- 1Energy-Rich Compounds Production by Photosynthetic Carbon Fixation Research Center, Shandong Key Lab of Applied Mycology, College of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 2Yan’an Tobacco (Group) Co., Ltd., Yan’an, China
- 3The Jinxiang County Agriculture and Rural Bureau, Jining, China
- 4Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
Introduction: Flue-cured tobacco is an important economic crop that is not tolerant of continuous cropping and can be influenced by planting soil conditions including rhizosphere microbial communities and soil physicochemical properties. The relationship between rhizosphere microbial communities and soil physicochemical properties under continuous cropping conditions is unclear.
Methods: This study investigated the succession of rhizosphere microbial community in continuous tobacco cropping soil for 1, 3, 5, 8, 10, 15, and 30 years. The physicochemical properties of the soil were measured, high-throughput sequencing was performed on the rhizosphere microbial community, and correlation analysis was conducted.
Results: The results suggested that continuous cropping could significantly enrich soil available nitrogen, available phosphorus, available potassium, and organic matter. Meanwhile, the alpha diversity of the bacterial community was significantly reduced with continuous cropping, indicating significant changes in the structure of bacterial and fungal communities. Based on linear discriminant analysis effect size (LEfSe), 173 bacterial and 75 fungal genera were identified with significant differences. The bacterial genera, Sphingomonas, Streptomyces, and Microvirga, were significantly positively correlated with continuous cropping years. The fungal genera, Tausonia, Solicocozyma, Pseudomycohila, and Fusarium, also showed significant positive correlation with continuous cropping years. Meanwhile, the fungal genera, Olpidium, Cephaliophora, and Cercophora, presented an opposite correlation. However, there are differences in the correlation between these bacterial and fungal genera related to continuous cropping years and other different soil physicochemical properties.
Discussion: In summary, this work could provide a reference for soil management and scientific fertilization of tobacco under continuous cropping conditions.
1 Introduction
As the main raw material of the tobacco industry, flue-cured tobacco has a long cultivation history all over the world, and its quality is very important in tobacco production (Ren et al., 2015). Continuous cropping refers to the cultivation method of repeatedly planting the same or similar crops in the same field, which is limited by the area of cultivated land and benefits from lower agricultural management costs compared to other cultivation methods, such as rotation, and is widely adopted in many places (Shipton, 1977; Chen et al., 2009). This long-term cropping pattern has inevitably resulted in great impacts on soil quality and field crop production. It usually leads to soil sickness, including deterioration of soil physicochemical properties, increase of pests and diseases, and accumulation of toxic substances. Moreover, long-term cropping can also increase the impact on the soil microbial community. All of these inevitably cause poor growth of crop plants, eventually resulting in yield and quality decline (Garbeva et al., 2004; Tan et al., 2021; Feng et al., 2022; Zhang et al., 2023). A recent study showed that the continuous cropping of sweet potato led to a 20%–30% reduction in yield, as well as a lack of production or even serious plant death (Gao et al., 2019). Continuous potato cropping caused a decline in soil fertility and aggravated the toxic effects of root secretions (Qin et al., 2017). Continuous cropping of chili led to the enrichment of pathogenic microbes (Chen et al., 2021), while continuous cropping of tomatoes significantly reduced soil enzyme activity, microbial biomass, and microbial metabolic activity, leading to changes in microbial community composition and structure, which caused changes in microbial community composition and structure (Fu et al., 2017). To sum up, there are three main factors associated with continuous cropping: imbalance of soil nutrients, shifts in microbial community composition, and autotoxicity of root exudates (Zhu et al., 2018). Tobacco, a type of plant in the Solanaceae family, is particularly intolerant to continuous cropping (Niu et al., 2017). The continuous cropping of Yunnan flue-cured tobacco has led to changes in environmental and biological factors in the soil (Yu et al., 2023). However, due to limited arable land, insufficient human resources, and a lack of scientific and advanced planting concepts, a large portion of tobacco planting still adopts continuous cropping.
The rhizosphere, as the narrow zone of soil that surrounds plant roots, is the habitat of countless microorganisms, and it can interact with plant roots (Philippot et al., 2013). The relationship between plant roots and rhizosphere microorganisms is also mutual: metabolites secreted by plant roots provide a favorable environment for the growth and reproduction of microorganisms (Edwards et al., 2015), and in return, rhizosphere microorganisms have significant effects on root traits and plant nutrition, growth, and development (Li et al., 2017b). As has been well documented, plants, soils, and microorganisms use the rhizosphere as the primary area for signal communication and material and energy exchange (Levy et al., 2017). Therefore, the study of the rhizosphere microbial community will be beneficial in addressing the issue of soil sickness caused by the continuous cropping of tobacco.
Soil microorganisms are critical to many processes including organic matter turnover, mineral nutrition cycling, soil structure maintenance, and toxin degradation (Blagodatskaya and Kuzyakov, 2013). Therefore, the variation of the soil microbial community is essential to the sustainability of soil ecosystem functions. On one hand, in agricultural production, soil microbial structure is determined by various factors, including farmland soil type, cropping pattern, and crop variety (Garbeva et al., 2004). On the other hand, soil productivity and sustainability depend on complex physicochemical and biological characteristics (Avidano et al., 2005). Therefore, the diversity and abundance of soil microbials significantly affect the productivity and sustainability of agricultural soil and thereby affect crop growth and quality (Nayyar et al., 2009). However, the general patterns of classification and functional characteristics of rhizosphere and microbial communities have largely remained unexplored. Understanding and managing the functions of rhizosphere microorganisms is crucial for supporting plant growth in different environmental conditions.
Additionally, because of its rapid response to environmental changes, the characteristics of microbial communities have been proven to be effective bio-indicators of soil conditions and environmental change (Welbaum et al., 2004; Chen et al., 2014). Recent studies have shown that bacterial communities presented huge differences when responding to various treatments, including abrupt flooding (Zhu et al., 2020), varying tillage depth in soil (Anderson et al., 2017), and climate changes (Dacal et al., 2022). The rhizosphere microbial community can also reflect the land use history (Li et al., 2019). Therefore, active management of soil microbial communities to improve soil conditions may be a promising method to improve crop productivity (Glick, 2010). However, specific populations, and the characteristics and relationships between plants and soil microbial communities must be further studied and understood.
In this study, we collected rhizosphere soil samples from seven different tobacco planting sites in vigorous growing stage: a place that had just planted flue-cured tobacco for 1 year (CC1Y), and places that had been continuously cropping flue-cured tobacco for 3 (CC3Y), 5 (CC5Y), 8 (CC8Y), 10 (CC10Y), 15 (CC15Y), and 30 (CC30Y) years, respectively. Soil physicochemical properties were analyzed, and high-throughput 16S rRNA and ITS rRNA gene sequencing was performed. The abundance, diversity, and composition of the microbial communities were analyzed. It was hypothesized that different continuous cropping treatments would lead to differences in soil physicochemical properties and recruit diverse soil microbial communities. There were two main objectives of this study: 1) to explore the effects of continuous tobacco cropping on soil microbial communities and physicochemical properties; and 2) to reveal the correlation between soil properties and soil microbial communities under continuous cropping systems.
2 Materials and methods
2.1 Study site description and soil sampling
The rhizosphere soil samples were collected from Yan’an city, Shanxi province, China (longitude 109° 32′E, latitude 36°07′N), at an altitude of 1,080 m, where the average annual rainfall is between 500 mm and the average yearly temperature is 8.1°C with a frost-free period of about 130 days. The soil type was basically cultivated loessial soil.
In this study, we selected seven different farmlands to gather our samples, including places that have been continuous cropped with tobacco for 3 (CC3Y), 5 (CC5Y), 8 (CC8Y), 10 (CC10Y), 15 (CC15Y), and 30 (CC30Y) years, as well as a place just cropped for 1 year (CC1Y). All treatments were with the same fertilizer and planting strategies, and all places have similar climatic conditions. Tobacco seedlings were cultivated in a greenhouse and then transplanted at the same time. The rhizosphere soil samples were collected on 15 May 2021, 1 month after transplantation. For rhizosphere soil collection, the five-point sampling method was adopted. Tobacco plant roots were carefully excavated from the soil, and visible particles were removed by gently shaking. Then, the fresh roots of each tobacco plant were collected into plastic bags, and the soil attached to the roots was shaken off tightly into the bags, then mixed thoroughly in equal proportions and stored. A part of the soil was taken to determine physicochemical properties, and high-throughput 16S rRNA and ITS rRNA gene sequencing was performed on the other soil samples. Each sample was analyzed in triplicate.
2.2 Analysis of soil properties
We used a glass electrode digital pH meter (PB-10, Sartorius, German) to determine the pH in a sample by mixing 10-g air-drying soil in 25 mL distilled water. Available potassium (AK) was extracted by ammonium acetate and measured by flame photometry (Hao et al., 2019). Soil organic matter (SOM) was determined using potassium dichromate oxidation-ammonium ferrous sulfate titration (Li et al., 2020b). Soil available nitrogen was measured using Automatic Kjeldahl Apparatus (FOSS), according to the Agricultural Industry Standards of the People’s Republic of China (NY/T1121.6-2006, LY/T1229-1999, and HJ7042014).
2.3 DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from fresh samples of 0.5 g with the Fast DNA® SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA). The V3-V4 region of bacterial 16S rRNA was amplified together with specific Illumina adapter and barcode sequences primer pair 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGA CTACHVGGGTWTCTAAT-3′). ITS1F (5′-CTTGGTCATTTAGAG GAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) were selected to target the fungal ITS1 region. PCR was conducted in 20 μL mixtures containing 1.0 μL (∼10 ng DNA) template, 4 μL 5 × FastPfu Buffer, 2 μL of forward/reverse primers, 0.4 μL FastPfu Polymerase, and 0.2 μL BSA, and dNase-rNase-Free deionized water was added to adjust the volume. The PCR procedure consisted of 95°C denaturing for 3 min; followed by 30 cycles of 95°C for 30 s, 54°C for 30 s, 72°C for 45 s, and a final step of 72°C extensions for 10 min. PCR product was purified and quantified with AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA), and the PCR product-constructed library sequencing was performed on the Illumina NovaSeq PE250 platform (Majorbio Technology Co., Ltd, Shang Hai, China). The raw data we obtained were in FASTQ format and had already cutoff adapter sequences. The raw data were submitted to the NCBI Sequence Read Archive (SRA) database under project number PRJNA982702.
