- 1Jiangxi Key Laboratory for Postharvest Technology and Nondestructive Testing of Fruits & Vegetables, College of Agronomy, Jiangxi Agricultural University, Nanchang, China
- 2College of Life Science and Technology, Honghe University, Mengzi, China
- 3Green Biotechnologies Research Centre of Excellence, University of Limpopo, Mankweng, South Africa
- 4College of Materials and Chemical Engineering, Pingxiang University, Pingxiang, China
Penicillium digitatum is the most severe pathogen that infects citrus fruits during storage. It can cause fruit rot and bring significant economic losses. The continuous use of fungicides has resulted in the emergence of drug-resistant strains. Consequently, there is a need to develop naturally and efficiently antifungal fungicides. Natural antimicrobial agents such as clove oil, cinnamon oil, and thyme oil can be extracted from different plant parts. They exhibited broad-spectrum antimicrobial properties and have great potential in the food industry. Here, we exploit a novel cinnamaldehyde (CA), eugenol (EUG), or carvacrol (CAR) combination antifungal therapy and formulate it into nanoemulsion form to overcome lower solubility and instability of essential oil. In this study, the antifungal activity evaluation and transcriptional profile of Penicillium digitatum exposed to compound nanoemulsion were evaluated. Results showed that compound nanoemulsion had a striking inhibitory effect on P. digitatum in a dose-dependent manner. According to RNA-seq analysis, there were 2,169 differentially expressed genes (DEGs) between control and nanoemulsion-treated samples, including 1,028 downregulated and 1,141 upregulated genes. Gene Ontology (GO) analysis indicated that the DEGs were mainly involved in intracellular organelle parts of cell component: cellular respiration, proton transmembrane transport of biological process, and guanyl nucleotide-binding molecular function. KEGG analysis revealed that metabolic pathway, biosynthesis of secondary metabolites, and glyoxylate and dicarboxylate metabolism were the most highly enriched pathways for these DEGs. Taken together, we can conclude the promising antifungal activity of nanoemulsion with multiple action sites against P. digitatum. These outcomes would deepen our knowledge of the inhibitory mechanism from molecular aspects and exploit naturally, efficiently, and harmlessly antifungal agents in the citrus postharvest industry.
Introduction
During the postharvest, handling, and transportation process, citrus usually suffers significant losses. Related studies show that citrus postharvest rot is mainly green mold caused by P. digitatum, accounting for more than 80% of the total crop losses (1). Currently, chemical agent application is the chief approach to combat this pathogen; notwithstanding it has a specific effect of prolonging the storage period of citrus, chemical agents have aroused concern about resistant strains, human health, and environmental pollution (2). Searching for exploitation of natural antifungal substances from plants as chemical compound alternatives has attracted more attention of researchers (3–7). Essential oil, a volatile aromatic compound, can be extracted from different parts of the plant (8). Essential oils have been recognized as GRAS (generally recognized as safe) by the US food and drug administration. Cinnamaldehyde and eugenol are the main components of cinnamon essential oil and clove oil, respectively. Carvacrol was found in the thyme oil or oregano oil and has been reported to possess antimicrobial activities (9–11). Cinnamaldehyde could effectively inhibit Aspergillus flavus through plasma membrane damage (5). Carvacrol has been demonstrated to have an antibacterial efficiency against Listeria monocytogenes and Escherichia coli (12); eugenol has an antifungal activity against Cryptococcus gattii and C. neoformans (13).
Antifungal drug combination against fungal growth is a reasonable strategy. Compared with individual fungicides, compound fungicides can reduce the effective dose. When two or more drugs act synergistically, it can reduce the cost and potential toxicity (14, 15). The combination can delay the evolution of fungal resistance. More importantly, they can make existing and approved drugs repurposed, bypassing the expensive and lengthy development of new antifungal agents (16, 17). Essential oils encapsulated in nanoform are a promising alternative for antimicrobial strategy (18, 19). Nanoemulsion can enhance an antibacterial or antifungal activity of essential oil against microorganisms and overcome the drawbacks of essential oils, such as low solubility, instable under light or oxygen (20, 21).
Transcriptomics is used to illuminate the transcriptional level of genes under different stress to elucidate its regulatory mechanism. In particular, a new high-throughput sequencing method (RNA-seq) has been widely used to study eukaryotic transcriptomes (22–24). RNA-seq technology has the advantages such as high-throughput, high-sensitivity, digital signal, accurate results, good reproducibility, low cost, and no species limitation. It can accurately explain the pathogenic mechanism of fungi and the inhibition mechanism of antifungal substances on microorganisms at the molecular level (25, 26).
In this study, the antifungal mechanism of a combination of eugenol, cinnamaldehyde, and carvacrol nanoemulsion on P. digitatum was determined with an RNA-Seq approach. P. digitatum transcriptome and the DEGs between nanoemulsion-treated and untreated samples were obtained, which provide a theoretical reference for improving the prevention and control effect of citrus postharvest diseases.
Materials and Methods
Reagent and Fungal Cultivation
Cinnamaldehyde, citral, and eugenol were purchased from Aladdin Co., Ltd. (Shanghai, China). First, a combination of cinnamaldehyde, citral, and eugenol in a ratio of 1:1:1 was dissolved in 2% Tween-80, then blended with a high-shear mixer (ULTRA TURRAX® T18 digital, IKA, Staufen, Germany), and finally passed through a microfluidizer (M-110P, Microfluidics Corporation, United States) to obtain a stock solution of 100 mg/ml. P. digitatum was isolated from green mold-infected citrus in our laboratory and maintained on PDA medium at 4°C. Fungal conidia from a 7-day-old culture were washed with sterilized water, filtered through four layers of gauze, and finally adjusted to a suspension of 1 × 107 conidia/mL by hemocytometer.
Antifungal Activity of Compound Nanoemulsion
The antifungal activity of compound nanoemulsion against P. digitatum was evaluated by the mycelial growth inhibition method (27) with some modifications. The stock nanoemulsions were added to the non-coagulated PDA to obtain the final concentration range (0.25, 0.125, 0.0625, 0.0313, 0.0156, and 0.0078 mg/mL). Two percent Tween-80 was added to PDA and taken as control. The plugs of mycelia (6 mm diameter) from the activated P. digitatum were transformed to the center of PDA plates and incubated at 28 ± 2°C for 7 days in the dark. The following formula calculated the growth inhibition rates of samples: mycelia growth inhibition rate = (control colony diameter-treated colony diameter) / (control colony diameter) × 100%. There are three replicates of each concentration, and the experiment was conducted twice.
Preparation of Nanoemulsion Treatment
About 1.5 ml spore suspension (1 × 107 conidia/ml) was added to 150 ml liquid culture medium (potato 200 g, glucose 20 g, 1 L distilled water) and cultured in an incubation shaker at 140 rpm for 48 h. Then, the mycelia were centrifuged at 3,000 × g for 20 min, followed by washing with phosphate buffer (pH 7.0) three times and resuspended in 100 ml PBS (pH 7.0). Afterward, the stock nanoemulsion was added to the flask (D-T) to the final concentration of 0.125 mg/mL, which is the minimum inhibitory concentration on P. digitatum, and then kept in an incubation shaker for 12 h; there was not nanoemulsion added, which was taken as the control (D-C). Finally, the mycelia, which removed phosphate buffer, were rapidly frozen in liquid nitrogen and kept in a −80°C refrigerator. Each treatment was performed three times.
Extraction, Quantification, and Qualification of RNA
Total RNA preparation, quality control, cDNA libraries construction, and RNA-seq were conducted by Shanghai Applied Protein Technology (APT) Co., Ltd. The TRIzol reagent was used (Invitrogen, United States) to isolate total RNA according to the instruction. One percent agarose gel electrophoresis was used to evaluate RNA degradation and contamination. RNA purity, concentration, and integrity were checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States), Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, United States), RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States) respectively. Each group were conducted for triple biological replicates.
Library Preparation for Transcriptome Sequencing
The input material is 3 μg RNA of each sample for the RNA preparations. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads by using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer's recommendations, and each sample was to attribute sequences by adding index codes. The divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X) were used to fragment. The fragments were used to synthesize the first-strand cDNA with random hexamer primer and M-MuLV Reverse Transcriptase (RNase H). The second-strand cDNA was transformed from the first-strand cDNA using RNase H and DNA polymerase I. Fragments of preferential lengths of about 250~300 bp were purified using the AMPure XP system (Beckman Coulter, Beverly, United States), end-repaired, and adapter-ligated. Then, 3 μl of USER Enzyme (NEB, United States) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and index (X) primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system.
De novo Transcriptome Assembly
After filtration of the lower-quality reads, obtained clean reads were used for the following analysis. The clean reads were mapped to the reference genome and assembled according to the previous method (Ouyang et al., 2016). The Trinity program was used to de novo assemble processed reads (28). The sequencing reads were used to construct a k-mer graph (k = 25). The seed k-mers were extended to both ends to form a contig. The overlapped contigs are clustered to form components, and each component becomes a set of possible representations of variable splicing isoform or homologous genes. Each component has a corresponding de Bruijn graph. The de Bruijn graph is simplified, and the path with continuous nodes is combined to form a more extended sequence, and the best path is found to obtain the transcriptional sequence.
Annotations of Unigenes
First, we were blasting the optimal transcript against the NCBI non-redundant protein sequences database (NR) and Swiss-Prot to obtain unigenes. To further annotate the unigenes, the Blast2GO program with Blast2GO default parameters was used to obtain Gene Ontology (GO) annotations. GO enrichment analyses of DEGs were implemented by the cluster profile package (version 3.4.4), in which gene length bias was corrected. GO terms with corrected P-value < 0.05 were considered significantly enriched by DEGs. We used the clusterProfiler package (version 3.4.4) to annotate the pathways of DEGs in KEGG pathways.
