Abstract
Sweet dessert watermelon (Citrullus lanatus) is one of the most important vegetable crops consumed throughout the world. The chemical composition of watermelon provides both high nutritional value and various health benefits. The present manuscript introduces a catalog of 1,679 small molecules occurring in the watermelon and their cheminformatics analysis for diverse features. In this catalog, the phytochemicals are associated with the literature describing their presence in the watermelon plant, and when possible, concentration values in various plant parts (flesh, seeds, leaves, roots, rind). Also cataloged are the chemical classes, molecular weight and formula, chemical structure, and certain physical and chemical properties for each phytochemical. In our view, knowing precisely what is in what we eat, as this catalog does for watermelon, supports both the rationale for certain controlled feeding studies in the field of precision nutrition, and plant breeding efforts for the development of new varieties with enhanced concentrations of specific phytochemicals. Additionally, improved and comprehensive collections of natural products accessible to the public will be especially useful to researchers in nutrition, cheminformatics, bioinformatics, and drug development, among other disciplines.
Introduction
Food is a complex mixture of chemical compounds, often numbering well over a thousand different compounds in any individual food item (1–3). That complexity expands when considering processing (4), the food matrix (5, 6), or byproducts, such as those derived from both human and microbial metabolism (7), as well as taints and off-flavors derived from degradation and packaging (8). Nonetheless, any catalog of the metabolites of food compounds coupled to research on their health effects ought to begin with knowledge of a chemical inventory of what is in the food, and in the forms in which it will be consumed. Here, we focus primarily on watermelon fruit, seed, and rind to provide a comprehensive, publicly accessible list of phytochemicals in watermelon.
Natural product databases generally are at the small end of the size spectrum of chemical databases when compared with the vastly larger PubChem (~110 million compounds) (9) and collections of synthetic compounds numbering in the billions (10). This necessitates a significant and genuine need to build resources for natural products. Existing natural product databases also suffer from missing links between the chemical structures and the organisms that produce them (3). These missing links often result from the standard practice that only newly elucidated structures are reported in scientific journals and then aggregated into public databases (11). Well-known metabolites identified in a newly studied organism or food are not reported. Hence, research programs in nutrition, cheminformatics, and drug development, among other disciplines, will benefit from natural product datasets that are comprehensive in scope and of a design that easily merges with other data.
Regarding human nutrition, knowledge of what is in a food is the basis by which to characterize the health benefits of that food. Those efforts support knowing what to eat to remain healthy (12) and assist in defining the “dark matter” or chemical complexity of nutrition (13, 14). In addition, comprehensive catalogs of the biochemicals present in a crop can stimulate projects in plant breeding and crop improvement, especially when coupled with genome sequencing and other such data streams (15, 16). Thus, to support and then fully implement projects in computational nutrition and cheminformatics research on natural products, and expand capabilities for dietary assessment, we sought to build a comprehensive catalog of compounds naturally occurring in watermelon.
Sweet dessert watermelon (Citrullus lanatus) is among the most important vegetable crops grown and consumed throughout the world, with global annual planting of more than 3 million hectares and production of over 100 million tons. China leads the world in watermelon production with an annual output of over 60 million tons. Other top watermelon producing countries are Turkey, India, Brazil, Algeria, Iran, Russia, United States, Egypt, Mexico, Kazakhstan, and Uzbekistan (with an annual production of 3.9, 2.5, 2.3, 2.2, 1.9, 1.8, 1.7, 1.6, 1.3, 1.3, and 1.2 million tons, respectively) (17).
Watermelon belongs to the xerophytic genus Citrullus, native to Africa (18). It was domesticated in Africa over 4000 years ago, while sweet dessert watermelons emerged in the Mediterranean region over 2000 years ago (19). It was introduced to India and China by the seventh and tenth centuries, respectively, and to Europe via Moorish Spain in the tenth century. There, watermelon has been cultivated successfully in the warmer Mediterranean regions of the continent. Watermelons were brought to the Americas by European colonists and with the slave trade from Africa during the sixteenth century (19). Today, watermelon is grown in 44 states in the USA, while most production is centered in Texas, Florida, Georgia, and California. Overall, sweet dessert watermelon varieties share a narrow genetic base, indicating a possible origin from a single founder population (20, 21). Those origins, their environments and the growth conditions of current production areas combined with detailed metabolomics will offer insight into origins of favored varieties as well as approaches to use levels of key compounds as quantitative traits for crop improvement.
Watermelon fruits contain a wide range of bioactive compounds, including glycosides, carotenoids, flavonoids, alkaloids, carbohydrates, fatty acids, and essential oils (22). Cucurbitacins, a rather broad family of bitter-tasting compounds in watermelon (23, 24), have drawn interest for their anti-oncogenic pharmacological properties (25). Through many years of evolution, domestication, and selection for desirable qualities, watermelon fruit has undergone significant changes in quality traits, mainly those associated with flesh color and texture, and nutrient and sugar content (26). Developing varieties with desirable fruit characteristics and high nutritional value is a top priority for watermelon breeding programs. Watermelon is a naturally rich source for the non-protein α-amino acid citrulline, which was reported to have antioxidant and vasodilatation activity (27). Citrulline was first isolated from watermelon by the Japanese researchers Yotaro Koga and Ryo Odake in 1914 (28) and further validated in 1930 (29). Lycopene was first reported in watermelon in 1930 (30), and like tomato, watermelon contains high levels of lycopene and other carotenoids with potential benefit for human health (31, 32). The health benefits of some of these compounds are known and continue to be the focus of nutrition research. Yet, interest is growing in documenting the chemical complexity of foods and assessing their impact on human health. The watermelon genome was sequenced, assembled, and annotated in 2019 (21), enabling exploration with bioinformatic tools to elucidate further its nutritive value and identify relevant biochemical pathways to tune the production of compounds of interest.
This manuscript presents a compilation of phytochemicals, linking chemically correct structures to the public resources where they were identified in watermelon and different parts of the plant. The 1,679 natural products that are part of this catalog underwent a curation process, their physicochemical properties were computed with cheminformatics tools, and all data are available at https://watermelon.naturalproducts.net. In this online database, users can freely browse and search for watermelon natural products.
Materials and Methods
Data Collection
Scientific articles on watermelon compounds or metabolomics were collected based on queries at PubMed and Agricola, 42 and 22 articles respectively, and supplemented with an additional 15 articles based on careful reading of other articles and 17 via personal communication. We also mined data from watermelon genome (CuCyC, genome v1) (33), and nutrition resources [Food Data Central (34); Phytochemical and Ethnobotanical Database at the USDA (35), PhytoHub (36)], and the LOTUS project (37). The latter aims to catalog documented pairs of natural products and the organisms producing them. Data collection was restricted to C. lanatus cultivars, varieties, and grafts. Expert knowledge of the authors directed the cataloging efforts to specific publications. We sought not to incorporate the compounds cataloged at FooDB (https://foodb.ca) and replicate that resource, but do include FooDB identifiers for compounds reported in other sources. Although essential for basic life processes, central metabolites, such as nucleotides, nucleosides, and ubiquitous coenzymes were excluded from the catalog, as these are shared by all living organisms.
Data Curation
After retrieving the literature, all collected information about the natural products was processed in Java with the Chemistry Development Kit (CDK) (38). For each molecule, the original SMILES were converted to unique and absolute SMILES, implicit hydrogens were tagged accordingly, compound aromaticity was corrected when appropriate, and tautomers and ionization states were standardized. Also, compounds of less than five heavy atoms were discarded. A structure-based compound unification was performed to prevent redundancies within the catalog. This was done using Tanimoto similarity with three different fingerprints, PubChem, Extended, and ECFP fingerprints, and a similarity threshold of 99% between two molecules for three of their fingerprints. Using three different fingerprints is necessary as they do not all perform well on all structure types, in particular for highly redundant monomeric structures like lipids and polysaccharides. The combined fingerprint comparison guarantees that two molecules with a Tanimoto similarity score over 99% with the three approaches are truly identical. The computer code for compound curation, unification, and calculation of features is available on GitHub (https://github.com/mSorok/Watermelon).
Content of the Catalog
The information on compounds found in watermelon is organized into tables pertinent to two distinct but overlapping disciplines: cheminformatics of natural products and human nutrition. All data are also available on the Watermelon Online website (https://watermelon.naturalproducts.net) with accompanying diverse search functionality. Data presented in these tables include common and alternative names in English, and compound identifiers in major chemical databases: CAS® (Chemical Abstracts Service), KEGG (Kyoto Encyclopedia of Genes and Genomes) (39), HMDB (Human Metabolome Database) (40), PubChem (9), ChEBI (Chemical Entities of Biological Interest) (41), FooDB (https://foodb.ca/), and LipidMaps (https://www.lipidmaps.org) (42). Additionally, provided for each molecule are the molecular formula and weight, together with classic structure representations, such as InChI, original (as from their source), canonical, and absolute SMILES, plus other representations such as Murcko scaffolds (43) (used generally for structure-activity relationship elucidation) and deepSMILES (an adaptation of SMILES for machine-learning purposes) (44). A wide range of molecular descriptors, such as AlogP, topological polar surface, atomic polarizabilities, Zagreb Index, Petitjean number, Kappa shape index, and the Lipinsky rule of five failures, have been computed with the CDK. Chemical pathways, superclasses, and classes were calculated with NPclassifier (https://npclassifier.ucsd.edu/) (45). This dataset is provided in Supplementary Table 1 and is available at https://watermelon.naturalproducts.net.
AFC Identifiers
Unique identifiers are a convenient means to refer to a compound without ambiguity. However, no single data repository has identifiers for all compounds cataloged here. Thus, we define the “AFC” identifier to represent Agricultural Research Service Food Compound and encourage its use in other catalogs. This has been assigned to all entries and serves as a bridge between data resources, the source literature, and across the two tables presented here.
