- State Key Laboratory of Oncogenes and Related Genes, Center for Single-Cell Omics, School of Public Health, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Colorectal cancer (CRC) is one of the most prevalent and life-threatening cancer types with limited therapeutic options worldwide. Gut microbiota has been recognized as the pivotal determinant in maintaining gastrointestinal (GI) tract homeostasis, while dysbiosis of gut microbiota contributes to CRC development. Recently, the beneficial role of postbiotics, a new concept in describing microorganism derived substances, in CRC has been uncovered by various studies. However, a comprehensive characterization of the molecular identity, mechanism of action, or routes of administration of postbiotics, particularly their role in CRC, is still lacking. In this review, we outline the current state of research toward the beneficial effects of gut microbiota derived postbiotics against CRC, which will represent the key elements of future precision-medicine approaches in the development of novel therapeutic strategies targeting gut microbiota to improve treatment outcomes in CRC.
1. Introduction
As one of the most prevalent malignant tumors in the world, colorectal cancer (CRC) is a major cause of the steadily rising cancer death. According to a global statistical analysis covering 35 major types of cancer, CRC was ranked as the 3rd after female breast cancer and lung cancer, and 2nd most cancer-related deaths after lung cancer (1, 2). Currently, tumor resection surgery, radiotherapy, and chemotherapy have been widely used as the most commonly recommended treatment option in CRC (3). Targeted therapy has also emerged as another practical option of anti-cancer therapy in the past few decades (4). However, the therapeutic efficacy of current strategies is usually compromised in the late-stage or metastatic CRCs (5) while overall responsive in early-stage CRCs with a low level of relapse (6). It was reported that early diagnosis of CRC was accompanied with effective interventions which would largely increase the survival rate and overall wellness of patients (7). Therefore, accurate diagnosis and intervention at the early stage is necessary for CRC treatment.
During CRC development and progression, a large number of intrinsic and extrinsic risk factors have been identified (8). The hereditary non-polyposis colorectal cancer (HNPCC), well-known as Lynch syndrome, is attributed to autosomal dominant genetic mutations in one of four DNA mismatch repair (MMR) genes (9, 10). The environmental factors such as low physical activity, exposure to toxicities, etc., were also reported to facilitate CRC progression (11). Dietary patterns are considered as another critical factor in regulating gut microbiota homeostasis, which is detrimental for both initiation and progression of CRC (12–15). Inflammation, especially chronic inflammation, plays an important role in CRC development, as revealed by the fact that patients with inflammatory bowel disease (IBD) have higher risks of CRC ranging from 8.3 to 18.4% (16–18).
Recently, gut microbiota has been demonstrated to be closely associated with human health with the advancement of high-throughput multi-omics technologies. It was revealed that the abundance of gut microbes or their derived molecules are closely related to CRC, emphasizing their potential as biomarkers in early CRC diagnosis (19). Specifically, bacterial species like Helicobacter pylori were found to drive CRC development (20). Fusobacterium nucleatum has also been shown both positively associated with and contribute to the carcinogenesis of CRC (21, 22). Porphyromonas species were shown positively correlated with the progression of CRC, suggesting a causal effect of Porphyromonas on CRC tumorigenesis (23). In contrast, certain microbes may have functional roles in preventing CRC progression, as revealed by the protective effects through gut microbial manipulation (24). Additionally, probiotics as beneficial microorganisms for human, has been proven to improve the chemotherapy effectiveness of CRC (25). In spite of the benefits, precise manipulation of gut microbiota by dietary intervention or probiotic administration is still challenging nowadays due to many technical limitations. Of note, individual adhesion to nutritional plans varies that would markedly affect the efficacy of dietary interventions (26, 27). Furthermore, transplantation of live microbes into human brought additional concerns regarding the inevitable safety and ethical issues, which further dampens its clinical utilization (28).
Now a growing number of researches are turning to the alternative, postbiotics, which are defined by the International Scientific Association of Probiotics and Prebiotics (ISSAP) as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” (2). It should be noticed the term of “paraprobiotics” was used in previous studies to describe non-living microbial cells or cell fractions that confer health benefit to the host, in separating from the past definition of postbiotics as “soluble products or metabolites with beneficial functions” (29). To comply with the most recent guideline of postbiotics and also to give a full picture of recent advances in related fields, we chose to use the broader definition of postbiotics by the ISSAP in the present review.
Distinct from the “prebiotics” defined as “a substrate that is selectively utilized by host microorganisms conferring a health benefit” (30), or the “probiotics” defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (31), the importance of postbiotics in regulating gut homeostasis remains at its infancy stage. As postbiotics are beneficial substances derived from microorganisms, they must be delivered at host surfaces through various routes to achieve their biological effects, such as oral cavity, gut, skin, urogenital tract, nasopharynx etc., (32). Previous studies have already uncovered the biological activities of gut microbe-derived metabolites in maintaining overall health and protecting against various diseases (33–35). In the context of CRC, the benefits of postbiotics have also been uncovered (36). Compared with live microorganisms (probiotics for instance), the molecular mechanisms of postbiotics as well as their biological effect are easier to be evaluated (28). A brief illustration of prebiotics, probiotics and postbiotics in the context of gut is displayed in Figure 1. Upon diet consumption, prebiotics can be released for gut microbial utilization. Outgrowth of gut commensals (or probiotics in the context of gut tissue), can exert multiple benefits on human health. Postbiotics, which are derived from both probiotics or many other kinds of microorganisms, can be naturally produced or directly applied on host surfaces for the improvement of human gut health and CRC prevention/treatment.
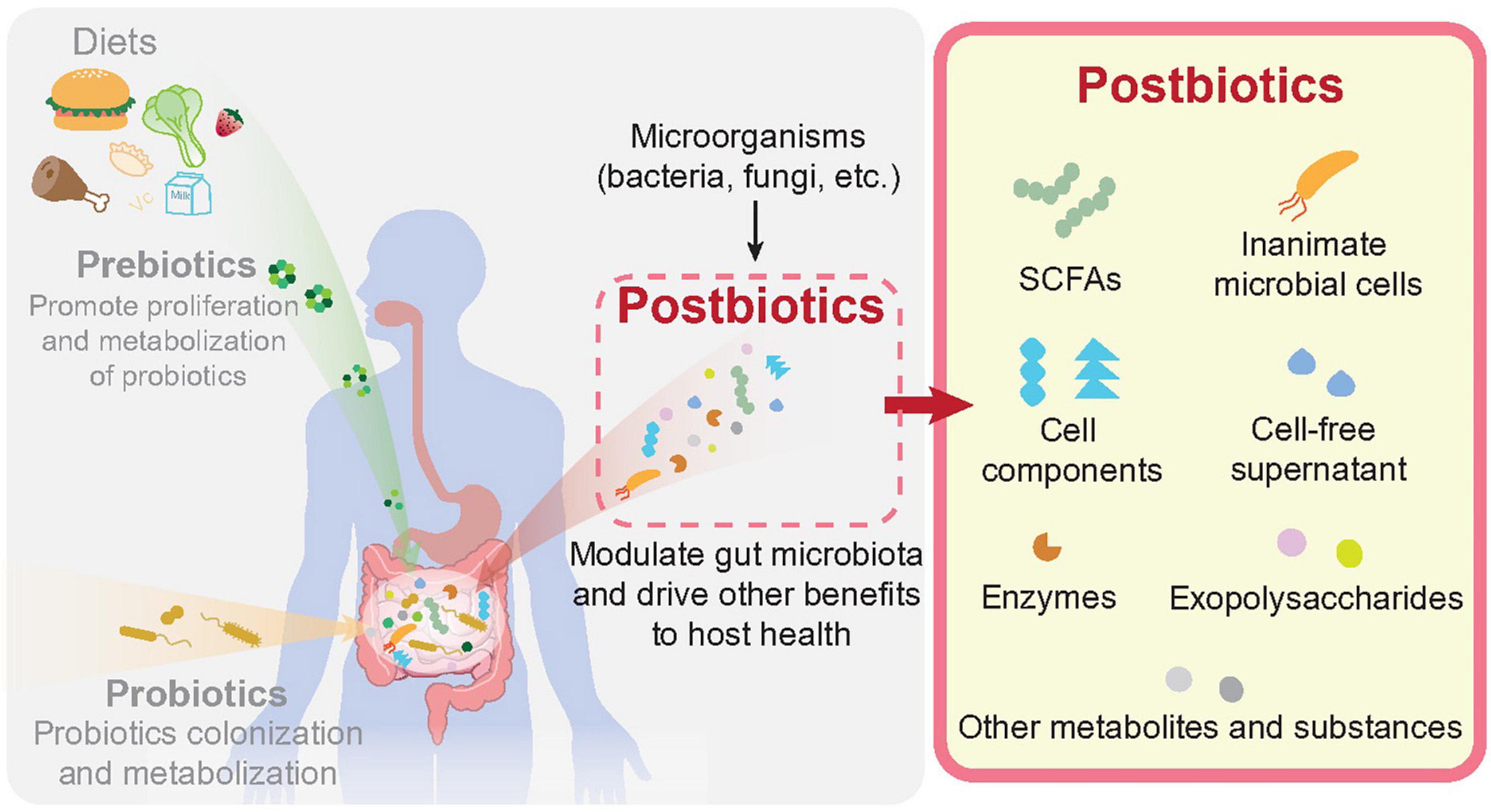
Figure 1. Prebiotics, probiotics, and postbiotics in human gut. As described in the main text, prebiotics are produced through consumption of daily intakes by our digest system, which are selectively utilized by host microorganisms conferring health benefits. Probiotics are defined as live microorganisms residing in human body, notably the intestinal lumen, which are beneficial for human health. Postbiotics are preparations of microbial derived substances that contribute to the health of host. Postbiotics can be derived from variable microbial origins, including cell components, cell-free supernatants, cell wall structures like exopolysaccharides (EPS), metabolites like short chain fatty acids as well as enzymes etc.
Although it is clear that the microbial sources or “sites of actions” of postbiotics are not limited to gut, the well-established relationship between gut microbiota and CRC make it a valuable topic to investigate the potential effects of gut microbial derived postbiotics in CRC progression. Here, we reviewed the current advances regarding the role of gut microbial derived postbiotics in CRC, including the identification, functional mechanism, and potential clinical applications.
2. Identification of postbiotics from multi-kingdom gut microbiota
Postbiotics are composed of various types of microbial preparations including microbial cells but also various peptides, vitamins, and structural components, etc., (37). Although it was proposed that chemically identified molecules from microorganisms were no longer suitable to be named as postbiotics (2), we prefer to include those molecules in this retrospective review as they were isolated from microorganisms and demonstrated to be effective against CRC, which would be insightful for future studies in related field.
Gut microbiota inhabited in the human gastrointestinal tract along with their derived metabolites are considered to be detrimental in human health and diseases (37). Growing evidence has demonstrated the close relationship between the abundance or activity of certain microbes and the incidence of various diseases. The importance of non-living microbial cells and their metabolites has also been recognized recently (34). Previous studies identified bioactive ingredients from gut microbiota by traditional biochemical approaches (centrifugation, ultrafiltration, metabolomics, etc.) (38). However, the biological functions of postbiotics in human health particularly the role of postbiotics in CRC initiation, progression, as well as treatment, are still poorly understood. Here described different types of postbiotics uncovered from bacteria, fungi as well as other microorganisms residing in the gut. The specific role of each type of postbiotics as well as the underlying mechanisms are illustrated in later sections. A concise list of different types of identified postbiotics, their source of identification, biological effects in CRC as well as the potential mechanisms are also summarized (Table 1).
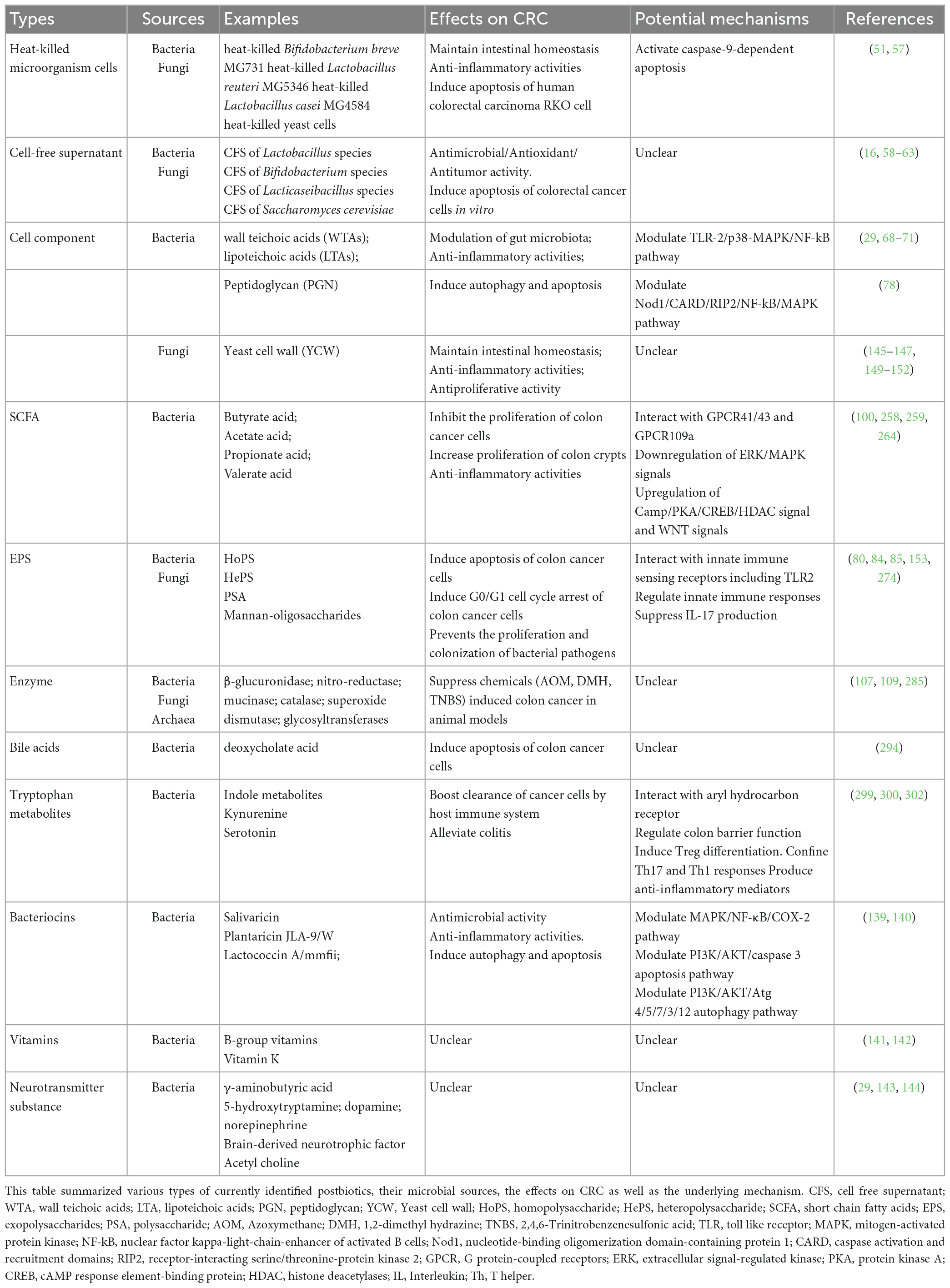
Table 1. List of identified postbiotics and beneficial effects for colorectal cancer (CRC) treatment.
2.1. Postbiotics from gut bacteria
2.1.1. Heat-killed bacterial cells
The heat-killed microbial cells had not been included into postbiotics until 2021, when the International Scientific Association of Probiotics and Prebiotics (ISAPP) incorporated the deliberately inanimate microbial cells, mostly heat-killed probiotic bacteria cells, into the definition of postbiotics (2, 39–41). To address the biological role of microbial cells, microorganisms were further inanimate by various methods like heating, chemical (e.g., formalin), gamma or ultraviolet rays, and sonication treatments (42–44). Lactobacillus and Bifidobacterium species, the mostly studied probiotics, were found to have beneficial functions in heat-killed formations (45–51). For instance, heat-killed Lactobacillus salivarius subsp. salicinius AP-32, L. rhamnosus CT-53, L. paracasei ET-66 could significantly inhibit the invasion of oral pathogens to improve oral health (45). Besides, heat-killed Lactobacillus and Bifidobacterium species are found to exhibit immunomodulatory effect. Heat-killed Bifidobacterium breve M-16V cells suppressed proinflammatory cytokine production in spleen cells and affected intestinal metabolism (46). Oral administration of heat-killed L. plantarum L-137 could enhance protection against IFV infection together with increased IFN-β production in the serum of infected mice at an early stage of infection (47). Lactobacillus rhamnosus GG (LGG) could ameliorate intestinal inflammation, and promote cytoprotective responses in the developing murine gut, thus playing a role in inflammatory bowel disease treatment (16, 48, 49). Li et al. found the heat-killed LGG could also decrease lipopolysaccharide (LPS)-induced proinflammatory mediators and increase anti-inflammatory mediators (50). Ueno et al. proved that administration of heat-killed Lactobacillus brevis SBC8803 could successfully maintain intestinal homeostasis and cure intestinal inflammation, which manifesting the potential for CRC treatment (51). Recently, Akkermansia muciniphila was found to be beneficial in the prevention of various diseases through maintaining the integrity of gut barrier (52). Implantation of live Akkermansia muciniphila exerted protective role against liver injury, colitis and CRC as well (53–55). Interestingly, Wang et al. showed that the pasteurized bacterium of Akkermansia muciniphila strikingly inhibit colitis associated tumorigenesis in mice, indicating the beneficial role of Akkermansia muciniphila derived components against CRC progression (56). Meanwhile, Kim et al. reported that heat-killed B. bifidum MG731, L. reuteri MG5346, and L. casei MG4584 significantly delayed tumor growth in colorectal carcinoma, which give direct evidence of heat-killed bacterial cells for CRC (57).
2.1.2. Cell-free supernatant (CFS)
Cell-free supernatant refers to the biologically active metabolites secreted by microorganisms. Here we mainly focus on the cell-free supernatant of bacteria, which are prepared after the bacterial cells were incubated, centrifuged, and removed (34). CFS has been reported to show antimicrobial, antioxidant, and antitumor activity (16, 58–63).
The antimicrobial activity refers to both antibacterial and antifungal activity (58, 59). The Saccharomyces cerevisiae CFS showed anti-biofilm activity against Staphylococcus aureus, which is the widely reported pathogen in both food and clinical environment (58, 64). The anti-biofilm property of S. cerevisiae CFS can be thus utilized to reduce the risk of infection by S. aureus in human (65). In addition, the cell-free supernatants derived from Lactobacillus paracasei subsp. Paracasei SM20 and Propionibacterium jensenii SM11 coculture all showed high antifungal activity against Candida pulcherrim and Rhodotorula mucilaginosa (59). Interestingly, CFS of B. animalis subsp. Lactis DSMZ 23032, L. acidophilus DSMZ 23033, L. rhamnosus SD11, and Lactobacillus brevis DSMZ 23034 exhibited strong antioxidant activity through DPPH radical scavenging and oxidative stress hypersensitivity in vivo (60). Pourramezan et al. found that there was a relationship between the antibacterial activity and antioxidant activity of CFS of Lactobacillus (61).
For Lactobacillus and Bifidobacterium species, their CFS were found to inhibit cancer progression (62). For example, L. casei and LGG cultures could prevent the invasion of colon cancer cells (62). In recent years, Lactobacillus cell-free supernatant (LCFS) has attracted more and more attention to its potential benefits on CRC prevention (63). The anti-cancer effects of L. fermentum CFS has been tested which proved that it could induce apoptotic cell death in three dimensional (3D) spheroids of colorectal cancer cells in vitro (63). Besides, the mRNA levels of apoptosis markers were dramatically induced after treating with LCFS in 3D system (63). Pahumunto et al. showed that CFS of Lacticaseibacillus paracasei SD1, Lacticaseibacillus rhamnosus SD4, Lacticaseibacillus rhamnosus SD11, and Lacticaseibacillus rhamnosus GG could inhibit pro-inflammatory cytokine expression in Caco-2 cells and inhibit the growth of Caco-2 cells, supporting its role in preventing CRC (16).