Quantification of Gene Expression Level
The FeatureCounts software (version 1.5.0-p3) was used to count the reads numbers mapped to each gene. Expression levels of unigenes were normalized and calculated as the values of fragments per kilobase of transcripts per million mapped fragments (FPKM). To select DEGs by DESeq2 package (version 1.16.1), DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values, which are based on the negative binomial distribution model, false discovery rate was calculated by Benjamini and Hochberg's approach. Genes with an adjusted P-value < 0.05 were assigned as differentially expressed.
Validation of RNA-Seq by qRT-PCR
Eight DEGs were selected randomly to confirm the RNA-Seq data by qRT-PCR. The total RNA was reverse-transcribed with a Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) (YEASEN Biotech Co, Ltd. Shanghai, China), according to the manufacturer's instructions. The qRT-PCR assay was performed on CFX Real-Time PCR Detection Systems (CFX96, BIO-RAD, United States). The actin gene was obtained as an internal reference (29), and other target gene primer pairs were designed by Primer-Blast of NCBI (National Centre of Biotechnology Information, Bethesda, MD, United States). The amplification program was as follows: one cycle at 95°C for 3 min, and 39 cycles at 95°C for 5 s, 60°C for 3 min. The relative gene expression was analyzed according to the method described in the study by Livak and Schmittgen (30). The primers for the qRT-PCR were synthesized by Tsingke Biotech (Beijing, China) and are presented in Table 1. Each reaction was conducted three times.
Determination of Oxidative Parameters
After the treatment with nanoemulsion, the mycelia were collected and used to determine the membrane lipid peroxidation-related parameters. The malondialdehyde (MDA) content was determined by following the previous method with minor modifications (31). According to the manufacturer's instructions, the H2O2 content was determined according to the method described in the study by Song et al. (32), using an H2O2 detection kit (Nanjing Jiancheng, Nanjing, China).
Determination of Soluble Protein Contents
Coomassie Bright Blue method was used to determine the change in soluble protein content in the mycelium of P. digitatum, and bovine serum protein was used as the standard curve. P. digitatum was incubated in a shaker at 27°C, 180 rpm for 48 h. The exact amounts of mycelia were suspended in 10 mL phosphoric acid buffer (pH 7.0), and nanoemulsion was added to make the final concentration of 0.125 mg/mL. The culture was kept in a shaker at 27°C, 180 rpm, for 0, 2, 4, 6, and 12 h. The mycelium was frozen by liquid nitrogen, and then the mycelia were grounded into a paste with 5 mL distilled water and centrifuged at 4°C, 12,000 rpm for 15 min. About 1.0 mL of supernatant was taken, and 5 mL of Coomassie Bright Blue solution was added. The solution was shaken thoroughly, then let stand for 5 min. The OD value was measured at 595 nm with distilled water as blank control. According to the standard curve, the soluble protein content of mycelia was calculated, and the result was expressed as the mass of soluble protein per gram of mycelia (mg/g). Each treatment has three biological replicates, and the experiment was conducted twice.
Statistical Analysis
Data were expressed as the mean ± SD, which was conducted triplicate, and were performed using SPSS 22.0 (SPSS Inc., United States). One-way ANOVA and Duncan's multiple range test were used to evaluate the significance (p < 0.05).
Results
Antifungal Activity of Nanoemulsion
As illustrated in Figure 1, nanoemulsion showed an increased inhibitory activity against P. digitatum with increasing concentration. At the concentration of 0.125 mg/mL, the mycelia germination inhibition rate was 66.05%; furthermore, while 0.25 mg/mL concentration significantly inhibited the mycelia growth, and the inhibition rate was 100.00% (p < 0.05) on the 7th day, the control did not show an inhibitory activity. These results indicated that nanoemulsion could inhibit the mycelia growth of P. digitatum in a dose-dependent manner.
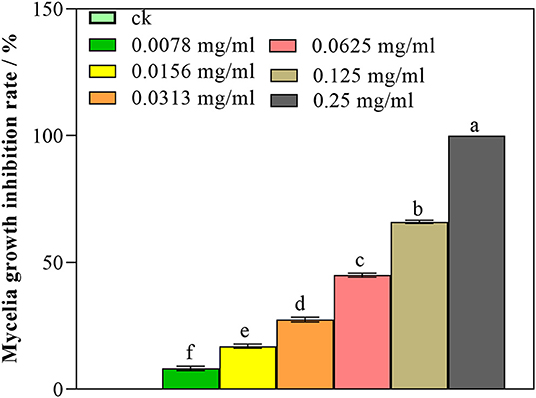
Figure 1. Antifungal activity of nanoemulsions against P. digitatum. Values are the mean ± standard deviation (SD) of three replicates. Different letters above the bars on columns indicate a significant difference (P < 0.05).
Transcriptome Sequencing Quality
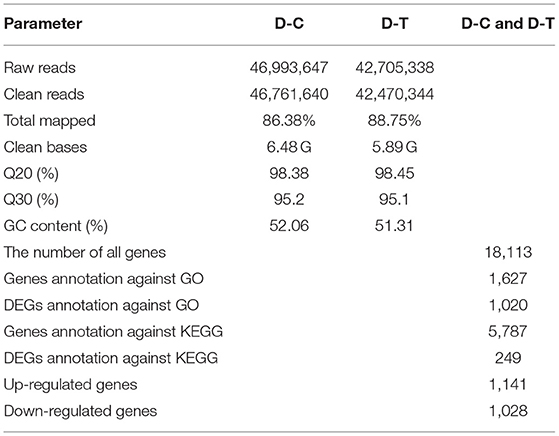
The quality inspection was conducted on all samples to meet the requirements of sequencing database construction. From the sequencing data shown in Table 2, RNA sequencing of nanoemulsion-treated and untreated P. digitatum generated 46.9 and 42.7 million raw reads, respectively. After filtering the adaptor sequences, 46.7 and 42.5 million clean reads were obtained. The GC content of each sample is above 50%, indicating that the base content is stable and there was no AT or GC separation; the Q30 base percentage of all samples is higher than 95%, which means obtained clean reads were accurate and can be used for subsequent analysis.
Transcriptional Stress Response of P. digitatum to Nanoemulsion
Figure 2 represents gene expression distribution differences and density distribution in control and nanoemulsion treatment samples. The gene expression distribution is different in three biological replicates from the same treatment, which is revealed in Figure 2A. Figure 2B indicates that most gene expression is in the lower level, while a few genes expression is in the higher level. Figure 3 shows 9,380 and 9,934 genes expressed in control and nanoemulsion treatment, of which 8,599 genes were co-expressed. The volcano plots revealed the difference in gene expression level and statistically significant difference in the DEGs. Two thousand one hundred and sixty-nine genes were differentially expressed in P. digitatum after nanoemulsion treatment: 1,141 genes showed upregulation tendency, whereas 1,028 genes were downregulated (Figure 4).
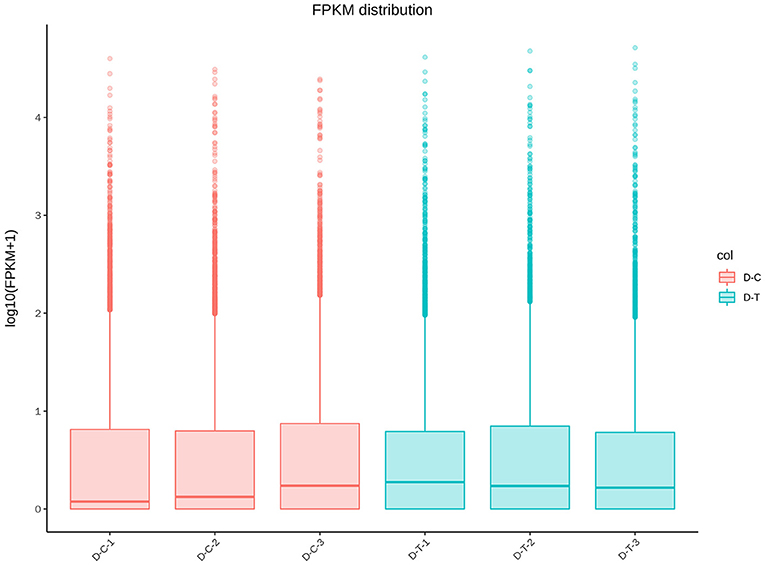
Figure 2. Gene expression-level comparison between control and nanoemulsion treatment: (A) FPKM distribution (Fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced) and (B) FPKM density distribution.
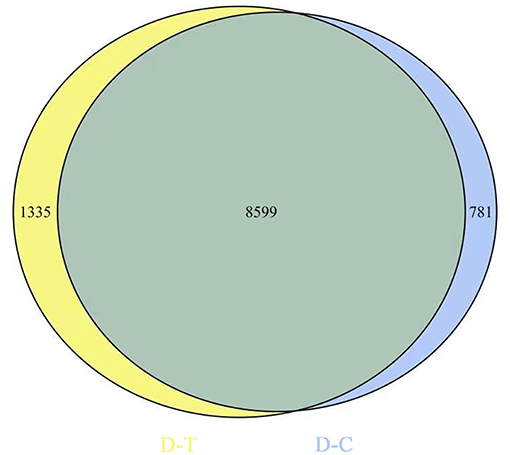
Figure 3. Venn diagram of gene expression. The number in the blue circle and yellow circle indicates the total number of genes expressed in the control and nanoemulsion treatment samples, respectively, and the green part of circles indicates that the gene is co-expressed in both samples.
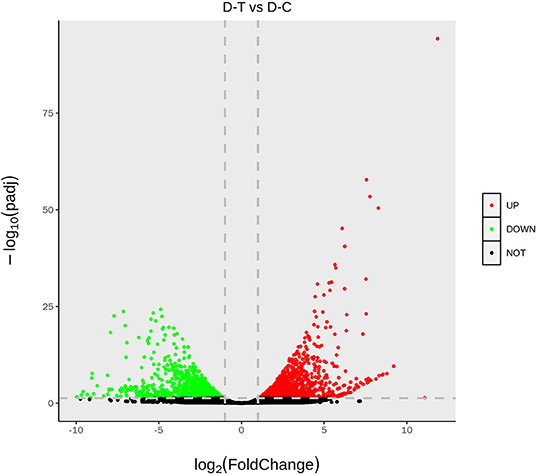
Figure 4. The volcano of differentially expressed genes. The abscissa indicates gene expression in different treatments; the ordinate indicates the statistical significance of gene expression. The scatter in the graph represents each gene, the black dot represents the gene with no significant difference, the red dot represents the upregulated gene with a significant difference, and the green dot represents the downregulated gene with a significant difference.