2.4 Bioinformatics, data analysis, and statistical analysis
After removing the adaptors and primer sequences, raw sequences were assembled for each sample according to the unique barcode using QIIME 1.9.1 (Caporaso et al., 2010). Flash 1.2.11 was used to merge the split sequences of each sample (Magoč and Salzberg, 2011). The valid labels of all samples were clustered using Uparse 11 (Edgar, 2013). Valuable sequences were divided into operational taxonomic units (OTUs) using USEARCH 11, with a 97% identity threshold as the cut-off point. For species annotation, representative bacterial sequences were matched against the SILVA 138 database (Quast et al., 2012), and the fungal representative sequences were classified using the UNITE 8.0 database (Kõljalg et al., 2013). Mothur 1.30.2 was used to analyze the alpha diversity. R language was used for redundancy analysis (RDA), principal coordinate analysis (pCoA), and unweighted paired group algorithm clustering. The unweighted-UniFrac index was used as the distance measure. The Functional Annotation of Prokaryotic Taxa database (FAPROTAX) was used to predict bacterial functional groups. BugBase (https://bugbase.cs.umn.edu/index.html/) database was used to perform phenotype prediction. All experimental data were analyzed using IBM SPSS 26.0 and Microsoft Excel 2019. Statistical significance was set at p < 0.05.
3 Results
3.1 Effects of long-term continuous cropping on soil physicochemical properties
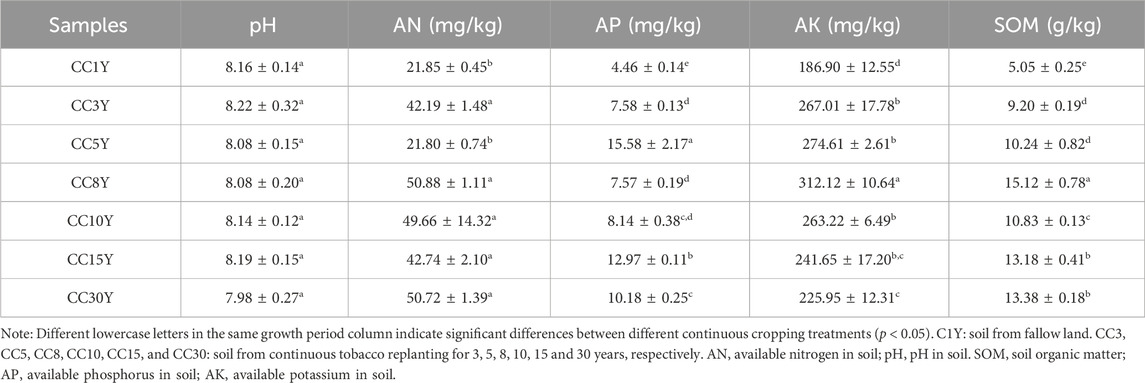
The physicochemical properties of tobacco rhizosphere soil varied greatly among the samples (Table 1). Results indicated that continuous cropping led to the accumulation of some soil nutrients, including available nitrogen (AN), available phosphorus (AP), available potassium (AK), and soil organic matter (SOM). The AN content in this study was between 21.80 mg/kg and 50.88 mg/kg, and compared to previous similar studies (24.20 mg/kg–97.30 mg/kg) is at a moderate level. The AK content was between 186.90 mg/kg and 312.12 mg/kg, and compared to previous studies (81.00 mg/kg–682.35 mg/kg) is at a moderate to high level. The AP content was relatively low compared to previous studies (10.30 mg/kg–93.90 mg/kg), with only 4.46 mg/kg to 15.58 mg/kg. The AP content was also relatively low compared to previous studies (15.61 mg/kg–98.10 mg/kg), with only 5.05 mg/kg to 15.12 mg/kg (Wang et al., 2020; Wang et al., 2022b; Wang et al., 2022c). The corresponding properties of the control sample CC1Y were basically at the lowest level. The available nitrogen content of CC1Y and CC5Y were significantly lower than that of other samples. CC1Y presented the lowest available phosphorus, available potassium, and soil organic matter contents among all samples. CC8Y had a high content of AN, AP, and SOM. The AP, AK, and SOM contents of samples with longer continuous cropping years, such as CC15Y and CC30Y, were at the middle level. Meanwhile, there were no significant differences observed in pH values of the soil samples. These results indicate that continuous cropping could result in soil nutrient accumulation to some extent.
3.2 Effects of continuous cropping on soil microbial diversity
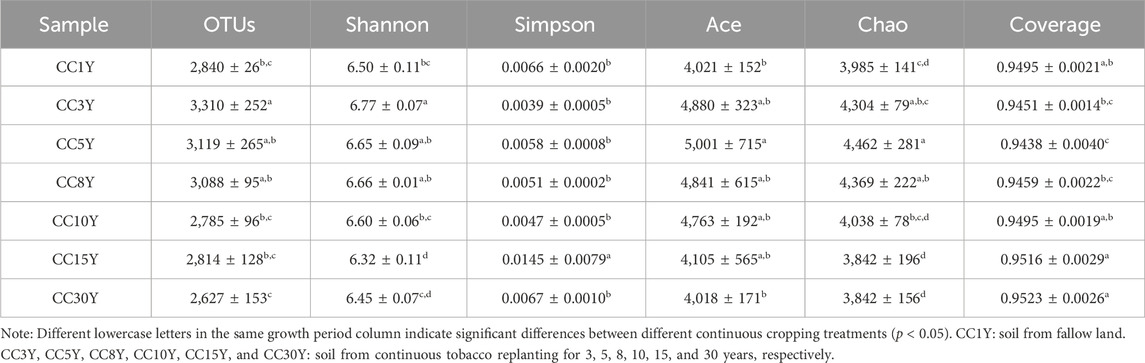
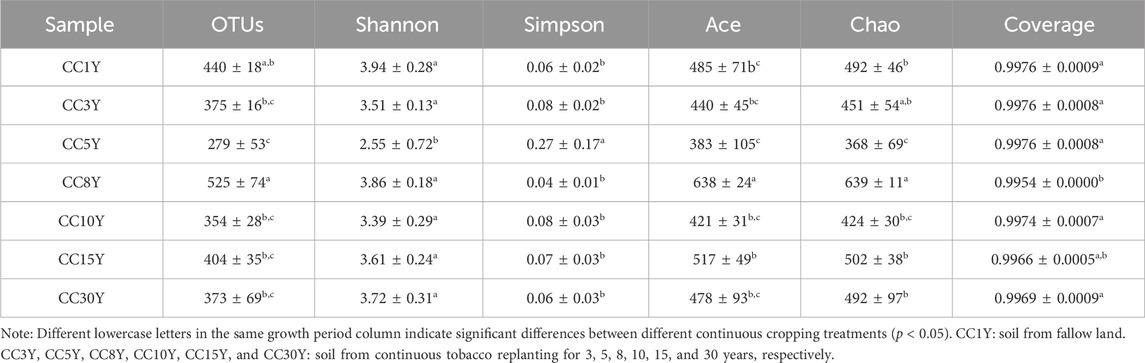
In our study, 946,516 high-quality clean tags from all bacterial samples and 1,027,944 high-quality clean tags from all fungal samples were collected after pair-end reads alignment and filtering. These were classified using a 97% sequence similarity criterion, and 4,648 operational taxonomic units (OTUs) were obtained in bacterial samples, and 1736 OTUs were obtained in fungal samples. Rarefaction curves tend to be flat in both kinds of samples (Supplementary Figure S1), indicating that the amount of sequencing data was sufficient to reflect the majority of bacterial and fungal diversity information and that the amounts of OTUs reached saturation for all the samples (Wang et al., 2019). Significant differences in alpha diversity among the seven samples were presented. The coverage index of all the samples were high and showed little difference, indicating that the sequencing results reflect the real situation of the bacterial community in the samples. The bacterial abundance of CC15Y and CC30Y was significantly lower than that of other samples (Table 2), indicated by the lower OTUs number (2,814 ± 128 and 2,627 ± 153) and Chao index (3,841 ± 196 and 3,842 ± 156). Interestingly, CC3Y and CC5Y samples showed significantly better species richness and diversity, where OTUs number and Chao index were 3,310 ± 252/3,119 ± 265 and 4,304 ± 79/4,462 ± 281, respectively. Meanwhile, the Ace index of CC3Y, CC5Y, and CC8Y were clearly higher than those of other samples. The OTUs number and Chao index of CC1Y sample were 2,840 ± 26 and 3,985 ± 141, respectively, implying a low level of bacterial abundance. Additionally, the fungal abundance of CC8Y was significantly higher than that of other samples, and that of CC5Y was significantly low, with no significant differences in other samples (Table 3). Taken together, these results demonstrate that long-term continuous cropping could significantly disturb the soil bacterial communities, but short-term continuous cropping might actually have a certain promoting effect on microbial community optimization. However, long-term continuous cropping did not indicate significant disturbances to soil fungal diversity.
3.3 Effects of continuous cropping on soil microbial community structure
The significant differences in microbial structural composition at the phylum level between different continuous soil samples are shown in Figures 1A, B. In all the samples, the dominant bacterial phyla did not change, and the first four types of dominant phyla were Actinobacteria, Proteobacteria, Acidobacteriota, and Chloroflexi. Moreover, the results indicated that the proportion of bacterial phyla was different, especially the four phyla of Chloroflexi, Firmicutes, Myxococcota, and Patescibacteria (Figure 1C). The proportion of Chloroflexi in CC1Y was 17.38% ± 2.86%, while it decreased to 13.60% ± 1.03%, 14.77% ± 0.85%, 14.57% ± 0.57%, 14.24% ± 1.30%, 12.57% ± 1.14%, and 12.50% ± 1.42%, respectively, along with the continuous cropping years. However, compared to previous studies, it was still at a higher level (Wang et al., 2022a; Wang et al., 2022c; Yu et al., 2023). The significance test of inter group differences indicated that Chloroflexi, Firmicutes, Myxococcota, and Patescibacteria had significant differences between different samples (Supplementary Figure S2A). Considering that continuous cropping does not significantly affect the dominant phylum of bacterial communities, it may be more effective to focus on phylum with significant differences, even if they are not dominant. Compared with the CC1Y group, the abundance of Patescibacteria bacteria in the multi-year continuous cropping tobacco soil decreased, which accounted for 1.07% ± 0.15% in CC1Y, at least double of the proportion in the other samples. Inversely, it was found that the abundances of Firmicutes and Myxococcota bacteria increased with the continuous cropping years. All these results suggested that continuous tobacco cultivation indeed interfered with the structure of the soil bacterial community.
