- 1Massachusetts Institute of Technology, Cambridge, MA, United States
- 2Metha Artificial Intelligence, Tel Aviv, Israel
Ruminant livestock production depends on microorganisms to ferment forages into valuable dairy and meat products. However, this process also generates enteric methane emissions, a significant contributor to anthropogenic greenhouse gases. Despite various strategies aimed at reducing methane emissions, success has been limited. In previous work, we developed an AI-driven model based on deep microbiome sequencing, which predicts the effect of feed additives on methane emissions. The model uses sequenced rumen samples from a given herd to construct microbiome networks to identify biomarkers associated with feed additive effectiveness in the reduction of methane emissions. In this study, we validated the model supplying a commercial methane-mitigating feed additive and performing hundreds of in-situ methane measurements across several commercial dairy farms. The results highlight the model’s robustness and precision, demonstrating its effectiveness in predicting enteric methane reductions and enhancing feed additive performance. Additionally, the model serves as a critical tool for data-driven decision-making, playing a pivotal role in advancing precision agriculture practices.
1 Introduction
Enteric methane emissions from ruminants, particularly dairy cattle, are a significant source of global greenhouse gases and play a critical role in climate change. These emissions arise as a byproduct of microbial fermentation in the rumen and constitute a considerable portion of the agriculture sector’s greenhouse gas emissions (Dillon et al., 2021; Mizrahi et al., 2021). Beyond its environmental impact, methane production can also signify a loss of dietary energy that could otherwise be directed toward livestock production such as milk, meat, etc. (Arndt et al., 2022; Tseten et al., 2022). Therefore, developing effective strategies to mitigate methane emissions in dairy cattle is crucial for promoting environmental sustainability and enhancing the efficiency of livestock production.
Among the diverse strategies under investigation, methane-mitigating feed additives have emerged as a focal point of interest. Natural compounds such as essential oils are particularly promising due to their bioactive properties and ability to alter rumen fermentation patterns, inhibit methanogenic archaea, and reduce methane production overall (Adesogan et al., 2013; Beauchemin et al., 2008; Falero et al., 2022; Permata et al., 2023). Additionally, essential oils improve feed efficiency, making them a practical and effective approach for methane mitigation (Durmic et al., 2014; Tseten et al., 2022).
Recent advances in artificial intelligence (AI) provide powerful tools for exploring, analyzing, and optimizing various agricultural processes, driving the advancement of precision agriculture (Negussie et al., 2019; Jeong et al., 2022). This study validates an innovative AI-driven model designed to predict the impact of methane-mitigating feed additives on enteric methane emissions in dairy cattle. By integrating rumen microbiome sequencing data from Israeli Holstein cows with hundreds of methane measurements across 13 commercial dairy herds, the model enables precise predictions of the impact of tested feed additive on methane emission outcomes.
The primary goal of this study was to validate the accuracy of the AI-driven model using an essential oil-based feed additive (Agolin Ruminant, Altech, Biere, Switzerland) recognized for its methane-reducing potential (Carrazco et al., 2020; Batley et al., 2024).
This research responds to the pressing demand for sustainable agricultural practices by targeting the reduction of the environmental footprint of livestock farming. By presenting an innovative method to predict the effects of feed additives on enteric methane emissions, the study advances global initiatives to mitigate climate change through precision agriculture.
2 Materials and methods
2.1 General study design
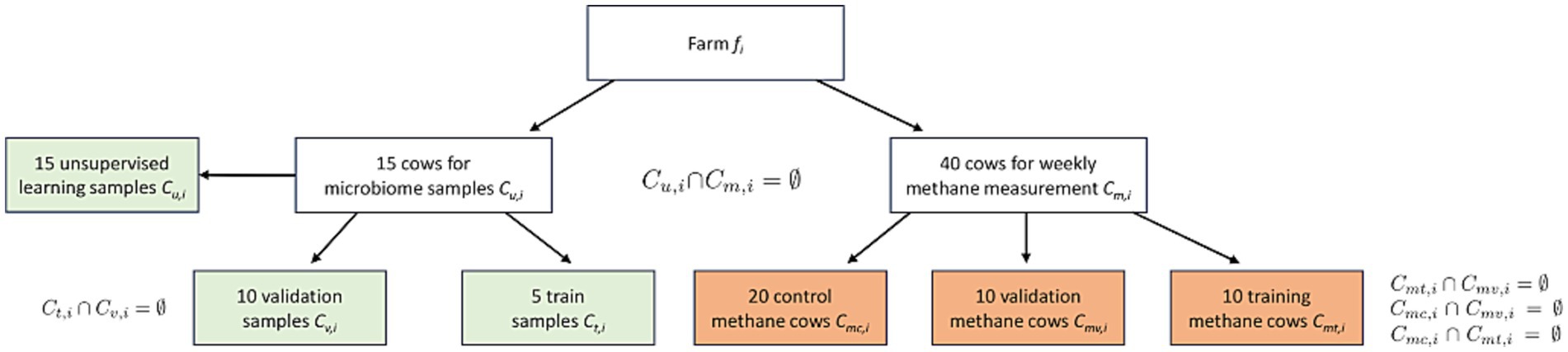
Previously we developed an AI-driven model based on a two-stage trial design targeting the prediction of the efficacy of methane-reducing additives using cows’ microbiome data. Briefly, the first stage involves an unsupervised machine-learning process trained on a diverse dataset of microbiome samples collected from cows across various farms. In the second stage, a smaller subset of cows, whose methane emissions have been periodically documented, is used to implement supervised learning (Figure 1). This stage aims to construct a predictive model that links microbiome profiles to the effectiveness of feed additives by generating a “predictive efficacy score” for each specific farm and feed additive tested (Altshuler et al., 2023; Altshuler et al., 2024).