Enrichment Analysis of GO and KEGG Pathways
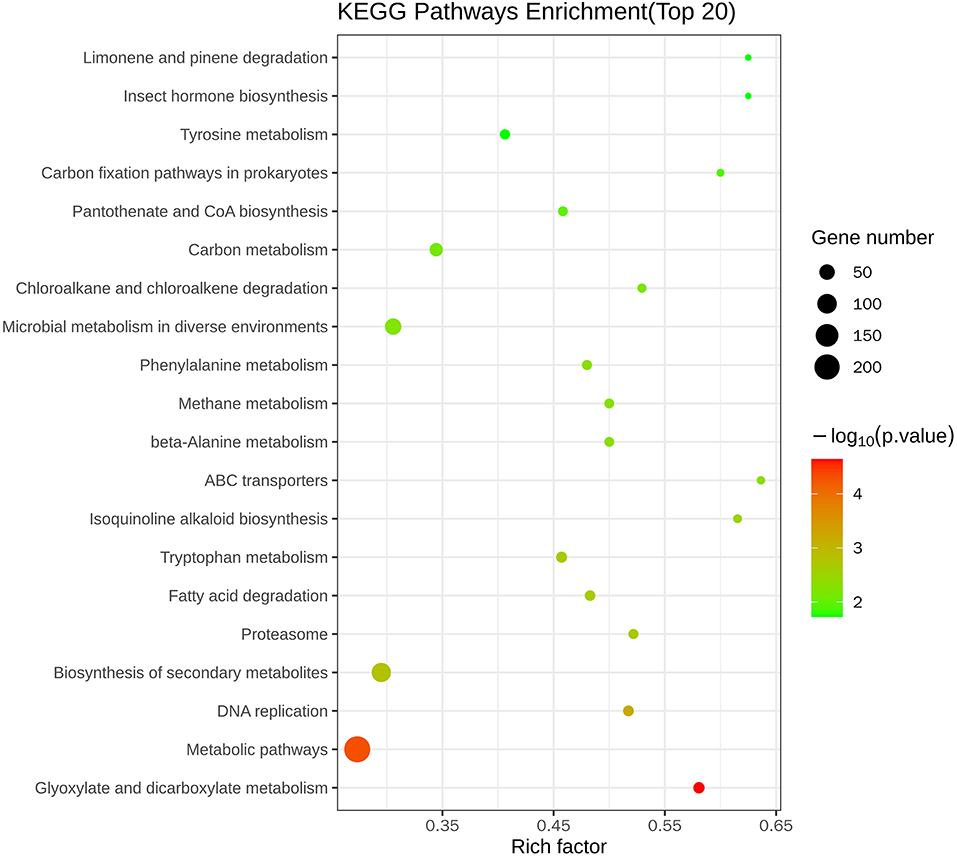
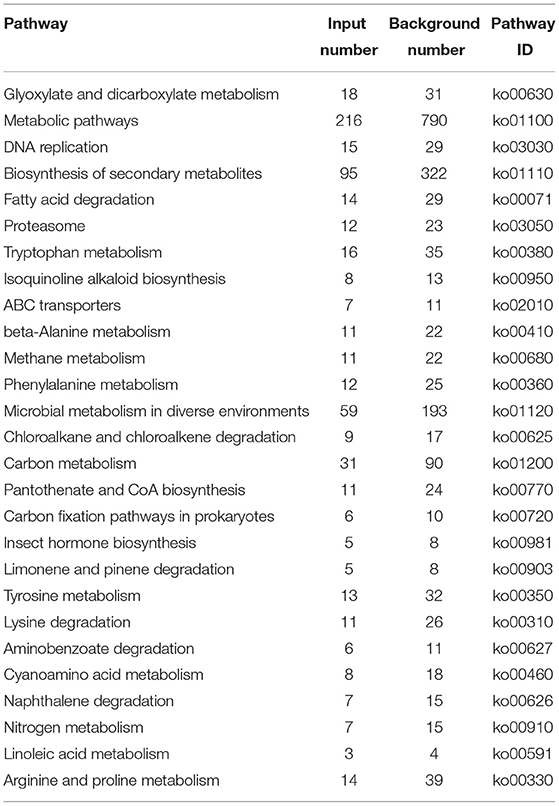
Figure 5 and Table 3 illustrate the most enriched biochemical pathways mapped by DEGs revealed by KEGG pathway analysis (P-value < 0.05). Therein, the significantly abundant DEGs (216) were enriched in the metabolic pathway (ko01100), 95 DEGs were enriched in the biosynthesis of secondary metabolites (ko01110), 18 DEGs were enriched in glyoxylate and dicarboxylate metabolism (ko00630), and 15 and 14 DEGs were enriched in DNA replication (ko03030) and fatty acid degradation (ko00071), respectively. Figures 6A,B shows the top 20 upregulated and downregulated DEGs in KEGG enrichment (P-value < 0.05). The results indicated that the downregulated pathways involved mainly belonged to the DNA replication, proteasome, chloroalkane, and chloroalkene degradation. The upregulated pathways involved are ABC transporters, alpha-linolenic acid metabolism, metabolic pathways, and biosynthesis of unsaturated fatty acids.
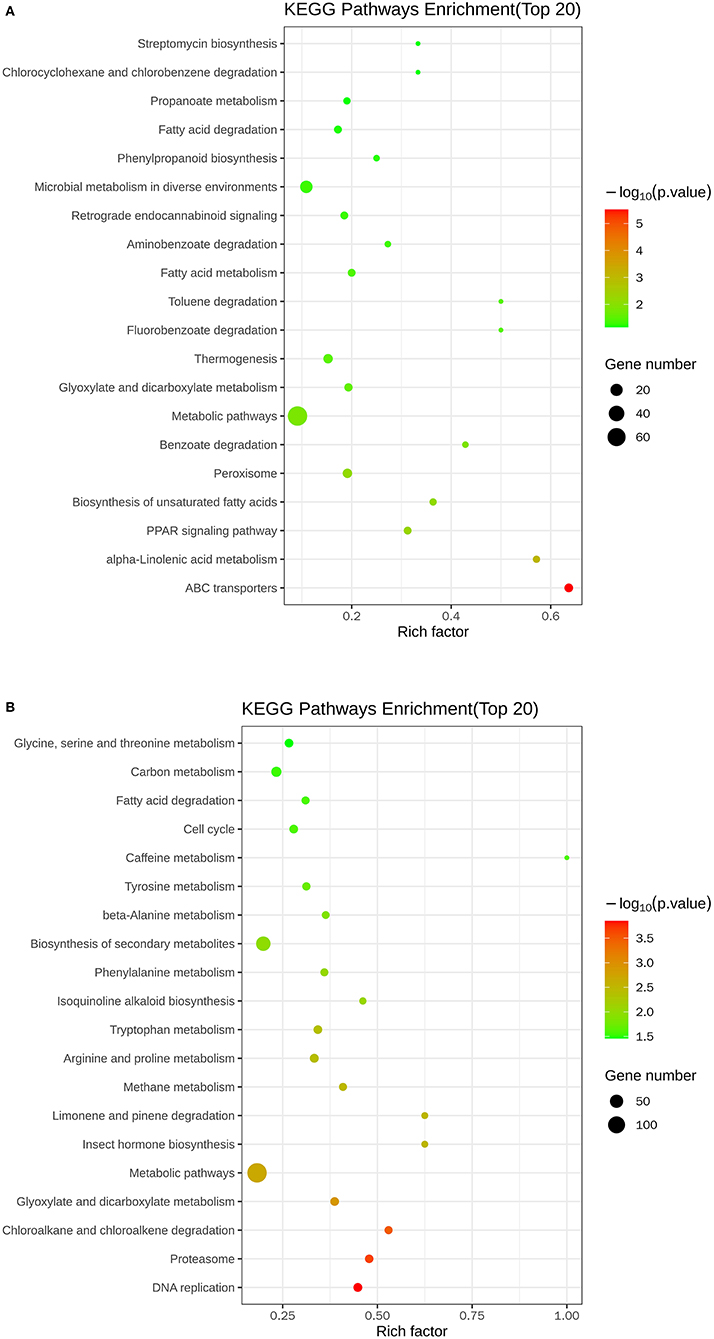
Figure 6. Scatter plots of the top 20 KEGG enrichment of upregulated (A) and downregulated (B) DEGs between control and nanoemulsion treatment.
