- 1Department of Experimental, Diagnostic and Specialty Medicine, Alma Mater Studiorum, University of Bologna, Bologna, Italy
- 2Department of Veterinary Medicine, Mediterranean Center for Disease Control (MCDC), University of Sassari, Sassari, Italy
The knowledge of disease determinants is a pre-requisite for disease prevention. Infectious diseases determinants can be classified in three ways, as: primary or secondary; intrinsic or extrinsic; and associated with host, agent, or environment. In the specific case of COVID-19 several of these determinants are currently unknown leading to difficulties in public health approach to this disease. In this paper, we attempt to address several of the current gaps on COVID-19 using a systematic analysis on recent findings and some preliminary knowledge on animal coronaviruses. A discussion on the impact of COVID-19 determinants in disease prevention and control will be based on the Environmental Change and Infectious Disease (EnVID) systemic framework to address several challenges that may affect the control of the SARS- CoV-2 pandemic spread both in industrialized and in developing Countries.
Introduction
The ongoing pandemic of a coronavirus-associated acute respiratory disease called coronavirus disease 19 (COVID-19), is the third documented spillover of an animal coronavirus to humans in only two decades (1). In 2011, Weiss and Leibovitz (2) concluded their Chapter on Coronavirus Pathogenesis by asking “Will SARS or another HCoV emerge from its reservoir? It seems like this could happen again given the identification of numerous bat SARS-like viruses and the finding of SARS-like virus in animal such as the civet.” This sentence sounded prophetic, especially now during the COVID-19 pandemic. Globally, COVID-19 is highlighting that preparedness toward pandemics is still not adequate to effectively deal with what is unknown. However, Asian countries that previously experienced similar epidemics (e.g., SARS) demonstrated a prompt and appropriate response aimed at containing viral spread and reducing disease impact.
Indeed, the rapid spreading of the virus across the world has exposed major gaps in the abilities of most countries to respond to a virulent new pathogen. The WHO-China Joint mission report (3) concludes that it is imperative to timely fill the knowledge gaps in the natural history of the disease to put in place effective control strategies (4) such as effective diagnostic tools, vaccines, and antivirals.
SARS-CoV- 2 is (only) the seventh coronavirus known to infect human in spite of the large coronavirus diversity already explored in animals. It is largely established that coronaviruses cause a large variety of diseases in wild and domestic species, both in livestock and companion animals, and in wildlife, leading to significant research in the last half of the Twentieth century (5). Given that SARS-CoV-2 has undoubtedly a zoonotic origin (6), a One Health approach is suggested in order to understand the origin and the causes of the pandemic (7).
This paper aims at analyzing some of the current unknowns on SARS-CoV-2 using recently published data and the available knowledge on coronaviruses infecting animals. Based on current scientific evidence, we elaborate the Environmental Change and Infectious Disease (EnvID) framework analysis (8) to facilitate the identification of relevant environmental and socio-economic factors that may affect disease burden and to provide links to interventions strategies in a One Health perspective.
The Pathogen
SARS-CoV-2 belongs to the Coronaviridae family that comprises enveloped, non-segmented, positive-sense RNA viruses (group 4 in Baltimore classification) named after their corona (crown) like surface, appreciable by electron microscopy, and formed by their largely protruding spike proteins.
According to the International Committee on Taxonomy of Viruses (ICTV) Coronaviridae study group (CSG) this family comprises 2 subfamilies (Table S1): Letovirinae, including the single species Microhyla letovirus 1, infecting the ornate chorus frog Microhyla fissipes (9), and Orthocoronavirinae, including 4 genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus) and 38 species. While all Alphacoronavirus and Betacoronavirus species infect mammals, Deltacoronavirus infects exclusively birds. Gammacoronavirus, a less diverse genus, includes 2 viral species, the Beluga whale coronavirus SW1, and Avian coronavirus, infecting, respectively, the beluga whale and birds. To date there is no evidence of coronavirus infection in reptiles, even if the presence of a Letovirinae coronavirus in amphibians encourage more coronavirus investigation in reptiles. Coronaviruses infecting human can cause mild respiratory symptoms and conjunctivitis (10, 11), such as Human coronavirus 229E and Human coronavirus NL63 (Alphacoronavirus), or mild enteric and respiratory disease, such as Betacoronavirus 1 OC43 and Human coronavirus HKU1 (Betacoronavirus); whereas Severe acute respiratory syndrome-related coronavirus (SARS) and Middle East respiratory syndrome-related coronavirus (MERS), both belonging to Betacoronavirus, can cause severe disease and are example of spillover of animal viruses to human. COVID-19 associated virus was recognized by CSG as forming a sister clade to the prototype human and bat severe acute respiratory syndrome coronaviruses (SARS-CoVs) of the species Severe acute respiratory syndrome-related coronavirus, and it was designated as SARS-CoV-2 (12).
