- 1School of Biotechnology, Jiangnan University, Wuxi, China
- 2Laboratory of Genomic and Precision Medicine, Wuxi School of Medicine, Jiangnan University, Wuxi, Jiangsu, China
- 3Wuxi Maternal and Child Healthcare Hospital, Wuxi, Jiangsu, China
Introduction: The activation of cerebral endothelial cells (CECs) has recently been reported to be the earliest acute neuroinflammation event in the CNS during sepsis-associated encephalopathy (SAE). Importantly, adenosine-to-inosine (A-to-I) RNA editing mediated by ADARs has been associated with SAE, yet its role in acute neuroinflammation in SAE remains unclear.
Methods: Our current study systematically analyzed A-to-I RNA editing in cerebral vessels, cerebral endothelial cells (CECs), and microglia sampled during acute neuroinflammation after treatment in a lipopolysaccharide (LPS)-induced SAE mouse model.
Results: Our results showed dynamic A-to-I RNA editing activity changes in cerebral vessels during acute neuroinflammation. Differential A-to-I RNA editing (DRE) associated with acute neuroinflammation were identified in these tissue or cells, especially missense editing events such as S367G in antizyme inhibitor 1 (Azin1) and editing events in lincRNAs such as maternally expressed gene 3 (Meg3), AW112010, and macrophage M2 polarization regulator (Mm2pr). Importantly, geranylgeranyl diphosphate synthase 1 (Ggps1) and another three genes were differentially edited across cerebral vessels, CECs, and microglia. Notably, Spearman correlation analysis also revealed dramatic time-dependent DRE during acute neuroinflammation, especially in GTP cyclohydrolase1 (Gch1) and non-coding RNA activated by DNA damage (Norad), both with the editing level positively correlated with both post-LPS treatment time and edited gene expression in cerebral vessels and CECs.
Discussion: The findings in our current study demonstrate substantial A-to-I RNA editing changes during acute neuroinflammation in SAE, underlining its potential role in the disease.
Introduction
Sepsis-related encephalopathy (SAE) is an acute progressive brain dysfunction caused by systemic inflammation without direct CNS infection (Kuperberg and Wadgaonkar, 2017). SAE is a common complication of sepsis, affecting up to 70% of patients, which can range from mild confusion to coma and is a significant predictor of mortality (Gofton and Young, 2012). Cerebrovascular dysfunction plays a vital role in the development of SAE. Sepsis triggers a cascade of inflammatory events that can damage the blood vessels in the brain, leading to impaired blood flow and oxygen delivery to brain tissue, and microglia activation predominates neuroinflammation (Xin et al., 2023). Animal studies on SAE have shown that the activation of cerebral endothelial cells (CECs) is the earliest event in the CNS during the onset of acute neuroinflammation, suggesting a substantial role for CECs in SAE pathogenesis.
Although the underlying mechanisms of SAE remain largely unclear, our previous study implicated a possible link between RNA editing and SAE (Zhang J.-Q. et al., 2022). The adenosine-to-inosine (A-to-I) editing mediated by the adenosine deaminase acting on RNA (ADAR) protein family (Gallo et al., 2017) in mammals plays an important function in neurodevelopment and neuropsychiatric diseases (Krestel and Meier, 2018; Gumpper et al., 2022), such as amyotrophic lateral sclerosis (ALS), developmental epileptic encephalopathy, and depression (Yang et al., 2021). Recent studies have implicated that ADAR could be involved in sepsis (Shangxun et al., 2020), and A-to-I RNA editing could possibly be linked to SAE, which could possibly be attributed to its importance in inflammation. ADAR expression is upregulated during acute inflammation (Yang et al., 2003). ADAR and its mediated A-to-I are involved in inflammation (Sun et al., 2021). In humans, ADAR deficiency in humans and mice could cause Aicardi–Goutières syndrome (AGS), which is a severe autoinflammatory disease (Rice et al., 2012; Nakahama et al., 2021). ADAR prevents autoinflammation by inhibiting the activator of apoptosis and necroptosis (de Reuver et al., 2022). Nevertheless, the role of RNA editing in SAE, especially during the onset of acute neuroinflammation, remains to be elucidated.
Herein, our current study conducted an epitranscriptomic analysis of A-to-I RNA editing and revealed dynamic RNA editing in cerebral vessels, CECs, and microglia during acute neuroinflammation in a mouse SAE model.
Materials and methods
RNA-Seq dataset retrieval
Raw RNA-Seq read data was downloaded from the NCBI Gene Expression Omnibus (GEO)1 database. The sequencing data of mouse cerebral vessels (GSE155516) used for RNA editing analysis contained a control group treated with PBS and treatment groups treated with 10 mg/kg lipopolysaccharide (LPS) for 15 min, 30 min, and 4 h (N = 3 for each group). The sequencing data of mouse CEC and microglia (GSE155517) contained a control group treated with PBS and treatment groups treated with LPS for 30 min, 1 h, and 2 h (N = 3 for each group) (Kodali et al., 2021).
RNA-Seq data processing and gene expression quantification
The raw sequencing data obtained above were analyzed following a pipeline as previously reported (Tao et al., 2021). In brief, Reads were aligned and mapped to the mouse genome (UCSC mm10) using RNA STAR (version 2.7.0e) (Dobin et al., 2013). Samtools (version 1.17) was used to filter the reads (Li et al., 2009). Base quality score recalibration was then performed with the resulting BAM files using GATK (version 4.1.3) and following the best practices workflows recommended by the documentation (Walker et al., 2018). Alignment files generated by RNA STAR were analyzed using FeatureCounts to obtain gene expression counts (Liao et al., 2014), and normalized gene expression levels (TPM) were calculated with edgeR (version 3.7) (Robinson et al., 2010).
Identification of high-confidence A-to-I RNA editing events
Single nucleotide variation (SNV) identification was performed using VarScan (version 2.4.4) (Koboldt et al., 2012) as previously reported (Zhang J.-Q. et al., 2022). Filtering criteria were set as base quality ≥25, sequencing depth ≥ 10, alternative allele depth ≥ 2, and frequency ≥ 1%, and false positive variants were filtered and removed using VarScan version 4.4 with default parameters. Further, high-confidence variants were retained, defined as those with editing levels ≥1% detected in at least two samples or annotated as known RNA editing variants in the REDIportal database (Mansi et al., 2021). SNVs were then annotated using the Ensembl Variant Effect Predictor (VEP)2 (McLaren et al., 2016).3
Enrichment analysis of gene functions and pathways
To understand the potential biological effects of RNA editing, Enrichr4 was used to analyze the gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathways enriched by edited genes and false discovery rate (FDR) < 0.05 was used as the cut-off (Kuleshov et al., 2016).
Statistical analysis
Comparison of RNA editing levels or gene expression levels between samples was performed using the ANOVA test or generalized linear model method and the likelihood ratio test to calculate the empirical p-values. Tukey’s Honest Significant Difference (HSD) test was used for post-hoc analysis between groups, and Benjamini-Hochberg correction was used for multiple comparisons. For RNA editing events with empirical GLM p < 0.05, an additional Fisher’s exact test was used to calculate Fisher’s P for intergroup comparisons of the total counts of the reference and alternative alleles among the four time-point groups. The Benjamini-Hochberg method was used to FDR adjustment of empirical p-values for multiple comparisons of RNA editing. RNA editing events were only considered to be differentially edited if both its (1) empirical GLM p < 0.05 and (2) GLM FDR < 0.05 or Fisher’s exact test FDR < 0.05. Principal component analysis (PCA) analysis was performed using the function Prcomp in R (version 3.6.3) software, and data visualization was performed using the ggplot2 (version 2.2.1) package (Tang et al., 2020). The correlation coefficient r and p-values were calculated using Spearman correlation analysis.