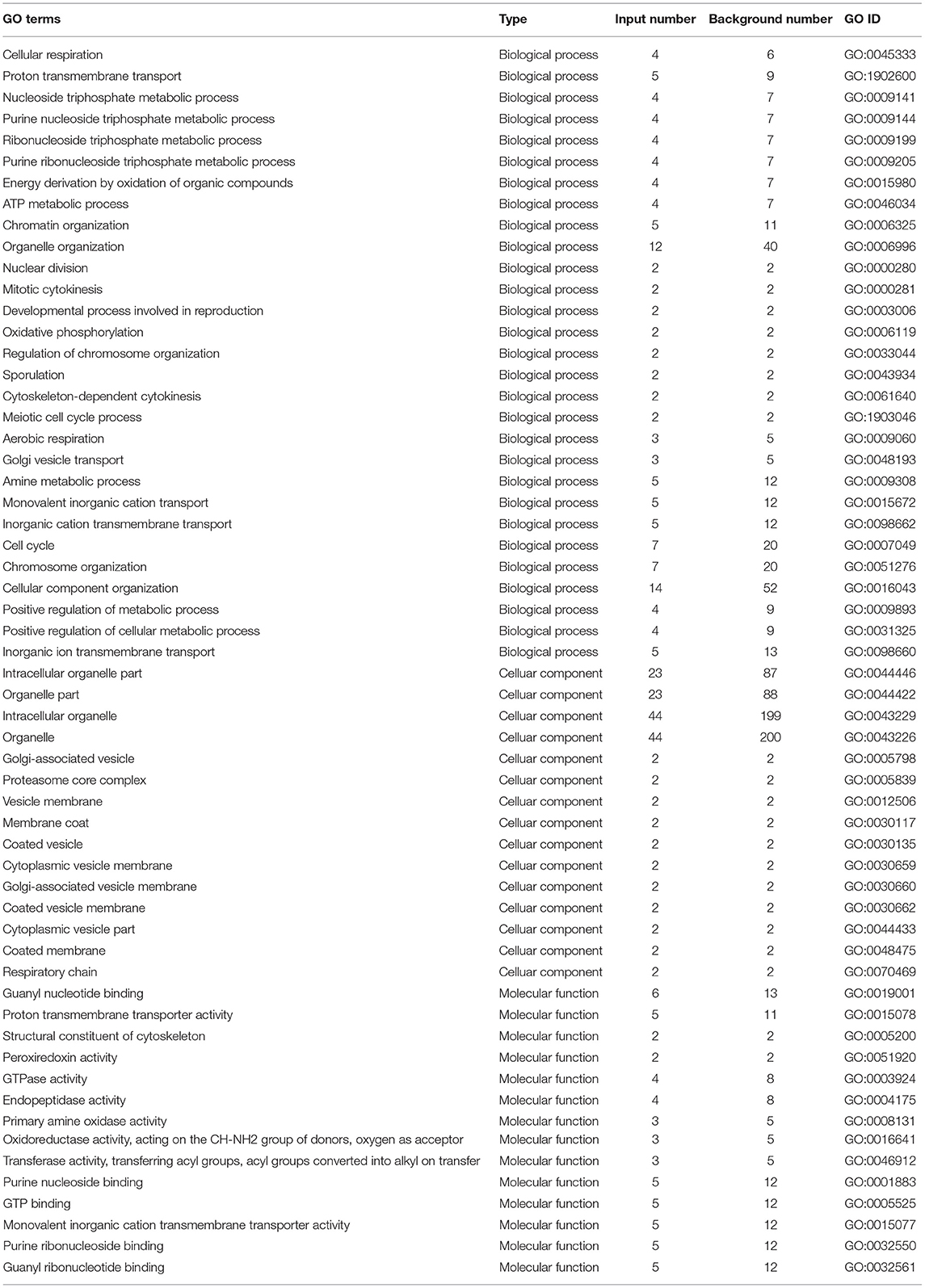
The most enriched GO in 2169 was DEGs (P-value < 0.05), which is indicated by Figure 7 and Table 4. In the biological process, the top three significant enrichment terms included cellular respiration (four DEGs), proton transmembrane transport (five DEGs), and nucleoside triphosphate metabolic process (four DEGs). The top three significant enrichment terms in cellular components were intracellular organelle part (23 DEGs), organelle part (23 DEGs), and intracellular organelle (44 DEGs). Furthermore, the top three significant enrichment terms in molecular function were mostly guanyl nucleotide-binding (six DEGs), proton transmembrane transporter activity (five DEGs), and structural constituents of the cytoskeleton (two DEGs). Figures 8A,B shows the top 20 upregulated and downregulated DEGs in GO enrichment (P-value < 0.05). The results indicated that the upregulated GO terms were cellular respiration, energy derivation by the oxidation of organic compounds, proton transmembrane transport of biological process; proton transmembrane transporter activity, monovalent inorganic cation transmembrane transporter activity and cytochrome-c oxidase activity of molecular function; mitochondrion, mitochondrial envelope, mitochondrial part of the cellular component of a cellular component. The downregulated GO terms were chromosome organization, cellular component organization, organelle organization of biological process; structural constituent of the cytoskeleton, structural molecular activity, purine nucleoside binding of molecular function, intracellular organelle, organelle, and intracellular organelle part of a cellular component.
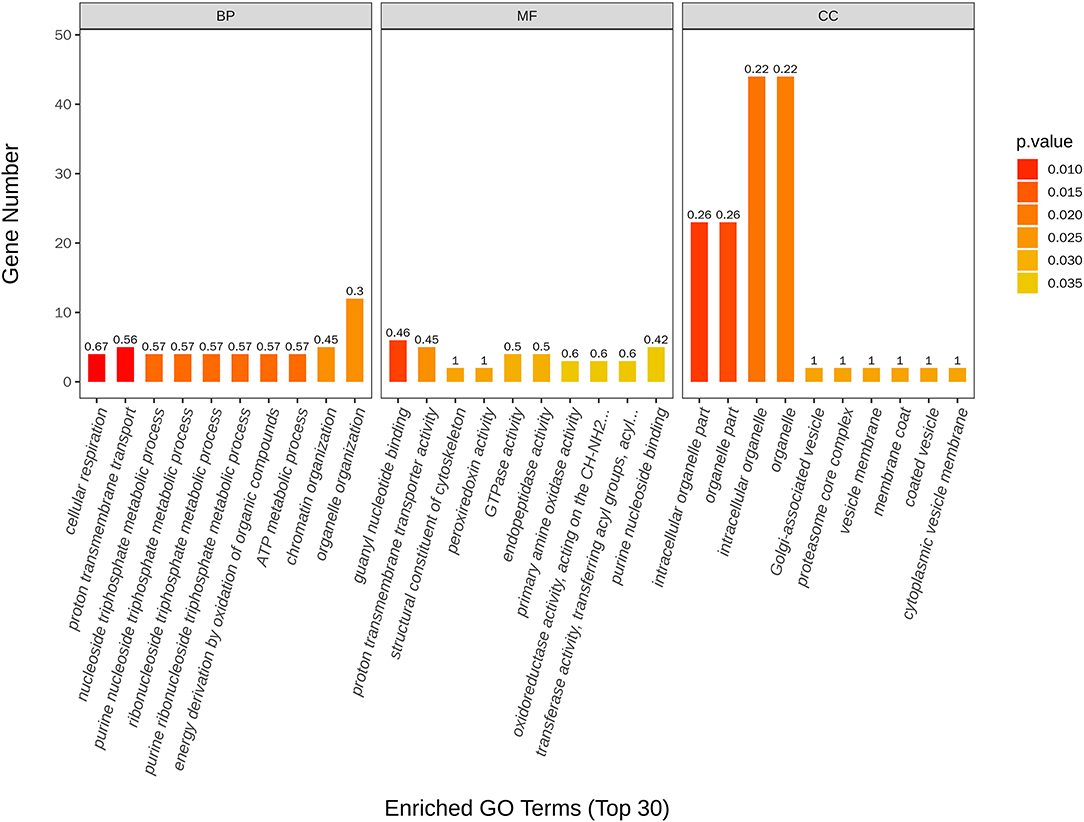
Figure 7. Top 30 GO terms of differentially expressed genes between control and nanoemulsion treatment.
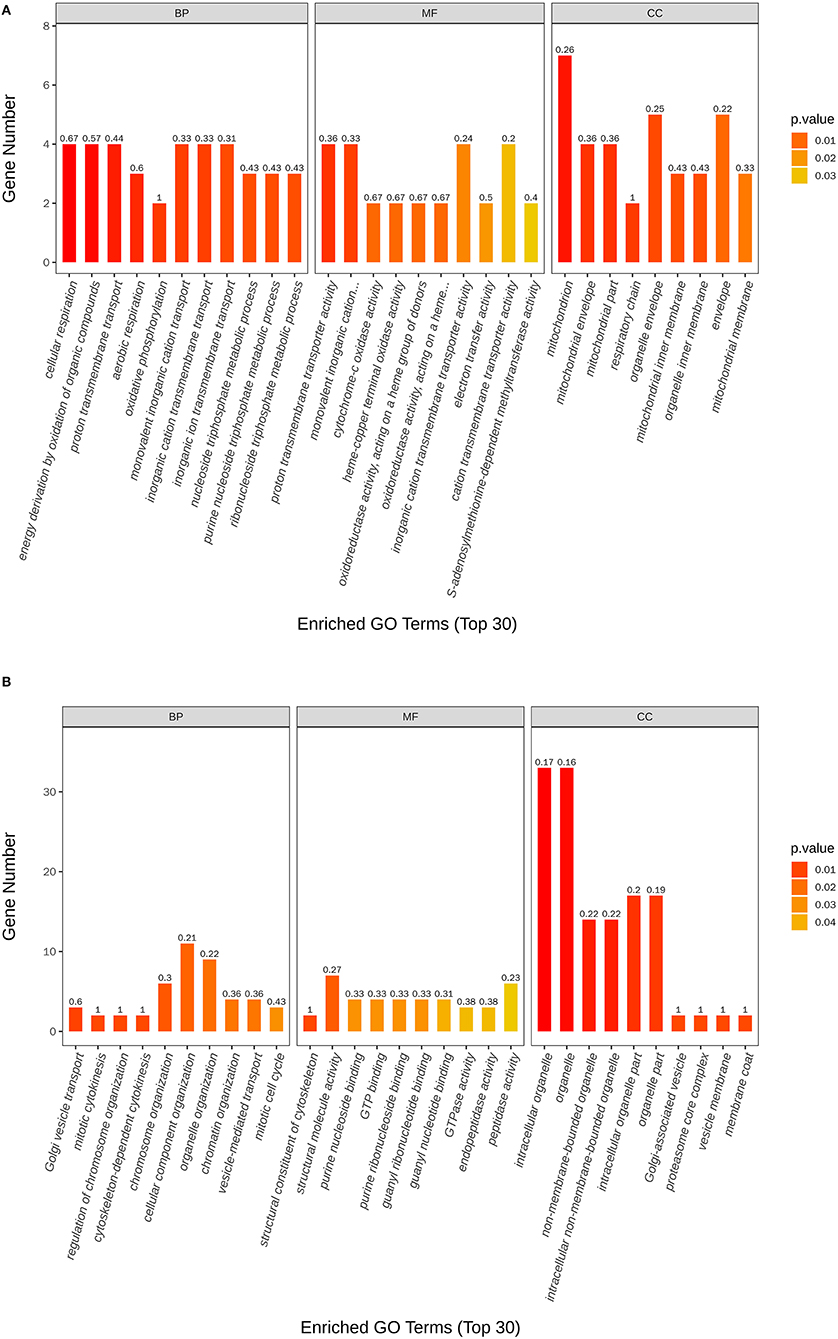
Figure 8. Scatter plots of the top 20 GO enrichment of upregulated (A) and downregulated (B) DEGs between control and nanoemulsion treatment.