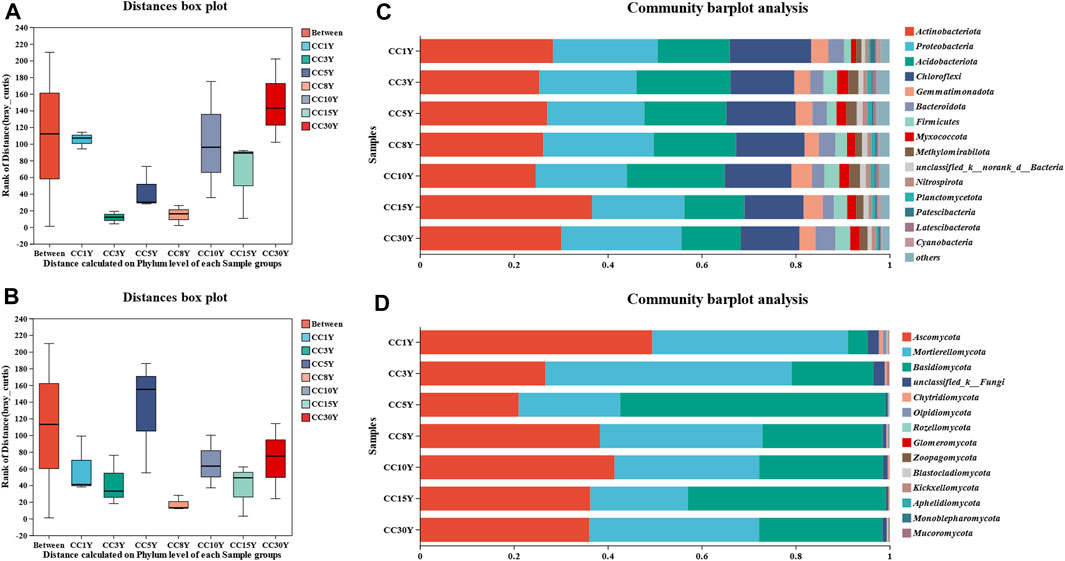
FIGURE 1. Microbiology community structure in the analysis of soil samples. (A) Rhizosphere bacteria sample grouping analysis in phylum level, (B) rhizosphere fungal sample grouping analysis in phylum level, (C) rhizosphere bacteria community structure in phylum level, (D) rhizosphere fungal community structure in phylum level.
The first three dominant fungal phyla were Ascomycota, Basidiomycota and Mortierellomycota (Figure 1D). In CC1Y samples, Ascomycota emerged as the predominant phylum, constituting 49.47% ± 10.12% of the total, indicating a moderate level. This contrasts with findings from previous studies (Wang et al., 2022b; Yu et al., 2023). On the other hand, Basidiomycota only accounted for 4.26% ± 1.59%, much lower than that of previous studies (Wang et al., 2022b; Yu et al., 2023). In continuous cropping samples, the abundance of Ascomycota was lower than that of CC1Y (21.05% ± 10.80%∼41.41% ± 10.61%), while the abundance of Basidiomycota was higher than that of CC1Y (17.42% ± 6.61%∼56.53% ± 22.06%), higher than in previous studies (Wang et al., 2022b; Yu et al., 2023). There were significant differences in the abundance of Mortierellomycota, Basidiomycota, unclassified_k__Fungi, and Glomeromycota in different fungal samples (Supplementary Figure S2B). This suggested that the continuous tobacco cultivation disturbed the fungal species abundance.
Moreover, PCoA analysis was performed at the family level (Figures 2A, B). For bacterial communities, PCoA analysis (PERMANOVA, R2 = 0.5528, p = 0.001) showed that there was a significant separation between CC1Y and continuous cropping samples. Of the differences at the family level, 31.45% could be explained by the PC1 axis, and 22.27% could be explained by the PC2 axis, indicating that continuous cropping caused significant changes in bacterial community structure (Figure 2A). Similarly, there was also significant separation between all samples of the fungal communities (Figure 2B). Of the differences at the family level, 39.58% could be explained by the PC1 axis, and 14.42% could be explained by the PC2 axis, and the differences were significant (R2 = 0.7128, p = 0.001).
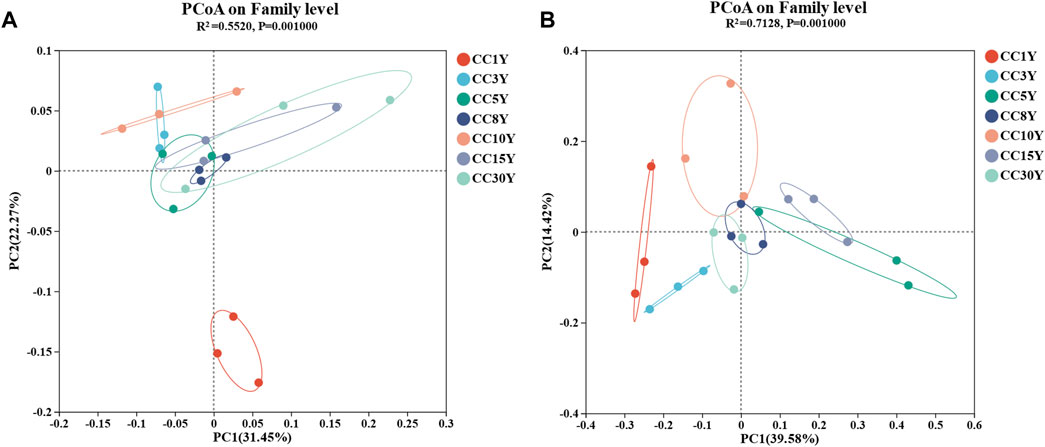
FIGURE 2. Microbiology community diversity in the analysis of soil samples. (A) Rhizosphere bacteria community principal co-ordinate analysis in family level (PERMANOVA, R2 = 0.5528, p = 0.001), (B) rhizosphere fungal community principal co-ordinate analysis in family level (PERMANOVA, R2 = 0.7128, p = 0.001).
In order to identify microbial communities with distinct characteristics in variable environments that are most responsive to continuous cropping, LEfSe analysis was conducted (Figures 3A, B). Among them, 173 taxa were identified at the genus level in the bacterial community (LDA value > 2, p value < 0.05), of which 41 were significantly enriched in the CC1Y samples. Meanwhile, 38, 11, 25, 13, 14, and 31 genera were significantly enriched in CC3Y, CC5Y, CC8Y, CC10Y, CC15Y, and CC30Y, respectively (Supplementary Table S2). At the genus level, 75 taxa were identified in the fungal community (LDA value > 2, p value < 0.05), of which 24 were significantly enriched in the CC1Y samples. Additionally, 13, 2, 6, 6, 2, and 10 genera were significantly enriched in the continuous cropping samples (Supplementary Table S3).
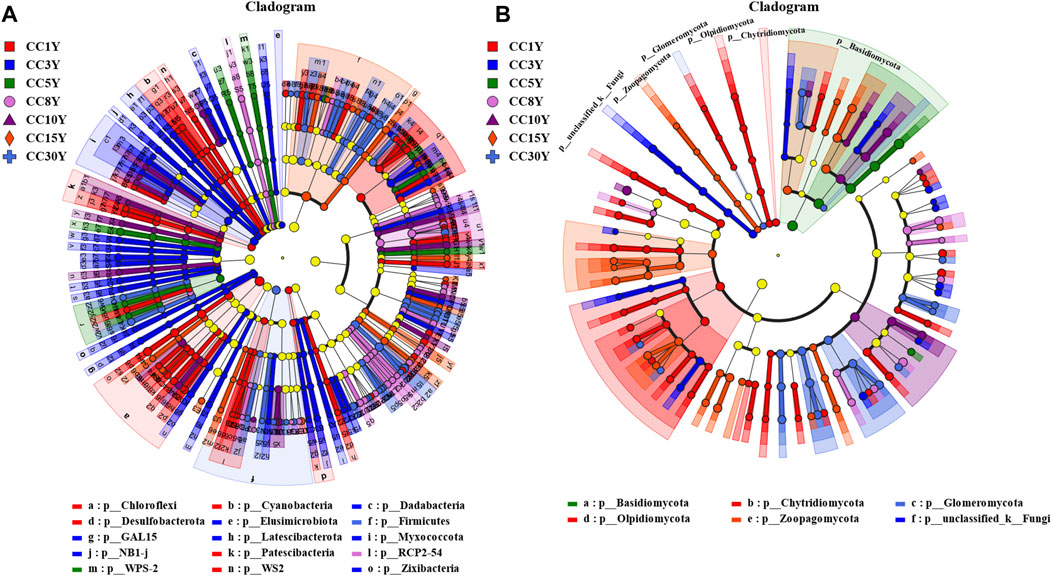
FIGURE 3. Microbiology community linear discriminant in the analysis of effect size (LEfSe) in soil samples. (A) Rhizosphere bacteria community LEfSe (LDA = 2), (B) rhizosphere fungal community LEfSe (LDA = 2).
3.4 Association of microbial communities with soil physicochemical properties
To elaborate the influence of physicochemical properties on the microbial community of tobacco soil, correlation analysis between the two aspects was conducted (Figures 4A, B). The results of RDA analysis demonstrated that the bacterial communities of CC15Y and CC30Y were more regulated by AN, AP, SOM, and continuous cropping years. pH was more moderated to the bacterial communities of CC3Y, CC5Y, and CC8Y. In addition, RDA1 and RDA2 explained a bacterial community variation of 25.33% and 12.06%, respectively (Figure 4A). Meanwhile, the fungal communities of CC1Y and CC3Y were more regulated by pH. That of CC15Y was more regulated by continuous cropping year, SOM, and AP. Meanwhile, the fungal communities of CC5Y were regulated by SOM and AP. RDA1 explained a fungal community variation of 52.96%, and RDA2 explained 9.22% (Figure 4B). To further clarify the effects of continuous cropping on the soil microbial communities, a regression analysis was carried out. With the increase of continuous cropping years, the Sobs index, which represents the number of observable OTUs, showed a significant downward trend (p = 0.0047) in bacterial samples (Figure 4C), but no significant differences were observed in fungal samples (Figure 4D). Considering the influence shown by the abundance of different species, the top 20 phyla were analyzed in association with physicochemical properties. It was clear that different phyla exhibited significant correlations with physicochemical properties. Proteobacteria phylum showed a significant positive correlation with available nitrogen solution, while Bacteroidota indicated a negative correlation with pH. Entotheonellaeota appeared significantly positively correlated with several factors, including continuous cropping years, SOM, and AP. However, Firmicutes only showed positive correlation with continuous cropping year, and Bdellovibrionota only showed positive correlation with AP. Moreover, Gemmatimonadota was significantly negatively correlated with AN and AK. Chloroflexi and Desulfobacterota were negatively correlated with continuous cropping year, SOM, and AP (Figure 5A). All these results suggested that continuous cropping is one important factor affecting the abundance of bacterial species. At the phylum level of fungal communities, among the top 10 dominant phyla, only Basidiomycota showed a significant positive correlation with continuous cropping year. Five phyla showed a significant negative correlation with continuous cropping year, including Ascomycota, Olpidiomycota, Rozellomycota, Chytridiomycota, and unclassified_ K_ Fungi (Figure 5B). There was a significant negative correlation between Basidiomycota and AP, while Ascomycota, Olpidiomycota, Rozellomycota, and Chytridiomycota were significantly positively correlated with AP. Ascomycota was highly significantly positively correlated with AN and SOM. Olpidiomycota was significantly positively correlated with pH. Among the phyla that were not significantly correlated with continuous cropping year, Mortierellomycota showed a highly significant negative correlation with AP, AN, and SOM. Glomeromycota showed a highly significant positive correlation with pH, AN, and SOM. Zoopagomycota showed a highly significant positive correlation with AK and AP. Moreover, there is a significant positive correlation between Aphelidiomycota and AK.