In this case study, the validation step was performed with a widely commercial feed additive, (Agolin Ruminant, Altech, Biere, Switzerland), which is composed of coriander seed oil, eugenol, geranyl acetate, and geraniol. This was followed by an extensive in-vivo methane emission procedure, which included biweekly methane measurements taken at the same time of day at each site over 3 months, resulting in at least seven time points. To avoid seasonal effects, and enhance methodological robustness, the model validation was conducted in independent cohorts across 13 different commercial farms located in various geographical regions of Israel, ranging from the cooler mountainous areas in the north to arid dessert regions in the south and template plains in the center of the country.
To assess the overall efficacy of the feed additive across all the participating farms, we conducted our data analysis using two simulated scenarios; in the first one (naive approach), we assume that the feed additive is supplied to all the participating farms; while in the second approach (optimized approach), we adopted a precision agriculture-oriented strategy, assuming that the feed additive is applied only to farms with a higher likelihood of benefiting from it based on the model’s predictions.
2.2 Animals and feed additive
Israeli Holstein cows from 13 commercial dairy farms in Israel participated in this study. The herds were maintained on a standard mixed diet with a 32/68 forage-to-concentrate ratio, while each farm followed its own nutritional regimen. As in many other countries, this could include by-products from the food industry, such as pulp, gluten meal, and citrus fruits. Due to the relatively small size of the farms (between 400 to 900 animals), cows within each herd were housed in the same barn. The participating farms were a mix of 70% Kibbutz-type, which are large, cooperatively owned and managed units, and 30% Moshav-type, which are smaller, family-owned farms. To ensure the welfare of the animals participating in the study, all experimental procedures adhered to the Guide for the Care and Use of Agricultural Animals in Research and Teaching (Ag Guide; Federation of Animal Science Societies (FASS), 2010). A qualified veterinarian closely supervised the rumen sampling procedure and all technical procedures to minimize potential stress or discomfort to the animals. Additionally, all animals were managed under standard farming practices without deviations from routine.
In this study, the selected feed additive (Agolin), was supplied by each local farm staff according to the producer’s instructions (1gram/cow/day for 60 days mixed with the concentrate part of the mix) as a top dressing to 20 randomly selected cows at each site (treatment group, 260 cows in total). Visual consumption tracking was conducted at each farm. The treated cows were identified by ear tags for future data collection. Additionally, another 20 cows were randomly selected at each farm to form the control group (260 cows in total), receiving the standard mixed diet without the feed additive. Periodic in-situ methane measurements were then taken to compare the model’s predictions with actual methane emissions.
2.3 Ruminal sampling and sequencing
The sample size was previously defined as at least 0.5% of the total herd size (Altshuler et al., 2023; Altshuler et al., 2024). However, to mitigate potential sequencing errors, the sampling process was set at 15 rumen samples per dairy farm. The samples were collected using a 300-cm polyvinyl stomach tube. The tube and speculum were thoroughly rinsed after each use to prevent cross-contamination (Supplementary material 1). To safeguard the samples from degradation during transport, the samples were treated with Zymo DNA/RNA shield buffer (Zymo Research, Cat. No. R1100-250) and transported at room temperature to a specialized facility for DNA extraction. Library preparation for paired-end sequencing was carried out using the Illumina DNA Prep kit (Illumina, Cat. no. 20060059) and the NovaSeq 6,000 S4 reagent kit v1.5 (300 cycles) (Illumina, Cat. no. 20028312) on an Illumina NovaSeq 6,000 instrument. The integrity of sequences was assessed by FASTQC, an standard trimming and filtering were performed by BBDUK software, discarding sequences shorter than 100 bp or having a Phred score below 35. Trimmed sequences are used in the model performance without the need for taxonomic or functional classification.
2.4 Enteric methane emission measurements
Enteric methane measurements used for model performance assessment and validation were obtained using the ATEX Gas Analyzer (Geotech SEM 5000), a specialized instrument for methane measurement. This instrument was selected after extensive comparisons with other well-established systems (Supplementary material 2). The results of these comparisons along with key advantages such as ease of use, non-intrusive operation, no need for animal training, and onsite performance, support the ATEX Gas Analyzer as a suitable tool for ruminant methane measurement (Altshuler et al., 2023; Altshuler et al., 2024). In addition, to these comparisons, the sensor used in this instrument has been previously recommended for cattle studies (Rey et al., 2019; Ribeiro et al., 2020).The ATEX device records continuous methane measurements every 3 to 5 s, generating hundreds of methane readings for animal. To ensure a single and consistent methane reading per cow for each visit, values lower than 5 ppm were removed, and then the median for each cow was obtained, establishing a dependable foundation for subsequent comparative analysis across various visits and cows. The threshold was set to 5 ppm based on our previous in-situ experience with the instrument; Field operators noted that when the instrument is positioned too far from cows, or even in open areas, background readings below 5 ppm are commonly observed.
Finally, model validation was conducted on each farm by comparing the model’s predictions with in-situ methane emissions. Methane measurements were consistently taken between 06:00 to 08:00 am.