Validation of the Expression of DEGs by qRT-PCR
As shown in Figure 9, eight DEGs were selected to validate the RNA-Seq results. The results of qRT-PCR experiments indicated that the expression of genes is in line with the RNA-Seq data and then confirmed the reliability of the RNA-Seq data.
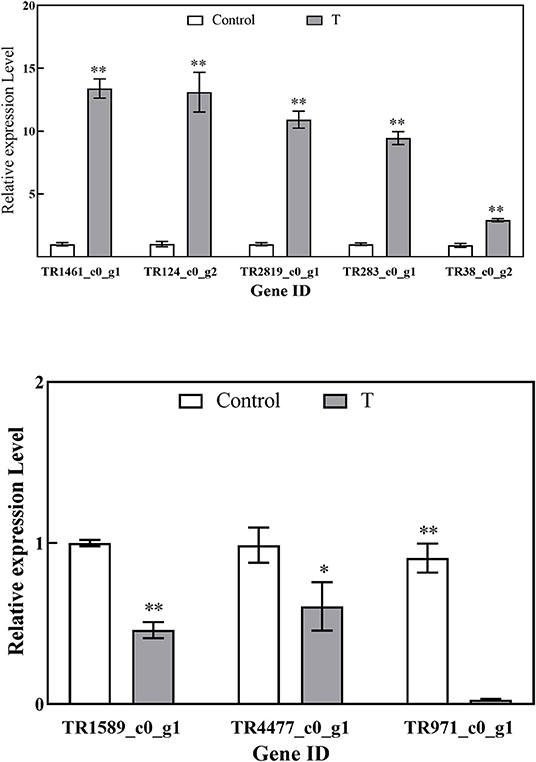
Figure 9. Relative expression levels of DEGs by RT-qPCR. Values are the mean ± standard deviation (SD) of three replicates. The “*”, “**” above the bars on columns indicate a significant difference (P < 0.05, P < 0.01) compared with control.
Effect of Nanoemulsion on Membrane Lipid Peroxidation
From Figures 10A,B, after nanoemulsion treatment, H2O2 accumulation could be significantly induced by nanoemulsion treatment in P. digitatum. Compared with the control group, the H2O2 content of the MIC treatment group increased more than 50% after 4 h. Similarly, the MDA content of mycelia increased significantly, while the content in the control group remained constant. With MIC, the MDA content reached a peak value when treated for 4 h. Afterward, the MDA content decreased gradually in MIC-treated sample, but it was still significantly higher than the control, which implies that the oxidative stress of P. digitatum refers to nanoemulsion.
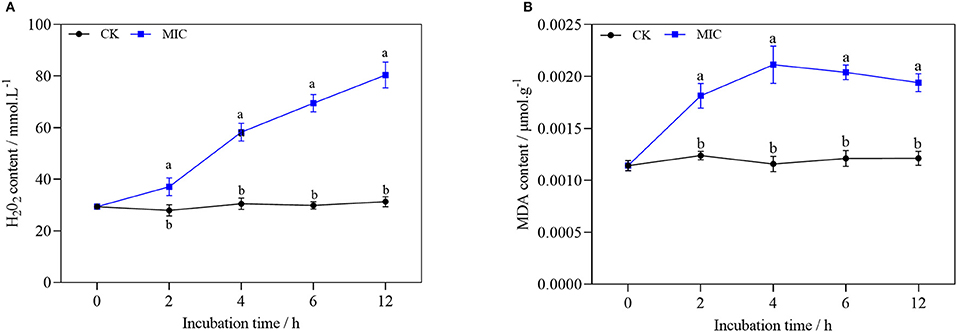
Figure 10. H2O2 (A) and MDA content (B) of P. digitatum treated without or with MIC nanoemulsion in different incubation times. Values are the mean ± standard deviation (SD) (n = 3), and the different letters indicate significant differences (P < 0.05).
According to previous reports, plant essential oil or constituents can induce eukaryotic cells to accumulate reactive oxygen species, leading to membrane lipid peroxidation damage (33). MDA is one of the essential products of membrane lipid peroxidation. H2O2 is a kind of reactive oxygen species, and it can be used as a molecular signal to improve cell defense ability and enhance cell tolerance at low concentrations. On the contrary, it can cause oxidative damage of lipid, protein, and nucleic acid molecules at high concentrations (34). After the treatment, the content of MDA and H2O2 increased sharply, indicating that the nanoemulsion could cause severe damage to membrane lipid peroxidation of the mycelia of P. digitatum.
Effect of Nanoemulsion Treatment on Mycelia Protein Synthesis
As shown in Figure 11, with the extension of treatment time, the soluble protein of mycelium decreased significantly in nanoemulsion samples, while the content of soluble protein in the control group showed a slowly increasing trend. After 2-h treatment, the difference was significant between the treatment and control group. The soluble protein content of nanoemulsion treatment was 1.86 mg/g at 12 h, which is 14.7 % lower than that of the control group, and the difference was significant.
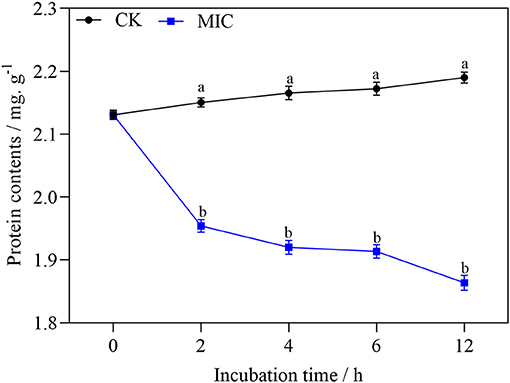
Figure 11. The protein content of P. digitatum treated without or with MIC nanoemulsion at different incubation times. Values are the mean ± standard deviation (SD) (n = 3), and the different letters indicate significant differences (P < 0.05).
Discussion
Under the influence of external stimuli, P. digitatum cells alleviate the adverse effects on growth by regulating gene expression patterns. Several studies have proved that the antimicrobial mechanism of essential oil includes alternating the distribution of fatty acids in the cell membrane, destroying cell walls, and reducing proton power, and the inactivation of ATPase, amylase, and protease was promoted (35–40). To deeply understand the action mechanism of nanoemulsion from molecular aspects and seek the pathway involved, we used RNA-Seq technology to profile the transcriptome of P. digitatum treated with nanoemulsion, which induced the expression of many stress response genes and the pathway involved to alleviate the adverse effect. Recent research by RNA-seq also observed different gene and pathway responses in P. digitatum cells treated by antifungal agent [(41, 42)].
Genes Related to Spore Germination and Cell Growth
Metabolic pathways are critical to cell growth and reproduction, and disruptions in metabolism can lead to cell death (43). KEGG enrichment analysis showed that most DEGs were enriched in the metabolic pathway, there were 144 down-regulated DEGs and 72 up-regulated DEGs respectively, indicating that the nanoemulsion inhibited the growth of P. digitatum by reducing its metabolic level. Fatty acids are the energy source for spore germination. The increase in fatty acid and protein contents can promote the germination of spores (44); as indicated by KEGG enrichment analysis, most downregulated DEGs were involved in fatty acid degradation and protein synthesis. Therefore, it can be inferred that nanoemulsion can inhibit spore germination through downregulation of lipid and amino acid metabolism.
Ribosomes are composed of many small subunits and large subunits, each containing a variety of ribosomal proteins and ribosomal RNA molecules. Protein biosynthesis is mainly carried out in ribosomes (45). According to GO enrichment analysis, most DEGs related to ribosome biogenesis in cellular components were downregulated. Four genes (TR4270_c0_g1, TR1973_c0_g1, TR2704_c0_g1, and TR4317_c0_g1) related to the ribosome protein were downregulated 2.73-, 1.80-, 1.71-, and 1.78-fold, respectively, by nanoemulsion compared with control. These genes illustrated that the nanoemulsion treatment destroyed the construction of the ribosome structure of P. digitatum.
Genes Related to Amino Acid Synthesis
Proteins play diverse functions with the cell and are essential for the vesicle trafficking of fungi (46). Protein synthesis is indispensable for spore germination and hypha formation (47). The KEGG enrichment pathway analysis revealed that there were abundant genes that regulate arginine and proline metabolism (13 DEGs); tryptophan metabolism (12 DEGs); phenylalanine metabolism (9 DEGs); tyrosine metabolism (10 DEGs); glycine, serine, and threonine metabolism (12 DEGs); lysine degradation (18 DEGs); and arginine biosynthesis (six DEGs), which were significantly downregulated by nanoemulsion compared with the control. The results are presented in Table 3. According to previous research, fungi usually activate the synthesis of some amino acids to maintain the vitality of cells under adverse stimuli (48, 49). Furthermore, as a signaling molecule, proline can trigger the expression of specific genes, regulate mitochondrial function, adjust osmotic pressure, act as a ROS scavenger, and affect cell proliferation or death (50). In our research, the amino acids and proline metabolism genes were significantly downregulated by nanoemulsion treatment compared with the control, suggesting that nanoemulsion treatment destroyed the osmotic balance ability of P. digitatum cells. The soluble protein contents of P. digitatum were found to be decreased with increasing treatment time (Figure 8), followed by the downregulation of gene expression of amino acids synthesis in P. digitatum revealed by RNA-seq data.
