- 1Wenzhou Vocational College of Science and Technology (Wenzhou Academy of Agricultural Sciences), Wenzhou, China
- 2Zhejiang Provincial Department of Agriculture and Rural Affairs, Hangzhou, China
Paddy–upland rotation systems are widely adopted to mitigate soil degradation in rice-based agroecosystems; however, their impacts on soil biota remain insufficiently understood. This study investigated the impacts of paddy continuous cropping (PA), upland continuous cropping (UP), and rice–loofah paddy–upland rotation (RO) on soil nematodes and microbial communities in southeastern China. Soil samples were collected prior to harvest at the end of the rice season and were analyzed for physicochemical properties, nematode communities via morphological identification, and microbial communities through high-throughput sequencing. The results showed that the RO system significantly increased soil pH, total phosphorus, available potassium, and available phosphorus, while reducing the abundance of the plant-parasitic nematode Hirschmanniella compared to the PA system. The total nematode abundance was highest in the UP system, where bacterivores predominated; the RO system was characterized by a higher proportion of algivores associated with flooded conditions, whereas the PA system was dominated by herbivores. The RO and PA system also improved nematode food web stability under flooded conditions, as indicated by higher maturity and structure indices relative to the UP system. Although microbial diversity did not differ significantly between systems, the community composition and predicted functional groups varied considerably. The relative abundance of Gemmatimonadota was significantly reduced in the PA system, while the abundance of Nitrospirota, Myxococcota, and Entorrhizomycota increased. Functional prediction revealed system-specific enrichment of bacterial metabolic groups associated with nitrogen cycling, carbon turnover, and redox-sensitive energy metabolism. Integration of soil physicochemical and biological indicators into a Soil Quality Index (SQI) ranked RO highest, underscoring its capacity to enhance soil ecological function and sustainability in rice-based systems.
1 Introduction
Rice is among the most important cereal crops and serves as the primary food source for over half of the global population. However, the continuous cropping of rice causes soil degradation and yield decline (Ren et al., 2020). Paddy–upland rotation is a farming system in which rice and upland crops are alternately planted in the same field over successive growing seasons. It is particularly common in South and East Asian countries such as Bangladesh, China, India, Nepal, and Pakistan (Zhou et al., 2014). This system is believed to improve soil quality, reduce methane emissions, enhance nitrogen availability, mitigate soil-borne pathogen pressures, and increase crop yields (Yin et al., 2025; Zhang D. et al., 2025). Paddy–upland rotation is characterized by seasonal water management and frequent alterations in wet and dry conditions, resulting in unique physicochemical properties and biological characteristics of the soil, which differ from those in dryland fields or continuous paddy fields (Zhou et al., 2014). Though paddy–upland rotation soils have been extensively studied in terms of soil nutrient cycling, organic carbon transformation, and soil fertility, research on soil biota in such systems remains limited.
Soil nematodes and microorganisms are ubiquitous in soils, constituting the essential components of terrestrial ecosystems (Bahram et al., 2018; van den Hoogen et al., 2019). They are frequently used as sensitive bioindicators in agroecosystems to assess soil quality and evaluate the impacts of various agricultural practices and fertilization regimes in soil environments (Bending et al., 2004; Martin and Sprunger, 2022; Wilhelm et al., 2023). Nematodes exhibit considerable taxonomic and functional diversity and are typically classified into trophic groups such as herbivores, bacterivores, fungivores, omnivores, and predators, each contributing to different aspects of soil ecological function (Yeates et al., 1993; Zhang et al., 2024). By decomposing organic matter and mineralizing nutrients, nematodes significantly contribute to nutrient cycling, energy flow, and ecosystem stability (van den Hoogen et al., 2019). In agricultural systems, plant-parasitic nematodes are major pathogenic organisms that pose a significant threat to plant health and productivity (Phani et al., 2021). Meanwhile, soil microorganisms influence crop growth by enhancing nitrogen fixation, promoting phosphate solubilization, facilitating mycorrhizal symbiosis, and inducing plant defense responses (Berg, 2009; Blundell et al., 2020; Timofeeva et al., 2023). Recent studies have also highlighted the critical role of soil microorganisms in regulating plant–nematode interactions (Zhang H. et al., 2025). However, soil microbial communities may also harbor plant pathogens or harmful rhizosphere microorganisms that can lead to disease outbreaks and yield losses (Mendes et al., 2013). Therefore, maintaining diverse and functionally beneficial nematode and microbial communities is essential for preserving soil health and ensuring sustainable agricultural productivity.
Soil quality is defined as the “the capacity of a soil to function within ecosystem and land-use boundaries to sustain biological productivity, maintain environmental quality, and promote plant and animal health” (Doran and Parkin, 1994). The soil quality index (SQI) integrates soil physicochemical and biological properties into a composite metric, providing a more comprehensive assessment than individual soil parameters and reducing interpretive ambiguity (Marion et al., 2022). Key soil properties—such as moisture, nutrient content, and pH—are known to strongly influence nematode and microbial diversity worldwide (Bahram et al., 2018; van den Hoogen et al., 2019). In paddy–upland rotations, alternating water regimes and changes in redox conditions can restructure soil food webs (Fang et al., 2024). Studies have shown that fluctuations between flooded and non-flooded states can suppress crop pests and pathogens (Ebihara et al., 2010). Differences in soil structure, nutrient availability, and management practices—including fertilization, pesticide application, and tillage—further shape soil biota (John et al., 2021; Matute and Anders, 2012; Zhang et al., 2023). Seasonal crop rotation also influences soil nematodes and microbial communities by modifying root exudates, plant residue inputs, and symbiotic interactions (Flower et al., 2019). Given these interacting drivers, a comprehensive assessment that integrates soil physicochemical and biological properties—and explicitly examines their interrelationships—is essential for evaluating the effects of paddy–upland rotation on soil quality and for elucidating the underlying mechanisms.
In this study, we investigated soil nematode and microbial communities under continuous paddy, continuous upland, and a locally practiced rice–loofah paddy–upland rotation system in southeastern China. Specifically, we aimed to: (1) evaluate how paddy–upland rotation affects the composition of soil nematode and microbial communities; (2) assess soil quality under different cropping systems by integrating physicochemical and biological indicators into an SQI framework; and (3) explore the relationships among soil properties, nematode communities, and microbial communities. We hypothesized that (1) paddy–upland rotation would significantly alter the structure of soil nematode and microbial communities through changes in water regime and nutrient management; (2) paddy–upland rotation would improve overall soil quality by increasing biodiversity, enhancing fertilization inputs, and reducing herbivore and pathogen pressures; and (3) soil properties, particularly moisture and nutrient levels, would serve as key drivers shaping the diversity and composition of soil biotic communities.
2 Materials and methods
2.1 Study area and experimental design
The experiment was established at an experiment station for paddy–upland rotation (27°28′10″–27°28′26″N, 120°21′20″–120°21′46″E) in Cangnan County, Zhejiang Province, China. This area has a subtropical marine monsoon climate, with an annual average temperature of 17.9°C and an annual average precipitation of 1753.9 mm. The soil at the site is classified as an Anthrosol with a clay loam texture.
The field experiment began in 2015 and included three treatments: a paddy continuous cropping system (PA), a paddy–upland rotation cropping system (RO), and an upland continuous cropping system (UP). Each treatment was replicated three times, with each plot measuring 60 m × 60 m (3,600 m2). All crops were managed according to local agricultural practices as described below. In the PA system, rice was planted in early July and harvested between late October and November. Chemical fertilizers were applied at rates of 180 kg N ha−1, 75 kg P₂O₅ ha−1, and 150 kg K₂O ha−1. The land remained fallow during the rest of the year. The RO system, promoted by local agricultural authorities to alleviate continuous cropping obstacles and enhance crop productivity, is widely practiced in both experimental and production fields across Cangnan County and surrounding areas. In this system, loofahs (Luffa aegyptiaca Mill.) were cultivated in plastic greenhouses from November to mid-May of the following year. Basal fertilization consisted of 70 kg N ha−1, 120 kg P₂O₅ ha−1, and 130 kg K₂O ha−1 in chemical fertilizers, along with 15,000 kg ha−1 of fully decomposed organic fertilizer. During fruit development, when the second batch of loofahs began to set, a high-potassium, low-nitrogen compound fertilizer was applied via drip irrigation at rates of 6 kg N ha−1, 4 kg P₂O₅ ha−1, and 23 kg K₂O ha−1. After the loofah harvest in June, the greenhouse film was removed. The rice was transplanted in July and harvested from late October to November. As soil fertility remained relatively high after loofah cultivation, no basal fertilizer was applied before rice planting. Instead, 75 kg ha−1 of urea was top-dressed 5–7 days after rice greening in order to promote tillering. In the UP system, loofahs were grown continuously in plastic greenhouses for two seasons per year. The fertilization regime followed that of the loofah stage in the RO system. Pest and disease control measures were implemented as needed to minimize yield loss.
Soil samples were collected prior to harvest at the end of the rice season in late October 2024. In each plot, ten soil cores (0–20 cm depth) were randomly taken approximately 20 cm from the crop root zone using a stainless steel auger (5 cm diameter) and composited into a homogeneous sample. Each soil sample was divided into three parts: one portion was stored at −80°C for microbial DNA sequencing, one was used for nematode extraction, and one was sent for soil physicochemical analysis.
2.2 Soil physicochemical analysis
The soil’s water content was determined by oven-drying the samples at 105°C for 48 h. Homogenized composite samples were air-dried, ground, and passed through a 0.15 mm sieve for further analysis. The soil’s pH was measured using a glass electrode in a 1:2.5 (v:v) soil-to-water suspension. Soil organic matter (SOM) was determined using the dichromate oxidation method. Total nitrogen (TN) was determined using the Kjeldahl method, while available nitrogen (AN) was determined using the alkaline hydrolysis diffusion method. Total phosphorus (TP) and available phosphorus (AP) were measured using the molybdenum-antimony colorimetric method. Total potassium (TK) was extracted by fusion with sodium hydroxide, and available potassium (AK) was extracted using ammonium acetate, before being quantified using flame photometry.