2.5 Model construction and performance
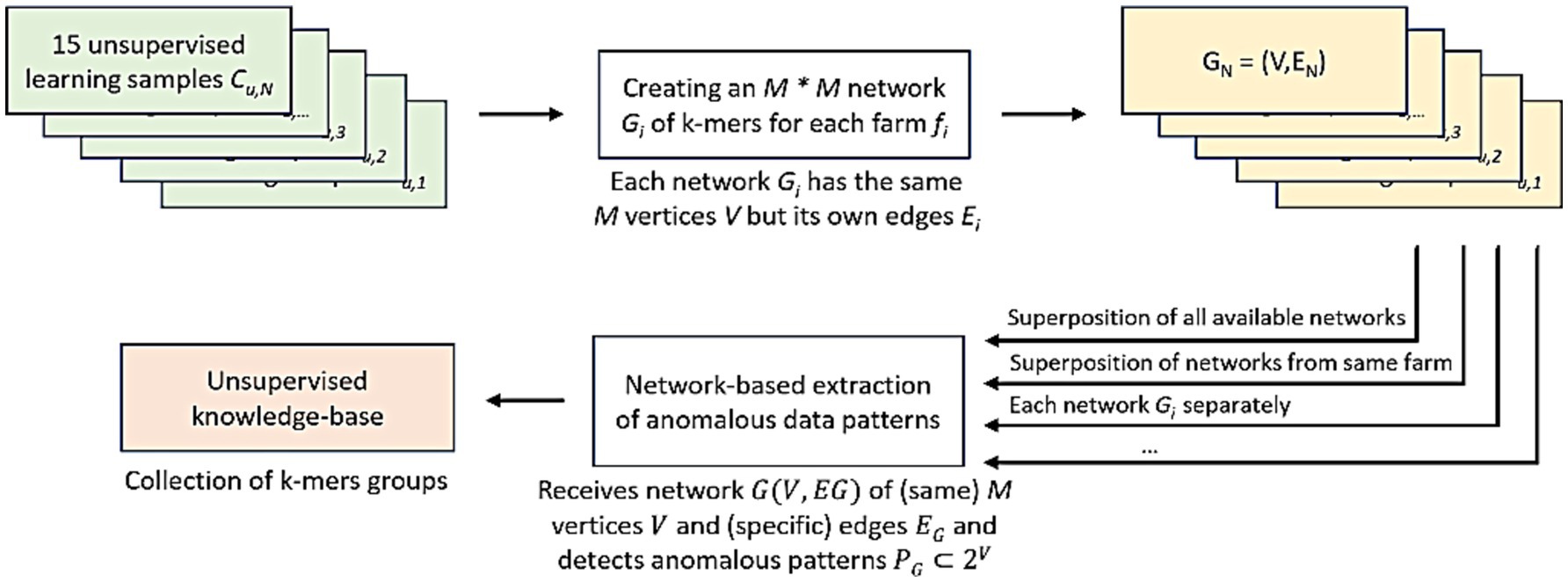
Initially, genomic sequences from a given rumen sample—consisting of 120 to 150 nucleotide strings—are filtered and analyzed. The AI-driven model processes these genomic sequences and generates subsequences of 30 nucleotides (k-mer 30). These subsequences are then displayed as a network of k-mers, where each node represents a unique k-mer, and the edges are formed by two k-mers derived from the same sequencing read. As a result, each network is unique and reflects the specific microbiome composition. The network structure reveals clusters of k-mers that frequently appear together (biomarkers); which can later be correlated with biological conditions such as methane emissions.
Next, networks from each rumen microbiome sample within a given farm are combined, creating a superposition network—a dense structure that provides a more comprehensive representation of farm-level features (Figure 2; Supplementary material 3). The use of a superposition network allows for the integration of new samples, and the model’s generic nature enables the analysis of biological targets or traits beyond feed additive efficacy, such as the effects of diet on milk or meat production, or susceptibility to diseases.
The generation of a superposition network for each farm facilitates the identification of subsequences (or biomarkers) statistically associated with methane emissions. Each microbiome marker from a given farm is then analytically validated to ensure it is unlikely to occur randomly in microbial genetic samples. A complete description of the model steps was previously published (Altshuler et al., 2023; Altshuler et al., 2024).
Finally, the identified biomarkers are grouped into two sets of DNA subsequences. The biomarkers positively associated with the desired biological condition (e.g., low methane emissions) are placed in the “top list” (or “positive list”), while those subsequences associated with a non-desired condition are grouped in the “bottom list” (or negative list). Both lists are used to analyze the farm’s overall response to a given feed additive, generating a predictive score ranging from 0 to 1 (Figure 3).