2.1.3. Cell components
According to the more recent definition of postbiotics by the ISSAP, cell components can be regarded as another major type of postbiotics. Two categories of cellular components were intensively investigated by far. Teichoic acids (TAs) are major constituent of bacterial cell walls, including both wall teichoic acids (WTAs) covalently linked to peptidoglycan and lipoteichoic acids (LTAs) anchored to the cytoplasmic membrane (66, 67). WTA was reported to be effective in modulating bacterial colonization (29), while LTA was discovered to exert immune-modulatory activity by recognizing Toll-like receptors (68). LTA could interact with TLR2 and TLR6 to active host immune response (69). In particular, LTA of Lactobacillus paracasei D3-5 could enhance mucin (Muc2) expression by modulating TLR-2/p38-MAPK/NF-kB pathway, which in turn reduces inflammation (70). In addition, LTA of Lactobacillus plantarum inhibited the phosphorylation of ERK and p38 kinase as well as the activation of NF-κB, resulting in decreased IL-8 production and inflammation suppression in porcine intestinal epithelial cells (71).
Peptidoglycan (PGN) is the conserved structure of Gram-negative bacterial which could be recognized by nucleotide-binding oligomerization domain-1 (NOD1) (72). Once activated by PGN, NOD1 could further activate the downstream NF-κB (72) and MAPK (73) pathway through interacting with a caspase activated and recruitment domain (CARD) and its adaptor protein, the receptor-interacting protein 2 (RIP2) (74–76). Besides, the PGN/Nod1/RIP2 pathway could induce autophagy and inflammatory signaling in response to bacterial infection (77). PGN of the Lactobacillus paracasei subsp. paracasei M5 strain exerted cytotoxic effects on HT-29 cells and induced a mitochondria-mediated apoptosis in colon cancer (78).
2.1.4. Exopolysaccharides (EPS)
Exopolysaccharides (EPS) is a subclass of polysaccharides that constitute the cell wall structure of bacteria and is characterized by less attachment to the cell surface compared with capsular polysaccharides (79). EPS can be structurally divided into two categories: homopolysaccharide (HoPS) and heteropolysaccharide (HePS). HoPS was composed of a single type of monosaccharides, such as D-glucose or D-fructose. Crosslinking of D-glucose through α- or β-glycosidic linkage resulted in α- or β- glucans, respectively, while connection of D-fructose through β-glycosidic linkage forms β-fructans (80). In contrast, HePS are composed of different types of monosaccharides, such as D-glucose, D-galactose, and L-Rhamnose (80). Besides, N-acetylation is also found to be presented on HePS, which further diversified their molecular structures (81).
Bacterial EPS has been evaluated as industrially important biopolymers with significant commercial potential (82). The effects of EPS on human health (and in particular, CRC) have also been recognized (80). EPS can be synthesized by a wide range of bacterial species such as Bifidobacteria sp. and Lactobacillus sp., which benefits the gut homeostasis (83). The protective role of EPS in CRC has been demonstrated previously. The EPS extracted from Lactobacillus acidophilus was found to inhibit the growth of Caco-2 cells in a dose-dependent manner (84). Similarly, Lactobacillus-derived EPS was found to suppress HT-29 cell growth by inducing the G0/G1 cell cycle arrest and apoptosis (85). In line with that, EPS produced by microalgae Chlorella pyrenoidosa, Scenedesmus sp., and Chlorococcum sp. was found to induce anti-tumor effect on both HCT116 and HCT8 cells (86), similar to EPS produced from Chlorella zofingiensis and Chlorella vulgaris (87). Taken together, these data suggest a universal protective effect of EPS against CRC independent of its sources. Nevertheless, the in vivo activity of EPS in CRC protection has not been fully clarified yet. It was revealed that EPS from Lactobacillus rhamnosus is critical in providing the “shield” for bacteria itself against the host immune defense, which is beneficial for the maintenance of gut microbiota homeostasis (88). Apart from that, EPS from Bifidobacterium longum subsp. Longum significantly alleviated DSS-induced colitis by maintaining the integrity of gut mucosal barrier (89), which was supported by the significantly altered level of EPS from Rhizopus nigricans in CRC mouse model (90). Taken together, these studies support the beneficial role of EPS against CRC.
2.1.5. Short chain fatty acids (SCFAs)
Short chain fatty acids (SCFAs) are a subset of saturated fatty acids with six or less carbon molecules including acetate, propionate, butyrate, pentatonic (valeric) acid, and hexanoic (caproic) acid (91). SCFAs are the main metabolic products of indigestible saccharides by anaerobic bacterial fermentation in human gut (91). Deficiency of SCFAs production can lead to the pathogenesis of many human diseases, such as autoimmune syndromes from allergies to asthma, metabolic diseases, neurological diseases, and cancers (92).
The protective effects of SCFAs against colorectal cancer were first discovered in the early 1980s. Whitehead et al. found that butyrate stimulation significantly inhibited the proliferation rate of human colorectal cancer cell line LIM1215 (93), accompanied by a similar finding that both propionate and butyrate suppressed the growth of human colorectal cancer HT-29 cells (94). In an independent study, it was shown that SCFA treatment, either alone or in combination, significantly increased the proliferation of colon crypts, suggesting a symbiotic effect on colorectal health, which eventually prevents the formation or progression of colorectal cancer (95). The anti-cancer effect of SCFAs is also supported by epidemiological studies revealing an inverse correlation between the level of dietary fibers and the incidence of human colorectal cancer (96). In line with this, the proportions of major SCFAs in enema samples of patients with colon polyps or colorectal cancer were strikingly different from healthy subjects, indicating the niche-dependent activity of SCFAs in response to CRC (97). Moreover, Gibson et al. investigated the fermentative production of butyrate in vivo as well as its protective effect against colorectal cancer in a rat model (98). It was found that the intake of natural fiber was associated with high concentration of butyrate in colon tissue and reduction of colorectal tumor formation, which highlighted the protective effect of butyrate in colorectal cancer. Propionate and valerate are also members of SCFAs effective in arresting cell growth or suppressing differentiation of human colon carcinoma cells (99). Although the in vivo activity of SCFAs in the context of CRC is still under investigation, the anti-inflammatory effects of SCFAs have been well-described and are essential for the inhibition of colitis and inflammatory bowel disease (IBD), which are both considered as the major risks for the development of CRC (100, 101). Of note, recent studies uncovered the physiological role of SCFA in preventing colitis-induced CRC in mice, as administration of SCFA mix strongly suppressed tumor burden in mice treated with Azoxymethane (AOM)/dextran sodium sulfate (DSS) which were commonly used for the induction of colitis-associated colorectal cancer (100). Colon inflammation as well as pathological score index was also reduced, consistent with the decreased secretion of pro-inflammatory cytokines including interlukin-17 (IL-17), IL-6, and tumor necrosis factor-α (TNF-α) (100, 102). Overall, these results highlighted the clinical potential of SCFAs in protection against CRC.
2.1.6. Enzyme
Enzymes are recognized as key regulators driving the metabolism of all organisms, with no exception for gut microbiome (103). In fact, enzymes encoded by microbes play essential roles in host-microbe interactions (103). Early studies on the epidemiology of CRC revealed that there was a negative correlation between the abundance of lactic acid-producing bacteria Bifidobacterium in human gut and the incidence of colorectal cancer (104). Lactic acid bacteria (LAB) are active in fermentation via synthesizing various enzymes, and the protective effects on colorectal cancer by dietary supplementation of different lactic acid-producing bacterial species have been well-established in murine studies (105–108). Therefore, the potential role of bacterial enzymes in inhibiting CRC development has also been indicated. In fact, it was shown that administration of Bifidobacterium longum and Lactobacillus acidophilus generated better outcome of AOM-induced colorectal cancer in rats, and was accompanied by increased β-glucuronidase and nitro-reductase activity in fecal samples (109). It was also demonstrated that the inhibitory effect of Bifidobacterium longum on colon carcinogenesis was induced by 2-Amino-3-methylimidazo [4,5-f] quinoline (IQ), a food mutagen (106). Consistently, supplementation of coconut cake was found to remarkably reduce the incidence of 1,2-dimethyl hydrazine (DMH)-induced colorectal cancer occurrence in a rat model, which was associated with the upregulation of β-glucuronidase and mucinase (107). In addition, oral administration of a catalase-producing Lactococcus lactis can prevent DMH-induced colorectal cancer in mice (108).
2.1.7. Other bacterial derived components
Other types of metabolites produced by gut microbes are also involved in regulating CRC progression, which include the microbially modified secondary bile acids (secondary BAs). Primary bile acids (primary BAs) are cholesterol derivatives that synthesized in the liver and then secreted into intestine for lipid absorption. Once utilized, most bile acids are recycled to liver via enterohepatic or systemic circulation. Meanwhile, a proportion of primary BAs is transported to colon where they are metabolized by gut microbiome to produce secondary BAs. The detailed signal transduction pathways of BA metabolism have been comprehensively reviewed (110). The relationship of diet, BAs, gut microbiota and CRC has been revealed in recent studies (110, 111). It was reported that the consumption of “unhealthy” western diet is associated with the increase of secondary BAs, especially deoxycholic acid (DCA) and lithocholic acid (LCA) in the gut, while the increased level of bile acids is tightly correlated with high CRC incidence (112–115). Therefore, secondary BAs are primarily considered as carcinogens in human gastrointestinal cancers (116). In addition, high-fat diets promote the synthesis and colonic delivery of bile acids, which stimulate the growth and activity of 7α-dehydroxylating bacteria to convert the primary BAs into secondary BAs (117). Animal studies also showed the effects of high fact diet in modulating the composition and activity of gut microbiota, which lead to increased BA secretion and colon tumorigenesis (118–120). Diet intervention has been implicated to be critical in preventing CRC through the regulation of BA levels in gut (121). These results indicate the biological role of gut microbial secreted BA in regulating the development of CRC. On the other side, ursodeoxycholic acid (UDCA), another member of secondary BAs (122), was effective in inducing apoptosis of colon cells in vitro (123), which is distinct from DCA (124). Consistently, another study uncovered the role of UDCA in inhibiting the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth (125). Mechanistically, it was revealed that UDCA suppressed the malignant progression of colorectal cancer by inhibiting the activation of Hippo/YAP pathway (126). Animal studies demonstrated the protective role of UDCA in chemical induced colon tumorigenesis (127, 128). According to a cross-sectional study, by analyzing the relationship between ursodiol use and colonic dysplasia (the precursor to colon cancer) in patients with ulcerative colitis and primary sclerosing cholangitis, it was revealed that the application of UCDA is associated with lower prevalence of colonic neoplasia (129). Although UDCA has been approved for the treatment of primary biliary cirrhosis by FDA (130), the mechanism of action of UCDA, the relationship of UCDA with gut microbiota, as well as the clinical use of UCDA in CRC treatment remains undetermined and therefore warrants further investigation.
Tryptophan is one of the essential amino acids supplied by the intake of dietary proteins (131). Despite the absorption of the majority of ingested proteins in small intestine, a small fraction of amino acids can reach colon where they can be absorbed by local microbes (132). Tryptophan metabolism is complicated and dependent on two different pathways: the kynurenine (Kyn) pathway and the indolic pathway (133). The significance of microbial tryptophan metabolites in human health has been investigated (134). In the context of CRC, it has been shown that Tryptophan metabolites derived from gut microbes regulate the homeostasis of gut immunity. The deficiency of Tryptophan secretion due to gut dysbiosis may cause exacerbated inflammatory response which eventually led to colorectal tumorigenesis (135). In fact, alteration of fecal tryptophan metabolism was correlated with shifted microbiota, which may be involved in the pathogenesis of colorectal cancer (136).
Bacteriocins are defined as ribosomally synthesized antimicrobial proteins or peptides. It could be divided into two categories: class I bacteriocins are modified by post-translational modifications; class II bacteriocins remain unaltered (137, 138). Given the fact that bacteriocins have antimicrobial activity, bacteriocins targeting CRC associated bacterial pathogens may have potential benefits against CRC. For example, Streptococcus salivarius DPC6993 which could secrete salivaricin against F. nucleatum in an ex vivo model of human colon, was shown to be effective in reducing the risk of CRC (139). Probiotics-derived bacteriocins, including plantaricin JLA-9, plantaricin W, lactococcin A, and lactococcin MMFII directly interact with CRC-promoting protein COX2 and modulate inflammasome complex NLRP3 and NF-kB pathways to reduce CRC-associated inflammation. They also have the potential of activating autophagy and apoptosis by regulating PI3K/AKT and caspase pathways in CRC (140).
In addition to the above-mentioned molecules, several other types of gut microbial derived molecules were also beneficial to human health. For instance, vitamins secreted by gut microbes, including B-group vitamins and vitamin K, were shown to be effective in preventing CRC (141, 142). Neurotransmitter substances synthesized by gut microbes, such as γ-aminobutyric acid (GABA), 5-hydroxytryptamine (5-HT), dopamine (DA), norepinephrine (NE), brain-derived neurotrophic factor (BDNF), and acetyl choline (29), were critical in regulating gut-brain axis (143, 144). However, their roles in CRC still awaits further investigation. In conclusion, a variety of probiotic bacteria-derived molecules have been identified, and the potential of these bacterial derived “postbiotics” in preventing/treating CRC have also been highlighted.
2.2. Postbiotics from gut fungi
2.2.1. Yeast cell wall (YCW) structural components
Yeast is one kind of highly adaptable microorganisms with proved benefits to human health. Yeast cell wall, particularly the mannan-oligosaccharides, were reported to be able to bind to fimbriated bacteria in the gut and prevent their attachment to gut mucosa which eventually prevents the proliferation and colonization of those bacterial pathogens (145). Furthermore, mannan-oligosaccharides (MOS) in the YCW have a strong binding affinity with type I-fimbriae in the gut (146, 147). The probiotic yeast S. boulardii, widely incorporated in dairy food, grains, fruit juices, chocolate, coffee, and tea, was demonstrated to ameliorate diarrheal conditions. Cell wall extracts of S. cerevisiae and S. boulardii could adsorb aflatoxin B1 (148), cholera toxin (149), and pathogenic E. coli (150) which are possibly by cell wall polysaccharides such as β-glucan, mannoprotein and chitin (148, 151). Furthermore, by adsorbing these, the cell wall extracts of S. boulardii could enhance immune responses in the intestinal mucosa (152). As for the relationship between YCW and CRC, the β-glucan of S. boulardii cell wall extract has been proven to prevent colon carcinogenesis in vitro (151). In addition, EPS was also found in probiotic yeast species like Kluyveromyces marxianus and Pichia kudriavzevii, indicating the complicated natural source of EPS (153). Similarly, EPS derived from yeast species Kluyveromyces marxianus and Pichia kudriavzevii showed a comparable anti-colorectal cancer activity (153). Collectively, these results highlight the potential of YCW as postbiotics against CRC.
2.2.2. Other compounds of fungi
The anaerobic gut fungal commensals (such as Anaeromyces robustus, Caecomyces churrovis, Neocallimastix californiae, and Piromyces finnis) are reported to encode diverse biosynthetic enzymes for natural products and antimicrobial peptides (AMP), including polyketide synthases (PKS), non-ribosomal peptide synthases (NRPS), etc., (154). A significant proportion of fungal products were detected in the fecal samples of gnotobiotic mice, which may affect the growth and differentiation of host epithelial and immune cells (155).
A postbiotic yeast cell wall-based blend containing hydrolyzed yeast cell wall of Saccharomyces cerevisiae, organic acids (n-butyric acid), vitamins (ascorbic acid), and essential oils can partially improve the health of pigs which were treated with multiple mycotoxins including aflatoxin B1 and deoxynivalenol (156). The potential of using S. boulardii-derived postbiotics (for instance, polyamines, organic acids, enzymes, etc.) as dietary supplements has also been suggested (157). However, the benefits of non-YCW fungal products in CRC remain unclear which awaits further investigation in the future.
2.3. Postbiotics from non-bacterial/fungal gut microbes
Methanogenic archaea are the most well-studied archaea in human gut. While archaea do not produce enzymes to break down complex carbohydrates for nutrition (which may rely on other symbiotic species in the gut), they do have special glycosyltransferases that can link monosaccharides to various ligands to form glycoconjugates (158). Furthermore, it was indicated that methanogens can downregulate gut trimethylamine (TMA) concentration through specific metabolism of trimethylamine depletion (159, 160), highlighting the probiotic effect of methanogens on human health. However, it remains largely unknown about the involvement of fungus or archaea-derived postbiotics in CRC progression/inhibition, which relies on comprehensive studies in the future.
2.4. Inter-kingdom interaction between postbiotics and CRC
Although the biological functions of individual postbiotics have been comprehensively investigated, the interactions between postbiotics derived from multi-kingdom gut microbiota as well as their roles in the development of CRC were also worthy to be evaluated. Coker et al. showed the ecological network between bacteria and fungi in healthy gut, which is disrupted in CRC patients, and the antagonized interkingdom interactions contribute to the progression of CRC (161). In another study, the interaction of bacteria and archaea has been shown important for CRC development, as healthy individuals have a positive association between bacterial and archaeal diversity, which disappeared in patients with CRC (162). Furthermore, Liu et al. recently investigated the four-kingdom microbiota alterations with CRC metagenomic datasets from 8 distinct geographical cohorts and finally demonstrated the feasibility of multi-kingdom markers as CRC diagnostic tools (163). Narunsky-Haziza et al. explored the mycobiome in 35 cancer types including colorectal cancer from 4 international cohorts. They discovered the widely fungal existence and the permissive co-occurring fungi-bacterial ecologies across diverse cancer types. Thus, prognostic and diagnostic capacities of the tissue and plasma mycobiomes together with bacteriomes were suggested (164). The role of inter-kingdom interactions of microorganisms in CRC has been uncovered, however, whether it affects the inter-kingdoms interaction of postbiotics and CRC progression still need to be investigated in the future.
3. Biological functions of postbiotics against colorectal cancer
3.1. In vitro biological activities of postbiotics
3.1.1. Inhibition of cancer cell proliferation
The current hallmarks of cancer embody eight hallmark capabilities and two enabling characteristics, include sustaining proliferative signaling, evading growth suppressors, avoiding immune destruction, enabling replicative immortality, tumor-promoting inflammation, activating invasion and metastasis, inducing or accessing vasculature, genome instability and mutation, resisting cell death and deregulating cellular metabolism (165, 166). With the growing evidence and our increasing understanding of cancer biology, emerging hallmarks and enabling characteristics are now introduced into the hallmarks of cancer, involving “unlocking phenotypic plasticity,” “non-mutational epigenetic reprogramming,” “polymorphic microbiomes,” and “senescent cells” (167). Among these, excessive cell proliferation mainly resulted from three types of mutations: mitogen or its signal transducing molecules to convey mitogen information, mutation of late-G1 cell-cycle checkpoints regulated by pRB, and uncontrolled expression of Myc (168–170). Therefore, one of the anti-cancer strategies is to block cancer cell proliferation, and that is how most postbiotics works against cancer (171). It was observed that the ratio of G0/G1 cells increased when treated with gemcitabine together with butyrate, a common postbiotic from intestinal bacteria. The percentage of cells in S-phase significantly decreased when treated with butyrate in PANC-1 cell, indicating that butyrate could inhibit the proliferation of human pancreatic cancer cells (172). Postbiotic metabolites produced by certain strains of L. plantarum exhibited selective cytotoxic activity via antiproliferation of human breast cancer cells, suggesting their potential against breast cancers (173). In addition, Lazarova et al. proposed that butyrate can induce cell cycle arrest which was dependent on the modulation of Wnt signal activity in CRC (174). When combined with chemotherapeutic agents like irinotecan, butyrate significantly inhibited cell proliferation which promoted the efficacy of chemotherapies for the treatment of colorectal cancer (175).
In addition to butyrate, varies of postbiotics derived from Lactobacillus also exhibited anti-proliferative activities on colorectal cancer cells. For instance, the cell-free culture supernatant of Lactobacillus has inhibitory effect on the proliferation of colorectal cancer cells (176). Another interesting study reported that cell free cultural supernatants of Lactobacillus conjugated with various linoleic acids significantly reduced the transcriptional level of genes required for colon cancer cell growth and proliferation, such as CDK1/2/6, PLK1, and SKP2, indicating the potential benefits of those metabolites in treating CRC (177).