The considerable diversity of coronaviruses is driven by their genetic and evolutionary features. Specifically, mutation and recombination appear to be particularly important in coronavirus evolution (13–16). As RNA viruses, CoVs tend to accumulate random mutations at a far higher rate than their hosts. In spite of the proofreading-repair activity of their polymerase, increasing copying accuracy up to 14-fold (17), coronaviruses form mutant spectra (mutant clouds, quasispecies); viral populations consisting of dynamic and complex mutant distributions, rather than unique genomic sequences (18). Quasispecies correlate to enhanced virulence and evolvability and render difficult to prevent and control viral diseases. The ability to exist as mutant clouds in an individual host have been described both in animal CoVs, such as the bovine enteric and respiratory coronaviruses (19) and in human SARS-CoV (20, 21). Furthermore, during CoV replication template switching favors homologous recombination among different CoVs lineages or with less related viruses. Cooperatively, CoVs circulation in multiple host species may increase recombination events (15). Among others, genetic recombination was demonstrated in animal viruses such as rodents CoVs (13, 22), porcine PEDV (23), cat and dogs CoVs (24, 25), Hedgehog CoVs (26), and bats (16, 27). Notably, recombination between Coronavirus and Orthoreovirus has been postulated in the case of Rousettus bat coronavirus GCCDC1 (28). Genetic recombination in Human CoVs, including, NL63, HKU1, OC43, SARS-CoV, and MERSCoV has also been documented (15, 16). Additionally, the large genome in CoV, and the presence of key mutational and recombination hotspots (15, 26, 29) account for extra plasticity in genome modifications, promoting intraspecies variability, host shifts, and novel CoVs to emerge (14, 15). Evidence for a rapid evolution of SARS-CoV-2 was recently shown by comparing 86 complete or near complete genomes from different parts of the world, depicting the great diversity in viral coding and non-coding regions (30).
The Hosts
It has been postulated that coronavirus evolution and dissemination is nourished by warm blooded flying vertebrates (bats and birds), ideal hosts for the coronavirus gene source; bats for Alphacoronavirus and Betacoronavirus, such as SARS-CoV and MERS-CoV, and birds for Gammacoronavirus and Deltacoronavirus (31, 32). Also, a rodent origin has been proposed for HCoV- OC43 and HKU1 (16).
SARS-CoV emerged in 2002 in southern China (Guangdong Province), as a novel clinical severe disease and rapidly spread to other 28 countries (31, 33). All early cases had a history of contact with living animals (in wet markets or restaurants). Molecular and serological data, and isolation, demonstrated that SARS-CoV originated from civet cats (Figure 1A), family Viverridae, in Guangdong market, and also that farmed civets did not play a role as reservoirs (31, 34, 35).
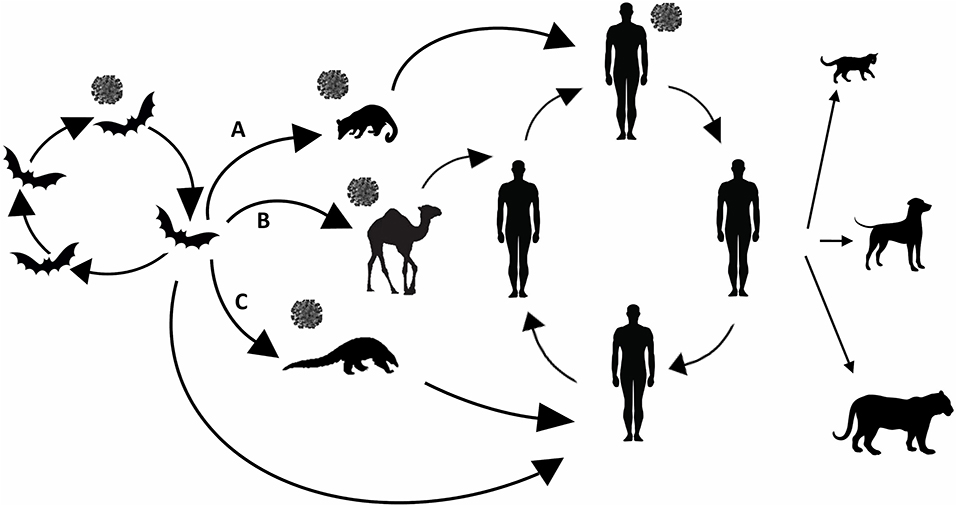
Figure 1. Animal origin of human coronaviruses. SARS- CoV (A) emerged from bats, infected civets and humans and adapted to these hosts before causing the SARS epidemic. MERS- CoV (B) likely spilled over from bats to dromedary camels. SARS-CoV-2 (C) emerged from bats spilled over to human either directly or by previous infecting pangolins. Reverse zoonosis transmission (from human to cats, dogs, and tiger) refers only to SARS-CoV-2.
Horseshoe bats family Rhinophidae host genetically diverse SARS-like coronaviruses, including ancestors of SARS-CoV and are considered the original source of SARS (31). Moreover, SARS and MERS related CoVs have been identified in Vespertilionidae and Molossidae bats (36).
Early cases of MERS in Saudi Arabia in 2012; (37) had contact with animals, in particular with dromedary camels (Figure 1B). Molecular and serological data indicated the presence of MERS viruses in dromedaries with high sequence similarities (>99%) to human MERS-CoV (38), and antibodies in camels could be traced back to the eighties (31, 39). More molecular data support that human and camel MERS-CoV isolates belong to the same coronavirus species, and that MERS-CoV originated from the bat gene pool (31). Ancestor analysis suggests that MERS-CoV could have spilled from bats to camels some 30 years ago in Africa, and it was subsequently introduced in the Arabian Peninsula by importing camels from the African continent (40).