2.3 Soil nematode extraction and identification
Soil nematodes were extracted from 100 g of fresh soil using the Baermann funnel method for 48 h and subsequently fixed in 4% formaldehyde. Nematodes were counted, and their abundance was expressed as the number of individuals per 100 g of dry soil. From each sample, 100 individuals (when possible) were randomly selected and identified to the genus level using a light microscope (BX53, Olympus, Japan). Identified genera were assigned to trophic groups following Yeates et al. (1993): bacterivores (Ba), fungivores (Fu), herbivores (He), and omnivores–predators (Op). In addition, we included a fifth category—“algivores” (Al)—to represent taxa that preferentially feed on algae and diatoms in flooded, algae-rich environments. This designation is not part of the classical trophic group framework but follows the ecological interpretation of Okada et al. (2011) for paddy soils. Specifically, Tobrilus and Rhabdolaimus were classified as algivores in this study; however, we acknowledge that these genera exhibit trophic plasticity and may also feed on bacteria or other resources under different environmental conditions (Nguyen et al., 2020; Liu et al., 2008). Nematodes were also allocated to five colonizer-persister (c-p) groups, ranging from colonizers with high fecundity and short generation times to persisters with long life cycles and higher sensitivity to disturbance (Bongers and Bongers, 1998).
Several ecological indices were calculated to evaluate the diversity and structure of nematode communities. These included richness (number of genus); Shannon diversity index (H′) = −∑ Pi (ln Pi), where Pi is the proportion of individuals in the ith taxon to the total number of nematodes; maturity index (MI) = ∑ vi fi, where vi is the c-p value of taxon i and fi is the frequency of that taxon in a sample (Bongers, 1990); enrichment index (EI) = 100 × (e / (e + b)) and structure index (SI) = 100 × (s / (b + s)), where e, b, s are the weighted proportions of the enriched component, basal component and structured component of the soil food web, respectively (Ferris et al., 2001).
2.4 Soil microbial DNA extraction and PCR amplification
Total genome DNA was extracted from 0.5 g of frozen soil using the TIANamp Soil DNA Kit (TianGen, China) according to the manufacturer’s instructions. The final DNA concentration and purity were evaluated on 1% agarose based on the absorbance ratios at 260/280 nm using the NanoDrop 2000 UV VIS spectrophotometer (Thermo Scientific, USA).
The V3-V4 regions of 16SrRNA gene were amplified using universal primers 341F (5’-CCTAYGGGRBGCASCAG-3′) and 806R (5′- GGACTACNNGGGTATCTAAT-3′) for bacteria and archaea. The ITS1 region of fungal rRNA was amplified using primers ITS-1F-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1-1F-R (5’-GCTGCGTTCTTCATCGATGC-3′). A polymerase chain reaction (PCR) was carried out with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs) that incorporated 2 μM of forward and reverse primers, as well as about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, before a final extension at 72°C for 5 min. PCR products were assessed using electrophoresis on 2% agarose gel. PCR products were mixed in equidensity ratios and the mixture of PCR products was purified using the Universal DNA Purification Kit (TianGen, China).
2.5 Soil microbial bioinformatics analysis
Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit (Illumina, USA) following the manufacturer’s recommendations; additionally, index codes were added. The library quality was assessed using Agilent 5,400 (Agilent Technologies Co Ltd., USA). The library was sequenced on an Illumina platform and 250 bp paired-end reads were generated.
Pair-end raw sequences were analyzed using the Quantitative Insights into Microbial Ecology (QIIME2) software. Demultiplexed sequences from each sample were quality filtered and trimmed, de-noised, and merged; then, the chimeric sequences were identified and removed using the DADA2 method in order to obtain the feature table of amplicon sequence variants (ASVs) (Callahan et al., 2016). Bacterial and fungal sequences were aligned against the SILVA 138.2 and UNITE+INSD database, respectively, for taxonomic annotation. Feature-level diversity indices, such as richness (number of observed ASVs) and the Shannon diversity index, were calculated to estimate the microbial diversity within an individual sample. Functional predictions were performed in the FRPROTAX (version 1.2.6) for bacteria (Langille et al., 2013). Soil fungal communities were functionally classified according to the FungalTraits database (Põlme et al., 2020).
2.6 Soil quality index analysis
The Soil Quality Index (SQI) based on soil properties, nematode and microbial communities was constructed using the minimum data set (MDS) to comprehensively assess soil quality. The calculation followed three main steps:
1 Selection of soil indicators for the MDS. The total data set (TDS) included soil properties (pH, SOM, TN, TP, TK, AN, AP, AK), nematode community metrics (Shannon diversity index, EI, SI, MI, relative abundance of herbivores, and relative abundance of omnivores–predators), and microbial diversity (bacterial and fungal Shannon diversity indices). SOM, TN, TP, TK, AN, AP, and AK were selected to represent nutrient availability; Shannon diversity indices for nematodes, bacteria, and fungi were used to reflect the soil’s ability to support biodiversity; the relative abundance of herbivores was used to indicate herbivore pressure, whereas the prevalence of omnivores–predators was associated with herbivore regulation. Generally, nematode communities characterized by high MI and SI values and low EI values indicate nutrient-enriched environments with well-structured food webs, representing healthy soils (Zhang et al., 2022). Principal component analysis (PCA) was applied to the standardized TDS to reduce dimensionality and identify the most informative indicators. Principle components with eigenvalues > 1.0 and at least explained 5% of the total variance were selected. For each selected component, variables with factor loadings within 10% of the highest loading were considered candidate MDS indicators. If more than one indicator were chosen in one principle component, Pearson’s correlation analysis was performed to reduce the redundancy. If indicators were significantly correlated (r ≥ 0.5) in the principle component, indicator with the highest factor loadings was retained in the MDS.
2 Scoring of selected indicators. Indicators in the MDS were transformed into unitless scores using min–max normalization (Qiao et al., 2025; Wang et al., 2024). Depending on their relationship with soil quality, the “more-is-better” function (Equation 1) was applied to pH, SOM, TN, TP, TK, AN, AP, AK, SI, MI, relative abundance of omnivores–predators, and the Shannon diversity indices of nematode, bacterial, and fungal communities. The “less-is-better” function (Equation 2) was applied to EI and the relative abundance of herbivores.
where f(x) is the scores of the indicator, Max and Min are the maximum and minimum measured values of the indicators.
3 Integration of indicator scores. The weight of the indicator was determined by variance explained by each principle component divided by the sum of the variance explained by all retained components (eigenvalues > 1.0). The final SQI was calculated as:
where Wi is the weight of indicator i and Si is its score.
2.7 Statistical analysis
A One-way ANOVA was used to test the effect of the cropping system on soil physicochemical properties, nematode ecological indices, microbial diversity indices and SQI. Tukey’s HSD post hoc test was used to evaluate the significance of differences between cropping systems. Data were log-transformed or arcsine square root-transformed (for proportional data) to improve normality and homogeneity when necessary. The nematode community composition was examined using principal coordinate analysis (PCoA) based on a Bray-Curtis distance matrix calculated from genus-level abundance data, while the bacterial and fungal communities were analyzed using the relative abundance at the ASV level. A permutational multivariate analysis of variance analysis (PERMANOVA) was conducted to quantitatively evaluate the effects of the cropping system on community composition. Linear discriminant analysis (LDA) was used to identify bacterial and fungal taxa that significantly differed between cropping systems (LDA > 3.5).
The partial least squares path model (PLS-PM) was built to examine the direct and indirect interactions among soil properties, nematodes communities, and bacterial and fungal communities. In the initial model, soil properties were represented by WC, pH, SOM, TN, TP, TK, AN, AP, and AK; nematode communities by the Shannon diversity index, EI, MI, SI, and PCoA 1; and bacterial and fungal communities by the Shannon diversity index and PCoA 1. Following model construction, variables with loadings < 0.7 were removed to improve model performance (Sanchez, 2013). Model quality was evaluated using the Goodness of Fit (GoF). All statistical analyses were performed using R 4.5.0.
3 Results
3.1 Soil physicochemical properties
We detected significant differences in soil water content (WC), pH, total phosphorus (TP), total potassium (TK), available nitrogen (AN), available phosphorus (AP), and available potassium (AK) between the three cropping systems (Table 1). The WC was significantly higher in the paddy–upland rotation (RO; 49.77%) and paddy continuous (PA; 48.77%) systems compared to the upland continuous system (UP; 31.87%). Soil pH was also significantly higher in the RO (6.02) system than in the PA (5.41) and UP (5.47) systems. TP, AK, and AP were significantly lower in the PA system. In contrast, AN and TK were highest in the UP system, intermediate in the PA system, and lowest in the RO system. No significant differences were observed in soil organic matter (SOM) and total nitrogen (TN) between the three systems.