Nutrition Data
Parallel to cataloging the natural products of watermelon and supporting nutrition research, effort was expended to assemble information, when available, on concentrations of compounds from different parts of the plant. The plant parts for which data are tabulated include (red) flesh, heart tissue, juice, seed, rind, peel, yellow flesh, seedling, leaf, root, other parts of the plant, and detected but plant part not reported. The collected data included the low value in the range, the high value in the range, deviation from those values, and units (assumed to be fresh or wet weight unless noted). This table (Supplementary Table 2) also provides for all compounds the citations to the literature and database sources. This information is archived at the USDA's Ag Data Commons (https://doi.org/10.15482/USDA.ADC/1522862), where updates will be provided.
Data Analysis
Simple statistical analyses and plots were made with ggplot in R, or Python 3 and the RDkit cheminformatic library for Python (46). The glycosylation analysis was performed with the Sugar Removal Utility (47) and RDkit. The graphical representation of the chemical space covered by the known watermelon natural products was performed with the t-distributed stochastic neighbor embedding (t-SNE), a dimensionality reduction method that captures a large fraction of the overall structural variance across the molecular set. t-SNE was performed with the scikit-learn Python 3 library and MACCS fingerprints.
Genome Mining
The C. lanatus genome (accession number GCA_000238415.2) was downloaded from the NCBI Genome on 1 Dec 2020. Online versions of plantiSMASH v.1 (48) and PRISM 3 (49) were used under default parameters to mine this genome for known biosynthetic gene clusters (BGCs) whose products synthesize small molecules such as non-ribosomal peptides (NRP) and polyketides.
Data Dictionary
The different terms and abbreviations are defined in Supplementary Table 3, and archived at https://doi.org/10.15482/USDA.ADC/1522862.
Results/Database Description
When writing this manuscript, the cheminformatics catalog of naturally occurring compounds in watermelon contains 1,679 curated molecules (Supplementary Table 1). This set does not include water, dissolved gases, minerals, salts, and common, central metabolism compounds, such as ubiquitous coenzymes (e.g., NADP, Coenzyme A) nor the nucleotides and their derivatives (e.g., ATP, ADP, AMP). As some of these compounds are nutrients, those are included in Supplementary Table 2.
General Characteristics
Molecules range in size (Figure 1A) from molecular weight 82.10 Da (dihydropyrimidine, AFC000168) to 2,286.8 Da for cold-adapted KDO2-lipid A (AFC001362). Grouping compounds into molecular weight bins of 25 units shows that molecular weight range 125–150 is the most populated with 161 compounds. The mean molecular weight in the catalog is 348.65 Da, and the median is 284.26 Da.
Figure 1
All molecules contain carbon except for the pyrophosphate ion (AFC000828). Oxalate (AFC00451) is the only compound that contains carbon and oxygen with no hydrogen atoms. There are 86 compounds that lack oxygen atoms, and of these, 58 are composed solely of carbon and hydrogen, ranging in molecular mass from ethenylbenzene (104.15 Da, AFC000284) to phytoene (544.94 Da, AFC000908). Additionally, 599, 333, and 149 compounds contain nitrogen, phosphorus, or sulfur atoms, respectively. Summary characteristics regarding the composition of watermelon natural products with these three atoms are presented in Figure 1B.
For each compound we determined the predicted partitioning between a hydrophobic and hydrophilic phase, using the Atomic logarithm of 1-octanol/water partition coefficient (AlogP) values (Figure 1C). This provides information on the solubility of a molecule based on its atomic constituents. A negative AlogP-value indicates a hydrophilic compound and a positive value is lipophilic. Of the natural products in this catalog, 925 (55.1%) are predicted to be hydrophilic and 754 (44.9%) lipophilic. The hydrophilicity of a compound has a direct impact on its distribution within cells and tissues and on its capacity to transit the cell membrane.
Each compound identified in watermelon is cross-referenced to the identifiers from seven different chemical compound databases (see section Materials and Methods). This information is provided to facilitate links between this resource and well-known, richly annotated databases of chemical compounds. Of the 1,679 compounds inventoried here, the range of representation spans from 1,591 (94.8%) with identifiers in PubChem (9) to 490 (29.2%) compounds found in the specialized LipidMaps (42) resource (Figure 1D). Because not all natural products cataloged here are found in the large databases and for ease of discussion, we created the AFC identifiers and assigned such to all cataloged compounds.
Chemical Classes and Known Bioactive Compounds
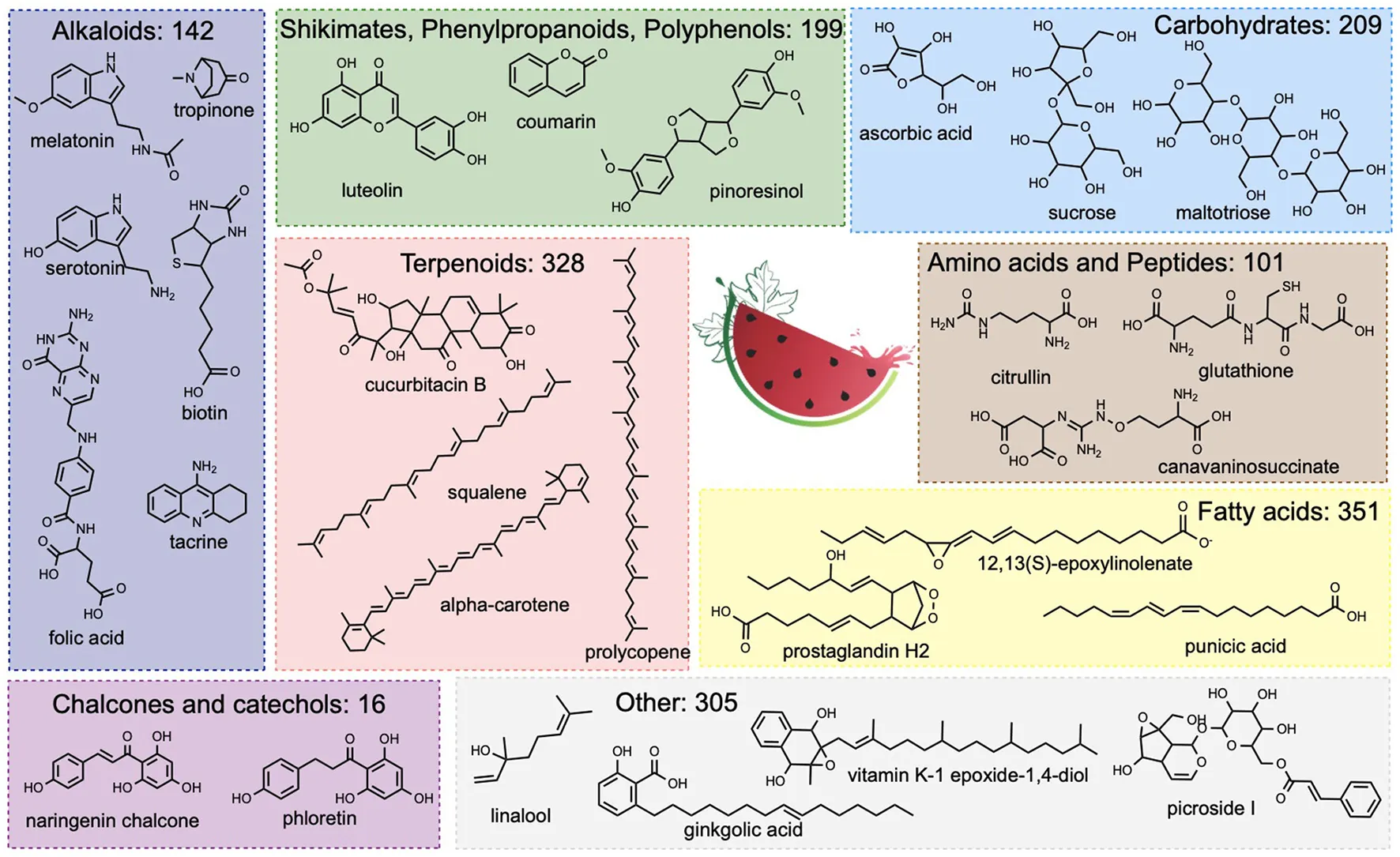
The classification of watermelon compounds with NPclassifier distributed them into seven distinct chemical classes: 351 compounds were identified as fatty acids (Figure 2), 328 as terpenoids (Figure 3), 209 as carbohydrates (Figure 4), 199 as shikimates, phenylpropanoids or polyphenols (Figure 5) 142 as alkaloids (Figure 6), 101 as amino acids, peptides, and NRPs (Figure 7), and 16 as type III plant polyketides (catechols, phloroglucinols, and chalcones—Figure 8). Among the latter, 28 compounds were classified in more than one category. Lastly, 305 compounds remained unclassified because of current limitations of NPclassifier.
Figure 2
Figure 3
Figure 4
Figure 5
Figure 6
Figure 7
Figure 8
Fatty Acids
Fatty acids form a very large group of natural compounds, and are the major components of lipids. Fatty acids can be classified variously: by saturation, number of carbons, or linearity. Dietary fatty acids also are important in human health and disease prevention (50). Three hundred fifty-one fatty acids were identified in the watermelon plant (Figure 2). Of particular interest are some known functional compounds, such as nervonic acid, which is beneficial to brain function (51), oleic acid, known to be a good general anti-inflammatory (52), and punicic acid, which has a wide range of biological properties, in particular antidiabetic and anti-obesity (53). Arachidic acid was found in the seeds and is one component of nanoparticles for drug delivery (54). Interestingly, two prostaglandins are found in the watermelon plant. H2 (55) regulates dilation of blood vessels, and stimulates platelet aggregation, and E2 is involved in modulating immune responses and has anti-inflammatory activity (56).