3.1.2. Induction of cancer cell apoptosis
Cell apoptosis is a programmed cell death with distinct morphological characteristics, including cell shrinkage, pyknosis, membrane blebbing, chromatin condensation and formation of cytoplasmic blebs (178, 179). Many diseases are related with apoptosis, such as AIDS (Acquired Immuno-Deficiency Syndrome), neurodegenerative diseases, autoimmune disorders, infectious disease, and cancer (180, 181). All somatic cells including cancer cells need to suppress apoptosis mainly by input of survival and trophic signals to survive (171). Besides, apoptosis also contributes to chemotherapy resistance (182). It’s a potential therapy to target the lesions that suppress apoptosis in tumor cells for cancer apart from inhibiting cancer cell proliferation (183–185). In recent years, postbiotics have been investigated to induce apoptosis in order to treat cancer (172, 173, 186). Postbiotic metabolites produced by six strains of L. plantarum induced apoptosis of MCF-7 breast cancer cells (173). Similarly, Kim et al. reported that heat-killed B. bifidum MG731, L. reuteri MG5346, and L. rhamnosus MG5200 induced apoptosis in human gastric cancer MKN1 cell to block tumor cell growth significantly. SCFAs, as one of the intensively investigated postbiotics, were also found to induce apoptosis in cancer treatment (186). SCFAs activated the mitochondrial apoptosis pathway and Fas death receptor apoptosis pathway in the breast cancer cell line 4T1 in vitro (186). Combining butyrate together with gemcitabine were found to decrease the percentage of live cells while increasing that of apoptotic cells in human pancreatic cancer cell lines (172). The underlying mechanism of butyrate’s anticarcinogenic effect is to modulate gene expression of key regulators in apoptosis and cell cycle such as cell cycle inhibitor p21 and proapoptotic protein BAK (99, 187–190).
Therefore, it is a potential way to harness postbiotics to treat CRC by inducing colorectal cancer cell apoptosis (183–185). The cultural supernatant of L. gallinarum significantly promoted apoptosis in CRC cell lines, HCT116 and LoVo, without impacting the cell growth of normal colonic epithelial cell line (NCM460), showing its specific beneficial potential against CRC (191). Consistently, Streptococcus thermophilus could secrete β-Galactosidase promoting CRC cells apoptosis and suppress the growth of CRC xenograft (192). Ma et al. discovered the induction of tumor apoptosis through PI3K/Akt signaling and caspase-3 activation in mice by the increasing production of beneficial metabolites including SCFAs when administrating sitosterol (193). Clostridium butyricum can suppress intestinal tumor development by modulating Wnt signaling and gut microbiota, including increasing the abundance of SCFAs-producing bacteria, especially butyrate-producing bacteria, thus suggest their potential efficacy against CRC (194). The in vitro activity of postbiotics on colorectal cancer cells is illustrated in Figure 2. On one side, postbiotics can efficiently inhibit colorectal cancer proliferation by disrupting the normal cell cycle of cancer cells; on the other side, postbiotics are effective in inducting cancer cell death through apoptotic pathways.
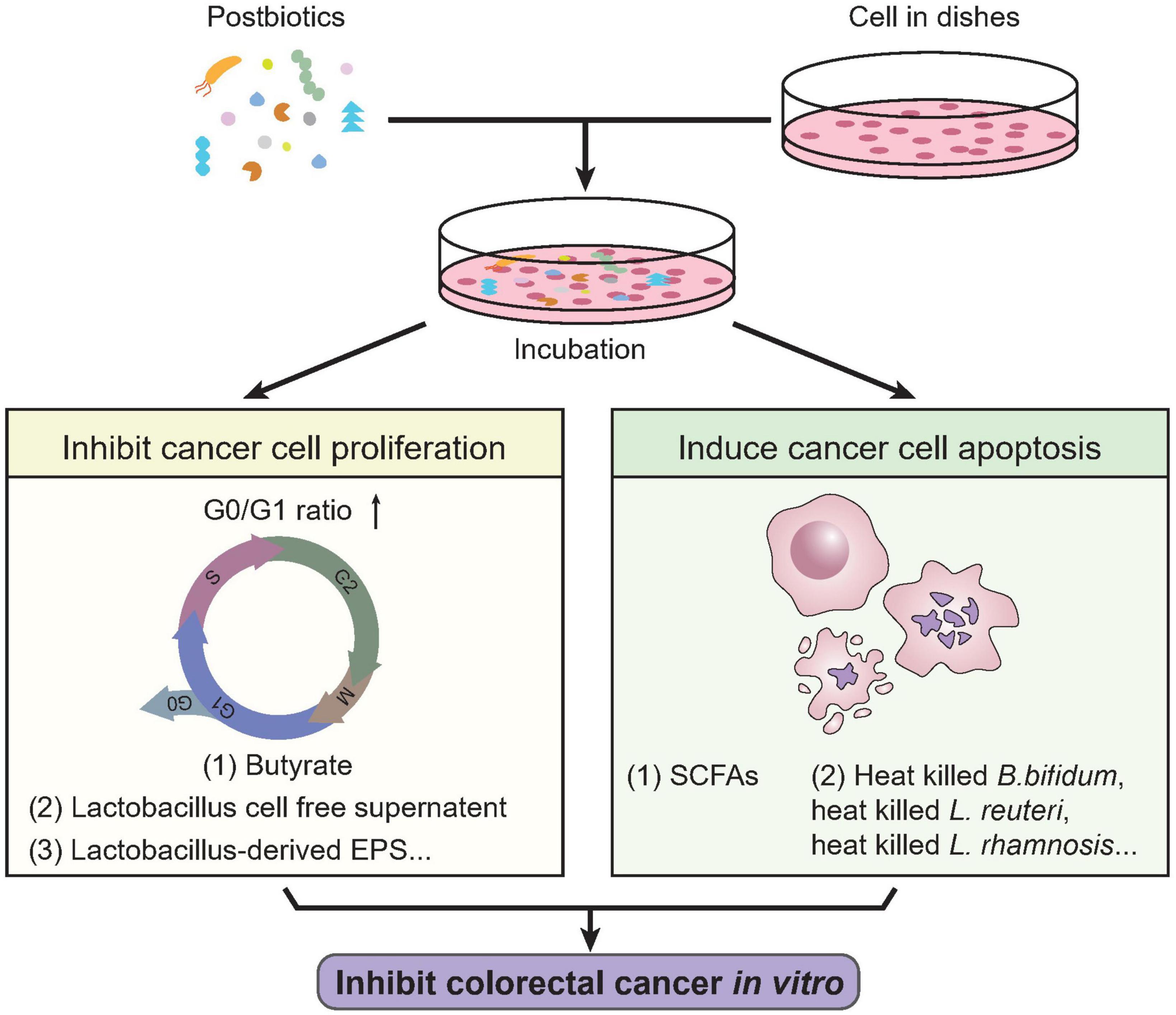
Figure 2. In vitro activities of postbiotics against colorectal cancer. Previous studies revealed multiple biological activities of postbiotics against colorectal cancer cells in vitro. Certain types of postbiotics like butyrate, lactobacillus cell free supernatant, Lactobacillus derived EPS, etc., were shown effective in inhibiting colorectal cancer cell proliferation, characterized by cell cycle arrest (increased G0/G1 ratio and decrease in S-phase). On the other hand, SCFAs, heat-killed B. Bifidum, L. reuteri, L. rhamnosis, etc., could induce apoptosis of colorectal cancer cells. These indicate the potential biological activity of postbiotics in treating CRC. SCFAs, short chain fatty acids.
3.2. In vivo biological activities of postbiotics
3.2.1. Modulation of gut microbial composition and abundance by postbiotics
The research on the effect of postbiotics on gut microbiota had been emerging in the past few years. It was found that postbiotics could modulate the composition and abundance of gut microbiota by promoting the growth of probiotics in the gastrointestinal tract of both healthy people and patients with different kinds of diseases (32). Terada et al. studied the effects of the consumption of heat-killed Enterococcus faecalis EC-12 on microbiota in healthy adults, and found that the levels of bifidobacteria and lactobacilli were significantly increased, while the abundance of lecithinase-positive clostridia including Clostridium perfringens, and Enterobacteriaceae were significantly decreased (195).
Postbiotics are reported to affect the gut microbiota of individuals with GI disorders, including inflammatory bowel disease (IBD). Sassone-Corsi et al. reported that microcins produced by Escherichia coli Nissle 1917 (EcN) could mediate inter and intra-species competition among the Enterobacteriaceae in the inflamed gut (196). The two major categories of IBD are Crohn’s disease (CD) and ulcerative colitis (197). In IBD patients, the pattern of gut microbiota dysbiosis is primarily characterized by a reduction in the abundance of bacterial species within the phylum of Firmicutes and Bacteroidetes and a relative increase of bacterial species belonging to the Enterobacteriaceae family, within the Phylum of Proteobacteria (198). Sokol et al. found that Faecalibacterium prausnitzii supernatant can effectively treat the intestinal microbiota disorders caused by CD and inhibit intestinal inflammation (197). Microbially modified bile acids were correlated with bacterial growth, activation of inflammation, and gut epithelium damage, suggesting an important role as components of intestinal antimicrobial defense (199, 200). It was revealed that cholic acid (primary bile acid) treatment significantly altered the composition of gut microbiota and promoted intestinal carcinogenesis, which is tightly correlated with the transformation of cholic acid to deoxycholic acid via bacterial 7α-dehydroxylation reaction, indicating the biological role of deoxycholic acid in gut microbiota homeostasis and CRC progression (201). Additionally, UCDA, which is mentioned earlier, was shown beneficial in improving the outcome of colitis via gut miciobiota modulation (202). It was also found that postbiotics could influence behavior in a mouse model of Citrobacter-induced colitis. A postbiotic derived from Limosilactobacillus fermentum and Lactobacillus delbrueckii shortened the small intestine and increased colon crypt depth in the mouse model, showing the protective effect of postbiotics on colitis (203).
The bacteriocin preparation fermented by lactic acid bacteria (LAB), lacidophilin tablets, has been extensively studied (204). Continuous intervention of lacidophilin tablets can reduce harmful bacteria and increase beneficial bacteria and normal bacteria. Lacidophilin tablets could adjust the composition and diversity of intestinal microbiota quickly in antibiotic-associated diarrhea (AAD) mice, and reconstruct the microbial communities which were mainly beneficial bacteria (205, 206). Specifically, lacidophilin-treated mice dramatically increased the abundance of Lactobacillus and decreased that of potential pathogens, such as Bacteroides, Parabacteroides, and Parasutterella, to nearly normal level (206). The interaction between postbiotics and anxiety and depressive disorder has also been studied. It showed that ADR-159 treatment, a heat-killed fermentate generated by two Lactobacillus strains, led to subtle but distinct changes in murine gut microbiota together with increased sociability and lower baseline corticosterone levels (stress hormone), supporting a beneficial effect on anxiety disorder and depressive disorder (207).
The in vivo function of postbiotics in regulating gut microbial composition in the context of CRC has not been fully elucidated. However, accumulating evidence showing the influence of gut microbial composition on CRC progression. Kimoto-Nira et al. investigated influence of long-term consumption of a Lactococcus lactis strain G50 on the intestinal flora of the senescence-accelerated mouse, showing that G50 could suppress the intestinal growth of H2S-producing bacteria (208). The H2S-producing intestinal bacteria include Salmonella sp., Shigella sp., and Citrobacter sp., all of which are associated with CRC (209). Gut microbiota dysbiosis was discovered in CRC patients. Clostridium butyricum supernatant was reported to decrease the relative abundance of pathogenic bacteria, such as Desulfovibrio, Odoribacter, and Helicobacter, which are related to CRC (194). Collectively, these results highlight the potential role of postbiotics in the prevention of CRC.
3.2.2. Modulation of gut commensalism by postbiotics
The first line of host defense against both native gut commensals and external pathogens is the intestinal mucosal barrier, which consists of tightly connected epithelial cells and is protected by two host-secreted mucous layers, inner mucus layer and outer mucus layer (210, 211). The intestinal mucus layer was composed of secreted mucin glycoproteins and other substances, which were secreted by goblet cell (212). It is found that mucin can not only provide adhesion sites for intestinal symbiotic bacteria, but can also separate the pathogens from Caco-2 cells (213). However, several intestinal pathogens could secrete enzymes to disrupt the mucous barrier or promote their invasion capability to facilitate their colonization and successful infection (214). Postbiotics could modulate gut commensalism by affecting the intestinal mucosal barrier. Bacterial SCFAs and other metabolites can induce mucus biosynthesis in germ-free mice (215, 216). For example, butyrate-treated human colorectal cells LS174T cells increased mucin protein expression, which enhanced adherence of probiotics Lactobacillus and Bifidobacterium and eventually inhibited the pathogenic Escherichia coli (E. coli) attachment (217, 218). Other probiotics supernatant, such as Lactobacillus rhamnosus GG culture supernatant, have been proved to promote mucin production to help the gut colonization of symbiotic bacteria and exert a preventive effect on most gut-derived pathogenic infections including E. coli K1 (218).
Adhesion is an important step for gut commensalism. If postbiotics provide adhesins such as fimbriae and lectins, they can compete with resident microorganisms for adhesion sites (219, 220). The isolated lectin domains of Llp1 and Llp2 exhibit significant inhibitory activity against biofilm formation of various pathogens, including clinical Salmonella species and pathogenic E. coli (220). EPS are extracellular polysaccharides attached to the bacterial cell wall which can affect adhesion by shielding cell surface adhesins or acting as ligands (221). It was found that EPS produced by LAB could block the adhesion of harmful bacteria to intestinal epithelial cells by inhibiting binding of adhesion factors to bacterial cell surface (222). Besides, different kinds of EPS could also modulate LAB’s efficiency of adhesion to intestinal epithelial cells in vitro (223). In addition to EPS, it was also found that heat-treated Lactobacillus acidophilus strain LB could inhibit the adhesion of entero-invasive pathogens to human intestinal Caco-2 cells (224). Thus, postbiotics may exert benefit in treating CRC by modulating gut commensalism through intestinal barrier remodulation (225).
3.2.3. Immunomodulatory activity of postbiotics
Postbiotics can modulate gut pathogenesis not only through competitive inhibition of receptor adhesion but also by changing the intestine barrier function or expression of specific regulatory genes. Tsilingiri et al. first confirmed the anti-inflammatory activity of the Lactobacillus paracasei supernatant (SN) after Salmonella infection of healthy tissue (226). They showed that the secretion of TNF-α, which is a proinflammatory cytokine, was increased after Salmonella infection. Proinflammatory cytokines are positive mediators of inflammation (227). But this effect was eliminated when SN was added together with Salmonella, manifesting an inflammatory potential of Salmonella (226). Mechanistically, SN can stimulate the epithelium cells resistant to Salmonella invasion rather than affecting Salmonella proliferation (226). Similarly, acetate was discovered to significantly increase resistance to enterohaemorrhagic E. coli O157:H7 infection in a mouse model (228). The could be attributed to the sealing properties of acetate on the intestinal barrier, eventually preventing lethal toxins from entering the general circulation (228). Ramakrishna et al. showed that some probiotics and their metabolites prevent pathogen invasion not only by blocking adherence sites but also by upregulating gene expression of MUC2 and antimicrobial peptides (229). Consistent with this is that Mack et al. showed that part of the beneficial effect of L. ramnosus and its supernatant was mediated by induction of mucin genes in intestinal epithelial cells (230).
The gut microbiota dysbiosis could promote the progression of CRC probably through pro-inflammatory responses (231–233). For example, the next generation sequencing analysis revealed that enrichment in Fusobacterium could promote inflammation in CRC (234). Inflammatory responses play an important role in different stages including initiation, malignant conversion, invasion, and metastasis of tumor such as CRC development (235). Some anti-inflammatory drugs like aspirin and sulindac were proved to prevent CRC and decrease side effects of inflammatory symptoms, hence bring new insights of CRC prevention and treatment (236, 237). Thus, the immunomodulatory activity of postbiotics can be harnessed in CRC prevention and treatment. For example, bile acids were reported to modulate gastrointestinal inflammation and play a role in CRC prevention (110). Niacin, which is also called nicotinic acid or vitamin B3, can be regarded as another kind of postbiotic because it could also be produced by intestinal microbes and exerts benefits to human health (238). It was reported that niacin can modulate immune responses through G protein-coupled receptor 109a (GPCR109a) to prevent colitis and colon cancer (239, 240). Apart from the anti-inflammation activity, postbiotics can also ameliorate CRC progression by inhibiting the enzymatic activity of pathogenic bacteria and reducing the amount and activity of virulence factors to attenuate gut pathogenesis (241, 242). However, certain microorganisms like Fusobacterium nucleatum, Escherichia coli NC101 and Bacteroides fragilis not only induce inflammation and ROS-mediated genotoxicity but also secrete toxins which induce DNA damage responses to promote CRC (243–245).
3.2.4. Modulation of the gut microbiota-host interactions by postbiotics
Gut microbiota-host interaction is critical for human health. Host inflammatory response is one of the most important signals modulating the homeostasis between gut microbiota and host. It was reported that inflammatory cell infiltration in adipose tissues, liver, and pancreatic islets is associated with an increase in LPS plasma levels, possibly derived from gut bacteria (246, 247). Moreover, the gut microbiota can regulate host inflammation by affecting cytokine secretion as well as the differentiation of inflammatory cell types, demonstrating the dynamic interplay between gut microbiota and the host immune (248, 249).
For instance, F. nucleatum promoted carcinogenesis by stimulating Wnt signaling pathway in CRC (250, 251). And the adhesion molecule FadA from F. nucleatum can bind to E cadherin, leading to activation of β-catenin and tumor development (252). Bacteroides fragilis was also known as the risk factor of CRC. Bacteroides fragilis toxins can bind to specific colonic epithelial receptor to activate Wnt and NF-κB signaling cascades, leading to an increased proinflammatory reactions including TH17 response and induction of DNA damage response, aggravating the colorectal tumorigenesis (253, 254). All the discussed in vivo biological roles of postbiotics in preventing CRC are illustrated in Figure 3. The beneficial roles of postbiotics in vivo are achieved via the following ways: postbiotics are effective in modulating gut microbial composition and the abundance of certain microbial species, which lead to homeostasis of gut microbiota; postbiotics can also benefit gut commensalism by promoting the integrity of intestinal mucosa tissue; postbiotics can specifically inhibit the invasion of pathogenic microbes in the gut; postbiotics are involved in the interaction between gut microbiota and host cells.
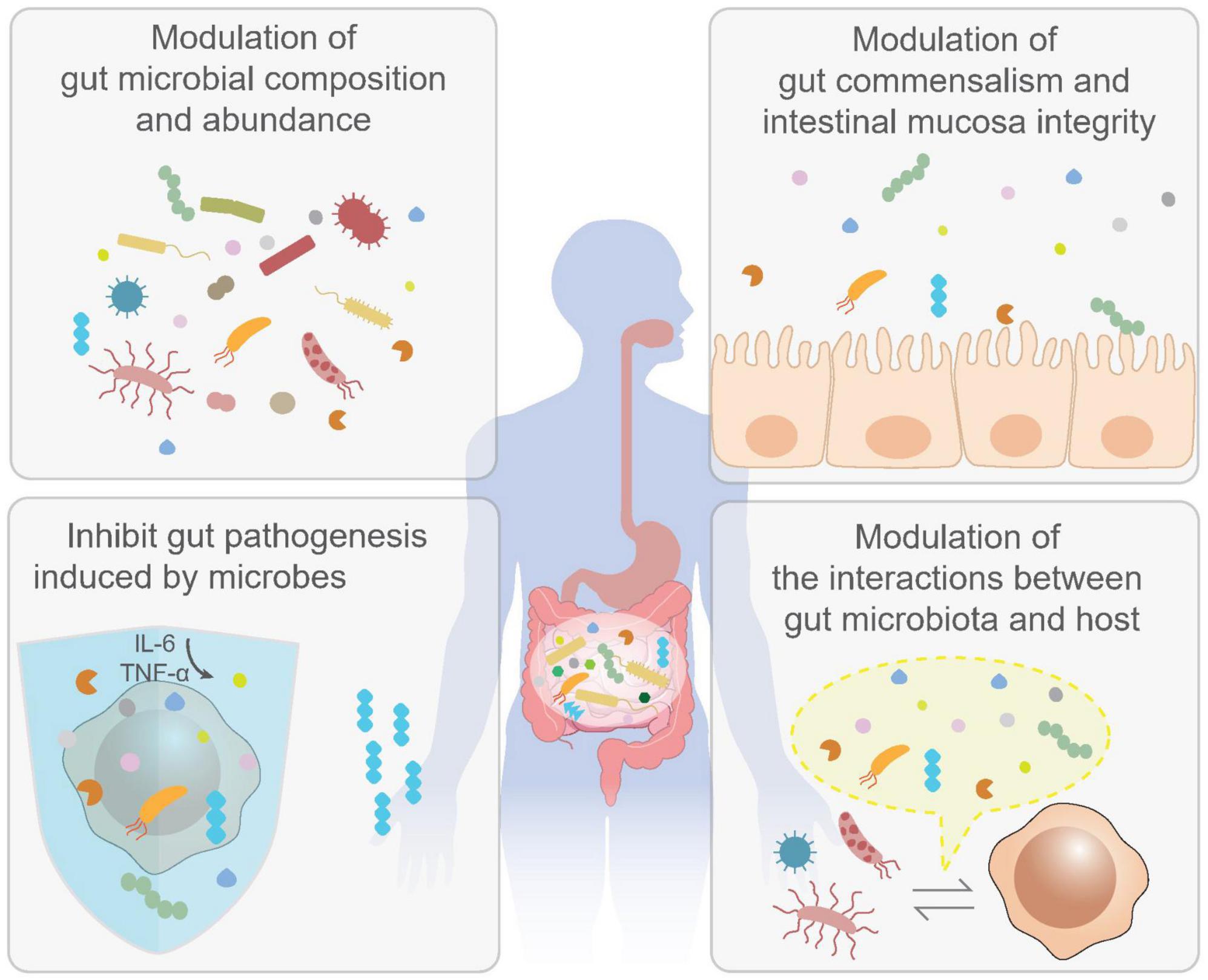
Figure 3. Biological activities of postbiotics in vivo. The benefits of postbiotics have also been well-documented by enormous in vivo studies. It has been shown that postbiotics are effective in the modulation of gut microbial composition and abundance (upper left); maintaining gut commensalism as well as intestinal mucosa integrity (upper right); inhibit gut pathogenesis induced by gut microbes (lower left) and modulation of interactions between gut microbiota and host and (lower right). These activities may contribute to the prevention and treatment of colorectal cancer (CRC).