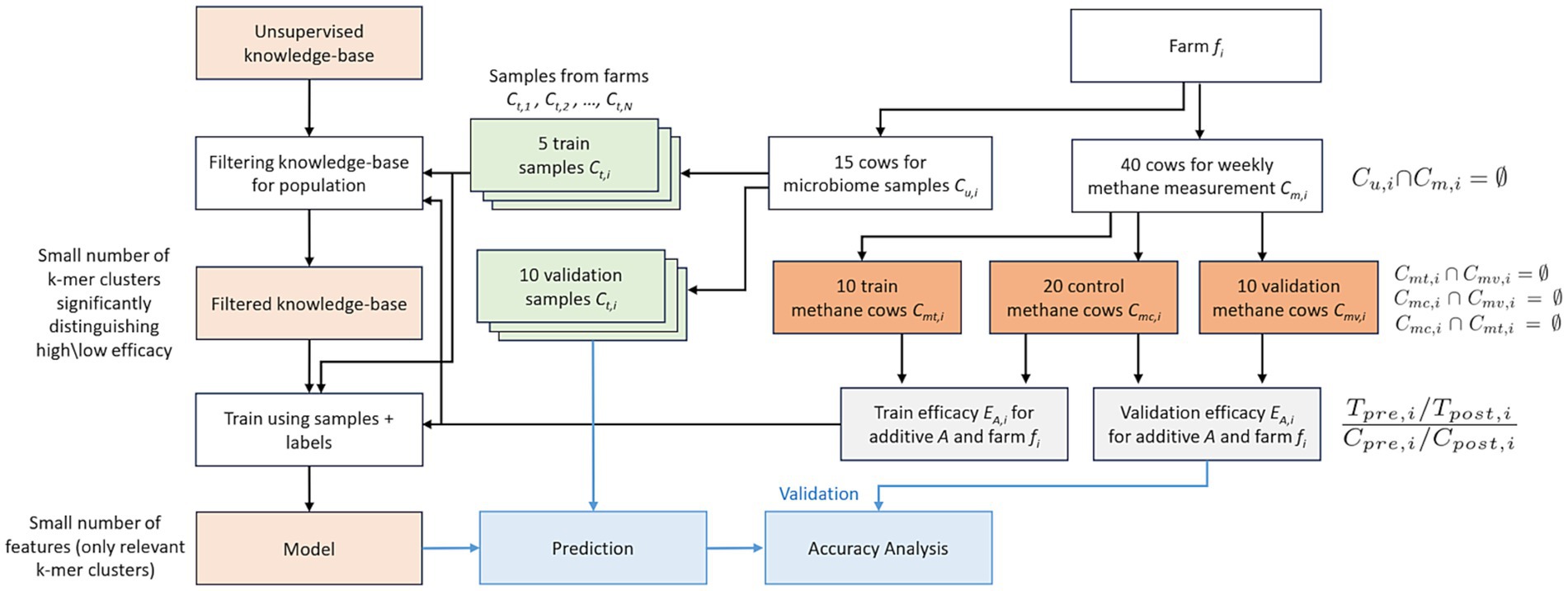
Figure 3. Schematic representation of the supervised learning phase paired with validation and prediction steps.
Briefly, the model computes the average score of individual cows within each farm, resulting in a global farm-level score ranging from −1 to 1. This score is then normalized through a simple mathematical process, yielding values between 0 (indicating expected low efficacy of the analyzed additive) and 1 (indicating expected high efficacy). Scores around 0.5 suggest insufficient information for an accurate prediction (Supplementary material 4).
This phase requires a model training step based on methane emissions, which utilizes 40 cows divided into three groups: training, control, and validation (Supplementary material 5).
3 Results
3.1 General feed additive efficacy
Figure 4 illustrates the efficacy analysis of the tested feed additive based on the obtained biomarkers (Supplementary material 6). Initially, efficacy is shown as a normal distribution under naive approach conditions, where no farm selection is applied. This is compared with the optimized or precision agriculture approach, where the feed additive is applied to only 50% of the farms with the higher chance of being positively impacted. The comparison demonstrates that the optimized strategy significantly enhances the global additive’s efficacy by focusing on farms where it is predicted to have the greatest impact. Thus, under the naive approach, the model predicts a general 8% reduction in methane emissions. In contrast, the optimized approach achieves an overall 14% reduction on enteric methane emissions.