Terpenoids
Terpenoids are the largest class of known natural products (57) and are characterized by their derivation from isoprene. Plant terpenoids are often used for their aromatic properties, but they also have notable pharmacological attributes. A total of 328 terpenoids have been described in watermelon, in particular cucurbitacins and carotenoids, and molecules representative of this class are shown in Figure 3. Cucurbitacins, also known as cucurbitane triterpenoids, have anti-inflammatory, antioxidant, and anticancer properties (58–60). Watermelon fruit is abundant in lycopene, which has significant antioxidant activity (61). Carotenoids as a group of phytochemicals are of intense interest for their overall benefits to human health. In particular their consumption is associated with lower risk of cardiovascular disease, cancer, and eye disease (61). Watermelon also contains squalene, a natural product with broad applications in nutrition, pharmacy, medicine, and cosmetics (62), erythrodiol, a vasorelaxant (63), and karounidiol which was observed to have anti-tumor effects (64). Distinct from terpenoids with pharmacological interest, the plant also contains pheophytin A (65), a beautiful molecule that can be used to estimate fruit ripening, and violaxanthin, which protects the plant from photooxidative damage (66).
Carbohydrates
The carbohydrate class of watermelon natural products is large, and despite the common association with a sweet taste, its molecules are not limited to this attribute. As expected, watermelon has a noted diversity of carbohydrates, including mono-, di-, and tri-saccharides, plus polysaccharides. Some examples of this group are illustrated in Figure 4. Only a few of these, such as glucose, fructose, and sucrose, impart a sweet taste to the fruit. The others, such as maltotriose, ajugose, and maltoheptaose, are synthesized and deposited in storage organs, such as seeds, during the maturation and ripening processes, which are then mobilized during early seed germination (67). In addition to typical carbohydrates, watermelon also contains chitotriose, an interesting carbohydrate-like molecule studied for antioxidant activity (68).
Shikimates, Phenylpropanoids, and Polyphenols
Shikimates, also known as shikimic acids, and the structurally similar phenylpropanoids, are a diverse family of natural products occurring in plants and synthesized from the aromatic amino acids phenylalanine and tyrosine. This family is also known for flavorful molecules, in particular flavonoids, but also coumarins and lignans. The watermelon plant contains 199 identified molecules from this chemical family (Figure 5). Among these, several have industrial or pharmacological interest, such as coumarin. Although toxic for humans in high concentrations, coumarin does add a pleasant odor in low concentrations, as is the case in watermelon. Coumarin also has a wide range of uses in industry, mainly related to its fragrance (69). Coumarin derivatives have demonstrated anti-inflammatory and antioxidant properties (70). Watermelon red flesh contains several flavonoids, polyphenols well-known for their pharmacological activities. These include luteolin, which has potential anti-cancer (71), anti-inflammatory, antioxidant, and anti-allergic activities (72), quercetin, with a vast range of activities, in particular antioxidant effects (73), and taxifolin, also recognized for its antioxidant properties (74). Watermelon also contains potent lignans, such as pinoresinol, with potential hepatoprotective effects (75) and thiosulfates like petivericin, involved in plant defense (76) with noted antibacterial and antifungal properties.
Alkaloids
Alkaloids are a class of natural products that contain at least one nitrogen atom and are produced by diverse organisms, with plants in particular. These molecules are known to have a wide spectrum of bioactivities, such as pharmacological applications, psychotropic, and stimulant use, and may be toxic. In general, alkaloids have a bitter taste for humans. The NPclassifier identified 142 alkaloids in the present watermelon natural products catalog, with selected examples shown in Figure 6. Among the alkaloids, particular attention is drawn to melatonin and serotonin, important for signaling and stress mitigation in plants (77), but also regulating mood, circadian cycles, and anxiety in mammals (78, 79). The watermelon fruit contains six of the eight types of water-soluble vitamin B: biotin (B7), folic acid (B9), thiamin (B1), riboflavin (B2), pantothenic acid (B5), and pyridoxine (B6). These compounds are involved in a wide range of metabolic processes in mammals and therefore are used for a broad spectrum of pharmacological applications (80). Also observed in the watermelon plant are xanthine and bilirubin, which have antioxidant effects (81, 82).
Amino Acids and Small Peptides
Over 100 non-proteinogenic amino acids and small peptides are reported in watermelon. The structures of selected examples are depicted in Figure 7. Among these, citrulline is most prominent, and watermelon remains its most important source known (27). Citrulline is used as a drug and in food supplements for its stimulating activity on protein synthesis in skeletal muscle (83), its cardioprotective and overall beneficial cardiovascular effects (84), and even for erectile dysfunction (85). In addition to citrulline, watermelon also contains high levels of glutathione and its derivatives (e.g., S-formylglutathione, glutathione disulfide, S-lactoglutathione), which show antioxidant activities (86). Four NRPs are reported (Figure 7) in the watermelon plant. However, as NRPs are known to be produced mainly by bacteria and fungi, caution is warranted as these NRPs might also be produced by a bacterium or fungus inside the plant or by the plant independent of bacterial or fungal infection.
Catechols, Phloroglucinols, and Chalcones
Sixteen natural products in the watermelon plant have been classified as catechols, phloroglucinols or chalcones, or type III polyketides produced only by plants. Representatives of this class are shown in Figure 8. Among these, two stand out for their recognized properties. Ginkgolic acid (AFC000953) is a natural product known for its anti-inflammatory (87) and neuroprotective (88) bioactivities. Phloretin (AFC001890) has various applications in medicine and cosmetics, derived from its broad and potent antioxidant activities (89).
Other Notable Molecules
Tannins are astringent polyphenolic biomolecules widely distributed in many plant species where they are mainly involved in protection against predation (90). It therefore is not surprising to find this molecule class in the watermelon plant. Interestingly, of five compounds reported to repel the malaria mosquito Anopheles gambiae (91), watermelon contains three: 2-nonanone, 6-methyl-5-hepten-2-one, and linalool. Another notable compound reported in watermelon is picroside I, a potent hepatoprotective antioxidant (92, 93). Structures of these molecules are shown in Figure 9.
Figure 9
Glycosylated Molecules
In addition to the aforementioned carbohydrates, watermelon contains 322 glycosylated molecules, i.e., non-carbohydrate molecules with glycosidic moieties attached. The glycosylation of a molecule positively affects its hydro-solubility and can increase or decrease its bioactivity. For example, in vitamin B6 in humans, glycosylation of the parent structure reduces its bioavailability (94).
In watermelon, luteolin, a flavonoid with potential anti-cancer, anti-inflammatory, antioxidant, and anti-allergic activities (72, 95), has five glycosylated derivatives. In Figure 10 the sugar moieties of these derivatives are marked in red, under the luteolin aglycon. Two studies demonstrated that glycosylation of luteolin at different positions is closely linked to the intensity and modulation of its antioxidant and anti-inflammatory effects (96, 97). Such glycosylation is catalyzed in vivo by glycosylases, enzymes that add sugar moieties to aglycons with various selectivity. Some glycosylases add only a specific type of sugar on a specific aglycon, while others add sugars less selectively, based on aglycon substructures. Further investigation is needed to elucidate watermelon glycosylase genes and to link those enzymes to their glycosylation capabilities. Doing so will support the eventual expansion of the current catalog with other, as yet uncharacterized glycosylated natural products.
Figure 10
Watermelon Chemical Space
t-SNE analysis, by design, converts a complex dataset of points in a high-dimensional space, such as chemical structures, and identifies an accurate representation of those data in lower-dimensional space, typically the flat 2-D version of paper or screen. Applying this tactic to the watermelon plant compound catalog shows great structural diversity (Figure 11A). The major compound classes are well-separated, as they are structurally distinct, although some map between chemical classes. The latter correspond to molecules that are hybrids, for example, glycosylated flavonoids. Compared to the chemical space occupied by all known natural products (Figure 11B, in gray), the watermelon natural products (Figure 11B, in red) cover a similar space, with only a few territories not represented. This indicates that the assembled NP catalog is relatively complete in terms of chemical diversity.
Figure 11
Human Nutrition
A major impetus for assembling this list of compounds that occur naturally in watermelon was to provide information on concentration in support of research in human nutrition and plant metabolism. Moreover, in consideration of reducing food waste, different parts of the watermelon plant are considered here as these are sources of livestock feed (98, 99). Although watermelon fruit is the most popular part of the plant consumed by people, the rind and seeds are not uncommon food items. Hence, collected in Supplementary Table 2 are the levels of different compounds from different parts of the plant, as parsed from the corresponding references. Values are presented for 300 different compounds in Supplementary Table 2 and derive from various experimental conditions, different cultivars, and varieties of melon, or measurement techniques. This table also lists 1,611 other compounds that have been detected in watermelon but not quantified, including dissolved gases, nucleotides, and nucleotide derivatives, and several incompletely characterized flavone glycosides, and the like. Altogether, this table provides useful information but is intended as a guide to the source literature and nutrition databases, the latter of which may be updated in the future. We note that no data are provided for nearly 85% of the natural products tabulated.
It is well-recognized that the diet feeds the metabolism of the gut microbiota, and those metabolites generated by the microbes can affect health in humans or act as biomarkers of intake of a specific food or food group. A diverse repertoire of natural products as present in watermelon underscores its potential as a prebiotic. For example, the oligosaccharide content of watermelon, including mannitol and 1-kestose, has suggested the fruit as a source of prebiotics (100). Of different fruit peels tested, yellow watermelon showed the highest probiotic activity on Lactobacillus rhamnosus and Bifidobacterium bifidum (101). In addition, supplementing the high-fat diets of obese male mice with different watermelon products improved serum insulin and fasting blood glucose levels, as well as the hepatic metabolite profile. Furthermore, supplementation with fiber-rich extracts of rind and skin showed added improvements in glucose metabolism and energy efficiency while shifting the microbiome composition (102). Although cataloging bacterially derived metabolites is beyond the scope of this work, the catalog of natural products presented here is a necessary component that supports such efforts.
