- 13D Structure Department, Fundación Instituto de Inmunología de Colombia, Bogotá, Colombia
- 2School of Medicine and Health Sciences Faculty, Universidad del Rosario, Bogotá, Colombia
- 3Medicine Faculty, Universidad de Ciencias Aplicadas y Ambientales, Bogotá, Colombia
- 4Medicine Faculty, Universidad Nacional de Colombia, Bogotá, Colombia
The 3D structural analysis of 62 peptides derived from highly pathogenic Plasmodium falciparum malaria parasite proteins involved in host cell invasion led to finding a striking association between particular β-turn types located in the N-terminal peripheral flanking residue region (preceding the polyproline II left-handed structures fitting into the HLA-DRβ* allele family) and modified immune protection-inducing protein structure induced long-lasting protective immunity. This is the first time association between two different secondary structures associated with a specific immunological function has been described: full, long-lasting protective immunity.
Introduction
In the search for a logical and rational methodology for vaccine development, we have proposed that in-depth, chemical, physical, structural and even mathematical approaches associated with an understanding of molecules' biological functions. We have thus consistently adopted such approach working with our prototype model, the threatening and scourging Plasmodium falciparum malaria parasite which afflicts around 200 million people, killing nearly 600,000 of them annually, mainly children below 5 years of age in sub-Saharan Africa (World Malaria Report, 20161).
This approach has involved molecular biology as well as functional analysis of the P. falciparum malaria parasite, enabling new molecules and their functions to be recognized (Wåhlin et al., 1984), as well as that of natural (human) and experimental (Aotus monkeys) host immune system molecules (Suárez et al., 2006, 2017; Lopez et al., 2014). This has facilitated the recognition of chemical, physical and structural rules regarding their interactions for tackling and resolving functional issues (Patarroyo et al., 2011). Here we show that vaccine component structural features are determinant regarding fully-protective, long-lasting protective immunity.
Vaccine components' 3D-structure can be assessed by 1H-NMR or X-ray diffraction; the protein structure work by many groups throughout the world has led to advances regarding knowledge about protein and peptide 3D structure and knowledge of the immune system molecules involved in major histocompatibility complex (http://www.cbs.dtu.dk/services/NetMHCIIpan/) Class I and Class II (HLA-DRβ*) antigen presentation to the T-cell receptor (TCR) to form the MHCII-peptide-TCR complex or immunological synapse to induce an appropriate immune response (Hennecke and Wiley, 2002; Rudolph et al., 2006). We have previously shown that immune protection-inducing protein structures (IMPIPS) (Patarroyo et al., 2015a) have a polyproline type II, left-handed-like (PPIIL-like) structure to enable fitting into the appropriate HLA-DRβ1* peptide binding region (PBR) (Patarroyo et al., 2012a, 2015a).
This manuscript is aimed at showing that, in the search for long-lasting, protective immunity (LLPI), these PPIIL-like structures must be preceded by specific and particular β-turn structures which, when preceding other structures like αR-helixes and/ or other β-turn types, could also activate immune system molecules whilst inducing short-lived protective immunity (SPI) structures. When associated with PPIIL amino acid sequences binding to the HLA-DRβ1* PBR having appropriate TCR contacting residue orientation, modified high activity binding peptides (mHABPs) can induce LLPI, which we have named IMPIPS (Patarroyo et al., 2015a; Alba et al., 2016). Specific preference for some of these β-turns and the complete absence of some others is also shown, as is the preferred association between some residues in both β-turns and PPIIL PBR sequences leading to high antibody titres and LLPI induction.
Materials and Methods
Ethics Statement, Animal Capture and Study Area
The current work was approved by the Fundación Instituto de Inmunología's ethics committee (FIDIC ethics committee). The capture, study, and scientific research of Aotus primates were authorized by the official Colombian environmental authority, CORPOAMAZONIA, (resolutions 0066/Sep/2006, 0028/May/2010, 0632/Jun/2010, 0042/Jan/2011 and 1209/Sep/2017). All animal-handling procedures were carried out according to the Guide for the Care and Use of Laboratory Animals, USA (National Research Council, 2011); such recommendations comply with Colombian regulations for biomedical research (resolution 8430/1993 and law 84/1989). CORPOAMAZONIA made a weekly visit to evaluate housing conditions, feeding regimens and the environmental enrichment of the monkeys captured. The monkeys were supervised by veterinarians and biologists; all individuals were released back into the Amazon jungle after the experimental procedures in optimal health conditions as determined by the Amazonian ethical committee and in the presence of CORPOAMAZONIA representatives.
Synthetic Peptides
Peptides were selected from several P. falciparum proteins as being relevant to the present study; cHABPs and mHABPs were thereby obtained by chemical synthesis. The solid-phase peptide synthesis (SPPS) method was used, following t-Boc strategy (Houghten, 1985). Cys-Gly residues were added to HABP C- and N-terminals during synthesis to enable polymerization and their complete characterization. Their purity was assessed by reverse-phase high-performance liquid chromatography (RP-HPLC) and their molecular mass was determined by mass spectrometry (MS-MALDI-TOF). Most have been synthesized in different studies throughout the last few years.
NMR Spectroscopy and Structural Models
This involved a protocol which has been used for several years for obtaining peptides' 3D structure by 1H-NMR. Native peptides (cHABPs) and their modified forms (mHABPs) from around 14 relevant proteins were prepared for 1H-NMR studies by dissolving ~10 mg monomer acetylated peptide in 600 μL TFE-d3/H2O 30:70% v/v. Spin systems were assigned by Double-Quantum-Filtered-Correlation SpectroscopY (DFQ-COSY) (Rance et al., 1983) and Total Correlation SpectroscopY (TOCSY) (Bax and Davis, 1985) experiments and Nuclear Overhauser Effect SpectroscopY (NOESY) (Jeener et al., 1979) 1H-1H 2D experiments. Standard spectrum procedure was used for sequential assignment. All NMR spectra were run on a Bruker DRX-600 spectrometer and processed on a computer equipped with TOPSPIN 1.3 software. All experiments were done at 295 K, except for temperature coefficients for predicting hydrogen bonds (−Δ∂HN/ΔT < 5) which were determined by TOCSY experiments, using 285, 295, 305, and 315 K temperatures.
