, Yamamoto A, Hata S, et al. Neoculin as a new taste-modifying protein occurring in the fruit of Curculigo latifolia. Biosci Biotechnol Biochem. (2004) 68:1403–7. doi: 10.1271/bbb.68.1403
20. Kurimoto E, Suzuki M, Amemiya E, Yamaguchi Y, Nirasawa S, et al. Curculin exhibits sweet-tasting and taste-modifying activities through its distinct molecular surfaces. J Biol Chem. (2007) 282:33252–6. doi: 10.1074/jbc.C700174200
21. Wel HVD, Larson G, Hladik A, Hladik CM, Hellekant G, Glaser D. Isolation and characterization of pentadin, the sweet principle of Pentadiplandra brazzeana Baillon. Chem Senses. (1989) 14:160–6. doi: 10.1093/chemse/14.1.75
22. Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. (2001) 106:381–90. doi: 10.1016/S0092-8674(01)00451-2
23. Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet umami taste. Proc Natl Acad Sci USA. (2002) 99:4692–6. doi: 10.1073/pnas.072090199
24. Ahmad R, Dalziel JE. G protein-coupled receptors in taste physiology and pharmacology. Front Pharmacol. (2020) 11:587664. doi: 10.3389/fphar.2020.587664
25. Maillet EL, Cui M, Jiang P, Mezei M, Hecht E, Quijada J, et al. Characterization of the binding site of aspartame in the human sweet taste receptor. Chem Senses. (2015) 40:577–6. doi: 10.1093/chemse/bjv045
26. Yang L, Zhu K, Yu H, Zhang X, Liu B. The flexible loop is a new sweetness determinant site of the sweet-tasting protein: characterization of novel sweeter mutants of the single-chain monellin (MNEI). Chem Senses. (2019) 44:607–14. doi: 10.1093/chemse/bjz057
27. Servant G, Kenakin T, Zhang L, Williams M, Servant N. The function and allosteric control of the human sweet taste receptor. Adv Pharmacol. (2020) 88:59–82. doi: 10.1016/bs.apha.2020.01.002
28. Temussi PA. Natural sweet macromolecules: how sweet proteins work. Cell Mol Life Sci. (2006) 63:1876–88. doi: 10.1007/s00018-006-6077-8
29. Temussi PA. Determinants of sweetness in proteins: a topological approach. J Mol Recognit. (2011) 24:1033–42. doi: 10.1002/jmr.1152
30. Yang L, Cui M, Liu B. Current progress in understanding the structure and function of sweet taste receptor. J Mol Neurosci. (2021) 72:234–44. doi: 10.1007/s12031-020-01642-4
31. Sung YH, Shin J, Chang HJ, Cho JM, Lee W. Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. Structural origin of sweetness. J Biol Chem. (2001) 276:19624–30. doi: 10.1074/jbc.M100930200
32. Assadi-Porter FM, Maillet EL, Radek JT, Quijada J, Markley JL, Max M. Key amino acid residues involved in multi-point binding interactions between brazzein, a sweet protein, and the T1R2-T1R3 human sweet receptor. J Mol Biol. (2010) 398:584–99. doi: 10.1016/j.jmb.2010.03.017
33. Lim JK, Jang JC, Kong JN, Kim MC, Kong KH. Importance of Glu53 in the C-terminal region of brazzein, a sweet-tasting protein. J Sci Food Agric. (2016) 96:3202–6. doi: 10.1002/jsfa.7501
34. Masuda T, Ohta K, Ojiro N, Murata K, Mikami B, Tani F, et al. A hypersweet protein: removal of the specific negative charge at Asp21 enhances thaumatin sweetness. Sci Rep. (2016) 6:20255. doi: 10.1038/srep20255
35. Zheng W, Yang L, Cai C, Ni J, Liu B. Expression, purification and characterization of a novel double-sites mutant of the single-chain sweet-tasting protein monellin (MNEI) with both improved sweetness and stability. Protein Expr Purif. (2017) 143:52–56. doi: 10.1016/j.pep.2017.10.010
36. Delfi M, Emendato A, Leone S, Lampitella EA, Porcaro P, Cardinale G, et al. A super stable mutant of the plant protein monellin endowed with enhanced sweetness. Life. (2021) 11:236. doi: 10.3390/life11030236
37. Tang N, Liu J, Cheng Y. Potential improvement of the thermal stability of sweet-tasting proteins by structural calculations. Food Chem. (2021) 345:128750. doi: 10.1016/j.foodchem.2020.128750
38. Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. (2009) 138:774–86. doi: 10.1016/j.cell.2009.07.038
39. Raman AS, White KI, Ranganathan R. Origins of allostery and evolvability in proteins: a case study. Cell. (2016) 166:468–80. doi: 10.1016/j.cell.2016.05.047
40. Zhou J, Wang Z, Mao Y, Wang L, Xiao T, Hu Y, et al. Proteogenomic analysis of pitaya reveals cold stress-related molecular signature. Peer J. (2020) 8:e8540. doi: 10.7717/peerj.8540
41. Nedrud D, Coyote-Maestas W, Schmidt D. A large-scale survey of pairwise epistasis reveals a mechanism for evolutionary expansion and specialization of PDZ domains. Proteins. (2021). doi: 10.1002/prot.26067. [Epub ahead of print].