In addition to its nutrition content, watermelon is known as a folk functional food, being offered, for example, as an ethnopharmacological diuretic (103). Rat models of urolithiasis demonstrated that watermelon pulp extract reduced calcium oxalate crystal count in kidney and urine, increased urinary pH and output, elevated serum creatinine clearance, and reduced urea and creatinine levels (104). In a rat model of diuresis, watermelon pulp extract produced diuresis, reduced serum chloride levels, and elevated urinary sodium and chloride levels, in addition to inhibiting aggregation of oxalate crystals (104). Sources for these benefits include citrate, antioxidants, steroids, and alkanes. Other folk medicine uses of watermelon were for erectile dysfunction in ancient Egypt (105), as a diuretic among Russlanddeutschen living in Germany (103), and to quench thirst and act as a diuretic according to traditional Chinese medicine practices. In many instances, results from folk medicine, molecular nutrition, and clinical studies agree, which underscores the healthy benefits of watermelon.
Genome Mining
The sequencing of the watermelon genome with its 11 chromosomes of different sizes is complete (21). plantiSMASH and PRISM with default parameters were used to identify eventual BGCs in each of the chromosomes (48, 49). PlantiSMASH predicted, spread across 10 of the 11 chromosomes, eight BGCs for terpene synthesis, six for saccharide synthesis, two for alkaloid synthesis, one for lignan synthesis, one for lignan-polyketide synthesis, one for saccharide-alkaloid synthesis, and three putative BGCs. Surprisingly, no NRP synthase clusters have been detected despite the documented presence of 121 NRPs in watermelon. Although the current version of PRISM is not adapted for plant genomes, it detected a total of 18 terpene BGCs across five chromosomes, overlapping significantly with plantiSMASH results for this compound category. A number of terpenes, alkaloids, NRPs, and polyketides are present in the watermelon NP catalog described here, but BGCs responsible for their synthesis were not detected by this analysis. Thus, these predictions are simply an initial glimpse of the biosynthesis capacities of watermelon. Deeper genome mining coupled with comparative genomics can lead to the discovery of other equally noteworthy natural products and the enzymes responsible for their biosynthesis.
Summary
This catalog is a unique resource that highlights the diversity of chemical compounds in watermelon. The information presented here will be useful in crop development research integrating metabolomics, phytochemical genomics, and plant breeding to improve nutritional values of watermelon. Such a curated list of compounds associated with a single food is a necessary component in building a comprehensive catalog of natural products in all foods and can serve as a reference set for testing automated methods to capture food-compound relationships. This catalog will support detailed analyses of watermelon and can be merged with other genomics data. Such analyses can identify loci for genes whose encoded proteins facilitate synthesis, transport or storage of specific compounds, and which then can be used for crop improvement with traditional plant breeding approaches and/or biotechnology methods, constructing new links between gene, protein, and compound, and expanding existing biochemical pathways.
Funding
LP and JO's work was funded in part by United States Department of Agriculture project number 8050–51000-107-00D, and this entity had no part in the design of this software, collection, analysis, and interpretation of data, nor in composing the manuscript. ED and GM's work was funded by Hatch project NC02724 and USDA-NIFA-SCRI 2016-51181-25404. AL was partially supported by USDA-NIFA-SCRI, Grant Award Number: 2020-51181-32139 (CucCAP), and by the National Watermelon Promotion Board (NWPB). MS was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–Project-ID 239748522–SFB 1127, ChemBioSys.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories: https://watermelon.naturalproducts.net and https://doi.org/10.15482/USDA.ADC/1522862, and in the Supplementary Material affiliated with this article.
Author contributions
MS, KM, ED, GM, PP-V, and LP: data extraction. MS, KM, and LP: data management and curation. MS and LP: data analysis. MS, AL, and LP: writing the manuscript. MS, KM, ED, GM, JO, PP-V, CS, AL, and LP: review and critical assessment of the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors acknowledge with gratitude the cooperative support of Sloane M. Zwanger for her assistance with database curation. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.729822/full#supplementary-material
References
1.
NavejaJJRico-HidalgoMPMedina-FrancoJL. Analysis of a large food chemical database: chemical space, diversity, and complexity. F1000Res. (2018) 7:Chem Inf Sci-993. 10.12688/f1000research.15440.2
2.
HootonFMenichettiGBarabásiAL. Exploring food contents in scientific literature with FoodMine. Sci Rep. (2020) 10:16191. 10.1038/s41598-020-73105-0
3.
SorokinaMMerseburgerPRajanKYirikMASteinbeckC. COCONUT online: collection of open natural products database. J Cheminform. (2021) 13:2. 10.1186/s13321-020-00478-9
4.
ZhangSJDe BruynFPothakosVTorresJFalconiCMoccandCet al. Following coffee production from cherries to cup: microbiological and metabolomic analysis of wet processing of Coffea arabica. Appl Environ Microbiol. (2019) 85:e02635–18. 10.1128/AEM.02635-18
5.
ChenCYMilburyPELapsleyKBlumbergJB. Flavonoids from almond skins are bioavailable and act synergistically with vitamins C and E to enhance hamster and human LDL resistance to oxidation. J Nutr. (2005) 135:1366–73. 10.1093/jn/135.6.1366
6.
MoughanPJ. Holistic properties of foods: a changing paradigm in human nutrition. J Sci Food Agric. (2020) 100:5056–63. 10.1002/jsfa.8997
7.
OliphantKAllen-VercoeE. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. (2019) 7:91. 10.1186/s40168-019-0704-8
8.
RidgwayKLalljieSPDSmithRM. Analysis of food taints and off-flavours: a review. Food Addit Contam A Chem Anal Control Expo Risk Assess. (2010) 27:146–68. 10.1080/19440040903296840
9.
KimSChenJChengTGindulyteAHeJHeSet al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. (2021) 49:D1388–95. 10.1093/nar/gkaa971
10.
RuddigkeitLvan DeursenRBlumLCReymondJR. Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J Chem Inf Model. (2012) 52:2864–75. 10.1021/ci300415d
11.
Shouchuang WangSAlseekhSFernieARLuoJ. The structure and function of major plant metabolite modifications. Mol Plant. (2019) 12:899–919. 10.1016/j.molp.2019.06.001
12.
RodgersGPCollinsFSPrecision nutrition - the answer to “What to eat to stay healthy”JAMA. (2020) 324:735–6. 10.1001/jama.2020.13601
13.
BarabásiALMenichettiGLoscalzoJ. The unmapped chemical complexity of our diet. Nature Food. (2020) 1:33–7. 10.1038/s43016-019-0005-1
14.
WestermanKEHarringtonSOrdovasJMParnellLD. PhyteByte: identification of foods containing compounds with specific pharmacological properties. BMC Bioinformatics. (2020) 21:238. 10.1186/s12859-020-03582-7
15.
PriceEJDrapalMPerez-FonsLAmahDBhattacharjeeRHeiderBet al. Metabolite database for root, tuber, and banana crops to facilitate modern breeding in understudied crops. Plant J. (2020) 101:1258–68. 10.1111/tpj.14649
16.
PourkheirandishMGoliczAABhallaPLSinghMB. Global role of crop genomics in the face of climate change. Front Plant Sci. (2020) 11:922. 10.3389/fpls.2020.00922
17.
Food Agriculture Organization of the United Nations. (2021). Available online at: http://faostat.fao.org/site/339/default.aspx (accessed February 22, 2021).
18.
ChomickiGRennerSS. Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytol. (2015) 205:526–32. 10.1111/nph.13163
19.
ParisHS. Origin and emergence of the sweet dessert watermelon, Citrullus lanatus. Ann Bot. (2015) 116:133–48. 10.1093/aob/mcv077
20.
LeviAThomasCEWehnerTCZhangX. Low genetic diversity indicates the need to broaden the genetic base of cultivated watermelon. HortScience. (2001) 36:1096–101. 10.21273/HORTSCI.36.6.1096
21.
WuSWangXReddyUSunHBaoKPatelTet al. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the US National Plant Germplasm System watermelon collection. Plant Biotechnol J. (2019) 17:2246–58. 10.1111/pbi.13136
22.
TliliIHdiderCLenucciMSIlahyRJebariHDalessandroG. Bioactive compounds and antioxidant activities during fruit ripening of watermelon cultivars. J Food Compos Anal. (2011) 24:923–8. 10.1016/j.jfca.2011.03.016
23.
MartinPAWBlackburnM. Inhibition of seed germination by extracts of bitter Hawkesbury watermelon containing cucurbitacin, a feeding stimulant for corn rootworm (Coleoptera: Chrysomelidae). J Econ Entomol. (2003) 96:441–5. 10.1603/0022-0493-96.2.441
24.
Davidovich-RikanatiRShalevLBaranesNMeirAItkinMCohenSet al. Recombinant yeast as a functional tool for understanding bitterness and cucurbitacin biosynthesis in watermelon (Citrullus spp.). Yeast. (2015) 32:103–14. 10.1002/yea.3049
25.
LinXFarooqiAA. Cucurbitacin mediated regulation of deregulated oncogenic signaling cascades and non-coding RNAs in different cancers: spotlight on JAK/STAT, Wnt/β-catenin, mTOR, TRAIL-mediated pathways. Semin Cancer Biol. (2020) 73:302–9. 10.1016/j.semcancer.2020.10.012
26.
YuanPHeNUmerMJZhaoSDiaoWZhuHet al. Comparative metabolomic profiling of Citrullus spp. fruits provides evidence for metabolomic divergence during domestication. Metabolites. (2021) 11:78. 10.3390/metabo11020078
27.
RimandoAMPerkins-VeaziePM. Determination of citrulline in watermelon rind. J Chromatogr A. (2005) 1078:196–200. 10.1016/j.chroma.2005.05.009
28.
KogaYOhtakeR. Study report on the constituents of squeezed watermelon. J Tokyo Chem Soc (Tokyo Kagaku Kaishi). (1914) 35:519–28 [translation provided in Fragkos KC, Forbes A. Was citrulline first a laxative substance? The truth about modern citrulline and its isolation. Nihon Ishigaku Zasshi [J Jpn History Med.]. (2011) 57:275–92].
29.
FragkosKCForbesA. Was citrulline first a laxative substance? The truth about modern citrulline and its isolation. Nihon Ishigaku Zasshi [J Jpn History Med.]. (2011) 57:275–92.
30.