Results
A-to-I RNA editing activity changes in cerebral vessels, CECs, and microglia during acute neuroinflammation in SAE
We first looked into the expression of RNA editing enzymes, including Adar and Adarb1, in the SAE mouse model. The most profound expression changes in ADARs during acute neuroinflammation were found in cerebral vessels (Figures 1A,B). In cerebral vessels, Adar expression slightly decreased at 30 min but dramatically increased at 4 h after LPS treatment, whereas Adarb1 expression rapidly increased at 15 min, and decreased at 4 h after LPS treatment. After LPS treatment, the average A-to-I RNA editing levels and the number of observed editing events in cerebral vessels increased rapidly at 15 min, followed by a substantial decrease at 30 min and 4 h (Figures 1C,D). Spearman correlation analysis revealed a significant correlation between the average RNA editing level and Adarb1 expression (Figure 1E).
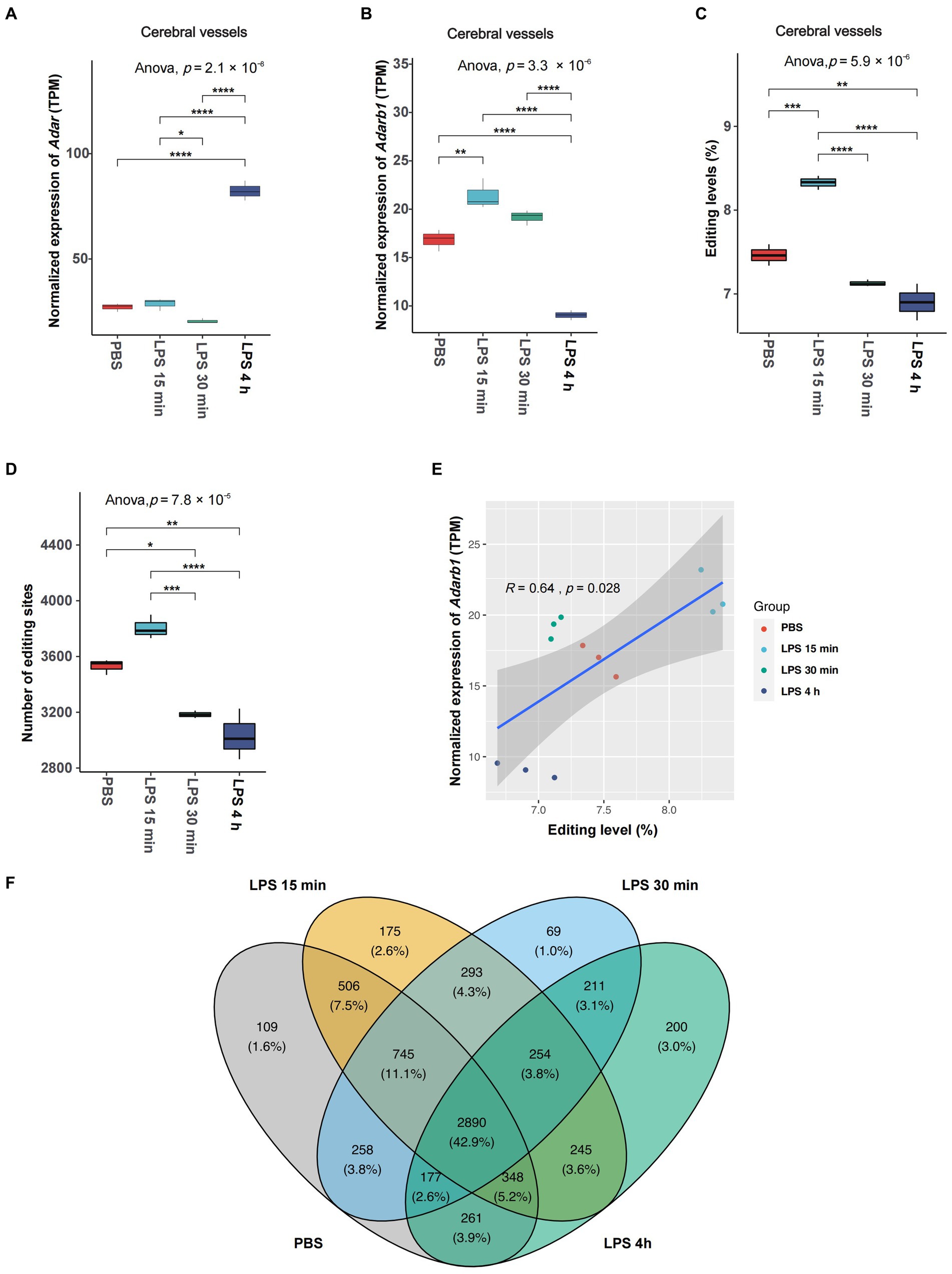
Figure 1. Overall changes in A-to-I RNA editing activities in cerebral vessels during acute neuroinflammation. (A,B) Expression of editing enzyme Adar and Adarb1 in cerebral vessels. (C) The average A-to-I RNA editing level and (D) number of editing events in cerebral vessels are shown. (E) Spearman correlation between the relative expression level and the avarge A-to-I RNA editing level in cerebral vessels. (F) Venn plot comparing A-to-I editing events detected in cerebral vessels among groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Although the expressions of Adar and Adarb1 or the number of observed editing events did not show significant changes in CECs during SAE (Supplementary Figure S1), the overall A-to-I RNA editing level significantly decreased 2 h after LPS treatment (Supplementary Figure S1C). Neither the expression of Adar and Adarb1 nor the average RNA editing level show significant changes in microglia during acute neuroinflammation (Supplementary Figures S2A–C).
The existence of individual RNA editing events was then compared among different time point groups. The Venn plot in Figure 1F shows that only 43.2% of the RNA editing events were observed across all groups in cerebral vessels, pointing to the high dynamics of RNA editing during SAE. Similar results were also observed in CECs (Supplementary Figure S1E) and microglia (Supplementary Figure S2E).
Annotation of A-to-I RNA editing events showed a relatively stable composition of functional categories during acute inflammation in SAE (Supplementary Figure S3). The largest proportion of A-to-I RNA editing events was 3′-untranslated region (UTR) editing in cerebral vessels and microglia and missense editing in CECs (Supplementary Figure S3).
Functional enrichment analysis also indicated dynamic changes of A-to-I RNA editing during acute neuroinflammation in SAE by identifying differential gene functions and pathways significantly enriched by edited genes in some time points but not others. The most differentially enriched biological processes and pathways in cerebral vessels are shown in Supplementary Figures S4A,B, such as vascular endothelial growth factor (VEGF) and receptor signaling, MAPK signaling, Apelin signaling, FoxO signaling, PI3K−Akt signaling, spliceosome, neuron projection development, and bacterial invasion of epithelial cells. The most differentially enriched biological processes and pathways in CECs are shown in Supplementary Figures S4C,D, such as protein localization to phagophore assembly site, positive regulation of protein dephosphorylation, regulation of cell cycle, positive regulation of mRNA metabolic process, protein K63−linked deubiquitination, cytoplasmic pattern recognition receptor signaling, Kaposi sarcoma−associated herpesvirus infection, B cell and T cell receptor signaling, and VEGF signaling. The most differentially enriched biological processes and pathways in microglia are shown in Supplementary Figures S4E,F, such as Huntington disease, pertussis, positive regulation of macroautophagy, cellular response to interleukin−6, mRNA catabolic process, Ras protein signal transduction, peroxisomal membrane transport, canonical Wnt signaling, protein K63−linked deubiquitination, mRNA surveillance, cytosolic DNA−sensing pathway, and RIG−I−like receptor signaling.