4. Molecular mechanisms of postbiotics against colorectal cancer
4.1. SCFA
Regarding the molecular mechanism of SCFAs in CRC development, it was shown that butyrate acid could suppress the expression of Neuropilin-1 (NRP-1) by inhibiting the activity of specificity protein 1 (Sp1), which eventually leading to the inhibition of angiogenesis and metastasis of colorectal cancer cells (255). In addition, SCFAs were shown to inhibit CRC cell proliferation by promoting the expression of endocan through extracellular signal-regulated kinases (ERK)/Mitogen-activated protein kinase (MAPK) signals (256). On the other hand, SCFAs induced apoptosis of colorectal cancer cells via the upregulation of Bcl-2-associated X protein (Bax) expression (257). G-protein coupled receptors (GPCR) 41/43 and GPCR109a were identified as the binding partners of SCFAs, suggesting the involvement of G protein as well as other downstream signals in mediating SCFAs’ biological functions on the host side (258, 259). Interestingly, it was revealed that deficiency of GPCR43 was associated with reduced abundance of gut commensal bacteria Bifidobacterium species and severe gut inflammation, meanwhile, restoration of Bifidobacterium sp. in GPCR43 knockout mice attenuated both inflammation and carcinogenesis (260, 261). In contrast, GPCR43 knockout mice exhibited a significantly higher abundance of Helicobacter hepaticus and Prevotellaceae family strains, both of which are tightly correlated with high intensity of GI inflammation and CRC development (261). These results imply the complicated interplay among SCFAs, gut microbiome, and CRC development. In an independent study, it was found that the lack of GPCR43 was associated with higher incidence of colonic tumor burden in Apcmin/+ mice [which is deficient in one allele of the adenomatous polyposis coli (Apc) gene and is highly vulnerable to the development of spontaneous CRC] (262). Mechanistic analysis further revealed that loss of GPCR43 leads to upregulation of cAMP/Protein Kinase A (PKA)/cAMP response element binding protein (CREBP)/histone deacetylase (HDAC) signaling axis, along with upregulated Wnt signaling and the downstream activation of nuclear β-catenin and c-Myc, suggesting the role of G protein-linked epigenetic modification in promoting CRC development (263). In contrast, the involvement of GPCR41 in SCFA-mediated signaling during CRC development has not been fully clarified, except for one study showing that the overexpression of GPCR41 depressed forskolin-stimulated cyclic AMP (cAMP) formation in response to SCFA treatment, similar to the endogenous function of GPCR41 in regulating Gαi/o activity (264). Overexpression of GPCR41 resulted in the inhibition of SCFA-induced histone acetylation, as well as reverse anti-proliferative and apoptotic effects of SCFAs (264). GPCR109a is another known receptor for SCFAs and is predominantly expressed in colon tissues (259). It was reported that GPCR109a is the receptor for butyrate (240, 259, 265). Interestingly, it was found that the expression level of GPCR109a is negatively correlated with colon tumor severity, which was significantly decreased in the normal colon tissue from Apcmin/+ mice as compared with that of wild-type littermates, and was nearly absent in the colon tumor tissues of Apcmin/+ mice (259). Loss of GPCR109a leads to the increased DNA methylation and upregulated activity of DNA methyltransferases, DNMT1 and DNMT3b (259). GPCR109a activation also resulted in elevated apoptosis through the inhibition of anti-apoptotic genes like B-cell lymphoma 2 (Bcl-2) and significant upregulation of pro-apoptotic effectors including Fas ligand (FasL), Fas associated death domain (FADD), and TNF receptor 1 (TNFR1). In addition, GPCR109a activation also led to the inhibition of nuclear transcription factor κB (NF-κB), which plays a central role in promoting colon inflammation, which may contribute to CRC development (259). Collectively, there are emerging lines of evidence illustrating the molecular mechanism of SCFA in modulating different host responses (240), however, the exact mechanism of SCFA in the context of CRC awaits further investigation.
In addition, recent studies revealed that the overall response of cancer patients following immune checkpoint blockade (ICB) treatments was affected by the composition of gut microbiota (266). Transplantation of fecal microbiota or commensal microbes significantly improved the efficacy of ICBs (267, 268). Notably, oral administration of SCFAs exhibits obvious benefits against cancers suggesting part of the regulatory mechanism of gut microbiota and host immune response (269). As accumulating immune therapeutic strategies are now approved in the clinical treatment of CRC, the role of SCFAs in colorectal cancer immunotherapies would be another interesting topic worth further investigation. Besides, SCFAs also protect the mucosal layer from damage by lowering the level of immune modulators, such as prostaglandins, formed by cyclooxygenase 2 (COX-2). It is reported that the level of COX-2 mRNA is increased in intestinal cancer tissues, and the level of prostaglandin E2, which contributes to inflammation and tumor growth, is also higher than normal tissues (270), while butyrate was found to reduce the expression of COX-2 in tumor tissues to prevent mucosal layer from prostaglandins (270).
4.2. EPS
Microorganism-derived components, including EPS, are generally recognized by host innate immune receptors named pattern recognition receptors (PRRs) including toll-like receptors (TLRs), nucleotide oligomerization domain-like receptors (NLRs), Retinoic acid-inducible gene (RIG)-I like receptors and lectin receptors, etc., (271). Notably, C-type lectin receptors (CLRs) can specifically recognize and bind to different kinds of bacteria-derived glycans, which induced receptor dimerization. Downstream signals involve the recruitment of immune tyrosine-based activation motif (ITAM) and activation of spleen tyrosine kinase (SYK), which further activating NF-κB and various immune responses (272). In addition, TLR2 was considered as another receptor of EPS. In 2005, Mazmanian et al. showed that polysaccharide (PSA) from gut commensal bacterium Bacteroides fragilis promoted the maturation of the host immune system. The “loss-of-function” mutant of B. fragilis in PSA production resulted in systemic T cell deficiencies and Th1/Th2 cell imbalance, indicating the role of PSA in host immune regulation (273). PSA derived from B. fragilis activated CD4+ T cells and the subsequent predominant Th1 response, which was diminished in TLR2–/– mice (274). TLR-2 pathway promoted the homeostasis of gut immunologic tolerance through sensing the B. fragilis derived PSA molecules, leading to activation of Foxp3+ regulatory T cells (275). A novel EPS from the biofilm of Thermus aquaticus YT-1 was proved to be effective in inducing macrophage activation through TLR-2 signal (276). Collectively, these results revealed the involvement of host innate immune system in sensing and transducing EPS-derived signals as well as their essential role in regulating host immune response.
In addition, accumulating evidence also suggested the immune regulatory activity of EPS on phagocytosis, immune activation, and antigen presentation (277–279). Previous studies demonstrated the capability of host immune system in distinguishing commensal bacteria-derived EPS from that of pathogens (80). It was further revealed that EPS could exhibit opposite immune regulating activities depending on the molecular property. For example, the negatively charged or small EPS can act as stimulators of immune cells, while neutral and large EPS would induce suppressive effects (280, 281). An in vivo study showed that EPS from Bacteroides fragilis protected animals from Helicobacter hepaticus-induced colitis by suppressing the production of IL-17, suggesting the anti-inflammatory activity of EPS and protective role against CRC (282). Recently, Kawaharada et al. identified epr3 gene in Lotus japonicus as the receptor of EPS derived from Mesorhizobium loti (283), the molecular structure of which was illustrated later (284). These results provided new insights into the functional mechanism of EPS as well as its role in plant-microbe symbiosis. However, the exact molecular mechanism of EPS in the mammalian system remains unclear. One possible explanation is the huge structural heterogeneity of EPS variants, which makes it difficult to access each host binding partner precisely (80). Overall, with ongoing intensive investigations, the potency of EPS as a promising type of postbiotic candidate in CRC treatment will be elucidated in the future.
4.3. Enzymes
To determine the biological role of bacterial enzymes in inhibiting CRC progression, LeBlanc et al. detected the existence of antioxidant enzymes, such as catalase (CAT) or superoxide dismutase (SOD) in the gut and their function in eliminating 2,4,6-Trinitrobenzenesulfonic acid (TNBS)-induced Crohn’s disease in mice, which was believed to be the major trigger of CRC (285). It was indicated that mice received engineered CAT or SOD-producing LAB showed a faster recovery of initial weight loss, increased enzymatic activities in the gut, and less intestinal inflammation compared to that with WT strain or blank controls, suggesting the protective role of antioxidant enzymes in CRC pathogenesis. Carmen et al. further demonstrated that the production of antioxidant enzymes by genetically modified LAB is effective against DMH-induced colorectal cancer in mice model (286). In addition, Lactobacillus strains with either the highest catalase activity or the highest dismutase-like activity were assessed for their protective effects on colorectal inflammation (287). It was found that Lactobacillus strains with dismutase-like activity were more effective in alleviating intestinal inflammation than strains producing catalase, indicating the crucial role of superoxide anion radical decomposition in this process (287). Collectively, the biological activity of bacterial enzymes in alleviating colorectal inflammation or preventing CRC has been uncovered, while the underlying mechanism remains unclear.
4.4. Other postbiotics
The relationship between bile acid transportation and gut microbial metabolism, as well as the implications on CRC development, has received more and more attention in recent years. Numerous studies have demonstrated the carcinogenic activity of bile acids, especially the secondary bile acids, in the development of colorectal cancer (288). It was revealed that bile acids disrupt colonic epithelial cells by inducing reactive oxygen species (ROS) production, genome destabilization as well as other effects, which eventually lead to colon tumorigenesis (289–291). Mechanistic studies further showed the involvement of MAPK, phosphoinositide 3-kinase (PI3K)/Akt and NF-κB in this process (292, 293). Despite this, other studies revealed the pro-apoptotic effect of the secondary bile acid deoxycholate acid, which is produced by gut microbiota, on colorectal cancer, as treatment with as low as 0.5 mM deoxycholate acid was able to induce apoptosis of colorectal cancer cells, which was comparable to the activity of SCFAs (294). However, the molecular mechanism remains unclear. A recent study illustrated different functions of primary and secondary bile acids in regulating host immune response against liver cancer (295). Specifically, primary bile acids produced by host metabolism increased CXCL16 expression, which could recognize and bind to CXCR6 expressed on natural killer T (NKT) cells. This interaction eventually led to hepatic tumor infiltration and tumor targeting of NKT cells. In contrast, secondary bile acids processed by gram-positive bacteria in the gut exhibited an opposite effect (295). Taken together, the exact role or molecular mechanism of bile acids, especially the microbial derived secondary bile acids in CRC development remain controversial, which relies on future studies to be better addressed.
For the working mechanism of tryptophan metabolites, it was found that indole metabolites and kynurenine can interact with aryl hydrocarbon receptor (AHR) to induce T regulatory cells differentiation, confine Th17 and Th1 responses and produce anti-inflammatory mediators (296, 297); kynurenine reduces tumor-infiltrating CD8+ T cells and mediates immune evasion of tumor cells (298), while indole metabolites alleviate colitis and protect against colorectal cancer (299, 300). Serotonin (5-HT) was also found to be critical in regulating GI inflammation, while the exact activity remains controversial. Evidence proved the anti-inflammatory role of serotonin in colitis, while others revealed the enhanced effect of serotonin in colorectal cancer progression (301). The interaction between tryptophan metabolites and host AHR is essential for regulating colon barrier function and inducing regulatory T cell differentiation (302). The association between fecal tryptophan metabolites and gut barrier functions was further validated by a clinical study, implying the involvement of tryptophan metabolism in the development of CRC (136). Figure 4 illustrated the presently identified working mechanisms of different postbiotics that are beneficial for CRC protection. Briefly, SCFAs (e.g., propionate and butyrate) can modulate cancer cell growth and promoting apoptosis of cancer cells via GPCR41/43/109a. Microbially derived secondary bile acid, notably deoxycholate acid, can efficiently induce cancer cell death via apoptotic pathway, which mimics SCFAs. EPS, notably PSA, can modulate Treg cell differentiation and activation through TLR2 sensing. Tryptophan metabolites are also effective in DC induced Treg differentiation via AHR sensing, which suppresses pro-inflammatory Th1/Th17 response. All these activities are implied in preventing CRC progression.
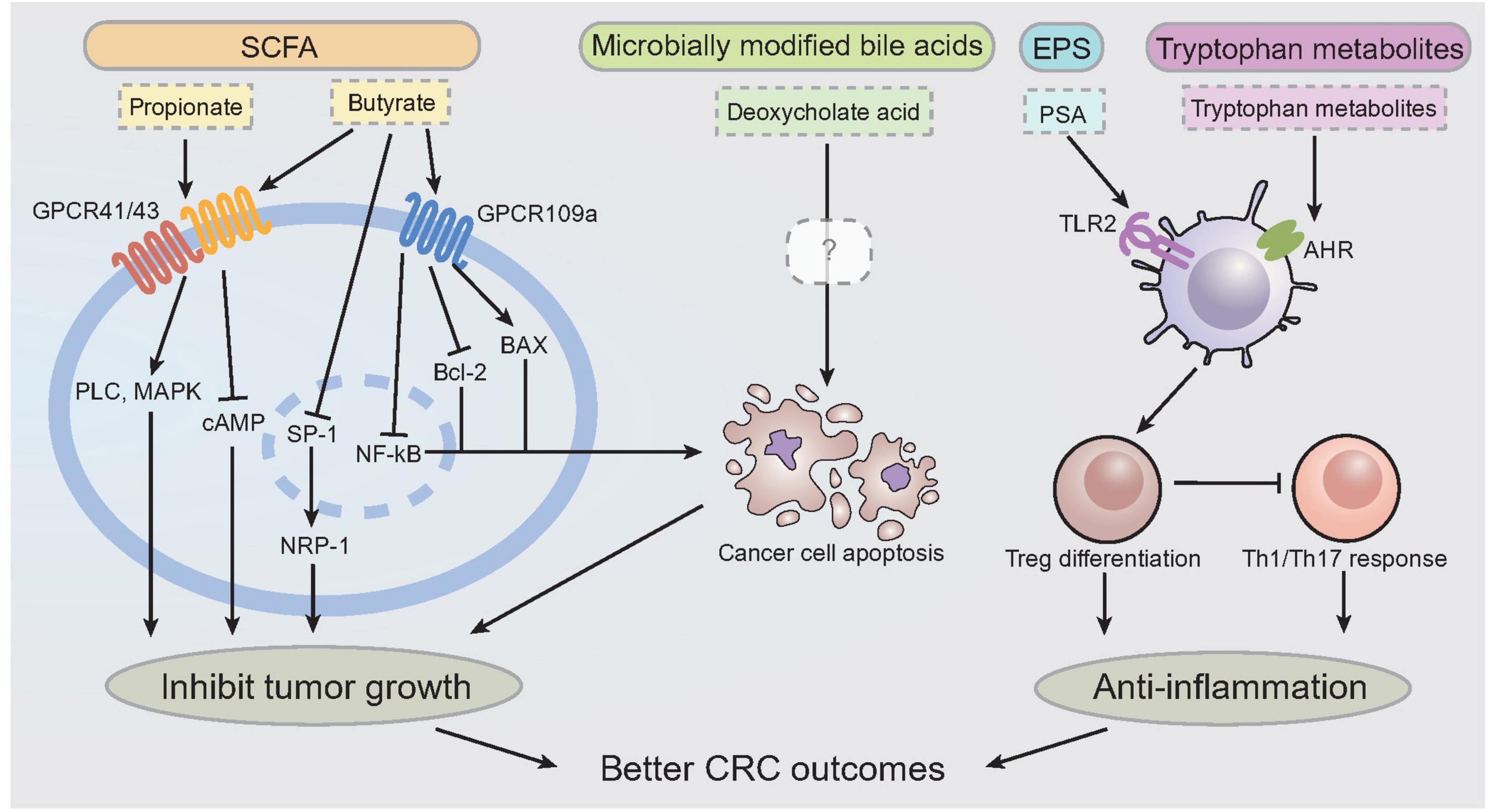
Figure 4. Molecular mechanism of postbiotics on colorectal cancer prevention and treatment. Working mechanisms of postbiotics against CRC. It has been shown that propionate and butyrate can bind to GPCR41/43 to promote the activation of PLC, MAPK, and signals intracellularly to inhibit tumor cell growth. Butyrate also binds to GPCR109a, which inhibits downstream intracellular signals including SP-1, NRP-1, to inhibit tumor cell growth, and also promote apoptosis via BAX and inhibition of Bcl-2, NF-kB activity. Deoxycholate acid, a type of microbially modified bile acids, can promote cancer cell apoptosis through unidentified mechanism. PSA interacts with TLR2 to promote the activation of DCs, which lead to the upregulation of T regulatory cells in GI environment. Tryptophan metabolites interact with AHR to exert the anti-inflammatory activity as well. GPCR, G-protein couple receptor; PLC, phospholipase C; MAPK, mitogen-activated protein kinase; cAMP, cyclic AMP; SP-1, Specific protein 1; NRP-1, Neuropilin-1; PSA, polysaccharide; TLR, toll like receptor; DC, dendritic cell; AHR, aryl hydrocarbon receptor; CRC, colorectal cancer.
5. Clinical potency of postbiotics in CRC
5.1. Description of analysis
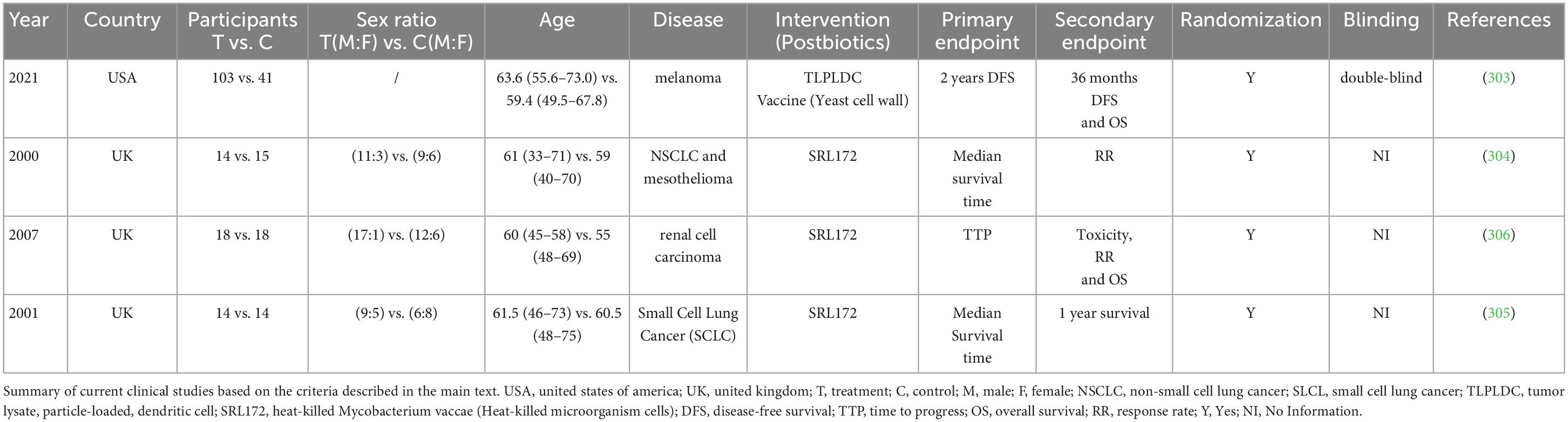
To obtain an complete picture of our current understanding regarding the clinical benefits of postbiotics, especially in cancer treatment, we went through multiple online databases [PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), Wanfang, etc.] to collect clinical studies published on or before the date of September 18, 2022 using following keywords: “postbiotics,” “Heat-killed bacterial cells,” “Cell-free supernatant,” “Teichoic acids,” “Exopolysaccharides,” “Short chain fatty acids,” “Enzyme,” “Yeast cell wall,” “Bile acid,” “Tryptophan,” “Bacteriocins,” “Vitamin,” “Neurotransmitter substance,” “Microbiota,” “therapy,” and “cancer,” without any other restrictions.