Distance constraints were extracted from NOESY spectra for obtaining structural models at 295 K temperature. All NOE intensities were converted into distance ranges as strong (1.8–2.8 Å), medium (2.8–3.5 Å) or weak (3.5–5.0 Å), along with a.rstrnt file. These, together with .inp, .car, .mdf input files and Insight II package (Accelrys, Inc, Software, USA), or .upl, .lol, .cya and .aco input files and Cyana software were used for calculating peptide structures. Distance geometry (DGII) software was used for building a set of conformers for 50 structures; these structures were then refined by using a simulated annealing protocol (Discover and Cyana software). Structures having low energy and NOE violations greater than 0.30 Å and no angle violations greater than ≥2.8° were then selected. The final distinctive low energy family, was assembled from consensus conformer lowest energy and superimposing the structured regions' backbones on structures obtained by 1H-NMR restrictions. The structure chosen have been indicated by our serial numbers and is shown after the dot (Figure 1).
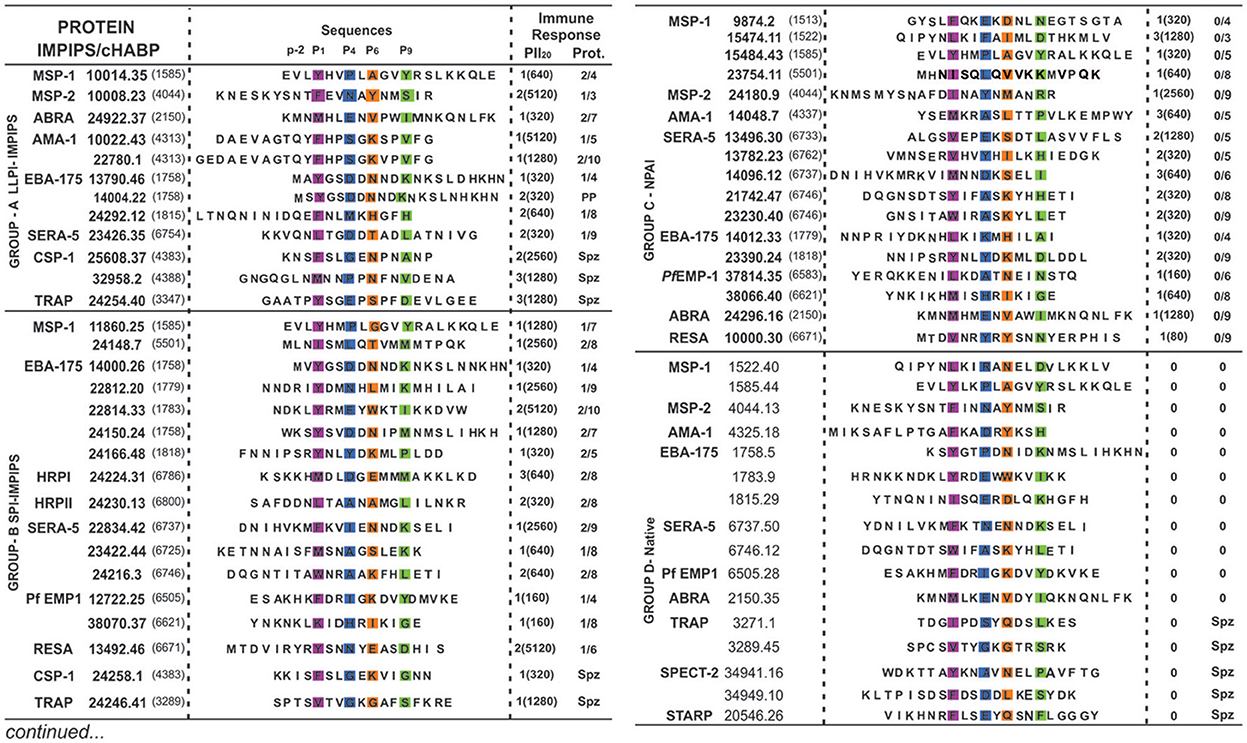
Figure 1. Modified and native cHABPs' chemical and immunological characteristics. P. falciparum merozoite (Mrz) or sporozoite (Spz)-derived proteins (in bold letters) where cHABPs (number in parenthesis) were identified, with their mHABP (in bold numbers with their associated conformed number after the dot) amino-acid sequences, highlighting PBR residues fitting into Pocket 1 (P1) in fuchsia, Pocket 4 (P4) in dark blue, Pocket 6 (P6) in orange and Pocket 9 (P9) in green, as well as the N-terminal PFR (–p2) preceding PBR residues essential in β-turn formation. Regarding immune response, the amount of monkeys producing immunofluorescence antibody (IFA) titres is shown in parenthesis, along with the amount of fully-protected monkeys (absolute absence of parasites in their blood during challenge). PP: partial protection. Group A (LLPI-IMPIPS), group B (SPI-IMPIPS), group C (NPAI) and group D (native) peptides. Spz peptide-induced protection could not be tested since there are no reliable Aotus monkey-adapted P. falciparum strains.
Classifying β-Turns
The N-terminal peripheral flanking region (PFR) adjacent to the PBR has been analyzed for many of the ~300 peptides studied by 1H-NMR; however, only 62 cHABPs and mHABPs have been included in this study due to the other peptides having been classified into different groups (Figure 2), having similarity regarding both their immunological and structural behavior. Peptides representative of each group were thus selected and are shown here. Chimera and Insight II software were used for structural analysis, measuring ψ and ϕ dihedral angles (Pettersen et al., 2004) and the distances between Cαi and Cαi+3 residues for each peptide (Reyes et al., 2017a). Different β-turns have been classified in our peptides according to recent work on dihedral angles (de Brevern, 2016), where ϕi+1, ψi+1, ϕi+2 and ψi+2 β-turns were taken into account: −60.00, −30.00, −90.00, 0.00 for type I turn, 60.00, 30.00, 90.00, 0.00 for type I', −60.00, 120.00, 80.00, 0.00 for type II, 60.00, −120.00, −80.00, 0.00 for type II', −120.0 130.0 55.0 41.0 for type IV1, −85.0 −15.0 −125.0 55.0 for type IV2, −71.0 −30.0 −72.0 −47.0 for type IV3, −97.0 −2.0 −117.0 −11.0 for type IV4, −60.00, 120.00, −90.00, 0.00 for VIa1, −120.00, −120.00, −60.00, 0.00 for VIa2 −135.00, 135.00, −75.00, 160.00 for VIb and −60.00, −30.00, −120.00, 120.00 for type VIII. It has been accepted that 3 angles are allowed to have ±30° deviation for β-type grouping and the fourth ±45°. β-turns which were found to be further apart were classified as distorted (marked by an asterisk *).