3.2 Soil nematode communities
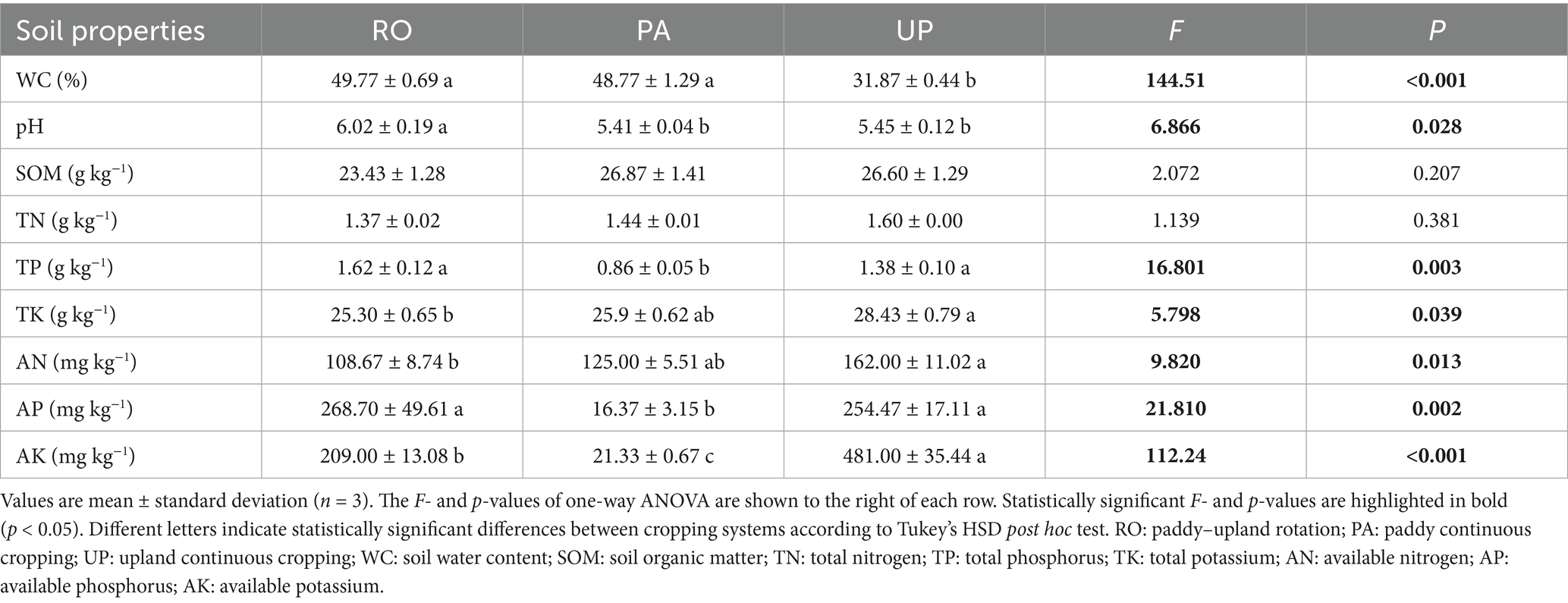
A total of 26 nematode genera were identified in the studied area (Supplementary Table S1). Mesodorylaimus (33.21%), Tobrilus (30.64%), and Rhabdolaimus (23.68%) were the dominant nematode genus in the RO system, while Hirschmanniella (38.34%), Rhabdolaimus (11.53%), and Tobrilus (11.36%) predominated in the PA system. Moreover, the UP system was dominated by Acrobeloides (66.67%) and Panagrolaimus (11.00%) (Figure 1A). The PCoA plot and PERMANOVA revealed that the cropping system had a significant effect on the community composition of soil nematodes (Figure 2A; R2 = 0.724; p = 0.003). The trophic structure also varied between cropping systems, whereby algivores were dominant in the RO system (55.15%), herbivores were dominant in the PA system (41.88%), and bacterivores dominated the UP system (93.67%) (Figures 3F–H). The relative abundance of fungivores and omnivores–predators did not significantly differ among the three systems (Figures 3I,J). The total nematode abundance was significantly higher in the UP system, intermediate in the PA system, and lowest in the RO system (Figure 3A), while the Shannon diversity was higher in the PA system, intermediate in the RO system, and lowest in the UP system (Figure 3B). There were no significant differences in nematode richness between cropping systems (Supplementary Table S2). The MI, EI, and SI were all significantly affected by the type of cropping system, being significantly lower in the UP system (Figure 3C–E).
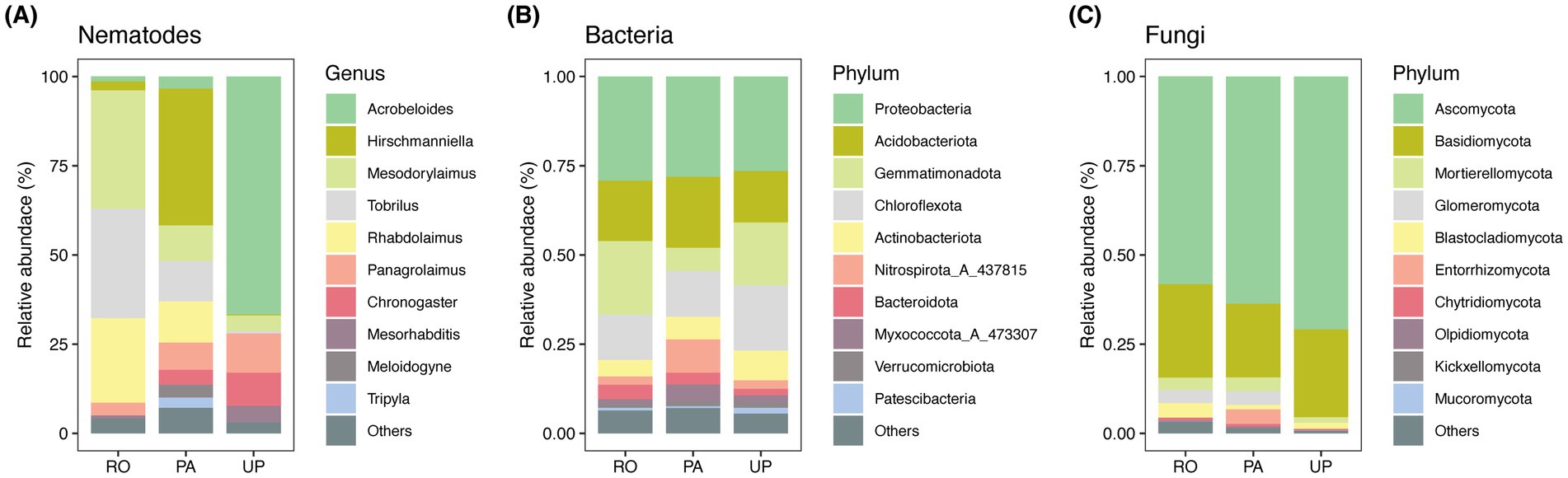
Figure 1. Relative abundance of soil nematode genus (A), bacterial phylum (B), and fungal phylum (C) in different cropping systems. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
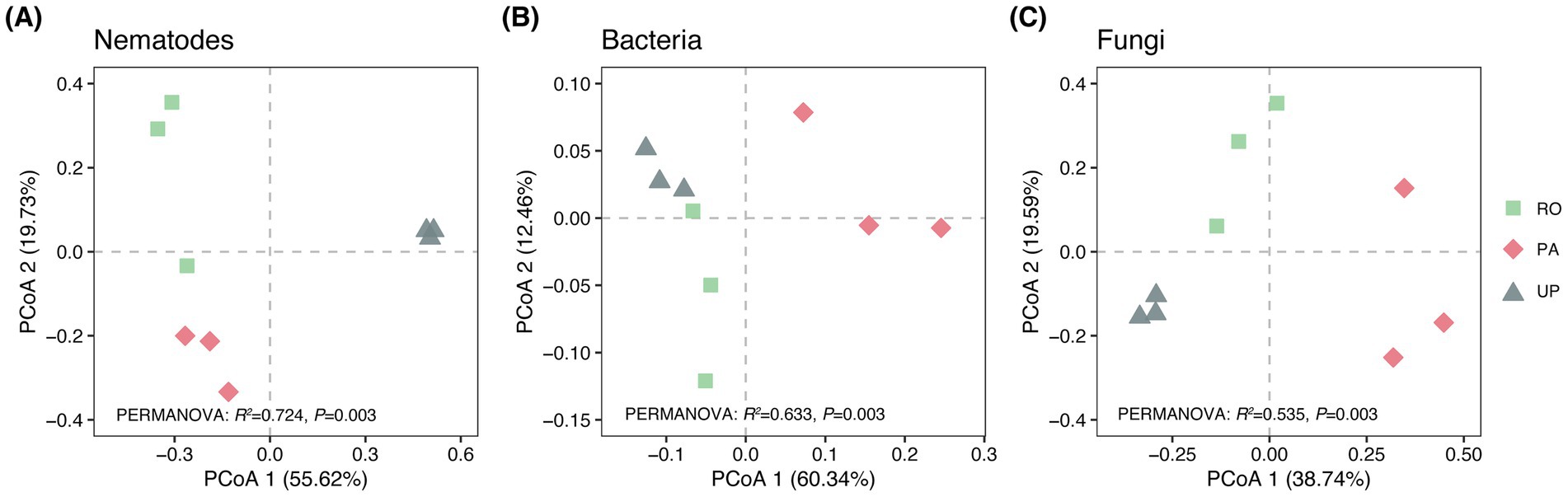
Figure 2. Principal coordinate analysis (PCoA) of the soil nematode (A), bacterial (B), and fungal (C) communities in different cropping systems. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
Figure 3. The ecological indices (A–E) and relative abundance of each trophic group (F–J) in different cropping systems. F-values and the significance of the ANOVA are depicted as follows: ***: p < 0.001; **: p < 0.01; *: p < 0.05; ns: not significant. Different letters indicate statistically significant differences between cropping systems according to Tukey’s HSD post hoc test. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
3.3 Soil microbial communities
Overall, Proteobacteria (26.51–29.22%), Gemmatimonadota (6.46–20.61%), Acidobacteriota (14.44–19.86%), and Chloroflexota (12.69–18.30%) were identified as the dominant bacterial phyla in the studied area (Figure 1B). The relative abundance of Gemmatimonadota was significantly reduced in the PA system (4.59%) compared to the RO (20.61%) and UP systems (17.60%), while the abundance of Nitrospirota and Myxococcota were reported to increased. Bacteroidota was less abundant in the UP system (1.79%) than in the RO (3.98%) and PA systems (3.35%) (Figure 1B). For fungal communities, Ascomycota (58.21–70.87%) and Basidiomycota (20.68–24.62%) were identified as the dominant phyla (Figure 1C). The relative abundance of Entorrhizomycota was significantly higher in the PA system (4.17%) compared to the RO (0.00%) and UP systems (0.09%) (Figure 1C).