A total of 538 literatures were systematically reviewed, and only a few original clinical studies investigating the effects of postbiotics on the outcome of cancer patients were selected for further analysis, which are listed in Table 2 and explicitly described as below (Table 2).
5.2. Major findings and interpretation
Among the selected studies, Vreeland et al. launched a randomized, double-blind, placebo-controlled trial in order to evaluate the clinical efficacy of “Tumor Lysate, Particle-Loaded, Dendritic Cell (TLPLDC)” vaccine as adjutant therapy to advanced-stage melanoma, which declared that patients enrolled in TLPLDC group had longer 24 months disease-free survival (62.9 vs. 34.8%, p = 0.041) (303). In addition, O’Brien et al. had demonstrated that traditional chemotherapy combined with heat-killed Mycobacterium vaccae SRL172 improved the response rate, median survival and 1 year survival by randomized trials in non-small cell lung cancer (NSCLC) and mesothelioma. Moreover, they also found that SRL172 could improve the situation of sleep (p = 0.08) and appetite (p = 0.01) of the patients (304). Assersohn et al. had applied traditional chemotherapy combined with heat-killed Mycobacterium vaccae SRL172 to Small Cell Lung Cancer (SCLC), which improved the median survival of the patients (305). Patel et al. suggested that patients with renal cell carcinoma treated with SRL172 plus IL-2 reduced adverse events in comparison to those treated with IL-2 alone (p < 0.001) (306). Unfortunately, none of those studies directly accessed the therapeutic effect of postbiotics on CRC patients.
Overall, the clinical effects of postbiotics in cancer prevention/treatment, in particular CRC, are still lacking as few studies have been drown by far. Additional evidence are still required to demonstrate the potential clinical effects of postbiotics in CRC prevention and treatment, which relied on well-designed randomized placebo-controlled clinical intervention trials.
6. “Pre-Pro-Post”: Comparison of different strategies in CRC treatment
6.1. Conceptual difference of prebiotics, probiotics, and postbiotics
With growing interest in uncovering the relationship between gut microbiota and human health in recent decades, different gut microbes are identified to be either beneficial or harmful in the pathogenesis of human diseases. Microbes that could promote human health or prevent disease are termed probiotics and emerged as novel strategies in clinical treatments (307). In 2014, the ISAPP defined probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (31). Dietary interventions, on the other hand, are more convenient in modulating gut microbiota without causing noticeable side effects. Effective components in such diets are called prebiotics and were also widely accepted as important supplementations by clinicians, researchers and consumers (308). In 2017, ISAPP further defined prebiotics as “a substrate that is selectively utilized by host microorganisms conferring a health benefit” (30). With the accumulation of research on probiotics, prebiotics, and microorganisms, it was found that inanimate microorganisms and their metabolites also behave beneficial activities. The first consensus definition of postbiotics was proposed in 2021, when ISAPP defined postbiotics as “preparation of inanimate microorganisms and/or their components that confers health benefits on the host” (31).
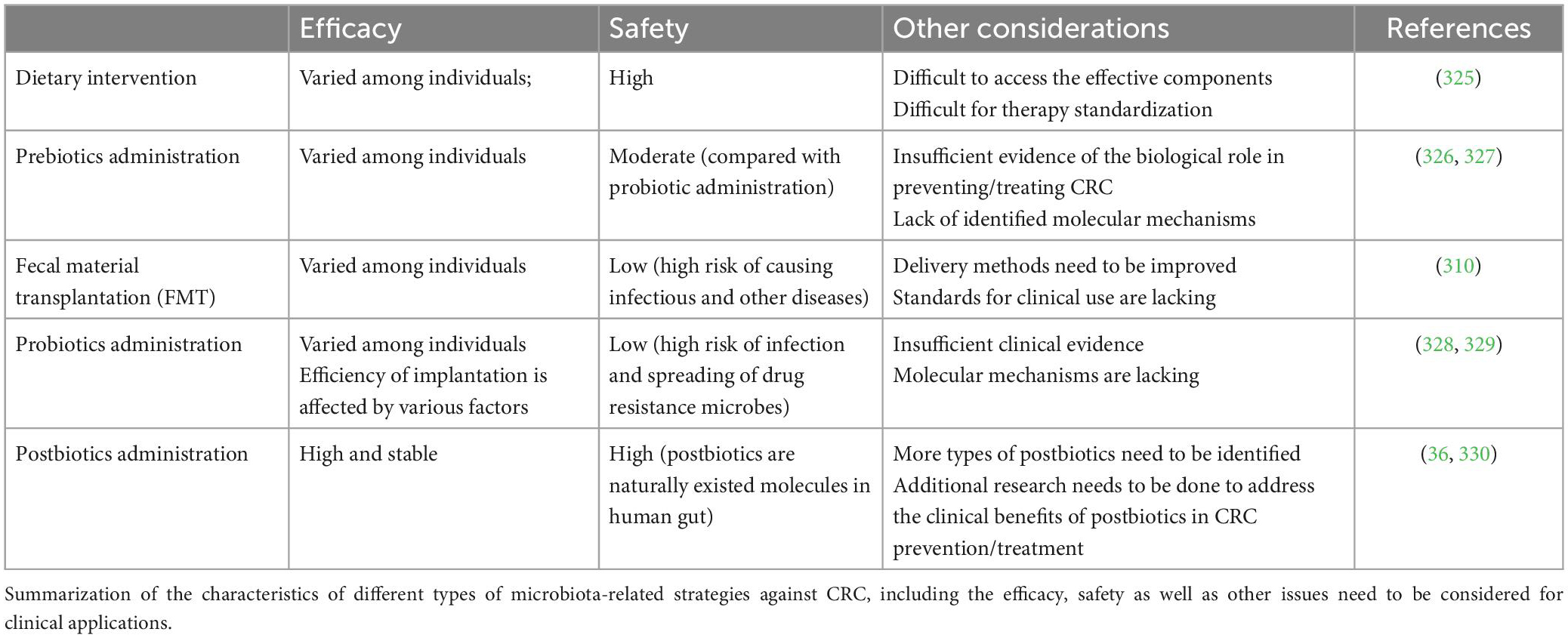
6.2. Efficacy/side-effects of different strategies on CRC treatments
Fecal microbiota transplantation (FMT), an emerging approach developed and used by clinicians recently, also achieved promising effects in treating multiple kinds of diseases including CRC (309, 310). While some studies provided mechanistic insights into how probiotic may confer benefits, they were largely performed in in vitro models. The clinical evidence for the efficacy of current probiotics or prebiotics as in medical use is still insufficient and inconsistent (311). In addition, many factors may affect the efficiency of exogeneous microbial implantation, including the colonization resistance by residing commensal species (312, 313). Individual response to probiotic colonization or dietary intervention also varies that further dampens the efficacy of “one strain fits all” strategy (314). FMT also introduces uncertainties from donor and fecal materials, which leads to unpredictable outcome in clinic regarding to efficiency or safety (315). Refined FMT strategy with defined consortium of cultured bacterial species seems to be able to overcome these limitations (316). Another major limitation of prebiotic or probiotic administration is the lack of clarity of defined functional mechanisms. Although previous studies have deciphered the molecular interplays among dietary intervention/probiotic administration, gut microbiota, and host response, a large proportion of studies are still observational or correlational. Thus, a comprehensive understanding of the causal interactions is urgent. Furthermore, the safety concerns of probiotic supplementation and FMT are also raised in pre-clinical studies (317).
In contrast, postbiotic administration is outperformed in both efficacy and safety to meet clinical requirements (28). As composed of derivatives from gut microbes, postbiotics refers to “chemicals” rather than “organisms.” With their natural existence in human gut, the safety issue in clinical use is feasible to be addressed through well-designed preclinical and clinical trials. Several studies found that probiotics could cause infectious risks. For example, Wagner et al. colonized athymic mice with human isolates of Lactobacillus reuteri, Lactobacillus acidophilus, Bifidobacterium animalis, or Lactobacillus rhamnosus GG (LGG). The result showed that colonization with the probiotics L. reuteri and LGG led to death in some athymic neonatal mice. This finding suggests that due to the nature of immune deficiency in neonates, it may put them at particularly high risk of sepsis when administrating probiotics (318). Besides, improper administration of probiotics may increase the risk of the spreading of drug resistant strains. For instance, Lactobacillus (such as Lactobacillus plantarum), which have plenty resistance factors, may become the transmission channel of antibiotic resistance genes (319). In terms of stability, researchers found that postbiotics are supposed to be more stable than the living bacteria they are derived from. That is, postbiotics are more suitable for long term use than probiotics. Venema et al. reported that peptides with antimicrobial properties, namely bacilysin and chlorotetaine, produced by Bacillus sp. strain CS93 are water soluble and active over a wide pH range, which could allow their application in a wide variety of food products (320, 321). Furthermore, for postbiotics, the characterization of molecular property, as well as elucidation of underlying mechanisms, would be much more straightforward through current approaches (28). In Table 3, we summarized the features, advantages and disadvantages of different strategies in treating CRC.
7. Conclusion and future perspectives
Over the last decades, the potency of postbiotics in treating various diseases has been uncovered by accumulating evidence. Compared with other microbiota manipulating strategies like dietary intervention, probiotic administration, or fecal material transplantation (FMT), postbiotic administration is superior in both safety and efficacy, which benefits from the clarification of their molecular identity and functional mechanism. However, there are still challenges in microbiome targeted therapy. First, only a limited categories of postbiotics are known, mainly restricted by the available techniques. Integrative methods including high-throughput multi-omics analysis, computational processing, large-scale screening and animal models are considered as the future direction for identification of new bioactive molecules from gut microbiota (322). Second, human microbiome may have unexpected interaction with supplemental metabolites, which may cause dysbiosis or transformation of metabolites into inactive or even toxic states. In addition, artificial upregulation of certain metabolites in human gut may affect the homeostasis of the endogenous metabolic feedback loop, which will cause the acquired deficiency in the production of metabolites by the human body (323). More importantly, low levels of a given metabolite in fecal sample do not precisely reflect its physiological roles in the gut where it exerts benefits. Multiple sampling sites would be an improvement in overcoming this limitation (324). Overall, it is necessary to systemically identify the existence of new bioactive compounds from gut microbes in the human body as well as their interaction between metabolite and resident microbiome prior to their utilization. Finally, commercialization of postbiotics should be controlled in a cost-effective way, which cannot be achieved by lab-based approaches and largely rely on the optimization of synthesis strategies by industries (35).
To systemically analyze the effects of different postbiotics conducted by different studies, appropriate classification and name of each kind of postbiotic is recommended in the future. Since the characterization of microbial source is a pre-requirement for a given postbiotic to be determined, it is an applicable way to name postbiotics by their microbial source and molecular class/characteristics (e.g., Bifidobacterium sp. derived cell free supernatant, or Lactobacillus sp. derived exopolysaccharides). Meanwhile, for those substances that can be found in multiple microbial species and the molecular structures have been identified (e.g., butyrate acid, propionate acid, etc.), the chemical name can be used instead for clearance.
Taken together, postbiotic administration is promising in preventing and treating CRC through the modulation of gut microbiota, which has been validated by a large number of preclinical studies. However, the clinical evidence of postbiotics in protecting against CRC is still insufficient. Given the fact that dietary intervention is critical in modulating gut microbiota and are considered as the major factor in CRC development, it is reasonable to expect a positive clinical outcome by precision nutrition. Therefore, large-scale, and double-blind clinical trials will be required in the future to promote the clinical utilization of postbiotics in CRC patients.
Author contributions
N-NL conceived and designed the manuscript. DS, XW, and YM wrote the manuscript and drafted the figures and tables. N-NL and HW revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by MOST Key R&D Program of China (2022YFC2304703 and 2020YFA0907200), National Natural Science Foundation of China (31900129, 32270202, 82030099, and 32200749), Natural Science Foundation of Shanghai (22ZR1435900), and Innovative research team of high-level local universities in Shanghai.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel R, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49.
2. Salminen S, Collado M, Endo A, Hill C, Lebeer S, Quigley E, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. (2021) 18:649–67.
3. Biller L, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. (2021) 325:669–85.
4. Hegde S, Sun W, Lynch J. Systemic and targeted therapy for advanced colon cancer. Exp Rev Gastroenterol Hepatol. (2008) 2:135–49.
5. Sartore-Bianchi A, Loupakis F, Argilés G, Prager G. Challenging chemoresistant metastatic colorectal cancer: therapeutic strategies from the clinic and from the laboratory. Ann Oncol. (2016) 27:1456–66. doi: 10.1093/annonc/mdw191
6. Norcic G. Liquid biopsy in colorectal cancer-current status and potential clinical applications. Micromachines. (2018) 9:300. doi: 10.3390/mi9060300
7. Joachim C, Macni J, Drame M, Pomier A, Escarmant P, Veronique-Baudin J, et al. Overall survival of colorectal cancer by stage at diagnosis: data from the martinique cancer registry. Medicine. (2019) 98:e16941. doi: 10.1097/MD.0000000000016941
8. Hull R, Francies F, Oyomno M, Dlamini Z. Colorectal cancer genetics, incidence and risk factors: in search for targeted therapies. Cancer Manag Res. (2020) 12:9869–82.
10. Steinke V, Engel C, Büttner R, Schackert H, Schmiegel W, Propping P. Hereditary Nonpolyposis Colorectal Cancer (HNPCC)/Lynch Syndrome. Dtsch Arztebl Int. (2013) 110:32–8.
11. Rattray N, Charkoftaki G, Rattray Z, Hansen J, Vasiliou V, Johnson C. Environmental influences in the etiology of colorectal cancer: the premise of metabolomics. Curr Pharmacol Rep. (2017) 3:114–25. doi: 10.1007/s40495-017-0088-z
12. Lipkin M, Reddy B, Newmark H, Lamprecht S. Dietary factors in human colorectal cancer. Annual Rev Nutr. (1999) 19:545–86.
13. O’Keefe S. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. (2016) 13:691–706.
14. Makki K, Deehan E, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. (2018) 23:705–15.
15. Wong S, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. (2019) 16:690–704.
16. Pahumunto N, Teanpaisan R. Anti-Cancer properties of potential probiotics and their cell-free supernatants for the prevention of colorectal cancer: an in vitro study. Probiotics Antimicrob Proteins. (2022) doi: 10.1007/s12602-022-09972-y [Epub ahead of print].
17. Eaden J, Abrams K, Mayberry J. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. (2001) 48:526–35.
18. Canavan C, Abrams K, Mayberry J. Meta-Analysis: colorectal and small bowel cancer risk in patients with crohn’s disease. Aliment Pharmacol Ther. (2006) 23:1097–104. doi: 10.1111/j.1365-2036.2006.02854.x
19. Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. (2018) 9:474–87. doi: 10.1007/s13238-018-0543-6
20. Shmuely H, Passaro D, Figer A, Niv Y, Pitlik S, Samra Z, et al. Relationship between Helicobacter pylori caga status and colorectal cancer. Am J Gastroenterol. (2001) 96:3406–10.
21. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium Nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. (2014) 33:1381–90. doi: 10.1007/s10096-014-2081-3
22. Rubinstein Mara R, Wang X, Liu W, Hao Y, Cai G, Han Yiping W. Fusobacterium Nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/β-catenin signaling via its fada adhesin. Cell Host Microbe. (2013) 14:195–206.
23. Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun. (2021) 12:5674. doi: 10.1038/s41467-021-25965-x
24. Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol. (2020) 11:615056. doi: 10.3389/fimmu.2020.615056
25. Pandey K, Naik S, Vakil B. Probiotics, prebiotics and synbiotics - a review. J Food Sci Technol. (2015) 52:7577–87.
27. Hearty A, McCarthy S, Kearney J, Gibney M. Relationship between attitudes towards healthy eating and dietary behaviour, lifestyle and demographic factors in a representative sample of irish adults. Appetite. (2007) 48:1–11. doi: 10.1016/j.appet.2006.03.329
28. Suez J, Elinav E. The path towards microbiome-based metabolite treatment. Nat Microbiol. (2017) 2:17075. doi: 10.1038/nmicrobiol.2017.75
29. Teame T, Wang A, Xie M, Zhang Z, Yang Y, Ding Q, et al. Paraprobiotics and postbiotics of probiotic lactobacilli, their positive effects on the host and action mechanisms: a review. Front Nutr. (2020) 7:570344. doi: 10.3389/fnut.2020.570344
30. Gibson G, Hutkins R, Sanders M, Prescott S, Reimer R, Salminen S, et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502.
31. Hill C, Guarner F, Reid G, Gibson G, Merenstein D, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14.
32. Abbasi A, Hajipour N, Hasannezhad P, Baghbanzadeh A, Aghebati-Maleki L. Potential in vivo delivery routes of postbiotics. Crit Rev Food Sci Nutr. (2020) 62:3345–69. doi: 10.1080/10408398.2020.1865260
33. Klemashevich C, Wu C, Howsmon D, Alaniz R, Lee K, Jayaraman A. Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Curr Opin Biotechnol. (2014) 26:85–90. doi: 10.1016/j.copbio.2013.10.006
34. Żółkiewicz J, Marzec A, Ruszczyński M, Feleszko W. Postbiotics-a step beyond pre- and probiotics. Nutrients. (2020) 12:2189. doi: 10.3390/nu12082189
35. Nataraj B, Ali S, Behare P, Yadav H. Postbiotics-Parabiotics: the new horizons in microbial biotherapy and functional foods. Microbial Cell Factor. (2020) 19:168. doi: 10.1186/s12934-020-01426-w
36. Rad A, Aghebati-Maleki L, Kafil H, Abbasi A. Molecular mechanisms of postbiotics in colorectal cancer prevention and treatment. Crit Rev Food Sci Nutr. (2021) 61:1787–803. doi: 10.1080/10408398.2020.1765310
37. Clemente Jose C, Ursell Luke K, Parfrey Laura W, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. (2012) 148:1258–70.
38. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
40. Aguilar-Toalá J, Garcia-Varela R, Garcia H, Mata-Haro V, González-Córdova A, Vallejo-Cordoba B, et al. Postbiotics: an evolving term within the functional foods field. Trends Food Sci Technol. (2018) 75:105–14.
41. Foo H, Loh T, Mutalib N, Rahim R. The Myth and Therapeutic Potentials Of postbiotics. Microbiome and Metabolome in Diagnosis, Therapy, and other Strategic Applications. Amsterdam: Elsevier (2019).
42. Deshpande G, Athalye-Jape G, Patole S. Para-Probiotics for Preterm Neonates-the Next Frontier. Nutrients. (2018) 10:871. doi: 10.3390/nu10070871
43. Taverniti V, Guglielmetti S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept). Genes Nutr. (2011) 6:261–74. doi: 10.1007/s12263-011-0218-x
44. Zorzela L, Ardestani S, McFarland L, Vohra S. Is there a role for modified probiotics as beneficial microbes: a systematic review of the literature. Benef Microbes. (2017) 8:739–54. doi: 10.3920/BM2017.0032
45. Chen Y, Hsieh P, Ho H, Hsieh S, Kuo Y, Yang S, et al. Antibacterial activity of viable and heat-killed probiotic strains against oral pathogens. Lett Appl Microbiol. (2020) 70:310–7.