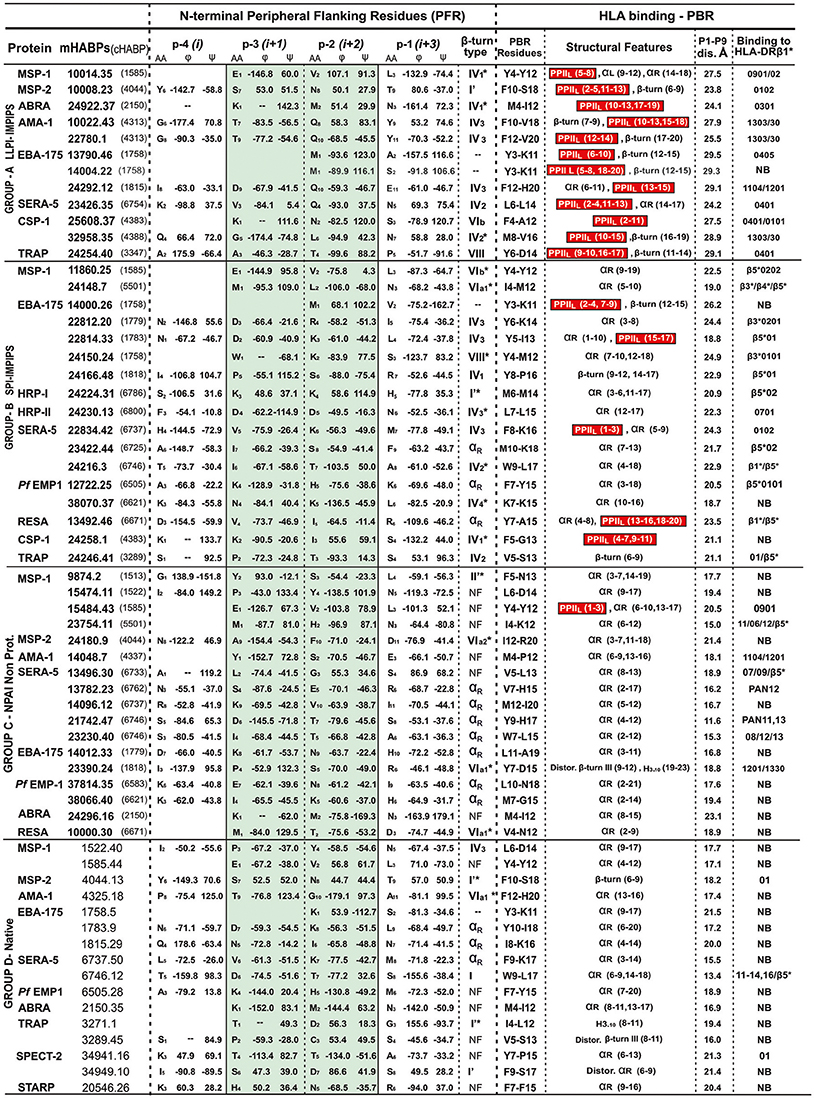
Figure 2. β-turn types and αR structures in the N-terminal PFR region preceding the PPIIL or other structures in PBR sequences. Previously classified by immunological results, A, B, C and D group peptides having residues located in the N-terminal PFR region (their amino acid and position shown in subscript in the central column) ϕ and ψ angles (in degrees °) classified according to (de Brevern, 2016) green highlighting relevant positions –p3 and –p2 in β-turn formation, αR or random structures (NF, not found). The next column describes the β-turn or NF structures, preceding residues fitting into PBR P1 to P9 of the HLA-DR molecules they bind to (last column). Red highlights (in parenthesis) residues in the structural features column forming PPIIL regions, followed by distance in Å between fittings into PBR P1 to P9. NB in the last column = not binding to any HLA-DR molecule, according to the netMHCIIpan 3.1 platform. Please note that all LLPI-IMPIPS (group A) PPIIL structures were preceded by specific β-turn types.
HLA-DRβ1* and IMPIPS
The NetMHCIIpan 3.1 algorithm (for predicting peptide binding to MHC-II molecules) was used for predicting HLADRβ* binding characteristics. This was based on the quantitative MHC class II binding capability of more than 100,000 peptides analyzed. Peptides binding to specific HLA-DRβ* alleles with high affinity (~95% specificity and 90% sensitivity) were accurately predicted (90%), as were correct HLA-DR peptide binding cores (previously determined by X-ray crystallography). This algorithm recognized peptides having very high theoretical binding to specific HLA-DRβ1* alleles and alternative β-chain isotypes, like HLA-DRβ3*, β4* and β5* alleles (Andreatta et al., 2015).
CLIP-binding HLA-DRβ1*0301 [PDB code: 1A6A, (Ghosh et al., 1995)] and HA-binding (hemagglutinin peptide) HLA-DRβ1*0401 [PDB code: 1J8H, (Hennecke and Wiley, 2002)] located in these molecules' PBR were used in this paper as templates for superimposing 32958.35 onto CLIP and 25608.37 onto HA. This was aimed at ascertaining the formation of H-bonds stabilizing the p-MHCII complex and highlighting interaction between α-chain-HLA-DRβ1* and the –p2 residue in the N-peripheral flanking region (β-turn fragment). The UCSF Chimera program was used for obtaining these measurements.
Immunization and Challenge
Immune response data and HLA-DR genotypes acquired from immunization studies of Aotus in groups of 5 to 8 spleen-intact monkeys for each peptide and each group have been previously described (Patarroyo et al., 2015a). Briefly, the Aotus monkeys were immunized with 250 μg polymerized peptide with Freund's complete adjuvant for the first dose and Freund's incomplete adjuvant for the second and third doses. Blood was used for immunological analysis on day 1 before (PI) the first immunization and 20 days after the first (I20), second (II20) and third (III20) immunizations.