ZechmeisterLTuzsonP. Der Farbstoff der Wasser-Melone. Berich Deutsch Chem Gesells A B. (1930) 63:2881–3. 10.1002/cber.19300631032
31.
Perkins-VeaziePCollinsJKDavisARRobertsW. Carotenoid content of 50 watermelon cultivars. J Agric Food Chem. (2006) 54:2593–7. 10.1021/jf052066p
32.
Perkins-VeaziePCollinsJKClevidenceBWuG. Watermelons and health. Acta Horticult. (2007) 731:121–8. 10.17660/ActaHortic.2007.731.17
33.
CuCyc. (2020). Available online at: http://cucyc.feilab.net/organism-summary?object=WATERMELON_GENOME (accessed July 2020–February 2021).
34.
FoodData Central,. (2021). Available online at: https://fdc.nal.usda.gov (accessed February 5, 2021).
35.
USDA. U.S. Department of Agriculture, Agricultural Research Service. 1992-2016.Dr. Duke's Phytochemical and Ethnobotanical Databases. (2020). Available online at: http://phytochem.nal.usda.gov/http://dx.doi.org/10.15482/USDA.ADC/1239279 (accessed October 27, 2020).
36.
PhytoHub. PhytoHub v1.4. (2021). Available online at:http://phytohub.eu on (accessed February 5, 2021).
37.
RutzASorokinaMGalgonekJMietchenDWillighagenEGrahamJet al. Open natural products research: curation and dissemination of biological occurrences of chemical structures through Wikidata. bioRxiv preprint. (2021) 2021.02.28.433265v1. 10.1101/2021.02.28.433265
38.
WillighagenELMayfieldJWAlvarssonJBergACarlssonLJeliazkovaNet al. The chemistry development kit (CDK) v20: atom typing, depiction, molecular formulas, and substructure searching. J Cheminform. (2017) 9:33. 10.1186/s13321-017-0220-4
39.
KanehisaMGotoSSatoYKawashimaMFurumichiMTanabeM. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. (2014) 42:D199–205. 10.1093/nar/gkt1076
40.
WishartDSFeunangYDMarcuAGuoACLiangKVázquez-FresnoRet al. HMDB 4.0: the human metabolome database for 2018.Nucleic Acids Res. (2018) 46:D608–17. 10.1093/nar/gkx1089
41.
HastingsJOwenGDekkerAEnnisMKaleNMuthukrishnanVet al. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. (2016) 44:D1214–9. 10.1093/nar/gkv1031
42.
FahyESudMCotterDSubramaniamS. LIPID MAPS online tools for lipid research. Nucleic Acids Res. (2007) 35:W606–12. 10.1093/nar/gkm324
43.
BemisGWMurckoMA. The properties of known drugs. 1. Molecular frameworks.J Med Chem. (1996) 39:2887–93. 10.1021/jm9602928
44.
O'BoyleNDalkeA. DeepSMILES: an adaptation of SMILES for use in machine-learning of chemical structures. ChemRxiv Preprint. (2018). 10.26434/chemrxiv.7097960.v1
45.
KimHWangMLeberCNothiasLFReherRKangKBet al. NPClassifier: a deep neural network-based structural classification tool for natural products. ChemRxiv Preprint. (2020). 10.26434/chemrxiv.12885494.v1
46.
RDKIT. Available online at: www.rdkit.org (accessed January 12, 2021).
47.
SchaubJZielesnyASteinbeckCSorokinaM. Too sweet: cheminformatics for deglycosylation in natural products. Cheminform. (2020) 12:67. 10.1186/s13321-020-00467-y
48.
KautsarSASuarez DuranHGBlinKOsbournAMedemaMH. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. (2017) 45:W55–63. 10.1093/nar/gkx305
49.
SkinniderMMerwinNJJohnstonCWMagarveyNAPRISM. 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. (2017) 45:W49–54. 10.1093/nar/gkx320
50.
KaurNChughVGuptaAK. Essential fatty acids as functional components of foods- a review. J Food Sci Technol. (2014) 51:2289–303. 10.1007/s13197-012-0677-0
51.
LiQChenJYuXGaoJM. A mini review of nervonic acid: source, production, and biological functions. Food Chem. (2019) 301:125286. 10.1016/j.foodchem.2019.125286
52.
Sales-CamposHReisde. Souza P, Crema Peghini B, Santana da Silva J, Ribeiro Cardoso C. An overview of the modulatory effects of oleic acid in health and disease. Mini Rev Med Chem. (2013) 13:201–10. 10.2174/138955713804805193
53.
ArunaPVenkataramanammaDSinghAKSinghRP. Health benefits of punicic acid: a review. Compr Rev Food Sci Food Saf. (2016) 15:16–27. 10.1111/1541-4337.12171
54.
TermsarasabUChoHJKimDHChongSChungSJShimCKet al. Chitosan oligosaccharide-arachidic acid-based nanoparticles for anti-cancer drug delivery. Int J Pharm. (2013) 441:373–80. 10.1016/j.ijpharm.2012.11.018
55.
HuangAKollerA. Endothelin and prostaglandin H2 enhance arteriolar myogenic tone in hypertension. Hypertension. (1997) 30:1210–5. 10.1161/01.hyp.30.5.1210
56.
KalinskiP. Regulation of immune responses by prostaglandin E2. J Immunol. (2012) 188:21–8. 10.4049/jimmunol.1101029
57.
FirnR. Nature's Chemicals: The Natural Products that Shaped Our World. New York, NY: Oxford University Press (2010) p. 250.
58.
AbdelwahabSIHassanLEASiratHMYagiSMAKokoWSMohanSet al. Anti-inflammatory activities of cucurbitacin E isolated from Citrullus lanatus var. citroides: role of reactive nitrogen species and cyclooxygenase enzyme inhibition. Fitoterapia. (2011) 82:1190–7. 10.1016/j.fitote.2011.08.002
59.
Tannin-SpitzTBergmanMGrossmanS. Cucurbitacin glucosides: antioxidant and free-radical scavenging activities. Biochem Biophys Res Commun. (2007) 364:181–6. 10.1016/j.bbrc.2007.09.075
60.
LeeDHIwanskiGBThoennissenNH. Cucurbitacin: ancient compound shedding new light on cancer treatment. Scientific World J. (2010) 10:413–8. 10.1100/tsw.2010.44
61.
KrinskyNIElizabethJ. Johnson EJ. Carotenoid actions and their relation to health and disease. Mol Aspects Med. (2005) 26:459–516. 10.1016/j.mam.2005.10.001
62.
SpanovaMDaumG. Squalene - biochemistry, molecular biology, process biotechnology, and applications. Eur J Lipid Sci. (2011) 113:1299–320. 10.1002/ejlt.201100203
63.
Rodríguez-RodríguezRHerreraMDPeronaJSRuiz-GutiérrezV. Potential vasorelaxant effects of oleanolic acid and erythrodiol, two triterpenoids contained in 'orujo' olive oil, on rat aorta. Br J Nutr. (2004) 92:635–42. 10.1079/bjn20041231
64.
AkihisaTTokudaHIchiishiEMukainakaTToriumiMUkiyaMet al. Anti-tumor promoting effects of multiflorane-type triterpenoids and cytotoxic activity of karounidiol against human cancer cell lines. Cancer Lett. (2001) 173:9–14. 10.1016/s0304-3835(01)00689-9
65.
AnderssonSCOlssonMEJohanssonE. Rumpunen K. Carotenoids in sea buckthorn (Hippophae rhamnoides L.) berries during ripening and use of pheophytin A as a maturity marker. J Agric Food Chem. (2009) 57:250–8. 10.1021/jf802599f
66.
HavauxMNiyogiKK. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci USA. (1999) 96:8762–7. 10.1073/pnas.96.15.8762
67.
DeyPM. 5 - Oligosaccharides. Methods Plant Biochem. (1990) 2:189–218. 10.1016/B978-0-12-461012-5.50011-2
68.
ChenASTaguchiTSakaiKKikuchiKWangMWMiwaI. Antioxidant activities of chitobiose and chitotriose. Biol Pharm Bull. (2003) 26:1326–30. 10.1248/bpb.26.1326
69.
EganDO'KennedyRMoranECoxDProsserEThornesRD. The pharmacology, metabolism, analysis, and applications of coumarin and coumarin-related compounds. Drug Metab Rev. (1990) 22:503–29. 10.3109/03602539008991449
70.
FylaktakidouKCHadjipavlou-LitinaDJLitinasKENicolaidesDN. Natural and synthetic coumarin derivatives with anti-inflammatory/antioxidant activities. Curr Pharm Des. (2004) 10:3813–33. 10.2174/1381612043382710
71.
LinYShiRWangXShenHM. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr Cancer Drug Targets. (2008) 8:634–46. 10.2174/156800908786241050
72.
SeelingerGMerfortISchemppCM. Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. (2008) 74:1667–77. 10.1055/s-0028-1088314
73.
BootsAWHaenenGRMMBastA. Health effects of quercetin: from antioxidant to nutraceutical. Eur J Pharmacol. (2008) 585:325–37. 10.1016/j.ejphar.2008.03.008
74.
GuptaMBBhallaTNGuptaGPMitraCRBhargavaKP. Anti-inflammatory activity of taxifolin. Jpn J Pharmacol. (1971) 21:377–82. 10.1254/jjp.21.377
75.
KimHYKimJKChoiJHJungJYOhWYKimDCet al. Hepatoprotective effect of pinoresinol on carbon tetrachloride-induced hepatic damage in mice. J Pharmacol Sci. (2010) 112:105–12. 10.1254/jphs.09234fp
76.
NwachukwuIDSlusarenkoAJGruhlkeMCH. Sulfur and sulfur compounds in plant defence. Nat Prod Commun. (2012) 7:395–400. 10.1177/1934578X1200700323
77.
ErlandLAEShuklaMRSinghASMurchSJSaxenaPK. Melatonin and serotonin: mediators in the symphony of plant morphogenesis. J Pineal Res. (2018) 64:12452. 10.1111/jpi.12452
78.