Differential A-to-I RNA editing during acute neuroinflammation in SAE
To identify the changes in RNA editing associated with acute neuroinflammation during SAE, the RNA editing levels of the sites among different time point groups were compared using the GLM method. 371 events in 295 genes in cerebral vessels (Supplementary Tables S1, S2), 355 events in 319 genes in CECs (Supplementary Tables S3, S4), and 85 events in 83 genes in microglia (Supplementary Tables S5, S6) showed DRE (Figures 2A–C). Importantly, most of the differentially edited genes showed differential expression during acute neuroinflammation, suggesting the potential effects of DRE on gene expression (Supplementary Tables S2, S4, S6). Moreover, the largest proportion of functional categories of these DRE events were 3′-UTR in cerebral vessels, CECs, and microglia, with a much higher proportion of intronic DRE in cerebral vessels than in CECs and microglia, as shown in Figure 2D. Notably, many DRE events in cerebral vessels were positively correlated with Adar and Adarb1 expressions (Figures 2E,F and Supplementary Tables S7, S8). PCA based on these DRE events showed that the samples of different time point groups could be well separated, with a large contribution rate of PC1 (cerebral vessels: 46.85%; CECs, 32.78%; and microglia, 33.03%, respectively) (Supplementary Figure S5).
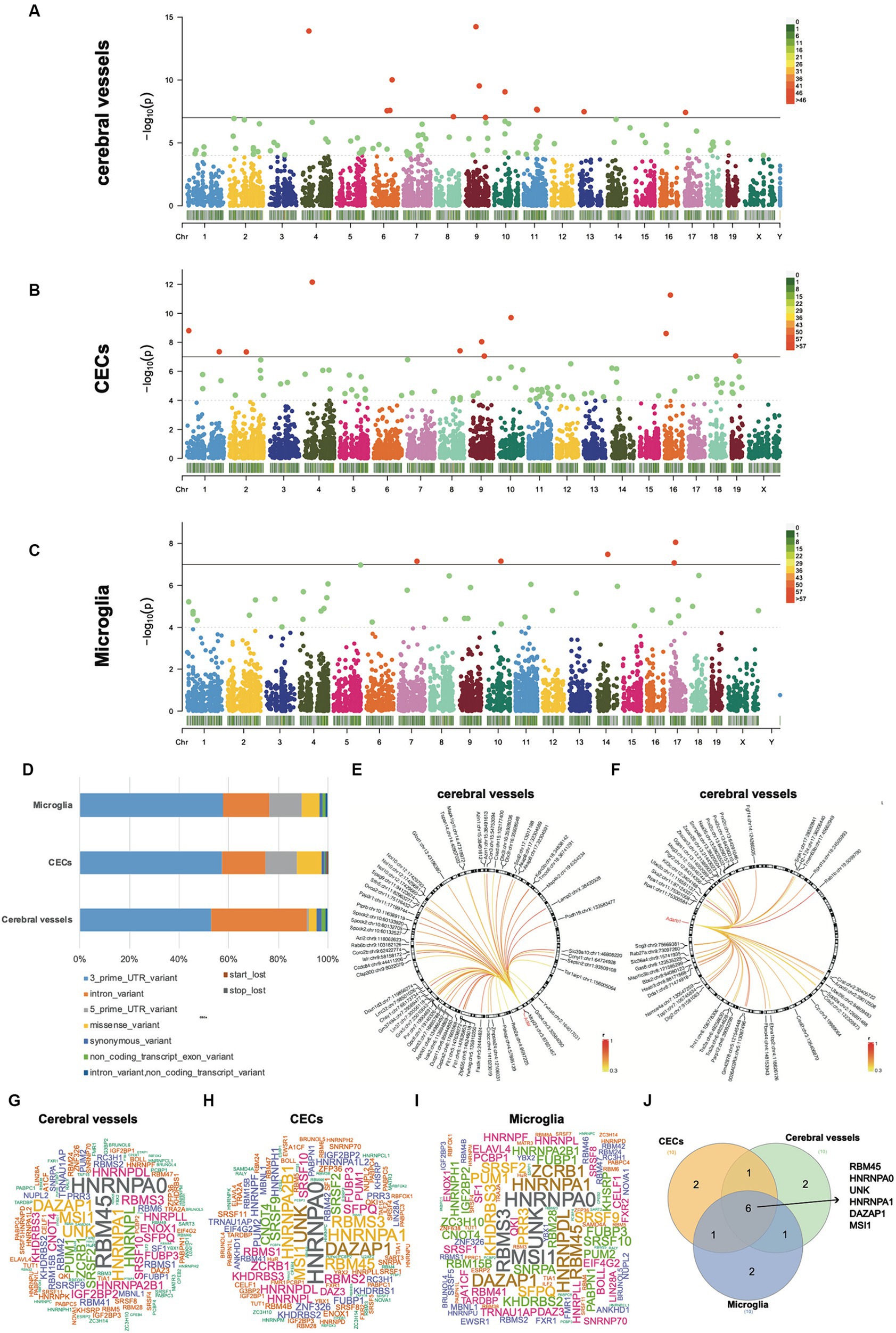
Figure 2. Differential A-to-I RNA editing during acute neuroinflammation. (A–C) Manhattan plots showing GLM p-values of individual A-to-I RNA editing events in cerebral vessels, CECs, and microglia. The solid horizontal line indicates p = 1.0 × 10−7, and the dashed horizontal line indicates p = 1.0 × 10−4. (D) Functional category distribution of DRE events in cerebral vessels, CECs, and microglia. (E,F) Differential RNA editing events associated with Adar and Adarb1 expressions in cerebral vessels. (G–I) Wordcloud plots showing the frequency of RBPs with binding sites overlapping with DRE sites. (J) Venn plot showing six RBPs shared by the top 10 frequent RBPs in the cerebral vessels, CECs, and microglia shown in panels (G–I). GLM, generalized linear model; DRE, differential RNA editing; RBPs, RNA binding proteins; CECs, cerebral endothelial cells.
Notably, several missense DRE events showed a more than 5% intergroup difference in editing levels during acute neuroinflammation (Table 1). Seven of these missense DRE events were found in cerebral vessels, including Azin1 p.S367G, Tmem63b p.Q619R, Grik5 p.K889R, Zfp771 p.K220R, Ube2o p.S93G, and Cacng8 p.S251G. Twelve were found in CECs, including Myo1e p.Q1009R, Sgpp1 p.T89A, Dhx36 p.E945G, Cdk13 p.Q103R and p.K96R, Cyfip2 p.K320E, Lrrc8b p.Q554R, Taf9b p.E40G, Plec p.H3003R, Ccni p.R75G, Akap1 p.S433G, and Lemd2 p.S226G. Only one, Tagap1 p.T187A, was found in Microglia. Notably, most of these differentially edited genes showed significantly differential expression (GLM FDR < 0.05) in the same tissue or cells during acute neuroinflammation. In addition, DRE was also observed in lincRNAs, such as maternally expressed gene 3 (Meg3) in cerebral vessels, AW112010 in CECs, and macrophage M2 polarization regulator (Mm2pr) in microglia.