LLPI-IMPIPS was assessed by re-challenging Aotus monkeys having high antibody titres and positive protection (defined as the complete absence of the parasite in a monkey's blood stream) 60 days after the end of the first trial in which a monkey proved fully-protected (Bermúdez et al., 2014; Alba et al., 2016).
Results and Discussion
Jardesky et al., using X-ray crystallography 20 years ago (Jardetzky et al., 1996), demonstrated that HLA-DRβ1*-associated endogenous peptides had a PPIIL structure (found later on in both antigenic and immunogenic peptides); these structures fit perfectly well into MHCII PBR (Dessen et al., 1997; Fremont et al., 1998).
Our group found that a specific group of immune protection-inducing protein structure (IMPIPS, involving LLPI and SPI) had or contained PPIIL-like structures in our search for a logical and rational methodology for malaria vaccine development (Patarroyo et al., 2012a,b, 2015a). Modified HABPs (mHABPs) had been developed against highly-virulent P. falciparum Aotus-adapted FVO strain lethal intravenous challenge; they were derived from conserved high activity binding peptides (cHABPs) from proteins directly involved in the parasite's invasion of a host (hepatocytes, endothelial or red blood) cells (Patarroyo et al., 2017). This was the first demonstration that chemically-synthesized, vaccine-induced immune protection was associated with a particular 3D structure; (Patarroyo et al., 2015a). As the ideal one should specifically bind to HLA-DRβ1* alleles, it was called Group A, inducing LLPI and having 26.5 ± 1.5 Å distance between residues 1 to 9 fitting into HLA-DRβ1* PBR (Alba et al., 2016), all having or containing PPIIL-like structures. This confirmed that PPIIL-like conformation is an absolute requirement for LLPI induction. Group B induced short-lived protective immunity (SPI) whose structure in the PBR binding region (HLA-DR residues 1 to 9) was ~3.5Å shorter, preferentially binding to HLA-DRβ3*, β4* or β5* haplotypes (Alba et al., 2016). Group C consisted of non-protective antibody-inducing (NPAI) mHABPs which were shorter in the PBR binding region and group D consisted of native cHABPs which did not induce antibody production or protection (Figure 1) or binding to any Class II molecule when used as immunogens, according to the netMHCIIpan 3.1. platform (Andreatta et al., 2015) (Figure 2).
Vey recently we demonstrated N-terminal peripheral flanking residue (PFR) (Reyes et al., 2017b) preference for amide (Q, N), sulfur-containing (M) and one having β-branched apolar (V) or large aliphatic (L) residues in IMPIPS position –p2. We have also shown preference for charged (E, K, R, D, H) and short polar (S, T) residues in SPI mHABP position –p2 (Reyes et al., 2017b).
This observation prompted us to look for an association between such immunological functions and particular 3D structures, applying Francis Cricks catch-phrase, “If you do not understand function, study structure and vice versa.” After having obtained ~300 3D structures for these peptides by powerful 1H-NMR spectroscopic analysis (600 MHz), we searched for an association between these characteristics and particular 3D structure conformation in our peptides (Patarroyo et al., 2011, 2012a,b, 2015a,b; Bermúdez et al., 2014; Alba et al., 2016; Reyes et al., 2017a).
Briefly, PPIIL are particular secondary structures in globular proteins and peptides, having left-handed geometry involving 3-5 amino acids, 9.3 Å pitch distance and three to five residues per turn, all amide bonds ideally having trans conformation (ω = 180°), an absence of internal H-bonds, side-chains almost perpendicular to a peptide's backbone and ϕ ~75° ± 25 and ψ ~145° ± 25° dihedral angles (Creamer, 1998; Stapley and Creamer, 1999; Adzhubei et al., 2013; Zondlo, 2013). Many of them contain prolines (~70%) (Chellgren and Creamer, 2004); however, ~40% of them do not (Kumar and Bansal, 2016) and a hexaproline peptide 3D structure confirmed the aforementioned physicochemical characteristics (Wilhelm et al., 2014).
β-turns must have a < 7.5Å distance between Cαi and Cαi+3 for residues involved in the turn (Hutchinson and Thornton, 1994), central residues are not helical and their ϕ and ψ angles define the β-turn type where, according to Brevern, a deviation of ± 30° from the canonical values is permitted for 3 of the 4 angles and ± 45° for a fourth one (Fuchs and Alix, 2005; de Brevern, 2016). Originally, Venkatachalam (1968) defined types I, II and III with their corresponding mirror images I', II' and III'; later on, Lewis added V and V' (Koch and Klebe, 2009) and Hutchinson et al., (Hutchinson and Thornton, 1994) divided VI into VIa1, VIa2 and VIb and precisely defined type VIII. Type VI is characterized by a cis-Pro in position i+2 and type VII is associated with a kink. The frequently occurring (~35% of all β-turns) and highly undefined type IV group was recently subdivided into types IV1, IV2, IV3, IV4, and IV miscellaneous (IVmisc) (de Brevern, 2016), observing some structural superimposition with previously-determined β-turn types (Madan et al., 2014).
Some turns, like type III' (1.5%), V (0.03%) and V' (0.02%), have recently been discarded due to their low frequency or their structural similarity with 310 helixes (e.g., type III and type I' similar to type III') (de Brevern, 2016).
We thus classified the PBR N-terminus peripheral flanking region groups as IMPIPS (LLPI, SPI), NPAI mHABPs and native cHABPs. It was striking that 23/29 (79%) SPIs or LLPI-IMPIPS exhibiting protection against challenge (Figure 1), had β-turn structures preceding HLA-DRβ1*, β3*, β4* or β5* Class II binding residues in their PBRs (Figure 2). It was observed that group B (SPI-IMPIPS) was much more diverse, having β-turn types IV1, IV2, IV3, IV4, VIa1, VIb, VIII (de Brevern, 2016) and three (3) αR structures, while group A was more selective regarding β-turn preference, having β-turns IV1 (similar to I'), IV2, IV3, VIb, VIII (Figure 2). Two EBA-175 (1758) cHABP-derived LLPI-IMPIPS in group A (13790.46 and 14004.22) were called short, since a perfect β-turn structure could not be assigned as only two amino acids preceded the PBR; this IMPIPS had M in position –p2 (Figure 2). It was striking that all IMPIPS (Group A) had β-turns structures. The preceding β-turn was very short for SPI-IMPIPS having PPIIL regions in one of them (14000.26); another was preceded by an αR region and another had the PBR in the αR region (Figure 2). It was clearly observed that most (7/17) NPAI (group C) had αR, type II'*, or β-structures (5/17), whilst another large group (6/17) had random structures (i.e. β-turn not found – NF) (Figure 2).