Evidence suggests that the SARS-CoV-2 emerged in late 2019 in a wet market in Wuhan, Hubei province, China (41, 42). However, retrospective analyses indicate that SARS-CoV-2 might have spilled somewhere else prior to December 2019 (6). Origin of this virus rapidly became one of the greatest concerns (Figure 1C). The idea of a laboratory-based origin is not plausible as there is no evidence showing that SARS-CoV-2 is a purposefully manipulated virus (11). Also, a snake origin of SARS-CoV-2 can be ruled out as no other coronaviruses have been found in reptiles, and there are not receptor signatures, or other strongly indicative molecular evidence (43) supporting this. It is now recognized that bats such as Rhinolophus affinis are natural viral reservoirs, and that the Malayan pangolin (Manis javanica) might be the SARS-CoV-2 intermediate host that brought the bat coronavirus to human hosts, even if some studies have proposed that the pangolin, illegally imported into southern China, may be a natural host rather than an intermediate host (44–46). SARS-CoV-2 infection has been demonstrated in two pet dogs in Hong Kong and two pet cats in Hong Kong and in Belgium (47, 48). The pet cases were in close contact with a confirmed COVID-19 human case. Also, a Malayan tiger in Bronx Zoo in New York City developed COVID-19 after exposure to an asymptomatically infected worker (49).
Transmission Pathways
Person-to-person transmission of SARS-CoV-2 has been documented as for the previously discovered SARS-CoV and MERS-CoV. All these coronaviruses seem predominantly transmitted by respiratory droplets that people cough, sneeze, or exhale over a relatively close distance.
A study conducted by Setti et al. (50) has shown that pollution may have played a role in the propagation of SARS-CoV-2. Researchers evidenced an association between the exceedances of the legal limits of the PM10 concentrations recorded during the 2 weeks preceding the first peak of COVID-19 cases in Northern Italy. This lead could be related to the conditions of airborne particulate matter pollution that may have exerted a boost action on transmission. Animal coronaviruses can replicate in the epithelial cells of both the respiratory and the enteric tracts (51). Enteric tropism was also reported for SARS-CoV-2, causing diarrhea in ~16–73% of patients in addition to respiratory symptoms (52). The transmission of SARS through water droplets from feces via air ventilation systems in Hong Kong was reported (53). Diarrhea and enteric symptoms were also reported in a significant proportion of COVID-19 patients. Recent reports show that SARS-CoV-2 has been detected in stool samples of COVID-19 cases (54–57).
According to WHO and Food Safety Authorities Network (INFOSAN) (58), more information on the potential for persistence of SARS-CoV-2 on foods traded internationally as well as the potential role of food in the transmission of the virus are needed. WHO suggests that the consumption of raw or undercooked animal products should be avoided. Raw meat, raw milk, or raw animal organs should be handled with care to avoid cross- contamination with uncooked foods. Results obtained with SARS-CoV surrogate, Bovine Coronavirus BCoV of the genus Betacoronavirus, showed that contaminated vegetables may serve as a vehicle for transmission through consumption. As an example, Mullis et al. (59) showed that BCoV on lettuce retained infectivity for at least 14 days under household refrigeration conditions. The ability of enteric coronavirus Porcine epidemic diarrhea virus (PEDv) to survive in specific feed ingredients, under modeled conditions simulating shipment, was shown suggesting that contaminated feed ingredients for pigs could serve as transboundary risk factors (60). Extended survival in soybean meal has been confirmed also for swine alphacoronavirus TGEV and deltacoronavirus PDCoV (61). These data lead to speculate that contaminated ready-to-consume produce may be a potential vehicle for zoonotic transmission of coronaviruses to humans. Awareness regarding the possible roles of water, fresh products, and fecal contamination in coronavirus transmission is required at times of human coronavirus outbreaks. The role of animals in SARS-CoV-2 transmission is still debated and need to be clarified. Experimental infection tests conducted in laboratory animals, suggest that SARS-CoV-2 replicates poorly in dogs, pigs, chickens, and ducks, but efficiently in ferrets and cats, with cats transmitting the virus via respiratory droplets (62). For these reasons, a possible role of animal hosts as reservoir and a further source of virus for humans cannot be ruled out, especially in hotspots of biodiversity, and in many developing countries characterized by close proximity among human, wild, and domestic animals.
Environmental and Socio-Economic Determinants
Urbanization can be considered an important distal environmental factor acting on COVID-19 transmission. According to FAO (63) cities, with their high population density, are vulnerable to the COVID-19 pandemic. The spread of the virus in crowded cities could have extensive morbidity and mortality consequences for urban populations. The very poor and those living in slums have extremely limited access to essential health and sanitation facilities, nutritious food and adequate infrastructure such as piped clean water and electricity. As during the SARS-CoV pandemic (64) SARS-CoV-2 was detected in wastewater collected at a major urban treatment facility in Massachusetts (65). SARS-CoV-2 was detected in sewage of 7 cities and the airport during the emergence of COVID-19 in the Netherlands. Sewage surveillance could be used to monitor the circulation of SARS-CoV-2 complementing current clinical surveillance (66).