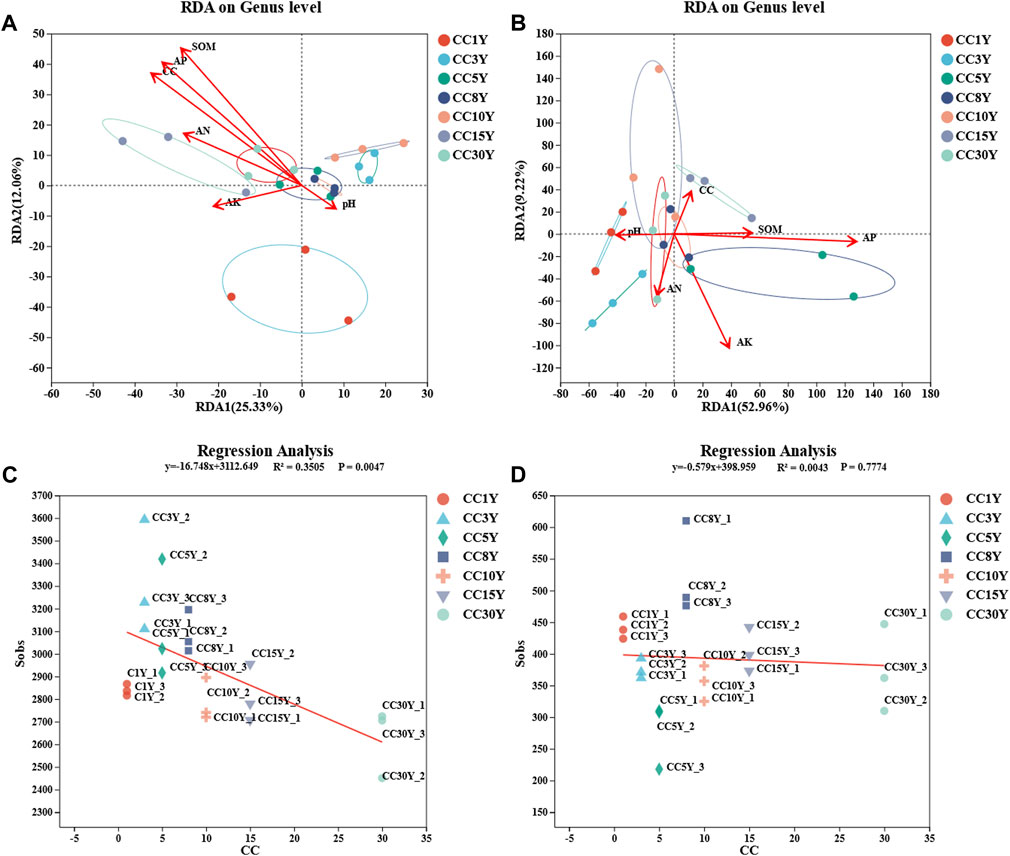
FIGURE 4. Correlation analysis of environmental factors with rhizosphere microbial diversity. (A) Redundancy analysis (RDA) of nutrient content (AN, pH, AK, AP, SOM), continuous cropping years, and rhizosphere bacteria (length = level), (B) redundancy analysis of nutrient content (AN, pH, AK, AP, SOM), continuous cropping years, and rhizosphere fungi (length = level), (C) linear regression of rhizosphere bacteria Sobs index and continuous cropping years, (D) linear regression of rhizosphere fungal Sobs index and continuous cropping years.
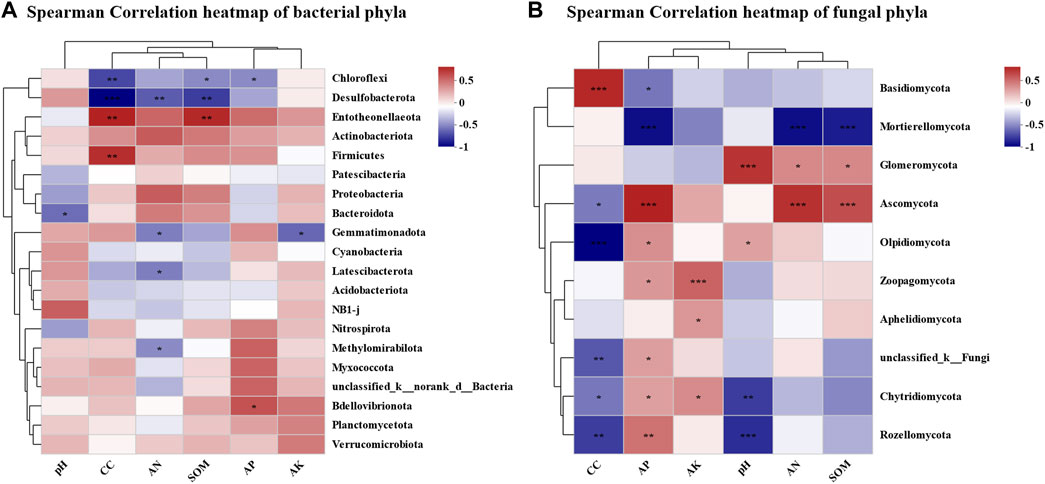
FIGURE 5. Correlation analysis of microbial community structure and environmental factors in continuous cropping tobacco-growing soil. (A) Heatmap of correlation analysis between rhizosphere bacteria and plant nutrient content (AN, pH, AK, AP, SOM) and continuous cropping years in phylum level (*p < 0.05, **p < 0.01, ***p < 0.001), (B) heatmap of correlation analysis between rhizosphere fungi and plant nutrient content (AN, pH, AK, AP, SOM), and continuous cropping years in phylum level (*p < 0.05, **p < 0.01, ***p < 0.001).
To determine the impact of microbial community differences between samples from different continuous cropping years, specific species screened through LEfSe analysis were combined to create an OTU set. The total OTU count of bacterial samples decreased from 9,894 to 4,399, and that of fungal samples decreased from 3,690 to 1,521 (Supplementary Table S1). The significance analysis of inter group differences demonstrated that among the top 15 dominant bacterial genera, the genera that can be identified and have significant differences are Sphingomonas, Nocardioides, Streptomyces, Bacillus, Marmoricola, and Microvirga (Figure 6A). Among the top 15 dominant fungal genera, the ones that can be identified and have significant differences are Tausonia, Solicoccozyma, Chaetomium, Pseudombrophila, Schizothecium, Fusarium, Paracylindrocarpon, and Cephaliophora (Figure 6B).
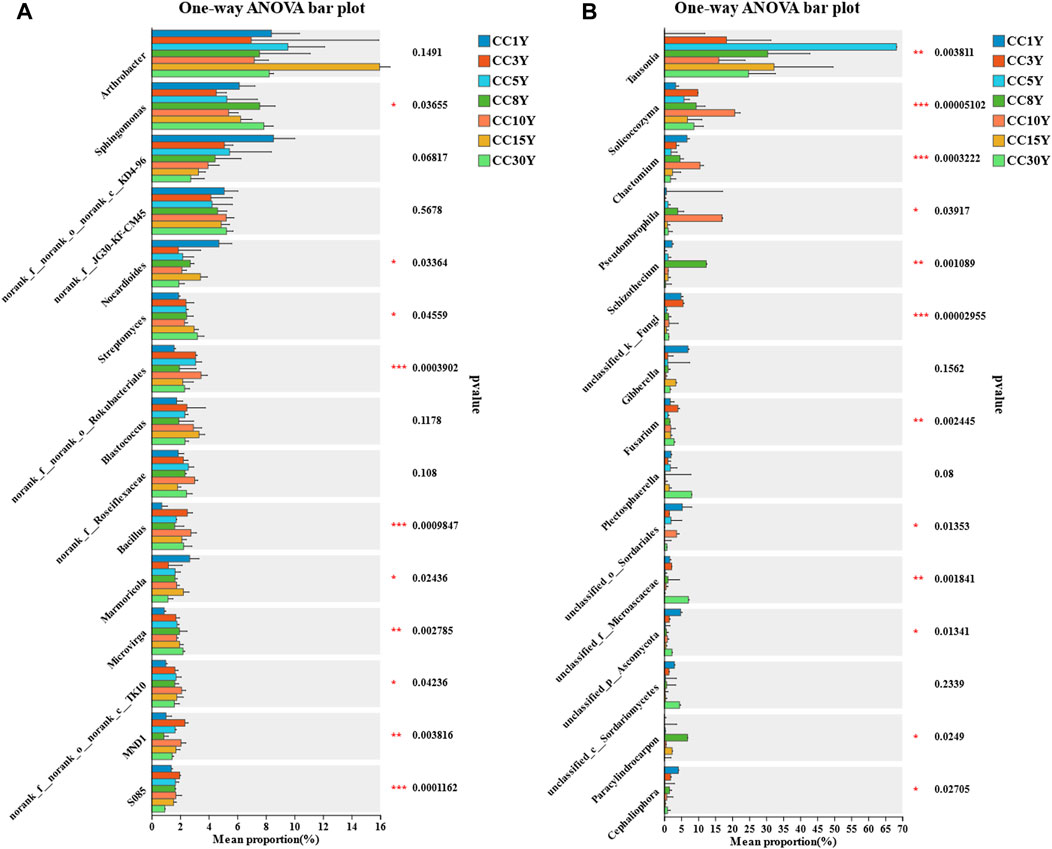
FIGURE 6. Analysis of genera differences in rhizosphere microbial diversity. (A) Significance test for inter group differences of rhizosphere bacteria community at the genus level (*p < 0.05, **p < 0.01, ***p < 0.001), (B) significance test for inter group differences of rhizosphere fungal community at the genus level (*p < 0.05, **p < 0.01, ***p < 0.001).