The situation in group D (native) was quite similar, as most native cHABPs had type I or I' (4/16), αR (3/16), VIa1 and IV3 β-turn (one each); interestingly, (6/16) had random structures (NF) in the preceding region, though sequence PBR binding by 1H-NMR had αR structures (theoretically, the netMHCpan 3.1 platform predicted that only 3/16 had some HLA-DR binding) (Figure 2). Such data clearly correlated 3D structural conformation with particular immunological outcomes where IMPIPS inducing LLPI had or contained PPIIL-like structures in the region fitting into the PBR preceded by specific β-turn types.
Interestingly β-turns I', IV, IVb, and VIII preceded PPIIL structures (Figure 2, highlighted in red) in most IMPIPS. This data showed LLPI-IMPIPS clear preference for type IV1 to IV3 and VIII β-turns while the most frequently occurring β-turns I (38.21%), II (11.81%), II' (2.51%), and IVmisc (~17%) accounted for ~70% of β-turns (Koch and Klebe, 2009; de Brevern, 2016) not being present, suggesting that β-turns I, II, II', and IVmisc are not associated with immune protection.
This was exceptional information since it showed the association of 2 different types of secondary structures to provide a defined functional role: LLPI induction (Figure 2, group A). We would like to stress that LLPI definition has been based on re-challenging Aotus monkeys having absolute protection (the complete absence of parasites in their blood) after immunization with these IMPIPs, the 1st and 2nd challenges mimicking what occurs in hyper-endemic areas where inhabitants may suffer as many as 18 infectious bites per night or repeated malaria episodes.
As previously stated (Reyes et al., 2017b), we have been increasing the amount of molecules analyzed. Group A (LLPI-IMPIPS) had a clear preference in –p2 for amide residues Q (4/12) and N (2/12), sulfur-containing M (3/12) and short alcohol (T), aliphatic amino acids (L) or β-branched apolar (V) amino acids, one per group. Group B (SPI-IMPIPS) had a clear preference in position−2 for charged residues, like K (5/17), R (1/17), H (1/17) and D (1/17), short alcohol S and T (2/17 each), β-branched I (2/17) and V (1/17) with contributions from L and M (1/17 each), whilst no single SPI-IMPIPS had amide (Q, N) residues in –p2. Group C preference in –p2 was clearly for T and S (3/17 each), with contributions from N and V (2/17 each), polar residues like Y, H, K, E and apolar residues like F, M, G (1/17 each) while p10 had a slight preference for E (4/17) and I (3/17) with contributions from M and R (2/18 each) and T, L, Y, V, A, and S (1/17 each). Group D native cHABP had no preference for residues in –p2 (Figure 2).
These results add quite interesting structure-function association-related information, since it has been suggested that IV3 and IV4 are very different regarding dihedral angle distribution but having similar amino acids composition. We found that this characteristic held true for position –p3 where D was present in most (4/7) IV3 in both IMPIPS groups; residues containing amides like Q and N were predominant in –p2 for LLPI-IMPIPS (6/12) (Figure 2, Group A), while positively-charged residues (K, H and R) were predominant in SPI-IMPIPS (7/17) (Figure 2, Group B). Proline was under-represented regarding β-turns in our mHABPs, since it has been suggested that type VI β-turns have cis-proline in position –p2 (we did not find this in our few type VI peptides). Type IV1 was closer to β I' than a β II turn; IV3 and IV4 were closer to β I turn, having very similar amino acid but very different structural conformation (de Brevern, 2016). It is striking that Q was only present in 4/12 group A (LLPI-IMPIPS) mHABPs in –p2 (Figure 2), while absent in the other 50 mHABPs from other groups, suggesting a very relevant role for these residues in LLPI-IMPIPS.
Despite similarities regarding dihedral angle distribution, our IMPIPS had very different amino acid composition to that described by de Brevern (2016), suggesting a different, distinctive amino acid composition but similar secondary structure conformation for peptides involved in protective immune responses.
Figure 3 shows the front view for β-turns in IMPIPS (LLPI and SPI) 3D structure (displayed in black sticks), most pointing toward the left-hand side, suggesting that once mHABPs have been anchored to HLA-DRβ1*, β3*, β4* or β5* PBR pocket 1 (P1), these specific β-turn sequences could interact with other specific residues located in Class II molecule α-chain. Group A LLPI-IMPIPS also had p2 pointing upwards and toward the right-hand side, while p3(cyan) also pointed upwards and toward the left-hand side (displaying a gauche+ orientation); exactly the opposite occurred in SPI-IMPIPS, confirming the critical role of these TCR-contacting residues' orientation in LLPI induction (Bermúdez et al., 2014; Alba et al., 2016).
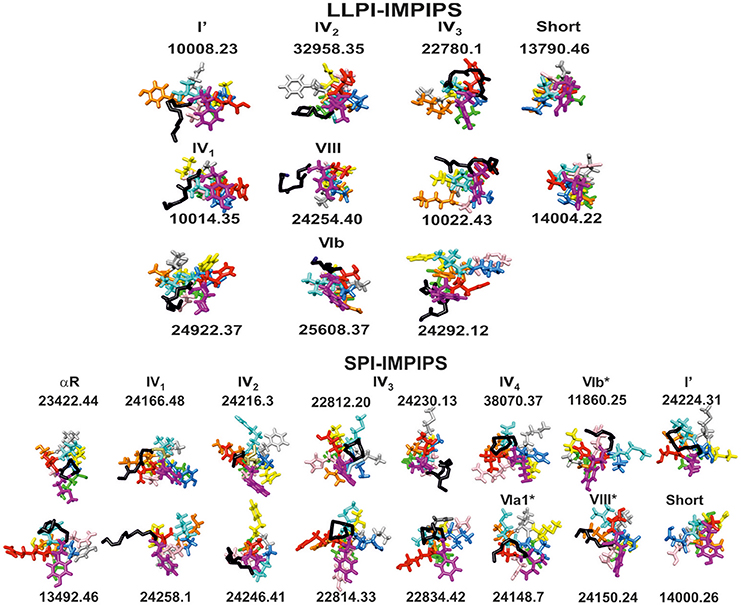
Figure 3. Front view of groups A (LLPI-IMPIPS) and B (SPI-IMPIPS); conformers determined by 1H-NMR. Color codes are the same as those for residues fitting into P1 (fuchsia), position p2, p3 (cyan), P4 (dark blue), p5 (pink), P6 (orange), p7 (gray), p8 (yellow), P9 (green). Residue positions (p) are those contacting the TCR. Black sticks highlights the backbone of residues forming β-turn types or αR structures preceding amino acid fitting into the PBR. Note that most N-terminal structures were left-hand orientated suggesting interaction with the HLA-DR α-chain to collaborate in stabilizing this complex. Note also the total absence of the most frequent β-turn types like I, II, II' and IVori, accounting for ~70% of β-turn types.