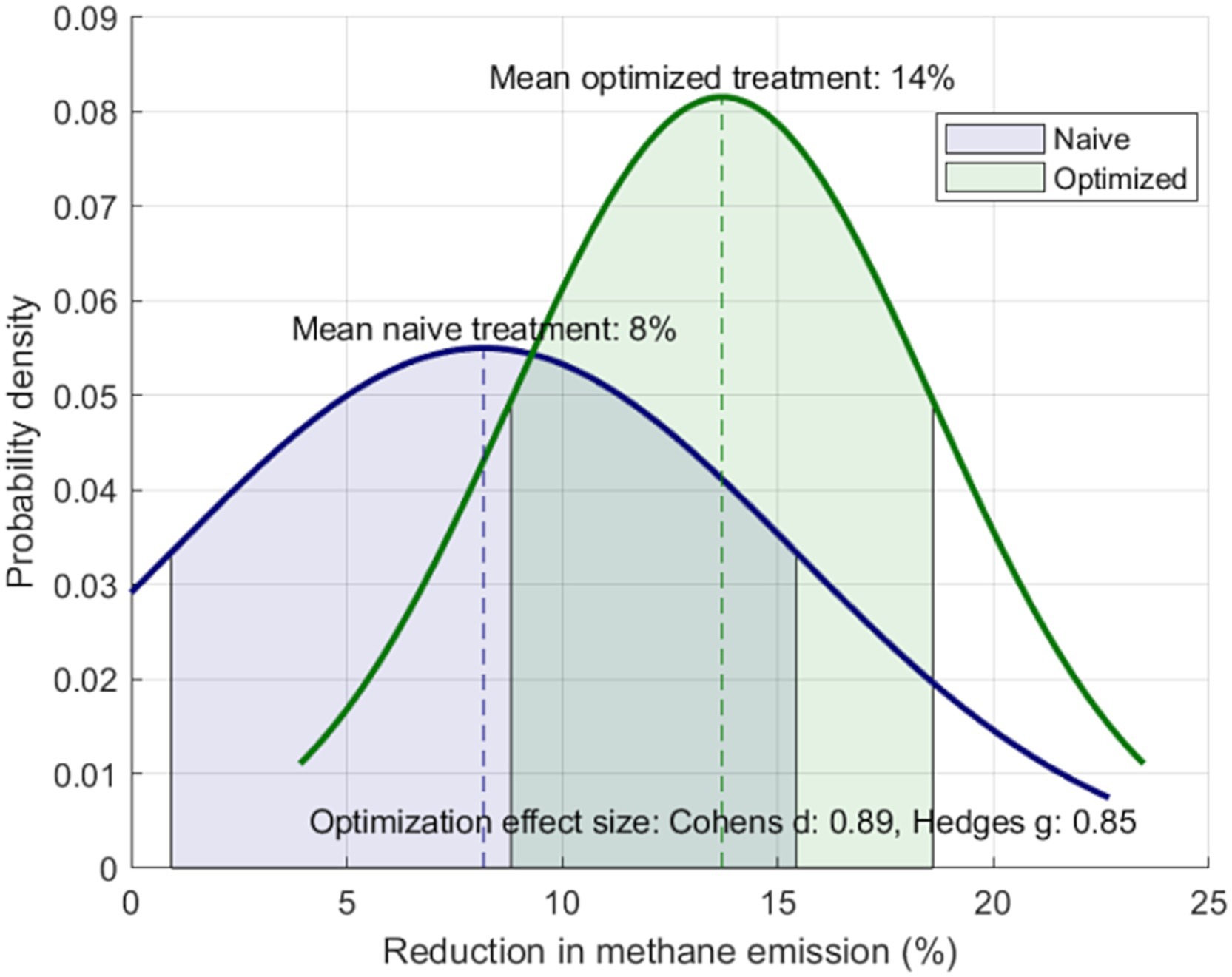
Figure 4. Normalized efficacy distribution for the feed additive, based on the identified k-mers markers, and regressed to fit a normal distribution (the dark Blue, denoted as ‘naive’). The Green chart, denoted as ‘optimized’, illustrates the additive’s efficacy across the 50% of farms predicted by our microbiome-based model to have the highest efficacy. The charts include the appropriate Cohen’s d and Hedge’s g metrics, indicating strong statistical significance of the observed effects under optimized conditions.
Despite both approaches achieving significant reductions in methane emissions, statistical analysis indicates that both approaches are significantly different (Cohen’s d = 0.89, Hedges’ g = 0.85), with a large effect size. In other words, the differences between the two scenarios are substantial and unlike to be due to random variation. Therefore, in the optimized scenario, the feed additive achieved a significant higher reduction in enteric methane emissions.
3.2 Prediction vs. real feed additive efficacy
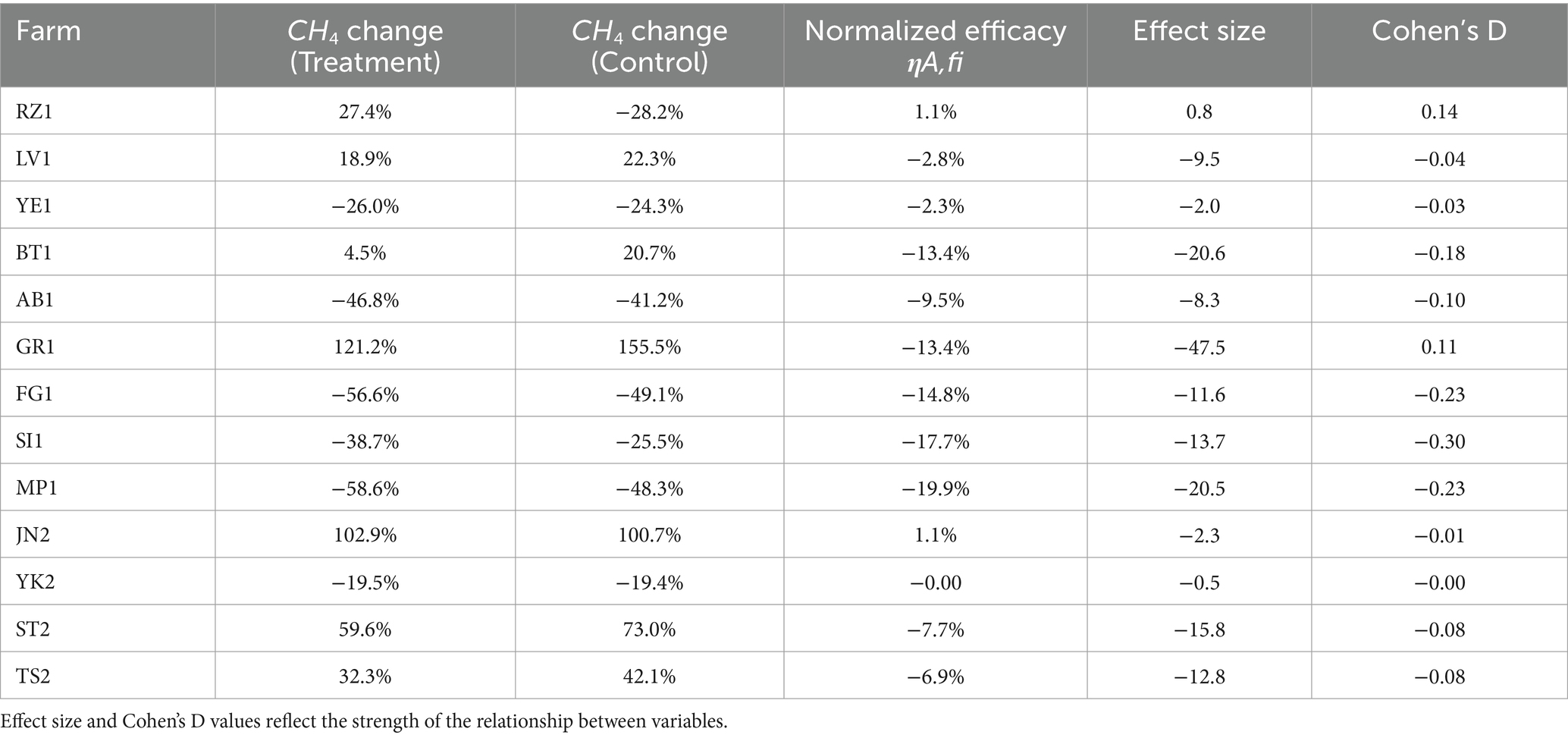
Methane emissions decreased significantly in the treated groups following the administration of the feed additive compared to the control groups; thus, the normalized efficacy column in Table 1 reflects the normalized mean percentage change in methane emissions, accounting for variations in control and treatment groups. Reductions in methane emissions were observed in 11 of 13 farms, with reductions ranging from 0.1 to 19%, with an overall reduction of 9.86%. In two cases, sites JN2 and RZ1 exhibited a slight increase of 1.1% in methane emission.