Furthermore, the ubiquitin-26S proteasome system is an important protein degradation system in cells (51) and plays the role of signal transduction, gene transcription, and programmed cell death. We found that 11 genes (TR1589_c0_g1, TR4378_c0_g1, TR4477_c0_g1, TR2443_c0_g1, TR10860_c0_g1, TR2507_c0_g1, TR2739_c0_g1, TR12942_c0_g1, TR1642_c0_g1, TR3076_c0_g1, and TR2492_c0_g1) encoding for 19S regulatory particle and 20S proteasome showed downregulation from 1.07- to 1.95-fold compared with nanoemulsion group with control in our study. Consequently, P. digitatum cells produced a huge quantity of damaged and erroneous protein after nanoemulsion treatment. Therefore, we supposed that nanoemulsions inhibit the cell growth by destroying proteins in the cytoplasm and nucleus.
Genes Related to Cell Integrity
Several studies have proved that the main targets of the antifungal activity of essential oils or their volatile components are cell walls, cell membranes, mitochondria, and intracellular genetic material (52, 53). The cell wall can maintain the cell morphology and control the material transportation and information transmission of the cell. Some proteins in the cell wall also have the function of disease prevention and stress resistance. Filamentous fungi cell walls contain chitin, which plays a pivotal role in the development and pathogenicity of fungi (54). Our results showed that several pathways involved in the cell wall formation, such as amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, were repressed by nanoemulsion. The gene TR1461_c0_g1 encoded for chitinase expression was upregulated, which might the more chitin was needed for cell wall synthesis, suggesting that the chitin content of P. digitatum may increase after the nanoemulsion treatment. The results are similar to the research reported previously (55).
Due to the lipophilicity of essential oils, researchers have proved that the plasma membrane was regarded as the active target of these antifungal agents (56). According to previous research, cells mediated the ratio of saturated fatty acids to unsaturated fatty acids, cis/trans unsaturated fatty acids, and unsaturated fatty acids response to external stress (57). Our data showed that some genes involved in cell membrane compositions were influenced after nanoemulsion treatment, such as biosynthesis of unsaturated fatty acids. Four DEGs in the biosynthesis of the unsaturated fatty acids pathway were all upregulated by 1.19- to 3.24-fold. The above results indicated that P. digitatum is responsible for maintaining the fluidity and permeability of the cell membrane by adjusting the composition and content of fatty acids. A similar antifungal mechanism was also reported by the study described by Hu et al. (39). Membrane is the target of Perilla frutescens essential oil against A. flavus.
Ergosterol is an essential component of the fungal cell membrane. Its primary function is to maintain the fluidity and permeability of the cell membrane. The decrease in ergosterol content usually leads to the disorder of cell permeability and the interruption of cell growth and proliferation (58). Some nanoemulsions encapsulated with antifungal agents showed a promising antifungal activity by inhibiting the ergosterol biosynthesis, leading to cell death (59, 60). In our study, the repressed tendency found in a gene (ERG2, ERG3, ERG4) encoding ergosterol biosynthesis was downregulated by 2.62-, 3.57-, and 2.71-fold, respectively. Previous research reported that ERG3 gene downregulation caused P. digitatum cells to lose the capacity to convert lanosterol to ergosterol (61).
Genes Related to Multidrug Resistance
Under abiotic stress, fungi can reduce intracellular drug levels by activating drug efflux transporters and enhance the ability of exogenous detoxification (62). Fungi drug resistance depends on significant facilitator superfamily transporters (MFS) and ATP-binding cassette (ABC), which can efflux exogenous drugs (63). The gene subfamilies include multidrug resistance (MDR/TAP, ABCB subfamily) and pleiotropic drug resistance (PDR, ABCG subfamily) (64). In our study, two ABCB subfamily genes showed an increase in expression in nanoemulsion-treated samples, which is 3.87- and 1.49-fold, respectively. Five genes belonging to the ABCG subfamily were also upregulated from 2.87- to 11.86-fold. Recent research reported that five transporter genes belong to the PDR network, which can efflux some hydrophobic molecules outside the cell (65), and the PDR proteins play a role in cell sterol uptake (66, 67) and quorum sensing in yeast (68). Therefore, it was speculated that P. digitatum could enhance the detoxification capacity and transport nanoemulsion out of the cells to minimize the injuries caused by nanoemulsion to P. digitatum cell.
Genes Related to Stress Response
Fungi could activate corresponding protection mechanisms by adjusting their gene expression under unfavorable external conditions. In our research, gene expression alterations related to stress response were influenced by nanoemulsion. Mitogen-activated protein kinases are present in many eukaryotes, including fungi. It plays an essential role in extracellular signal transduction and cell development and differentiation (69). Research has been reported that there are three classes of MAP kinases, namely, Fus3/Kss1, Hog1, and Slt2 in yeast and filamentous (70), and in the P. digitatum, three mitogen-activated protein kinases regulate osmotic pressure, growth and conidiation, cell development, and virulence (71). In our research, one gene, TR3246_c0_g1 encoding MAPK, was downregulated 2.46-fold by nanoemulsion compared with control. These results indicate that nanoemulsion can effectively inhibit P. digitatum to pathogenicity-associated MAPK cascades. Research also reported that ΔPdSlt2 mutants generated much fewer conidia than the wild type, indicating that PdSlt2 positively controls conidia formation in P. digitatum (72).
Reactive oxygen species is a byproduct of normal oxygen metabolism and decisive in cell signal transmission. Nevertheless, excessive levels of reactive oxygen species can damage cell and gene structure (73). ROS cause damage to biomacromolecules, resulting in devastating damage. DNA may break, mutate, and change its thermal stability after oxidative damage, which seriously affects the standard transcription and translation of genetic information (74). Generally, cells can alleviate ROS damage to cells through the action of enzymes (superoxide dismutase, catalase). Some small molecules, such as glutathione, also play critical cellular antioxidants (75). In our research, the gene (TR16620_c0_g1) relative to peroxiredoxin activity was upregulated by 3.06-fold. The ROS-mediated response has been previously observed for nanoemulsion treatment (76, 77). Similar results showed that genes (CAT1, SOD1, SOD4, SOD5, AOX2, and YHB1) showed upregulated tendency after peptide MAF-1A treatment, involved in oxidative stress response (78).
Cells can produce a large quantity of energy to neutralize the stress from external stimuli to maintain their vitality. Mitochondria could produce ATP and regulate cell metabolism by oxidative phosphorylation (79). In this research, most genes are related to carbon metabolism, while glyoxylate and dicarboxylate genes related to metabolism were repressed after nanoemulsion treatment. The gene (TR3140_c0_g1) relative to acetyl-CoA acetyltransferase in the tricarboxylic acid cycle and oxidative phosphorylation pathway was downregulated by 2.19-fold, indicating that nanoemulsion treatment might influence the intracellular respiration of P. digitatum, which is followed by the most enriched item in the biological process revealed by GO function enrichment analysis of RNA-seq data.
Conclusions
In conclusion, through RNA-seq technology, we analyzed the effect of nanoemulsion on the P. digitatum from the transcriptional level. In-depth RNA-seq analysis revealed that DEGs mainly involved in cell integrity (cell wall and membrane), amino acid synthesis, proteasome, glyoxylate and dicarboxylate metabolism, ribosomes biogenesis, and mitogen-activated protein kinases were notably affected by nanoemulsion treatment. The results indicated that nanoemulsion triggered gene expression variation and induced multiple pathways involvement. A deep understanding of the transcriptomic view mechanism further demonstrated the applicability of nanoemulsion as a natural origination, environment-friendly, and safe approach to combat against the green mold of citrus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: SubmissionID: SUB9953553; BioProject ID: PRJNA745208; http://www.ncbi.nlm.nih.gov/bioproject/745208.