A tempting hypothesis is that IMPIPS mHABPs interact via their PPIIL preceding β-turn structures with Pheα51, Alaα52, Pheα54 and Trpα43 residues located in the MHCII molecule 310-helix (Yin and Stern, 2013) establishing H-bonds between residues in –p2 backbone atoms and Trpα43 and Serα53 side-chain and backbone atoms or interactions between the aforementioned peptide and α-chain-MHCII side-chain hydrogens. NetMHCPan 3.1 (Andreatta et al., 2015) predicted 32958.35 IMPIPS binding to HLA-DRβ1*1303; however, we were able to superimpose 32958.35 onto HLA-DRβ1*0301 (Ghosh et al., 1995) due to sequence and haplotype similarity; this complex has been shown to have interactions between L6:O and Hγ:Serα53 and L6:Hδ1 and HZ2:Trpα43 (Yin and Stern, 2013) and 25608.37 has been shown to have H-bonds between N2:O and Hγ:Serα53 and 32958.35 (Figure 4). They collaborate in stabilizing peptide binding to the PBR, specifically when fitting into P1 where peptides having low binding affinity and kinetic instability are highly susceptible to HLA-DM-mediated peptide exchange (Wieczorek et al., 2017). The opposite has been clearly demonstrated, as epitope selection is constrained by favoring the presentation of peptides having longer HLA-DM-mediated half-lives (Yin et al., 2012).
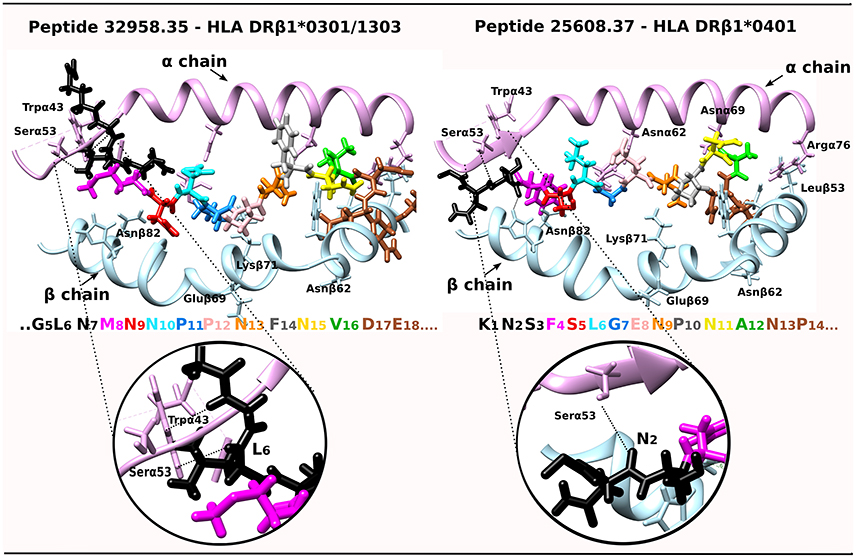
Figure 4. IMPIPS 32958.35 interaction with HLA-DRβ1*0301 and IMPIPS 25608.37 interaction with HLA-DRβ1*0401. DRβ1*' α-chain contact with N-terminal peripheral flanking residues (–p2 in black) are shown by a dotted line. The colors of residues in each IMPIPS are taken from the code for Figure 3 and MHC II α (pink ribbon) and β-chain residues (blue).
Two more facts should be highlighted. All these LLPI-IMPIPS (Group A), with one (10008.23, having 23.8 Å) exception, had 24.1Å to 29.1 Å distance (26.5 Å ± 2.5 Å) between residues fitting into P1 to P9 of the HLA-DRβ1* PBR of the allele to which they specifically bound. This was different to what occurs with SPIs-IMPIPS (Group B) having a 18.7 Å to 24.4Å distance between the same residues (21.6 Å ± 2.5 Å), preferentially binding to β3*, β4*, β5* allelic families (Figure 2) (Alba et al., 2016).
This distance was shorter (11.6 Å to 21.4 Å) in NPAI mHABPs (group C), some binding to HLA-DRβ1* alleles but mainly proceeded by α-helix structures (7/17), II'*, , β-turns (4/17), or not found (NF) structures (6/17). The distance was even shorter in native cHABPs (Group D), very few binding to some HLA-DR alleles (3/16) (Figure 2), thereby confirming the critical role of distance between residues fitting into P1 to P9 in the HLA-DRβ1*PBR.
Another striking physicochemical and immunological characteristic was found in IMPIPS mHABPs; no particular β-turn type was found to be associated with any HLA-DR allele or allele family (Figure 2).
Polar residues like Q, N or T, or large aliphatic residues like L in –p2 were associated with the highest antibody titres, while those having V or M in this position had lower antibody levels, suggesting a preference for such amino acid in this position (–p2) for stabilizing this complex and antibody production. There was a strong association between residues fitting into P1, like F and L with Q and N in position –p2, while Y in P1 was mainly associated with V, M or T in –p2.
It has been clearly shown here that type IV1 had more classical characteristics, being closer to type I' β-turns, that type IV2 and VIII were structurally very close, having high amino acid sequence similarity, that IV2 seemed to be a less extended form of VIII and that type IV3 seemed to be a half-turn, as shown here. This is why these β -turns have been grouped in Figure 3.