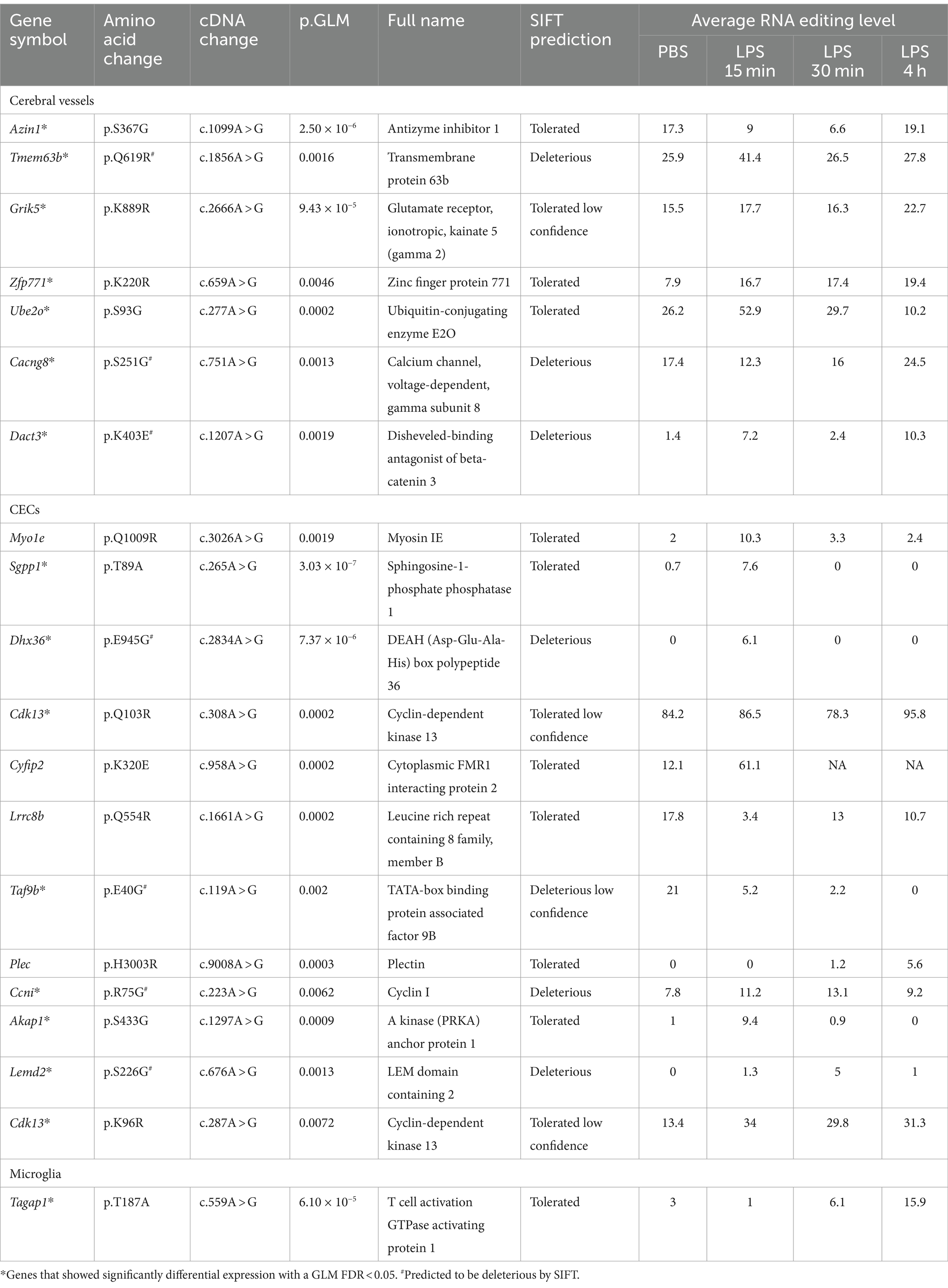
Table 1. Differential missense A-to-I editing with at least 5% intergroup editing level difference in cerebral vessels, CECs, and microglia during acute neuroinflammation.
The RBPmap tool was then used to predict how DRE events overlapped with and potentially affected RBP binding sites (Figures 2G–J). Our results showed that six RBPs, including RBM45, HNRNPA0, UNK, HNRNPA1, DAZAP1, and MSI1, were shared among the top 10 frequent RBPs with binding sites potentially affected by DRE. Interestingly, all these six RBP genes showed significant differential expression in cerebral vessels with empirical GLM p < 0.05. Dazap1, Hnrnpa0, and Unk were also differentially expressed in CECs, whereas only Hnrnpa1 was differentially expressed in microglia with empirical GLM p < 0.05 (Supplementary Table S9).
Four genes were differentially edited across cerebral vessels, CECs, and microglia (Figure 3A), including DnaJ Heat Shock Protein Family (Hsp40) member C18 (Dnajc18), OVCA2 Serine Hydrolase Domain Containing (Ovca2), Geranylgeranyl Diphosphate Synthase 1 (Ggps1) and Microfibril Associated Protein 1a (Mfap1a). Notably, 49 genes were differentially edited in both cerebral vessels and CECs, much more than those in both cerebral vessels (nine) or CECs (13) and microglia, pointing to a larger similarity of DRE between cerebral vessels and CECs. Gene ontology enrichment analysis further showed that these genes edited in both cerebral vessels and CECs were mainly involved in biological processes related to isoprenoid biosynthesis and response to misfolded protein and retinoic acid (Figure 3B).
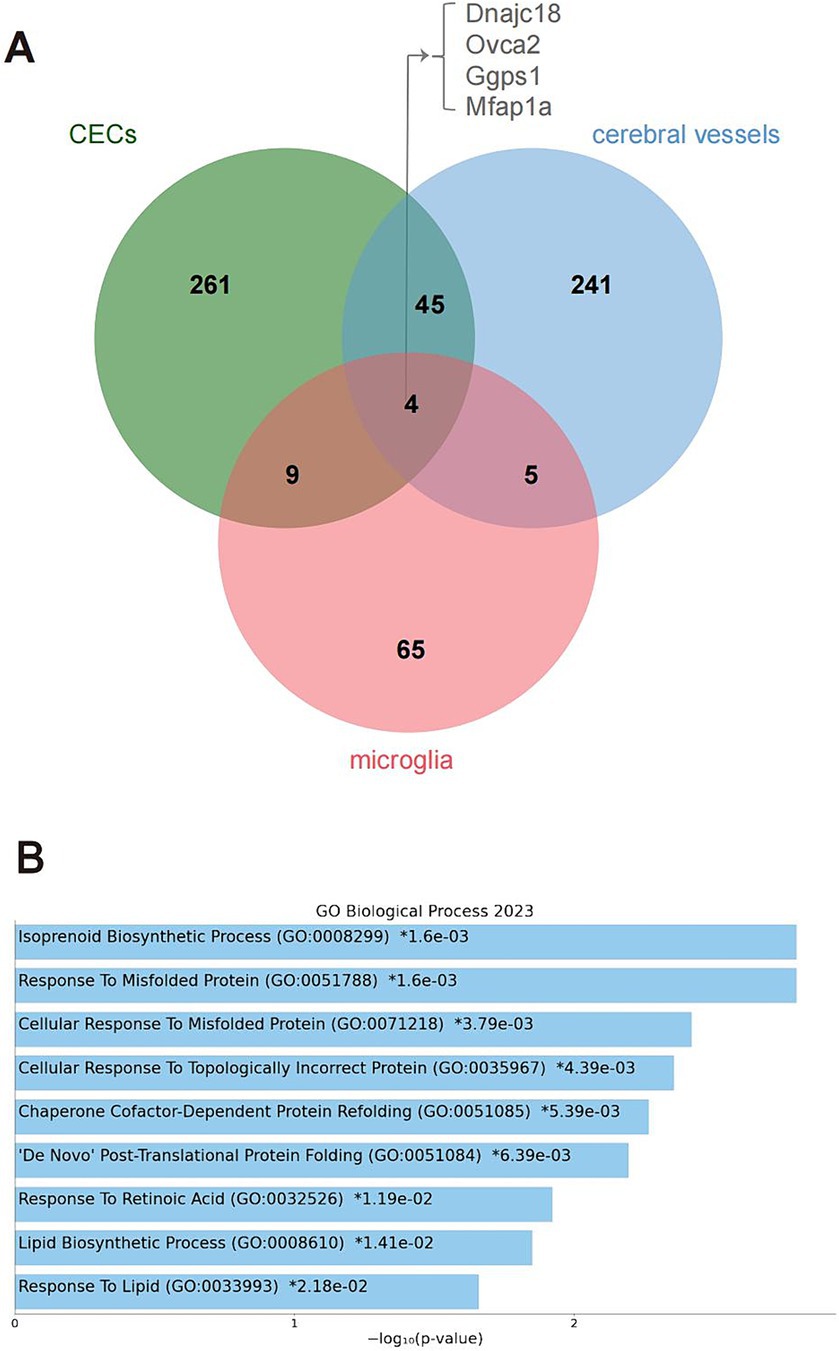
Figure 3. Genes differentially edited across cerebral vessels, CECs, and microglia during acute neuroinflammation. (A) Venn plot showing four common differentially edited genes in the tree tissues or cell types. (B) GO analysis showing biological processes significantly enriched by the four common differentially edited genes. CECs, cerebral endothelial cells; GO, gene ontology.