Climate is a further distal environmental factor that might indirectly influence SARS-CoV-2 transmission. The WHO assumption is that COVID-19 spread will not ameliorate during summer period. The effect of seasons on transmission of COVID-19 is still unknown. A negative correlation between warmer climate and COVID-19 spread has been suggested by a number of pre-print sources (67–69) and news. However, Caspi and collaborators conclude that their findings, on possible reduced spread during warm season, should be cautiously interpreted and need to be validated as an association between warmer climate and reduced COVID-19 spread might be due to local patterns of transmission rather than by climate.
It has been speculated that the warm months of summer in the northern hemisphere might not necessarily reduce transmission below the value of unity as they do for influenza A given the fact that SARS-CoV-2 R0 was estimated 2–3 (1). SARS-CoV-2 spreads in Countries characterized by tropical climates, such as Singapore. For this reason, winter is not considered a necessary condition for SARS-CoV-2 diffusion and persistence (70). The independence of viral spread from high temperature has been already proved for other coronaviruses such as the SARS- CoV surrogate animal virus transmissible gastroenteritis virus (TGEV) a diarrheal pathogen of swine and mouse hepatitis virus MHV a respiratory and enteric pathogen of mice that showed the ability to survive on surfaces for days at 20°C and wide range of Relative Humidity (RH) levels (20–60%). The animal surrogates showed to be more resistant to inactivation on surfaces than previously studied human coronaviruses, such as 229E. Several studies demonstrated that SARS-CoV and MERS-CoV have the capacity to survive on dry surfaces for a sufficient duration to facilitate onward transmission. SARS-CoV and MERS-CoV were shown to be able to contaminate the environment and fomites, promoting viral access to mucous membranes of the nose, eyes or mouth through individual self-inoculation by hands (71). In fact, SARS-CoV related mathematical and animal models, and intervention studies suggested that contact with contaminated environment is the most important route in some scenarios, such as in health care facilities (72).
SARS-CoV and its surrogates can also survive in environmental reservoirs such as water, foods, and in sewage for extended periods (71, 73–75). SARS-CoV and probably MERS-CoV are shed into the environment at concentrations exceeding the infective dose and they can survive for considerable time on surfaces. The surface survival of SARS/MERS-CoV was shown to be greater than that of other respiratory viruses such as influenza virus. In particular, infective MERS-CoV could still be recovered after 48 h at the 20°C−40% RH condition, whereas the virus remained viable for 8 and 24 h at 30°C−80% RH and 30°C 30% RH, respectively. Instead, H1N1 influenza virus, known to have a seasonal spread pattern, cannot be recovered after 4 h at the same environmental conditions (73).
Economic development should be considered as a further distal factor, FAO (63) stated that there is a particularly high risk of infection for the 1.2 billion people living in the congested and overcrowded informal urban settlements where conditions are already unsafe and unhealthy for human living.
Discussion
Climate change, urbanization and subsequent loss of natural habitats, changes in human habits and behavior, collectively leading to human and animals living in close proximity, have been identified as the main drivers for the emergence of viral diseases (76).
To some extent COVID-19 outbreak may be considered as an indirect consequence of global environmental changes (77). In fact, multiscale environmental changes encompassing social processes, such as over-crowded urban settings where human and wildlife can come into close contact (e.g., wet markets), trading of exotic animals (6), large-scale population migration, such as those linked to the Chinese New Year celebration (78), were identified as possible causes of the emergence of COVID-19 and of its pandemic spread.
To develop plans and policies for intervention strategies, a preliminary knowledge of disease determinants is necessary (79). COVID-19 was immediately defined as a contact-transmissible infectious disease, spread via direct contact between individuals. Outbreak control measures were thus aimed at reducing the amount of mixing in the population to delay the peak and reduce the final size of the epidemic (80). However, the course of the COVID-19 epidemic is defined by a series of further key factors, some of which are still poorly understood (1). The amount of scientific data produced during the COVID-19 pandemic is amazingly huge, and this can be crucial to translate research into effective public health policies and practices.
According to recent data, COVID-19 is not only linked to person to person transmission, as indirect transmission may also occur. Recent scientific evidence seems to suggest that environmental factors and changes may act as extrinsic determinants in the epidemiology of COVID-19 and of other human and animal coronaviruses.
Based on the available scientific literature, we applied the EnvID framework (8) to facilitate the identification of the environment-disease relationships and connections that may impact on disease burden (Figure 2). The EnvID framework encompasses 3 interlocking components including: environment, transmission and disease, and it defines three transmission groups: group I, including directly transmitted diseases; group II, including vector-borne diseases; and group III, including environmentally mediated diseases with non-human host. At first, based on the available data, we attempted to attribute COVID-19 to one of these three groups.