The PCoA plot and PERMANOVA revealed that the type of cropping system had a significant effect on the bacterial (R2 = 0.633; p = 0.003) and fungal (R2 = 0.535; p = 0.003) community composition at the ASV level (Figures 2B,C). The ASV richness and Shannon diversity of bacterial and fungal communities did not differ significantly between cropping systems.
The LEfSe analysis revealed significant changes in the bacterial and fungal communities at different taxonomic levels (Figure 4). Most taxa of bacteria were enriched in PA, including Bryobacterales (Bryobacteraceae, Palsa-187), Thermoanaerobaculales (Thermoanaerobaculaceae, RBG_13_68_16), UBA6911 (Gp18_AA60), UBA10030 (UBA6906, CAADGV01), Bacteroidota, RPRB01, Haliangiales_463188 (Haliangiaceae_463188, Haliangium_463188), Thermodesulfovibrionia (Thermodesulfovibrionales, UBA6898, GW_Nitrospira_1), VBDM01, and Sideroxydans. The relative abundance of Gemmatimonadaceae (SCN_70_22, AG11) and Thiobacillaceae (Thiobacillus) was significantly increased in the RO system. The relative abundance of Saccharimonadia (Saccharimonadales, UBA10212), Pseudolabrys_502538, and Trinickia_580244 was higher in the UP system. Meanwhile, for fungi, the RO system was enriched in Penicillium, Trichosphaeriales (Trichosphaeriaceae, Nigrospora), and Blastocladiomycota (Blastocladiomycetes, Blastocladiales, Physodermataceae, Paraphysoderma). Ascobolaceae (Ascobolus), Pezizomycetes (Pezizales), Glomerellaceae (Colletotrichum), Volutella, and Lophotrichus were more abundant in the UP system. Leotiomycetes (Thelebolales, Pseudeurotiaceae, Pseudeurotium), Stachybotryaceae (Stachybotrys), Ophiostomatales (Ophiostomataceae, Leptographium), Strophariaceae (Psilocybe), and Funneliformis were reported to be significantly increased in the PA system.
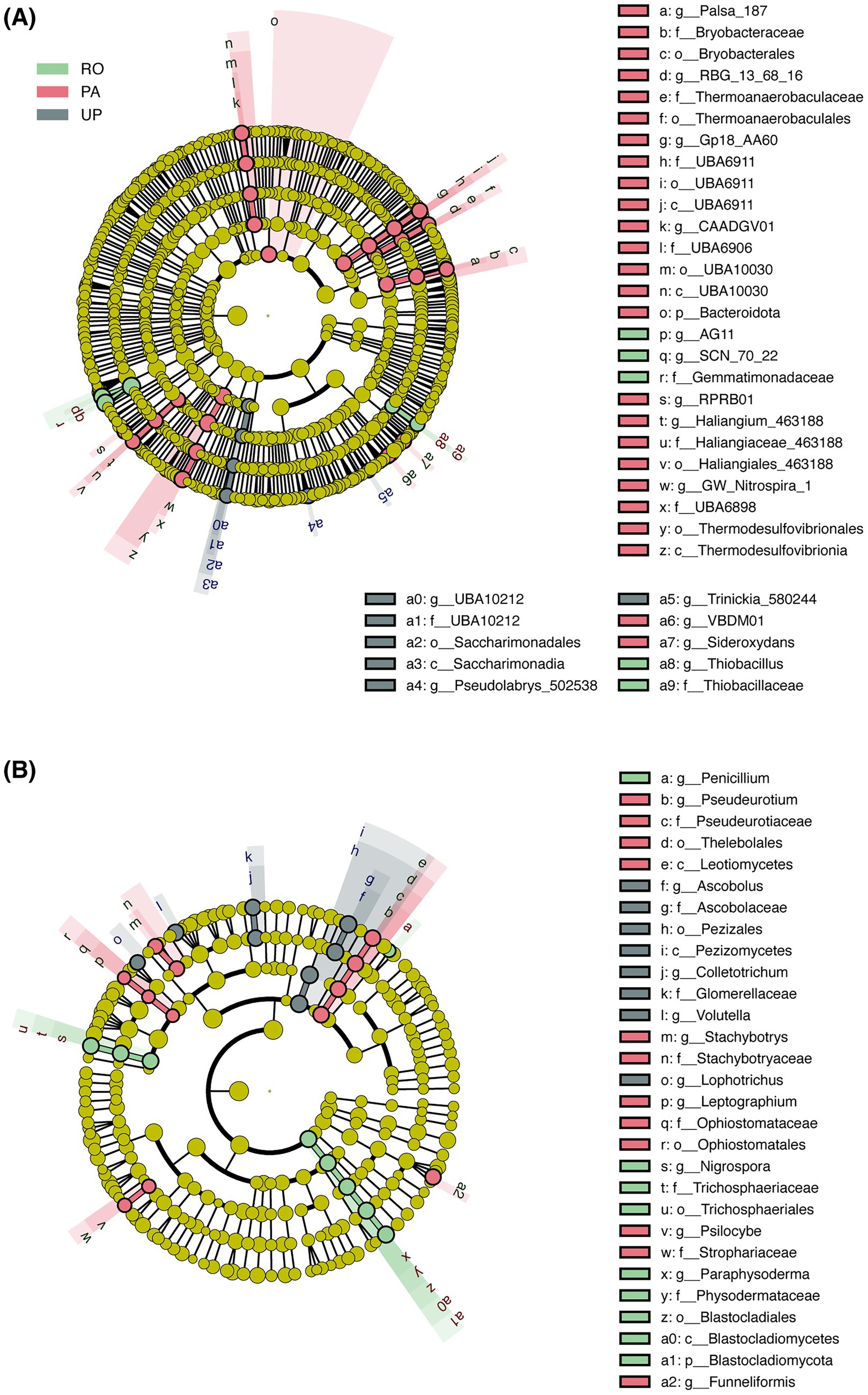
Figure 4. LEfSe cladograms showing significantly enriched bacterial (A) and fungal (B) taxa between different cropping systems (LDA score >3.5). Colored dots indicate taxa with significantly different relative abundances. Concentric circles represent phylogenetic levels from the phylum to the genus. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
We used FAPROTAX to predict the ecological functions of bacteria (Figure 5 and Supplementary Table S3). The relative abundance of bacterial taxa that are associated with nitrogen fixation and predatory or exoparasitic lifestyles was significantly higher in the PA system, while those involved in cellulolysis and ureolysis were significantly lower. In the RO system, functional groups linked to dark oxidation, nitrate reduction, nitrate respiration, nitrogen respiration and photoautotrophy were more abundant. Functional groups related to aerobic ammonia oxidation and aerobic chemoheterotrophy were significantly more abundant in the UP system. For fungi, we used the FungalTraits database to classify taxa into 19 functional groups; however, no significant differences in any functional group were observed between the cropping systems (Supplementary Table S4).
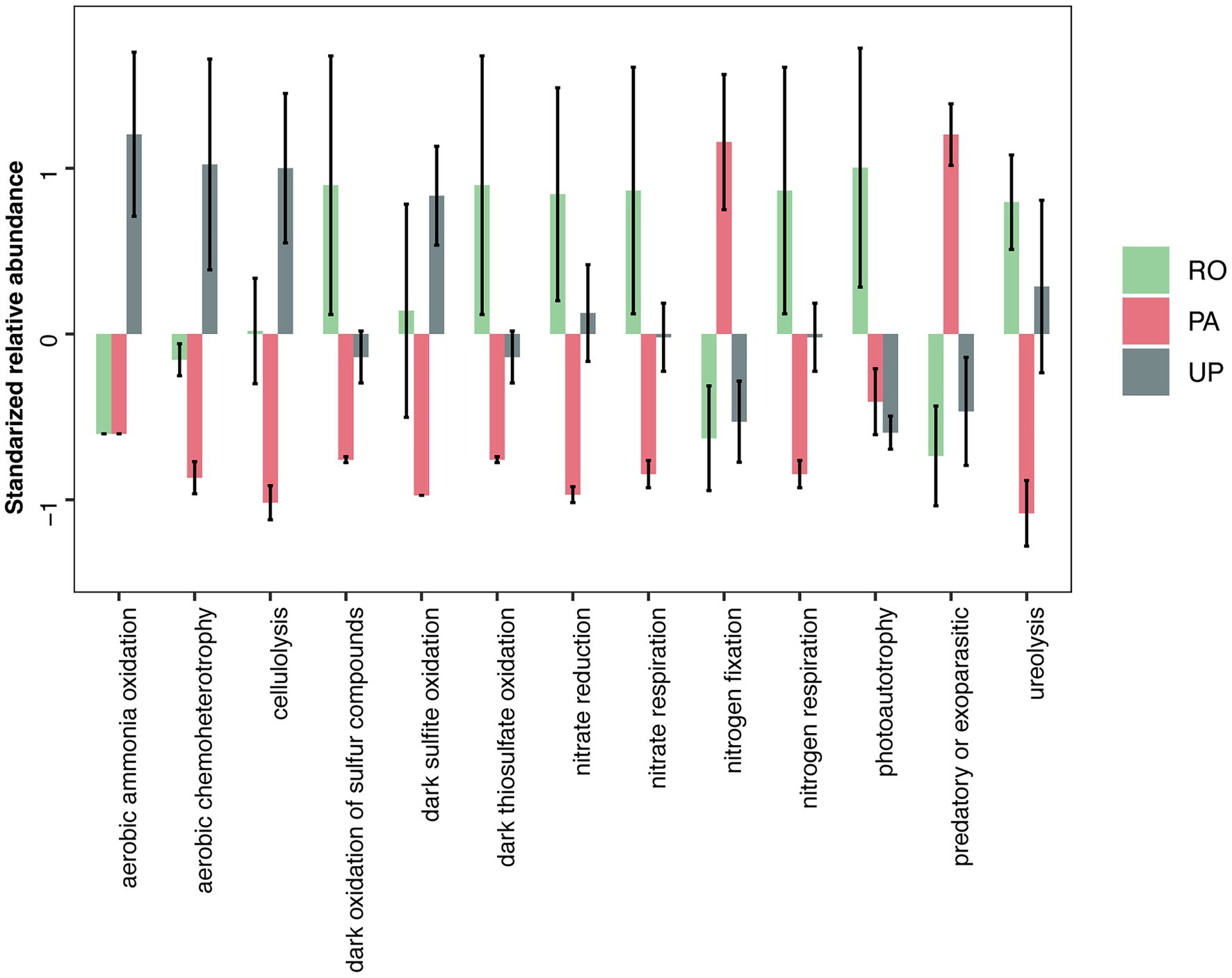
Figure 5. Effects of cropping system on the relative abundance of bacterial functional groups. Relative abundance values were standardized to improve visualization. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
3.4 Soil quality index
To comprehensively evaluate the effects of cropping systems on soil quality, we integrated soil physicochemical properties with nematode and microbial community metrics. Principal component analysis (PCA) was applied to the total data set, and four principal components (PCs) with eigenvalues > 1.0 and explaining at least 5% of the total variance were retained. Together, these four PCs accounted for 91.0% of the total variance (Supplementary Table S5). For PC1, the factor loading matrix identified four variables—structure index (SI), available potassium (AK), maturity index (MI), and available nitrogen (AN)—within 10% of the highest factor loading (SI = 0.806). Because these variables were highly correlated with total nitrogen (TN) (r ≥ 0.5; Supplementary Table S6), SI was retained as the representative indicator for this component. Similarly, total phosphorus (TP) was retained for PC2, TN for PC3, and the Shannon diversity index of fungal communities (SF) for PC4. The Soil Quality Index (SQI) was calculated using the weighted sum of these four indicators: SQI = 0.453 × SI + 0.312 × TP + 0.159 × TN + 0.076 × SF. The SQI values ranked the cropping systems as RO > PA > UP, indicating that the paddy–upland rotation system supported the highest overall soil quality (Figure 6).