What we have shown here is that PPIIL-like structures must be preceded by specific β-turn types to induce an LLPI-IMPIPS, speeding up the long-sought-after process of vaccine design for humankind's health and welfare.
Author Contributions
AB and MEP: conceived and supervised the study; AB, MV, and MPA: analyzed data; AB, MEP, and MPA: wrote the manuscript; MEP and MAP: made manuscript revisions. All authors contributed to the discussion of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by The Colombian Science, Technology and Innovation Department (COLCIENCIAS) through contract RC#0309-201. We would like to thank Mr Jason Garry for making appropriate corrections to the use of the English language.
Abbreviations
MHC, Major histocompatibility complex; TCR, T-cell receptor; HLA, Human leukocyte antigen; IMPIPS, Immune protection-inducing protein structure; PPIIL, Polyproline type II, left-handed; PBR, Peptide binding region; LLPI, Long-lasting, protective immunity; SPI, Short-lived protective immunity; cHABP, Conserved high activity binding peptide; mHABP, Modified high activity binding peptide; NPAI, Non-protective antibody-inducing; PFR, Peripheral flanking residue; Mrz, Merozoite; Spz, Sporozoite; IFA, Immunofluorescence assay.
Footnotes
References
Adzhubei, A. A., Sternberg, M. J., and Makarov, A. A. (2013). Polyproline-II helix in proteins: structure and function. J. Mol. Biol. 425, 2100–2132. doi: 10.1016/j.jmb.2013.03.018
Alba, M. P., Suarez, C. F., Varela, Y., Patarroyo, M. A., Bermudez, A., and Patarroyo, M. E. (2016). TCR-contacting residues orientation and HLA-DRbetas* binding preference determine long-lasting protective immunity against malaria. Biochem. Biophys. Res. Commun. 477, 654–660. doi: 10.1016/j.bbrc.2016.06.115
Andreatta, M., Karosiene, E., Rasmussen, M., Stryhn, A., Buus, S., and Nielsen, M. (2015). Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67, 641–650. doi: 10.1007/s00251-015-0873-y
Bax, A., and Davis, D. G. (1985). MLEV-17 based two dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65, 355–360. doi: 10.1016/0022-2364(85)90018-6
Bermúdez, A., Calderon, D., Moreno-Vranich, A., Almonacid, H., Patarroyo, M. A., Poloche, A., et al. (2014). Gauche(+) side-chain orientation as a key factor in the search for an immunogenic peptide mixture leading to a complete fully protective vaccine. Vaccine 32, 2117–2126. doi: 10.1016/j.vaccine.2014.02.003
Chellgren, B. W., and Creamer, T. P. (2004). Short sequences of non-proline residues can adopt the polyproline II helical conformation. Biochemistry 43, 5864–5869. doi: 10.1021/bi049922v
Creamer, T. P. (1998). Left-handed polyproline II helix formation is (very) locally driven. Proteins 33, 218–226. doi: 10.1002/(SICI)1097-0134(19981101)33:2<218::AID-PROT6>3.0.CO;2-E
de Brevern, A. G. (2016). Extension of the classical classification of beta-turns. Sci. Rep. 6:33191. doi: 10.1038/srep33191
Dessen, A., Lawrence, C. M., Cupo, S., Zaller, D. M., and Wiley, D. C. (1997). X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity 7, 473–481. doi: 10.1016/S1074-7613(00)80369-6
Fremont, D. H., Monnaie, D., Nelson, C. A., Hendrickson, W. A., and Unanue, E. R. (1998). Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity 8, 305–317. doi: 10.1016/S1074-7613(00)80536-1
Fuchs, P. F., and Alix, A. J. (2005). High accuracy prediction of beta-turns and their types using propensities and multiple alignments. Proteins 59, 828–839. doi: 10.1002/prot.20461
Ghosh, P., Amaya, M., Mellins, E., and Wiley, D. C. (1995). The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature 378, 457–462.
Hennecke, J., and Wiley, D. C. (2002). Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J. Exp. Med. 195, 571–581. doi: 10.1084/jem.20011194
Houghten, R. A. (1985). General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc. Natl. Acad. Sci. U.S.A. 82, 5131–5135. doi: 10.1073/pnas.82.15.5131
Hutchinson, E. G., and Thornton, J. M. (1994). A revised set of potentials for beta-turn formation in proteins. Protein Sci. 3, 2207–2216. doi: 10.1002/pro.5560031206
Jardetzky, T. S., Brown, J. H., Gorga, J. C., Stern, L. J., Urban, R. G., Strominger, J. L., et al. (1996). Crystallographic analysis of endogenous peptides associated with HLA-DR1 suggests a common, polyproline II-like conformation for bound peptides. Proc. Natl. Acad. Sci. U.S.A. 93, 734–738. doi: 10.1073/pnas.93.2.734
Jeener, J. M. B., Backman, P., and Ernst, R. R. (1979). Investigation of enhance processes by two dimensional NMR spectroscopy. J. Chem. Phys. 71, 4546–4553. doi: 10.1063/1.438208
Koch, O., and Klebe, G. (2009). Turns revisited: a uniform and comprehensive classification of normal, open, and reverse turn families minimizing unassigned random chain portions. Proteins 74, 353–367. doi: 10.1002/prot.22185
Kumar, P., and Bansal, M. (2016). Structural and functional analyses of PolyProline-II helices in globular proteins. J. Struct. Biol. 196, 414–425. doi: 10.1016/j.jsb.2016.09.006
Lopez, C., Suarez, C. F., Cadavid, L. F., Patarroyo, M. E., and Patarroyo, M. A. (2014). Characterising a microsatellite for DRB typing in Aotus vociferans and Aotus nancymaae (Platyrrhini). PLoS ONE 9:e96973. doi: 10.1371/journal.pone.0096973
Madan, B., Seo, S. Y., and Lee, S. G. (2014). Structural and sequence features of two residue turns in beta-hairpins. Proteins 82, 1721–1733. doi: 10.1002/prot.24526
National Research Council (2011). Guide for the Care and Use of Laboratory Animals. Washington: DC: National Academies Press.