Time-dependent DRE during acute neuroinflammation
Spearman correlation analysis was used to identify time-dependent DRE events using time after LPS treatment as an independent continuous variable. Our results showed 304, 302, and 340 time-dependent DRE events in cerebral vessels (Supplementary Table S10), CECs (Supplementary Table S11), and microglia (Supplementary Table S12), respectively (all Spearman p < 0.05). Moreover, 11 time-dependent DRE events were shared by cerebral vessels and CECs, 11 were shared by CECs and microglia, and 5 were shared by cerebral vessels and microglia (Figure 4A). Among these time-dependent DRE events shared by cerebral vessels and CECs, two showed cis-regulatory effects on the edited gene expression, including 3′-UTR editing in GTP cyclohydrolase1 (Gch1) Gch1:chr14:47155050 and lincRNA editing in non-coding RNA activated by DNA damage (Norad) (Norad:chr2:156390419) (Figures 4D–K and Supplementary Tables S2, S4). The top 25 time-dependent events correlated with time in CECs and cerebral vessels, as shown in Figures 4B,C, most of which positively correlated with the edited gene expression. Notably, the correlation coefficient (r) was 0.95 and 0.81 for Gch1:chr14:47155050 in cerebral vessels (Figure 4E) and CECs (Figure 4G), respectively. Notably, the Gch1 3′-UTR RNA editing was observed only after LPS treatment but not in PBS controls. RBPmap prediction showed that Gch1:chr14:47155050 might affect the binding of PCBP to Gch1 mRNA (Supplementary Figure S6), an RBP highly expressed in the nervous system. Both the editing and expression levels of Norad decreased in cerebral vessels (Figures 4H,I) and CECs (Figures 4J,K), respectively.
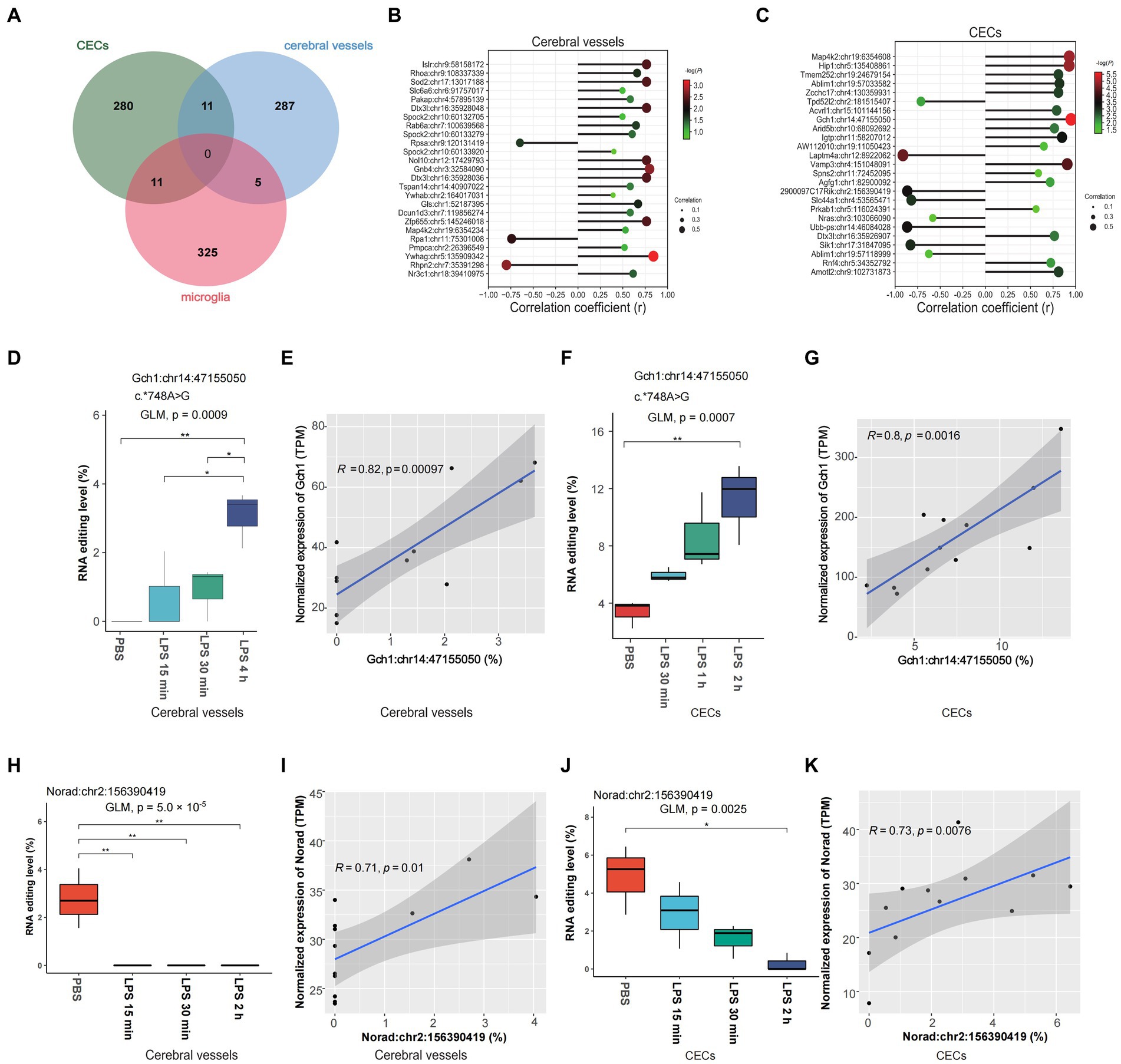
Figure 4. Time-dependent A-to-I RNA editing in cerebral vessels, CECs, and microglia during acute neuroinflammation. (A) Venn plot comparing the A-to-I editing events that highly correlated with time after LPS treatment (Spearman p < 0.05) in cerebral vessels, CECs, and microglia. (B,C) Spearman correlation of the top 25 differentially edited events strongly correlated with time in CECs and cerebral vessels. (D) RNA editing of Gch1:chr14:47155050 in cerebral vessels. (E) Spearman correlation between the editing level of Gch1:chr14:47155050 and Gch1 expression in cerebral vessels. (F) RNA editing of Gch1:chr14:47155050 in CECs. (G) Spearman correlation between the editing level of Gch1:chr14:47155050 and Gch1 expression in CECs. (H) RNA editing level of Norad:chr2:156390419 in cerebral vessels. (I) Correlation between levels of Norad:chr2:156390419 editing and Norad expression in cerebral vessels. (J) RNA editing level of Norad:chr2:156390419 in CECs. (K) The correlation between editing efficiency at Norad:chr2:156390419 editing and Norad expression in CECs. CECs, cerebral endothelial cells; Gch1, GTP cyclohydrolase1; Norad, non-coding RNA actived by DNA damage.