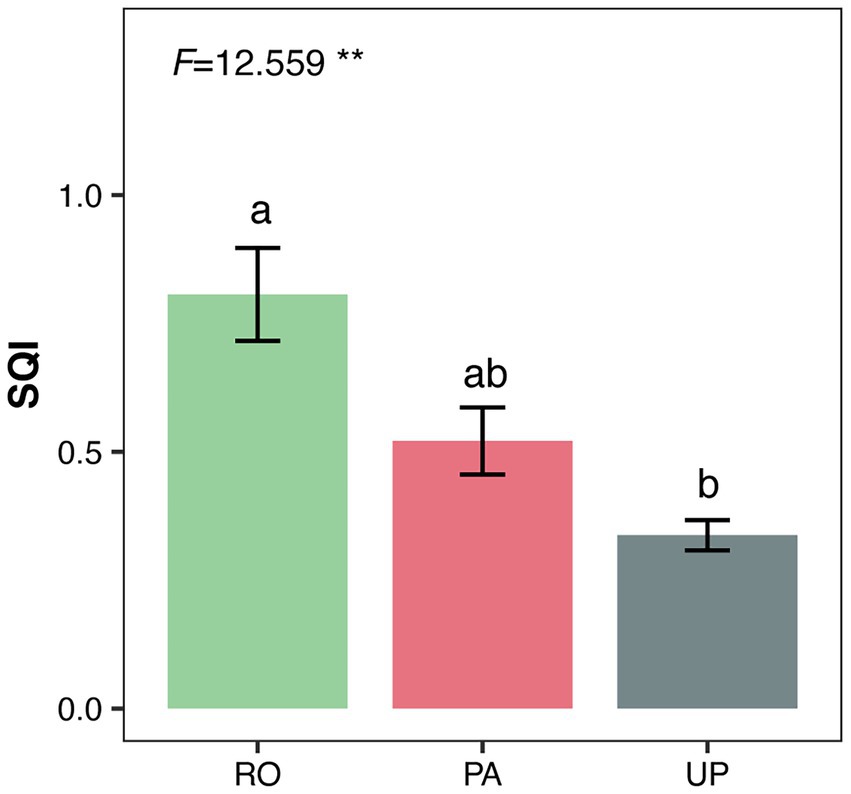
Figure 6. Soil quality index in different cropping systems. F-values and the significance of ANOVA are shown as follows: **: p < 0.01; *: p < 0.05. Different letters indicate statistically significant differences between cropping systems according to Tukey’s HSD post hoc test. RO: paddy–upland rotation; PA: paddy continuous cropping; UP: upland continuous cropping.
3.5 PLS-PM
The PLS-PM showed a good overall fit (GoF = 0.801) and effectively described the interaction pathways among soil properties, and bacterial, fungal, and nematode communities (Figure 7). Soil properties exerted a significant effect on nematode communities and a marginally significant effect on bacterial and fungal communities (p = 0.072). In addition, bacterial communities significantly influenced fungal communities. In contrast, nematode communities were not significantly affected by bacterial or fungal communities.
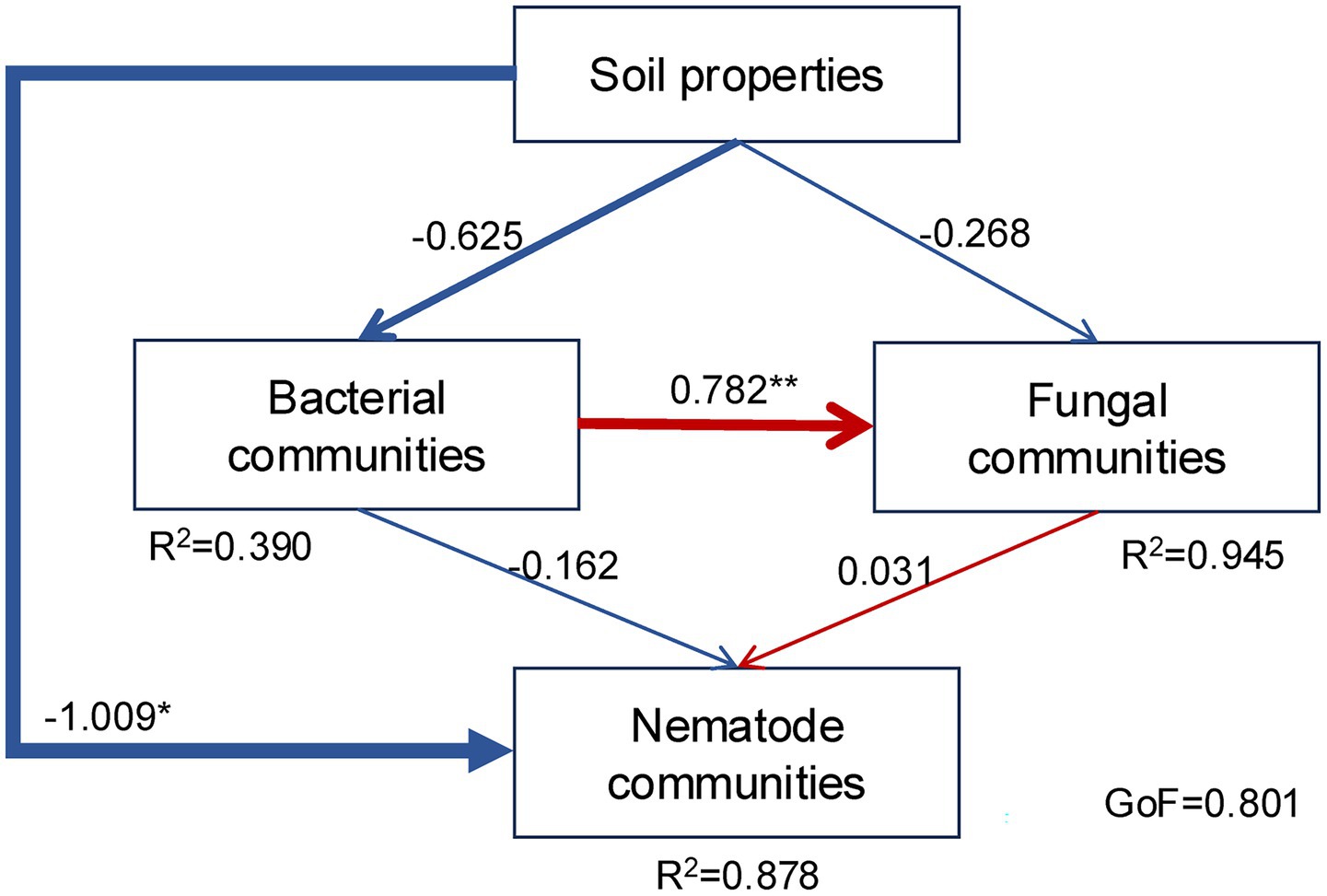
Figure 7. Partial least squares path model (PLS-PM) describing the direct and indirect interactions among soil properties, nematode communities, and bacterial and fungal communities. Soil properties were represented by WC, TK, AN, and AK; bacterial communities by PCoA 1; fungal communities by PCoA 1; and nematode communities by PCoA 1, EI, SI, and MI. Numbers on arrows indicate standardized path coefficients, with arrow widths proportional to their magnitudes. R2 adjacent to the latent variables are the coefficients of determination. The GoF is the goodness of fit. Blue and red represent negative and positive correlations, respectively. **p < 0.01, *p < 0.05.