46. Sugahara H, Yao R, Odamaki T, Xiao J. Differences between Live and Heat-Killed Bifidobacteria in the Regulation of Immune Function and the Intestinal Environment. Benef Microbes. (2017) 8:463–72. doi: 10.3920/BM2016.0158
47. Maeda N, Nakamura R, Hirose Y, Murosaki S, Yamamoto Y, Kase T, et al. Oral Administration of Heat-Killed Lactobacillus Plantarum L-137 Enhances Protection against Influenza Virus Infection by Stimulation of Type I Interferon Production in Mice. Int Immunopharmacol. (2009) 9:1122–5. doi: 10.1016/j.intimp.2009.04.015
48. Donato K, Gareau M, Wang Y, Sherman P. Lactobacillus Rhamnosus Gg Attenuates Interferon-{Gamma} and tumour necrosis factor-alpha-induced barrier dysfunction and pro-inflammatory signalling. Microbiology. (2010) 156:3288–97. doi: 10.1099/mic.0.040139-0
49. Lin P, Nasr T, Berardinelli A, Kumar A, Neish A. The probiotic Lactobacillus Gg may augment intestinal host defense by regulating apoptosis and promoting cytoprotective responses in the developing murine gut. Pediatr Res. (2008) 64:511–6. doi: 10.1203/PDR.0b013e3181827c0f
50. Li N, Russell W, Douglas-escobar M, Hauser N, Lopez M, Neu J. Live and heat-killed Lactobacillus rhamnosus Gg: effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatr Res. (2009) 66:203–7. doi: 10.1203/PDR.0b013e3181aabd4f
51. Ueno N, Fujiya M, Segawa S, Nata T, Moriichi K, Tanabe H, et al. Heat-Killed Body of Lactobacillus Brevis Sbc8803 Ameliorates Intestinal Injury in a Murine Model of Colitis by Enhancing the Intestinal Barrier Function. Inflamm Bowel Dis. (2011) 17:2235–50. doi: 10.1002/ibd.21597
52. Cani P, Depommier C, Derrien M, Everard A, de Vos W. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol. (2022) 19:625–37. doi: 10.1038/s41575-022-00631-9
53. Raftar S, Ashrafian F, Abdollahiyan S, Yadegar A, Moradi H, Masoumi M, et al. The Anti-Inflammatory effects of Akkermansia muciniphila and its derivates in hfd/ccl4-induced murine model of liver injury. Sci Rep. (2022) 12:2453. doi: 10.1038/s41598-022-06414-1
54. Liu Q, Lu W, Tian F, Zhao J, Zhang H, Hong K, et al. Akkermansia muciniphila Exerts Strain-Specific Effects on Dss-Induced Ulcerative Colitis in Mice. Front Cell Infect Microbiol. (2021) 11:698914. doi: 10.3389/fcimb.2021.698914
55. Fan L, Xu C, Ge Q, Lin Y, Wong C, Qi Y, et al. A. Muciniphila Suppresses Colorectal Tumorigenesis by Inducing Tlr2/Nlrp3-Mediated M1-Like Tamsa. Muciniphila Induces M1-Like Tams That Suppress Crc. Cancer Immunol Res. (2021) 9:1111–24. doi: 10.1158/2326-6066.CIR-20-1019
56. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, et al. A Purified Membrane Protein from <Em>Akkermansia muciniphila</Em> or the Pasteurised Bacterium Blunts Colitis Associated Tumourigenesis by Modulation of Cd8<Sup>+</Sup> T Cells in Mice. Gut. (2020) 69:1988–97. doi: 10.1136/gutjnl-2019-320105
57. Kim S, Kang C, Kim G, Cho H. Anti-Tumor effects of heat-killed L. Reuteri Mg5346 and L. Casei Mg4584 against human colorectal carcinoma through caspase-9-dependent apoptosis in xenograft model. Microorganisms. (2022) 10:533. doi: 10.3390/microorganisms10030533
58. Kim Y, Yu H, Park Y, Lee N, Paik H. Anti-Biofilm Activity of Cell-Free Supernatant of Saccharomyces cerevisiae against Staphylococcus aureus. J Microbiol Biotechnol. (2020) 30:1854–61.
59. Schwenninger S, Lacroix C, Truttmann S, Jans C, Spörndli C, Bigler L, et al. Characterization of Low-Molecular-Weight Antiyeast Metabolites Produced by a Food-Protective Lactobacillus-Propionibacterium Coculture. J Food Prot. (2008) 71:2481–7. doi: 10.4315/0362-028x-71.12.2481
60. Amaretti A, di Nunzio M, Pompei A, Raimondi S, Rossi M, Bordoni A. Antioxidant Properties of Potentially Probiotic Bacteria: In Vitro and in Vivo Activities. Appl Microbiol Biotechnol. (2013) 97:809–17.
61. Pourramezan Z, Kasra Kermanshahi R, Oloomi M, Aliahmadi A, Rezadoost H. In Vitro Study of Antioxidant and Antibacterial Activities of Lactobacillus Probiotic Spp. Folia Microbiol. (2018) 63:31–42. doi: 10.1007/s12223-017-0531-x
62. Escamilla J, Lane M, Maitin V. Cell-Free Supernatants from Probiotic Lactobacillus Casei and Lactobacillus Rhamnosus Gg Decrease Colon Cancer Cell Invasion in Vitro. Nutr Cancer. (2012) 64:871–8. doi: 10.1080/01635581.2012.700758
63. Lee J, Lee J, Kim S, Kang D, Yoo H. Evaluating Cell Death Using Cell-Free Supernatant of Probiotics in Three-Dimensional Spheroid Cultures of Colorectal Cancer Cells. J Vis Exp. (2020) 160:E61285–302. doi: 10.3791/61285
64. Tong S, Davis J, Eichenberger E, Holland T, Fowler V Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin Microbiol Rev. (2015) 28:603–61.
65. Haghi Ghahremanloi Olia A, Ghahremani M, Ahmadi A, Sharifi Y. Comparison of biofilm production and virulence gene distribution among community- and hospital-acquired Staphylococcus aureus isolates from Northwestern Iran. Infect Genet Evol. (2020) 81:104262. doi: 10.1016/j.meegid.2020.104262
66. Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen R, et al. The extracellular biology of the Lactobacilli. FEMS Microbiol Rev. (2010) 34:199–230.
67. Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in gram-positive physiology and host interactions. Nat Rev Microbiol. (2008) 6:276–87.
68. Lovering A, Lin L, Sewell E, Spreter T, Brown ED, Strynadka N. Structure of the bacterial teichoic acid polymerase Tagf provides insights into membrane association and catalysis. Nat Struct Mol Biol. (2010) 17:582–9. doi: 10.1038/nsmb.1819
69. Mohamadzadeh M, Pfeiler E, Brown J, Zadeh M, Gramarossa M, Managlia E, et al. Regulation of Induced Colonic Inflammation by Lactobacillus Acidophilus Deficient in Lipoteichoic Acid. Proc Natl Acad Sci U.S.A. (2011) 108(Suppl. 1):4623–30.
70. Wang S, Ahmadi S, Nagpal R, Jain S, Mishra S, Kavanagh K, et al. Lipoteichoic Acid from the Cell Wall of a Heat Killed Lactobacillus Paracasei D3-5 Ameliorates Aging-Related Leaky Gut, Inflammation and Improves Physical and Cognitive Functions: From C. Elegans to Mice. Geroscience. (2020) 42:333–52.
71. Noh S, Kang S, Yun C, Han S. Lipoteichoic acid from Lactobacillus Plantarum Inhibits Pam2csk4-Induced Il-8 Production in Human Intestinal Epithelial Cells. Mol Immunol. (2015) 64:183–9. doi: 10.1016/j.molimm.2014.11.014
72. Girardin S, Boneca I, Carneiro L, Antignac A, Jéhanno M, Viala J, et al. Nod1 Detects a Unique Muropeptide from Gram-Negative Bacterial Peptidoglycan. Science. (2003) 300:1584–7.
73. Allison C, Kufer T, Kremmer E, Kaparakis M, Ferrero R. Helicobacter Pylori Induces Mapk Phosphorylation and Ap-1 Activation Via a Nod1-Dependent Mechanism. J Immunol. (2009) 183:8099–109. doi: 10.4049/jimmunol.0900664
74. Girardin S, Tournebize R, Mavris M, Page A, Li X, Stark G, et al. Card4/Nod1 Mediates Nf-Kappab and Jnk Activation by Invasive Shigella flexneri. EMBO Rep. (2001) 2:736–42. doi: 10.1093/embo-reports/kve155
75. Inohara N, Koseki T, Lin J, del Peso L, Lucas P, Chen F, et al. An induced proximity model for Nf-Kappa B Activation in the Nod1/Rick and Rip Signaling Pathways. J Biol Chem. (2000) 275:27823–31. doi: 10.1074/jbc.M003415200
76. Hasegawa M, Fujimoto Y, Lucas P, Nakano H, Fukase K, Núñez G, et al. A critical role of Rick/Rip2 Polyubiquitination in Nod-Induced Nf-Kappab Activation. EMBO J. (2008) 27:373–83. doi: 10.1038/sj.emboj.7601962
77. Irving A, Mimuro H, Kufer T, Lo C, Wheeler R, Turner L, et al. The Immune Receptor Nod1 and Kinase Rip2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. (2014) 15:623–35. doi: 10.1016/j.chom.2014.04.001
78. Wang S, Han X, Zhang L, Zhang Y, Li H, Jiao Y. Whole peptidoglycan extracts from the Lactobacillus paracasei Subsp. Paracasei M5 strain exert anticancer activity in Vitro. Biomed Res Int. (2018) 2018:2871710. doi: 10.1155/2018/2871710
79. Xiao R, Zheng Y. Overview of microalgal extracellular polymeric substances (EPS) and Their Applications. Biotechnol Adv. (2016) 34:1225–44. doi: 10.1016/j.biotechadv.2016.08.004
80. Oerlemans M, Akkerman R, Ferrari M, Walvoort M, de Vos P. Benefits of bacteria-derived exopolysaccharides on gastrointestinal microbiota, immunity and health. J Funct Foods. (2021) 76:104289. doi: 10.1016/j.foodres.2022.111445
81. Badel S, Bernardi T, Michaud P. New Perspectives for Lactobacilli exopolysaccharides. Biotechnol Adv. (2011) 29:54–66.
82. Freitas F, Alves V, Reis M. Advances in bacterial exopolysaccharides: from production to biotechnological applications. Trends Biotechnol. (2011) 29:388–98.
83. Salazar N, Gueimonde M, de Los Reyes-Gavilán C, Ruas-Madiedo P. Exopolysaccharides Produced by Lactic Acid Bacteria and Bifidobacteria as Fermentable Substrates by the Intestinal Microbiota. Crit Rev Food Sci Nutr. (2016) 56:1440–53.
84. Deepak V, Ram Kumar Pandian S, Sivasubramaniam S, Nellaiah H, Sundar K. Optimization of anticancer exopolysaccharide production from probiotic Lactobacillus acidophilus by response surface methodology. Prep Biochem Biotechnol. (2016) 46:288–97. doi: 10.1080/10826068.2015.1031386
85. Di W, Zhang L, Yi H, Han X, Zhang Y, Xin L. Exopolysaccharides Produced by Lactobacillus Strains Suppress Ht−29 Cell Growth Via Induction of G0/G1 Cell Cycle Arrest and Apoptosis. Oncol Lett. (2018) 16:3577–86. doi: 10.3892/ol.2018.9129
86. Zhang J, Liu L, Ren Y, Chen F. Characterization of exopolysaccharides produced by microalgae with antitumor activity on human colon cancer cells. Int J Biol Macromol. (2019) 128:761–7. doi: 10.1016/j.ijbiomac.2019.02.009
87. Zhang J, Liu L, Chen F. Production and characterization of exopolysaccharides from chlorella zofingiensis and chlorella vulgaris with anti-colorectal cancer activity. Int J Biol Macromol. (2019) 134:976–83. doi: 10.1016/j.ijbiomac.2019.05.117
88. Lebeer S, Claes I, Verhoeven T, Vanderleyden J, De Keersmaecker S. Exopolysaccharides of Lactobacillus rhamnosus Gg form a protective shield against innate immune factors in the intestine. Microb Biotechnol. (2011) 4:368–74. doi: 10.1111/j.1751-7915.2010.00199.x
89. Yan S, Yang B, Zhao J, Zhao J, Stanton C, Ross R, et al. A ropy exopolysaccharide producing strain Bifidobacterium longum Subsp. Longum Ys108r alleviates dss-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. (2019) 10:1595–608. doi: 10.1039/c9fo00014c
90. Lu Y, Wang J, Ji Y, Chen K. Metabonomic variation of exopolysaccharide from rhizopus nigricans on Aom/Dss-Induced colorectal cancer in mice. Onco Targets Ther. (2019) 12:10023–33. doi: 10.2147/OTT.S226451
91. Hernández-Granados M, Franco-Robles E. Postbiotics in human health: Possible new functional Ingredients? Food Res Int. (2020) 137:109660. doi: 10.1016/j.foodres.2020.109660
92. Tan J, McKenzie C, Potamitis M, Thorburn A, Mackay C, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. (2014) 121:91–119.
93. Whitehead R, Young G, Bhathal P. Effects of short chain fatty acids on a new human colon carcinoma cell line (Lim1215). Gut. (1986) 27:1457–63. doi: 10.1136/gut.27.12.1457
94. Gamet L, Daviaud D, Denis-Pouxviel C, Remesy C, Murat J. Effects of short-chain fatty acids on growth and differentiation of the human colon-cancer cell line Ht29. Int J Cancer. (1992) 52:286–9.
95. Scheppach W, Bartram P, Richter A, Richter F, Liepold H, Dusel G, et al. Effect of short-chain fatty acids on the human colonic mucosa in vitro. JPEN J Parenter Enteral Nutr. (1992) 16:43–8.
96. Masrul M, Nindrea R. Dietary fibre protective against colorectal cancer patients in asia: a meta-analysis. Open Access Maced J Med Sci. (2019) 7:1723–7. doi: 10.3889/oamjms.2019.265
97. Weaver G, Krause J, Miller T, Wolin M. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut. (1988) 29:1539–43. doi: 10.1136/gut.29.11.1539
98. McIntyre A, Gibson P, Young G. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut. (1993) 34:386–91. doi: 10.1136/gut.34.3.386
99. Hinnebusch B, Meng S, Wu J, Archer S, Hodin R. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. (2002) 132:1012–7. doi: 10.1093/jn/132.5.1012
100. Tian Y, Xu Q, Sun L, Ye Y, Ji G. Short-chain fatty acids administration is protective in colitis-associated colorectal cancer development. J Nutr Biochem. (2018) 57:103–9. doi: 10.1016/j.jnutbio.2018.03.007
101. Parada Venegas D, De la Fuente M, Landskron G, González M, Quera R, Dijkstra G. Short Chain Fatty Acids. (Scfas)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.00277
102. Hu Y, Le Leu R, Christophersen C, Somashekar R, Conlon M, Meng X, et al. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis. (2016) 37:366–75. doi: 10.1093/carcin/bgw019
103. Jia B, Han X, Kim K, Jeon C. Discovery and mining of enzymes from the human gut microbiome. Trends Biotechnol. (2022) 40:240–54.
104. Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly T, et al. The amount of bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am J Pathol. (2018) 188:2839–52. doi: 10.1016/j.ajpath.2018.08.015
105. Goldin B, Gorbach S. Effect of Lactobacillus acidophilus dietary supplements on 1,2-dimethylhydrazine dihydrochloride-induced intestinal cancer in rats. J Natl Cancer Inst. (1980) 64:263–5. doi: 10.1093/jnci/64.2.263
106. Reddy B, Rivenson A. Inhibitory effect of bifidobacterium longum on colon, mammary, and liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Cancer Res. (1993) 53:3914–8.
107. Nalini N, Manju V, Menon V. Effect of coconut cake on the bacterial enzyme activity in 1,2-dimethyl hydrazine induced colon cancer. Clin Chim Acta. (2004) 342:203–10. doi: 10.1016/j.cccn.2004.01.001
108. de Moreno de LeBlanc A, LeBlanc J, Perdigón G, Miyoshi A, Langella P, Azevedo V, et al. Oral Administration of a Catalase-Producing Lactococcus lactis can prevent a chemically induced colon cancer in mice. J Med Microbiol. (2008) 57:100–5. doi: 10.1099/jmm.0.47403-0
109. Lee W. Effects of lactic acid bacteria on intestinal microbial enzyme activity and composition in rats treated with azoxymethane. J Microbiol. (2001) 39:154–61.
110. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. (2018) 15:111–28.
111. Roberts D, Chidambaram S, Kinross J. The role of the colonic microbiota and bile acids in colorectal cancer. Curr Opin Gastroenterol. (2022) 38:179–88.
112. Cai J, Sun L, Gonzalez F. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. (2022) 30:289–300. doi: 10.1016/j.chom.2022.02.004
113. Ocvirk S, Wilson A, Appolonia C, Thomas T, O’Keefe S. Fiber, fat, and colorectal cancer: new insight into modifiable dietary risk factors. Curr Gastroenterol Rep. (2019) 21:62. doi: 10.1007/s11894-019-0725-2
114. Liu Y, Zhang S, Zhou W, Hu D, Xu H, Ji G. Secondary bile acids and tumorigenesis in colorectal cancer. Front Oncol. (2022) 12:813745. doi: 10.3389/fonc.2022.813745
115. Nagengast F, Grubben M, van Munster I. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. (1995) 31:1067–70.
116. Bernstein H, Bernstein C, Payne C, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. (2005) 589:47–65.
117. Ocvirk S, O’Keefe S. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet – gut microbiota interactions. Curr Nutr Rep. (2017) 6:315–22. doi: 10.1007/s13668-017-0219-5
118. Fu T, Coulter S, Yoshihara E, Oh T, Fang S, Cayabyab F, et al. FXR regulates intestinal cancer stem cell proliferation. Cell. (2019) 176:1098–112.e18.
119. Schulz M, Atay C, Heringer J, Romrig F, Schwitalla S, Aydin B, et al. High-Fat-Diet-Mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. (2014) 514:508–12. doi: 10.1038/nature13398
120. Islam K, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. (2011) 141:1773–81.
121. Wolf P, Gaskins H, Ridlon J, Freels S, Hamm A, Goldberg S, et al. Effects of taurocholic acid metabolism by gut bacteria: a controlled feeding trial in adult african american subjects at elevated risk for colorectal cancer. Contemp Clin Trials Commun. (2020) 19:100611. doi: 10.1016/j.conctc.2020.100611
122. Winston J, Rivera A, Cai J, Patterson A, Theriot C. Secondary bile acid ursodeoxycholic acid alters weight, the gut microbiota, and the bile acid pool in conventional mice. PLoS One. (2021) 16:e0246161. doi: 10.1371/journal.pone.0246161
123. Im E, Martinez J. Ursodeoxycholic Acid (Udca) Can Inhibit Deoxycholic Acid (Dca)-Induced Apoptosis Via Modulation of Egfr/Raf-1/Erk Signaling in Human Colon Cancer Cells. J Nutr. (2004) 134:483–6. doi: 10.1093/jn/134.2.483
124. Shiraki K, Ito T, Sugimoto K, Fuke H, Inoue T, Miyashita K, et al. Different effects of bile acids, ursodeoxycholic acid and deoxycholic acid, on cell growth and cell death in human colonic adenocarcinoma cells. Int J Mol Med. (2005) 16:729–33.
125. Kim E, Cho J, Kim E, Kim Y. Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One. (2017) 12:e0181183. doi: 10.1371/journal.pone.0181183
126. Zhang H, Xu H, Zhang C, Tang Q, Bi F. Ursodeoxycholic acid suppresses the malignant progression of colorectal cancer through Tgr5-Yap Axis. Cell Death Discovery. (2021) 7:207. doi: 10.1038/s41420-021-00589-8
127. Kohno H, Suzuki R, Yasui Y, Miyamoto S, Wakabayashi K, Tanaka T. Ursodeoxycholic acid versus sulfasalazine in colitis-related colon carcinogenesis in mice. Clin Cancer Res. (2007) 13:2519–25. doi: 10.1158/1078-0432.CCR-06-2727
128. Wali R, Stoiber D, Nguyen L, Hart J, Sitrin M, Brasitus T, et al. Ursodeoxycholic acid inhibits the initiation and postinitiation phases of azoxymethane-induced colonic tumor development. Cancer Epidemiol Biomarkers Prev. (2002) 11:1316–21.
129. Tung B, Emond M, Haggitt R, Bronner M, Kimmey M, Kowdley K, et al. Ursodiol use is associated with lower prevalence of colonic neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis. Ann Intern Med. (2001) 134:89–95. doi: 10.7326/0003-4819-134-2-200101160-00008
130. Ikegami T, Matsuzaki Y. Ursodeoxycholic acid: mechanism of action and novel clinical applications. Hepatol Res. (2008) 38:123–31.