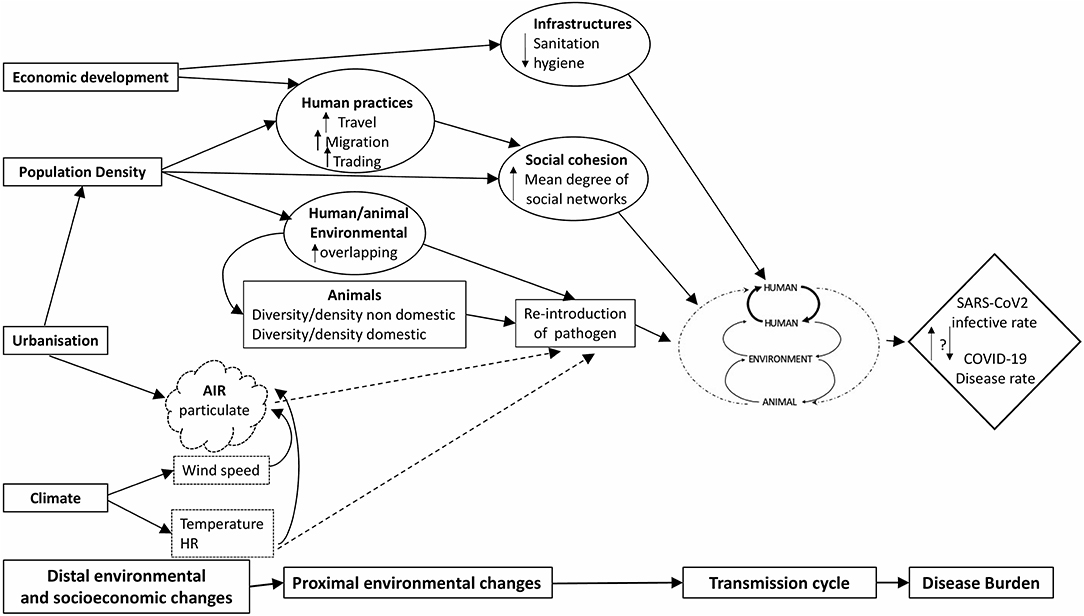
Figure 2. Causal EnvID diagram showing the relationship between distal and proximal environmental changes and COVID-19 disease burden. Solid lines indicate actual influence and scattered lines represent potential influence.
In agreement with the prevalent scientific evidence, COVID-19 might be classified as group I, as it can be transmitted person to person being mainly affected by social processes, such as over-crowding. Social distancing has in fact shown to prevent transmission from symptomatic and non-symptomatic cases, hence flattening the epidemic and delaying the peak.
However, SARS-CoV-2 is also quite resistant in the environment, and the role of non-human hosts as reservoir in infected areas cannot be overlooked. For this reason, it may be also attributed to group III, including those diseases for which transmission can be affected by the modification of human exposure to contaminated environment, media including water, and possibly food and infected animals. Besides indirect contact with contaminated environment, a direct transmission from human to animals and vice-versa cannot be ruled out.
In Figure 2, we represented this transmission dynamic with dashed arrows, as it requires to be better investigated to provide more scientific evidence, in a true One Health approach, where human and veterinary medicine need to collaborate thoroughly to clearly define the whole causal relationships of disease transmission.
According to Brierley et al. (81), transmission routes with environmental components (e.g., fecal–oral or food borne transmission) would be associated with higher virulence than direct, contact-based transmission. EnvID analysis shows that distal environmental changes, such as those related to urbanization and climate, need to be considered as they may act through multiple intermediate steps on COVID-19 transmission. As an example, pollution linked either to urbanization and to climate factors may play a role. It is known in fact that a prolonged exposure to air pollution leads to a chronic inflammatory stimulus, even in young and healthy subjects (82). For this reason, a contribute of air pollution and particulate cannot be neglected as an environmental factor that may directly or indirectly influence the transmission cycle of SARS-CoV-2. Regarding climate, it has been shown that SARS-CoV and MERS-CoV are quite resistant even at a temperature of 30°C when RH is not exceeding 30%. It has been shown for influenza virus that at low RH, evaporation of water from exhaled bioaerosols would occur rapidly, leading to the formation of droplet nuclei; conversely, at high RH, small respiratory droplets would take on water, increase in size and settle more quickly out of the air (83). If SARS-CoV-2 will also show similar resistance patterns, as the other CoVs, and its transmission is not impaired at 30°C, this may suggest that contact-based spread may predominate in the tropics where RH is generally high, whereas aerosol transmission may play a larger role in temperate climates. Even if person to person transmission is considered the main transmission pathway, our proposed transmission pattern underlines that human, environment and animal, might all play a role in the potential spread and persistence of SARS-CoV-2 with a different weight in different geographical and social contexts. In particular, environmental contamination and human/animal habitat overlapping cannot be overlooked especially in developing countries. Coronaviruses exist and can maintain their viability in sewage and wastewater, originating from the fecal discharge of infected patients, highlighting the importance of sanitation to protect public health (84). In developing countries, where water and sanitation systems are often insufficient or ineffective, it is necessary to consider that SARS-CoV-2 transmission might be amplified through water contamination in cities.