Environmental factor correlation analysis presented three genera in bacterial community showing a significant positive correlation with continuous cropping year (Figure 7A)—Sphingomonas, Streptomyces, and Microvirga. These three genera were all significantly positively correlated with SOM. Sphingomonas was significantly positively correlated with AN, and significantly negatively correlated with pH. Streptomyces was significantly positively correlated with AP. Among the bacterial genera that were not significantly correlated with continuous cropping year, Lysobacter showed significant positive correlation with AN and significant negative correlation with AP. Blastococcus showed significant positive correlation with AP. Bryobacter showed a significant negative correlation with pH. Four out of the top 20 dominant genera in the fungal community, namely, Tausonia, Solicocozyma, Pseudomycohila, and Fusarium, showed a significant positive correlation with continuous cropping year (Figure 7B). Five genera, namely, Olpidium, Cephaliophora, Cercophora, Unclassified_O_Sordariales, and unclassified_ K_ Fungi, showed a significant negative correlation with continuous cropping year. Among them, Tausonia was significantly negatively correlated with AP. Solicocozyma was significantly negatively correlated with AP and AN. Fusarium was significantly positively correlated with SOM. Olpidium was significantly positively correlated with pH and AP. Among the fungal genera that were not significantly correlated with continuous cropping year, Mortierella showed a highly significant negative correlation with AN, AP, and SOM. Schizotocium showed a highly significant positive correlation with SOM, AP, and AK. Plectosphaerella showed a highly significant positive correlation with AN and SOM, and showed a significant positive correlation with pH and AP. Alternaria showed a significant positive correlation with AP and AN. Gibberella was significantly positively correlated with SOM.
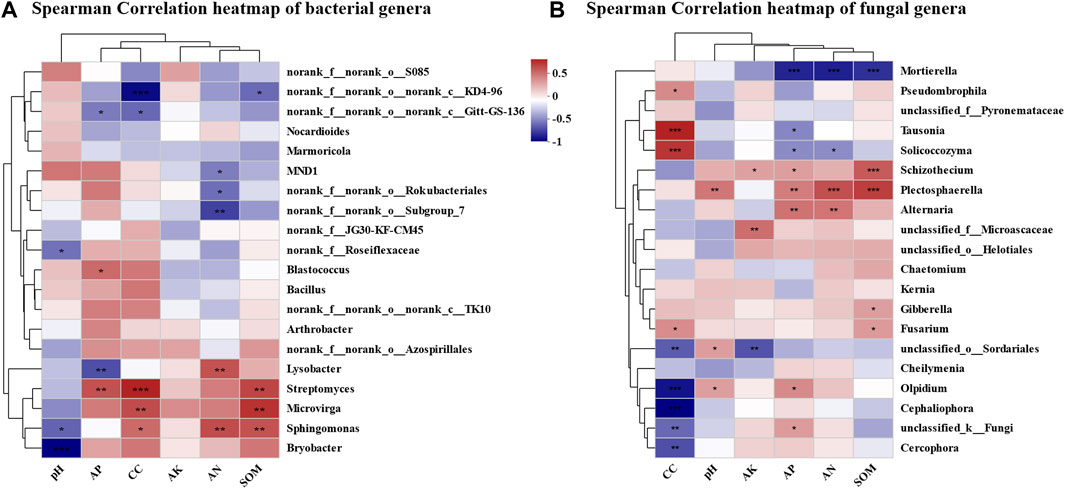
FIGURE 7. Correlation analysis of microbial community structure and environmental factors in continuous cropping tobacco-growing soil. (A) Heatmap of correlation analysis between rhizosphere bacteria and plant nutrient content (AN, pH, AK, AP, SOM) and continuous cropping years in genus level (*p < 0.05, **p < 0.01, ***p < 0.001), (B) heatmap of correlation analysis between rhizosphere fungi and plant nutrient content (AN, pH, AK, AP, SOM), and continuous cropping years in genus level (*p < 0.05, **p < 0.01, ***p < 0.001).
3.5 Effects of continuous cropping on bacterial community function
To further explore the effects of long-term continuous cropping on rhizosphere bacterial community function in tobacco soil, the FAPROTAX database was used (Supplementary Figure S3). According to the results, chemoheterotrophy, aerobic chemoheterotrophy, and ureolysis functions of long-term continuous cropping rhizosphere bacterial communities were significant increased (Figure 8A). The functions of aromatic compound degradation, photoautotrophy, nitrogen respiration, nitrate respiration, and oxygenic photoautotrophy were significant decreased. These results indicated that the specific species changes in bacterial community structure in continuous cropping samples may be more significantly related to the functions that affect material cycling.
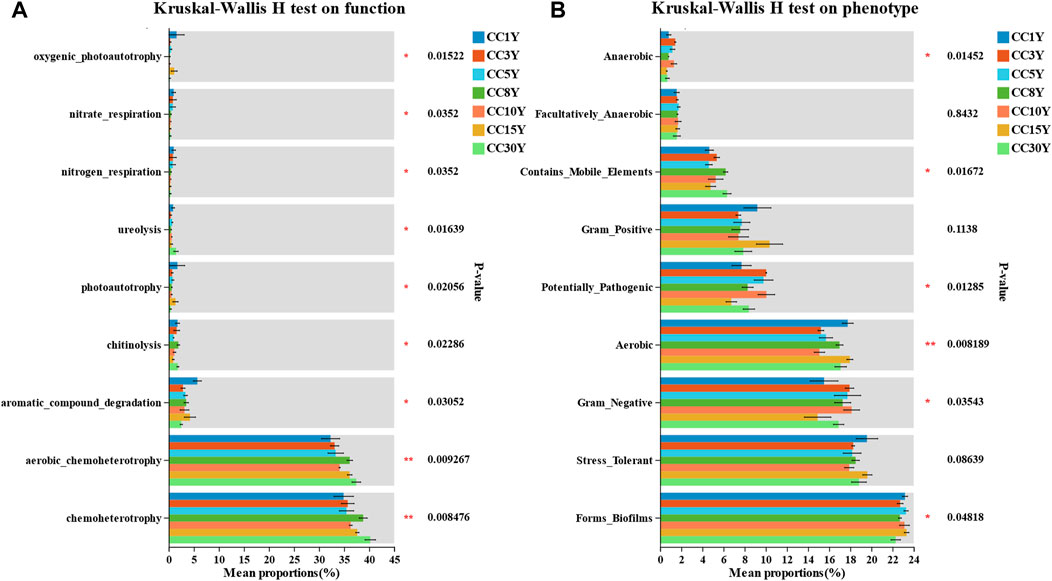
FIGURE 8. Functional and phenotype prediction of microbial community in continuous cropping tobacco-growing rhizosphere soil. (A) Significance test for inter group differences of FAPROTAX functions prediction of rhizosphere bacteria diversity in tobacco plants (*p < 0.05, **p < 0.01, ***p < 0.001), (B) significance test for inter group differences of BugBase phenotype prediction of rhizosphere bacteria diversity in tobacco plants (*p < 0.05, **p < 0.01, ***p < 0.001).
Moreover, BugBase database analysis was applied to predict the phenotypes of bacterial communities in continuous cropping samples (Figure 8B), with significant differences in biofilm formation, and significant differences in whether samples were gram negative, aerobic, potentially pathogenic, contained mobile elements, or were anaerobic. The proportion of phenotypes forming biofilms in CC1Y samples was significantly higher than that in continuous cropping samples. This indicated that specific species changes in continuous cropping samples may have a greater impact on the diversity and stability of bacterial community phenotypes. Among them, Sphingomonas is one of the main contributors of phenotypes of biofilm formation, and in phenotypes that are gram negative and aerobic (Supplementary Figures S3A–C).
4 Discussion
In this study, it was found that as the continuous cropping years of tobacco increased, soil physicochemical properties changed, soil quick-available nutrients such as nitrogen, phosphorus, and potassium accumulated, soil organic matter also increased. Soil available nutrients are important indicators for evaluating soil fertility and are the direct source of the three basic elements required by plants, which have a significant impact on crop yield and quality (Oldroyd and Leyser, 2020). In previous studies, the impact of continuous cropping on soil properties of different crops was not the same; it has been shown that long-term continuous cropping of crops such as peanuts and chili leads to an increase in the content of available potassium, available phosphorus, available nitrogen, and soil organic matter (Chen et al., 2021; Li et al., 2022a).
Our sampling method was similar to earlier investigations of continuous tobacco cropping by Yu, involving samples obtained within the same year. Nonetheless, while Yu’s sampling primarily emphasized distinctions between continuous and non-continuous cropping, our sampling method was adopted based on the variations across continuous cropping years (Yu et al., 2023). In Yu’s study, continuous cropping could significantly increase the available nitrogen content and significantly decrease the available potassium of tobacco grown in Yunnan. But in Yan’s study, continuous cropping significantly reduced the soil organic matter, nitrate nitrogen, and available potassium content of Liangshan tobacco, but significantly increased the content of available phosphorus (Yan et al., 2021). The results showed that tobacco cultivation for more than 3 years led to an accumulation of a large amount of available nutrients in the soil, including available potassium and available phosphorus. On one hand, this might be relative to the long-term fertilization during continuous cropping process, and it may also be related to the preferential absorption of plants (Chen et al., 2020). However, in previous studies, soil sickness brought about by continuous cropping could not be prevented by only correcting the imbalanced physicochemical factors in soil (Wilson et al., 2004).
Soil microorganisms have been shown to play important roles, being able to participate in the circulation of elements such as carbon, nitrogen, phosphorus, and sulfur (Yi et al., 2019; Li et al., 2022b). Continuous planting of the same crop is a convenient way to manage farmland, but the cultivation of a single crop affects the soil texture and the structure and function of the root microbial community of the crop (Niu et al., 2016; Wei and Yu, 2018; Li et al., 2020a; Dang et al., 2020). Our study indicated that long-term continuous cropping significantly reduced the diversity and stability of the soil bacterial community, and this is consistent with previous studies (Tan et al., 2021). However, continuous cropping did not show a significant impact on the diversity and stability of fungal community, in agreement with previous studies (Yu et al., 2023). The healthy growth of plants and the formation and maintenance of disease-resistant soils benefit from high diversity in the soil microbiome (Yuan et al., 2014). Therefore, the loss of rhizosphere bacterial richness and diversity may be one of the factors leading to poor growth of tobacco. In this study, the abundance of Chloroflexi and Patescibacteria in soil decreased along with continuous planting years. As reported, the most abundant bacteria belonging to Chloroflexi were involved in various organic transformations and sulfate metabolism, and they could reduce the potential for membrane contamination (Miura et al., 2007; Hug et al., 2013; Narsing Rao et al., 2022). The decrease in Chloroflexi abundance may be one of the reasons for the accumulation of soil organic matter. After years of continuous cropping, the abundance of the Patescibacteria phylum declined. Patescibacteria has been identified as potentially the large autotrophic taxa involved in the nitrogen, sulfur, and iron cycles (Herrmann et al., 2019). A decrease in the abundance of the Patescibacteria phylum indicated the decrease of some part of the microbial communities involved in the circulation of elements. These reductions in the abundance of beneficial microorganisms leading to a huge impact on the rhizosphere environment may be one of the reasons for nutrient imbalances. Ascomycota, Basidiomycota, and Mortierellomycota were dominant fungal phyla, consistent with previous studies (Wang et al., 2020). Ascomycota is a saprophytic soil fungus that plays a major ecological role as a decomposer in nature and is influenced by plant species and planting methods. In this study, there was a significant correlation between Ascomycota and AN, AP, and SOM, and the decrease of Ascomycota abundance may be one of the reasons for the enrichment of various available nutrients. As reported, Mortierella could dissolve mineral phosphorus in soil by synthesizing and secreting oxalic acid (Spaepen et al., 2007), and it can also grow rapidly in iron-rich soil and use its mycelium to facilitate the growth of beneficial bacteria such as Bacillus (Jeewani et al., 2021). Olpidiomycota has also been reported to be significantly enriched in non-continuous cropping sugar beets (Cui et al., 2022). However, an opposite result was shown, where the abundance of Chytridiomycota increased significantly in the continuous cropping of sweet potato (Gao et al., 2019). Together, these comparisons indicated that the microbial community were affected by various factors, among which crop variety played a key role. Additionally, a significant positive correlation between Basidiomycota and continuous cropping year indicated that continuous cropping significantly enriches Basidiomycota. This is in contrast to the results of many previous studies (Beule et al., 2019; Beule et al., 2021; Pang et al., 2021; Xu et al., 2022) but similar in a study of continuous cropping in grape (Liu et al., 2021). Basidiomycota has a wide variety of fungal species, making it one of the most advanced groups of fungi, with extremely wide distribution and complex functions (He et al., 2022). The impact of changes in Basidiomycota abundance requires more detailed research. Above all, the balance of rhizosphere microecology is an important measure for maintaining the balance of soil and plant microbiota, as a good rhizosphere microecological environment can not only promote the absorption and utilization of nutrients by plants but also help plants resist the invasion of diseases (Gonzalez et al., 2018; Zhou et al., 2020; Ling et al., 2022). The disruption of the rhizosphere microecological balance could lead to a decrease in sustainable soil productivity (Berendsen et al., 2012).