To test accuracy, we compare the enteric methane emissions with the prediction obtained by the AI-driven model (Figure 5). Thus, the model accurately predicted the effect of the feed additive at each site. When values in the efficacy score (axis x) were higher than 0.6, a higher impact of the feed additive in methane reduction (axis y) was observed. Thus, sites GR1, AB1, FG1, MP1, and SI1 obtained higher values for the efficacy score and were the most positively impacted by the feed additive, exhibiting reductions on enteric methane emissions of 13.4, 9.5, 14.8, 19.9, and 17.7%, respectively (Table 1).
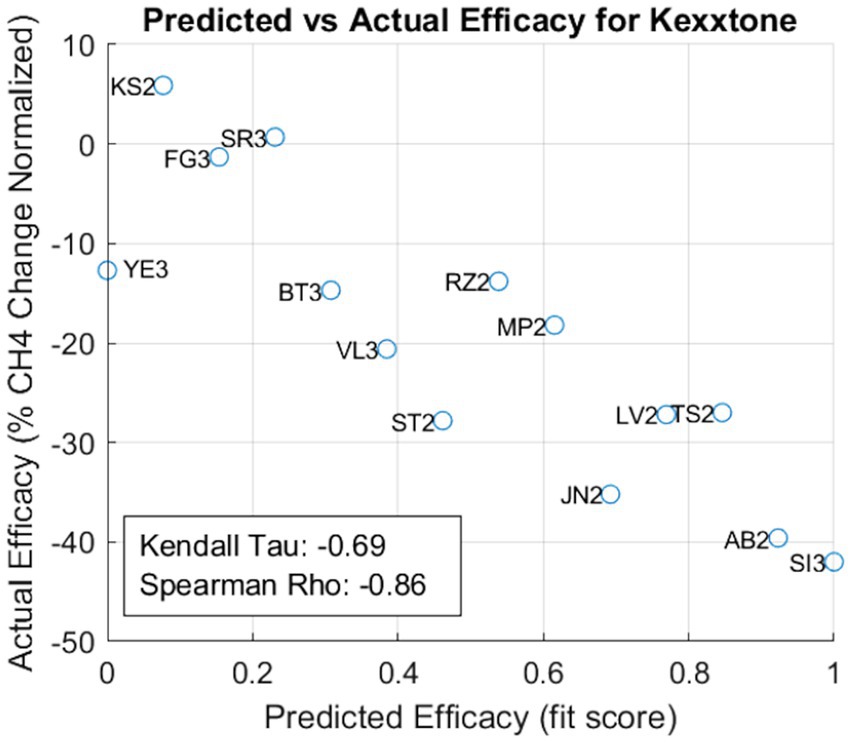
Figure 5. Predictive model accuracy for the tested feed additive. The scatter plot ranks the farms by their predicted efficacy (x-axis) against their actual, measured efficacy (y-axis). Spearman’s ρ and Kendall’s τ are displayed, serving as statistical measures of correlation between the predicted and actual efficacies.
The statistical analysis of all the sites analyzed corroborates the accuracy of the model. Both Kendall’s Tau and the Spearman’s Rho metrics (−0.73 and − 0.89 respectively) indicate a strong negative monotonic relationship between the model’s predictions and the actual methane emissions. In other words, when the predictive score was higher for a given site, real enteric methane emissions were significantly reduced. Conversely, when the predictive score was low, minimal reductions in enteric methane emissions were observed, thereby validating the accuracy of the model.