Discussion
Our previous study implicated the association of brain A-to-I RNA editing with sepsis, yet its role in SAE remains to be further investigated. Through transcriptome-wide analyses, our study investigated the temporal dynamics of A-to-I RNA editing in cerebral vessels, CECs, and microglia during acute neuroinflammation in septic mice, underlining the importance of such epigenetic changes in the disease.
Existing studies have shown widespread A-to-I RNA editing among genes expressed in the CNS (Behm and Öhman, 2016), which could play potentially important roles in neurodevelopment and brain function. Inflammation and immune Moreover, A-to-I RNA editing was also associated with the pathogenesis of neurological and psychiatric disorders, including amyotrophic lateral sclerosis, epilepsy, developmental epileptic encephalopathy, depression, and schizophrenia (Yang et al., 2021). In line with such a role, our current study highlighted the role of A-to-I RNA editing during acute neuroinflammation in SAE. Neuroinflammation, vascular dysfunction, and BBB disruption were thought to play an important role in the progression of SAE (Kikuchi et al., 2019). Activation of CECs resulted in compromised BBB and brain dysfunction. Microglia also contributed to neuroinflammation. In this study, dynamic A-to-I RNA editing changes observed in different tissues or cell types might contribute to an in-depth understanding of the pathogenesis during acute neuroinflammation in SAE.
Notably, functional enrichment analysis suggested the involvement of A-to-I RNA editing in common important gene functions and pathways dynamically altered during acute neuroinflammation in SAE. The most commonly altered biological functions and pathways were related to inflammation and immune response, in line with the neuroinflammatory characteristic of SAE. Two of the common pathways that showed changes between cerebral vessels and CECs were related to VEGF and VEGFR signaling pathway and MAPK signaling pathway. The VEGF family and VEGFRs play a crucial role in the health and function of cerebral vessels and their endothelial cells. Studies suggest VEGF signaling through VEGFR2 might have neuroprotective effects, promoting neuronal survival under hypoxic conditions (Wittko-Schneider, et al., 2013; Silva-Hucha et al., 2021), and brain expression of VEGFB is associated with cognitive aging and Alzheimer’s disease (Mahoney et al., 2021).
Our analysis revealed numerous missense A-to-I RNA editing associated with acute neuroinflammation in various tissues or cell types, suggesting a potential role of A-to-I RNA editing in recoding proteins in SAE. As suggested by the SIFT prediction, these missense editing events could have functional impacts on the encoded protein. Notably, the differential Azin1 S367G editing in cerebral vessels is conserved between humans and mice, which has been found to enhance cancer cell stemness, promote tumor angiogenesis, and might drive metastasis (Han et al., 2015; Wei et al., 2022). Our finding thus added to the biological functions of Azin1 S367G editing by showing its involvement during acute neuroinflammation in SAE. In addition, our findings also suggested a potential involvement of lincRNA editing during acute neuroinflammation in SAE. Meg3 could promote Nlrp3-mediated inflammation in microglia, and its elevated expression could induce endothelial dysfunction (Jiang et al., 2022). LncRNA Aw112010 has recently been found to be a key modulator of inflammation. AW112010 could promote inflammatory T-cell differentiation by suppressing IL-10 expression through demethylation of H3K4, and its elevated expression might increase during monocyte aging (Barman et al., 2022). Intriguing, Mm2pr, a lincRNA differentially edited in microglia during acute neuroinflammation in our current study, has been recently identified as an essential modulator for M2 macrophage polarization with a potential role in macrophage-promoted tumorigenesis (Cao et al., 2019). Such differential lincRNA editing might modulate the acute neuroinflammation process in SAE, which warrants further investigation.
Our current study identified genes differentially edited across cerebral vessels, CECs, and microglia, which could be hotspots of A-to-I RNA editing during acute neuroinflammation. Dnajc18 encodes a protein of the DNAJ family, which is highly expressed in the brain and plays a key role in neurodegenerative disorders, such as Parkinson’s Disease, and may also be involved in ubiquitin-dependent ERAD pathway and cellular response to misfolded protein (Huang et al., 2019; Zhang K. et al., 2022). In addition, Dnajc18 has recently been identified to be involved in congenital and structural heart disorders and cardiomyopathy in both humans and mice (Spielmann et al., 2022). Ovca2 encodes a serine hydrolase and is downregulated and degraded during retinoid-induced apoptosis (Prowse et al., 2002), with its role in SAE unknown. Ggps1 encodes a protein belonging to the prenyltransferase family, and its knockout ameliorates ventilator-induced lung injury by regulating TLR2/4-AP-1 signaling (Wan et al., 2020). Ggps1 is also involved in muscular dystrophy and promotes rab37-mediated autophagy (Tucker et al., 2020; Kaiyrzhanov et al., 2022; Wang et al., 2022).
In addition, our study revealed time-dependent dynamic changes in A-to-I RNA editing during acute neuroinflammation, especially Gch1: chr14:47155050. Gch1 encodes a member of the GTP cyclohydrolase family, which could be upregulated in the vascular wall during inflammation and in endothelial cells stimulated by cytokines such as IL-6, hs-CRP, and LPS (Antoniades et al., 2011). Increased GCH1 expression leads to greater tetrahydrobiopterin (BH4) production, which acts as a defense mechanism against systemic inflammation by helping maximize eNOS function and maintaining endothelial function. Moreover, GCH1 also plays a role in inflammatory and peripheral neuropathic pain (Nasser et al., 2013; Xiao et al., 2023). Gch1 variants have recently been associated with the risk and age of onset of Parkinson’s disease (Pan et al., 2020). The impact of A-to-I RNA editing at Gch1:chr14:47155050 on SAE remained unanswered, but it could possibly be involved in BH4 production by regulating Gch1 expression. In addition, NORAD in humans encodes a lincRNA reported to inhibit vascular endothelial cell senescence and apoptosis (Bian et al., 2020) and has a potential role in neurological diseases (Wang B. et al., 2020). The findings of time-dependent DRE in Gch1 and Norad warranted further study on the role of RNA editing in cerebral vessels and CECs in SAE.
In conclusion, our current study demonstrates dynamic alterations in A-to-I RNA editing in cerebral vessels, CECs, and microglia in a mouse model during acute neuroinflammation in SAE. Therefore, our findings provided new insight into understanding the role played by A-to-I RNA editing in SAE.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo under accession IDs GSE155516 and GSE155517.
Ethics statement
The requirement of ethical approval was waived by the Ethics Committee of Jiangnan University for the studies involving animals because the study was a reanalysis of publicly available datasets. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
Y-NL: Data curation, Formal analysis, Writing – original draft. Y-PL: Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. J-QZ: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. NL: Conceptualization, Methodology, Writing – review & editing. Z-YW: Methodology, Software, Writing – review & editing. YR: Writing – review & editing. J-HC: Conceptualization, Data curation, Supervision, Writing – review & editing. Y-YJ: Formal analysis, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by grants from the National Natural Science Foundation of China (No. 31671311, 82302492), the “Six Talent Peak” Plan of Jiangsu Province (No. SWYY-127), the Innovative and Entrepreneurial Talents of Jiangsu Province, the Program for High-Level Entrepreneurial and Innovative Talents of Jiangsu Province, Natural Science Foundation of Guangdong Province/Guangdong Basic and Applied Basic Research Foundation (2019A1515012062), Taihu Lake Talent Plan, and Fundamental Research Funds for the Central Universities (JUSRP51712B and JUSRP1901XNC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer QL declared a past co-authorship with the author J-HC to the handling editor.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2024.1435185/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | RNA editing activities in CECs during acute neuroinflammation. (A,B) Expression of editing enzyme Adar and Adarb1 in CECs and (C,E) A-to-I RNA editing events identified from mouse CECs. (C) A-to-I RNA editing levels and (D) number of editing sites in cerebral vessels are shown. (E) Venn plot comparing the A-to-I editing sites detected in 2 or more samples among groups.