Recent studies have suggested that land use history affects crop rhizosphere microbiomes and plant physiology (Li et al., 2019). Soils with specific microbial community structure characteristics can also reflect soil changes and health levels (Shao et al., 2019). In the correlation analysis with soil properties in bacterial samples, Streptomyces, Sphingomonas, Microvirga, and Bryobacter showed a significant positive correlation with continuous cropping years. Streptomyces is one of the most important bacterial communities in soil, with great potential for the bioremediation of toxic compounds (Álvarez et al., 2012), and plays an important role in the decomposition of organic matter that is beneficial for crop production (Qin et al., 2011). The results of this study are consistent with a study of the continuous cropping of sugarcane (Pang et al., 2021) but in contrast to a study of continuous cropping in soybean (Xu et al., 2022), implying the diversity between different crops. Sphingomonas has the function of promoting plant growth and decomposing aromatic compounds (Khan et al., 2014; Zhou et al., 2016). A previous study used it as an inoculum for soil herbicide removal experiments (Li et al., 2017a). Microvirga has the ability to decompose the pyridinecarboxamide compound, and it is used as a bioremediation agent (Zhao et al., 2021). In fungal samples, it was found that Tausonia was the first dominant genus in Basidiomycota taxa, and its abundance increased with continuous cropping years, with significant differences; this partially explained the significant increase in Basidiomycota. Environmental factor correlation analysis at the genus level showed that Tausonia and Solicocozyma under the Basidiomycota phylum were significantly positively correlated with continuous cropping years, consistent with previous studies on continuous cropping of sugar beet (Cui et al., 2022). Previous studies have shown that under the reduced nitrogen fertilizer environment of tobacco, the abundance of Tausonia significantly increased (Shen et al., 2022). Tausonia may participate in regulating tobacco sugar content and may also play a role in plant tolerance to low temperature and fermentation processes (Kim et al., 2020; Wang et al., 2022b). Tausonia pullulans has been reported to be able to delay the mycorrhizal colonization of beneficial fungi in tomato roots (Mestre et al., 2022). As a Solanaceae crop, the tobacco plant may also affect the colonization of beneficial fungi due to the enrichment of the Tausonia genus. Similarly, the Fusarium and Pseudomycohila genera under the Ascomycota phylum also showed a significant positive correlation with the duration of continuous cropping. Fusarium genera have various pathogenic fungi, among which Fusarium oxysporum can directly cause root rot. Therefore, the increase in Fusarium abundance might greatly increase the possibility of soil disease. The changes in these specific genera with different functions during continuous cropping could provide a reference for the exploration of microbial agent products, which could be beneficial in solving soil sickness caused by continuous cropping.
The cycling of nitrogen in soil is influenced by both plant and microbial functional genes (Morse et al., 2018). Previous studies have shown that an increase in nitrogen inhibits microbial activity, such as microbial respiration, and a decrease in microbial biomass carbon (Liu and Greaver, 2010; Hu et al., 2021). In this study, there was a significant decrease in nitrogen metabolism functions and a significant increase in respiratory function in bacterial communities, with significant differences in continuous cropping. This is in contrast to previous studies, indicating that differential microorganisms may not fully explain functional changes. A previous study showed that differential microorganisms can serve as important indicators of environmental change (Berard et al., 2004), but the function of soil microbial communities is not entirely determined by bacteria, and the function of fungi is equally important. In Yu’s research, the correlation between allelochemicals, microbial communities, and tobacco continuous cropping is evident, with allelochemicals leading to a more significant influence than microbial communities. In Yu’s findings, however, soil physicochemical properties did not show a direct correlation with the continuous cropping of tobacco (Yu et al., 2023). Therefore, the exploration of microbial communities should extend beyond surface-level assessments of species richness and diversity. A deeper and more comprehensive examination into their functional roles and interactions should be performed. It should be noted that the bacterial community function in this study was predicted using only the FAPROTAX database, rather than being validated through experiments as other studies have done, which could have led to differences from the actual situation (Tao et al., 2020; Jin et al., 2023; Zhou et al., 2023). Therefore, further experimental verification is needed to assess the impact of microbial community changes on microbial community function. It should also be noted that the sampling method adopted in this study, which involves selecting different plots in the same year, present some limitations to a certain extent. A more accurate method for continuous cropping study would be to sample the same plot for multiple consecutive years. Consecutive sampling would play a crucial role in decreasing the impact of environmental changes on the dynamic shifts within planting soil and microbial communities. It facilitates a clearer comprehension of the change trends in soil and microbial communities. Furthermore, consecutive sampling contributes to evaluating the relative stability of microbial communities, enhancing the reliability of study data. Nevertheless, the consecutive sampling method poses specific challenges for practical implementation.
Of course, considering the complexity of plant microbial and microbial interactions, optimizing soil microbial partnerships to improve soil quality is a challenging task (Brodeur, 2012; Toju et al., 2018; Sessitsch et al., 2019). Sometimes the change of a single microorganism could lead to great changes in the entire community (Tao et al., 2020), indicating the regulatory complexity of microbial community structure. However, the soil shape obtained by changing the rhizosphere microbial community is more durable than changing the fertilization method (Zhou et al., 2023). Therefore, for soil sickness caused by continuous cropping in farmland production, we recommend reducing the application of fertilizers, and instead balancing the microbial community. This is similar to previous research that confirmed that reducing nitrogen inputs could benefit tobacco (Shen et al., 2021). Changing the application of fertilizers to other new types of fertilizers, such as chitin fertilizers (Fan et al., 2022) and organic fertilizers (Chen et al., 2018), is also an effective strategy. Furthermore, changing farming methods would also be an effective way (Liu et al., 2020). These proposed strategies could help to restore the balance of the rhizosphere microecological environment.
5 Conclusion
This study analyzed the changes in the microbial community structure of tobacco rhizosphere under different continuous cropping years. The relationship between soil microbial community and soil physicochemical properties was revealed. Research has demonstrated that continuous cropping significantly impacted soil physicochemical properties during the long-term continuous cropping process, leading to the enrichment of some soil nutrients. The bacterial and fungal communities in the rhizosphere soil of continuous cropping tobacco underwent significant compositional changes, with some genera significantly associated with continuous cropping. Together, these results could be of benefit for providing a theoretical reference for the proper management and scientific fertilization of tobacco planting soil under continuous cropping conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/982702.
Author contributions
Conceptualization, JR, LY, ZW, and JY; experiment implementation, GL, ZW, PG, JZ, LW, PD, YJ, YZ, and XZ; data processing, GL, ZW, PG, YZ, and JR; writing–original draft preparation, GL and ZW; writing–review and editing, XS, JR, and JY; funding acquisition, JY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the science and technology planning project of Shaanxi Tobacco Company Yan’an Company (Grant No. 2020-130), the technology development project of Inner Mongolia Kunming Cigarette Limited Liability Company (Grant No. 202315010534-JS-062), the “First class grassland science discipline” program in Shandong Province, and the Talents of High Level Scientific Research Foundation (Grant No. 6651121004) of Qingdao Agricultural University.
Acknowledgments
The authors would like to thank the “Laboratory for Agricultural Molecular Biology” of Qingdao Agricultural University for providing laboratory apparatus.