4 Discussion
In this study, we evaluated the community structure and functional profiles of soil nematodes, bacteria, and fungi under three cropping systems: continuous paddy (PA), continuous upland (UP), and paddy–upland rotation (RO). Our findings reveal that although microbial diversity remained stable, significant shifts in nematode and microbial community composition and ecological function occurred, driven by water regime and nutrient redistribution. Integration of soil physicochemical and biological indicators into the Soil Quality Index (SQI) demonstrated that the RO system maintained the highest overall soil quality, highlighting its potential to enhance ecosystem functions and sustainability in this agroecosystem.
Previous studies have shown that rotating rice with upland crops improves the physical and chemical properties of paddy soil, oxidizes reductive substances produced during rice cultivation, neutralizes soil acidity, and enhances crop yield (Kögel-Knabner et al., 2010; Zhang D. et al., 2025). In our study, the elevated soil water content observed in the RO and PA systems was primarily attributed to flooded rice cultivation, which involved prolonged waterlogging and elevated groundwater levels. Anaerobic conditions in flooded soils can lead to phosphorus fixation in less-bioavailable forms, such as Fe–P or Al–P complexes, and promote potassium loss through leaching (Lee et al., 2004; Maranguit et al., 2017), and subsequently resulting in significantly lower levels of TP and AP in the RO system. Crop rotation has been reported to alter soil pH (Godsey et al., 2007). In our study, the RO system exhibited significantly higher pH values compared to the PA and UP systems, likely due to the alternating aerobic and anaerobic conditions that enhanced redox balance and limited the accumulation of acidic by-products (Pan et al., 2014; Yin et al., 2025). The UP system, characterized by two annual loofah cropping cycles, likely involved greater fertilizer inputs and more intensive management, contributing to the highest levels of AN and TK observed in this study. Paddy–upland rotation has been shown to improve nutrient use efficiency and reduce nutrient loss (Chen et al., 2018). In our rotation system, rice can also be employed to make more efficient use of the residual soil nutrients resulting from the high fertilizer inputs applied during the loofah cultivation phase.
Water management, soil fertility and crop type are considered key factors in shaping soil nematode communities in paddy–upland systems (Li et al., 2024; Liu et al., 2008; Okada et al., 2011). In the RO and PA systems, the flooded conditions during the rice-growing stage promoted the proliferation of algae and diatoms, leading to the dominance of Mesodorylaimus, Tobrilus, and Rhabdolaimus, which are known to feed on these organisms (Okada et al., 2011). Notably, the plant-feeding nematode Hirschmanniella appears to be particularly well adapted to the anaerobic conditions of flooded soils and is commonly found parasitizing the roots of submerged crops such as rice and lotus (Beesa et al., 2021; Uematsu et al., 2020). In our study, the paddy–upland rotation effectively reduced the dominance of Hirschmanniella during the rice stage, possibly due to fluctuations between flooded and non-flooded conditions, as well as the absence of host plants during the upland phase, which may substantially suppress the population of this nematode. In the UP system, bacterial-feeding nematodes were overwhelmingly dominant. The aerobic soil conditions appear to be more favorable for the growth and reproduction of Acrobeloides and Panagrolaimus in our study. These bacterial-feeding nematodes, which are characterized by low c-p values, are also capable of rapidly responding to fertilizer inputs, resulting in increased population densities (Bongers and Bongers, 1998). This may partly explain the higher nematode abundance observed in the UP system. The higher maturity (MI) and structure (SI) indices observed in the paddy fields of the RO and PA systems indicate a more structured and stable soil food web under flooded conditions (Bongers and Ferris, 1999; Okada et al., 2011). However, given the generally lower total nematode abundance in paddy soils, these environments may be less suitable habitats than upland soils due to reduced oxygen availability. These findings are consistent with previous studies reporting greater MI and EI values in paddy fields compared to upland fields (Okada et al., 2011).
Numerous studies have shown that cropping systems are major drivers of microbial community structure; however, the effect of crop rotation on soil microbial diversity can be context-dependent (Liu et al., 2023; Venter et al., 2016). Although crop rotation can alter soil properties and nutrient availability, the high adaptability of microorganisms, combined with interspecific competition and food web interactions, may buffer short-term shifts in microbial diversity. For example, Wang et al. (2017) reported that crop rotation increased microbial biomass and activity but did not affect bacterial diversity. Similarly, our study did not observe significant differences in either bacterial or fungal diversity. However, more pronounced changes were found in microbial community composition, which can have important implications for crop productivity and soil functions including organic matter decomposition and nutrient cycling (Sun et al., 2024; Wang et al., 2017; Zuber et al., 2018). Our findings revealed that the relative abundance of Gemmatimonadota was significantly reduced in the PA system, whereas Nitrospirota, Myxococcota, and Entorrhizomycota were more abundant. Gemmatimonadota are involved in nitrogen and phosphorus cycling and suggested to prefer aerobic, drier soil conditions (Mujakić et al., 2022). The anaerobic environment created by the prolonged flooding in the PA system likely suppressed their activity, resulting in a reduced relative abundance. Nitrospirota are key nitrite-oxidizing bacteria involved in the nitrification process, converting nitrite to nitrate (Daims et al., 2015; Liu et al., 2022). Myxococcota are known for their predatory behavior, feeding on other bacteria and contributing to the regulation of microbial communities (Wang et al., 2020). The anaerobic conditions in flooded paddy fields may favor the growth of Nitrospirota and Myxococcota species which are adapted to such environments, leading to their proliferation (Fang et al., 2024). Entorrhizomycota is a phylum of fungi that includes root-associated parasites forming galls on plant roots, particularly in the Cyperaceae and Juncaceae families (Bauer et al., 2015; Riess et al., 2019). The flooded conditions in paddy fields may favor the proliferation of Entorrhizomycota, possibly due to the presence of suitable host plants and the anaerobic environment that supports their life cycle (Ellouze et al., 2014). In contrast, the absence of these conditions in upland and rotation systems may restrict their occurrence. LEfSe analysis revealed a greater number of bacterial taxa enriched in the PA system. This enrichment pattern suggests these bacteria are specifically adapted to, and functionally active within, the long-term anaerobic environment characteristic of flooded rice monoculture. Many of these enriched taxa are likely involved in complex redox reactions and nutrient cycling processes essential under oxygen-limited conditions, such as Sideroxydans (iron-oxidizing bacteria) (Zhou et al., 2022), Thiobacillus (sulfur-oxidizing bacteria) (Sasaki et al., 1998), and Thermodesulfovibrionia (sulfate-reducing bacteria) (Kushkevych et al., 2021).
Functional prediction based on FAPROTAX further revealed shifts in bacteria function groups, suggesting potential impacts on ecosystem services. The increased abundance of taxa involved in nitrogen fixation and predation in the PA system may be linked to the higher relative abundance of the phyla Nitrospirota and Myxococcota, as previously discussed. Fluctuations in redox potential due to paddy water management strongly influence microbial community structure and function, thereby affecting short-term biogeochemical processes such as decomposition and nutrient cycling (Kögel-Knabner et al., 2010). Under flooded conditions, an anoxic environment typically slows decomposition rates (Kögel-Knabner et al., 2010; Zhou et al., 2014). Cellulolytic bacteria play a key role in decomposing plant residues (Bautista-Cruz et al., 2024), while ureolytic bacteria convert urea into ammonia, making it available for plant uptake or transformation through the nitrogen cycle (Hasan, 2000). The reduced abundance of these functional groups in the PA system suggests the presence of alterations in carbon and nitrogen cycling under flooded conditions. In contrast, the well-aerated soils of the UP system provide favorable conditions for aerobic ammonia oxidation and aerobic chemoheterotrophy, enhancing nitrification and organic matter decomposition (Zhang et al., 2018). Notably, in the RO system, bacterial functional groups linked to dark oxidation, nitrate reduction, nitrate respiration, nitrogen respiration, and photoautotrophy were more abundant. These functions are closely tied to energy metabolism and nitrogen cycling processes that are highly sensitive to oxygen availability (Pett-Ridge and Firestone, 2005). The seasonal alternation between anaerobic and aerobic phases in the RO system generates fluctuating redox conditions, promoting a broader range of metabolic pathways than in either continuous paddy or continuous upland systems. Such functional versatility likely supports more efficient nitrogen turnover, sustained nutrient availability, and greater microbial adaptability to environmental fluctuations. It should be emphasized, however, that functional predictions based solely on taxonomic assignments cannot confirm ecosystem-level processes. Future work could validate these inferences by targeting key microbial groups through functional gene quantification and by measuring enzyme activities to establish direct links between microbial community composition, nutrient cycling, and plant health outcomes (Li et al., 2018).
Apart from the cropping system itself, differences in fertilization regimes and microclimatic conditions across treatments may have contributed to variation in soil nutrient availability and biological communities. In particular, the RO and UP systems involved the use of plastic greenhouses during the loofah-growing phase, which likely created distinct microclimatic environments (e.g., increased temperature and humidity) relative to the open-field paddy system (PA). These greenhouse conditions can accelerate organic matter decomposition, modify microbial activity, and affect nematode dynamics by altering soil temperature and moisture regimes (Bandopadhyay et al., 2018). Additionally, the RO and UP systems received higher fertilization inputs—particularly during loofah cultivation—which likely contributed to elevated nutrient levels and may have stimulated copiotrophic microbial taxa and bacterial-feeding nematodes (Ferris et al., 2001; Leff et al., 2015). Such differences in nutrient inputs and environmental conditions complicate direct attribution of biological responses solely to the cropping system itself.