SUPPLEMENTARY FIGURE S2 | RNA editing activities in microglia during acute neuroinflammation. (A,B) Expression of editing enzyme Adar and Adarb1 in microglia and (C–E) A-to-I RNA editing events identified from mouse microglia. (C) A-to-I RNA editing levels and (D) number of editing sites in cerebral vessels are shown. (E) Venn plot comparing the A-to-I editing sites detected in 2 or more samples among groups.
SUPPLEMENTARY FIGURE S3 | A-to-I RNA editing variants during acute neuroinflammation. Results are shown for (A) cerebral vessels, (B) CECs, and (C) microglia.
SUPPLEMENTARY FIGURE S4 | The most differentially enriched biological processes and pathways during acute neuroinflammation. Results of GO biological processes and KEGG pathways are shown for (A,B) cerebral vessels, (C,D) CECs, and (E,F) microglia.
SUPPLEMENTARY FIGURE S5 | Principle component analysis of the A-to-I RNA editing events differentially edited between different groups during acute neuroinflammation. Results are shown for (A) cerebral vessels, (B) CECs, and (C) microglia.
SUPPLEMENTARY FIGURE S6 | The results of overlapping RBP binding sites of Gch1:chr14:47155050 using the RBPmap.
Footnotes
1. ^ https://www.ncbi.nlm.nih.gov/geo/
2. ^ https://www.ensembl.org/vep
References
Antoniades, C., Cunnington, C., Antonopoulos, A., Neville, M., Margaritis, M., Demosthenous, M., et al. (2011). Induction of vascular GTP-Cyclohydrolase I and endogenous tetrahydrobiopterin synthesis protect against inflammation-induced endothelial dysfunction in human atherosclerosis. Circulation 124, 1860–1870. doi: 10.1161/CIRCULATIONAHA.111.029272
Barman, P. K., Shin, J. E., Lewis, S. A., Kang, S., Wu, D., Wang, Y., et al. (2022). Production of MHCII-expressing classical monocytes increases during aging in mice and humans. Aging Cell 21:e13701. doi: 10.1111/acel.13701
Behm, M., and Öhman, M. (2016). RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 32, 165–175. doi: 10.1016/j.tig.2015.12.005
Bian, W., Jing, X., Yang, Z., Shi, Z., Chen, R., Xu, A., et al. (2020). Downregulation of LncRNA NORAD promotes ox-LDL-induced vascular endothelial cell injury and atherosclerosis. Aging 12, 6385–6400. doi: 10.18632/aging.103034
Cao, J., Dong, R., Jiang, L., Gong, Y., Yuan, M., You, J., et al. (2019). LncRNA-MM2P identified as a modulator of macrophage M2 polarization. Cancer Immunol. Res. 7, 292–305. doi: 10.1158/2326-6066.CIR-18-0145
de Reuver, R., Verdonck, S., Dierick, E., Nemegeer, J., Hessmann, E., Ahmad, S., et al. (2022). ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607, 784–789. doi: 10.1038/s41586-022-04974-w
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Gallo, A., Vukic, D., Michalík, D., O’Connell, M. A., and Keegan, L. P. (2017). ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 136, 1265–1278. doi: 10.1007/s00439-017-1837-0
Gofton, T. E., and Young, G. B. (2012). Sepsis-associated encephalopathy. Nat. Rev. Neurol. 8, 557–566. doi: 10.1038/nrneurol.2012.183
Gumpper, R. H., Fay, J. F., and Roth, B. L. (2022). Molecular insights into the regulation of constitutive activity by RNA editing of 5HT2C serotonin receptors. Cell Rep. 40:111211. doi: 10.1016/j.celrep.2022.111211
Han, L., Diao, L., Yu, S., Xu, X., Li, J., Zhang, R., et al. (2015). The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 28, 515–528. doi: 10.1016/j.ccell.2015.08.013
Huang, Y., Arora, K., Mun, K. S., Yang, F., Moon, C., Yarlagadda, S., et al. (2019). Targeting DNAJB9, a novel ER luminal co-chaperone, to rescue ΔF508-CFTR. Sci. Rep. 9, 9808. doi: 10.1038/s41598-019-46161-4
Jiang, Y., Zhu, H., Chen, H., Yu, Y.-C., Xu, Y.-T., Liu, F., et al. (2022). Elevated expression of lncRNA MEG3 induces endothelial dysfunction on HUVECs of IVF born offspring via epigenetic regulation. Front. Cardiovasc. Med. 8:717729. doi: 10.3389/fcvm.2021.717729
Kaiyrzhanov, R., Perry, L., Rocca, C., Zaki, M. S., Hosny, H., Araujo Martins Moreno, C., et al. (2022). GGPS1-associated muscular dystrophy with and without hearing loss. Ann. Clin. Transl. Neurol. 9, 1465–1474. doi: 10.1002/acn3.51633
Kikuchi, D. S., Campos, A. C. P., Qu, H., Forrester, S. J., Pagano, R. L., Lassègue, B., et al. (2019). Poldip2 mediates blood-brain barrier disruption in a model of sepsis-associated encephalopathy. J. Neuroinflammation 16:241. doi: 10.1186/s12974-019-1575-4
Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D., Lin, L., et al. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. doi: 10.1101/gr.129684.111
Kodali, M. C., Chen, H., and Liao, F.-F. (2021). Temporal unsnarling of brain’s acute neuroinflammatory transcriptional profiles reveals panendothelitis as the earliest event preceding microgliosis. Mol. Psychiatry 26, 3905–3919. doi: 10.1038/s41380-020-00955-5
Krestel, H., and Meier, J. C. (2018). RNA editing and retrotransposons in neurology. Front. Mol. Neurosci. 11:163. doi: 10.3389/fnmol.2018.00163
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377
Kuperberg, S. J., and Wadgaonkar, R. (2017). Sepsis-associated encephalopathy: the blood-brain barrier and the sphingolipid rheostat. Front. Immunol. 8:597. doi: 10.3389/fimmu.2017.00597
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Mahoney, E. R., Dumitrescu, L., Moore, A. M., Cambronero, F. E., De Jager, P. L., Koran, M. E. I., et al. (2021). Brain expression of the vascular endothelial growth factor gene family in cognitive aging and Alzheimer’s disease. Mol. Psychiatry 26, 888–896. doi: 10.1038/s41380-019-0458-5
Mansi, L., Tangaro, M. A., Lo Giudice, C., Flati, T., Kopel, E., Schaffer, A. A., et al. (2021). REDIportal: millions of novel A-to-I RNA editing events from thousands of RNAseq experiments. Nucleic Acids Research 49, D1012–D1019. doi: 10.1093/nar/gkaa916
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R. S., Thormann, A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17:122. doi: 10.1186/s13059-016-0974-4
Nakahama, T., Kato, Y., Shibuya, T., Inoue, M., Kim, J. I., Vongpipatana, T., et al. (2021). Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutières-syndrome-like encephalopathy. Immunity 54, 1976–1988.e7. doi: 10.1016/j.immuni.2021.08.022