Conflict of interest
Authors PG, LW, PD, YJ, and LY were employed by Yan’an Tobacco (Group) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1251938/full#supplementary-material
References
Álvarez, A., Yañez, M. L., Benimeli, C. S., and Amoroso, M. J. (2012). Maize plants (Zea mays) root exudates enhance lindane removal by native Streptomyces strains. Int. Biodeterior. Biodegrad. 66 (1), 14–18. doi:10.1016/j.ibiod.2011.10.001
Anderson, C., Beare, M., Buckley, H. L., and Lear, G. (2017). Bacterial and fungal communities respond differently to varying tillage depth in agricultural soils. PeerJ 5, e3930. doi:10.7717/peerj.3930
Avidano, L., Gamalero, E., Cossa, G. P., and Carraro, E. (2005). Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30 (1), 21–33. doi:10.1016/j.apsoil.2005.01.003
Berard, A., Rimet, F., Capowiez, Y., and Leboulanger, C. (2004). Procedures for determining the pesticide sensitivity of indigenous soil algae: a possible bioindicator of soil contamination? Arch. Environ. Contam. Toxicol. 46 (1), 24–31. doi:10.1007/s00244-003-2147-1
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 (8), 478–486. doi:10.1016/j.tplants.2012.04.001
Beule, L., Arndt, M., and Karlovsky, P. (2021). Relative abundances of species or sequence variants can Be misleading: soil fungal communities as an example. Microorganisms 9 (3), 589. doi:10.3390/microorganisms9030589
Beule, L., Lehtsaar, E., Corre, M. D., Schmidt, M., Veldkamp, E., and Karlovsky, P. (2019). Poplar rows in temperate agroforestry croplands promote bacteria, fungi, and denitrification genes in soils. Front. Microbiol. 10, 3108. doi:10.3389/fmicb.2019.03108
Blagodatskaya, E., and Kuzyakov, Y. (2013). Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol. Biochem. 67, 192–211. doi:10.1016/j.soilbio.2013.08.024
Brodeur, J. (2012). Host specificity in biological control: insights from opportunistic pathogens. Evol. Appl. 5 (5), 470–480. doi:10.1111/j.1752-4571.2012.00273.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi:10.1038/nmeth.f.303
Chen, H., Hou, R., Gong, Y., Li, H., Fan, M., and Kuzyakov, Y. (2009). Effects of 11 years of conservation tillage on soil organic matter fractions in wheat monoculture in Loess Plateau of China. Soil Tillage Res. 106 (1), 85–94. doi:10.1016/j.still.2009.09.009
Chen, M., Li, X., Yang, Q., Chi, X., Pan, L., Chen, N., et al. (2014). Dynamic succession of soil bacterial community during continuous cropping of peanut (Arachis hypogaea L.). PLoS One 9 (7), e101355. doi:10.1371/journal.pone.0101355
Chen, P., Wang, Y.-z., Liu, Q.-z., Zhang, Y.-t., Li, X.-y., Li, H.-q., et al. (2020). Phase changes of continuous cropping obstacles in strawberry (Fragaria × ananassa Duch.) production. Appl. Soil Ecol. 155, 103626. doi:10.1016/j.apsoil.2020.103626
Chen, W., Guo, X., Guo, Q., Tan, X., and Wang, Z. (2021). Long-term chili monoculture alters environmental variables affecting the dominant microbial community in rhizosphere soil. Front. Microbiol. 12, 681953. doi:10.3389/fmicb.2021.681953
Chen, W., Teng, Y., Li, Z., Liu, W., Ren, W., Luo, Y., et al. (2018). Mechanisms by which organic fertilizer and effective microbes mitigate peanut continuous cropping yield constraints in a red soil of south China. Appl. Soil Ecol. 128, 23–34. doi:10.1016/j.apsoil.2018.03.018
Cui, R., Geng, G., Wang, G., Stevanato, P., Dong, Y., Li, T., et al. (2022). The response of sugar beet rhizosphere micro-ecological environment to continuous cropping. Front. Microbiol. 13, 956785. doi:10.3389/fmicb.2022.956785
Dacal, M., García-Palacios, P., Asensio, S., Wang, J., Singh, B. K., and Maestre, F. T. (2022). Climate change legacies contrastingly affect the resistance and resilience of soil microbial communities and multifunctionality to extreme drought. Funct. Ecol. 36 (4), 908–920. doi:10.1111/1365-2435.14000
Dang, H., Zhang, T., Li, G., Mu, Y., Lv, X., Wang, Z., et al. (2020). Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol. 20 (1), 291. doi:10.1186/s12866-020-01977-3
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 (10), 996–998. doi:10.1038/nmeth.2604
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U. S. A. 112 (8), E911–E920. doi:10.1073/pnas.1414592112
Fan, Y., Liu, J., Liu, Z., Hu, X., Yu, Z., Li, Y., et al. (2022). Chitin amendments eliminate the negative impacts of continuous cropping obstacles on soil properties and microbial assemblage. Front. Plant Sci. 13, 1067618. doi:10.3389/fpls.2022.1067618
Feng, Y., Zhang, H., Song, X., Ge, T., Zhu, J., Zhou, C., et al. (2022). Microalgae as a potential conditioner for continuous cropping obstacles for taro (Colocasia esculenta L. Schott) production. J. Clean. Prod. 369, 133356. doi:10.1016/j.jclepro.2022.133356
Fu, H., Zhang, G., Zhang, F., Sun, Z., Geng, G., and Li, T. (2017). Effects of continuous tomato monoculture on soil microbial properties and enzyme activities in a solar greenhouse. Sustainability 9 (2), 317. doi:10.3390/su9020317
Gao, Z., Han, M., Hu, Y., Li, Z., Liu, C., Wang, X., et al. (2019). Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front. Microbiol. 10, 2269. doi:10.3389/fmicb.2019.02269
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). MICROBIAL diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi:10.1146/annurev.phyto.42.012604.135455
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28 (3), 367–374. doi:10.1016/j.biotechadv.2010.02.001
Gonzalez, E., Pitre, F. E., Pagé, A. P., Marleau, J., Guidi Nissim, W., St-Arnaud, M., et al. (2018). Trees, fungi and bacteria: tripartite metatranscriptomics of a root microbiome responding to soil contamination Microbiome. 6 (1), 53. doi:10.1186/s40168-018-0432-5
Hao, X., Zhu, P., Zhang, H., Liang, Y., Yin, H., Liu, X., et al. (2019). Mixotrophic acidophiles increase cadmium soluble fraction and phytoextraction efficiency from cadmium contaminated soils. Sci. Total Environ. 655, 347–355. doi:10.1016/j.scitotenv.2018.11.221
He, M.-Q., Zhao, R.-L., Liu, D.-M., Denchev, T. T., Begerow, D., Yurkov, A., et al. (2022). Species diversity of Basidiomycota. Fungal Divers. 114, 281–325. doi:10.1007/s13225-021-00497-3
Herrmann, M., Wegner, C. E., Taubert, M., Geesink, P., Lehmann, K., Yan, L., et al. (2019). Predominance of cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 10, 1407. doi:10.3389/fmicb.2019.01407
Hu, Y., Chen, M., Yang, Z., Cong, M., Zhu, X., and Jia, H. (2021). Soil microbial community response to nitrogen application on a swamp meadow in the arid region of central asia. Front. Microbiol. 12, 797306. doi:10.3389/fmicb.2021.797306
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1 (1), 22. doi:10.1186/2049-2618-1-22
Jeewani, P. H., Van Zwieten, L., Zhu, Z., Ge, T., Guggenberger, G., Luo, Y., et al. (2021). Abiotic and biotic regulation on carbon mineralization and stabilization in paddy soils along iron oxide gradients. Soil Biol. Biochem. 160, 108312. doi:10.1016/j.soilbio.2021.108312
Jin, X., Rahman, M. K. U., Ma, C., Zheng, X., Wu, F., and Zhou, X. (2023). Silicon modification improves biochar's ability to mitigate cadmium toxicity in tomato by enhancing root colonization of plant-beneficial bacteria. Ecotoxicol. Environ. Saf. 249, 114407. doi:10.1016/j.ecoenv.2022.114407
Khan, A. L., Waqas, M., Kang, S. M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52 (8), 689–695. doi:10.1007/s12275-014-4002-7
Kim, M. J., Lee, H. W., Kim, J. Y., Kang, S. E., Roh, S. W., Hong, S. W., et al. (2020). Impact of fermentation conditions on the diversity of white colony-forming yeast and analysis of metabolite changes by white colony-forming yeast in kimchi. Food Res. Int. 136, 109315. doi:10.1016/j.foodres.2020.109315
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22 (21), 5271–5277. doi:10.1111/mec.12481
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2017). Genomic features of bacterial adaptation to plants. Nat. Genet. 50 (1), 138–150. doi:10.1038/s41588-017-0012-9
Li, H., Li, C., Song, X., Liu, Y., Gao, Q., Zheng, R., et al. (2022a). Impacts of continuous and rotational cropping practices on soil chemical properties and microbial communities during peanut cultivation. Sci. Rep. 12 (1), 2758. doi:10.1038/s41598-022-06789-1
Li, J., Wang, Y. K., Guo, Z., Li, J. B., Tian, C., Hua, D. W., et al. (2020a). Effects of conservation tillage on soil physicochemical properties and crop yield in an arid loess plateau, China. Sci. Rep. 10 (1), 4716. doi:10.1038/s41598-020-61650-7
Li, R., Dorfler, U., Munch, J. C., and Schroll, R. (2017a). Enhanced degradation of isoproturon in an agricultural soil by a Sphingomonas sp. strain and a microbial consortium. Chemosphere 168, 1169–1176. doi:10.1016/j.chemosphere.2016.10.084
Li, X., Jousset, A., de Boer, W., Carrion, V. J., Zhang, T., Wang, X., et al. (2019). Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 13 (3), 738–751. doi:10.1038/s41396-018-0300-0
Li, X., Shi, X. S., Lu, M. Y., Zhao, Y. Z., Guo, R. B., and Peng, H. (2020b). Improved nitrogen conservation capacity during composting of dairy manure amended with oil shale semi-coke as the porous bulking agent. J. Hazard Mater 388, 121742. doi:10.1016/j.jhazmat.2019.121742
Li, Y., Han, C., Dong, X., Sun, S., and Zhao, C. (2022b). Soil microbial communities of dryland legume plantations are more complex than non-legumes. Sci. Total Environ. 822, 153560. doi:10.1016/j.scitotenv.2022.153560
Li, Y., Li, Z., Arafat, Y., Lin, W., Jiang, Y., Weng, B., et al. (2017b). Characterizing rhizosphere microbial communities in long-term monoculture tea orchards by fatty acid profiles and substrate utilization. Eur. J. Soil Biol. 81, 48–54. doi:10.1016/j.ejsobi.2017.06.008
Ling, N., Wang, T., and Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions Nat. Commun. 13 (1), 836. doi:10.1038/s41467-022-28448-9
Liu, L., and Greaver, T. L. (2010). A global perspective on belowground carbon dynamics under nitrogen enrichment. Ecol. Lett. 13 (7), 819–828. doi:10.1111/j.1461-0248.2010.01482.x
Liu, Q., Wang, S., Li, K., Qiao, J., Guo, Y., Liu, Z., et al. (2021). Responses of soil bacterial and fungal communities to the long-term monoculture of grapevine. Appl. Microbiol. Biotechnol. 105 (18), 7035–7050. doi:10.1007/s00253-021-11542-1
Liu, Z., Liu, J., Yu, Z., Yao, Q., Li, Y., Liang, A., et al. (2020). Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 197, 104503. doi:10.1016/j.still.2019.104503