Veenstra-VanderWeeleJAndersonGMCookEHJr. Pharmacogenetics and the serotonin system: initial studies and future directions. Eur J Pharmacol. (2000) 410:165–81. 10.1016/s0014-2999(00)00814-1
79.
MalhotraSSawhneyGPandhiP. The therapeutic potential of melatonin: a review of the science. MedGenMed. (2004) 6:46.
80.
Fernandez-MejiaC. Pharmacological effects of biotin. J Nutr Biochem. (2005) 16:424–7. 10.1016/j.jnutbio.2005.03.018
81.
AzamSHadiNKhanNUHadiSM. Antioxidant and prooxidant properties of caffeine, theobromine and xanthine. Med Sci Monit. (2003) 9:BR325–30.
82.
VitekL. The protective role of the heme catabolic pathway in hepatic disorders. Antioxid Redox Signal. (2021). 10.1089/ars.2021.0080
83.
BahriSZerroukNAusselCMoinardCCrennPCurisEet al. Citrulline: from metabolism to therapeutic use. Nutrition. (2013) 29:479–84. 10.1016/j.nut.2012.07.002
84.
RomeroMJPlattDHCaldwellRBCaldwellRW. Therapeutic use of citrulline in cardiovascular disease. Cardiovasc Drug Rev. (2006) 24:275–90. 10.1111/j.1527-3466.2006.00275.x
85.
CormioLDe SiatiMLorussoFSelvaggioOMirabellaLSanguedolceFet al. Oral L-citrulline supplementation improves erection hardness in men with mild erectile dysfunction. Urology. (2011) 77:119–22. 10.1016/j.urology.2010.08.028
86.
PompellaAVisvikisAPaolicchiADe TataVCasiniAF. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol. (2003) 66:1499–503. 10.1016/s0006-2952(03)00504-5
87.
GerstmeierJSeegersJWittFWaltenbergerBTemmlVRollingerJMet al. Ginkgolic acid is a multi-target inhibitor of key enzymes in pro-inflammatory lipid mediator biosynthesis. Front Pharmacol. (2019) 10:797. 10.3389/fphar.2019.00797
88.
MangoDWeiszFNisticòR. Ginkgolic acid protects against Aβ-induced synaptic dysfunction in the hippocampus. Front Pharmacol. (2016) 7:401. 10.3389/fphar.2016.00401
89.
BehzadSSuredaABarrecaDNabaviSFRastrelliLNabaviSM. Health effects of phloretin: from chemistry to medicine. Phytochem Rev. (2017) 16:527–33. 10.1007/s11101-017-9500-x
90.
BarbehennRVConstabelCP. Tannins in plant-herbivore interactions. Phytochemistry. (2011) 72:1551–65. 10.1016/j.phytochem.2011.01.040
91.
MengerDJVan LoonJJATakkenW. Assessing the efficacy of candidate mosquito repellents against the background of an attractive source that mimics a human host. Med Vet Entomol. (2014) 28:407–13. 10.1111/mve.12061
92.
ChanderRKapoorNKDhawanBN. Picroliv, picroside-I and kutkoside from Picrorhiza kurrooa are scavengers of superoxide anions. Biochem Pharmacol. (1992) 44:180–3. 10.1016/0006-2952(92)90054-m
93.
AnsariRAAswalBSChanderRDhawanBNGargNKKapoorNKet al. Hepatoprotective activity of kutkin - the iridoid glycoside mixture of Picrorhiza kurrooa. Indian J Med Res. (1988) 87:401–4.
94.
NakanoHMcMahonLGGregory IIIJF. Pyridoxine-5'-beta–glucoside exhibits incomplete bioavailability as a source of vitamin B-6 and partially inhibits the utilization of co-ingested pyridoxine in humans. J Nutr. (1997) 127:1508–13. 10.1093/jn/127.8.1508
95.
HussainYCuiJHKhanHAschnerMBatihaGEJeandetP. Luteolin and cancer metastasis suppression: focus on the role of epithelial to mesenchymal transition. Med Oncol. (2021) 38:66. 10.1007/s12032-021-01508-8
96.
ChoiJSIslamMNAliMYKimYMParkHJSohnHSet al. The effects of C-glycosylation of luteolin on its antioxidant, anti-Alzheimer's disease, anti-diabetic, and anti-inflammatory activities. Arch Pharm Res. (2014) 37:1354–63. 10.1007/s12272-014-0351-3
97.
OdontuyaGHoultJRSHoughtonPJ. Structure-activity relationship for antiinflammatory effect of luteolin and its derived glycosides. Phytother Res. (2005) 19:782–6. 10.1002/ptr.1723
98.
ErhirhieEEkeneN. Medicinal values on Citrullus lanatus (watermelon): pharmacological review. Int J Res Pharma Biomed Sci. (2013) 4:1305–12.
99.
WadhwaMBakshiMPS. Utilization of Fruit and Vegetable Wastes as Livestock Feed and as Substrates for Generation of Other Value-Added Products. Food and Agriculture Organization of the United Nations (2013).
100.
Jovanovic-MalinovskaRKuzmanovaSWinkelhausenE. Oligosaccharide profile in fruits and vegetables as sources of prebiotics and functional foods. Int J Food Prop. (2014) 17:949–65. 10.1080/10942912.2012.680221
101.
HaoCLEsahEMTajarudinHAAkterBSallehRM. Effect of potential prebiotics from selected fruits peel on the growth of probiotics. J Food Process Preserv. (2021) 2021:e15581. 10.1111/jfpp.15581
102.
BecraftARSturmMLMendezRLParkSHLeeSIShayNF. Intake of watermelon or its byproducts alters glucose metabolism, the microbiome, and hepatic proinflammatory metabolites in high-fat-fed male C57BL/6 J mice. J Nutr. (2020) 150:434–42. 10.1093/jn/nxz267
103.
PieroniAGrayC. Herbal and food folk medicines of the Russlanddeutschen living in Künzelsau/Taläcker, South-Western Germany. Phytother Res. (2008) 22:889–901. 10.1002/ptr.2410
104.
SiddiquiWAShahzadMShabbirAAhmadA. Evaluation of anti-urolithiatic and diuretic activities of watermelon (Citrullus lanatus) using in vivo and in vitro experiments. Biomed Pharmacother. (2018) 97:1212–21. 10.1016/j.biopha.2017.10.162
105.
Haimov-KochmanRSciaky-TamirYHurwitzA. Reproduction concepts and practices in ancient Egypt mirrored by modern medicine. Eur J Obstet Gynecol Reprod Biol. (2005) 123:3–8. 10.1016/j.ejogrb.2005.03.022
106.
DavisARKingSR. MSW-28, a full-flavor crisp watermelon line with high lycopene and medium brix. HortScience. (2007) 42:1715–6. 10.21273/HORTSCI.42.7.1715
107.
NolteAJvon LoeseckeHW. Characteristics and composition of watermelon seed oil (Cuban Queen variety). J Agric Food Chem. (1939) 61:889–91. 10.1021/ja01873a034
108.
YaoYLiuWZhouHZhangDLiRLiCet al. The relations between minor components and antioxidant capacity of five fruits and vegetable seed oils in China. J Oleo Sci. (2019) 68:625–35. 10.5650/jos.ess19005
109.
TabiriBAgbenorheviJKWireko-ManuFDOmpoumaEI. Watermelon seeds as food: nutrient composition, phytochemicals and antioxidant activity. Int J Nutr Food Sci. (2016) 5:139–44. 10.11648/j.ijnfs.20160502.18
110.
TliliIHdiderCLenucciMSRiadhIJebariHDalessandroG. Bioactive compounds and antioxidant activities of different watermelon (Citrullus lanatus (Thunb. Mansfeld) cultivars as affected by fruit sampling area.J Food Compost Anal. (2011) 24:307–14. 10.1016/j.jfca.2010.06.005
111.
KimYGoodnerKL. Bioactive Compounds in Watermelon Flesh and Rind. Synergy Flavors Sensus Technical Note (2009) 1–6. Available online at: https://www.synergytaste.com/sites/synergytaste.com/files/SEN-TN-0021-Bioactive_Compounds_in_Watermelon_Flesh_and_Rind.pdf (accessed January 2021).
112.
KimYCChoiDZhangCLiuH. Lee S. Profiling cucurbitacins from diverse watermelons (Citrullus spp.).Hortic Environ Biotechnol. (2018) 59:557–66.
113.
DavisARPerkins-VeaziePCollinsJLeviA. LSW-177 and LSW-194: Red-fleshed watermelon lines with low-total soluble solids. HortScience. (2008) 43:538–9. 10.21273/HORTSCI.43.2.538
114.
NgiefuCKPaquotCVieuxA. Oil-bearing plants of Zaire. II Botanical families providing oils of medium unsaturation. Oleagineux. (1976) 31:545–7.
115.
KamelBSDawsonHKakudaY. Characteristics and composition of melon and grape seed oils and cakes. J Am Oil Chem Soc. (1985) 62:881–3. 10.1007/BF02541750
116.
ZhongYShiJZhengZNawazMAChenCChengFet al. NMR-based fruit metabonomic analysis of watermelon grafted onto different rootstocks under two potassium levels. Sci Hortic. (2019) 258:108793. 10.1016/j.scienta.2019.108793
117.
Mendoza-EnanoMLStanleyRFrankD. Linking consumer sensory acceptability to volatile composition for improved shelf-life: a case study of fresh-cut watermelon (Citrullus lanatus). Postharvest Biol Technol. (2019) 154:137–47. 10.1016/j.postharvbio.2019.03.018
118.
GrossKCAcostaPB. Fruits and vegetables are a source of galactose: implications in planning the diets of patients with galactosaemia. J Inherit Metab Dis. (1991) 14:253–8. 10.1007/BF01800599
119.
DavisARCollinsJFishWWTadmorYWebber IIICLPerkins-VeazieP. Rapid method for total carotenoid detection in canary yellow-fleshed watermelon. J Food Sci. (2007) 72:S319–23. 10.1111/j.1750-3841.2007.00381.x
120.