42. Templeton CM, Ostovar pour S, Hobbs JR, Blanch EW, Munger SD, Conn GL. Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II Helix. Chem Senses. (2011) 36:425–34. doi: 10.1093/chemse/bjr007
43. Leone S, Pica A, Merlino A, Sannino F, Temussi PA, Picone D. Sweeter and stronger: enhancing sweetness and stability of the single chain monellin MNEI through molecular design. Sci Rep. (2016) 6:34045. doi: 10.1038/srep34045
44. Assadi-Porter FM, Abildgaard F, Blad H, Markley JL. Correlation of the sweetness of variants of the protein brazzein with patterns of hydrogen bonds detected by NMR spectroscopy. J Biol Chem. (2003) 278:31331–9. doi: 10.1074/jbc.M302663200
45. Singarapu KK, Tonelli M, Markley JL, Assadi-Porter FM. Structure-function relationships of brazzein variants with altered interactions with the human sweet taste receptor. Protein Sci. (2016) 25:711–9. doi: 10.1002/pro.2870
46. Lee JW, Cha JE, Jo HJ, Kong KH. Multiple mutations of the critical amino acid residues for the sweetness of the sweet-tasting protein, brazzein. Food Chem. (2013) 138:1370–3. doi: 10.1016/j.foodchem.2012.10.140
47. Dittli SM, Rao H, Tonelli M, Quijada J, Markley JL, Max M, et al. Structural role of the terminal disulfide bond in the sweetness of brazzein. Chem Senses. (2011) 36:821–30. doi: 10.1093/chemse/bjr057
48. Zhao M, Xu X, Liu B. Structure basis of the improved sweetness and thermostability of a unique double-sites single-chain sweet-tasting protein monellin (MNEI) mutant. Biochimie. (2018) 154:156–63. doi: 10.1016/j.biochi.2018.08.010
49. Liu X, Wang Y, Gutierrez JS, Damsker JM, Nagaraju K, Hoffman EP, et al. (2020) Disruption of a key ligand-H-bond network drives dissociative properties in vamorolone for Duchenne muscular dystrophy treatment. Proc Natl Acad Sci USA. 117:24285–93. doi: 10.1073/pnas.2006890117
50. Saldaño TE, Tosatto SCE, Parisi G, Fernandez-Alberti S. Network analysis of dynamically important residues in protein structures mediating ligand-binding conformational changes. Eur Biophys J. (2019) 48:559–68. doi: 10.1007/s00249-019-01384-1
51. de Vries RMJM, Meijer FA, Doveston RG, Leijten-van de Gevel IA, Brunsveld L. Cooperativity between the orthosteric and allosteric ligand binding sites of RORγt. Proc Natl Acad Sci USA. (2021) 118:e2021287118. doi: 10.1073/pnas.2021287118
52. Kant R. Sweet proteins-potential replacement for artificial low calorie sweeteners. Nutr J. (2005) 4:5. doi: 10.1186/1475-2891-4-5
Keywords: sweet-tasting proteins, intramolecular interaction forces, protein sector, network, sweetness, stability
Citation: Zhao X, Wang C, Zheng Y and Liu B (2021) New Insight Into the Structure-Activity Relationship of Sweet-Tasting Proteins: Protein Sector and Its Role for Sweet Properties. Front. Nutr. 8:691368. doi: 10.3389/fnut.2021.691368
Received: 06 April 2021; Accepted: 18 May 2021;
Published: 18 June 2021.
Edited by:
Jinsong Bao, Zhejiang University, ChinaReviewed by:
Denis Baranenko, ITMO University, RussiaJosé Pinela, Polytechnic Institute of Bragança (IPB), Portugal
Copyright © 2021 Zhao, Wang, Zheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Liu, ZXJ0cmRmZ2dAcWx1LmVkdS5jbg==
 Xiangzhong Zhao1
Xiangzhong Zhao1 Bo Liu
Bo Liu