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi:10.1093/bioinformatics/btr507
Mestre, M. C., Tamayo Navarrete, M. I., and García Garrido, J. M. (2022). Exploring the yeast-mycorrhiza-plant interaction: Saccharomyces eubayanus negative effects on arbuscular mycorrhizal formation in tomato plants. Plant Soil 479 (1-2), 529–542. doi:10.1007/s11104-022-05538-7
Miura, Y., Watanabe, Y., and Okabe, S. (2007). Significance of Chloroflexi in performance of submerged membrane bioreactors (MBR) treating municipal wastewater. Environ. Sci. Technol. 41 (22), 7787–7794. doi:10.1021/es071263x
Morse, N., Payne, E., Henry, R., Hatt, B., Chandrasena, G., Shapleigh, J., et al. (2018). Plant-microbe interactions drive denitrification rates, dissolved nitrogen removal, and the abundance of denitrification genes in stormwater control measures. Environ. Sci. Technol. 52 (16), 9320–9329. doi:10.1021/acs.est.8b02133
Narsing Rao, M. P., Luo, Z. H., Dong, Z. Y., Li, Q., Liu, B. B., Guo, S. X., et al. (2022). Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles. Environ. Res. 209, 112888. doi:10.1016/j.envres.2022.112888
Nayyar, A., Hamel, C., Lafond, G., Gossen, B. D., Hanson, K., and Germida, J. (2009). Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 43 (1), 115–121. doi:10.1016/j.apsoil.2009.06.008
Niu, J., Chao, J., Xiao, Y., Chen, W., Zhang, C., Liu, X., et al. (2017). Insight into the effects of different cropping systems on soil bacterial community and tobacco bacterial wilt rate. J. Basic Microbiol. 57 (1), 3–11. doi:10.1002/jobm.201600222
Niu, J., Rang, Z., Zhang, C., Chen, W., Tian, F., Yin, H., et al. (2016). The succession pattern of soil microbial communities and its relationship with tobacco bacterial wilt. BMC Microbiol. 16 (1), 233. doi:10.1186/s12866-016-0845-x
Oldroyd, G. E. D., and Leyser, O. (2020). A plant's diet, surviving in a variable nutrient environment. Science 368 (6486), eaba0196. doi:10.1126/science.aba0196
Pang, Z., Dong, F., Liu, Q., Lin, W., Hu, C., and Yuan, Z. (2021). Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 12, 627569. doi:10.3389/fmicb.2021.627569
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11 (11), 789–799. doi:10.1038/nrmicro3109
Qin, S., Xing, K., Jiang, J. H., Xu, L. H., and Li, W. J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89 (3), 457–473. doi:10.1007/s00253-010-2923-6
Qin, S., Yeboah, S., Cao, L., Zhang, J., Shi, S., and Liu, Y. (2017). Breaking continuous potato cropping with legumes improves soil microbial communities, enzyme activities and tuber yield. PLoS One 12 (5), e0175934. doi:10.1371/journal.pone.0175934
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi:10.1093/nar/gks1219
Ren, X., He, X., Zhang, Z., Yan, Z., Jin, H., Li, X., et al. (2015). Isolation, identification, and autotoxicity effect of allelochemicals from rhizosphere soils of flue-cured tobacco. J. Agric. Food Chem. 63 (41), 8975–8980. doi:10.1021/acs.jafc.5b03086
Sessitsch, A., Pfaffenbichler, N., and Mitter, B. (2019). Microbiome applications from lab to field: facing complexity. Trends Plant Sci. 24 (3), 194–198. doi:10.1016/j.tplants.2018.12.004
Shao, P., Liang, C., Lynch, L., Xie, H., and Bao, X. (2019). Reforestation accelerates soil organic carbon accumulation: evidence from microbial biomarkers. Soil Biol. Biochem. 131, 182–190. doi:10.1016/j.soilbio.2019.01.012
Shen, M. C., Shi, Y. Z., Bo, G. D., and Liu, X. M. (2022). Fungal inhibition of agricultural soil pathogen stimulated by nitrogen-reducing fertilization. Front. Bioeng. Biotechnol. 10, 866419. doi:10.3389/fbioe.2022.866419
Shen, M. C., Zhang, Y. Z., Bo, G. D., Yang, B., Wang, P., Ding, Z. Y., et al. (2021). Microbial responses to the reduction of chemical fertilizers in the rhizosphere soil of flue-cured tobacco. Front. Bioeng. Biotechnol. 9, 812316. doi:10.3389/fbioe.2021.812316
Shipton, P. J. (1977). Monoculture and soilborne plant pathogens. Annu. Rev. Phytopathology 15 (1), 387–407. doi:10.1146/annurev.py.15.090177.002131
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31 (4), 425–448. doi:10.1111/j.1574-6976.2007.00072.x
Tan, G., Liu, Y., Peng, S., Yin, H., Meng, D., Tao, J., et al. (2021). Soil potentials to resist continuous cropping obstacle: three field cases. Environ. Res. 200, 111319. doi:10.1016/j.envres.2021.111319
Tao, C., Li, R., Xiong, W., Shen, Z., Liu, S., Wang, B., et al. (2020). Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 8 (1), 137. doi:10.1186/s40168-020-00892-z
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., et al. (2018). Core microbiomes for sustainable agroecosystems. Nat. Plants 4 (5), 247–257. doi:10.1038/s41477-018-0139-4
Wang, J., Li, M., Zhou, Q., and Zhang, T. (2022a). Effects of continuous cropping Jiashi muskmelon on rhizosphere microbial community. Front. Microbiol. 13, 1086334. doi:10.3389/fmicb.2022.1086334
Wang, M., Zhang, L., He, Y., Huang, L., Liu, L., Chen, D., et al. (2022b). Soil fungal communities affect the chemical quality of flue-cured tobacco leaves in Bijie, Southwest China. Sci. Rep. 12 (1), 2815. doi:10.1038/s41598-022-06593-x
Wang, P., Yan, S., Zhang, W., Xie, X., Li, M., Ren, T., et al. (2022c). Effect of soil management systems on the rhizosphere bacterial community structure of tobacco: continuous cropping vs. paddy-upland rotation. Front. Plant Sci. 13, 996858. doi:10.3389/fpls.2022.996858
Wang, S., Cheng, J., Li, T., and Liao, Y. (2020). Response of soil fungal communities to continuous cropping of flue-cured tobacco. Sci. Rep. 10 (1), 19911. doi:10.1038/s41598-020-77044-8
Wang, Z., Yang, Y., Xia, Y., Wu, T., Zhu, J., Yang, J., et al. (2019). Time-course relationship between environmental factors and microbial diversity in tobacco soil. Sci. Rep. 9 (1), 19969. doi:10.1038/s41598-019-55859-4
Wei, Z., and Yu, D. (2018). Analysis of the succession of structure of the bacteria community in soil from long-term continuous cotton cropping in Xinjiang using high-throughput sequencing. Arch. Microbiol. 200 (4), 653–662. doi:10.1007/s00203-018-1476-4
Welbaum, G. E., Sturz, A. V., Dong, Z., and Nowak, J. (2004). Managing soil microorganisms to improve productivity of agro-ecosystems. Crit. Rev. Plant Sci. 23 (2), 175–193. doi:10.1080/07352680490433295
Wilson, S., Andrews, P., and Nair, T. S. (2004). Non-fumigant management of apple replant disease. Sci. Hortic. 102 (2), 221–231. doi:10.1016/j.scienta.2004.01.001
Xu, X., Luo, Q., Wei, Q., Jiang, S., Dong, C., Faruque, M. O., et al. (2022). The deterioration of agronomical traits of the continuous cropping of stevia is associated with the dynamics of soil bacterial community. Front. Microbiol. 13, 917000. doi:10.3389/fmicb.2022.917000
Yan, L., Zhang, W., Duan, W., Zhang, Y., Zheng, W., and Lai, X. (2021). Temporal bacterial community diversity in the nicotiana tabacum rhizosphere over years of continuous monocropping. Front. Microbiol. 12, 641643. doi:10.3389/fmicb.2021.641643
Yi, X., Yi, K., Fang, K., Gao, H., Dai, W., and Cao, L. (2019). Microbial community structures and important associations between soil nutrients and the responses of specific taxa to rice-frog cultivation. Front. Microbiol. 10, 1752. doi:10.3389/fmicb.2019.01752
Yu, F., Yan, Y., Dong, Q., Jiang, C., Zu, C., and Shen, J. (2023). The changes in rhizosphere metabolome and microbiota are the main direct obstacles to continuous cropping in tobacco (nicotiana tabacum L.). Agronomy 13 (4), 964. doi:10.3390/agronomy13040964
Yuan, S., Wang, L., Wu, K., Shi, J., Wang, M., Yang, X., et al. (2014). Evaluation of Bacillus-fortified organic fertilizer for controlling tobacco bacterial wilt in greenhouse and field experiments. Appl. Soil Ecol. 75, 86–94. doi:10.1016/j.apsoil.2013.11.004
Zhang, C., Zhang, Q., Luo, M., Wang, Q., and Wu, X. (2023). Bacillus cereus WL08 immobilized on tobacco stem charcoal eliminates butylated hydroxytoluene in soils and alleviates the continuous cropping obstacle of Pinellia ternata. J. Hazard Mater 450, 131091. doi:10.1016/j.jhazmat.2023.131091
Zhao, Y. X., Guo, L., Wang, L., Jiang, N. D., Chen, K. X., and Dai, Y. J. (2021). Biodegradation of the pyridinecarboxamide insecticide flonicamid by Microvirga flocculans and characterization of two novel amidases involved. Ecotoxicol. Environ. Saf. 220, 112384. doi:10.1016/j.ecoenv.2021.112384
Zhou, L., Li, H., Zhang, Y., Han, S., and Xu, H. (2016). Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Braz J. Microbiol. 47 (2), 271–278. doi:10.1016/j.bjm.2016.01.001
Zhou, Y., Coventry, D. R., Gupta, V. V. S. R., Fuentes, D., Merchant, A., Kaiser, B. N., et al. (2020). The preceding root system drives the composition and function of the rhizosphere microbiome Genome Biol. 21 (1), 89. doi:10.1186/s13059-020-01999-0
Zhou, X., Zhang, J., Khashi, U. R. M., Gao, D., Wei, Z., Wu, F., et al. (2023). Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes. Mol. Plant 16 (5), 849–864. doi:10.1016/j.molp.2023.03.009
Zhu, S., Wang, Y., Xu, X., Liu, T., Wu, D., Zheng, X., et al. (2018). Potential use of high-throughput sequencing of soil microbial communities for estimating the adverse effects of continuous cropping on ramie (Boehmeria nivea L. Gaud). PLoS One 13 (5), e0197095. doi:10.1371/journal.pone.0197095
Keywords: tobacco, long-term continuous cropping, physicochemical properties, microbiota, correlation
Citation: Li G, Gong P, Zhou J, Wang L, Song X, Ding P, Jin Y, Zhang Y, Zhou X, Yang J, Wang Z, Yun L and Ren J (2024) The succession of rhizosphere microbial community in the continuous cropping soil of tobacco. Front. Environ. Sci. 11:1251938. doi: 10.3389/fenvs.2023.1251938
Received: 03 July 2023; Accepted: 14 December 2023;
Published: 19 January 2024.
Edited by:
Meihua Deng, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Paul Daly, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaPaula Pérez-Rodríguez, University of Vigo, Spain
Yanning Zheng, Chinese Academy of Sciences (CAS), China
Xingang Zhou, Northeast Agricultural University, China
Copyright © 2024 Li, Gong, Zhou, Wang, Song, Ding, Jin, Zhang, Zhou, Yang, Wang, Yun and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Yun, MjQ0ODM4Njc5QHFxLmNvbQ==; Jie Ren, cmVuamllQGNhYXMuY24=
 Guitong Li1
Guitong Li1 Yuzhen Zhang
Yuzhen Zhang Jianming Yang
Jianming Yang Zhaobao Wang
Zhaobao Wang Jie Ren
Jie Ren