LewinsohnESitritYBarEAzulayYMeirAZamirDet al. Carotenoid pigmentation affects the volatile composition of tomato and watermelon fruits, as revealed by comparative genetic analyses. J Agric Food Chem. (2005) 53:3142–8. 10.1021/jf047927t
121.
WehnerTCNaegeleRPPerkins-VeazieP. Heritability and genetic variance components associated with citrulline, arginine, and lycopene content in diverse watermelon cultigens. HortScience. (2017) 52:936–40. 10.21273/HORTSCI11255-16
122.
Vázquez-ManjarrezNUlaszewskaMGarcia-AloyMMattiviFPraticòGDragstedLOet al. Biomarkers of intake for tropical fruits. Genes Nutr. (2020) 15:11. 10.1186/s12263-020-00670-4
123.
TabataMTanakaSChoHJUnoCShimakuraJItoMet al. Production of an anti-allergic triterpene bryonolic acid, by plant cell cultures. J Nat Prod. (1993) 56:165–74. 10.1021/np50092a001
124.
MoraisDRRottaEMSargiSCBonafeEGSuzukiRMSouzaNEet al. Proximate composition, mineral contents and fatty acid composition of the different parts and dried peels of tropical fruits cultivated in Brazil. J Braz Chem Soc. (2017) 28:308–18. 10.5935/0103-5053.20160178
125.
KasoteDMJayaprakashaGKOngKCrosbyKMPatilBS. Hormonal and metabolites responses in Fusarium wilt-susceptible and -resistant watermelon plants during plant-pathogen interactions. BMC Plant Biol. (2020) 20:481. 10.1186/s12870-020-02686-9
126.
LiuCHZhang HY DaiZYLiuXLiuYDengXXet al. Volatile chemical and carotenoid profiles in watermelons [Citrullus vulgaris (Thunb) Schrad (Cucurbitaceae)] with different flesh colors. Food Sci Biotechnol. (2012) 21:531–41. 10.1007/s10068-012-0068-3
127.
JacobAGEtongDITijjaniA. Proximate, mineral and anti-nutritional compositions of melon (Citrullus lanatus) seeds. Br J Res. (2015) 2:142–51.
128.
LvPLiNLiuHGuHZhaoWE. Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars. Food Chem. (2015) 174:52–9. 10.1016/j.foodchem.2014.11.022
129.
ZhuQGaoPLiuSZhuZAmanullahSDavisARet al. Comparative transcriptome analysis of two contrasting watermelon genotypes during fruit development and ripening. BMC Genomics. (2017) 18:3. 10.1186/s12864-016-3442-3
130.
Costa de ContoLLefevre GragnaniMAMausDIfanger AmbielHSChiuMCGrimaldiRet al. Characterization of crude watermelon seed oil by two different extraction methods. J Am Oil Chem Soc. (2011) 88:1709–14. 10.1007/s11746-011-1850-8
131.
MahlaHRRathoreSSVenkatesanKSharmaR. Analysis of fatty acid methyl esters and oxidative stability of seed purpose watermelon (Citrullus lanatus) genotypes for edible oil. J Food Sci Technol. (2018) 55:1552–61. 10.1007/s13197-018-3074-5
132.
OuassorIAqilY. BelmaghraouiW, El Hajjaji S. Characterization of two Moroccan watermelon seeds oil varieties by three different extraction methods. OCL. (2020) 27:13. 10.1051/ocl/2020010
133.
ImbsABPhamLQ. Lipid composition of ten edible seed species from North Vietnam. J Am Oil Chem Soc. (1995) 72:957–61. 10.1007/BF02542074
134.
HarnlyJMDohertyRFBeecherGRHoldenJMHaytowitzDB. Flavonoid content of US fruits, vegetables, and nuts. J Agric Food Chem. (2006) 54:9966–77. 10.1021/jf061478a
135.
TadmorYKingSLeviADavisARMeirAWassermanBet al. Comparative fruit coloration in watermelon and tomato. Food Res Int. (2005) 38:837–41. 10.1016/j.foodres.2004.07.011
136.
ChoHJTanakaSKamisakoWTabataM. Biosynthesis of bryonolic acid in cultured cells of watermelon. Phytochemistry. (1993) 33:1407–13. 10.1016/0031-9422(93)85100-6
137.
FeizyJJahaniMAhmadiS. Antioxidant activity and mineral content of watermelon peel. J Food Bioprocess Eng. (2020) 3:35–40. 10.22059/jfabe.2020.75811
138.
JawadUMGaoLGebremeskelHSafdarLBYuanPZhaoSet al. Expression pattern of sugars and organic acids regulatory genes during watermelon fruit development. Sci Hortic. (2020) 265:109102. 10.1016/j.scienta.2019.109102
139.
SaitoKYokoyamaHNojiMMurakoshiI. Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J Biol Chem. (1995) 270:16321–6. 10.1074/jbc.270.27.16321
140.
MushtaqMSultanaBBhattiHN. Asghar M. RSM based optimized enzyme-assisted extraction of antioxidant phenolics from underutilized watermelon (Citrullus lanatus Thunb.) rind. J Food Sci Technol. (2015) 52:5048–56. 10.1007/s13197-014-1562-9
141.
HildebrandDFAfitlhileMFukushigeH. Regulation of oxylipin synthesis. Biochem Soc Trans. (2000) 28:847–9. 10.1042/bst0280847
142.
BianchiGProvenziL. Rizzolo A. Evolution of volatile compounds in ‘Cuoredolce®’ and ‘Rugby’ mini- watermelons (Citrullus lanatus (Thunb) Matsumura and Nakai) in relation to ripening at harvest. J Sci Food Agric. (2020) 100:945–52. 10.1002/jsfa.10023
143.
ShadungKGMashelaPWMphosiMS. Suitable drying temperature from preserving cucurbitacins in fruit of wild cucumber and wild watermelon. Am Soc Hortic Sci. (2016) 26:816–9. 10.21273/HORTTECH03400-16
144.
FanJParkEZhangLEdirisingheIBurton-FreemanBSandhuAK. Pharmacokinetic parameters of watermelon (rind, flesh, and seeds) bioactive components in human plasma: a pilot study to investigate the relationship to endothelial function. J Agric Food Chem. (2020) 68:7393–403. 10.1021/acs.jafc.0c02756
145.
SulaimanFAhmad AzamAAhamad BustamamMSFakuraziSAbasFLeeYXet al. Metabolite profiles of red and yellow watermelon (Citrullus lanatus) cultivars using a 1 H-NMR metabolomics approach. Molecules. (2020) 25:3235. 10.3390/molecules25143235
146.
LiuYHeCSongHL. Comparison of SPME versus SAFE processes for the analysis of flavor compounds in watermelon juice. Food Anal Methods. (2018) 11:1677–89. 10.1007/s12161-018-1153-x
147.
FadimuGJGhafoorKBabikerEEAl-JuhaimiFAbdulraheemRAAdenekanMK. Ultrasound-assisted process for optimal recovery of phenolic compounds from watermelon (Citrullus lanatus) seed and peel. J Food Meas Charact. (2020) 14:1784–93. 10.1007/s11694-020-00426-z
148.
TripodiGCondursoCCincottaFMerlinoMVerzeraA. Aroma compounds in mini watermelon fruits from different grafting combinations. J Sci Food Agric. (2020) 100:1328–35. 10.1002/jsfa.10149
149.
AsgharMNShahzadMTNadeemIAshrafCM. Phytochemical and in vitro total antioxidant capacity analysis of peel extracts of different cultivars of Cucumis melo and Citrullus lanatus. Pharm Biol. (2013) 51:226–32. 10.3109/13880209.2012.717228
150.
BangHDavisARKimSLeskovarDIKingSR. Flesh color inheritance and gene interactions among canary yellow, pale yellow, and red watermelon. J Am Soc Hortic Sci. (2010) 135:362–8. 10.21273/JASHS.135.4.362
151.
HuangYZhaoLKongQChengFNiuMXieJet al. Comprehensive mineral nutrition analysis of watermelon grafted onto two different rootstocks. Hortic Plant J. (2016) 2:105–13. 10.1016/j.hpj.2016.06.003
152.
YajimaISakakibaraHIdeJYanaiTHayashK. Volatile flavor components of watermelon (Citrullus vulgaris). Agric Biol Chem. (1985) 49:3145–50. 10.1080/00021369.1985.10867246
153.
HalderTGadgilVN. Fatty acids of callus tissues of six species of cucurbitaceae. Phytochemistry. (1983) 22:1965–7. 10.1016/0031-9422(83)80024-7
154.
SongQJoshiMDiPiazzaJJoshiV. Functional relevance of citrulline in the vegetative tissues of watermelon during abiotic stresses. Front Plant Sci. (2020) 11:512. 10.3389/fpls.2020.00512
155.
ZhangBTolstikovVTurnbullCHicksLMOliverF. Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc Natl Acad Sci USA. (2010) 107:13532–7. 10.1073/pnas.0910558107
156.
GaoLZhaoSLuXHeNLiuW. ‘SW’, a new watermelon cultivar with a sweet and sour flavor. HortScience. (2018) 53:895–6. 10.21273/HORTSCI12857-18
157.
OgbujiKMcCutcheonGSSimmonsAMSnookMEHarrisonHFLeviA. Partial leaf chemical profiles of a desert watermelon species (Citrullus cococynthis) and heirloom watermelon cultivars (Citrullus lanatus var. lanatus). Am Soc Hortic Sci. (2012) 47:580–4. 10.21273/hortsci.47.5.580
158.
ayaprakashaGKPatilBSA metabolomics approach to identify and quantify the phytochemicals in watermelons by quantitative (1)HNMR. Talanta. (2016) 153:268–77. 10.1016/j.talanta.2016.02.060
159.
BeaulieuJCLeaJM. Characterization and semiquantitative analysis of volatiles in seedless watermelon varieties using solid phase microextraction. J Agric Food Chem. (2006) 54:7789–93. 10.1021/jf060663l
160.