Attribution of the burden of disease to environmental risks, highlights in fact the importance of environmental protection for people's health and can inform priority setting for targeted management of environmental determinants (85). The public health measures to contain COVID-19 spreading are being based on the scientific data currently available that mainly derive from studies on the infection's dynamics observed in industrialized countries. An important bias on the large amount of data produced in China and other Asian countries and the lower contributions from Europe and North America is evident. This is clearly due to the timing and geographic spread of the epidemic. As suggested by the EnvID framework, distal and proximal environmental factors, transmission dynamics and consequent morbidity and lethality may differ in different socio-economic contexts. It is thus essential to collect data on the determinants of infection even in developing countries where the capacities of the healthcare infrastructures, diagnostic and research capacities are very limited. In fact, country socio-economic profiles were shown to have an influence on the growth rate of epidemics so that R0 might differ in different geographic areas (70). As an example, FAO warns that the policies to limit the effects of the virus, successfully applied in many industrialized Countries, and based on lockdowns and physical distancing, can spell disaster for the livelihood of individuals and families in developing Countries, leading to food insecurity and deficient nutrition.
Tackling with environmental risk factors always entails intersectoral collaborations and a One Health approach. Research inputs from both human and animal health sides, including many other scientific and non-scientific stakeholders, will be needed to facilitate a systemic way to effectively deal with COVID-19 emergency. A One Health perspective is foreseen, especially in the Global South, to design and implement research programs and policies in which multiple sectors communicate and work together to achieve better public health outcomes.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found in the EnvID model which is presented as a figure and discussed in the text.
Author Contributions
AS shaped the conceptualization. AS and AA each contributed to research, critical analysis, and writing of the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2020.00302/full#supplementary-material
References
1. Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. (2020) 21:931–4. doi: 10.1016/S0140-6736(20)30567-5
2. Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. (2011) 81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2
3. World Health Organization (2020). Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). Available online at: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
4. Bedford J, Enria D, Giesecke J, Heymann DL, Ihekweazu C, Kobinger G, et al. COVID-19: towards controlling of a pandemic. Lancet. (2020) 395:1015–8. doi: 10.1016/S0140-6736(20)30673-5
5. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
6. Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. (2020) 181:223–7. doi: 10.1016/j.cell.2020.03.035
7. El Zowalaty ME, Järhult JD. From SARS to COVID-19: a previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans – call for a one health approach. One Health. (2020). 9:100124. doi: 10.1016/j.onehlt.2020.100124
8. Eisenberg JNS, Desai MA, Levy K, Bates SJ, Liang S, Naumoff K, et al. Environmental determinants of infectious disease: a framework for tracking causal links and guiding public health research. Environ Health Perspect. (2007) 115:1216–23. doi: 10.1289/ehp.9806
9. Bukhari K, Mulley G, Gulyaeva AA, et al. Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology. (2018) 524:160–71. doi: 10.1016/j.virol.2018.08.010
10. Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. (2018) 100:163–88. doi: 10.1016/bs.aivir.2018.01.001
11. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. (2020) 26:450–2. doi: 10.1038/s41591-020-0820-9
12. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. (2020) 5:536–44. doi: 10.1038/s41564-020-0695-z
13. Lai MM, Baric RS, Makino S, Keck JG, Egbert J, Leibowitz JL, et al. Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol. (1985) 56:449–56. doi: 10.1128/JVI.56.2.449-456.1985
14. Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. (2009) 234:1117–27. doi: 10.3181/0903-MR-94
15. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. (2016) 24:490–502. doi: 10.1016/j.tim.2016.03.003
16. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
17. Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. (2011) 8:270–9. doi: 10.4161/rna.8.2.15013
18. Domingo E. Quasispecies and the development of new antiviral strategies. Prog Drug Res. (2003) 60:133–58. doi: 10.1007/978-3-0348-8012-1_4
19. Zhang X, Hasoksuz M, Spiro D, Halpin R, Wang S, Vlasova A, et al. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology. (2007) 363:1–10. doi: 10.1016/j.virol.2007.03.018
20. Xu D, Zhang Z, Chu F, Li Y, Jin L, Zhang L, et al. Genetic variation of SARS coronavirus in Beijing Hospital. Emerg Infect Dis. (2004) 10:789–94. doi: 10.3201/eid1005.030875
21. Xu HF, Wang M, Zhang ZB, Zou XZ, Gao Y, Liu XN, et al. An epidemiologic investigation on infection with severe acute respiratorY syndrome coronavirus in wild animals traders in Guangzhou. Zhonghua Yu Fang Yi Xue Za Zhi. (2004) 38:81–3.