5 Conclusion
This study demonstrates that paddy–upland rotation significantly reshapes soil nematode and microbial communities while achieving a higher Soil Quality Index (SQI) compared to continuous paddy or upland cropping systems. The rotation system influenced nematode trophic structure, reduced the abundance of plant-parasitic nematodes such as Hirschmanniella, and enhanced food web stability, as reflected by the higher maturity and structure indices. While the microbial diversity remained stable, the community composition and functional profiles were markedly altered. In microbial communities, the relative abundance of the functional groups involved in nitrogen cycling, carbon turnover, and redox-sensitive energy metabolism varied. These shifts reflect the profound influence of cropping systems on belowground biodiversity and ecosystem functioning. Our findings support the ecological benefits of paddy–upland rotation and provide valuable insights for improving soil health in rice-based agroecosystems.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author(s).
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SP: Funding acquisition, Formal analysis, Methodology, Conceptualization, Writing – review & editing, Software, Investigation, Writing – original draft. YW: Investigation, Resources, Writing – review & editing. JC: Writing – review & editing, Investigation, Methodology. SW: Formal analysis, Writing – review & editing, Funding acquisition. XC: Validation, Funding acquisition, Writing – review & editing, Data curation. PW: Investigation, Writing – review & editing. XW: Data curation, Writing – review & editing. CY: Supervision, Writing – review & editing, Funding acquisition. YZ: Writing – review & editing, Funding acquisition, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Wenzhou Basic Scientific Research Project [N2024004], Cangnan Industrial Research Institute of Modern Agriculture [2023CNYJY13], ‘Three Rural and Nine Directions’ Science and Technology Collaboration Plan [2023SNJF036], Wenzhou Agricultural Harvest Fund [FSJH202307], Promotion Project of Science Technology Strengthening Agriculture Industry in Pingyang County [2023PYII04] and the Doctoral Research Start-up Fund of Wenzhou Vocational College of Science and Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2025.1646192/full#supplementary-material
References
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Bandopadhyay, S., Martin-Closas, L., Pelacho, A. M., and DeBruyn, J. M. (2018). Biodegradable plastic mulch films: impacts on soil microbial communities and ecosystem functions. Front. Microbiol. 9:819. doi: 10.3389/fmicb.2018.00819
Bauer, R., Garnica, S., Oberwinkler, F., Riess, K., Weiß, M., and Begerow, D. (2015). Entorrhizomycota: a new fungal phylum reveals new perspectives on the evolution of fungi. PLoS One 10:e0128183. doi: 10.1371/journal.pone.0128183
Bautista-Cruz, A., Aquino-Bolaños, T., Hernández-Canseco, J., and Quiñones-Aguilar, E. E. (2024). Cellulolytic aerobic bacteria isolated from agricultural and forest soils: an overview. Biology 13:102. doi: 10.3390/biology13020102
Beesa, N., Sasnarukkit, A., Jindapunnapat, K., Tivet, F., Bellafiore, S., and Chinnasri, B. (2021). Species characterization and population dynamics of Hirschmanniella mucronata in lowland rice fields managed under conservation agriculture in Cambodia. J. Saudi Soc. Agric. Sci. 20, 137–145. doi: 10.1016/j.jssas.2020.12.009
Bending, G. D., Turner, M. K., Rayns, F., Marx, M.-C., and Wood, M. (2004). Microbial and biochemical soil quality indicators and their potential for differentiating areas under contrasting agricultural management regimes. Soil Biol. Biochem. 36, 1785–1792. doi: 10.1016/j.soilbio.2004.04.035
Berg, G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Blundell, R., Schmidt, J. E., Igwe, A., Cheung, A. L., Vannette, R. L., Gaudin, A. C. M., et al. (2020). Organic management promotes natural pest control through altered plant resistance to insects. Nat Plants 6, 483–491. doi: 10.1038/s41477-020-0656-9
Bongers, T. (1990). The maturity index: an ecological measure of environmental disturbance based on nematode species composition. Oecologia 83, 14–19. doi: 10.1007/BF00324627
Bongers, T., and Bongers, M. (1998). Functional diversity of nematodes. Appl. Soil Ecol. 10, 239–251. doi: 10.1016/S0929-1393(98)00123-1
Bongers, T., and Ferris, H. (1999). Nematode community structure as a bioindicator in environmental monitoring. Trends Ecol. Evol. 14, 224–228. doi: 10.1016/S0169-5347(98)01583-3
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, S., Liu, S., Zheng, X., Yin, M., Chu, G., Xu, C., et al. (2018). Effect of various crop rotations on rice yield and nitrogen use efficiency in paddy–upland systems in southeastern China. Crop J. 6, 576–588. doi: 10.1016/j.cj.2018.07.007
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Doran, J. W., and Parkin, T. B. (1994). Defining and assessing soil quality. In Defining Soil Quality for a Sustainable Environment. eds J. W. Doran, D. C. Coleman, D. F. Bezdicek, and B. A. Stewart), Soil Science Society of America, Madison, 3–21. doi: 10.2136/sssaspecpub35.c1
Ebihara, Y., Uematsu, S., and Nomiya, S. (2010). Control of Verticillium dahliae at a strawberry nursery by paddy-upland rotation. J. Gen. Plant Pathol. 76, 7–20. doi: 10.1007/s10327-009-0205-x
Ellouze, W., Esmaeili Taheri, A., Bainard, L. D., Yang, C., Bazghaleh, N., Navarro-Borrell, A., et al. (2014). Soil fungal resources in annual cropping systems and their potential for management. Biomed. Res. Int. 2014:531824. doi: 10.1155/2014/531824
Fang, Y., Qiu, J., and Li, X. (2024). Mechanisms of irrigation water levels on nitrogen transformation and microbial activity in paddy fields. Water 16:3021. doi: 10.3390/w16213021
Ferris, H., Bongers, T., and Goede, R. G. M. D. (2001). A framework for soil food web diagnostics: extension of the nematode faunal analysis concept. Appl. Soil Ecol. 18, 13–29. doi: 10.1016/S0929-1393(01)00152-4
Flower, K. C., Hüberli, D., Collins, S. J., Thomas, G., Ward, P. R., and Cordingley, N. (2019). Progression of plant-parasitic nematodes and foliar and root diseases under no-tillage with different crop rotations. Soil Tillage Res. 191, 18–28. doi: 10.1016/j.still.2019.03.010
Godsey, C. B., Pierzynski, G. M., Mengel, D. B., and Lamond, R. E. (2007). Changes in soil pH, organic carbon, and extractable aluminum from crop rotation and tillage. Soil Sci. Soc. Am. J. 71, 1038–1044. doi: 10.2136/sssaj2006.0170
Hasan, H. A. H. (2000). Ureolytic microorganisms and soil fertility: a review. Commun. Soil Sci. Plant Anal. 31, 2565–2589. doi: 10.1080/00103620009370609
John, K., Zaitsev, A. S., and Wolters, V. (2021). Soil fauna groups respond differentially to changes in crop rotation cycles in rice production systems. Pedobiologia 84:150703. doi: 10.1016/j.pedobi.2020.150703
Kögel-Knabner, I., Amelung, W., Cao, Z., Fiedler, S., Frenzel, P., Jahn, R., et al. (2010). Biogeochemistry of paddy soils. Geoderma 157, 1–14. doi: 10.1016/j.geoderma.2010.03.009
Kushkevych, I., Dordević, D., Vítězová, M., and Rittmann, S. K. M. R. (2021). Environmental impact of sulfate-reducing bacteria, their role in intestinal bowel diseases, and possible control by bacteriophages. Appl. Sci. 11:735. doi: 10.3390/app11020735
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lee, C. H., Park, C. Y., Park, K. D., Jeon, W. T., and Kim, P. J. (2004). Long-term effects of fertilization on the forms and availability of soil phosphorus in rice paddy. Chemosphere 56, 299–304. doi: 10.1016/j.chemosphere.2004.02.027
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Li, G., Li, X., and Liu, T. (2024). Reassessing soil nematode diversity under fertilization in a paddy–upland rotation system. Appl. Soil Ecol. 195:105216. doi: 10.1016/j.apsoil.2023.105216
Li, D., Zhang, X., Green, S. M., Dungait, J. A., Wen, X., Tang, Y., et al. (2018). Nitrogen functional gene activity in soil profiles under progressive vegetative recovery after abandonment of agriculture at the Puding karst critical zone observatory, SW China. Soil Biol. Biochem. 125, 93–102. doi: 10.1016/j.soilbio.2018.07.004
Liu, M., Chen, X., Qin, J., Wang, D., Griffiths, B., and Hu, F. (2008). A sequential extraction procedure reveals that water management affects soil nematode communities in paddy fields. Appl. Soil Ecol. 40, 250–259. doi: 10.1016/j.apsoil.2008.05.001
Liu, S., Coyne, M. S., Grove, J. H., and Flythe, M. D. (2022). Nitrite oxidizing bacteria, Nitrobacter and Nitrospira, are differently influenced by season, fertilizer, and tillage in long-term maize culture. Appl. Soil Ecol. 177:104530. doi: 10.1016/j.apsoil.2022.104530
Liu, Q., Zhao, Y., Li, T., Chen, L., Chen, Y., and Sui, P. (2023). Changes in soil microbial biomass, diversity, and activity with crop rotation in cropping systems: a global synthesis. Appl. Soil Ecol. 186:104815. doi: 10.1016/j.apsoil.2023.104815