131. Richard D, Dawes M, Mathias C, Acheson A, Hill-Kapturczak N, Dougherty D. L-Tryptophan: basic metabolic functions, behavioral research and therapeutic indications. Int J Tryptophan Res. (2009) 2:45–60. doi: 10.4137/ijtr.s2129
132. Roager H, Licht T. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:3294.
133. Wyatt M, Greathouse K. Targeting dietary and microbial tryptophan-indole metabolism as therapeutic approaches to colon cancer. Nutrients. (2021) 13:1189. doi: 10.3390/nu13041189
134. Platten M, Nollen E, Röhrig U, Fallarino F, Opitz C. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discovery. (2019) 18:379–401. doi: 10.1038/s41573-019-0016-5
135. Zhang H, Zhang A, Miao J, Sun H, Yan G, Wu F, et al. Targeting regulation of tryptophan metabolism for colorectal cancer therapy: a systematic review. RSC Adv. (2019) 9:3072–80. doi: 10.1039/c8ra08520j
136. Sun X, Zhao D, Zhou Y, Wang Q, Qin G, Yao S. Alteration of fecal tryptophan metabolism correlates with shifted microbiota and may be involved in pathogenesis of colorectal cancer. World J Gastroenterol. (2020) 26:7173–90. doi: 10.3748/wjg.v26.i45.7173
137. Cotter P, Ross R, Hill C. Bacteriocins - a Viable Alternative to Antibiotics? Nat Rev Microbiol. (2013) 11:95–105.
138. Simons A, Alhanout K, Duval R. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. (2020) 8:639. doi: 10.3390/microorganisms8050639
139. Lawrence G, McCarthy N, Walsh C, Kunyoshi T, Lawton E, O’Connor P, et al. Effect of a Bacteriocin-Producing Streptococcus salivarius on the Pathogen Fusobacterium nucleatum in a Model of the Human Distal Colon. Gut Microbes. (2022) 14:2100203. doi: 10.1080/19490976.2022.2100203
140. Patra S, Sahu N, Saxena S, Pradhan B, Nayak S, Roychowdhury A. Effects of probiotics at the interface of metabolism and immunity to prevent colorectal cancer-associated gut inflammation: a systematic network and meta-analysis with molecular docking studies. Front Microbiol. (2022) 13:878297. doi: 10.3389/fmicb.2022.878297
141. Hill M. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev. (1997) 6(Suppl. 1):S43–5.
142. LeBlanc J, Milani C, de Giori G, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. (2013) 24:160–8. doi: 10.1016/j.copbio.2012.08.005
144. Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. (2018) 1693:128–33.
145. Suganuma K, Hamasaki T, Hamaoka T. Effect of dietary direct-fed microbial and yeast cell walls on cecal digesta microbiota of layer chicks inoculated with nalidixic acid resistant Salmonella enteritidis. Poult Sci. (2021) 100:101385. doi: 10.1016/j.psj.2021.101385
146. Agunos A, Ibuki M, Yokomizo F, Mine Y. Effect of Dietary B 1–4 Mannobiose in the Prevention of Salmonella enteritidis Infection in Broilers. Br Poultry Sci. (2007) 48:331–41.
147. Ganner A, Stoiber C, Uhlik J, Dohnal I, Schatzmayr G. Quantitative Evaluation of E. Coli F4 and Salmonella typhimurium Binding Capacity of Yeast Derivatives. AMB Express. (2013) 3:62. doi: 10.1186/2191-0855-3-62
148. Pereyra C, Gil S, Cristofolini A, Bonci M, Makita M, Monge M, et al. The production of yeast cell wall using an agroindustrial waste influences the wall thickness and is implicated on the aflatoxin B1 adsorption process. Food Res Int. (2018) 111:306–13. doi: 10.1016/j.foodres.2018.05.026
149. Czerucka D, Rampal P. Diversity of Saccharomyces boulardii Cncm I-745 Mechanisms of Action against Intestinal Infections. World J Gastroenterol. (2019) 25:2188–203. doi: 10.3748/wjg.v25.i18.2188
150. Lenka S, Singh D, Paul S, Gayen A, Chandra MS. Boulardii fails to hold its cell wall integrity against nonpathogenic E. Coli: Are Probiotic Yeasts Losing the Battle? ACS Infect Dis. (2021) 7:733–45. doi: 10.1021/acsinfecdis.0c00413
151. Fortin O, Aguilar-Uscanga B, Vu K, Salmieri S, Lacroix M. Cancer chemopreventive, antiproliferative, and superoxide anion scavenging properties of Kluyveromyces marxianus and Saccharomyces cerevisiae Var. Boulardii Cell Wall Components. Nutr Cancer. (2018) 70:83–96. doi: 10.1080/01635581.2018.1380204
152. Liu M, Liu W, Zhang W, Yao J, Mo X. Ultrasound-Assisted extraction of boulardii yeast cell wall polysaccharides: characterization and its biological functions on early-weaned lambs. Food Sci Nutr. (2021) 9:3617–30. doi: 10.1002/fsn3.2318
153. Rahbar Saadat Y, Yari Khosroushahi A, Movassaghpour A, Talebi M, Pourghassem Gargari B. Modulatory Role of Exopolysaccharides of Kluyveromyces marxianus and Pichia kudriavzevii as probiotic yeasts from dairy products in human colon cancer cells. J Funct Foods. (2020) 64:103675.
154. Swift C, Louie K, Bowen B, Olson H, Purvine S, Salamov A, et al. Anaerobic gut fungi are an untapped reservoir of natural products. Proc Natl Acad Sci U.S.A. (2021) 118:e2019855118. doi: 10.1073/pnas.2019855118
155. Pettersen V, Dufour A, Arrieta M. Metaproteomic profiling of fungal gut colonization in gnotobiotic mice. Anim Microb. (2022) 4:14.
156. Holanda D, Yiannikouris A, Kim S. Investigation of the efficacy of a postbiotic yeast cell wall-based blend on newly-weaned pigs under a dietary challenge of multiple mycotoxins with emphasis on deoxynivalenol. Toxins. (2020) 12:504. doi: 10.3390/toxins12080504
157. Chan M, Liu S. Fortifying foods with synbiotic and postbiotic preparations of the probiotic yeast, Saccharomyces boulardii. Curr Opin Food Sci. (2022) 43:216–24.
158. Nkamga V, Henrissat B, Drancourt M. Archaea: essential inhabitants of the human digestive microbiota. Hum Microb J. (2017) 3:1–8.
159. Gaci N, Borrel G, Tottey W, O’Toole P, Brugère J. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. (2014) 20:16062–78. doi: 10.3748/wjg.v20.i43.16062
160. Brugère J, Borrel G, Gaci N, Tottey W, O’Toole P, Malpuech-Brugère C. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes. (2014) 5:5–10. doi: 10.4161/gmic.26749
161. Coker O, Nakatsu G, Dai R, Wu W, Wong S, Ng S, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. (2019) 68:654–62. doi: 10.1136/gutjnl-2018-317178
162. Coker O, Wu W, Wong S, Sung J, Yu J. Altered gut archaea composition and interaction with bacteria are associated with colorectal cancer. Gastroenterology. (2020) 159:1459–70.e5. doi: 10.1053/j.gastro.2020.06.042
163. Liu N, Jiao N, Tan J, Wang Z, Wu D, Wang A, et al. Multi-Kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat Microbiol. (2022) 7:238–50. doi: 10.1038/s41564-021-01030-7
164. Narunsky-Haziza L, Sepich-Poore G, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. (2022) 185:3789–806.e17. doi: 10.1016/j.cell.2022.09.005
168. Hunter T. Signaling–2000 and Beyond. Cell. (2000) 100:113–27. doi: 10.1016/s0092-8674(00)81688-8
169. Harbour J, Dean D. The Rb/E2f Pathway: expanding roles and emerging paradigms. Genes Dev. (2000) 14:2393–409. doi: 10.1101/gad.813200
171. Evan G, Vousden K. Proliferation, cell cycle and apoptosis in cancer. Nature. (2001) 411:342–8.
172. Panebianco C, Villani A, Pisati F, Orsenigo F, Ulaszewska M, Latiano T, et al. Butyrate, a postbiotic of intestinal bacteria, affects pancreatic cancer and gemcitabine response in in vitro and in vivo models. Biomed Pharmacother. (2022) 151:113163. doi: 10.1016/j.biopha.2022.113163
173. Chuah L, Foo H, Loh T, Mohammed Alitheen N, Yeap S, Abdul Mutalib N, et al. Postbiotic metabolites produced by lactobacillus plantarum strains exert selective cytotoxicity effects on cancer cells. BMC Complement Altern Med. (2019) 19:114. doi: 10.1186/s12906-019-2528-2
174. Lazarova D, Bordonaro M, Carbone R, Sartorelli A. Linear relationship between WNT activity levels and apoptosis in colorectal carcinoma cells exposed to butyrate. Int J Cancer. (2004) 110:523–31. doi: 10.1002/ijc.20152
175. Encarnação J, Pires A, Amaral R, Gonçalves T, Laranjo M, Casalta-Lopes J, et al. Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J Nutr Biochem. (2018) 56:183–92. doi: 10.1016/j.jnutbio.2018.02.018
176. Zhang R, Zhou Z, Ma Y, Du K, Sun M, Zhang H, et al. Anti-Gastric Cancer Activity of the Cell-Free Culture Supernatant of Serofluid Dish and Lactiplantibacillus plantarum Yt013. Front Bioeng Biotechnol. (2022) 10:898240. doi: 10.3389/fbioe.2022.898240
177. Peng M, Lee S, Rahaman S, Biswas D. Dietary probiotic and metabolites improve intestinal homeostasis and prevent colorectal cancer. Food Funct. (2020) 11:10724–35. doi: 10.1039/d0fo02652b
178. Kerr J, Wyllie A, Currie A. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. (1972) 26:239–57.
180. Thompson C. Apoptosis in the pathogenesis and treatment of disease. Science. (1995) 267:1456–62.
181. Song D, Meng J, Cheng J, Fan Z, Chen P, Ruan H, et al. Pseudomonas aeruginosa quorum-sensing metabolite induces host immune cell death through cell surface lipid domain dissolution. Nat Microbiol. (2019) 4:97–111. doi: 10.1038/s41564-018-0290-8
182. Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s achilles’. Heel. Cancer Cell. (2008) 13:472–82.
183. Evan G, Wyllie A, Gilbert C, Littlewood T, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by C-Myc Protein. Cell. (1992) 69:119–28.
184. Evan G, Littlewood TA. Matter of life and cell death. Science. (1998) 281:1317–22. doi: 10.1126/science.281.5381.1317
185. Schmitt C, Lowe S. Apoptosis and therapy. J Pathol. (1999) 187:127–37. doi: 10.3390/cancers11060822
186. Ma W, Zhang Y, Yu M, Wang B, Xu S, Zhang J, et al. Corrigendum to ‘in-Vitro and in-Vivo anti-breast cancer activity of synergistic effect of berberine and exercise through promoting the apoptosis and immunomodulatory effects’. Int Immunopharmacol. (2020) 88:106899.
187. Davie J. Inhibition of histone deacetylase activity by butyrate. J Nutr. (2003) 133(7 Suppl.):2485s–93s.
188. Chirakkal H, Leech S, Brookes K, Prais A, Waby J, Corfe B. Upregulation of Bak by Butyrate in the Colon Is Associated with Increased Sp3 Binding. Oncogene. (2006) 25:7192–200. doi: 10.1038/sj.onc.1209702
189. Dashwood R, Myzak M, Ho E. Dietary Hdac Inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. (2006) 27:344–9. doi: 10.1093/carcin/bgi253
190. Hamer H, Jonkers D, Venema K, Vanhoutvin S, Troost F, Brummer R. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. (2008) 27:104–19.
191. Sugimura N, Li Q, Chu E, Lau H, Fong W, Liu W, et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut. (2021) 71:2011–21. doi: 10.1136/gutjnl-2020-323951
192. Li Q, Hu W, Liu W, Zhao L, Huang D, Liu X, et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting B -Galactosidase. Gastroenterology. (2021) 160:1179–93.e14. doi: 10.1053/j.gastro.2020.09.003
193. Ma H, Yu Y, Wang M, Li Z, Xu H, Tian C, et al. Correlation between microbes and colorectal cancer: tumor apoptosis is induced by sitosterols through promoting gut microbiota to produce short-chain fatty acids. Apoptosis. (2019) 24:168–83. doi: 10.1007/s10495-018-1500-9
194. Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating wnt signaling and gut microbiota. Cancer Lett. (2020) 469:456–67. doi: 10.1016/j.canlet.2019.11.019
195. Terada A, Bukawa W, Kan T, Mitsuoka T. Effects of the consumption of heat-killed enterococcus faecalis ec-12 preparation on microbiota and metabolic activity of the faeces in healthy adults. Microbial Ecol Health Dis. (2004) 16:188–94.
196. Sassone-Corsi M, Nuccio S, Liu H, Hernandez D, Vu C, Takahashi A, et al. Microcins Mediate Competition among Enterobacteriaceae in the Inflamed Gut. Nature. (2016) 540:280–3. doi: 10.1038/nature20557
197. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán L, Gratadoux J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci U S A. (2008) 105:16731–6. doi: 10.1073/pnas.0804812105
198. Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. (2015) 37:47–55.
199. Inagaki T, Moschetta A, Lee Y, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U.S.A. (2006) 103:3920–5.
200. Begley M, Gahan C, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. (2005) 29:625–51.
201. Wang S, Dong W, Liu L, Xu M, Wang Y, Liu T, et al. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol Carcinog. (2019) 58:1155–67.
202. He Q, Wu J, Ke J, Zhang Q, Zeng W, Luo Z, et al. Therapeutic role of ursodeoxycholic acid in colitis-associated cancer via gut microbiota modulation. Mol Therapy. (2022) 31:585–98.
203. Warda A, de Almeida Bettio P, Hueston C, Di Benedetto G, Clooney A, Hill C. Oral administration of heat-treated lactobacilli modifies the murine microbiome and reduces citrobacter induced colitis. Front Microbiol. (2020) 11:69. doi: 10.3389/fmicb.2020.00069
204. De Vuyst L, Leroy F. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol. (2007) 13:194–9.
205. Li Z, Wang W, Liu D, Guo Y. Effects of Lactobacillus acidophilus on gut microbiota composition in broilers challenged with Clostridium perfringens. PLoS One. (2017) 12:e0188634. doi: 10.1371/journal.pone.0188634
206. Yang N, Zhan Y, Wan J, Li Y, Hu X, Liu W. Effects of lacidophilin tablets, yogurt, and bifid triple viable capsules on the gut microbiota of mice with antibiotic-associated diarrhea. Can J Infect Dis Med Microbiol. (2022) 2022:6521793. doi: 10.1155/2022/6521793
207. Warda A, Rea K, Fitzgerald P, Hueston C, Gonzalez-Tortuero E, Dinan T, et al. Heat-Killed Lactobacilli alter both microbiota composition and behaviour. Behav Brain Res. (2019) 362:213–23. doi: 10.1016/j.bbr.2018.12.047
208. Kimoto-Nira H, Mizumachi K, Okamoto T, Sasaki K, Kurisaki J. Influence of long-term consumption of a Lactococcus lactis strain on the intestinal immunity and intestinal flora of the senescence-accelerated mouse. Br J Nutr. (2009) 102:181–5. doi: 10.1017/S0007114508143574
209. Huycke M, Gaskins H. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med. (2004) 229:586–97. doi: 10.1177/153537020422900702
210. Turner J. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. (2009) 9:799–809.
212. Martens E, Neumann M, Desai M. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. (2018) 16:457–70. doi: 10.1038/s41579-018-0036-x
213. Pelaseyed T, Bergström J, Gustafsson J, Ermund A, Birchenough G, Schütte A, et al. The Mucus and Mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. (2014) 260:8–20. doi: 10.1111/imr.12182
214. McGuckin M, Lindén S, Sutton P, Florin T. Mucin Dynamics and Enteric Pathogens. Nat Rev Microbiol. (2011) 9:265–78.
215. Wrzosek L, Miquel S, Noordine M, Bouet S, Joncquel Chevalier-Curt M, Robert V. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. (2013) 11:61. doi: 10.1186/1741-7007-11-61
216. Johansson M, Jakobsson H, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro A, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. (2015) 18:582–92. doi: 10.1016/j.chom.2015.10.007
217. Jung T, Park J, Jeon W, Han K. Butyrate Modulates Bacterial Adherence on Ls174t Human Colorectal Cells by Stimulating Mucin Secretion and Mapk Signaling Pathway. Nutr Res Pract. (2015) 9:343–9. doi: 10.4162/nrp.2015.9.4.343
218. He X, Zeng Q, Puthiyakunnon S, Zeng Z, Yang W, Qiu J, et al. Lactobacillus rhamnosus Gg Supernatant Enhance Neonatal Resistance to Systemic Escherichia Coli K1 Infection by Accelerating Development of Intestinal Defense. Sci Rep. (2017) 7:43305. doi: 10.1038/srep43305
219. Tytgat H, Douillard F, Reunanen J, Rasinkangas P, Hendrickx A, Laine P, et al. Lactobacillus Rhamnosus Gg Outcompetes Enterococcus faecium Via Mucus-Binding Pili: evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol. (2016) 82:5756–62. doi: 10.1128/AEM.01243-16
220. Petrova M, Imholz N, Verhoeven T, Balzarini J, Van Damme E, Schols D, et al. Lectin-Like Molecules of Lactobacillus Rhamnosus Gg Inhibit Pathogenic Escherichia Coli and Salmonella Biofilm Formation. PLoS One. (2016) 11:e0161337. doi: 10.1371/journal.pone.0161337
221. Barnett A, Roy N, McNabb W, Cookson A. The Interactions between Endogenous Bacteria, Dietary Components and the Mucus Layer of the Large Bowel. Food Funct. (2012) 3:690–9. doi: 10.1039/c2fo30017f
222. Korcz E, Kerényi Z, Varga L. Dietary fibers, prebiotics, and exopolysaccharides produced by lactic acid bacteria: potential health benefits with special regard to cholesterol-lowering effects. Food Funct. (2018) 9:3057–68. doi: 10.1039/c8fo00118a
223. Ruas-Madiedo P, Gueimonde M, Margolles A, de los Reyes-Gavilán C, Salminen S. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J Food Prot. (2006) 69:2011–5. doi: 10.4315/0362-028x-69.8.2011
224. Coconnier M, Bernet M, Kernéis S, Chauvière G, Fourniat J, Servin A. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 Cells by Lactobacillus acidophilus Strain Lb Decreases Bacterial Invasion. FEMS Microbiol Lett. (1993) 110:299–305. doi: 10.1111/j.1574-6968.1993.tb06339.x
225. Martin T, Jiang W. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. (2009) 1788:872–91.
226. Tsilingiri K, Barbosa T, Penna G, Caprioli F, Sonzogni A, Viale G, et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut. (2012) 61:1007–15. doi: 10.1136/gutjnl-2011-300971
228. Fukuda S, Toh H, Taylor T, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. (2012) 3:449–54. doi: 10.4161/gmic.21214
229. Ramakrishna B. Probiotic-Induced changes in the intestinal epithelium: implications in gastrointestinal disease. Trop Gastroenterol. (2009) 30:76–85.
230. Mack D, Ahrne S, Hyde L, Wei S, Hollingsworth M. Extracellular Muc3 mucin secretion follows adherence of lactobacillus strains to intestinal epithelial cells in vitro. Gut. (2003) 52:827–33. doi: 10.1136/gut.52.6.827
231. Hung R, Ulrich C, Goode E, Brhane Y, Muir K, Chan A, et al. Cross cancer genomic investigation of inflammation pathway for five common cancers: lung, ovary, prostate, breast, and colorectal cancer. J Natl Cancer Inst. (2015) 107:djv246. doi: 10.1093/jnci/djv246
232. Chai E, Siveen K, Shanmugam M, Arfuso F, Sethi G. Analysis of the intricate relationship between chronic inflammation and cancer. Biochem J. (2015) 468:1–15.
233. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi M. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. (2017) 18:197.
234. Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. (2013) 66:462–70.
236. Suh O, Mettlin C, Petrelli N. Aspirin use, cancer, and polyps of the large bowel. Cancer. (1993) 72:1171–7.
237. Rigau J, Piqué J, Rubio E, Planas R, Tarrech J, Bordas J. Effects of long-term sulindac therapy on colonic polyposis. Ann Intern Med. (1991) 115:952–4. doi: 10.7326/0003-4819-115-12-952
238. Fang Y, Yan C, Zhao Q, Xu J, Liu Z, Gao J, et al. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. (2021) 12:720–35.
239. Zeman M, Vecka M, Perlík F, Staòková B, Hromádka R, Tvrzická E, et al. Pleiotropic effects of niacin: current possibilities for its clinical use. Acta Pharm. (2016) 66:449–69. doi: 10.1515/acph-2016-0043
240. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. (2014) 40:128–39. doi: 10.1016/j.immuni.2013.12.007
241. Homayoni Rad A, Akbarzadeh F, Mehrabany E. Which are more important: prebiotics or probiotics? Nutrition. (2012) 28:1196–7.
242. Vernazza C, Rabiu B, Gibson G. Human Colonic Microbiology and the Role of Dietary Intervention: Introduction to Prebiotics. In: Gibson GR, Rastall RA editors. Prebiotics: Development & Application. Chichester: John Wiley & Sons, Ltd. (2006).