Patarroyo, M. E., Alba, M. P., Rojas-Luna, R., Bermudez, A., and Aza-Conde, J. (2017). Functionally relevant proteins in Plasmodium falciparum host cell invasion. Immunotherapy 9, 131–155. doi: 10.2217/imt-2016-0091
Patarroyo, M. E., Bermudez, A., and Alba, M. P. (2012a). The high immunogenicity induced by modified sporozoites' malarial peptides depends on their phi (varphi) and psi (psi) angles. Biochem. Biophys. Res. Commun. 429, 81–86. doi: 10.1016/j.bbrc.2012.10.088
Patarroyo, M. E., Bermudez, A., Alba, M. P., Vanegas, M., Moreno-Vranich, A., Poloche, L. A., et al. (2015a). IMPIPS: the immune protection-inducing protein structure concept in the search for steric-electron and topochemical principles for complete fully-protective chemically synthesised vaccine development. PLoS ONE 10:e0123249. doi: 10.1371/journal.pone.0123249
Patarroyo, M. E., Bermudez, A., and Patarroyo, M. A. (2011). Structural and immunological principles leading to chemically synthesized, multiantigenic, multistage, minimal subunit-based vaccine development. Chem. Rev. 111, 3459–3507. doi: 10.1021/cr100223m
Patarroyo, M. E., Moreno-Vranich, A., and Bermudez, A. (2012b). Phi (Phi) and psi (Psi) angles involved in malarial peptide bonds determine sterile protective immunity. Biochem. Biophys. Res. Commun. 429, 75–80. doi: 10.1016/j.bbrc.2012.10.089
Patarroyo, M. E., Patarroyo, M. A., Pabon, L., Curtidor, H., and Poloche, L. A. (2015b). Immune protection-inducing protein structures (IMPIPS) against malaria: the weapons needed for beating Odysseus. Vaccine 33, 7525–7537. doi: 10.1016/j.vaccine.2015.09.109
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Rance, M., Sorensen, O. W., Bodenhausen, G., Wagner, G., Ernst, R. R., and Wuthrich, K. (1983). Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117, 479–485. doi: 10.1016/0006-291X(83)91225-1
Reyes, C., Moreno-Vranich, A., and Patarroyo, M. E. (2017a). The role of pi-interactions and hydrogen bonds in fully protective synthetic malaria vaccine development. Biochem. Biophys. Res. Commun. 484, 501–507. doi: 10.1016/j.bbrc.2017.01.077
Reyes, C., Rojas-Luna, R., Aza-Conde, J., Tabares, L., Patarroyo, M. A., and Patarroyo, M. E. (2017b). Critical role of HLA-DRbeta* binding peptides' peripheral flanking residues in fully-protective malaria vaccine development. Biochem. Biophys. Res. Commun. 489, 339–345. doi: 10.1016/j.bbrc.2017.05.123
Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466. doi: 10.1146/annurev.immunol.23.021704.115658
Stapley, B. J., and Creamer, T. P. (1999). A survey of left-handed polyproline II helices. Protein Sci. 8, 587–595. doi: 10.1110/ps.8.3.587
Suárez, C. F., Pabon, L., Barrera, A., Aza-Conde, J., Patarroyo, M. A., and Patarroyo, M. E. (2017). Structural analysis of owl monkey MHC-DR shows that fully-protective malaria vaccine components can be readily used in humans. Biochem. Biophys. Res. Commun. 491, 1062–1069. doi: 10.1016/j.bbrc.2017.08.012
Suárez, C. F., Patarroyo, M. E., Trujillo, E., Estupinan, M., Baquero, J. E., Parra, C., et al. (2006). Owl monkey MHC-DRB exon 2 reveals high similarity with several HLA-DRB lineages. Immunogenetics 58, 542–558. doi: 10.1007/s00251-006-0127-0
Venkatachalam, C. M. (1968). Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers 6, 1425–1436. doi: 10.1016/0005-2795(68)90174-8
Wåhlin, B., Wahlgren, M., Perlmann, H., Berzins, K., Bjorkman, A., Patarroyo, M. E., et al. (1984). Human antibodies to a Mr 155,000 Plasmodium falciparum antigen efficiently inhibit merozoite invasion. Proc. Natl. Acad. Sci. U.S.A. 81, 7912–7916. doi: 10.1073/pnas.81.24.7912
Wieczorek, M., Abualrous, E. T., Sticht, J., Alvaro-Benito, M., Stolzenberg, S., Noe, F., et al. (2017). Major Histocompatibility Complex (MHC) Class, I., and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 8:292. doi: 10.3389/fimmu.2017.00292
Wilhelm, P., Lewandowski, B., Trapp, N., and Wennemers, H. (2014). A crystal structure of an oligoproline PPII-helix, at last. J. Am. Chem. Soc. 136, 15829–15832. doi: 10.1021/ja507405j
Yin, L., Calvo-Calle, J. M., Dominguez-Amorocho, O., and Stern, L. J. (2012). HLA-DM constrains epitope selection in the human CD4 T cell response to vaccinia virus by favoring the presentation of peptides with longer HLA-DM-mediated half-lives. J. Immunol. 189, 3983–3994. doi: 10.4049/jimmunol.1200626
Yin, L., and Stern, L. J. (2013). HLA-DM focuses on conformational flexibility around P1 pocket to catalyze peptide exchange. Front. Immunol. 4:336. doi: 10.3389/fimmu.2013.00336
Keywords: malaria-vaccine, β-turns, structure-function, IMPIPS, LLPI, PPIIL
Citation: Bermudez A, Alba MP, Vanegas M, Patarroyo MA and Patarroyo ME (2018) Specific β-Turns Precede PPIIL Structures Binding to Allele-Specific HLA-DRβ1* PBRs in Fully-Protective Malaria Vaccine Components. Front. Chem. 6:106. doi: 10.3389/fchem.2018.00106
Received: 22 December 2017; Accepted: 22 March 2018;
Published: 06 April 2018.
Edited by:
Julio A. Camarero, University of Southern California, United StatesReviewed by:
Alexander Shekhtman, University at Albany (SUNY), United StatesBogdan Olenyuk, Proteogenomics Research Institute for Systems Medicine, United States
Copyright © 2018 Bermudez, Alba, Vanegas, Patarroyo and Patarroyo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel E. Patarroyo, bWVwYXRhcnJAZ21haWwuY29t
 Adriana Bermudez1,2
Adriana Bermudez1,2 Manuel A. Patarroyo
Manuel A. Patarroyo Manuel E. Patarroyo
Manuel E. Patarroyo