Nasser, A., Bjerrum, O. J., Heegaard, A.-M., Møller, A. T., Larsen, M., Dalbøge, L. S., et al. (2013). Impaired behavioural pain responses in hph-1 mice with inherited deficiency in GTP cyclohydrolase 1 in models of inflammatory pain. Mol. Pain 9:5. doi: 10.1186/1744-8069-9-5
Pan, H.-X., Zhao, Y.-W., Mei, J.-P., Fang, Z.-H., Wang, Y., Zhou, X., et al. (2020). GCH1 variants contribute to the risk and earlier age-at-onset of Parkinson’s disease: a two-cohort case-control study. Transl. Neurodegener. 9:31. doi: 10.1186/s40035-020-00212-3
Prowse, A. H., Vanderveer, L., Milling, S. W. F., Pan, Z.-Z., Dunbrack, R. L., Xu, X.-X., et al. (2002). OVCA2 is downregulated and degraded during retinoid-induced apoptosis. Int. J. Cancer 99, 185–192. doi: 10.1002/ijc.10334
Rice, G. I., Kasher, P. R., Forte, G. M. A., Mannion, N. M., Greenwood, S. M., Szynkiewicz, M., et al. (2012). Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248. doi: 10.1038/ng.2414
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Shangxun, Z., Junjie, L., Wei, Z., Yutong, W., Wenyuan, J., Shanshou, L., et al. (2020). ADAR1 alleviates inflammation in a murine sepsis model via the ADAR1-miR-30a-SOCS3 axis. Mediat. Inflamm. 2020, 9607535–9607515. doi: 10.1155/2020/9607535
Silva-Hucha, S., Pastor, A. M., and Morcuende, S. (2021). Neuroprotective effect of vascular endothelial growth factor on motoneurons of the oculomotor system. Int. J. Mol. Sci. 22:814. doi: 10.3390/ijms22020814
Spielmann, N., Miller, G., Oprea, T. I., Hsu, C.-W., Fobo, G., Frishman, G., et al. (2022). Extensive identification of genes involved in congenital and structural heart disorders and cardiomyopathy. Nat. Cardiovasc. Res. 1, 157–173. doi: 10.1038/s44161-022-00018-8
Sun, T., Yu, Y., Wu, X., Acevedo, A., Luo, J.-D., Wang, J., et al. (2021). Decoupling expression and editing preferences of ADAR1 p150 and p110 isoforms. Proc. Natl. Acad. Sci. U. S. A. 118:e2021757118. doi: 10.1073/pnas.2021757118
Tang, S., Jing, H., Huang, Z., Huang, T., Lin, S., Liao, M., et al. (2020). Identification of key candidate genes in neuropathic pain by integrated bioinformatic analysis. J. Cell. Biochem. 121, 1635–1648. doi: 10.1002/jcb.29398
Tao, J., Ren, C.-Y., Wei, Z.-Y., Zhang, F., Xu, J., and Chen, J.-H. (2021). Transcriptome-wide identification of G-to-a RNA editing in chronic social defeat stress mouse models. Front. Genet. 12:680548. doi: 10.3389/fgene.2021.680548
Tucker, E. J., Rius, R., Jaillard, S., Bell, K., Lamont, P. J., Travessa, A., et al. (2020). Genomic sequencing highlights the diverse molecular causes of Perrault syndrome: a peroxisomal disorder (PEX6), metabolic disorders (CLPP, GGPS1), and mtDNA maintenance/translation disorders (LARS2, TFAM). Hum. Genet. 139, 1325–1343. doi: 10.1007/s00439-020-02176-w
Walker, M. A., Pedamallu, C. S., Ojesina, A. I., Bullman, S., Sharpe, T., Whelan, C. W., et al. (2018). GATK PathSeq: a customizable computational tool for the discovery and identification of microbial sequences in libraries from eukaryotic hosts. Bioinformatics 34, 4287–4289. doi: 10.1093/bioinformatics/bty501
Wan, B., Xu, W.-J., Chen, M.-Z., Sun, S.-S., Jin, J.-J., Lv, Y.-L., et al. (2020). Geranylgeranyl diphosphate synthase 1 knockout ameliorates ventilator-induced lung injury via regulation of TLR2/4-AP-1 signaling. Free Radic. Biol. Med. 147, 159–166. doi: 10.1016/j.freeradbiomed.2019.12.024
Wang, Z., Chen, M., Pan, X., Wang, L., Yin, C., Lin, Q., et al. (2022). Knockout of GGPPS1 restrains rab37-mediated autophagy in response to ventilator-induced lung injury. Hum. Cell 35, 871–884. doi: 10.1007/s13577-022-00692-7
Wang, B., Xu, L., Zhang, J., Cheng, X., Xu, Q., Wang, J., et al. (2020). LncRNA NORAD accelerates the progression and doxorubicin resistance of neuroblastoma through up-regulating HDAC8 via sponging miR-144-3p. Biomed. Pharmacother. 129:110268. doi: 10.1016/j.biopha.2020.110268
Wei, Y., Zhang, H., Feng, Q., Wang, S., Shao, Y., Wu, J., et al. (2022). A novel mechanism for A-to-I RNA-edited AZIN1 in promoting tumor angiogenesis in colorectal cancer. Cell Death Dis. 13, 1–12. doi: 10.1038/s41419-022-04734-8
Wittko-Schneider, I. M., Schneider, F. T., and Plate, K. H. (2013). Brain homeostasis: VEGF receptor 1 and 2—two unequal brothers in mind. Cell Mol. Life Sci. 70, 1705–1725. doi: 10.1007/s00018-013-1279-3
Xiao, Y., Yuan, Y., Yang, Y., Liu, B., Ding, Z., Luo, J., et al. (2023). GCH1 reduces LPS-induced alveolar macrophage polarization and inflammation by inhibition of ferroptosis. Inflamm. Res. 72, 1941–1955. doi: 10.1007/s00011-023-01785-1
Xin, Y., Tian, M., Deng, S., Li, J., Yang, M., Gao, J., et al. (2023). The key drivers of brain injury by systemic inflammatory responses after sepsis: microglia and neuroinflammation. Mol. Neurobiol. 60, 1369–1390. doi: 10.1007/s12035-022-03148-z
Yang, J.-H., Nie, Y., Zhao, Q., Su, Y., Pypaert, M., Su, H., et al. (2003). Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278, 45833–45842. doi: 10.1074/jbc.M308612200
Yang, Y., Okada, S., and Sakurai, M. (2021). Adenosine-to-inosine RNA editing in neurological development and disease. RNA Biol. 18, 999–1013. doi: 10.1080/15476286.2020.1867797
Zhang, J.-Q., Pan, J.-Q., Wei, Z.-Y., Ren, C.-Y., Ru, F.-X., Xia, S.-Y., et al. (2022). Brain Epitranscriptomic analysis revealed altered A-to-I RNA editing in septic patients. Front. Genet. 13:887001. doi: 10.3389/fgene.2022.887001
Keywords: RNA editing, sepsis-associated encephalopathy, acute neuroinflammation, cerebral vessels, cerebral endothelial cells, microglia
Citation: Li Y-N, Liang Y-P, Zhang J-Q, Li N, Wei Z-Y, Rao Y, Chen J-H and Jin Y-Y (2024) Dynamic A-to-I RNA editing during acute neuroinflammation in sepsis-associated encephalopathy. Front. Neurosci. 18:1435185. doi: 10.3389/fnins.2024.1435185
Edited by:
Zhexing Wen, Emory University School of Medicine, United StatesReviewed by:
Jie Zhang, Third People’s Hospital of Zhongshan City, ChinaBo Yan, Fudan University, China
Qiyang Li, Southern Medical University, China
Copyright © 2024 Li, Liang, Zhang, Li, Wei, Rao, Chen and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yijian Rao, cmFveWlqaWFuQGppYW5nbmFuLmVkdS5jbg==; Jian-Huan Chen, Y2poX2Jpb0Bob3RtYWlsLmNvbQ==; Yun-Yun Jin, amlueXVueXVuQGppYW5nbmFuLmVkdS5jbg==
†These authors have contributed equally to this work
 Yu-Ning Li1
Yu-Ning Li1 Jian-Huan Chen
Jian-Huan Chen