Author Contributions
CW and JC involved in conceptualization, resources, supervision, and project administration. RY, XC, and CC involved in methodology. RY and XC involved in software and formal analysis. RY and CC involved in validation. RY, XC, and QH involved in investigation. RY and CW involved in data curation and visualization. RY involved in writing the original draft preparation. CW and KR involved in writing the review and editing. CW involved in funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31760598) and the Natural Science Foundation of Jiangxi Province of China (20181BCB24005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.724419/full#supplementary-material
References
1. Costa JH, Bazioli JM, Moraes Pontes JGd, Fill TP. Penicillium digitatum infection mechanisms in citrus: what do we know so far? Fungal Biol UK. (2019) 123:584–93. doi: 10.1016/j.funbio.2019.05.004
2. Pétriacq P, López A, Luna E. Fruit decay to diseases: can induced resistance and priming help? Plants Basel. (2018) 7:77. doi: 10.3390/plants7040077
3. Manoharan RK, Lee JH, Lee J. Antibiofilm and anti-hyphal activities of cedar leaf essential Oil, camphor, and fenchone derivatives against Candida albicans. Front Microbiol. (2017) 8:1476. doi: 10.3389/fmicb.2017.01476
4. Lyles JT, Kim A, Nelson K, Bullard-Roberts AL, Hajdari A, Mustafa B, et al. The chemical and antibacterial evaluation of St John's Wort Oil Macerates used in Kosovar traditional medicine. Front Microbiol. (2017) 8:1639. doi: 10.3389/fmicb.2017.01639
5. Qu S, Yang KL, Chen L, Liu M, Geng QR, He XN, et al. Cinnamaldehyde, a promising natural preservative against Aspergillus flavus. Front Microbiol. (2019) 10:2895. doi: 10.3389/fmicb.2019.02895
6. Wan CP, Kahramanoglu I, Okatan V. Application of plant natural products for the management of postharvest diseases in fruits. Folia Hortic. (2021) 33:203–15. doi: 10.2478/fhort-2021-0016
7. Meneguetti BT, Machado LdS, Oshiro KGN, Nogueira ML, Carvalho CME, Franco OL. Antimicrobial peptides from fruits and their potential use as biotechnological tools-a review and outlook. Front Microbiol. (2017) 7:2136. doi: 10.3389/fmicb.2016.02136
8. Burt S. Essential oils: their antibacterial properties and potential applications in foods—a review. Int J food microbiol. (2004) 94:223–53. doi: 10.1016/j.ijfoodmicro.2004.03.022
9. Ribes S, Fuentes A, Talens P, Barat JM. Prevention of fungal spoilage in food products using natural compounds: a review. Crit Rev Food Sci. (2018) 50:2002–16. doi: 10.1080/10408398.2017.1295017
10. Rao JJ, Chen BC, McClements DJ. Improving the efficacy of essential oils as antimicrobials in foods: mechanisms of action. Annu Rev Food Sci T. (2019) 10:365–87. doi: 10.1146/annurev-food-032818-121727
11. Montagu A, Joly-Guillou ML, Rossines E, Cayon J, Kempf M, Saulnier P. Stress Conditions induced by carvacrol and cinnamaldehyde on Acinetobacter baumannii. Front Microbiol. (2016) 7:01133. doi: 10.3389/fmicb.2016.01133
12. Braschi G, Patrignani F, Siroli L, Lanciotti R, Schlüter O, Fröhling A. Flow cytometric assessment of the morphological and physiological changes of Listeria monocytogenes and Escherichia coli in response to natural antimicrobial exposure. Front Microbiol. (2018) 9:2783. doi: 10.3389/fmicb.2018.02783
13. Alves JCO, Ferreira GF, Santos JR, Silva LCN, Rodrigues JFS, Neto WR, et al. Eugenol induces phenotypic alterations and increases the oxidative burst in Cryptococcus. Front Microbiol. (2017) 8:2419. doi: 10.3389/fmicb.2017.02419
14. Spitzer M, Robbins N, Wright GD. Combinatorial strategies for combating invasive fungal infections. Virulence. (2017) 8:169–85. doi: 10.1080/21505594.2016.1196300
15. Trifan A, Luca SV, Greige-Gerges H, Miron A, Gille E, Aprotosoaie A, et al. Recent advances in tackling microbial multidrug resistance with essential oils: combinatorial and nano-based strategies. Crit Rev Microbiol. (2020) 46:338–57. doi: 10.1080/1040841X.2020.1782339
16. May HC, Yu JJ, Guentzel MN, Chambers JP, Cap AP, Arulanandam BP. Repurposing auranofin, ebselen, and PX-12 as antimicrobial agents targeting the thioredoxin system. Front Microbiol. (2018) 9:336. doi: 10.3389/fmicb.2018.00336
17. Smani Y, Miró canturri A, Ayerbe algaba R. Drug repurposing for the treatment of bacterial and fungal infections. Front Microbio. (2018) 10:41. doi: 10.3389/fmicb.2019.00041
18. Das S, Singh VK, Dwivedy AK, Chaudhari AK, Deepika, Dubey NK. Eugenol loaded chitosan nanoemulsion for food protection and inhibition of Aflatoxin B-1 synthesizing genes based on molecular docking. Carbohyd Polym. (2021). 255:117339. doi: 10.1016/j.carbpol.2020.117339
19. Guo MM, Zhang LJ, He Q, Ali Arabi S, Zhao HH, Chen WJ, et al. Synergistic antibacterial effects of ultrasound and thyme essential oils nanoemulsion against Escherichia coli O157:H7. Ultrason Sonochem. (2020) 66:104988. doi: 10.1016/j.ultsonch.2020.104988
20. Zheng BJ, McClements J. Formulation of more efficacious Curcumin delivery systems using colloid science: enhanced solubility, stability, and bioavailability. Molecules. (2020) 25:2791. doi: 10.3390/molecules25122791
21. Zhao CC, Wei LP, Yin BB, Liu FG, Li JY, Liu XB, et al. Encapsulation of lycopene within oil-in-water nanoemulsions using lactoferrin: impact of carrier oils on physicochemical stability and bioaccessibility. Int J Biol Macromol. (2020) 153:912–20. doi: 10.1016/j.ijbiomac.2020.03.063
22. Li XD, Feng GR, Wang WJ, Yi LH, Deng LL, Zeng KF. Effects of peptide C12-OOWW-NH2 on transcriptome and cell wall of the postharvest fungal pathogen Penicillium digitatum. Front Microbiol. (2020) 11:574882. doi: 10.3389/fmicb.2020.574882
23. Cortes BW, Naditz AL, Anast JM, Schmitz-Esser S. Transcriptome sequencing of Listeria monocytogenes reveals major gene expression changes in response to lactic acid stress exposure but a less pronounced response to oxidative stress. Front Microbiol. (2020) 10:3110. doi: 10.3389/fmicb.2019.03110
24. Shen WH, Wang DH, Wei LL, Zhang Y. Fungal elicitor-induced transcriptional changes of genes related to branched-chain amino acid metabolism in Streptomyces natalensis HW-2. Appl Microbiol Biot. (2020) 104:4471–82. doi: 10.1007/s00253-020-10564-5
25. Zhang ZB, Zhao PC, Zhang PA, Su LY, Jia HR, Wei XK, et al. Integrative transcriptomics and metabolomics data exploring the effect of chitosan on postharvest grape resistance to Botrytis cinerea. Postharvest Biol Tec. (2020) 167:111248. doi: 10.1016/j.postharvbio.2020.111248
26. Samaras A, Ntasiou P, Myresiotis C, Karaoglanidis G. Multidrug resistance of Penicillium expansum to fungicides: whole transcriptome analysis of MDR strains reveals overexpression of efflux transporter genes. Int J Food Microbiol. (2020) 335:108896. doi: 10.1016/j.ijfoodmicro.2020.108896
27. Yan YF, Yang CJ, Shang XF, Zhao ZM, Liu YQ, Zhou R, et al. Bioassay-guided isolation of two antifungal compounds from Magnolia officinalis, and the mechanism of action of honokiol. Pestic Biochem Phys. (2020) 170:104705. doi: 10.1016/j.pestbp.2020.104705
28. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. (2013) 8:1494–512. doi: 10.1038/nprot.2013.084
29. OuYang QL, Tao NG, Jing GX. Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genom. (2016) 17:599. doi: 10.1186/s12864-016-2943-4
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT Method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
31. Yang Q, Wang J, Zhang P, Xie SN, Yuan XL, Hou XD, et al. In vitro and in vivo antifungal activity and preliminary mechanism of cembratrien-diols against Botrytis cinerea. Ind Crop Prod. (2020) 154:112745. doi: 10.1016/j.indcrop.2020.112745
32. Song XS, Gu KX, Gao J, Wang JX, Ding SC, Zhou MG. Ethylenediaminetetraacetic acid disodium salt acts as an antifungal candidate molecule against Fusarium graminearum by inhibiting DON biosynthesis and chitin synthase activity. Toxins. (2021) 13:17. doi: 10.3390/toxins13010017
33. Ji ZH, Zhang YX, Tian J, Wang FB, Song MY, Li H. Oxidative stress and cytotoxicity induced by tetrachlorobisphenol A in Saccharomyces cerevisiae cells. Ecotox Environ Safe. (2018) 161:1–7. doi: 10.1016/j.ecoenv.2018.05.070
34. Zhang ZQ, Chen J, Li BQ, He C, Chen Y, Tian SP. Influence of oxidative stress on biocontrol activity of Cryptococcus laurentii against blue mold on peach fruit. Front Microbiol. (2017) 8:151. doi: 10.3389/fmicb.2017.00151
35. Wang Y, Feng KW, Yang HH, Yuan YH, Yue TL. Antifungal mechanism of cinnamaldehyde and citral combination against Penicillium expansum based on FT-IR fingerprint, plasma membrane, oxidative stress and volatile profile. Rsc Adv. (2018) 8:5806–15. doi: 10.1039/C7RA12191A
36. Guo FY, Chen QP, Liang Q, Zhang M, Chen WX, Chen HM, et al. Antimicrobial activity and proposed action mechanism of Linalool against Pseudomonas fluorescens. Front Microbiol. (2021) 12:562094. doi: 10.3389/fmicb.2021.562094
37. Cao YF, Zhou AL, Zhou DG, Xiao XL, Yu YG, Li XF. Cronobacter sakazakii CICC 21544 responds to the combination of carvacrol and citral by regulating proton motive force. LWT-Food Sci Technol. (2020) 122:109040. doi: 10.1016/j.lwt.2020.109040
38. Scariot FJ, Pansera MS, Delamare APL, Echeverrigaray S. Citral and geraniol induce necrotic and apoptotic cell death on Saccharomyces cerevisiae. World J Microb Biot. (2021) 37:42. doi: 10.1007/s11274-021-03011-8
39. Hu ZY, Yuan K, Zhou Q, Lu C, Du LH, Liu F. Mechanism of antifungal activity of Perilla frutescens essential oil against Aspergillus flavus by transcriptomic analysis. Food Control. (2021) 123:107703. doi: 10.1016/j.foodcont.2020.107703