4 Discussion
This study successfully validated a novel AI-driven model that uses rumen microbiome data to accurately predict the efficacy of a commercial feed additive (Agolin) in reducing enteric methane emissions. The robustness of this validation is demonstrated by various factors addressed in the study, such as the wide geographical and climatological diversity across Israel, the different types of dairy farms analyzed (Kibbutz and Moshav, or extensive vs. familiar), and the varied diets involved on each farm. Additionally, our model overcomes traditional biases related to taxonomical read assignments and limitations of culture-based methods, which are often restricted to a limited number of cow rumen microorganisms.
The model’s prediction accuracy validated in this study is consistent with validations performed in parallel using different types of feed additives (Altshuler et al., 2023; Altshuler et al., 2024). Thus, this model can provide a valuable tool for herd management and methane mitigation strategies, as it quantifies the impact of feed additives on enteric methane emissions. Therefore, integrating our model with any feed additive could help farmers make personalized, faster and more informed decisions. Moreover, the model’s nature allows for the integration of new data and adaptation to the changing conditions in any dairy. Another significant advantage of the tested model is its to be it can also be widely adapted to diverse data sources, such as cows, sheep, soil, or even humans. It can also be modified to assess other critical microbiome-related outcomes, such as disease risk or productivity, facilitating early interventions and improving livestock health and operational efficiency.
The model’s effectiveness was supported by its consistent performance across different herds, suggesting that the patterns observed from reads and k-mers follow a power-law distribution—a relationship where one quantity changes as a fixed power of another, indicating consistent patterns across different scales (see Altshuler et al., 2023, section 5.8.3), which suggests that our results follow a more universal principle, and they are not result of coincidence.
The feed additive here assessed exhibits an important capacity to reduce enteric methane emissions, reaching in some farm reductions close to 20%, which is higher than the previously reported reductions on methane emissions (Becker et al., 2023; Bach et al., 2023); however also it was detected in some cases in which the effect was negligible or even slightly higher than the control group. This point is extremely important because our model predicted this phenomenon, generating very low (close to zero) predictive scores for these sites, despite the lack of clear differences observed in-situ between these and the other diaries.
Despite these promising results, the study presents some limitations. Efforts to reduce bias by categorizing cows based on age, lactation period, and milk yield remain crucial, as individual differences in diet, and environment may still influence microbiome composition and methane production. Additionally, potential mislabeling during the supervised learning phase could affect the model’s accuracy, highlighting the need for precise data collection. Future research should address these issues more thoroughly. Additionally, it is evident that the results obtained here apply only to the tested feed additive; therefore, the model needs to undergo further validation with other widely used feed additives.
5 Conclusion
This AI-driven model effectively predicted the impact of the tested feed additive on enteric methane emissions, even in cases where the effect was minimal. As the model undergoes further validation with additional methane-mitigating feed additives, it will enable the development of customized feed additive strategies at the farm level, helping farmers optimize both environmental sustainability and productivity. Moreover, the model’s flexibility extends its application to forecasting other critical microbiome-related outcomes, such as disease risk and productivity. Therefore, this model holds significant potential for global impact by helping farmers and nutritionists optimize personalized strategies in response to emerging methane reduction policies.
Data availability statement
Microbiome data used in this research was collected from privately owned dairy farms. To respect the confidentiality agreements established with these farms and to protect the privacy of the farm owners, access to the raw data is restricted and will be made available upon request.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because all animal procedures were conducted following the Agricultural Animals in Research and Teaching (Ag Guide, FASS 2010) and supervised by a qualified veterinarian. The staff responsible for methane measurements were carefully trained to use the methane measuring instrument before performing in-situ measurements.
Author contributions
YA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing. TC: Project administration, Writing – review & editing. SC: Investigation, Methodology, Writing – review & editing. JG: Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The study received funding from Metha Artificial Intelligence, which had previously raised capital from financial investors. None of these investors had any role or influence in the study design, data collection, analysis, interpretation, manuscript preparation, or the decision to submit the work for publication. The funding covered operational expenses only—such as equipment, feed additives, logistics, and sample collection.
Acknowledgments
We sincerely thank the farmers from the participating Kibbutz and Moshav dairy farms in Israel for their cooperation and support throughout this study. Their participation and assistance were crucial to the success of this research.
Conflict of interest
YA, TC, SC, and JG were employed by the company Metha Artificial Intelligence, Israel.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2025.1548223/full#supplementary-material
References
Adesogan, T., Yang, W., Lee, C., Gerber, P. J., Henderson, B., and Tricarico, J. M. (2013). Mitigation of methane and nitrous oxide emissions from animal—special topics. J. Anim. Sci. 91, 5045–5069.
Altshuler, Y., Calvao Chebach, T., and Cohen, S. (2024). “From microbes to methane: AI-based predictive modeling of feed additive efficacy in dairy cows” in Applied swarm intelligence. ed. Y. Altshuler (Florida: CRC Press).
Altshuler, Y., Chebach, T. C., and Cohen, S. (2023). From microbes to methane: AI-based predictive modeling of feed additive efficacy in dairy cows. arXiv.