243. Boleij A, Hechenbleikner E, Goodwin A, Badani R, Stein E, Lazarev M, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. (2015) 60:208–15. doi: 10.1093/cid/ciu787
244. McCoy A, Araújo-Pérez F, Azcárate-Peril A, Yeh J, Sandler R, Keku T. Fusobacterium is associated with colorectal adenomas. PLoS One. (2013) 8:e53653. doi: 10.1371/journal.pone.0053653
246. Cani P. Human gut microbiome: hopes, threats and promises. Gut. (2018) 67:1716–25. doi: 10.1136/gutjnl-2018-316723
247. Wait R, Chiesa G, Parolini C, Miller I, Begum S, Brambilla D, et al. Reference Maps of Mouse Serum Acute-Phase Proteins: Changes with Lps-Induced Inflammation and Apolipoprotein a-I and a-Ii Transgenes. Proteomics. (2005) 5:4245–53. doi: 10.1002/pmic.200401292
248. Schirmer M, Smeekens S, Vlamakis H, Jaeger M, Oosting M, Franzosa E, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. (2016) 167:1897.
249. Parolini C. Effects of Fish N-3 Pufas on Intestinal Microbiota and Immune System. Mar Drugs. (2019) 17:374.
250. Badi I, Cinquetti R, Frascoli M, Parolini C, Chiesa G, Taramelli R, et al. Intracellular Ankrd1 Protein Levels Are Regulated by 26s Proteasome-Mediated Degradation. FEBS Lett. (2009) 583:2486–92. doi: 10.1016/j.febslet.2009.07.001
251. Rubinstein M, Baik J, Lagana S, Han R, Raab W, Sahoo D, et al. Fusobacterium Nucleatum Promotes Colorectal Cancer by Inducing Wnt/B -Catenin Modulator Annexin A1. EMBO Rep. (2019) 20:e47638. doi: 10.15252/embr.201847638
252. Rubinstein M, Wang X, Liu W, Hao Y, Cai G, Han Y. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/B -Catenin Signaling Via Its Fada Adhesin. Cell Host Microbe. (2013) 14:195–206.
253. Goodwin A, Destefano Shields C, Wu S, Huso D, Wu X, Murray-Stewart T, et al. Polyamine Catabolism Contributes to Enterotoxigenic Bacteroides fragilis-Induced Colon Tumorigenesis. Proc Natl Acad Sci U.S.A. (2011) 108:15354–9. doi: 10.1073/pnas.1010203108
254. Wu S, Rhee K, Albesiano E, Rabizadeh S, Wu X, Yen H, et al. A human colonic commensal promotes colon tumorigenesis via activation of T Helper Type 17 T Cell Responses. Nat Med. (2009) 15:1016–22. doi: 10.1038/nm.2015
255. Yu D, Waby J, Chirakkal H, Staton C, Corfe B. Butyrate suppresses expression of neuropilin i in colorectal cell lines through inhibition of Sp1 Transactivation. Mol Cancer. (2010) 9:276. doi: 10.1186/1476-4598-9-276
256. Zuo L, Lu M, Zhou Q, Wei W, Wang Y. Butyrate suppresses proliferation and migration of RKO colon cancer cells though regulating endocan expression by Mapk Signaling Pathway. Food Chem Toxicol. (2013) 62:892–900.
257. Ruemmele F, Schwartz S, Seidman E, Dionne S, Levy E, Lentze M. Butyrate Induced Caco-2 Cell Apoptosis Is Mediated Via the Mitochondrial Pathway. Gut. (2003) 52:94–100.
258. Brown A, Goldsworthy S, Barnes A, Eilert M, Tcheang L, Daniels D, et al. The Orphan G Protein-Coupled Receptors Gpr41 and Gpr43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J Biol Chem. (2003) 278:11312–9. doi: 10.1074/jbc.M211609200
259. Thangaraju M, Cresci G, Liu K, Ananth S, Gnanaprakasam J, Browning D, et al. Gpr109a Is a G-Protein-Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Res. (2009) 69:2826–32. doi: 10.1158/0008-5472.CAN-08-4466
260. Sivaprakasam S, Gurav A, Paschall A, Coe G, Chaudhary K, Cai Y, et al. An Essential Role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis. (2016) 5:e238. doi: 10.1038/oncsis.2016.38
261. Macia L, Tan J, Vieira A, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors Gpr43 and Gpr109a facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. (2015) 6:6734. doi: 10.1038/ncomms7734
262. Pan P, W.Skaer C, Wang H, Oshima K, Huang Y, Yu J. Loss of free fatty acid receptor 2 enhances colonic adenoma development and reduces the chemopreventive effects of black raspberries in Apcmin/+ Mice. Carcinogenesis. (2016) 38:86–93. doi: 10.1093/carcin/bgw122
263. Chang S, Shen M, Liu C, Pu C, Hu J, Huang CJA. Gut butyrate-producing bacterium butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol Lett. (2020) 20:327. doi: 10.3892/ol.2020.12190
264. Wu J, Zhou Z, Hu Y, Dong S. Butyrate-Induced Gpr41 activation inhibits histone acetylation and cell growth. J Genet Genomics. (2012) 39:375–84. doi: 10.1016/j.jgg.2012.05.008
265. Lukasova M, Malaval C, Gille A, Kero J, Offermanns S. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor Gpr109a expressed by immune cells. J Clin Invest. (2011) 121:1163–73. doi: 10.1172/JCI41651
266. Dai Z, Zhang J, Wu Q, Fang H, Shi C, Li Z, et al. Intestinal microbiota: a new force in cancer immunotherapy. Cell Commun Signal. (2020) 18:90.
267. Gopalakrishnan V, Spencer C, Nezi L, Reuben A, Andrews M, Karpinets T, et al. Gut Microbiome Modulates Response to Anti Pd-1 Immunotherapy in Melanoma Patients. Science. (2018) 359:97–103.
268. Routy B, Chatelier E, Derosa L, Duong C, Alou M, Daillère R, et al. Gut Microbiome Influences Efficacy of Pd-1 based immunotherapy against epithelial tumors. Science. (2018) 359:91–7.
269. Hersi F, Elgendy S, Al Shamma S, Altell R, Sadiek O, Omar H. Cancer immunotherapy resistance: the impact of microbiome-derived short-chain fatty acids and other emerging metabolites. Life Sci. (2022) 300:120573. doi: 10.1016/j.lfs.2022.120573
270. Lim H, Joo H, Choi J, Yi J, Yang M, Cho D, et al. Increased expression of cyclooxygenase-2 protein in human gastric carcinoma. Clin Cancer Res. (2000) 6:519–25.
272. Geijtenbeek T, Gringhuis S. Signalling through C-Type lectin receptors: shaping immune responses. Nat Rev Immunol. (2009) 9:465–79. doi: 10.1038/nri2569
273. Mazmanian S, Liu C, Tzianabos A, Kasper D. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
274. Wang Q, McLoughlin R, Cobb B, Charrel-Dennis M, Zaleski K, Golenbock D, et al. A bacterial carbohydrate links innate and adaptive responses through Toll-Like Receptor 2. J Exp Med. (2006) 203:2853–63.
275. Round J, Lee S, Li J, Tran G, Jabri B, Chatila T, et al. The Toll-Like Receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. (2011) 332:974–7. doi: 10.1126/science.1206095
276. Lin M, Yang Y, Chen Y, Hua K, Lu C, Sheu F, et al. A novel exopolysaccharide from the biofilm of Thermus aquaticus Yt-1 Induces the Immune Response through Toll-Like Receptor 2. J Biol Chem. (2011) 286:17736–45. doi: 10.1074/jbc.M110.200113
277. Liu C, Tseng K, Chiang S, Lee B, Hsu W, Pan T. Immunomodulatory and Antioxidant Potential of Lactobacillus Exopolysaccharides. J Sci Food Agric. (2011) 91:2284–91.
278. Hickey A, Stamou P, Udayan S, Ramón-Vázquez A, Esteban-Torres M, Bottacini F, et al. Bifidobacterium Breve Exopolysaccharide Blocks Dendritic Cell Maturation and Activation of Cd4+ T Cells. Front Microbiol. (2021) 12:653587. doi: 10.3389/fmicb.2021.653587
279. Matsuzaki C, Hayakawa A, Matsumoto K, Katoh T, Yamamoto K, Hisa K. Exopolysaccharides Produced by Leuconostoc mesenteroides Strain Ntm048 as an immunostimulant to enhance the mucosal barrier and influence the systemic immune response. J Agric Food Chem. (2015) 63:7009–15. doi: 10.1021/acs.jafc.5b01960
280. Hidalgo-Cantabrana C, López P, Gueimonde M, de Los Reyes-Gavilán C, Suárez A, Margolles A. Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob Proteins. (2012) 4:227–37. doi: 10.1007/s12602-012-9110-2
281. Laiño J, Villena J, Kanmani P, Kitazawa H. Immunoregulatory effects triggered by lactic acid bacteria exopolysaccharides: new insights into molecular interactions with host cells. Microorganisms. (2016) 4:27. doi: 10.3390/microorganisms4030027
282. Mazmanian S, Round J, Kasper DLA. Microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5.
283. Kawaharada Y, Nielsen M, Kelly S, James E, Andersen K, Rasmussen S, et al. Differential Regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun. (2017) 8:14534. doi: 10.1038/ncomms14534
284. Muszyński A, Heiss C, Hjuler C, Sullivan J, Kelly S, Thygesen M, et al. Structures of exopolysaccharides involved in receptor-mediated perception of mesorhizobium loti by lotus Japonicus. J Biol Chem. (2016) 291:20946–61.
285. LeBlanc J, del Carmen S, Miyoshi A, Azevedo V, Sesma F, Langella P, et al. Use of Superoxide Dismutase and Catalase Producing Lactic Acid Bacteria in Tnbs Induced Crohn’s Disease in Mice. J Biotechnol. (2011) 151:287–93. doi: 10.1016/j.jbiotec.2010.11.008
286. Del Carmen S, de Moreno de LeBlanc A, Levit R, Azevedo V, Langella P, Bermúdez-Humarán L. Anti-Cancer Effect of Lactic Acid Bacteria Expressing Antioxidant Enzymes or Il-10 in a Colorectal Cancer Mouse Model. Int Immunopharmacol. (2017) 42:122–9.
287. Tomusiak-Plebanek A, Heczko P, Skowron B, Baranowska A, Okoń K, Thor P, et al. Lactobacilli with superoxide dismutase-like or catalase activity are more effective in alleviating inflammation in an inflammatory bowel disease mouse model. Drug Des Devel Ther. (2018) 12:3221–33. doi: 10.2147/DDDT.S164559
288. Nguyen T, Ung T, Kim N, Jung Y. Role of bile acids in colon carcinogenesis. World J Clin Cases. (2018) 6:577–88.
289. Ignacio Barrasa J, Olmo N, Pérez-Ramos P, Santiago-Gómez A, Lecona E, Turnay J, et al. Deoxycholic and chenodeoxycholic bile acids induce apoptosis via oxidative stress in human colon adenocarcinoma cells. Apoptosis. (2011) 16:1054–67. doi: 10.1007/s10495-011-0633-x
290. Booth L, Gilmore I, Bilton R. Secondary Bile Acid Induced DNA Damage in Ht29 Cells: Are Free Radicals Involved? Free Radic Res. (1997) 26:135–44.
291. Bernstein C, Holubec H, Bhattacharyya A, Nguyen H, Payne C, Zaitlin B, et al. Carcinogenicity of Deoxycholate, a Secondary Bile Acid. Arch Toxicol. (2011) 85:863–71.
292. Shant J, Cheng K, Marasa B, Wang J, Raufman J. Akt-Dependent Nf-Kappab Activation Is Required for Bile Acids to Rescue Colon Cancer Cells from Stress-Induced Apoptosis. Exp Cell Res. (2009) 315:432–50. doi: 10.1016/j.yexcr.2008.11.003
293. Baek M, Park J, Park J, Kim M, Kim H, Bae W, et al. Lithocholic acid upregulates upar and cell invasiveness Via MAPK and AP-1 Signaling in Colon Cancer Cells. Cancer Lett. (2010) 290:123–8. doi: 10.1016/j.canlet.2009.08.030
294. Hague A, Elder D, Hicks D, Paraskeva C. Apoptosis in Colorectal Tumour Cells: Induction by the Short Chain Fatty Acids Butyrate, Propionate and Acetate and by the Bile Salt Deoxycholate. Int J Cancer. (1995) 60:400–6. doi: 10.1002/ijc.2910600322
295. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut Microbiome–Mediated Bile Acid Metabolism Regulates Liver Cancer Via Nkt Cells. Science. (2018) 360:eaan5931.
296. Chen W, Liang X, Peterson A, Munn D, Blazar B. The Indoleamine 2,3-Dioxygenase Pathway Is Essential for Human Plasmacytoid Dendritic Cell-Induced Adaptive T Regulatory Cell Generation. J Immunol. (2008) 181:5396–404. doi: 10.4049/jimmunol.181.8.5396
297. Mezrich J, Fechner J, Zhang X, Johnson B, Burlingham W, Bradfield C. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. J Immunol. (2010) 185:3190–8.
298. Campesato L, Budhu S, Tchaicha J, Weng C, Gigoux M, Cohen I, et al. Blockade of the Ahr Restricts a Treg-Macrophage Suppressive Axis Induced by L-Kynurenine. Nat Commun. (2020) 11:4011. doi: 10.1038/s41467-020-17750-z
299. Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, et al. Commensal Bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. (2013) 8:e80604. doi: 10.1371/journal.pone.0080604
300. Busbee P, Menzel L, Alrafas H, Dopkins N, Becker W, Miranda K, et al. Indole-3-Carbinol Prevents Colitis and Associated Microbial Dysbiosis in an Il-22-Dependent Manner. JCI Insight. (2020) 5:e127551. doi: 10.1172/jci.insight.127551
301. Kannen V, Bader M, Sakita J, Uyemura S, Squire J. The dual role of serotonin in colorectal cancer. Trends Endocrinol Metab. (2020) 31:611–25. doi: 10.1016/j.tem.2020.04.008
302. Scott S, Fu J, Chang P. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci U.S.A. (2020) 117:19376–87.
303. Vreeland T, Clifton G, Hale D, Chick R, Hickerson A, Cindass J, et al. A Phase Iib Randomized Controlled Trial of the Tlpldc Vaccine as Adjuvant Therapy after Surgical Resection of Stage Iii/Iv Melanoma: a primary analysis. Ann Surg Oncol. (2021) 28:6126–37. doi: 10.1245/s10434-021-09709-1
304. O’Brien M, Watkins D, Ryan C, Priest K, Corbishley C, Norton A, et al. A Randomised Trial in Malignant Mesothelioma (M) of Early (E) Versus Delayed (D) Chemotherapy in Symptomatically Stable Patients: The Med Trial. Ann Oncol. (2006) 17:270–5. doi: 10.1093/annonc/mdj073
305. Assersohn L, Souberbielle B, O’Brien M, Archer C, Mendes R, Bass R, et al. A Randomized Pilot Study of Srl172 (Mycobacterium Vaccae) in Patients with Small Cell Lung Cancer (Sclc) Treated with Chemotherapy. Clin Oncol. (2002) 14:23–7. doi: 10.1053/clon.2001.0030
306. Patel P, Sim S, O’Donnell D, Protheroe A, Beirne D, Stanley A, et al. An Evaluation of a Preparation of Mycobacterium Vaccae (Srl172) as an Immunotherapeutic Agent in Renal Cancer. Eur J Cancer. (2008) 44:216–23. doi: 10.1016/j.ejca.2007.11.003
307. Reid G, Jass J, Sebulsky M, McCormick J. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. (2003) 16:658–72.
308. Stratton R, Elia MA. Review of reviews: a new look at the evidence for oral nutritional supplements in clinical practice. Clin Nutr Suppl. (2007) 2:5–23.
309. Kim K, Gluck M. Fecal microbiota transplantation: an update on clinical practice. Clin Endosc. (2019) 52:137–43.
310. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: current status and perspectives. Int J Cancer. (2019) 145:2021–31. doi: 10.1002/ijc.32003
311. Washburn R, Sandberg D, Gazdik Stofer M. Supplementation of a single species probiotic does not affect diversity and composition of the healthy adult gastrointestinal microbiome. Hum Nutr Metab. (2022) 28:200148.
312. Yoon Y, Suh J, Kang E, Kim J. Efficacy and safety of fecal microbiota transplantation for decolonization of intestinal multidrug-resistant microorganism carriage: beyond Clostridioides difficile Infection. Ann Med. (2019) 51:379–89. doi: 10.1080/07853890.2019.1662477
313. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microb features. Cell. (2018) 174:1388–405.e21. doi: 10.1016/j.cell.2018.08.041
314. Landy J, Walker A, Li J, Al-Hassi H, Ronde E, English N, et al. Variable alterations of the microbiota, without metabolic or immunological change, following faecal microbiota transplantation in patients with chronic pouchitis. Sci Rep. (2015) 5:12955.
315. Basson A, Zhou Y, Seo B, Rodriguez-Palacios A, Cominelli F. Autologous fecal microbiota transplantation for the treatment of inflammatory bowel disease. Transl Res. (2020) 226:1–11.
316. Pereira F, Wasmund K, Cobankovic I, Jehmlich N, Herbold C, Lee K, et al. rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile Colonization. Nat Commun. (2020) 11:5104. doi: 10.1038/s41467-020-18928-1
317. Merrick B, Allen L, Masirah M, Forbes B, Shawcross D, Goldenberg S. Regulation, risk and safety of faecal microbiota transplant. Infect Prev Pract. (2020) 2:100069.
318. Wagner R, Warner T, Roberts L, Farmer J, Balish E. Colonization of congenitally immunodeficient mice with probiotic bacteria. Infect Immun. (1997) 65:3345–51. doi: 10.1128/iai.65.8.3345-3351.1997
319. Toomey N, Bolton D, Fanning S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from irish pork and beef abattoirs. Res Microbiol. (2010) 161:127–35. doi: 10.1016/j.resmic.2009.12.010
320. Venema K. Foreword: prebiotics that modulate the endogenous microbiota are also very important. Benef Microbes. (2013) 4:1–2. doi: 10.3920/BM2013.x001
321. Phister T, O’Sullivan D, McKay L. Identification of Bacilysin, Chlorotetaine, and Iturin a Produced by Bacillus Sp. Strain Cs93 Isolated from Pozol, a Mexican Fermented Maize Dough. Appl Environ Microbiol. (2004) 70:631–4. doi: 10.1128/AEM.70.1.631-634.2004
322. Wang Q, Wang K, Wu W, Giannoulatou E, Ho J, Li L. Host and microbiome multi-omics integration: applications and methodologies. Biophys Rev. (2019) 11:55–65.
323. Hitchings R, Kelly L. Predicting and understanding the human microbiome’s impact on pharmacology. Trends Pharmacol Sci. (2019) 40:495–505.
324. Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B, et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. (2020) 10:151. doi: 10.3389/fcimb.2020.00151
325. Vernia F, Longo S, Stefanelli G, Viscido A, Latella G. Dietary factors modulating colorectal carcinogenesis. Nutrients. (2021) 13:143.
326. Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. (2020) 39:4925–43.
327. Mahdavi M, Laforest-Lapointe I, Massé E. Preventing colorectal cancer through prebiotics. Microorganisms. (2021) 9:1325.
328. Sivamaruthi B, Kesika P, Chaiyasut C. The role of probiotics in colorectal cancer management. Evid Based Complement Alternat Med. (2020) 2020:3535982.
329. Doron S, Snydman D. Risk and safety of probiotics. Clin Infect Dis. (2015) 60(Suppl. 2):S129–34.
Keywords: postbiotics, CRC, gut microbiota, nutritional intervention, immune regulation
Citation: Song D, Wang X, Ma Y, Liu N-N and Wang H (2023) Beneficial insights into postbiotics against colorectal cancer. Front. Nutr. 10:1111872. doi: 10.3389/fnut.2023.1111872
Received: 30 November 2022; Accepted: 21 February 2023;
Published: 10 March 2023.
Edited by:
Nancy D. Turner, Michigan State University, United StatesReviewed by:
Guangqiang Wang, University of Shanghai for Science and Technology, ChinaJacques Izard, University of Nebraska-Lincoln, United States
Julio Plaza-Diaz, Children’s Hospital of Eastern Ontario (CHEO), Canada
Copyright © 2023 Song, Wang, Ma, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning-Ning Liu, bGl1bmluZ25pbmdAc2hzbXUuZWR1LmNu; Hui Wang, aHVpd2FuZ0BzaHNtdS5lZHUuY24=
†These authors have contributed equally to this work
 Dingka Song
Dingka Song Xiuli Wang†
Xiuli Wang† Ning-Ning Liu
Ning-Ning Liu Hui Wang
Hui Wang