40. Geweely NS, Afifi HA, Ibrahim DM, Soliman MM. Inhibitory effect of essential oils on growth and physiological activity of deteriorated fungal species isolated from three archeological objects, Saqqara excavation, Egypt. Geomicrobiol J. (2020) 37:520–33. doi: 10.1080/01490451.2020.1731021
41. Feng GR, Li XD, Wang WJ, Deng LL, Zeng KF. Effects of peptide thanatin on the growth and transcriptome of Penicillium digitatum. Front Microbiol. (2020) 11:606482. doi: 10.3389/fmicb.2020.606482
42. Guo MX, Liu J, Xu ZL, Wang J, Li TT, Lei HT, et al. 2-Methoxy-1,4-naphthoquinone induces metabolic shifts in Penicillium digitatum revealed by high-dimensional biological data. J Arg Food Chem. (2020) 68:9697–706. doi: 10.1021/acs.jafc.0c03396
43. Green DR, Galluzzi L, Kroemer G. Metabolic control of cell death. Science. (2014) 345:1250256. doi: 10.1126/science.1250256
44. Cristina de CS, Vila T, Vinícius de MB, Ishida K. New targets for the development of antifungal agents. Encyclopedia Mycol. (2021) 1:456–67. doi: 10.1016/B978-0-12-809633-8.21026-1
45. Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. (2003) 313:17–42. doi: 10.1016/S0378-1119(03)00629-2
46. Patiño-Medina JA, Valle-Maldonado MI, Maldonado-Herrera G, Pérez-Arques C, Jácome-Galarza IE, Díaz-Pérez C, et al. Role of Arf-like proteins (Arl1 and Arl2) of Mucor circinelloides in virulence and antifungal susceptibility. Fungal Genet Biol. (2019) 129:40–51. doi: 10.1016/j.fgb.2019.04.011
47. Liu JY, Men JL, Chang MC, Feng CP, Yuan LG. iTRAQ-based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress. J Proteomics. (2017) 156:75–84. doi: 10.1016/j.jprot.2017.01.009
48. Lai TF, Wang Y, Fan YY, Zhou YY, Bao Y, Zhou T. The response of growth and patulin production of postharvest pathogen Penicillium expansum to exogenous potassium phosphite treatment. Int J Food Microbiol. (2017) 244:1–10. doi: 10.1016/j.ijfoodmicro.2016.12.017
49. Wang QH, Ji YP, Qu YY, Qi YK, Li DW, Liu ZY, et al. The response strategies of Colletotrichum gloeosporioides ss due to the stress caused by biological control agent Bacillus amyloliquefaciens deciphered by transcriptome analyses. Biol Control. (2020) 150:104372. doi: 10.1016/j.biocontrol.2020.104372
50. Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. (2010) 15:89–97. doi: 10.1016/j.tplants.2009.11.009
51. Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. (2004) 55:555–90. doi: 10.1146/annurev.arplant.55.031903.141801
52. Krishnamoorthy R, Gassem MA, Athinarayanan J, Periyasamy VS, Prasad S, Alshatwi AA. Antifungal activity of nanoemulsion from Cleome viscosa essential oil against food-borne pathogenic Candida albicans. Saudi J Biol Sci. (2021) 28:286–93. doi: 10.1016/j.sjbs.2020.10.001
53. Tariq S, Wani S, Rasool W, Bhat MA, Prabhakar A, Shalla AH, et al. A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens. Microb Pathogenesis. (2019) 134:103580. doi: 10.1016/j.micpath.2019.103580
54. Gandía M, Harries E, Marcos JF. Identification and characterization of chitin synthase genes in the postharvest citrus fruit pathogen Penicillium digitatum. Fungal Biol UK. (2012) 116:654–64. doi: 10.1016/j.funbio.2012.03.005
55. Jiang CM, Li ZZ, Shi YH, Guo D, Pang B, Chen XQ, et al. Bacillus subtilis inhibits Aspergillus carbonarius by producing iturin A, which disturbs the transport, energy metabolism, and osmotic pressure of fungal cells as revealed by transcriptomics analysis. Int J Food Microbiol. (2020) 330:108783. doi: 10.1016/j.ijfoodmicro.2020.108783
56. Jugreet BS, Suroowan S, Rengasamy RRK, Mahomoodally MF. Chemistry, bioactivities, mode of action and industrial applications of essential oils. Trends Food Sci Tech. (2020) 101:89–105. doi: 10.1016/j.tifs.2020.04.025
57. Bajerski F, Wagner D, Mangelsdorf K. Cell membrane fatty acid composition of Chryseobacterium frigidisoli PB4T, isolated from antarctic glacier forefield soils, in response to changing temperature and pH conditions. Front microbiol. (2017) 8:677. doi: 10.3389/fmicb.2017.00677
58. Bibi M, Murphy S, Benhamou RI, Rosenberg A, Ulman A, Bicanic T, et al. Combining colistin and fluconazole synergistically increases fungal membrane permeability and antifungal cidality. ACS Infect Dis. (2021) 7:377–89. doi: 10.1021/acsinfecdis.0c00721
59. Kalagatur NK, Ghosh OSN, Sundararaj N, Mudili V. Antifungal activity of chitosan nanoparticles encapsulated with Cymbopogon martinii essential oil on plant pathogenic fungi Fusarium graminearum. Front Pharmacol. (2018) 9:610. doi: 10.3389/fphar.2018.00610
60. Chaudhari AK, Singh VK, Das S, Deepika, Prasad J, Dwivedy AK, et al. Improvement of in vitro and in situ antifungal, AFB (1) inhibitory and antioxidant activity of Origanum majorana L. essential oil through nanoemulsion and recommending as novel food preservative. Food Chem Toxicol. (2020) 143:111536. doi: 10.1016/j.fct.2020.111536
61. OuYang QL, Liu YM, Oketch OR, Zhang ML, Shao XF, Tao NG. Citronellal exerts its antifungal activity by targeting ergosterol biosynthesis in Penicillium digitatum. J Fungi. (2021) 7:60432. doi: 10.3390/jof7060432
62. Morschhäuser J. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet Biol. (2010) 47:94–106. doi: 10.1016/j.fgb.2009.08.002
63. Paul S, Moye-Rowley WS. Multidrug resistance in fungi: regulation of transporter-encoding gene expression. Front Physiol. (2014) 5:143. doi: 10.3389/fphys.2014.00143
64. Kovalchuk A, Driessen AJM. Phylogenetic analysis of fungal ABC transporters. BMC Genom. (2010) 11:177. doi: 10.1186/1471-2164-11-177
65. Balzi E, Goffeau A. Yeast multidrug resistance: The PDR network. J Bioenerg Biomembr. (1995) 27:71–6. doi: 10.1007/BF02110333
66. Wilcox LJ, Balderes DA, Wharton B, Tinkelenberg AH, Rao G, Sturley SL. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. (2002) 277:32466–72. doi: 10.1074/jbc.M204707200
67. Moreno A, Banerjee A, Prasad R, Falson P. PDR-like ABC systems in pathogenic fungi. Res Microbiol. (2019) 170:417–25. doi: 10.1016/j.resmic.2019.09.002
68. Hlaváček O, Kučerová H, Harant K, Palková Z, Váchová L. Putative role for ABC multidrug exporters in yeast quorum sensing. FEBS Lett. (2009) 583:1107–13. doi: 10.1016/j.febslet.2009.02.030
69. Martínez-Soto D, Ruiz-Herrera J. Functional analysis of the MAPK pathways in fungi. Revista Iberoamericana de Micología. (2017) 34:6. doi: 10.1016/j.riam.2017.02.006
70. Jiang C, Zhang X, Liu HQ, Xu JR. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. (2018) 14:1006875. doi: 10.1371/journal.ppat.1006875
71. Gandía M, Garrigues S, Hernanz-Koers M, Manzanares P, Marcos J. F. Differential roles, crosstalk and response to the antifungal protein AfpB in the three mitogen-activated protein kinases (MAPK) pathways of the citrus postharvest pathogen Penicillium digitatum. Fungal Genet Biol. (2019) 124:17–28. doi: 10.1016/j.fgb.2018.12.006
72. De Ramón-Carbonell M, Sánchez-Torres P. PdSlt2 Penicillium digitatum mitogen-activated-protein kinase controls sporulation and virulence during citrus fruit infection. Fungal Biol UK. (2017) 121:1063–74. doi: 10.1016/j.funbio.2017.09.004
73. Hamann A, Brust D, Osiewacz HD. Apoptosis pathways in fungal growth, development and ageing. Cell. (2008) 16:276–83. doi: 10.1016/j.tim.2008.03.003
74. Lv QZ, Ni TJH, Li LP, Li T, Zhang DZ, Jiang YY. A new antifungal agent (4-phenyl-1, 3-thiazol-2-yl) hydrazine induces oxidative damage in Candida albicans. Front Cell Infect Mi. (2020) 10:578956. doi: 10.3389/fcimb.2020.578956
75. Devi SM, Raj N, Sashidhar RB. Efficacy of short-synthetic antifungal peptides on pathogenic Aspergillus flavus. Pestic Biochem Phys. (2021) 174:104810. doi: 10.1016/j.pestbp.2021.104810
76. Shanmugapriya K, Kim H, Lee YW, Kang HW. Cellulose nanocrystals/nanofibrils loaded astaxanthin nanoemulsion for the induction of apoptosis via ROS-dependent mitochondrial dysfunction in cancer cells under photobiomodulation. Int J Biol Macromol. (2020) 149:165–77. doi: 10.1016/j.ijbiomac.2020.01.243
77. Liu Q, Gao Y, Fu X, Chen W, Yang JH, Chen ZY, et al. Preparation of peppermint oil nanoemulsions: investigation of stability, antibacterial mechanism and apoptosis effects. Colloid Surface B. (2021) 201:111626. doi: 10.1016/j.colsurfb.2021.111626
78. Wang T, Xiu JF, Zhang YC, Wu JW, Ma XL, Wang Y, et al. Transcriptional responses of Candida albicans to antimicrobial peptide MAF-1A. Front Microbiol. (2017) 8:894. doi: 10.3389/fmicb.2017.00894
Keywords: citrus, RNA-seq, Penicillium digitatum, nanoemulaion, antifungal
Citation: Yang R, Chen X, Huang Q, Chen C, Rengasamy KRR, Chen J and Wan C (2021) Mining RNA-Seq Data to Depict How Penicillium digitatum Shapes Its Transcriptome in Response to Nanoemulsion. Front. Nutr. 8:724419. doi: 10.3389/fnut.2021.724419
Received: 13 June 2021; Accepted: 18 August 2021;
Published: 14 September 2021.
Edited by:
Satyanarayan R. S. Dev, Florida Agricultural and Mechanical University, United StatesReviewed by:
Paloma Sánchez-Torres, Institute of Agrochemistry and Food Technology (IATA), SpainAnand Kumar Chaudhari, Banaras Hindu University, India
Copyright © 2021 Yang, Chen, Huang, Chen, Rengasamy, Chen and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunpeng (Craig) Wan, Y2h1bnBlbmd3YW5AanhhdS5lZHUuY24=; Jinyin Chen, amlueWluY2hlbkAxMjYuY29t
†These authors have contributed equally to this work
 Ruopeng Yang
Ruopeng Yang Xiu Chen
Xiu Chen Qiang Huang
Qiang Huang Chuying Chen
Chuying Chen Kannan R. R. Rengasamy
Kannan R. R. Rengasamy Jinyin Chen
Jinyin Chen Chunpeng (Craig) Wan
Chunpeng (Craig) Wan