Arndt, C., Hristov, A., Price, W., McClelland, S., Pelaez, A., Cueva, S., et al. (2022). Full adoption of the most effective strategies to mitigate methane emissions by ruminants can help meet the 1.5 °C target by 2030 but not 2050. PNAS 119:e2111294119. doi: 10.1073/pnas.2111294119
Bach, A., Elcoso, G., Escartin, E., Spengler, K., and Jouve, A. (2023). Modulation of milking performance, methane emissions, and rumen microbiome on dairy cows by dietary supplementation of a blend of essential oils. Animal 17:100825. doi: 10.1016/j.animal.2023.100825
Batley, R., Romanzini, E., da Silva, K., de Souza, W., Quigley, S., Harper, K., et al. (2024). The essential oil blend Agolin ruminant L reduces methane production in vitro and in vivo when included in the drinking water of cattle. J. Anim. Sci. 102. doi: 10.1093/jas/skae315
Beauchemin, K. A., Kreuzer, M., O’mara, F., and McAllister, T. A. (2008). Nutritional management for enteric methane abatement: a review. Aust. J. Exp. Agric. 48, 21–27. doi: 10.1071/EA07199
Becker, F., Spengler, K., Reinicke, F., and Heider-van Diepen, C. (2023). Impact of essential oils on methane emissions, milk yield, and feed efficiency and resulting influence on the carbon footprint of dairy production systems. Environ. Sci. Pollut. Res. 30, 48824–48836. doi: 10.1007/s11356-023-26129-8
Carrazco, A. V., Peterson, C. B., Zhao, Y., Pan, Y., McGlone, J. J., DePeters, E. J., et al. (2020). The impact of essential oil feed supplementation on enteric gas emissions and production parameters from dairy cattle. Sustain. For. 12:10347. doi: 10.3390/su122410347
Dillon, J., Stackhouse-Lawson, K., Thoma, G., Gunter, S., Rotz, C., Kebreab, E., et al. (2021). Current state of enteric methane and the carbon footprint of beef and dairy cattle in the United States. Anim. Front. 11, 57–68. doi: 10.1093/af/vfab043
Durmic, Z., Moate, P., Eckard, R., Revell, D., Williams, R., and Vercoe, P. (2014). In vitro screening of selected feed additives, plant essential oils and plant extracts for rumen methane mitigation. J. Sci. Food Agric. 94, 1191–1196. doi: 10.1002/jsfa.6396
Falero, E., Santana, Á., Diez, H., Longueira, E., Hernández, D., Repetto, J., et al. (2022). 336 Anavrin®, monensin or both on the diet of finishing steers fed TMR? Effect on performance and rumen fermentation. J. Anim. Sci. 100, 163–164. doi: 10.1093/jas/skac247.304
Federation of Animal Science Societies (FASS) (2010). Guide for the care and use of agricultural animals in research and teaching. 3rd Edn. Champaign, IL: FASS.
Jeong, S., Fisher, M., Breunig, H., Marklein, A., Hopkins, F., and Biraud, S. (2022). Artificial intelligence approach for estimating dairy methane emissions. Environ. Sci. Technol. 56, 4849–4858. doi: 10.1021/acs.est.1c08802
Mizrahi, I., Wallace, R. J., and Moraıs, S. (2021). The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Negussie, E., Gonzales, O., Haas, Y., Gengler, N., Soyeurt, H., Peiren, N., et al. (2019). “Machine learning ensemble algorithms in predictive analytics of dairy cattle methane emission using imputed versus non-imputed datasets.” Greenhouse gas and animal agriculture conference – 7th GGAA, Brazil.
Permata, D., Komang, G., and Anuraga, J. (2023). Evaluation of essential oil supplementation as a feed additive on rumen fermentation characteristics and methane mitigation in ruminants: a meta-analysis. Vet. Integrative Sci. 22, 463–473. doi: 10.12982/VIS.2024.032
Rey, J., Atxaerandio, R., Ruiz, R., Ugarte, E., Gonzalez-Recio, O., Garcia-Rodriguez, A., et al. (2019). Comparison between noninvasive methane measurement techniques in cattle. Animals 9:563. doi: 10.3390/ani9080563
Ribeiro, A., Pinedo, L., Codognoto, L., Cavali, J., Porto, M., Dos Santos, B., et al. (2020). Comparison of methods to measure enteric methane emissions from ruminants: an integrative review. Res. Soc. Dev. 9:e8259118143. doi: 10.33448/rsd-v9i11.8143
Keywords: enteric methane emissions, dairy, AI model, predictive model, precision agriculture, feed additive
Citation: Altshuler Y, Chebach TC, Cohen S and Gatica J (2025) AI-driven precision agriculture for enteric methane mitigation: cross-farm validation with an essential oil-based feed additive. Front. Sustain. Food Syst. 9:1548223. doi: 10.3389/fsufs.2025.1548223
Edited by:
Rajiv Kumar Srivastava, Texas A&M University, United StatesReviewed by:
Faheem Ahmed Khan, National Research and Innovation Agency (BRIN), IndonesiaJoana Lima, Scotland’s Rural College, United Kingdom
Selim Esen, Republic of Turkey Ministry of Agriculture and Forestry, Türkiye
Subrat Kumar Behera, Orissa University of Agriculture and Technology, India
Preeti Lakhani, Lala Lajpat Rai University of Veterinary and Animal Sciences, India
Bożena Króliczewska, Wroclaw University of Environmental and Life Sciences, Poland
Copyright © 2025 Altshuler, Chebach, Cohen and Gatica. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaniv Altshuler, eWFuaXZhbEBtaXQuZWR1
 Yaniv Altshuler
Yaniv Altshuler Tzruya Calvao Chebach2
Tzruya Calvao Chebach2