FuLXuBTXuXRGanRYZhangYXiaEQet al. Antioxidant capacities and total phenolic contents of 62 fruits. Food Chem. (2011) 129:345–50. 10.1016/j.foodchem.2011.04.079
161.
JeffreyJLTurnerNDKingSR. Carotenoid bioaccessibility from nine raw carotenoid-storing fruits and vegetables using an in vitro model. J Sci Food Agric. (2012) 92:2603–10. 10.1002/jsfa.5768
162.
RankoffGPopowA. Untersuchungen über das Öl aus den Samen von Wassermelonen. Fette Seifen. (1941) 48:489–91. 10.1002/lipi.19410480802
163.
FredesASalesCBarredaMValcárcelMRosellóSBeltránJ. Quantification of prominent volatile compounds responsible for muskmelon and watermelon aroma by purge and trap extraction followed by gas chromatography-mass spectrometry determination. Food Chem. (2016) 190:689–700. 10.1016/j.foodchem.2015.06.011
164.
TarachiwinLMasakoOFukusakiE. Quality evaluation and prediction of Citrullus lanatus by 1H NMR-based metabolomics and multivariate analysis. J Agric Food Chem. (2008) 56:5827–35. 10.1021/jf800418u
165.
ShibaharaAYamamotoKNakayamaTKajimotoG. cis-Vaccenic acid in pulp lipids of commonly available fruits. J Am Oil Chem Soc. (1987) 64:397–401. 10.1007/BF02549303
166.
Abu-ReidahIMArráez-RománDSegura-CarreteroAFernández-GutiérrezA. Profiling of phenolic and other polar constituents from hydro-methanolic extract of watermelon (Citrullus lanatus) by means of accurate-mass spectrometry (HPLC–ESI–QTOF–MS). Food Res Int. (2013) 51:354–62. 10.1016/j.foodres.2012.12.033
167.
KastingRDelwicheCC. Ornithine, citrulline, and arginine metabolism in watermelon seedlings. Plant Physiol. (1958) 33:350–4. 10.1104/pp.33.5.350
168.
ChoHJTanakaSFukuiHTabataM. Formation of bryonolic acid in cucurbitaceous plants and their cell cultures. Phytochemistry. (1992) 31:3893–6. 10.1016/S0031-9422(00)97548-4
169.
Perkins-VeaziePCollinsJK. Carotenoid changes of intact watermelons after storage. J Agric Food Chem. (2006) 54:5868–74. 10.1021/jf0532664
170.
KantorMLeviAThiesJGunerNKantorCParnhamSet al. NMR analysis reveals a wealth of metabolites in root-knot nematode resistant roots of Citrullus amarus watermelon plants. J Nematol. (2018) 50:303–16. 10.21307/jofnem-2018-030
171.
GulerZCandirEYetisirHKaracaFSolmazI. Volatile organic compounds in watermelon (Citrullus lanatus) grafted onto 21 local and two commercial bottle gourd (Lagenaria siceraria) rootstocks. J Hortic Sci Biotechnol. (2014) 89:448–52. 10.1080/14620316.2014.11513105
172.
ZhongYChenCNawazMAJiaoYZhengZShiXet al. Using rootstock to increase watermelon fruit yield and quality at low potassium supply: a comprehensive analysis from agronomic, physiological and transcriptional perspective. Sci Hortic. (2018) 241:144–51. 10.1016/j.scienta.2018.06.091
173.
CondursoCVerzeraADimaGTripodiGCrinòPParatoreAet al. Effects of different rootstocks on aroma volatile compounds and carotenoid content of melon fruits. Sci Hortic. (2012) 148:9–16. 10.1016/j.scienta.2012.09.015
174.
LiuSGaoPWangXDavisARBalochAMLuanF. Mapping of quantitative trait loci for lycopene content and fruit traits in Citrullus lanatus. Euphytica. (2015) 202:411–26. 10.1007/s10681-014-1308-9
175.
RaziqSAnwarFMahmoodZShahidSA. Nadeem R. Characterization of seed oils from different varieties of watermelon [Citrullus lanatus (Thunb)] from Pakistan. Gras Aceit. (2012) 63:365–72. 10.3989/gya.022212
176.
DavisARWebber IIICLFishWWWehnerTCKingSPerkins-VeazieP. L-citrulline levels in watermelon cultigens tested in two environments. HortScience. (2011) 46:1572–5. 10.21273/HORTSCI.46.12.1572
177.
Tarazona-DíazMPViegasJMoldao-MartinsMAguayoE. Bioactive compounds from flesh and by-product of fresh-cut watermelon cultivars. J Sci Food Agric. (2011) 91:805–12. 10.1002/jsfa.4250
178.
AslamAZhaoSAzamMLuXHeNLiBet al. Comparative analysis of primary metabolites and transcriptome changes between ungrafted and pumpkin-grafted watermelon during fruit development. PeerJ. (2020) 8:e8259. 10.7717/peerj.8259
179.
JarretRLLevyIJ. Oil and fatty acid contents in seed of Citrullus lanatus Schrad. J Agric Food Chem. (2012) 60:5199–204. 10.1021/jf300046f
180.
IlahyRTliliISiddiquiMWHdiderCLenucciMS. Inside and beyond color: comparative overview of functional quality of tomato and watermelon fruits. Front Plant Sci. (2019) 10:769. 10.3389/fpls.2019.00769
181.
RicoXGullónBAlonsoJLYáñezR. Recovery of high value-added compounds from pineapple, melon, watermelon and pumpkin processing by-products: an overview. Food Res Int. 132:109086. 10.1016/j.foodres.2020.109086
182.
GrassiSPiroGLeeJMZhengYFeiZJDalessandroGet al. Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genomics. (2013) 14:781. 10.1186/1471-2164-14-781
183.
SilouTKissotokene-NtinouOMvoula TsieriMOuambaJMKiakouamaS. Contribution a l'etude des corps gras de graines de quatre espèces des cucurbitacees cultivées au Congo. J Soc Chim Tunisie. (1990) 2:13–21.
184.
KikuchiTIkedayaATodaAIkushimaKYamakawaTOkadaRet al. Pyrazole alkaloids from watermelon (Citrullus lanatus) seeds. Phytochem Lett. (2015) 12:94–7. 10.1016/j.phytol.2015.02.017
185.
Estévez-SantiagoROlmedilla-AlonsoBFernández-JalaoI. Bioaccessibility of provitamin A carotenoids from fruits: application of a standardised static in vitro digestion method. Food Funct. (2016) 7:1354–66. 10.1039/c5fo01242b
186.
NoeFFFowdenL. α-amino-β-(pyrazolyl-N) propionic acid: a new amino-acid from Citrullus vulgaris (water melon). Nature. (1959) 184:BA69–70. 10.1038/184069a0b
187.
FredesARosellóSBeltránJCebolla-CornejoJPérez-de-CastroAGisbertCet al. Fruit quality assessment of watermelons grafted onto citron melon rootstock. J Sci Food Agric. (2017) 97:1646–55. 10.1002/jsfa.7915
188.
KongQYuanJGaoLLiuPCaoLHuangYet al. Transcriptional regulation of lycopene metabolism mediated by rootstock during the ripening of grafted watermelons. Food Chem. (2017) 214:406–11. 10.1016/j.foodchem.2016.07.103
189.
WangCQiaoAFangXSunLGaoPDavisARet al. Fine mapping of lycopene content and flesh color related gene and development of molecular marker-assisted selection for flesh color in watermelon (Citrullus lanatus). Front Plant Sci. (2019) 10:1240. 10.3389/fpls.2019.01240
190.
SaftnerRLuoYMcEvoyJAbbottJAVinyardB. Quality characteristics of fresh-cut watermelon slices from non-treated and 1-methylcyclopropene- and/or ethylene-treated whole fruit. Postharvest Biol Technol. (2007) 44:71–9. 10.1016/j.postharvbio.2006.11.002
191.
MilczarekRRSedejI. Enhancing nutritional quality of spray-dried concentrated watermelon juice using watermelon by-product carrier blends. J Food Qual. (2020).
192.
Al-KhalifaAS. Physicochemical characteristics, fatty acid composition, and lipoxygenase activity of crude pumpkin and melon seed oils. J Agric Food Chem. (1996) 44:964–6. 10.1021/jf950519s
193.
GaoLZhaoSLuXHeNZhuHDouJet al. Comparative transcriptome analysis reveals key genes potentially related to soluble sugar and organic acid accumulation in watermelon. PLoS ONE. (2018) 13:e0190096. 10.1371/journal.pone.0190096
194.
RippergerHSeifertK. Die Cucurbitacine von Citrullus lanatus var. citroides Tetrahedron. (1975) 31:1561–3. 10.1016/0040-4020(75)87012-8
Summary
Keywords
food chemistry, natural products, natural compounds, watermelon, phytochemicals
Citation
Sorokina M, McCaffrey KS, Deaton EE, Ma G, Ordovás JM, Perkins-Veazie PM, Steinbeck C, Levi A and Parnell LD (2021) A Catalog of Natural Products Occurring in Watermelon—Citrullus lanatus. Front. Nutr. 8:729822. doi: 10.3389/fnut.2021.729822
Received
23 June 2021
Accepted
09 August 2021
Published
14 September 2021
Volume
8 - 2021
Edited by
José M. Alvarez-Suarez, Universidad San Francisco de Quito, Ecuador
Reviewed by
Jacob Mashilo, University of KwaZulu-Natal, South Africa; Emmanuel Oladeji Alamu, International Institute of Tropical Agriculture, Zambia
Updates
Copyright
© 2021 Sorokina, McCaffrey, Deaton, Ma, Ordovás, Perkins-Veazie, Steinbeck, Levi and Parnell.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Sorokina maria.sorokina@uni-jena.deLaurence D. Parnell laurence.parnell@usda.gov
This article was submitted to Nutrition and Food Science Technology, a section of the journal Frontiers in Nutrition
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.