22. Wang W, Lin XD, Guo WP, Zhou RH, Wang MR, Wang CQ, et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. (2015) 1:19–27. doi: 10.1016/j.virol.2014.10.017
23. Tian PF, Jin YL, Xing G, Qv LL, Huang YW, Zhou JY. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis. (2014) 20:1735–8. doi: 10.3201/eid2010.140338
24. Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, et al. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. J Virol. (2009) 83:1532–7. doi: 10.1128/JVI.01937-08
25. Terada Y, Matsui N, Noguchi K, Kuwata R, Shimoda H, Soma T, et al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE. (2014) 9:e106534. doi: 10.1371/journal.pone.0106534
26. Lau SKP, Luk HKH, Wong ACP, Fan RYY, Lam CSF, Li KSM, et al. Identification of a novel betacoronavirus (merbecovirus) in amur hedgehogs from China. Viruses. (2019) 11:980. doi: 10.3390/v11110980
27. Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. (2016) 10:609–20. doi: 10.1038/ismej.2015.138
28. Huang C, Liu WJ, Xu W, Jin T, Zhao Y, Song J, et al. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. (2016) 12:e1005883. doi: 10.1371/journal.ppat.1005883
29. Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission J Virol. (2010) 84:3134–46. doi: 10.1128/JVI.01394-09
30. Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Gen Evol. (2020) 81:104260. doi: 10.1016/j.meegid.2020.104260
31. Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. (2015) 12:221. doi: 10.1186/s12985-015-0422-1
32. Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as thegene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11
33. Zhong NS, Zheng BJ, Li YM, Poon Xie ZH, Chan KH, Li PH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. (2003) 362:1353–8. doi: 10.1016/S0140-6736(03)14630-2
34. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. (2003) 10:276–8. doi: 10.1126/science.1087139
35. Tu C, Crameri G, Kong X, Chen J, Sun Y, Yu M, et al. Antibodies to SARS coronavirus in civets. Emerg Infect. Dis. (2004) 10:2244–8. doi: 10.3201/eid1012.040520
36. Lecis R, Mucedda M, Pidinchedda E, Pittau M, Alberti A. Molecular identification of Betacoronavirus in bats from Sardinia (Italy): first detection and phylogeny. Virus Genes. (2019) 55:60–7. doi: 10.1007/s11262-018-1614-8
37. Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. (2012) 17:20290. doi: 10.1101/2020.02.19.950253
38. Yusof MF, Eltahir YM, Serhan WS, Hashem FM, Elsayed EA, Marzoug BA, et al. Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi Emirate. UAE Virus Genes. (2015) 50:509–13. doi: 10.1007/s11262-015-1174-0
39. Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. (2013) 13:859–66. doi: 10.1016/S1473-3099(13)70164-6
40. Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. (2014) 88:11297–303. doi: 10.1128/JVI.01498-14
41. Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. (2020) 6:14. doi: 10.1038/s41421-020-0153-3
42. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
43. Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. Proteome Res. (2020) 19:1351–60. doi: 10.1021/acs.jproteome.0c00129
44. Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. (2020) 30:1346–51. doi: 10.1016/j.cub.2020.03.022
45. Liu P, Jiang JZ, Hua Y, Wang X, Hou F, Wan XF, et al. Are pangolins the intermediate host of the 2019 novel coronavirus (2019-nCoV)? PLOS Pathogens. (2020) 16:e1008421. doi: 10.1371/journal.ppat.1008421
46. Wong MC, Cregeen SJJ, Ajami NJ, Petrosino JF. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. (2020). doi: 10.1101/2020.02.07.939207
47. OIE–World Animal Health Organization (2020). Available online at: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=33892
48. OIE–World Animal Health Organization (2020). Available online at: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=33832
49. OIE–World Animal Health Organization (2020). Available online at: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=33885
50. Setti L, Passarini F, De Gennaro G, Barbieri P, Pallavicini A, Ruscio M, et al. Searching for SARS-COV-2 on particulate matter: a possible early indicator of COVID-19 epidemic recurrence. Int J Environ Res Public Health. (2020) 17:2986. doi: 10.3390/ijerph17092986
51. Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Investig. (2003) 111:1605–9. doi: 10.1172/JCI18819
52. Han C, Duan C, Zhang S, Spiegel B, Shi H, Wang W, et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. (2020) 115:916–23. doi: 10.14309/ajg.0000000000000664
53. McKinney KR, Gong YY, Lewis TG. Environmental transmission of SARS at Amoy gardens. J Environ Health. (2006) 68:26–30.
54. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
55. Yeo C. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. (2020) 5:335–7. doi: 10.1016/S2468-1253(20)30048-0
56. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. (2020) 158:1831–3.e3. doi: 10.1053/j.gastro.2020.02.055
57. Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. (2020) 26:502–5. doi: 10.1038/s41591-020-0817-4
58. World Health organization and Food Safety Authorities Network (2020). Coronavirus Disease 2019 (COVID-19). Situation Report – 32. Available online at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200221-sitrep-32-covid-19.pdf?sfvrsn=4802d089_2
59. Mullis L, Saif LJ, Zhang Y, Zhang X, Azevedo MS. Stability of bovine coronavirus on lettuce surfaces under household refrigeration conditions. Food Microbiol. (2012) 30:180–6. doi: 10.1016/j.fm.2011.12.009
60. Dee S, Neill C, Singrey A, Clement T, Cochrane R, Jones S, et al. Modeling the transboundary risk of feed ingredients contaminated with porcine epidemic diarrhea virus. BMC Vet Res. (2016) 12:51. doi: 10.1186/s12917-016-0674-z
61. Trudeau MP, Verma H, Sampedro F, Urriola PE, Shurson GC, Goyal SM, et al. Environmental persistence of porcine coronaviruses in feed and feed ingredients. PLoS ONE. (2017) 12:e0178094. doi: 10.1371/journal.pone.0178094
62. Shi J, Wen Z, Zhong G, Yang H, Wang C, Liu R, et al. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. Science. (2020) 368:1016–20. doi: 10.1126/science.abb7015
63. Food and Agriculture Organisation. Urban Food Systems and COVID-19: The Role of Cities and Local Governments in Responding to the Emergency. (2020). Available online at: http://www.fao.org/3/ca8600en/CA8600EN.pdf
64. Wang XW, Li J, Guo T, Zhen B, Kong Q, Yi B, et al. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People's Liberation Army. Water Sci Technol. (2005) 52:213–21. doi: 10.2166/wst.2005.0266
65. Wu F, Xiao A, Zhang J, Gu X, Lee L, Kauffman K, et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv [preprint]. (2020). doi: 10.1101/2020.04.05.20051540
66. Medema G, Heijnen L, Elsinga G, Italiaander R. Presence of SARS-coronavirus-2 in sewage. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.29.20045880
67. Araujo MB, Naimi B. Spread of SARS-CoV-2 Coronavirus likely to be constrained by climate. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.12.20034728
68. Ficetola GF, Rubolini D. Climate affects global patterns of COVID-19 early outbreak dynamics. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.23.20040501
69. Caspi G, Shalit U, Kristensen SL, Aronson D, Caspi L, Rossenberg O, et al. Climate effect on COVID-19 spread rate: an online surveillance tool. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.26.20044727
70. Neher RA, Dyrdak R, Druelle V, Hodcroft EB, Albert J. Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. Swiss Med Wkly. (2020) 150:w20224. doi: 10.4414/smw.2020.20224
71. Casanova L, Rutala WA, Weber DJ, Sobsey MD. Survival of surrogate coronaviruses in water. Water Res. (2009) 43:1893–8. doi: 10.1016/j.watres.2009.02.002
72. Otter JA, Donskey C, Yezli S, Douthwaite S, Goldenberg SD, Weber DJ. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. (2016) 92:235–50. doi: 10.1016/j.jhin.2015.08.027
73. van Doremalen N, Bushmaker T, Munster VJ. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. (2013) 18:20590. doi: 10.2807/1560-7917.ES2013.18.38.20590
74. Chan KH, Peiris JS, Lam SY, Poon LL, Yuen KY, Seto WH. The effects of temperature and relative humidity on the viability of the SARS Coronavirus. Adv Virol. (2011) 2011:734690. doi: 10.1155/2011/734690
75. Yepiz-Gomez MS, Gerba CP, Bright KR. Survival of respiratory viruses on fresh produce. Food Environ Virol. (2013) 5:150–6. doi: 10.1007/s12560-013-9114-4
76. Murray KA, Daszak P. Human ecology in pathogenic landscapes: two hypotheses on how land use change drives viral emergence. Curr Opin Virol. (2013) 3:79–83. doi: 10.1016/j.coviro.2013.01.006
77. Chakraborty I, Maity P. COVID-19 outbreak: migration, effects on society, global environment and prevention. Sci Total Environ. (2020) 728:138882. doi: 10.1016/j.scitotenv.2020.138882
78. Wang Q, Su M. A preliminary assessment of the impact of COVID-19 on environment - a case study of China. Sci Total Environ. (2020) 728:138915. doi: 10.1016/j.scitotenv.2020.138915
79. Shrivastava SR, Shrivastava PS, Ramasamy J. Emerging and re-emerging infectious diseases: public health perspective. Int J Prev Med. (2013) 4:736–7.
80. Prem K, Liu Y, Russell TW, Kucharski AJ, Eggo RM, Davies N, et al. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. Lancet Public Health. (2020) 5:e260–71. doi: 10.1101/2020.03.09.20033050
81. Brierley L, Pedersen AB, Woolhouse MEJ. Tissue tropism and transmission ecology predict virulence of human RNA viruses. PLoS Biol. (2019) 17:e3000206. doi: 10.1371/journal.pbio.3000206
82. Conticini E, Frediani B, Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ Pollut. (2020) 26:114465. doi: 10.1016/j.envpol.2020.114465
83. Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. (2007) 3:1470–6. doi: 10.1371/journal.ppat.0030151
84. Naddeo V, Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ Sci Water Res Technol. (2020) 6:1213–6. doi: 10.1039/D0EW90015J
Keywords: one health, COVID-19, SARS-CoV-2, disease determinants, public health, vertebrate hosts, epidemics
Citation: Scagliarini A and Alberti A (2020) COVID-19: An Appeal for an Intersectoral Approach to Tackle With the Emergency. Front. Public Health 8:302. doi: 10.3389/fpubh.2020.00302
Received: 22 April 2020; Accepted: 04 June 2020;
Published: 16 June 2020.
Edited by:
Zisis Kozlakidis, International Agency for Research on Cancer (IARC), FranceReviewed by:
Zohar Lederman, The University of Hong Kong, Hong KongJaquelline Germano De Oliveira, Oswaldo Cruz Foundation (Fiocruz), Brazil
Copyright © 2020 Scagliarini and Alberti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandra Scagliarini, YWxlc3NhbmQuc2NhZ2xpYXJpbmlAdW5pYm8uaXQ=
 Alessandra Scagliarini
Alessandra Scagliarini Alberto Alberti
Alberto Alberti