Maranguit, D., Guillaume, T., and Kuzyakov, Y. (2017). Effects of flooding on phosphorus and iron mobilization in highly weathered soils under different land-use types: short-term effects and mechanisms. Catena 158, 161–170. doi: 10.1016/j.catena.2017.06.023
Marion, L. F., Schneider, R., Cherubin, M. R., Colares, G. S., Wiesel, P. G., da Costa, A. B., et al. (2022). Development of a soil quality index to evaluate agricultural cropping systems in southern Brazil. Soil Tillage Res. 218:105293. doi: 10.1016/j.still.2021.105293
Martin, T., and Sprunger, C. D. (2022). Soil food web structure and function in annual row-crop systems: how can nematode communities infer soil health? Appl. Soil Ecol. 178:104553. doi: 10.1016/j.apsoil.2022.104553
Matute, M., and Anders, M. (2012). Influence of rice rotation systems on soil nematode trophic groups in Arkansas. J. Agric. Sci. 4, 11–20. doi: 10.5539/jas.v4n2p11
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mujakić, I., Piwosz, K., and Koblížek, M. (2022). Phylum gemmatimonadota and its role in the environment. Microorganisms 10:151. doi: 10.3390/microorganisms10010151
Nguyen, S. V., Nguyen, P. T. K., Araki, M., Perry, R. N., Ba Tran, L., Minh Chau, K., et al. (2020). Effects of cropping systems and soil amendments on nematode community and its relationship with soil physicochemical properties in a paddy rice field in the Vietnamese Mekong Delta. Appl. Soil Ecol. 156:103683. doi: 10.1016/j.apsoil.2020.103683
Okada, H., Niwa, S., Takemoto, S., Komatsuzaki, M., and Hiroki, M. (2011). How different or similar are nematode communities between a paddy and an upland rice fields across a flooding–drainage cycle? Soil Biol. Biochem. 43, 2142–2151. doi: 10.1016/j.soilbio.2011.06.018
Pan, Y., Koopmans, G., Bonten, L., Song, J., Luo, Y., Temminghoff, E., et al. (2014). Influence of pH on the redox chemistry of metal (hydr)oxides and organic matter in paddy soils. J. Soils Sediments 14, 1713–1726. doi: 10.1007/s11368-014-0919-z
Pett-Ridge, J., and Firestone, M. (2005). Redox fluctuation structures microbial communities in a wet tropical soil. Appl. Environ. Microbiol. 71, 6998–7007. doi: 10.1128/AEM.71.11.6998-7007.2005
Phani, V., Khan, M. R., and Dutta, T. K. (2021). Plant-parasitic nematodes as a potential threat to protected agriculture: current status and management options. Crop Protect. 144:105573. doi: 10.1016/j.cropro.2021.105573
Põlme, S., Abarenkov, K., Henrik Nilsson, R., Lindahl, B. D., Clemmensen, K. E., Kauserud, H., et al. (2020). FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 105, 1–16. doi: 10.1007/s13225-020-00466-2
Qiao, Y., Gao, D., Fan, Y., Zhang, L., Yan, Q., Guo, L., et al. (2025). Deep plowing enhanced soil nematode communities and soil quality in dryland wheat fields on the loess plateau of China. Agric. Ecosyst. Environ. 387:109625. doi: 10.1016/j.agee.2025.109625
Ren, X., Chen, F., Ma, T., and Hu, Y. (2020). Soil quality characteristics as affected by continuous rice cultivation and changes in cropping systems in South China. Agriculture 10:443. doi: 10.3390/agriculture10100443
Riess, K., Schön, M. E., Ziegler, R., Lutz, M., Shivas, R. G., Piątek, M., et al. (2019). The origin and diversification of the Entorrhizales: deep evolutionary roots but recent speciation with a phylogenetic and phenotypic split between associates of the Cyperaceae and Juncaceae. Org. Divers. Evol. 19, 13–30. doi: 10.1007/s13127-018-0384-4
Sasaki, K., Tsunekawa, M., Ohtsuka, T., and Konno, H. (1998). The role of sulfur-oxidizing bacteria Thiobacillus thiooxidans in pyrite weathering. Colloids Surf. Physicochem. Eng. Aspects 133, 269–278. doi: 10.1016/S0927-7757(97)00200-8
Sun, Y., Yang, X., Elsgaard, L., Du, T., Siddique, K. H. M., Kang, S., et al. (2024). Diversified crop rotations improve soil microbial communities and functions in a six-year field experiment. J. Environ. Manag. 370:122604. doi: 10.1016/j.jenvman.2024.122604
Timofeeva, A. M., Galyamova, M. R., and Sedykh, S. E. (2023). Plant growth-promoting soil bacteria: nitrogen fixation, phosphate solubilization, siderophore production, and other biological activities. Plants 12:4074. doi: 10.3390/plants12244074
Uematsu, S., Yabu, T., Yao, M., Kurihara, T., and Koga, H. (2020). Ultrastructure of Hirschmanniella diversa early-stage infection in browning rhizomes of Indian lotus. J. Nematol. 52, 1–9. doi: 10.21307/jofnem-2020-055
van den Hoogen, J., Geisen, S., Routh, D., Ferris, H., Traunspurger, W., Wardle, D. A., et al. (2019). Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198. doi: 10.1038/s41586-019-1418-6
Venter, Z. S., Jacobs, K., and Hawkins, H.-J. (2016). The impact of crop rotation on soil microbial diversity: a meta-analysis. Pedobiologia 59, 215–223. doi: 10.1016/j.pedobi.2016.04.001
Wang, G., Dong, Y., Rong, L., Yang, W., and Duan, X. (2024). The influence of vegetation restoration on soil quality in abandoned farmlands in the Yuanjiang dry-hot valley. Catena 242:108109. doi: 10.1016/j.catena.2024.108109
Wang, Y., Ji, H., Wang, R., Guo, S., and Gao, C. (2017). Impact of root diversity upon coupling between soil C and N accumulation and bacterial community dynamics and activity: result of a 30year rotation experiment. Geoderma 292, 87–95. doi: 10.1016/j.geoderma.2017.01.014
Wang, W., Luo, X., Ye, X., Chen, Y., Wang, H., Wang, L., et al. (2020). Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land. Appl. Soil Ecol. 146:103365. doi: 10.1016/j.apsoil.2019.103365
Wilhelm, R. C., Amsili, J. P., Kurtz, K. S. M., van Es, H. M., and Buckley, D. H. (2023). Ecological insights into soil health according to the genomic traits and environment-wide associations of bacteria in agricultural soils. ISME Commun. 3:1. doi: 10.1038/s43705-022-00209-1
Yeates, G. W., Bongers, T., De Goede, R. G., Freckman, D. W., and Georgieva, S. S. (1993). Feeding habits in soil nematode families and genera-an outline for soil ecologists. J. Nematol. 25, 315–331.
Yin, X., Li, Q., Zhang, X., Wang, Z., Li, Q., Wei, L., et al. (2025). Paddy–upland rotation improves rice growth and reduces greenhouse gas emissions in winter paddy fields. Agronomy 15:349. doi: 10.3390/agronomy15020349
Zhang, H., Guo, D., Lei, Y., Lozano-Torres, J. L., Deng, Y., Xu, J., et al. (2025). Cover crop rotation suppresses root-knot nematode infection by shaping soil microbiota. New Phytol. 245, 363–377. doi: 10.1111/nph.20220
Zhang, X., Hu, B. X., Ren, H., and Zhang, J. (2018). Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 633, 518–528. doi: 10.1016/j.scitotenv.2018.03.158
Zhang, H., Luo, G., Wang, Y., Fei, J., Xiangmin, R., Peng, J., et al. (2023). Crop rotation-driven change in physicochemical properties regulates microbial diversity, dominant components, and community complexity in paddy soils. Agric. Ecosyst. Environ. 343:108278. doi: 10.1016/j.agee.2022.108278
Zhang, D., Sun, J., Peng, S., Wang, Y., Hua, Q., Wu, P., et al. (2025). Paddy-upland rotation combined with manure application: an optimal strategy for enhancing soil multifunctionality. J. Environ. Manag. 373:123788. doi: 10.1016/j.jenvman.2024.123788
Zhang, C., Wright, I. J., Nielsen, U. N., Geisen, S., and Liu, M. (2024). Linking nematodes and ecosystem function: a trait-based framework. Trends Ecol. Evol. 39, 644–653. doi: 10.1016/j.tree.2024.02.002
Zhang, C., Xue, W., Xue, J., Zhang, J., Qiu, L., Chen, X., et al. (2022). Leveraging functional traits of cover crops to coordinate crop productivity and soil health. J. Appl. Ecol. 59, 2627–2641. doi: 10.1111/1365-2664.14264
Zhou, N., Keffer, J. L., Polson, S. W., and Chan, C. S. (2022). Unraveling Fe(II)-oxidizing mechanisms in a facultative Fe(II) oxidizer, Sideroxydans lithotrophicus strain ES-1, via culturing, transcriptomics, and reverse transcription-quantitative PCR. Appl. Environ. Microbiol. 88:e0159521. doi: 10.1128/aem.01595-21
Zhou, W., Lv, T.-F., Chen, Y., Westby, A. P., and Ren, W.-J. (2014). Soil physicochemical and biological properties of paddy-upland rotation: a review. Sci. World J. 2014:856352. doi: 10.1155/2014/856352
Keywords: paddy-upland rotation, soil nematodes, microbial communities, soil biodiversity, functional group, sustainable agriculture
Citation: Pan S, Wu Y, Chen J, Wang S, Cai X, Wu P, Wang X, Yan C and Zheng Y (2025) Paddy-upland rotation improves soil quality by reshaping soil nematode and microbial communities. Front. Sustain. Food Syst. 9:1646192. doi: 10.3389/fsufs.2025.1646192
Edited by:
Liming Ye, Ghent University, BelgiumReviewed by:
Andrey S. Zaitsev, Senckenberg Museum of Natural History Görlitz, GermanyRodolfo Lizcano Toledo, Tolima University, Colombia
Gen Li, Nanjing Agricultural University, China
Copyright © 2025 Pan, Wu, Chen, Wang, Cai, Wu, Wang, Yan and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengjin Yan, eWNqMDY4NUAxNjMuY29t; Yongli Zheng, eW9uZ2xpemhlbmdAeWVhaC5uZXQ=
 Sufeng Pan
Sufeng Pan Yonghan Wu1
Yonghan Wu1 Yongli Zheng
Yongli Zheng